Abstract
Phytic acid, a phosphorylated derivative of myo-inositol, functions as the major storage form of phosphorus in plant seeds. Myo-inositol phosphates, including phytic acid, play diverse roles in plants as signal transduction molecules, osmoprotectants, and cell wall constituents. d-myo-inositol-3-phosphate synthase (MIPS EC 5.5.1.4) catalyzes the first step in de novo synthesis of myo-inositol. A soybean (Glycine max) MIPS cDNA (GmMIPS1) was isolated by reverse transcriptase-PCR using consensus primers designed from highly conserved regions in other plant MIPS sequences. Southern-blot analysis and database searches indicated the presence of at least four MIPS genes in the soybean genome. Northern-blot and immunoblot analyses indicated higher MIPS expression and accumulation in immature seeds than in other soybean tissues. MIPS was expressed early in the cotyledonary stage of seed development. The GmMIPS1 expression pattern suggested that it encodes a MIPS isoform that functions in seeds to generate d-myo-inositol-3-phosphate as a substrate for phytic acid biosynthesis.
Nutrient reserves accumulate during seed development for remobilization during germination and early seedling growth. Phosphorus in seeds is stored primarily in the form of phytic acid (phytate, myo-inositol hexakisphosphate, InsP6), which is a derivative of inositol. During seed development phytic acid is deposited in spherical inclusions known as globoids or as complexes with seed storage proteins in protein bodies (Prattley and Stanley, 1982; Lott et al., 1995). The stored phytate is hydrolyzed by the activity of phytase enzymes during germination to provide inorganic phosphate and myo-inositol to the growing seedling.
As a component of animal feed, phytate from seeds compromises the availability of dietary phosphorus because non-ruminants lack the digestive enzymes to hydrolyze phytic acid. The negatively charged phytic acid molecule also chelates other mineral nutrients, which lowers their bioavailablity. In addition to the nutritional effects of phytate in animal diets, there are also environmental consequences. Undigested phytic acid is excreted in manure, which leads to elevated soil phosphorus levels when manure is applied repeatedly as fertilizer in areas of livestock production. High soil phosphorus levels, coupled with the potential for run-off, can lead to environmental phosphorus pollution and eutrophication (Sharpley et al., 1994).
Myo-inositol is a precursor to compounds in plants that function not only in phosphorus storage, but also in signal transduction, stress protection, hormonal homeostasis, and cell wall biosynthesis (for review, see Morré et al., 1990; Loewus and Murthy, 2000). Production of the second messenger InsP3, a derivative of inositol, leads to release of intracellular calcium in cellular signaling. Methylated forms of myo-inositol, ononitol and pinitol, accumulate in plants in response to salinity stress to function in osmotic adjustment. Plant cells contain a number of conjugates of indole-3-acetic acid, including indole-3-acetic acid-myo-inositol, which allow storage of excess amounts of auxin. Oxidation of myo-inositol to d-GlcUA plays a role in the biosynthesis of compounds that are constituents of plant cells walls.
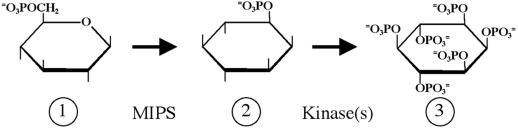
The first step in the synthesis of myo-inositol is the conversion of d-Glc-6-P to d-myo-inositol-3-phosphate by the isomerase d-myo-inositol-3-phosphate synthase (MIPS). The proposed phytic acid biosynthetic pathway (Loewus and Murthy, 2000) also includes phosphorylation steps catalyzed by one or more kinases that have not been well studied in plants (Fig. 1). The MIPS coding sequence has been cloned and characterized from a number of prokaryotic and eukaryotic sources. The first reported MIPS gene was isolated from Saccharomyces cerevisiae using a genetic complementation strategy (Johnson and Henry, 1989). A sequence similar to the yeast MIPS gene was identified in Spirodela polyrrhiza, an aquatic angiosperm (Smart and Fleming, 1993). MIPS sequences have been reported from Arabidopsis (Johnson, 1994), Citrus paradisii (Abu-Abied and Holland, 1994), common ice plant (Ishitani et al., 1996), and tobacco (Hara et al., 2000), and are highly conserved at the nucleotide level.
Figure 1.
Phytate biosynthesis. d-Glc-6-P (1) is converted to d-myo-inositol-3-phosphate (2) by the activity of MIPS. d-myo-inositol-3-phosphate is further phosphorylated to yield myo-inositol hexakisphosphate (3) by several kinase steps.
