Abstract
Whereas remarkable advances have uncovered mechanisms that drive nervous system assembly, the processes responsible for the lifelong maintenance of nervous system architecture remain poorly understood. Subsequent to its establishment during embryogenesis, neuronal architecture is maintained throughout life in the face of the animal’s growth, maturation processes, the addition of new neurons, body movements, and aging. The Caenorhabditis elegans protein SAX-7, homologous to the vertebrate L1 protein family of neural adhesion molecules, is required for maintaining the organization of neuronal ganglia and fascicles after their successful initial embryonic development. To dissect the function of sax-7 in neuronal maintenance, we generated a null allele and sax-7S-isoform-specific alleles. We find that the null sax-7(qv30) is, in some contexts, more severe than previously described mutant alleles and that the loss of sax-7S largely phenocopies the null, consistent with sax-7S being the key isoform in neuronal maintenance. Using a sfGFP::SAX-7S knock-in, we observe sax-7S to be predominantly expressed across the nervous system, from embryogenesis to adulthood. Yet, its role in maintaining neuronal organization is ensured by postdevelopmentally acting SAX-7S, as larval transgenic sax-7S(+) expression alone is sufficient to profoundly rescue the null mutants’ neuronal maintenance defects. Moreover, the majority of the protein SAX-7 appears to be cleaved, and we show that these cleaved SAX-7S fragments together, not individually, can fully support neuronal maintenance. These findings contribute to our understanding of the role of the conserved protein SAX-7/L1CAM in long-term neuronal maintenance and may help decipher processes that go awry in some neurodegenerative conditions.
Keywords: neuronal maintenance, lifelong, L1, sax-7, Ig, cleavage
Introduction
An important yet poorly understood question of neurobiology is how the organization of neural circuits is maintained over a lifetime to ensure their proper function. Largely established during embryogenesis, the architecture of the nervous system needs to persist throughout life in the face of the animal’s growth, the addition of new neurons, maturation processes, body movements, and aging. Whereas significant progress has been made in understanding the processes driving neuronal development, little is known about the mechanisms ensuring lifelong maintenance of nervous system architecture and function.
Research using Caenorhabditis elegans has uncovered a number of immunoglobulin (Ig) superfamily molecules required for the long-term maintenance of neuronal architecture (Benard and Hobert 2009). These include the large extracellular protein DIG-1 (Benard et al. 2006; Johnson and Kramer 2012), the small two-Ig domain proteins ZIG-3, ZIG-4, and ZIG-10 (Aurelio et al. 2002; Benard and Hobert 2009; Benard et al. 2012; Cherra and Jin 2016), the ectodomain of the FGF receptor EGL-15 (Bülow et al. 2004), as well as SAX-7/L1CAM (Zallen et al. 1999; Sasakura et al. 2005; Wang et al. 2005; Pocock et al. 2008; Zhou et al. 2008). Here, we further the investigation of SAX-7’s role in the lifelong maintenance of neuronal architecture.
SAX-7 is an evolutionary conserved transmembrane cell adhesion molecule homologous to mammalian L1CAM (Hortsch 2000; Chen et al. 2001; Hortsch et al. 2014). In C. elegans, SAX-7 exists as two main isoforms, a long isoform SAX-7L and a short isoform SAX-7S. These two isoforms are identical for their intracellular tail, transmembrane domain (TM), and most of their extracellular region including five identical fibronectin type III domains (FnIII), and four Ig-like domains. They differ in the N-terminal extracellular region, where SAX-7S has four Ig domains (Ig 3–6), whereas SAX-7L has six Ig domains (Ig 1–6). Transgenes of SAX-7S, but not of SAX-7L, rescue the defects of sax-7 loss-of-function mutants, indicating that the SAX-7S isoform is central to sax-7 functions (Sasakura et al. 2005; Wang et al. 2005; Pocock et al. 2008; Ramirez-Suarez et al. 2019). Vertebrate proteins of the SAX-7/L1CAM family include L1CAM, NrCAM, CHL1, and Neurofascin (Brummendorf and Rathjen 1996; Brummendorf et al. 1998; Hortsch 2000; Haspel and Grumet 2003; Hortsch et al. 2014).
sax-7/L1CAM is well known to contribute to the development of distinct neurons in C. elegans. It is involved in dendrite development and axon guidance (Zhao et al. 1998; Heiman and Pallanck 2011; Schafer and Frotscher 2012; Dong et al. 2013; Salzberg et al. 2013; Diaz-Balzac et al. 2015; 2016; Zhu et al. 2017; Yip and Heiman 2018; Chen et al. 2019; Ramirez-Suarez et al. 2019; Cebul et al. 2020; Sherry et al. 2020). In flies and mammals, homologs of sax-7 function in neuronal migration, axon guidance, and synaptogenesis (Bieber et al. 1989; Hall and Bieber 1997; Sonderegger et al. 1998; Rougon and Hobert, 2003; Godenschwege et al. 2006). In humans, mutations in L1CAM severely impair neuronal development, leading to disorders collectively referred to L1 or CRASH syndrome for corpus callosum hypoplasia, mental retardation, aphasia, spastic paraplegia, and hydrocephalus (Fransen et al. 1997; Hortsch et al. 2014).
Besides their roles in neuronal development, SAX-7/L1CAM family members also function in the mature nervous system to preserve neuronal organization. In C. elegans, sax-7 is required for maintaining neuronal organization well after development is completed, as specific neuronal structures that initially develop normally in sax-7 mutant animals, later become disorganized. For instance, in sax-7 mutants, a subset of axons within the ventral nerve cord, which developed normally during embryogenesis, become displaced to the contralateral fascicle during the first larval stage; and neurons within embryonically established ganglia become progressively disorganized by late larval stages and adulthood in sax-7 mutants (Zallen et al. 1999; Sasakura et al. 2005; Wang et al. 2005; Pocock et al. 2008; Zhou et al. 2008). Such postdevelopmental neuronal disorganization displayed by sax-7 mutant animals can be prevented if animals are paralyzed (Sasakura et al. 2005; Pocock et al. 2008), indicating that the mechanical stress from body movements contributes to perturbing neuronal architecture in these mutants. In mammals, roles for L1 family members in the adult nervous system have been revealed as well, through the study of conditional knockouts. Adult-specific knockout of neurofascin affects rats behavior and alters the axon initial segment in mice (Kriebel et al. 2011; Zonta et al. 2011); knockout of L1CAM specifically in the adult mouse brain leads to behavioral deficits and synaptic transmission changes (Law et al. 2003); and CHL1 conditional depletion in a subtype of forebrain neurons in mice leads to defects in working memory duration (Kolata et al. 2008). Thus, L1CAM family of neural adhesion proteins contributes to preserving the functionality of the mammalian adult nervous system.
Despite the evolutionarily conserved importance of SAX-7/L1CAM, its role in the long-term maintenance of the neuronal architecture remains unclear. In order to better understand how SAX-7/L1CAM participates in neuronal maintenance, here we have generated and characterized a null allele of sax-7, tested the temporal requirements for sax-7S neuronal maintenance function, determined the endogenous expression pattern of SAX-7S, and assessed the function of SAX-7S cleavage products in neuronal maintenance. Our results further our understanding of the roles of the evolutionarily conserved molecule SAX-7/L1CAM in the lifelong persistence of neuronal organization and function.
Materials and methods
Nematode strains and genetics
Nematode cultures were maintained in an incubator at 20°C (unless otherwise noted) on Nematode growth medium agar plates (NGM) plates seeded with Escherichia coli OP50 bacteria as described (Brenner 1974). Alleles used in this study are listed in Table 1. Strains were constructed using standard genetic procedures and are listed in Table 2. Genotypes were confirmed by genotyping PCR or by sequencing when needed. Primers used to build strains are listed in Table 3. All the mutant alleles and reporter strains are outcrossed with the Bristol N2 wild-type strain at least three times prior to use for analysis or strain building.
Table 1.
List of sax-7 mutant alleles used
| Allele | Nature of alleles | Location on cosmid C18F3 | Reference |
|---|---|---|---|
| qv31 |
732-bp insertion sfGFP::SAX-7S construct |
After 12,809 | This study |
| qv30 |
19,972-bp deletion Total loss of function |
8,364–2,8335 | This study |
| qv25 |
47-bp insertion, creates an ORF frameshift and a stop codon in sax-7S signal peptide |
After 12,785 | This study |
| qv26 |
36-bp in frame insertion, but disrupts sax-7S signal peptide |
After 12,785 | This study |
| eq2 | 648-bp deletion | 8,041–8,688 | Wang et al. (2005) |
| nj53 | 724-bp deletion | 8,122–8,845 | Sasakura et al. (2005) |
| nj48 | 582-bp deletion | 12,457–13,038 | Sasakura et al. (2005) |
| tm1448 | 1,727-bp deletion | 22,599–24,325 |
Mitani lab at NBRP C. elegans |
| eq1 | 2,020-bp deletion | 26,591–28,605 | Wang et al. (2005) |
Table 2.
List of strains used
| Name | Genotype | Transgene | Reference |
|---|---|---|---|
| Wild-type reference strains | |||
| N2 | Brenner (1974) | ||
| hdIs29 V | Psra-6::DsRed2; Podr-2::cfp | Schmitz et al. (2008) | |
| OH4589 | bwIs2 otIs133 II | Pflp-1::gfp, Pttx-3::rfp | Pocock et al. (2008) |
| VQ51 | oyIs14 V | Psra-6::gfp | Sarafi-Reinach et al. (2001) |
| sax-7S knock-ins | |||
| VQ1290 | sax-7(qv31) IV | [sfgfp::sax-7S] | This study |
| TH502 | unc-119(ed3) III; ddIs290 | [sax-7::ty1::egfp::3FLAG] | Sarov et al. (2012) |
| sax-7 mutants | |||
| VQ1047 | sax-7(qv30) IV | This study | |
| VQ1058 | sax-7(qv30) IV; hdIs29 V | This study | |
| VQ1057 | sax-7(qv30) IV; bwIs2 otIs133 II | This study | |
| VQ1000 | sax-7(qv30) IV; oyIs14 V | This study | |
| OH4587 | sax-7(nj48) IV | Sasakura et al. (2005) | |
| OH7984 | sax-7(nj48) IV; oyIs14 V | Pocock et al. (2008) | |
| VQ397 | sax-7(nj48) IV; hdIs29 V | This study | |
| OH4588 | sax-7(nj48) IV; bwIs2 otIs133 II | Pocock et al. (2008) | |
| VQ976 | sax-7(qv25) IV | This study | |
| VQ1011 | sax-7(qv25) IV; oyIs14 V | This study | |
| VQ1269 | sax-7(qv25) IV; hdIs29 V | This study | |
| VQ1259 |
sax-7(qv25) IV; bwIs2 otIs133 II |
This study | |
| VQ977 | sax-7(qv26) IV | This study | |
| VQ1012 | sax-7(qv26) IV; oyIs14 V | This study | |
| LH2 | sax-7(eq2) IV | Wang et al. (2005) | |
| OH6028 | sax-7(eq2) IV; oyIs14 V | Benard et al. (2012) | |
| LH81 | sax-7(eq1) IV | Wang et al. (2005) | |
| OH8904 | sax-7(eq1) IV; oyIs14 V | Benard et al. (2012) | |
| IK637 | sax-7(nj53) IV | Sasakura et al. (2005) | |
| OH9002 | sax-7(nj53) IV; oyIs14 V | Benard et al. (2012) | |
| sax-7(tm1448) IV | Mitani lab at NBRP; Wang et al. (2005) | ||
| Transgenic lines | |||
| VQ1357 | sax-7(qv30) IV; hdIs29 V; qvEx377 | pCB189, Plgc-11::gfp, pBSK+. Line #1 | This study |
| VQ1358 | sax-7(qv30) IV; hdIs29 V; qvEx378 | pCB189, Plgc-11::gfp, pBSK+. Line #2 | This study |
| VQ1359 | sax-7(qv30) IV; hdIs29 V; qvEx379 | pCB189, Plgc-11::gfp, pBSK+. Line #3 | This study |
| VQ1566 | sax-7(qv30) IV; hdIs29 V; qvEx476 | pCB428, Punc-122::rfp, pBSK+, Line #1 | This study |
| VQ1587 | sax-7(qv30) IV; hdIs29 V; qvEx485 | pCB428, Punc-122::rfp, pBSK+, Line #2 | This study |
| VQ1465 | sax-7(qv30) IV; oyIs14 V; qvEx234 | pCB191, Punc-122::rfp, Pttx-3::mCherry, pBSK+ | This study |
| VQ1375 | sax-7(qv30) IV; hdIs29 V; qvEx391 | pCB195, Plgc-11::gfp, pBSK+. Line #1 | This study |
| VQ1377 | sax-7(qv30) IV; hdIs29 V; qvEx393 | pCB195, Plgc-11::gfp, pBSK+. Line #2 | This study |
| VQ1583 | sax-7(qv30) IV; hdIs29 V; qvEx481 | pCB430, Punc-122::rfp, pBSK+. Line #1 | This study |
| VQ1588 | sax-7(qv30) IV; hdIs29 V; qvEx486 | pCB430, Punc-122::rfp, pBSK+. Line #2 | This study |
| VQ1590 | sax-7(qv30) IV; hdIs29 V; qvEx488 | pCB430, Punc-122::rfp, pBSK+. Line #3 | This study |
| VQ1584 | sax-7(qv30) IV; hdIs29 V; qvEx482 | pCB429, Punc-122::rfp, pBSK+. Line #1 | This study |
| VQ1586 | sax-7(qv30) IV; hdIs29 V; qvEx484 | pCB429, Punc-122::rfp, pBSK+. Line #2 | This study |
| VQ1589 | sax-7(qv30) IV; hdIs29 V; qvEx487 | pCB429, Punc-122::rfp, pBSK+. Line #3 | This study |
| VQ1449 | sax-7(qv30) IV; hdIs29 V; qvEx441 | pCB426, Punc-122::rfp, Pttx-3::mCherry, pBSK+. Line #1 | This study |
| VQ1116 | sax-7(qv30) IV; oyIs14 V; qvEx309 | pCB224, Punc-122::rfp, Pttx-3::mCherry, pBSK+. Line #2 | This study |
| VQ1117 | sax-7(qv30) IV; oyIs14 V; qvEx310 | pCB224, Punc-122::rfp, Pttx-3::mCherry, pBSK+. Line #3 | This study |
| VQ1582 | sax-7(qv30) IV; hdIs29 V; qvEx480 | pCB431, Punc-122::rfp, pBSK+. Line #1 | This study |
| VQ1594 | sax-7(qv30) IV; oyIs14 V; qvEx489 | pCB431, Punc-122::rfp, pBSK+. Line #2 | This study |
| VQ1112 | sax-7(qv30) IV; oyIs14 V; qvEx305 | pCB401, Punc-122::rfp, Pttx-3::mCherry, pBSK+. Line #1 | This study |
| VQ1113 | sax-7(qv30) IV; oyIs14 V; qvEx306 | pCB401, Punc-122::rfp, Pttx-3::mCherry, pBSK+. Line #2 | This study |
| VQ1114 | sax-7(qv30) IV; oyIs14 V; qvEx307 | pCB401, Punc-122::rfp, Pttx-3::mCherry, pBSK+. Line #3 | This study |
| VQ1585 | sax-7(qv30) IV; hdIs29 V; qvEx483 | pCB432, Punc-122::rfp, pBSK+. Line #1 | This study |
| VQ1596 | sax-7(qv30) IV; hdIs29 V; qvEx490 | pCB432, Plgc-11::gfp, pBSK+. Line #2 | This study |
| VQ1597 | sax-7(qv30) IV; hdIs29 V; qvEx491 | pCB432, Plgc-11::gfp, pBSK+. Line #3 | This study |
| VQ1059 | sax-7(qv30) IV; hdIs29 V; qvEx243 | pCB219, Pceh-22::gfp, pBSK+ | This study |
| VQ1062 | sax-7(qv30) IV; hdIs29 V; qvEx246 | pCB213, Punc-122::rfp, pBSK+ | This study |
| VQ1118 | sax-7(qv30) IV; hdIs29 V; qvEx311 | pCB402, Pceh-22::gfp, pBSK+. Line #1 | This study |
| VQ1119 | sax-7(qv30) IV; hdIs29 V; qvEx312 | pCB402, Pceh-22::gfp, pBSK+. Line #2 | This study |
| VQ1121 | sax-7(qv30) IV; hdIs29 V; qvEx314 | pCB212, Punc-122::rfp, pBSK+. Line #1 | This study |
| VQ1120 | sax-7(qv30) IV; hdIs29 V; qvEx313 | pCB212, Punc-122::rfp, pBSK+. Line #2 | This study |
| VQ1787 |
sax-7(qv30) IV; hdIs29 V; qvEx577 |
pCB471, Plgc-11::gfp, pBSK+. Line #1 | This study |
| VQ1788 |
sax-7(qv30) IV; hdIs29 V; qvEx578 |
pCB473, Plgc-11::gfp, pBSK+. Line #3 | This study |
| VQ1789 |
sax-7(qv30) IV; hdIs29 V; qvEx579 |
pCB472, Punc-122::gfp, pBSK+. Line #6 | This study |
| VQ1065 | sax-7(qv30) IV; hdIs29 V; qvEx243; qvEx246 | pCB219, Pceh-22::gfp, pBSK+ and pCB213, Punc-122::rfp, pBSK+ | This study |
| VQ1123 | sax-7(qv30) IV; hdIs29 V; qvEx311; qvEx314 | pCB402, Pceh-22::gfp, pBSK+. Line #1 and pCB212, Punc-122::rfp, pBSK+. Line #1 | This study |
| VQ1122 | sax-7(qv30) IV; hdIs29 V; qvEx312; qvEx313 | pCB402, Pceh-22::gfp, pBSK+. Line #2 and pCB212, Punc-122::rfp, pBSK+. Line #2 | This study |
| VQ1129 | sax-7(qv30) IV; hdIs29 V; qvEx311; qvEx246 | pCB402, Pceh-22::gfp, pBSK+. Line #1 and pCB213, Punc-122::rfp, pBSK+ | This study |
| VQ1803 |
sax-7(qv30) IV; hdIs29 V; qvEx577; qvEx579 |
pCB471, Plgc-11::gfp, pBSK+. Line #1 and pCB472, Punc-122::gfp, pBSK+. Line #6 |
This study |
| VQ1804 |
sax-7(qv30) IV; hdIs29 V; qvEx578; qvEx579 |
pCB473, Plgc-11::gfp, pBSK+. Line #3 and pCB472, Punc-122::gfp, pBSK+. Line #6 | This study |
Table 3.
