Abstract
We present 3D virtual pancreatography (VP), a novel visualization procedure and application for non-invasive diagnosis and classification of pancreatic lesions, the precursors of pancreatic cancer. Currently, non-invasive screening of patients is performed through visual inspection of 2D axis-aligned CT images, though the relevant features are often not clearly visible nor automatically detected. VP is an end-to-end visual diagnosis system that includes: a machine learning based automatic segmentation of the pancreatic gland and the lesions, a semi-automatic approach to extract the primary pancreatic duct, a machine learning based automatic classification of lesions into four prominent types, and specialized 3D and 2D exploratory visualizations of the pancreas, lesions and surrounding anatomy. We combine volume rendering with pancreas- and lesion-centric visualizations and measurements for effective diagnosis. We designed VP through close collaboration and feedback from expert radiologists, and evaluated it on multiple real-world CT datasets with various pancreatic lesions and case studies examined by the expert radiologists.
Index Terms—: Visual diagnosis, pancreatic cancer, automatic segmentation, lesion classification, planar reformation
1. Introduction
Pancreatic cancer (PC) is among the most aggressive cancers with less than 10% survival rate over a 5-year period [1]. Such low rate is partially attributed to the asymptomatic nature of the disease, leading to most cases remaining undetected until late stage. Early detection and characterization of distinctive precursor lesions can significantly improve the prognosis of PC. However, accurate characterization of lesions on CT scans is challenging since the relevant morphological and shape features are not clearly visible in conventional 2D views. These features also overlap between lesion types making a manual assessment subject to human errors and variability in diagnosis: the diagnostic accuracy of experienced radiologists is within 67–70% range [2]. A designated system with tools and 3D visualizations developed specifically for the detection, segmentation and analysis of the pancreas and lesions, which are inherently 3D, could provide additional diagnostic insights, such as the relationship between the lesions and pancreatic ducts which often is difficult to establish on traditional 2D views (Fig. 1). To the best of our knowledge, there are currently no such visualization systems.
Fig. 1.
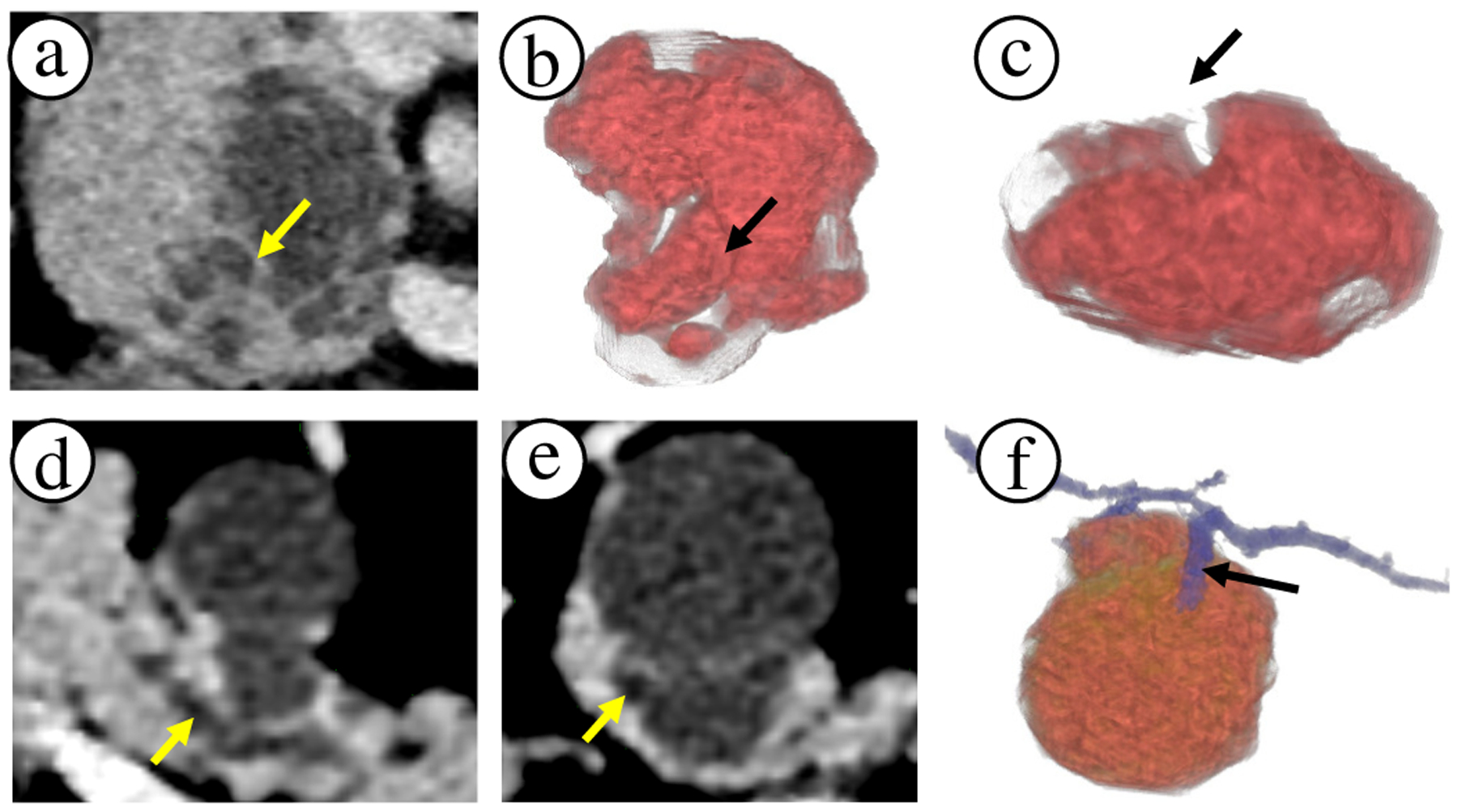
Comparison of 2D slice views to 3D visualizations. (a) Axial slice of a lesion on pancreas head with apparent internal lesion septation wall (see arrow). (b)-(c) The same region can be external crevice rather than an internal septation wall. Thus, 3D visualizations reveal important shape and size information of the lesion cystic components, which can impact the diagnosis. (d) Axial slice of a lesion and primary duct (arrow) in the pancreas body. (e) Secondary duct that connects to the lesion appears very subtle on the slice view (arrow). (f) 3D visualization clearly shows the branching secondary duct connecting with the lesion. 3D visualizations can draw attention to such subtleties.
Pancreatography is an invasive endoscopic procedure to diagnose the pancreas. We have developed instead a non-invasive computer-assisted procedure using a CT scan, hence the term virtual pancreatography (VP). It employs a comprehensive visualization system that automatically segments the pancreas and pancreatic lesions, and automatically classifies the segmented lesions being one of four prominent types. It incorporates tools for user-assisted extraction of the primary duct, and provides effective exploratory visualizations of the pancreas, lesions, and related features. VP combines 3D volume rendering and 2D visualizations constructed through multi-planar reformation (MPR) and curved planar reformation (CPR) to provide better mappings between 3D visualization and raw CT intensities. Such mappings allow radiologists to effortlessly inspect and verify 3D regions of interest using familiar 2D CT reconstructions. VP was designed through close collaboration and feedback from collaborating radiologists, which we discuss throughout the paper. Our contributions include:
To the best of our knowledge, VP is the first end-to-end visualization system for diagnosis of pancreatic lesions, including: automatically segmenting pancreatic structures, automatically classifying lesions, exploratory 3D/2D visualizations, and automatic/manual measurements.
Novel and efficient model for the automatic segmentation of diseased pancreas and pancreatic lesions and a universal training procedure, which speeds up the convergence, including for small targets, such as lesions.
Semi-automatic approach for extraction of the pancreatic duct; and novel approach to modeling the pancreas centerline by weighting both the duct and pancreas geometry.
Enhancement and visualization of internal lesion features (e.g., cystic and solid sacs, septation, calcification).
3D and 2D curved planar reformation (CPR) views for the visualization of duct-lesion relationship.
Evaluation through case studies of real-world diseased pancreas datasets and expert radiologists’ feedback.
2. Related Work
Visual Diagnosis Systems.
Technological progress in image acquisition, processing, and visualization has enabled fundamentally new medical virtual diagnosis and virtual endoscopy [3]. For instance, virtual colonoscopy (VC) [4–6] is a well-established technique for evaluation of colorectal cancer that are highly effective using advanced visualization methods, such as electronic biopsy [7], colon flattening [8, 9], and synchronized display of two (e.g., prone, supine) scans [10]. Similarly, virtual endoscopy has been applied to bronchoscopy [11, 12], angioscopy [13, 14], and sinus cavity [15, 16]. Prostate cancer visualization [17, 18] and mammography screening are examples where rendering [19, 20] and machine learning techniques [21] assist clinicians in decision-making. Specialized visualizations have also been used in surgical [22] and therapy planning [23]. 3D visualization was utilized in contrast enhanced rat pancreas [24], and MR microscopy for micro scale features in mouse pancreas [25]. However, to our knowledge, there are currently no comprehensive visual diagnosis systems for non-invasive screening and analysis of pancreatic lesions in humans. VP is an end-to-end visualization system that is designed for ease of use and effective diagnosis of PC.
Visual Reformation and Illustrations.
Visual reformations such as volumetric re-sectioning, CPR [26–28], and volumetric reformations [29–32] introduce deliberate simplifications to spatial data that reduce visual complexity, overcome occlusion, and minimize required interactions. Centerlines are well suited to describe tube-like anatomical structures such as ducts, vessels, nerves, the colon, and elongated muscles [33], and can be leveraged to deform 3D structures into planar views [10, 26, 34]. The encoded local volumetric information can help in detecting abnormalities in vascular structures, such as stenosis [35] and aneurysms [36]. VP introduces pancreas-specific novel visualizations (e.g., duct-centric CPR and re-sectioning views), built upon principles of such reformation and exploratory techniques.
Pancreas Segmentation.
Segmentation of the pancreas and its lesions is critical for supporting ready-to-use, clutter-free 3D visualizations for diagnosis due to its deep-seated position in the abdomen. It also supports quantitative assessment such as lesion classification and measurements. Most of recent works on automatic pancreas segmentation can be classified into (1) models based on probabilistic atlases [37–39] and statistical shape models [40], which heavily rely on the computationally expensive and often imperfect volumetric registration step; and (2) machine learning-based models utilizing coarse-to-fine segmentation pipelines [41–44] or subsequent conditional random field frameworks for output refinement [45]. However, these methods were only evaluated on healthy pancreata. Thus, they might be unsuitable for segmenting diseased pancreas and lesions, considering the associated challenges, such as severe intra-patient variability in appearance, size, and shape. Moreover, the small footprint of pancreatic lesions on CT scans complicates the segmentation problem further as traditional techniques are sensitive to target size. A few works acknowledges these challenges, proposing intricate and highly tailored solutions for lesion segmentation [46].
Unlike previous works, which propose complex, multi-stage, and meticulously tailored segmentation solutions, we describe an efficient ConvNet-based segmentation model, a neural network with convolutional layers, with an unorthodox training procedure which alleviates the detrimental effects of small footprints of target structures.
Lesion Classification.
The use of computer-aided diagnosis (CAD) for lesion classification may not only assist radiologists but also reduces the subjectivity of lesion differentiation. Although many CAD systems have been proposed for benign and malignant masses in various organs, the number of pancreas-specific solutions is limited. Some focus only on classifying pancreatic cystic neoplasms, utilizing off-the-shelf ConvNets [47] and support vector machine (SVM) classifiers [48], while others use complex ConvNets based on lesion CT appearance [49]. Our VP integrates a highly reliable module for classification of the four most common pancreatic lesion types [50, 51]. A Bayesian combination of a probabilistic random forest and a custom ConvNet leverages the multi-source data, analyzing patients’ demographics and the raw CT, to obtain a holistic picture of the case and to make the final diagnosis [50].
3. Application Background
The pancreas is an elongated abdominal organ oriented horizontally towards the abdomen upper left side. It is surrounded by the stomach, spleen, liver, and intestine. It secretes important digestive juices for food assimilation and produces hormones for regulating blood sugar levels. The pancreas is broadly divided into three regions: head, body, and tail (Fig. 2a). The primary duct passes approximately along the central axis of the elongated organ, and can be visible on a CT scan due to dilation. Pancreatic lesions are frequently encountered due to the increased usage of high-quality cross-sectional imaging. While weight loss, nausea, jaundice, and abdominal pain are common symptoms, the majority of cases are diagnosed incidentally. The most common types of pancreatic lesion are intraductal papillary mucinous neoplasm (IPMN), mucinous cystic neoplasm (MCN), serous cystadenoma (SCA), and solid-pseudopapillary neoplasm (SPN). These lesions vary in degrees of their malignancy and aggressiveness; IPMN and MCN are considered precursors to PC and offer the potential for early disease identification, whereas SCA and SPN have low malignancy potential [52]. VP is an end-to-end visual diagnostic aid to clinicians in making an accurate PC diagnosis.
Fig. 2.
