Abstract
Recurrent loss-of-function mutations of spliceosome gene, ZRSR2, occur in myelodysplastic syndromes (MDS). Mutation/loss of ZRSR2 in human myeloid cells primarily causes impaired splicing of the U12-type introns. In order to further investigate the role of this splice factor in RNA splicing and hematopoietic development, we generated mice lacking ZRSR2. Unexpectedly, Zrsr2-deficient mice developed normal hematopoiesis with no abnormalities in myeloid differentiation evident in either young or ≥1-year old knockout mice. Repopulation ability of Zrsr2-deficient hematopoietic stem cells was also unaffected in both competitive and non-competitive reconstitution assays. Myeloid progenitors lacking ZRSR2 exhibited mis-splicing of U12-type introns, however, this phenotype was moderate compared to the ZRSR2-deficient human cells. Our investigations revealed that a closely related homolog, Zrsr1, expressed in the murine hematopoietic cells, but not in human cells contributes to splicing of U12-type introns. Depletion of Zrsr1 in Zrsr2 KO myeloid cells exacerbated retention of the U12-type introns, thus highlighting a collective role of ZRSR1 and ZRSR2 in murine U12-spliceosome. We also demonstrate that aberrant retention of U12-type introns of MAPK9 and MAPK14 leads to their reduced protein expression. Overall, our findings highlight that both ZRSR1 and ZRSR2 are functional components of the murine U12-spliceosome, and depletion of both proteins is required to accurately model ZRSR2-mutant MDS in mice.
Introduction
Mutations in RNA splicing factors constitute the leading class of genetic alterations in myelodysplastic syndromes (MDS).1-4 Somatic mutations in spliceosome genes, SF3B1, SRSF2, U2AF1 and ZRSR2 are observed in >50% of MDS.2,3 These mutations are early events during disease development and largely occur mutually exclusive of each other.1-5 Intense efforts in the last few years have enhanced our understanding of the impact of spliceosome mutations on RNA splicing and defined a mis-splicing pattern for each mutation.6-17 Animal models expressing mutant hotspots of splice factor genes - SRSF2 (P95H), SF3B1 (K700E) and U2AF1 (S34F) - have enabled elucidation of consequences of these genetic lesions on RNA splicing and hematopoietic development.6,7,12,15,16,18-20 Unlike mutational hotspots observed in SF3B1, SRSF2 and U2AF1, alterations of ZRSR2 are truncating mutations spread throughout the transcript. ZRSR2 is located on human chromosome X and somatic mutations are primarily observed in males, suggesting its lossof- function in MDS.1 It is involved in 3’ splice site recognition and interacts with the U2AF2/U2AF1 heterodimer and SRSF2 during pre-spliceosome assembly.21 ZRSR2 is recruited in an ATP-dependent fashion to the U12-type intron splice site, and is required for the formation of the spliceosome complex.22 We have previously illustrated that either truncating mutations of ZRSR2 in MDS or its silencing in acute myeloid leukemia (AML) cells impair predominantly splicing of U12-type introns, excision of which is mediated by minor spliceosome assembly.8 However, animal models of ZRSR2 deficiency have not been reported and an in-depth understanding of its function is lacking.
In this study, we have generated the first mouse model of ZRSR2 deficiency to further uncover its function in splicing and to evaluate the effect of its loss on normal and malignant hematopoiesis. Although deletion of Zrsr2 induces aberrant splicing of U12-type introns in mouse myeloid precursors, myeloid differentiation is largely unaffected in young, as well as ≥1-year old Zrsr2 knockout (KO) male mice. ZRSR2-deficient hematopoietic stem cells (HSC) retain multilineage reconstitution ability, suggesting limited function of ZRSR2 in mouse hematopoiesis. We further investigated its closely related homolog, ZRSR1, and uncovered that it compensates for the loss of ZRSR2 in mouse hematopoietic cells, and concurrent deficiency of both proteins leads to more profound defects in splicing of U12-type introns. Hence, murine models lacking both ZRSR2 and ZRSR1 are necessary to replicate faithfully mis-splicing caused by lossof- function mutations of ZRSR2 in MDS.
Methods
Generation of Zrsr2 knockout mice
C57BL/6N embryonic stem (ES) cells (Zrsr2tm1(KOMP)Vlcg) with targeted deletion of mouse Zrsr2 were obtained from UC Davis KOMP Repository. Mice with a constitutive deletion of Zrsr2 were generated at the transgenic facility of CSI Singapore. Briefly, ES cells were microinjected into BALB/cJInv blastocysts and resulting chimera were mated with C57BL/6 mice. Black offspring were genotyped, and mice carrying the Zrsr2 null allele were used to establish a colony of Zrsr2 knockout (KO) mice. Subsequently, mice were crossed with CMV-Cre strain to remove the neomycin selection cassette. In this study, Zrsr2 KO mice refer to those before or after Cre-mediated excision, as they were phenotypically identical in all experiments. Primers used for genotyping are listed in the Online Supplementary Table S1.
All animal experiments were approved by the Institutional Animal Care and Use Committee of the National University of Singapore, Singapore.
Flow cytometric analysis and fluorescence-activated cell sorting
Single cell suspensions from bone marrow (BM), spleen and thymus were stained with fluorochrome-conjugated antibodies for 30 minutes. The lineage cocktail was comprised of antibodies targeting murine CD19, CD3ε, CD11b, Gr1 and TER119. Cells were washed with 2% fetal bovine serum in phosphate buffered saline and resuspended in SYTOX Blue Dead Cell Stain (ThermoFisher Scientific). Cells were acquired on FACS LSR II (BD Biosciences) and data were analyzed using FACSDIVA software (BD Biosciences). Cells were sorted on FACSAria cell sorter (BD Biosciences).
RNA sequencing
Total RNA was extracted from sorted myeloid precursors (common myeloid precursors [CMP], granulocyte monocyte precursors [GMP] and megakaryocyte erythroid precursors [MEP]), murine embryonic fibroblasts [MEF] and ex vivo cultured Lin−Kit+ BM cells using either RNeasy Micro or Mini Kits (Qiagen). Libraries of polyA-selected RNA were prepared using either TruSeq sample preparation kit (CMP, GMP, MEP and MEF) or NEBNext Ultra RNA Library Prep Kit (Lin−Kit+ BM cells). Libraries were sequenced on HiSeq 4000, and paired-end reads were mapped to either mouse reference transcriptome (GRCm38/mm10; Ensemble version 84) or human hg38 reference genome using the STAR aligner23 with params ‘-- outSAMstrandField intronMotif --alignSJDBoverhangMin 6 -- alignIntronMax 299999 --outFilterMultimapNmax 4 -- scoreGapATAC -4’.
