Abstract
Bladder cancer (BC) is one of the most frequent cancer in the world, and its incidence is rising worldwide, especially in developed countries. Urine metabolomics is a powerful approach to discover potential biomarkers for cancer diagnosis. In this study, we applied an ultra-performance liquid chromatography coupled to mass spectrometry (UPLC-MS) method to profile the metabolites in urine from 29 bladder cancer patients and 15 healthy controls. The differential metabolites were extracted and analyzed by univariate and multivariate analysis methods. Together, 19 metabolites were discovered as differently expressed biomarkers in the two groups, which mainly related to the pathways of phenylacetate metabolism, propanoate metabolism, fatty acid metabolism, pyruvate metabolism, arginine and proline metabolism, glycine and serine metabolism, and bile acid biosynthesis. In addition, a subset of 11 metabolites of those 19 ones were further filtered as potential biomarkers for BC diagnosis by using logistic regression model. The results revealed that the area under the curve (AUC) value, sensitivity and specificity of receiving operator characteristic (ROC) curve were 0.983, 95.3% and 100%, respectively, indicating an excellent discrimination power for BC patients from healthy controls. It was the first time to reveal the potential diagnostic markers of BC by metabolomics, and this will provide a new sight for exploring the biomarkers of the other disease in the future work.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-022-09318-5.
Keywords: Bladder cancer, Urinary metabolomics, UPLC-MS, Potential biomarker, Diagnosis
Introduction
Bladder cancer (BC), also known as urinary bladder cancer, is the tenth most frequent cancer in the world (sixth in men and seventeenth in women), and its incidence is steadily rising worldwide, especially in developed countries, with approximately 550,000 new cases annually [1, 2]. Prolonged exposure to environmental and occupational chemicals could result in the tumorigenesis of BC. Among them, tobacco smoke is the main known cause, which is a possible explanation that greater tobacco smoke in men leads to the 4-fold gender discrepancy in BC incidence [1, 3, 4]. In addition, BC is a heterogeneous disease and possesses a high risk of morbidity and recurrence [5]. Among BC patients, it has primary and recurrent bladder cancer, and the stages of BC could be classified into T1, T2, T3, T4, Ta, etc [6]. The current BC diagnoses are mainly based on urinary cytology, cystoscopy and radiological imaging [6–8]. Cystoscopy is invasive, painful and costly, and it has low sensitivity for diagnosing high-grade superficial tumors. Particularly, it may lead to a high psychological burden for some patients once coupled with biopsy [7, 8]. Urinary cytology is a noninvasive test with high specificity, but poor sensitivity [9]. Therefore, it is urgent to seek more new noninvasive, sensitive and less expensive methods for BC diagnosis.
The reported biomarkers of bladder cancer mainly focused on the gene expression, such as CDK1, MAGEA3, etc [10]. Some of them lacked experimental validations. Meanwhile, gene markers might be failed since they could be regulated by the other proteins or signals. In recent years, metabolomics has proved to be a powerful technique for investigating the variation of endogenous small molecules during life activities in a high-throughput mode [10, 11]. Metabolites have played important roles in biological systems that diseases cause the disruption of biochemical pathways, and the metabolites changes observed in patients as primary indicators have been an important part of clinical practice [12]. Nowadays, metabolomics has been recognized as the preferred approach for biomarker identification, early disease diagnosis and searching related pathways [10, 13, 14]. For example, with the help of urine metabolomics, a marker discovery pipeline selected six putative markers from the metabolomic profiles, which could be employed for the discrimination of BC samples from hernia samples [15].
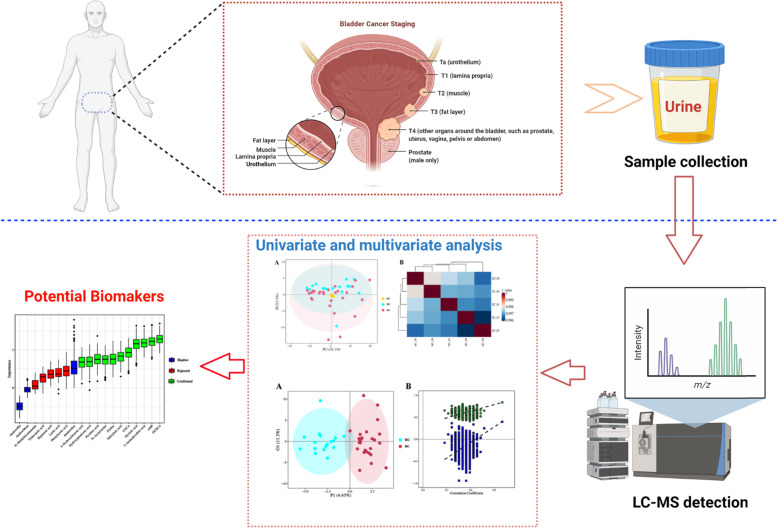
Mass spectrometry (MS) is a generally used platform for metabolomics analysis, and it is always coupled with advanced separation techniques such as gas chromatography (GC-MS), liquid chromatography (LC-MS) and/or others [16–18]. However, GC-MS is only suitable for analyzing volatile metabolites, resulting in the limited application. On the contrary, LC-MS has been widely used for metabolomics analysis benefitting from its high separation power and resolution [19, 20]. Therefore, in this study, a method by ultra-performance liquid chromatography coupled to mass spectrometry (UPLC-MS) was developed and applied to detect endogenous metabolites in urine from BC and healthy control groups. Multivariate statistical analysis methods were employed to identify significantly differential metabolites and potential biomarkers. The pattern recognition analytical techniques, including principal components analysis (PCA), partial least squares discriminant analysis (PLS-DA) and orthogonal partial least squares discriminant analysis (OPLS-DA), were used to comprehensively evaluate the metabolites that were present in any given biological case or that were connected to a specific disease condition (Fig. 1). As a result, a combinatorial biomarker panel, with high sensitivity and specificity, was explored and defined as core indicators in BC diagnosis.
Fig. 1.
The workflow of urine biomarker discovery in bladder cancer
Materials and methods
Chemicals and reagents
Formic acid was of analytical grade and obtained from Sigma-Aldrich (St. Louis, MO, USA). Methanol (Optima LC-MS grade), acetonitrile (Optima LC-MS grade), and isopropanol (Optima LC-MS grade) were purchased from Thermo Fisher Scientific (FairLawn, NJ, USA). Sable isotope-labeled internal standards and the derivatization regents, 3-nitrophenylhydrazine (3-NPH) and N-(3-(dimethylamino)propyl)-N′-ethylcarbodiimide (EDC)·HCl were purchased from Sigma-Aldrich (St. Louis, MO, USA). Ultra-pure water was produced by a Milli-Q system equipped with a LC-MS Pak filter (Millipore, Billerica, MA, USA).
