Abstract
A new amino acid transporter was identified from the Arabidopsis expressed sequence tag cDNAs by expressing the cDNA in a yeast amino acid transport mutant. Transport analysis of the expressed protein in yeast showed that it transports aromatic and neutral amino acids, as well as arginine. This transporter (ANT1, aromatic and neutral transporter) also transports indole-3-acetic acid and 2,4-dichlorophenoxyacetic acid. The cDNA is 1.6 kb in length with an open reading frame that codes for a protein with 432 amino acids and a calculated molecular mass of 50 kD. Hydropathy analysis showed ANT1 is an integral membrane protein with 11 putative membrane-spanning domains. Southern analysis and a BLAST search of the Arabidopsis genome database suggests that ANT1 is part of a small gene family containing at least five members. Phylogenetic comparisons with other known amino acid transporters in plants suggests that ANT1 represents a new class of amino acid transporter. RNA gel-blot analysis showed that this transporter is expressed in all organs with highest abundance in flowers and cauline leaves.
Amino acids are the principal form of nitrogen available to the heterotrophic tissues of the plant (Bush, 1999). They are essential precursors not only for protein biosynthesis, but also in such diverse molecules as pigments, growth factors, lignin, nucleic acids, phytohormones, and phytoalexins. Amino acids are known as the “currency of nitrogen exchange” (Bush, 1999) because they are transported from sites of primary nitrogen assimilation to import-dependent tissues and organs by way of the plants' vascular system. Transport into plant cells is generally mediated by proton motive force coupled symporters (Bush, 1993; Fischer et al., 1998; Ortiz-Lopez et al., 2000).
Several amino acid transporters have been isolated from Arabidopsis by functional complementation of yeast amino acid transport mutants. AAP1 was the first isolated by functional complementation (Frommer et al., 1993; Hsu et al., 1993) and it was subsequently shown to be part of a small gene family of related transporters that are differentiated by substrate specificity and expression patterns (Fischer et al., 1995, 1998). Additional amino acid transporters have been isolated from Arabidopsis, including a cationic amino acid transporter (CAT1; Frommer et al., 1995), which is related to a previously described animal amino acid transporter (Yoshimoto et al., 1991; Christensen, 1992) and two Pro transporters (ProT1 and ProT2; Rentsch et al., 1996). Chen and Bush (1997) recently described a Lys and His transporter (LHT1). Orthologs of these transporters have been identified in other plants (Williams et al., 1992, 1996; Lalanne et al., 1995).
Amino acid transport activity is involved in all processes associated with nitrogen allocation during plant growth. Therefore, identifying novel amino acid transporter genes that participate in resource allocation is essential for understanding this fundamental process. One of the approaches that we have used (Chen and Bush, 1997) to identify additional amino acid transporters has been to survey the Arabidopsis expressed sequence tags (ESTs) cDNA database (Newman et al., 1994) for sequences that exhibit some similarity to AAP1 using the BLAST search protocol (Altschul et al., 1990). Several of the ESTs identified using this strategy included previously described clones, whereas others represented novel cDNAs that had small regions of sequence similarity. We obtained the novel EST cDNAs and tested them for amino acid transport activity by expressing them in yeast amino acid transport mutants. In the results reported here we describe a new class of amino acid transporter identified using this bioinformatic approach.
