Introduction
Cancer burden has raised from 16.8 million to 18.1 million new cases with 9.6 million cancer deaths in 2018 alone.1, 2 Leukemia is the thirteenth most common cancer with 4.37 lakh new cases, and 2.11 lakh related deaths globally.2 Based on the origin of the predominant cell type (lymphoid or myeloid) and the rate of disease progression (acute or chronic), leukemia is categorized into four major subtypes: acute lymphoid leukemia (ALL), chronic lymphocytic leukemia (CLL), acute myeloid leukemia (AML), and chronic myelogenous leukemia (CML).3, 4
Of which, the incidence of ALL and AML are 1.08 lakh and 1.40 lakh, respectively.4 ALL is a common cancer in children and is caused by uncontrolled production of bone marrow hematopoietic precursor cells.5 Advancement in the therapeutics has increased the overall survival rate but influencing factors such as lineage switching and the rise of mixed lineages at relapses or with chemotherapy often changes the prognosis of the illness. Lineage switching is a rare phenomenon in which ALL transforms from lymphoid to myeloid lineage or vice versa. This situation rarely occurs especially in adults, and the prognosis is highly variable. The specific causative factors, central mechanisms involved in these phenomena are still unclear and yet to be identified. In the present case report, we are presenting a rare case of lineage switching of ALL to AML in a young adult of 19 years during chemotherapy.
Case presentation
A 19-year-old male was presented at our hematology center due to weakness, weight loss, and pancytopenia. Upon physical examination, he was pale, with bone pain, and referred asthenia. Other physical examinations have shown no other abnormalities. Initial laboratory tests were performed. From complete blood count (CBC) decreased total leukocyte count was observed and using peripheral blood smear (PBS) blast cells were identified (Figure 1B and D). Bone and marrow aspirate (BMA) and flow cytometric analysis disclosed CD19 (+), CD7 (+), HLA-DR (+), and CD38 (+) in suggestive of B-ALL (Table 1 and Figure 1A). Fluorescence in situ Hybridization (FISH) test for BCR/ABL;t(9:22), MLL gene rearrangement; t(11q23), E2A gene rearrangement, TEL/AML1 ES; t(12;21) was performed using TEL/AML1 ES: t(12;21) (p13;q22): (ETV6/RUNX1) dual color translocation probe, VYSIS (Abbott), Zytolight SPEC BCR/ABL1 dual color dual fusion probe, E2A gene rearrangement:t(19p13); dual color break apart probe, Cytotest, t(11q23): LSI MLL dual color, break apart rearrangement probe, and metasystems. Reports from FISH analysis was revealed to be negative (Figure 1E). RT- PCR for BCR-ABL rearrangements tests was not done. Baseline cytogenetics was normal. Augmented Berlin-Frankfurt-Munster (aBFM)-90 regimen was initiated. Induction phase was non-eventful. However, Post induction marrow MRD by flow cytometry was negative.
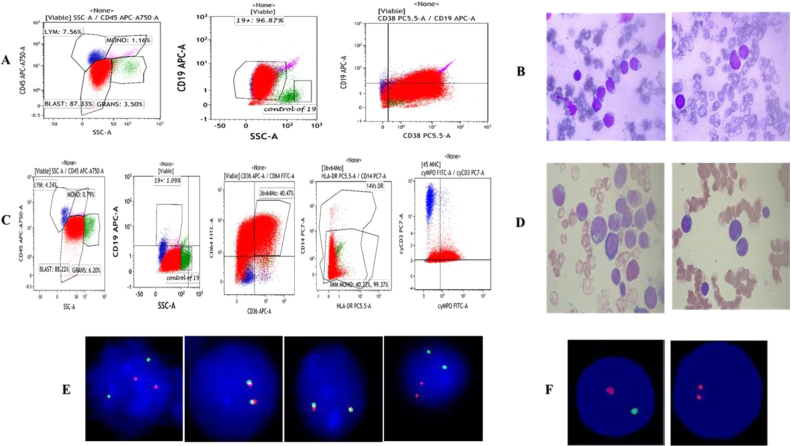
Figure 1.