All of the standards were purchased from TRC Chemicals (Toronto, ON, Canada), Sigma-Aldrich (St. Louis, MO, USA) and Steraloids Inc. (Newport, RI, USA). They were accurately weighed and dissolved in appropriate solutions to obtain individual stock solutions at the concentration of 5.0 mg mL− 1. Appropriate amount of each stock solution was mixed to get stock calibration solutions.
Apparatus
An ultra-performance liquid chromatography coupled to tandem mass spectrometry (ACQUITY UPLC-Xevo TQ-S, Waters Corp., Milford, MA, USA) with an electrospray ionization (ESI) source was operated under positive and negative ion modes for the quantitation of metabolites. The UPLC-MS system was controlled by MassLynx 4.1 software. The chromatographic separations were carried out by an ACQUITY BEH C18 column (100 mm × 2.1 mm, 1.7 μm) (Waters, Milford, MA) at a flow rate of 0.4 mL min− 1. The mobile phases were consisted of 0.1% formic acid in water (solvent A) and acetonitrile/isopropanol (70:30, v:v) (solvent B), and a gradient elution program was set as follows: 0–1 min, 5% B; 1–5 min, 5–30% B; 5–9 min, 30–50% B; 9–12 min, 50–79% B; 12–15 min, 78–95% B; 15–16 min, 95–100% B; 16–18 min, 100% B. The main parameters of ESI source were optimized and adopted as follows: 1.2 kV (ESI−) and 3.2 kV (ESI+) of capillary voltage, 150 °C of source temperature, 550 °C of desolvation temperature, and 1200 L h− 1 of desolvation gas flow (N2). Collision-induced dissociation (CID) activation was used for the MS/MS fragmentation with an isolation width of m/z 3.0.
Clinical samples
A total of 44 subjects, including 29 BC patients (BCs) and 15 healthy controls (HCs), were recruited at the First Affiliated Hospital, Zhejiang University School of Medicine. Among the collected BC patients, 19 were classified into high stage and 10 were low stage. The detailed information was showed in the supplementary materials Table S1. The experiment was approved by Zhejiang University Institutional Review Board, and informed consent forms were obtained from all participants. The diagnosis, staging and other information of BCs were obtained from the database for inpatients of the First Affiliated Hospital. The midstream urine was freshly collected in the morning after overnight fasting, then transferred into an Eppendorf tube, which was stored at − 80 °C before use.
Urine sample preparation
Metabolomics analysis on urine samples was conducted by using the Q300 Metabolite Assay Kit (Human Metabolomics Institute, Inc., Shenzhen, Guangdong, China), referring to reported method with some modifications [21]. In brief, samples were firstly thawed on the ice-bath to reduce sample degradation. Then, 25 μL of urine was added to a 96-well plate, which was loaded to the Biomek 4000 workstation (Biomek 4000, Beckman Coulter, Brea, California, USA) [21]. The cold methanol containing partial internal standards was automatically added to each sample, and the samples were subsequently vortexed for 5 min [22]. After centrifugation for 30 min at 4000×g (Allegra X-15R, Beckman Coulter, Indianapolis, IN, USA), 30 μL of supernatant and 20 μL of fresh derivative reagents (200 mM 3-NPH in 75% methanol and 96 mM EDC-6% pyridine solution in methanol) were added to each well of a new clean 96-well plate [22]. After derivatization at 30 °C for 60 min, each sample was diluted by 350 μL of cold 50% methanol and stored at − 20 °C for 20 min. After centrifugation with the conditions of 4000×g and 4 °C for 30 min, 135 μL of supernatant and 15 μL of internal standards were added to each well on a new 96-well plate. And the remaining wells were filled with serial diluted derivatized stock standards. At last, the sample plate was sealed for the subsequent UPLC-MS analysis.
Quality control approach for metabolomic analysis
Periodic analysis of real samples together with quality control (QC) samples was applied in this study to ensure the excellent quality of metabolic profiling [12]. In detail, five injections of QC samples were put in the analytical platform in the first instance. Next, before inserting 5 samples, one QC sample was breathed into the sample set in order. The QC samples were prepared by a mixture of BCs and HCs samples with the same volumes.
Data analysis and statistical analysis
The raw MS data files were processed by Targeted Metabolome Batch Quantification (TMBQ) software (v1.0, Human Metabolomics Institute, Inc., Shenzhen, Guangdong, China) to perform peak integration, calibration, and quantitation for each metabolite [23]. Identified metabolites were further annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/) and Human Metabolome Database (HMDB) (http://www.hmdb.ca/) [24]. Metaboanalyst (https://www.metaboanalyst.ca/) was employed to perform the metabolic pathway enrichment of differential metabolites. PCA and OPLS-DA were carried out by metaX software [25]. Univariate analysis (t-test) was employed to calculate the statistical significance (P-value) [26]. The metabolites with variable importance in the projection (VIP) > 1, P-value < 0.05 and |fold change (FC)| >0 were regarded as differential metabolites [27].
Results
Clinical information of participants
An untargeted metabolomics method was used to study urine samples from 29 BCs and 15 HCs. The participants’ clinical information is summarized in Table 1. These participants aged from 48 to 92 years old with an average age of 68.2. In the BCs, 21 (72.4%) were male and 8 (27.6%) were female. In the HCs, 12 (80.0%) were male and 3 (20.0%) were female.
Table 1.
Characteristics of enrolled patients
| Groups | No. of subjects | Gender | |
|---|---|---|---|
| Male | Female | ||
| Sample number | 44 | 33 | 11 |
| BC patient | 29 | 21 | 8 |
| HC control | 15 | 12 | 3 |
| Among patients | |||
| Age range | 68.2 (48–92) | ||
| Stage | |||
| Ta | 11 | 7 | 4 |
| T1 | 6 | 6 | 0 |
| T2 | 5 | 2 | 3 |
| T3 | 4 | 4 | 0 |
| T4 | 3 | 2 | 1 |
| MIBC | 12 | 8 | 4 |
| NMIBC | 17 | 13 | 4 |
| High grade | 19 | 12 | 7 |
| Low grade | 10 | 9 | 1 |
| Primary | 18 | 14 | 4 |
| Recurrence | 11 | 7 | 4 |
QC sample analysis
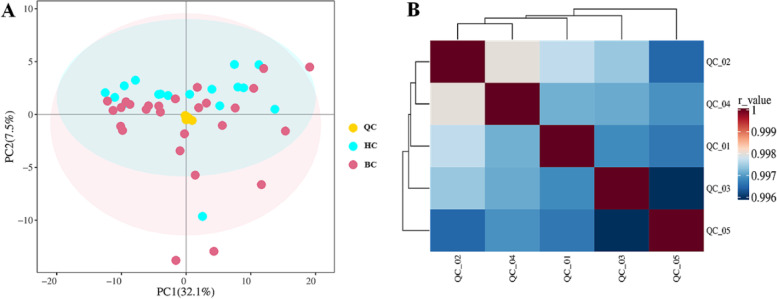
PCA analysis was performed based on QC samples and tested samples. The PCA score plot is shown in Fig. 2A. The result indicated that QC samples formed a cluster without any obvious drift during metabonomic profiling. In addition, the pearson correlation (calculated by Pearson Correlation Coefficient) of any two QC samples was within 0.996 and 1 (Fig. 2B). These results demonstrated the current metabolomics data had good stability and reproducibility.
