Abstract
Background
Viral infections are often treated with empiric antibiotics due to suspected bacterial coinfections, leading to antibiotic overuse. We aimed to describe antibiotic resistance (ABR) trends and their association with the influenza season in ambulatory and inpatient settings in the United States.
Methods
We used the BD Insights Research Database to evaluate antibiotic susceptibility profiles in 30-day nonduplicate bacterial isolates collected from patients >17 years old at 257 US healthcare institutions from 2011 to 2019. We investigated ABR in Gram-positive (Staphylococcus aureus and Streptococcus pneumoniae) and Gram-negative (Enterobacterales [ENT], Pseudomonas aeruginosa [PSA], and Acinetobacter baumannii spp [ACB]) bacteria expressed as the proportion of isolates not susceptible ([NS], intermediate or resistant) and resistance per 100 admissions (inpatients only). Antibiotics included carbapenems (Carb), fluoroquinolones (FQ), macrolides, penicillin, extended-spectrum cephalosporins (ESC), and methicillin. Generalized estimating equations models were used to evaluate monthly trends in ABR outcomes and associations with community influenza rates.
Results
We identified 8 250 860 nonduplicate pathogens, including 154 841 Gram-negative Carb-NS, 1 502 796 Gram-negative FQ-NS, 498 012 methicillin-resistant S aureus (MRSA), and 44 131 NS S pneumoniae. All S pneumoniae rates per 100 admissions (macrolide-, penicillin-, and ESC-NS) were associated with influenza rates. Respiratory, but not nonrespiratory, MRSA was also associated with influenza. For Gram-negative pathogens, influenza rates were associated with the percentage of FQ-NS ENT, FQ-NS PSA, and Carb-NS ACB.
Conclusions
Our study showed expected increases in rates of ABR Gram-positive and identified small but surprising increases in ABR Gram-negative pathogens associated with influenza activity. These insights may help inform antimicrobial stewardship initiatives.
Keywords: antibiotic resistance, hospitals, influenza, seasonality, United States
Between 2011–2019, we identified associations in the US between increased antibiotic resistance and influenza rates for both Gram-positive and -negative pathogens, specifically fluoroquinolone-not susceptible (NS) Enterobacterales, fluoroquinolone-NS Pseudomonas aeruginosa, and carbapenem-NS Acinetobacter. These associations may inform antimicrobial stewardship initiatives.
Yearly influenza season invokes an interplay of many different respiratory viruses, including metapneumovirus and respiratory syncytial virus, which may incite secondary bacterial infections, particularly in patients at high risk due to structural lung disease, chronic conditions such as diabetes, or an immunocompromised state [1–4]. In addition, influenza infection may directly facilitate bacterial infections due to increased bacterial adherence, influenza-associated immunosuppression, and other mechanisms [3, 5]. Across various studies, approximately 11% to 35% of patients with laboratory-confirmed influenza have a bacterial coinfection [6], and patients with influenza and bacterial coinfections who develop community-acquired pneumonia (CAP) have more severe disease and higher mortality rates than CAP patients without coinfections [7]. Even in the absence of bacterial coinfection, many patients with respiratory conditions receive empiric treatment with antibiotics. In one study, 41% of ambulatory patients treated with empiric antibiotics due to respiratory symptoms did not have a diagnosis for which antibiotics were indicated [8]. The higher rates of both bacterial infections and antibiotic use during influenza seasons are thought to play a role in seasonal antibiotic resistance (ABR), particularly in respiratory isolates [9].
Much of the current literature has evaluated changes in resistance patterns for the pathogen to which a vaccine is directed (eg, Streptococcus pneumoniae resistance and the pneumococcal vaccine) or more commonly isolated Gram-positive respiratory copathogens such as Staphylococcus aureus. However, there are minimal data on seasonal influences on resistance in nonrespiratory and Gram-negative pathogens. A previous study based on the BD Insights Research Database (Franklin Lakes, NJ) detected seasonal trends in ABR Gram-negative pathogens isolated from inpatients, but it did not explore possible associations with these patterns or evaluate seasonality in ambulatory patients [10]. Aggregated facility-level resistance trends may be of value in choosing empiric therapies for potential secondary bacterial coinfections, particularly if vaccines are championed further upstream to help mitigate the inciting viral infection. Insights on increased isolation of resistant pathogens associated with respiratory virus season my help spearhead vaccination campaigns, which are increasingly needed. Furthermore, epidemiological knowledge of resistant pathogens that peak during respiratory virus surges may help forewarn and inform infection prevention and control and antimicrobial stewardship programs. The goal of our study was to explore US trends in antibiotic-resistant respiratory and nonrespiratory bacteria and the relationship of these trends with influenza rates derived from facilities in the BD Insights Research Database, which includes approximately 13% of all-cause annual hospital admissions in the United States [11].
