Summary
Angiotensin-converting enzyme 2 (ACE2) was identified as a molecule that mediates the cellular entry of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Several membrane molecules of the host cell must cooperate in this process. While ACE2 serves in a membrane receptor-mediating interaction with the surface spike (S) glycoprotein of SARS-CoV-2 located on the virus envelope, enzyme A disintegrin and metalloproteinase 17 (ADAM17) regulates ACE2 availability on the membrane and transmembrane protease serine 2 (TMPRSS2) facilitates virus-cell membrane fusion. Interestingly, ACE2, ADAM17 and TMPRSS2 show a daily rhythm of expression in at least some mammalian tissue. The circadian system can also modulate COVID-19 progression via circadian control of the immune system (direct, as well as melatonin-mediated) and blood coagulation. Virus/ACE2 interaction causes ACE2 internalization into the cell, which is associated with suppressed activity of ACE2. As a major role of ACE2 is to form vasodilatory angiotensin 1–7 from angiotensin II (Ang II), suppressed ACE2 levels in the lung can contribute to secondary COVID-19 complications caused by up-regulated, pro-inflammatory vasoconstrictor Ang II. This is supported by the positive association of hypertension and negative COVID-19 prognosis although this relationship is dependent on numerous comorbidities. Hypertension treatment with inhibitors of renin-angiotensin system does not negatively influence prognosis of COVID-19 patients. It seems that tissue susceptibility to SARS-CoV-2 shows negative correlation to ACE2 expression. However, in lungs of infected patient, a high ACE2 expression is associated with better outcome, compared to low ACE2 expression. Manipulation of soluble ACE2 levels is a promising COVID-19 therapeutic strategy.
Keywords: Hypertension, ACE, Circadian, Gender, Ontogenesis
Introduction
Coronavirus disease 2019 (COVID-19) is caused by the coronavirus-2 (SARS-CoV-2) and represents a challenge for the health system and societies all over the world. The COVID-19 dashboard of the Center for Systems Science and Engineering at Johns Hopkins University features daily updated global information about COVID-19 cases and deaths from 192 countries (Dong et al. 2020). There were more than 202 million COVID-19 cases worldwide, with more than 4.29 million deaths as of 9 August 2021. The unexpected quick progression of SARS-CoV-2 resulted from its rapid spread via respiratory droplets from an infected person and by direct contact with contaminated surfaces (Vardoulakis et al. 2020).
Extensive research has been conducted on the characteristics of virus and the mechanisms of its penetration into cells. As a result of the enormous effort of the scientific community, major accomplishments were reported in an extremely short time. During one year, we learned more about risk factors for mortality, but also about positive factors, which can help patients successfully survive COVID-19. Despite this, additional knowledge on the pathogenesis of the disease is needed for effective treatment and to minimize its negative consequences; this must be the additional approach of massive vaccination programs.
Dorjee et al. (2020) revealed that the case fatality risks of the prevalence of death in risk groups were: cardiovascular disease (52 %), chronic obstructive pulmonary disease (51 %), chronic kidney disease (48 %), chronic liver disease (39 %), age ≥60 years (35 %), hypertension (28 %), smoking history (27 %), male gender (26 %) and diabetes (24 %). The major cause of mortality from COVID-19 is represented by respiratory failure, but in addition to the lungs, the cardiovascular system is also of critical importance, as outlined by extensive epidemiological evidence. Large epidemiological studies implicate hypertension as an important risk factor of the complicated form of COVID-19 disease. A strong association has been found between COVID-19-related deaths and high blood pressure in the OpenSAFELY platform study. The hypertension is associated with a higher risk of COVID-19 mortality in patients up to the age of 70 years. The adjusted HR for COVID-19 death for the age group of 18–39 years old patients with hypertension is 3.10 (1.69–5.70), 40–49 olds have HR 2.73 (1.96–3.81), patients in age between 50–59 years have HR 2.07 (1.73–2.47) and in age group between 60–69 years the adjusted HR is 1.32 (1.17–1.50) of COVID-19 death. The risk in patients above the age of 70 years is lower (adjusted HR 0.94 (0.86–1.02) and adjusted HR 0.73 (0.69–0.78) for age groups 70–79 olds and over 80 years old patients, respectively). The hypertension hazard ratio adjusted not only for age and sex, but also for diabetes and obesity decreased from 1.09 (1.05–1.14) to HR 0.97 (0.92–1.01) (Williamson et al. 2020). The severity of COVID-19 was higher in hypertensive patients, showing a hazard ratio (HR) of 2.27 (95 % confidence interval [CI]: 1.8–2.86), compared to non-hypertensive patients. In addition, hypertensive patients exhibited a HR of 3.48 (95 % CI: 1.72–7.08) of dying from COVID-19, compared to non-hypertensive patients (Zhang et al. 2020).
The role of ACE2, as a part of renin-angiotensin system, in COVID-19 progression
Hypertension is frequently associated with an up-regulated renin-angiotensin system (RAS), which is a critical regulator of the cardiovascular system with strong impacts on blood pressure, water balance and vascular function. RAS functioning is based on multistep enzymatic processing, leading to the conversion of precursors to effectors in a way that allows negative feedback to eliminate aberrations within the RAS. Among enzymes involved in the RAS is a zinc-dependent metalloprotease (ACE2) discovered in 2000 that shows a strong influence on the levels of major RAS effector molecules angiotensin II (Ang II) and angiotensin 1–7 (Ang1–7) (Hooper et al. 2020).
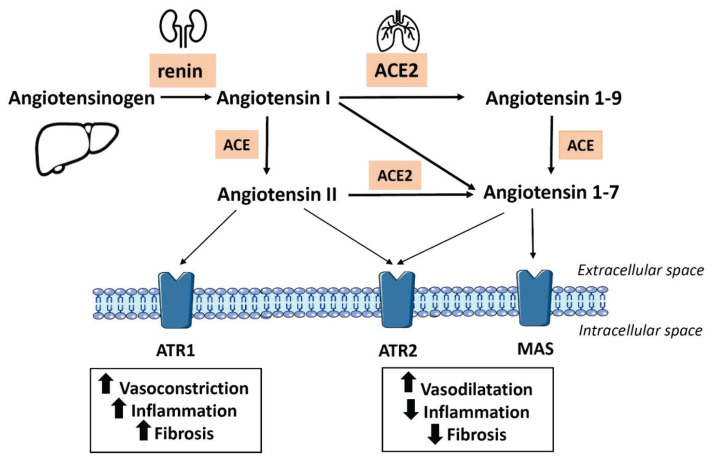
The precursor of Ang II, angiotensin I (Ang I), is formed from angiotensinogen, originating in the liver, by renal peptidase renin released by the juxtaglomerular cells in the kidney in response to blood pressure decrease (Herichova and Szantoova 2013, for a review). Conversion of Ang I to Ang II is executed by angiotensin-converting enzyme (ACE), which strongly resembles ACE2 by its structure. This step is followed by a rise in Ang II levels, which causes an increase in blood pressure and shows a strong pro-inflammatory influence, mediated mostly via angiotensin receptor type 1 (ATR1). Unlikely ACE, ACE2 converts both, Ang I and Ang II, to angiotensin 1–9 (Ang1–9) and Ang1–7, respectively. Ang1–7 is the primary vasodilatory ACE2 product and is known to counterbalance the effects of Ang II. It binds to the MAS receptor, exerts anti-inflammatory and anti-fibrotic actions and reduces blood pressure (Fig. 1) (Schmieder et al. 2007). Therefore, ACE2 activity is considered to be a preventive factor of cardiovascular pathologies associated with up-regulated Ang II levels. Ang II can be metabolized to angiotensin IV, which together with Ang II, plays a significant role in thrombosis development (Mogielnicki et al. 2005). In addition to the systemic RAS, most of its components, including ACE2, are also locally formed in different organs, such as the heart, brain, kidney and gastrointestinal tract, creating a so-called peripheral RAS (Paul et al. 2007).
Fig. 1.
Renin-angiotensin system cascade. Angiotensinogen is converted to the decapeptide angiotensin I by renin. This step is followed by the generation of angiotensin II (Ang II) from angiotensin I by angiotensin-converting enzyme (ACE). Ang II binds to transmembrane angiotensin type 1 (ATR1) or type 2 (ATR2) receptors, which are associated with different physiological responses. Effects of Ang II can be terminated by its conversion to angiotensin 1–7 with use of the angiotensin-converting enzyme 2 (ACE2). ACE2 can also convert angiotensin I to form angiotensin 1–9, which can be modified by ACE to generate angiotensin 1–7. Angiotensin 1–7 can exert its anti-inflammatory and other beneficial counterbalancing effects of Ang II after binding to MAS receptor.
Unfortunately, despite ACE2 function as an Ang II-counterbalancing the RAS component, ACE2 is also related to COVID-19 progression. This feature of ACE2 was discovered due to the similarity of SARS-CoV-2 to SARS-CoV, its close relative. The SARS-CoV virus was responsible for an epidemic of severe acute respiratory syndrome (SARS) in 2002 in China. It penetrates the cell membrane by binding to ACE2 via the receptor binding domain (RBD), which is a part of the transmembrane spike (S) glycoprotein. The S protein of SARS-CoV-2 shares approximately 75 % of the identity of SARS-CoV.
High-affinity interaction between SARS-CoV-2 and ACE2 was confirmed under in vitro conditions. Binding frequency between SARS-CoV-2 and ACE2 increased exponentially with prolonged contact time (Yang et al. 2020). The RBD of the SARS-CoV-2 S protein has similar or lower binding capacity compared to SARS-CoV (Shang et al. 2020) but SARS-CoV-2 shows 10- to 20-fold higher affinity to ACE2, possibly explaining the faster spread of COVID-19 worldwide (Wrapp et al. 2020). Therefore, not surprisingly, ACE2 was quickly recognized as a functional receptor for SARS-CoV-2 and ACE2 role in facilitation of SARS-CoV infection was confirmed (Saponaro et al. 2020).