MIPS sequences comprise gene families in some plant species. In maize, seven sequences hybridizing to a MIPS probe were mapped to different chromosomes (Larson and Raboy, 1999). In Arabidopsis, two distinct MIPS genes have been identified (Johnson, 1994; Johnson and Burk, 1995; Johnson and Sussex, 1995). The existence of multiple MIPS genes in plants may permit differential MIPS expression for many different physiological functions; however, MIPS gene family expression has not been well characterized to date.
We report the isolation of a MIPS cDNA from developing soybean (Glycine max) seeds. The genomic organization of soybean MIPS genes and the structure of the gene corresponding to the isolated MIPS cDNA were examined. At least four different MIPS sequences were identified from the soybean expressed sequence tag (EST) database and Southern hybridization results. Patterns of MIPS expression were analyzed in developing soybean seeds and other plant tissues. Together, these data suggest that we have isolated a member of a MIPS gene family that is abundantly expressed in developing soybean seeds to function in phytic acid synthesis.
RESULTS AND DISCUSSION
Isolation and Analysis of a MIPS cDNA
A MIPS cDNA (GmMIPS1) was isolated from developing soybean seeds using reverse transcriptase- (RT) PCR (Fig. 2). The 1,791-bp cDNA sequence contained a 1,533-bp open reading frame that could encode a protein of 510 amino acids. The putative soybean MIPS protein has a predicted molecular mass of 56.5 kD. BLAST database searches were performed with the MIPS cDNA sequence and predicted protein sequence (Altschul et al., 1997). The searches revealed a high degree of sequence identity between GmMIPS1 and MIPS genes from plants and other organisms. The highest scoring match was to the tobacco MIPS sequence (Hara et al., 2000), which showed 80.6% identity to GmMIPS1 at the nucleotide level and 92.2% identity at the amino acid. Sequence identity to yeast was 40.2% at the nucleotide level and 41.5% at the amino acid level (Johnson and Henry, 1989), further confirming the high degree of conservation among MIPS sequences from different sources.
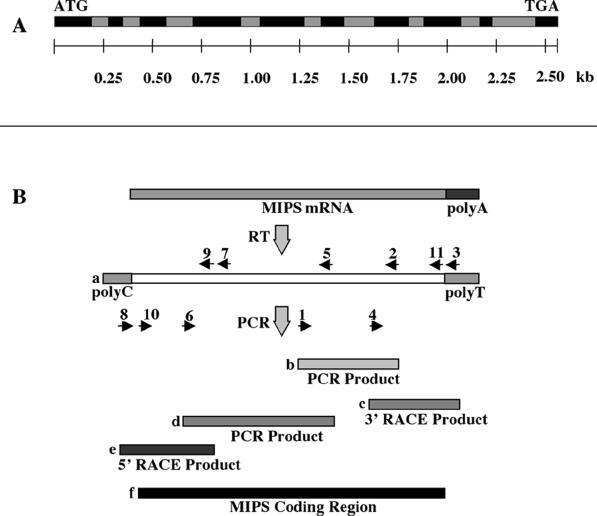
Figure 2.
GmMIPS1 gene structure and cDNA cloning strategy. A, The sequence of a genomic PCR fragment was compared with the cloned MIPS cDNA sequence to define intron-exon borders. Introns are represented in gray and exons are represented in black. Scale indicates size in kilobases. B, Cloning of GmMIPS1 cDNA sequences. Total RNA from developing seeds was used as template for cDNA synthesis. Locations of PCR primers on the MIPS sequence are indicated (a). Primers 1 and 2 from conserved MIPS regions were used to amplify an internal fragment (b). The 3′ end of the MIPS cDNA was amplified by 3′-RACE using anchored primer 3 and gene-specific primer 4 (c). A second internal PCR product was amplified using primer 5 from a 5′-conserved region in conjunction with a primer 6 from the first PCR product (d). The 5′ end of the coding sequence and untranslated sequence were obtained by using primer 7 for cDNA synthesis and adapter-anchored primer 8 and gene-specific primer 9 for amplification by 5′-RACE (e). A fragment containing the full-length coding sequence was amplified by high-fidelity PCR using primers 10 and 11 (f).
