Abstract
We investigated the influence of light of different wavelengths on the expression of the psbA gene, which encodes the D1 protein of the photosystem II and the psaE gene, which encodes the subunit Psa-E of the photosystem I, in Synechocystis sp PCC 6803. In an attempt to differentiate between a light-sensory and a redox-sensory signaling processes, the effect of orange, blue, and far-red light was studied in the wild-type and in a phycobilisome-less mutant. Transferring wild-type cells from one type of illumination to another induced changes in the redox state of the electron transport chain and in psbA and psaE expression. Blue and far-red lights (which are preferentially absorbed by the photosystem I) induced an accumulation of psbA transcripts and a decrease of the psaE mRNA level. In contrast, orange light (which is preferentially absorbed by the photosystem II) induced a large accumulation of psaE transcripts and a decrease of psbA mRNA level. Transferring mutant cells from blue to orange light (or vice versa) had no effect either on the redox state of the electron transport chain or on the levels of psbA and psaE mRNAs. Thus, light quality seems to regulate expression of these genes via a redox sensory mechanism in Synechocystis sp PCC 6803 cells. Our data suggest that the redox state of one of the electron carriers between the plastoquinone pool and the photosystem I has opposite influences on psbA and psaE expression. Its reduction induces accumulation of psaE transcripts, and its oxidation induces accumulation of psbA mRNAs.
Cyanobacteria, like algae and higher plants, have developed a large number of mechanisms to allow optimal use of absorbed light energy and to avoid oxidative damage induced by excessive excitation (photoinhibition; for review, see Prasil et al., 1992; Aro et al., 1993; Keren and Ohad, 1998). Short-term regulatory responses involve modifications of the organization of the existent photosynthetic apparatus. These processes are required to redistribute the energy absorbed by the photosystems I and II (PSI and PSII) (state transitions; for review, see Allen, 1992a, 1995; Keren and Ohad, 1998) and to decrease the number of photons arriving at the reaction centers (photoprotection; for review, see Niyogi, 1999). Changes in the redox state of the electron transport chain and in the transthylakoid proton gradient induce these mechanisms. In green algae and higher plants the mechanism of state transitions involves redox-dependent phosphorylation/dephosphorylation reactions of the PSII antenna, inducing a movement of part of the antenna between the PSII and the PSI. These processes are accompanied by changes in the fluorescence emission of both photosystems (for review, see Williams and Allen, 1987; Biggins and Bruce, 1989; Allen, 1992b). The mechanisms involved in state transition in cyanobacteria, which contains phycobilisomes as PSII antenna, are not as well characterized as in higher plants. It has been proposed that a lateral diffusion of the phycobilisomes leads to changes in the cross-section of PSII and PSI antennae (Mullineaux et al., 1986, 1997; Mullineaux and Holzwarth, 1990). However, results from other laboratories suggested that the mobile elements are the photosystems (Olive et al., 1986, 1997; Schluchter et al., 1996; El Bissati et al., 2000).
Long-term responses involve changes in the expression of genes encoding proteins related to photosynthesis. This regulation serves to replace the damaged proteins during light stress and to adapt the stoichiometry of the photosystems to the different light conditions. Plants have evolved multiple light-sensing (photoreceptors) and redox-sensing systems for the perception of the level and quality of light on the surrounding environment. There are at least three types of photoreceptors: the red/far-red-absorbing phytochromes; the blue/UV-A-absorbing cryptochromes; and the UV-B receptors (for review, see Rüdiger, 1992; Quail et al., 1995; Furuya and Schafer, 1996). In cyanobacteria, the recent sequencing of the Synechocystis sp PCC 6803 chromosome revealed six genes (slr0473, sll1124, sll1473, sll0821, slr1393, slr1212) showing different degrees of similarity to the chromophore binding site of plant phytochrome (Kaneko et al., 1996). The ORF slr0473, cph1, shows the largest homology to plant phytochromes. It encodes a polypeptide that is able, in vitro, to autocatalytically attach the linear tetrapyrrole chromophores, phycocyanobilin and phytochromobilin, and to exhibit a red/far-red photochemistry similar to plant phytochromes (Hughes et al., 1997; Lamparter et al., 1997; Yeh et al., 1997). Its role in vivo is still unknown. The ORF sll1124, plpA, which also shows homology to the chromophore binding site of phytochromes and His kinases, encodes a protein that is needed for blue light growth of Synechocystis sp PCC 6803 cells (Wilde et al., 1997).
Depending on the redox state of the photosynthetic electron transport chain, redox-sensing molecules modulate the expression of genes (Allen, 1993; Allen et al., 1995). Light energy is transduced into chemical energy by the PSII and the PSI complexes operating in series. Water oxidation and quinone reduction occur in PSII and plastocyanin oxidation and NADP+ reduction in PSI. The electron transport from water to NADP+ is coupled to a proton translocation across the membrane, generating a proton motive force for ATP synthesis. NADPH (reducing power) formation is related only to linear electron transfer between the PSII and the PSI, whereas ATP (chemical energy) is also synthesized during the cyclic electron transfer around the PSI. The redox state of the different components of the photosynthetic electron transport chain is modulated by light intensity and quality. An emerging idea is that in mature plants, the redox-sensing processes are very important on the regulation of photosynthetic genes (Durnford and Falkowski, 1997). It has been reported that in chloroplasts, the redox state of the plastoquinone (PQ) pool is involved in the transcriptional regulation of cab genes, encoding the family of light-harvesting complex proteins (Escoubas et al., 1995), and the genes encoding PSI and PSII proteins (Pfannschmidt et al., 1999). On the other hand, the translation of transcripts of the chloroplast psbA gene, encoding the D1 protein of the reaction center of PSII, appears to be regulated by the redox state of thiol proteins (like thioredoxine) (Danon and Mayfield, 1994a, 1994b).
In the past we have studied the light regulation of two photosynthetic genes, the psbA and the psaE genes, in a strain of cyanobacteria, Synechocystis sp PCC 6803. As stated above, the psbA gene encodes the D1 protein that, together with the D2 protein, forms the core of PSII. The heterodimer binds all the cofactors involved in primary charge separation and stabilization leading to water oxidation and reduction of plastoquinone. The psaE gene encodes the subunit Psa-E of the PSI involved in ferredoxin binding (Rousseau et al., 1993).
In cyanobacteria, light essentially modulates the transcription of the psbA genes (for review, see Golden, 1995). In Synechocystis sp PCC 6803 and PCC 6714 there are three psbA genes: psbA1, psbA2, and psbA3 (Ajlani et al., 1989; Mohamed and Jansson, 1989). The psbA1 copy is silent and never expressed. The psbA2 and psbA3 copies share 99% of nucleotide identity and encode an identical D1 protein (Mohamed and Jansson, 1989; Bouyoub et al., 1993). The level of expression of the two copies however, is different. Approximately 80% to 90% of the psbA transcripts originate from psbA2, whereas only 10% to 20% originate from psbA3 in cells grown at low-light or high-light conditions (Bouyoub et al., 1993; Mohamed et al., 1993). Shifting cells to higher irradiances increases the steady-state levels of both psbA2 and psbA3 transcripts (Mohamed et al., 1993). This rise is due to an increase in transcription activity (Constant et al., 1997). Light also modulates the transcription of the psaE gene that is present as a unique copy in the chromosome of Synechocystis sp PCC 6803 (Constant et al., 1997; Alfonso et al., 1999, 2000).
