Abstract
An abscisic acid (ABA)-induced cDNA fragment encoding a putative protein phosphatase 2C (PP2C) was obtained by means of differential reverse transcriptase-polymerase chain reaction approach. The full-length clone was isolated from a cDNA library constructed using mRNA from ABA-treated beechnut (Fagus sylvatica) seeds. This clone presents all the features of plant type PP2C and exhibits homology to members of this family such as AthPP2CA (61%), ABI1 (48%), or ABI2 (47%), therefore it was named FsPP2C1. The expression of FsPP2C1 is detected in dormant seeds and increases after ABA treatment, when seeds are maintained dormant, but it decreases and tends to disappear when dormancy is being released by stratification or under gibberellic acid treatment. Moreover, drought stress seems to have no effect on FsPP2C1 transcript accumulation. The FsPP2C1 transcript expression is tissue specific and was found to accumulate in ABA-treated seeds rather than in other ABA-treated vegetative tissues examined. These results suggest that the corresponding protein could be related to ABA-induced seed dormancy. By expressing FsPP2C1 in Escherichia coli as a histidine tag fusion protein, we have obtained direct biochemical evidence supporting Mg2+-dependent phosphatase activity of this protein.
Seed dormancy, an adaptative mechanism to ensure plant survival, can be overcome by chilling, light, or plant hormone treatment (Schneider and Gifford, 1994). This phenomenon is one of the least understood in seed biology (Hilhorst, 1995). Research has focused on the physiological differences of dormant and non-dormant seeds and on the role that abscisic acid (ABA) plays in the induction and maintenance of seed dormancy (Bewley and Black, 1994; Nicolás et al., 1997).
ABA modulates different events of plant growth and development, including seed maturation, dormancy, and germination (Chandler and Robertson, 1994; Leung and Giraudat, 1998). The signal pathways of this regulation are still unknown, but phosphorylation/dephosphorylation have been suggested to be involved in these processes (Trewavas, 1988; Verhey and Walker-Simmons, 1997).
Many extracellular stimuli elicit diverse intracellular responses through phosphorylation/dephosphorylation cascades. The phosphorylation state of a protein is controlled by protein kinases and protein phosphatases. The protein Ser/Thr phosphatases involve two different families, one of them including protein phosphatases 1, 2A, and 2B (PP1, PP2A, and PP2B), which are homologous to one another, and a second group of protein phosphatases 2C (PP2C), which are structurally distinct proteins. PP2Cs are not evolutionary related to the other families of Ser/Thr protein phosphatases; they are Mg2+- or Mn2+-dependent, insensitive to okadaic acid (strong inhibitor of the other groups of protein phosphatases), and lack a regulatory subunit as compared with other types of phosphatases (for review, see Rodríguez, 1998). In eukaryotes, PP2Cs have been implicated as regulatory factors in protein kinase cascades that become activated as a result of stress (Moore et al., 1991). For instance, in both fission yeast and budding yeast, PP2C enzymes have an important role as negative regulators of a mitogen-activated protein (MAP) kinase pathway activated after osmotic stress (Maeda et al., 1994; Shiozaki and Russell, 1995; Meskiene et al., 1998).
Since ABA is a key signal that modulates plant responses to environmental stress and seed dormancy, several genes and proteins regulated by this hormone have been identified and studied in relation to these processes. By using ABA response mutants (ABA insensitive, abi), which produce non-dormant seeds (Koornneef et al., 1984) as a genetic approach, various components that participate in the ABA signal transduction pathway involved in seed dormancy have been recognized, including two highly homologous members of the plant type-PP2C family, ABI1 (Leung et al., 1994; Meyer et al., 1994), and ABI2 (Leung et al., 1997; Rodríguez et al., 1998a). ABI1 and ABI2 have been shown to be negative regulators of ABA signaling pathways as proved by mutational effects of PP2Cs on ABA-inducible transcription (Sheen, 1998) and by genetic analysis of abi1-1 revertant mutants (Gosti et al., 1999). Some other protein kinase and phosphatase genes regulated by ABA have also been reported to be involved in seed dormancy or stress responses, including PKABA1 (Anderberg and Walker-Simmons, 1992), FsPK1, and FsPK2 (Lorenzo et al., 2000) or AtP2C-HA (Rodríguez et al., 1998b).
Our work is focused on the mechanism of ABA action in the induction and maintenance of dormancy in beechnut (Fagus sylvatica) seeds and in the expression of specific genes involved in this process. Although beechnut lacks the genetic tractability of Arabidopsis, it represents a suitable model to study seed dormancy of woody plants where little is known about the mechanism involved in this process. Furthermore, in previous work we have shown that beechnuts seeds exhibit a specially deep degree of dormancy, maintained by ABA, and overcome by stratification or gibberellic acid (GA3) treatment (during 6 and 3 weeks, respectively), by regulating the expression of some dormancy-related genes (Nicolás et al., 1996, 1997, 1998).