MIPS Gene Structure and Organization
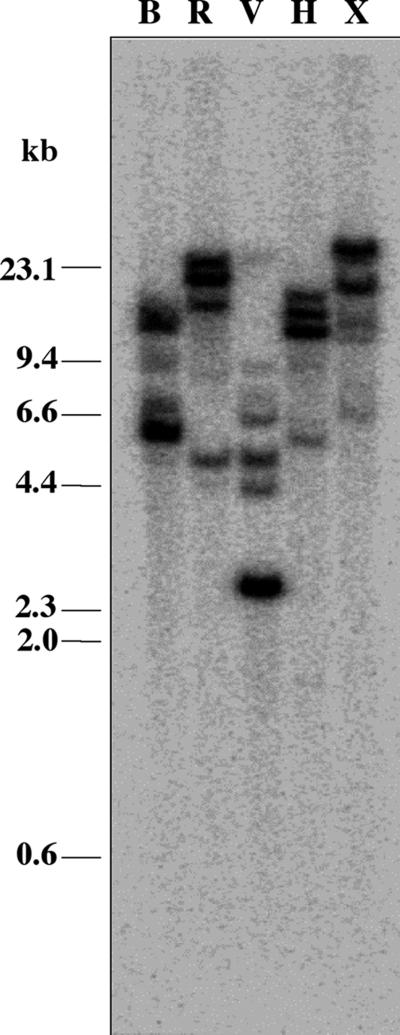
An estimate of MIPS sequence copy number was determined by Southern-blot analysis. Under stringent conditions, multiple high-Mr bands were detected using the full-length cDNA as the MIPS hybridization probe (Fig. 3). The data indicated that the soybean genome contains four or more loci with MIPS homology.
Figure 3.
Southern-blot analysis of soybean genomic DNA. A full-length MIPS cDNA fragment was [32P] labeled to probe a blot of soybean genomic DNA at high stringency. DNA samples (10 μg) were digested with the following restriction endonucleases: BglII (B), EcoRI (R), EcoRV (V), HindIII (H), and XbaI (X), respectively. HindIII-digested bacteriophage λ DNA was used as a size marker.
A MIPS genomic sequence corresponding to GmMIPS1 was generated by PCR amplification and was compared with the cDNA sequence for assignment of exons and introns. The region encompassing the start (ATG) to stop (TGA) codons in the genomic PCR product consisted of 2,607 bp containing nine introns (Fig. 2A). Sequence analysis revealed that restriction sites for EcoRI, HindIII, and XbaI were absent in the genomic sequence. Genomic restriction fragments generated by the three enzymes were larger than the size of the genomic PCR product, consistent with the lack of these sites in the sequence (Fig. 3). Due to the high degree of sequence similarity among plant MIPS genes, high stringency hybridization did not reveal which fragment corresponded to the cloned GmMIPS1 sequence.
Analysis of Soybean cDNAs with Homology to MIPS
The GmMIPS1 cDNA sequence was compared with soybean ESTs that have been identified as putative MIPS sequences in a soybean database, SoyBase (R. Shoemaker, U.S. Department of Agriculture-Agricultural Research Service, Iowa State University). Analysis of the large number of soybean cDNA sequences with MIPS homology facilitated assignment of the ESTs to four highly similar, yet distinct MIPS sequences (GmMIPS1–GmMIPS4) as shown in Table I. (The complete GmMIPS1 sequence and alignment with other MIPS family members from the EST database, as well as EST accession nos., are shown in online supplementary material.) The GmMIPS1 cDNA isolated from immature seeds in these studies was identical to the EST sequences obtained from several immature cotyledon libraries. The majority of ESTs from GmMIPS2 originated primarily from root libraries, but also included sequences from shoot, flower, and seed coat libraries. GmMIPS1 and GmMIPS2 showed 96% identity over a span of 1,465 nucleotides and shared a very similar 3′-untranslated region. The EST sequences corresponding to GmMIPS3 were derived predominantly from flower libraries, but were also found in leaves, buds, and germinated cotyledons. Only two ESTs were assigned to GmMIPS4 (from buds and young pods). GmMIPS4 sequences were similar to GmMIPS3 in coding and 3′-untranslated sequences. In addition to the soybean cDNAs that were assigned to GmMIPS1 through GmMIPS4, there were several individual cDNAs in the EST database that could not be placed with any other sequences.
Table I.
Classification of soybean ESTs with MIPS homology
| Library Source | No. of EST Sequences Encoding
|
|||
|---|---|---|---|---|
| GmMIPS1 | GmMIPS2 | GmMIPS3 | GmMIPS4 | |
| Immature cotyledons | 16 | 0 | 0 | 0 |
| Roots | 2 | 7 | 0 | 0 |
| Immature seed coats | 1 | 3 | 0 | 0 |
| Shoot tips | 0 | 1 | 0 | 0 |
| Flowers | 0 | 1 | 5 | 0 |
| Immature flowers | 0 | 1 | 3 | 0 |
| Germinated cotyledons | 0 | 0 | 2 | 0 |
| Leaves | 0 | 0 | 2 | 0 |
| Buds | 0 | 0 | 2 | 1 |
| Young pods | 0 | 0 | 0 | 1 |
No. of EST sequences and library source of cDNA for each MIPS family member are indicated.