Differential splicing analysis
A list of valid introns was extracted from the Gencode24 gene transfer format file post removing transcripts with biotype ‘retained_intron’. Introns overlapping with an exon at their junctions were removed using pybedtools25,26 and gffutils (https://github.com/daler/gffutils). They were subsequently classified as either U2-type or U12-type based on position weight matrices from splicerack using gimmemotifs.27,28
MSI (mis-splicing index) values were calculated for each intron as described before.8 Only those introns with a coverage of at least one read at each of their junctions and with total coverage of the two junctions higher than four were considered. Also, introns which did not have a coverage of at least one for 95% of their length were filtered out. In order to identify differentially spliced introns, differences in MSI values (DMSI) were calculated as DMSI=MSIknockout−MSIwildtype. Significance of this difference was computed using a Fisher’s test, and the obtained Pvalues were corrected for multiple testing.
The python and R scripts used for analyses are available at https://github.com/pd321/intron-retention-scripts.
Accession codes
RNA sequencing data were deposited in Gene Expression Omnibus database repository under accession numbers GSE151470 and GSE152432.
Results
Aberrant retention of U12-type introns in ZRSR2-deficient murine hematopoietic cells
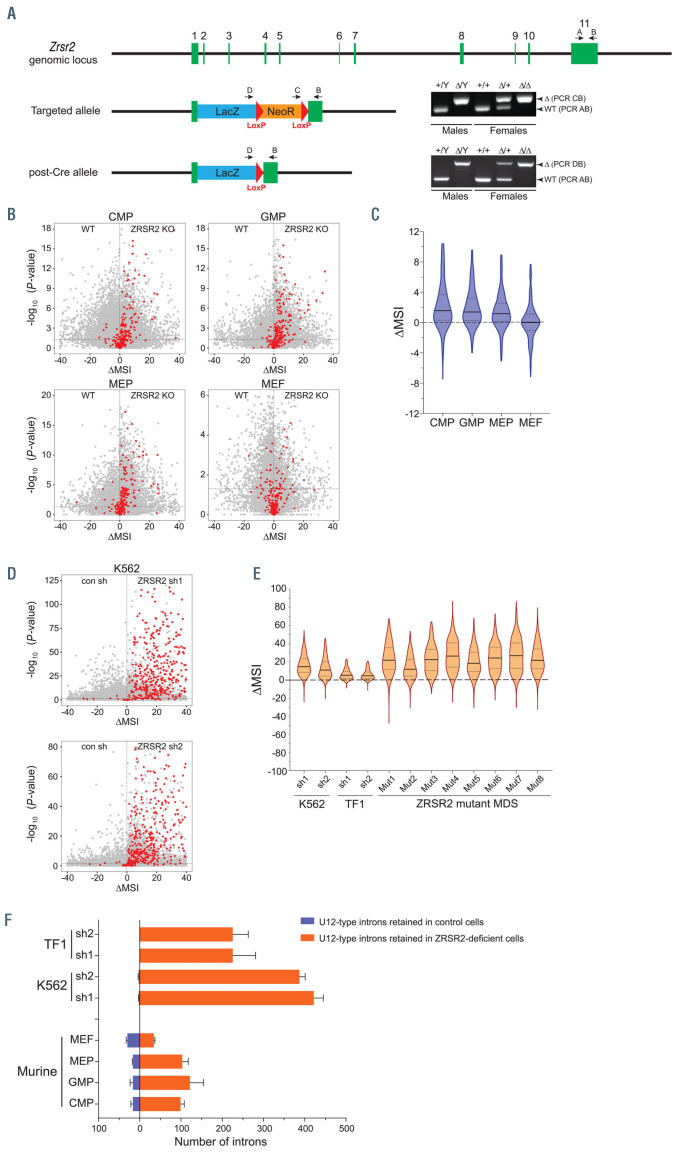
Mice lacking the entire Zrsr2 coding sequence were obtained by germline transmission of the targeted allele (Figure 1A). Following this, neomycin selection cassette was removed through mating with CMV-Cre transgenic mice, in effect replacing the Zrsr2 coding sequence with β- galactosidase gene (Figure 1A). Complete lack of Zrsr2 transcripts was evident in quantitative polymerase chain reaction (qPCR) analysis of BM, spleen and thymus cells, as well as RNA sequencing of targeted ES cells (Online Supplementary Figure S1A and B). Loss of Zrsr2 did not alter expression of its homolog, Zrsr1, in hematopoietic cells (Online Supplementary Figure S1B).
In order to assess the effect of ZRSR2 deficiency on RNA splicing, we performed RNA sequencing on sorted wild-type (WT) and KO myeloid precursor populations from BM: CMP (Lin−Kit+Sca1−CD34+FcγRII/IIIlo), GMP (Lin−Kit+Sca1−CD34+FcγRII/IIIhi), MEP (Lin−Kit+Sca1−CD34−FcγRII/III−), as well as MEF from male WT and Zrsr2 KO embryos. Aberrant retention of U12- type introns was observed in all three ZRSR2-deficient myeloid progenitors, but was not evident in MEF (Figure 1B and C; Online Supplementary Table S2). Despite a trend towards mis-splicing of U12-type introns in murine hematopoietic cells, we noted that the effect of ZRSR2 deficiency on splicing (as indicated by DMSI values) was significantly lower than that previously observed by us in ZRSR2 mutant human MDS BM and ZRSR2-deficient human AML cell lines, TF1 and K562 (Figure 1B and E; Online Supplementary Table S3).8 The number of aberrantly retained U12-type introns in Zrsr2 KO murine myeloid precursors was notably lower than ZRSR2 knockdown K562 and TF1 cells (Figure 1F). Only a modest effect on splicing was unexpected, especially given a complete loss of ZRSR2 expression in our mouse model. Moreover, U12-type introns are highly conserved between human and mouse genomes (Online Supplementary Figure S2), so a similar effect of ZRSR2 deficiency would be expected in the two species. Overall, our findings indicated reduced dependence of the U12-spliceosome on ZRSR2 in murine myeloid cells.
Figure 1.
