Abstract
Lithospermum erythrorhizon produces red naphthoquinone pigments that are shikonin derivatives. They are accumulated exclusively in the roots of this plant. The biosynthesis of shikonin is strongly inhibited by light, even though other environmental conditions are optimized. Thus, L. erythrorhizon dark-inducible genes (LeDIs) were isolated to investigate the regulatory mechanism of shikonin biosynthesis. LeDI-2, showing the strict dark-specific expression, was further characterized by use of cell suspension cultures and hairy root cultures as model systems. Its mRNA accumulation showed a similar pattern with that of shikonin. In the intact plants LeDI-2 expression was observed solely in the root, and the longitudinal distribution of its mRNA was also in accordance to that of shikonin. LeDI-2 encoded a very hydrophobic polypeptide of 114 amino acids that shared significant similarities with some root-specific polypeptides such as ZRP3 (maize) and RcC3 (rice). Reduction of LeDI-2 expression by its antisense DNA in hairy roots of L. erythrorhizon decreased the shikonin accumulation, whereas other biosynthetic enzymes, e.g. p-hydroxybenzoic acid:geranyltransferase, which catalyzed a critical biosynthetic step, showed similar activity as the wild-type clone. This is the first report of the gene that is involved in production of secondary metabolites without affecting biosynthetic enzyme activities.
Light plays an important role in being the essential energy source for photosynthesis, as well as an environmental signal for plants (Thompson, 1991). When plant seedlings are exposed to light, dramatic physiological changes are observed, e.g. repression of hypocotyl elongation, opening cotyledons, greening, etc. In secondary metabolic research it has been extensively shown that light functions as not only a stimulatory element for the production of many phenolic compounds (Ohl et al., 1990; Weisshaar and Jenkins, 1998), but also a critical inducing factor for flavonoid and terpenoid biosyntheses in some plant species (Hahlbrock and Scheel, 1989; Yamaura et al., 1991). In those studies it was demonstrated that the expressions of genes encoding biosynthetic enzymes were strongly activated by light irradiation, and light-dependent transcriptional regulators responsible for the positive regulation were also identified (Feldbruegge et al., 1994; Procissi et al., 1997). Contrary to those light-activated secondary metabolisms, there are several plants in which the production of secondary metabolites are inhibited by light irradiation, e.g. shikonin and anthraquinones (Tabata et al., 1974; Igbavboa et al., 1985). In fact, those compounds are specifically accumulated in the root or rhizome of those intact plants. However, there are few molecular genetic studies on genes involved in plant secondary metabolism preferentially expressed in darkness.
The production system of shikonin, a red naphthoquinone derivative in Lithospermum erythrorhizon, is a model cell culture system suitable for biochemical and molecular biological studies on dark-inducible secondary metabolism, i.e. the light inhibition is very strong, but reversible (Tabata et al., 1974; Heide et al., 1989), the product is visible, the biosynthetic pathway is well characterized (Li et al., 1998; Yazaki et al., 1999), and it is transformable with an appropriate binary vector (Yazaki et al., 1998). The productivity of shikonin by the cell suspension cultures was very high and it was the first example of industrial production of plant-derived pharmaceutical (Tabata and Fujita, 1985). To clarify the negative regulation mechanism of the secondary metabolism we isolated cDNAs strongly expressed in darkness in the cell culture, which were designated L. erythrorhizon dark-inducible (LeDI) genes (Yazaki et al., 1999). In this study we report the structure of LeDI-2 whose expression showed the exclusive preference to darkness, and we also report on the functional analyses of the gene product to demonstrate its involvement in the secondary metabolism specific to the root.
RESULTS
Cloning and Structure Analysis of LeDI-2
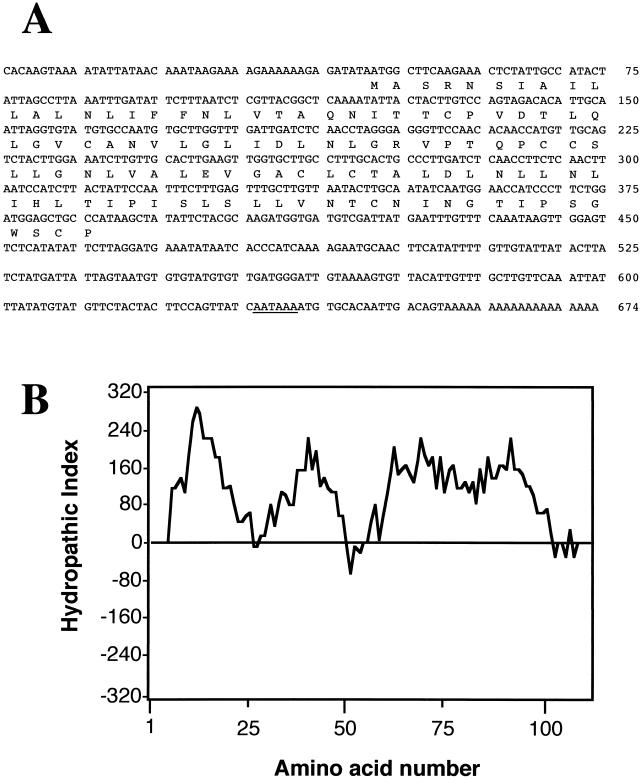
A series of the cDNA fragments whose expressions were preferentially induced in the dark were isolated by subtractive hybridization in a previous study (Yazaki et al., 1995a). One of those genes, LeDI-2, showed the strongest dark inducibility. Using the cDNA fragment as a probe, a near full-length clone of LeDI-2 (674 bp) was isolated from a lambda-ZAP cDNA library (accession no. D45901). Sequence analysis showed that it had an open reading frame of 114 amino acids (Fig. 1A). The deduced amino acid sequence was rich in Leu (22%) and showed high hydrophobicity (Fig. 1B), suggesting that it encoded a membrane-associated protein. Swiss-Prot database search revealed significant sequence similarities with several plant proteins known for specific expression in the root tissues such as ZRP3 of maize (John et al., 1992), RCc2 of rice (Xu et al., 1995), PVR5 of bean (Choi et al., 1996), and AIR1 of Arabidopsis (Neuteboom et al., 1999; Fig. 2). Although some proteins of this family, especially carrot (DC2.15, Aleith and Richter, 1990) or oilseed rape (SAC51, Coupe et al., 1993) proteins were reported as Pro-rich proteins due to Pro-rich domain in the N-terminal region, LeDI-2 lacked this characteristic sequence. However, LeDI-2 possessed eight Cys residues that were conserved among this family (Fig. 2). These results suggest that LeDI-2 also belongs to this family and has a similar secondary structure through disulfide linkage, although the role of the Pro-rich domain and the function of those polypeptides in plant cells have not been confirmed.