Fig. 2.
A PCA score plot for QC samples and tested samples. Yellow dots denote QC samples, blue dots are HC samples and red dots represent BC samples. B correlation heat map for QC samples
Urine metabolomics data analysis
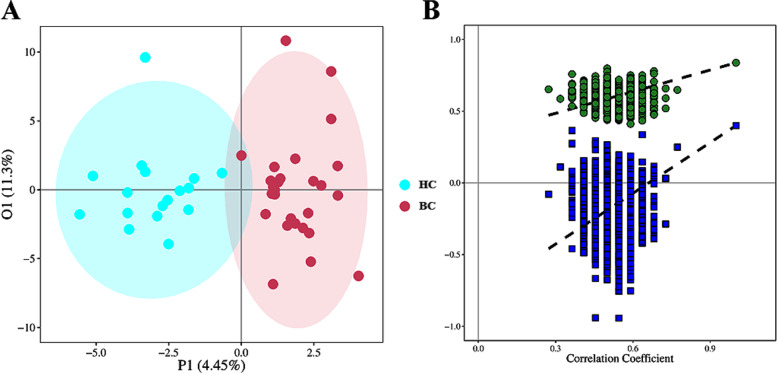
Based on the untargeted metabolomics technique, a total of 208 metabolites were identified in the urine samples. To evaluate the discriminating power of the obtained 208 metabolites, we performed OPLS-DA analysis for the urine samples from 29 BCs and 15 HCs (Fig. 3A). The OPLS-DA model was constructed by performing 7-fold cross-validation, and the result showed satisfactory modeling and prediction with 1 predictive component and 2 orthogonal components (R2Xcum = 0.157, R2Ycum = 0.837, Q2 cum = 0.399). To avoid model over-fitting, the model was further validated with a permutation multivariate analysis of variance (PERMANOVA), and the result indicated that the probability of this model randomly occurring was less than 0.001 (Fig. 3B). From these satisfactory results, the metabolic profiling of BC patients showed significantly discriminative potential from that of HCs.
Fig. 3.
A OPLS-DA score plot for HC and BC groups. Blue circles and red circles represent data for HC and BC samples, respectively. (B) The correlation coefficient (R2) distribution plot of the permutation test for the OPLS-DA model
Identification of metabolic biomarkers
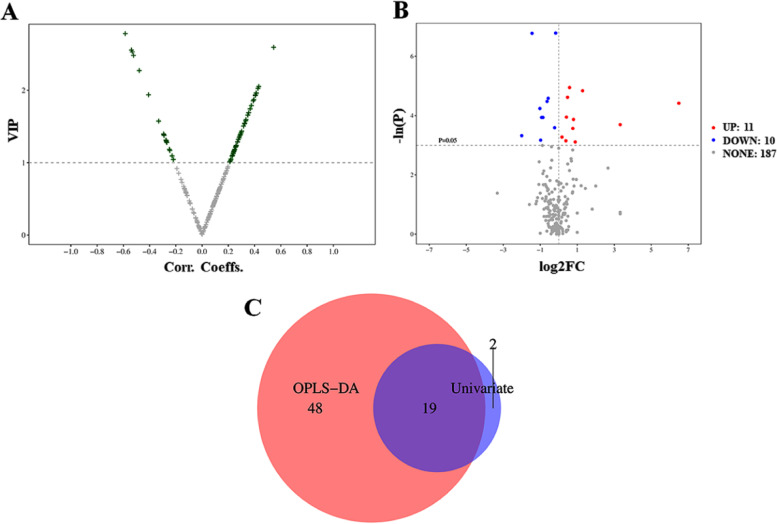
In the current study, we applied two types of analysis to identify the significantly changed metabolites in BC patients and explore potential biomarkers for diagnosis of BC. Firstly, VIP scores of obtained 208 metabolites were extracted from the OPLS-DA model. The volcano plot of VIP scores for these metabolites is shown in Fig. 4A. The green crosses in the volcano plot indicated 67 significantly changed metabolites with VIP > 1. Secondly, t-test was employed to calculate the P-value and fold change (FC), which was shown in the volcano plot (Fig. 4B). As a result, 21 metabolites with P value < 0.05 and |FC|>0 were highlighted as differential metabolites, including 11 upregulation (marked in red) and 10 downregulation (marked in blue). In consideration of stage and gender influence, we carefully analyzed the data (Fig. S1 and Fig. S2). The top 10 metabolites were listed in Table S2 and S3. There were common metabolites, such as AMP, GUDCA, etc. Meanwhile, different metabolites between low grade and high grade were also found, such as N-Methylnicotinamide. There was some difference based on stage or gender analysis. However, due to the limitation of samples, more samples should be collected to make a direction. All the metabolites were presented in the top 21 metabolites of changed metabolites in BC patients.
Fig. 4.