METHODS
Study Design
We conducted a retrospective, ecological analysis to evaluate possible associations between community influenza and ABR rates using real-world data collected between 2011 and 2019 from patients >17 years old at US acute healthcare institutions in the BD Insights Research Database. This database includes small and large acute healthcare facilities in urban and rural areas with geographical representation across the United States [10, 12, 13]. The outcomes of interest were the percentage of ABR pathogens for both inpatients and ambulatory patients and the rates of ABR pathogens per 100 admissions (inpatients only). Outcomes were analyzed by setting (ambulatory or inpatient) and source (respiratory vs nonrespiratory [urine, blood, skin/wound, intraabdominal, and other sites]). Geographic localization was based on US Department of Health and Human Services (HHS) regions. Associations between ABR bacteria and community influenza rates were evaluated as described below. The influenza status of individual patients with positive bacterial cultures was not assessed.
Patient Consent
Outcome studies using this retrospective, deidentified dataset were approved, and informed consent was waived by the New England Institutional Review Board (Wellesley, MA; institutional review board no. 120180023).
Laboratory Analyses
Susceptibility results and pathogen identification were based on facility reports from healthcare facilities in the BD Insights Research Database. Microbiology results likely associated with surveillance or environmental cultures were excluded using previously described methodology [14].
Antibiotic resistance was based on a test result of resistance (for methicillin-resistant S aureus [MRSA]) or not susceptible ([NS] defined as intermediate or resistant) for specified pathogens and antibiotics as defined by local testing facilities. The following pathogen/antibiotic resistance patterns were assessed: (1) MRSA - S aureus with a test result of resistant to methicillin; (2) S pneumoniae - NS to macrolides, penicillin, or extended-spectrum cephalosporins (ESC); (3) Enterobacterales (ENT) and Pseudomonas aeruginosa (PSA) - NS to carbapenems (Carb) or fluoroquinolones (FQ); (4) Acinetobacter baumannii spp (AC) - NS to Carb; (5) Stenotrophomonas maltophilia - NS to FQ.
Influenza polymerase chain reaction and antigen laboratory data were used to determine facility-level influenza rates (positive results/100 tests). Although other respiratory viruses also display seasonal variations, influenza accounts for the majority of cases in adults [15] and was therefore chosen as the focus of respiratory virus evaluations.
Statistical Analysis
Influenza and ABR rates were calculated for each year-quarter Q1 2011 through Q4 2019 to best encapsulate the concept of a “season”. Sensitivity analyses were conducted at the monthly level(s) to test the extent of any potential misclassification, with year-quarters accurately capturing seasonal fluctuations in the influenza rate. Graphical representations were based on monthly levels to best highlight these seasonal trends.
Generalized estimating equations and logistic regression were used to evaluate the monthly trends for percentage of ABR isolates and the effect of the influenza season on ABR. Generalized linear mixed models were used to model the resistance rate per 100 admissions and explore associations between influenza and ABR rates. The time series data were viewed as repeated measures and acute healthcare facilities were modeled as random effect. These analyses were run separately by pathogen group while controlling for setting, source of isolates, and healthcare facility-level variables (region, bed size, and other characteristics). These analyses were then stratified by setting type and by source of isolates (respiratory vs nonrespiratory). The rates of ABR were graphically evaluated and superimposed to examine visual and statistical associations between the ABR time series and the influenza incidence time series. Results for the association between ABR and influenza are presented as β coefficients reflecting the degree of change in antimicrobial resistance for every 1-unit change in the influenza rate. Positive coefficients reflect positive associations.
We stratified the analyses by pathogen type and evaluated which pathogens were significantly associated with influenza rates, and whether these relationships varied by pathogen type (Gram negative vs Gram positive). By stratifying the analysis, we were able to assess the effect of other variables such as setting type and source of isolates on relationships between influenza season and ABR. All statistical analyses were conducted using R V 4.0.3 (R Core Team 2020) and the R geepack package. P < .05 was considered statistically significant.
RESULTS
A total of 257 acute healthcare facilities with 38 619 461 admissions contributed data to this study. Approximately 40% (39.3%) of healthcare facilities were in HHS Region 4 (South/Southeast) and 72% were in urban locations (Table 1). Between 2011 and 2019, we identified 8 250 860 confirmed 30-day nonduplicate pathogens and 3 510 459 influenza tests across 38 619 461 admissions (Supplementary Table 1). The most common pathogens were ENT (6 316 806 [76.6%]) and PSA (749 832 [9.1%]). Data on influenza rates (percent of positive tests) tracked well with influenza data from the Centers for Disease Control and Prevention [16] (Supplementary Figure 1).
Table 1.