RESULTS
An EST cDNA Complements a Yeast Amino Acid Transport Mutant
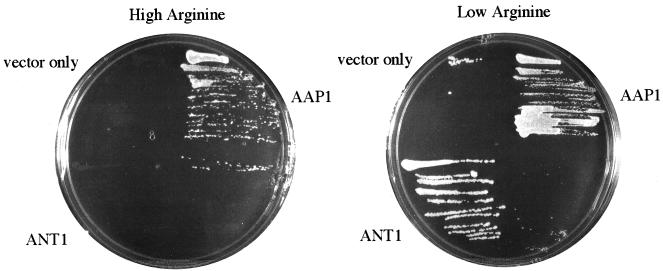
We selected several Arabidopsis EST cDNAs as possible amino acid transporters based on their low level of sequence similarity to AAP1. These ESTs were first analyzed by restriction digestion for pattern and length, and those that were sufficiently long to potentially contain a complete open reading frame were subcloned into a yeast expression vector. We expressed these in JT16 to test them for amino acid transport activity and initially identified LHT1 based on its ability to complement growth and mediate active transport (Chen and Bush, 1997). The other ESTs we screened did not complement growth in our initial analysis. However, hydropathy analysis of the partial amino acid sequences that were available suggested one of the non-complementing ESTs (EST42) contained several transmembrane domains that would be consistent with a transport protein. If EST42 encodes an amino acid transporter, we reasoned that its inability to complement JT16 may be unrelated to its ability to transport His. We noted that the high affinity transporter for Arg (CAN1: Ahmad and Bussey, 1986) is also deleted in JT16, and consequently the growth medium is supplemented with high Arg (6 mm). We hypothesized that EST42 may encode an Arg transporter that causes toxic levels of Arg to accumulate or that Arg acts as a competitive inhibitor of His transport under our growth conditions. We tested this notion by reducing the Arg concentration to 0.6 mm and found that EST42 then allowed for growth on low His (Fig. 1). These results suggested the expressed protein transports Arg and His.
Figure 1.
Arg levels altered the ability of ANT1 to complement JT16, a His auxotroph. The ANT1 cDNA was subcloned into λYES, a yeast expression vector, and transformed into JT16 for functional complementation of His transport. ANT1 did not complement JT16 under His-limiting conditions (130 μm His) in the presence of high Arg (6 mm). When the Arg concentration was reduced to 0.6 mm (low Arg), ANT1 allowed for growth. The positive control for this experiment was AAP1-expressing cells. AAP1 transports His, but not Arg (Frommer et al., 1993, Hsu et al., 1993). The negative control was insert-free vector.
Transport Properties of ANT1
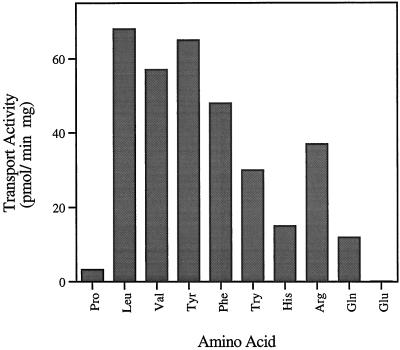
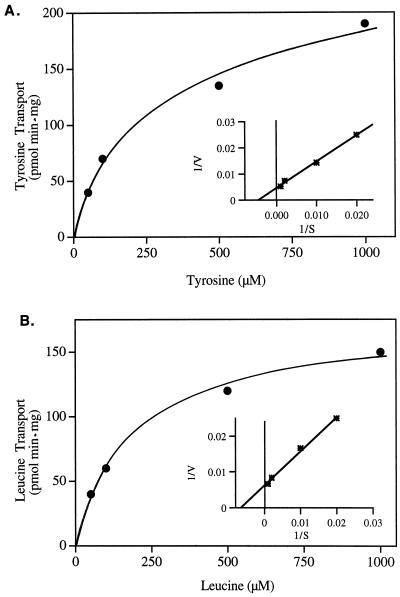
We measured the uptake of several amino acids into JT16-expressing EST42 to determine the substrate specificity of this transporter (Fig. 2). The expressed protein was able to transport a variety of amino acids with particularly high rates of flux for aromatic and neutral amino acids. We named this gene product ANT1, for aromatic and neutral amino acid transporter. As predicted from the complementation experiments (Fig. 1), the transporter also exhibited reasonable rates of His and Arg transport. Leu and Tyr exhibited saturable uptake kinetics with apparent Km values of 163 and 240 μm, respectively (Fig. 3).
Figure 2.
Transport in ANT1-expressing cells. Ten amino acids were tested as potential substrates in ANT1 expressing JT16 (A). Each amino acid was tested at 100 μm. The endogenous transport activity was subtracted using transformed JT16 with insert-free vector to measure background transport.