A Volcano plot of VIP scores from OPLS-DA model. The green crosses represent the metabolites with VIP>1 and the grey crosses represent the metabolites with VIP ≤ 1. B Volcano plot with the univariate statistical test (−ln P) and the magnitude of the change (log2FC) of metabolites. Red points represent the metabolites with P-value < 0.05 and FC>0. Blue points represent the metabolites with P-value < 0.05 and FC < 0. Grey points represent the metabolites with P-value>0.05. C Venn diagram integrating results from volcano-plots of OPLS-DA model and univariate statistical test
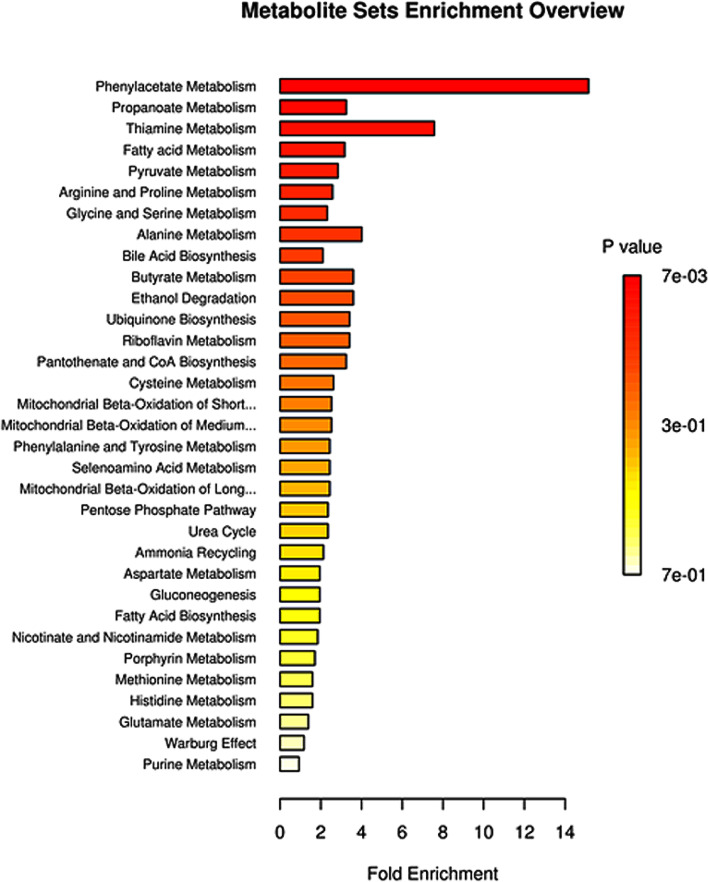
Finally, a Venn diagram was plotted to integrate the selected results from the OPLS-DA model and univariate analysis (Fig. 4C). In the overlapped area of the Venn diagram, a total of 19 metabolites could meet the following criteria of VIP > 1 and P-value < 0.05 and |FC|>0, which were regarded as metabolic biomarkers. Their detailed information is listed in Table 2. Based on the relative abundance of differential metabolites, pathway enrichment results showed that 33 metabolic pathways were identified in Small Molecule Pathway Database (SMPDB). Among them, 7 pathways, including phenylacetate metabolism, propanoate metabolism, fatty acid metabolism, pyruvate metabolism, arginine and proline metabolism, glycine and serine metabolism, and bile acid biosynthesis, were significantly enriched with at least 2 annotated metabolites (Fig. 5). The detailed pathway enrichment results are displayed in Table 3.
Table 2.
19 differential metabolites annotated in KEGG or HMDB database
| Metabolite | Class | HMDB | KEGG | P-value | FC | VIP |
|---|---|---|---|---|---|---|
| Hydroxypropionic acid | Organic Acids | HMDB0000700 | C01013 | 0.0195 | 0.5273 | 1.2951 |
| AMP | Nucleotides | HMDB0000045 | C00020 | 0.0079 | 2.4444 | 1.1625 |
| Lactic acid | Organic Acids | HMDB0000190 | C00186 | 0.0446 | 1.8547 | 1.6818 |
| Picolinic acid | Pyridines | HMDB0002243 | C10164 | 0.0102 | 0.6731 | 1.2883 |
| 4-Hydroxybenzoic acid | Benzoic Acids | HMDB0000500 | C00156 | 0.0114 | 0.6455 | 1.3049 |
| Phenylacetic acid | Benzenoids | HMDB0000209 | C07086 | 0.0429 | 1.3069 | 1.9231 |
| Salicyluric acid | Benzoic Acids | HMDB0000840 | C07588 | 0.0144 | 0.4935 | 1.378 |
| Proline | Amino Acids | HMDB0000162 | C00148 | 0.0209 | 1.7364 | 1.0882 |
| N-Acetylserine | Amino Acids | HMDB0002931 | NA | 0.042 | 0.5078 | 1.0862 |
| 5-Aminolevulinic acid | Amino Acids | HMDB0001149 | C00430 | 0.0011 | 0.3679 | 2.5489 |
| N-Methylnicotinamide | Pyridines | HMDB0003152 | NA | 0.0283 | 1.6952 | 1.783 |
| Heptanoic acid | Fatty Acids | HMDB0000666 | C17714 | 0.0378 | 1.129 | 2.0465 |
| GUDCA | Bile Acids | HMDB0000708 | NA | 0.0121 | 90.0 | 1.9545 |
| CDCA | Bile Acids | HMDB0000518 | C02528 | 0.0099 | 1.3894 | 2.5883 |
| GCDCA | Bile Acids | HMDB0000637 | C05466 | 0.0071 | 1.5 | 1.4274 |
| Tridecanoic acid | Fatty Acids | HMDB0000910 | C17076 | 0.0276 | 0.8571 | 2.2681 |
| Myristic acid | Fatty Acids | HMDB0000806 | C06424 | 0.0011 | 0.8913 | 2.5243 |
| 3-Hydroxylisovalerylcarnitine | Carnitines | NA | NA | 0.0195 | 0.5544 | 1.3705 |
| Palmitoylcarnitine | Carnitines | HMDB0000222 | C02990 | 0.0249 | 10.0 | 1.3128 |
Fig. 5.
Differential metabolite pathway analysis. The color depth and column length indicate the disturbance degree of the pathway
Table 3.