Salient features of the immunophenotype, at diagnosis: (A) blasts (red) co-express CD19/CD34 and CD38 antigens, whereas they do not express CD10, CD20, CD33 or CD117 antigens. The expressions of other myeolid and lymphoid antigens of the T lineage was not detected. (B) Bone marrow aspiration images at diagnosis.
Salient features of the immunophenotype, at relapse: (C) blasts (red) now co-express CD36/CD64/Cy MPO and CD117 while they do not express the CD19 antigen. Color indicators: red color – blasts, green – granulocytes, blue – lymphocytes, and violet – monocytes. (D) Bone marrow aspiration images at relapse.
(E) FISH study at diagnosis (left to right): interphase cell showing 2 orange, 2 green signals indicating BCR/ABL: Ph negative status.
Signal pattern showing 2 fusion (yellow) signals indicative of negative status for E2A gene rearrangement/translocation.
Signal pattern showing 2 fusion (yellow) signals indicative of negative status for MLL; t(11q23) gene rearrangement/translocation.
Interphase cell showing 2 orange, 2 green signals indicating TEL/AML1 ES: t(12;21) (p13;q22):negative status.
(F) FISH study at replapse: on the left – cell showing one orange and one green signals, indicating positive for loss of chromosome 17p13, locus (90%), nuc ish (TP53, NF1) × 1[180/200]; on the right – cell showing two orange signals, indicating negative for deletion of the 20q12 locus, nuc ish (D20S108 × 3) [20/200].
Table 1.
Clinical and laboratory features of the ALL at diagnosis and AML at transformation.
| Clinical data | At diagnosis | Follow up | At relapse |
|---|---|---|---|
| Bone marrow blast count | 96% lymphoblasts | Less than 3% blast/hematogones | 88% myeloblast |
| Flow cytometry | HLA-DR+, CD7+ (dim), CD19+, CD38+, MPO−, CD13−, CD15−, CD33−, and CD34− | MRD undetectable | MPO+, CD13+ (dim), CD15+, CD33+, CD38+, CD64+, CD 36+, CD19-, CD22−, and CD117− |
| Cytogenetics | Baseline cytogenetics normal | Not tested | t(15 17) (q21 q22), Inv 16 (p13 q22), and t(8 21) (q22 q22) negative |
| FISH | FISH for ALL: BCR/ABL: t(9:22) – normal MLL gene rearrangement; t(11q23) – normal E2A gene rearrangement – normal TEL/AML1 ES; t(12;21) – normal | Not tested | 17p13 loss positive |
| Lumbar puncture | Negative | Not tested | Negative |
So the patient was proceeded with the consolidation phase, with time the patient has developed severe pain in legs and body aches. Once again, all the tests such as CBC, PBS, flow cytometry, cytogenetics and FISH were performed. Where CBC has shown an increase in total leukocyte count with peripheral blood smear showing blasts. Flow cytometry results have shown myeloid and monocytic markers CyMPO, CD13, CD33, CD117, CD36, CD64, CD10, CD15, CD38, CD4 and CD7confirming AML with monocytic differentiation (Table 1 and Figure 1C). Whereas cytogenetics leukemia translocation panel was negative for t(15 17) (q21 q22), Inv 16 (p13 q22),t(8 21) (q22 q22) and FISH was positive for loss of the TP 53(17p 13), confirming the lineage switch (Table 1 and Figure 1F). FLAG-IDA chemotherapy regimen followed by allogeneic hematopoietic stem cell transplantation (HSCT) was planned. As on date, the patient is on chemotherapy with regular follow-ups.
Discussion
Lineage switch from ALL to AML in adolescents and adults are very rare. We present a young adult patient case in which de novo leukemia underwent lineage switch from ALL to AML. The pathogenesis for lineage switch in acute leukemia (AL) is currently unknown. Three hypotheses have been proposed to explain this unusual event. These include mechanisms of a common progenitor, trans-differentiation, and dedifferentiation.6 A lineage switch may represent either the emergence of a new leukemic clone or high plasticity attributes or a relapse of the original clone with heterogeneity at the morphological level.5, 7 It has been noted that in many patients, lineage switching occurs at relapse.5, 8, 9
With respect to our patient, initial bone marrow aspirate findings were in favor of AL. Immunophenotyping by flow cytometry of bone marrow sample was also in suggestive of B-ALL. FISH for ALL was negative and baseline cytogenetics were normal. Over period of time, patient was relapsed during chemotherapy. Peripheral blood smear was performed and reappearance of blasts was observed. Immunophenotyping studies were performed, suggesting AML with monocytic differentiation. Further reports from AML translocation panel was negative, whereas cytogenetic studies were positive for loss of tp 53 (17p 13) locus (90%), confirming the lineage switching, as shown in Table 1.