List of primers used to genotype the gene sax-7 when build strains
| Allele | Primer | Sequence | PCR product(s) (bp) | Cosmid coordinates |
|---|---|---|---|---|
| qv30 | Mutant specific | |||
| oCB747 | tctctcaaaattcttcgcaagc | 326 | C18F3 8252…8273, forward | |
| oCB1025 | cgggaagaaatgaaacagga | C18F3 28531…28550, reverse | ||
| Wild-type specific | ||||
| oCB1022 | tggtggtagcgatggtgtag | 609 | C18F3 12312…12331, forward | |
| oCB1023 | agttcgatgttctcggctgt | C18F3 12901…12920, reverse | ||
| qv25 | oCB1022 | tggtggtagcgatggtgtag | 656 (mt), | C18F3 12312…12331, forward |
| oCB1023 | agttcgatgttctcggctgt | 609 (wt) | C18F3 12901…12920, reverse | |
| qv26 | oCB1022 | tggtggtagcgatggtgtag | 645 (mt), | C18F3 12312…12331, forward |
| oCB1023 | agttcgatgttctcggctgt | 609 (wt) | C18F3 12901…12920, reverse | |
| nj48 | oCB1022 | tggtggtagcgatggtgtag | 257 (mt), | C18F3 12312…12331, forward |
| oCB208 | gagttattggggtattttagcg | 825 (wt) | C18F3 13115…13136, reverse |
RT-PCR for sax-7 alleles
This analysis was performed with wild-type [N2], sax-7L-specific mutants [sax-7(eq2) and sax-7(nj53)], hypomorphic mutants of both isoforms [sax-7(nj48) and sax-7(tm1448)], and intracellular sax-7 mutant [sax-7(eq1)] strains. Total RNA was extracted from worm samples using Trizol (Invitrogen) according to the manufacturer’s instructions. RNA (500 ng) was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and random primers. PCR reactions were carried out with 1st strand cDNA template, and 0.25 μM of each primer for sax-7 cDNA amplification in 10 mM Tris pH 8.3, 1.5 mM MgCl2, 50 mM KCl, 0.2 mM deoxynucleotides, and 1 U Phusion DNA polymerase for 30 cycles of 94°C for 10 s, 55°C for 20 s, and 72°C for 45 s. Primers used to detect sax-7 transcript are as following: oCB985 (CGATTTGCAACTCAACAGGA), oCB986 (TGGTGCTCATGAAGATCAG), oCB987 (GTGTCCCGAACTGATTCGAT), oCB988 (TTTGTGGAACGTATTGACC), oCB989 (GGAACGTATTGACCTGAAACAG), oCB990 (TTGATCGTCCTGTCCGTGTA), and oCB991 (GACCACCGAATACCACAACC).
Primers oCB992 (TCGCTTCAAATCAGTTCAGC) and oCB993 (GCGAGCATTGAACAGTGAAG) were used for the control gene Y45F10D.4 (Hoogewijs et al. 2008) cDNA amplification.
Generation of sax-7 null allele by CRISPR-Cas9 (knockout)
gRNA plasmids (pCB392 and pCB393):
The gRNAs plasmids were made as previously described (Arribere et al. 2014). To obtain a deletion of the entire locus of sax-7, we used two target sequences, one on the 1st exon of the sax-7 long isoform (gtggccagtgagtaacaag reverse target sequence, pCB392) and the other one on the last exon of sax-7 corresponding to exon 17 and 14 of long and short isoform, respectively (ccggcatcaagctcttttg reverse target sequence, pCB393).
pCB392: Forward and reverse oligonucleotides (oCB1511: AAACcttgttactcactggccacC and oCB1510: TCTTGgtggccagtgagtaacaag, respectively), containing the 5ʹ target sequence and overhangs compatible with BsaI sites in plasmid pRB1017 (Arribere et al. 2014), were annealed and ligated into pRB1017 cut with BsaI to create the gRNA plasmid pCB392.
pCB393: Forward and reverse oligonucleotides (oCB1513: AAACcaaaagagcttgatgccggC and oCB1512: TCTTGccggcatcaagctcttttg, respectively), containing the 3ʹ target sequence and overhangs compatible with BsaI sites in plasmid pRB1017 (Arribere et al. 2014), were annealed and ligated into pRB1017 cut with BsaI to create the gRNA plasmid pCB393.
Plasmids were confirmed by sequencing with M13 reverse primer.
The repair donor ssDNA oligonucleotide (repair template):
We designed the repair donor simple-strand DNA oligonucleotide and ordered to Integrated DNA Technologies (IDT) (oCB1514: GATTCTAGATCACGTCGAAAGACCACCATCATGAGGAGCTTCATATTTCTAGCTTGATGCCGGCCGAACGGCCCGAGAAAGGATCAACGTCGACGTTTG, forward). The donor sequence starts with 50 nucleotides corresponding to the 5ʹ homology arm of sax-7L at the 5ʹ target site, followed by 49 nucleotides corresponding to the 3ʹ homology arm of sax-7 at the 3ʹ target site.
sax-7 deletion is located from 8373 to 28,330 bp on cosmid C18F3, deletion of 19,957 bp (Table 1, Supplementary Figure S1A).
Microinjection:
DNA mixture was prepared in injection buffer (20 mM potassium phosphate, 3 mM potassium citrate, 2% PEG, pH 7.5). The injection mix contained the Cas9 plasmid (pDD162; Dickinson et al. 2013) at 50 ng/µL, the gRNA plasmids pCB392 and pCB393 at 50 ng/µL each, the ssDNA donor oCB1514 at 20 ng/µL, the gRNA plasmid pJA58 (dpy-10 target; Arribere et al. 2014) at 50 ng/µL, and the ssDNA repair template for dpy-10 (dpy-10(cn64); Arribere et al. 2014) at 20 ng/µL. Mutations in the dpy-10 gene were used as CRISPR coconversion marker.
Screening:
F1 progeny were screened for Rol and Dpy phenotypes 3–4 days after injection. Rol or Dpy F1 animals were singled and the F2 progeny were screened by PCR for the absence of sax-7 gene with two couples of primers. The first couple of primers outside sax-7, oCB747 (TCTCTCAAAATTCTTCGCAAGC, forward) and oCB1025 (CGGGAAGAAATGAAACAGGA, reverse), giving a band when sax-7 is knockout works. Second couple of primers inside sax-7, oCB212 (GAAATACACACAAATACGAGTGC, forward) and oCB723 (TAGTTGATTAAAATGTTTCAAGATTG, reverse) giving a band in wild type (no knockout of sax-7).
Identification:
The strain resulting from this genome editing is identified as sax-7(qv30) (Tables 1–3) and verified by sequencing the deletion junctions (Supplementary Figure S1A) and also failed to amplify any product by several PCR reactions with primers targeting most of sax-7 exons.
Generation of sax-7(qv25) and sax-7(qv26), sax-7S-specific alleles by CRISPR-Cas9
Two insertion–deletion mutants, namely sax-7(qv25) and sax-7(qv26) (Tables 1–3, Supplementary Figure S1, B and C), were obtained during our efforts to insert sfgfp in the sax-7S-specific locus by CRIPSR-Cas9, described below.
Generation sfGFP::SAX-7S by CRISPR-Cas9 (knock-in)
We chose the protein marker sfGFP as a gene tag because it encodes a GFP variant that folds robustly even when fused to poorly folded proteins and its modified structure resists to the acidic extracellular environment (Pedelacq et al. 2006).
gRNA plasmids (pCB394 and pCB395):
The gRNAs plasmids were made as previously described (Arribere et al. 2014). Two target sequences were selected at the end of the exon 1 of sax-7S-specific locus (sax-7S/C18F3.2a, d), located in the predicted sax-7S signal peptide [ggatgtctactgttccttg forward target sequence (pCB394) and tgaaatgaaactaaccaca reverse target sequence (pCB395)].
pCB394: Forward and reverse oligonucleotides (oCB1515: TCTTGggatgtctactgttccttg and oCB1516: AAACcaaggaacagtagacatccC, respectively), containing the target sequence and overhangs compatible with BsaI sites in plasmid pRB1017 (Arribere et al. 2014), were annealed and ligated into pRB1017 cut with BsaI to create the gRNA plasmid pCB394.
pCB395: Forward and reverse oligonucleotides (oCB1518: AAACtgtggttagtttcatttcaC and oCB1517: TCTTGtgaaatgaaactaaccaca, respectively), containing the target sequence and overhangs compatible with BsaI sites in plasmid pRB1017 (Arribere et al. 2014), were annealed and ligated into pRB1017 cut with BsaI to create the gRNA plasmid pCB395.
Plasmids were confirmed by sequencing with M13 reverse primer.
The repair donor PCR amplicon (repair template):
We decided to design the repair donor DNA in order that the new gene insertion takes place directly at the end of the exon 1 of sax-7S, in sax-7S signal peptide. The end of the sax-7S signal peptide is at beginning of the exon 2 of sax-7S. Thus, it was necessary to add this signal sequence part localized downstream the insertion area (TCGGATCGCTACTACACA at the beginning of exon 2) at the end of exon 1, along with the gene sfgfp to be inserted, so as to ensure the presence of the entire signal peptide (Figure 4B, Supplementary Figure S1D).
Figure 4.
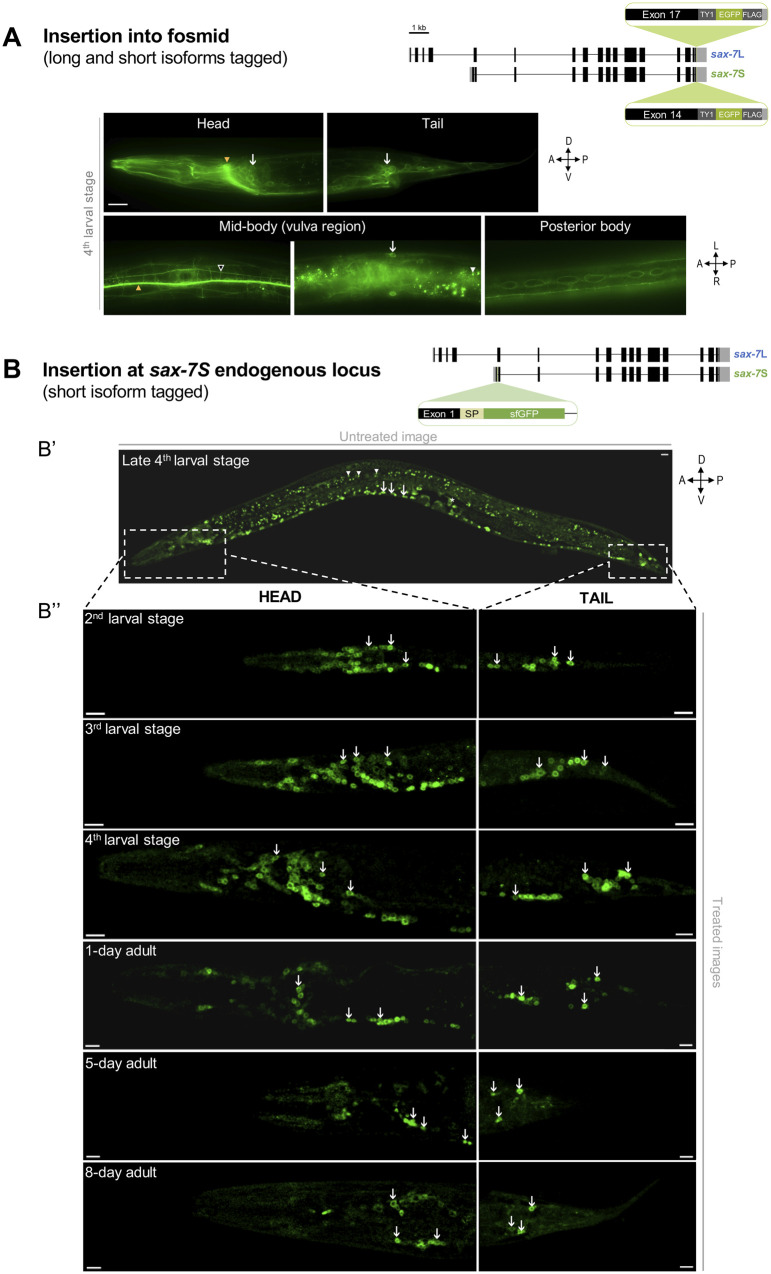
SAX-7S is expressed in virtually all neurons throughout life. (A) Images of SAX-7::GFP expression reporting both SAX-7L and SAX-7S. As shown on the schematics, in this previously published transgene (Sarov et al. 2012), the gene coding for EGFP was inserted into the gene sax-7 by fosmid recombineering in such a way that both SAX-7S and SAX-7L isoforms were tagged, making it impossible to distinguish between them. SAX-7::GFP is broadly expressed in neurons and epidermal cells (vulval cells, seam cells). (B) Confocal images showing sfGFP::SAX-7S expression. As shown on the schematics, the gene coding for sfGFP was inserted by CRISPR-Cas9 at the end of exon 1 of sax-7S in order to specifically tag SAX-7S (see Supplementary Figure S1D; qv31 in Table 1). “sfGFP,” superfolderGFP; “SP,” export signal peptide sequence part of sax-7S inserted along with sfgfp. (Bʹ) Untreated confocal image of a late 4th larval stage worm. Arrows indicate neurons of ventral nerve cord and arrowheads point to examples of background green auto-fluorescence due to gut granules. Dotted boxes indicate the body region (head or tail) analyzed in B″. (B″) Images of animals at the indicated larval stages and days of adulthood examined by confocal microscopy followed by unmixing. Aged worms (>5-day old) have notably increased background auto-fluorescence. Arrows indicate sfGFP::SAX-7S expression in neurons of the head (left) or tail (right) ganglia. n ≥ 20 animals examined by confocal microscopy for each stage. z-stack projections. Scale bar, 10 µm.
The repair donor DNA was amplified by PCR using first, primers oCB1525 (GTGTCGGATCGCTACTACACAATGAGCAAAGGAGAAGAAC, forward) and oCB1527 (ATGTGCCCTAAAAAGAAAAATGAAATGAAACTAACTTTGTAGAGCTCATCCATGC, reverse) and a plasmid containing the sequence of sfGFP as template. Primers oCB1525 contains 18 bases in 5ʹ upstream sfgfp corresponding to the missing sax-7S signal peptide sequence part and oCB1527 contains 35 bases corresponding to 3ʹ homology arms of sax-7S at the target site. A second PCR was amplified on the previous products with primers oCB1526 (TCATATTCCTGCTAGGATGTCTACTGTTCCTTGTGTCGGATCGCTAC, forward) and oCB1527 (ATGTGCCCTAAAAAGAAAAATGAAATGAAACTAACTTTGTAGAGCTCATCCATGC, reverse). Primer oCB1526 contains 35 bases corresponding to 5ʹ homology arms of sax-7S at the target site. sfgfp with signal peptide part were inserted immediately following amino acid 29 of SAX-7S.
Microinjection:
DNA mixture was prepared in injection buffer (20 mM potassium phosphate, 3 mM potassium citrate, 2% PEG, pH 7.5). The injection mix contained the Cas9 plasmid (pDD162; Dickinson et al. 2013) at 50 ng/µL, the gRNA plasmids pCB394 and pCB395 at 25 ng/µL each, the 5ʹarm::sp::sfgfp::3ʹarm donor PCR (containing the signal peptide, sp) at 100 ng/µL, the gRNA plasmid pJA58 (dpy-10 target; Arribere et al. 2014) at 50 ng/µL, and the ssDNA repair template for dpy-10 (dpy-10(cn64); Arribere et al. 2014) at 20 ng/µL. Mutations in the dpy-10 gene were used as CRISPR coconversion marker.
Screening:
F1 progeny were screened for Rol and Dpy phenotypes 3–4 days after injection. Rol or Dpy F1 animals were singled and the F2 progeny were screened by PCR for the presence of sax-7S signal peptide and sfgfp in the sax-7S locus with primers oCB1022 (TGGTGGTAGCGATGGTGTAG, forward) and oCB818 in sfgfp (TTCAGCACGCGTCTTGTAGG, reverse) for the 5ʹ insertion side and oCB1427 in sfgfp (AAAAGCGTGACCACATGGTCC, forward) and oCB1023 (AGTTCGATGTTCTCGGCTGT, reverse) for the 3ʹ insertion side.
Identification:
The new strain resulting from this genome editing is identified as sax-7(qv31[sfgfp::sax-7S]) (Tables 1 and 2), which is abbreviated as sfgfp::sax-7S. The modified locus was verified by sequencing of the entire region (Supplementary Figure S1D).
Microinjection to generate transgenic animals
Transgenic animals were generated by standard microinjection techniques (Mello and Fire 1995). Each construct was injected at 0.5 ng/μL (pCB471), 1 ng/μL (pCB191), 5 ng/μL (pCB219, pCB213, pCB402, and pCB212), 10 ng/μL (pCB224 and pCB426), or 25 ng/μL (pCB428, pCB189, pCB195, pCB430, pCB429, pCB431, pCB401, pCB432, pCB472, and pCB473), along with one or two coinjection markers to select transgenics, including Pceh-22::gfp (50 ng/μL) and Plgc-11::gfp (50 ng/μL) labeling the pharynx in green, Pttx-3::mCherry (50 ng/μL) labeling AIY neurons in red, and Punc-122::rfp (50 ng/μL) labeling coelomocytes in red. As needed, pBSK+ was used to increase the total DNA concentration of the injection mixes to 200 ng/µL. For details on transgenic strains and their injection mix composition, see Table 2.