Figure 1.
A, Nucleotide sequence of cDNA and deduced amino acid sequence of LeDI-2. Putative polyadenylation signal is underlined. B, Hydrophobicity profile of LeDI-2 polypeptide. Kyte-Doolittle hydrophobicity index appears in the left margin. Positive values indicate hydrophobic regions. Amino acid numbers are shown beneath the plot.
Figure 2.
Multiple alignment of amino acid sequences of LEDI-2 and the orthologs. The origins of each molecular species are as follows: RcC2 and RcC3 (rice), ZRP3 (maize), DC2.15 (carrot), SAC51 (oilseed rape), CR14KD (Catharanthus roseus; Hotze et al., 1994), PVR5 (Phaseolus vulgaris), and AIR1 (Arabidopsis). Conserved Cys residues are highlighted with asterisks.
Expression of LeDI-2 in Cell Suspension Cultures
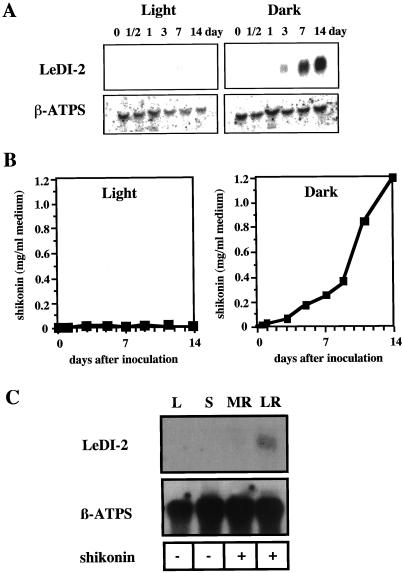
As described in the “Introduction,” the induction of shikonin biosynthesis by darkness in L. erythrorhizon cell cultures is well established (Heide et al., 1989; Yazaki et al., 1999). Thus, the gene expression of LeDI-2 was analyzed in cultured cells grown in the dark or in the light. Figure 3A illustrates the expression pattern of LeDI-2 in L. erythrorhizon cell suspension cultures in the dark or under illumination by northern hybridization. RNA-blot analyses showed that LeDI-2 mRNA is highly accumulated in the dark, whereas continuous light clearly suppressed its accumulation. This expression pattern of LeDI-2 mRNA during culture also corresponded to the accumulation pattern of shikonin derivatives, as shown in Figure 3B.
Figure 3.
A, Northern-blot hybridization of LeDI-2 in cell suspension cultures of L. erythrorhizon in M9 medium. Left, Gel blots of illuminated cultures, and those of dark-cultured cells are shown on the right. β-ATPS indicates a β-subunit of ATP synthase used as a load control (Boutry and Chua, 1985). B, Time course of shikonin production in those cell cultures agitated under illumination or in the dark. C, Organ-specific expression of LeDI-2 in the intact plant of L. erythrorhizon. L, Leaves; S, stems; MR, main root; LR, lateral roots. Shikonin accumulation monitored with the plant materials are shown beneath the blot: (+), shikonin derivatives are detected; (-), shikonin derivatives are not detectable.
Since there was positive correlation between LeDI-2 mRNA accumulation and shikonin accumulation, we further characterized the organ specific accumulation of LeDI-2 mRNA in the intact plant of L. erythrorhizon. RNAs were isolated from leaf, stem, lateral root, and main root, which were then probed with LeDI-2 cDNA. Again, LeDI-2 mRNA was only detected in the roots, where shikonin derivatives exclusively accumulated (Fig. 3C).
Expression of LeDI-2 in Hairy Root Cultures
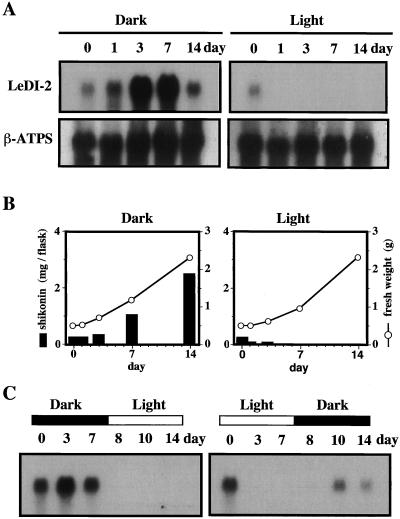
L. erythrorhizon is transformable with an aid of Agrobacterium rhizogenes and the hairy root system obtained by this method shows a similar shikonin productivity to the cell cultures (Shimomura et al., 1991). This transformation system provides a useful tool to characterize the function of LeDI-2 because an antisense DNA can be introduced into the L. erythrorhizon genome with a binary vector (Yazaki et al., 1998). Before the transformation with LeDI-2-antisense, the endogenous expression of LeDI-2 gene in hairy root cultures were determined. As shown in Figure 4A, LeDI-2 expression in cultured hairy roots was very similar to that in cell suspension cultures, i.e. light completely inhibited the LeDI-2 mRNA accumulation, as well as shikonin biosynthesis, without affecting the cell growth (Fig. 4B), whereas both accumulations were induced in the dark. One minor difference was that LeDI-2 mRNA increased earlier in hairy root cultures than in cell suspension cultures.
Figure 4.