Based on previous finding a low ACE2 expression should be beneficial and inhibit spread of SARS-CoV-2. As virus/ACE2 interaction causes ACE2 internalization into the cell, SARS-CoV-2 infection is associated with suppressed expression and activity of ACE2 (Saponaro et al. 2020, Xu et al. 2021). However, experimental data do not support the hypothesis that low ACE2 during COVID-19 disease is beneficial for patients. A lack of ACE2 was associated with increased lung injury after viral infection and this detrimental effect was markedly ameliorated in mice genetically lacking lung epithelial ATR1a (Xu et al. 2021). The protective role of ACE2 in acute lung injury was also evidenced by experiments using animal Ace2−/− knockout models, in which ACE2 deficiency was associated with worse lung pathology (Gagliardi et al. 2020). As ACE2 deficiency can boost the vasoconstrictor and pro-inflammatory arm of the RAS (Hooper et al. 2020) several authors attribute the organ damage accompanying the difficult form of COVID-19 to up-regulated Ang II (Gagliardi et al. 2020, Hanff et al. 2020), implicating the beneficial role of ACE2 in lung tissue.
So decrease in ACE2 expression may attenuate the SARS-CoV-2 infection, however, it can slow-down Ang II conversion and cause an increased activation of ATR1 and subsequent tissue damage. On the other hand, abundant ACE2 expression mediates increased virus penetration, but less tissue damage (Zores and Rebeaud 2020). ACE2 expression can be up-regulated by antihypertensive drugs, e.g. ACE inhibitors or ATR1 blockers that are both recommended as first-line medications for hypertension treatment. However, it was confirmed that treatment with an ACE inhibitor or ATR1 blockers is not associated with greater susceptibility to SARS-CoV-2 infection or worse disease course (Lee et al. 2020, Zores and Rebeaud 2020, Xu et al. 2021).
To summarize, ACE2 expression influences COVID-19 progression in two ways. There is a negative effect, as ACE2 mediates virus internalization into the cell and beneficial effect, as ACE2 counterbalances Ang II induced hypertension and inflammation. Beneficial effect of ACE2 seems to be prevailing as increased ACE2 expression in nasopharyngeal epithelium was associated with reduced respiratory distress in COVID-19 patients (Rossi et al. 2021).
Molecular mechanism of ACE2 receptor role in COVID-19 infection and alternative pathways
The molecular mechanism of SARS-CoV-2 penetration into the human body is well understood. Host cell receptor recognition, cell attachment and membrane fusion are executed after binding of surface S glyco-proteins located on the virus envelope to host cell membrane. The trimeric S protein consists of an extracellular N-terminal domain, transmembrane (TM) domain, and intracellular C-terminal domain. Each monomer of the S protein contains two functional subunits, the receptor-binding spike S1 subunit and membrane-fusing spike S2 subunit. The S1 subunit includes a N-terminal domain and RBD. The S2 subunit includes a fusion peptide (FP), heptapeptide repeat sequence 1 (HR1), heptapeptide repeat sequence 2 (HR2), TM domain and cytoplasm domain (Shang et al. 2020). The distal S1 subunit of the S protein is responsible for receptor binding, while the TM S2 subunit mediates fusion between the viral envelope and target cell membrane (Hoffmann et al. 2020).
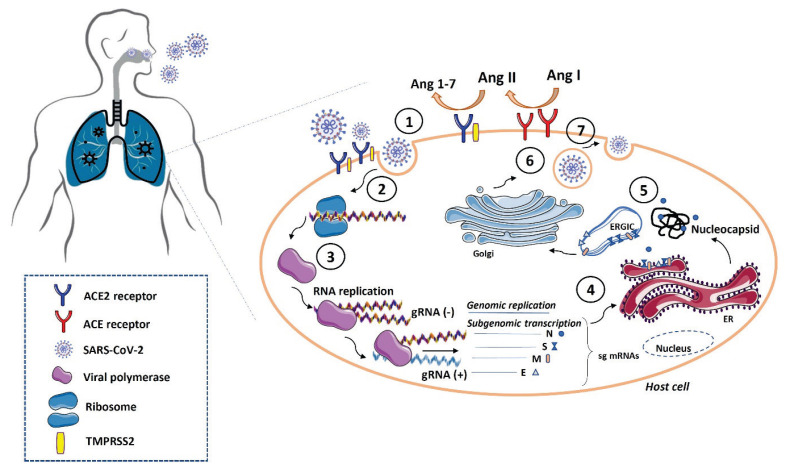
The S protein requires proteolytic activation, which allows the fusion of the viral and cellular membranes. Host proteases, including the transmembrane serine protease-2 (TMPRSS2) and cathepsin, are required for dissociation of S1 subunit and structural changes of subunit S2. During the viral infection, binding of RBD to a target membrane receptor allows an insertion of a FP into the cell membrane and fusion of virus/cell membranes, which is followed by the release of the viral genome into the host cell cytoplasm (Li et al. 2005, Ou et al. 2020). Subsequently, the open reading frames (ORF1a and ORF1ab) are translated and polyproteins PP1A and PP1AB, respectively, are produced. These polyproteins are post-translationally modified into the non-structural proteins that form the RNA replicase-transcriptase complex. Translated proteins are inserted into the endoplasmic reticulum (ER) and move from the ER to the Golgi intermediate compartment (ERGIC), where the viral genome interacts with the N protein and is incorporated into the ERGIC membrane. Here, the mature virions are formed and released from the infected cell by exocytosis (V’kovski et al. 2021) (Fig. 2).
Fig. 2.
SARS-CoV-2 replication in the host cells. Firstly, SARS-CoV-2 virus detects ACE2 receptors and binds to the host cell by its transmembrane spike (S) protein (1). Both ACE2 and TMPRSS2 are co-expressed at the cell surface and act as a point for coronavirus infection. Subsequently, the viral genome is released into the host cell (2). Viral genome replication involves several host and viral proteins that control RNA polymerization, proofreading and final virus assembly (3). Translated proteins (nucleocapsid N, spike S, membrane M, and envelope E) are inserted into the endoplasmic reticulum (ER) (4) and move from the ER to the Golgi intermediate compartment (ERGIC). Here, the viral genome interacts with the N protein and is incorporated into the ERGIC membrane (5). Finally, the mature virions are formed (6) and released from the infected cell by exocytosis (7). Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; ACE, angiotensin-converting enzyme; ACE2, angiotensin-converting enzyme 2; TMPRSS2, transmembrane serine protease-2; E, envelope; M, membrane; N, nucleocapsid; S, spike.
Previously, the role of TMPRSS2 was demonstrated in the entry of influenza virus, SARS-CoV and MERS-CoV virus. Recently, the role of TMPRSS2 was also described in SARS-CoV-2 infection (Dai et al. 2020, Hoffmann et al. 2020). In general, ACE2 and TMPRSS2 are co-expressed at the cell surface and act as a point for coronavirus infection (Fig. 2). The TMPRSS2 protease is essential in S protein priming, which is required for viral entry into the host cells and subsequent spreading in target cells (Hooper et al. 2020). Camostat mesylate, a TMPRSS2 inhibitor, is able to block SARS-CoV-2 virus entry into the Caco-2 and Vero-TMPRSS2 cells, indicating protease-dependent virus entry. Its combination with cathepsin B and L inhibitor amplifies the effect of camostat mesylate, which indicates that not only TMPRSS2, but also other proteases, are important in S protein priming during virus entry (Hoffmann et al. 2020).
The entry of SARS-CoV-2 into the host cells can be regulated by interferon-inducible transmembrane protein 3 (IFITM3) (Dai et al. 2020), which can inhibit virus replication and invasion. Generally, interferon-induced transmembrane proteins play an important role in the immune system by regulating virus fusion with endocytic vesicles; thereby, controlling the spread of the virus (Spence et al. 2019). The expression level of IFITM3 mRNA and protein vary in human tissues and organs. Abundant mRNA expression of IFITM3 was confirmed in all organs, however, mainly in lung, liver and blood cells. Interestingly, at protein level, IFITM3 was expressed more in gastrointestinal tract and endocrine tissues, and expression was reduced in lung, liver and blood cells (Dai et al. 2020). These results may explain the severity of lung disease observed in COVID-19 patients. The antiviral effect of IFITM3 may be amplified by valproic acid, which was found to enhance its expression in human bronchial epithelium at the transcription level (Hachim et al. 2020).
Several studies have confirmed that coronavirus infection can be enhanced by the cell surface neuropilin receptors, NRP1 (known as the vascular endothelial cell growth factor 165 receptor [VEGF165R]) and NRP2 vascular endothelial cell growth factor 165 receptor 2 (VEGF165R2) (Mayi et al. 2021). In general, NRP receptors participate in the regulation of angiogenesis, neurogenesis, and tumor progression (Pellet-Many et al. 2008). NRP1 expression was found in various tissues of the body, including abundant expression in endothelial and vascular smooth muscle cells, as well as neurons, retinal vasculature, and adipose tissue macrophages (Niland and Eble 2019). Interaction with neuropilin receptors requires the presence of a unique amino acid sequence at the carboxy terminus of the protein or peptide, named C-end rule (CendR motif). In SARS-CoV-2 infection, proteolytic cleavage of the S1 protein by the host cell protease furin is responsible for the generation of the CendR motif that interacts with NRP1 receptors (Daly et al. 2020). NRP1 knockout in HeLa cells expressing ACE2 caused a reduction in SARS-CoV-2 infection, compared to wild-type cells expressing ACE2 (Daly et al. 2020).
NRP1 receptors are abundantly expressed in the olfactory epithelium, suggesting that these receptors can represent ACE2-independent pathways of SARS-CoV-2 infection (Ramani et al. 2020) and may potentiate infection into the neurons and olfactory cells resulting in olfactory sense disruption. NRP1 and NRP2 show higher expressions in human lung tissue and human olfactory epithelium, compared to ACE2. Therefore, it is hypothesized that SARS-CoV-2-related anosmia in humans is potentiated by the NRP1 receptors in olfactory cells and neurons (Mayi et al. 2021).
Additionally, diabetes is associated with up-regulated expression of NRP1. This observation can explain the increased risk of SARS-CoV-2 infection in patients with diabetes (Cantuti-Castelvetri et al. 2020).