Distribution of Study Healthcare Facilities
| Characteristic | n | % |
|---|---|---|
| Overall | 257 | |
| HHS Region | ||
| Region 1 (CT, ME, MA, NH, RI, VT) | 14 | 5.5 |
| Region 2 (NJ, NY) | 3 | 1.2 |
| Region 3 (DE, DC, MD, PA, VA, WV) | 10 | 3.9 |
| Region 4 (AL, FL, GA, KY, MS, NC, SC, TN) | 101 | 39.3 |
| Region 5 (IL, IN, MI, MN, OH, WI) | 35 | 13.6 |
| Region 6 (AR, LA, NM, OK, TX) | 39 | 15.2 |
| Region 7 (IA, KS, MO, NE) | 16 | 6.2 |
| Region 8 (CO, MT, ND, SD, UT, WY) | 13 | 5.1 |
| Region 9 (AZ, CA, HI, NV) | 24 | 9.3 |
| Region 10 (AK, ID, OR, WA) | 2 | 0.8 |
| Urban/Rural | ||
| Urban | 185 | 72.0 |
| Rural | 72 | 28.0 |
| Teaching Status | ||
| Nonteaching | 217 | 84.4 |
| Teaching | 40 | 15.6 |
| Bed Size | ||
| < 100 | 74 | 28.8 |
| 100–300 | 99 | 38.5 |
| > 300 | 84 | 32.7 |
Abbreviations: AL, Alabama; AR, Arizona; AK, Arkansas; CA, California; CO, Colorado; CT, Connecticut; DE, Delaware; DC, District of Columbia; FL, Florida; GA, Georgia; HHS, US Department of Health and Human Services; HI, Hawaii; IA, Iowa; ID, Idaho; IL, Illinois; IN, Indiana; KY, Kentucky; KS, Kansas; LA, Louisiana; MA, Massachusetts; MD, Maryland; ME, Maine; MI, Michigan; MN, Minnesota; MO, Missouri; MS, Mississippi; MT, Montana; NE, Nebraska; OH, Ohio; OK, Oklahoma; NC, North Carolina; ND, North Dakota; NH, New Hampshire; NJ, New Jersey; NM, New Mexico; NV, Nevada; NY, New York; OR, Oregon; PA, Pennsylvania; RI, Rhode Island; SC, South Carolina; SD, South Dakota; TN, Tennessee; TX, Texas; VA, Virginia; VT, Vermont; WA, Washington; WI, Wisconsin; WV, West Virginia; WY, Wyoming.
Antibiotic Resistance in Gram-Positive and Gram-Negative Pathogens
Analyses of Gram-positive pathogens revealed that slightly over half of S aureus isolates tested (498 012 of 989 796 [50.3%]) were MRSA. High levels of ABR were also observed in the 74 554 S pneumoniae isolates tested: 28 421 (38.1%) were NS to macrolides, 13 240 (17.8%) were NS to penicillin, and 2470 (3.3%) were NS to ESC (Table 2).
Table 2.
Distribution of Gram-Positive Antibiotic-resistant Pathogens by Setting, Culture Source, and Quarter
| Characteristic | Staphylococcus aureus | Streptococcus pneumoniae | ||||
|---|---|---|---|---|---|---|
| Tested | MRSA (%) | Tested | Macrolide-NS (%) | Penicillin-NS (%) | ESC-NS (%) | |
| Overall | 989 796 | 498 012 (50.3%) | 74 554 | 28 421 (38.1%) | 13 240 (17.8%) | 2470 (3.3%) |
| Setting | ||||||
| Ambulatory | 501 644 | 236 435 (47.1%) | 25 666 | 10 303 (40.1%) | 4993 (19.5%) | 959 (3.7%) |
| Inpatient | 488 152 | 261 577 (53.6%) | 48 888 | 18 118 (37.1%) | 8247 (16.9%) | 1511 (3.1%) |
| Culture Source | ||||||
| Respiratory | 106 170 | 56 691 (53.4%) | 34 554 | 15 384 (44.5%) | 7437 (21.5%) | 1373 (4.0%) |
| Nonrespiratory | 883 626 | 441 321 (49.7%) | 40 000 | 13 037 (31.2%) | 5803 (17.5%) | 1097 (3.1%) |
| Quarter | ||||||
| Q1 | 230 928 | 116 857 (50.6%) | 24 286 | 9328 (38.4%) | 4271 (17.6%) | 825 (3.4%) |
| Q2 | 239 865 | 121 466 (50.6%) | 18 190 | 6871 (37.8%) | 3224 (17.7%) | 579 (3.2%) |
| Q3 | 266 029 | 133 085 (50.0%) | 11 764 | 4587 (39.0%) | 2300 (19.6%) | 451 (3.8%) |
| Q4 | 252 974 | 126 604 (50.1%) | 20 314 | 7635 (37.6%) | 3445 (17.0%) | 615 (3.0%) |
Abbreviations: ESC, extended-spectrum cephalosporins; MRSA, methicillin-resistant S aureus; NS, not susceptible.