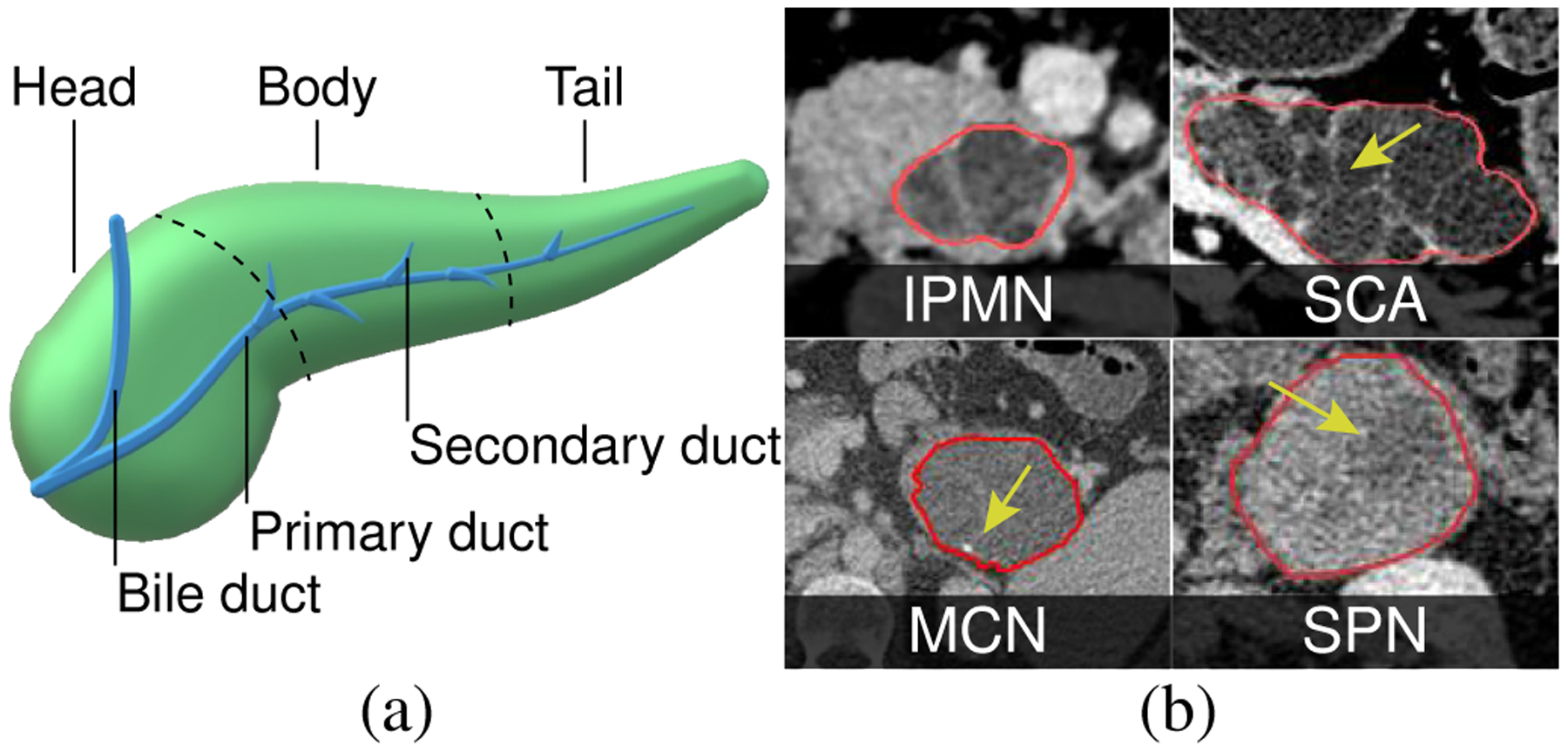
(a) The pancreas. (b) Pancreatic lesions with classical radiological features: peripheral calcification (MCN), macrocystic components with septation (SCA), cyst encapsulated by solid component (SPN), multiple cystic components connected to pancreatic duct (IPMN).
The initial diagnosis of pancreatic lesions often starts with the age and gender of a patient, along with lesion location within the pancreas gland. The diagnosis is reinforced with the imaging characteristics identified on CT scans [52, 53]. Visible characteristics of lesions on CT images include features such as: (a) calcifications - calcium deposits that appear as bright high intensity specks; (b) cystic components - small (microcystic) and large (macrocystic) sacs typically filled with fluids that appear as dark approximately spherical regions; (c) septations - relatively brighter wall-like structures usually separating the cystic components; (d) solid components - solid light gray regions within the lesions; (e) duct dilation and communication - dilation of the primary duct and how it communicates with the lesion. The lesion morphological appearance based on these visible features can help in characterizing them. However, making a correct diagnosis is challenging since some diagnostic features overlap between different lesion types. Examples of the appearance of these lesions in CT are depicted in Fig. 2b. The main diagnostic features of these lesions are:
IPMN is common in both males and females, typically with a mean age of 69, and can appear anywhere in the pancreas. Radiologically, IPMN has a microcystic appearance and always communicates with the pancreatic duct. It typically does not have calcification.
MCN is normally diagnosed in perimenopausal women with a mean age of 50 and is often located in the body or tail of the pancreas. It typically contains a single cystic component (macrocystic) and might have peripheral calcification and thick septation.
SCA is frequent in women with a mean age of 65 and typically arises in the pancreatic head. CT appearance is usually a lobulated mass with a honeycomb-like (micro- or macro-cystic) form, which is often calcified.
SPN mostly occurs in young women with a mean age of 25. These lesions typically appear as cystic components encapsulated by solid mass that may be calcified.
4. Design Overview
Conventional Workflow.
Current diagnostic workflow consists of several tasks (T). CT volumes are reviewed merely in the axial, coronal, and sagittal 2D planes using conventional radiological systems. After the initial 2D scroll through the CT data, the radiologists locate the pancreas and evaluate the pancreas volume for presence of lesions (T1). If found, they study the lesion morphology (T2) by scrolling back and forth in the 2D views, mentally reconstructing its 3D anatomy and internal features (as in Sec. 3). By scrolling again through the images, radiologists try to determine if the lesion is connected to the duct (T3), which can often be visible only if well dilated. When making the final diagnosis (T4), the lesion location within the pancreas and patient’s age and gender are also considered, and no CAD system is used. Radiologists use linear measurements on subjectively chosen 2D slices to calculate lesion size (T5).
Design Motivations.
Interpretation of lesion morphology (T2) on 2D views is challenging. For example, lesion walls with complex non-convex shape may appear similar to internal septation (Fig. 1a–c). Similarly, examining the duct (T3) is cumbersome as it splits into multiple components through cross sections (Fig. 1d–f). A typical span of the pancreas can be 150–200 axial slices. Mental reconstruction of 3D structure of lesion, duct, and duct-lesion relationship requires significant scrolling and mental effort. Additionally, the variability in radiologists’ expertise, lack of specialized tools for lesion detection (T1), analysis (T2, T4), and accurate measurement (T5), introduce diagnosis inconsistencies. Based on these time and quality challenges, we introduce specialized 3D tools. This is also motivated by previous works [54, 55], that emphasize importance of 3D for augmenting diagnosis, treatment, and surgery. Our system further aims to maximize the CT scans information and to introduce more consistency into the characterization of pancreatic lesions through exploratory 3D/2D visualization accompanied with machine learning-based automatic lesion classification. Dr. Matthew Barish (M.B., co-author) and Dr. Kevin Baker (K.B.), each with more than 10 years of experience, served as our expert radiologists. From conversations with them about the tasks and limitations of their current workflow, we have outlined the following:
Develop an automatic algorithm for pancreas and lesion segmentation (supports T1).
Provide 3D and 2D visualizations of the pancreas and lesions with inter-linked views to explore and acquire information from different views. Provide presets modes to visualize the lesion and enhance its characteristic internal features (supports T2).
Develop a method for semi-automatic extraction of the pancreatic duct, and support 3D visualization and 2D reformed viewpoints for detailed analysis of the relationship between the duct and lesions. These include (a) an orthogonal cutting plane that slides along the duct centerline (centerline-guided re-sectioning) to improve visual coherence in tracking the footprint (cross-section) of the duct across its length, and (b) a CPR of the pancreas to visualize the entire duct and the lesion in a single 2D viewpoint (supports T3).
Employ simpler local 1D transfer functions (TFs) by leveraging the already segmented structures, and provide a simplified interface for TF manipulation to keep the visualizations intuitive and avoid misinterpretation of data. While being more extensive, this tool performs similar to grayscale window/level adjustment commonly used by radiologists (supports T2-T3).
Incorporate a module for the automatic classification of lesions, based on a detailed analysis of the demographic and radiological features (supports T4).
Create a tool for automatic and manual assessment of the size of the lesions (supports T5).
VP is intended to provide comprehensive visualization tools for qualitative analysis using the radiologists’ expertise in identifying characteristic malignant features, while augmenting the diagnostic process with quantitative analysis, such as automatic classification and measurements.
5. VP Pipeline
The VP system pipeline is illustrated in Fig. 3. Broadly, it employs the following modules: (1) automatic segmentation of the pancreas and lesions; (2) semi-automatic extraction of primary pancreatic duct, followed by centerline and CPR computation; (3) automatic lesion classification into four common types; and (4) a full featured user-interface for exploratory visualizations and measurement tools. The input abdominal CT scan is first passed through the automatic segmentation module, which segments the pancreas and pancreatic lesions, if present. The segmentations are then passed to a Duct Extraction Window where the radiologist isolates the primary pancreatic duct interactively through a semi-automatic process. The radiologist may skip this step if a duct is invisible. Here, the user is provided an opportunity to manually correct any errors in the segmentation masks through automatic invocation of 3D Slicer [56]. Finalizing the segmentation results then triggers the computation of the pancreas and duct centerlines, CPR, and automatic classification of the segmented lesions. The input CT volume, segmented pancreas, lesions, duct, computed centerlines, and the classification probabilities are then loaded into the main visualization interface. In the following subsections, we describe the VP components that support tasks T1-T5, along with discussions and feedback from radiologists as appropriate. These have been expressed after using VP on multiple real-world patient scans.
Fig. 3.
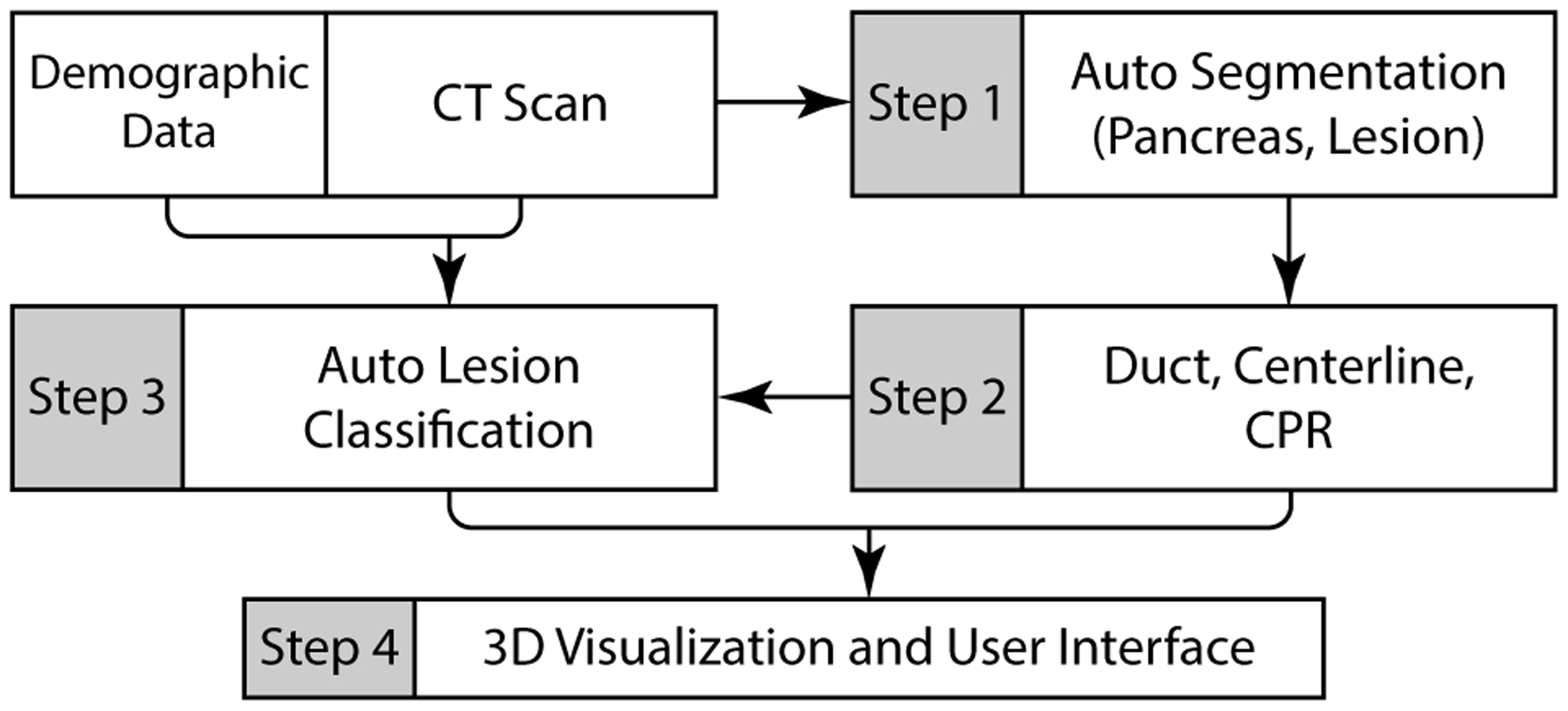
A schematic overview of the VP system pipeline.
5.1. Segmentation of Pancreas Anatomy
Being deeply seated in the retroperitoneum, the pancreas is quite difficult to visualize. Its close proximity to obstructing surrounding organs (stomach, spleen, liver, intestine), further complicates its visual analysis and diagnosis. Segmentation of important components (pancreas, lesions, duct) can provide better control on visualization design and interactions due to explicit knowledge of structure boundaries (supports T1, T2).
5.1.1. Automatic Pancreas and Lesion Segmentation
Architecture.
Our segmentation method was designed with computational efficiency and universality in mind. We use the same architecture and training procedure to perform segmentations for both the pancreas and lesions, which demonstrates the generality of our approach. Additionally, it also simplifies the implementation due to the common pipeline for both structures. The novel architecture is based on a multi-scale 3D ConvNet for processing 3D sub-volumes, which has two branches: (1) to detect whether or not the sub-volume contains the target structure for segmentation (healthy tissue of the pancreatic gland or tissue of a lesion), and (2) to predict a set of voxels that constitutes the target structure. In addition to improving convergence [57], such a two-branch model speeds up the inference by avoiding predicting a full-resolution segmentation mask for a sub-volume if it does not contain the target structure, speeding up the inference process, on average, by 34.8%.
The input to our network is a 3D sub-volume of the original CT scan, and the outputs are a binary label and a binary mask of the target segmentation tissue. Similar to classical segmentation approaches, our model consists of decoder and encoder paths. However, unlike classical models, these paths do not mirror each other. The overall architecture is illustrated in Fig. 4. More formally, the encoder includes three consequently connected convolutional layers of various kernel sizes and strides, each followed by a Leaky ReLU activation function and batch normalization layers, proceeded by three ResNet blocks [58]. The decoder includes three ResNet blocks, followed by two convolutional layers, where the last layer ends with a sigmoid function. Additionally, an auxiliary branch is connected to the encoder to perform a binary classification of the input for the presence of the target structure. This auxiliary branch is composed of one convolutional layer of 32 kernels of 1×1×1 size, followed by a max-pooling and reshaping layers and three fully-connected layers, each connected by the Leaky ReLU function, while the last layer ends with a sigmoid.