In this report we describe and characterize a new cDNA clone (FsPP2C1) coding for a Ser/Thr PP2C. It is up-regulated by ABA and specifically expressed in ABA-treated seeds; thus it seems to be correlated with the ABA-induced seed dormancy. Expression of this cDNA clone in Escherichia coli as a His tag fusion protein shows Mg2+-dependent phosphatase activity.
RESULTS
Isolation and Characterization of a cDNA Clone from Beechnut Seeds, Coding for a Type PP2C
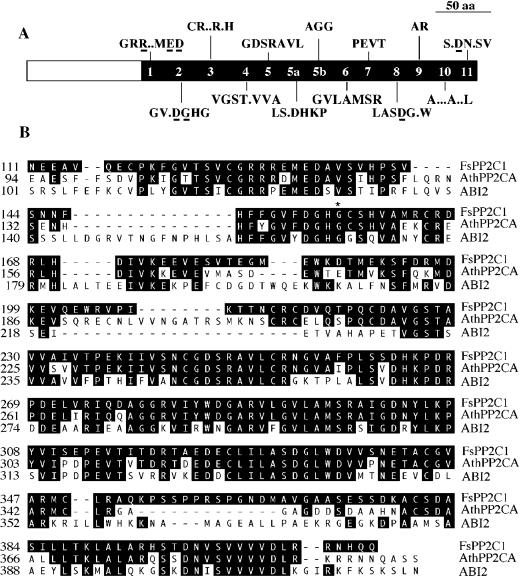
In a previous report by Nicolás et al. (1996) we found that ABA is involved in beechnut dormancy while GA3 is able to release it. As PP2Cs are key elements in ABA signal transduction and play a crucial role as regulators of seed dormancy (Leung et al., 1994; Meyer et al., 1994), we attempted to identify PP2Cs induced by ABA, putatively involved in ABA signal transduction, and related to seed dormancy. A differential reverse transcriptase (RT)-PCR approach was used starting from mRNA extracted of either ABA-treated or GA3-treated seeds, and using degenerate oligonucleotides corresponding to conserved motifs 2 (DGHGGS) and 8 (LASDGLWD) present in all Ser/Thr PP2Cs (Rodríguez, 1998). Some cDNA fragments, encoding partial gene products with homology to PP2Cs, were selected and amplified from cDNA of ABA-treated seeds as template, but absent when the template used was cDNA of GA3-treated seeds. One of them was used as a probe to screen a cDNA library constructed from mRNA of ABA-treated seeds (Nicolás et al., 1997) and the corresponding full-length clone, named FsPP2C1, was found and sequenced (accession no. AJ277743). It had 1,383 bp and contained an open reading frame of 1,242-bp-long. The ATG start codon was preceded by an in-frame stop codon. The length of FsPP2C1 cDNA correlates well with the size of the mRNA determined by northern blot (1.4 kb approximately). The deduced sequence of the corresponding protein contains 413 amino acids and has a predicted molecular mass of 45.3 kD. As shown in Figure 1A, FsPP2C1 protein includes the 11 conserved catalytic subdomains present in all PP2Cs (Rodríguez, 1998) with the residues putatively involved in the coordination of the phosphate and metal ions (Das et al., 1996).
Figure 1.
A, Schematic diagram of FsPP2C1 showing the conserved domains within the PP2C family. Residues putatively involved in the coordination of the phosphate and metal ions are underlined. B, Alignment of the derived amino acid sequences from FsPP2C1 with ABI2 and AthPP2CA from Arabidopsis. The Gly residue that is substituted by Asp in the AthPP2CA and abi2 mutant proteins is indicated by an asterisk.
FsPP2C1 deduced amino acid sequence was compared with EMBL databases and revealed homology with different proteins of the PP2C family. FsPP2C1 shares homology with plant type-PP2C such as AthPP2CA (61% identity) (Kuromori and Yamamoto, 1994) and with the catalytic domain of ABI1 (48% identity) (Leung et al., 1994; Meyer et al., 1994) and ABI2 (47% identity) of Arabidopsis (Leung et al., 1997; Rodríguez et al., 1998a) (Fig. 1B). The structural organization of FsPP2C1 follows the plant pattern as described by Rodríguez (1998); however, the short N-terminal extension is unrelated to ABI1/ABI2 but similar to that of the AthPP2CA protein.