SMPDB pathway enrichment
| Pathway | Total | Expected | Hits | P-value | Adjust P-value | FDR | Enriched compounds |
|---|---|---|---|---|---|---|---|
| Phenylacetate Metabolism | 9 | 0.132 | 2 | 0.0068 | 0.666 | 0.666 | AMP; Phenylacetic acid |
| Propanoate Metabolism | 42 | 0.615 | 2 | 0.123 | 1 | 1 | AMP; Hydroxypropionic acid |
| Fatty acid Metabolism | 43 | 0.63 | 2 | 0.128 | 1 | 1 | AMP; Palmitoylcarnitine |
| Pyruvate Metabolism | 48 | 0.703 | 2 | 0.153 | 1 | 1 | AMP; Lactic acid |
| Arginine and Proline Metabolism | 53 | 0.776 | 2 | 0.18 | 1 | 1 | AMP; Proline |
| Glycine and Serine Metabolism | 59 | 0.864 | 2 | 0.212 | 1 | 1 | AMP; 5-Aminolevulinic acid |
| Bile Acid Biosynthesis | 65 | 0.952 | 2 | 0.246 | 1 | 1 | CDCA; GCDCA |
| Thiamine Metabolism | 9 | 0.132 | 1 | 0.125 | 1 | 1 | AMP |
| Alanine Metabolism | 17 | 0.249 | 1 | 0.223 | 1 | 1 | AMP |
| Butyrate Metabolism | 19 | 0.278 | 1 | 0.246 | 1 | 1 | AMP |
| Ethanol Degradation | 19 | 0.278 | 1 | 0.246 | 1 | 1 | AMP |
| Ubiquinone Biosynthesis | 20 | 0.293 | 1 | 0.258 | 1 | 1 | 4-Hydroxybenzoic acid |
| Riboflavin Metabolism | 20 | 0.293 | 1 | 0.258 | 1 | 1 | AMP |
| Pantothenate and CoA Biosynthesis | 21 | 0.308 | 1 | 0.269 | 1 | 1 | AMP |
| Cysteine Metabolism | 26 | 0.381 | 1 | 0.322 | 1 | 1 | AMP |
| Mitochondrial Beta-Oxidation of Short Chain Saturated Fatty Acids | 27 | 0.396 | 1 | 0.332 | 1 | 1 | AMP |
| Mitochondrial Beta-Oxidation of Medium Chain Saturated Fatty Acids | 27 | 0.396 | 1 | 0.332 | 1 | 1 | AMP |
| Phenylalanine and Tyrosine Metabolism | 28 | 0.41 | 1 | 0.342 | 1 | 1 | AMP |
| Selenoamino Acid Metabolism | 28 | 0.41 | 1 | 0.342 | 1 | 1 | AMP |
| Mitochondrial Beta-Oxidation of Long Chain Saturated Fatty Acids | 28 | 0.41 | 1 | 0.342 | 1 | 1 | AMP |
| Pentose Phosphate Pathway | 29 | 0.425 | 1 | 0.352 | 1 | 1 | AMP |
| Urea Cycle | 29 | 0.425 | 1 | 0.352 | 1 | 1 | AMP |
| Ammonia Recycling | 32 | 0.469 | 1 | 0.381 | 1 | 1 | AMP |
| Aspartate Metabolism | 35 | 0.513 | 1 | 0.409 | 1 | 1 | AMP |
| Gluconeogenesis | 35 | 0.513 | 1 | 0.409 | 1 | 1 | Lactic acid |
| Fatty Acid Biosynthesis | 35 | 0.513 | 1 | 0.409 | 1 | 1 | Myristic acid |
| Nicotinate and Nicotinamide Metabolism | 37 | 0.542 | 1 | 0.426 | 1 | 1 | AMP |
| Porphyrin Metabolism | 40 | 0.586 | 1 | 0.452 | 1 | 1 | 5-Aminolevulinic acid |
| Methionine Metabolism | 43 | 0.63 | 1 | 0.477 | 1 | 1 | AMP |
| Histidine Metabolism | 43 | 0.63 | 1 | 0.477 | 1 | 1 | AMP |
| Glutamate Metabolism | 49 | 0.718 | 1 | 0.523 | 1 | 1 | AMP |
| Warburg Effect | 58 | 0.85 | 1 | 0.586 | 1 | 1 | Lactic acid |
| Purine Metabolism | 74 | 1.08 | 1 | 0.678 | 1 | 1 | AMP |
Potential biomarkers for BC diagnosis
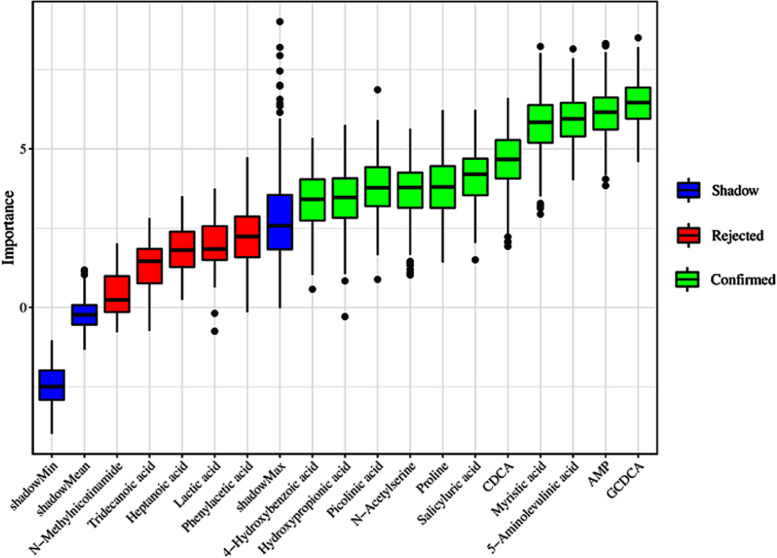
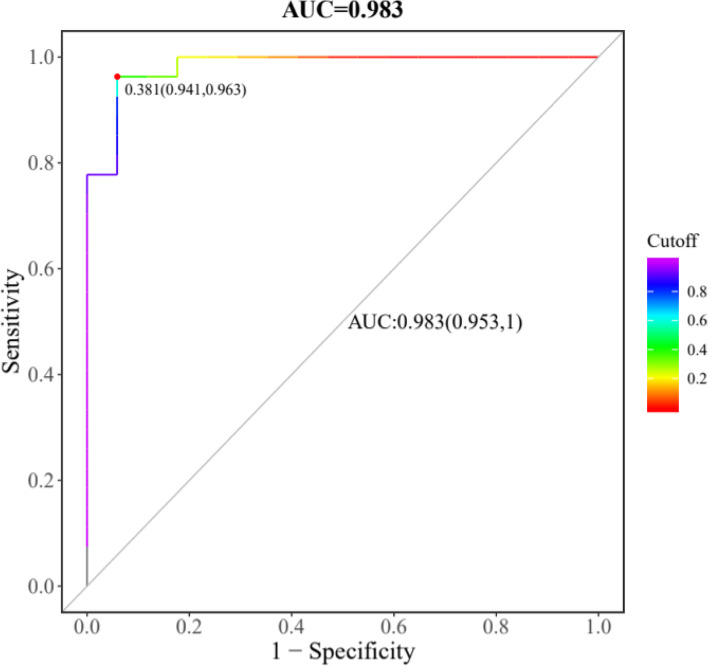
In order to find out candidate biomarkers from 19 identified differential metabolites, we carried out random forest (RF), support vector machine (SVM) and boruta analysis in sequence. First, we got union set between top 10 metabolites from RF and top 10 metabolites from SVM, which were employed to carry out further selection of potential biomarkers using boruta analysis. In this study, the result of the boruta algorithm for selecting the most important metabolites is shown in Fig. 6, and a total of 11 metabolites, namely glycochenodeoxycholic acid (GCDCA), adenosine monophosphate (AMP), 5-Aminolevulinic acid, myristic acid, chenodeoxycholic acid (CDCA), salicyluric acid, proline, N-Acetylserine, picolinic acid, hydroxypropionic acid and 4-Hydroxybenzoic acid, were labeled as “Confirmed”, which could be used for model building and prediction. These 11 selected potential biomarkers were further combined by logistic regression (LR) model to build the biomarker panel, and the final receiving operator characteristic (ROC) curve is shown in Fig. 7. It can be observed that the biomarker panel had an area under the curve (AUC) of 0.983 and the values of sensitivity and specificity reached 95.3% and 100% at the best cut-off points.