Molecular cloning
For sax-7 constructs, the sax-7 cDNA was subcloned under the control of promoters rab-3 (Nonet et al. 1997), unc-14 (Ogura et al. 1997), or heat-shock promoter hsp-16.2 that express in neurons and other tissues (Jones et al. 1986; Fire et al. 1990; Stringham et al. 1992). The gene-coding sequences of sax-7/C18F3.2b and sax-7/C18F3.2a were used for the long and short isoform, respectively (available on WormBase). All inserts were verified by sequencing.
Phsp16.2::sax-7S (pCB191):
Vector pRP100 (Punc-14::sax-7S; Pocock et al. 2008) was digested with HindIII and BamHI to release Punc-14 and ligated with insert of Phsp-16.2 digested out of pPD49.78 (was a gift from Andrew Fire; Addgene plasmid # 1447; RRID: Addgene_1447) with the same restriction enzymes.
Prab-3::sax-7S (pCB428):
Punc-14::sax-7S::Myc (pCB189):
Cloned by Gibson assembly. For this plasmid, we used the vector pRP100 (Punc-14::sax-7S; Pocock et al. 2008). The FLAG::sax-7S::Myc construct was made by amplifying the 5ʹ end of the sax-7S cDNA from pRP100, carrying a BamHI site, with two nested PCR reactions adding FLAG tag sequence (GATTACAAGGATGACGACGATAAG) right after the signal peptide sequence in the exon 2 and, by amplifying the 3ʹ end of sax-7S cDNA from pRP100, carrying NcoI site, with two nested PCR reactions adding Myc tag sequence (GAGCAGAAACTCATCTCTGAAGAGGATCTG) right before the stop codon, in the exon 14. The vector pRP100 was digested with BamHI and NcoI enzymes to release nontagged sax-7S cDNA in order to clone the synthesized fragment FLAG::sax-7S::Myc into it with the same restriction enzymes. As a note, western blot experiments with several anti-FLAG antibodies were done in the attempt of detecting the N-terminus part of SAX-7, but failed.
Punc-14::sax-7L (pCB195):
Cloned through Gibson assembly. The HA::SAX-7L::V5 construct was made by amplifying the 5ʹ end of the sax-7L cDNA from Punc-17::sax-7L construct, carrying BamHI site, with two nested PCR reactions adding HA tag sequence (TACCCATACGACGTCCCAGACTACGCT) after the signal peptide sequence (exon 1) in the exon 2 (between 60 and 61 sax-7L cDNA bases). Also, by amplifying the 3ʹ end of sax-7L cDNA carrying NcoI site, with two nested PCR reactions adding V5 tag sequence (GGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACG) right before the stop codon, in the exon 17. The vector pRP100 was digested with BamHI and NcoI enzymes to release nontagged sax-7S cDNA in order to clone the synthesized fragment HA::SAX-7L::V5 into it with the same restriction enzymes.
Punc-14::sax-7SΔIg3–4 (pCB430):
Punc-14::sax-7SΔIg5–6 (pCB429):
Punc-14::sax-7SΔFnIII#3 (pCB224):
Punc-14::sax-7SΔFnIII#3::Myc (pCB426):
Vector pCB189 (Punc-14::FLAG::sax-7S::Myc) was digested with PstI and SalI restriction enzymes to release the sax-7S cDNA fragment containing the FnIII#3 domain and ligated with insert of sax-7S cDNA fragment without the FnIII#3 domain, digested out of pCB224 (Punc-14::sax-7SΔFnIII#3; Pocock et al. 2008) with the same restriction enzymes. As a note, western blot experiments with several anti-FLAG antibodies were done in the attempt of detecting the N-terminus part of SAX-7, but failed.
Punc-14::sax-7SΔFnIII (pCB431):
Punc-14::sax-7SΔAnkyrin (pCB401):
The vector Pttx-3::sax-7SΔAnkyrin (gift from H.E. Bülow) was digested with HindIII and BamHI restriction enzymes to release the fragment Pttx-3 and ligated with insert of Punc-14, digested out of pCB174 (Punc-14::sax-7LΔ11, Pocock et al. 2008) with the same restriction enzymes.
Punc-14::sax-7SΔICD (pCB432):
Punc-14::sax-7S Ig3 to serine protease cleavage site (RWKR) (Fragment A) (pCB219):
From pCB189 (Punc-14::FLAG::sax-7S::Myc), the sax-7S cDNA fragment FLAG::Ig3 to serine protease cleavage site (RWKR) (amino acid 745) was amplified with primers oCB798 (CATGATgctagcATGGGGTTACGAGAGACGATGG, forward) and oCB799 (ATCATGccatggCTATCTCTTCCATCTGAACTTTC, reverse) to add on NheI and NcoI restriction sites, respectively. Vector pCB195 (Punc-14::HA::sax-7L::V5) was digested with NheI and NcoI and ligated with the insert of sax-7S cDNA fragment using the same restriction enzymes. As a note, western blot experiments with several anti-FLAG antibodies were done in the attempt of detecting the N-terminus part of SAX-7, but failed.
Punc-14::sax-7S serine protease cleavage site (RWKR) to PDZ::Myc (Fragment B) (pCB213):
From pCB189 (Punc-14::FLAG::sax-7S::Myc), the sax-7S cDNA fragment serine protease cleavage site (RWKR) (amino acid 742) to PDZ::Myc was amplified with primers oCB811 (ACTGGCCACATATCATCAGGCAGCATAGATTGGTCAGCGAGATGGAAGAGATCAATTCG, forward) and oCB801 (ATCATGccatggCTACAGATCCTCTTCAGAGATG, reverse) to add the sax-7L signal peptide sequence and an NcoI restriction site, respectively. This first nest product was then amplified with primers oCB812 (CATGATgctagcATGAGGAGCTTCATATTCCTCTTGTTACTCACTGGCCACATATCATCAGG, forward) and oCB801 (ATCATGccatggCTACAGATCCTCTTCAGAGATG, reverse) to add the NheI restriction site. Then, vector pCB195 (Punc-14::HA::sax-7L::V5) was digested with NheI and NcoI and ligated with the insert of sax-7S cDNA fragment using the same restriction enzymes.
Punc-14::sax-7S Ig3 to proximal-transmembrane cleavage site (Fragment C) (pCB402):
From pCB189 (Punc-14::FLAG::sax-7S::Myc), the sax-7S cDNA fragment Ig3 to proximal-transmembrane cleavage site (amino acid 1024) was amplified with primers oCB798 (CATGATgctagcATGGGGTTACGAGAGACGATGG, forward) and oCB807 (ATCATGccatggCTAACGAGAACTCGTTCCCGTCG, reverse) to add NheI and NcoI restriction sites, respectively. Then, vector pCB195 (Punc-14::HA::sax-7L::V5) was digested with NheI and NcoI and ligated with the insert of sax-7S cDNA fragment using the same restriction enzymes.
Punc-14::sax-7S proximal-transmembrane cleavage site to PDZ::Myc (Fragment D) (pCB212):
From pCB189 (Punc-14::FLAG::sax-7S::Myc), the sax-7S cDNA fragment from proximal-transmembrane cleavage site (amino acid 1024) to PDZ::Myc was amplified with primers oCB813 (TCACTGGCCACATATCATCAGGCAGCATAGATTGGTCAGCGGAAGAAATGTCTATCTTTTG, forward) and oCB801 (ATCATGccatggCTACAGATCCTCTTCAGAGATG, reverse) to add the sax-7L signal peptide sequence and NcoI restriction site, respectively. This first nested product was then amplified with primers oCB812 (CATGATgctagcATGAGGAGCTTCATATTCCTCTTGTTACTCACTGGCCACATATCATCAGG, forward) and oCB801 (ATCATGccatggCTACAGATCCTCTTCAGAGATG, reverse) to add on NheI restriction site. Then, vector pCB195 (Punc-14::HA::sax-7L::V5) was digested with NheI and NcoI and ligated with the insert of sax-7S cDNA fragment using the same restriction enzymes.
Pdpy-7::sax-7S-Fragment A (From Ig3 to serine protease cleavage site RWKR), (pCB471):
pCB219 was digested with HindIII and BamHI to release the 1424 bp sax-7S-fragment A, which was ligated to a 248 bp HindIII and BamHI restriction fragment containing Pdpy-7.
Prab-3::sax-7S-Fragment A (From Ig3 to serine protease cleavage site RWKR), (pCB473):
pCB428 was digested with BamHI and ApaI to remove sax-7S and ligated to a 3161 bp BamHI and ApaI restriction fragment from pCB219 containing sax-7S-fragment A.
Prab-3::sax-7S-Fragment B (From serine protease cleavage site RWKR to PDZ::Myc), (pCB472):
pCB428 was digested with BamHI and ApaI to remove sax-7S and ligated to a 2204 bp BamHI and ApaI restriction fragment from pCB213 containing sax-7S-fragment B.
Immunoblot analysis
The analysis of Figure 1D was performed with strains carrying the reporter oyIs14, either wild-type or sax-7S-specific mutants. For each strain, either (1) large populations of worms were collected or (2) 100 L4-stage animals were collected in M9 solution and bacteria were washed off. Worms were fed and grown on plates at 20°C for at least three generations before collecting. Because the amount of SAX-7 protein was too low to detect all the protein forms on the analysis above, large pellets of thousands of mixed-stage worm populations were collected by washing plates, mostly devoid of bacteria, with M9 solution.
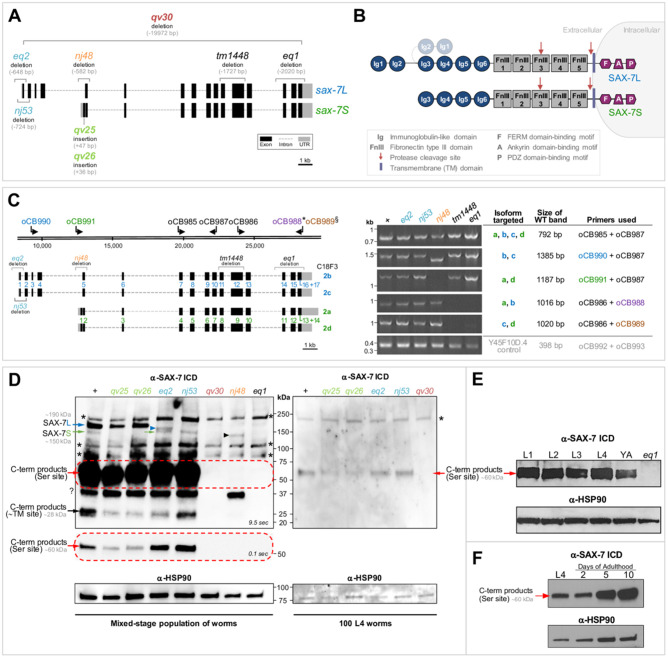
Figure 1.
Analysis of sax-7 mutant alleles. (A) Schematics of the gene structure for the sax-7 short (C18F3.2a) and long (C18F3.2b) isoforms. The mutant alleles used in this study are indicated, including the newly generated the null allele qv30 and sax-7S-specific alleles qv25 and qv26 (see Supplementary Figure S1, A–C for sequence information). Alleles nj48, tm1448, and eq1 affect both isoforms, and alleles eq2 and nj53 are sax-7L-specific (see Table 1 for allele information). (B) Schematics of the protein structure of SAX-7L and SAX-7S. Red arrows indicate cleavage sites: serine protease cleavage site in FnIII#3, or cleavage site proximal to the transmembrane (TM) domain. The two N-terminal Ig domains Ig1 and Ig2 may fold at the hinge region onto Ig3 and Ig4 indicated in gray (Pocock et al. 2008). (C) Schematics of the four encoded sax-7 isoforms. Isoforms a and d, and isoforms b and c, are nearly identical, except for a short sequence of 9 extra nucleotides at the beginning of exons 17 and 14 in isoforms c and d, respectively. sax-7 mutant alleles and primers used for RT-PCR analysis are indicated. Primer oCB990 (blue) was used to detect the long isoforms (b and c). Primer oCB991 (green) was used to detect the short isoforms (a and d). Primer oCB989§ (brown) specifically targets isoforms c and d, as its 3ʹ-end sequence primes on the 9 extra nucleotides of isoforms c and d. Conversely, primer oCB988* (violet) specifically targets isoforms a and b, as it was designed to prime at the exon junction lacking the 9 extra nucleotides. To the right, detection of sax-7 transcripts by RT-PCR. All RT-PCR products were confirmed by sequencing and correspond to the predicted sax-7 sequences. In mutant nj48 , transcripts are detected. The sax-7 long isoforms (b and c) RT-PCR product amplified with the primers oCB990/oCB987 is shorter than in the wild type, in accordance with the nj48 deletion where exon 5 of sax-7L is deleted. As expected (primer oCB991 falls within the nj48 deletion), no transcript for short isoforms (a and d) were detected. In mutants nj53 and eq2, the 5ʹ UTR and exon 1 of the sax-7 long isoforms (b and c) are deleted, and for eq2, part of exon 2 is deleted as well. Yet, sax-7 long (b and c) transcripts are detected in nj53 and eq2. Finally, in mutants tm1448 and eq1, both long (b and c) and short (a and d) transcripts are detected. Y45F10D.4 is a housekeeping gene used as an RT-PCR control (Hoogewijs et al. 2008). (D) Western blot analyses of SAX-7 protein. An antibody specific to the intracellular domain (ICD) of SAX-7 was used, which detects a region in the C-terminus of SAX-7S and SAX-7L (Chen et al. 2001). anti-HSP90 was used as a loading control. “+” indicates wild-type strain. Representative membranes of at least three independent repeats each. Asterisks (*) denote nonspecific bands, which are unrelated to SAX-7 as they are present in extracts of (1) sax-7(qv30) null mutants, where the entire sax-7 genomic locus is deletedand (2) eq1 mutant, where the entire the region coding for the epitope targeted by this antibody is deleted. “?” indicates an unknown form of SAX-7, which is detected in both wild-type and sax-7 mutants, except for the null qv30 and the epitope-control eq1. Left panel: >5000 mixed-stages worm populations were loaded per well. The band corresponding to full-length SAX-7L (∼190 kDa) is indicated by a blue arrow; SAX-7L is detected in the wild type and in mutants qv25 and qv26, but not in eq2 and nj53, as expected. The band corresponding to full-length SAX-7S (∼150 kDa) is indicated by a green arrow; SAX-7S is detected in the wild type and in mutants eq2 and nj53 (indicated by a green arrow), but not in qv25 and qv26, as expected. A presumably truncated mutant version of SAX-7L is detected in eq2 (blue arrowhead), which is not detected in wild type or eq1 and qv30 controls. Also, a truncated mutant version of SAX-7 is detected in nj48 (black arrowhead, unclear whether it corresponds to a truncated SAX-7S and/or SAX-7L). Two cleavage products are also detected: a highly abundant band at ∼60 kDa, indicated by a red arrow, corresponds to the C-terminal product resulting from cleavage at the serine protease site; a less abundant band at ∼28 kDa, indicated by a black arrow, corresponds to the C-terminal product from the cleavage site near to the TM and appears to run as a double band. An exposure of 9.5 s is required to see the bands of full-length SAX-7L and SAX-7S (arrows); however, at this exposure, the ∼60 kDa cleavage product saturates the area indicated by the red dotted box. To better distinguish level differences among mutants, the same ∼60 kDa membrane area was exposed for 0.1 s and is shown underneath. In SAX-7S-specific mutants qv25 and qv26, the 60 kDa C-terminal product (resulting from cleavage at the serine protease site) is detected at a lower level of compared to wild type; however, in SAX-7L-specific mutants eq2 and nj53 the level of this 60 kDa C-terminal-serine protease cleavage products is comparable to wild type, suggesting that most of this cleavage product may be derived from full-length SAX-7S. On the other hand, the 28 kDa C-terminal cleavage (resulting product from cleavage site near to TM) appears to be less abundant in eq2. Right panel: 100 L4 worms (4th larval stage) were loaded per well. While not all protein forms can be detected with this lower protein amount, the 60 kDa C-terminal product from cleavage at the serine protease site is again clearly detected at lower levels in the sax-7S-specific mutants qv25 and qv26, compared wild type. (E) Western blot analyses of SAX-7 protein at the 1st, 2nd, 3rd, and 4th larval stages (L1, L2, L3, and L4, respectively) of wild-type N2 animals. The most abundant SAX-7 product from cleavage at the serine protease site is detected throughout development. Anti-HSP90 was used as a loading control. (F) Western blot analyses of SAX-7 protein in 100 wild-type N2 animals at the 4th larval stage (L4), and at 2, 5, and 10 days of adulthood (devoid of embryos). The most abundant SAX-7 product from cleavage at the serine protease site is detected throughout adulthood. Anti-HSP90 was used as a loading control.
For Figure 1E, N2 animals were grown in large amounts in liquid culture, synchronized by bleaching and hatching in M9 buffer, and then grown on solid media up to the larval stages L1, L2, L3, and L4, when they were collected.
For Figure 1F, 100 wild-type (N2) animals of the L4 stage, or of 2-, 5-, or 10-day of adulthood were collected. To collect adult animals devoid of embryos, all animals were forced to lay their embryos by being treated with serotonin (50 μL of 30 mg/mL serotonin in M9 buffer added per plate for 1 h) and were then collected.
For Figure 5B, strains used were in the hdIs29 background carrying different sax-7S protein fragments. Worms were allowed to grow for ∼2 generations by feeding with ∼30 transgenic worms (or nontransgenic for wild type). Because these assays require large pellets of thousands of worms, rather than picking transgenic animals, worms were collected by washing populations on plates. We estimate that around ∼50% of animals carry the various extrachromosomal transgenes: unstable nonintegrated extrachromosomal arrays are lost during cell divisions and over generations, so that by the time that the worms were collected from plates, only a proportion of the animals are transgenics (verified by visual inspection). Mixed-stage worm populations from plates devoid of bacteria were collected in M9 solution.