Regulatory Effect of ABA on the Expression of FsPP2C1 in Dormant Beechnuts
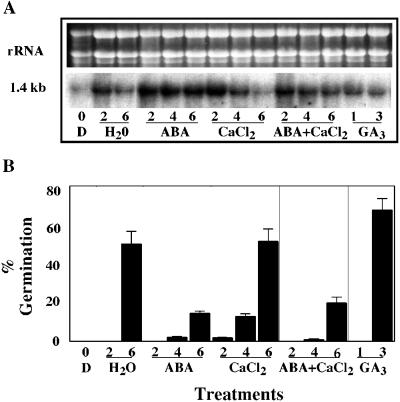
The effect of ABA on the accumulation of FsPP2C1 mRNA in beechnut seeds was tested by northern blot. Expression of FsPP2C1 (Fig. 2A) was inversely correlated with the germination after the indicated treatments (Fig. 2B). Transcript levels are initially low in dormant seeds and increase for all the treatments after 2 weeks imbibition. During stratification at 4°C in water (treatment previously reported to be efficient in breaking dormancy in these seeds) expression decreased as the stratification period proceeded and dormancy is dissipated, reaching 50% germination after 6 weeks (Nicolás et al., 1996). In the presence of GA3, which increases the percentage of seed germination and produces a fast release from dormancy after only 3 weeks of treatment (Nicolás et al., 1996), the transcript levels are even lower than those found in the control in water. The addition of ABA maintains the dormant state in beechnut, decreasing the percentages of germination to 20% compared with the control in water. This hormone induces the expression of the FsPP2C1 transcript to higher levels than the rest of treatments. Although calcium is important for the expression of other beechnut PP2Cs (O. Lorenzo, G. Nicolás, C. Nicolás, D. Rodríguez, and P.L. Rodríguez, unpublished data), the addition of calcium does not seem to have any effect on the levels of expression of this clone as compared with the stratification treatment. However, calcium treatment appears to antagonize the ABA-induced maintenance of FsPP2C1 expression.
Figure 2.
A, Northern-blot analysis of total RNA isolated from beechnut dormant seeds (D) and dormant seeds incubated from 1 to 6 weeks at 4°C in water, 100 μm ABA, 1 mm CaCl2, 100 μm ABA + 1 mm CaCl2, and 100 μm GA3. RNA (10 μg per lane) was used and hybridized with FsPP2C1 cDNA probe. Top, Ethidium bromide-stained gel showing rRNAs. B, Germination percentages of beechnut seeds obtained with the indicated treatments.
Since ABA has been involved in stress responses, other stress conditions were checked in relation to the expression of this clone. A water deficit was imposed by imbibing stratified seeds in the presence of polyethylene glycol (PEG; 30%) at 15°C (optimal temperature for germination of beechnuts), but FsPP2C1 transcript levels were not affected by this treatment. No differences were found in either water or PEG at 15°C, the transcript levels being almost undetectable and even lower than those observed after 6 weeks of stratification (data not shown). PEG-treated seeds were alive and viable after the treatment, as shown by transferring them to water and observing the seedling growth.
Transcripts Tissue Specificity
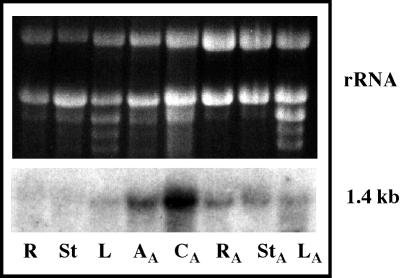
Expression of FsPP2C1 clone in different parts of beechnut seedlings (6 weeks old) and seeds was analyzed for tissue specificity. The FsPP2C1 transcript was found to accumulate in the ABA-treated seeds, mainly in cotyledons but also in embryonic axes, whereas the level of expression in other ABA-treated and untreated tissues like leaves, stems, and roots was almost undetectable (Fig. 3).
Figure 3.
Expression pattern of FsPP2C1 transcripts in different beechnut seed (2 weeks imbibed in ABA) and seedling (6 weeks old) untreated tissues: roots (R), stems (St), and leaves (L); ABA-treated tissues: embryonic axes (AA), cotyledons (CA), roots (RA), stems (StA), and leaves (LA). RNA (10 μg per lane) was used and hybridized with FsPP2C1 cDNA probe. Top, Ethidium bromide-stained gel showing rRNAs.
Phosphatase Activity of Recombinant FsPP2C1 Protein
To confirm that FsPP2C1 protein does show phosphatase activity, the coding fragment of the clone was expressed in E. coli as His tag fusion protein (Fig. 4). Cells carrying the recombinant plasmid were grown and the production of recombinant proteins was induced by the addition of isopropyl β-d-thiogalactopyranoside. The fusion proteins were recovered from E. coli cells that had been grown at 30°C and purified by Ni2+ affinity chromatography according to the manufacturer's instruction (Novagen, Madison, WI). The purified, recombinant protein has an apparent molecular mass (approximately 50 kD) larger than that expected from the amino acid sequence due to the His tag (approximately 3 kD) added to the in vitro expressed protein.