Heparan sulphate proteoglycans (HSPGs) are a family of cell-surface or extracellular matrix glycoproteins containing one or more heparan sulphate (HS) chains. The function of HSPGs depends on the location in the cell (e.g. basement membranes, secretory vesicles, or extracellular matrix). In general, HSPGs can facilitate cell-cell interactions and motility, act as co-receptors for various growth factor receptors, and regulate the distribution and activity of proteases (Sarrazin et al. 2011).
The interaction of SARS-CoV-2 S protein and cell surface HS is mediated by the RBD in the S1 subunit. In virus-cell attachment, both cellular HS and ACE2 are required, indicating that HS acts as a co-receptor in coronavirus infection. Secondly, HS increases virus recruitment to the cell surface; thus, potentiating its entry into cells. Consistent with this view, the HS-S protein complex represents a therapeutic target in COVID-19 prevention. In human adenocarcinoma cell lines, A549 and H1299, hepatoma Hep3B cells and melanoma A375 cells, enzymatic degradation of HS significantly reduced virus binding. Interestingly, the intensity of HS expression in cell lines did not correlate with the binding of S protein (Clausen et al. 2020), which can reflect differences in HS structure and expression levels of ACE2.
Sialic acid was also described as an attachment factor for different viruses, including alpha- and beta-coronaviruses. Coronaviruses interact with sialic acids, which are terminal residues of glycoconjugates located on the cell surface, before binding to receptors (Cuervo and Grandvaux 2020).
Regulation of ACE2 expression in tissues and involved control factors
Comparative ACE2 expression
The structure of ACE2 differs among species and this also affects its capacity to serve as a SARS-CoV-2 receptor for entry into host cells. Among species with an ACE2 structure allowing SARS-CoV-2 infection are those of the bat (Rhinolophus sinicus), pangolin (Manis javanica), cat (Felis catus), dog (Canis lupus), pig (Sus scrofa), human (Homo sapiens), monkey (Macaca mulatta), rabbit (Oryctolagus cuniculus) and goat (Capra hircus). On the other hand, differences in ACE2 structure causes the hen (Gallus gallus), mouse (Mus musculus) and rat (Rattus norvegicus) to have lower susceptibilities to SARS-CoV-2 entry. Unlikely other authors, Cao et al. (2020) suspected rat (Rattus norvegicus) to be a potential host of SARS-CoV-2. It was assumed that the species’ specific differences in vulnerability to SARS-CoV-2 infection were based on the short sequence in the RBD region that influenced the ability of ACE2 to facilitate viral entry (Zhang et al. 2021a). However, the newly identified Y217 residue with a strong impact on virus entry via ACE2 does not interact with the RBD; it rather influences ACE2 transport into the cell (Zhang et al. 2021a). Therefore, it is likely there is more than one way in which structure of ACE2 receptor influences its capacity to deliver SARS-CoV-2 into the cell.
In spite of species-dependent differences in capacity to convey SARS-CoV-2 propagation, ACE2 shows high homology in its DNA sequence among wildlife and laboratory species and is highly conserved. The human ACE2 protein sequence shows 98.1 % identity with orangutan (Pongo abeli), 85.2 % with cat (Felis catus), 83.5 % with civet (Paguma larvata), 81.1 % with cattle (Bos taurus), 82.5 % with rat (Rattus norvegicus) and 82.1 % with mouse (Mus musculus) (Cao et al. 2020).
In humans, the ACE2 gene is located on chromosome Xp22 and contains 18 exons and 20 introns. Recently, six ACE2 transcript variants and several ACE2 isoforms were identified. The best-known isoforms are full-length ACE2 (isoform 1) and soluble ACE2 generated by the enzymatic shedding of the full-length ACE2 from the membrane (Hooper et al. 2020). There are three additional isoforms that differ in the presence of the N-terminal signal peptide domain (SP) and length of ectodomain with peptidase activity (ED) and/or collectrin homology domain (CHD). The ACE2 isoform 2 has a shorter CHD than isoform 1, and the ACE2 isoform 3 contains shorter ED and CHD than isoform 1. The ACE2 isoform 4 (delta ACE2) lacks SP and the first 357 residues of ED and this structural change disables SARS-CoV-2 entry into cells and carboxypeptidase activity (Badawi and Ali 2021, Onabajo et al. 2020).
Transcription factors
There are several transcription factors known to influence ACE2 expression and it is believed that ACE2 belongs to the group of interferon-stimulated genes (Cuervo and Grandvaux 2020). A cAMP-response element binding protein responsive element was also described in its promoter (Clarke et al. 2014). Bioinformatic research of available human data implicated the roles of signal transducer and activator of transcription 1 and 3, interferon regulatory factor 1 and 8, interferon (INF)-gamma and -delta in the regulation of ACE2 expression (Salka et al. 2021). Pro-inflammatory cytokines tumor necrosis factor (TNF), INF-beta, interleukin (IL)-6 and -4 cause down-regulation of ACE2 expression (Oz and Lorke 2021); however, IL-1beta up-regulates ACE2 expression (Clarke et al. 2014). Recently, the regulation of ACE2 expression by interferons has been challenged by the discovery of an interferon-responsive delta ACE2 isoform that cannot facilitate SARS-CoV-2 entry into cells (Onabajo et al. 2020).
A functional p53 binding site was discovered in an ACE2 gene promoter and validated by luciferase assay. Knockout of TP53 caused an increase in ACE2 mRNA expression in porcine primary kidney fibroblast cells. On the other hand, a tissue- and sex-specific influence of p53 deficiency on ACE2 mRNA expression was described in the kidney and small intestine of pigs of both sexes (Zhang et al. 2021b).
It was also suggested that ACE2 expression can be influenced by sirtuin 1 (SIRT1), as ChIP-sequencing has demonstrated SIRT1 binding to the ACE2 promoter. Therefore, it was hypothesized that under energy stress conditions in cooperation with specific co-factors, SIRT1 initiates ACE2 transcription (Clarke et al. 2014).
In addition to above-mentioned ways ACE2 expression can also be regulated by methylation, glycosylation and phosphorylation (for review see Saponaro et al. 2020).
Shedding from the membrane
ACE2 can serve as a SARS-CoV-2 receptor, only while it is bound to the cell membrane. However, ACE2 can be detached from the membrane by the activity of several enzymes that contribute to a generation of soluble forms of ACE2 (Hooper et al. 2020) that possess biological activity (Jia et al. 2009) but cannot facilitate SARS-CoV-2 infection. Shedding of ACE2 from the membrane is catalyzed mainly via ADAM17. ADAM17 belongs to a group of modular transmembrane proteases that are involved in many signaling pathways by shedding membrane-anchored proteins. ADAM17 is also known as a tumor necrosis factor-alpha converting enzyme (TACE), as it was first identified as a sheddase that cleaves and activates membrane-bound tumor necrosis factor α (TNF-α) (Lorenzen et al. 2016). It is now known that ADAM17 cleaves more than 80 substrates, and among them are epidermal growth factor receptor (EGFR) and ACE2 (Lorenzen et al. 2016, Zipeto et al. 2020).
ADAM17 cleaves ACE2 in the juxtamembrane region of the ectodomain and in this way increases the level of soluble ACE2 and inhibits binding of SARS-CoV-2 S1 protein to its receptor (Lambert et al. 2005).
The regulation of ADAM17 availability is facilitated by the RAS, as Ang II induces ADAM17 expression and its membrane translocation via ATR1 receptors. A higher availability of ADAM17 results in ACE2 shedding from the cytoplasmatic membrane and as a result, higher levels of ACE2 in the plasma (Patel et al. 2014).
ACE2 expression in tissues
The expression of ACE2 mRNA in human tissues was investigated soon after its discovery (Harmer et al. 2002) and this analysis confirmed its ubiquitous expression, with highest levels in the testis, following by the gastrointestinal tract, lungs, kidney and heart. The lowest ACE2 mRNA expression was observed in the spleen, brain, ovary and myometrium. These results were partially confirmed by RNA sequencing in the study of Fu et al. (2020), demonstrating high levels of ACE2 mRNA expression in the human small intestine, kidney and heart. In mice, ACE2 showed the highest mRNA levels in the lung, breast, retina and kidney (Fu et al. 2020). The highest expression of ACE2 in pigs was observed in the kidney and small intestine (Zhang et al. 2021a). One of the latest GTEx (The Genotype-Tissue Expression) database searches confirmed high human ACE2 expression in the testis, small intestine, kidney, heart, breast and colon and low expression in the brain and spleen. According to a search of this study, ACE2 mRNA expression in the liver and lung was similar (Hu et al. 2021), which differs from above mentioned studies.
Taken together, available results implicate that ACE2 expression in the gastrointestinal tract and the kidney is high and similar in humans, mice and pigs.
Expression of mRNA and protein levels of ACE2 differ in some cases. The highest expression of ACE2 protein was demonstrated in the cardiovascular system, kidney and intestine (Saponaro et al. 2020). The expression of ACE2 was detected by immunohisto-chemistry in the endothelial cells of arteries and veins in all investigated tissues, including the lung, skin, gastrointestinal tract, kidney, brain, and spleen, among others. Similarly, ACE2 expression has been detected in smooth muscle cells of the gastrointestinal tract (Hamming et al. 2004).
Ubiquitous ACE2 expression in human tissues explains the wide range of COVID-19 symptoms, such as nausea, vomiting, diarrhea, and myocarditis, among others (Hanff et al. 2020).
The expression of ACE2 in specific cell types, such as type II alveolar cells, endothelial, myocardial cells, proximal tubule cells of the kidney, ileum and esophagus epithelial cells, and bladder urothelial cells, seems to parallel the vulnerability of organs to COVID-19 infection (Zou et al. 2020).
ACE2 expression in the respiratory tract
ACE2 protein expression was also demonstrated in the lungs including lung epithelium, which is suspected to serve as a site of SARS-CoV-2 entry. However, a detailed investigation of respiratory system, in respect to ACE2 expression, reshaped this view. In humans, there is a gradient in ACE2 expression, with the highest levels of expression detected in the upper respiratory tract and lower expression in the lungs. Abundant ACE2 protein expression was detected in human paranasal sinus specimens. Therefore, the sinonasal epithelium has recently been considered a major entry route for SARS-CoV-2 (Soni et al. 2021).