Figure 3.
Kinetics of amino acid transport in ANT1-expressing cells. Transport of Tyr (A) and Leu (B) were measured at a range of 50 to 1,000 μm. Inset, Lineweaver-Burk plot of the same data. The Km values for Tyr and Leu were 240 and 160 μm, respectively.
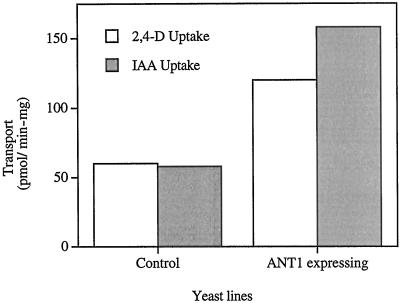
This is the first report of a plant amino acid transporter with significant rates of aromatic amino acid transport. That observation raised the possibility that this carrier may also be capable of transporting auxin because earlier biochemical and molecular investigations provided evidence that auxin uptake may be mediated by amino acid permeases (Jones, 1994; Bennett et al., 1996). This hypothesis is not an untenable idea given the structural similarity between indole-3-acetic acid (IAA) and Trp. Preliminary experiments examining this question showed that IAA was an effective competitor of Tyr uptake (data not shown). Therefore, we measured radiolabeled IAA and 2,4-dichlorophenoxyacetic acid (2,4-D) accumulation in NAT1 expressing cells and showed that both were transported (Fig. 4).
Figure 4.
IAA and 2,4-D transport into control and ANT1-expressing cells. IAA and 2,4-D were tested at 100 μm. Control transport represents yeast cells transformed with insert-free vectors. ANT1 was expressed using λYES.
ANT1 Belongs to a New Class of Transport Protein
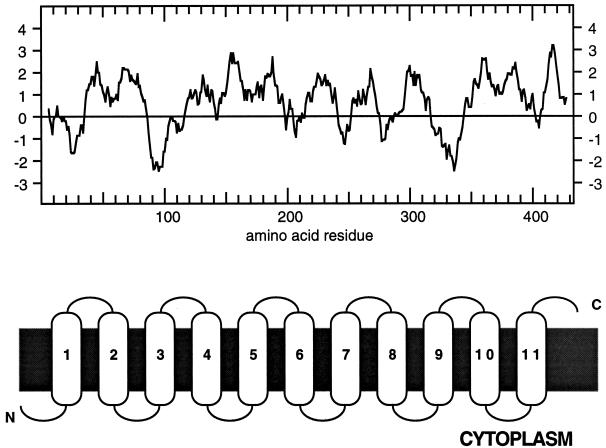
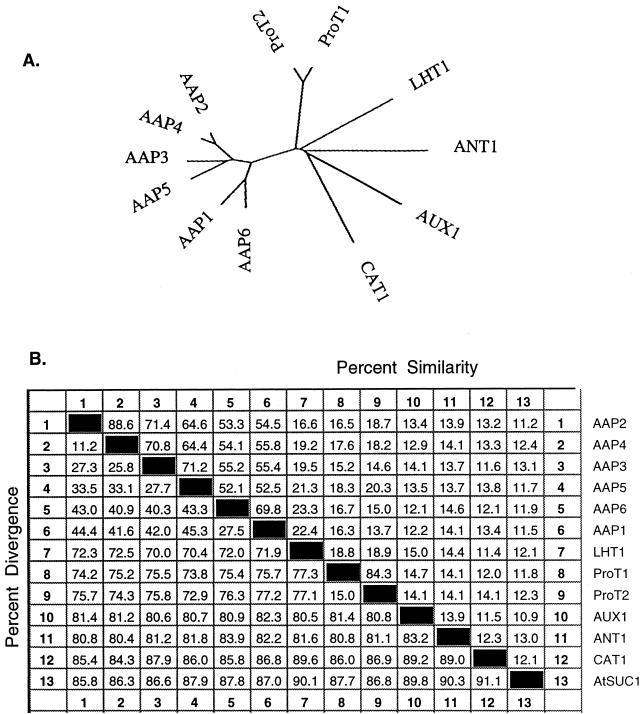
The ANT1 cDNA contains 1,636 bp (accession no. U39783) with an open reading frame that codes for a protein of 432 amino acid residues and a predicted molecular mass of 50 kD. The deduced amino acid sequence is highly hydrophobic with 11 putative transmembrane domains (Kyte and Doolittle, 1982; Fig. 5). Gapped BLAST results with scores greater than 100 (Altschul et al., 1997) from the current protein databases include a variety of putative amino acid transporters and hypothetical proteins from Arabidopsis, fruit fly, Saccharomyces cerevisiae, and Caenorhabditis elegans (data not shown). ANT1 also shares regions of similarity with a vesicular γ-amino-butyrate transporter associated with neurotransmission in rats (McIntire et al., 1997: 43% similar). Sequence alignments of the known families of plant amino acid and auxin transporters using Clustal W suggests ANT1 belongs to a new class of amino acid transporter (Fig. 6A). ANT1 is 