Abstract
Accurate assessment and monitoring of the Plasmodium falciparum Kelch 13 (pfk13) gene associated with artemisinin resistance is critical to understand the emergence and spread of drug-resistant parasites in malaria-endemic regions. In this study, we evaluated the genomic profile of the pfk13 gene associated with artemisinin resistance in P. falciparum in Nigerian children by targeted sequencing of the pfk13 gene. Genomic DNA was extracted from 332 dried blood (DBS) spot filter paper samples from three Nigerian States. The pfk13 gene was amplified by nested polymerase chain reaction (PCR), and amplicons were sequenced to detect known and novel polymorphisms across the gene. Consensus sequences of samples were mapped to the reference gene sequence obtained from the National Center for Biotechnology Information (NCBI). Out of the 13 single nucleotide polymorphisms (SNPs) detected in the pfk13 gene, five (F451L, N664I, V487E, V692G and Q661H) have not been reported in other endemic countries to the best of our knowledge. Three of these SNPs (V692G, N664I and Q661H) and a non-novel SNP, C469C, were consistent with late parasitological failure (LPF) in two States (Enugu and Plateau States). There was no validated mutation associated with artemisinin resistance in this study. However, a correlation of our study with in vivo and in vitro phenotypes is needed to establish the functional role of detected mutations as markers of artemisinin resistance in Nigeria. This baseline information will be essential in tracking and monitoring P. falciparum resistance to artemisinin in Nigeria.
Introduction
Malaria, one of the major global health challenges, has co-existed with humans for over 40 centuries. Control of this ancient disease heavily relies upon the use of antimalarial drugs [1]. Artemisinin-based combination therapies (ACTs) [e.g., artemether-lumefantrine (AL) and artesunate-amodiaquine (AA)] are the current line of treatment for malaria. These drugs were recommended by the World Health Organization (WHO) as first-line treatment for uncomplicated falciparum malaria in 2001 and have since been widely adopted to treat falciparum malaria globally [2]. Artemisinin (ART)-resistant P. falciparum has been confirmed to have emerged from the Greater Mekong Subregion (GMS) Thai-Cambodian border [3, 4] and has spread to other malaria endemic regions [5–8].
Several point mutations in the pfk13 gene (F446I, N458Y, M476I, Y493H, R539T, I543T, P553L, R561H, P574L and C580Y) have been validated to correlate with clinical ART resistance in Southeast Asia and South America [2, 9] and confirmed to confer elevated survival rates based on Ring-stage Survival Assays (RSA)0–3 h [10–12]. Furthermore, mutations and increased copy numbers of genes such as P. falciparum multidrug resistant gene-1 (pfmdr1) and P. falciparum chloroquine resistance transporter (pfcrt) gene have been linked to resistance in artemisinin (and derivatives) partner drugs such as lumefantrine, amodiaquine, mefloquine and piperaquine [13–18]. Due to these mutations, the efficacy of ACTs may be compromised [19]. Recently for the first time in Africa, two of the validated mutations (R561H and P574L) on the pfk13 gene have been reported in Rwanda to be associated with in vitro resistance to ACTs [7, 8]. However, in other sub-Saharan African countries such as Nigeria, no association between ART-resistant parasites and the ten validated mutations has been found [20–23].
Despite the need to regularly survey for the emergence of these pfk13 mutant alleles in different malaria-endemic regions of Nigeria, only a small number of systematic molecular epidemiological studies on field P. falciparum isolates have so far been conducted [2]. A few studies have reported cases with delayed response to ACTs [22, 23], and a sporadic scan for amino acid mutations in the pfk13 gene identified three nonsynonymous mutations (G592R, Q613H, and G665S) and other synonymous mutations [9].
This study aims to describe the potential emergence and spread of ART-resistance alleles in the pfk13 gene in three Nigerian States. The study was designed to inform malaria policymakers and research scientists in accordance with the objectives of the therapeutic efficacy study (TES) of the National Malaria Elimination Program (NMEP) of the Federal Republic of Nigeria recommended by the WHO.
Materials and methods
Study site
This is a retrospective, cross-sectional, community-based study which is part of the TES for monitoring antimalarial efficacies of Artesunate-amodiaquine (AA), Artemether-lumefantrine (AL) and Dihydroartemisinin-piperaquine (DHP) in the treatment of uncomplicated P. falciparum infections in children aged 6–96 months old in Nigeria. A cohort of 586 children was enrolled from three sentinel sites of the 2018 antimalarial drug efficacy testing and monitoring of the NMEP of the Federal Ministry of Health, namely; Kura (n = 200), Barkin Ladi (n = 185) and Agbani (n = 201), in Kano, Plateau and Enugu States respectively. Full description of study sites is available online at https://www.health.gov.ng/doc/2018-TES-FINAL-REPORT.pdf
Enrolment and sample collection
Children were enrolled if they were 6–96 months old. Children were eligible for enrolment if they had symptoms compatible with acute uncomplicated malaria, P. falciparum mono-infection with parasite count ranging from 2,000–200,000 asexual forms/μl by microscopy and body (axillary) temperature of ≥ 37.5 °C or a history of fever in the 24 hours preceding presentation. Children with severe malaria, severe malnutrition, serious underlying diseases (renal, cardiac, or hepatic diseases), and known allergies to the study drugs were excluded. Follow-up clinical and parasitological evaluations were done daily on days 1 to 3 and then on days 7, 14, 21, 28, 35, and 42.