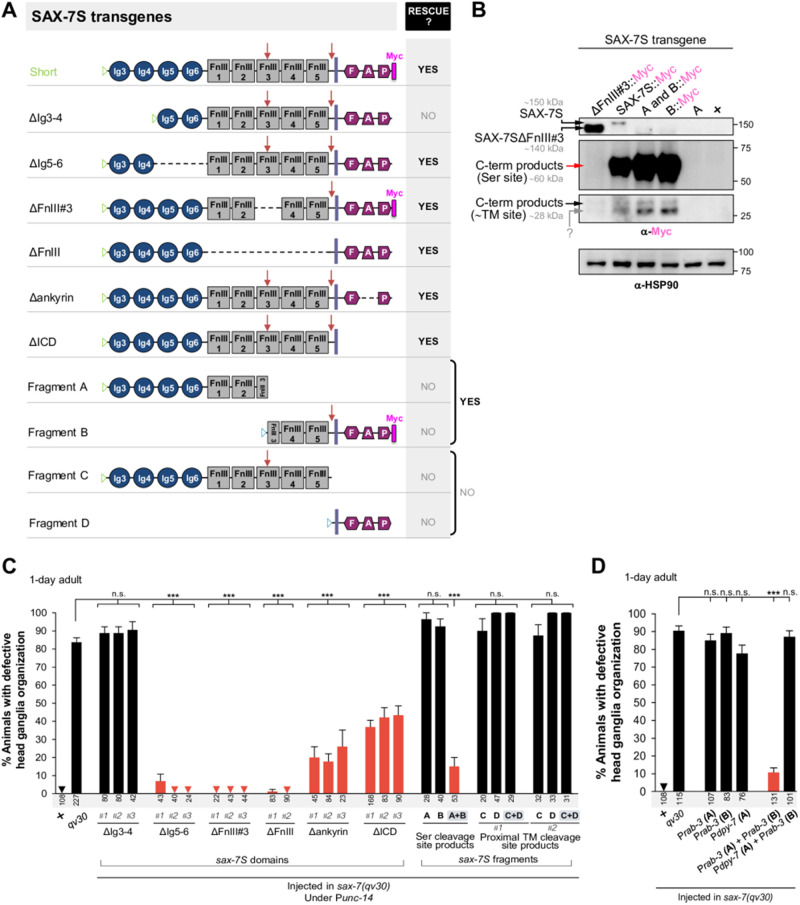
Figure 5.
The two SAX-7S cleavage products derived from the serine protease cleavage site, together, can mediate the maintenance of neuronal architecture. (A) Schematics of full-length and recombinant transgenic versions of SAX-7S used. Blue triangles indicate the signal peptide of SAX-7L. Green triangles indicate the signal peptide of SAX-7S. “ΔIg3–4” contains the entire SAX-7S protein except for the two first Ig domains. “ΔIg5–6” contains the entire SAX-7S protein except for the Ig5 and 6 domains. In “ΔFnIII#3,” SAX-7S::Myc lacks the 3rd FnIII domain. In “ΔFnIII,” SAX-7S lacks all FnIII domains. In “Δankyrin,” SAX-7S lacks the intracellular ankyrin-binding domain. In “ΔICD,” SAX-7S lacks the intracellular domain. “Fragment A” contains the SAX-7S protein region from Ig3 to the serine protease cleavage site (RWKR). “Fragment B” contains the SAX-7S::Myc protein region from the serine protease cleavage site (RWKR) to PDZ::Myc. “Fragment C” contains the SAX-7S protein region from Ig3 to the proximal-transmembrane cleavage site. “Fragment D” contains the SAX-7S protein region from the proximal-transmembrane cleavage site to PDZ. “Ig,” Immunoglobulin-like domain; “FnIII,” Fibronectin type III domain; “F,” FERM domain-binding motif; “A,” Ankyrin binding motif; “P,” PDZ domain-binding motif; bold violet line indicates the TM; red arrows indicate serine protease cleavage site in FnIII#3 or, cleavage site close to the TM. (B) Western blot analysis of wild-type animals (+), sax-7(qv30) null mutants expressing transgenes for various Myc-tagged SAX-7S fragments. N-terminal and C-terminal fragments of SAX-7S proteins were detected with anti-Myc antibody. Mixed-stage populations of >5000 worms were loaded per well, including a variable proportion of animals that actually carry the extrachromosomal array (and therefore are transgenic), as the array gets lost randomly upon cell divisions and generations; this comparison is only qualitative. As expected, in lysates of worms with transgene SAX-7SΔFnIII#3, an uncleaved band smaller than the full-length SAX-7S is detected. “C-term products” indicates C-terminal cleavage product, “Ser site” indicates serine protease cleavage site, “∼TM site” indicates cleavage site near to TM. “?” indicates an unknown form of SAX-7S. The three top anti-Myc panels correspond to the same membrane but at different exposure times in order to facilitate the observation of bands that are largely different in abundance (as was done in Figure 1D). α-HSP90 was used as a loading control. (C, D) The relative positioning of the soma and nerve ring axons of chemosensory neurons ASH/ASI was evaluated using the reporter Psra-6::DsRed2. Wild-type control and qv30 mutants, along with distinct SAX-7S recombinant transgenic animals, were examined as 1-day adults. In panel C, SAX-7S transgenes are expressed under the control of the Punc-14 promoter (domain analyses are shown on the left, and fragment analyses on the right). The simultaneous absence of Ig3 and 4 of SAX-7S fails to rescue qv30 mutants, while other domain deletions remain fully or largely functional. Two separate sets of independent extrachromosomal arrays for fragments C and D were tested (C#1+D#1, and C#2+D#2), and failed to rescue. In panel D, SAX-7S fragments are expressed under the control of different promoters (epidermal promoter Pdpy-7 and pan-neuronal promoter Prab-3, as indicated). SAX-7S fragments A and B failed to rescue the defects of sax-7 mutant animals when expressed under separate promoters (D), while they rescued when expressed in the same cells, under either the Punc-14 or the Prab-3 promoters (C and D, respectively), indicating that the two SAX-7S protein fragments together somehow reconstitute function. Sample sizes are indicated under each column of the graph. Error bars are the standard error of the proportion. Asterisks denote significant difference: ***P ≤ 0.001 (z-tests, P-values were corrected by multiplying by the number of comparisons, Bonferroni correction). “+,” indicates wild-type strain; n.s., not significant.
NETI (NaCl, EDTA, Tris, IGEPAL) buffer and protease inhibitors (Roche #11836153001) were added to worm pellets with 2X Laemmli sample buffer (Bio-Rad #161-0737) and 5% β-mercaptoethanol (v/v), and immediately frozen in liquid nitrogen. Samples were boiled for 5 min at 95°C and centrifuged for 10 min at 10,000 rpm prior to loading with capillary tips. Proteins were separated by SDS-PAGE on a 4–15% Mini-PROTEAN® TGX Stain-Free™ gel (Bio-Rad #456-8084) and transferred with the Trans-Blot® Turbo™ RTA Transfer Kit (Bio-Rad #170-4275) to an LF (low fluorescence) PVDF membrane using the Trans-Blot® Turbo™ Transfer System (Bio-Rad). Membranes were blocked in 5% BSA (VWR #0175) and 5% nonfat milk. To detect SAX-7, blots were incubated in 1:8000 rabbit anti-SAX-7, an affinity-purified antibody generated against the SAX-7 cytoplasmic tail [gift of Chen et al. (2001)] and 1:5000 goat anti-rabbit HRP secondary antibody (Bio-Rad #170-5046). To detect Myc tagged recombinant versions of SAX-7, blots were incubated in 1:500 mouse anti-Myc (CST #2276) and 1:3000 goat anti-mouse HRP secondary antibody (Jackson ImmunoResearch #115-035-003). For the loading control, membranes were incubated in 1:1000 rabbit anti-HSP90 antibody (CST #4874) and 1:5000 goat anti-rabbit HRP secondary antibody (Bio-Rad #170-5046). Signal was revealed using Clarity Max™ Western ECL Substrate (Bio-Rad #170-5062) and imaged using the ChemiDoc™ System (Bio-Rad). This analysis was performed three times for each set of experiments.
Microscopy and imaging
Worms were grown at 20°C for at least three generations prior to analysis. Worm stages are indicated in the figures. Tweny-four-hour post-L4 stage is considered “1st day of adulthood,” 24 h after that is considered “day 2 of adulthood,” and so on.
Neuroanatomical observations
Neuroanatomy was examined in wild-type and mutant animals using specific reporters. Worms were anesthetized with 75 mM of sodium azide (NaN3) and mounted on 5% agarose pads on glass slides. Animals were observed with Nomarski or fluorescence microscopy (Carl Zeiss Axio Scope.A1 or Axio Imager.M2), and images were acquired using the AxioCam camera (Zeiss) and processed using AxioVision (Zeiss), with 60× oil immersion objective (expected for PVQ/PVP axons: 100× oil immersion objective).
Analysis of ASH/ASI cell body positioning with respect to the nerve ring:
Cell body pairs of ASH/ASI chemosensory neurons and the nerve ring (neuropil of the worm), positioned in the head ganglia of the worm, were visualized using hdIs29. Animals were analyzed in a lateral orientation. Normally, both the two ASH and the two ASI soma are located posterior to the nerve ring. Animals were counted as mutant when at least one ASH or ASI soma was touching, on top of, or anterior to the nerve ring. Animals were counted as wild type when all ASH/ASI soma were positioned posterior to the nerve ring.
Analysis of AVK/AIY soma:
Cell body pairs of AVK/AIY interneurons were visualized using bwIs2 to label AVK in green, and otIs133 to label AIY in red. Animals were analyzed when in a ventral orientation. Cell bodies of AVK/AIY localized to the head ganglia of the worm in the retrovesicular ganglion. Normally, both neuron pairs AVKL/AIYL (left) and AVKR/AIYR (right) adhere to each other (White et al. 1986b; Pocock et al. 2008). Animals were counted as mutant when one or two AVK/AIY pairs were detached. Animals were counted as wild type when both of the AVK/AIY soma pairs were in contact.
Analysis of PHA/PHB soma:
Cell body pairs of PHA/PHB chemosensory neurons were visualized using DiI (1,1ʹ-Dioctadecyl-3,3,3ʹ,3ʹ-Tetramethylindocarbocyanine Perchlorate) staining procedure (Hedgecock et al. 1985). This is a lipophilic fluorescent stain for labeling cell membranes and hydrophobic structures, providing an alternative for labeling cells and tissues. In our case, it allows us to stain and visualize by a pink fluorescence the ciliated amphid (ADL, ASH, ASI, ASJ, ASK, and AWB) and phasmid (PHA and PHB) neurons (Collet et al. 1998) that are exposed to the outside environment. Animals were analyzed in a ventral orientation. Cell bodies of PHA/PHB localized to the tail ganglia of the worm in the lumbar ganglion. Normally, both neuron pairs PHAL/PHBL (left) and PHAR/PHBR (right) adhere to each other (White et al. 1986a). Animals were counted as mutant when any of the PHA/PHB pairs were detached from one another. Animals were counted as wild type when both of the PHA/PHB soma pairs were in contact.
Analysis of PVQ/PVP axons:
PVQ and PVP axons were visualized in animals using oyIs14 (PVQ labeled in green) or hdIs29 (PVQ labeled in red and PVP in green). Animals were analyzed in a ventral orientation. The axons of the PVQL/PVPL and the PVQR/PVPR neurons are normally located within the left and right fascicles of the ventral nerve cord, respectively. Animals were counted as having an axon flip-over defect when one of the PVQ/PVP axons was flipped to the opposite fascicle at any point along the ventral nerve cord, as previously described (Benard et al. 2006).
Other phenotypic observations
Analysis of embryonic lethality:
Embryos were picked and spread onto a new plate kept at 20°C. After ∼16 h, the number of larvae and dead embryos were counted. This experiment was repeated three times.
Analysis of brood size:
L4 worms were singled on a new plate independently. The number of embryos laid was counted each day of adulthood until day 4 of adulthood and the total amount of laid embryos during 4 days was calculated. This was done at least seven times.
Analysis of egg-laying:
Ten L4-stage worms were put on one plate and their ability to lay embryos normally was examined each day from day 1 to 5 of adulthood. Worms deficient in embryo laying retain them inside their bodies and display an Egl phenotype (Trent et al. 1983; Desai and Horvitz 1989). When counted defective they were removed from the plate. This was done 10 times.
Expression pattern analysis of sfGFP::SAX-7S
Fluorescence images of sax-7::ty1::egfp::3FLAG strain (Figure 4A; Sarov et al. 2012) were captured by fluorescence microscopy (Carl Zeiss Axio Imager.M2), and images were acquired using the AxioCam camera (Zeiss) and processed using AxioVision (Zeiss), with 60× oil immersion objective.
Fluorescent images of qv31, the sfgfp::sax-7S strain (Figure 4B), were captured using a Nikon A1R confocal microscope and processed using ImageJ. For each stage, at least eight worms were examined in detail. Nematodes were immobilized in 75 mM of NaN3 and mounted on 5% agarose pads on glass slides. All fluorescence images for sfgfp::sax-7S strain were obtained with the same settings using a Nikon Ti-e spinning disk confocal with 60× oil immersion objective. Images were three-dimensionally unmixed with NIS-Elements image acquisition and analysis software. The green fluorescent background is commonly seen in worms (gut granules), which disturbs the analysis of green fluorescent fusion proteins. In this study, we took advantage of a microscopy technique which “unmixes” overlapping spectral emissions after acquisition. Thanks to highly sensitive GaAsP-detectors, signals can be distinguished by the process called “spectral unmixing” (Ackermann 2017).
For this, we acquired images for wild-type N2 animals and determined an region of interest (ROI) in the pharynx in the head of the worm, giving a spectral profile defined as “background” green auto-fluorescence the worm. Then, with the sfgfp::sax-7S CRISPR-Cas9 strain, which expresses “real” green fluorescence, we acquired images and determined an ROI to the soma part of one neuron in the head of the worm, giving a spectral profile defined as “real” green fluorescence in the case of the sfGFP fluorophore. Finally, the “background” profile was subtracted from the “real” green fluorescence profile, keeping the real green fluorescence emission coming from sfGFP for the entire animal. ND2 files generated with NIS-Elements were imported into Fiji for analysis. Maximum intensity projections were generated by selecting stacks that had both ventral and dorsal signals.
Heat-shock inducible expression of sax-7S(+)
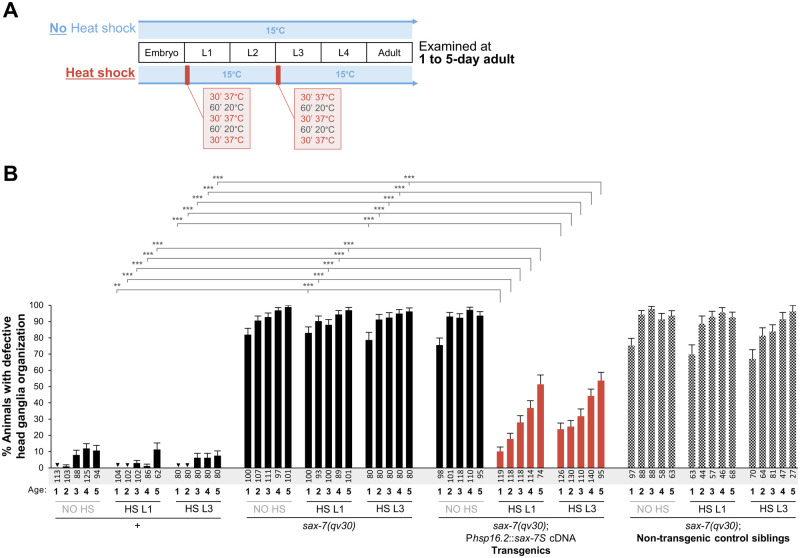
This analysis was performed with wild type [oyIs14], null mutant [sax-7(qv30); oyIs14], and null mutant transgenic animals carrying sax-7S cDNA under heat-shock promoter hsp16.2 that express in neurons and other tissues (Jones et al. 1986; Fire et al. 1990; Stringham et al. 1992). Worms were maintained in the incubator at 15°C for at least two generations prior to analysis. Plates were fed with a lot of gravid adult hermaphrodites and left at 15°C overnight. Embryos were then picked on a new plate and kept for ∼6 h at 15°C, after which any remaining unhatched embryos were removed from the plates leaving only freshly hatched L1s (on average 3.5-h old) on the plate. Animals were either heat shocked immediately as freshly L1s, or as L3s (∼42-h posthatch). Heat shock treatment consisted of three cycles of 30 min at 37°C with a 60 min recovery period at 20°C between each cycle, after which plates were put back at 15°C until analysis as adults (Figure 3A). All experiments were repeated at least twice. Neuroanatomical analysis of ASH/ASI cell body positioning with respect to the nerve ring was performed on animals as 1-, 2-, 3-, 4-, and 5-day-old adults.
Figure 3.