Figure 4.
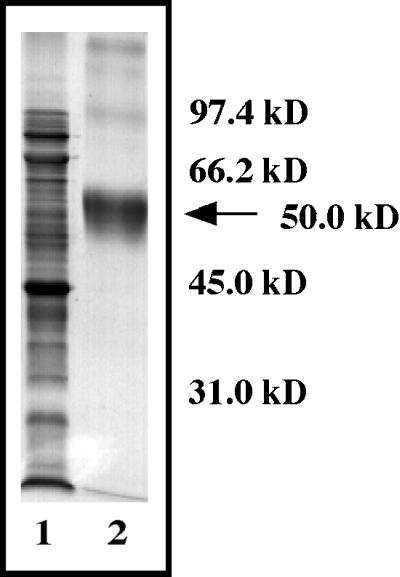
Expression and purification of fusion protein pET-FsPP2C1. Soluble extracts of E. coli (lane 1) and purified protein (lane 2) were resolved by 12% (w/v) SDS-PAGE and stained with Coomassie Brilliant Blue. Position of the fusion protein is indicated by an arrow.
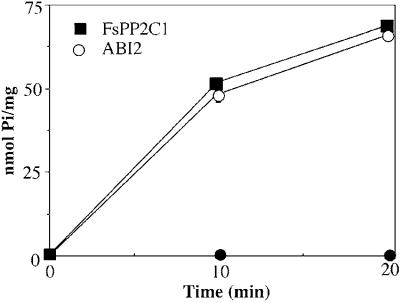
To determine whether FsPP2C1 fusion protein exhibits Mg2+-dependent in vitro phosphatase activity and its ability to dephosphorylate labeled casein (a commonly used artificial substrate for measuring PP2C activity), we carried out a time-dependent phosphatase activity assay and ABI2 from Arabidopsis was used as PP2C activity control. Both fusion proteins exhibit Mg2+-dependent phosphatase activity since no detectable activity was present in the absence of the divalent cation Mg2+ (Fig. 5).
Figure 5.
PP2C activity of FsPP2C1 (▪) and ABI2 (○) proteins. Reaction mixture (90 μL) contained 40 ng of each fusion protein in 50 mm Tris-HCl, pH 7.5, 0.1% (v/v) 2-mercaptoethanol, and 10 mm magnesium acetate. The reactions were incubated with 32P-labeled casein at 30°C for the indicated time periods. Phosphatase activity in the abscense of Mg2+ ions (●). Values have been subtracted from the amount of 32Pi (inorganic phosphate) released in control samples without protein. Each value represents the average of duplicate assays and phosphatase activity is expressed as nmol of 32Pi released per mg of protein.
DISCUSSION
Dormancy constitutes an intrinsic block to germination. Beechnut seeds display an embryo dormancy that can be released by cold treatment (stratification) at 4°C or by application of GA3 (Nicolás et al., 1996). In previous work we reported that the addition of ABA reverses the effect of stratification on the breaking of dormancy (Nicolás et al., 1996). We have cloned and characterized some specific genes encoding for a Gly-rich protein (Nicolás et al., 1997), a GTP-binding protein (Nicolás et al., 1998), and two protein kinases (FsPK1 and FsPK2) (Lorenzo et al., 2000) that are up-regulated by ABA in hydrated growth-arrested seeds. A possible involvement of all these genes in dormancy is suggested by their ABA induction in seeds.
ABA has been shown to play an important role in many of the processes related to the formation, germination, and dormancy of seeds (Bewley, 1997). Our understanding of ABA-mediated processes involving protein phosphorylation/dephosphorylation events during seed development and maturation is just emerging. In developing seeds ABA levels peak at the onset of dessication, resulting in the induction of many ABA-response genes. This increase in ABA levels has been associated with acquisition of dessication tolerance and dormancy in seeds (for review, see Bewley and Black, 1994). During embryo maturation, cell division cycle control is crucial and must be regulated by the ABA signal pathway and thereby converge with other growth-inhibitory and -stimulatory signaling cascades (Iten et al., 1999). Involvement of protein phosphorylation/dephosphorylation processes in these ABA-mediated events are indicated by the identification of ABA regulated protein kinases (PKs) and by genetic evidences suggesting that protein phosphatases are involved in ABA responses (for review, see Verhey and Walker-Simmons, 1997; Campalans et al., 1999; Iten et al., 1999).