Two to three drops of finger-pricked blood samples were blotted on 3mm Whatman filter paper (Whatman International Limited, Maidstone, United Kingdom) before treatment initiation (Day 0) and post-treatment initiation on days 7, 14, 21, 28, 35 and 42. Blood samples impregnated on filter papers were allowed to air-dry appropriately at room temperature and kept in airtight envelopes with silica gel at room temperature until analysed. A total of 586 DBS filter paper samples from children were sent to the African Centre of Excellence for Genomics of Infectious Diseases (ACEGID), Redeemer’s University, for molecular analysis. Samples were collected during the intense malaria season (August-October) of 2018.
Ethical declaration
The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the National Health Research Ethics Committee, Federal Ministry of Health (FMOH), Abuja, Nigeria. Written informed consent was obtained from parents/guardians for children prior to enrollment in this study. Child assent was obtained from children aged 84–96 months.
Assessment of treatment outcome
Response to drug treatment was evaluated using the following treatment indices:
Adequate clinical and parasitological response (ACPR), early treatment failure (ETF), late clinical failure (LCF) and late parasitological failure (LPF) [Full description of WHO TES study protocol available at https://www.who.int/docs/default-source/documents/publications/gmp/methods-for-surveillance-of-antimalarial-drug-efficacy.pdf?sfvrsn=29076702_2].
Asexual parasite reduction ratio (PRR) one or two days after treatment initiation (PRRD1 or PRRD2) defined as the ratio of asexual parasitaemia pre-treatment initiation and that on day one or two, respectively.
Asexual parasite positivity on day 1 (APPD1), 2 (APPD2) or 3 (APPD3) is defined as the proportion of children with residual asexual parasitaemia one, two or three days after treatment initiation, respectively Asexual parasite clearance time (PCT) defined as the time elapsing between drug administration and absence of microscopic detection of viable asexual parasitaemia.
Parasite genomic DNA extraction
A total of 300 pre-treatment (Day 0) DBS filter paper samples from all three sites were utilised in this study. Of the 300 samples, 32 had LPF making a final total of 332 samples selected for analysis. Parasite genomic DNA was extracted from 332 DBS samples using the Zymo Quick-DNA Miniprep Plus kit (17062 Murphy Avenue Irvine, California 192614, United States of America) according to the manufacturer’s protocol.
Amplification of pfk13 gene and targeted sequencing
The pfk13 propeller domain was amplified by nested PCR using PuReTaq Ready-To-Go PCR beads (GE Healthcare UK Limited) according to the manufacturer’s protocol. We used primers kelch-outer-F 5′-gggaatctggtggtaacagc-3′ and kelch-outer-R 5′-cggagtgaccaaatctggga- 3′ for the primary PCR, and kelch-inner-F 5′-gccttgttgaaagaagcaga-3′ and kelch-inner-R 5′- gccaagctgccattcatttg-3′ for the nested PCR. The nested PCR product was 849 bp and corresponds to nucleotide sequence 1279–2127 (representing codons 427–709) of PF3D7_1343700 K13 propeller domain, which included mutations correlated with delayed parasite clearance [10].
The ready-to-go PCR beads were reconstituted to a final volume of 20 μL in the primary reaction, 2 μL of DNA was amplified with 0.5 μM of each primer. Cycling conditions were 95 °C for 1 minute, followed by 35 cycles at 95°C for 20 seconds, 58 °C for 20 seconds, and 60 °C for 1 minute, with a final extension at 60 °C for 3 minutes.
One microlitre of the primary reaction was further amplified with 0.5 μM of each primer in the nested PCR reaction. Cycling conditions were 95 °C for 1 minute, followed by 35 cycles at 95 °C for 20 seconds, 56 °C for 20 seconds, and 60 °C for 1 minute, with a final extension at 60 °C for 3 minutes. Nested amplicons were analysed by electrophoresis on a 2% agarose gel to confirm amplification. We purified the nested amplicons using ExoSAP-IT (Affymetrix, Santa Clara, CA, USA), and we sequenced them using BigDye Terminator v.1.1 (Life Technologies, Carlsbad, CA, USA) and the same primers as for nested PCR (kelch-inner-F and kelch-inner-R). We carried out sequencing using an Applied Biosystems 3500 XL series Genetic Analyser at ACEGID, Redeemer’s University, Ede, Osun State, Nigeria.
Data analysis
Identification of polymorphisms
Chromatograms of the individual sequences were viewed using Geneious v2020.0.4 [24, 25] and manual base calling was carried out as needed for some of the sequences. Consensus sequences were generated for all samples. The consensus sequences were generated by first aligning the forward and reverse reads for individual samples using Geneious alignment. A consensus was reached from the resulting alignment by choosing the highest quality base and also carrying out manual base calling for regions of ambiguities. To detect polymorphisms in the sequences encoding the pfk13 propeller gene, we obtained the reference nucleotide sequence of the pfk13 gene from the NCBI database (PF3D7_1343700 sequence region spanning region 1,724,817–1,726,997 bp of chromosome 13). We mapped the consensus sequences of the samples to the reference gene sequence using Geneious v2020.0.4.
Statistical analysis of treatment outcome
Discrete variables (such as proportions of frequencies) were compared by calculating χ2 using Yates’ correction, Fisher’s exact or Mantel Haenszel tests. Normally distributed, continuous data were analysed by Student’s t-test or analysis of variance (ANOVA) as it is applicable. Mann–Whitney U tests or Kruskal Wallis tests (or by Wilcoxon ranked sum test) was used to compare data that did not conform to normal distribution. P values of <0.05 were taken to indicate significant differences.