Fig. 4.
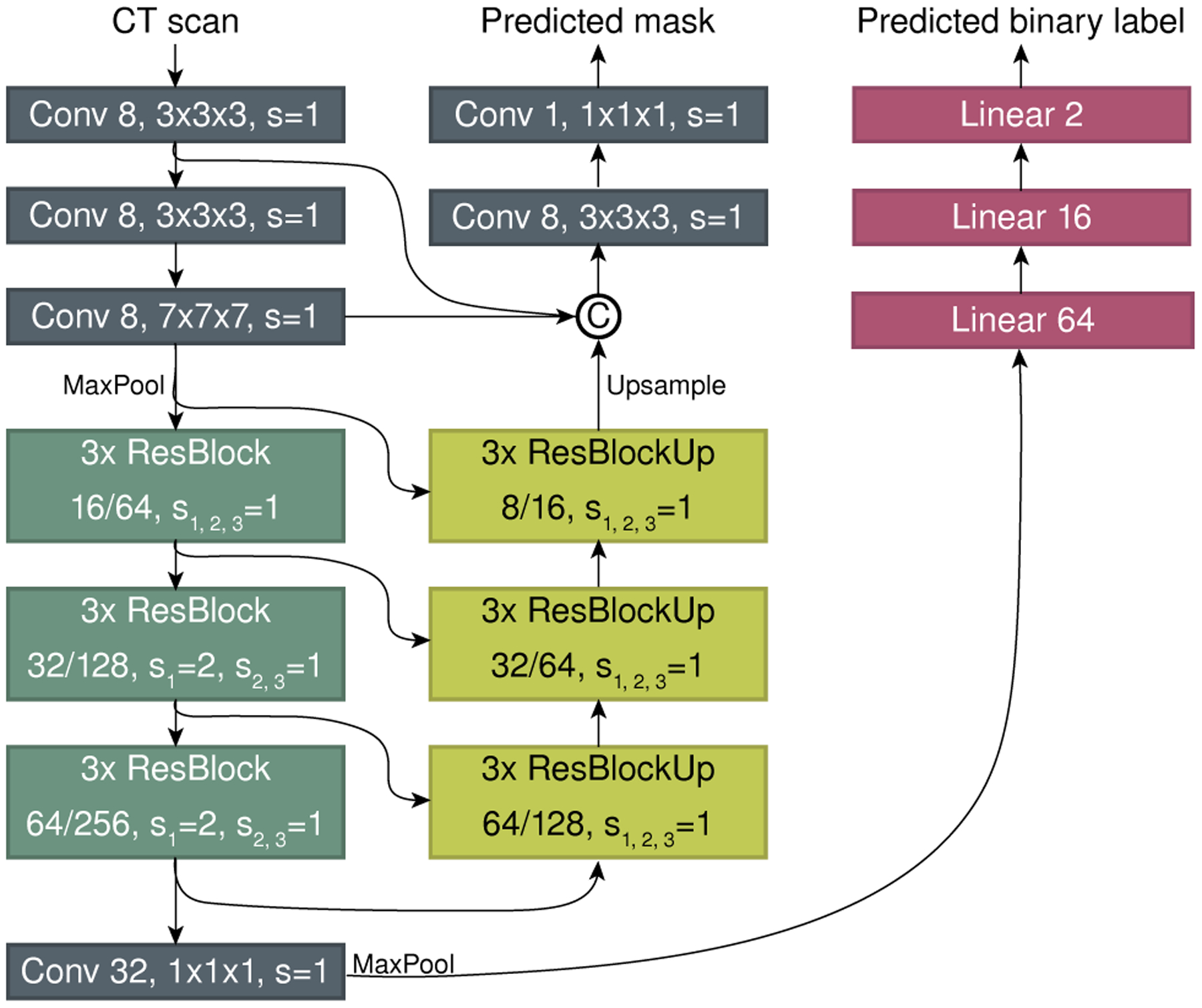
A schematic view of the segmentation model.
Training Methodology.
The proposed network was trained to minimize a joint loss, which is a summation of a binary voxel-wise cross-entropy from the auxiliary classifier and a Dice coefficient (DC)-based loss from the decoder part of the model. However, the latter loss function is known for being challenging to optimize when the target occupies only a small portion of the input due to the learning process getting trapped in the spurious local minima without predicting any masks. Several workarounds have been proposed, including for methods targeting segmentation of the pancreas and pancreatic lesions [59, 60]. However, these methods were tailored for segmenting a particular structure, and cannot be easily extended for other structures. Contrarily, we propose a novel training methodology for minimizing the DC-based loss function which can be applied for the segmentation of any structure, regardless of its size, using any base model. Specifically, we describe an optimization technique which is based on the following iterative process. To alleviate the obstructive issues of the target structure being too small with regards to the overall input size, we begin training a model on the smaller sub-volumes extracted from the center of the original sub-volumes and upsampled to the target input size. This allows us to pretrain a model where the target structure can occupy a significant portion of the input sub-volume. By gradually increasing the size of the extracted sub-volumes we can fine-tune the model weights without it getting stuck in a local minima. Eventually, we fine-tune the model on the original target resolution, which is 32 × 256 × 256 for both pancreas and its lesion segmentation. Our system was developed and validated using 134 abdominal contrast-enhanced CT scans imaged with a Siemens SOMATOM scanner (Siemens Medical Solutions, Malvern, PA).
5.1.2. Semi-Automatic Primary Duct Extraction
We have developed a semi-automatic approach for duct segmentation (supports T3), based on a multi-scale vesselness filter [61], using an adjustable threshold parameter. Vesselness filters are designed to enhance vascular structures through estimation of local geometry. The eigenvalues of a locally computed Hessian matrix are used to estimate vesselness (or cylindricity) of each voxel x ∈ P, where P is the segmented pancreas. Higher values indicate higher probability of a vessel at that voxel. We compute normalized vesselness response values R(x). The user then adjusts the threshold parameter t to compute a response:
| (1) |
Note that this is not a binarization, but a thresholding that results in multiple connected components of R′(x) if zero values are considered as empty space. Default value for t is set to 0.0015. A total vesselness value is calculated for each resulting connected component Ci as the sum of response values within Ci:
| (2) |
The pancreatic duct is selected by the user as the first n connected components with largest values. In other words, the list of connected components is sorted by values (largest value first), and the first n components are chosen as the duct segmentation mask. Integer value of n is chosen by user through a spin button. The default n = 1 is sufficient in most cases. We found that our metric (Eq. 2) is more reliable than simply picking n largest components by volume, since it provides a more balanced composition of both size and vesselness probability of a component. While the vesselness enhancement filter [61] is well-known, our contribution lies in applying Eq. 2 to eliminate occlusion due to noisy regions and tailoring the filter to extract the pancreatic duct with minimal user interaction.
Expert Feedback.
The radiologists appreciated the automatic segmentation of the pancreas and lesions, which significantly improves and simplifies the workflow in comparison to manual or semi-automatic approaches [62]. They also expressed their liking of the duct segmentation, which was easy to extract adjusting a single slider. While the overlap between the produced and manual outlines was not 100% accurate, the radiologists noted that it only slightly deviated from the true boundaries, which is sufficient for practical use when studying duct-lesion relationship.
5.2. Centerline Computation
Once the duct has been segmented and the Duct Extraction Window has been closed, the two centerlines: the duct centerline and pancreas centerline (see Fig. 5), are computed (supports T3). Both the pancreas and duct centerlines create navigation paths for viewing 3D rendering compared with orthogonal 2D raw CT sections of the pancreas volume (Sec. 5.4.3). The duct centerline provides a strict re-sectioning along the duct volume, which can be used for closer inspection of duct boundary and cross-section in the raw CT data. It is also used for computing the CPR view (Sec. 5.4.4). On the other hand, the pancreas centerline provides smoother re-sectioning of the entire pancreas volume as it is geometrically less twisted, and is also used by the automatic lesion classification module for determining the location of the lesion within the pancreas.
Fig. 5.
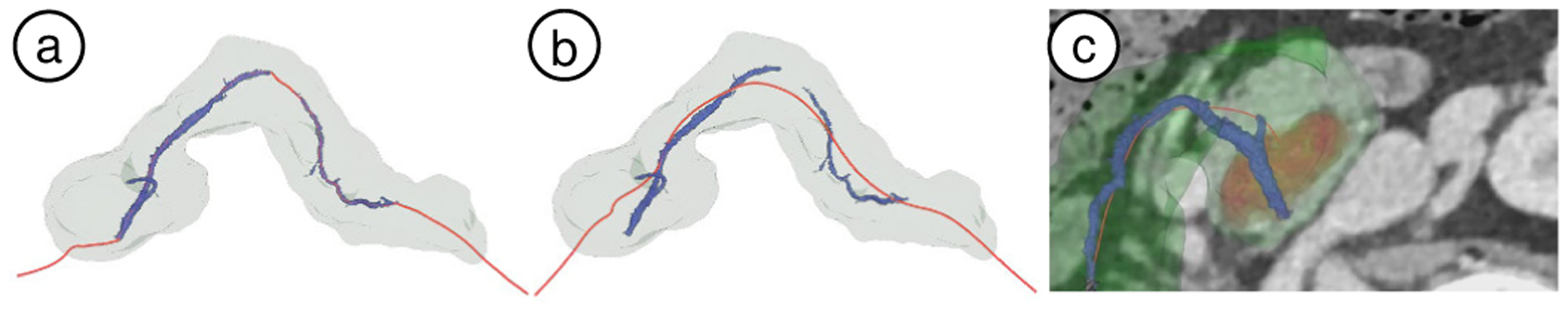
(a) Duct centerline (red curve) on case VP002. It passes through the primary duct (blue) two components, following the pancreas elongated shape, while approximately maintaining constant distance from the pancreas surface. (b) Pancreas centerline (red curve) on VP002. (c) Centerline-guided re-sectioning view embedded in the 3D view for VP011 (red: lesion volume, green: pancreas surface, blue: duct volume).
Pancreas Centerline.
The pancreas centerline is computed using our penalized distance algorithm [63]. This approach uses a graph to represent paths in a grid of voxels. Penalty values are assigned to every voxel (graph node) x in pancreas P, based on that voxel’s distance from the pancreas surface. The path between the pancreas extreme ends with the minimum cost in the graph, is calculated as the centerline. A distance field d(x) is computed on each node as the shortest distance to the pancreas surface. Since we want the centerline to pass through the pancreas inner-most voxels, we want the penalty values to be higher for voxels closer to the surface. Thus, we substract each voxel distance value from the maximum distance dmax:
| (3) |
This results in higher penalty values for the voxels closer to the pancreas surface, as desired. Note that d′(x) is always positive, as d(x) ≤ dmax. We then use the distance field d′ as penalty values and compute the pancreas centerline [63].
Duct Centerline.
Due to the noise in CT scans and variability in dilation, the extracted duct volume may be fragmented into multiple connected components. Since the pancreas itself is an elongated object, we model the duct centerline to follow the entire pancreas length, as well as pass through every connected component of the extracted duct. The centerline follows the pancreas shape wherever the primary duct is disconnected. The penalized distance algorithm [63] focuses on a single object boundary for computing skeleton curves. When computing our duct centerline, we face the problem of handling multiple objects (multiple connected components of the duct and pancreas). A naive approach could be taken where individually computed centerlines of duct components could be connected with each other and to the pancreas extreme ends using the penalty field d′ (Eq. 3). However, this leads to excessive curve bending as it enters and exits the duct components due to abrupt change in penalty field (see Fig. 6). Our contribution is to intuitively model a curve that passes through the primary duct components and also respects the pancreas geometry without excessive correction as it enters and exits the duct components. The duct centerline uses the same end-points (x0, x1) as the pancreas centerline, and passes through all of the extracted duct fragments. In local regions where the primary duct is not contiguous, the duct centerline follows the pancreas shape. This is achieved by a trade-off between finding the shortest path and maintaining a constant distance to the pancreas surface. The trade-off is modelled through a modification of the penalized distances used in the algorithm as described below (see Fig. 7).
Fig. 6.
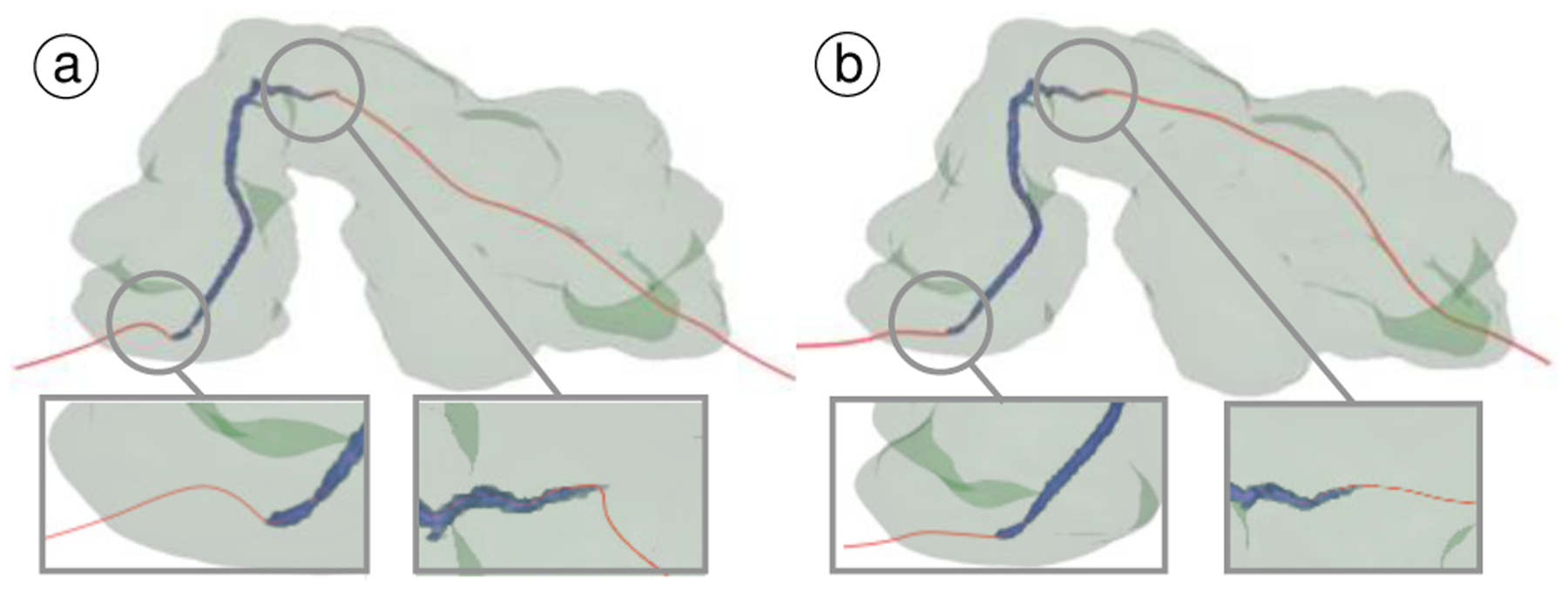
Comparing benefits of our approach to compute duct centerline: (a) computed naively using penalty field, and (b) our approach. Note the reduction in bending at entry and exit points. Smoother centerline is necessary for constructing CPR and re-sectioning views.