15% similar to any of the previously described plant amino acid transporters and AUX1 (Fig. 6B), the putative auxin transporter (Bennett et al., 1996). In contrast, amino acid transporters that are considered to be part of well-defined gene families, such as the AAPs and ProTs, range from 52% to 87% similar. Likewise, transporters from unrelated families of carriers such as CAT1 (a basic amino acid transporter) and AtSUC1 (a proton-Suc symporter) range from 11% to 14% similar (Fig. 6B). We conclude from these data that ANT1 belongs to a new class of plant amino acid transporter, but we cannot rule out evolutionary links to other families of transporters.
Figure 5.
Hydropathy profile of ANT1. Hydropathy analysis of ANT1 (accession no. U39783) using the Kyte and Doolittle (1982) algorithm was performed by DNA Strider 1.1 (A) or Gunnar von Heijne's algorithm performed by TopPred II (B). The latter analysis used Kyte and Doolittle hydrophobicity values, as well as the positive inside rule and analysis of loop distances on both sides of the membrane.
Figure 6.
Phylogenetic comparison of ANT1 with other amino acid transporters. A phylogenetic analysis of ANT1 with other plant transporters was generated with the Clustal W method of sequence alignments using a PAM250 residue weight table (Higgins and Sharp, 1989). A, An unrooted tree comparing ANT1 with other plant amino acid transporters. B, Sequence pair distances using the same method. Accession numbers: AAP1, L16240; AAP2, X71787; AAP3, X77499; AAP4, X77500; AAP5, X77501; AAP6, X95736; ProT1, X995737; ProT2, X995738; LHT1, U39782; ANT1, U39783; AUX1, X98772; CAT1, X77502; and AtSUC1, X75365.
ANT1 May Be Part of a Small Gene Family in the Arabidopsis Genome and It Is Preferentially Expressed in Flowers and Cauline Leaves
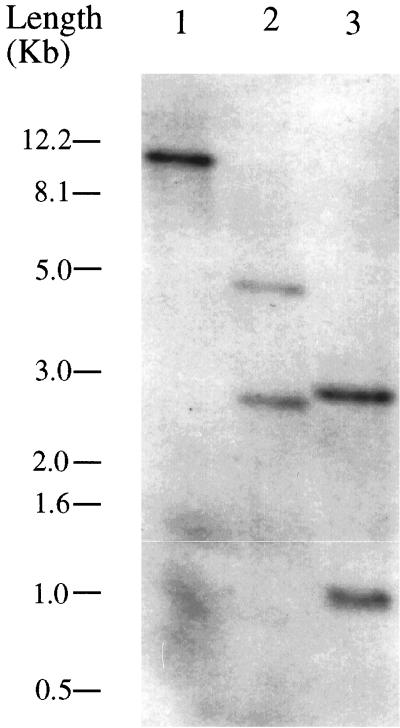
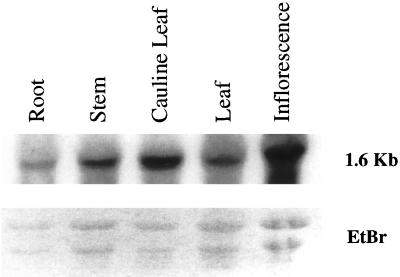
Southern hybridization suggests that ANT1 is a single copy gene in the Arabidopsis genome (Fig. 7). However, a BLAST search (on August, 28, 2000; Altschul et al., 1997) of the Arabidopsis nucleotide database identified four other genomic sequences with 41% to 55% similarity, suggesting ANT1 is a member of a small gene family. Tissue-specific northern analysis showed that ANT1 is transcribed as a 1.6-kb message that is expressed in all tissues with maximum abundance in flowers and cauline leaves (Fig. 8).