Results
Demographics
Of the 300 participants’ samples analysed, 165 (55%) were male. The mean age of all children was 58.42±25.71 months (95% confidence interval (CI) 55.71–61.52). The geometric mean of the asexual parasitemia was 17,765 μL-1 (95%CI 15498–20363) (Table 1). Children enrolled in Kano State were significantly (p<0.0001) younger and had significantly (p = 0.046) lower enrollment asexual parasitaemia compared to children enrolled in other States (Table 1).
Table 1. Demographic characteristics of children at enrollment.
| Parameter | Enugu | Kano | Plateau | All | P value |
|---|---|---|---|---|---|
| n | 100 | 100 | 100 | 300 | |
| Gender | |||||
| M: F | 55:45 | 60:40 | 50:50 | 165:135 | 0.36 |
| Age (Months) | |||||
| Mean | 57.69 | 51.7 | 66.54 | 58.62 | <0.0001 |
| 95% CI | 52.33–63.05 | 46.6–56.8 | 62.24–70.85 | 55.71–61.52 | |
| ≤ 60months | 60 | 70 | 47 | 177 | 0.004 |
| Asexual Parasitemia | |||||
| Geometric mean | 17098 | 14526 | 22984 | 17765 | 0.046 |
| 95% CI | 13060–22384 | 12130–17395 | 17801–29676 | 15498–20363 | |
| ≥ 100,000 | 10 | 3 | 16 | 29 | 0.004 |
Study treatment outcome
The overall ACPR_c values for AA, AL and DHP in Enugu, Kano and Plateau States were 100%, 99.3% and 100% respectively. The ACPR_c values for AA in Kano and Plateau States were 100% respectively while the ACPR_c values for AL in Enugu, Kano and Plateau States were 98%, 100% and 100% respectively. Only Enugu State tested DHP and recorded an ACPR_c value of 100%. Of the 32 presented with LPF, 14 occurred in Enugu, 11 in Kano and 8 in Plateau State (Table 2).
Table 2. Summary of treatment outcome by State and drug.
| State | Treatment Outcome | Drug | Total | ||
|---|---|---|---|---|---|
| AA | AL | DHP | |||
| Enugu | Number of samples | 50 | 50 | 100 | |
| ETF | 0 | 0 | 0 | ||
| LCF | 0 | 0 | 0 | ||
| LPF | 11 | 2 | 14 | ||
| ACPR_u | 39 | 48 | 87 | ||
| ACPR_c | 49 | 50 | 99 | ||
| %ACPR_c | 98 | 100 | 99 | ||
| Kano | Number of samples | 50 | 50 | 100 | |
| ETF | 0 | 0 | 0 | ||
| LCF | 0 | 0 | 0 | ||
| LPF | 2 | 9 | 11 | ||
| ACPR_u | 48 | 41 | 89 | ||
| ACPR_c | 50 | 50 | 100 | ||
| %ACPR_c | 100 | 100 | 100 | ||
| Plateau | Number of samples | 50 | 50 | 100 | |
| ETF | 0 | 0 | 0 | ||
| LCF | 0 | 0 | 0 | ||
| LPF | 1 | 7 | 8 | ||
| ACPR_u | 49 | 43 | 92 | ||
| ACPR_c | 50 | 50 | 100 | ||
| %ACPR_c | 100 | 100 | 100 | ||
| Total | Number of samples | 100 | 150 | 50 | 300 |
| ETF | 0 | 0 | 0 | 0 | |
| LCF | 0 | 0 | 0 | 0 | |
| LPF | 3 | 27 | 2 | 32 | |
| ACPR_u | 97 | 123 | 47 | 267 | |
| ACPR_c | 100 | 149 | 50 | 299 | |
| %ACPR_c | 100 | 99.3 | 100 | 99.7 | |
Crude ACPR (ACPR_u); PCR-corrected ACPR (ACPR_c).
WHO protocol for parasite genotyping to differentiate recrudescence from new infections available at http://whqlibdoc.who.int/publications/2008/9789241596305_eng.pdf.
Pfk13 gene mutations
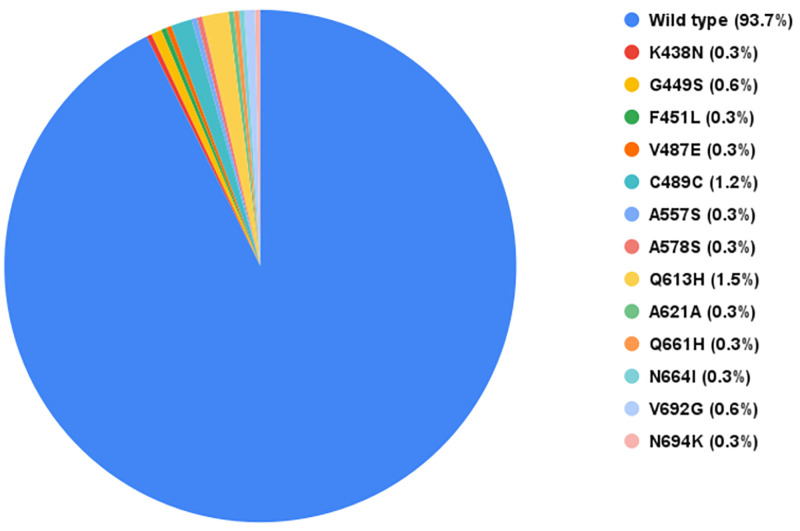
We amplified and sequenced the pfk13 gene from all the 332 samples (300 pre-treatment samples and 32 LPF samples). Thirteen pfk13 gene mutations were detected in 21 out of the 332 sequences analysed in this study (Table 3). The prevalence of parasites with mutations on chromosome 13 in the propeller region of the pfk13 protein was 6.3%, 93.7% did not have mutations (Fig 1). The highest occurring mutation Q613H (1.5%) was detected in five samples (three from Plateau State and two from Enugu State). The C469C mutation with 1.2% was detected in four samples from Enugu State. Mutations V692G and G449S were detected in 0.6% of samples (G449S in Kano State; V692G in Enugu and Plateau States). We summarized mutations detected in this study and previous Nigerian studies in Table 4.