Fig. 7.
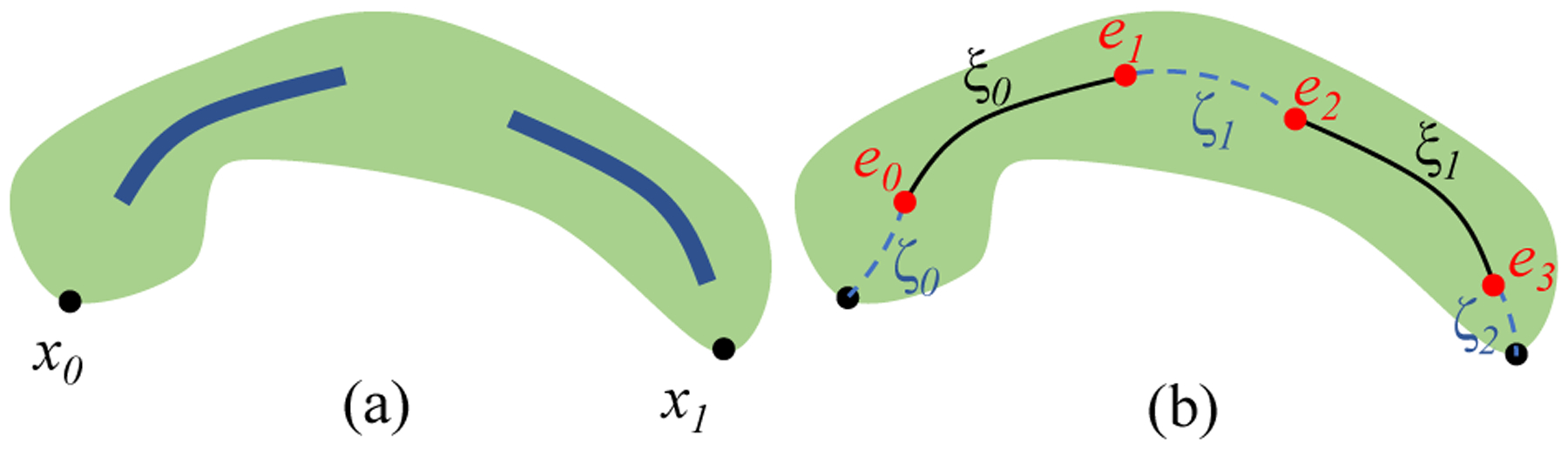
Duct centerline algorithm. (a) Pancreas with two duct components (blue). x0, x1 are end-points of pancreas centerline. (b) ξ0, ξ1 are detached duct component centerlines. ζ0, ζ1, ζ2 are connecting curves. Together, the path ζ0, ξ0, ζ1, ξ1, ζ2 forms the entire duct centerline.
We first calculate the centerlines {ξi} for each connected component of the duct volume independently, using the same process as described for the pancreas centerline. The centerline fragments are then oriented and sorted correctly to align along the length of the pancreas. Each pair of consecutive fragments (ξk, ξk+1) are then connected by computing the shortest penalized path {ζi} between their closest end-points. This includes connections to the global end points x0 and x1. When computing a connecting curve ζ0 between end points e1 and e2, the previously computed pancreas distance field d′(x) from Eq. 3 is transformed as:
| (4) |
where l = (d′(e1) + d′(e2))/2 is the average value between e1 and e2. The modified distance field d″(x) is used as penalty values in the voxel graph for computing the connecting curve ζ1. Together, alternating curves {ζ1} and {ξi} form a single continuous duct centerline.
5.3. Automatic Lesion Classification
We utilize our previously developed CAD algorithm for classification of pancreatic cystic lesions [50], which uses patient demographic information and the CT images as input (supports T4). The classifier is comprised of two main modules: (1) a probabilistic random forest (RF) classifier, trained on a set of manually selected features; and (2) a CNN for analyzing the high-level radiological features. To automatically estimate the lesion location within the pancreas, we divide the centerline of the pancreas into three even segments, representing its head, body, and tail, and determine the closest segment to the segmented lesion center of mass. The final classification probabilities for the four most common lesion types are generated by the Bayesian combination of the RF and CNN, where the classifiers predictions were weighted using the prior knowledge in a form of confusion matrices. The overall scheme of our CAD algorithm is depicted in Fig. 8 and the visualization of the classification results in the user interface is shown in Fig. 14f. Our previously developed lesion classification pipeline [50] has been tested using a blinded study [51]. The model achieves an overall accuracy of 91.7%. For more details, see our previous work [50].
Fig. 8.
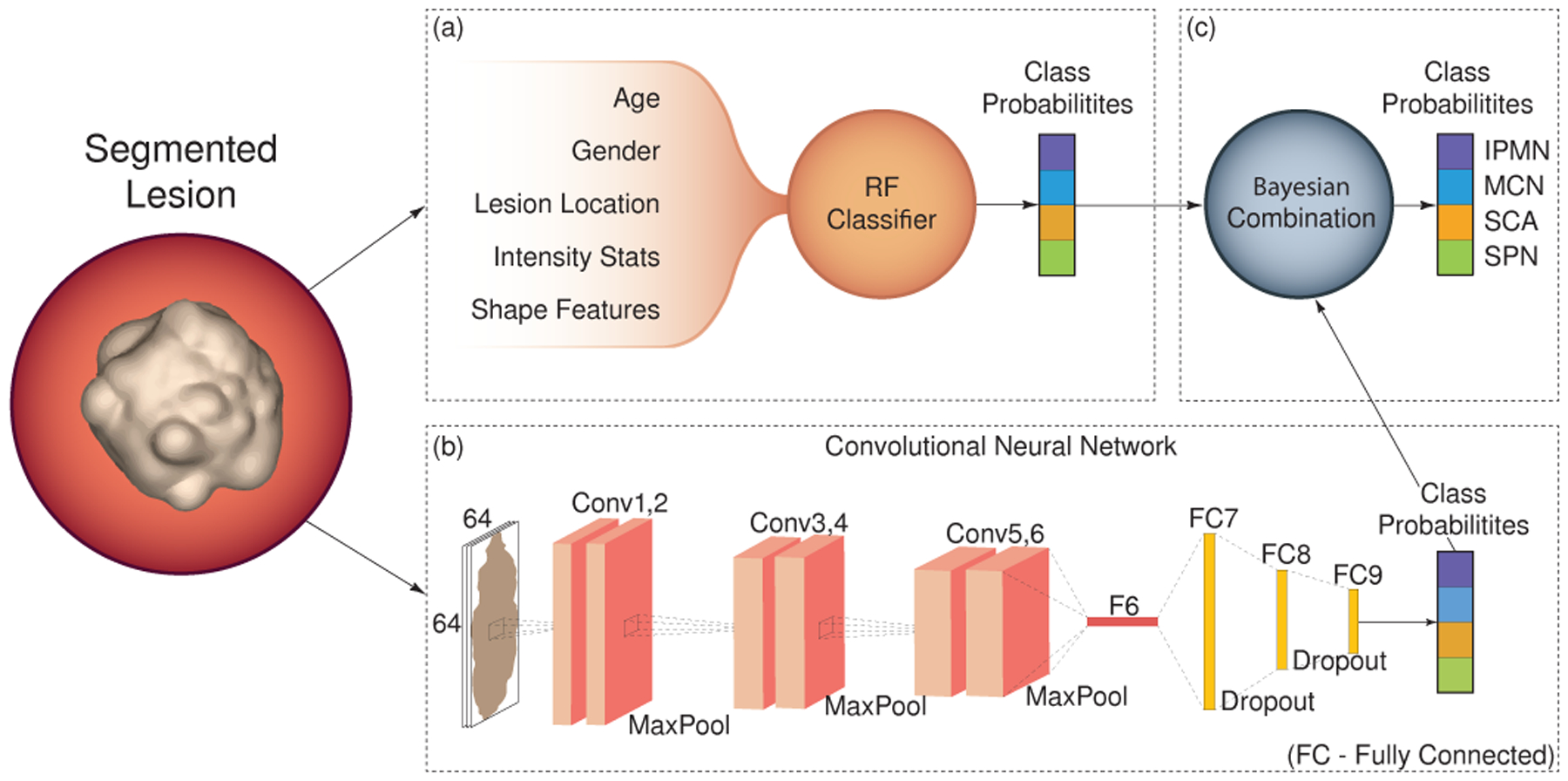
A schematic view of our classification model.
Fig. 14.
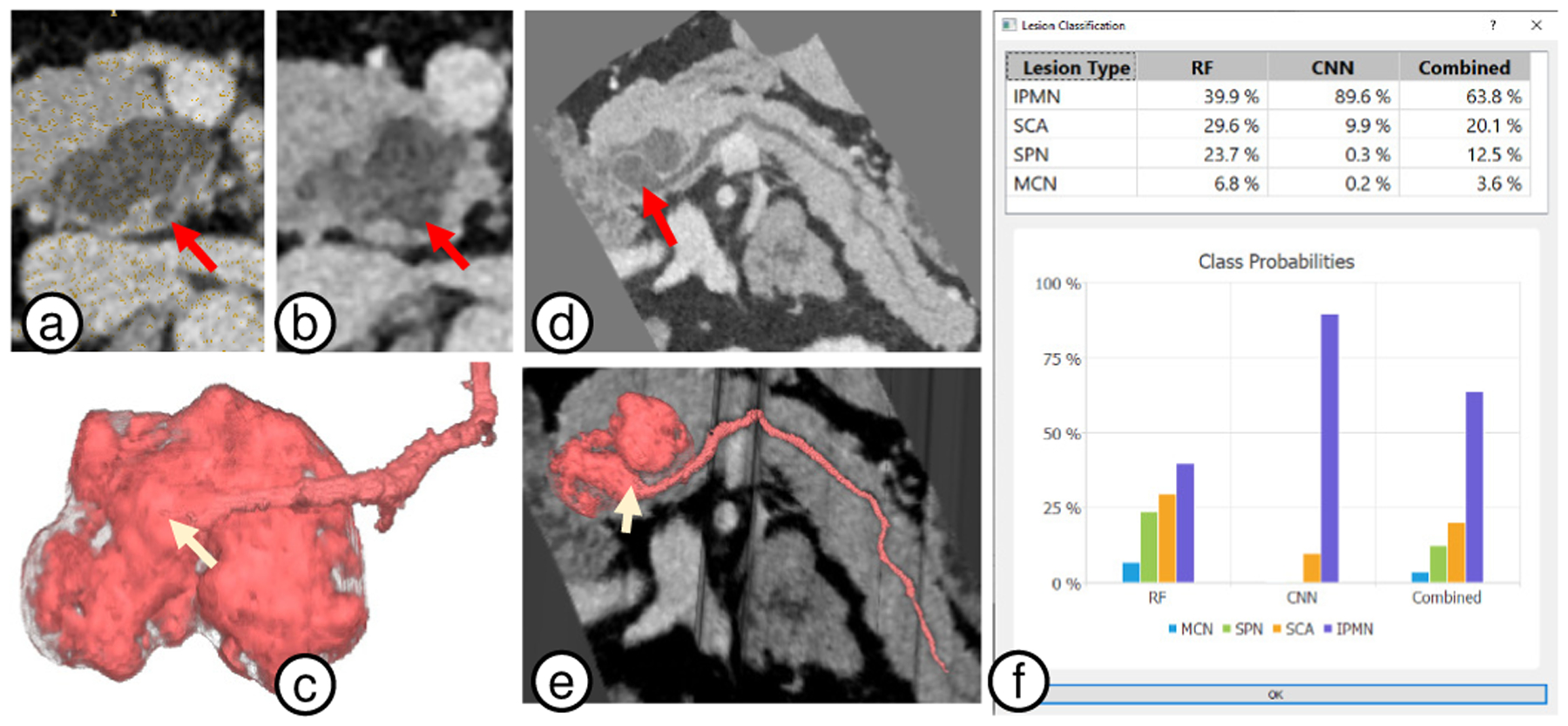
Case VP011. (a) 2D view with duct footprint (red arrow) next to grey lesion. (b) 2D lesion view as the duct (red arrow) merges into the lesion. It is difficult to visualize the separation between the duct and lesion, and thus notice the duct geometry. (c) 3D visualizations show a clear fusiform dilatation (white arrow) of the main duct as it connects with the lesion. (d-e) 2D and 3D CPR views provide additional views to confirm the diagnosis. (f) Finally, lesion classification independently classifies the lesion as an IPMN (overall probability 63.8%).
Expert Feedback.
The lesion automatic classification was very appreciated, with the radiologists noting that it can provide a crucial assistance during the diagnosis process, especially to radiologists who may have had little exposure to pancreatic pathology. They particularly commented on how viewing the RF and CNN classification probabilities separately before they are combined allowed them to weight the importance of each and refine the final diagnosis.
5.4. Visualizations and User Interface
Once the pancreas, lesions, and duct are segmented, the centerlines are computed, and the classification probabilities are generated, they are available in the user interface for visual diagnosis. A snapshot of the user interface is shown in Fig. 9. The screen space is broadly divided into seven frames. To the left is a searchable table with the patient information and CT image metadata extracted from the CT Scan DICOM header. Below the patient information table is a pancreas CPR 2D view that can switch between Centerline-Guided Re-sectioning (Sec. 5.4.3) and Duct-Centric CPR (Sec. 5.4.4) view. In the top center, a viewport displays the Pancreas-Centric 3D view for all our 3D visualizations. To the right, the three conventional planar views: axial, coronal, and sagittal views that are common in radiology applications, support the interactions common in radiological interfaces, including panning, zooming, and window/level adjustment.