The mutation or loss of p53 gene is most frequently seen in solid tumors and many other cancer types.10 However, their frequency in hematological malignancies is relatively very low. p53 gene demonstrated a pivotal role in disease development, progression, regulation of cell proliferation, apoptosis, maintenance of genome integrity, chromosomal stability, resistance to DNA-damaging drugs, responding, and repairing the damaged DNA. Any mutation or loss of p53 function will cause genomic instability, chemotherapy resistance, and cell cycle deregulation.10, 11 Nevertheless, a strong correlation was found to be associated with mutation or loss of p53 and its related unfavorable prognostic factors and resistance to chemotherapy in hematological malignancies.12, 13 Rossi et al. presented 7 cases of lineage switching from B-ALL to AML at relapse. In all these 7 cases, 11q23/MLL gene abnormalities were detected.14 In another study, 18 pediatric lineage switch cases were reported because of chromosomal aberrations involving 11q rearrangements.5
Over 25% of leukemia patients suffer relapses and among them lineage switching was observed to be very rare. Criteria for such lineage switching might be morphological, immunophenotypic and cytogenetic alterations in the same original cell lineage (lymphoid or myeloid).5, 15, 16, 17 In cytogenetic alterations, very low incidence of p53 mutations (5%) were reported in infants and young adults with AML/ALL.18, 19, 20 No studies have conformed the possible reasons for p53 mutations. In many instances, p53 mutated AML patients were observed to be having a significantly inferior complete remission duration, lower response rate, and overall survival.18
Despite tremendous progress and advancement in the knowledge and technology in understanding the AL, still many questions to be addressed about the mechanisms driving this lineage switching. Ultimately this phenomenon leads to poor prognosis and resistance to therapy. Hence much more research has to be done in deciphering, understanding and unfolding the underlying mechanisms of these gene functions, interrelationships and their role in these cases for further better management.
Financial disclosure
The authors declared that this study has received no financial support.
Conflict of interest
The authors have no conflicts of interest to declare.
Informed consent
Written informed consent was obtained from the patient.
Acknowledgements
The authors would like to thank Dr. Yasam Venkata Ramesh from HCG Manavata Cancer Centre, Centre for Difficult Cancers (CDC), Nashik, India, for his medical writing assistance.
References
- 1.World Health Organization (WHO). Latest global cancer data. https://www.who.int/cancer/PRGlobocanFinal.pdf [cited 2020 January 10].
- 2.Global Burden of Disease Cancer Collaboration Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: a systematic analysis for the global burden of disease study. JAMA Oncol. 2019;5(12):1749–1768. doi: 10.1001/jamaoncol.2019.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.https://www.wcrf.org/dietandcancer/cancer-trends/worldwide-cancer-data [cited 10.01.20].