Table 3. Pfk13 polymorphisms observed in Enugu, Kano and Plateau States.
| S/N | K13 Amino Acid Locus | Nucleotide Locus | Reference Allele | Mutant Allele | Mutation Type | Country Previously Observed | Nigerian State Observed |
|---|---|---|---|---|---|---|---|
| 1. | K438N | 1314 | A | T | Non-synonymous | SE Asian Countries | Kano |
| 2. | G449S | 1345 | G | A | Non-synonymous | Mali | Kano |
| 3. | F451L | 1353 | T | - | Non-synonymous | - | Kano |
| 4. | C469C | 1407 | C | T | Synonymous | Kenya, Malawi, Senegal, Niger, Congo, DRC | Enugu |
| 5. | V487E | 1460 | T | A | Non-synonymous | - | Enugu |
| 6. | A557S | 1669 | G | T | Non-synonymous | Congo, DRC, Côte d’Ivoire | Enugu |
| 7. | A578S | 1732 | G | T | Non-synonymous | Uganda, Kenya, DRC, Gabon, Mali, Ghana, Cameroon, Cambodia, India | Enugu |
| 8. | Q613H | 1839 | A | T | Non-synonymous | Senegal, Ghana, Tanzania | Enugu & Plateau |
| 9. | A621A | 1863 | T | A | Synonymous | - | Enugu |
| 10. | Q661H | 1983 | A | - | Non-synonymous | - | Enugu |
| 11. | N664I | 1991 | A | T | Non-synonymous | - | Plateau |
| 12. | V692G | 2075 | T | G | Non-synonymous | - | Enugu & Plateau |
| 13. | N694K | 2082 | T | A | Non-synonymous | Angola, Cote d’Ivoire | Kano |
Fig 1. A chart showing the frequency distribution of pfk13 gene polymorphisms observed in this study.
Table 4. Molecular surveillance of Pfk13 propeller polymorphisms in Nigeria.
| S/N | Mutations Previously observed | Year observed | Reference |
|---|---|---|---|
| 1 | H136N | 2015, 2016 | [26] |
| 2 | K189T | 2015, 2016 | [26] |
| 3 | E433G | 2018 | [27] |
| 4 | F434I | 2018 | [27] |
| 5 | F434S | 2018 | [27] |
| 6 | K438N | 2018 | Observed in this study |
| 7 | P441S | 2017, 2018 | [28] |
| 8 | F442F | 2018 | [27] |
| 9 | G449S | 2018 | Observed in this study |
| 10 | F451L | 2018 | Observed in this study |
| 11 | D464N | 2010, 2011 | [29] |
| 12 | C469C* | 2017 | [28] |
| 13 | V487E | 2018 | Observed in this study |
| 14 | F492F | 2018 | [27] |
| 15 | G496G | 2014 | [30] |
| 16 | V510V | 2016 | [20] |
| 17 | P553P | 2016 | [20] |
| 18 | A557S | 2018 | Observed in this study |
| 19 | A578S* | 2010, 2011 | [29] |
| 20 | V589V | 2016 | [28] |
| 21 | K610R | 2014 | [30] |
| 22 | Q613H* | 2010, 2011, 2015, 2016 | [29, 26] |
| 23 | A621A* | 2014 | [31] |
| 24 | A626T | 2014 | [30] |
| 25 | A627A | 2014 | [30] |
| 26 | V650F | 2016 | [28] |
| 27 | Q661H | 2018 | Observed in this study |
| 28 | N664I | 2018 | Observed in this study |
| 29 | N664N | 2017 | [28] |
| 30 | G665C | 2016 | [20] |
| 31 | V666V | 2016 | [20] |
| 32 | A676A | 2016 | [28] |
| 33 | I684N | 2018 | [27] |
| 34 | I684T | 2018 | [27] |
| 35 | E688K | 2018 | [27] |
| 36 | V692G | 2018 | Observed in this study |
| 37 | N694K | 2018 | Observed in this study |
*Mutations observed in previous studies and our study.
Treatment outcome of children infected with mutated P. falciparum
Characteristics of the children with P. falciparum with pfk13 mutations are shown in Table 5. Children infected with the mutated parasites are relatively older children (mean age: 53.7 ± 22 months, 14 of 21 were aged ≥48 months). They were also characterized by low enrollment asexual parasitaemia (geometric mean parasitaemia: 17,530 μL-1; 16 of 21 children had enrollment asexual parasitaemia < 50,000μL-1), relatively slow clearance time (two-third of the cohort cleared parasitaemia by after day 1). Following artemisinin-based combination treatment of the uncomplicated infection, 7 of 21 children (33%) had recurrent parasitaemia within 21–42 days of follow-up post treatment initiation (mean time to recurrence: 29±6.3 days), of these seven, only one child from Enugu State had recurrent parasitaemia due to recrudescence while four children from Enugu State and two from Plateau State had recurrent parasitaemia due to reinfection. It appears LPF was consistent with C469C (predominant in Enugu State) and V692G (observed in Enugu and Plateau States). Also, the Q613H mutation occurring in these States had ACPR phenotype.