We have previously studied the light regulation of psbA and psaE gene expression by changing the fluence of white light and/or by addition of inhibitors of the photosynthetic electron transport (Constant et al., 1997; Alfonso et al., 1999, 2000). We have shown that changes in the redox state of different components of the photosynthetic and respiratory electron carriers induce modifications in psbA and psaE mRNAs production. Transcription of both genes increased when Synechocystis cells that were incubated for long time in darkness (low reducing conditions) were illuminated (more reducing conditions). Addition of chemicals that prevent the reduction of the electron transport chain between the PQ pool and the PSI and the production of NADPH, inhibited this light-induced increase. However, psbA and psaE expression did not always respond in the same way under different stimuli. Transfer of cells from low intensities of white light to saturating intensities induced the accumulation of psbA mRNAs and the decrease of psaE mRNA level. A similar response was observed when chemicals like 3-(3, 4-dichlorophenyl)-1,1- dimethylurea (DCMU) or 2,5–dibromo-3-methyl-6–isopropyl-benzoquinone (DBMIB) were added to cells incubated at low intensities of white light. Both, DCMU and DBMIB, prevent the reduction of the electron carriers between the PQ pool and PSI and induced the closure of PSII centers. Although the existence of redox control of the expression of psbA and psaE genes was clearly demonstrated by our results, we could not exclude the role of photoreceptors responding to the changing light quality. This possibility became more realistic with the discovery of phytochrome-like photoreceptors in Synechocystis cells that may absorb blue, red, and far-red light. Moreover, it has been suggested that in the cyanobacterium Synechoccocus PCC 7942, a blue-light receptor regulates the expression of the different copies of psbA (Tsinoremas et al., 1994). However, other research groups have proposed that a redox signal via the excitation pressure of the PSII (Campbell et al., 1995) or via the cellular thiol redox state (Sippola and Aro, 1999) are involved in the light regulation of psbAII and psbAIII expression in Synechoccocus PCC 7942.
In the present report, we describe the influence of light of different wavelengths on the expression of psbA and psaE genes. In an attempt to differentiate between a light-sensory to a redox-sensory signaling process involved in the regulation of the expression of these two genes, the influence of blue, orange, and far-red illumination in the transcript levels of psbA and psaE has been also studied in a phycobilisome-less mutant (PAL mutant; Ajlani and Vernotte, 1998). This mutant derives from a phycocyanin-less mutant (Elmorjani et al., 1986) in which the apcE gene (encoding Lcm, the core-membrane linker) and the apcAB genes (encoding APa and b subunits) were inactivated. The mutant is totally devoid of phycobiliproteins, has intact photosystems, and is enriched in PSII (Ajlani and Vernotte, 1998).
Here we provide evidence that the differential expression of the PSII and the PSI genes under blue and far-red light (preferentially absorbed by PSI) and orange light (preferentially absorbed by PSII) is regulated via a redox-sensing pathway rather than by a photoreceptor.
RESULTS
Short-Term Responses to Changes in Light Quality: State Transitions followed by Fluorescence Measurements
The use of a mutant, in which the effect of orange and blue illumination on the redox state of the photosynthetic electron transport chain will be different to that observed in the wild type, should be a good tool to differentiate between a light-sensory to a redox-sensory mechanism involved in the regulation of gene expression. In Synechocystis cells, the energy collection of PSII is mainly done by the phycobilisomes, large extramembrane complexes formed by the blue phycobiliproteins, phycocyanin, and allophycocyanin, whereas chlorophyll (Chl) a is the major energy collector pigment in PSI. In wild-type cells, orange light, which is absorbed principally by the phycocyanin, induces a larger oxygen evolving activity than blue light, principally absorbed by chlorophyll molecules. In contrast, in the PC/APC-deficient mutant (PAL mutant), blue and orange lights induce similar oxygen-evolving activities (data not shown). Thus, one can expect that transfer of mutant cells from orange to blue light (or vice versa) will not induce mechanisms depending on changes in the redox state of the intersystem electron carriers. One of these mechanisms is state transitions that regulates the redistribution of the absorbed energy between PSII and PSI. As in higher plants and green algae, exposure of Synechocystis wild-type cells to light absorbed predominantly by PSII (light 2 is orange light) causes a relative decrease of the PSII fluorescence yield. Illumination with light absorbed preferentially by PSI (light 1 is blue light or far-red light), inversely, induces a relative increase of the PSII fluorescence yield. The redox state of the PQ pool regulates these variations in PSII fluorescence yield that are related to changes in the organization of the photosynthetic apparatus (Mullineaux and Allen, 1990).
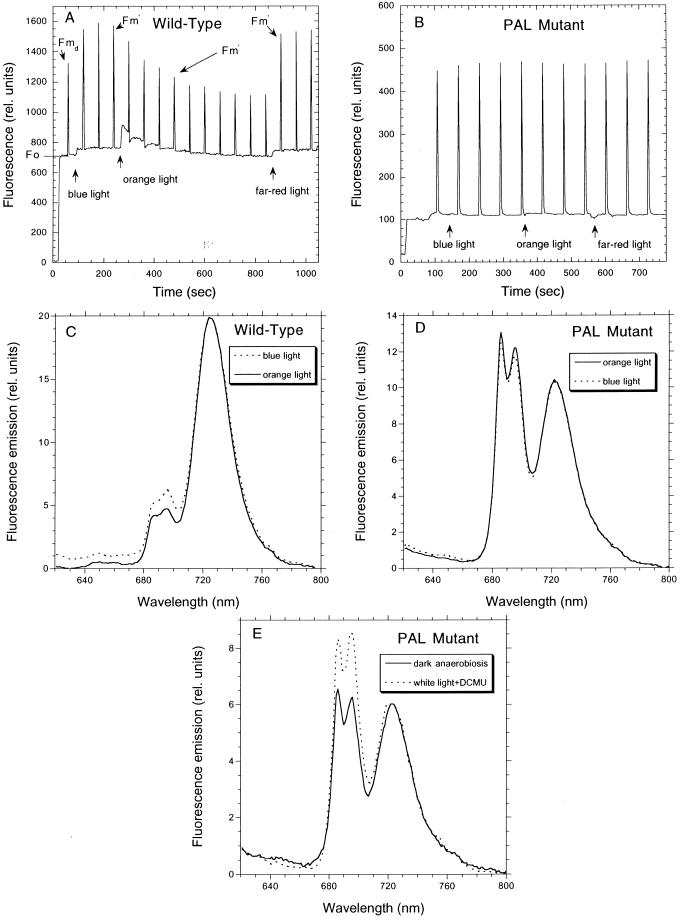
Fluorescence changes related to state transitions could be followed by fluorescence measurements at room temperature or at 77 K (Fig. 1). At room temperature, most of the observed fluorescence emission is related to PSII. A typical room temperature fluorescence trace in Synechocystis wild-type cells is shown in Figure 1A (see also “Materials and Methods”). Dark-adapted cells present a low Fmd level characteristic of cyanobacteria. Upon illumination by low-intensity blue light, exciting preferentially PSI, a maximal level of Fm′ is reached (state 1). This level is similar to Fm, obtained in the presence of light plus DCMU (data not shown). Then, orange illumination, preferentially exciting PSII, induced a large quenching of Fm′ (state 2). The quenching was suppressed by illumination with far-red light that also preferentially excites PSI. In contrast, illumination of PAL mutant cells with orange, blue, or far-red light did not induce any change in the PSII fluorescence level (Fig. 2B) even when the incubation under each type of illumination was prolonged until 40 min (data not shown).