Fig. 6.
Relative importance (RI) of metabolites calculated by boruta algorithm
Fig. 7.
Receiver operating characteristic (ROC) curve of a logistic regression model for distinguishing BCs from HCs by using 11 potential biomarkers conjunctively
Discussion
Bladder cancer is one of the most prevalent malignancies of the urinary system, which is related to multiple factors, such as genetic susceptibility, environmental exposure as well as unhealthy lifestyles [5]. In recent years, there is a trend of increased incidence and mortality worldwide for BC [28, 29]. As is known, metabolomics has proved to be a powerful technique for investigating the variation of endogenous small molecules during life activities in a high-throughput mode [10], which could help us to identify potential metabolic biomarkers and study possible mechanisms for BC. Because bladder is a temporary storage of urine and the urinary metabolic characteristics could closely reveal changes that occur during pathological conditions, urinary metabolomics has some advantages over other biological fluids, such as noninvasive collection and convenient storage. In the present study, a nontargeted UPLC-MS method was applied to profile the metabolites of urine from 29 BC patients and 15 healthy subjects. A group of 19 metabolites was discovered as differently expressed biomarkers, which mainly related to phenylacetate metabolism, propanoate metabolism, fatty acid metabolism, pyruvate metabolism, etc. (Table 3). In addition, the combination of 11 potential biomarkers showed an excellent discrimination capability of BCs from HCs, which could be potentially used for clinical diagnosis, prognosis monitoring, and early detection of BC patients.
Significantly altered metabolites and related pathways
Among the 19 differential metabolites, the changes of AMP, glycoursodeoxycholic acid (GUDCA) and palmitoylcarnitine were the most dramatic ones with FC>2. Adenosine monophosphate, known as AMP, is a nucleotide that is found in RNA. AMP can be produced during adenosine triphosphate (ATP) synthesis by the enzyme adenylate kinase urine (Human Metabolome Database). The AMP-activated protein kinase (AMPK) is sensitive to cellular AMP/ATP ratio, in which a high AMP or low ATP level activates AMPK [30, 31]. In the present research, the content of AMP in BCs was higher than that in HCs. AMPK activation could provide a growth advantage to tumor cells by regulating cellular metabolic plasticity, thus providing tumor cells the flexibility to adapt to metabolic stress [32]. The role of core enzyme (AMPK) was analyzed by using Onconmine and cBIO portal. As showed in the following Fig. S3, it was found that the survival probability of Protein Kinase AMP-Activated Catalytic Subunit Alpha 2 (PRKAA2, one sub-type of AMPK) altered patients was relatively lower than those without alteration. Higher expression of AMP could result in the worse survival conditions.
The potential biomarkers of GUDCA, CDCA and GCDCA were also belonging to the group of bile acids, and they were all up-regulated in urine from BC patients as compared to the HC group. Bile acids are physiological detergent molecules, so are highly cytotoxic. Meanwhile, it is found that conjugated bile acids can activate the sphingosine 1-phosphate receptor 2 that activates intracellular ERK1/2 and AKT signaling to promote the invasive growth of cholangiocarcinoma, which is commonly associated with chronic cholestasis [33], and overexpression of bile acids in urine might result in worse condition of urinary bladder, even cancer. It was reported that bile acids were implicated as etiologic agents in cancer of the gastrointestinal tract, including cancers of the esophagus, stomach, small intestine, liver, biliary tract, pancreas and colon [34]. The bile acids could generate cellular reactive oxygen species and induce multiple stresses on cells including DNA damage, endoplasmic reticulum stress and mitochondrial damage, which lead to genomic instability, apoptosis, necrosis, autophagic cell death, etc. [35–38].
Palmitoylcarnitine is a well-known intermediate in mitochondrial fatty acid oxidation, which naturally exists in blood, feces, saliva and urine. In this study, the content of palmitoylcarnitine on bladder cancer patients was 10 times higher than that in healthy subjects. Some studies have reported the accumulation of palmitoy lcarnitine in diabetes mellitus type II, obesity and kidney cancer [39–41]. It was found that the increased contents of most acylcarnitines in the urine of cancer patients together with high cancer grades in those patients, and higher chain length acylcarnitines, such as palmitoylcarnitine, showed inhibitory effects on nuclear factor kappa-B (NF-kB) activation, indicating an immune modulatory effect [41]. Besides, it was reported that palmitoylcarnitine might represent a potential biomarker of the metabolic dysfunction associated with prostate cancer [42]. The result showed that, at physiological levels of palmitoylcarnitine, there were no effects on the prostate cancer cells; however, at high levels of palmitoylcarnitine, it drove tumor development through inducing key inflammatory cytokines and gene expression associated with glycolysis.
Interestingly, salicylic acid was a characteristic index presented in the urine samples of BC. Through analyzing the drug administration, we found that some of BC patients often used salicylic acid-related drugs, such as aspirin. This indicated that overdose of salicylic acid-related drugs might contribute to the tumorigenesis of BC. Due to the small samples in our work, more subjects were needed to be collected to validate this speculation.
Diagnosis of BCs from HCs
The AUC was 0.983 for BC diagnosis, with 95.3% sensitivity and 100% specificity, showing satisfactory discrimination power by the eleven-biomarker panel. The results suggested that the eleven-biomarker panel could be used for the diagnosis of BCs from HCs. In addition, besides N-Acetylserine, all other potential biomarkers could individually achieve a high AUC value higher than 0.7 with satisfactory sensitivity and specificity. It should be noted that, because these 11 differential metabolites were chosen based on boruta algorithm using 29 BCs and 15 HCs, this predictive accuracy may be biased upward. Thus, the additional verification set should include a wider range of subjects, and their ages and gender should be carefully matched in the further study.
Conclusion
In this study, we established and applied a UPLC-MS based metabolomics to investigate the metabolite difference in urine from 29 bladder cancer patients and 15 healthy subjects. An obvious discrimination was obtained by OPLS-DA analysis based on the identified metabolites between bladder cancer patients and healthy controls. In addition, 19 metabolites were discovered as differently expressed biomarkers in the two groups. Based on boruta analysis, 11 of them were selected and combined to test the potentiality of diagnosis of BC by using LR model. The AUC, sensitivity and specificity of ROC curve were 0.983, 95.3% and 100%, respectively, showing an excellent discrimination power for BC diagnosis.