Single-cell analysis of non-diseased human lung by detecting promoter inactivity/inactivity state by open chromatin regions sequencing revealed the highest ACE2 gene accessibility in alveolar type 2 cells (AT2) compared to other cell types, although gene body chromatin accessibility was also prominent in other epithelial cell types of the lung. In spite of that, ACE2 expression in AT2 cells was not high. The highest expression of ACE2 was detected in goblet cells, followed by AT2/club-like, AT1/AT2-like and AT2 cells. Generally, lung epithelial cells showed a higher expression of ACE2, compared to mesenchymal, endothelial and hematopoietic cell types (Wang et al. 2020). After infection spreads in the body, ciliated and AT2 cells are the most infected cell types in the human lung (Hou et al. 2020). In the human nasal epithelium, ACE2 expression exerted the highest level in goblet and ciliated cells (Sungnak et al. 2020).
A high expression of ACE2 protein has been detected in rat nasal mucosa, as well as in lung (Soni et al. 2021). In particular, ACE2 immunoreactivity was detected predominantly in the alveolar epithelium, bronchiolar epithelium, endothelium and smooth muscle cells of the pulmonary vessels (Xie et al. 2006). On the other hand, in mice, basal ACE2 protein expression in the lung was undetectable, but expression was inducible by hyperoxia or bacterial infection (Soni et al. 2021).
Ontogenesis
Age is strongly associated with risk of COVID-19 mortality. Epidemiological data implicate a much lower incidence and severity of SARS-CoV-2 in children, compared to adult and older persons (Stokes et al. 2020), indicating that with increasing age, the risk of COVID-19 mortality is higher. People aged 60–69 years have a HR of 2.4 (95 % CI: 2.16–2.66) for COVID-19-related death compared to those 50–59 years old. People aged 70–79 years have more than a six-fold higher risk of COVID-19 mortality than those 50–59-years old. People aged ≥80 years have a HR of 20.60 (95 % CI: 18.70–22.68) for COVID-19 death compared to 50–59-year-olds (Williamson et al. 2020). A comprehensive systematic review and meta-analysis of 77 studies (n=38906 hospitalized patients) revealed an overall prevalence of all COVID-19-hospitalized deaths of 20 % and severe disease of 28 % of patients. Nearly 50 % of the hospitalized patients were older than 60 years (Dorjee et al. 2020).
The significance of the comorbidities in COVID-19 progression is also age dependent, however, in some cases, it shows an opposite pattern, as expected (e.g. hypertension is more relevant comorbidity for young people than for older patients). A meta-analysis of 18 studies found that COVID-19 patients with hypertension who were younger than 50 years had a HR of 6.43 (95 % CI: 3.40–12.17) of death due to COVID-19 in comparison with the normotensive COVID-19 patients of the same age. Interestingly, hypertensive patients older than 50 years of age were at risk of death with a HR of 2.66 (95 % CI: 1.27–5.57) compared to age matched COVID-19 control (Zhang et al. 2020). This inverse relationship was confirmed when hypertension in younger patients (18–39 years of age) was associated with higher COVID-19 mortality, compared to older hypertensive patients with HR of 3.10 (95 % CI: 1.69–5.70) (Williamson et al. 2020).
As there are obvious age-dependent factors influencing COVID-19 disease in patients, several research groups focused on the ontogenetic aspects of ACE2 expression in the respiratory tract. Bunyavanich et al. (2020) investigated ACE2 gene expression in four groups of probands in different age categories: aged <10 years, 10–17 years, 18–24 years and >24 years. The authors reported increasing trend in ACE2 gene expression correlating with age, with slightly lower expression in children <10 years of age. This finding is in accordance with age depend changes of ACE2 levels in serum exerting higher levels in older probands compared to children (Swärd et al. 2020, Pavel et al. 2021). Single-cell screening revealed a higher ACE2 expression in AT2 cells in 30-year-old adults, compared to 3-year-old children. Quantitative analysis of ACE2 expression within ACE2-expressing cells did not show age-dependent changes (Wang et al. 2020). Similarly, in cohort of patients from 18 to 70 years a positive correlation between ACE2 expression in lung and age was revealed (Bao et al. 2021). A positive correlation between ACE2 expression in nasopharyngeal epithelium and age was observed also in patients diagnosed with COVID-19 (Rossi et al. 2021). On the contrary, lower ACE2 mRNA expression in nasal epithelia of young patients (0–17 years) compared to adults (18–39 years) and older adults (40–66) was reported by Plaas et al. (2021). Changes in ACE2 expression dependent on COVID-19 progression were also reported. According Baker et al. (2020) ACE2 expression is responsive to duration of terminal phase before death in COVID-19 patients. Change in ACE2 expression was positively associated with age as old patients (60–69 years) showed highest expression of ACE2 that exerted decrease by 14 % in each preceding decade. So there are several factors that can influence ACE2 expression which may explain differences in results of experimental studies.
Considering the receptor role of ACE2 in COVID-19 infection, prevailing evidence implicating the age-dependent difference in ACE2 levels in the respiratory tract and circulation are consistent with the observation that the course of COVID-19 disease is milder in children. However, ACE2 levels are not likely to be the most important factor causing an age-dependent susceptibility to COVID-19. It seems that other factors showing age-dependent changes, such as endothelial damage, susceptibility to coagulation, differences in immune system response to SARS-CoV-2 and incidence of comorbidities, should be considered (Zimmerman and Curtis 2020).
Effect of the circadian system on ACE2 facilitated SARS-CoV-2 spread and associated factors
The circadian system generates and regulates the genetically determined endogenous rhythms in physiology and behavior, with periods close to 24 h and allows humans to be synchronized to external environmental cycles. The dominant synchronizing factor in this process is external light (L):dark (D) cycle, but other factors can influence the circadian system functioning as well. In this way, organisms can prepare in advance for known challenges (Gibbs et al. 2014).
The circadian system is composed from central and peripheral oscillators. While the central oscillator is located in the bilateral hypothalamic nuclei called suprachiasmatic nuclei (because of their position above optic chiasm), peripheral oscillators are localized in all other tissues and cells of the human body. Central and peripheral oscillators are connected via network of neural and humoral pathways that allow efficient coordination of endogenous rhythms inside the organism. A key molecular mechanism involved in generation of circadian oscillations is based on expression of so called “clock genes” that are able to regulate their own expression. A regulatory region playing crucial role in this process is E-box (CACGTG) that is present in promoter region of clock genes PER and CRY. Expression of per and cry genes is via E-box induced by heterodimer BMAL1:CLOCK and inhibited by clock gene protein products PER and CRY. Coordinated expression of transcription factors BMAL1 and CLOCK and clock genes PER and CRY creates transcription-translation feedback loop (TTFL) whose one complete cycle lasts circa 24 h. As E-box is frequently employed regulatory region in transcriptome TTFL can impose circadian rhythmicity on many genes. In this way the circadian system influences all body organ systems and most of regulatory pathways, including the RAS (Honma 2018).
Daily rhythm in ACE2, ADAM17 and TMPRSS2 expression
All three main members of molecular machinery involved in SARS-CoV-2 entry into cells (ACE2, ADAM17 and TMPRSS2) are, to some extent, controlled by the circadian system. Firstly, a low amplitude daily rhythm of ACE2 mRNA expression with the highest levels at the transition from the active to passive phase of the L:D cycle was detected in the hearts of rats. The rhythm of the expression of ACE mRNA was not detected, but the ACE/ACE2 ratio showed a clear daily rhythm with maximum levels in the active phase of the L:D cycle. ACE/ACE2 rhythmicity was significantly attenuated in hypertensive rats (Herichova et al. 2013). The subsequent study demonstrated the rhythmic expression of ACE2 mRNA in rat aorta, with higher levels during the passive phase of the L:D cycle (Herichova et al. 2014).
Actually, several other components of the RAS show circadian rhythmicity as well. Aldosterone levels shows a clear-cut rhythm with maximum before beginning of active phase of LD cycle in serum of rats while ACE exerts a daily rhythm with peak at the beginning of light phase (Lemmer et al. 2000). In human a daily rhythm of ACE activity with peak at the end of light phase was detected in plasma (Veglio et al. 1987). Similarly, a daily rhythm of aldosterone and plasma renin activity show a daily rhythm in human plasma with maximal levels before beginning of active phase of 24 h cycle (Cugini et al. 1992). Plasma levels of Ang I and Ang II display a daily rhythm with highest concentrations closely before beginning of active phase of 24 h cycle in rat (Schiffer et al. 2001). Expression ATR1 mRNA exerts a daily rhythm with the peak at the active phase of 24 h cycle in the rat heart (Herichova et al. 2013). Unfortunately, there are no data on the circadian rhythmicity of ACE2 expression in the respiratory system in humans. However, above mentioned findings substantiate a chronopharmacological approach to the treatment of hypertension.
According to the Circadian Expression Profiles DataBase (Pizarro et al. 2013), mRNA of ADAM17 shows a significant daily rhythm in the lung, kidney and liver in mice. Maximum of rhythmic ADAM17 expression in the lung and kidney was observed at the transition from the active to passive phase of the L:D cycle, while its expression in the liver peaked in the middle of the active phase. In humans, ADAM17 exerted a rhythmic pattern in the visceral fat tissue at the transition from active to passive phase of 24 h cycle. The existence of a clear-cut rhythm in ADAM17 expression is in accordance with the presence of a regulatory site for CLOCK and Aryl hydrocarbon receptor nuclear translocator-like protein 1 (ARNTL, BMAL1) in its sequence (Dreos et al. 2017).
TMPRSS2 also shows a rhythmic profile in mouse pituitary and suprachiasmatic nuclei, with maximal levels at the beginning of the active phase of the L:D cycle (Pizarro et al. 2013).
Therefore, a rhythmic expression of the factors regulating ACE2 availability for SARS-CoV-2 entry should be also taken into consideration when the role of ACE2 in COVID-19 progression is evaluated.