Figure 1.
Fluorescence changes during state transitions. A and B, Measurements of fluorescence yield by a PAM fluorometer during different types of illumination in wild-type (A) and PAL mutant (B) cells. Dark-adapted cells for 5 min were successively illuminated with blue (30 μE m−2 s−1), orange (30 μE m−2 s−1), and far-red (30 μE m−2 s−1) light. Saturating pulses (3,200 μE m−2 s−1, 800-ms duration) were applied to assess Fm dark (Fmd) and Fm′. The pulses were separated by 100 s. Chl concentration 2 μg mL−1. For details see “Materials and Methods.” C and D, Fluorescence emission spectra at 77 K of dark-adapted (5 min) wild-type (C) and PAL (D) cells illuminated 30 min with orange light (solid line) and then 30 min with blue light (dashed line) at room temperature. E, Fluorescence emission spectra at 77 K of PAL cells incubated 30 min in darkness under anaerobic conditions (solid line) or incubated 30 min under white illumination in the presence of DCMU (20 μm; dashed line). The excitation wavelength was 440 nm. The spectra were normalized to the PSI fluorescence peak at 725 nm. Chl concentration, 2 μg mL−1.
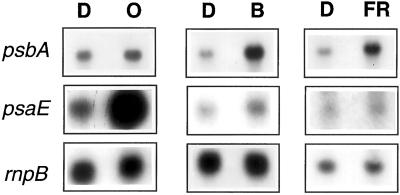
Figure 2.
Northern-blot analysis of psbA and psaE transcript levels in dark-adapted (15 min) cells (15 μg Chl mL−1) that were incubated for 10 min in blue (30 μE m−2 s−1), orange (30 μE m−2 s−1), or far-red (30 μE m−2 s−1) light. The rnpB RNA was always used as loading control.
Figure 1, C and D, shows the 77-K fluorescence emission spectra of dark-adapted wild-type and mutant cells that were successively illuminated by orange (30 min) and blue light (30 min). In the fluorescence spectra generated by excitation at 440 nm that is mainly absorbed by chlorophyll molecules, PSII (685 and 695 nm) and PSI (720 nm) fluorescence emissions were observed in wild-type and mutant cells (Fig. 1, C and D). The mutant presented a greater PSII/PSI emission ratio due to an increased number of PSII centers relative to PSI centers (Ajlani and Vernotte, 1998). In the wild type, the ratio of PSII/PSI fluorescence emission was smaller under orange (or darkness) than under blue illumination (Fig. 1C), indicating a transition to state 1, upon transfer of cells from orange to blue light. In the PAL mutant, no change in the value of the ratio was detected under these different illuminations, indicating that blue and orange light were unable to induce state transitions in PAL cells. However, when intersystem electron transport carriers were reduced by cell incubation in darkness under anaerobic conditions or oxidized by cell incubation in the presence of DCMU under white illumination, changes in the PSII/PSI fluorescence emission ratio were induced (Fig. 1E). Under oxidizing conditions, the ratio PSII/PSI was larger than under reducing conditions, indicating that the mutant cells were able to induce state transitions even in the absence of phycocyanin, allophycocyanin, and the Lcm protein when the redox state of the PQ pool changes.
The PAL mutant thus appears as a good tool to discriminate between a light sensory mechanism and a redox control of gene expression. In wild-type cells, blue and orange lights provoke changes in the redox state of the photosynthetic electron transport chain inducing redox-controlled mechanisms, as state transitions, whereas in PAL cells, blue and orange light do not induce these mechanisms.
Behavior of the psbA and psaE Transcripts upon Transfer of Wild-Type Cells from Light 1 to Light 2 and Vice Versa
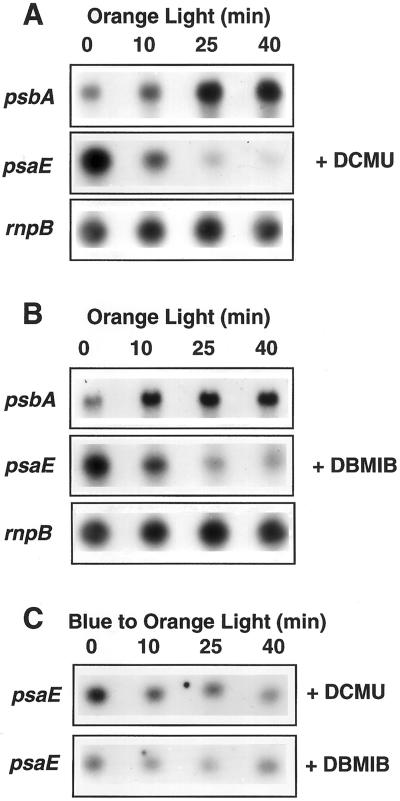
It has been already shown that transfer of Synechocystis wild-type cells from darkness into white light induces an increase in the level of various mRNAs including psbA and psaE mRNAs (Alfonso et al., 2000). To test the effect of light quality on the expression of these two genes, dark-adapted cells (for 30 min) were transferred to orange, blue or far-red illumination with an intensity of 15 μE m−2 s−1 for 10 min. Cell exposure to blue and far-red illumination induced an increase in psbA transcript levels (300%–400%). Cell exposure to orange light had no effect on the level of this transcript (Fig. 2). In contrast, orange illumination induced an increase in psaE transcript level (500%–600%), whereas far-red and blue illumination had no effect on the level of this transcript (Fig. 2).
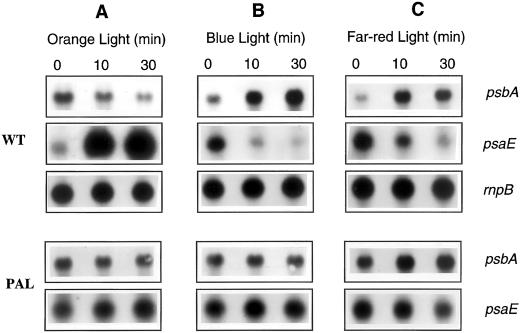
We further studied the effect of light 1 and light 2 illumination on the expression of these genes by incubating wild-type Synechocystis cells under one type of illumination and then transferring them to the other type of illumination. When blue light-adapted cells (for 30 min) were transferred to orange light, a decrease in psbA mRNA levels and an increase in the level of psaE transcript were observed (Fig. 3A). The opposite effect was observed, when orange light-adapted cells were transferred to blue or far-red illumination (Fig. 3, B and C), a decrease of psaE mRNA and an increase of psbA mRNA levels. The effect of light quality was greater on psaE (700%–800%) than on psbA gene (300%–400%) expression. These results suggested that genes coding for PSII and PSI proteins respond in a different way to light 1 and light 2 illumination: light 1 affects the expression of PSII encoding genes and light 2 increases the expression of PSI encoding genes.
Figure 3.
Northern-blot analysis of psbA and psaE transcript levels in blue light-adapted wild-type and PAL mutant Synechocystis sp PCC 6,803 cells (30 min; 15 μg Chl mL−1) transferred to orange light (A); orange light-adapted cells transferred to blue light (B); and orange light-adapted cells transferred to far-red light (C). The rnpB RNA level was used as loading control. The light intensities were the same used in Figure 2 (30 μE m−2 s−1).