Effects of light on LeDI-2 expression and shikonin production in hairy root cultures of L. erythrorhizon in M9 medium. A, Left panels show the accumulation of LeDI-2 mRNA in hairy roots cultured in the dark, and right ones are those under illumination. B, Cell growth and accumulation of shikonin in hairy roots cultured in the dark (left) or under illumination (right). The culture was started by inoculating 0.5 g of hairy roots that had been cultured in Murashige and Skoog liquid medium in the dark. C, Light-induced up- and down-regulation of LeDI-2 expression during the culture period. Hairy root cultures were transferred from a dark to light (left) or light- to-dark (right) condition 7 d after inoculation.
Figure 4C depicts a more detailed inhibitory effect of light on LeDI-2 mRNA accumulation. When the flask was transferred to light after cultivation in darkness for 7 d, the LeDI-2 mRNA level dropped down to undetectable level only 1 d after the irradiation (left). According to the quick decrease of LeDI-2 mRNA level, shikonin production immediately ceased under illumination (data not shown). LeDI-2 expression was, however, induced when the flask under illumination is transferred to darkness, i.e. mRNA of LeDI-2 started to accumulate at least 2 d after the transfer (right), and shikonin production was also recovered in the dark condition (data not shown).
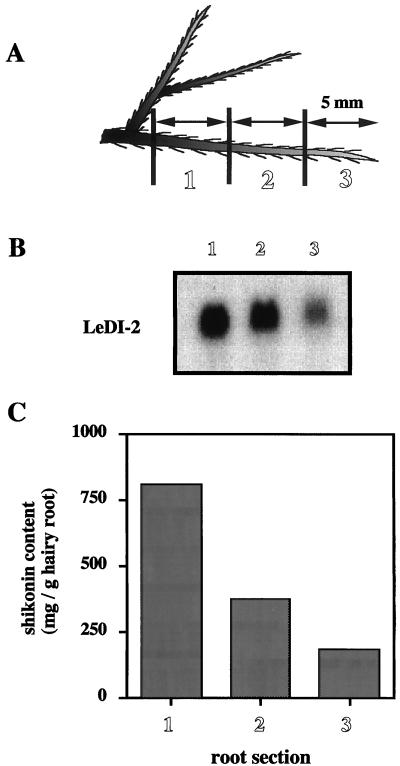
Because consecutive stages of development can be observed in a single longitudinal root axis, the accumulation pattern of LeDI-2 mRNA at different stages of development was detected by northern-blot analysis (Fig. 5). Figure 5B shows the longitudinal distribution of LeDI-2 mRNA throughout the hairy root in liquid cultures. The highest accumulation was observed in section 1, as the mRNA levels decrease to the region of the root tip. The accumulation of shikonin in the hairy root showed a high correlation with the mRNA level of LeDI-2 as revealed in Figure 5C. Nearly the same results were obtained for all transformants tested.
Figure 5.
Longitudinal distribution of LeDI-2 mRNA throughout the primary root of L. erythrorhizon (clone no. 5). Root tissues were cut to three sections of 5 mm in length as shown in A. Northern-blot analysis of RNA isolated from each segment of the hairy root using LeDI-2 as a hybridization probe (B). Shikonin contents in each root segment are shown in C.
LeDI-2 Expression in Transgenic Hairy Roots and Shikonin Production
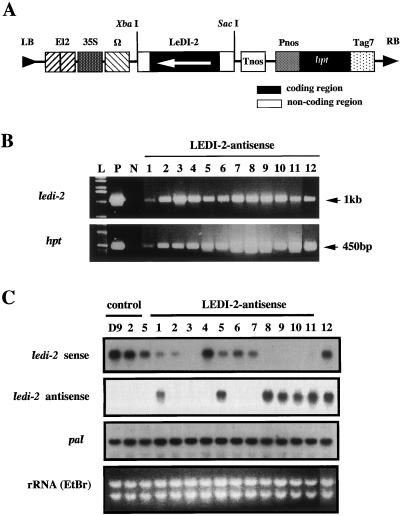
To characterize the function of the LeDI-2 polypeptide in vivo we constructed a binary vector containing the full-length cDNA of LeDI-2 in the antisense orientation (Fig. 6A). We used the enhancer-equipped cauliflower mosaic virus 35S promoter (El2 promoter) to drive LeDI-2 antisense, which had a strong promoter activity in L. erythrorhizon hairy roots (Yazaki et al., 1998), and the hygromycin resistant (hpt) gene was utilized as the selection marker. T-DNA region of the binary vector is co-integrated with the T-DNAs derived from Ri plasmid harbored by A. rhizogenes. To confirm the co-integrated T-DNA of the binary vector, hpt and LeDI-2 gene were amplified by PCR using the genomic DNA of each hairy root clone as the template and specific primers for introduced genes. In the case of this experiment about 45% of the hairy root generated by the infection survived on the hygromycin plate, and the hpt gene, as well as LeDI-2 antisense driven by El2 promoter, has been detected in more than 90% of those hygromycin-resistant clones (Fig. 6B). Total RNA was prepared from transformants of 7-d cultures where LeDI-2 expression was the highest, and hybridized with a single-strand RNA probe, sense or antisense of LeDI-2 (Fig. 6C). Northern-blot analyses indicated that several clones, e.g. clones 8 through 11, showed strongly reduced LeDI-2 mRNA accumulation accompanied with the overexpression of antisense LeDI-2 mRNA, whereas clone 3 showed strong reduction of LeDI-2 mRNA without the accumulation of LeDI-2 antisense. Clones 1 and 5 revealed modest suppression with the accumulation of antisense LeDI-2 mRNA. The mRNA level of Phe ammonia-lyase (PAL) involved in shikonin biosynthesis (Yazaki et al., 1997) as an internal standard of secondary metabolic activity showed, however, a nearly constant level of PAL mRNA in all the clones. This suggests that the introduced LeDI-2 antisense gene does not interfere in the general secondary metabolic activity, but rather is specific for LeDI-2 expression.
Figure 6.