The data from the soybean EST project facilitated the generation of an “electronic northern,” which served as a predictive first step in determining the expression patterns of MIPS gene family members (Table I). The frequency of GmMIPS1 EST sequences and the derivation of these GmMIPS1 sequences from immature cotyledons suggests that GmMIPS1 is abundantly expressed early in seed development, perhaps to serve in phytic acid biosynthesis.
Soybean MIPS RNA Expression
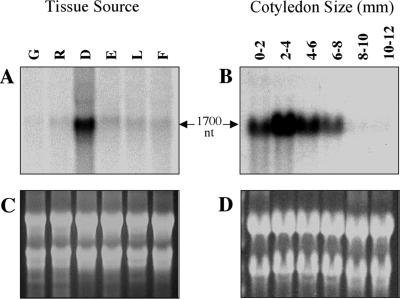
Northern-blot analyses were performed with total RNA from different soybean tissues and various stages of seed development using the GmMIPS1 cDNA as the hybridization probe (Fig. 4). Steady-state RNA levels were higher in developing seeds than in other soybean tissues, including flowers, leaves, roots, germinating cotyledons, and somatic embryos (Fig. 4A). MIPS transcript was observed in cotyledons at the earliest developmental stages analyzed, 0 to 2 mm (corresponding in size up to approximately stage C; Meinke et al., 1981). Maximal levels of MIPS RNA were observed in 2- to 4-mm seeds (equivalent to stages D–F; Fig. 4B). As development progressed, MIPS transcript levels decreased to nearly undetectable levels by 8- to 10-mm seeds (stage J and beyond). Although differences in the relative abundance of MIPS RNA were easily detected using GmMIPS1 as a probe, the high degree of sequence similarity between gene family members prevented us from definitively concluding that the observed patterns were gene specific. However, coupled with the relative abundance of GmMIPS1 in the EST database and the origin of GmMIPS1 sequences primarily from immature seeds, these results strongly suggest that the high level of MIPS transcript in soybean seeds may be primarily due to GmMIPS1 expression.
Figure 4.
Analysis of MIPS expression patterns in soybean tissues. After transfer to membranes, soybean RNA samples were probed using a [32P]-labeled MIPS cDNA. A and C, RNA samples from soybean tissues. G, Germinated cotyledons; R, young roots; D, developing seeds; E, globular-stage somatic embryos; L, young leaves; F, flowers. B and D, RNA samples from different stages of seed development: 0 to 2, 2 to 4, 4 to 6, 6 to 8, 8 to 10, and 10 to 12 mm. A and B are autoradiographs; C and D are the corresponding ethidium bromide-stained samples to visualize RNA quantity and quality.
Immunodetection of Soybean MIPS
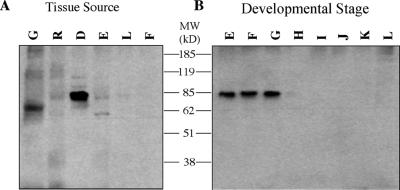
Western-blot analyses were performed to further corroborate the pattern of MIPS expression. To produce an antibody for immunodetection studies, a MIPS fusion protein construct was generated by insertion of a PCR product into an expression vector containing a His tag. After demonstrating that the fusion protein was successfully expressed in bacterial cells, the vector was used for large-scale protein expression and antibody production (Styer, 2000). The generation of a polyclonal antibody raised against the MIPS fusion protein has allowed the examination of MIPS protein expression and will provide a useful tool for further immunocytochemical studies. Immunoblot analysis of protein extracts from developing seeds, flowers, roots, somatic embryos, leaves, and germinated seeds indicated that developing seeds contained the highest levels of MIPS protein (Fig. 5A). The faint immunoreactive bands observed in other tissues may represent additional MIPS isoforms. The accumulation of the MIPS protein was observed in early seed development at stages E, F, and G (2.6–5.4 mm) as seen from the developmental series of immature cotyledons (Fig. 5B).
Figure 5.
Immunodetection of soybean MIPS protein. Antibody produced against the soybean MIPS fusion protein was used to examine protein expression patterns in soybean protein extracts. A, Protein extracts from soybean tissues. G, Germinating cotyledons; R, young roots; D, developing seeds; E, globular somatic embryos; L, young leaves; F, flowers. B, Lanes E through L represent protein extracts from soybean cotyledons at sequential stages of development as classified by Meinke et al. (1981). Size markers used were the BenchMark Prestained Protein Ladder (Life Technologies).