Behavior of the psbA and psaE Transcripts after Transfer of PAL Mutant Cells from Light 2 to Light 1 Illumination and Vice Versa and from Low- to High-Light Intensities
The fact that blue and far-red light had similar effects on the expression of these genes and opposite effects to those induced by orange light suggested that the redox state of at least one electron carrier of the photosynthetic electron transport chain is involved in this regulation. To test this hypothesis we followed the changes in psaE and psbA mRNA levels induced by orange and blue illumination in PAL mutant cells. Similar regulation of psbA and psaE expression in wild-type and PAL cells would suggest involvement of photoreceptors, whereas different behavior of psbA and psaE mRNAs would suggest redox control.
Transfer of PAL cells from blue to orange (Fig. 3A) or from orange to blue light (Fig. 3B) had no effect on psbA and psaE transcript levels. Far-red light, that even in PAL cells is absorbed more by the PSI than by the PSII, had an effect on the levels of the mRNAs: psbA mRNA level slightly increased and psaE mRNA level slightly decreased (Fig. 3C). These results suggested that the variations in mRNA levels observed under low-light intensities of blue and orange light in Synechocystis cells are induced by changes in the redox state of the electron transport chain rather than by the activation of different photoreceptors.
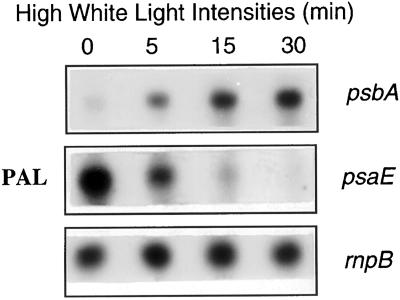
Transfer of Synechocystis wild-type cells from low to high white light intensities induces an activation of psbA expression and an inhibition of psaE transcription (Mohamed and Jansson, 1989; Mohamed et al., 1993; Constant et al., 1997). When PAL mutant cells were transferred from 30 μE m−2 s−1 to 1,800 μE m−2 s−1 intensity of white light the level of psbA mRNA largely increased during the first 15 min of high-light incubation, whereas the level of psaE mRNA rapidly decreased (Fig. 4). We conclude that whereas the PAL mutant has lost the gene expression regulation by orange and blue light, it maintains that involving light intensity.
Figure 4.
Northern-blot analysis of psbA and psaE transcript levels in PAL mutant cells. PAL cells (15 μg Chl m−1) were pre-incubated under low intensities of white light for 15 min (30 μE m−2 s−1) and then transferred to high-light intensities (1,800 μE m−2 s−1).
Effect of the Photosynthetic Inhibitors, DBMIB and DCMU, on the Expression of psbA and psaE under Different Light Conditions
To elucidate which electron carrier is involved in the regulation of the expression of the psbA and psaE genes we studied the effect of the electron transport inhibitors, DCMU and DBMIB, on psbA and psaE mRNA accumulation under different light conditions in wild-type cells.
Under illumination, in the presence of DCMU, an inhibitor of the electron transport between the primary (QA) and secondary (QB) electron acceptor quinones of the PSII, the PQ pool, and cyt b6f cannot be further reduced by PSII; the intersystem electron carriers are mainly oxidized. On the other hand, addition of DCMU increases the lifetime of QA− by inhibiting the electron transfer from QA− to QB. As a consequence, the concentration of QA− (i.e. closed centers) increases under a given light regime (Trebst, 1980).
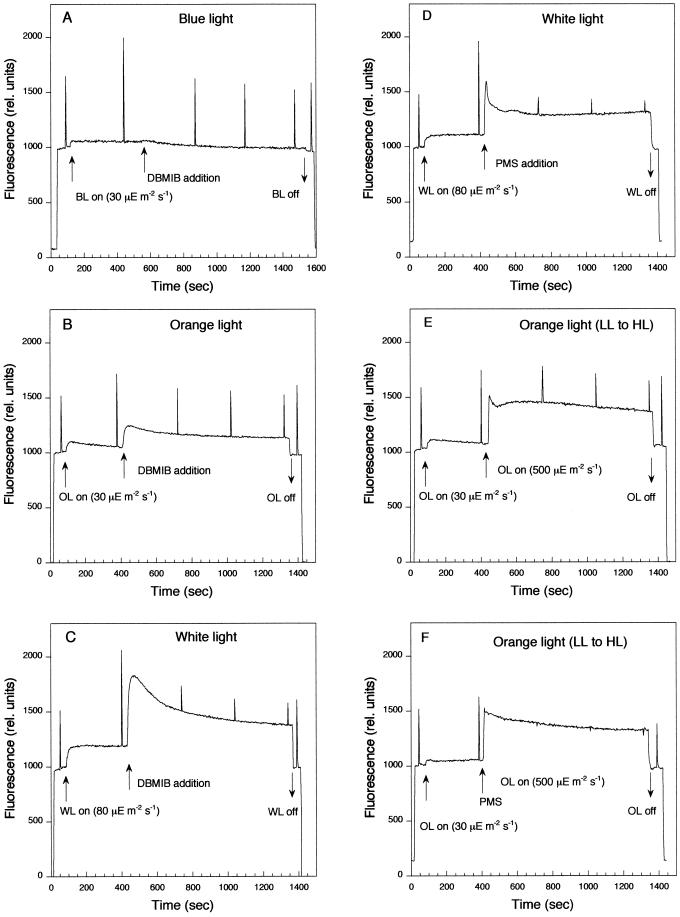
DBMIB largely inhibits linear and cyclic electron transports as well as respiration. It prevents plastoquinone re-oxidation by its binding to the Qo site of the cyt b6f. In its presence, while the plastoquinone pool is mainly reduced, the other electron carriers (including cyt b6f) are mostly oxidized. DBMIB can also induce the accumulation of QA− (closed centers) depending on light conditions (Trebst, 1980). The level of closed centers can be monitored by the fluorescence level measured in a PAM fluorometer. At room temperature, the yield of fluorescence depends on the oxido-reduction state of QA. When QA is oxidized, a minimum level of fluorescence is observed since the excitons are efficiently trapped by open centers. When QA is fully reduced (maximal concentration of closed centers), the fluorescence level reaches a maximum since the centers are closed and thus unable to trap excitons. Figure 5 shows the changes on the yield of fluorescence upon addition of DBMIB under blue (Fig. 5A), orange (Fig. 5B), and white illumination (Fig. 5C). Under orange and white illumination, addition of DBMIB induces an increase on the yield of fluorescence (Fs′ level) and on the number of closed centers. After 15 min of cell incubation in the presence of DBMIB, 35% of PSII centers were closed in orange light and 70% in white light. In contrast, under blue illumination, addition of DBMIB had almost no effect on the closure of PSII centers. Under all light conditions, DBMIB induced a decrease of the Fm′ level, indicating a transition to state 2 related to the reduction of the PQ pool.
Figure 5.
Measurements of fluorescence yield by a PAM fluorometer during different types of illumination in wild-type cells. Dark-adapted cells for 5 min were illuminated with blue (30 μE m−2 s−1) (A), orange (30 μE m−2 s−1) (B, E, and F), or white (80 μE m−2 s−1) (C and D) light. After 5-min incubation, 15 μm DBMIB (A–C) or 50 μm PMS (D and F) was added. E and F, The cells after 5 min of incubation under low-light intensities of orange light (30 μE m−2 s−1) were transferred to higher intensities of orange light (500 μE m−2 s−1). PMS was added just before the cell transfer. Saturating pulses (3,200 μE m−2 s−1, 800-ms duration) were applied to assess Fm′. The pulses were separated by 100 s. Chl concentration, 15 μg mL−1.