Table 5. Clinical features of children with mutated P falciparum in this study.
| Study site | Sample ID | Gender | Age (Month) | Enrollment asexual parasitaemia (uL-1) | Mutation Position | Parasite clearance time (day) | Antimalarial Treatment | Treatment outcome |
|---|---|---|---|---|---|---|---|---|
| Enugu | 33 | M | 48 | 30571 | A578S | 1 | AL | ACPR |
| 89 | F | 60 | 12817 | Q613H | 2 | AL | ACPR | |
| 106 | F | 36 | 16290 | Q613H | 2 | AL | ACPR | |
| 134 | M | 72 | 57850 | A621A | 2 | AL | ACPR | |
| 154 | M | 60 | 3424 | V487E | 2 | AL | ACPR | |
| 179 | M | 36 | 61320 | A557S | 2 | DHP | ACPR | |
| 197 | M | 48 | 2055 | C469C | 2 | DHP | LPF (D42) | |
| 228+ | F | 24 | 91680 | C469C | 2 | AL | LPF (D21) | |
| 236 | M | 36 | 9794 | C469C | 2 | AL | LPF (D28) | |
| 239 | M | 72 | 6714 | V692G | 1 | AL | LPF (D28) | |
| Q661H | ||||||||
| 254 | M | 96 | 29229 | C469C | 2 | AL | LPF (D28) | |
| Kano | ||||||||
| 14 | M | 12 | 15923 | N694K | 2 | AA | ACPR | |
| 26 | F | 24 | 37880 | F451L | 1 | AL | ACPR | |
| 51 | F | 84 | 26523 | K438N | 2 | AA | ACPR | |
| 59 | M | 36 | 16888 | G449S | 2 | AL | ACPR | |
| 77 | M | 48 | 2250 | G449S | 1 | AA | ACPR | |
| Plateau | 31 | F | 84 | 26069 | Q613H | 1 | AA | ACPR |
| 58 | M | 60 | 16304 | Q613H | 1 | AL | ACPR | |
| 62 | F | 72 | 61080 | Q613H | 2 | AL | ACPR | |
| 77 | F | 60 | 5428 | N664I | 1 | AL | LPF (D28) | |
| 155 | M | 60 | 54739 | V692G | 2 | AA | LPF (D28) | |
| Mean (SD) | 53.71 (22) | 17, 530* | 1.67 (0.48) | 29 (6.3)# |
*Geometric mean;
#Mean LPF;
+Only sample with recrudescence recurrent parasitaemia.
Comparison of responsiveness indices following treatment initiation in children with and without mutated Pfk13
As a result of the small number of samples with mutations in our study (n = 21), samples with mutation were matched for age, gender, enrolment asexual parasitaemia, same day presentation and same treatment with those without. Following treatment intiation, treatment indices such as APPD1-2 and PRRD1-2 were similar in the 2 groups (P>0.2). None of the children with mutant parasites had persistent asexual parasitaemia two or three days after treatment initiation. In comparison, 28.6% (6 of 21) of children and 4.8% (1 of 21) of children of the cohort infected with non-mutant parasites had persistent asexual parasitaemia two and three days after treatment initiation respectively. PCT was significantly longer in children infected with non-mutant parasites than those infected with mutant parasites (2.3±1.2 days versus 1.7±0.5 days, respectively; P = 0.03).
Discussion
The knowledge of mutations in the pfk13 gene associated with slow clearance of artemisinin derivatives provides the ability to track the emergence and prevent the spread of resistant parasites and assess the effectiveness of control measures.
In this study, we detected a total of 13 SNPs (out of which 11 were non-synonymous) in the pfk13 gene of P falciparum obtained from Nigerian children. None of them was among the ten SNPs (F446I, N458Y, M476I, Y493H, R539T, I543T, P553L, R561H, P574L and C580Y) which have been validated to be associated with artemisinin resistance in Southeast Asia, South America and Rwanda [2, 6–9]. The absence of the validated mutations in our study is in line with many recent studies carried out in Africa [32–40] and in Nigeria [20, 27, 29, 34, 41, 42].
In addition, eight (A578S, C469C, Q613H, K438N, A621A, N694K, G499S and A557S) of the 13 SNPs observed in our study have also been observed in Southeast Asia and other sub-Saharan African countries [10, 28, 30, 31, 33, 34, 36, 39, 40, 43–47] as represented in Table 4. The Q613H was the most prominent SNP observed in our study and has been reported in recent studies in Nigeria [26, 29]. However, A578S is the most prominent SNP observed among sub-Saharan African countries like Ghana, Kenya, Gabon, DRC, Uganda, Cameroon and Mali [34, 39, 46]. This mutation (A578S) represents a change from a non-polar to a polar amino acid and can alter the shape of the pfk13 protein in the regions it has been observed in [35]. Therefore, more attention must be paid to this mutation as it is emerging to be the most prominent mutation observed in the pfk13 gene in sub-Saharan Africa [34, 39, 36].
We detected five novel SNPs (F451L, N664I, V487E, V692G and Q661H), which to the best of our knowledge, have not yet been described elsewhere. Three of these SNPs (V692G, N664I and Q661H) and a non-novel SNP, C469C were associated with LPF in two States (Enugu and Plateau States) (Table 5). More studies would have to be carried out to understand these mutations better and unravel their effects or importance.
