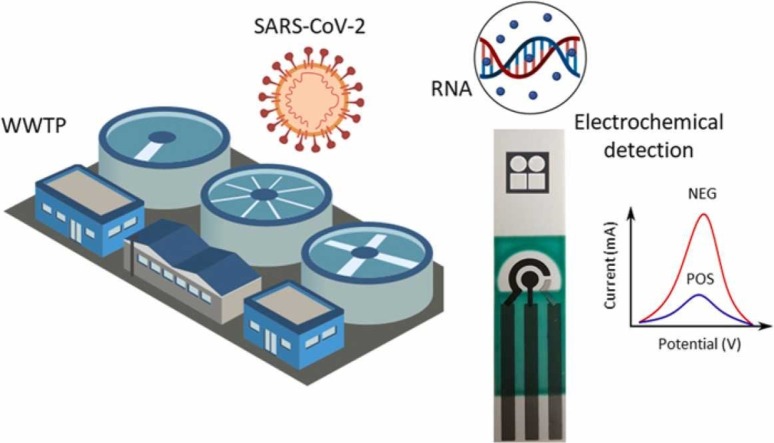
Abstract
The current pandemic COVID-19 caused by the coronavirus SARS-CoV-2, has generated different economic, social and public health problems. Moreover, wastewater-based epidemiology could be a predictor of the virus rate of spread to alert on new outbreaks. To assist in epidemiological surveillance, this work introduces a simple, low-cost and affordable electrochemical sensor to specifically detect N and ORF1ab genes of the SARS-CoV-2 genome. The proposed sensor works based on screen-printed electrodes acting as a disposable test strip, where the reverse transcription loop-mediated isothermal amplification (RT-LAMP) reaction takes place. Electrochemical detection relies upon methylene blue as a redox intercalator probe, to provide a diffusion-controlled current encoding the presence and concentration of RT-LAMP products, namely amplicons or double-stranded DNA. We test the performance of the sensor by testing real wastewater samples using end-point and time course measurements. Results show the ability of the electrochemical test strip to specifically detect and quantify RT-LAMP amplicons below to ~ 2.5 × 10−6 ng/μL exhibiting high reproducibility. In this sense, our RT-LAMP electrochemical sensor is an attractive, efficient and powerful tool for rapid and reliable wastewater-based epidemiology studies.
Keywords: SARS-CoV-2, RT-LAMP, Electrochemical sensor, Redox intercalator, Wastewater, Epidemiology
Graphical Abstract
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viruses, has rapidly spread worldwide bringing serious consequences for human life and the global economy [1]. Since the outbreak, SARS-CoV-2 pandemic has infected more than 239 million people worldwide resulting in death of about 4.8 million people (https://covid19.who.int/). Hence, curbing the spread of infection is paramount at present.
Testing is a key starting point to contain COVID-19 transmission and accurate SARS-CoV-2 detections. Nevertheless, clinical testing is more expensive and time consuming to detect new outbreaks than testing of wastewater for the presence of SARS-CoV-2 [2], [3], [4], [5]. Currently, for detecting and quantifying SARS-CoV-2 one can find, rapid antigen detection (RAD) [6], rapid detection of antibodies (RDA) [7] and molecular detection [8]. This latter is the most reliable method to detect the presence SARS-CoV-2 due to its specificity [9], [10]. On the other hand, testing of wastewater for the presence of SARS-CoV-2 nucleic acid as a surveillance and management tool is essential to slow the spread of the virus [11]. SARS-CoV-2 may enter wastewater systems from pathogen shedding in human waste, resulting in a potentially fecal-oral transmission with a serious health consequence [12], [13], [14]. Recently, several groups in different countries isolated and detected the genetic material of SARS-CoV-2 in wastewater using reverse transcription quantitative polymerase chain reaction (RT-qPCR), as a gold standard technique [15], [16], [17]. However, one important problem is that RT-qPCR still requires expensive laboratory infrastructure and skilled technicians or scientists to complete the assay. Furthermore, more efforts are needed to develop rapid and accurate detection tools for wastewater surveillance and management of the SARS-CoV-2 spread using molecular diagnostics in limited-resources settings [18].
In this context, it results imperative the application of versatile and affordable tools to detect viral or microbiological pathogens in environmental samples in a fast and sensitive manner [19]. The nucleic acid based isothermal amplification methods have been extensively deployed as a sensitive and straightforward techniques [20]. Specifically, loop-mediated isothermal amplification with simultaneous reverse-transcription (RT-LAMP) allows for rapid and analytically sensitive detection of nucleic acids within one hour that requires only a heat source [21]. Several groups are currently developing LAMP-based protocols for the detection of SARS-CoV-2 in clinical samples [22], [23], [24], [25]. However, scarce information is reported about RT-LAMP technique as a cheaper and faster option for monitoring the genetic material of SARS-CoV-2 in wastewater-based epidemiology [26], and its integration with attractive sensing schemes.
Among the wide variety of transduction mechanisms for detecting nucleic acid amplification, one can find electrochemical-based sensors [27], [28], optical devices [29], colorimetric assays [30], luminescence-based sensors [31], and surface plasmon resonance apparatus [32], to mention only a few. Particularly, electrochemical transduction has demonstrated its ability to provide a cost-effective alternative to circumvent manufacturing and integration processes to robust devices. Moreover, electrochemical sensors exhibit several advantages such as low-cost, portability, miniaturization and high reliability, ideal for in-situ measurements [33], [34]. Nevertheless, common electrochemical biosensors need a labeled receptor to be immobilized on the sensitive element or electrode [35], [36]. Due to the COVID-19 pandemic, several investigations have been devoted to simplify the experimental processes and methods to provide affordable and versatile platforms suitable for a easy-to-develop sensors in resource-limited or field settings [37]. For instance, the work in [38] shows the trends in electrochemical sensors for rapid detection of SARS-CoV-2 from human samples focusing on viral nucleic acid, immunoglobulin, antigen, and the entire viral particles.