Fig. 9.
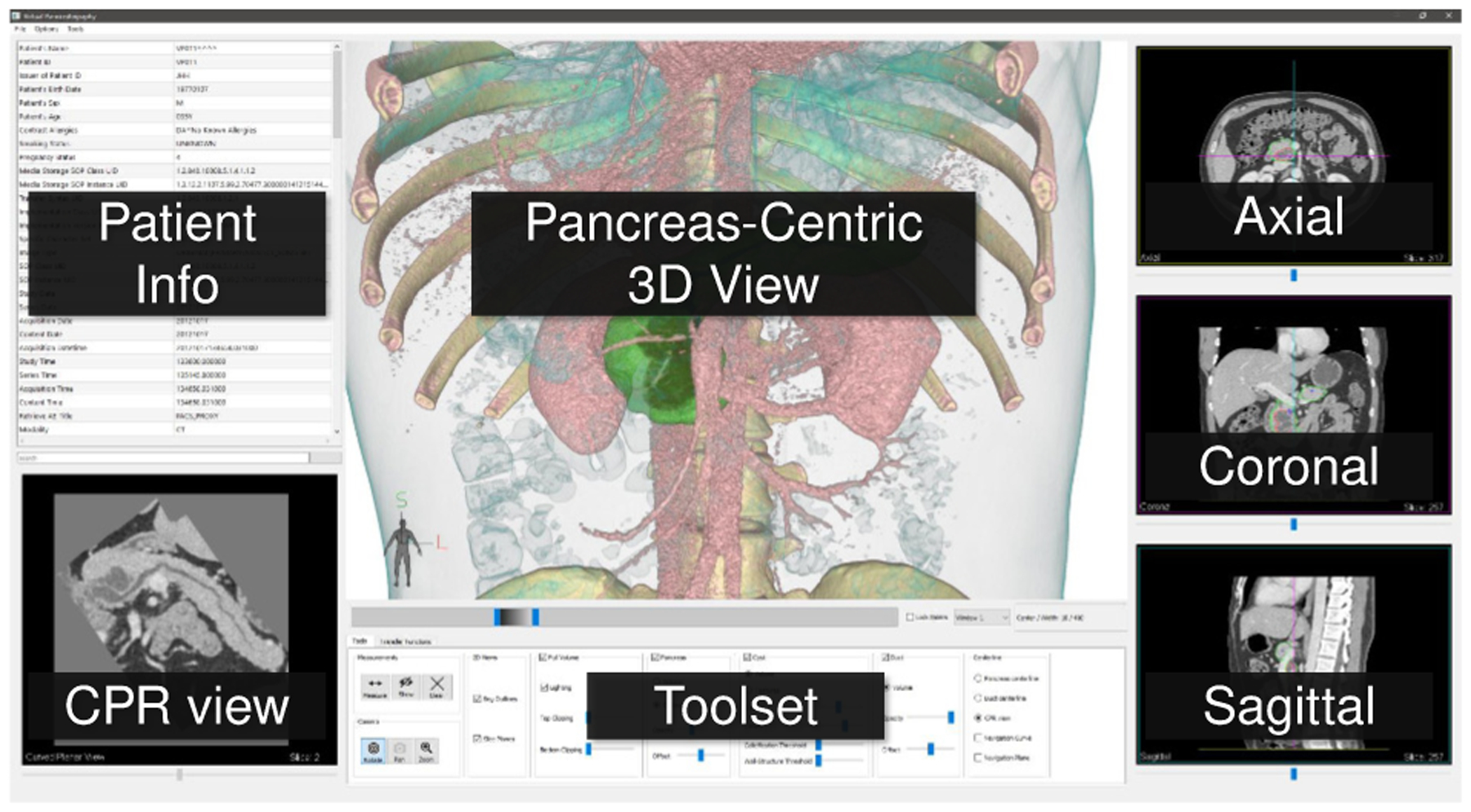
The VP user interface.
Linking of rendering canvases such as viewports and embedded cutting planes/surfaces is often used for correlation of features across viewpoints in diagnostic and visual exploration systems [64, 65]. In our system, the 3D and 2D views are linked to facilitate correlation of relevant pancreas and lesion features across viewpoints. A point selected on any of the 2D views will automatically navigate the other 2D views to the clicked voxel. Additionally, a 3D cursor highlights the position of the selected voxel in the 3D viewpoint. Similarly, the user can also directly select a point in 3D by clicking from two different camera positions (perspectives). Each time the user clicks the 3D view, a ray is cast identifying a straight line passing through the 3D scene. Two such clicks uniquely identify a 3D point. The 2D views are automatically changed to the selected voxel. This linking of views allows comparison between features in 3D visualizations with CT raw intensities in 2D.
Finally, at the screen bottom, we place the user tools/options to control the 3D and 2D visualizations and their parameters. These include visibility (show/hide) and opacity of segmented structures, clipping planes, and window/level setting (grayscale color map) common for all the 2D views. Please refer to the supplementary video for the VP visualization and user interface in action.
5.4.1. Pancreas-Centric 3D View
The Pancreas-Centric 3D view renders the segmented anatomical structures (pancreas, lesions, duct) individually or in different combinations, as shown in Fig. 10b–d (supports T1, T2). The context around the pancreas can also be visualized in the form of the entire CT volume. The user is provided with two clipping planes (top and bottom) to restrict this context view (see Fig. 10a). This is useful to reduce occlusions and focus on the region of interest.
Fig. 10.
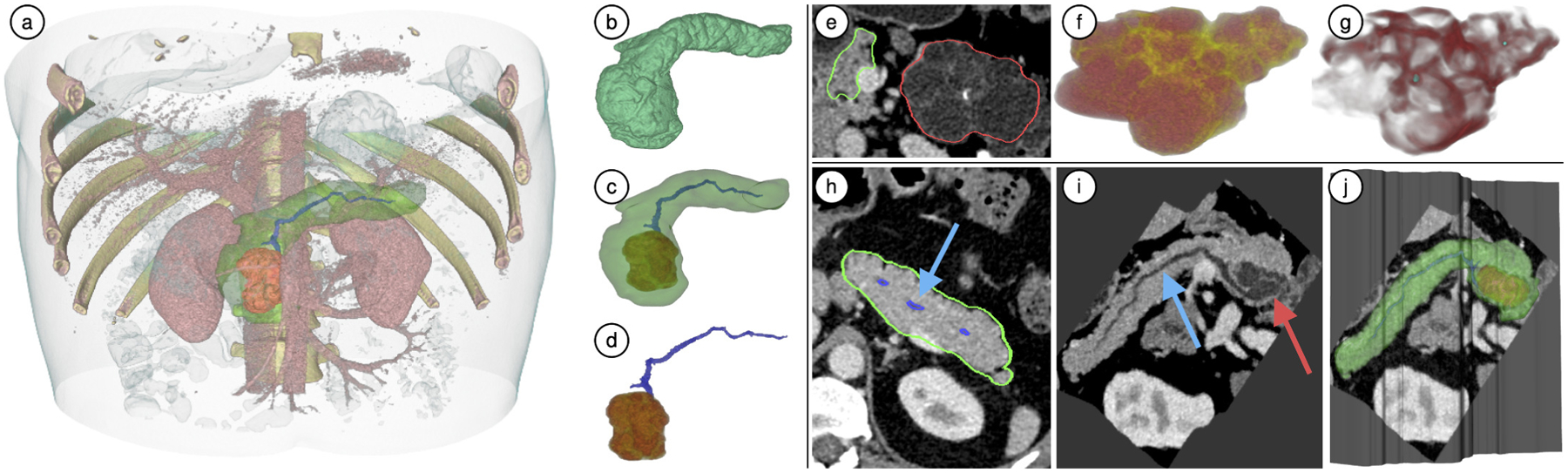
(a-d) Pancreas-centric 3D visualizations of case VP011: (a) Context volume around the pancreas clipped from the top and bottom for focused visualization; (b-d) The segmented anatomical structures and the CT volume rendered in different combinations. (e-g) Enhanced lesion visualization for VP001: (e) Axial view of the lesion (red) and pancreas (green); (f) DLR view shows septation (yellow) and cystic components (red); (g) EFR view: calcifications (cyan blobs) and septation (red) are enhanced through a Hessian-based objectness filter to provide clearer view of internal lesion features. (h-j) CPR views of VP011: (h) Axial 2D view of pancreas: pancreatic primary duct does not lie in a single plane and appears broken; (i) Duct-centric CPR view of the pancreas: the primary duct is completely visible in a single plane along with a lesion sectional view; (j) CPR surface embedded in 3D view: incorporating such a view along with 3D visualization is helpful in understanding how the CPR view was constructed.
The TF is pre-designed for the context volume (CT volume surrounding the pancreas) and does not require manual editing as CT intensities have a similar range across patients. Similarly, the TFs used for the pancreas, lesions, and duct volume are also pre-designed and are re-scaled to the scalar intensity ranges within the segmented structures for every patient. We provide simplified controls over the optical properties of our 3D visualizations. They can be modified through a simplified interface using two sliders: opacity and offset; rather than editing a multi-node polyline which can have significantly more degrees of freedom. The opacity slider controls the global opacity of the segmented structure and the offset slider applies a negative/positive offset to the TF to control how the colors are applied to the segment. We provide an advanced tab in the toolset where the 1D TFs can be edited as a multi-node polyline, if desired.
Expert Feedback.
The radiologists tested our 3D visualizations using multiple real-world cases. They noted that the 3D pancreas-centric visualizations combining pancreas outline along with lesion and duct volumes are very useful to assess the duct and lesion relationship. For example, in case of an IPMN it is critical to investigate if the lesion arises from the duct. They were also able to identify the shape of duct dilation and further analyze the IPMN type (main duct vs side branch communication) through our 3D visualization, which not only helps in lesion characterization, but can also inform the decision making in surgery. Such analysis would be much harder in 2D planar views as the shape of duct dilation and connectivity with side branches can be misleading in 2D slices. K.B. also suggested that the 3D visualization and measurement capabilities of the system could be potentially useful for a pre-operative planning since the surgeons can visualize the structures in 3D before the actual procedure. The radiologists also found our simplified two-slider TF editing interface very simple and intuitive as compared to editing a multi-node polyline.
5.4.2. Lesion Visualization
As described in Sec. 3, the morphology and appearance of the pancreatic lesions (Fig. 2) on CT scans are paramount in their diagnosis. This includes an assessment of visible characteristics of the lesions, such as septation, central and peripheral calcification, and presence of cystic and solid components (task T2). To this end, we visualize the internal features of lesions in two different modes, namely: direct lesion rendering (DLR) and enhanced features rendering (EFR).
The DLR visualization mode performs a lesion direct volume rendering using a two-color (red, yellow) preset TF that applies a higher opacity red color for darker regions (e.g., cystic components), and a lower opacity yellow color for relatively brighter ones (e.g., septation, solid components). This allows the radiologist to easily find the correct boundary between features (e.g., septation, cystic components). Another example of DLR rendering is in Fig. 10f.
Similar to a previous curvature-based approach [66] to enhance features in volume rendering, the EFR mode visualizes the lesion through enhancement of local geometry using a Hessian-based enhancement filter (Fig. 10a–d). A locally computed Hessian at multiple scales can quantify the probability of a local structure, such as a blob, plate, or a vessel. We apply the plate enhancement for wall structures and blob enhancement along with thresholding for calcifications. Both volumes are then combined as a 2-channel (red-green) 3D texture for simultaneous volume rendering of enhanced features (see Fig. 10g).
Expert Feedback.
When testing the 3D lesion visualization capabilities, the radiologists noticed the visible septation, cystic components, and calcifications. They were able to understand relative sizes of the cystic components, particularly in cases of SCA and SPN that have macro-cystic appearance. Through the lesion 3D shape, they were also able to characterize IPMN lesions between main duct and side branch IPMNs, since side branch IPMNs have a more regular spherical shape as compared to main duct IPMNs.
5.4.3. Centerline-Guided Re-sectioning
In VP, we incorporate an exploratory visualization that combines 3D visualization with a 2D slice view of raw CT intensities. An orthogonal intersecting plane slides along either the pancreas or duct centerline and renders the raw CT intensities in grayscale (see Fig. 5c). Simultaneously, all 3D anatomical structures (pancreas, lesions, primary duct, surrounding CT volume), can be combined in the visualization through visibility controls. The sliding plane represents a centerline-guided re-sectioning of the pancreas volume, and can be viewed in the 3D view and separately as a 2D slice view. This sectional view provides coherent tracking of the primary duct along its length and closely studying its relationship with the lesion (task T3). Radiologists can take a closer look at how the 3D anatomical structures, such as lesions and duct, correspond with raw CT values (tasks T2, T3) by directly overlapping the 3D rendering with the intersecting plane. This approach instills greater confidence in the radiologists during diagnosis as they can correlate 3D structures to 2D images that they are traditionally interpreting. It also provides a better comparison than switching context across multiple viewports.
Expert Feedback.
The radiologists interacted with the centerline-guided re-sectioning view embedded in the 3D pancreas-centric view. They very much appreciated the idea of being able to verify 3D features by directly correlating them with the 2D intersection plane of raw CT intensities. Through our implemented linked views and point selection, the radiologists were able to relate corresponding points between 3D and 2D views.
5.4.4. Duct-Centric CPR View
The primary duct size and its relationship with pancreatic lesions is important for visual diagnosis, as only IPMN lesions communicate with the primary duct. CPR techniques reconstruct the longitudinal cross-sections of vessels, which enables visualization of the entire vessel length in a single projected viewpoint. Points on the vessel centerline are swept along an arbitrary direction to construct a developable surface along the vessel length. The surface samples the intersecting volume at every point and is flattened without distortion into a 2D image. Using the computed duct centerline as a guide curve, we generate pancreas duct-centric CPR views in Fig. 10h–j (task T3). The direction in which the centerline is swept CPR surface construction is chosen by the user through a widget that shows the surface rendering of the pancreas, lesion, and centerline curve. The sweeping direction is always normal to the screen. This widget allows to choose an appropriate direction to generate the desired CPR viewpoint, thus helping in visualizing the duct-lesion relationship in a single viewpoint.