The pattern of MIPS expression observed in developing soybean seeds was consistent with the expectation that high levels of MIPS expression should precede and/or accompany synthesis and accumulation of phytic acid. In rice, recent studies demonstrated a clear relationship between patterns of MIPS expression and accumulation of phytic acid (Yoshida et al., 1999). MIPS RNA expression was detected by in situ hybridization in globular stage rice embryos, prior to the formation of phytate-containing globoids. In maturing rice seeds, abundant levels of the MIPS transcript were observed in the scutellum and aleurone layer.
Raboy and Dickinson (1987) measured phytic acid and phosphate levels throughout soybean development. Separation of seed extracts using anion-exchange chromatography showed that phytic acid levels increased steadily until late in seed maturation (18.7–33.6 μg per seed per day). At seed maturity, very little free phosphate remained in the seed because it had been incorporated into phytic acid. Although Raboy and Dickinson (1987) reported accumulation of phytic acid throughout seed maturation, our examination of MIPS protein levels during seed development indicated high level expression only in early cotyledonary stages. This suggests that conversion of d-Glc-6-P to d-myo-inositol-3-phosphate occurs earlier in seed development than accumulation of the final product, phytic acid. Identification of kinases and their expression patterns will provide a more detailed picture of the steps in phytic acid biosynthesis.
Potential for Seed-Specific MIPS Isoforms
Further evidence for seed-specific MIPS expression is provided by studies on low phytic acid (lpa) mutants in maize. Reduction in phytate levels by 50% to 95% has been achieved in lpa mutants following ethyl methanesulfonate mutagenesis (Larson and Raboy, 1999; Raboy et al., 2000). The lpa mutants are viable and several appeared identical to wild type for traits other than the low phytic acid phenotype. One copy of the maize MIPS gene mapped to the same location as the lpa-1 trait on chromosome 1S. It is possible that maize lpa-1 represents a mutation affecting seed-specific MIPS gene expression, resulting in decreased MIPS activity and lower phytate accumulation in seeds.
The association between the MIPS gene and the lpa phenotype is less clear in other cereals. The lpa-1 mutations in barley and rice mapped to chromosomes 2H and 2L, respectively (Larson et al., 1998, 2000). Only a single locus with identifiable MIPS homology has been located in barley and rice, neither of which mapped to the same location as the lpa phenotype (Larson and Raboy, 1999; Larson et al., 2000). This raises the possibility that barley and rice may contain other cryptic copies of genes encoding MIPS activity, or alternatively, that the lpa phenotype in barley and rice consists of mutations in regulatory genes affecting MIPS expression or enzyme activity.
In Arabidopsis, two distinct MIPS sequences were identified (Johnson, 1994; Johnson and Burk, 1995) and subsequently verified by their presence in EST and genome sequencing projects. No specific functions have been assigned to either gene in Arabidopsis. The two copies may simply be redundant or may perform different physiological functions within the plant. Additional copies of the MIPS gene in other plants may allow for further specialization of MIPS isozymes.
Compared with Arabidopsis, soybean has a greater capacity for nutrient storage in the seed. The amount of phytate stored in soybean seeds may exceed levels required for normal seed function. Raboy et al. (1985) decreased the phytic acid content of soybean seeds to one-third the normal amount by growing maternal plants in low phosphate conditions. Germination and seedling viability were not compromised in the resulting low phytate soybeans. Our data suggest that large-seeded plants such as maize and soybean may utilize a seed-abundant isoform for high level accumulation of phytic acid. Future comparisons of MIPS function in Glycine soja, a small-seeded relative of soybean, and teosinte, a wild relative of maize, may address this hypothesis.
Alteration of the MIPS gene is a potential approach for the development of low-phytic acid soybeans (Hitz and Sebastian, 1998). Mutants with decreased MIPS activity in soybean seeds contained significantly less seed phytic acid, as well as reduced levels of the inositol derivatives, raffinose and stachyose. The MIPS sequence from the mutant soybeans contained a single substitution at nucleotide 1,188 of the coding sequence (G to T), resulting in a change from Lys in the wild type to Asn in the mutant sequence. Other strategies for decreasing MIPS gene expression such as antisense suppression may provide additional routes for the future development of soybeans with low-phytic acid phenotypes to improve phosphorus availability.