Monitoring and detecting SARS-CoV-2, however, remain a challenging task, even more when testing environmental samples due to its complex structure. To overcome the difficulties encountered in classical benchtop equipment and methods, herein we report the development and potentiality of an electrochemical sensor for detecting SARS-CoV-2 in wastewater samples. To the best of our knowledge, the sensor innovates in the following aspects:
-
•
By working around a RT-LAMP reaction, it is a cost-effective and less-time consuming alternative to the classical RT-PCR amplification, without losing specificity.
-
•
It works around screen-printed electrodes (SPEs) and minimal instrumentation, which is a current trend for field-deployable and low-cost detection systems.
-
•
The sensor is primarily devoted to retrieve end-point results for detecting RT-LAMP amplicons, and additionally, shows promising results for real-time quantification.
-
•
Its potentiality is demonstrated by measuring real wastewater samples for a current sanitary problem, which is an underestimated application for electrochemical detection devices.
Summarizing, the proposed device acts as a test strip to detect, previously concentrated and extracted, nucleic acid fragments of SARS-CoV-2 by monitoring the diffusion-controlled current, promoted by the amplification reaction and a redox intercalating probe. As a whole, the device aims to show how easily affordable technologies can be versatile tools for environmental surveillance in water analysis laboratories with the minimum infrastructure to perform the concentration and extraction of nucleic acids.
The rest of the paper is organized as follows. In Section 2 we introduce the methods for sample collection, RNA concentration and extraction, as well as the monitoring of RT-LAMP reaction with electrochemical transduction. Section 3 shows the experimental results to assess the performance of the proposal alongside a thorough discussion. Finally, Section 4 is devoted to the conclusions.
2. Materials and methods
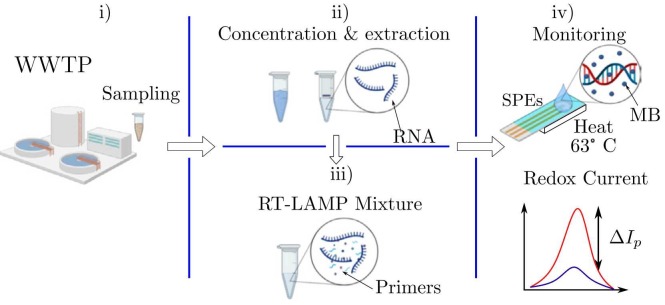
Fig. 1 shows the device workflow comprising four main stages: i) wastewater sampling, ii) RNA concentration, iii) RT-LAMP mixture, and iv) the electrochemical monitoring of RT-LAMP reaction. First, the samples are collected and then the nucleic acids are extracted and concentrated using a custom-developed method [18]. Together these methodologies take a time of 1 h 30 min in the laboratory with minimum infrastructure. Afterwards, the RNA is mixed with the RT-LAMP primers and methylene blue (MB) as a redox intercalator for the electrochemical transduction. Hence, a micro-volume sample is drop cast over the surface of custom fabricated screen-printed electrodes (SPEs), wherein the RT-LAMP reaction takes place by controlling the local temperature at 63 ∘C. Thereby, the resultant diffusion-controlled current, promoted by the redox process, is monitored by a portable potentiostat to provide a measure of the RT-LAMP reaction. The amplification and monitoring take approximately 30 min. Finally, the peak current change %ΔI p encodes the concentration of the nucleic acids according to the sensor model. Jointly, the turnaround time for a complete experiment is 2 h. To validate the electrochemical monitoring performance, a colorimetric assay was simultaneously performed on the evaluated wastewater samples.
Fig. 1.
Workflow of RT-LAMP based electrochemical sensor in wastewater samples. i) Sampling from wastewater treatment plant. ii) Nucleic acid extraction and concentration. iii) RT-LAMP mixtures for genetic amplification. iv) Electrochemical monitoring of the RT-LAMP products via redox current.
2.1. Wastewater samples
The sampling was carried out at two wastewater treatment plants (WTTP) in the metropolitan area of the City of Queretaro, Mexico (see Table 1). In South and Santa Rosa plants, the influent was sampled. Samples were collected from the period between May 31 and June 7, 2021. The influent samples (500 mL) were collected during the morning (9–11 am) and kept at 4 ∘C until their use.
Table 1.
Samples taken from wastewater treatment plants (WWTP) alongside its capability and location coordinates.
| Sample | WWTP | Sampling date | Plant capability (L/s) | Location |
|---|---|---|---|---|
| L9 | South | May 31th | 400 | 20∘ 33’ 19.9" N, 100∘ 25’ 49.0" W |
| IPS | South | June 7th | 400 | 20∘ 33’ 19.9" N, 100∘ 25’ 49.0" W |
| ISR | Santa Rosa | June 7th | 30 | 20∘ 44’ 4.6" N, 100∘ 27’ 4.1" W |
2.2. Concentration and extraction of RNA
Samples were concentrated the same day of sampling, using the electronegative membrane method owing its detection limit for SARS-CoV-2 genes [18]. Briefly, the pH of samples was adjusted to 3.5 with 2 N HCl and then were filtered through a negatively charged nitrocellulose membrane (0.45 μm pore diameter, Millipore, Netherlands). According to the manufacturer’s instructions, the membranes were cut and used directly in the RNeasy Power Microbiome extraction kit (Qiagen, Germany) for RNA extraction. RNA was stored at − 20∘C until its use.