Respiratory sources accounted for 10.7% of S aureus isolates and approximately half (46.3%) of S pneumoniae isolates. The ABR rates in respiratory isolates were slightly higher than in nonrespiratory isolates (Table 2). We also observed higher ABR rates for inpatients versus ambulatory patients for MRSA but not for S pneumoniae. With respect to seasonal trends, percentages of MRSA showed little variation across quarters. The highest rates of ABR in S pneumoniae were recorded in Q3 for all 3 of the antibiotic classes evaluated.
For Gram-negative pathogens, the majority of isolates tested were ENT (6 316 806 of 7 186 510 [87.9%]). Although not analyzed in this study, previous analyses with this database indicate that approximately 56% of ENT inpatient isolates in our database are Escherichia coli, followed by Klebsiella pneumoniae and Proteus mirabilis [13]. Overall, 154 841 (2.2%) of Gram-negative pathogens were Carb-NS and 1 502 796 (20.9%) were FQ-NS (Table 3). As observed for Gram-positive pathogens, ABR rates were higher in respiratory versus nonrespiratory isolates. However, only 5.3% of Gram-negative isolates had a respiratory source (2.6% of ENT, 23.1% of PSA, 25.8% of ACB, and 47.4% of S maltophilia). The ABR rates were higher for inpatients versus ambulatory patients for all pathogens and antibiotics evaluated. The percentages of resistant isolates were generally highest in Q1 with the exception of Carb-NS ENT, which was highest in Q4.
Table 3.
Distribution of Gram-Negative Antibiotic-Resistant Pathogens by Setting, Culture Source, and Quarter
| Characteristic | Enterobacterales | Pseudomonas aeruginosa | Acinetobacter spp | Stenotrophomonas maltophilia | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tested | Carb-NS (%) | FQ-NS (%) | Tested | Carb-NS (%) | FQ-NS (%) | Tested | Carb-NS (%) | Tested | FQ-NS (%) | |
| Overall | 6 316 806 | 43 272 (0.7%) | 1 310 150 (20.7%) | 749 382 | 96 961 (12.9%) | 183 956 (24.6%) | 48 172 | 14 608 (30.3%) | 72 150 | 8690 (12.0%) |
| Setting | ||||||||||
| Ambulatory | 4 040 147 | 15 837 (0.4%) | 750 020 (18.5%) | 329 996 | 31 159 (9.4%) | 73 309 (22.2%) | 16 808 | 2616 (15.6%) | 25 471 | 2932 (11.5%) |
| Inpatient | 2 276 659 | 27 435 (1.2%) | 560 130 (24.6%) | 419 386 | 65 802 (15.7%) | 110 647 (26.4%) | 31 364 | 11 992 (38.2%) | 46 679 | 5758 (12.3%) |
| Culture source | ||||||||||
| Respiratory | 165 252 | 6319 (3.8%) | 34 438 (20.8%) | 173 086 | 40 459 (23.4%) | 54 696 (31.6%) | 12 420 | 5670 (45.7%) | 34 224 | 4562 (13.3%) |
| Nonrespiratory | 6 151 554 | 36 953 (1.2%) | 1 275 712 (20.1%) | 576 296 | 56 502 (9.9%) | 129 260 (18.8%) | 35 752 | 8938 (26.0%) | 37 926 | 4128 (10.4%) |
| Quarter | ||||||||||
| Q1 | 1 437 289 | 9661 (0.7%) | 311 796 (21.6%) | 172 887 | 23 040 (13.3%) | 44 119 (25.6%) | 10 671 | 3674 (34.4%) | 16 407 | 2046 (12.5%) |
| Q2 | 1 521 108 | 10 517 (0.7%) | 320 811 (21.2%) | 177 877 | 23 521 (13.2%) | 44 966 (25.2%) | 11 452 | 3 627 (31.7%) | 17 314 | 2108 (12.2%) |
| Q3 | 1 697 602 | 11 514 (0.7%) | 338 012 (19.9%) | 200 624 | 25 214 (12.6%) | 47 774 (23.8%) | 13 666 | 3661 (26.8%) | 19 261 | 2223 (11.5%) |
| Q4 | 1 660 807 | 11 580 (0.7%) | 339 531 (20.5) | 197 994 | 25 214 (12.7%) | 47 097 (23.8%) | 12 383 | 3646 (29.4%) | 19 168 | 2313 (12.1%) |
Abbreviations: Carb, carbapenem; FQ, fluoroquinolones; NS, not susceptible.