Under orange illumination, addition of DBMIB or DCMU induced a large decrease of psaE mRNA level (Fig. 6, A and B). Moreover, the accumulation of psaE transcripts usually observed after cell transfer from blue to orange illumination was inhibited by the presence of DCMU or DBMIB (Fig. 6C). In contrast, addition of one or other of these chemicals, induced an increase of the level of psbA mRNAs in orange light (Fig. 6, A and B). Under blue illumination, addition of DBMIB had no effect on the levels of psaE and psbA transcripts (data not shown); the level of psbA mRNA remained high and that of psaE mRNA remained low. However, addition of DCMU induced a slight increase on the level of psbA transcripts without effect on psaE mRNA levels (data not shown). Our hypothesis is that under blue light, conditions in which the level of the psbA mRNA was already high, the closure of PSII centers induced by addition of DCMU (but not by DBMIB, see Fig. 5) gave a second activation signal.
Figure 6.
Effect of DCMU and DBMIB on psbA and psaE transcript levels under different light conditions. A and B, Wild-type cells were pre-incubated for 30 min in orange light before 20 μm DCMU (A) or 15 μm DBMIB (B) was added. Time 0 (addition of DCMU or DBMIB) coincide with 30 min of orange incubation. C, Wild-type cells were incubated 30 min in blue light and then transferred to orange light. DBMIB or DCMU was added just before cell transfer to orange light.
We have already demonstrated that in Synechocystis cells, addition of DCMU or DBMIB does not modified the stability of the psbA mRNA during the first 40 min after the addition of DCMU or DBMIB, suggesting that the increase in the psbA mRNA levels is due to an activation of transcription (Alfonso et al., 1999).
The results presented in this section indicated that the redox state of the PQ pool itself is not involved in the regulation of psbA and psaE expression as is the case in higher plants (Pfannschmidt et al., 1999). The addition of the chemicals, DCMU or DBMIB, which has opposite effects on the redox state of the PQ pool, induces the same effect on the levels of psbA and psaE mRNAs: i.e. an increase of the level of psbA mRNAs and a decrease of psaE mRNA level.
Accumulation of psbA mRNA after Cell Transfer to Higher Light Intensities
We have shown that the level of psbA mRNA increases when wild-type cells are transferred from orange to blue light. This transfer induces an oxidation of the cyt b6f complex that may be the signal for psbA mRNA accumulation. It has already been shown that cell transfer to higher intensities of white light also induces an activation of psbA expression. Questions arise about the signal involved in this case. Under saturating light conditions, the oxidation of PQH2 by the cyt b6f is the overall rate-limiting step. This rate limitation is related to the electron transfer between the PQH2 and the cyt f. Murakami and Fujita (1991) have demonstrated by analysis of flash-induced oxidation-reduction of cyt f in Synechocystis sp PCC 6803 cells that the level of reduced cyt f decreased under a light intensity greater than that required for light saturation of photosynthesis (Murakami and Fujita, 1991). A low PSI/PSII ratio is observed under these conditions. They proposed that there is a strong correlation between the rapid reduction of flash oxidized cyt f (and/or the level of reduced cyt b6) and the stimulation of PSI formation (Murakami and Fujita, 1991, 1993).
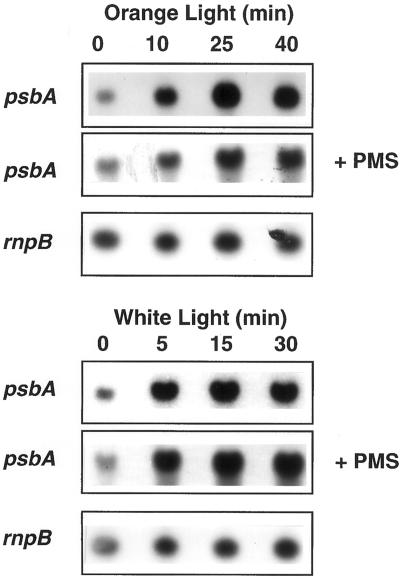
To test whether psbA mRNA accumulation under high-light intensities is related to a decrease in the concentration of reduced cyt f, we prevented cyt f oxidation by adding phenazine methosulfate (PMS; 50 μm) just before cell transfer from low- to high-light intensities. PMS is a fast electron donor to P700+ (reduced PMS to P700+ electron donation 6 ms at 50 μm) (Vassiliev et al., 1998) and it artificially mediates cyclic electron transport around the PSI (Trebst, 1974). It operates by picking up an electron from the reducing side of PSI and then rapidly giving up the electron to reduced P700+. In its presence, light induces the reduction of all the electron transport chain up to P700 (including the cyt f), whereas it prevents NADP+ reduction. Addition of PMS under white illumination (Fig. 5D) or just before cell transfer from low to high intensities of white (data not shown) or orange light (Fig. 5E versus Fig. 5F) induced a further increase of Fs′ level and a decrease of the Fm′ level. Thus the presence of PMS provoked the transition to state 2 and a further closure of PSII centers, indicating that this chemical induced the reduction of the electron transport chain. Figure 7 shows that transfer of Synechocystis cells from low to high intensities not only of white light but also of orange light, induced accumulation of psbA mRNAs. The presence of PMS did not inhibit this increase, suggesting that the oxidation of the cyt f is not the signal involved in the accumulation of psbA mRNAs upon cell transfer to higher light intensities.
Figure 7.
Northern-blot analysis of psbA transcript level in wild-type cells under high-light intensities of orange or white light in the absence or presence of PMS. A, Cells were pre-incubated for 30 min in orange light (30 μE m−2 s−1) and then transferred to 500 μE m−2 s−1 of orange light. B, Cells were pre-incubated for 15 min in white light (30 μE m−2 s−1) and then transferred to 1,800 μE m−2 s−1 of white light. The level of the rnpB RNA remained constant.
DISCUSSION
Redox Control Versus Involvement of Photoreceptors
The use of a phycobiliprotein-less mutant of Synechocystis sp PCC 6803 (PAL mutant) allowed us to differentiate between redox control and the involvement of photoreceptors in the regulation of the expression of psaE and psbA genes by different qualities of light in cyanobacteria. Transfer of wild-type cells from orange to blue light (or vice versa) induced changes in the redox state of the photosynthetic electron transport chain and in the expression of psbA and psaE genes. In contrast, transfer of PAL cells from one light to the other did not provoke changes in the redox state of the electron carriers and did not induce changes in psbA and psaE transcript accumulation. However, in this mutant psbA and psaE genes are still regulated by light intensities. Moreover, addition of DCMU under white illumination induced an accumulation of psbA transcripts (data not shown) like in wild-type cells. Under these conditions, the electron carriers between the two photosystems were oxidized and the cells were in state 1. These results indicate that blue and orange light regulate psbA and psaE gene expression via a redox control and suggest that photoreceptors are not involved in this regulation, at least in Synechosystis cells.
The Redox State of Which Component of the Electron Transport Chain Regulates psaE and psbA Expression?