Figure 7.
Southern analysis of ANT1. Arabidopsis genomic DNA was digested with BamHI (lane 1), EcoRI (lane 2), and HindIII (lane 3). The ANT1 cDNA has an internal EcoRI and HindIII site.
Figure 8.
RNA gel-blot analysis of ANT1 expression. A, A single band at 1.6 kb hybridized with the ANT1 probe. B, Equal loading of total RNA (40 μg) was demonstrated for root, stem, cauline leaf, and leaf RNA using ethidium bromide staining. Inflorescence RNA was slightly more abundant.
DISCUSSION
In the results presented here we have described a new class of amino acid transporter based on substrate specificity and phylogenetic comparisons. ANT1 is the first plant amino acid transporter that preferentially moves aromatic amino acids in addition to neutral amino acids and Arg. It is interesting that ANT1 also transports auxin. BLAST searches of the Arabidopsis genome suggest that ANT1 is part of a small gene family and RNA-gel bot analysis showed this transporter is expressed in the major organs of the plant.
Analysis of ANT1 transport activity showed that it translocates aromatic and neutral amino acids, as well as Arg (Fig. 2). In yeast, two aromatic amino acid transporters have been identified and one of them, TAT1/VAP1, also transports neutral amino acids (Andre, 1995). The ability of ANT1 to transport Trp raised the possibility that it may also move auxin and ANT1 expressing yeast exhibited IAA and 2,4-D transport. It is important to note, however, that there was significant accumulation of these molecules in the insert-free vector controls (Fig. 4). This suggests a native carrier in yeast is capable of significant IAA and 2,4-D conductance or that these molecules also exhibit substantial rates of simple diffusion cross the plasma membrane. The latter hypothesis is consistent with experiments using purified liposomes that are capable of ΔpH-dependent IAA accumulation in the absence of any protein conducting pathways (D.R. Bush, unpublished data). Nonetheless, these results show that ANT1 possess some auxin transport activity. Given the complexity of auxin action in plant growth and development, and the broad substrate specificity of ANT1, the physiological significance of auxin transport activity will most easily be evaluated in future experiments characterizing the physiology of ANT1 knock-outs.
Hydropathy analysis of ANT1 suggests it has 11 putative transmembrane domains. Chang and Bush (1997) recently determined the topology of AAP1 and they showed that transporter has 11 transmembrane domains with the N terminus in the cytoplasm and C terminus facing outside the cell. Although ANT1 has low similarity to AAP transporter family, it is intriguing that the hydropathy profile of ANT1 also predicts 11 membrane-spanning domains.
The third putative transmembrane domains of PheP and AroP, two aromatic amino acid transporters in Escherichia coli, are rich in aromatic amino acids, and mutations in some of these residues abolish transport function (Pi et al., 1993). These non-polar amino acid residues are most likely facing the lipid phase of the bilayer and the charged residues in this helix could be facing a hydrophilic channel, thus participating in the transport reaction. This would be consistent with the loss of function observed for mutations in these residues (Pi et al., 1993). There are four aromatic amino acids within the putative third transmembrane domain of ANT1 (E, F, L, I, F, T, A, Q, C, G, G, S, V, A, Y, L, V, F, I, G). Future experiments exploring the role of these residues in the transport reaction should be revealing.