Deficiency of ZRSR2 causes aberrant retention of U12- type introns in murine myeloid cells. (A) Generation of Zrsr2 knockout (KO) mice. Mice carrying the targeted Zrsr2 allele were crossed with CMV-Cre mice to excise the neomycin resistance cassette (post-Cre allele). Polymerase chain reaction analysis (right) shows genotyping of deleted Zrsr2 alleles in male and female mice. (B) Dot plots show intron retention in Zrsr2-deficent murine myeloid precursors (CMP, GMP and MEP) and MEF compared to wild-type (WT) cells. U12- type introns are depicted as red circles. (C) Range of DMSI values for retention of U12-type introns in murine cells. Outliers were removed from the plot using Gout method (Q=1). (D) Intron retention in ZRSR2 knockdown K562 cells (expressing either short hairpin RNA [shRNA] shRNA1 or shRNA2) compared to control transduced cells. (E) Range of DMSI values for intron retention (U12-type introns) in TF1, K562 and myelodysplastic syndromes (MDS) bone marrow cells. Outliers were removed from the plot using Gout method (Q=1). RNA sequencing data of ZRSR2- deficient TF1 cells and ZRSR2 mutant MDS used in this analysis has been previously published.8 (F) Number of U12-type introns retained in murine and human cells lacking ZRSR2 (P<0.05; Fisher's exact test). CMP: common myeloid precursors; GMP: granulocyte monocyte precursors; MEP: megakaryocyte erythroid precursors; MEF: murine embryonic fibroblasts; MSI: mis-splicing index. Difference in MSI values (DMSI) was calculated as DMSI=MSIknockout−MSIwild-type.
Normal myeloid development in mice lacking ZRSR2
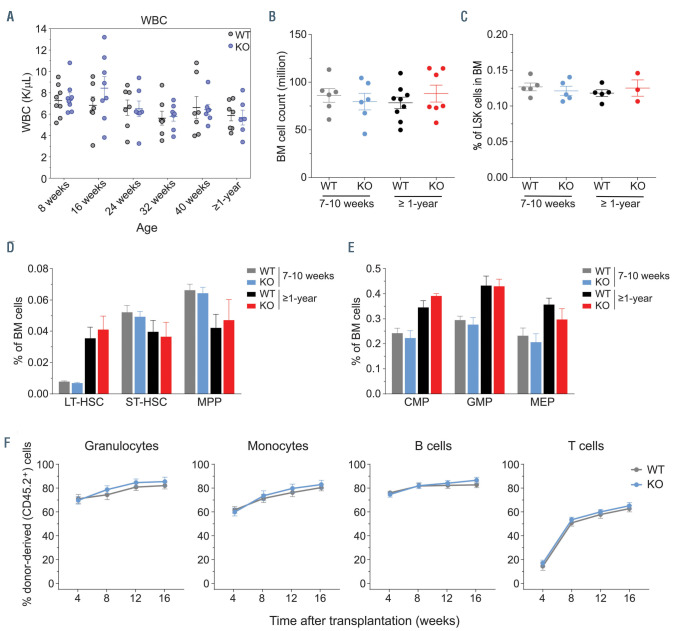
We analyzed hematopoietic compartment in ZRSR2- deficient (Zrsr2D/Y) compared to WT (Zrsr2+/Y) male mice. No significant difference was observed in peripheral blood counts of young mice of both genotypes; and age-dependent defects were also not apparent (Figure 2A; Online Supplementary Figure S3). Initial analysis of young males (7-10 weeks old) showed that deficiency of ZRSR2 neither affected the BM cellularity nor the frequency of Lin−Sca1+Kit+ (LSK) cells (Figure 2B and C). Proportion of HSC (CD34–Flt3– LSK cells) and progenitors also remained unaltered in young male mice (Figure 2D). MDS is primarily a disease of elderly, and in order to understand if loss of ZRSR2 manifested its effect with aging, BM of ≥1-year old male mice were examined. Surprisingly, old ZRSR2- deficient males also exhibited normal BM cellularity and frequencies of HSC and multipotent progenitors (Figure 2B to D). Moreover, proportion of myeloid precursors, CMP, GMP and MEP, and stages of erythroid development were also largely unaltered in the BM of young and old ZRSR2- deficient male mice (Figure 2E; Online Supplementary Figure S4A). Normal myeloid differentiation was also evident by unchanged frequencies of granulocytes in spleen and BM of Zrsr2 KO mice (Online Supplementary Figure S4B).
In order to evaluate the repopulation ability of Zrsr2 KO HSC, competitive repopulation assays were performed. ZRSR2-deficient HSC reconstituted both myeloid and lymphoid lineages in recipient mice as efficiently as WT cells (Figure 2F), suggesting that repopulation potential of HSC is maintained in the absence of ZRSR2. Further, in non-competitive repopulation assays, loss of ZRSR2 did not affect the peripheral blood cell counts in recipient mice even 1 year after transplantation (Online Supplementary Figure S5). Collectively, our comprehensive analyses of hematopoietic development in Zrsr2 KO mice and reconstitution ability of ZRSR2-deficient HSC demonstrated that ZRSR2 is not essential for hematopoietic development in mice.