Rhythms in immunity
The host circadian system also influences the pathogenicity of viral infections via the direct regulation of viral replication within the target cells, and indirectly via circadian rhythms in innate and adaptive immune responses (Meira et al. 2020). When mice were infected with herpes virus at various time-points of the day, significant differences in the severity of the effects were found and ascribed to the circadian clock (Matsuzawa et al. 2018). The time of infection may predict survival, as shown after influenza infection, because mice infected at the beginning of their active phase showed higher mortality than those infected at the onset of the rest phase. These temporal differences were abolished in mice with knockout of clock gene Bmal1 (Gibbs et al. 2014).
It is well known that the immune system exhibits a distinct circadian rhythmicity (Scheiermann et al. 2013), which may affect vulnerability to pathogens. For example, it has been shown that an endogenous circadian clock within lung epithelial cells modulates neutrophil recruitment through the chemokine ligand CXCL5 (Pick et al. 2019). Therefore, it is possible that a weakened circadian organization may increase the severity of COVID-19 disease. This might be important because lighting conditions in intensive care units, required for continued observation of patients, suppress the L:D signal, which entrains the circadian system. As a result, the damped circadian rhythmicity may interfere with the protective role of the immune system.
Recent research shows that patients infected with SARS-CoV-2 exhibited endothelial dysfunction due to endothelial inflammation (Paces et al. 2020, Varga et al. 2020). The damaged endothelium facilitates coagulation and thrombus formation in large vessels, but also in small arterioles and capillaries. This process is also influenced by the circadian system. The most studied rhythmically expressed coagulation regulator is plasminogen activator inhibitor 1 (PAI-1). Gene coding PAI-1 is under control of transcription factors nuclear receptor subfamily 1 group D member 1 and RAR-related orphan receptor alpha that are regulated by basic feed-back loop of the circadian oscillator. Consequently PAI-1 levels in plasma exert a distinct daily rhythm with highest levels at the beginning of active phase and nadir at the beginning of passive phase in humans (Carmona et al. 2020). In addition to PAI-1 the circadian system influences much more factors involved in regulation of coagulation cascade and clotting function that are reviewed elsewhere (Haspel et al. 2021).
Melatonin
Another link between the circadian system and COVID-19 could be hormone melatonin (Cardinali et al. 2020). Melatonin biosynthesis follows distinct circadian rhythms, with high levels during the night and low during the day in both diurnal and nocturnal species. In this way, plasma concentrations of melatonin represent a reliable biomarker of circadian clock function, and its suppressed levels indicate a disturbed circadian organization. Moreover, melatonin was repeatedly reported to have anti-inflammatory and immune-protective effects (da Cunha Pedrosa et al. 2010). Because of its low toxicity, melatonin has been recommended as an adjuvant for COVID-19 treatment (Tan and Hardeland 2020, Cardinali et al. 2020).
Gender-dependent differences in ACE2-COVID-19 interactions
In addition to hypertension and age, it was shown that gender is an important risk factor for the severe COVID-19 form and mortality (Gagliardi et al. 2020).
According to the COVID-19 gender-disaggregated data tracker, at the global level, more females are COVID-19 tested (57 % of tested persons). The COVID-19 diagnosis equally affects both genders (50 % of cases), but females are less likely to be hospitalized (48 % of hospitalizations). Only 31 % of intensive care unit COVID-19 admissions and 43 % of COVID-19 deaths were females, compared to males (Hawkes et al. 2021). The data from the OpenSAFELY platform support the gender-based differences in COVID-19 mortality. Men exhibited a higher risk of COVID-19 mortality, fully adjusted on comorbidities and another risk factors, in comparison to women (HR, 1.59 [95 % CI: 1.53–1.65] (Williamson et al. 2020). Males have a significantly higher COVID-19 fatality rate than females ≥30 years of age (Scully et al. 2020) and comprise nearly 59 % of the hospitalized patients with COVID-19 (Dorjee et al. 2020).
Gender-dependent differences in COVID-19 incidence and severity are not completely elucidated. Concerning the association of gender-dependent susceptibility to COVID-19 and ACE2, no conclusive evidence proving that higher male mortality issues from the differences in ACE2 expression in the respiratory tract or ACE2 levels in the circulation exist, although these ideas were proposed and there are indications that support them.
Firstly, ACE2 gene is located on the X chromosome region that is not subjected to X chromosome inactivation, and ACE2 transcription from both X chromosomes can lead to ACE2 overexpression (Tukiainen et al. 2017). Unfortunately, data regarding gender-dependent expression of ACE2 do not support this prediction (Liu et al. 2010).
ACE2 expression is influenced by male, as well as female, sex hormones. ACE2 expression has been shown to be inducible by androgens (Deng et al. 2021) and this was proposed as a reason for higher susceptibility to COVID-19 in males. However, estradiol (E2) was also reported to induce ACE2 expression (Chen et al. 2020). On the other hand, it was shown that E2 inhibits ACE2 expression in mice kidneys (Liu et al. 2010).
Available experimental results indicate that males have higher levels of circulating ACE2 compared to females (Sama et al. 2020, Swärd et al. 2020, Pavel et al. 2021). Similarly, ACE2 expression in lungs is higher in males compared to females (Bao et al. 2021). In mice a higher expression of ACE2 was observed in the kidney, but not in the heart or lung of males compared to females (Liu et al. 2010). Unlikely above mentioned studies Rossi et al. (2021) did not observe sex-dependent difference in ACE2 expression in nasopharyngeal epithelium of COVID-19 patients. Therefore, in spite of some diversity in results, there is prevailing evidence that ACE2 shows higher levels in men compared to females.
ACE2 as a therapeutic target
Several strategies to reduce SARS-CoV-2 binding to ACE2 are being intensively investigated. Firstly, small molecules (Huentelman et al. 2004) or peptides (Huang et al. 2003) interacting with ACE2 are being tested. Efficiency of N-(2-aminoethyl)-1 aziridine-ethanamine (NAAE) in the protection against SARS-CoV infection based on the inhibition of S protein interactions with ACE2 has been reported (Huentelman et al. 2004). Another approach is based on the use of monoclonal antibodies isolated from B-cells of SARS-CoV-2 infected persons, which can compete with ACE2 for binding to the RBD. In general, neutralizing monoclonal antibodies recognize some parts of the S protein (e.g. RBD and N-terminal domain), which interact with ACE2. Of many RBD-specific monoclonal antibodies, three (P2C-1F11, P2C-1A3 and P2B-2F6) were identified as the most promising in the neutralization of SARS-CoV-2 activity through ACE2 receptor mimicry. The most potent is the P2C-1F11 antibody, which shares the highest RBD-ACE2 homology (Ge et al. 2021).
Exogenous administration of ACE2 includes soluble recombinant human ACE2 (rhACE2), which has already been tested in the first and second phase of clinical trials (Khan et al. 2017). Recombinant human ACE2 successfully inhibits virus attachment to the host cells, preventing the entry of SARS-CoV-2, as well as reducing SARS-CoV-2 infections under in vitro conditions (Monteil et al. 2020). Interestingly, the use of mouse recombinant-soluble ACE2 did not successfully inhibit SARS-CoV-2 infection.
The efficiency of SARS-CoV-2 entry into host cells may also be influenced by ACE2 shedding. Therefore, various molecules enhancing ACE2 shedding have been developed, including agonist phorbol 12-myristate 13-acetate (PMA) and ionomycin, or chemokines and cytokines (Jia et al. 2021).
Conclusions
The role of ACE2 in COVID-19 progression is ambiguous. ACE2 facilitates SARS-CoV-2 entry into the cell and it seems that tissue susceptibility to SARS-CoV-2 shows a negative correlation to ACE2 expression before infection. However, when an organism is infected, a high ACE2 expression is more beneficial, compared to low expression, as ACE2 counterbalances the pro-inflammatory effect of Ang II by its cleaving and via the independent mechanism mediated by MAS receptors. Therefore, new therapeutic strategies are focused mainly on membrane bound ACE2, in order to preserve circulating ACE2 that continues to possess some enzymatic activity. Hypertension treatment by ACE inhibitors or ATR1 blockers that induce ACE2 expression does not cause worsening of COVID-19 course. Epidemiological data referring about sex-dependent difference in COVID-19 susceptibility are in line with evidence implicating higher ACE2 expression in males compared to females. Age dependent changes in COVID-19 incidence are most probably not related to ACE2 expression. ACE2, ADAM17 and TMPRSS2 show a daily rhythm of expression in at least some mammalian tissue. The circadian system can also modulate COVID-19 progression via its effect on the immune system. Because of the fundamental role of ACE2, ADAM17 and TMPRSS2 in the COVID-19 course, pathogenesis of the circadian regulation of membrane-bound ACE2 can be important for the evaluation of the risk of infection by SARS-CoV-2.
Acknowledgements
Supported by grant APVV-16-0209.
Abbreviations
- ACE2
angiotensin-converting enzyme 2
- ADAM17
enzyme A disintegrin and metalloproteinase 17
- Ang 1–7
angiotensin 1–7
- Ang 1–9
angiotensin 1–9
- Ang I
angiotensin I
- Ang II
angiotensin II
- ARNTL
aryl Hydrocarbon Receptor Nuclear Translocator-Like
- ATR1
angiotensin receptor type 1
- ATR1a
angiotensin receptor type 1a
- AT2
alveolar type 2 cells
- ATR2
angiotensin receptor type 2
- CendR motif
C-end rule motif
- CHD
collectrin homology domain
- CI
confidence interval
- COVID-19
coronavirus disease 2019
- ED
ectodomain
- EGFR
epidermal growth factor receptor
- ER
endoplasmatic reticulum
- ERGIC
endoplasmic reticulum Golgi intermediate compartment
- FP
fusion peptide
- gRNA
guide RNA
- sgRNA
single guide RNA
- GTEx
the genotype-tissue expression
- HR
hazard ratio
- HR1
heptapeptide repeat sequence 1
- HR2
heptapeptide repeat sequence 2
- HS
heparan sulphate
- HSPGs
heparan sulphate proteoglycans
- IFITM3
interferon-induced transmembrane protein 3
- IL
interleukin
- INF
interferon
- L:D cycle
light:dark cycle
- MAS
MAS receptor
- MERS
Middle East Respiratory Syndrome Coronavirus
- NRP1
neuropilin receptor 1
- NRP2
neuropilin receptor 2
- ORF1a
open reading frame 1a
- ORF1ab
open reading frame 1ab
- PMA
phorbol 12-myristate 13-acetate
- PP1A
polyprotein 1A
- PP1AB
polyprotein 1AB
- RAS
renin-angiotensin system
- RBD
receptor binding domain
- S
spike glycoproteins
- SARS
severe acute respiratory syndrome
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- SIRT1
sirtuin 1
- SP
signal peptide
- TM
transmembrane domain
- TMPRSS2
transmembrane protease serine 2
- TNF
tumor necrosis factor
- VEGF165R
vascular endothelial cell growth factor 165 receptor 2
Footnotes
Conflict of Interest
There is no conflict of interest.