Pfannschmidt et al. (1999) have proposed that in higher plants lights 1 and 2 regulate the expression of genes encoding PSI and PSII proteins via the redox state of the PQ pool. Li and Sherman (2000) recently have analyzed the long-term effects (6 h) of DCMU and DBMIB on transcript levels of PSII and PSI genes in growing cultures of Synechocystis sp PCC 6803. They also concluded that expression of PSI and PSII genes is under redox control. As we have shown for the psbA and psaE transcripts, they observed that oxidizing conditions induced an increase of the mRNA levels of PSII genes and a decrease of those of PSI genes, whereas under reducing conditions in the presence of Glc, the steady-state levels of PSII transcripts decreased and those of PSI slightly increased. However, whereas we have observed a clear effect of DBMIB inducing an increase of the psbA mRNA levels and a decrease of psaE mRNA level, they showed that in the presence of DBMIB, the mRNA levels remained almost unchanged. The lack of effect of DBMIB on PS gene expression observed by these authors could be explained by the fact that DBMIB is relatively unstable and is easily converted to other compounds. We have already shown that DBMIB (20 μm) inhibits 90% of oxygen evolution activity, 10 min after its addition to Synechocystis sp PCC 6803 cells (30 μg Chl mL−1) and only 40% 60 min after its addition (Alfonso et al., 2000). In our hands, addition of DCMU or DBMIB inhibits the accumulation of psaE transcripts and increases the levels of psbA mRNAs. Moreover, the presence of DBMIB inhibits the increase on the level of psaE mRNA when cells are transferred from blue to orange light. Thus, our results do not support the hypothesis that the redox state of the PQ pool regulates the expression of genes encoding PSI and PSII subunits.
In Synechococcus PCC 7942 cells, the redox state of one of the electron acceptors of the PSI seems to play an important role in the regulation of psbA expression (Sippola and Aro, 1999). An up-regulation of psbAII/psbAIII genes is induced by the reducing power generated by PSI (Sippola and Aro, 1999). In Synechocystis cells, although the presence of reducing power is required for psbA (and psaE) expression (Alfonso et al., 2000), the redox state of the electron carriers after the PSI seems not to be the principal signal for the regulation of psbA (and psaE) expression. We have already shown that the presence of methylviologen, which prevents the reduction of PSI electron acceptors, had no effect on psaE and psbA expression (Alfonso et al., 2000). In the present work, we have shown that the presence of PMS or DBMIB, that also avoids the reduction of PSI electron acceptors, does not prevent the accumulation of psbA mRNAs upon cell transfer from low- to high-light intensities. Moreover, the addition of DBMIB, instead of inducing a decrease of the levels of psbA mRNAs, provokes an increase of these levels. Finally, the oxidation of the electron acceptors of PSI, generated by the cell transition from light 1 to light 2, and that induced by DBMIB addition had opposite effects on the accumulation of psaE transcripts.
Although our results do not allow to determine which redox component is involved in the expression of psbA and psaE genes, they clearly demonstrate that in Synechocystis cells, light quality regulates the expression of photosynthetic genes via a redox control that seems to involve the oxido-reduction reactions occurring in the cyt b6f complex. We propose, as a working hypothesis, that the occupancy of the Qox site in the cyt b6f complex, rather than the redox state of cyt f (as proposed by Murakami and Fujita, 1991; Fujita, 1997) plays a role in the regulation of psaE and psbA gene expression. This hypothesis is based in the effect of DBMIB (that inhibits the re-oxidation of the PQ pool by binding at the Qox site) on psbA and psaE expression and in the fact that addition of PMS (that prevents the oxidation of the cyt f) had no effect on psbA mRNA accumulation.
We cannot exclude from our results the possibility that under high-light intensities other signals are involved in the regulation of psbA gene expression such as the redox state of PSII the reducing power of the cell or oxygen radicals. Cell transfer from light 2 to light 1 or from low- to high-light intensities has opposite effects on the redox state of the intersystem electron transports (with the exception of cyt f) (Murakami and Fujita, 1991; Fujita, 1997), whereas both conditions induce an accumulation of psbA mRNAs. We have already proposed that under white light illumination an increase in the concentration of closed PSII centers could be a signal for the accumulation of psbA mRNAs (Alfonso et al., 1999). Here we have described some results that support this working hypothesis. The effect of DBMIB depended on light conditions. Under blue light, conditions in which this chemical did not induce the closure of centers, its addition had no effect, whereas addition of DCMU slightly increased the already high level of psbA mRNAs. In addition, cell transfer from low to high intensities of orange light also induced an accumulation of psbA mRNAS that PMS or DBMIB did not inhibit it. Although our results are in favor of a different signal for psbA accumulation when the cells are transferred to higher light intensities or to light 1, further experiments are needed to confirm this hypothesis.
MATERIALS AND METHODS
Cyanobacterium and Culture Conditions
Wild-type and PAL mutant Synechocystis sp PCC 6803 cells were grown under photoautotrophic conditions in the mineral medium described by Herdman et al. (1973) containing twice the concentration of sodium nitrate. Cells were shaken in a rotatory shaker (120 rpm) at 30°C under a 5% (v/v) CO2-enriched atmosphere and continuous illumination from fluorescent white lamps giving a total intensity of 80 μE m−2 s−1.
The Chl a concentration was spectrophotometrically determined in methanol extracts using the extinction coefficient at 665 nm of 74.5 mL mg−1 cm−1.
Light and Chemical Treatments of the Cells
Wild-type and mutant Synechocystis sp PCC 6803 cells were harvested in the exponential phase of growth by centrifugation and resuspended in fresh culture medium containing 50 mm HEPES (pH 6.8) at a final concentration of 15 μg Chl mL−1. The cells were pre-incubated for 15 min in darkness at 30°C in a thermo-stated glass cuvette (4-cm diameter) under gentle stirring and then they were illuminated by far-red, blue, or orange light (30 μE m−2 s−1). Far-red and blue light (PSI light) were obtained by filtering white light through a Schott RG filter (cut-on 690 nm) for far-red light or through a blue filter (350–530 nm; max 450 nm; 195 filter, LEE filters; Andover, Hampshire, UK) for blue light. Illumination of cells with orange light (PSII light) was obtained from white light filtered through two filters: an orange filter (cut-on 580 nm; 021 filter, LEE filters; Andover) and a cut-off 650 nm filter (Oriel, Courtaboeuf, France). For low to high light experiments, the cells (15 μg Chl mL−1) were pre-incubated under low light 80 μE m−2 s−1 for 15 min and then transferred to high-light intensity of approximately 1,800 μE m−2 s−1. Samples were collected for various analyses at different times during the different light and inhibitory treatments as indicated in “Results.”
Isolation of RNA and Northern Blotting
Total RNA was isolated from 15 mL of Synechocystis sp PCC 6803 (15 μg Chl mL−1) cells using hot phenol and LiCl as described by Mohamed and Jansson (1989). RNA samples were denatured for 3 min at 68°C and separated by electrophoresis using 1.2% (w/v) agarose gel containing formaldehyde as denaturing agent. The gel was transferred into a charged nylon membranes (Hybond N+, Amersham, Buckinghamshire, UK) by capillary blotting and fixed to the membrane by 5-min UV exposure and 2 h at 80°C. Hybridizations were performed at 42°C with the different radioactive probes for psbA, psaE, and rnpB genes. Northern blots were exposed to x-ray film (Kodak, Rochester, NY) to obtain autoradiograms. The analysis of the autoradiograms was performed by combination of a scanner (studio Iisi, AGFA) and a Macintosh Power 7100/80 computer using the public software domain NIH Image program (developed at U.S. National Institutes of Health and available from the Internet (zippy.nimh.nih.gov) or on floppy disc from the National Technical Information Service, Springfield, VA, part no. PB95–500195GEI).