Multivariate Association Between Influenza and Antibiotic Resistance Rates
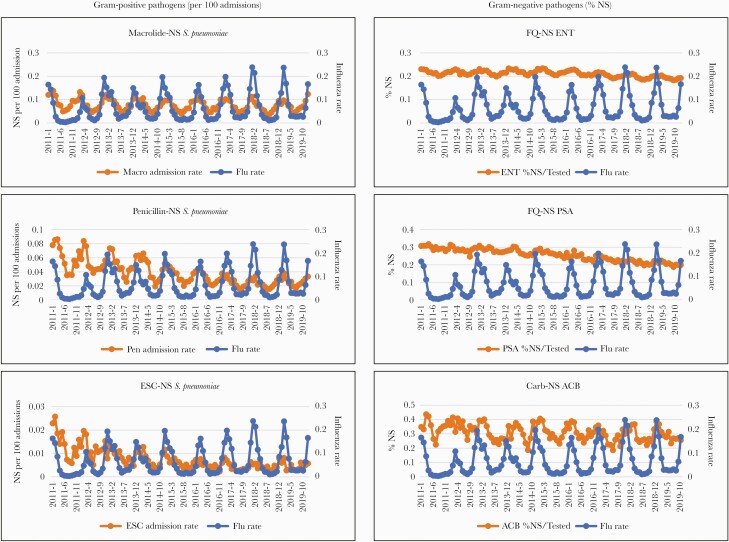
Respiratory MRSA rates per 100 admissions were statistically associated with influenza rates, but overall rates of MRSA (both respiratory and nonrespiratory sources) were not (Table 4). The rate per 100 admissions of ABR S pneumoniae showed significant associations with influenza rates for both respiratory isolates and overall isolates for all 3 antibiotic classes evaluated (macrolide-, penicillin-, and ESC-NS) (Table 4 and Figure 1). Macrolide-NS S pneumoniae had the strongest association (β coefficient of 0.464 overall and 0.253 for respiratory isolates; P < .001 for both associations). Associations between percentage of resistant Gram-positive pathogens and influenza rates were not observed.
Table 4.
Summary of Multivariate Regression Analysesa for the Association Between ABR and Influenza Rates by Source and Setting
| ABR | β Coefficient (P Value) | ||||
|---|---|---|---|---|---|
| Overall | Source | Setting | |||
| Respiratory | Nonrespiratory | Ambulatory | Inpatient | ||
| Gram-Positive ABR per 100 Admissions | |||||
| MRSA | 0.060 (0.615) | 0.066 (0.028) | −0.087 (0.065) | NA | NA |
| Macrolide-NS Streptococcus pneumoniae | 0.464 (<0.001) | 0.253 (<0.001) | 0.068 (0.376) | NA | NA |
| Penicillin-NS S pneumoniae | 0.062 (0.011) | 0.056 (0.046) | 0.044 (0.103) | NA | NA |
| ESC-NS S pneumoniae | 0.033 (0.036) | 0.032 (0.012) | 0.018 (0.073) | NA | NA |
| Gram-Negative ABR Percent NS | |||||
| FQ-NS ENT | 0.041 (<0.001) | 0.130 (<0.001) | 0.031 (0.030) | 0.018 (0.043) | 0.048 (<0.001) |
| FQ-NS PSA | 0.039 (0.015) | 0.022 (0.036) | 0.020 (0.087) | 0.032 (0.172) | 0.044 (0.015) |
| Carb-NS ACB | 0.205 (<0.001) | 0.379 (<0.001) | 0.134 (0.040) | 0.123 (0.077) | 0.255 (<0.001) |
Abbreviations: ABR antibiotic resistance; ACB, Acinetobacter spp; Carb, carbapenem; ENT, Enterobacterales; ESC, extended-spectrum cephalosporin; FQ, fluoroquinolone; MRSA, methicillin-resistant Staphylococcus aureus; NA, not applicable; NS, not susceptible; PSA, Pseudomonas aeruginosa.
Data were adjusted for region, teaching status, urban/rural location, bed size, and season.
NOTE: Statistically significant findings are shown in bold.
Figure 1.
Significant associations between antibiotic resistance and influenza rates from 2011 to 2019. Influenza rate is expressed as positive tests per 100. ACB, Acinetobacter spp; Carb, carbapenem; ENT, Enterobacterales; ESC, extended-spectrum cephalosporin; FQ, fluoroquinolone; MRSA, methicillin-resistant Staphylococcus aureus; NS, not susceptible; PSA, Pseudomonas aeruginosa.