Expert Feedback.
One of the radiologists was already familiar with the general concept of CPR reconstruction, and appreciated that we tailored it to the pancreas by including the primary duct and lesion in a single view. Through the 3D embedded CPR view showing both the CPR surface and 3D rendering of the duct and lesion, the radiologists found it helpful to verify that the duct is connected to the lesion, and also to identify how the duct dilates as it connects to the lesion. The duct dilation geometry would not be clearly visible in 2D views. Visualizing this dilation can also help in surgical decisions such as the location of potential resection to remove the malignant mass.
5.4.5. 2D and 3D Measurements
Measurement is an integral part of visual diagnosis as it helps to quantify lesion size and cystic components (T5). We provide the capability to interactively measure in all our 2D planar views, by clicking and drawing measurements. We also provide automatic lesion 3D measurements of the lesions as the dimensions of its best fitting oriented bounding box. Additionally, automatic volume measurements of segmented structures (pancreas, lesions, duct) are also available. Please refer to the video for these tools in action.
Expert Feedback.
In the current workflow, measurements are performed by subjectively choosing an axis-aligned 2D slice. The radiologists explained that the choice of this plane can drastically affect measurement values. They agreed that estimating the volume of the segmented lesions and using a best fitting box for calculating size is a significantly more reliable approach to measurement.
6. Results and Evaluation
We evaluated our VP visualization system on real-world patient CT scans that were accompanied with ground truth masks of the pancreas and lesions outlined by a radiologist, and ground truth histopathological diagnosis confirmed by a pathologist after specimen resection. Due to the challenges associated with the manual segmentation of the duct, manual duct outlines were acquired for only four cases. The size of each volume is Z × 512 × 512, where Z is the number of axial slices ranging from 418 to 967.
Segmentation.
To develop and test the automatic segmentation module, we randomly divided the dataset into training and testing sets using a ratio of 90%/10%. Qualitative evaluation of the performance was done by comparing the predicted masks Ŷ against the ground truth masks Y using the common DC-based metric .
Our model achieves an accuracy of 83.75% ± 12.76% and 80.07% ± 4.03% for pancreas and lesion segmentation, respectively, outperforming the current state-of-the-art solution [46] by 4.52% and 16.63%, respectively.
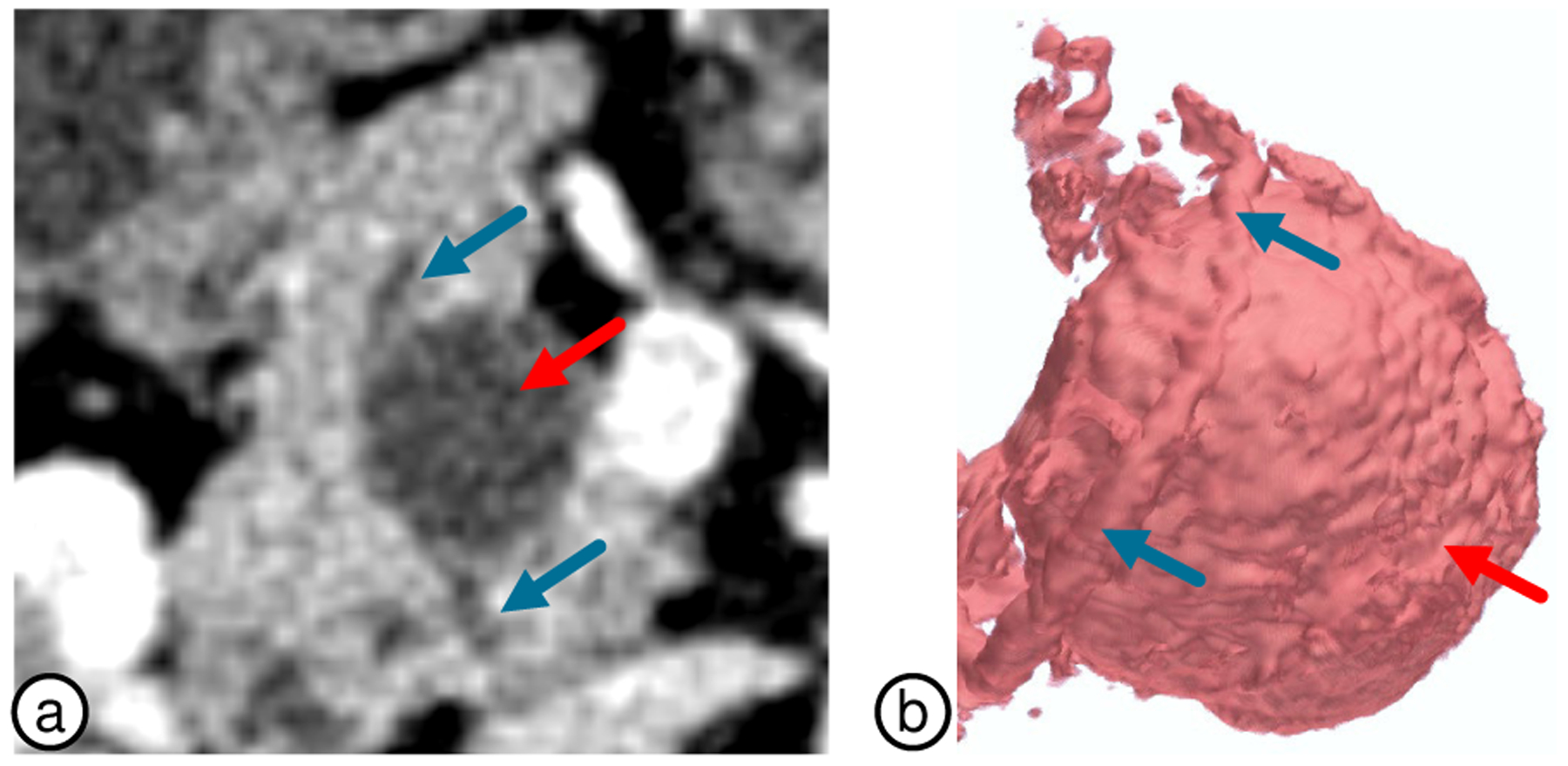
Duct and Centerline Extraction.
A qualitative comparison of our duct extraction approach with manual segmentations is shown in Fig. 12b–c, which is able to overcome the noise and occlusions caused by noisy voxels, that are mistakenly enhanced with the vesselness filter (Fig. 12a). Additionally, our duct centerline approach reduces the excessive bending as it enters and exists the duct by utilizing a penalty model which weights both duct and pancreas geometry when computing the curve (Fig. 6).
Fig. 12.
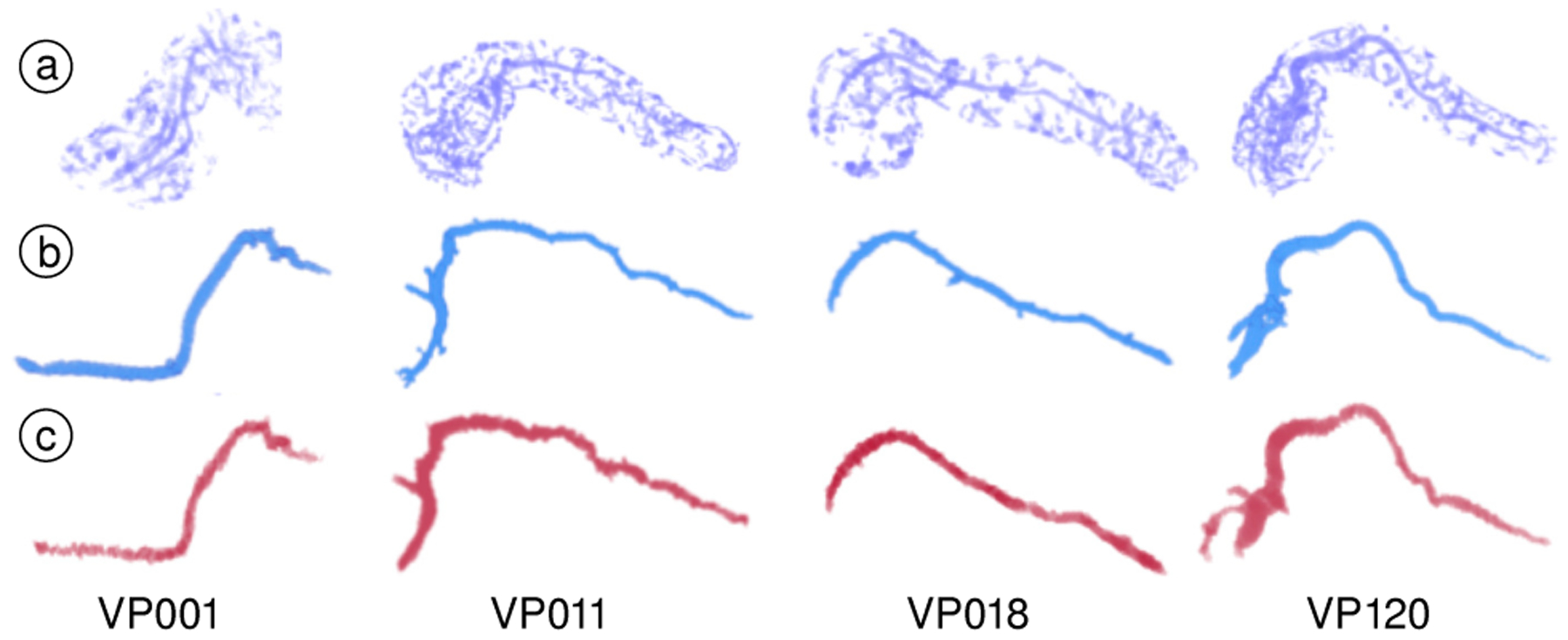
Duct volume rendering. (a) Direct application of the vesselness filter. (b) Extraction using our approach. (c) Manual segmentations.
CPR and Lesion Visualization.
Examples of the 3D pancreas-centric and CPR views are shown in Fig. 11 for two patients with dilated ducts. These can assist in determining the presence of connection between the duct and the lesion in particularly difficult cases when the conventional 2D axial views are inadequate. Examples of the DLR and EFR visualizations of the lesions are shown in Fig. 13. These facilitate better understanding of lesions shape and structure in comparison to viewing them in 2D images, by enhancing the characteristic lesion structures, such as external wall, calcification, internal septation, solid and cystic components.
Fig. 11.
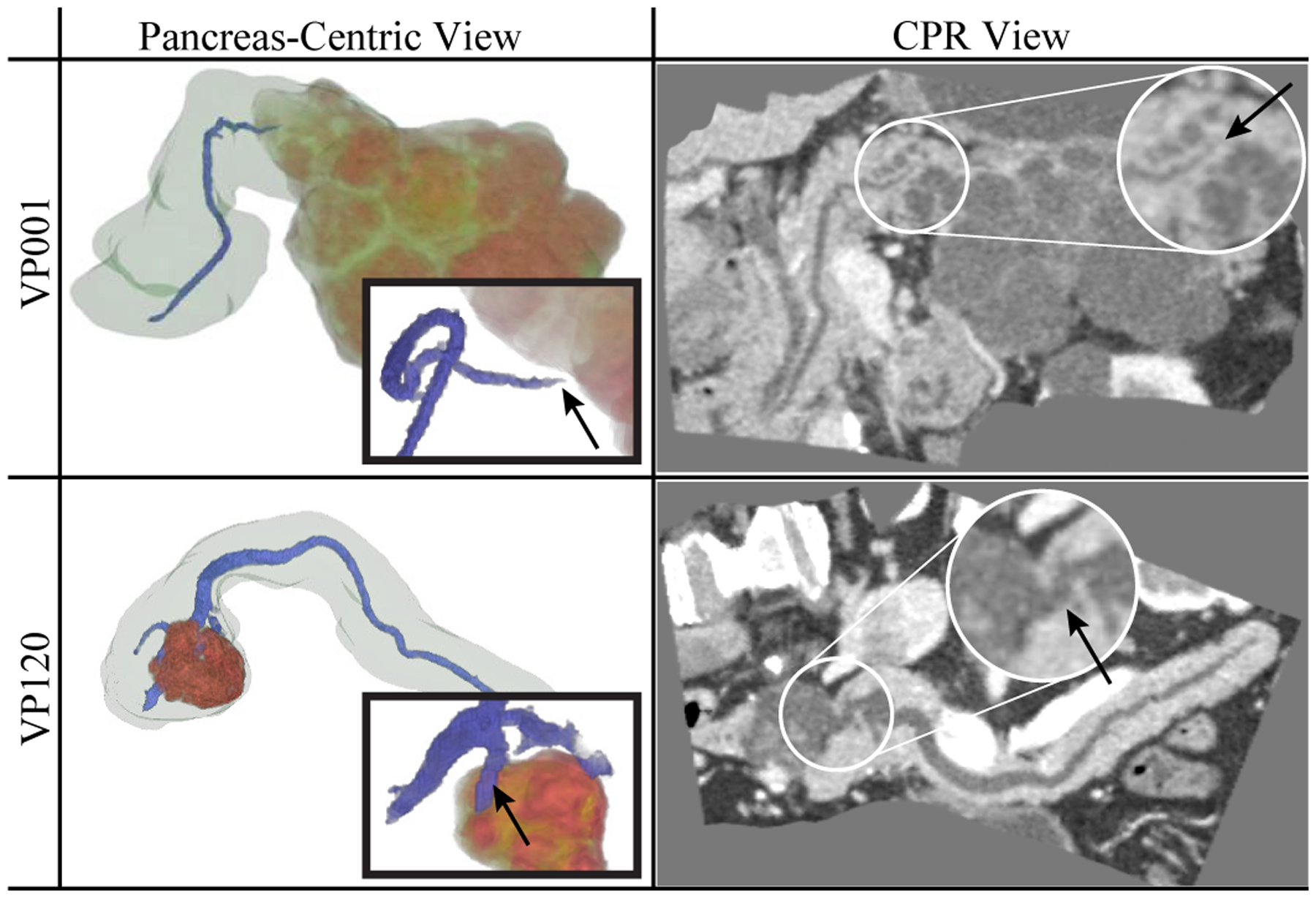
Pancreas-centric and CPR views, assisting in confirming presence (VP120, IPMN) or absence (VP001, SCA) of duct-lesion connection (see arrows).
Fig. 13.