Murine Zrsr1 is a putative functional copy of Zrsr2
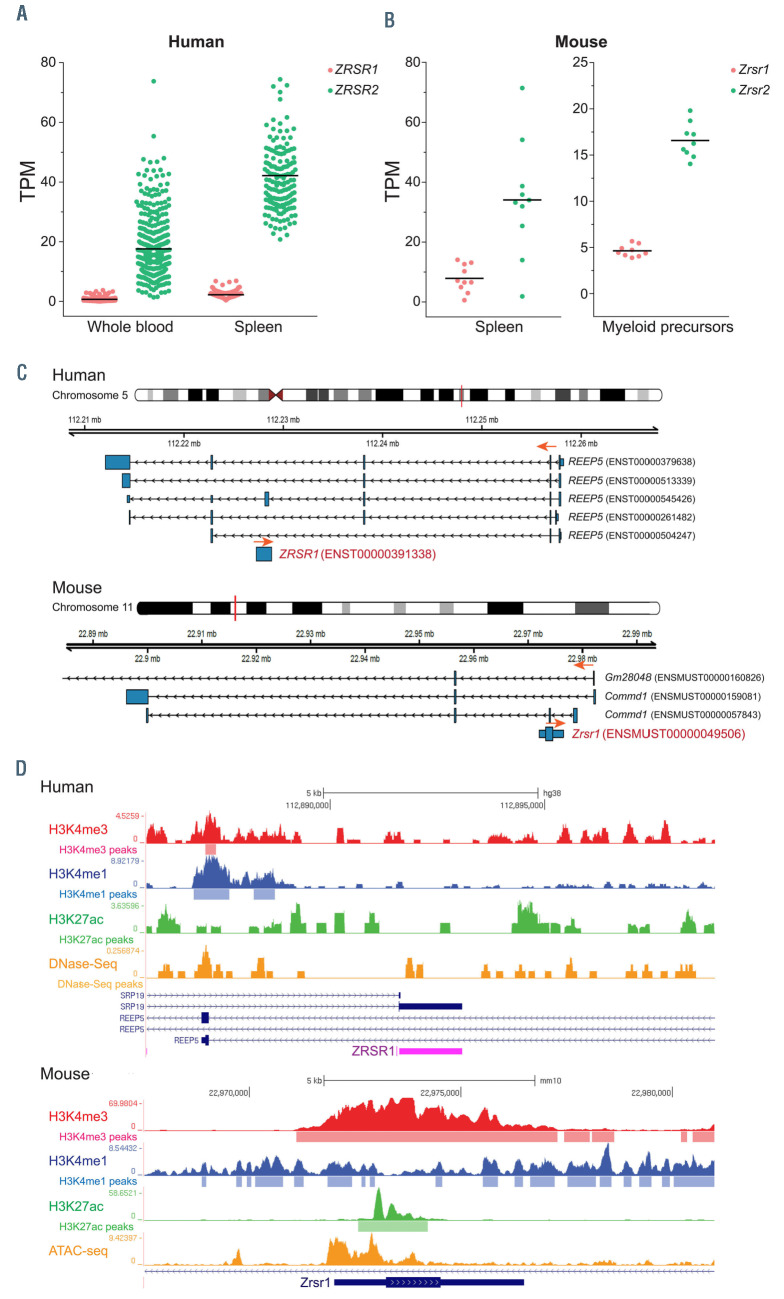
Given our unexpected observations that deletion of Zrsr2 in mice did not impact hematopoietic development and modestly affected splicing of U12-type introns, we postulated that other spliceosome protein(s) might compensate for its absence. ZRSR1, a closely-related homolog, is a single exon gene formed by retrotransposition of ZRSR2 cDNA sequence. This autosomal copy of X-linked ZRSR2 gene is highly similar to the parent gene in coding sequence (95% and 77% identical in human and mouse, respectively) and amino acid sequences (92% and 75% identical in human and mouse, respectively) with a conserved open reading frame. While ZRSR2 is ubiquitously expressed, human ZRSR1 is designated as a pseudogene with negligible transcript levels in human tissues (https://gtexportal.org/home/), including the hematopoietic cells (Figure 3A). In contrast, the mouse Zrsr1 gene is expressed in hematopoietic cells, albeit at levels lower than Zrsr2 (Figure 3B). This difference in transcriptional regulation of murine and human ZRSR1 genes is possibly caused by their location in distinct, nonorthologous genomic loci (Figure 3C). Human ZRSR1 is located in the intron of the REEP5 gene on chromosome 5q while the mouse gene localizes to the first intron of the Commd1 gene on chromosome 11 (Figure 3C), a genomic locus not syntenic to human chromosome 5. This demonstrates that retrotransposition of ZRSR2 gene occurred independently in rodents and primates, after the evolutionary divergence between the two mammalian orders.
Furthermore, we inspected the chromatin structure and presence of histone marks associated with transcriptional activation at the ZRSR1 locus in human and murine cells, using available DNase-seq/ATAC-seq and ChIP-Seq data, respectively. While no ZRSR1 locus-specific signal was observed for histone marks associated with gene activation (H3K4me3, H3K4me1 and H3K27ac) in human common myeloid progenitors, significant ChIP-Seq peaks for all three histone modifications were detected in murine hematopoietic cells (Figure 3D). Correspondingly, accessible chromatin was evident the murine but not human ZRSR1 locus (Figure 3D). Based on epigenetic profiles and expression levels, we hypothesized that murine ZRSR1 encodes for a functional protein that could possibly regulate splicing of the U12-type introns.
Loss of ZRSR1 impairs splicing of U12-type introns in the absence of ZRSR2
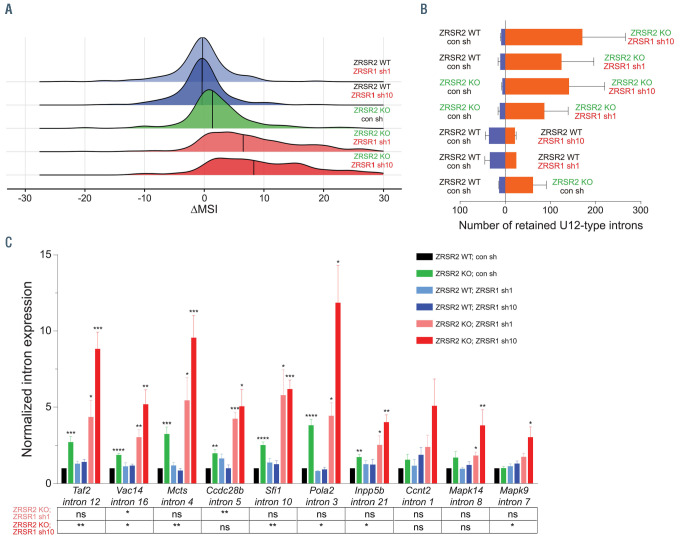
In order to investigate if ZRSR1 functionally compensates for deficiency of ZRSR2 in splicing of U12-type introns, we silenced expression of Zrsr1 in murine myeloid precursors using short hairpin RNA (shRNA) (Online Supplementary Figure S6A). Stable knockdown of Zrsr1 in WT Lin−Kit+ BM cells did not notably alter splicing of the U12-type introns, although it resulted in a reduced number of myeloid colonies in the methylcellulose medium (Figure 4A; Online Supplementary Figures S6B and S7A; Online Supplementary Table S4). Zrsr2 KO myeloid cells exhibited sizable retention of the U12-type introns (Figure 4A and B; Online Supplementary Figure S7A; Online Supplementary Table S4), similar to that previously described for sorted CMP, GMP and MEP populations (Figure 1B and C). Notably, knockdown of Zrsr1 in ZRSR2-deficient myeloid cells further exacerbated missplicing of the U12-type introns (Figure 4A and B; Online Supplementary Figure; Online Supplementary Table S4), suggesting that ZRSR1 can also regulate their splicing. Comparison of Zrsr2 single KO with Zrsr2/Zrsr1 doubledeficient Lin−Kit+ BM cells showed two subclasses of retained introns in Zrsr2/Zrsr1 double deficient cells. The first subclass comprises those introns which are retained in Zrsr2 KO cells, and their DMSI increases significantly in double-deficient cells; while the second category includes introns which are retained only when both ZRSR1 and ZRSR2 are lacking (Online Supplementary Figure S7B).