Expression of sax-7S(+) during larval stages is sufficient for sax-7S to function in the maintenance of the neuronal organization. (A) Summary of the heat-shock experiments performed. Animals were kept at 15°C at all times except during heat shock at 37°C (red boxes). Heat shock was done at either the 1st (L1) or the 3rd (L3) larval stage. Animals were later analyzed at days 1, 2, 3, 4, and 5 of adulthood. (B) Quantification of the relative position between the soma of ASH/ASI and the nerve ring (as in Figure 2A), visualized using the reporter oyIs14 (Psra-6::gfp), at days 1, 2, 3, 4, and 5 of adulthood (age indicated under each bar of the graph). Transgenic animals carry a transgene of sax-7S(+) expressed under the control of a heat-shock promoter [Phsp-16.2::sax-7S(+)]. Controls include the wild type, sax-7(qv30) mutants, and nontransgenic siblings of the transgenic animals, which are derived from the same mothers and grew on the same plates, but which do not carry the extrachromosomal array harboring the transgene. Additionally, for all of the four genetic conditions, neuroanatomical analyses were done in the absence of heat shock so as to ensure that no transgene expression occurred in the absence of heat shock. The defects normally displayed by adult sax-7(qv30) mutants are profoundly rescued by heat-shock-induced expression of sax-7S(+) during larval stages, as seen in heat-shocked sax-7(qv30) adult mutants carrying the transgene (orange bars). Nontransgenic siblings, however, are severely defective, indicating that the rescue of defects is dependent on the expression of sax-7S(+) upon heat shock. “+” indicates wild type; “NO HS,” no heat-shock; “HS L1,” heat shock was performed at the 1st larval stage; “HS L3,” heat shock was performed at the 3rd larval stage. Sample sizes are indicated under each bar of the graph. Error bars are the standard error of the proportion. Asterisks denote significant difference: ***P ≤ 0.001 (z-tests, P-values were corrected by multiplying by the number of comparisons, Bonferroni correction).
Statistical analysis
z-tests and Student’s t-tests, as indicated in each case, were performed in MS Office Excel. P-values were corrected by multiplying by the number of comparisons using the Bonferroni correction. Error bars in bar graphs represent standard error of proportion (S.E.P.).
Data availability
Mutant and genome engineered strains will be available at the Caenorhabditis Genetics Center, and all strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplementary material is available at GENETICS online.
Results
Molecular analysis of previous sax-7 mutant alleles
The interpretation of previous structure–function analyses for sax-7 was limited by the lack of a clear null mutation for the gene. In particular, the existence of gene product in sax-7(nj48), an allele reported to be a complete loss of function of the gene sax-7, has not been fully assessed. We examined sax-7 transcripts by RT-PCR for nj48, as well as for other sax-7 mutant alleles, including the sax-7L-specific alleles eq2 and nj53, and two alleles that affect both sax-7 isoforms, tm1448 and eq1 (Figure 1, A and B). We detected transcripts corresponding to all isoforms of sax-7 in all mutants tested (Figure 1C; all RT-PCR products were verified by sequencing), except when the primer targets a sequence that is deleted by a given mutation. In particular, transcript was detected in nj48 mutants, using four different primer pairs (Figure 1C), indicating that nj48 is not a null allele.
We also carried out western blots to characterize the expression of the protein SAX-7 in sax-7(nj48) and other mutant alleles (Figure 1D). To detect SAX-7, we used a purified antibody generated against the cytoplasmic tail of SAX-7 (Chen et al. 2001). In wild-type extracts, we detect five protein bands of ∼190, 150, 60, 40, and 28 kDa that are absent in the control eq1, in which the epitope-containing region of SAX-7 is deleted, as well as in a new deletion allele qv30 that we generated, in which the entire locus of sax-7 is absent (see below). The 190 kDa band (Figure 1D, blue arrow) and the 150 kDa band (Figure 1D, green arrow) correspond to the predicted SAX-7L and SAX-7S full-length protein, respectively, as previously reported (Chen et al. 2001; Sasakura et al. 2005; Wang et al. 2005). Two bands at 60 and 28 kDa appear to be cleavage products. The 60 kDa band (Figure 1D, red arrow) is likely the C-terminal fragment resulting from proteolytic cleavage of SAX-7 at the serine protease site in the 3rd FnIII domain (Figure 1B). This cleavage site is conserved in vertebrate L1 proteins (Faissner et al. 1985; Sadoul et al. 1988; Hortsch 1996, 2000; Nayeem et al. 1999; Silletti et al. 2000; Mechtersheimer et al. 2001; Haspel and Grumet 2003; Kalus et al. 2003; Matsumoto-Miyai et al. 2003; Xu et al. 2003; Schafer and Altevogt 2010; Lutz et al. 2012, 2017). The 28 kDa band (Figure 1D, black arrow), which runs as a doublet, is likely the predicted C-terminal fragment resulting from the proteolytic cleavage of SAX-7 at the proximal-transmembrane extracellular site (TM site, Figure 1B). Similar metalloprotease cleavage sites have been reported in vertebrate L1CAM proteins (Sadoul et al. 1988; Beer et al. 1999; Nayeem et al. 1999; Mechtersheimer et al. 2001; Gutwein et al. 2003; Haspel and Grumet 2003; Kalus et al. 2003; Matsumoto-Miyai et al. 2003; Xu et al. 2003; Naus et al. 2004; Maretzky et al. 2005; Riedle et al. 2009; Jafari et al. 2010; Schafer and Altevogt 2010; Kiefel et al. 2012; Zhou et al. 2012; Tatti et al. 2015; Linneberg et al. 2019; Maten et al. 2019). Finally, a 40 kDa band is detected in the wild type, but is absent in the controls (Figure 1D), suggesting yet another form of SAX-7 (also recently indicated in WormBase). Noteworthy, we find that the level of the full-length SAX-7L protein is higher than the full-length SAX-7S and that most SAX-7 is detected as a cleaved form. In particular, the serine protease cleavage product (∼80%) is most abundant (only the C-terminal fragment of the serine protease cleavage can be detected, as epitope located in the C-terminus). In contrast, the proximal-TM cleavage site product is less abundant (Figure 1D). The 60 kDa-C-terminal-serine protease cleavage product is detected throughout larval stages and adulthood (Figure 1, E and F). Importantly, in extracts of nj48 mutants, two forms of SAX-7 protein were detected, which were absent in controls: a 40 kDa band (Figure 1D, indicated by a question mark), and a 140 kDa band, likely a truncated form of SAX-7 protein (Figure 1D, black arrowhead). Thus, sax-7 transcript and SAX-7 protein are detected in extracts of sax-7(nj48) mutants, revealing that nj48 is not a null allele.
Generation of a sax-7 null and sax-7S-specific mutant alleles
To generate a null allele of sax-7, we used CRISPR-Cas9 technology and deleted the entire locus of the sax-7 gene. Two targets were used, one on the 1st exon of sax-7L and one on the last exon of sax-7 (exon 17 and 14 of the long and short isoform, respectively), resulting a 19,972-bp deletion (Figure 1A, Supplementary Figure S1A). This new mutant, named sax-7(qv30), is a clear null allele of sax-7 and was verified by multiple PCRs, sequencing, and western blot (Figure 1D). sax-7(qv30) null mutants are viable and have a somewhat reduced brood size, but their egg-laying and embryonic viability are normal (Supplementary Figure S2).
We also generated sax-7S-isoform-specific alleles, as this isoform has been found to be functionally important. Using CRISPR-Cas9 technology, we targeted the 1st exon of sax-7S specifically (in a region corresponding to an intron in sax-7L) and obtained two small sax-7S-specific insertion alleles, qv25 and qv26, both predicted to be strong loss-of-function alleles of sax-7S. qv25 has a 47-bp insertion and qv26, a 36-bp insertion (Supplementary Figure S1, B and C). Both alleles disrupt the sax-7S export signal peptide sequence, likely affecting SAX-7S protein synthesis. As a further consequence of the qv25 insertion, a stop codon is generated in the open reading frame of sax-7S (Supplementary Figure S1B). At the protein level, using the antibody against the SAX-7 cytoplasmic tail (Chen et al. 2001), as expected we detected no full-length SAX-7S in extracts of these mutants, while full-length SAX-7L was detected (190 kDa band; Figure 1D). Noteworthy, it appears that when SAX-7S is affected, as in qv25 and qv26, the 60 kDa-C-terminal-serine protease cleavage product is less abundant than in wild type or sax-7L-specific mutants eq2 and nj53 (60 kDa band; Figure 1D). This was consistently observed in all of the western blots done using either mixed worm populations or samples containing 100 L4 worms (≥3 independent repeats in each case). It thus appears that a large proportion of the C-terminal-serine protease cleavage product may originate from the cleavage of SAX-7S protein specifically.
Phenotypic characterization of new sax-7 mutants
We characterized the phenotypic consequences of the complete loss of sax-7 function in sax-7(qv30) mutants in neuronal maintenance. As a measure of head ganglia organization, we examined two pairs of head chemosensory neurons (ASH and ASI) from the 2nd larval stage to adulthood, as previously done for other mutants (Benard et al. 2006, 2009, 2012). The soma of these neurons are located in the lateral head ganglia and their axons project into the nerve ring. We visualized these four neurons using the fluorescent Psra-6::gfp or Psra-6::DsRed2 and noted the relative position of the ASH/ASI cell bodies with respect to the nerve ring. We found that head ganglia organization is normal in 2nd larval stage qv30 null mutants, but becomes progressively disorganized by the 4th larval stage, worsening into adulthood (Figure 2A). Similar disorganization of ASH/ASI has been described in nj48 mutant adults (Benard et al. 2012).
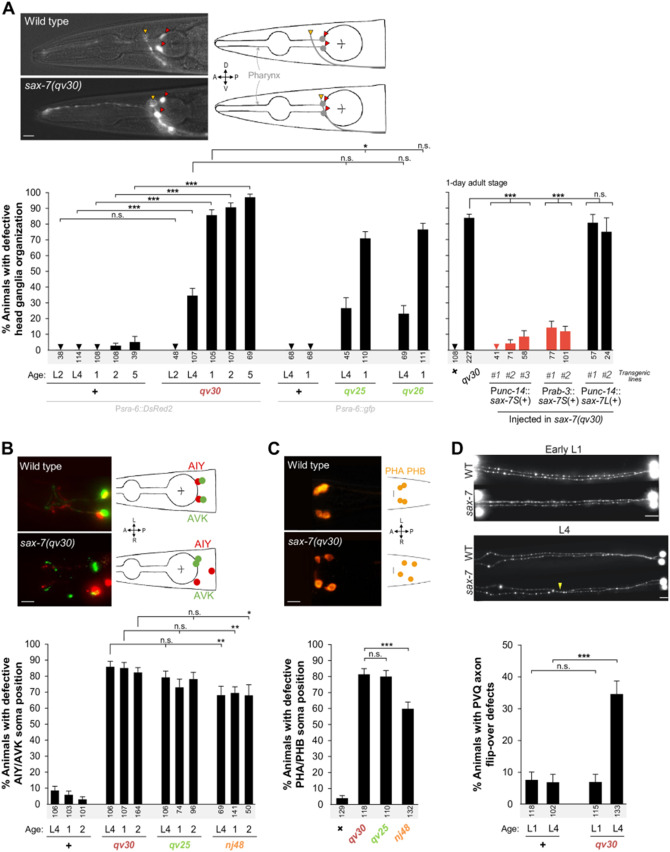
Figure 2.
Neuronal maintenance defects in the sax-7 mutant alleles qv30, qv25, and qv26. (A) sax-7S is required to maintain head ganglia organization postdevelopmentally. Fluorescence images of the head region of 1-day-old adults, where the soma and axons of the chemosensory neurons ASH and ASI are visualized using reporter Psra-6::DsRed2. Drawings illustrate microscopy images. Reporters Psra-6::DsRed2 (hdIs29) and Psra-6::gfp (oyIs14) give comparable results for all genotypes tested. In the wild type, the soma of neurons ASH/ASI (red arrowheads) are positioned posteriorly relative to the nerve ring (yellow arrowhead) throughout stages. In sax-7 mutants, the relative positioning between the soma of neurons ASH/ASI and the nerve ring is initially normal (soma posterior to nerve ring), but becomes progressively defective in late larvae onwards (soma can either overlap with or become anterior to the nerve ring). Quantification of the relative positioning between the ASH/ASI soma and the nerve ring in wild type, null mutant qv30, and sax-7S-specific mutants qv25 and qv26. Animals were examined at the 2nd (L2) and 4th (L4) larval stages, as well as at days 1, 2, or 5 of adulthood. Rescue of qv30 null mutant defects by expression of sax-7S(+) in the nervous system using the heterologous promoters Punc-14 and Prab-3. “#1, #2, #3” indicate independent transgenic lines. Statistical comparisons are with qv30 mutant. (B) sax-7S is required to maintain the retrovesicular ganglion organization. Fluorescence images showing the soma of two pairs of interneurons AVK and AIY on either sides of the 1-day-old adult animal visualized using reporters Pflp-1::gfp and Pttx-3::DsRed2. In the wild-type animals, the soma of AVK (green) and AIY (red) are adjacent with each other. In sax-7 mutants, one or both of the AVK and AIY neuron pairs become separate from one another. Quantification of animals showing separate pairs of AVK and AIY soma in wild type, qv30, qv25, and nj48, at the 4th larval stage (L4) and days 1 and 2 of adulthood. (C) sax-7S functions to maintain tail ganglia organization. Fluorescence images of the chemosensory neurons PHA and PHB in L4 larvae, visualized using DiI staining, whose soma are located in the lumbar ganglia on each side of the animal. In the wild type, the PHA and PHB soma are adjacent to each other. In sax-7 mutants, one or both of the PHA/PHB pairs are separated from one another. Quantification of disorganized soma position in wild type, qv30, qv25, and nj48, at L4. (D) sax-7S is required to maintain axon position within the ventral nerve cord. Fluorescence images of the interneurons PVK (visualized using oyIs14). PVQ axons extend ipsilaterally along the ventral nerve cord during embryogenesis. In the wild type, each PVQ axon remains on the ipsilateral side of the ventral nerve cord. In sax-7 mutants, while PVQ axons develop normally and are positioned like wild type at birth (early 1st larval stage), they later become displaced to the opposite side of the ventral nerve cord. Quantification of displaced PVQ axons in wild type and qv30, at hatching (early L1 stage) and the 4th larval stage (L4). Scale bars, 10 μm. Sample sizes are indicated under each column of the graph. Error bars are the standard error of the proportion. Asterisks denote significant difference: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (z-tests, P-values were corrected by multiplying by the number of comparisons, Bonferroni correction). “+” indicates wild-type strain; n.s., not significant.
We also examined the precise axon position of a pair of bilateral interneurons (PVQ) in the ventral nerve cord, labeled by the reporter Psra-6::gfp (oyIs14). Like in the wild type, these axons are normally positioned in freshly hatched 1st stage larvae of qv30 mutants, indicating that they had extended normally along the ventral nerve cord during embryogenesis (Figure 2D). However, compared to wild type where the PVQ axons maintain their normal position, PVQ axons inappropriately flip-over to the other side of the ventral nerve cord in 35% of sax-7(qv30) mutants (Figure 2D). We further examined the axon position of another pair of bilateral interneurons (PVP) in the ventral nerve cord of 4th larval stage animals, labeled by the reporter Podr-2::cfp (in hdIs29; this strain also harbors Psra-6::DsRed2 enabling the visualization of the PVQ interneurons). In the wild type, both PVP and PVQ axons remain well-positioned in virtually all animals (94%, n = 117), but in 37.5% (n = 80) of sax-7(qv30) mutants, the PVP and PVQ axons flip-over to the other side of the ventral nerve cord in, which is similar to nj48 mutants (Pocock et al. 2008; Benard et al. 2012).
Other aspects of the neuroanatomy of qv30 mutants were more severe than nj48 mutants. For instance, we observed retrovesicular ganglia organization by visualizing the neurons AIY and AVK (using reporters Pttx-3::mCherry and Pflp-1::gfp, respectively) and found that 85% of 1-day adult qv30 mutant animals display disjointed AIY and AVK soma, compared to 70% in nj48 mutants (Figure 2B). Also, using DiI staining, we found that the position of the soma of PHA and PHB in the tail ganglia is defective in 81% of qv30 mutants, compared to 60% of nj48 mutants, at the 4th larval stage (Figure 2C). Thus, while nj48 is a strong allele displaying similar penetrance to the null allele qv30 in some neuronal contexts, its loss of function is partial and less severe than the null qv30 in other neuronal contexts.
SAX-7S is required for neuronal maintenance
sax-7S, but not sax-7L, has previously been found to be sufficient to rescue neuronal maintenance defects in sax-7(nj48) mutants (Sasakura et al. 2005; Pocock et al. 2008). We verified whether sax-7S is also sufficient to rescue such defects in the sax-7(qv30) null mutants, by generating transgenic qv30 null mutant animals carrying wild-type copies of sax-7S(+) expressed neuronally [using the transgenes Prab-3::sax-7S(+) or Punc-14::sax-7S(+); as a note Punc-14 drives expression pan-neuronally as well as in some hypodermal cells]. We found that qv30 transgenic animals were profoundly rescued for head ganglia disorganization (Figure 2A). On the other hand, wild-type sax-7L(+) did not rescue qv30 transgenic mutant animals [transgene Punc-14::sax-7L(+); Figure 2A], similar to findings using the allele nj48 (Sasakura et al. 2005; Pocock et al. 2008). This is consistent with the absence of defects in sax-7L-specific mutants eq2 and nj53 (Sasakura et al. 2005; Benard et al. 2012). Altogether, these results further demonstrate that sax-7S mediates neuronal maintenance function.
To directly assess the phenotypic consequences of specifically disrupting sax-7S, we analyzed neuronal maintenance defects of the newly generated sax-7S-specific mutants qv25 and qv26 (Figures 1A and 2A). We found that the penetrance of their defects is similar to qv30 null mutant animals. For instance, the head ganglia of qv25 and qv26 animals become disorganized from the 4th larval stage onwards, similar to the qv30 null mutants (Figure 2A). Also, the soma of retrovesicular ganglion neurons AIY and AVK become disorganized from the 4th larval stage in qv25 mutants, similar to qv30 mutants (Figure 2B). Finally, the soma of tail neurons PHA and PHB get displaced from the 4th larval stage onwards in qv25 mutants, similar to qv30 mutants (Figure 2C). Thus, the specific disruption of sax-7S leads to neuronal maintenance defects that are similar to those resulting from the complete loss of sax-7 (deleting both sax-7S and sax-7L), confirming the key role of SAX-7S in the maintenance of neuronal architecture.