Associations were observed between the percentage of ABR Gram-negative pathogens and influenza rates but not between ABR rates per 100 admissions and influenza rates (Table 4 and Figure 1). The influenza rate was significantly associated with the percentages of FQ-NS ENT (β coefficient of 0.041; P < .001), FQ-NS PSA (β coefficient of 0.039; P = .015), and Carb-NS ACB (β coefficient of 0.205; P < .001). Associations were observed for both respiratory and nonrespiratory sources for Carb-NS ACB and FQ-NS ENT, but only for respiratory FQ-NS PSA (Table 4). An association for FQ-NS ENT was observed in both ambulatory patients and inpatients, whereas associations for Carb-NS ACB and FQ-NS PSA were observed in inpatients only.
DISCUSSION
Our study, based on a large nationwide database, demonstrates clear seasonality to ABR that follows trends in influenza rates after controlling for setting, source of isolates, and healthcare facility characteristics. These associations do not imply causality, because the causes of ABR are multifactorial [11]. However, they provide important insights into ABR trends that may help health systems strategically allocate resources, including vaccination drives and antimicrobial stewardship initiatives [17]. We have also documented associations between influenza rates and increased inpatient antibiotic use for key antibiotic classes used to treat respiratory infections, including macrolides, ESC, and FQ (unpublished observations, KC Yu et al., 2022). Together, these studies provide insights into both bacterial pathogen transmission and antibiotic use at the hospital level that may be appropriate for targeted interventions and for informing policies that facilitate reduced ABR.
In Gram-positive pathogens, we identified significant associations between influenza rates and the rate per 100 admissions for drug-resistant S pneumoniae and respiratory MRSA. Staphylococcus aureus and S pneumoniae are the most common causes of secondary bacterial infections associated with influenza [6]. Antibiotic resistance in these key respiratory pathogens has the potential to complicate treatment of influenza coinfections, including CAP, and potentially impair patient outcomes [18]. An increased number of ABR admissions during influenza season may further increase the burden on hospitals. Based on β coefficients, a 20% increase in the rate of influenza would be associated with approximately 90 more patients with macrolide-NS pneumoniae, 12 more patients with penicillin-NS S pneumoniae, and 7 more patients with ESC-NS S pneumoniae per 1000 admissions.
For Gram-negative pathogens, the percentage of Carb-NS ACB, FQ-NS ENT, and FQ-NS PSA were significantly associated with influenza rates. Based on β coefficients, a 20% increase in the rate of influenza would be associated with an approximately 1% increase in FQ-NS ENT and PSA isolates and a 4% increase in Carb-NS ACB isolates. This result is somewhat surprising because Gram-negative bacteria are less commonly involved in influenza coinfections [6]. Other studies that explored seasonality, but did not specifically evaluate associations with influenza rates, are consistent with our observations, including studies reporting increased ciprofloxacin and beta-lactam resistance in E coli during winter months compared with summer months [19, 20], and a previous study using our database that found higher rates of Carb-NS ACB in Q1 versus Q3 [10]. The associations we observed between Gram-negative resistance and influenza rates suggest that empiric antibiotic use associated with influenza season may have collateral effects on nonrespiratory pathogens and add corroborative evidence that potential bacterial pathogens of public health interest may be important targets for infection prevention practices in a post-COVID-19 era [21]. Given the association of these infections with poor outcomes and increased costs [22, 23], increases in ABR observed in our study can have meaningful impacts on healthcare systems.
Our study provides practical insights for healthcare facilities to incorporate into their stewardship policies and programs to help guide more appropriate antimicrobial selection in patients prone to respiratory infections based on season and severity of influenza rates in local communities. Information on resistant pathogens that peak during surge capacity of respiratory virus season may help forewarn infection prevention and control programs. Antimicrobial stewardship programs, which are now mandated by Centers for Medicare & Medicaid Services (CMS) as a Condition of Participation [24], can also be informed by these trends. The results from this aggregated facility-level analysis support the expansion of influenza vaccine coverage further upstream to help mitigate the inciting viral infection and thereby reduce secondary bacterial coinfections and antibiotic use [25–28]. We concur with the World Health Organization Action Framework on leveraging vaccines to reduce antibiotic use and prevent ABR [17] that vaccines and vaccination should be considered core components of stewardship policies and strategies.
Although some studies have reported a substantial lag time between increased antibiotic use during influenza season and ABR [29], other studies have shown an approximately 1-month lag period [19], which is more in line with the data reported here. A modeling study of seasonal antibiotic use suggested that due to a strong stabilizing force, resistance fluctuates in phase with usage [30].