Sequence comparisons among the known amino acid transporters in plants suggest that ANT1 is not part of a previously described family of carriers (Fig. 6). Thus, ANT1 represents a new class of amino acid transporter in plants. A recent search of the Arabidopsis genome (August 28, 2000) identified four additional sequences with 41% to 55% similarity, suggesting ANT1 is a member of a small gene family in Arabidopsis. Moreover, sequence comparisons with the current protein databases identified a variety of putative transporters and hypothetical proteins from Arabidopsis, fruit fly, Rattus norvegicus, C. elegans, and S. cerevisiae that exhibit at least 45% similarity with ANT1, suggesting it is part of a eukaryotic gene family. It is noteworthy that no prokaryotic sequences were identified in this search.
Amino acids are the currency of nitrogen exchange in plants and their transporters play key roles in all aspects of amino acid allocation (Bush, 1999). Given the diverse pathways of nitrogen assimilation and the different tissues involved in amino acid partitioning, it isn't surprising that several gene families that code for a variety of amino acid transporters have been identified. These genes are differentiated by tissue-specific expression patterns that may also be influenced by developmental and/or environmental signals (Ortiz-Lopez et al., 2000). ANT1 is expressed in every organ of the plant (Fig. 8), suggesting a possible role in the systemic distribution of amino acids. This pattern would also be consistent with a role in auxin transport, perhaps through the plant vascular system. Future experiments determining the cell-specific expression pattern of ANT1 and the phenotype of ANT1 knockouts should provide additional insight into its role in nitrogen metabolism and auxin function.
MATERIALS AND METHODS
Growth Conditions
Arabidopsis ecotype Columbia plants were grown on soil in growth chambers with 12 h of light at 150 to 250 μE m−2 s−1 at 20°C, 70% relative humidity, and a 12-h dark period at 18°C, 70% relative humidity for 3 to 4 weeks. Tissues were harvested, frozen in liquid nitrogen, and stored at −80°C until further use.
Cloning Strategy for EST cDNA
EST cDNAs were obtained from the Arabidopsis Stock Center. Each EST was analyzed by restriction digestion and those with sufficiently long inserts were subcloned into λYES (Elledge et al., 1991) and pYES2 (Invitrogen, Carlsbad, CA). All digestions, ligations, and transformation were as described by Sambrook et al. (1989).
Yeast Transformation and Complementation
The Saccharomyces cerevisiae strain used in this study was JT16 (MATa hip1-614 his4-401 ura3-52 ino1 can1; Tanaka and Fink 1985). JT16 was maintained in the S1 medium (Hsu et al., 1993) supplemented with 0.2% (w/v) Ura. Electroporation was used to introduce plasmids into JT16 (Gietz et al., 1992) and Ura+ transformants were selected on S1 medium containing 1 m sorbitol (Hsu et al., 1993). Positive transformants were maintained in S1 medium. Complementation of JT16 was tested with His-limiting medium as described, except that the Arg content was lowered to 0.01% (w/v; Hsu et al., 1993). Insert-free vectors were run as negative controls in all relevant experiments.
Transport Measurements
Yeast transport activity was measured as described by Hsu et al. (1993). In brief, cells were grown to midlogarithmic phase, collected by centrifugation, and suspended at 200 to 300 mg cells per milliliter in transport buffer that contained His-limiting medium minus His and Arg (pH adjusted to 5.0 with KOH). Ten microliters of cells was used in a 400-μL transport reaction that contained 0.2 to 1.0 μCi of 14C-labeled amino acid and unlabeled amino acid to the desired final concentration. At predetermined time points, 180 μL was removed from the reaction buffer and the cells were collected and washed on a micropore filter. JT16 cells transformed with insert-free vectors were used to measure background transport activity in all transport experiments and results are reported as net transport (i.e. ANT1-expressing cell accumulation minus accumulation by insert-free vector controls). Each experiment was repeated at least three times and the se never exceeded 14% of the averaged results.