2.3. RT-LAMP reactions
In a two step RT-LAMP assay, RNA (5μL) was reverse transcribed with QuantiTect Reverse Transcription Kit (Qiagen, Germany) following manufacturer’s instructions. 50 μL LAMP reaction contained: 5 μL Buffer Bst (NEB), 3 μL MgSO4 (NEB), 5 μL of 2 mM dNTPs (Thermo Scientific), 2 μL of 10X primer’s core mix, 2 μL of 10X primer’s loop mix, 10 U of Bst 2.0 DNA polymerase (NEB), 4 μL cDNA and nuclease free water to make up 50 μL of reaction volume. The specific primers were designed by [39] for the SARS-CoV-2 N and ORF1ab genes, and were validated using PrimerExplorer V5 (https://primerexplorer.jp/e/) as shown in Table 2. RT-LAMP reactions were monitored by the electrochemical sensor using MB at 6 μM at 63∘C in uniform temperature for 30 min. Also, the reactions of RT-LAMP assays were checked on 1% agarose gel stained with SYBR Safe DNA Gel Stain for the presence of ladder pattern, and the products were also verified by sequencing. Afterwards, the concentration of the RT-LAMP amplified products was estimated using a Nanodrop™ spectrophotometer. Finally, to test the specificity of the amplification, the products of the RT-LAMP reactions were validated by sequencing (Genbank OM522662).
Table 2.
RT-LAMP primers to detect SARS-CoV-2 genome [39] in wastewater samples.
| Gene | Primer | Sequence (5’ to 3’) | Concentration (μM) |
|---|---|---|---|
| ORF1ab Amplicon: 203 bp | F3 | TGCTTCAGTCAGCTGATG | 0.2 |
| B3 | TTAAATTGTCATCTTCGTCCTT | 0.2 | |
| FIP | TCAGTACTAGTGCCTGTGCC- CACAATCGTTTTTAAACGGGT | 1.6 | |
| BIP | TCGTATACAGGGCTTTTGACATCTA- TCTTGGAAGCGACAACAA | 1.6 | |
| Loop F | CTGCACTTACACCGCAA | 0.8 | |
| Loop B | GTAGCTGGTTTTGCTAAATTCC | 0.8 | |
| N Amplicon: 165 bp | F3 | CGGCAGTCAAGCCTCTTC | 0.2 |
| B3 | TTGCTCTCAAGCTGGTTCAA | 0.2 | |
| FIP | TCCCCTACTGCTGCCTGGAG- CGTTCCTCATCACGTAGTCG | 1.6 | |
| BIP | TTCTCCTGCTAGAATGGCTGGC- TCTGTCAAGCAGCAGCAAAG | 1.6 | |
| Loop B | AATGGCGGTGATGCTGCTCT | 0.8 |
2.4. Electrochemical monitoring
For electrochemical monitoring, MB was added to the RT-LAMP mixture as an electroactive intercalator [40], exhibiting strong and specific binding ability to dsDNA amplicon without inhibiting the RT-LAMP process. A negative control template (NTC) was composed by RT-LAMP master mix and MB without genome owing the well-know performance of the RNA concentration and extraction [18]. The detection strategy relies upon the measurement of a change in the faradaic current, promoted by the free-to-diffuse state of the MB as the RT-LAMP progresses. Thus, the current amplitude is attenuated because the MB becomes less active to an electron exchange following the complex formation with DNA amplicon compared with its free counterpart. As the sensing element, a three electrode electrochemical cell based on screen-printed electrodes (SPEs) was used. This configuration allowed to be treated as a disposable test strip. Therein, the working and counter electrodes were fabricated using carbon paste; whereas, the reference electrode was made of Ag/AgCl ink. The test strips have an active area of 12.56 mm2 over a flexible substrate of 33 mm width, 10 mm length and 1 mm thickness, which allows to measure samples of 50 μL volume. Before electrochemical monitoring, each SPEs-based test strip was prepared by performing a cyclic voltammetry (CV) in the range from 1.0 to − 0.6 V and a scan rate of 100 mV/s scan rate using a Tris acetate buffer (10 mM, pH 7.4). Afterwards, the SPEs were rinsed with deionized water (DI) and dried in a stream of N2. On the other hand, owing the working temperature of RT-LAMP reaction, a polyamide-based substrate heater was placed behind the electrochemical test strip. A digital controller was adjusted to provide a temperature of 63 ± 0.5∘C for the SPEs thus enabling enough stability for the isothermal amplification process. The electrochemical experiments were carried out using a custom-made portable potentiostat [41]. Due to its high sensitivity and reliable performance, square-wave voltammetry (SWV) was selected as the analytical technique to monitor RT-LAMP reactions. For this purpose, measurements were performed in the potential range from 0 V to - 0.5 V, with a step amplitude of 5 mV and frequency of 15 Hz. Each measurement was repeated five times in a biological triplicate to assess the variability of the sensor.