A, Binary vector constructs (pBinHygLeDI-2as) for transformation of L. erythrorhizon. A full-length cDNA of LeDI-2 was subcloned under the control of El2 promoter. The orientation of the coding region of LeDI-2 cDNA is shown with an arrow, and the flanking regions of non-coding sequences are indicated as white. El2 has two enhancer sequences in tandem attached upstream of the cauliflower mosaic virus 35S promoter. B, Genomic PCR to detect exogenous LeDI-2 and hpt gene integrated in plant genome. P, Positive control in which plasmid DNA (pBinHygLeDI-2as) was used as the template; N, negative control using genomic DNA of control hairy root (clone 5) as the template; L, DNA ladder as size marker. C, Northern-blot analyses of LeDI-2 mRNA level in transformants. LeDI-2 sense shows the level of sense strand detected by antisense probe, and LeDI-2 antisense shown was probed with the sense probe. Three control hairy root clones that were not transformed with a binary vector were employed as control. The mRNA level of PAL was also detected with a consensus sequence of PAL cDNA fragment (Yazaki et al., 1997).
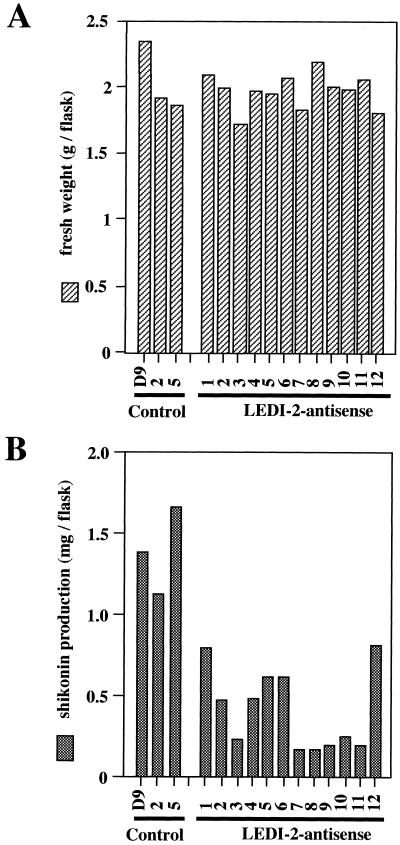
Figure 7 shows the growth and the accumulation of shikonin derivatives in those hairy root clones in which LeDI-2 expression is suppressed by the transformation of antisense DNA, compared with control hairy root cultures. The growth of antisense-induced hairy roots was not affected by the suppression of endogenous LeDI-2 expression, as shown in Figure 7A. Shikonin levels in LeDI-2 antisense clones were, however, clearly lower than those of control hairy roots, and there was no antisense clone whose shikonin production exceeded the level of shikonin in the control group (Fig. 7B). In particular, numbers 3 and 7 through 11 showed only low level of shikonin accumulation, which had an appreciable correlation with the mRNA level of LEDI-2 detected by RNA probe. Because shikonin is localized in small granules and secreted out of the cells, the shikonin amount recovered from the medium and the shikonin content in root tissues were separately measured, but the ratio was mostly constant (data not shown).
Figure 7.
A, Fresh weight of hairy root clones 14 d after inoculation. Each clone was cultured in 30 mL of medium (inoculum size 0.5g). B, Shikonin productivity of each hairy root clone. Shikonin derivatives were extracted from culture media and from hairy root tissues after 14 d of culture kept in the dark.
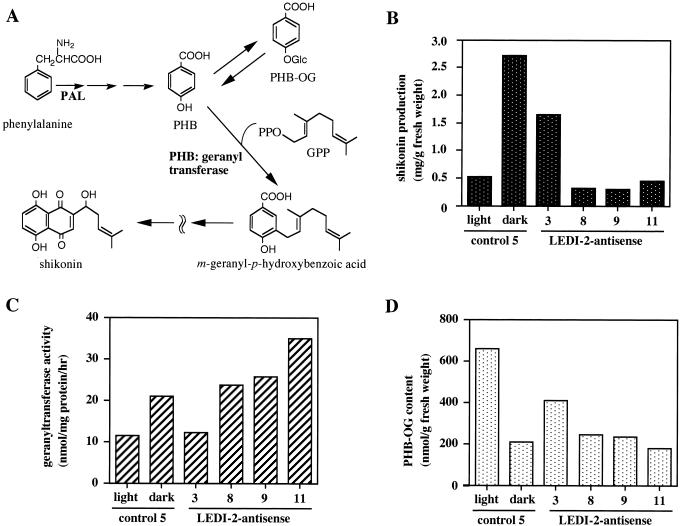
A critical regulatory reaction step in shikonin biosynthesis is p-hydroxybenzoic acid (PHB):geranyltransferase, which is very sensitive to ammonium ion, 2,4-D, and light, by which shikonin production is dramatically inhibited (Fig. 8A). This crucial enzyme activity was measured with three antisense clones that produced only small amounts of shikonin. Figure 8C shows that the enzyme activities of PHB:geranyltransferase in antisense clones are almost the same level as the control, whereas accumulation of shikonin is suppressed (Fig. 8B). It is remarkable that shikonin production is inhibited where geranyltransferase is as active as the control. The amount of PHB-O-glucoside was also analyzed and the result is shown in Figure 8D. There is usually a clear negative correlation with geranyltransferase because the glucoside is a storage form of the shikonin precursor, PHB, in the cells. Therefore, when geranyltransferase is suppressed by light, for instance, the level of PHB-O-glucoside increases. In antisense clones, however, their levels are almost within the same range as the control hairy root, indicating that the geranylation step is not influenced by LeDI-2 antisense.
Figure 8.
A, Biosynthetic pathway of shikonin. When the expression of geranyltransferase is inhibited by light, the excess amount of PHB is accumulated as its glucoside form. PHB-OG, O-glucoside of PHB; GPP, geranylpyrophosphate. Shikonin production and PHB:geranyltransferase activity in control and LeDI-antisense hairy root clones are indicated in B and C, respectively. The accumulation level of PHB-OG in the root tissues of those clones are also monitored in D. Cells were cultured in the dark or under continuous light (80 μE/m−2 s−1) with fluorescent lamps.