A limitation of our study is that it did not include data after 2019 and therefore does not reflect significant changes in antibiotic treatment patterns in response to severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) [31, 32], which may have also influenced ABR rates [33]. Future research will be required to evaluate the influence of pandemic alterations, including antibiotic prescribing, on ABR [34]. Earlier studies have shown that non-SARS-CoV-2 coronaviruses have a seasonality pattern similar to influenza [15]; additional research will be required to delineate whether SARS-CoV-2 will also ultimately have an influenza-like seasonality, which could further impact temporal trends in antibiotic use and ABR. It is possible that variations in local influenza vaccination rates may have affected the associations we observed. Additional respiratory viruses were not evaluated in this study, and they may also influence associations with ABR. Future evaluations of associations between other respiratory viruses and ABR should include the pediatric population because the majority of adenovirus, parainfluenza virus, and respiratory syncytial virus cases occur in children under the age of 18, a population that was not included in our study [15]. Further research is needed to explore potential associations between influenza rates and ABR at the individual level.
CONCLUSIONS
In conclusion, our study generates insights on the small but significant seasonal changes in Gram-negative resistance, particularly in the inpatient setting, and corroborates the increased occurrence of ABR Gram-positive respiratory pathogens during the influenza season. Our data may help inform targeted antimicrobial stewardship initiatives in preparation for and during influenza season and provide support for vaccination programs to decrease secondary bacterial coinfections and resulting antibiotic use, particularly in patients with chronic underlying conditions who are at risk of hospitalization during the influenza season.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
The study conception, design, and analysis plan was codeveloped by researchers from Sanofi Pasteur and Becton, Dickinson & Company. We thank Dr. Sharon L. Cross (Fusion MD Medical Science Network, Inc.) for providing manuscript support with funding from Becton, Dickinson & Company.
Financial support . This study was funded by a grant from Sanofi Pasteur to Becton, Dickinson & Company. Medical writing was supported by Becton, Dickinson & Company.
Potential conflicts of interest . V. G., K. C. Y., and J. A. W. are employees of Becton, Dickinson & Company, which was contracted by Sanofi Pasteur to conduct the study. K. C. Y. and V. G also own stock in Becton, Dickinson & Company. H. K. and A. A. are employees of Sanofi Pasteur and may hold shares and/or stock options in the company. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Shieh WJ, Blau DM, Denison AM, et al. 2009 pandemic influenza (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am J Pathol 2010; 177:166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Metersky ML, Masterton RG, Lode H, File TM Jr, Babinchak T.. Epidemiology, microbiology, and treatment considerations for bacterial pneumonia complicating influenza. Int J Infect Dis 2012; 16:e321–31. [DOI] [PubMed] [Google Scholar]
- 3. Metzger DW, Sun K.. Immune dysfunction and bacterial coinfections following influenza. J Immunol 2013; 191:2047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morris DE, Cleary DW, Clarke SC.. Secondary bacterial infections associated with influenza pandemics. Front Microbiol 2017; 8:1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rowe HM, Meliopoulos VA, Iverson A, Bomme P, Schultz-Cherry S, Rosch JW.. Direct interactions with influenza promote bacterial adherence during respiratory infections. Nat Microbiol 2019; 4:1328–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klein EY, Monteforte B, Gupta A, et al. The frequency of influenza and bacterial coinfection: a systematic review and meta-analysis. Influenza Other Respir Viruses 2016; 10:394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lim YK, Kweon OJ, Kim HR, Kim TH, Lee MK.. Impact of bacterial and viral coinfection in community-acquired pneumonia in adults. Diagn Microbiol Infect Dis 2019; 94:50–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Havers FP, Hicks LA, Chung JR, et al. Outpatient antibiotic prescribing for acute respiratory infections during influenza seasons. JAMA Netw Open 2018; 1:e180243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martinez EP, Cepeda M, Jovanoska M, et al. Seasonality of antimicrobial resistance rates in respiratory bacteria: a systematic review and meta-analysis. PLoS One 2019; 14:e0221133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gupta V, Ye G, Olesky M, Lawrence K, Murray J, Yu K.. Trends in resistant Enterobacteriaceae and Acinetobacter species in hospitalized patients in the United States: 2013–2017. BMC Infect Dis 2019; 19:742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2019. Available at: www.cdc.gov/DrugResistance/Biggest-Threats.html. Accessed 16 September 2021.
- 12. Gupta V, Yu KC, Schranz J, Gelone SP.. A multicenter evaluation of the US prevalence and regional variation in macrolide-resistant S. pneumoniae in ambulatory and hospitalized adult patients in the US. Open Forum Infect Dis 2021; 8:ofab063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCann E, Srinivasan A, DeRyke CA, et al. Carbapenem-nonsusceptible Gram-negative pathogens in ICU and non-ICU settings in US hospitals in 2017: a multicenter study. Open Forum Infect Dis 2018; 5:ofy241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brosette SE, Hacek DM, Gavin PJ, et al. A laboratory-based, hospital-wide, electronic marker for nosocomial infection: the future of infection control surveillance? Am J Clin Pathol 2006; 125:34–9. [PubMed] [Google Scholar]
- 15. Choe YJ, Smit MA, Mermel LA.. Comparison of common respiratory virus peak incidence among varying age groups in Rhode Island, 2012-2016. JAMA Netw Open 2020; 3:e207041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Centers for Disease Control and Prevention. Weekly U.S. influenza surveillance report. Available at: https://www.cdc.gov/flu/weekly/index.htm. Accessed 10 August 2021.