In the present study, we isolated and characterized a FsPP2C1 clone using a differential RT-PCR approach to identify PP2C whose levels of expression increased after ABA treatment. Furthemore, we also provide evidence that it corresponds to the plant type-PP2C family. These proteins seem to have a crucial role in different processes of plant growth, including seed dormancy (Himmelbach et al., 1998). The FsPP2C1 deduced protein shows high similarity to Arabidopsis PP2Cs (Fig. 1B), such as AthPP2CA (Kuromori and Yamamoto, 1994), ABI1 (Leung et al., 1994; Meyer et al., 1994), and ABI2 (Leung et al., 1997; Rodríguez et al., 1998a). It contains the PP2C core with the 11 catalytic subdomains present in all Ser/Thr PP2C (Fig. 1A) (Rodríguez, 1998) including the residues binding the phosphate and metal ions (Das et al., 1996). However, the N-terminal domain is different from ABI1 and ABI2 but shows some stretches of amino acid sequence identity with AtPP2CA. These differences in the N-terminal extension have been suggested to facilitate the interaction with different substrates (Rodríguez, 1998). Moreover, these PP2Cs have been involved in the ABA signal tranduction cascade (Himmelbach et al., 1998; Rodríguez, 1998), acting in some cases as negative regulators of ABA responses (Gosti et al., 1999), as recently demonstrated by mutational analysis of ABI1 and AthPP2CA (Sheen, 1998). It is interesting that FsPP2C1 protein sequence also has the two characteristic Gly residues G151 and G157 flanking the DGH154–156 active site uniquely conserved in ABI1, ABI2, and AthPP2CA (Sheen, 1998). Mutations in G180D of abi1, G145D of AthPP2CA, and G168D found in abi2 confer dominant ABA-insensitive effect (Leung et al., 1997; Sheen, 1998) and could have a similar effect in G157 of FsPP2C1.
FsPP2C1 encodes a functional PP2C as demonstrated by the expression of FsPP2C1 in E. coli as His tag fusion protein (Fig. 4). This protein exhibits Mg2+-dependent phosphatase activity as shown in the in vitro phosphatase activity assay. In the absence of the divalent cation Mg2+ no detectable activity was present (Fig. 5). This Mg2+-dependent activity was comparable with another plant PP2C, ABI2 of Arabidopsis (Rodríguez et al., 1998a). The reaction mechanism catalyzed by PP2Cs has been proposed by Das et al. (1996) and it is well established that PP2C depends on either Mg2+ or Mn2+ ions for catalytic activity.
The expression of FsPP2C1 was determined in the seed over 6 weeks of imbibition under the different treatments that maintain or eliminate dormancy (Fig. 2), as well as in different tissues of the young seedling (Fig. 3).
FsPP2C1 expression is specifically induced in seeds upon ABA treatment but not by drought stress (Fig. 2), while stratification or gibberellin treatment decrease the level of transcripts. Therefore, expression of FsPP2C1 negatively correlates with germination and is abolished by treatments that break seed dormancy. Furthermore, this gene is specifically expressed in ABA-treated dormant seeds (Fig. 3) and as far as we know, FsPP2C1 is the first plant type-PP2C expressed preferably in seeds, since other previously reported PP2Cs are also expressed in vegetative tissues.
Other PP2Cs similar to FsPP2C1, such as ABI1 and ABI2, have been involved in different stress responses (Leung et al., 1997). However, FsPP2C1 transcripts were not affected by water deficit and they do not show the expression profile described for other ABA-induced dessication-related proteins. Besides, its expression increases after 2 weeks imbibition for all treatments (Fig. 2A), which negatively correlates to dessication, suggesting an important role for this gene during early weeks of stratification. At longer periods the expression decreases except in the presence of ABA, supporting our proposal of an specific up-regulation by ABA.
Taken together, these results suggest that FsPP2C1 might play a role in ABA-induced seed dormancy. However, it is not clear whether FsPP2C1 expression is a cause or effect of non-germination in ABA-treated seeds and genetic evidence is necessary to firmly establish whether FsPP2C1 is a positive regulator of seed dormancy. As transgenic work is not feasible in beechnut, we have initiated the construction of Arabidopsis plants that overexpress FsPP2C1. We will also evaluate the ABA response of plants with altered levels of AthPP2CA, the best match of FsPP2C1 in the Arabidopsis genome.
MATERIALS AND METHODS
Plant Material and Germination Conditions
Beechnut (Fagus sylvatica) seeds were obtained from the Danish State Forestry Tree Improvement Station. Seeds were dried to a moisture content of 10% and stored at −4°C in sealed jars. The pericarp was manually removed, and seeds were previously sterilized in 1% (w/v) sodium hypoclorite before imbibition in sterile water containing 100 μm ABA, 1 mm CaCl2, 100 μm ABA + 1 mm CaCl2 or 100 μm GA3. Seeds were maintained in the different media at 4°C from 1 to 6 weeks. Drought stress conditions were generated by imbibing seeds (maintained during 6 weeks at 4°C in water) from 2 to 6 d in 30% (w/v) PEG at 15°C.