DISCUSSION
Light is an essential controlling factor in the development and metabolism of higher plants. In response to light, a large number of genes are not only up-regulated, but are also down-regulated in their expression. Dark-inducible gene expression was intensively studied in the research field of plant senescence (Shimada et al., 1998; Scheumann et al., 1999), amino acid synthesis (Tsai and Coruzzi, 1990; Yamagata et al., 1998), and induction of pathogenesis-related proteins (Eyal et al., 1992; Sessa et al., 1995). The light sensing mechanisms have been also intensively studied by several groups (Quail, 1994; Tobin and Kehoe, 1994; Neuhaus et al., 1997; Cashmore et al., 1999). In the field of secondary metabolism, however, very little is known about the down-regulation mechanisms of metabolite production by light at molecular level. Shikonin production in L. erythrorhizon is a suitable model to study the negative regulation of secondary metabolism (for review, see Yazaki et al., 1999).
Characterization of LeDI-2
In this paper we have demonstrated the isolation of a dark-inducible cDNA, LeDI-2, from the cell cultures of L. erythrorhizon, and its putative involvement in shikonin production, which is dark-specific secondary metabolism in L. erythrorhizon, by alteration of LeDI-2 expression using antisense DNA. Its expression in the cultured cells, as well as the hairy roots, preceded the accumulation of shikonin derivatives in the dark and was strongly inhibited under illumination. The accumulation of LeDI-2 in hairy root cultures started earlier compared with that of cell cultures, but this might be because the cells in hairy roots were differentiated and already expressing the biosynthetic enzymes involved in shikonin biosynthesis and LeDI-2, as well. The localization of LeDI-2 mRNA in the intact plant was restricted to root tissue, which is the sole organ for shikonin accumulation. The predicted amino acid sequence shares significant similarities with several plant polypeptides of unknown function, but some of which are reported to be specifically expressed in the roots. In the case of ZRP3 of maize (John et al., 1992) and PVR-5 in bean (Choi et al., 1996), the highest expression was observed in the cortical ground meristem of the roots where maximum cell division occurred. Because these small polypeptides seem to be membrane associated due to the high hydrophobicity, those authors presumed that they may play a role in the cell differentiation or in transport of substances necessary for cell division in plant cells (John et al., 1992; Choi et al., 1996). LeDI-2, however, showed almost no influence on the cell growth in L. erythrorhizon, even if the expression is strongly suppressed by the antisense DNA (Fig. 7), suggesting that LeDI-2 polypeptide plays no important role in cell division.
In hairy roots of L. erythrorhizon, higher levels of LeDI-2 mRNA were detected in the older part of the tissues (Fig. 5). This was consistent with the accumulation site of shikonin derivatives in the roots, i.e. higher accumulations were observed in older tissue of the root for both. Another characteristic for LeDI-2 amino acid sequence is that the Pro-rich domain, which occurs in most of the family members of this gene in other plant species, is lacking in LeDI-2 polypeptide, although the function of this domain in those orthologs is still an open question.
The Phenotype of Transgenic Hairy Roots
To analyze the involvement of the LeDI-2 polypeptide in shikonin production, hairy root clones transformed with full-length LeDI-2 sense and antisense constructs were prepared. We obtained a number of antisense transformants in which the accumulation of LeDI-2 mRNA was strongly suppressed, but sense constructs gave no strain overexpressing LeDI-2 (data not shown), thus we analyzed LeDI-2 antisense transformants in further detail. Shikonin production in those antisense clones was shown to be significantly lower than the control (Fig. 6), whereas no clear suppression of the growth or a remarkable morphological change in those hairy roots was observed. Then the influence of LeDI-2 suppression on the biosynthetic enzymes was analyzed.
Shikonin is synthesized in vivo by the coupling of two molecules: PHB derived from the shikimate pathway through PAL (Yazaki et al., 1997) and geranyl diphosphate, which is a product of mevalonate pathway (Li et al., 1998). This reaction is catalyzed by PHB:geranyltransferase, a critical regulatory enzyme in shikonin biosynthesis (Heide et al., 1989; Mühlenweg et al., 1998), i.e. this enzyme activity shows a direct correlation to the production of shikonin under various culture conditions (Heide et al., 1989; Yazaki et al., 1998). However, we could not find a significant decrease in the geranyltransferase activity in the antisense transformants, and the expression level of PAL was also unchanged (Fig. 6). To our knowledge it is noteworthy that shikonin production was strongly suppressed in those LeDI-2 antisense clones in spite of the high geranyltransferase activity as the control.
Putative Function of LeDI-2 Family in Plants
It is presumed that the biosynthetic enzymes after the prenylation step are localized in membrane vesicles derived from endoplasmic reticulum, and finally shikonin is secreted out of the cells with lipid to form red granules attached on the cell walls (Tsukada and Tabata, 1984). The intracellular vesicles observed in shikonin-producing cells, where the putative shikonin precursors are located, are covered by a phospholipid monolayer (Tabata, 1996). A well-studied example of such vesicles covered by a lipid monolayer in the plant cell is the oil body, which is further covered with a small hydrophobic polypeptide, oleosin. LeDI-2 polypeptide has no significant sequence similarity with the oleosin family, but it may localize in the membrane of such vesicles in which a large amount of lipid including shikonin is compartmented. By use of a green fluorescent protein-fusion protein with LeDI-2 we have observed the green fluorescence in the intracellular vesicles that seemed to be derived from endoplasmic reticulum (data not shown), but the identification by the electron microscopic experiment of these vesicles with those specific for shikonin will be needed.
As there are many orthologs in various plant species that do not have shikonin productivity, the function of LeDI-2 polypeptide in vivo may not be specific for the accumulation of shikonin. An Arabidopsis ortholog AIR1 protein, which is the most similar one to LeDI-2 (Fig. 2), is expected to be located outside the cell or at the plasma membrane (Neuteboom et al., 1999). There may be a similar transport mechanism between AIR1 protein in Arabidopsis and LeDI-2 polypeptide in L. erythrorhizon that may affect the shikonin accumulation.
Although the exact function of LeDI-2 in L. erythrorhizon is still unclear, the previous knowledge about shikonin production and the results obtained in this study suggest that the gene product of LeDI-2 is involved in the productivity of shikonin, probably by a sort of indirect effect on efficiency of shikonin production, e.g. affecting the stability or transport of the intracellular vesicles where shikonin biosynthesis takes place. This hypothesis may be supported by the identification of LeDI-2 polypeptide in shikonin-specific membrane vesicles by immunogold electron microscopic studies using an appropriate antibody against LeDI-2 protein. Further studies on the analysis of shikonin precursors in those LeDI-2-localized vesicles, and on the pulse-labeling of LeDI-2 polypeptide will offer clues to understand how LeDI-2 affects the production of the lipophilic secondary metabolites such as shikonin derivatives.