Supplementary Information
Acknowledgements
We thank the financial support from the department of R&D in Yaneng Bioscience, Co., Ltd.
Authors’ contributions
Shujun Zhou and Chaojun Wang made substantial contributions to the concept and design of the work. Xu Zhang, Huaixing Kang and Rui Wang analyzed and interpreted the data and were major contributors in writing the manuscript, and Hongling Wang substantively revised it. Qing Nie performed the sample collection and major technical work. All authors read and approved the final manuscript.
Funding
This work was financially supported by the department of R&D in Yaneng Bioscience, Co., Ltd and Xiangtan Science and Technology Foundation (SF-YB20201005).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The fresh urine samples in this study were obtained from adult patients at the First Affiliated Hospital, Zhejiang University School of Medicine, which was approved by Zhejiang University Institutional Review Board.
Informed consent was obtained from all of participants with both sex above 18 years old.
This research was approved by Zhejiang University Institutional Review Board including all experimental protocols and all methods in accordance with the relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rui Wang, Huaixing Kang and Xu Zhang contributed equally to this work.
Contributor Information
Rui Wang, Email: 2290461273@qq.com.
Huaixing Kang, Email: kanghuaixin@163.com.
Xu Zhang, Email: 11618324@zju.edu.cn.
Qing Nie, Email: nieqing@yanengbio.com.
Hongling Wang, Email: whongling2009@163.com.
Chaojun Wang, Email: wangchaojundf@hotmail.com.
Shujun Zhou, Email: zhoushujun@yanengbio.com.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Richters A, Aben KKH, Kiemeney LALM. The global burden of urinary bladder cancer: an update. World J Urol. 2020;38(8):1895–1904. doi: 10.1007/s00345-019-02984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boffetta P. Tobacco smoking and risk of bladder cancer. Scand J Urol Nephrol. 2008;42:45–54. doi: 10.1080/03008880802283664. [DOI] [PubMed] [Google Scholar]
- 4.Soria F, Marra G, Capoun O, Soukup V, Gontero P. Prevention of bladder cancer incidence and recurrence: tobacco use. Curr Opin Urol. 2018;28(1):80–87. doi: 10.1097/Mou.0000000000000453. [DOI] [PubMed] [Google Scholar]
- 5.Burger M, Catto JWF, Dalbagni G, Grossman HB, Herr H, Karakiewicz P, Kassouf W, Kiemeney LA, La Vecchia C, Shariat S, et al. Epidemiology and risk factors of urothelial bladder Cancer. Eur Urol. 2013;63(2):234–241. doi: 10.1016/j.eururo.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 6.Issaq HJ, Nativ O, Waybright T, Luke B, Veenstra TD, Issaq EJ, Kravstov A, Mullerad M. Detection of bladder cancer in human urine by metabolomic profiling using high performance liquid chromatography/mass spectrometry. J Urology. 2008;179(6):2422–2426. doi: 10.1016/j.juro.2008.01.084. [DOI] [PubMed] [Google Scholar]
- 7.Putluri N, Shojaie A, Vasu VT, Vareed SK, Nalluri S, Putluri V, Thangjam GS, Panzitt K, Tallman CT, Butler C, et al. Metabolomic profiling reveals potential markers and bioprocesses altered in bladder Cancer progression. Cancer Res. 2011;71(24):7376–7386. doi: 10.1158/0008-5472.Can-11-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasikanti KK, Esuvaranathan K, Ho PC, Mahendran R, Kamaraj R, Wu QH, Chiong E, Chan ECY. Noninvasive urinary Metabonomic diagnosis of human bladder Cancer. J Proteome Res. 2010;9(6):2988–2995. doi: 10.1021/pr901173v. [DOI] [PubMed] [Google Scholar]
- 9.Witjes JA, Lebret T, Comperat EM, Cowan NC, De Santis M, Bruins HM, Hernandez V, Espinos EL, Dunn J, Rouanne M, et al. Updated 2016 EAU guidelines on muscle-invasive and metastatic bladder Cancer. Eur Urol. 2017;71(3):462–475. doi: 10.1016/j.eururo.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 10.Wishart DS. Metabolomics for investigating physiological and pathophysiological processes. Physiol Rev. 2019;99(4):1819–1875. doi: 10.1152/physrev.00035.2018. [DOI] [PubMed] [Google Scholar]
- 11.Rinschen MM, Ivanisevic J, Giera M, Siuzdak G. Identification of bioactive metabolites using activity metabolomics. Nat Rev Mol Cell Bio. 2019;20(6):353–367. doi: 10.1038/s41580-019-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Hou H, Chen H, Luo Y, Zhang L, Zhang Y, Liu H, Zhang F, Liu Y, Wang A, et al. Urinary metabolomics for discovering metabolic biomarkers of laryngeal cancer using UPLC-QTOF/MS. J Pharm Biomed Anal. 2019;167:83–89. doi: 10.1016/j.jpba.2019.01.035. [DOI] [PubMed] [Google Scholar]
- 13.Guijas C, Montenegro-Burke JR, Warth B, Spilker ME, Siuzdak G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat Biotechnol. 2018;36(4):316–320. doi: 10.1038/nbt.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blazenovic I, Kind T, Ji J, Fiehn O. Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites. 2018;8(2):31–54. doi: 10.3390/metabo8020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao CH, Chen CL, Lin JY, Chen CJ, Fu SH, Chen YT, Chang YS, Yu JS, Tsui KH, Juo CG, et al. Metabolite marker discovery for the detection of bladder cancer by comparative metabolomics. Oncotarget. 2017;8(24):38802–38810. doi: 10.18632/oncotarget.16393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beale DJ, Pinu FR, Kouremenos KA, Poojary MM, Narayana VK, Boughton BA, Kanojia K, Dayalan S, Jones OAH, Dias DA. Review of recent developments in GC-MS approaches to metabolomics-based research. Metabolomics. 2018;14(11):1–31. doi: 10.1007/s11306-018-1449-2. [DOI] [PubMed] [Google Scholar]
- 17.Papadimitropoulos MEP, Vasilopoulou CG, Maga-Nteve C, Klapa MI. Untargeted GC-MS metabolomics. Methods Mol Biol. 1738;2018:133–147. doi: 10.1007/978-1-4939-7643-0_9. [DOI] [PubMed] [Google Scholar]