Seedlings were obtained from 4 weeks-stratified seeds sown in a controlled environment chamber under a 12-h-light and 12-h-dark cycle at 15°C in moist vermiculite and harvested after 6 weeks. ABA-treated seedlings were watered every 2 d and misted daily with a solution of 100 μm ABA, and the corresponding tissues were collected 6 d afterward. Then, treated or untreated seedlings were separated into roots, leaves, and stems. All collected tissues were frozen in liquid nitrogen and stored at −80°C.
Differential RT-PCR Approach
Total RNA from either ABA-treated or GA3-treated seeds was extracted using the Qiagen pack-500 cartridge (Qiagen USA, Valencia, CA) following the manufacturer's protocol. Poly(A+) RNA was purified from each preparation of total RNA by affinity chromatography in oligo(dT)-cellulose columns using the mRNA Purification Kit (Pharmacia Biotech, Piscataway, NJ). cDNA was synthesized from 1 μg of poly(A+) RNA prepared either from ABA-treated or GA3-treated seeds using the 1st Strand cDNA Synthesis kit for RT-PCR AMV (Roche Diagnostics, Mannheim, Germany) with oligo-p(dT) used as a primer and following the manufacturer's instruction. Each cDNA was used as a template for a PCR reaction with degenerate oligonucleotides corresponding to two subdomains conserved among the Ser/Thr protein phosphatases (Rodríguez, 1998). The forward primer consisted of a 18-mer of the sequence 5′-GA(T,C) GG(T,C) CATGG(A,C) GG(C,T)(T,C)(C,A) T-3′ encoding the DGHGGS amino acid sequence (subdomain 2) and the reverse primer was a 24-mer of the sequence 5′-ATCCCAAA(G,C)(A,C) CC(A,G) TCACT(G,T,C) GC(T,C) AA-3′ corresponding to the LASDGLWD amino acid sequence (subdomain 8). The conditions of PCR were as follows: 1 min 94°C, 2 min 45°C, 2 min 72°C for 30 cycles, and 10 min 72°C, using [α-35S] ATP. PCR products were fractionated on a 6% (w/v) acrylamide gel, dried, and exposed to autoradiographic films. A DNA band of approximately 550 bp was amplified using as a template cDNA prepared from ABA-treated seeds. In contrast, this DNA band was absent when cDNA prepared from GA3-treated seeds was used as a template. The 550-bp DNA band was excised from the gel, re-amplified under the same PCR conditions, cloned into the pCR 2.1 vector (Original TA Cloning kit, Invitrogen, Carlsbad, CA), and sequenced. As the predicted gene product encoded by this clone revealed homology to PP2C, we named it FsPP2C1.
Isolation of the cDNA Clone
The full-length cDNA clone was isolated from a cDNA library constructed using poly(A+) RNA from seeds imbibed in 100 μm ABA for 2 weeks as a template (Nicolás et al., 1997) with FsPP2C1 PCR fragment as a probe. Double stranded cDNA was synthesized using a cDNA kit (Stratagene, La Jolla, CA) following the manufacturer's instructions and inserted between the EcoRI and XhoI sites of the Uni-ZAP XR vector (Stratagene). The recombinant cDNA of the clones selected for further analysis (FsPP2C1) was excised from the phage in pBluescript SK+ (Stratagene) using the biological rescue recommended by the supplier.
DNA Sequencing
Plasmid DNA templates were isolated by the Wizard Plus Minipreps DNA Purification System (Promega, Madison, WI). Determination of the nucleotide sequence of the cDNA clone was performed on a ABI 377 sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA) using the Taq DyeDeoxy Terminator Cycle Sequencing kit. The DNA and deduced protein sequences were compared to other sequences in the EMBL databases (GenBank and SwissProt, respectively), using the FASTA algorithm (Pearson and Lipman, 1988).