MATERIALS AND METHODS
Plant Materials
Cell suspension cultures of Lithospermum erythrorhizon (strain M18TOM derived from M18) were maintained in Linsmaier and Skoog medium (1965) supplemented with indoleacetic acid (10−6 M) and kinetin (10−5 M). They produced shikonin derivatives when cultured in M9 medium (Fujita et al., 1981) in darkness (Yazaki et al., 1997). Hairy roots of L. erythrorhizon obtained by infection with Agrobacterium rhizogenes (strain 15834) have been subcultured in Murashige and Skoog medium without plant hormones, whereas they were cultured also in M9 medium for shikonin production (Yazaki et al., 1998). Unless otherwise stated, the experiments were done with a representative hairy root clone number 5 cultured in M9 medium.
cDNA Cloning
A subtracted probe in which dark-expressing cDNAs of L. erythrorhizon were enriched was used as a hybridization probe to screen a lambda-ZAP cDNA library (Yazaki et al., 1995a). In the first screening, approximately 360 signals were detected out of approximately 6,000 plaques from which redundancy was checked by random cross-hybridization experiments to obtain six subsets of cDNAs. The dark-selectivity was then checked by RNA-blot hybridization where total RNAs of the dark-grown cells and of illuminated cells were immobilized on a nylon membrane Hybond N+ (Amersham, Buckinghamshire, UK), which was hybridized with the individual cDNA. One of them, LeDI-2, which was the smallest subset (two clones), showed the strongest dark selectivity. The nucleotide sequence data reported in this paper will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases (with the accession no. D45901).
Extraction and Analysis of RNA
Total RNA was extracted from cultured cells or hairy roots by use of Plant RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Poly(A)+ RNA was prepared by QuickPrep Micro mRNA Purification Kit (Pharmacia Biotech, Piscataway, NJ). Fifteen micrograms of total RNA or 3 μg of mRNA (per lane) was separated in a formamide-containing 1% (w/v) agarose gel and capillary blotted onto a nylon membrane Hybond N+. The membrane was hybridized according to the standard procedure. The hybridization probe was a full-length cDNA of LeDI-2, unless otherwise stated. The 18S rDNA of broad bean or the DNA fragment of the β-subunit of ATP synthase from Nicotiana plumbaginitolia was used for the load control in the northern analyses. Intact plants of L. erythrorhizon were a generous gift from the Botanical Garden of Takeda Chemical Industries, which were cultivated in pots for 2 years. Each tissue shown in the organ-specific northern hybridization experiment was excised and immediately frozen in liquid nitrogen.
Vector Construction
Because we found only a weak suppression of LeDI-2 in the previous study by introducing the coding region of LeDI-2 as the antisense DNA (Yazaki et al., 1999), full-length cDNA was used in this study. Full-length LeDI-2 cDNA was amplified by PCR with a primer pair that annealed on the T3 and T7 promoter region by which BglII site and SacI site were introduced on 5′ and 3′ ends, respectively. The PCR product was subcloned down stream of El2 promoter in pUC118 by the restriction sites BamHI and SacI. The El2 promoter consists of the 35S promoter having two enhancers in tandem and an omega sequence. The entire expression cassette between EcoRI and HindIII sites was then transferred into a binary vector (pBiHyg-HSE), a pBin19 derivative (Gatz et al., 1992) whose EcoRI and HindIII sites had been inverted. The selection marker for plant transformation was the hygromycin resistant-gene, htp. This completed plasmid (pBinHygLeDI-2as) was introduced to A. rhizogenes (strain 15834) by electroporation.
Transformation of L. erythrorhizon
Transformation of L. erythrorhizon and the selection of transformants was done according to the method established in our laboratory (Yazaki et al., 1998), except for plant material. In this study cultured shoots were utilized for A. rhizogenes-mediated transformation instead of the seedlings. Axenic shoot cultures of L. erythrorhizon applied for A. rhizogenes infection have been kept on hormone-free Linsmaier and Skoog medium of one-half salt concentration. After removing the root tissue, shoots of 5 to 8 cm were used for the infection with A. rhizogenes. Detection of the foreign gene integrated into the plant genome was carried out by PCR in which genomic DNA was used as the template. Genomic DNA was prepared from 100 mg of each L. erythrorhizon hairy root clone using the DNA extraction kit Phytopure (Scotlab, UK). In the PCR, two sets of primer pairs, which were designed to detect the LeDI-2 from the binary vector or the hpt gene, were used to amplify the 1-kb and 450-bp fragments, respectively. The primer sequences for hpt detection were previously reported (Yazaki et al., 1998). Those for antisense LeDI-2 were GATATCTCCACTGACGTAAGG (forward primer) and CCCATCTCATAAATAACGTC (reverse primer), which annealed to the 35S promoter and nopaline synthase terminator regions, respectively. PCR was done by the standard three-step program and then 5 μL of the reaction mixture was loaded on an agarose gel (1.5%, w/v) for electrophoresis.
Chemical Analysis of Shikonin and PHB-O-Glucoside
Quantitative analyses of shikonin produced by cell cultures and hairy root cultures were done 2 weeks after inoculation. Cultured cells or hairy roots were harvested by filtration through Miracloth (Calbiochem, San Diego). Because PHB-O-glucoside is localized in the vacuoles of the cells (Yazaki et al., 1995b), its content in the transformants was determined by HPLC analysis of the cell-free extract prepared as reported elsewhere (Tani et al., 1993). The solvent system consisted of H2O:CH3OH:acetic acid (74:5:1), column: ODS-120A (Tosoh, Japan), detection: A254.