References
- BADAWI S, ALI BR. ACE2 nascence, trafficking, and SARS-CoV-2 pathogenesis: The saga continues. Hum Genomics. 2021;15:8. doi: 10.1186/s40246-021-00304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAKER SA, KWOK S, BERRY GJ, MONTINE TJ. Angiotensin-converting enzyme 2 (ACE2) expression increases with age in patients requiring mechanical ventilation. PLoS One. 2021;16:e0247060. doi: 10.1371/journal.pone.0247060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAO W, ZHANG X, JIN Y, HAO H, YANG F, YIN D, CHEN X, XUE Y, HAN L, ZHANG M. Factors associated with the expression of ACE2 in human lung tissue: Pathological evidence from patients with normal FEV1 and FEV1/FVC. J Inflamm Res. 2021;14:1677–1687. doi: 10.2147/JIR.S300747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUNYAVANICH S, DO A, VICENCIO A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA. 2020;323:2427–2429. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CANTUTI-CASTELVETRI L, OJHA R, PEDRO LD, DJANNATIAN M, FRANZ J, KUIVANEN S, Van der MEER F, KALLIO K, KAYA T, ANASTASINA M, SMURA T, LEVANOV L, SZIROVICZA L, TOBI A, KALLIO-KOKKO H, ÖSTERLUND P, JOENSUU M, MEUNIER FA, BUTCHER SJ, WINKLER MS, ET AL. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAO Y, SUN Y, TIAN X, BAI Z, GONG Y, QI J, LIU D, LIU W, LI J. Analysis of ACE2 gene-encoded proteins across mammalian species. Front Vet Sci. 2020;7:457. doi: 10.3389/fvets.2020.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARDINALI DP, BROWN GM, PANDI-PERUMAL SR. Can melatonin be a potential “silver bullet” in treating COVID-19 patients? Diseases. 2020;8:44. doi: 10.3390/diseases8040044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN J, JIANG Q, XIA X, LIU K, YU Z, TAO W, GONG W, HAN JJ. Individual variation of the SARS-CoV-2 receptor ACE2 gene expression and regulation. Aging Cell. 2020;19:e13168. doi: 10.1111/acel.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLARKE NE, BELYAEV ND, LAMBERT DW, TURNER AJ. Epigenetic regulation of angiotensin-converting enzyme 2 (ACE2) by SIRT1 under conditions of cell energy stress. Clin Sci (Lond) 2014;126:507–516. doi: 10.1042/CS20130291. [DOI] [PubMed] [Google Scholar]
- CLAUSEN TM, SANDOVAL DR, SPLIID CHB, PIHL J, PERRETT HR, PAINTER CHD, NARAYANAN A, MAJOWICZ SA, KWONG EM, McVICAR RN, THACKER BE, GLASS CHA, YANG Z, TORRES JL, GOLDEN GJ, ET AL. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057. doi: 10.2139/ssrn.3657535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CUERVO NZ, GRANDVAUX N. ACE2: Evidence of role as entry receptor for SARS-CoV-2 and implications in comorbidities. Elife. 2020;9:e61390. doi: 10.7554/eLife.61390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CUGINI P, LUCIA P, Di PALMA L, RE M, CANOVA R, GASBARRONE L, CIANETTI A. Effect of aging on circadian rhythm of atrial natriuretic peptide, plasma renin activity, and plasma aldosterone. J Gerontol. 1992;47:B214–B219. doi: 10.1093/geronj/47.6.B214. [DOI] [PubMed] [Google Scholar]
- Da CUNHA PEDROSA AM, WEINLICH R, MOGNOL GP, ROBBS BK, VIOLA JP, CAMPA A, AMARANTE-MENDES GP. Melatonin protects CD4+ T cells from activation-induced cell death by blocking NFAT-mediated CD95 ligand upregulation. J Immunol. 2010;184:3487–3494. doi: 10.4049/jimmunol.0902961. [DOI] [PubMed] [Google Scholar]
- DAI Y-J, ZHANG W-N, WANG W-D, HE S-Y, LIANG CH-C, WANG D-W. Comprehensive analysis of two potential novel SARS-CoV-2 entries, TMPRSS2 and IFITM3, in healthy individuals and cancer patients. Int J Biol Sci. 2020;16:3028–3036. doi: 10.7150/ijbs.51234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DALY JL, SIMONETTI B, KLEIN K, CHEN K-E, WILLIAMSON MK, ANTON-PLAGARO C, SHOEMARK DK, SIMON-GRACIA L, BAUER M, HOLLANDI R, GREBER UF, HORVATH P, SESSIONS RB, HELENIUS A, HISCOX JA, TEESALU T, MATTHEWS DA, DAVIDSCON AD, COLLINS BM, CULLEN PJ, YAMAUCHI Y. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DENG Q, RASOOL RU, RUSSELL RM, NATESAN R, ASANGANI IA. Targeting androgen regulation of TMPRSS2 and ACE2 as a therapeutic strategy to combat COVID-19. iScience. 2021;24:102254. doi: 10.1016/j.isci.2021.102254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DONG E, HONGRU D, GARDNER L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DORJEE K, KIM H, BONOMO E, DOLMA R. Prevalence and predictors of death and severe disease in patients hospitalized due to COVID-19: A comprehensive systematic review and meta-analysis of 77 studies and 38,000 patients. PLoS One. 2020;15:e0243191. doi: 10.1371/journal.pone.0243191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DREOS R, AMBROSINI G, GROUX R, CAVIN PÉRIER R, BUCHER P. The eukaryotic promoter database in its 30th year: focus on non-vertebrate organisms. Nucleic Acids Res. 2017;45:D51–D55. doi: 10.1093/nar/gkw1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FU J, ZHOU B, ZHANG L, BALAJI KS, WEI C, LIU X, CHEN H, PENG J, FU J. Expressions and significances of the angiotensin-converting enzyme 2 gene, the receptor of SARS-CoV-2 for COVID-19. Mol Biol Rep. 2020;47:4383–4392. doi: 10.1007/s11033-020-05478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAGLIARDI MC, TIERI P, ORTONA E, RUGGIERI A. ACE2 expression and sex disparity in COVID-19. Cell Death Discov. 2020;6:37. doi: 10.1038/s41420-020-0276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GE J, WANG R, JU B, ZHANG Q, SUN J, CHEN P, ZHANG S, TIAN Y, SHAN S, CHENG L, ZHOU B, SONG S, ZHAO J, HAIYAN W, SHI X, DING Q, LIU L, ZHAO J, ZHANG Z, WANG X, ZHANG L. Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry. Nat Commun. 2021;12:250. doi: 10.1038/s41467-020-20501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIBBS J, INCE L, MATTHEWS L, MEI J, BELL T, YANG N, SAER B, BEGLEY N, POOLMAN T, PARIOLLAUD M, FARROW S, DEMAYO F, HUSSELL T, WORTHEN GS, RAY D, LOUDON A. An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat Med. 2014;20:919–926. doi: 10.1038/nm.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HACHIM MY, HEIALY SA, HACHIM IY, HALWANI R, SENOK AC, MAGHAZACHI AA, HAMID Q. Interferon-induced transmembrane protein (IFITM3) is upregulated explicitly in SARS-CoV-2 infected lung epithelial cells. Front Immunol. 2020;11:1372. doi: 10.3389/fimmu.2020.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAMMING I, TIMENS W, BULTHUIS ML, LELY AT, NAVIS G, Van GOOR H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANFF TC, HARHAY MO, BROWN TS, COHEN JB, MOHAREB AM. Is there an association between COVID-19 mortality and the renin-angiotensin system? A call for epidemiologic investigations. Clin Infect Dis. 2020;71:870–874. doi: 10.1093/cid/ciaa329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARMER D, GILBERT M, BORMAN R, CLARK KL. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532:107–110. doi: 10.1016/S0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- HAWKES S, TANAKA S, PANTAZIS A, GAUTAM A, KIWUWA-MUYINGO S, BUSE K, PURDIE A. Recorded but not revealed: exploring the relationship between sex and gender, country income level, and COVID-19. Lancet Glob Health. 2021;9:e751–e752. doi: 10.1016/S2214-109X(21)00170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HERICHOVA I, SZANTOOVA K. Renin-angiotensin system: upgrade of recent knowledge and perspectives. Endocr Regul. 2013;47:39–52. doi: 10.4149/endo_2013_01_39. [DOI] [PubMed] [Google Scholar]
- HERICHOVA I, SOLTESOVA D, SZANTOOVA K, MRAVEC B, NEUPAUEROVA D, VESELA A, ZEMAN M. Effect of angiotensin II on rhythmic per2 expression in the suprachiasmatic nucleus and heart and daily rhythm of activity in Wistar rats. Regul Pept. 2013;186:49–56. doi: 10.1016/j.regpep.2013.06.016. [DOI] [PubMed] [Google Scholar]
- HERICHOVA I, ZSOLDOSOVA K, VESELA A, ZEMAN M. Effect of angiotensin II infusion on rhythmic clock gene expression and local renin-angiotensin system in the aorta of Wistar rats. Endocr Regul. 2014;48:144–151. doi: 10.4149/endo_2014_03_144. [DOI] [PubMed] [Google Scholar]
- HOFFMANN M, KLEINE-WEBER H, SCHROEDER S, KRÜGER N, HERRLER T, ERICHSEN S, SCHIERGENS TS, HERRLER G, WU N-H, NITSCHE A, MÜLLER MA, DROSTEN CH, PÖHLMANN S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HONMA S. The mammalian circadian system: a hierarchical multi-oscillator structure for generating circadian rhythm. J Physiol Sci. 2018;68:207–219. doi: 10.1007/s12576-018-0597-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOOPER NM, LAMBERT DW, TURNER AJ. Discovery and characterization of ACE2 - a 20-year journey of surprises from vasopeptidase to COVID-19. Clin Sci. 2020;134:2489–2501. doi: 10.1042/CS20200476. [DOI] [PubMed] [Google Scholar]