Northern-Blot Analysis
In northern-blot analysis, 10 μg of total RNA isolated from treated seeds or seedlings were fractionated in denaturing formaldehyde agarose gels and transferred to nylon membranes (Hybond N+, Amersham, Buckinghamshire, UK) with 25 mm sodium-phosphate buffer (pH 6.8). Blotted membranes were hybridized with a FsPP2C1 probe labeled with 32P using the Random primed kit (Roche Diagnostics, Mannheim, Germany) at 42°C overnight in 5× SSC (1× SSC is 0.15 m NaCl, 15 mm Tri-sodium citrate), 1% (w/v) SDS, 5× Denhardt's solution (1× Denhardt's solution is 0.02% [w/v] bovine serum albumin, 0.02% [w/v] Ficoll 400, and 0.02% [w/v] polyvinylpyrrolidone) plus 50% (w/v) deionized formamide. Membranes were washed at 42°C twice with 2× SSC, 0.1% (w/v) SDS for 5 min each and once with 0.5× SSC, 0.1% (w/v) SDS for 15 min. They were then exposed to X-Omat films (Kodak, Rochester, NY).
Expression and Purification of FsPP2C1 Recombinant Protein
The coding region of FsPP2C1 was amplified by PCR with primers 5′-CATATGGCTGGGATTTGCTGT-3′ (which contained the NdeI site underlined and the translation-initiation codon in bold) and 5′-CTCGAGCTATTGTTGATGATT-3′ (which contained the XhoI site underlined and the stop codon in bold), subcloned in frame into the NdeI and XhoI sites of the pET28a(+) vector (Novagen), and verified by DNA sequencing. FsPP2C1 protein was expressed in Escherichia coli BL21(DE3) as His-tag fusion protein, cells carrying the recombinant plasmid were grown at 30°C in 2× YT until A600 reached 0.6 units, and recombinant protein was induced by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 1 mm. Cells were harvested by centrifugation at 5,000g for 5 min after 2 h of induction and lysed using Bug Buster protein extraction reagent (Novagen). Fusion protein was affinity-purified using Ni2+-resin (Novagen) according to the manufacturer's instruction. Proteins were analyzed by SDS-PAGE and Coomassie Blue staining.
The pMalc2-ABI2 construct was provided by Dr. P.L. Rodríguez and fusion protein was purified with amylose resin (New England Biolabs, Beverly, MA) following the manufacturer's protocol.
Protein Determination
Protein concentration was measured using a Bio-Rad Laboratories protein assay kit based on the method of Bradford (1976) with bovine serum albumin as standard.
Assay for Phosphatase Activity
For the PP2C activity assays 40 ng of the fusion proteins were used in a 90-μL reaction mixture (Rodríguez et al., 1998a), containing 50 mm Tris-HCl, pH 7.5, 0.1% (v/v) 2-mercaptoethanol, and 10 mm magnesium acetate. The reactions were incubated with 32P-labeled casein (McGowan and Cohen, 1988) at 30°C. Aliquots (30 μL) were removed at the indicated time periods, and the reaction was stopped by addition of 100 μL of 20% (w/v) trichloroacetic acid. Samples were centrifuged, 100 μL of the upper phase were mixed with 1 mL of Ready Safe Liquid Scintillation Cocktail (Beckman Instruments, Fullerton, CA) and radioactivity determined in a Tri-Carb 2100 TR Liquid Scintillation Analyzer (Packard Instruments, Meriden, CT).
Footnotes
This work was supported by the Dirección General de Investigación Científica y Técnica (Spain; grant no. PB96–1313), by Junta de Castilla y León (grant no. SA60/99), and by a research fellowship from Universidad de Salamana (Salamanca, Spain; to O.L.).
LITERATURE CITED
- Anderberg RJ, Walker-Simmons MK. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinase. Proc Natl Acad Sci USA. 1992;89:10183–10187. doi: 10.1073/pnas.89.21.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Black M. Dormancy and the control of germination. In: Bewley JD, Black M, editors. Seeds: Physiology of Development and Germination. Ed 2. New York: Plenum Press; 1994. pp. 199–271. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Campalans A, Messeguer R, Goday A, Pagès M. Plant responses to drought, from ABA signal transduction events to the action of the induced proteins. Plant Physiol Biochem. 1999;37:327–340. [Google Scholar]
- Chandler PM, Robertson M. Gene expression regulated by abscisic acid and its relation to stress tolerance. Annu Rev Plant Physiol. 1994;45:113–141. [Google Scholar]
- Das AK, Helps NR, Cohen PTW, Barford D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 Å resolution. EMBO J. 1996;15:6798–6809. [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AAR, Vartanian N, Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signalling. Plant Cell. 1999;11:1897–1909. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilhorst HWM. A critical update on seed dormancy I: primary dormancy. Seed Sci Res. 1995;5:61–73. [Google Scholar]