Enzyme Assay
Hairy roots (0.5 g) were inoculated in 30 mL of M9 medium supplemented with 3 mL of liquid paraffin for extracting shikonin derivatives from the cell surface to trap the pigments in the paraffin layer. Seven days after inoculation, the harvested hairy roots were homogenized and cell-free extract was prepared by the method of Tani et al. (1993). PHB:geranyltransferase activity was measured according to Heide et al. (1989). The productivity of shikonin was also monitored by measuring the shikonin content in the lipid phase. An aliquot of the paraffin layer (10 μL) was partitioned with 1.2 mL of 2.5% (w/v) KOH, and absorbance of the aqueous phase was measured at 620 nm.
ACKNOWLEDGMENTS
We thank Dr. I. Furusawa of Kyoto University and Dr. C. Gatz of the Institut fuer Genbiologische Forschung (Berlin) for the generous gifts of El2 promoter and pBin19 derivative, respectively. We are also grateful to Dr. L. Brigham of the University of Arizona for the critical reading of the manuscript. The intact L. erythrorhizon plant was a generous gift from the Kyoto Botanical Garden of Takeda Chemical Industries.
Footnotes
This work was supported in part by the Japanese Society for Promotion of Science (grant to A.B.) and by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (to K.Y.).
LITERATURE CITED
- Aleith F, Richter G. Gene expression during induction of somatic embryogenesis in carrot cell suspensions. Planta. 1990;183:17–24. doi: 10.1007/BF00197562. [DOI] [PubMed] [Google Scholar]
- Boutry M, Chua N-H. A nuclear gene encoding the beta subunit of the mitochondria ATP synthase in Nicotiana plumbaginifolia. EMBO J. 1985;4:2159–2165. doi: 10.1002/j.1460-2075.1985.tb03910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D. Cryptochromes: blue light receptors for plant and animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- Choi DW, Song JY, Kwon YM, Kim SG. Characterization of a cDNA encoding a proline-rich 14-kDa protein in developing cortical cells of the roots of bean (Phaseolus vulgaris) seedling. Plant Mol Biol. 1996;30:973–982. doi: 10.1007/BF00020808. [DOI] [PubMed] [Google Scholar]
- Coupe SA, Taylor JE, Isaac PG, Roberts JA. Identification and characterization of a proline-rich mRNA that accumulates during pod development in oil seed rape (Brassica napus L.) Plant Mol Biol. 1993;23:1223–1232. doi: 10.1007/BF00042355. [DOI] [PubMed] [Google Scholar]
- Eyal Y, Sagee O, Fluhr R. Dark-induced accumulation of a basic pathogenesis-related (PR-1) transcript and a light requirement for its induction by ethylene. Plant Mol Biol. 1992;19:589–599. doi: 10.1007/BF00026785. [DOI] [PubMed] [Google Scholar]
- Feldbruegge M, Sprenger M, Dinkelbach M, Yazaki K, Harter K, Weisshaar B. Functional analysis of a light-responsive plant bZIP transcriptional regulator. Plant Cell. 1994;6:1607–1621. doi: 10.1105/tpc.6.11.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hara Y, Suga C, Morimoto T. Production of shikonin derivatives by cell suspension cultures of Lithospermum erythrorhizon: a new medium for the production of shikonin derivatives. Plant Cell Rep. 1981;1:61–63. doi: 10.1007/BF00269273. [DOI] [PubMed] [Google Scholar]
- Gatz C, Frohberg C, Wendenburg R. Stringent repression and homogeneous de-repression by tetracycline of a modified CaMV 35S promoter in intact transgenic tobacco plants. Plant J. 1992;2:397–404. doi: 10.1111/j.1365-313x.1992.00397.x. [DOI] [PubMed] [Google Scholar]
- Hahlbrock K, Scheel D. Physiology and molecular biology of phenylpropanoid metabolism. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:347–369. [Google Scholar]
- Heide L, Nishioka N, Fukui H, Tabata M. Enzymatic regulation of shikonin biosynthesis in Lithospermum erythrorhizon cell cultures. Phytochemistry. 1989;28:1873–1877. [Google Scholar]
- Hotze M, Waitz A, Schröder J. cDNA for a 14-kilodalton polypeptide from Madagascar periwinkle (Catharanthus roseus) Plant Physiol. 1994;104:1097–1098. doi: 10.1104/pp.104.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igbavboa U, Sieweke H-J, Leistner E, Röwer I, Hüsemann W, Barz W. Alternative formation of anthraquinones and lipoquinones in heterotrophic and photoautotrophic cell suspension cultures of Morinda lucida Benth. Planta. 1985;166:537–544. doi: 10.1007/BF00391279. [DOI] [PubMed] [Google Scholar]
- John I, Wang H, Held BM, Wurtele ES, Colbert JT. An mRNA that specifically accumulates in maize root delineates a novel subset of developing cortical cells. Plant Mol Biol. 1992;20:821–831. doi: 10.1007/BF00027153. [DOI] [PubMed] [Google Scholar]
- Li S-M, Hennig S, Heide L. Shikonin: a geranyl diphosphate-derived plant hemiterpenoid formed via the mevalonate pathway. Tetrahedron Lett. 1998;39:2721–2724. [Google Scholar]
- Linsmaier EM, Skoog F. Organic growth factor requirements of tobacco tissue cultures. Physiol Plant. 1965;18:100–127. [Google Scholar]
- Mühlenweg A, Melzer M, Li S-M, Heide L. 4-Hydroxybenzoate 3-geranyltransferase from Lithospermum erythrorhizon: purification of a plant membrane-bound prenyltransferase. Planta. 1998;205:407–413. doi: 10.1007/s004250050337. [DOI] [PubMed] [Google Scholar]