Hybridization Probes
psbA Probe
A KpnI- KpnI fragment of 0.7 kb containing the psbA2 gene region of Synechocystis sp PCC 6714 encoding the 3′ half of the gene was used as homologous probe (Ajlani et al., 1989). This probe recognized the two expressed psbA copies, psbA2 and psbA3. Because the psbA2 and psbA3 coding regions present a high degree of homology at the nucleotide level with an identity of >99%, it was impossible to construct specific probes for each copy.
psaE Probe
A 0.35-kb AvaI-Eco24 I fragment corresponding to the full-length psaE sequence of Synechocystis sp PCC 6803 was used as a probe. This fragment was obtained from the plasmid pBSPsaE (gift from Dr. B. Lagoutte) coding the PSA-E subunit of PSI (Rousseau et al., 1993).
rnpB Probe
A 0.38-kb fragment encoding the sequence of the rnpB gene of Anacystis nidulans (Synechocystis sp PCC 6301; gift from A. Vioque, Sevilla, Spain) was used as a probe (Vioque, 1992). This gene, that is also present in Synechocystis sp PCC 6803 (Vioque, 1992), encodes the constitutively expressed RNA component of the ribonuclease P.
Fluorescence Measurements
The yield of Chl fluorescence was continuously monitored in a modulated fluorometer (PAM chlorophyll fluorometer; Walz, Effelrich, Germany) adapted to a DW1 Hansatech oxygen electrode as previously described (Arsalane et al., 1994). Cell suspensions (2 μg Chl mL−1) were placed in a stirred cuvette (30°C). Cells adapted to dark for 5 min were brought to state 1 by illumination with blue light (450 nm, Corning 4.96) at 30 μE m−2 s−1 and to state 2 by illumination with orange light (601 nm, filter: Balzer B-40 599 10) at 30 μE m−2 s−1. See Figure 1A for illustration of a fluorescence trace.
The minimal fluorescence level (F0) was determined by illuminating dark-adapted cells with a low intensity of red-modulated light. The minimal fluorescence level in the light-adapted state (F0′) was determined by briefly interrupting the continuous blue or orange light. Maximum fluorescence level of the dark-adapted (Fmd) or light adapted cells (Fm′) was measured by a 800-ms high-intensity white pulse (3,200 μE m−2 s−1). Application of such pulses of intense light, which transiently close all PSII centers and removes photochemical quenching, serves to distinguish photochemical from non-photochemical quenching at any time. The saturating multiple turnover white pulses were produced by an electronic shutter (Uniblitz, Vincent, Rochester, NY; opening time of 2 ms) put in front of a KL-1,500 quartz-iodine lamp (Schott, Mainz, Germany) and controlled by the accessory module PAM-103. The maximal level of fluorescence (Fm) cannot be determined in darkness because in cyanobacteria a large non-photochemical quenching is present under dark conditions. As a consequence, Fm was determined in the presence of 10 μm DCMU and white or blue light. This inhibitor blocks the electron transport from QA− to QB, causing reaction center closure and cancels all the photochemical quenching.
The fluorescence emission spectra at 77 K were recorded on a fluorescence spectrophotometer (F-3010, Hitachi, Tokyo). Excitation was done at 440 nm. Emission was scanned from 620 to 800 nm. One milliliter of cell suspension (2 μg Chl mL−1) was illuminated by blue or orange light for 30 min. It was then quickly filtered, and the filter was immediately plunged into liquid nitrogen.
ACKNOWLEDGMENTS
We express our gratitude to Dr. G. Ajlani for providing us the PAL mutant, which was indispensable for this study, and for critical reading of the manuscript. We thank Dr. B. Lagoutte and Dr. A. Vioque for giving us the psaE and rnpB DNA probes. We also thank Dr. B. Lagoutte and Dr. A.W. Rutherford for critical reading of the manuscript and Dr. J.-C. Duval and Dr. C. Richaud for helpful discussions. Finally, we acknowledge I. Perewoska for technical assistance.
LITERATURE CITED
- Ajlani G, Kirilovsky D, Picaud M, Astier C. Molecular analysis of psbA mutations responsible for various herbicide resistance phenotypes in Synechocystis 6714. Plant Mol Biol. 1989;13:469–479. doi: 10.1007/BF00027307. [DOI] [PubMed] [Google Scholar]
- Ajlani G, Vernotte C. Construction and characterization of a phycobiliprotein-less mutant of Synechocystis sp. PCC 6803. Plant Mol Biol. 1998;37:577–580. doi: 10.1023/a:1005924730298. [DOI] [PubMed] [Google Scholar]
- Alfonso M, Perewoska I, Constant S, Kirilovsky D. Redox control of psbA expression in cyanobacteria Synechocystis strains. J Photochem Photobiol B Biol. 1999;48:104–113. [Google Scholar]
- Alfonso M, Perewoska I, Kirilovsky D. Redox control of psbA gene expression in the cyanobacterium Synechocystis PCC 6803: involvement of the cytochrome b6f complex. Plant Physiol. 2000;122:505–516. doi: 10.1104/pp.122.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF. How does protein phosphoylation regulate photosynthesis? Trends Biochem Sci. 1992a;17:12–17. doi: 10.1016/0968-0004(92)90418-9. [DOI] [PubMed] [Google Scholar]
- Allen JF. Protein phosphorylation in regulation of photosynthesis. Biochim Biophys Acta. 1992b;1098:275–335. doi: 10.1016/s0005-2728(09)91014-3. [DOI] [PubMed] [Google Scholar]
- Allen JF. Redox control of gene expression and the function of chloroplast genomes: an hypothesis. Photosynth Res. 1993;36:95–102. doi: 10.1007/BF00016274. [DOI] [PubMed] [Google Scholar]
- Allen JF. Thylakoid protein phosphorylation, state 1-state 2 transitions, and photosystem stoichiometry adjustment: redox control at multiple levels of gene expression. Physiol Plant. 1995;93:196–205. [Google Scholar]
- Allen JF, Alexciev K, Hakansson G. Regulation by redox signalling. Curr Biol. 1995;5:869–872. doi: 10.1016/s0960-9822(95)00176-x. [DOI] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B. Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta. 1993;1143:113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- Arsalane W, Rousseau B, Duval J. Influence of the pool size of the xanthophyll cycle on the effects of light stress in a diatom: competition between photoprotection and photoinhibition. Photochem Photobiol. 1994;60:237–243. [Google Scholar]
- Biggins J, Bruce D. Regulation of excitation energy transfer in organisms containing phycobilins. Photosynth Res. 1989;20:1–34. doi: 10.1007/BF00028620. [DOI] [PubMed] [Google Scholar]
- Bouyoub A, Vernotte C, Astier C. Functional analysis of the two homologous psbA gene copies in Synechocystis PCC 6714 and PCC 6803. Plant Mol Biol. 1993;21:249–258. doi: 10.1007/BF00019941. [DOI] [PubMed] [Google Scholar]