- Himmelbach A, Iten M, Grill E. Signalling of abscisic acid to regulate plant growth. Phil Trans R Soc Lond B Biol Sci. 1998;353:1439–1444. doi: 10.1098/rstb.1998.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iten M, Hoffman T, Grill E. Receptors and signalling components of plant hormones. J Recept Signal Transduct Res. 1999;19:41–58. doi: 10.3109/10799899909036636. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant. 1984;61:377–383. [Google Scholar]
- Kuromori T, Yamamoto M. Cloning of cDNAs from Arabidopsis thaliana that encode putative protein phosphatase 2C and a human Dr1-like protein by transformation of a fission yeast mutant. Nucleic Acid Res. 1994;22:5296–5301. doi: 10.1093/nar/22.24.5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J. Abscisic acid signal transduction. Annu Rev Plant Physiol. 1998;19:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Rodríguez D, Nicolás C, Nicolás G. Characterization and expression of two protein kinase and an EIN3-like genes, which are regulated by ABA and GA3 in dormant Fagus sylvatica seeds. In: Black M, Bradford KJ, Vazquez-Ramos J, editors. Seed Biology: Advances and Applications. Wallingford, UK: CAB International; 2000. pp. 329–340. [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito M. A two component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- McGowan CH, Cohen P. Protein phosphatase-2C from rabbit skeletal muscle and liver: an Mg2+-dependent enzyme. Methods Enzymol. 1988;159:416–426. doi: 10.1016/0076-6879(88)59041-9. [DOI] [PubMed] [Google Scholar]
- Meskiene I, Bogre L, Glaser W, Balog J, Brandstotter M, Zwerger K, Ammerer G, Hirt H. MP2C, a plant protein phosphatase 2C, function as a negative regulator of mitogen activated protein kinase pathways in yeast and plants. Proc Natl Acad Sci USA. 1998;95:1938–1943. doi: 10.1073/pnas.95.4.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- Moore F, Weekes J, Hardie DG. Evidence that AMP triggers phosphorylation as well as direct allosteric activation of rat liver AMP-activated protein kinase. Eur J Biochem. 1991;199:691–697. doi: 10.1111/j.1432-1033.1991.tb16172.x. [DOI] [PubMed] [Google Scholar]
- Nicolás C, Nicolás G, Rodríguez D. Antagonistic effects of abscisic acid and gibberellic acid on the breaking of dormancy of Fagus sylvatica seeds. Physiol Plant. 1996;96:244–250. [Google Scholar]
- Nicolás C, Nicolás G, Rodríguez D. Transcripts of a gene, encoding a small GTP-binding protein from Fagus sylvatica, are induced by ABA and accumulated in the embryonic axis of dormant seeds. Plant Mol Biol. 1998;36:487–491. doi: 10.1023/a:1005906601446. [DOI] [PubMed] [Google Scholar]
- Nicolás C, Rodríguez D, Poulsen F, Eriksen EN, Nicolás G. The expression of an abscisic acid-responsive glycine-rich protein coincides with the level of seed dormancy in Fagus sylvatica. Plant Cell Physiol. 1997;38:1303–1310. doi: 10.1093/oxfordjournals.pcp.a029122. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence analysis. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez PL. Protein phosphatases 2C (PP2C) function in higher plants. Plant Mol Biol. 1998;38:919–927. doi: 10.1023/a:1006054607850. [DOI] [PubMed] [Google Scholar]
- Rodríguez PL, Benning G, Grill E. ABI2, a second protein phosphatase 2C involved in ABA signal transduction in Arabidopsis. FEBS Lett. 1998a;421:185–190. doi: 10.1016/s0014-5793(97)01558-5. [DOI] [PubMed] [Google Scholar]
- Rodríguez PL, Leube MP, Grill E. Molecular cloning in Arabidopsis thaliana of a new protein phosphatase 2C (PP2C) with homology to ABI1 and ABI2. Plant Mol Biol. 1998b;38:879–883. doi: 10.1023/a:1006012218704. [DOI] [PubMed] [Google Scholar]
- Schneider WL, Gifford DJ. Loblolly pine seed dormancy I: the relationship between protein synthesis and the loss of dormancy. Physiol Plant. 1994;90:246–252. [Google Scholar]
- Sheen J. Mutational analysis of protein phosphatases 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci USA. 1998;95:975–980. doi: 10.1073/pnas.95.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K, Russell P. Counteractive roles of protein phosphatase 2C (PP2C) and a MAP kinase kinase homolog in the osmoregulation of fission yeast. EMBO J. 1995;14:492–502. doi: 10.1002/j.1460-2075.1995.tb07025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas AJ. Timing and memory processes in seed embryo dormancy: a conceptual paradigm for plant development questions. Bioessays. 1988;6:87–92. [Google Scholar]
- Verhey SD, Walker-Simmons MK. Abscisic acid-mediated responses in seeds involving protein kinases and phosphatases. In: Ellis RM, Black M, Murdoch AJ, Hong TD, editors. Basic and Applied Aspects of Seed Biology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 225–233. [Google Scholar]