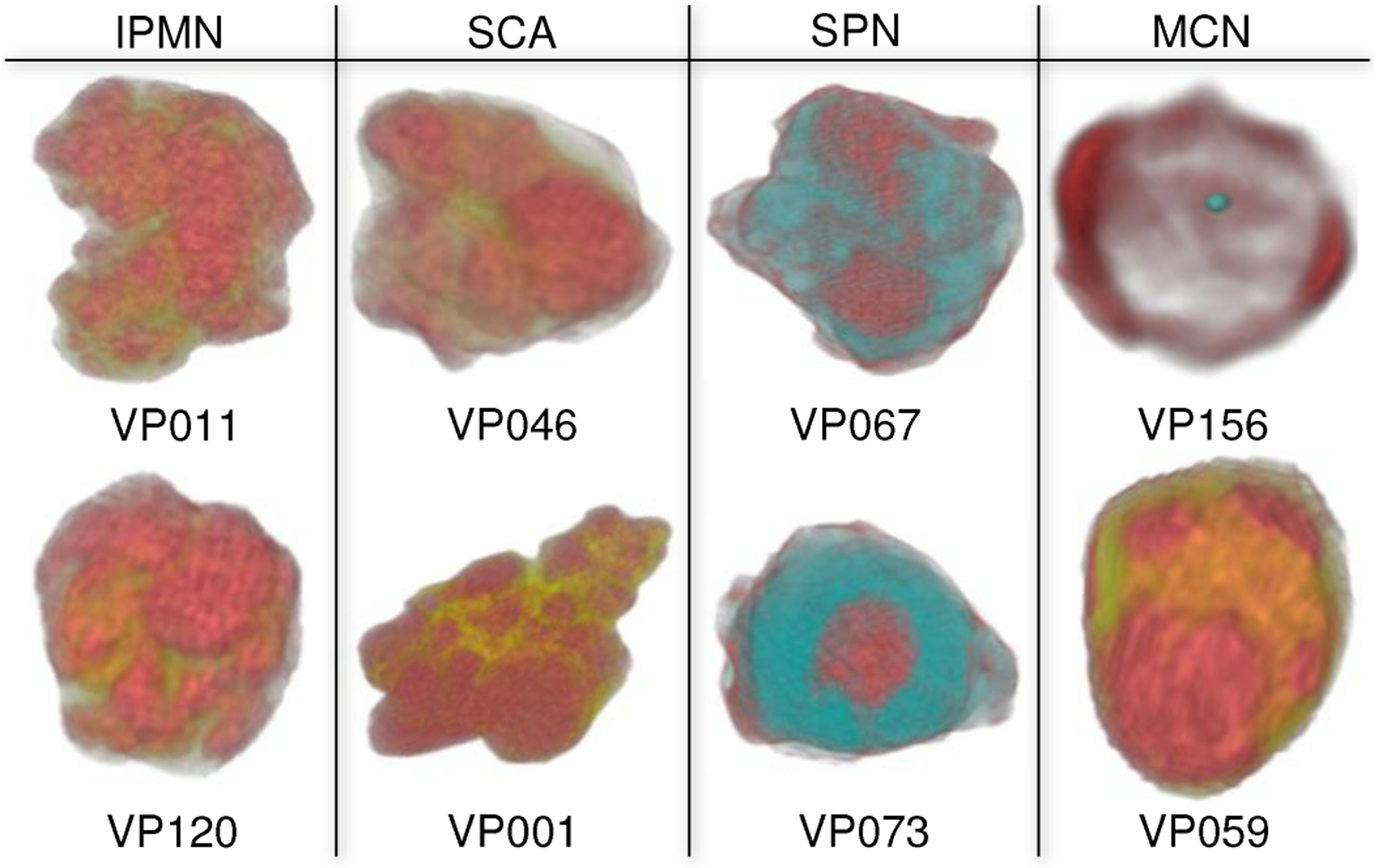
3D visualization of lesions enhancing the important characteristics that can inform their visual classification. VP156 in EFR mode to render wall structures (red) and calcification (cyan). Others are in DLR mode: cystic components (red), septation / soft tissue (yellow), and solid components (cyan) that is an SPN characteristic feature.
Case Studies.
Our expert radiologists: M.B. and K.B., used our VP system on multiple real-world datasets before providing their suggestions and feedback on VP components. The qualitative feedback regarding individual VP components of our system has been discussed throughout Sec. 5. We describe two real-world case studies conducted by M.B. with VP that demonstrate the effectiveness and utility of VP. He provided his analysis of the cases as below.
VP011 is a 35 years old male. Fig. 14 shows different visualizations used for the analysis. The 3D visualization (Fig. 14a) clearly shows the primary duct connected to a lesion through a fusiform (spindle shape) dilatation. M.B. didn’t notice the fusiform shape in the conventional 2D axial view, but immediately noticed it in 3D. He independently confirmed the shape through different viewpoints of 2D and 3D embedded CPR views (Fig. 14b). Third confirmation came from the automatic lesion classification (Fig. 14c) of IPMN probability of 39.9% from RF, 89.6% from CNN, and 63.8% combined. Based on multiple sources, including 3D visualizations showing main duct fusiform dilatation and the lesion shape, M.B. classified the lesion as a mixed main-duct and side-branch IPMN.
VP018 is a 72 years old female. Fig. 15a shows a 2D axial view of the primary duct and the lesion. M.B. expressed difficulty to confirm whether the primary duct enters the lesion or is simply adjacent to it. Using the 3D visualization (Fig. 15b) he inferred that the duct is touching the lesion, passing around without entering it. Noticing this information, along with the fact that the lesion shows a unilocular (spherical) shape, the radiologist concluded that this is consistent with the characteristic of side-branch IPMN. This conclusion is confirmed by the automatic classification that characterizes the lesion as IPMN (RF: 25%, CNN: 73%, combined: 48%), which concurs with the ground truth. M.B. agreed that such characterizations would have been harder in the traditional workflow that uses only axis-align 2D views, without the 3D and CPR visualizations.
Fig. 15.
Case VP018. (a) 2D axial view of the main duct (blue arrows) and the lesion (gray circular region, red arrow). It was hard for the radiologist to discern whether the duct is adjacent or enters the lesion. (b) 3D visualization clearly shows the main duct (blue arrows) touching the lesion (red arrow), but passes around without entering it.
Updated Workflow.
The radiologists were asked to compare the new VP system with their conventional workflow and how they would change their workflow. In the conventional workflow (as described in Sec. 4), the radiologists rely heavily on raw 2D axis-aligned views and often don’t use even familiar tools such as de-noising filters, cutting planes, and maximum intensity projections. Depending on the case, a radiologist could spent significant time scrolling back and forth for understanding the relationship between lesions and duct, when a typical span of the pancreas can be 150–200 axial slices. Based on the demonstrated datasets and analyzed real-world cases, the radiologists agreed that the 3D visualizations of VP could improve the diagnosis process effectiveness and provide additional data points for decision making. Based on our novel VP, they suggested an updated VP workflow as follows: (1) overview a case and inspect the lesions using 2D axis-aligned views and provided segmentation outlines; (2) visualize the lesion and its internal features in 3D to further understand the lesion morphological structure; (3) study the lesion and duct relationship in 3D, CPR view, and duct-centric re-sectioning views; and (4) finally confirm own diagnosis and characterization with the automatic classification results. Since our automatic classification (step (4)) is at least 91.7% accurate [51], and the other steps can further enhance this diagnosis accuracy, we anticipate that the radiologists’ accuracy using VP will improve substantially from the current 67–70% [2].
7. Conclusion and Future Work
We have presented 3D Virtual Pancreatography (VP), a comprehensive visualization application and its system that enables quantitative and qualitative visual diagnosis and characterization of pancreatic lesions. VP was developed in close collaboration between computer scientists and expert radiologists who are experienced in diagnosing pancreatic lesions with a goal to understand their requirements and cater VP to their needs. We developed a fully automatic segmentation pipeline for the pancreas and lesions, and a semi-automatic approach for duct extraction. We presented combined 3D and 2D exploratory visualizations for effective diagnosis and characterization of lesions. Our novel approach to compute the pancreas centerline that incorporates the pancreas geometry as well as the connected components of the extracted duct is utilized to construct exploratory visualizations, such as duct-centric pancreas re-sectioning and CPR view. We incorporated feature enhancement visualizations for lesions and their internal features, such as septation, calcifications, and cystic/solid components. Quantitative analysis is supported through automatic lesion classification and automatic/manual measurements.
We have demonstrated the utility of our VP system and visualizations through real-world case-studies and expert radiologists’ feedback. In the future, we plan to perform a larger quantitative user study with multiple radiologists to concretely evaluate VP benefits and shortcomings for further improvements and suitability for deployment in the real world. As future enhancements, our radiologists also suggested that we add explicit tools to track lesion growth in a patient over time. Additionally, we plan to evaluate our segmentation model and the training procedure on other organs, potentially using a non-Dice-based metric.
Supplementary Material
Acknowledgments
We thank our collaborators at Stony Brook Medicine, including Dr. Kevin Baker, for evaluating our system and providing feedback, and at Johns Hopkins, Drs. Ralph Hruban and Elliot Fishman, for the pancreas CT datasets. This work has been partially supported by NSF grants CNS1650499, OAC1919752, ICER1940302; Marcus Foundation; and National Heart, Lung, and Blood Institute of NIH under Award U01HL127522; New York State Center for Advanced Technology in Biotechnology; Stony Brook University; Cold Spring Harbor; Brookhaven National; Feinstein Institute for Medical Research; and New York State Economic Development Department, Contract C14051. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Biographies

Shreeraj Jadhav is a PhD candidate in the Department of Computer Science at Stony Brook University. He received his B.E. in Mechanical Engineering from the University of Pune, India in 2006 and his M.S. in Computing from the School of Computing, University of Utah in 2012. His research interests include computer graphics, visualization, VR, and analysis techniques for medical diagnosis.

Konstantin Dmitriev received his PhD in Computer Science from Stony Brook University in 2020. He did his Bachelor of Computer Science and Applied Mathematics from St. Petersburg Electrotechnical University, Russia. His research interests include medical imaging, visualization, machine learning, and computer vision.

Joseph Marino is a Postdoctoral Research Fellow. He received his PhD in Computer Science (2012), and BS in Computer Science and Applied Mathematics & Statistics (2006) both from Stony Brook University. His research focuses on visualization and computer graphics for medical imaging, including visualization and analysis for pancreatic and prostate cancer screening, and enhancements for virtual colonoscopy. He is a recipient of the 2016 Long Island Technology Hall of Fame Patent Award (2016).

Matthew Barish is an Associate Professor of Clinical Radiology, Vice-Chair, Radiology Operations, Management and Informatics, Co-Director of Body Imaging at Stony Brook University Hospital since 2007, graduated from Boston University and Fellowship from Yale-New Haven Hospital. He specializes in Abdominal Imaging, world-renown expert in Virtual Colonoscopy (VC), and co-leads the Division of Abdominal Radiology. He is a Fellow of the American College of Radiology and serves as Director of the American College of Radiology Education Center for VC. He has published extensively on MR Cholangiopanc Reatography, VC, 3D Imaging and Image Processing.

Arie E. Kaufman is a Distinguished Professor of Computer Science, Director of Center of Visual Computing, and Chief Scientist of Center of Excellence in Wireless and Information Technology at Stony Brook University. He served as Chair of Computer Science Department, 1999–2017. He has conducted research for >40 years in visualization and graphics and their applications, and published >350 refereed papers. He was the founding Editor-in-Chief of IEEE TVCG, 1995–98. He is an IEEE Fellow, ACM Fellow, National Academy of Inventors Fellow, recipient of IEEE Visualization Career Award (2005), and inducted into Long Island Technology Hall of Fame (2013) and IEEE Visualization Academy (2019). He received his PhD in Computer Science from Ben-Gurion University, Israel (1977).
Contributor Information
Shreeraj Jadhav, Computer Science Department, Stony Brook University..
Konstantin Dmitriev, Computer Science Department, Stony Brook University..
Joseph Marino, Computer Science Department, Stony Brook University..
Matthew Barish, Department of Radiology, Stony Brook Medicine..
Arie E. Kaufman, Computer Science Department, Stony Brook University..