Sequencing
Dideoxynucleotide sequencing (Sanger et al., 1977) was adapted from the Sequenase booklet using Sequenase 2.0 version (Amersham, Buckinghamshire, UK) and double-strand DNA as templates. Plasmids for sequencing were isolated using QIAprep column (Qiagen, Valencia, CA). Sequencing reactions were performed according to the manufacturer's description.
Southern Hybridization
Arabidopsis DNA was isolated according to Dellaporta et al. (1983). The DNA was digested with different restriction enzymes, resolved in 0.8% (w/v) agarose gel in Tris-acetate EDTA buffer, and depurinated with 25 n HCl. DNA was transferred onto Hybond N+ membrane (Amersham) using 0.4 n NaOH, and UV cross-linked (Stratagene, La Jolla, CA). The probe was radiolabeled by random primer (Megaprimed System, Amersham) and purified with either Bio-Spin column (Bio-Rad, Hercules, CA) or Qiaquick nucleotide removal kit (Qiagen). The hybridization and wash conditions followed the suggestions of the membrane manufacturer (Amersham). Low stringency washes consisted of two 10-min washes with 2× SSPE (Sambrook et al., 1989) and 0.1% (w/v) SDS at room temperature. The high stringency wash included an additional wash with 1× SSPE and 0.1% (w/v) SDS at 65°C for 15 min and two washes of 0.1× SSPE and 0.1% (w/v) SDS at 65°C for 30 min.
RNA Gel Blot
Total RNA was isolated with TriZol following the manufacturer's instructions (Gibco-BRL, Cleveland). Forty micrograms of total RNA was size fractionated in a formaldehyde gel (Zielinski, 1987), transferred to a nylon membrane (Nytran, Schleicher & Schuell, Keene, NH), UV-cross linked, and prehybridized in buffer (7% [w/v] SDS, 0.25 m Na2HPO4, pH 7.4, 1 mm EDTA, pH 8, and 1% [w/v] bovine serum albumin) for 2 h at 65°C. The denatured radiolabeled probe was diluted into the buffer (50 ng DNA/mL) and hybridized overnight. The filters were washed twice for 15 min at room temperature in 2× SSC, 0.1% (w/v) SDS and was then washed for 15 min at room temperature in 0.1× SSC, 0.1% (w/v) SDS. The α32P-labeled probe was generated using random primers (Megaprimed System, Amersham). The specific activity of the DNA probe was routinely 107 cpm/μg. The filters were exposed for 7 d with enhancing screens. Staining with ethidium bromide showed approximately equal amounts of total RNA in each lane.
LITERATURE CITED
- Ahmad M, Bussey H. Yeast arginine permease: nucleotide sequence of the CAN1 gene. Curr Genet. 1986;10:587–592. doi: 10.1007/BF00418125. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre B. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast. 1995;11:1575–1611. doi: 10.1002/yea.320111605. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Bush DR. Proton-coupled sugar and amino acid transporters in plants. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:513–542. [Google Scholar]
- Bush DR. Amino Acid Transport. In: Singh BK, editor. Plant Amino Acids: Biochemistry and Biotechnology. New York: Marcel Dekker; 1999. pp. 305–318. [Google Scholar]
- Chang H-C, Bush DR. Topology of NAT2, a prototypical example of a new family of amino acid transporters. J Biol Chem. 1997;48:30552–30557. doi: 10.1074/jbc.272.48.30552. [DOI] [PubMed] [Google Scholar]
- Chen L, Bush DR. LHT1, a lysine and histidine-specific amino acid transporter. Plant Physiol. 1997;115:1127–1134. doi: 10.1104/pp.115.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen HN. A retrovirus uses cationic amino acid transporter as a cell surface receptor. Nutr Rev. 1992;50:47–48. doi: 10.1111/j.1753-4887.1992.tb02512.x. [DOI] [PubMed] [Google Scholar]
- Dellaporta ST, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;4:19–21. [Google Scholar]
- Elledge SJ, Mulligan JT, Ramer S, Spottswood M, Davis RW. λYES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc Natl Acad Sci USA. 1991;88:1731–1735. doi: 10.1073/pnas.88.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer WN, Andre B, Rentsch D, Krolkiewicz S, Tegeder M, Breitkreuz K, Frommer WB. Amino acid transport in plants. Trends Plant Sci. 1998;3:188–195. [Google Scholar]