2.5. Colorimetric assays
The assay was performed in a 50 μL reaction mixture containing 4 μL of 10x primer mix of 16 μM (each) of Forward Inner Primer (FIP) and Backward Inner Primer (BIP), 2 μM (each) of F3 and B3 primers, 4 μM (each) of Forward Loop (LoopF) and Backward Loop (LoopB) primers, 20 μL of WarmStart™ Colorimetric Lamp 2X Master Mix (M1800, New England BioLabs INC.) 10 μL of DNAse, RNAase free water, and 5 μl of RNA template. The reaction mixture was performing at 63 ∘C for 30 min on a dry bath. Finally, the concentration of the RT-LAMP amplified products were estimated using a Nanodrop™ spectrophotometer.
3. Results and discussion
3.1. Performance and specificity
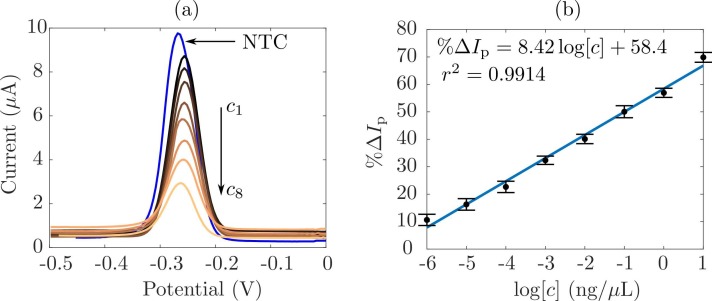
As the first step, we characterized the electrochemical sensor by using well-know concentrations of double-stranded DNA. Fig. 2(a) shows the square-wave voltammograms retrieved for a NTC, as the base signal and, for eight concentrations (c 1 = 0.001, c 2 = 0.01, c 3 = 0.1, c 4 = 1, c 5 = 10, c 6 = 100, c 7 = 1000 and c 4 = 10000 × 10−3 ng/μL). Therein, one can see how the peak current is located around the MB formal potential, − 0.25 V. Moreover, it is worth to notice a decrease in the peak current as the concentration grows up. This situation is due to the MB acting as a redox intercalating probe. For the NTC sample, there are no amplicons, and hence, the electroactive molecules tend to diffuse onto the surface of the working electrode, thus giving a high peak current signal. On the other hand, as the concentration increases due to the RT-LAMP reaction, more dsDNA was synthesized and the MB intercalated, hence the amount of free electroactive molecules decrease. As a result, it causes a drop in the peak amplitude of the diffusion-controlled current. To quantify such phenomenon, one can compute the change in the peak current (%ΔI p) due to the concentration as [42].
| (1) |
where I m and I 0 are the measured peak currents in the presence of amplicons and for the NTC, respectively. Following this rationale, Fig. 2(b) shows the calibration curve of the proposed sensor. The plot illustrates the experimental data (black dots), the uncertainty (vertical lines) and the linear model that best fits them (solid line). As expected, the output of the sensor, %ΔI p, exhibits an increasing trend as the concentration c grows up. Indeed, a large amount of amplicons implies less amount of free MB at the electrode surface, and hence, the current ratio I m∕I 0, in 1, diminishes. Thereby, the sensor sensitivity can be computed as ; whereas, r 2 determines a highly linear behavior and a goodness of the fit ~ 99%, thus leading to a limit-of-detection (LoD) of 0.038 × 10−3 ng∕μL in the concentration range from 0.001 to 10,000 × 10−3 ng/μL, which is in the same order of magnitude as previous reports [43].
Fig. 2.
Calibration results for sensing SARS-CoV-2 amplicons in wastewater samples. (a) Square-wave voltammograms of the base signal (NTC) and four concentrations from c1 to c4. (b) Calibration curve of the ratio of change in the peak current % ΔIp as a function of the concentration. The sensitivity is and the goodness of the fit is r2.
3.2. RT-LAMP monitoring
Once the performance of the sensor was assessed, we performed experiments in two scenarios to verify the ability of the sensor to monitor RT-LAMP reactions. First, we considered an end-point measurement to retrieve information about SARS-CoV-2 positive samples. Lastly, we monitored the time course of the RT-LAMP reaction for a positive sample at different concentrations to assess the dynamic performance of the sensor.