Data reported in this study suggests that all the thirteen (13) mutations observed in our study are less likely to be associated with a delayed parasite clearance phenotype as evidenced by similar parasite reduction ratios and proportion with persistent asexual parasitaemia following treatment initiation. In addition, significantly longer asexual PCT in children infected with non-mutant pfk13 parasites indicate that the mutants identified in the parasites circulating in Nigeria do not confer resistance to artemisinin derivatives.
As some of these mutations seem to be indigenous to African parasites, it is imperative that in vivo and in vitro studies are conducted to validate their possible roles in the emergence and spread of ART reduced susceptibility/resistance in Nigeria and sub-Saharan Africa as a whole as observed in previous studies [48, 49].
Although these polymorphisms reported in this study have not been associated with ART resistance, recent studies have shown that two of the validated mutations (R561H and P574L) associated with ART-resistance have been observed in Rwanda, a country which like Nigeria is in the Sub-saharan African Region [7, 8]. This is worrisome because it is only a matter of time before such mutant malaria parasites are introduced into Nigeria due to migration patterns between these two countries. With the emergence of these mutations in the pfk13 gene, there is a need for constant and routine monitoring to avoid bad surprises and to have in place control strategies should resistant parasites emerge.
Conclusion
There was no validated mutation associated with ART resistance in this study. However, we observed novel and other established mutations reported to be circulating in Nigeria and other African countries. Correlation of mutation data obtained in this study with in vivo and in vitro phenotypes is needed to establish the functional role of detected mutations as markers of artemisinin resistance in Nigeria.
Acknowledgments
The authors thank all the patients, their parents or guardians for volunteering to participate in the study. We acknowledge technical support from colleagues at the ACEGID. We also acknowledge the principal investigators (PIs) in each of the three sentinel locations considered in this study and the National Malaria Elimination Program of the Federal Ministry of Health in Nigeria.
Data Availability
All sequences from this study are available on GenBank with accession numbers MT113570 - MT113901.
Funding Statement
This work is made possible by support from Flu Lab and a cohort of generous donors through TED’s Audacious Project, including the ELMA Foundation, MacKenzie Scott, the Skoll Foundation, and Open Philanthropy. This work was supported by grants from the National Institute of Allergy and Infectious Diseases (https://www.niaid.nih.gov), NIH-H3Africa (https://h3africa.org) (U01HG007480 and U54HG007480 to C.T.H), the World Bank grant (worldbank.org) (ACE IMPACT project) to C.T.H. The U.S President’s Malaria Initiative (USPMI) funded the primary drug efficacy study from which samples were obtained for the current study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kim Y, Schneider KA. Evolution of Drug Resistance in Malaria Parasite Populations. Nat Ed Knowledge 2013;4(8): 6. [Google Scholar]
- 2.World Health Organization, Report on antimalarial drug efficacy, resistance and response: 10years of surveillance (2010–2019), World Health Organization, 2020.
- 3.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361(5): 455–67. doi: 10.1056/NEJMoa0808859 Erratum in: N Engl J Med. 2009;361(17): 1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Artemisinin Resistance in Cambodia 1 (ARC1) Study Consortium. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008. Dec 11;359(24): 2619–20. doi: 10.1056/NEJMc0805011 [DOI] [PubMed] [Google Scholar]
- 5.Mishra N, Bharti RS, Mallick P, Singh OP, Srivastava B, Rana R, et al. Emerging polymorphisms in falciparum Kelch 13 gene in Northeastern region of India. Malar J. 2016. Dec 3;15(1):583. doi: 10.1186/s12936-016-1636-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathieu LC, Cox H, Early AM, Mok S, Lazrek Y, Paquet JC, et al. Local emergence in Amazonia of Plasmodium falciparum k13 C580Y mutants associated with in vitro artemisinin resistance. Elife. 2020. May 12;9: e51015. doi: 10.7554/eLife.51015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uwimana A, Legrand E, Stokes BH, Ndikumana JM, Warsame M, Umulisa N, et al. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat Med. 2020. Oct;26(10): 1602–1608. doi: 10.1038/s41591-020-1005-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uwimana A, Legrand E, Stokes BH, Ndikumana JM, Warsame M, Umulisa N, et al. Author Correction: Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat Med. 27(6): 1113–1115. doi: 10.1038/s41591-021-01365-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daily JP. K13-Propeller Mutations and Malaria Resistance. N Engl J Med. 2016. Jun 23;374(25): 2492–3. doi: 10.1056/NEJMe1604520 [DOI] [PubMed] [Google Scholar]
- 10.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014. Jan 2;505(7481): 50–5. doi: 10.1038/nature12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol. 2014. Aug;32(8): 819–21. doi: 10.1038/nbt.2925 [DOI] [PubMed] [Google Scholar]
- 12.Straimer J, Gnädig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, et al. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015. Jan 23;347(6220): 428–31. doi: 10.1126/science.1260867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000. Feb 24;403(6772): 906–909. doi: 10.1038/35002615 [DOI] [PubMed] [Google Scholar]
- 14.Sanchez CP, Rotmann A, Stein WD, Lanzer M. Polymorphisms within PfMDR1 alter the substrate specificity for anti-malarial drugs in Plasmodium falciparum. Mol Microbiol. 2008. Nov;70(4): 786–98. doi: 10.1111/j.1365-2958.2008.06413.x [DOI] [PubMed] [Google Scholar]
- 15.Veiga MI, Dhingra SK, Henrich PP, Straimer J, Gnädig N, Uhlemann AC, et al. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun. 2016. May 18;7: 11553. doi: 10.1038/ncomms11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, et al. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis. 2016. Mar;16(3): 357–65. doi: 10.1016/S1473-3099(15)00487-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mbaye A, Dieye B, Ndiaye YD, Bei AK, Muna A, Deme AB, et al. Selection of N86F184D1246 haplotype of Pfmrd1 gene by artemether-lumefantrine drug pressure on Plasmodium falciparum populations in Senegal. Malar J. 2016. Aug 25;15(1): 433. doi: 10.1186/s12936-016-1490-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kayode A, Ajogbasile F, Akano K, Uwanibe J, Oluniyi P, Eromon P, et al. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter (Pfcrt) and multidrug-resistant gene 1 (Pfmdr-1) in Nigerian children 10 years post-adoption of artemisinin-based combination treatments. International Journal for Parasitology. 2021. March; 51(4): 301–310. doi: 10.1016/j.ijpara.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization, Status report on Artemisinin and artemisinin-based combination therapy resistance (2017), World Health Organization, 2017.