Postdevelopmental expression of sax-7S is sufficient for maintaining the neuronal architecture
Although the ventral nerve cord and head ganglia assemble during embryogenesis, sax-7(qv30) null mutants manifest ventral nerve cord flip-over defects during larval development, and head ganglia become disorganized by late larval stages, progressively worsening into adulthood. The appearance of defects in sax-7 mutants could theoretically result from either (1) undetected embryonic neuronal development defects that later worsen as the animal grows and moves or (2) deficient neuronal maintenance during larval and adult stages. To distinguish between these possibilities, we carried out rescue assays of qv30 null mutants with wild-type sax-7S(+) copies expressed under the control of an inducible heat-shock promoter, which drives expression in neurons and other tissues (Jones et al. 1986; Fire et al. 1990; Stringham et al. 1992). For this, we generated transgenic qv30 animals carrying the transgene Phsp16.2::sax-7S(+) as an extrachromosomal array. All animals were kept at 15°C [a colder temperature to prevent expression of Phsp16.2::sax-7S(+)], except during heat-shock treatments (Figure 3A). The organization of the ASI and ASH head ganglia neurons was examined in 1-, 2-, 3-, 4-, and 5-day-old adults. We controlled for head ganglia organization in the strains grown continuously at 15°C being indeed (1) normal in wild-type animals; (2) defective in qv30 mutants; (3) not rescued in the absence of heat shock, in transgenic qv30 animals carrying the transgene [Phsp-16.2::sax-7S(+)], indicating that the transgene is not expressed without heat shock; and (4) defective in qv30 nontransgenic control siblings under the same conditions (Figure 3B).
To determine the temporal requirement in sax-7 function, we heat shocked 1st (L1) or 3rd (L3) larval stage animals that had otherwise been grown at 15°C and examined head ganglia organization at days 1, 2, 3, 4, and 5 of adulthood (Figure 3A). Wild-type animals, qv30 mutants, transgenic qv30 animals carrying the transgene [Phsp-16.2::sax-7S(+)], and their qv30 nontransgenic siblings were analyzed in parallel. An additional control consisted of heat-shock treatment alone, in the absence of the transgene, which did not modify the defects of sax-7 mutants (head ganglia are similarly disorganized in qv30 animals whether heat shocked or not; Figure 3B). Also, heat shock did not alter head ganglia organization in wild-type animals (Figure 3B). In contrast, when transgenic qv30 animals carrying the transgene Phsp16.2::sax-7S(+) were heat shocked at L1, or even as late as L3, their neuronal organization was profoundly rescued. This rescue by heat shock-induced expression of sax-7S(+) is dependent on the presence of the transgene, as nontransgenic control siblings (which grew side by side with qv30 transgenics) were not rescued (Figure 3B). Together, these results indicate that wild-type activity of sax-7S provided postembryonically is sufficient for it to function in the maintenance of neuronal architecture. Heat shock-induced expression of sax-7S(+) had a more profound rescue effect when triggered during the 1st larval stage than at the 3rd larval stage. This indicates that sax-7S function during larval stages contributes to neuronal maintenance, which is consistent with the observation that head ganglia disorganization in sax-7(qv30) mutant animals starts being detected at the 4th larval stage (L4; Figure 2A). That heat shock-induced expression of sax-7S(+) as late as the 3rd larval stage could strongly rescue the defect highlights sax-7S(+) postembryonic function. Moreover, we found that the rescue of qv30 mutants following induction of sax-7S(+) is more profound in younger adults (days 1–3), as compared to older adults (days 4 and 5, Figure 3B). By day 5 of adulthood, more than 6 days have passed after heat shock-induced expression of Phsp16.2::sax-7S(+), suggesting that de novo expression of sax-7S may be required to sustain its maintenance function during adulthood. In sum, sax-7S can function postdevelopmentally to maintain the organization of embryonically developed neuronal architecture.
Endogenous sax-7S is expressed in neurons
The sax-7 gene is expressed strongly and broadly across the nervous system, as visualized with a fosmid (Sarov et al. 2012) where both the short and long isoforms are tagged with gfp (Figure 4A; Ramirez-Suarez et al. 2019). To elucidate the expression pattern of sax-7S, we used CRISPR-Cas9 technology to tag the sax-7S isoform specifically with sfgfp at its endogenous genomic locus (Figure 4B). We targeted the end of the 1st exon of sax-7S in a precise region that corresponds to intron 4 of sax-7L, and inserted sfgfp, preceded by sax-7S-signal peptide coding sequence (sfGFP::SAX-7S; Supplementary Figure S1D, Figure 4B). This knock-in allele of sax-7, named qv31, which was verified by sequencing (Supplementary Figure S1D), does not affect overall growth, morphology, or behavior, and head ganglia organization of qv31 animals is normal (n = 84, 2% defects, examined at 1-day adult by DiI staining, similar to wild type with 0% defects, n = 76).
To characterize the temporal and spatial expression pattern of qv31 sfGFP::SAX-7S, we used conventional and confocal fluorescence microscopy with spectral unmixing. sfGFP::SAX-7S is seen most predominantly and abundantly across the nervous system, where it is observed in virtually all neurons of the head and tail ganglia, the ventral nerve cord, as well as in isolated neurons located along the body wall (e.g., HSN near the vulva, and the PVM postdeirid neuron). Expression of sfGFP::SAX-7S in neurons is first observed in embryos (Supplementary Figure S3A) and persists throughout larval stages and adulthood, including in 5- and 8-day adults (Figure 4B). The sfGFP::SAX-7S signal appears less bright in aged adults, perhaps due to a decline in expression/stability of sax-7S or due to an increased autofluorescence of aging worms. While virtually all neurons express SAX-7S, differences in the level of expression are observed among neurons. sfGFP::SAX-7S is also occasionally detected in other cell types, such as in epidermal cells of the developing vulva and the uterus at the L4 stage, but not in adults (Supplementary Figure S3B). In sum, SAX-7S appears to be transiently and weakly expressed in developing cells of the epidermis, but its expression is strongest and sustained in virtually all neurons from embryogenesis to adulthood.
As a note, previous reports where sax-7 (both L and S indistinctly) was tagged intracellularly reported SAX-7 protein signal in axons, dendrites, or the plasma membrane (Chen et al. 2001; Wang et al. 2005; Ramirez-Suarez et al. 2019). Here, the sfGFP::SAX-7S signal in qv31 animals appears to be perinuclear in neuronal cell bodies, which is surprising for a transmembrane protein, and is likely artifactual. Indeed, in our effort to exclusively tag SAX-7S with sfGFP by CRISPR-Cas9, the only option was to insert the sfGFP very close to the predicted signal peptide of SAX-7S, which possibly affects the cleavage of the signal peptide and/or targeting of the protein. Nonetheless, there appears to be sufficient functional SAX-7S in qv31 animals as they are phenotypically wild type; it is plausible that sufficient SAX-7S is cleaved away from sfGFP to sustain its functions. Thus, whereas qv31 does not reliably inform about the subcellular localization of the protein SAX-7S, it yields valuable information about the spatio-temporal expression pattern of sax-7S.
Domains Ig3–4 of SAX-7S are necessary for its function in neuronal maintenance
L1 family members play diverse roles via homophilic interactions through their extracellular domains which mediate homophilic cell adhesion (Brummendorf and Rathjen 1996; Brummendorf et al. 1998; Hortsch 2000; Haspel and Grumet 2003), and mutating different extracellular Ig-like domains of vertebrate L1 perturbs its homophilic and/or heterophilic binding in in vitro assays (Holm et al. 1995; Zhao and Siu 1995; Montgomery et al. 1996; Felding-Habermann et al. 1997; Blaess et al. 1998; Kunz et al. 1998; De Angelis et al. 1999; Oleszewski et al. 1999; Haspel et al. 2000; Castellani et al. 2002; De Angelis et al. 2002) and neurite outgrowth (Appel et al. 1993). In C. elegans, neuronal expression of an SAX-7S recombinant version lacking Ig5–6 domains rescued AIY/AVK neuronal soma position defects of sax-7(nj48) mutants, whereas a recombinant version lacking Ig3–4 domains does not (Pocock et al. 2008). We asked whether such SAX-7S recombinant versions lacking specific Ig domains could rescue head ganglia organization in qv30 null mutant animals. We found that the transgene Punc-14::sax-7SΔIg5–6 rescued the position of the soma of the neurons ASH and ASI relative to the nerve ring in qv30 null mutants, but that the transgene Punc-14::sax-7SΔIg3–4 did not rescue (Figure 5C). This indicates that only Ig domains 3 and 4 of SAX-7S are required for its role in the maintenance head ganglia organization.
FnIII domains of L1 family members play diverse roles in neurite outgrowth, homophilic binding, and interactions with various partners (Holm et al. 1995; Silletti et al. 2000; Haspel and Grumet 2003; Kalus et al. 2003; Koticha et al. 2005; Maten et al. 2019). We asked whether FnIII domains are necessary for sax-7S function in C. elegans to maintain head ganglia organization. The recombinant transgene Punc-14::sax-7SΔFnIII#3::Myc lacks the third FnIII (FnIII#3) domain, which harbors the serine protease cleavage site (Figure 5A, ΔFnIII#3). In extracts of qv30 transgenic animals carrying this transgene, a ∼140 kDa band is detected with anti-Myc antibodies (Figure 5B), which is the expected size for uncleaved SAX-7S minus the FnIII#3 domain (full-length SAX-7S would be ∼150 kDa). We found that this transgene rescues head ganglia organization defects of qv30 mutant animals (Figure 5C), indicating that uncleaved SAX-7SΔFnIII#3 can function in neuronal maintenance, at least in a transgenic overexpression situation. We next tested a transgene that lacks all five FnIII domains, Punc-14::sax-7SΔFnIII, and found that it also rescues the head ganglia organization defects of qv30 null mutants (Figure 5C), indicating that the FnIII domains are dispensable for sax-7S to function in neuronal maintenance. Such transgene lacking all FnIII domains also rescued AIY and AVK soma position in nj48 mutants (Pocock et al. 2008), as well as AIY position and branching in sax-7(dz156) mutants (Diaz-Balzac et al. 2015).
The intracellular region (ICD ) of SAX-7/L1CAM shows a strong homology between vertebrates and invertebrates, and mutations in the cytoplasmic domain lead to X-linked hydrocephalus in humans (Wong et al. 1995b). This intracellular part contains motifs (FERM, ankyrin, and PDZ binding-domain motifs; Figure 1B), which mediate interactions with intracellular components and cytoskeletal proteins (Davis and Bennett 1994; Wong et al. 1995a; Koroll et al. 2001; Schaefer et al. 2002; Gil et al. 2003; Falk et al. 2004; Davey et al. 2005; Dirks et al. 2006; Gunn-Moore et al. 2006; Herron et al. 2009). We tested whether a transgene of sax-7S lacking the ankyrin-binding motif (Punc-14::sax-7SΔankyrin) could function to maintain head ganglia organization and found that qv30 null mutants were significantly rescued by this transgene (Figure 5C). Thus, the ankyrin-binding motif does not appear to be necessary for SAX-7S function in the maintenance of head ganglia. We next asked if SAX-7S could function in neuronal maintenance without its intracellular domain (Punc-14::sax-7SΔICD) and found a partial but significant rescue of the qv30 defects in head ganglia organization in animals expressing this transgene (Figure 5C). Yet, 40% of the qv30 animals neuronally expressing the SAX-7S transgene devoid of the intracellular domain display maintenance defects; this incomplete rescue may reflect that the intracellular domain is important for SAX-7S function but that its requirement can be partially alleviated in an overexpression situation, or alternatively it may be due to mosaicism or overexpression of the plasmid, which could possibly interfere with intermolecular interactions.
Serine protease SAX-7S fragments can, together, function in neuronal maintenance
The most abundant detected form of SAX-7 appears to be a serine protease cleavage product (Figure 1D), which splits the molecule within the third FnIII domain (Figure 1B). Other detected cleavage products result from a cleavage site proximal to the TM (Figure 1D). We tested whether the protein fragments predicted to result from serine protease-site cleavage, or the TM-proximal cleavage, could function in maintaining neuronal organization, similarly to full-length SAX-7S. For this, we constructed four separate transgenes encoding each of the four predicted fragments of the protein SAX-7S, from cleavage at either the serine protease or the TM proximal sites. Some of these transgenes encode versions of SAX-7S with a C-terminal Myc tag, which can then be examined by immunoblots (Figure 5, A and B). Cleavage of SAX-7S at the serine protease site within the third FnIII domain results in N- and C-terminal fragments, which we named “SAX-7S-fragment-A” and “SAX-7S-fragment-B,” respectively (Figure 5A). Another cleavage event proximal to the TM results in N- and C-terminal fragments, which we named “SAX-7S-fragment-C” and “SAX-7S-fragment-D,” respectively (Figure 5A). We tested each of these fragments alone, or in reciprocal combinations, for their ability to rescue the neuronal maintenance defects of sax-7(qv30) mutants when expressed under the promoter Punc-14. Neither “SAX-7S-fragment-A” alone (Punc-14::sax-7S-[N-terminal] Ig3 up to serine protease site) nor “SAX-7S-fragment-B alone” (Punc-14::sax-7S-serine protease site up to PDZ [C-terminal]) could rescue head ganglia organization defects of qv30 mutant animals (Figure 5C). Similarly, neither “SAX-7S-fragment-C” alone (Punc-14::sax-7S-[N-terminal] Ig3 up to TM site) nor “SAX-7S-fragment-D” alone (Punc-14::sax-7S-TM site up to PDZ [C-terminal]) could rescue head ganglia organization defects of qv30 mutant animals (Figure 5C).
We next tested whether the serine protease cleavage N- and C-terminal SAX-7S fragments together, i.e., “SAX-7S-fragment-A” and “SAX-7S-fragment-B” together, or whether the TM-proximal cleavage N- and C-terminal SAX-7S fragments together, i.e., “SAX-7S-fragment-C” and “SAX-7S-fragment-D” together, could rescue neuronal maintenance defects of sax-7(qv30) mutant animals. To generate doubly transgenic animals harboring the two respective transgenes, we avoided simultaneously comicroinjecting the two transgenes, as DNA recombination events between two transgenes could potentially reconstitute a full-length gene. Rather, we used genetic crosses to generate doubly transgenic animals (harboring two independent extrachromosomal arrays). We obtained a doubly transgenic strain carrying two extrachromosomal arrays, one for “SAX-7S-fragment-C” and one for “SAX-7S-fragment-D.” We found that the combination of SAX-7S fragments C and D did not rescue sax-7(qv30) mutant phenotype (Figure 5C, “C + D”; two independent sets of extrachromosomal arrays were tested and failed to rescue). Thus, SAX-7S fragments C and D predicted to result from cleavage proximal to the TM domain, even when present simultaneously, cannot fulfill sax-7S function in neuronal maintenance.
In a similar manner, we crossed the strain carrying the extrachromosomal array for “SAX-7S-fragment-A” with the second transgenic strain carrying the extrachromosomal array for “SAX-7S-fragment-B.” The resulting strain has animals carrying both extrachromosomal arrays, which we tested for rescue of qv30 head ganglia organization defects. The simultaneous presence of both fragments A and B profoundly rescued the head ganglia organization in sax-7(qv30) mutant animals (Figure 5C, “A + B”). This finding indicates that the two serine protease cleavage products together can mediate neuronal maintenance. As an important control, we verified that the rescue observed in the doubly transgenic strain depends on the simultaneous presence of both transgenes (for fragments A and B). Indeed, each of the two rederived in singly transgenic lines, each carrying only one of the two extrachromosomal arrays no longer rescued the sax-7(qv30) defects (fragment A alone: n = 28, 82% defects, or fragment B alone: n = 35, 91% defects; at 1-day adult), further confirming that only when the two SAX-7S fragments A and B are present together in an animal can then mediate neuronal maintenance (Figure 5C, “A” and “B”).
We analyzed protein extracts of doubly transgenic animals carrying both extrachromosomal arrays for fragments A and B, and as expected, no full-length SAX-7S is detected (Figure 5B), confirming that the distinct SAX-7S fragments A and B, together, can fulfill the role of SAX-7S in neuronal maintenance.
To test whether the two serine protease cleavage products (SAX-7S fragments A and B) need to be coexpressed within the same cell to function, we drove the expression of these fragments either in the same cell or in two different cell types. For this, we made three transgenes encoding SAX-7S fragment A under the neuronal promoter Prab-3 (Prab-3::SAX-7S-fragment-A) or under the epidermal promoter Pdpy-7 (Pdpy-7::SAX-7S-fragment A), and SAX-7S fragment B under Prab-3 (Prab-3::SAX-7S-fragment B). We generated independent transgenic strains each one carrying one of these transgenes and found that neither SAX-7S fragment A or B alone was able to rescue the neuronal maintenance defects of sax-7(qv30) mutants (Figure 5D), as above. We used genetic crosses to generate doubly transgenic animals harboring two independent extrachromosomal arrays to avoid DNA recombination and assess whether the SAX-7S fragments A and B in combination, but expressed from different cells, could function in neuronal maintenance. We found that when SAX-7S fragments A and B are both expressed in neurons, they could fully rescue neuronal maintenance defects (Figure 5D). However, when SAX-7S fragments A and B were expressed from different cells (i.e., fragment A expressed in the epidermis and fragment B expressed in neurons), then these fragments were not able to rescue the neuronal maintenance defects. These results might suggest that SAX-7S fragments A and B may somehow reassemble in a complex, intracellularly within neurons, and be secreted together to fulfill their function in neuronal maintenance. As a note, such reassembly would be noncovalent as no full-length SAX-7S protein is detected on denaturing western blots (Figure 5B). Alternatively, it is possible that epidermally expressed SAX-7S fragment A may not reach the neuron surface, where fragment B is located (the extracellular matrix needs to be traversed). In any case, cleaved fragments are detected by immunoblot analysis in wild-type animals throughout life and can provide function when expressed simultaneously in neurons. Together, our findings show that while the cleavage at the serine protease site is not absolutely necessary for SAX-7S function in an over-expression situation, the two cleaved fragments A and B resulting from it, functionally complement to mediate normal SAX-7S function for the maintenance of neuronal architecture in C. elegans.