Retention of U12-type introns in the Zrsr2/Zrsr1 double- deficient cells was validated by quantitative real-time PCR (qRT-PCR) analysis of 10 introns, which illustrated a modest effect of loss of ZRSR2 alone on splicing of U12- type introns, while concomitant deficiency of ZRSR1 and ZRSR2 significantly enhanced intron retention compared to either WT or single KO cells (Figure 4C).
Further, in order to verify the compensatory role of murine ZRSR1 in splicing of U12-type introns, CRISPR/Cas9 technique was used to generate myeloid cells (32D) lacking either one or both ZRSR proteins. Firstly, the whole Zrsr2 coding sequence was deleted using two guide RNA targeting exons 2 and 11 of Zrsr2 gene (Online Supplementary Figure S8A and B). Two single cell clones of Zrsr2 KO 32D cells were generated (Online Supplementary Figure S8B and C). Subsequently, we deleted the N-terminus portion of the Zrsr1 gene to generate single or double KO 32D cells (Online Supplementary Figure S8A, B, D and E). qRT-PCR analysis demonstrated aberrant retention of U12-type introns in Zrsr1/Zrsr2 double-deficient cells compared to control cells (Online Supplementary Figure S8F).
Figure 2.
Hematopoietic development and reconstitution potential are unperturbed upon deletion of Zrsr2. (A) Peripheral blood white blood cell (WBC) counts in wild-type (WT) and Zrsr2 knockout (KO) mice. (B) Total bone marrow (BM) leukocyte counts (femurs + tibias) in young (7-10 weeks) and old (≥1-year old) WT and Zrsr2-deficient mice. (C) Proportion of LSK cells in the BM of young and old WT and Zrsr2 KO mice. (D and E) Frequencies of LT-HSC, ST-HSC and MPP (D) (Young mice: five WT and five KO; Old mice: five WT and three KO) and myeloid precursors (CMP, GMP, MEP) (E) (Young mice: five WT and five KO; Old mice: six WT and four KO). (F) Donor-derived B cells, T cells, granulocytes and monocytes in peripheral blood of recipient mice in competitive repopulation assays (six recipient mice/group). HSC: hematopoietic stem cells; CMP: common myeloid precursors; GMP: granulocyte monocyte precursors; MEP: megakaryocyte erythroid precursors.
Figure 3.
Human and mouse ZRSR1 are located in non-orthologous genomic loci. (A) Levels of ZRSR1 and ZRSR2 transcripts in human whole blood and spleen cells. Expression data are collated from Genotype-Tissue Expression (GTEx) portal. (B) Expression levels of Zrsr1 and Zrsr2 transcripts in murine spleen cells (Bgee database) and myeloid precursors (in-house RNA sequencing of sorted CMP, GMP and MEP cells). TPM: transcripts per million reads. (C) Genomic location of human and murine ZRSR1 genes. Orange arrows denote the direction of transcription for each gene. (D) H3K4me3, H3K4me1 and H3K27ac ChIP-seq signals and regions of open chromatin at human and murine ZRSR1 locus. All data were downloaded from the ENCODE Consortium. Human ChIP-seq and DNase-seq data are for common myeloid progenitors, while murine data are from following sources - H3K4me3: CD1 embryonic erythroblasts; H3K4me1: CD1 embryonic megakaryocytes; H3K27ac: bone marrow macrophages; ATAC-seq: adult erythroblasts; CMP: common myeloid precursors; GMP: granulocyte monocyte precursors; MEP: megakaryocyte erythroid precursors.
Figure 4.
Loss of ZRSR1 exacerbates retention of U12-type introns in ZRSR2-deficient murine hematopoietic cells. (A) Distribution of DMSI values for retention of U12-type introns in Lin−Kit+ bone marrow (BM) cells lacking either ZRSR1 (sh1 or sh10) or ZRSR2 or both compared to control cells. (B) Number of U12-type introns retained (P<0.05; Fisher's exact test) in various pair-wise comparisons of Lin−Kit+ BM cells deficient in either one or both ZRSR proteins. (C) Normalized expression of representative U12-type introns in Lin−Kit+ BM cells determined using quantitative real-time polymerase chain reaction. Expression of U12-type introns was measured relative to expression of flanking exons. Data are from at least three replicates and represented as mean ± standard error of the mean. P-values for each group compared to the ‘ZRSR2 WT; con sh’ group are depicted in the plot. Statistical difference between the Zrsr2/Zrsr1-double deficient and Zrsr2 KO cells are shown below the graph. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; ns: not significant; MSI: mis-splicing index: Difference in MSI values (DMSI) was calculated as DMSI=MSIknockout−MSIwildtype.
In order to verify that human ZRSR1 is a non-functional copy of ZRSR2, we generated K562 cells lacking either ZRSR1 or ZRSR2 or both proteins together. DZRSR1 cells were first generated using CRISPR/Cas9 technique followed by silencing of ZRSR2 using shRNA. As expected, ZRSR2 deficiency alone impaired splicing of U12-type introns. However, loss of ZRSR1, either alone or when combined with ZRSR2, did not significantly alter the splicing of U12-type introns in K562 cells (Online Supplementary Figure S9). Taken together, our experiments highlight a functional role of murine ZRSR1 in splicing of the U12-type introns.