3.2.1. End-point measurements
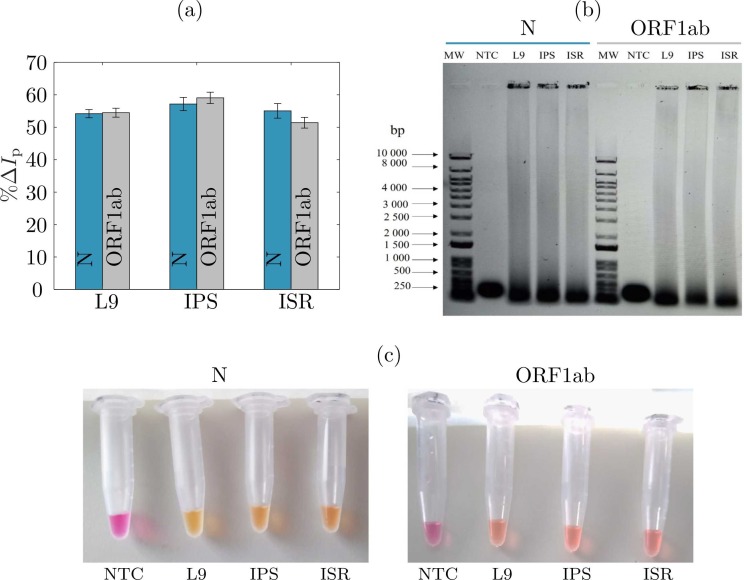
To test the reliability of our sensor in the basis of MB/RT-LAMP reaction, we performed standard RT-LAMP (without MB) and MB-LAMP in eppendorf tubes while also measuring a similar MB/RT-LAMP reaction with our electrochemical test strip. To guarantee reproducible results, we used the same target concentrations, for three positive samples labeled as L9, IPS and ISR (see Table 1) and verified the results using gel electrophoresis. Firstly, electrochemical measurements were performed following an end-point procedure. That is, the voltammograms were measured after 30 min of the RT-LAMP reaction. Subsequently, we computed the peak current change % ΔI p as in 1. Fig. 3(a) depicts the results retrieved by our sensor for these three samples. One can see, the peak current change decreased almost 55%, which reflects the success of the RT-LAMP reaction. It makes sense, the intercalation of MB to double-stranded amplicons significantly reduced the concentration of free MB at the electrode surface, and hence, diminished the peak current with respect to the negative control sample. To verify that the end-point measurements were reliable we performed an electrophoresis test on a 1% agarose gel for RT-LAMP N and ORF1ab reaction products, respectively. Fig. 3(b) shows the agarose gel for molecular weight markers (MW), negative control samples (NTC) and the three tested samples (L9, IPS and ISR). From Fig. 3(b), one can observe that only the positive reactions resulted in a ladder pattern, while the NTCs did not show any detectable amplicons. Ultimately, by using the calibration curve shown in Fig. 2(a), we computed the estimated concentrations by our sensor for two SARS-CoV-2 genes, N and ORF1ab, in three samples. To validate the electrochemical sensor, we concurrently performed a colorimetric assay as shown in Fig. 2(c). Therein, one can see the negative reactions indicated in pink, and how the positive reactions change the color to yellow. As expected, this effect is due to the presence of phenol red within the RT-LAMP reaction mix, which allows a straightforward differentiation among positive and negative samples. Finally, the concentration of both, electrechmical and colorimetric assays, was verified with the Nanodrop™ spectrophotometer. Table 3 summarizes the concentration results given by its mean value and uncertainty. These results thereby allow us to confirm that the proposed sensor reproduces well the Nanodrop™ measurements and agree with the colorimetric readings. Thus, the sensor accuracy is above 90%, with the largest error for the sample IPS, which is the one more concentrated. End-point measurements allowed to measure the concentration of the amplicons for the N and ORF1ab genes after the amplification process promoted by the RT-LAMP reaction. One should keep in mind that, those concentrations are not the number of copies in the total RNA isolated from the wastewater samples. Hence, this experiment was useful to validate the sensor to only detect the presence of SARS-CoV-2 genome, and to corroborate the concentration of the dsDNA products given by the redox current change due to the RT-LAMP amplification.
Fig. 3.
Results of sensing SARS-CoV-2 in wastewater samples. a) Ratio of change in the peak current % ΔIp for genes N and ORF1ab of three samples labeled as L9, IPS and ISR. (b) Loop-mediated amplification visualized in a 1% agarose gel stained with SYBR Safe DNA Gel Stain, showing characteristic RT-LAMP amplicon profiles in positive samples (L9, IPS, ISR) and no amplification in non-template controls (NTC). (c) Colorimetric assays for determining the presence genes N and ORF1b after the RT-LAMP reaction.
Table 3.
Comparison of the mean value and uncertainty for measured concentrations using the electrochemical sensor, the Nanodrop™ spectrophotometer and the colorimetric assays.
| Sample | Gene | Concentration (ng/μL) |
||
|---|---|---|---|---|
| Sensor | Nanodrop | Colorimetry | ||
| L9 | N | 2.42 ± 0.26 | 2.29 ± 0.17 | 2.15 ± 0.38 |
| ORF1ab | 2.60 ± 0.28 | 2.77 ± 0.13 | 2.84 ± 0.18 | |
| IPS | N | 3.15 ± 0.42 | 3.47 ± 0.04 | 2.98 ± 0.32 |
| ORF1ab | 3.45 ± 0.86 | 3.69 ± 0.03 | 3.01 ± 0.14 | |
| ISR | N | 2.71 ± 0.47 | 2.82 ± 0.13 | 2.90 ± 0.50 |
| ORF1ab | 1.95 ± 0.34 | 1.99 ± 0.05 | 2.03 ± 0.45 | |
3.2.2. Time course measurements
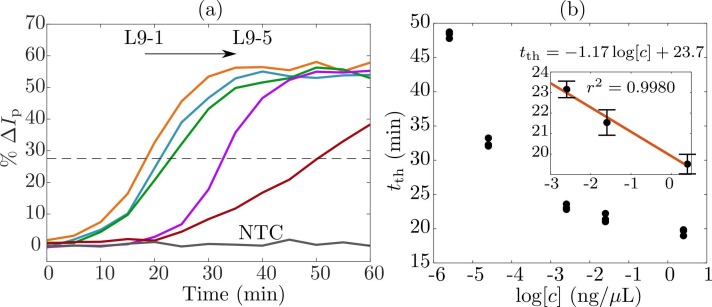
As the last experiment, we measured the time course of the RT-LAMP reaction. For this purpose, we used five different initial nucleic acid concentrations using the sample L9 (L9–1 =251.8, L9–2 =25.18, L9–3 =2.518, L9–4 =0.02518 and L9–5 =0.002518 × 10−3 ng/μL), to specifically detect the fragment N of SARS-CoV-2 genome. Electrochemical measurements were carried out every 5 min up to 60 min. That is, at each time instant, we collected the voltammogram retrieved by SWV, and computed the peak current change with respect to a negative control sample. Fig. 4(a) shows the mean value for a biological triplicate of the peak current change % ΔI p as a function of RT-LAMP reaction time. Therein, one can see the amplification time course described by sigmoidal-like curves as in classical genetic amplification processes [44]. From left to right, it is possible to see the effect of the concentration in the RT-LAMP time course. Interestingly, for concentrations L9–1, L9–2 and L9–3, the amplification traces show an exponential phase occurring starting from 15 up to 20 min. This situation could be attributed to the relatively high concentration of the samples, such that the RT-LAMP amplicons can be easily generated within a short-time period. On the other hand, for highly diluted concentrations, the RT-LAMP reaction takes more time to start generating amplicons, starting at approximately 30 and 50 min for samples L9–4 and L9–5, respectively. Therefore, MB remains free at the surface of the electrodes for a relatively long time until amplicons start generating.