- 20.Oboh MA, Ndiaye D, Antony HA, Badiane AS, Singh US, Ali NA, et al. Status of Artemisinin Resistance in Malaria Parasite Plasmodium falciparum from Molecular Analyses of the Kelch13 Gene in Southwestern Nigeria. Biomed Res Int. 2018. Oct 3; 2018:2305062. doi: 10.1155/2018/2305062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madamet M, Kounta MB, Wade KA, Lo G, Diawara S, Fall M, et al. Absence of association between polymorphisms in the K13 gene and the presence of Plasmodium falciparum parasites at day 3 after treatment with artemisinin derivatives in Senegal. Int J Antimicrob Agents. 2017. Jun;49(6): 754–756. doi: 10.1016/j.ijantimicag.2017.01.032 [DOI] [PubMed] [Google Scholar]
- 22.Ajayi NA, Ukwaja KN. Possible artemisinin-based combination therapy-resistant malaria in Nigeria: a report of three cases. Rev Soc Bras Med Trop. 2013. Jul-Aug;46(4): 525–7. doi: 10.1590/0037-8682-0098-2013 [DOI] [PubMed] [Google Scholar]
- 23.Wundermann GS, Osiki A. Currently Observed Trend in the Resistance of Malaria to Artemisinin Based Combination Therapy in Nigeria—A Report of 5 Cases. IJTDH [Internet]. 3 Feb. 2017. [cited 18Apr.2021];21(2):1–5. Available from: https://www.journalijtdh.com/index.php/IJTDH/article/view/20337. [Google Scholar]
- 24.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012. Jun 15;28(12): 1647–9. doi: 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geneious Prime 2020.0.4. www.geneious.com.
- 26.Idowu AO, Oyibo WA, Bhattacharyya S, Khubbar M, Mendie UE, Bumah VV, et al. Rare mutations in Pfmdr1 gene of Plasmodium falciparum detected in clinical isolates from patients treated with anti-malarial drug in Nigeria. Malar J. 2019. Sep 18;18(1): 319. doi: 10.1186/s12936-019-2947-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abubakar UF, Adam R, Mukhtar MM, Muhammad A, Yahuza AA, Ibrahim SS. Identification of Mutations in Antimalarial Resistance Gene Kelch13 from plasmodium falciparum Isolates in Kano, Nigeria. Trop Med Infect Dis. 2020. May 27;5(2): 85. doi: 10.3390/tropicalmed5020085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Ruan W, Zhou S, Huang F, Lu Q, Feng X, et al. Molecular surveillance of Pfcrt and k1 propeller polymorphisms of imported Plasmodium falciparum cases to Zhejiang Province, China between 2016 and 2018. Malar J. 2020. Feb 4;19(1):59. doi: 10.1186/s12936-020-3140-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Igbasi U, Oyibo W, Omilabu S, Quan H, Chen SB, Shen HM, et al. Kelch 13 propeller gene polymorphism among Plasmodium falciparum isolates in Lagos, Nigeria: Molecular Epidemiologic Study. Trop Med Int Health. 2019. Aug;24(8): 1011–1017. doi: 10.1111/tmi.13273 [DOI] [PubMed] [Google Scholar]
- 30.Yang C, Zhang H, Zhou R, Qian D, Liu Y, Zhao Y, et al. Polymorphisms of Plasmodium falciparum k13-propeller gene among migrant workers returning to Henan Province, China from Africa. BMC Infect Dis. 2017. Aug 10;17(1): 560. doi: 10.1186/s12879-017-2634-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prosser C, Meyer W, Ellis J, Lee R. Resistance screening and trend analysis of imported falciparum malaria in NSW, Australia (2010 to 2016). PLoS One. 2018. May 29;13(5): e0197369. doi: 10.1371/journal.pone.0197369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conrad MD, Bigira V, Kapisi J, Muhindo M, Kamya MR, Havlir DV, et al. Polymorphisms in K13 and falcipain-2 associated with artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan children. PLoS One. 2014. Aug 21;9(8): e105690. doi: 10.1371/journal.pone.0105690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torrentino-Madamet M, Fall B, Benoit N, Camara C, Amalvict R, Fall M, et al. Limited polymorphisms in k13 gene in Plasmodium falciparum isolates from Dakar, Senegal in 2012–2013. Malar J. 2014. Dec 4;13: 472. doi: 10.1186/1475-2875-13-472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamau E, Campino S, Amenga-Etego L, Drury E, Ishengoma D, Johnson K, et al. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis. 2015. Apr 15;211(8): 1352–5. doi: 10.1093/infdis/jiu608 Erratum in: J Infect Dis. 2016 Jan 15;213(2): 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouattara A, Kone A, Adams M, Fofana B, Maiga AW, Hampton S, et al. Polymorphisms in the K13-propeller gene in artemisinin-susceptible Plasmodium falciparum parasites from Bougoula-Hameau and Bandiagara, Mali. Am J Trop Med Hyg. 2015. Jun;92(6): 1202–6. doi: 10.4269/ajtmh.14-0605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, et al. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in Sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis. 2015. Mar 1;211(5): 680–8. doi: 10.1093/infdis/jiu467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eboumbou Moukoko CE, Huang F, Nsango SE, Kojom Foko LP, Ebong SB, Epee Eboumbou P, et al. K-13 propeller gene polymorphisms isolated between 2014 and 2017 from Cameroonian Plasmodium falciparum malaria patients. PLoS One. 2019. Sep 3;14(9): e0221895. doi: 10.1371/journal.pone.0221895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ocan M, Akena D, Nsobya S, Kamya MR, Senono R, Kinengyere AA, et al. K13-propeller gene polymorphisms in Plasmodium falciparum parasite population in malaria affected countries: a systematic review of prevalence and risk factors. Malar J. 2019. Mar 7;18(1): 60. doi: 10.1186/s12936-019-2701-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.WWARN K13 Genotype-Phenotype Study Group. Association of mutations in the Plasmodium falciparum Kelch13 gene (Pf3D7_1343700) with parasite clearance rates after artemisinin-based treatments-a WWARN individual patient data meta-analysis. BMC Med. 2019. Jan 17;17(1): 1. doi: 10.1186/s12916-018-1207-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaye A, Sy M, Ndiaye T, Siddle KJ, Park DJ, Deme AB, et al. Amplicon deep sequencing of kelch13 in Plasmodium falciparum isolates from Senegal. Malar J. 2020. Mar 30;19(1): 134. doi: 10.1186/s12936-020-03193-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dokunmu T, Olasehinde G, Oladejo D, Adjekukor C, Akinbohun A, Onileere O, et al. Evaluation of Plasmodium falciparum K13 gene polymorphism and susceptibility to dihydroartemisinin in an endemic area. BMRAT [Internet]. 2018. Sep 24;5(9): 2651–7. Available from: http://www.bmrat.org/index.php/BMRAT/article/view/474. [Google Scholar]
- 42.Oyebola KM, Aina OO, Idowu ET, Olukosi YA, Ajibaye OS, Otubanjo OA, et al. A barcode of multilocus nuclear DNA identifies genetic relatedness in pre- and post-Artemether/Lumefantrine treated Plasmodium falciparum in Nigeria. BMC Infect Dis. 2018. Aug 13;18(1): 392. doi: 10.1186/s12879-018-3314-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boussaroque A, Fall B, Madamet M, Camara C, Benoit N, Fall M, et al. Emergence of Mutations in the K13 Propeller Gene of Plasmodium falciparum Isolates from Dakar, Senegal, in 2013–2014. Antimicrob Agents Chemother. 2015. Oct 26;60(1): 624–7. doi: 10.1128/AAC.01346-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talundzic E, Ndiaye YD, Deme AB, Olsen C, Patel DS, Biliya S, et al. Molecular Epidemiology of Plasmodium falciparum kelch13 Mutations in Senegal Determined by Using Targeted Amplicon Deep Sequencing. Antimicrob Agents Chemother. 2017. Feb 23;61(3): e02116–16. doi: 10.1128/AAC.02116-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Li N, Siddiqui FA, Xu S, Geng J, Zhang J, et al. In vitro susceptibility of Plasmodium falciparum isolates from the China-Myanmar border area to artemisinins and correlation with K13 mutations. Int J Parasitol Drugs Drug Resist. 2019. Aug; 10: 20–27. doi: 10.1016/j.ijpddr.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Shi Y, Zhang W, Yan H, Lin K, Wei S, et al. K13-propeller gene polymorphisms of Plasmodium falciparum and the therapeutic effect of artesunate among migrant workers returning to Guangxi, China (2014–2017). Malar J. 2019. Oct 16;18(1): 349. doi: 10.1186/s12936-019-2984-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mensah BA, Aydemir O, Myers-Hansen JL, Opoku M, Hathaway NJ, Marsh PW, et al. Antimalarial Drug Resistance Profiling of Plasmodium falciparum Infections in Ghana Using Molecular Inversion Probes and Next-Generation Sequencing. Antimicrob Agents Chemother. 2020. Mar 24;64(4): e01423–19. doi: 10.1128/AAC.01423-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bustamante C, Folarin OA, Gbotosho GO, Batista CN, Mesquita EA, Brindeiro RM, et al. In vitro-reduced susceptibility to artemether in P. falciparum and its association with polymorphisms on transporter genes. J Infect Dis. 2012. Aug 1;206(3): 324–32. doi: 10.1093/infdis/jis359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mbaye A, Gaye A, Dieye B, Ndiaye YD, Bei AK, Affara M, et al. Ex vivo susceptibility and genotyping of Plasmodium falciparum isolates from Pikine, Senegal. Malar J. 2017. Jun 14;16(1): 250. doi: 10.1186/s12936-017-1897-6 [DOI] [PMC free article] [PubMed] [Google Scholar]