Mis-splicing of MAPK9 and MAPK14 impacts their protein expression in murine and human cells
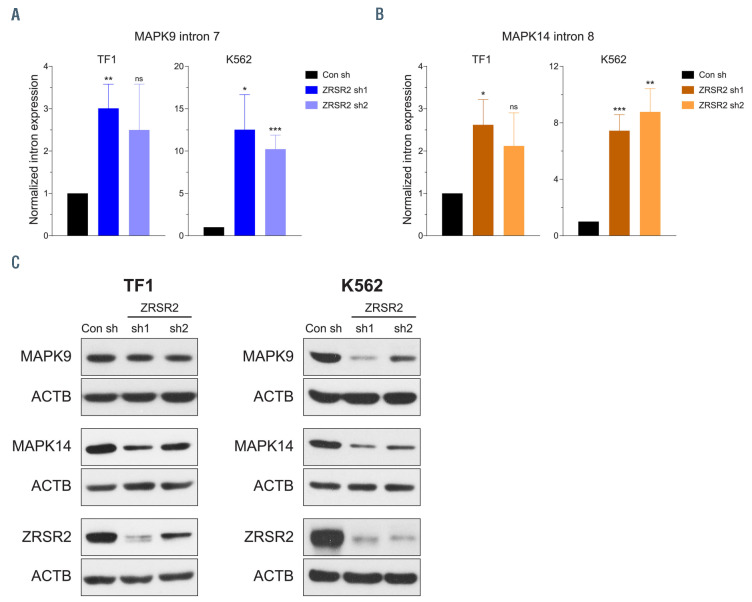
Several crucial genes harbour U12-type introns including members of the mitogen-activated protein kinase (MAPK) family of serine-threonine kinases. MAPK proteins play a crucial role in a wide range of biological processes including hematopoiesis.29 We focused on splicing of two members, MAPK9 (JNK2) and MAPK14 (p38α), as aberrant retention of their U12-type introns was verified in murine myeloid precursors deficient in both Zrsr1 and Zrsr2 (Figure 4C). Intron retention of both MAPK9 and MAPK14 resulted in the generation of aberrant transcripts containing a premature stop codon. In order to investigate if the retention of U12-type introns in these two genes affects their protein expression, western blotting was performed with anti-MAPK9 and anti- MAPK14 antibodies. Protein levels of both MAPK9 and MAPK14 were reduced, albeit modestly, in murine myeloid precursors, and MAPK14 levels were also reduced in 32D cells lacking ZRSR2 and ZRSR1 (Online Supplementary Figure S10A and B). Retention of U12-type introns of MAPK9 and MAPK14 was also observed in ZRSR2 knockdown K562 and TF1 cells (Figure 5A and B). This was accompanied by significantly reduced MAPK9 and MAPK14 protein expression in human cells with silencing of ZRSR2 (Figure 5C). Amongst the two cell lines, K562 cells exhibited increased mis-splicing of both MAPK9 and MAPK14, which correlated with a more profound effect on their protein levels, compared to the TF1 cells (Figure 5A to C). These observations illustrate that mis-splicing of MAPK9 and MAPK14 results in lower levels of these proteins, potentially impacting their function in hematopoietic development.
Figure 5.
Aberrant retention of U12-type introns in human MAPK9 and MAPK14 genes impacts their protein expression. (A and B) Normalized expression of U12-type introns of MAPK9 (A) and MAPK14 (B) in TF1 and K562 cells stably expressing short hairpin RNA (shRNA) targeting human ZRSR2. The expression of U12-type introns was measured relative to the expression of flanking exons using quantitative polymerase chain reaction (qPCR). Data are from five PCR experiments and represented as mean ± standard error of the mean. *P<0.05, **P<0.01, ***P<0.001, ns: not significant. (C) Protein levels of human MAPK9 and MAPK14 in TF1 and K562 cells transduced with shRNA targeting ZRSR2.
Discussion
Mouse models have been effective in deciphering key splicing and hematopoietic defects caused by mutations of spliceosome genes. Hematopoietic cells from mice expressing either mutant SF3B1, SRSF2 or U2AF1 recapitulated the patterns of splicing changes observed in mutant MDS samples.6,12,15,16,18-20 Although the splice factors are evolutionary conserved, intronic sequences are divergent between human and mouse. These differences in intronic splicing elements and regulatory motifs between the two species possibly lead to a divergent set of mis-spliced transcripts, which can contribute to phenotypic differences in hematopoietic development. This presents a conundrum about the utility of mouse models to identify genes whose splicing is altered by spliceosome mutation in MDS. We have previously demonstrated that ZRSR2 mutations/deficiency impairs splicing of the U12- type intron in human cells.8,17 U12-type introns are highly conserved during evolution, and the tissue-specific expression of transcripts harboring this class of introns is largely preserved between mouse and human.30,31 Therefore, we envisaged that a murine model would faithfully replicate the loss of ZRSR2 in leukemic human cells. Indeed, murine ZRSR2 KO myeloid cells exhibited aberrant retention of U12-type introns. Unexpectedly, splicing of the U12-type introns was unaffected in ZRSR2-deficient MEF, suggesting tissue-specific effects of ZRSR2-deficiency on splicing. A previous study also demonstrated a more pronounced mis-splicing of U12- type introns in blood cells compared to fibroblast and amniocytes in Taybi–Linder syndrome (TALS) cases harboring germline mutations of a U12 spliceosome-specific component, RNU4ATAC.32
One limitation of our constitutive KO mouse model is that complete absence of ZRSR2 caused by germline loss of ZRSR2 might promote functional compensation during development. Therefore, findings from our KO mice may not reflect the consequences of acute loss of ZRSR2 in myeloid cells. Hematopoietic cell-specific inducible models of ZRSR2 deficiency are needed to further elucidate the effect of conditional depletion of ZRSR2 during adult hematopoiesis, akin to what is observed in ZRSR2 mutant MDS.
Mice with constitutive ZRSR2 deletion developed normally with no overt hematopoietic defects and functionally competent hematopoietic stem cells. This is in contrast with mice expressing mutant SF3B1, SRSF2 or U2AF1, which displayed a range of hematopoietic phenotypes. 6,12,15,16,18-20,33 Isolated loss-of-function mutations of ZRSR2 have been associated with macrocytic anaemia in patients with myeloid diseases.34 However, our Zrsr2 KO mice did not display any significant changes in size and counts of blood cells. Moreover, while ZRSR2 deficiency affected splicing of U12-type introns in murine hematopoietic cells, magnitude of splicing defects was modest compared to either MDS BM cells or human leukemia cell lines. This led us to investigate whether ZRSR1 can functionally compensate for the lack of ZRSR2.
ZRSR1 is a retrotransposed copy of ZRSR2, which originated via independent transposition events in rodents and primates. Murine Zrsr1 is an imprinted gene and expressed from the unmethylated, paternal allele.35-40 Unlike the human counterpart, which is designated as a pseudogene, the murine gene is expressed in hematopoietic cells, albeit at lower levels compared with Zrsr2. Our analyses revealed open chromatin and enrichment of epigenetic marks associated with transcriptional activation only at the murine Zrsr1 locus, supporting evidence for transcription of the murine retrogene.