- Neuhaus G, Bowler C, Hiratsuka K, Yamagata H, Chua N-H. Phytochrome-regulated repression of gene expression requires calcium and cGMP. EMBO J. 1997;15:2554–2564. doi: 10.1093/emboj/16.10.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuteboom LW, Ng JMY, Kuyper M, Clijdesdale OR, Hooykaas PJJ, van der Zaal BJ. Isolation and characterization of cDNA clones corresponding with mRNAs that accumulated during auxin-induced lateral root formation. Plant Mol Biol. 1999;39:273–287. doi: 10.1023/a:1006104205959. [DOI] [PubMed] [Google Scholar]
- Ohl S, Hedrick SA, Chory J, Lamb CJ. Functional properties of a phenylalanine ammonia-lyase promoter from Arabidopsis. Plant Cell. 1990;2:837–848. doi: 10.1105/tpc.2.9.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procissi A, Dolfini S, Ronchi A, Tonelli C. Light-dependent spatial and temporal expression of pigment regulatory genes in developing maize seeds. Plant Cell. 1997;9:1547–1557. doi: 10.1105/tpc.9.9.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. Photosensory perception and signal transduction in plants. Curr Opin Genet Dev. 1994;4:652–61. doi: 10.1016/0959-437x(94)90131-l. [DOI] [PubMed] [Google Scholar]
- Scheumann V, Schoch S, Rudiger W. Chlorophyll b reduction during senescence of barley seedlings. Planta. 1999;209:364–370. doi: 10.1007/s004250050644. [DOI] [PubMed] [Google Scholar]
- Sessa G, Yang X-Q, Raz V, Eyal Y, Fluhr R. Dark induction and subcellular localization of the pathogenesis-related PRB-1b protein. Plant Mol Biol. 1995;28:537–534. doi: 10.1007/BF00020400. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Wu GJ, Watanabe A. A protein encoded by din1, dark-inducible and senescence-associated gene of radish, can be imported by isolated chloroplasts and has sequence similarity to sulfide dehydrogenase and other small stress proteins. Plant Cell Physiol. 1998;39:139–142. doi: 10.1093/oxfordjournals.pcp.a029350. [DOI] [PubMed] [Google Scholar]
- Shimomura K, Sudo H, Saga H, Komada H. Shikonin production and secretion by hairy root cultures of Lithospermum erythrorhizon. Plant Cell Rep. 1991;10:282–285. doi: 10.1007/BF00193142. [DOI] [PubMed] [Google Scholar]
- Tabata M. The mechanism of shikonin biosynthesis in Lithospermum cell cultures. Plant Tissue Culture Lett. 1996;13:117–125. [Google Scholar]
- Tabata M, Fujita Y. Production of shikonin by plant cell cultures. In: Zaitlin M, Day P, Hollaender A, editors. Biotechnology in Plant Science. Orlando, FL: Academic Press; 1985. pp. 207–218. [Google Scholar]
- Tabata M, Mizukami H, Hiraoka N, Konoshima M. Pigment formation in callus cultures of Lithospermum erythrorhizon. Phytochemistry. 1974;13:927–932. [Google Scholar]
- Tani M, Takeda K, Yazaki K, Tabata M. Effects of oligogalacturonides on biosynthesis of shikonin in Lithospermum cell cultures. Phytochemistry. 1993;34:1285–1290. [Google Scholar]
- Thompson WF. Physiological and molecular studies of light-regulated genes in higher plants. Annu Rev Plant Physiol. 1991;42:423–466. [Google Scholar]
- Tobin EM, Kehoe DM. Phytochrome regulated gene expression. Semin Cell Biol. 1994;5:335–346. doi: 10.1006/scel.1994.1040. [DOI] [PubMed] [Google Scholar]
- Tsai FY, Coruzzi GM. Dark-induced and organ-specific expression of two asparagine synthetase genes in Pisum sativum. EMBO J. 1990;9:323–332. doi: 10.1002/j.1460-2075.1990.tb08114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Tabata M. Intracellular localization and secretion of naphthoquinone pigments in cell cultures of Lithospermum erythrorhizon. Planta Med. 1984;50:338–341. doi: 10.1055/s-2007-969725. [DOI] [PubMed] [Google Scholar]
- Weisshaar B, Jenkins GI. Phenylpropanoid biosynthesis and its regulation. Curr Opin Plant Biol. 1998;1:251–257. doi: 10.1016/s1369-5266(98)80113-1. [DOI] [PubMed] [Google Scholar]
- Xu Y, Buchholz WG, DeRose RT, Hall TC. Characterization of a rice gene family encoding root-specific proteins. Plant Mol Biol. 1995;27:237–248. doi: 10.1007/BF00020180. [DOI] [PubMed] [Google Scholar]
- Yamagata H, Nakajima A, Bowler C, Iwasaki T. Molecular cloning and characterization of a cDNA encoding asparagine synthetase from soybean (Glycine max L.) cell cultures. Biosci Biotechnol Biochem. 1998;62:148–150. doi: 10.1271/bbb.62.148. [DOI] [PubMed] [Google Scholar]
- Yamaura T, Tanaka S, Tataba M. Participation of phytochrome in the photoregulation of terpenoid synthesis in thyme seedlings. Plant Cell Physiol. 1991;32:603–608. [Google Scholar]
- Yazaki K, Bechthold A, Tabata M. Nucleotide sequence of a cDNA from Lithospermum erythrorhizon homologous to PR-1 of parsley. Plant Physiol. 1995a;108:1331–1332. doi: 10.1104/pp.108.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K, Inushima K, Kataoka M, Tabata M. Intracellular localization of UDPG: p-hydroxybenzoate glucosyltransferase and its reaction product in Lithospermum cell cultures. Phytochemistry. 1995b;38:1127–1130. [Google Scholar]
- Yazaki K, Kataoka M, Honda G, Severin K, Heide L. cDNA cloning and gene expression of phenylalanine ammonia-lyase in Lithospermum erythrorhizon. Biosci Biotechnol Biochem. 1997;61:1995–2003. doi: 10.1271/bbb.61.1995. [DOI] [PubMed] [Google Scholar]
- Yazaki K, Matsuoka H, Ujihara T, Sato F. Shikonin biosynthesis in Lithospermum erythrorhizon: light-induced negative regulation of secondary metabolism. Plant Biotechnol. 1999;16:335–342. [Google Scholar]
- Yazaki K, Tanaka S, Matsuoka H, Sato F. Stable transformation of Lithospermum erythrorhizon with Agrobacterium rhizogenes and shikonin production of the transformants. Plant Cell Rep. 1998;18:214–219. doi: 10.1007/s002990050559. [DOI] [PubMed] [Google Scholar]