- 4.Hao T., Li T., Buck M.A., Buck A., Chen W. An emerging trend of rapid increase of leukemia but not all cancers in the aging population in the United States. Sci Rep. 2019;9:12070. doi: 10.1038/s41598-019-48445-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acosta D.E., Pelayo R. Lineage switching in acute leukemias: a consequence of stem cell plasticity? Bone Marrow Res. 2012;2012:406796. doi: 10.1155/2012/406796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu B., Jug R., Luedke C., Su P., Rehder C., McCall C., et al. Lineage switch between B-lymphoblastic leukemia and acute myeloid leukemia intermediated by “occult” myelodysplastic neoplasm: two cases of adult patients with evidence of genomic instability and clonal selection by chemotherapy. Am J Clin Pathol. 2017;148(2):136–147. doi: 10.1093/ajcp/aqx055. [DOI] [PubMed] [Google Scholar]
- 7.Krawczuk R.M., Zak J., Jaworowska B. A lineage switch from AML to ALL with persistent translocation t(4;11) in congenital leukemia. Med Pediatr Oncol. 2003;41:95–96. doi: 10.1002/mpo.10276. [DOI] [PubMed] [Google Scholar]
- 8.Cobaleda C., Jochum W., Busslinger M. Conversion of mature B-cells into T-cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 9.Park M., Koh K.N., Kim B.E., Im H.J., Jang S., Park C.J., et al. Lineage switch at relapse of childhood acute leukemia: a report of four cases. J Korean Med Sci. 2011;26:829–831. doi: 10.3346/jkms.2011.26.6.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris C.C. Structure and function of the p53 tumor suppressor gene: clues for rational cancer therapeutic strategies. J Natl Cancer Inst. 1996;88:1442–1455. doi: 10.1093/jnci/88.20.1442. [DOI] [PubMed] [Google Scholar]
- 11.Kirsch D.G., Kastan M.B. Tumor-suppressor p53: implications for tumor development and prognosis. J Clin Oncol. 1998;16:3158–3168. doi: 10.1200/JCO.1998.16.9.3158. [DOI] [PubMed] [Google Scholar]
- 12.Krug U., Ganser A., Koeffler H.P. Tumor suppressor genes in normal and malignant hematopoiesis. Oncogene. 2002;21:3475–3495. doi: 10.1038/sj.onc.1205322. [DOI] [PubMed] [Google Scholar]
- 13.Preudhomme C., Fenaux P. The clinical significance of mutations of the p53 tumor suppressor gene in haematological. Br J Haematol. 1997;98:502–511. doi: 10.1046/j.1365-2141.1997.2403057.x. [DOI] [PubMed] [Google Scholar]
- 14.Rossi J.G., Bernasconi A.R., Alonso C.N., Rubio P.L., Gallego M.S., Carrara C.A., et al. Lineage switch in childhood acute leukemia: an unusual event with poor outcome. Am J Hematol. 2012;87:890–897. doi: 10.1002/ajh.23266. [DOI] [PubMed] [Google Scholar]
- 15.Park B.G., Park C.J., Jang S., Seo E.J., Chi H.S., Lee J.H. Erythroleukemia relapsing as precursor B-cell lymphoblastic leukemia. Korean J Lab Med. 2011;31:81–85. doi: 10.3343/kjlm.2011.31.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grammatico S., Vitale A., Starza R.L., Gorello P., Angelosanto N., Negulici A.D., et al. Lineage switch from pro-B acute lymphoid leukemia to acute myeloid leukemia in a case with t(12;17)(p13;q11)/TAF15–ZNF384 rearrangement. Leuk Lymphom. 2013;54:1802–1805. doi: 10.3109/10428194.2012.753450. [DOI] [PubMed] [Google Scholar]
- 17.Chung H.J., Park C.J., Jang S., Chi H.S., Seo E.J., Seo J.J. A case of lineage switch from acute lymphoblastic leukemia to acute myeloid leukemia. Korean J Lab Med. 2007;27:102–105. doi: 10.3343/kjlm.2007.27.2.102. [DOI] [PubMed] [Google Scholar]
- 18.Kadia T.M., Jain P., Ravandi F., Garcia-Manero G., Andreef M., Takahashi K., et al. TP53 mutations in newly diagnosed acute myeloid leukemia: clinicomolecular characteristics, response to therapy, and outcomes. Cancer. 2016;122:3484–3491. doi: 10.1002/cncr.30203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Megonigal M.D., Rappaport F.E., Nowell P.C., Lange B.J., Felix C.A. Potential role for wild-type p53 in leukemias with MLL gene translocations. Oncogene. 1998;16:1351–1356. doi: 10.1038/sj.onc.1201637. [DOI] [PubMed] [Google Scholar]
- 20.Kawamura M., Ohnishi H., Guo S.X., Sheng X.M., Minegishi M., Hanada R., et al. Alterations of the p53, p21, p16, p15 and RAS genes in childhood T-cell acute lymphoblastic leukemia. Leukemia. 1999;23:115–126. doi: 10.1016/s0145-2126(98)00146-5. [DOI] [PubMed] [Google Scholar]