Discussion
After the initial establishment of the nervous system, neuronal maintenance molecules function to actively preserve neuronal structural organization and integrity. One such molecule is C. elegans SAX-7, homologous to vertebrate L1 proteins, whose developmental roles have been studied (Dong et al. 2013; Salzberg et al. 2013), but whose roles in the long-term maintenance of nervous system organization remain unclear. Here we have generated and characterized a complete loss-of-function allele of sax-7, examined the endogenous expression pattern of SAX-7S, tested the temporal requirements for sax-7S, and assessed the function of SAX-7S cleavage products in the maintenance of neuronal architecture.
New sax-7 alleles: a complete null and two sax-7S-specific alleles
The sax-7(nj48) allele, previously considered to be a null, has detectable sax-7 transcripts and proteins (Figure 1). A new mutant, sax-7(qv30), deletes the entire sax-7 genomic locus, resulting in the complete loss of function of the gene (Figure 1, A and D). This null allele facilitates the interpretation of experiments without the caveat of potential truncated protein products present in hypomorphic alleles, which is especially important for rescue assays with transgenes encoding protein fragments.
qv30 mutant animals display defects that are in some cases stronger than previously studied alleles, for instance in the maintenance of some ganglia organization (e.g., AIY and AVK neuron pairs in the retrovesicular ganglion; PHA and PHB neuron pairs in the tail ganglion; Figure 2). sax-7S(+), but not sax-7L(+), can rescue the defects of null mutants sax-7(qv30) (Figure 2A), supporting that sax-7S is the key isoform in the maintenance of neuronal architecture, as previously described (Sasakura et al. 2005; Pocock et al. 2008). This is in accordance with previous reports that sax-7L-specific mutant alleles (eq2 and nj53) do not lead to neuronal maintenance defects (Pocock et al. 2008; Benard et al. 2012) and that sax-7L(+) cannot rescue neuronal maintenance defects of mutants nj48 and ky146, where both sax-7 isoforms are affected (Pocock et al. 2008).
Our analysis of two new sax-7 short-specific alleles, qv25 and qv26, further supports the notion that sax-7S is the important isoform in neuronal maintenance. Mutant animals for each of these two sax-7S alleles display defects similar to the null qv30 (Figure 2). As a note, other sax-7S-specific alleles with different molecular lesions have recently been isolated (Chen et al. 2019; Rahe et al. 2019). Together, our new findings and previous results unequivocally establish that SAX-7S is the important isoform mediating maintenance of neuronal architecture (Sasakura et al. 2005; Wang et al. 2005; Pocock et al. 2008; Benard et al. 2012; Diaz-Balzac et al. 2015).
Postembryonic expression of sax-7S is sufficient to maintain head ganglia organization
Expression of sax-7S(+) during larval stages, which is well after the embryonic assembly of neuronal ganglia, is sufficient to function in maintaining ganglia organization (Figure 3). Indeed, driving sax-7S expression (under the control of a heat-shock promoter) at the 1st larval stage, or as late as the 3rd larval stage, was sufficient to profoundly rescue neuronal maintenance defects in qv30 null mutant animals. While the rescue is profound, it is not complete, possibly due to the mosaicism of the extrachromosomal array bearing the transgene and the failure to recapitulate normal sax-7S(+) expression levels. Nonetheless, larval expression profoundly rescues the null mutants, pointing to the fact that sax-7S(+) functions postdevelopmentally to ensure the maintenance of the neuronal organization. This finding rules out the possibility that the neuronal maintenance defects of sax-7 mutants are a result of an undetected embryonic defect that is amplified by the growth and movement of the animal. Instead, our result is consistent with an active requirement for sax-7 postembryonically to maintain the organization of an already established nervous system structure.
Thus far, only a handful of molecules have been identified that function to maintain specific aspects of the nervous system. This likely is a reflection of the difficulty associated with determining an adult role for molecules that also play critical roles during development. A postembryonic neuronal role for sax-7, the C. elegans homolog of the mammalian L1CAM family, is a conserved property of this gene family. Indeed, loss of L1CAM specifically from the adult mouse brain led to an increase in basal excitatory synaptic transmission and behavioral alterations (Law et al. 2003). In rats, postdevelopmental nervous system knockdown of Neurofascin severely compromised the already established composition of the axon initial segment and led to an onset of motor deficits (Kriebel et al. 2011; Zonta et al. 2011). Postnatal disruption of CHL1 in excitatory neurons of the mouse forebrain affected the duration of working memory (Kolata et al. 2008). Thus, the continued importance of L1 family members in the adult nervous system is conserved from worm to mammals, suggesting that our findings in C. elegans will likely have implications in other organisms.
SAX-7S is robustly expressed across the nervous system
Transgenic expression of sax-7S(+) under different tissue-specific promoters has been used to test for function (this study; Sasakura et al. 2005; Pocock et al. 2008; Zhou et al. 2008; Benard et al. 2012; Dong et al. 2013; Salzberg et al. 2013; Diaz-Balzac et al. 2015; Zhu et al. 2017; Ramirez-Suarez et al. 2019). Here we have generated sfgfp insertion specifically in the sax-7S locus and characterized its endogenous expression pattern. sfGFP::SAX-7S is robustly expressed in virtually all neurons (Figure 4), consistent with the role of SAX-7S in the C. elegans nervous system. Indeed, transgenic wild-type copies of sax-7S(+) expressed pan-neuronally (Punc-14::sax-7S, Prab-3::sax-7S) rescue sax-7 mutant defects including head ganglia disorganization (Figure 2A), PVQ axon flip-over, AIY and AVK neuronal soma displacement (Pocock et al. 2008), AIY soma position and branching (Diaz-Balzac et al. 2015), AFD neuronal soma position (Sasakura et al. 2005), and PVD length or defasiculation (Ramirez-Suarez et al. 2019), as well as neuronal SAX-7 expression rescues dendrite retrograde extension (Cebul et al. 2020). Interestingly, we observed that sfGFP::SAX-7S expression levels vary among specific neurons in a given animal, and these neuron-specific differences appear to be reproducible across animals. Future studies will address the functional relevance of such SAX-7S expression-level signatures.
Transgenic expression of sax-7S(+) in the hypodermis [using the epidermal promoter in Pdpy-7::sax-7(+) transgene] rescues the PVD dendrite defects of sax-7 mutants (Dong et al. 2013; Salzberg et al. 2013; Zhu et al. 2017; Chen et al. 2019). Despite our careful analyses of animals at all developmental stages, including with unmixing confocal microscopy, we did not observe sfGFP::SAX-7S expression in the body wall epidermis (hyp 7 cells). This suggests that either (1) the endogenous level of SAX-7S in the epidermis is too low to be detected or (2) the functional form of SAX-7S, in this context is the C-terminal-serine protease cleavage product (fragment B), which cannot be seen with the qv31 sfGFP::SAX-7S knock-in, as the fluorescent protein is fused N-terminally (Figure 5A). Consistent with this idea, PVD dendrites can be rescued with SAX-7S constructs lacking N-terminal domains Ig3–4 or Ig5–6 (Dong et al. 2013; Salzberg et al. 2013). Tagging the intracellular domain of SAX-7 may allow for visualization of epidermal expression, but such a construct cannot be specific to the short isoform (SAX-7S), if done at the endogenous genomic locus, as both isoforms share the entire intracellular C-terminal region. Previous immuno-histochemistry analyses using an antibody generated against the C-terminal cytoplasmic tail of SAX-7 reported expression of SAX-7 in multiple tissues, including robust signal in neuronal cell bodies, as well as in the nerve ring (major bundle of axons) and the ventral nerve cord (Chen et al. 2001; Wang et al. 2005).
SAX-7S cleavage products in neuronal maintenance
SAX-7S and SAX-7L proteins could be reliably distinguished on immunoblots thanks to robust controls: the mutant allele eq1, where the sequence coding for the intracellular domain of sax-7 containing the epitope recognized by the antibody is deleted (Chen et al. 2001), and the null qv30 where the entire sax-7 locus is deleted. We observed that in wild-type animals, (1) full-length SAX-7S is less abundant than the full-length SAX-7L; (2) the vast majority of SAX-7 protein is cleaved throughout developmental stages and adulthood, as an abundant ∼60 kDa cleavage product, seemingly derived from the serine protease cleavage site; and (3) another less abundant cleavage product of ∼28 kDa may result from cleavage at a site near the transmembrane (Figure 1D). In the sax-7S-specific alleles qv25 and qv26, where SAX-7S is absent, the abundance of full-length SAX-7L is similar to wild type, and the ∼60 kDa serine protease cleavage product is less abundant compared to the wild type, suggesting that the SAX-7S protein may be preferentially cleaved compared to SAX-7L. Also, in the sax-7L-specific alleles eq2 and nj53, the ∼60 kDa serine protease cleavage product appears more abundant than the wild type, perhaps revealing that the SAX-7S cleavage may be favored, resulting in a lower level of full-length SAX-7S vs full-length SAX-7L.
When the serine protease cleavage site is deleted (sax-7S-ΔFnIII#3), the resulting recombinant protein is functional in head ganglia maintenance (Figure 5, B and C), indicating that the cleavage is not essential for function in the maintenance of neural architecture, at least with a highly expressed transgene. Consistent with this, motor neuron axon outgrowth defects upon knockdown of l1camb in zebrafish can be rescued by expression of a noncleavable form of L1cam (Linneberg et al. 2019). However, this may be context-specific as Reelin-mediated cleavage of L1CAM in the mouse brain is important for neurodevelopment (Lutz et al. 2017). Furthermore, we find sax-7S-ΔFnIII#3 is primarily detected as full length via western blot (Figure 5B), consistent with recent work which shows that mutating the cleavage site within the third FnIII domain of L1CAM leads to detectable full-length protein with no FnIII-domain-mediated cleavage products detected (Kleene et al. 2020). This suggests that the third FnIII has conserved importance in SAX-7/L1CAM processing.
Although the two serine protease cleavage products (SAX-7S-fragment-A and -B) cannot function individually in neuronal maintenance, we find that their simultaneous expression in neurons fulfills sax-7S neuronal maintenance function. The soluble ectodomain of L1cam similarly cannot solely restore l1cam knockdown-mediated defects in motor neuron axon outgrowth in zebrafish L1cam (Linneberg et al. 2019). It is possible that, in vivo, serine protease cleavage fragments A and B exist (as suggested by our immunoblot analysis) and may interact together to maintain neuronal architecture. In support of this, furin-mediated cleavage products of Tractin (the L1CAM homolog in leech) can interact in vitro, and these fragments together, not individually, can mediate adhesion in an in vitro S2 cell aggregation assay (Xu et al. 2003). As we know that SAX-7S can also promote homophilic adhesion in an in vitro cell aggregation assay (Sasakura et al. 2005), this points to the intriguing possibility that SAX-7S fragments together in vivo may have adhesive neural-maintenance-promoting properties. Future studies will help to address whether these SAX-7S fragments similarly function together or whether their function in neural maintenance is through other interacting factors.
Supplementary Material
Acknowledgments
The authors thank Maria Doitsidou for comments on the manuscript; Denis Flipo for assistance with confocal microscopy and unmixing; Andrea Thackeray for help with the project; the following researchers for sharing reagents: Max Heiman (for a plasmid containing the sfgfp gene), Lihsia Chen (for anti-C-terminal SAX-7 antibodies), Hannes Bülow and Roger Pocock for plasmids, as well as the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440), and WormBase.
Funding
This work was supported by funds from the USA National Institutes of Health (R01 AG041870-01A), Canadian Institutes of Health Research (Project Grant 707700), Natural Sciences and Engineering Research Council of Canada (NSERC, Discovery Grant 2017-06553), and Fond de Recherche du Québec-Santé (FRQS Chercheur-boursier Jr2 32719 and Sr 284221) to C.Y.B.; and graduate studies scholarships to V.E.D. by the Université du Québec à Montréal and the CERMO-FC Research Center and to M.N. by the Fond de Recherche du Québec-Santé.
Conflicts of interest
None declared.
Literature cited
- Ackermann C. 2017. https://www.uni-heidelberg.de/nic/a1r.html
- Appel F, Holm J, Conscience JF, Schachner M.. 1993. Several extracellular domains of the neural cell adhesion molecule L1 are involved in neurite outgrowth and cell body adhesion. J Neurosci. 13:4764–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, et al. 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 198:837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurelio O, Hall DH, Hobert O.. 2002. Immunoglobulin-domain proteins required for maintenance of ventral nerve cord organization. Science. 295:686–690. [DOI] [PubMed] [Google Scholar]
- Beer S, Oleszewski M, Gutwein P, Geiger C, Altevogt P.. 1999. Metalloproteinase-mediated release of the ectodomain of L1 adhesion molecule. J Cell Sci. 112:2667–2675. [DOI] [PubMed] [Google Scholar]
- Benard C, Hobert O.. 2009. Looking beyond development: maintaining nervous system architecture. Curr Top Dev Biol. 87:175–194. [DOI] [PubMed] [Google Scholar]
- Benard C, Tjoe N, Boulin T, Recio J, Hobert O.. 2009. The small, secreted immunoglobulin protein ZIG-3 maintains axon position in Caenorhabditis elegans. Genetics. 183:917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard CY, Blanchette C, Recio J, Hobert O.. 2012. The secreted immunoglobulin domain proteins ZIG-5 and ZIG-8 cooperate with L1CAM/SAX-7 to maintain nervous system integrity. PLoS Genet. 8:e1002819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard CY, Boyanov A, Hall DH, Hobert O.. 2006. DIG-1, a novel giant protein, non-autonomously mediates maintenance of nervous system architecture. Development. 133:3329–3340. [DOI] [PubMed] [Google Scholar]
- Bieber AJ, Snow PM, Hortsch M, Patel NH, Jacobs JR, et al. 1989. Drosophila neuroglian: a member of the immunoglobulin superfamily with extensive homology to the vertebrate neural adhesion molecule L1. Cell. 59:447–460. [DOI] [PubMed] [Google Scholar]
- Blaess S, Kammerer RA, Hall H.. 1998. Structural analysis of the sixth immunoglobulin-like domain of mouse neural cell adhesion molecule L1 and its interactions with alpha(v)beta3, alpha(IIb)beta3, and alpha5beta1 integrins. J Neurochem. 71:2615–2625. [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummendorf T, Kenwrick S, Rathjen FG.. 1998. Neural cell recognition molecule L1: from cell biology to human hereditary brain malformations. Curr Opin Neurobiol. 8:87–97. [DOI] [PubMed] [Google Scholar]
- Brummendorf T, Rathjen FG.. 1996. Structure/function relationships of axon-associated adhesion receptors of the immunoglobulin superfamily. Curr Opin Neurobiol. 6:584–593. [DOI] [PubMed] [Google Scholar]
- Bülow HE, Boulin T, Hobert O.. 2004. Differential functions of the C. elegans FGF receptor in axon outgrowth and maintenance of axon position. Neuron. 42:367–374. [DOI] [PubMed] [Google Scholar]
- Castellani V, De Angelis E, Kenwrick S, Rougon G.. 2002. Cis and trans interactions of L1 with neuropilin-1 control axonal responses to semaphorin 3A. EMBO J. 21:6348–6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebul ER, McLachlan IG, Heiman MG.. 2020. Dendrites with specialized glial attachments develop by retrograde extension using SAX-7 and GRDN-1. Development. 147:dev180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Hsu HW, Chang YH, Pan CL.. 2019. Adhesive L1CAM-Robo signaling aligns growth cone F-actin dynamics to promote axon-dendrite fasciculation in C. elegans. Dev Cell. 48:215–228.e5. [DOI] [PubMed] [Google Scholar]
- Chen L, Ong B, Bennett V.. 2001. LAD-1, the Caenorhabditis elegans L1CAM homologue, participates in embryonic and gonadal morphogenesis and is a substrate for fibroblast growth factor receptor pathway-dependent phosphotyrosine-based signaling. J Cell Biol. 154:841–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherra SJ, Jin Y.. 2016. A two-immunoglobulin-domain transmembrane protein mediates an epidermal-neuronal interaction to maintain synapse density. Neuron. 89:325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet J, Spike CA, Lundquist EA, Shaw JE, Herman RK.. 1998. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics. 148:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey F, Hill M, Falk J, Sans N, Gunn-Moore FJ.. 2005. Synapse associated protein 102 is a novel binding partner to the cytoplasmic terminus of neurone-glial related cell adhesion molecule. J Neurochem. 94:1243–1253. [DOI] [PubMed] [Google Scholar]
- Davis JQ, Bennett V.. 1994. Ankyrin binding activity shared by the neurofascin/L1/NrCAM family of nervous system cell adhesion molecules. J Biol Chem. 269:27163–27166. [PubMed] [Google Scholar]
- De Angelis E, MacFarlane J, Du JS, Yeo G, Hicks R, et al. 1999. Pathological missense mutations of neural cell adhesion molecule L1 affect homophilic and heterophilic binding activities. EMBO J. 18:4744–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis E, Watkins A, Schafer M, Brummendorf T, Kenwrick S.. 2002. Disease-associated mutations in L1 CAM interfere with ligand interactions and cell-surface expression. Hum Mol Genet. 11:1–12. [DOI] [PubMed] [Google Scholar]