Summary
Our MIPS RNA and protein expression data suggest that GmMIPS1 expression is under the control of a strong, developmentally regulated promoter in immature seeds. This is further supported by the fact that the GmMIPS1 ESTs are abundant and derived primarily from cDNA libraries from immature cotyledons. The observation of at least four bands by high stringency Southern-blot analysis is consistent with the presence of four loci containing highly similar MIPS sequences in the soybean genome. It is likely that GmMIPS1 is expressed most strongly in seeds and plays a critical role in phytic acid biosynthesis, whereas the other three soybean MIPS genes may be involved in other aspects of inositol metabolism. Differential regulation of multiple genes would allow specific MIPS expression during periods of high demand for inositol, such as would be expected during phytic acid accumulation in seed development. Closer examination of the expression of these sequences in plant tissues using sensitive methods such as differential RT-PCR will aid in elucidating the multiple roles for MIPS in plant cells.
MATERIALS AND METHODS
Plant Material
Soybean (Glycine max L. Merr. cv Williams 82) plants were grown in an environmental chamber (Conviron, Pembina, ND) in 24 h of light until approximately 8 weeks of age and were then transferred to a 16-h light/8-h dark cycle to induce flowering. Somatic embryos were initiated from immature soybean cotyledons and were maintained on D20 medium (Finer, 1988). Plant tissues were harvested and stored at −80°C.
DNA and RNA Extraction
DNA for restriction endonuclease digestion and Southern-blot analysis was isolated as described by Dellaporta et al. (1983). Total RNA was isolated from tissue samples (100 mg) using the RNAeasy kit (Qiagen, Valencia, CA). RNA used for RT-PCR amplification was isolated from seeds ranging in size from 2 to 10 mm, which were pooled since the precise timing of MIPS expression in seeds was unknown. For northern analysis, the seeds were divided into groups based on length as measured from apical end to basal end of the seed (0–2, 2–4, 4–6, 6–8, 8–10, and 10–12 mm).
Isolation and Analysis of cDNA and Genomic Sequences
A soybean MIPS cDNA was isolated from developing seeds using a PCR approach that was based on conservation of MIPS coding sequences among plant species. A multiple sequence alignment of several previously reported plant MIPS cDNA sequences from GenBank (Arabidopsis, Phaseolus vulgaris, oilseed rape, Spirodela polyrrhiza, and common ice plant) was performed using the Clustal method (Thompson et al., 1994) in the MegAlign program (Lasergene Software, DNAstar, Madison, WI). Oligonucleotides for PCR amplification are included in online supplementary data.
Oligo-dT-primed cDNA synthesis was performed with 5 μg of total RNA from pooled seeds using Superscript II reverse transcriptase (3′-RACE system, Life Technologies, Rockville, MD). PCR reactions (50 μL) were assembled with Taq Master Mix according to the manufacturer's recommendations (Qiagen) and amplification was performed with a Robocycler Gradient 40 thermocycler (Stratagene, La Jolla, CA). An initial 496-bp MIPS PCR product was cloned into the SmaI site of pTZ19R (Fermentas, Hanover, MD). Dideoxy sequencing was performed using the Sequitherm Excel II kit (Epicenter Technologies, Madison, WI).
RACE was used to amplify the 5′ and 3′ ends of the soybean MIPS cDNA sequence (Life Technologies). A 412-bp PCR product from the 3′ end of the MIPS cDNA was cloned into the SmaI site of pTZ19R and was sequenced as described above. The 5′ end of the MIPS cDNA was amplified in two rounds of 5′-RACE. PCR products of 836 and 295 bp were cloned into the SmaI site of pTZ19R and were sequenced. The sequences from the overlapping MIPS cDNA fragments were assembled into a contiguous sequence using the program SeqMan (DNAstar). A 1,551-bp product containing the full-length MIPS cDNA was amplified by high-fidelity PCR (Pfu polymerase; Stratagene) and cloned into the SmaI site of pTZ19R. The MIPS sequence was submitted to GenBank (accession no. AF293970). DNA from soybean hypocotyls was utilized as a template for amplifying a genomic product spanning the MIPS coding region and was sequenced at the Virginia Polytechnic Institute and State University Sequencing Facility.
DNA- and RNA-Blot Analyses
DNA samples (10 μg) were digested with restriction enzymes, separated by agarose gel electrophoresis, and transferred onto a nylon membrane (Schleicher & Schuell, Keene, NH). For use as a hybridization probe, a DNA fragment containing the full-length MIPS coding sequence was gel purified and labeled with α-[32P]dATP by random priming (Random Primers Kit, Life Technologies). High stringency DNA hybridizations and subsequent washes were performed according to the manufacturer's specifications (Ambion, Austin, TX). The membrane was exposed to a storage phosphor screen for 2 to 24 h, which was scanned by a Storm 860 imager (Molecular Dynamics, Sunnyvale, CA).