- 17. World Health Organization. Leveraging vaccines to reduce antibiotic use and prevent antimicrobial resistance. An action framework and annexe to immunization agenda 2030. Available at: https://www.who.int/publications/m/item/leveraging-vaccines-to-reduce-antibiotic-use-and-prevent-antimicrobial-resistance. Accessed 5 October 2021.
- 18. Liu Y, Ling L, Wong SH, et al. Outcomes of respiratory viral-bacterial co-infection in adult hospitalized patients. EClinicalMedicine 2021; 37:100955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun L, Klein EY, Laxminarayan R.. Seasonality and temporal correlation between community antibiotic use and resistance in the United States. Clin Infect Dis 2012; 55:687–94. [DOI] [PubMed] [Google Scholar]
- 20. Ramsey EG, Royer J, Bookstaver PB, et al. Seasonal variation in antimicrobial resistance rates of community-acquired Escherichia coli bloodstream isolates. Int J Antimicrob Agents 2019; 54:1–7. [DOI] [PubMed] [Google Scholar]
- 21. Yu K. PACCARB virtual public meeting: intersection of COVID-19 and AMR. Available at: https://www.youtube.com/watch?v=bQd4aBXhqr8&feature=youtu.be&t=4111. Accessed 4 August 2021.
- 22. American Society for Microbiology. CMS final rule on antibiotic stewardship programs. Available at: https://asm.org/Articles/Policy/CMS-Final-Rule-on-Antibiotic-Stewardship-Programs. Accessed 8 August 2021.
- 23. Nelson RE, Stayton RB, Stevens VW, et al. Attributable mortality of healthcare-associated infections due to multidrug-resistant Gram-negative bacteria and methicillin-resistant Staphylococcus. Infect Control Hosp Epidemiol 2017; 38:848–56. [DOI] [PubMed] [Google Scholar]
- 24. Bartsch SM, McKinnell JA, Mueller LE, et al. Potential economic burden of carbapenem-resistant Enterobacteriaceae (CRE) in the United States. Clin Microbiol Infect 2017; 23:48.e9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klugman KP, Black S.. Impact of existing vaccines in reducing antibiotic resistance: primary and secondary effects. Proc Natl Acad Sci U S A 2018; 115:12896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith AM, Huber VC.. The unexpected impact of vaccines on secondary bacterial infections following influenza. Viral Immunol 2018; 31:159–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bloom DE, Black S, Salisbury D, Rappuoli R.. Antimicrobial resistance and the role of vaccines. Proc Natl Acad Sci 2018; 115:12868–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klein EY, Schueller E, Tseng KK, Morgan DJ, Laxminarayan, Nandi A.. The impact of influenza vaccination on antibiotic use in the United States, 2010–2017. Open Forum Infect Dis 2020; 7:ofaa223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Choe YJ, Smit MA, Mermel LA.. Seasonality of respiratory viruses and bacterial pathogens. Antimicrob Resist Infect Control 2019; 8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blanquart F, Lehtinen S, Fraser C.. An evolutionary model to predict the frequency of antibiotic resistance under seasonal antibiotic use, and an application to Streptococcus pneumoniae. Proc Biol Sci 2017; 284:20170679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Puzniak L, Bauer KA, Yu KC, et al. Effect of inadequate empiric antibacterial therapy on hospital outcomes in SARS-CoV-2-positive and -negative US patients with a positive bacterial culture: a multicenter evaluation from March to November 2020. Open Forum Infect Dis 2021; 8:ofab232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Puzniak L, Finelli L, Yu KC, et al. A multicenter analysis of the clinical microbiology and antimicrobial usage in hospitalized patients in the US with or without COVID-19. BMC Infect Dis 2021; 21:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gaspar GG, Ferreira L, Feliciano CS, et al. Pre- and post-COVID-19 evaluation of antimicrobial susceptibility for healthcare-associated infections in the intensive care unit of a tertiary hospital. Rev Soc Bras Med Trop 2021; 54:e00902021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nieuwlaat R, Mbuagbaw L, Mertz D, et al. Coronavirus disease 2019 and antimicrobial resistance: parallel and interacting health emergencies. Clin Infect Dis 2021; 72:1657–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.