References
- [1].Cancer Facts & Figures. American Cancer Society, 2019. [Google Scholar]
- [2].Sahani DV, Sainani NI, Blake MA, Crippa S, Mino-Kenudson M, and del Castillo CF, “Prospective evaluation of reader performance on MDCT in characterization of cystic pancreatic lesions and prediction of cyst biologic aggressiveness,” Am J Roentgenol, 197(1):W53–W61, 2011. [DOI] [PubMed] [Google Scholar]
- [3].Bartz D, “Virtual endoscopy in research and clinical practice,” Comput Graph Forum, 24(1):111–126, 2005. [Google Scholar]
- [4].Hong L, Kaufman A, Wei Y-C, Viswambharan A, Wax M, and Liang Z, “3D virtual colonoscopy,” IEEE Biomed Vis, 26–32, 1995. [Google Scholar]
- [5].Hong L, Muraki S, Kaufman A, Bartz D, and He T, “Virtual voyage: Interactive navigation in the human colon,” SIGGRAPH, pp. 27–34, 1997. [Google Scholar]
- [6].Vilanova A, König A, and Gröller E, “VirEn: A virtual endoscopy system,” Machine Graphics and Vision, vol. 8, pp. 469–487, 1999. [Google Scholar]
- [7].Wan M, Dachille F, Kreeger K, Lakare S, Sato M, Kaufman AE, Wax MR, and Liang Z, “Interactive electronic biopsy for 3D virtual colonscopy,” SPIE Medical Imaging, 4321:483–489, 2001. [Google Scholar]
- [8].Hong W, Gu X, Qiu F, Jin M, and Kaufman A, “Conformal virtual colon flattening,” Proc of ACM SPM, pp. 85–93, 2006. [Google Scholar]
- [9].Bartrolí AV, Wegenkittl R, König A, Gröller E, and Sorantin E, “Virtual colon flattening,” Data Visualization, pp. 127–136, 2001. [Google Scholar]
- [10].Zeng W, Marino J, Gurijala KC, Gu X, and Kaufman A, “Supine and prone colon registration using quasi-conformal mapping,” IEEE TVCG, vol. 16, no. 6, pp. 1348–1357, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wegenkittl R, Vilanova A, Hegedust B, Wagner D, Freund MC, and Gröller E, “Mastering interactive virtual bronchioscopy on a low-end pc,” IEEE Vis, pp. 461–464, 2000. [Google Scholar]
- [12].Rodenwaldt J, Kopka L, Roedel R, Margas A, and Grabbe E, “3D virtual endoscopy of the upper airway: optimization of the scan parameters in a cadaver phantom and clinical assessment,” Computer Assisted Tomography, vol. 21, no. 3, pp. 405–411, 1997. [DOI] [PubMed] [Google Scholar]
- [13].Bartz D, Straßer W, Skalej M, and Welte D, “Interactive exploration of extra-and interacranial blood vessels,” IEEE Vis, pp. 389–547, 1999. [Google Scholar]
- [14].Gobbetti E, Pili P, Zorcolo A, and Tuveri M, “Interactive virtual angioscopy,” IEEE Vis, pp. 435–438, 1998. [Google Scholar]
- [15].Yagel R, Stredney D, Wiet GJ, Schmalbrock P, Rosenberg L, Sessanna DJ, and Kurzion Y, “Building a virtual environment for endoscopic sinus surgery simulation,” Computers & Graphics, vol. 20, no. 6, pp. 813–823, 1996. [Google Scholar]
- [16].Rogalla P, “Virtual endoscopy of the nose and paranasal sinuses,” Virtual Endoscopy and Related 3D Techniques, pp. 17–37, 2001. [Google Scholar]
- [17].Marino J and Kaufman A, “Prostate cancer visualization from MR imagery and MR spectroscopy,” Comput Graph Forum, vol. 30, no. 3, pp. 1051–1060, 2011. [Google Scholar]
- [18].Raidou RG, Casares-Magaz O, Amirkhanov A, Moiseenko V, Muren LP, Einck JP, Vilanova A, and Gröller ME, “Bladder Runner: Visual analytics for the exploration of RT-induced bladder toxicity in a cohort study,” Comput Graph Forum, 37(3):205–216, 2018. [Google Scholar]
- [19].Ghaderi MA, Heydarzadeh M, Nourani M, Gupta G, and Tamil L, “Augmented reality for breast tumors visualization,” Proc IEEE Eng Med Biol Soc, pp. 4391–4394, 2016. [DOI] [PubMed] [Google Scholar]
- [20].Webb LJ, Samei E, Lo JY, Baker JA, Ghate SV, Kim C, Soo MS, and Walsh R, “Comparative performance of multiview stereoscopic and mammographic display modalities for breast lesion detection,” Med Phys, 38(4):1972–19820, 2011. [DOI] [PubMed] [Google Scholar]
- [21].Wang J, Yang X, Cai H, Tan W, Jin C, and Li L, “Discrimination of breast cancer with microcalcifications on mammography by deep learning,” Scientific Reports, vol. 6, p. 27327, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Smit N, Lawonn K, Kraima A, DeRuiter M, Sokooti H, Bruckner S, Eisemann E, and Vilanova A, “Pelvis: Atlas-based surgical planning for oncological pelvic surgery,” IEEE TVCG, vol. 23, no. 1, pp. 741–750, 2016. [DOI] [PubMed] [Google Scholar]
- [23].Raidou RG, Casares-Magaz O, Muren LP, Van der Heide UA, Rørvik J, Breeuwer M, and Vilanova A, “Visual analysis of tumor control models for prediction of radiotherapy response,” Comput Graph Forum, 35(3):231–240, 2016. [Google Scholar]
- [24].Yin T, Coudyzer W, Peeters R, Liu Y, Cona MM, Feng Y, Xia Q, Yu J, Jiang Y, Dymarkowski S, Huang G, Chen F, Oyen R, and Ni Y, “Three-dimensional contrasted visualization of pancreas in rats using clinical MRI and CT scanners,” Contrast Media & Molecular Imaging, vol. 10, no. 5, pp. 379–387, 2015. [DOI] [PubMed] [Google Scholar]
- [25].Grippo PJ, Venkatasubramanian PN, Knop RH, Heiferman DM, Iordanescu G, Melstrom LG, Adrian K, Barron MR, Bentrem DJ, and Wyrwicz AM, “Visualization of mouse pancreas architecture using MR microscopy,” The American Journal of Pathology, vol. 179, no. 2, pp. 610–618, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kanitsar A, Wegenkittl R, Fleischmann D, and Gröller ME, “Advanced curved planar reformation: Flattening of vascular structures,” IEEE Vis, pp. 43–50, 2003. [Google Scholar]
- [27].Kok P, Baiker M, Hendriks EA, Post FH, Dijkstra J, Lowik CW, Lelieveldt BP, and Botha CP, “Articulated planar reformation for change visualization in small animal imaging,” IEEE TVCG, vol. 16, no. 6, pp. 1396–1404, 2010. [DOI] [PubMed] [Google Scholar]
- [28].Brambilla A, Angelelli P, Andreassen Ø, and Hauser H, “Comparative visualization of multiple time surfaces by planar surface reformation,” IEEE Pacific Visualization Symposium, pp. 88–95, 2016. [Google Scholar]
- [29].Auzinger T, Mistelbauer G, Baclija I, Schernthaner R, Köchl A, Wimmer M, Gröller ME, and Bruckner S, “Vessel visualization using curved surface reformation,” IEEE TVCG, 19(12):2858–2867, 2013. [DOI] [PubMed] [Google Scholar]
- [30].Lampe OD, Correa C, Ma K-L, and Hauser H, “Curve-centric volume reformation for comparative visualization,” IEEE TVCG, vol. 15, no. 6, pp. 1235–1242, 2009. [DOI] [PubMed] [Google Scholar]
- [31].Williams D, Grimm S, Coto E, Roudsari A, and Hatzakis H, “Volumetric curved planar reformation for virtual endoscopy,” IEEE TVCG, vol. 14, no. 1, pp. 109–119, 2008. [DOI] [PubMed] [Google Scholar]
- [32].Kretschmer J, Soza G, Tietjen C, Suehling M, Preim B, and Stamminger M, “ADR-anatomy-driven reformation,” IEEE TVCG, vol. 20, no. 12, pp. 2496–2505, 2014. [DOI] [PubMed] [Google Scholar]
- [33].Fridman Y, Pizer SM, Aylward S, and Bullitt E, “Extracting branching tubular object geometry via cores,” Med Im Anal, vol. 8, no. 3, pp. 169–176, 2004. [DOI] [PubMed] [Google Scholar]
- [34].Nadeem S, Marino J, Gu X, and Kaufman A, “Corresponding supine and prone colon visualization using eigenfunction analysis and fold modeling,” IEEE TVCG, vol. 23, no. 1, pp. 751–760, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sorantin E, Halmai C, Erdohelyi B, Palágyi K, Nyúl LG, Ollé K, Geiger B, Lindbichler F, Friedrich G, and Kiesler K, “Spiral-CT-based assessment of tracheal stenoses using 3-D-skeletonization,” IEEE TMI, vol. 21, no. 3, pp. 263–273, 2002. [DOI] [PubMed] [Google Scholar]
- [36].Straka M, Cervenansky M, Cruz AL, Kochl A, Sramek M, Gröller E, and Fleischmann D, “The VesselGlyph: Focus context visualization in CT-angiography,” IEEE Vis, pp. 385–392, 2004. [Google Scholar]
- [37].Chu C, Oda M, Kitasaka T, Misawa K, Fujiwara M, Hayashi Y, Nimura Y, Rueckert D, and Mori K, “Multi-organ segmentation based on spatially-divided probabilistic atlas from 3D abdominal CT images,” MICCAI, pp. 165–172, 2013. [DOI] [PubMed] [Google Scholar]
- [38].Wolz R, Chu C, Misawa K, Mori K, and Rueckert D, “Multi-organ abdominal CT segmentation using hierarchically weighted subject-specific atlases,” MICCAI, pp. 10–17, 2012. [DOI] [PubMed] [Google Scholar]
- [39].Karasawa K, Oda M, Kitasaka T, Misawa K, Fujiwara M, Chu C, Zheng G, Rueckert D, and Mori K, “Multi-atlas pancreas segmentation: atlas selection based on vessel structure,” Med Image Anal, vol. 39, pp. 18–28, 2017. [DOI] [PubMed] [Google Scholar]
- [40].Okada T, Linguraru MG, Hori M, Summers RM, Tomiyama N, and Sato Y, “Abdominal multi-organ segmentation from CT images using conditional shape–location and unsupervised intensity priors,” Med Image Anal, 26(1):1–18, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhu Z, Xia Y, Shen W, Fishman EK, and Yuille AL, “A 3D coarse-to-fine framework for volumetric medical image segmentation,” arXiv preprint arXiv:1712.00201, 2017. [Google Scholar]
- [42].Yu Q, Xie L, Wang Y, Zhou Y, Fishman EK, and Yuille AL, “Recurrent saliency transformation network: Incorporating multi-stage visual cues for small organ segmentation,” IEEE Conference on Computer Vision and Pattern Recognition, pp. 8280–8289, 2018. [Google Scholar]
- [43].Roth HR, Lu L, Farag A, Shin H-C, Liu J, Turkbey EB, and Summers RM, “Deeporgan: Multi-level deep convolutional networks for automated pancreas segmentation,” MICCAI, pp. 556–564, 2015. [Google Scholar]
- [44].Roth HR, Oda H, Hayashi Y, Oda M, Shimizu N, Fujiwara M, Misawa K, and Mori K, “Hierarchical 3D fully convolutional networks for multi-organ segmentation,” arXiv preprint arXiv:1704.06382, 2017. [Google Scholar]
- [45].Cai J, Lu L, Xie Y, Xing F, and Yang L, “Pancreas segmentation in MRI using graph-based decision fusion on convolutional neural networks,” MICCAI, pp. 674–682, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhou Y, Xie L, Fishman EK, and Yuille AL, “Deep supervision for pancreatic cyst segmentation in abdominal CT scans,” MICCAI, pp. 222–230, 2017. [Google Scholar]
- [47].Hu H, Li K, Guan Q, Chen F, Chen S, and Ni Y, “A multi-channel multi-classifier method for classifying pancreatic cystic neoplasms based on ResNet,” Proc of ICANN, pp. 101–108, 2018. [Google Scholar]
- [48].Wei R, Lin K, Yan W, Guo Y, Wang Y, Li J, and Zhu J, “Computer-aided diagnosis of pancreas serous cystic neoplasms: A radiomics method on preoperative MDCT images,” Technology in Cancer Research & Treatment, vol. 18, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Li H, Lin K, Reichert M, Xu L, Braren R, Fu D, Schmid R, Li J, Menze B, and Shi K, “Differential diagnosis for pancreatic cysts in CT scans using densely-connected convolutional networks,” arXiv preprint arXiv:1806.01023, 2018. [DOI] [PubMed] [Google Scholar]
- [50].Dmitriev K, Kaufman A, Javed AA, Hruban RH, Fishman EK, Lennon AM, and Saltz JH, “Classification of pancreatic cysts in computed tomography images using a random forest and convolutional neural network ensemble,” MICCAI, 150–158, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Dmitriev K, Marino J, Baker K, and Kaufman AE, “Visual analysis of a computer-aided diagnosis system for pancreatic lesions,” IEEE TVCG, 2019. [DOI] [PubMed] [Google Scholar]
- [52].Lennon AM and Wolfgang C, “Cystic neoplasms of the pancreas,” J Gastrointest Surg, vol. 17, no. 4, pp. 645–653, 2013. [DOI] [PubMed] [Google Scholar]
- [53].Cho H-W, Choi J-Y, Kim M-J, Park M-S, Lim JS, Chung YE, and Kim KW, “Pancreatic tumors: Emphasis on CT findings and pathologic classification,” Korean J Radiol, 12(6):731–739, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Rowe SP and Fishman EK, “Image processing from 2D to 3D,” Multislice CT, pp. 103–120, 2019. [Google Scholar]
- [55].Sakas G, “Trends in medical imaging: from 2D to 3D,” Computers & Graphics, vol. 26, no. 4, pp. 577–587, 2002. [Google Scholar]
- [56].Pieper S, Halle M, and Kikinis R, “3D Slicer,” Proc IEEE Int Symp Biomed Imaging, pp. 632–635, 2004. [Google Scholar]
- [57].Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, and Rabinovich A, “Going deeper with convolutions,” Proc of IEEE ICCV, pp. 1–9, 2015. [Google Scholar]
- [58].He K, Zhang X, Ren S, and Sun J, “Deep residual learning for image recognition,” Proc of IEEE CVPR, pp. 770–778, 2016. [Google Scholar]
- [59].Zhou Y, Xie L, Shen W, Wang Y, Fishman EK, and Yuille AL, “A fixed-point model for pancreas segmentation in abdominal CT scans,” MICCAI, pp. 693–701, 2017. [Google Scholar]
- [60].Cai J, Lu L, Xie Y, Xing F, and Yang L, “Improving deep pancreas segmentation in CT and MRI images via recurrent neural contextual learning and direct loss function,” arXiv preprint arXiv:1707.04912, 2017. [Google Scholar]
- [61].Sato Y, Nakajima S, Atsumi H, Koller T, Gerig G, Yoshida S, and Kikinis R, “3D multi-scale line filter for segmentation and visualization of curvilinear structures in medical images,” CVRMed-MRCAS, pp. 213–222, 1997. [DOI] [PubMed] [Google Scholar]
- [62].Dmitriev K, Gutenko I, Nadeem S, and Kaufman A, “Pancreas and cyst segmentation,” SPIE Medical Imaging, 9784:97842C, 2016. [Google Scholar]
- [63].Bitter I, Kaufman AE, and Sato M, “Penalized-distance volumetric skeleton algorithm,” IEEE TVCG, 7(3):195–206, 2001. [Google Scholar]
- [64].Kohlmann P, Bruckner S, Kanitsar A, and Gröller ME, “Contextual picking of volumetric structures,” IEEE Pacific Visualization Symposium, pp. 185–192, 2009. [Google Scholar]
- [65].Mirhosseini S, Gutenko I, Ojal S, Marino J, and Kaufman A, “Immersive virtual colonoscopy,” IEEE TVCG, 25(5):2011–21, 2019. [DOI] [PubMed] [Google Scholar]
- [66].Kindlmann G, Whitaker R, Tasdizen T, and Moller T, “Curvature-based transfer functions for direct volume rendering: Methods and applications,” IEEE VIS, pp. 513–520, 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