Fig. 4.
RT-LAMP time course measurements for sensing SARS-CoV-2 in wastewater samples. (a) Retrieved peak current change %ΔIp for five concentrations L9–1 = 251.8, L9–2 = 25.18, L9–3 = 2.518, L9–4 = 0.02518 and L9–5 = 0.002518 × 10−3 ng/μL as a function of the reaction time. (b) Correlation between the time-to-threshold tth and concentration c in a logarithmic scale.
As shown in Fig. 4(a), the dashed horizontal line indicates the threshold value of the peak current change at which RT-LAMP exhibits an exponential transition. Hence, for each concentration the time-to-threshold value t th indicates the needful time for the amplification to succeed. To quantitatively assess the RT-LAMP performance, Fig. 4(b) depicts the relationship among the time-to-threshold and the concentration in a logarithmic scale. From there, one can deduce that the sensor operates in two regimes.
-
•
For concentrations below 10−3 ng/μL, the required amplification time is above to 30 min.
-
•
For highly concentrated samples, greater than 10−3 ng/μL, the time-to-threshold of RT-LAMP reaction takes less than 25 min.
Though this behavior was to be expected, it is convenient to focus on the last three samples. Therefore, the inset of Fig. 4(b) confirms an inverse correlation between the time-to-threshold and the concentration, with a sensitivity of 1.17 min/, and linearity of approximately 99%. This result shows how the electrochemical sensor could also serve as an alternative method for quantitative molecular tests with enough sensitivity. Also, the time course experiment confirmed that the electrochemical test strip was able to detect concentrations as low as 2.5 × 10−6 ng/μL. However, it is worth to notice that, for highly diluted samples, the isothermal reaction requires more time to reach the plateau. This could be acceptable for classical molecular tests; nonetheless, for our purposes, it could be problematic as we are interested in fast measurements for applications in limited-resources settings.
3.3. Towards sensor-based wastewater surveillance
The use of wastewater surveillance as an epidemiological tool has raised more interest in detecting the increment of the viral load, which could be related to disease spread in the population. Another approach though, is the near-source tracking, applying the surveillance on a small spatial scale, in vulnerable or higher risk groups, like people in prisons, schools, hospitals and factories. Therein, the only detection of the virus in their wastewater or sewage system is a valuable tool to prevent a local outbreak, followed by targeted clinical tests [45]. For instance, using a conventional RT-qPCR approach monitoring the sewage system for a prison, Carrillo-Reyes et al. [18] were able to detect the presence of the SARS-CoV-2 previous the report of clinical cases by the local health authorities. Following the current trends, the proposed electrochemical sensor showed promising results for detecting SARS-CoV-2 in real wastewater samples. The main advantages of the device are that, it does not require sophisticated infrastructure to succeed; and the electrochemical detection preserve acceptable sensitivity compared with optical methods, but its instrumentation is cost-effective for field-deployable devices. Following this rationale, the described approach is at least 5000 USD cheaper than a classical setup by replacing the thermocycler and the optical detection apparatus. In further studies, the proposed sensor can be modified to integrate all the methods, such as concentration and extraction stages, in a single device. This improvement could be a significant advance towards point-of-collection devices for automated analysis in wastewater surveillance for near-source tracking to rapidly identify SARS-CoV-2, as well as other pathogens.
4. Conclusions
In this work, a low-cost, affordable and accurate electrochemical sensor for the sensitive detection of SARS-CoV-2 nucleic acids was successfully developed and evaluated. The results demonstrated the ability of the sensor to perform measurements in real wastewater samples, and were validated with a colorimetric assay and a commercial apparatus. The proposed sensor was primarily devoted to detect the presence of SARS-CoV-2 genome by means of end-point measurements with a detection limit of 38 × 10−6 ng/μL. Moreover, the sensing device was also able to track the time course of the RT-LAMP reaction for concentrations as low as 2.5 × 10−6 ng/μL. Though is a promising result for quantitative assays, it requires further validation with RT-qPCR experiments. The versatility and features of the electrochemical RT-LAMP-based sensor make it as an attractive alternative to detect SARS-CoV-2 in low-resource settings for surveillance the COVID-19 spread in environmental scenarios. Finally, the device could be further improved to be an integrated system for stand-alone measurements, and can be extended for detecting other pathogens.