- 18.Theodoridis G, Gika HG, Wilson ID. LC-MS-based methodology for global metabolite profiling in metabonomics/metabolomics. Trac-Trend Anal Chem. 2008;27(3):251–260. doi: 10.1016/j.trac.2008.01.008. [DOI] [Google Scholar]
- 19.Becker S, Kortz L, Helmschrodt C, Thiery J, Ceglarek U. LC-MS-based metabolomics in the clinical laboratory. J Chromatogr B. 2012;883:68–75. doi: 10.1016/j.jchromb.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 20.Zhou B, Xiao JF, Tuli L, Ressom HW. LC-MS-based metabolomics. Mol BioSyst. 2012;8(2):470–481. doi: 10.1039/c1mb05350g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie G, Wang L, Chen T, Zhou K, Zhang Z, Li J, Sun B, Guo Y, Wang X, Wang Y, et al. A metabolite Array technology for precision medicine. Anal Chem. 2021;93(14):5709–5717. doi: 10.1021/acs.analchem.0c04686. [DOI] [PubMed] [Google Scholar]
- 22.Tang G, Du Y, Guan H, Jia J, Zhu N, Shi Y, et al. Butyrate ameliorates skeletal muscle atrophy in diabetic nephropathy by enhancing gut barrier function and FFA2-mediated PI3K/Akt/mTOR signals. Br J Pharmacol. 2021. 10.1111/bph.15693. [DOI] [PubMed]
- 23.Qiu J, Yan J, Liu W, Liu X, Lin J, Du Z, Qi L, Liu J, Xie G, Liu P, et al. Metabolomics analysis delineates the therapeutic effects of Huangqi decoction and astragalosides on alpha-naphthylisothiocyanate (ANIT) -induced cholestasis in rats. J Ethnopharmacol. 2021;268:113658. doi: 10.1016/j.jep.2020.113658. [DOI] [PubMed] [Google Scholar]
- 24.Wishart DS, Mandal R, Stanislaus A, Ramirez-Gaona M. Cancer Metabolomics and the Human Metabolome Database. Metabolites. 2016;6(1). 10.3390/metabo6010010. [DOI] [PMC free article] [PubMed]
- 25.Cao Z, Xia W, Zhang X, Yuan H, Guan D, Gao L. Hepatotoxicity of nutmeg: a pilot study based on metabolomics. Biomed Pharmacother. 2020;131:110780. doi: 10.1016/j.biopha.2020.110780. [DOI] [PubMed] [Google Scholar]
- 26.Vinaixa M, Samino S, Saez I, Duran J, Guinovart JJ, Yanes O. A guideline to univariate statistical analysis for LC/MS-based untargeted metabolomics-derived data. Metabolites. 2012;2(4):775–795. doi: 10.3390/metabo2040775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu L, Lai Q, Feng Q, Li Y, Feng J, Xu B. Serum metabolic profiling analysis of chronic gastritis and gastric Cancer by untargeted metabolomics. Front Oncol. 2021;11:636917. doi: 10.3389/fonc.2021.636917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009;374(9685):239–249. doi: 10.1016/S0140-6736(09)60491-8. [DOI] [PubMed] [Google Scholar]
- 29.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the global burden of disease study 2010. Lancet. 2012;380(9859):2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W, Guan KL. AMP-activated protein kinase and cancer. Acta Physiol. 2009;196(1):55–63. doi: 10.1111/j.1748-1716.2009.01980.x. [DOI] [PubMed] [Google Scholar]
- 31.Rehman G, Shehzad A, Khan AL, Hamayun M. Role of AMP-activated protein kinase in cancer therapy. Arch Pharm. 2014;347(7):457–468. doi: 10.1002/ardp.201300402. [DOI] [PubMed] [Google Scholar]
- 32.Faubert B, Vincent EE, Poffenberger MC, Jones RG. The AMP-activated protein kinase (AMPK) and cancer: many faces of a metabolic regulator. Cancer Lett. 2015;356(2):165–170. doi: 10.1016/j.canlet.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 33.Li T, Apte U. Bile acid metabolism and signaling in cholestasis, inflammation, and Cancer. Adv Pharmacol. 2015;74:263–302. doi: 10.1016/bs.apha.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernstein H, Bernstein C, Payne CM, Dvorak K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J Gastroentero. 2009;15(27):3329–3340. doi: 10.3748/wjg.15.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernstein C, Bernstein H, Payne CM, Beard SE, Schneider J. Bile salt activation of stress response promoters in Escherichia coli. Curr Microbiol. 1999;39(2):68–72. doi: 10.1007/s002849900420. [DOI] [PubMed] [Google Scholar]
- 36.Bernstein H, Payne CM, Bernstein C, Schneider J, Beard SE, Crowley CL. Activation of the promoters of genes associated with DNA damage, oxidative stress, ER stress and protein malfolding by the bile salt, deoxycholate. Toxicol Lett. 1999;108(1):37–46. doi: 10.1016/S0378-4274(99)00113-7. [DOI] [PubMed] [Google Scholar]
- 37.Payne CM, Crowley C, Washo-Stultz D, Briehl M, Bernstein H, Bernstein C, Beard S, Holubec H, Warneke J. The stress-response proteins poly (ADP-ribose) polymerase and NF-kappa B protect against bile salt-induced apoptosis. Cell Death Differ. 1998;5(7):623–636. doi: 10.1038/sj.cdd.4400395. [DOI] [PubMed] [Google Scholar]
- 38.Goldman A, Condon A, Adler E, Minnella M, Bernstein C, Bernstein H, Dvorak K. Protective effects of glycoursodeoxycholic acid in Barrett's esophagus cells. Dis Esophagus. 2010;23(2):83–93. doi: 10.1111/j.1442-2050.2009.00993.x. [DOI] [PubMed] [Google Scholar]
- 39.Adams SH, Hoppel CL, Lok KH, Zhao L, Wong SW, Minkler PE, Hwang DH, Newman JW, Garvey WT. Plasma Acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J Nutr. 2009;139(6):1073–1081. doi: 10.3945/jn.108.103754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mihalik SJ, Goodpaster BH, Kelley DE, Chace DH, Vockley J, Toledo FGS, DeLany JP. Increased levels of plasma Acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity. 2010;18(9):1695–1700. doi: 10.1038/oby.2009.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ganti S, Taylor SL, Kim K, Hoppel CL, Guo LN, Yang J, Evans C, Weiss RH. Urinary acylcarnitines are altered in human kidney cancer. Int J Cancer. 2012;130(12):2791–2800. doi: 10.1002/ijc.26274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Al-Bakheit A, Traka M, Saha S, Mithen R, Melchini A. Accumulation of Palmitoylcarnitine and its effect on pro-inflammatory pathways and calcium influx in prostate Cancer. Prostate. 2016;76(14):1326–1337. doi: 10.1002/pros.23222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.