- Desai C, Horvitz HR.. 1989. Caenorhabditis elegans mutants defective in the functioning of the motor neurons responsible for egg laying. Genetics. 121:703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Balzac CA, Lazaro-Pena MI, Ramos-Ortiz GA, Bulow HE.. 2015. The adhesion molecule KAL-1/anosmin-1 regulates neurite branching through a SAX-7/L1CAM-EGL-15/FGFR receptor complex. Cell Rep. 11:1377–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Balzac CA, Rahman M, Lazaro-Pena MI, Martin Hernandez LA, Salzberg Y, et al. 2016. Muscle- and skin-derived cues jointly orchestrate patterning of somatosensory dendrites. Curr Biol. 26:2379–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B.. 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 10:1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks P, Thomas U, Montag D.. 2006. The cytoplasmic domain of NrCAM binds to PDZ domains of synapse-associated proteins SAP90/PSD95 and SAP97. Eur J Neurosci. 24:25–31. [DOI] [PubMed] [Google Scholar]
- Dong X, Liu OW, Howell AS, Shen K.. 2013. An extracellular adhesion molecule complex patterns dendritic branching and morphogenesis. Cell. 155:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faissner A, Teplow DB, Kubler D, Keilhauer G, Kinzel V, et al. 1985. Biosynthesis and membrane topography of the neural cell adhesion molecule L1. EMBO J. 4:3105–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J, Thoumine O, Dequidt C, Choquet D, Faivre-Sarrailh C.. 2004. NrCAM coupling to the cytoskeleton depends on multiple protein domains and partitioning into lipid rafts. Mol Biol Cell. 15:4695–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felding-Habermann B, Silletti S, Mei F, Siu CH, Yip PM, et al. 1997. A single immunoglobulin-like domain of the human neural cell adhesion molecule L1 supports adhesion by multiple vascular and platelet integrins. J Cell Biol. 139:1567–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Harrison SW, Dixon D.. 1990. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 93:189–198. [DOI] [PubMed] [Google Scholar]
- Fransen E, Van Camp G, Vits L, Willems PJ.. 1997. L1-associated diseases: clinical geneticists divide, molecular geneticists unite. Hum Mol Genet. 6:1625–1632. [DOI] [PubMed] [Google Scholar]
- Gil OD, Sakurai T, Bradley AE, Fink MY, Cassella MR, et al. 2003. Ankyrin binding mediates L1CAM interactions with static components of the cytoskeleton and inhibits retrograde movement of L1CAM on the cell surface. J Cell Biol. 162:719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godenschwege TA, Kristiansen LV, Uthaman SB, Hortsch M, Murphey RK.. 2006. A conserved role for Drosophila neuroglian and human L1-CAM in central-synapse formation. Curr Biol. 16:12–23. [DOI] [PubMed] [Google Scholar]
- Gunn-Moore FJ, Hill M, Davey F, Herron LR, Tait S, et al. 2006. A functional FERM domain binding motif in neurofascin. Mol Cell Neurosci. 33:441–446. [DOI] [PubMed] [Google Scholar]
- Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Gast D, et al. 2003. ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. FASEB J. 17:292–294. [DOI] [PubMed] [Google Scholar]
- Hall SG, Bieber AJ.. 1997. Mutations in the Drosophila neuroglian cell adhesion molecule affect motor neuron pathfinding and peripheral nervous system patterning. J Neurobiol. 32:325–340. [PubMed] [Google Scholar]
- Haspel J, Friedlander DR, Ivgy-May N, Chickramane S, Roonprapunt C, et al. 2000. Critical and optimal Ig domains for promotion of neurite outgrowth by L1/Ng-CAM. J Neurobiol. 42:287–302. [PubMed] [Google Scholar]
- Haspel J, Grumet M.. 2003. The L1CAM extracellular region: a multi-domain protein with modular and cooperative binding modes. Front Biosci. 8:s1210–1225. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Thomson JN, Perkins LA.. 1985. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev Biol. 111:158–170. [DOI] [PubMed] [Google Scholar]
- Heiman MG, Pallanck L.. 2011. Neurons at the extremes of cell biology. Mol Biol Cell. 22:721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron LR, Hill M, Davey F, Gunn-Moore FJ.. 2009. The intracellular interactions of the L1 family of cell adhesion molecules. Biochem J. 419:519–531. [DOI] [PubMed] [Google Scholar]
- Holm J, Appel F, Schachner M.. 1995. Several extracellular domains of the neural cell adhesion molecule L1 are involved in homophilic interactions. J Neurosci Res. 42:9–20. [DOI] [PubMed] [Google Scholar]
- Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR.. 2008. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol. 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortsch M. 1996. The L1 family of neural cell adhesion molecules: old proteins performing new tricks. Neuron. 17:587–593. [DOI] [PubMed] [Google Scholar]
- Hortsch M. 2000. Structural and functional evolution of the L1 family: are four adhesion molecules better than one? Mol Cell Neurosci. 15:1–10. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Nagaraj K, Mualla R.. 2014. The L1 family of cell adhesion molecules: a sickening number of mutations and protein functions. Adv Neurobiol. 8:195–229. [DOI] [PubMed] [Google Scholar]
- Jafari G, Burghoorn J, Kawano T, Mathew M, Morck C, et al. 2010. Genetics of extracellular matrix remodeling during organ growth using the Caenorhabditis elegans pharynx model. Genetics. 186:969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RP, Kramer JM.. 2012. Neural maintenance roles for the matrix receptor dystroglycan and the nuclear anchorage complex in Caenorhabditis elegans. Genetics. 190:1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D, Russnak RH, Kay RJ, Candido EP.. 1986. Structure, expression, and evolution of a heat shock gene locus in Caenorhabditis elegans that is flanked by repetitive elements. J Biol Chem. 261:12006–12015. [PubMed] [Google Scholar]
- Kalus I, Schnegelsberg B, Seidah NG, Kleene R, Schachner M.. 2003. The proprotein convertase PC5A and a metalloprotease are involved in the proteolytic processing of the neural adhesion molecule L1. J Biol Chem. 278:10381–10388. [DOI] [PubMed] [Google Scholar]
- Kiefel H, Bondong S, Hazin J, Ridinger J, Schirmer U, et al. 2012. L1CAM: a major driver for tumor cell invasion and motility. Cell Adh Migr. 6:374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene RLutz DLoers GBork UBorgmeyer U, et al. . 2021. Revisiting the proteolytic processing of cell adhesion molecule L1. J Neurochem. 157:1102–1117. 10.1111/jnc.15201 32986867 [DOI] [PubMed] [Google Scholar]
- Kolata S, Wu J, Light K, Schachner M, Matzel LD.. 2008. Impaired working memory duration but normal learning abilities found in mice that are conditionally deficient in the close homolog of L1. J Neurosci. 28:13505–13510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroll M, Rathjen FG, Volkmer H.. 2001. The neural cell recognition molecule neurofascin interacts with syntenin-1 but not with syntenin-2, both of which reveal self-associating activity. J Biol Chem. 276:10646–10654. [DOI] [PubMed] [Google Scholar]
- Koticha D, Babiarz J, Kane-Goldsmith N, Jacob J, Raju K, et al. 2005. Cell adhesion and neurite outgrowth are promoted by neurofascin NF155 and inhibited by NF186. Mol Cell Neurosci. 30:137–148. [DOI] [PubMed] [Google Scholar]
- Kriebel M, Metzger J, Trinks S, Chugh D, Harvey RJ, et al. 2011. The cell adhesion molecule neurofascin stabilizes axo-axonic GABAergic terminals at the axon initial segment. J Biol Chem. 286:24385–24393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz S, Spirig M, Ginsburg C, Buchstaller A, Berger P, et al. 1998. Neurite fasciculation mediated by complexes of axonin-1 and Ng cell adhesion molecule. J Cell Biol. 143:1673–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JW, Lee AY, Sun M, Nikonenko AG, Chung SK, et al. 2003. Decreased anxiety, altered place learning, and increased CA1 basal excitatory synaptic transmission in mice with conditional ablation of the neural cell adhesion molecule L1. J Neurosci. 23:10419–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linneberg C, Toft CLF, Kjaer-Sorensen K, Laursen LS.. 2019. L1cam-mediated developmental processes of the nervous system are differentially regulated by proteolytic processing. Sci Rep. 9:3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz D, Sharaf A, Drexler D, Kataria H, Wolters-Eisfeld G, et al. 2017. Proteolytic cleavage of transmembrane cell adhesion molecule L1 by extracellular matrix molecule Reelin is important for mouse brain development. Sci Rep. 7:15268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz D, Wolters-Eisfeld G, Joshi G, Djogo N, Jakovcevski I, et al. 2012. Generation and nuclear translocation of sumoylated transmembrane fragment of cell adhesion molecule L1. J Biol Chem. 287:17161–17175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretzky T, Schulte M, Ludwig A, Rose-John S, Blobel C, et al. 2005. L1 is sequentially processed by two differently activated metalloproteases and presenilin/gamma-secretase and regulates neural cell adhesion, cell migration, and neurite outgrowth. Mol Cell Biol. 25:9040–9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maten MV, Reijnen C, Pijnenborg JMA, Zegers MM.. 2019. L1 cell adhesion molecule in cancer, a systematic review on domain-specific functions. Int J Mol Sci. 20:4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Miyai K, Ninomiya A, Yamasaki H, Tamura H, Nakamura Y, et al. 2003. NMDA-dependent proteolysis of presynaptic adhesion molecule L1 in the hippocampus by neuropsin. J Neurosci. 23:7727–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechtersheimer S, Gutwein P, Agmon-Levin N, Stoeck A, Oleszewski M, et al. 2001. Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J Cell Biol. 155:661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C, Fire A.. 1995. DNA transformation. Methods Cell Biol. 48:451–482. [PubMed] [Google Scholar]
- Montgomery AM, Becker JC, Siu CH, Lemmon VP, Cheresh DA, et al. 1996. Human neural cell adhesion molecule L1 and rat homologue NILE are ligands for integrin alpha v beta 3. J Cell Biol. 132:475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naus S, Richter M, Wildeboer D, Moss M, Schachner M, et al. 2004. Ectodomain shedding of the neural recognition molecule CHL1 by the metalloprotease-disintegrin ADAM8 promotes neurite outgrowth and suppresses neuronal cell death. J Biol Chem. 279:16083–16090. [DOI] [PubMed] [Google Scholar]
- Nayeem N, Silletti S, Yang X, Lemmon VP, Reisfeld RA, et al. 1999. A potential role for the plasmin(ogen) system in the posttranslational cleavage of the neural cell adhesion molecule L1. J Cell Sci. 112:4739–4749. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, et al. 1997. Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci. 17:8061–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K, Shirakawa M, Barnes TM, Hekimi S, Ohshima Y.. 1997. The UNC-14 protein required for axonal elongation and guidance in Caenorhabditis elegans interacts with the serine/threonine kinase UNC-51. Genes Dev. 11:1801–1811. [DOI] [PubMed] [Google Scholar]
- Oleszewski M, Beer S, Katich S, Geiger C, Zeller Y, et al. 1999. Integrin and neurocan binding to L1 involves distinct Ig domains. J Biol Chem. 274:24602–24610. [DOI] [PubMed] [Google Scholar]
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS.. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 24:79–88. [DOI] [PubMed] [Google Scholar]
- Pocock R, Benard CY, Shapiro L, Hobert O.. 2008. Functional dissection of the C. elegans cell adhesion molecule SAX-7, a homologue of human L1. Mol Cell Neurosci. 37:56–68. [DOI] [PubMed] [Google Scholar]
- Rahe D, Carrera I, Cosmanescu F, Hobert O.. 2019. An isoform-specific allele of the sax-7 locus. MicroPubl Biol.10.17912/micropub.biology.000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Suarez NJ, Belalcazar HM, Salazar CJ, Beyaz B, Raja B, et al. 2019. Axon-dependent patterning and maintenance of somatosensory dendritic arbors. Dev Cell. 48:229–244.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedle S, Kiefel H, Gast D, Bondong S, Wolterink S, et al. 2009. Nuclear translocation and signalling of L1-CAM in human carcinoma cells requires ADAM10 and presenilin/gamma-secretase activity. Biochem J. 420:391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougon G, Hobert O.. 2003. New insights into the diversity and function of neuronal immunoglobulin superfamily molecules. Annu Rev Neurosci. 26:207–238. [DOI] [PubMed] [Google Scholar]
- Sadoul K, Sadoul R, Faissner A, Schachner M.. 1988. Biochemical characterization of different molecular forms of the neural cell adhesion molecule L1. J Neurochem. 50:510–521. [DOI] [PubMed] [Google Scholar]
- Salzberg Y, Diaz-Balzac CA, Ramirez-Suarez NJ, Attreed M, Tecle E, et al. 2013. Skin-derived cues control arborization of sensory dendrites in Caenorhabditis elegans. Cell. 155:308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafi-Reinach TR, Melkman T, Hobert O, Sengupta P.. 2001. The lin-11 LIM homeobox gene specifies olfactory and chemosensory neuron fates in C. elegans. Development. 128:3269–3281. [DOI] [PubMed] [Google Scholar]
- Sarov M, Murray JI, Schanze K, Pozniakovski A, Niu W, et al. 2012. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell. 150:855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakura H, Inada H, Kuhara A, Fusaoka E, Takemoto D, et al. 2005. Maintenance of neuronal positions in organized ganglia by SAX-7, a Caenorhabditis elegans homologue of L1. EMBO J. 24:1477–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AW, Kamei Y, Kamiguchi H, Wong EV, Rapoport I, et al. 2002. L1 endocytosis is controlled by a phosphorylation-dephosphorylation cycle stimulated by outside-in signaling by L1. J Cell Biol. 157:1223–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer MK, Altevogt P.. 2010. L1CAM malfunction in the nervous system and human carcinomas. Cell Mol Life Sci. 67:2425–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer MK, Frotscher M.. 2012. Role of L1CAM for axon sprouting and branching. Cell Tissue Res. 349:39–48. [DOI] [PubMed] [Google Scholar]
- Schmitz C, Wacker I, Hutter H.. 2008. The Fat-like cadherin CDH-4 controls axon fasciculation, cell migration and hypodermis and pharynx development in Caenorhabditis elegans. Dev Biol. 316:249–259. [DOI] [PubMed] [Google Scholar]
- Sherry T, Handley A, Nicholas HR, Pocock R.. 2020. Harmonization of L1CAM expression facilitates axon outgrowth and guidance of a motor neuron. Development. 147:dev193805. [DOI] [PubMed] [Google Scholar]
- Silletti S, Mei F, Sheppard D, Montgomery AM.. 2000. Plasmin-sensitive dibasic sequences in the third fibronectin-like domain of L1-cell adhesion molecule (CAM) facilitate homomultimerization and concomitant integrin recruitment. J Cell Biol. 149:1485–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonderegger P, Kunz S, Rader C, Buchstaller A, Berger P, et al. 1998. Discrete clusters of axonin-1 and NgCAM at neuronal contact sites: facts and speculations on the regulation of axonal fasciculation. Prog Brain Res. 117:93–104. [DOI] [PubMed] [Google Scholar]
- Stringham EG, Dixon DK, Jones D, Candido EP.. 1992. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol Biol Cell. 3:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatti O, Gucciardo E, Pekkonen P, Holopainen T, Louhimo R, et al. 2015. MMP16 mediates a proteolytic switch to promote cell-cell adhesion, collagen alignment, and lymphatic invasion in melanoma. Cancer Res. 75:2083–2094. [DOI] [PubMed] [Google Scholar]
- Trent C, Tsuing N, Horvitz HR.. 1983. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics. 104:619–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kweon J, Larson S, Chen L.. 2005. A role for the C. elegans L1CAM homologue lad-1/sax-7 in maintaining tissue attachment. Dev Biol. 284:273–291. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S.. 1986a. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 314:1–340. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S.. 1986b. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 314:1–340. [DOI] [PubMed] [Google Scholar]
- Wong EV, Cheng G, Payne HR, Lemmon V.. 1995a. The cytoplasmic domain of the cell adhesion molecule L1 is not required for homophilic adhesion. Neurosci Lett. 200:155–158. [DOI] [PubMed] [Google Scholar]
- Wong EV, Kenwrick S, Willems P, Lemmon V.. 1995b. Mutations in the cell adhesion molecule L1 cause mental retardation. Trends Neurosci. 18:168–172. [DOI] [PubMed] [Google Scholar]
- Xu YZ, Ji Y, Zipser B, Jellies J, Johansen KM, et al. 2003. Proteolytic cleavage of the ectodomain of the L1 CAM family member Tractin. J Biol Chem. 278:4322–4330. [DOI] [PubMed] [Google Scholar]
- Yip ZC, Heiman MG.. 2018. Ordered arrangement of dendrites within a C. elegans sensory nerve bundle. eLife. 7:e35825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen JA, Kirch SA, Bargmann CI.. 1999. Genes required for axon pathfinding and extension in the C. elegans nerve ring. Development. 126:3679–3692. [DOI] [PubMed] [Google Scholar]
- Zhao X, Siu CH.. 1995. Colocalization of the homophilic binding site and the neuritogenic activity of the cell adhesion molecule L1 to its second Ig-like domain. J Biol Chem. 270:29413–29421. [DOI] [PubMed] [Google Scholar]
- Zhao X, Yip PM, Siu CH.. 1998. Identification of a homophilic binding site in immunoglobulin-like domain 2 of the cell adhesion molecule L1. J Neurochem. 71:960–971. [DOI] [PubMed] [Google Scholar]
- Zhou L, Barao S, Laga M, Bockstael K, Borgers M, et al. 2012. The neural cell adhesion molecules L1 and CHL1 are cleaved by BACE1 protease in vivo. J Biol Chem. 287:25927–25940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Opperman K, Wang X, Chen L.. 2008. unc-44 Ankyrin and stn-2 gamma-syntrophin regulate sax-7 L1CAM function in maintaining neuronal positioning in Caenorhabditis elegans. Genetics. 180:1429–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Liang X, Wang XM, Shen K.. 2017. Dynein and EFF-1 control dendrite morphology by regulating the localization pattern of SAX-7 in epidermal cells. J Cell Sci. 130:4063–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonta B, Desmazieres A, Rinaldi A, Tait S, Sherman DL, et al. 2011. A critical role for Neurofascin in regulating action potential initiation through maintenance of the axon initial segment. Neuron. 69:945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mutant and genome engineered strains will be available at the Caenorhabditis Genetics Center, and all strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplementary material is available at GENETICS online.