- Fischer WN, Kwart M, Hummel S, Frommer WB. Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis. J Biol Chem. 1995;27:16315–16320. doi: 10.1074/jbc.270.27.16315. [DOI] [PubMed] [Google Scholar]
- Frommer WB, Hummel S, Riesmeier JW. Expression cloning in yeast of a cDNA encoding a broad specificity amino acid permease from Arabidopsis thaliana. Proc Natl Acad Sci USA. 1993;90:5944–5948. doi: 10.1073/pnas.90.13.5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer WB, Hummel S, Unseld M, Ninnemann O. Seed and vascular expression of a high-affinity transporter for cationic amino acids in Arabidopsis. Proc Natl Acad Sci USA. 1995;92:12036–12040. doi: 10.1073/pnas.92.26.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH. Improved methods for high efficiency transformation of intact yeast cells. Nucleic Acid Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM. Fast and sensitive multiple sequence alignments on a microcomputer. CABIOS. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- Hsu LC, Chiou TJ, Chen L, Bush DR. Cloning a plant amino acid transporter by functional complementation of a yeast amino acid transport mutant. Proc Natl Acad Sci USA. 1993;90:7441–7445. doi: 10.1073/pnas.90.16.7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM. Auxin-binding proteins. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:393–420. [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lalanne E, Vedel F, de Paepe R. Isolation of a Nicotiana sylvestris cDNA (Accession no. U31932) encoding an amino acid transporter homologous to Arabidopsis thaliana amino acid permeases. Plant Physiol. 1995;109:722. [Google Scholar]
- McIntire SL, Reimer RJ, Schuske K, Edwards RH, Jorgensen EM. Identification and characterization of the vesicular GABA transporter. Nature. 1997;389:870–876. doi: 10.1038/39908. [DOI] [PubMed] [Google Scholar]
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Lopez A, Chang HC, Bush DR. Amino acid transporters in plants. Biochim Biophys Acta. 2000;1465:275–280. doi: 10.1016/s0005-2736(00)00144-9. [DOI] [PubMed] [Google Scholar]
- Pi J, Wookey PJ, Pittard AJ. Site-directed mutagenesis reveals the importance of conserved charged residues for the transport activity of the PheP permease of Escherichia coli. J Bacteriol. 1993;175:7500–4. doi: 10.1128/jb.175.22.7500-7504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Hirner B, Schmelzer E, Frommer WB. Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. Plant Cell. 1996;8:1437–1446. doi: 10.1105/tpc.8.8.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka J, Fink GR. The histidine permease gene (HIP1) of Saccharomyces cerevisiae. Gene. 1985;38:205–214. doi: 10.1016/0378-1119(85)90219-7. [DOI] [PubMed] [Google Scholar]
- Williams LE, Bick JA, Neelam A, Weston KN, Hall JL. Biochemical and molecular characterization of sucrose and amino acid carriers in Ricinus communis. J Exp Bot. 1996;47:1211–1216. doi: 10.1093/jxb/47.Special_Issue.1211. [DOI] [PubMed] [Google Scholar]
- Williams LE, Nelson SJ, Hall JL. Characterization of solute/proton cotransport in plasma membrane vesicles from Ricinus cotyledons, and a comparison with other tissues. Planta. 1992;186:541–550. doi: 10.1007/BF00198034. [DOI] [PubMed] [Google Scholar]
- Yoshimoto T, Yoshimoto E, Meruelo D. Molecular cloning and characterization of a novel human gene homologoud to the murineecotropic retroviral receptor. Virology. 1991;185:10–17. doi: 10.1016/0042-6822(91)90748-z. [DOI] [PubMed] [Google Scholar]
- Zielinski RE. Calmodulin mRNA in barley (Hordeum vulgare L.), apparent regulation by cell proliferation and light. Plant Physiol. 1987;84:937–943. doi: 10.1104/pp.84.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]