In our study, silencing of Zrsr1 had no evident effect on the splicing of U12-type introns in Zrsr2 WT myeloid cells. In contrast, deficiency of ZRSR1 clearly exacerbated the mis-splicing of U12-type introns in cells lacking ZRSR2, thereby underlining that both ZRSR1 and ZRSR2 collectively contribute to splicing of U12-type introns in mouse hematopoietic cells. While RNA splicing was not studied in Zrsr1 KO mice,41 recent studies have shown that expression of truncated Zrsr1 impacts splicing of the U12-type introns in testis and hypothalamus.42,43 Interestingly, Zrsr1-mutant mice exhibit defects in erythrocyte maturation and fewer peripheral red blood cells, with apparent morphological abnormalities.42 The differences observed in impact of ZRSR1 deficiency on splicing of U12-type introns in our study (myeloid precursors) versus studies investigating mutant Zrsr1-expressing spermatocytes/ hypothalamus could possibly arise because of relative levels of ZRSR2 and ZRSR1 in different cell types. Another possibility is that the truncated ZRSR1 can impair recruitment of ZRSR2 to U12-type intron splice site, thereby perturbing its splicing function.
Signal transduction mediated by MAP kinase pathway plays a vital role in numerous biological processes via phosphorylation of several downstream substrates.44 Genes encoding several members of MAP kinase family harbor U12-type introns, hence their splicing is dependent on ZRSR2 activity. We identified two candidates, MAPK9 and MAPK14, where aberrant splicing resulted in decreased protein levels both in human and murine cells. The effect of ZRSR2/ZRSR1 double deficiency on MAPK9 and MAPK14 protein levels in murine cells was less pronounced compared to human cells lacking ZRSR2, which corresponds with milder effect on splicing of U12- type introns in murine cells. While MAPK9 regulates Tcell apoptosis and proliferation, and MAPK14 is indispensable for definitive erythropoiesis in mice,45,46 these proteins have not been directly implicated in pathogenesis of MDS. Although we experimentally validated mis-splicing of just two of the MAPK members, a broader effect on splicing of multiple components can be envisaged. Moreover, in murine myeloid precursors, we also validated aberrant retention of U12-type intron of MCTS1, which modulates MAPK pathway by promoting phosphorylation of MAPK1 and MAPK3.47 Given an essential role of MAPK proteins in regulating hematopoiesis,29 collective decrease in their protein levels can potentially be detrimental to myeloid/erythroid differentiation and expansion, thereby contributing to the disease phenotype in MDS.
Taken together, unlike human ZRSR1 pseudogene, Zrsr1 in mice is a functional autosomal copy of Zrsr2 and contributes to splicing of U12-type introns. This is also supported by a recent study which demonstrated that expression of at least one copy of either maternal Zrsr2 or paternal Zrsr1 is necessary for viability of murine embryos.48 Additionally, our study highlights that splicing of U12-type introns in murine cells depends conceivably on the balance between expression levels of ZRSR2 and ZRSR1. Hence, deficiency of ZRSR2 alone is insufficient to impact extensively RNA splicing in mice, and further studies with concurrent deficiency of ZRSR1 and ZRSR2 are warranted to replicate complete loss of ZRSR activity. Notably, germline expression of truncated Zrsr1 and Zrsr2 alleles showed that double mutant mice are non-viable, with Zrsr1/Zrsr2 double mutant embryos exhibiting defects in early preimplantation development.48 Hence, conditional KO alleles of both Zrsr1 and Zrsr2 are required to investigate their combined loss in adult mice.
Supplementary Material
Acknowledgements
We thank the staff of Comparative Medicine, NUS for their support in maintaining mouse colonies. We also acknowledge expert help and support from the FACS facility at CSI, Singapore.
Funding Statement
Funding: This work was funded by the Leukemia and Lymphoma Society, the Singapore Ministry of Health’s National Medical Research Council (NMRC) under its Singapore Translational Research (STaR) Investigator Award to HPK (NMRC/STaR/0021/2014), the NMRC Center Grant awarded to the National University Cancer Institute of Singapore (NMRC/CG/012/2013) and the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centers of Excellence initiatives. This research is also supported by the RNA Biology Center at the Cancer Science Institute of Singapore, NUS, as part of funding under the Singapore Ministry of Education’s Tier 3 grants, grant number MOE2014-T3-1-006. We thank the Melamed Family for their generous support.
References
- 1.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64-69. [DOI] [PubMed] [Google Scholar]
- 2.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, Cazzola M, Boultwood J, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365(15):1384-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damm F, Kosmider O, Gelsi-Boyer V, et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood. 2012;119(14):3211-3218. [DOI] [PubMed] [Google Scholar]
- 6.Kim E, Ilagan JO, Liang Y, et al. SRSF2 Mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27(5):617-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komeno Y, Huang YJ, Qiu J, et al. SRSF2 Is Essential for hematopoiesis, and its myelodysplastic syndrome-related mutations dysregulate alternative pre-mRNA splicing. Mol Cell Biol. 2015;35(17):3071-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madan V, Kanojia D, Li J, et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat Commun. 2015;6:6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Lieu YK, Ali AM, et al. Diseaseassociated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci U S A. 2015;112(34):E4726-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ilagan JO, Ramakrishnan A, Hayes B, et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2015;25(1):14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okeyo-Owuor T, White BS, Chatrikhi R, et al. U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia. 2015;29(4):909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirai CL, Ley JN, White BS, et al. Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell. 2015;27(5):631-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alsafadi S, Houy A, Battistella A, et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun. 2016;7:10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darman RB, Seiler M, Agrawal AA, et al. Cancer-Associated SF3B1 hotspot mutations induce cryptic 3' splice site selection through use of a different branch point. Cell Rep. 2015;13(5):1033-1045. [DOI] [PubMed] [Google Scholar]