- HOU YJ, OKUDA K, EDWARDS CE, MARTINEZ DR, ASAKURA T, DINNON KH, KATO T, LEE RE, YOUNT BL, MASCENIK TM, CHEN G, OLIVIER KN, GHIO A, TSE LV, LEIST SR, GRALINSKI LE, SCHÄFER A, DANG H, GILMORE R, NAKANO S, ET AL. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HU RW, LIU C, GONG JP, CAO ZX. Differential expression and immune correlation analysis of COVID-19 receptor ACE2 and TMPRSS2 genes in all normal and tumor tissues. Eur Rev Med Pharmacol Sci. 2021;25:1724–1731. doi: 10.26355/eurrev_202102_24882. [DOI] [PubMed] [Google Scholar]
- HUANG L, SEXTON DJ, SKOGERSON K, DEVLIN M, SMITH R, SANYAL I, PARRY T, KENT R, ENRIGHT J, WU Q-L, CONLEY G, DEOLIVEIRA D, MORGANELLI L, DUCAR M, WESCOTT CHR, LADNER RC. Novel peptide inhibitors of angiotensin-converting enzyme 2. J Biol Chem. 2003;278:15532–15540. doi: 10.1074/jbc.M212934200. [DOI] [PubMed] [Google Scholar]
- HUENTELMAN MJ, ZUBCEVIC J, PRADA JAH, XIAO X, DIMITROV DS, RAIZADA MK, OSTROV DA. Structure-based discovery of a novel angiotensin-converting enzyme 2 inhibitor. Hypertension. 2004;44:903–906. doi: 10.1161/01.HYP.0000146120.29648.36. [DOI] [PubMed] [Google Scholar]
- JIA H, NEPTUNE E, CUI H. Targeting ACE2 for COVID-19 therapy: Opportunities and challenges. Am J Respir Cell Mol Biol. 2021;64:416–425. doi: 10.1165/rcmb.2020-0322PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIA HP, LOOK DC, TAN P, SHI L, HICKEY M, GAKHAR L, CHAPPELL MC, WOHLFORD-LENANE C, McCRAY PB., JR Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am J Physiol Lung Cell Mol Physiol. 2009;297:L84–L96. doi: 10.1152/ajplung.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHAN A, BENTHIN C, ZENO B, ALBERTSON TE, BOYD J, CHRISTIE JD, HALL R, POIRIER G, RONCO JJ, TIDSWELL M, HARDES K, POWLEY WM, WRIGHT TJ, SIEDERER SK, FAIRMAN DA, LIPSON DA, BAYLIFFE AI, LAZAAR AL. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit Care. 2017;21:234. doi: 10.1186/s13054-017-1823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAMBERT DW, YARSKI M, WARNER FJ, THORNHILL P, PARKIN ET, SMITH AI, HOOPER NM, TURNER AJ. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2) J Biol Chem. 2005;280:30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE MMY, DOCHERTY KF, SATTAR N, MEHTA N, KALRA A, NOWACKI AS, SOLOMON SD, VADUGANATHAN M, PETRIE MC, JHUND PS, McMURRAY JJV. Renin-angiotensin system blockers, risk of SARS-CoV-2 infection and outcomes fromCoViD-19: systematic review and meta-analysis. Eur Heart J Cardiovasc Pharmacother. 2020:pvaa138. doi: 10.1093/ehjcvp/pvaa138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI F, LI W, FARZAN M, HARRISON SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- LEMMER B, WITTE K, SCHÄNZER A, FINDEISEN A. Circadian rhythms in the renin-angiotensin system and adrenal steroids may contribute to the inverse blood pressure rhythm in hypertensive TGR (mREN-2)27 rats. Chronobiol Int. 2000;17:645–658. doi: 10.1081/CBI-100101071. [DOI] [PubMed] [Google Scholar]
- LIU J, JI H, ZHENG W, WU X, ZHU JJ, ARNOLD AP, SANDBERG K. Sex differences in renal angiotensin converting enzyme 2 (ACE2) activity are 17β-oestradiol-dependent and sex chromosome-independent. Biol Sex Differ. 2010;1:6. doi: 10.1186/2042-6410-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LORENZEN I, LOKAU J, KORPYS Y, OLDEFEST M, FLYNN CM, KÜNZEL U, GARBERS C, FREEMAN M, GRÖTZINGER J, DÜSTERHÖFT S. Control of ADAM17 activity by regulation of its cellular localisation. Sci Rep. 2016;6:35067. doi: 10.1038/srep37364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATSUZAWA T, NAKAMURA Y, OGAWA Y, ISHIMARU K, GOSHIMA F, SHIMADA S, NAKAO A, KAWAMURA TJ. Differential day-night outcome to HSV-2 cutaneous infection. J Invest Dermatol. 2018;138:233–236. doi: 10.1016/j.jid.2017.07.838. [DOI] [PubMed] [Google Scholar]
- MAYI BS, LEIBOWITZ JA, WOODS AT, AMMON KA, LIU AE, RAJA A. The role of neuropilin-1 in COVID-19. PLoS Pathog. 2021;17:e1009153. doi: 10.1371/journal.ppat.1009153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEIRA ECM, MIYAZAWA M, GOZAL D. Putative contributions of circadian clock and sleep in the context of SARS-CoV-2 infection. Eur Respir J. 2020;55:2001023. doi: 10.1183/13993003.01023-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOGIELNICKI A, CHABIELSKA E, PAWLAK R, SZEMRAJ J, BUCZKO W. Angiotensin II enhances thrombosis development in renovascular hypertensive rats. Thromb Haemost. 2005;93:1069–1076. doi: 10.1160/TH04-10-0701. [DOI] [PubMed] [Google Scholar]
- MONTEIL V, KWON H, PRADO P, HAGELKRÜYS A, WIMMER RA, STAHL M, LEOPOLDI A, GARRETA E, HURTADO DEL POZO C, PROSPER F, ROMERO JP, WIRNSBERGER G, ZHANG H, SLUTSKY AS, CONDER R, MONTSERRAT N, MIRAZIMI A, PENNINGER JM. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NILAND S, EBLE JA. Neuropilins in the context of tumor vasculature. Int J Mol Sci. 2019;20:639. doi: 10.3390/ijms20030639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ONABAJO OO, BANDAY AR, STANIFER ML, YAN W, OBAJEMU A, SANTER DM, FLOREZ-VARGAS O, PIONTKIVSKA H, VARGAS JM, RING TJ, KEE C, DOLDAN P, TYRRELL DL, MENDOZA JL, BOULANT S, PROKUNINA-OLSSON L. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat Genet. 2020;52:1283–1293. doi: 10.1038/s41588-020-00731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OU X, LIU Y, LEI X, LI P, MI D, REN L, GUO L, GUO R, CHEN T, HU J, XIANG Z, MU Z, CHEN X, CHEN J, HU K, JIN Q, WANG J, QIAN Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OZ M, LORKE DE. Multifunctional angiotensin converting enzyme 2, the SARS-CoV-2 entry receptor, and critical appraisal of its role in acute lung injury. Biomed Pharmacother. 2021;136:111193. doi: 10.1016/j.biopha.2020.111193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PACES J, STRIZOVA Z, SMRZ D, CERNY J. COVID-19 and the immune system. Physiol Res. 2020;69:379–388. doi: 10.33549/physiolres.934492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PATEL VB, CLARKE N, WANG Z, FAN D, PARAJULI N, BASU R, PUTKO B, KASSIRI Z, TURNER AJ, OUDIT GY. Angiotensin II induced proteolytic cleavage of myocardial ACE2 is mediated by TACE/ADAM-17: a positive feedback mechanism in the RAS. J Mol Cell Cardiol. 2014;66:167–176. doi: 10.1016/j.yjmcc.2013.11.017. [DOI] [PubMed] [Google Scholar]
- PAUL M, MEHR AP, KREUTZ R. Physiology of local renin-angiotensin systems. Physiol Rev. 2006;86:747–803. doi: 10.1152/physrev.00036.2005. [DOI] [PubMed] [Google Scholar]
- PAVEL AB, WU J, RENERT-YUVAL Y, Del DUCA E, GLICKMAN JW, MILLER RL, PALLER AS, KRUEGER JG, GUTTMAN-YASSKY E. SARS-CoV-2 receptor ACE2 protein expression in serum is significantly associated with age. Allergy. 2021;76:875–878. doi: 10.1111/all.14522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PELLET-MANY C, FRANKEL P, JIA H, ZACHARY I. Neuropilins: structure, function and role in disease. Biochem J. 2008;411:211–226. doi: 10.1042/BJ20071639. [DOI] [PubMed] [Google Scholar]
- PICK R, HE W, CHEN CS, SCHEIERMANN C. Time-of-day-dependent trafficking and function of leukocyte subsets. Trends Immunol. 2019;40:524–537. doi: 10.1016/j.it.2019.03.010. [DOI] [PubMed] [Google Scholar]