The expression of the MIPS gene in developing seeds and other tissues was analyzed by northern-blot hybridization. RNA (10 μg) from staged developing seeds and other soybean tissues (germinated cotyledons, young roots, somatic embryos, young leaves, and flowers) was separated by formaldehyde agarose gel electrophoresis (1.2% [w/v] agarose) prior to transfer to nylon membranes (Schleicher & Schuell). Duplicate sets of lanes were loaded and one set was stained with ethidium bromide to confirm sample integrity and equal loading of samples. The blots were probed and washed as described for Southern hybridizations. Detection of hybridizing RNA bands was performed using the phosphorimager as described above.
Protein Expression and Immunoblot Analyses
For high-level bacterial expression and subsequent protein purification, a fragment containing the majority of the MIPS coding sequence was fused in frame to a 6× His sequence in the expression plasmid pET-32a (+) (Novagen, Madison, WI). A 1,425-bp portion of the MIPS cDNA encoding amino acid residues 24 to 498 was amplified by high-fidelity PCR from a MIPS cDNA and was cloned into the pET-32a (+) vector. The resulting plasmid was introduced into the bacterial strain BL21 (DE3) pLysS (Novagen). The constructs were provided to the laboratory of Dr. Glenda Gillaspy (Virginia Polytechnic Institute and State University) for production and purification of the MIPS fusion protein (Styer, 2000), which was shipped to a commercial facility (Cocalico Biologicals, Reamstown, PA) for polyclonal antibody production.
Total protein was extracted from developing seeds, flowers, germinating cotyledons, leaves, roots, and somatic embryos. Immature seed samples were grouped by developmental stages as described previously (Meinke et al., 1981). Stages collected included: E = 2.6 to 3.4 mm, F = 3.6 to 4.4 mm, G = 4.6 to 5.4 mm, H = 5.6 to 6.4 mm, I = 6.6 to 7.4 mm, J = 7.6 to 8.4 mm, K = 8.6 to 9.4 mm, and L = 9.6 to 10.4 mm. Protein was extracted with homogenization buffer (0.15 m KCl, 50 mm Tris-HCl, and 5 mm EDTA) in the presence of a plant protease inhibitor cocktail (P-9599, Sigma, St. Louis) and 0.1% (v/v) Triton X-100. Protein was quantified according to a bovine serum albumin standard curve using a protein assay kit (Bio-Rad, Hercules, CA).
For immunodetection of MIPS, protein extracts were subjected to SDS-PAGE and were subsequently transferred to membranes according to manufacturer's specifications (Bio-Rad). Membranes were incubated with primary antibody and secondary (horseradish peroxidase conjugated goat-anti-rabbit; Sigma) antibody at 1:10,000 dilutions. Protein bands were detected using the chemiluminescent enhanced chemiluminescence Plus Western Blotting Detection System according to the manufacturer's specifications (Amersham Pharmacia Biotech, Piscataway, NJ).
ACKNOWLEDGMENTS
We thank Regina Hanlon for excellent technical assistance, and John McDowell and Craig Nessler for critical review of the manuscript.
Footnotes
This work was supported in part by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (grant no. 97–35504–4997).
LITERATURE CITED
- Abu-Abied M, Holland D. The gene c-ino1 from Citrus paradisi is highly homologous to tur1 and ino1 from yeast and Spirodela encoding for myo-inositol phosphate synthase. Plant Physiol. 1994;106:1689. doi: 10.1104/pp.106.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Bio Rep. 1983;1:19–21. [Google Scholar]
- Finer JJ. Apical proliferation of embryogenic tissue of soybean (Glycine max L. Merrill) Plant Cell Rep. 1988;7:238–241. doi: 10.1007/BF00272532. [DOI] [PubMed] [Google Scholar]
- Hara K, Yagi M, Koizumi N, Kusano T, Sano H. Screening of wound-responsive genes identifies an immediate-early expressed gene encoding a highly charged protein in mechanically wounded tobacco plants. Plant Cell Physiol. 2000;41:684–691. doi: 10.1093/pcp/41.6.684. [DOI] [PubMed] [Google Scholar]
- Hitz WD, Sebastian SA, inventors. April 7, 1998. Soybean plant producing seeds with reduced levels of raffinose saccharides and phytic acid. International Patent Application, WO 98/45448, pp 1–57