- 15.Mupo A, Seiler M, Sathiaseelan V, et al. Hemopoietic-specific Sf3b1-K700E knock-in mice display the splicing defect seen in human MDS but develop anemia without ring sideroblasts. Leukemia. 2017;31(3):720-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obeng EA, Chappell RJ, Seiler M, et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell. 2016;30(3):404-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madan V, Li J, Zhou S, et al. Distinct and convergent consequences of splice factor mutations in myelodysplastic syndromes. Am J Hematol. 2020;95(2):133-143. [DOI] [PubMed] [Google Scholar]
- 18.Fei DL, Zhen T, Durham B, et al. Impaired hematopoiesis and leukemia development in mice with a conditional knock-in allele of a mutant splicing factor gene U2af1. Proc Natl Acad Sci U S A. 2018;115(44):E10437-E10446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kon A, Yamazaki S, Nannya Y, et al. Physiological Srsf2 P95H expression causes impaired hematopoietic stem cell functions and aberrant RNA splicing in mice. Blood. 2018;131(6):621-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smeets MF, Tan SY Xu JJ, et al. Srsf2(P95H) initiates myeloid bias and myelodysplastic/ myeloproliferative syndrome from hemopoietic stem cells. Blood. 2018;132(6): 608-621. [DOI] [PubMed] [Google Scholar]
- 21.Tronchere H, Wang J, Fu XD. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature. 1997;388(6640): 397-400. [DOI] [PubMed] [Google Scholar]
- 22.Shen H, Zheng X, Luecke S, Green MR. The U2AF35-related protein Urp contacts the 3' splice site to promote U12-type intron splicing and the second step of U2- type intron splicing. Genes Dev. 2010;24(21):2389-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frankish A, Diekhans M, Ferreira AM, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766-D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dale RK, Pedersen BS, Quinlan AR. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics. 2011;27(24):3423-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheth N, Roca X, Hastings ML, Roeder T, Krainer AR, Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34(14):3955-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Heeringen SJ, Veenstra GJ. GimmeMotifs: a de novo motif prediction pipeline for ChIP-sequencing experiments. Bioinformatics. 2011;27(2):270-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geest CR, Coffer PJ. MAPK signaling pathways in the regulation of hematopoiesis. J Leukoc Biol. 2009;86(2):237-250. [DOI] [PubMed] [Google Scholar]
- 30.Lin CF, Mount SM, Jarmolowski A, Makalowski W. Evolutionary dynamics of U12-type spliceosomal introns. BMC Evol Biol. 2010;10:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olthof AM, Hyatt KC, Kanadia RN. Minor intron splicing revisited: identification of new minor intron-containing genes and tissue- dependent retention and alternative splicing of minor introns. BMC Genomics. 2019;20(1):686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cologne A, Benoit-Pilven C, Besson A, et al. New insights into minor splicing-a transcriptomic analysis of cells derived from TALS patients. RNA. 2019;25(9):1130-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu JJ, Smeets MF, Tan SY, Wall M, Purton LE, Walkley CR. Modeling human RNA spliceosome mutations in the mouse: not all mice were created equal. Exp Hematol. 2019;70:10-23. [DOI] [PubMed] [Google Scholar]
- 34.Fleischman RA, Stockton SS, Cogle CR. Refractory macrocytic anemias in patients with clonal hematopoietic disorders and isolated mutations of the spliceosome gene ZRSR2. Leuk Res. 2017;61:104-107. [DOI] [PubMed] [Google Scholar]
- 35.Hatada I, Sugama T, Mukai T. A new imprinted gene cloned by a methylationsensitive genome scanning method. Nucleic Acids Res. 1993;21(24):5577-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayashizaki Y, Shibata H, Hirotsune S, et al. Identification of an imprinted U2af binding protein related sequence on mouse chromosome 11 using the RLGS method. Nat Genet. 1994;6(1):33-40. [DOI] [PubMed] [Google Scholar]
- 37.Hatada I, Kitagawa K, Yamaoka T, et al. Allele-specific methylation and expression of an imprinted U2af1-rs1 (SP2) gene. Nucleic Acids Res. 1995;23(1):36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shibata H, Yoshino K, Sunahara S, et al. Inactive allele-specific methylation and chromatin structure of the imprinted gene U2af1-rs1 on mouse chromosome 11. Genomics. 1996;35(1):248-252. [DOI] [PubMed] [Google Scholar]
- 39.Feil R, Boyano MD, Allen ND, Kelsey G. Parental chromosome-specific chromatin conformation in the imprinted U2af1-rs1 gene in the mouse. J Biol Chem. 1997;272(33):20893-20900. [DOI] [PubMed] [Google Scholar]
- 40.Nabetani A, Hatada I, Morisaki H, Oshimura M, Mukai T. Mouse U2af1-rs1 is a neomorphic imprinted gene. Mol Cell Biol. 1997;17(2):789-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sunahara S, Nakamura K, Nakao K, Gondo Y, Nagata Y, Katsuki M. The oocyte-specific methylated region of the U2afbp-rs/U2af1- rs1 gene is dispensable for its imprinted methylation. Biochem Biophys Res Commun. 2000;268(2):590-595. [DOI] [PubMed] [Google Scholar]
- 42.Horiuchi K, Perez-Cerezales S, Papasaikas P, et al. Impaired spermatogenesis, muscle, and erythrocyte function in U12 intron splicingdefective Zrsr1 mutant mice. Cell Rep. 2018;23(1):143-155. [DOI] [PubMed] [Google Scholar]
- 43.Alen F, Gomez-Redondo I, Rivera P, et al. Sex-dimorphic behavioral alterations and altered neurogenesis in U12 intron splicingdefective Zrsr1 mutant mice. Int J Mol Sci. 2019;20(14):3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75(1):50-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura K, Sudo T, Senftleben U, Dadak AM, Johnson R, Karin M. Requirement for p38alpha in erythropoietin expression: a role for stress kinases in erythropoiesis. Cell. 2000;102(2):221-231. [DOI] [PubMed] [Google Scholar]
- 46.Sabapathy K, Kallunki T, David JP, Graef I, Karin M, Wagner EF. c-Jun NH2-terminal kinase (JNK)1 and JNK2 have similar and stage-dependent roles in regulating T cell apoptosis and proliferation. J Exp Med. 2001;193(3):317-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu HL, Choy CO, Kasiappan R, et al. MCT-1 oncogene downregulates p53 and destabilizes genome structure in the response to DNA double-strand damage. DNA Repair (Amst). 2007;6(9):1319-1332. [DOI] [PubMed] [Google Scholar]
- 48.Gomez-Redondo I, Ramos-Ibeas P, Pericuesta E, Fernandez-Gonzalez R, Laguna-Barraza R, Gutierrez-Adan A. Minor splicing factors Zrsr1 and Zrsr2 are essential for early embryo development and 2-celllike conversion. Int J Mol Sci. 2020;21(11):4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.