- PIZARRO A, HAYER K, LAHENS NF, HOGENESCH JB. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Res. 2013;41:D1009–D1013. doi: 10.1093/nar/gks1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAMANI A, MÜLLER L, OSTERMANN PN, GABRIEL E, ABIDA-ISLAM P, MÜLLER-SCHIFFMANN A, MARIAPPAN A, GOUREAU O, GRUELL H, WALKER A, ANDRÉE M, HAUKA S, HOUWAART T, DILTHEY A, WOHLGEMUTH K, OMRAN H, KLEIN F, WIECZOREK D, ADAMS O, TIMM J, KORTH C, SCHAAL H, GOPALAKRISHNAN J. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020;39:e106230. doi: 10.15252/embj.2020106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSSI ÁD, De ARAÚJO JLF, De ALMEIDA TB, RIBEIRO-ALVES M, De ALMEIDA VELOZO C, De ALMEIDA JM, De CARVALHO LEITÃO I, FERREIRA SN, Da SILVA OLIVEIRA J, ALVES HJ, CHEI S. Association between ACE2 and TMPRSS2 nasopharyngeal expression and COVID-19 respiratory distress. Sci Rep. 2021;11:9658. doi: 10.1038/s41598-021-88944-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALKA K, ABUTALEB K, CHORVINSKY E, THIRUVENGADAM G, ARROYO M, GOMEZ JL, GUTIERREZ MJ, PILLAI DK, JAISWAL JK, NINO G. IFN Stimulates ACE2 expression in pediatric airway epithelial cells. Am J Respir Cell Mol Biol. 2021;64:515–518. doi: 10.1165/rcmb.2020-0352LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAMA IE, RAVERA A, SANTEMA BT, Van GOOR H, TER MAATEN JM, CLELAND JGF, RIENSTRA M, FRIEDRICH AW, SAMANI NJ, NG LL, DICKSTEIN K, LANG CC, FILIPPATOS G, ANKER SD, PONIKOWSKI P, METRA M, Van VELDHUISEN DJ, VOORS AA. Circulating plasma concentrations of angiotensin-converting enzyme 2 in men and women with heart failure and effects of renin-angiotensin-aldosterone inhibitors. Eur Heart J. 2020;41:1810–1817. doi: 10.1093/eurheartj/ehaa373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAPONARO F, RUTIGLIANO G, SESTITO S, BANDINI L, STORTI B, BIZZARRI R, ZUCCHI R. ACE2 in the era of SARS-CoV-2: Controversies and novel perspectives. Front Mol Biosci. 2020;7:588618. doi: 10.3389/fmolb.2020.588618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SARRAZIN S, LAMANNA WC, ESKO JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. 2011;3:a004952. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHIFFER S, PUMMER S, WITTE K, LEMMER B. Cardiovascular regulation in TGR(mREN2)27 rats: 24h variation in plasma catecholamines, angiotensin peptides,and telemetric heart rate variability. Chronobiol Int. 2001;18:461–474. doi: 10.1081/CBI-100103969. [DOI] [PubMed] [Google Scholar]
- SCULLY EP, HAVERFIELD J, URSIN RL, TANNENBAUM C, KLEIN SL. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat Rev Immunol. 2020;20:442–447. doi: 10.1038/s41577-020-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHANG J, WAN Y, LUO CH, YE G, GENG Q, AUERBACH A, LI F. Cell entry mechanism of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHEIERMANN C, KUNISAKI Y, FRENETTE PS. Circadian control of the immune system. Nat Rev Immunol. 2013;13:190–198. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHMIEDER RE, HILGERS KF, SCHLAICH MP, SCHMIDT BMW. Renin-angiotensin system and cardiovascular risk. Lancet. 2007;369:1208–1219. doi: 10.1016/S0140-6736(07)60242-6. [DOI] [PubMed] [Google Scholar]
- SONI S, JIANG Y, TESFAIGZI Y, HORNICK JL, ÇATALTEPE S. Comparative analysis of ACE2 protein expression in rodent, non-human primate, and human respiratory tract at baseline and after injury: A conundrum for COVID-19 pathogenesis. PLoS One. 2021;16:e0247510. doi: 10.1371/journal.pone.0247510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPENCE JS, HE R, HOFFMANN H-H, DAS T, THINON E, RICE CHM, PENG T, CHANDRAN K, HANG HC. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat Chem Biol. 2019;15:259–268. doi: 10.1038/s41589-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STOKES EK, ZAMBRANO LD, ANDERSON KN, MARDER EP, RAZ KM, El BURAI FELIX S, TIE Y, FULLERTON KE. Coronavirus Disease 2019 Case Surveillance - United States, January 22–May 30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:759–765. doi: 10.15585/mmwr.mm6924e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUNGNAK W, HUANG N, BECAVIN C, BERG M, QUEEN R, LITVINUKOVA M, TALAVERA-LOPEZ C, MAATZ H, REICHART D, SAMPAZIOTIS F, WORLOCK KB, YOSHIDA M, BARNES JL. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SWÄRD P, EDSFELDT A, REEPALU A, JEHPSSON L, ROSENGREN BE, KARLSSON MK. Age and sex differences in soluble ACE2 may give insights for COVID-19. Crit Care. 2020;24:221. doi: 10.1186/s13054-020-02942-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAN DX, HARDELAND R. Potential utility of melatonin in deadly infectious diseases related to the overreaction of innate immune response and destructive inflammation: Focus on COVID-19. Melatonin Res. 2020;3:120–143. doi: 10.32794/mr11250052. [DOI] [Google Scholar]
- TUKIAINEN T, VILLANI AC, YEN A, RIVAS MA, MARSHALL JL, SATIJA R, AGUIRRE M, GAUTHIER L, FLEHARTY M, KIRBY A, CUMMINGS BB, CASTEL SE, KARCZEWSKI KJ, AGUET F, BYRNES A. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VARDOULAKIS S, SHEEL M, LAL A, GRAY D. COVID-19 environmental transmission and preventive public health measures. Aust N Z J Public Health. 2020;44:333–335. doi: 10.1111/1753-6405.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VARGA Z, FLAMMER AJ, STEIGER P, HABERECKER M, ANDERMATT R, ZINKERNAGEL AS, MEHRA MR, SCHUEPBACH RA, RUSCHITZKA F, MOCH H. Endothelial cell infections and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VEGLIO F, PIETRANDREA R, OSSOLA M, VIGNANI A, ANGELI A. Circadian rhythm of the angiotensin converting enzyme (ACE) activity in serum of healthy adult subjects. Chronobiologia. 1987;14:21–25. [PubMed] [Google Scholar]
- V’KOVSKI P, KRATZEL A, STEINER S, STALDER H, THIEL V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG A, CHIOU J, POIRION OB, BUCHANAN J, VALDEZ MJ, VERHEYDEN JM, HOU X, KUDTARKAR P, NARENDRA S, NEWSOME JM, GUO M, FADDAH DA, ZHANG K, YOUNG RE, BARR J, SAJTI E, MISRA R, HUYCK H, ROGERS L, POOLE C, WHITSETT JA, PRYHUBER G, XU Y, GAULTON KJ, PREISSL S, SUN X. Single-cell multiomic profiling of human lungs reveals cell-type-specific and age-dynamic control of SARS-CoV2 host genes. Elife. 2020;9:e62522. doi: 10.7554/eLife.62522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLIAMSON EJ, WALKER AJ, BHASKARAN K, BACON S, BATES C, MORTON CE, CURTIS HJ, MEHRKAR A, EVANS D, INGLESBY P, COCKBURN J, McDONALD HI, MacKENNA B, TOMLINSON L, DOUGLAS IJ, RENTSCH CT, MATHUR R, WONG AYS, GRIEVE R, HARRISON D, ET AL. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WRAPP D, WANG N, CORBETT KS, GOLDSMITH JA, HSIEH CH-L, ABIONA O, GRAHAM BS, McLELLAN JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIE X, CHEN J, WANG X, ZHANG F, LIU Y. Age- and gender-related difference of ACE2 expression in rat lung. Life Sci. 2006;78:2166–2171. doi: 10.1016/j.lfs.2005.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XU F, GAO J, BERGMANN S, SIMS AC, ASHBROOK DG, BARIC RS, CUI Y, JONSSON CB, LI K, WILLIAMS RW, SCHUGHART K, LU L. Genetic dissection of the regulatory mechanisms of Ace2 in the infected mouse lung. Front Immunol. 2021;11:607314. doi: 10.3389/fimmu.2020.607314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XU J, TENG Y, SHANG L, GU X, FAN G, CHEN Y, TIAN R, ZHANG S, CAO B. The effect of prior angiotensin-converting enzyme inhibitor and angiotensin receptor blocker treatment on coronavirus disease 2019 (COVID-19) susceptibility and outcome: A systematic review and meta-analysis. Clin Infect Dis. 2021;72:e901–e913. doi: 10.1093/cid/ciaa1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG J, PETITJEAN SJL, KOEHLER M, ZHANG Q, DUMITRU AC, CHEN W, DERCLAYE S, VINCENT SP, SOUMILLION P, ALSTEENS D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat Commun. 2020;11:4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG HL, LI YM, SUN J, ZHANG YY, WANG TY, SUN MX, WANG MH, YANG YL, HU XL, TANG YD, ZHAO J, CAI X. Evaluating angiotensin-converting enzyme 2-mediated SARS-CoV-2 entry across species. J Biol Chem. 2021a;296:100435. doi: 10.1016/j.jbc.2021.100435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG J, WU J, SUN X, XUE H, SHAO J, CAI W, JING Y, YUE M, DONG C. Association of hypertension with the severity and fatality of SARS-CoV-2 infection: A meta-analysis. Epidemiol Infect. 2020;148:e106. doi: 10.1017/S095026882000117X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG Y, NIU G, FLISIKOWSKA T, SCHNIEKE A, FLISIKOWSKI K. A tissue- and gender-specific regulation of the SARS-CoV-2 receptor ACE2 by p53 in pigs. Biochem Biophys Res Commun. 2021b;553:25–29. doi: 10.1016/j.bbrc.2021.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZIMMERMANN P, CURTIS N. Why is COVID-19 less severe in children? A review of the proposed mechanisms underlying the age-related difference in severity of SARS-CoV-2 infections. Arch Dis Child. 2021;106:429–439. doi: 10.1136/archdischild-2020-320338. [DOI] [PubMed] [Google Scholar]
- ZIPETO D, PALMEIRA JDF, ARGANARAZ GA, ARGANARAZ ER. ACE2/ADAM17/TMPRSS2 interplay may be the main risk factor for COVID-19. Front Immunol. 2020;11:576745. doi: 10.3389/fimmu.2020.576745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZOU X, CHEN K, ZOU J, HAN P, HAO J, HAN Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZORES F, REBEAUD ME. COVID and the renin-angiotensin system: Are hypertension or its treatments deleterious? Front Cardiovasc Med. 2020;7:71. doi: 10.3389/fcvm.2020.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]