- Ishitani M, Majumder AL, Bornhouser A, Michalowski CB, Jensen RG, Bohnert HJ. Coordinate transcriptional induction of myo-inositol metabolism during environmental stress. Plant J. 1996;9:537–548. doi: 10.1046/j.1365-313x.1996.09040537.x. [DOI] [PubMed] [Google Scholar]
- Johnson MD. The Arabidopsis thaliana myo-inositol 1-phosphate synthase (EC 5.5.1.4) Plant Physiol. 1994;105:1023–1024. doi: 10.1104/pp.105.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD, Burk D. Isozyme of 1L-myo-inositol-1-phosphate synthase from Arabidopsis (accession no. U30250; PGR95-067) Plant Physiol. 1995;109:721. doi: 10.1104/pp.107.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD, Henry SA. Biosynthesis of inositol in yeast: primary structure of myo-inositol-1-phosphate synthase (EC 5.5.1.4) and functional analysis of its structural gene, the INO1 locus. J Biol Chem. 1989;264:1274–1283. [PubMed] [Google Scholar]
- Johnson MD, Sussex IM. 1-L-myo-inositol 1-phosphate synthase from Arabidopsis thaliana. Plant Physiol. 1995;107:613–619. doi: 10.1104/pp.107.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson SR, Raboy V. Linkage mapping of maize and barley myo-inositol 1-phosphate synthase DNA sequences: correspondence with a low phytic acid mutation. Theor Appl Genet. 1999;99:27–36. [Google Scholar]
- Larson SR, Rutger JN, Young KA, Raboy V. Isolation and genetic mapping of a non-lethal rice (Oryza sativa L.) low phytic acid mutant. Crop Sci. 2000;40:1397–1405. [Google Scholar]
- Larson SR, Young KA, Cook A, Blake TK, Raboy V. Linkage mapping of two mutations that reduce phytic acid content of barley grain. Theor Appl Genet. 1998;97:141–146. [Google Scholar]
- Loewus FA, Murthy PPN. Myo-Inositol metabolism in plants. Plant Sci. 2000;150:1–19. [Google Scholar]
- Lott JNA, Greenwood JS, Batten GD. Mechanisms and regulation of mineral nutrient storage during seed development. In: Kigel J, Galili G, editors. Seed Development and Germination. New York: Marcel Dekker; 1995. pp. 215–235. [Google Scholar]
- Meinke DW, Chen J, Beachy RN. Expression of storage-protein genes during soybean seed development. Planta. 1981;153:130–139. doi: 10.1007/BF00384094. [DOI] [PubMed] [Google Scholar]
- Morré DJ, Boss WF, Loewus FA. Inositol Metabolism in Plants. New York: Wiley-Liss; 1990. [Google Scholar]
- Prattley CA, Stanley DW. Protein-phytate interactions in soybeans: I. Localization of phytate in protein bodies and globoids. J Food Biochem. 1982;6:243–253. [Google Scholar]
- Raboy V, Dickinson DB. The timing and rate of phytic acid accumulation in developing soybean seeds. Plant Physiol. 1987;85:841–844. doi: 10.1104/pp.85.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboy V, Gerbasi PF, Young KA, Stoneberg SD, Pickett SG, Bauman AT, Murthy PPN, Sheridan WF, Ertl DS. Origin and seed phenotype of maize low phytic acid 1-1 and low phytic acid 2-1. Plant Physiol. 2000;124:355–368. doi: 10.1104/pp.124.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboy V, Hudson SJ, Dickinson DB. Reduced phytic acid content does not have an adverse effect on germination of soybean seeds. Plant Physiol. 1985;79:323–325. doi: 10.1104/pp.79.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpley AN, Chapra SC, Wedepohl R, Sims JT, Daniel TC, Reddy KR. Managing agricultural phosphorus for protection of surface waters: issues and options. J Environ Qual. 1994;23:437–451. [Google Scholar]
- Smart CC, Fleming AJ. A plant gene with homology to D-myo-inositol-3-phosphate synthase is rapidly and spatially up-regulated during an abscisic-acid-induced morphogenic response in Spirodela polyrrhiza. Plant J. 1993;4:279–293. doi: 10.1046/j.1365-313x.1993.04020279.x. [DOI] [PubMed] [Google Scholar]
- Styer J. Regulating inositol biosynthesis in plants: myo-inositol phosphate synthase and myo-inositol monophosphatase. MS thesis. 2000. Virginia Polytechnic Institute and State University, Blacksburg. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida KT, Wada T, Koyama H, Mizobuchi-Fukuoka R, Naito S. Temporal and spatial patterns of accumulation of the transcript of myo-inositol-1-phosphate synthase and phytin-containing particles during seed development in rice. Plant Physiol. 1999;119:65–72. doi: 10.1104/pp.119.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]