CRediT authorship contribution statement
Roberto G. Ramírez-Chavarría: Conceptualization, Methodology, Funding acquisition, Writing – original draft. Elizabeth Castillo-Villanueva: Investigation, Methodology, Writing – original draft. Bryan E. Alvarez-Serna: Investigation, Methodology, Software, Writing – review & editing. Julián Carrillo-Reyes Investigation, Methodology, Validation, Writing – review & editing. Rosa María Ramírez-Zamora: Writing – review & editing, Validation, Resources. Germán Buitrón Writing – review & editing, Validation, Resources. Luis Alvarez-Icaza Writing – review & editing, Validation, Resources.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the grant DGAPA-UNAM-PAPIIT TA100221.
Editor: V. Victor
References
- 1.Mofijur M., Fattah I.R., Alam M.A., Islam A.S., Ong H.C., Rahman S.A., Najafi G., Ahmed S., Uddin M.A., Mahlia T. Impact of COVID-19 on the social, economic, environmental and energy domains: lessons learnt from a global pandemic. Sustain. Prod. Consum. 2021;26:343–359. doi: 10.1016/j.spc.2020.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lotfi M., Hamblin M.R., Rezaei N. COVID-19: transmission, prevention, and potential therapeutic opportunities. Clin. Chim. Acta. 2020;508:254–266. doi: 10.1016/j.cca.2020.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 6.Scohy A., Anantharajah A., Bodéus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26(6):845–848. doi: 10.1371/journal.pone.0240502. [DOI] [PubMed] [Google Scholar]
- 8.Jalandra R., Yadav A.K., Verma D., Dalal N., Sharma M., Singh R., Kumar A., Solanki P.R. Strategies and perspectives to develop SARS-CoV-2 detection methods and diagnostics. Biomed. Pharmacother. 2020;129 doi: 10.1016/j.biopha.2020.110446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arena F., Pollini S., Rossolini G.M., Margaglione M. Summary of the available molecular methods for detection of SARS-CoV-2 during the ongoing pandemic. Int. J. Mol. Sci. 2021;22(3):1298. doi: 10.3390/ijms22031298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumura Y., Shimizu T., Noguchi T., Nakano S., Yamamoto M., Nagao M. Comparison of 12 molecular detection assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2021;23(2):164–170. doi: 10.1016/j.jmoldx.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kataki S., Chatterjee S., Vairale M.G., Sharma S., Dwivedi S.K. Concerns and strategies for wastewater treatment during COVID-19 pandemic to stop plausible transmission. Resour. Conserv. Recycl. 2021;164 doi: 10.1016/j.resconrec.2020.105156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lahrich S., Laghrib F., Farahi A., Bakasse M., Saqrane S., ElMhammedi M. Review on the contamination of wastewater by COVID-19 virus: Impact and treatment. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.142325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pulicharla R., Kaur G., Brar S.K. A year into the COVID-19 pandemic: Rethinking of wastewater monitoring as a preemptive approach. J. Environ. Chem. Eng. 2021 doi: 10.1016/j.jece.2021.106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langone M., Petta L., Cellamare C., Ferraris M., Guzzinati R., Mattioli D., Sabia G. SARS-CoV-2 in water services: presence and impacts. Environ. Pollut. 2021;268 doi: 10.1016/j.envpol.2020.115806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., et al. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hata A., Hara-Yamamura H., Meuchi Y., Imai S., Honda R. Detection of SARS-CoV-2 in wastewater in japan during a COVID-19 outbreak. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrillo-Reyes J., Barragán-Trinidad M., Buitrón G. Surveillance of SARS-CoV-2 in sewage and wastewater treatment plants in Mexico. J. Water Process. Eng. 2021;40 doi: 10.1016/j.jwpe.2020.101815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pilevar M., Kim K.T., Lee W.H. Recent advances in biosensors for detecting viruses in water and wastewater. J. Hazard. Mater. 2021;410 doi: 10.1016/j.jhazmat.2020.124656. [DOI] [PubMed] [Google Scholar]
- 20.Augustine R., Hasan A., Das S., Ahmed R., Mori Y., Notomi T., Kevadiya B.D., Thakor A.S. Loop-mediated isothermal amplification (LAMP): a rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology. 2020;9(8) doi: 10.3390/biology9080182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12) doi: 10.1093/nar/28.12.e63. e63–e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim B., Ratcliff J., Nawrot D.A., Yu Y., Sanghani H.R., Hsu C.-C., Peto L., Evans S., Hodgson S.H., Skeva A., et al. Clinical validation of optimised RT-LAMP for the diagnosis of SARS-CoV-2 infection. Sci. Rep. 2021;11(1):1–11. doi: 10.1038/s41598-021-95607-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang W.E., Lim B., Hsu C.-C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020;13(4):950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamb L.E., Bartolone S.N., Ward E., Chancellor M.B. Rapid detection of novel coronavirus/severe acute respiratory syndrome coronavirus 2 (sars-cov-2) by reverse transcription-loop-mediated isothermal amplification. PLoS One. 2020;15(6):1–15. doi: 10.1371/journal.pone.0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., Chen W.-H., Yin X. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin. Chem. 2020;66(7):975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amoah I.D., Mthethwa N.P., Pillay L., Deepnarain N., Pillay K., Awolusi O.O., Kumari S., Bux F. RT-LAMP: a cheaper, simpler and faster alternative for the detection of SARS-CoV-2 in wastewater. Food Environ. Virol. 2021:1–10. doi: 10.1007/s12560-021-09489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farzin L., Sadjadi S., Sheini A., Mohagheghpour E. A nanoscale genosensor for early detection of COVID-19 by voltammetric determination of RNA-dependent RNA polymerase (RdRP) sequence of SARS-CoV-2 virus. Microchim. Acta. 2021;188(4):1–12. doi: 10.1007/s00604-021-04773-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng Y., Pan Y., Sun Z., Li J., Yi Y., Yang J., Li G. An electrochemical biosensor for sensitive analysis of the SARS-CoV-2 RNA. Biosens. Bioelectron. 2021;186 doi: 10.1016/j.bios.2021.113309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmad M., Sharma P., Kamai A., Agrawal A., Faruq M., Kulshreshtha A. HRPZyme assisted recognition of SARS-CoV-2 infection by optical measurement (HARIOM) Biosens. Bioelectron. 2021;187 doi: 10.1016/j.bios.2021.113280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moon J., Kwon H.-J., Yong D., Lee I.-C., Kim H., Kang H., Lim E.-K., Lee K.-S., Jung J., Park H.G., et al. Colorimetric detection of SARS-CoV-2 and drug-resistant ph1n1 using crispr/dcas9. ACS Sens. 2020;5(12):4017–4026. doi: 10.1021/acssensors.0c01929. [DOI] [PubMed] [Google Scholar]
- 31.Fan Z., Yao B., Ding Y., Zhao J., Xie M., Zhang K. Entropy-driven amplified electrochemiluminescence biosensor for RdRp gene of SARS-CoV-2 detection with self-assembled DNA tetrahedron scaffolds. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang C.-H., Wu T.-H., Chang C.-C., Lo H.-Y., Liu H.-W., Huang N.-T., Lin C.-W. Biosensing amplification by hybridization chain reaction on phase-sensitive surface plasmon resonance. Biosensors. 2021;11(3) doi: 10.3390/bios11030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill A., Tait S., Baillie C., Virdis B., McCabe B. Microbial electrochemical sensors for volatile fatty acid measurement in high strength wastewaters: a review. Biosens. Bioelectron. 2020 doi: 10.1016/j.bios.2020.112409. [DOI] [PubMed] [Google Scholar]
- 34.Pilevar M., Kim K.T., Lee W.H. Recent advances in biosensors for detecting viruses in water and wastewater. J. Hazard. Mater. 2020 doi: 10.1016/j.jhazmat.2020.124656. [DOI] [PubMed] [Google Scholar]
- 35.Lu D., Zhu D.Z., Gan H., Yao Z., Fu Q., Zhang X.J. Prospects and challenges of using electrochemical immunosensors as an alternative detection method for SARS-CoV-2 wastewater-based epidemiology. Sci. Total Environ. 2021 doi: 10.1016/j.scitotenv.2021.146239. [DOI] [Google Scholar]
- 36.Mao K., Zhang H., Yang Z. An integrated biosensor system with mobile health and wastewater-based epidemiology (iBMW) for COVID-19 pandemic. Biosens. Bioelectron. 2020;169 doi: 10.1016/j.bios.2020.112617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suleman S., Shukla S.K., Malhotra N., Bukkitgar S.D., Shetti N.P., Pilloton R., Narang J., NeeTan Y., Aminabhavi T.M. Point of care detection of covid-19: advancement in biosensing and diagnostic methods. Chem. Eng. J. 2021;414 doi: 10.1016/j.cej.2021.128759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar N., Shetti N.P., Jagannath S., Aminabhavi T.M. Electrochemical sensors for the detection of sars-cov-2 virus. Chem. Eng. J. 2022;430 doi: 10.1016/j.cej.2021.132966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Tholoth M., Bau H.H., Song J. A single and two-stage, closed-tube, molecular test for the 2019 novel coronavirus (COVID-19) at home, clinic, and points of entry. ChemRxiv. 2020 doi: 10.26434/chemrxiv.11860137. 10.26434/chemrxiv.11860137.v1. [DOI] [Google Scholar]
- 40.Martin A., Bouffier L., Grant K.B., Limoges B., Marchal D. Real-time electrochemical lamp: a rational comparative study of different dna intercalating and non-intercalating redox probes. Analyst. 2016;141:4196–4203. doi: 10.1039/C6AN00867D. [DOI] [PubMed] [Google Scholar]
- 41.Ramírez-Chavarría R.G., Sánchez-Pérez C., Romero-Ornelas L., Ramón-Gallegos E. Time-constant-domain spectroscopy: an impedance-based method for sensing biological cells in suspension. IEEE Sens. J. 2021;21(1):185–192. doi: 10.1109/JSEN.2020.3014569. [DOI] [Google Scholar]
- 42.Ahmed M.U., Nahar S., Safavieh M., Zourob M. Real-time electrochemical detection of pathogen DNA using electrostatic interaction of a redox probe. Analyst. 2013;138:907–915. doi: 10.1039/C2AN36153A. [DOI] [PubMed] [Google Scholar]
- 43.Kumblathan T., Liu Y., Uppal G.K., Hrudey S.E., Li X.-F. Wastewater-based epidemiology for community monitoring of SARS-CoV-2: progress and challenges. ACS Environ. 2021 doi: 10.1021/acsenvironau.1c00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swillens S., Dessars B., Housni H.E. Revisiting the sigmoidal curve fitting applied to quantitative real-time pcr data. Anal. Biochem. 2008;373(2):370–376. doi: 10.1016/j.ab.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 45.Hassard F., Lundy L., Singer A.C., Grimsley J., DiCesare M. Innovation in wastewater near-source tracking for rapid identification of COVID-19 in schools. Lancet Microbe. 2021;2(1):e4–e5. doi: 10.1016/S2666-5247(20)30193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]