- Campbell D, Zhou G, Gustafsson P, Öquist G, Clarke AK. Electron transport regulates exchange of two forms of photosystem II D1 protein in the cyanobacterium Synechococcus. EMBO J. 1995;14:5457–5466. doi: 10.1002/j.1460-2075.1995.tb00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constant S, Perewoska I, Alfonso M, Kirilovsky D. Expression of the psbA gene during photoinhibition and recovery in Synechocystis PCC 6714: inhibition and damage of transcriptional and translational machinery prevent the restoration of photosystem II activity. Plant Mol Biol. 1997;34:1–13. doi: 10.1023/a:1005754823218. [DOI] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. ADP-dependent phosphorylation regulates RNA-binding in vitro: implications in light-modulated translation. EMBO J. 1994a;13:2227–2235. doi: 10.1002/j.1460-2075.1994.tb06500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. Light-regulated translation of chloroplast messenger RNAs through redox potential. Science. 1994b;266:1717–1719. doi: 10.1126/science.7992056. [DOI] [PubMed] [Google Scholar]
- Durnford DG, Falkowski PG. Chloroplast redox regulation of nuclear gene-transcription during photoacclimation. Photosynth Res. 1997;53:229–241. [Google Scholar]
- El Bissati K, Delphin E, Murata N, Etienne A-L, Kirilovsky D. Photosystem II fluorescence quenching in the cyanobacterium Synechocystis PCC 6803: involvement of two different mechanisms. Biochim Biophys Acta. 2000;1457:229–242. doi: 10.1016/s0005-2728(00)00104-3. [DOI] [PubMed] [Google Scholar]
- Elmorjani K, Thomas JC, Sebban P. Phycobilisomes of wild type and pigment mutants of the cyanobacterium Synechocystis PCC 6803. Arch Microbiol. 1986;146:186–191. [Google Scholar]
- Escoubas JM, Lomas M, Laroche J, Falkowski PG. Light-intensity regulation of cab gene-transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA. 1995;92:10237–10241. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y. A study on the dynamic features of photosystem stoichiometry: accomplishments and problems for future studies. Photosynth Res. 1997;53:83–93. [Google Scholar]
- Furuya M, Schafer E. Photoperception and signalling of induction reactions by different phytochromes. Trends Plant Sci. 1996;1:301–307. [Google Scholar]
- Golden SS. Light-responsive gene expression in cyanobacteria. J Bacteriol. 1995;177:1651–1654. doi: 10.1128/jb.177.7.1651-1654.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdman M, Delaney SF, Carr NG. A new medium for the isolation and growth of auxotrophic mutants of the blue-green alga Anacystis nidulans. J Gen Microbiol. 1973;79:233–237. [Google Scholar]
- Hughes J, Lamparter T, Mittmann F, Hartmann E, Gärtner W, Wilde A, Börner T. A prokaryotic phytochrome. Nature. 1997;386:663. doi: 10.1038/386663a0. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803: II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Keren N, Ohad I. State transition and photoinhibition. In: Rochaix JD, Goldschmidt-Clermont M, Merchants S, editors. The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 569–596. [Google Scholar]
- Lamparter T, Mittmann F, Gaertner W, Boerner T, Hartmann E, Hughes J. Characterization of recombinant phytochrome from the cyanobacterium Synechocystis. Proc Natl Acad Sci USA. 1997;94:11792–11797. doi: 10.1073/pnas.94.22.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Sherman L. A redox-responsive regulator of photosynthesis gene expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2000;182:4268–4277. doi: 10.1128/jb.182.15.4268-4277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed A, Eriksson J, Osiewacz HD, Jansson C. Differential expression of the psbA genes in the cyanobacterium Synechocystis 6803. Mol Gen Genet. 1993;238:161–168. doi: 10.1007/BF00279543. [DOI] [PubMed] [Google Scholar]
- Mohamed A, Jansson C. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol Biol. 1989;13:693–700. doi: 10.1007/BF00016024. [DOI] [PubMed] [Google Scholar]
- Mullineaux CW, Allen JF. State 1-State 2 transitions in the cyanobacterium Synechococcus 6301 are controlled by the redox state of electron carriers between photosystems I and II. Photosynth Res. 1990;23:297–311. doi: 10.1007/BF00034860. [DOI] [PubMed] [Google Scholar]
- Mullineaux CW, Boult M, Sanders CE, Allen JF. Fluorescence induction transients indicate altered absorption cross-section during light-state transitions in the cyanobacterium Synechococcus 6301. Biochim Biophys Acta. 1986;851:147–150. [Google Scholar]
- Mullineaux CW, Holzwarth AR. A proportion of photosystem II core complexes are decoupled from the phycobilisome in light-state 2 in the cyanobacterium Synechococcus 6301. FEBS Lett. 1990;260:245–248. [Google Scholar]
- Mullineaux CW, Tobin MJ, Jones GR. Mobility of photosynthetic complexes in thylakoid membranes. Nature. 1997;390:421–424. [Google Scholar]
- Murakami A, Fujita Y. Regulation of photosystem stoichiometry in the photosynthetic system of the cyanophyte Synechocystis PCC 6714 in response to light-intensity. Plant Cell Physiol. 1991;32:223–230. [Google Scholar]
- Murakami A, Fujita Y. Regulation of stoichiometry between PS I and PS II in response to light regime for photosynthesis observed with Synechocystis PCC 6714: relationship between redox state of cit b6f complex and regulation of PS I formation. Plant Cell Physiol. 1993;34:1175–1180. [Google Scholar]
- Niyogi KK. Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- Olive J, Ajlani G, Astier C, Recouvreur M, Vernotte C. Ultrastructure and light adaptation of phycobilisome mutants of Synechocystis PCC 6803. Biochim Biophys Acta. 1997;1319:275–282. [Google Scholar]
- Olive J, M'Bina I, Vernotte C, Astier C, Wollman FA. Randomization of the EF particles in thylakoid membranes of Synechocystis 6714 upon transition from state I to state II. FEBS Lett. 1986;208:308–312. [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628. [Google Scholar]
- Prasil O, Adir N, Ohad I. Dynamics of photosystem II: mechanism of photoinhibition and recovery processes. In: Barbers J, editor. The Photosystems: Structure, Function and Molecular Biology. Amsterdam: Elsevier Science Publishers; 1992. pp. 295–348. [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D. Phytochromes: photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Rousseau F, Sétif P, Lagoutte B. Evidence for the involvement of PSI-E in the reduction of ferredoxin by photosystem I. EMBO J. 1993;12:1755–1765. doi: 10.1002/j.1460-2075.1993.tb05823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdiger W. Events in the phytochrome molecule after irradiation. Photochem Photobiol. 1992;56:803–809. doi: 10.1111/j.1751-1097.1992.tb02236.x. [DOI] [PubMed] [Google Scholar]
- Schluchter W, Shen G, Zhao J, Bryant D. Characterization of psaI and psaL mutants of Synechococcus sp strain PCC 7002: a new model for state transitions in cyanobacteria. Photochem Photobiol. 1996;64:53–66. doi: 10.1111/j.1751-1097.1996.tb02421.x. [DOI] [PubMed] [Google Scholar]
- Sippola K, Aro EM. Thiol redox state regulates expression of psbA genes in Synechococcus sp. PCC 7942. Plant Mol Biol. 1999;41:425–433. doi: 10.1023/a:1006347808475. [DOI] [PubMed] [Google Scholar]
- Trebst A. Energy conservation in photosynthetic electron transport of chloroplasts. In: Briggs W, Green P, Joness R, editors. Annual Review of Plant Physiology. Palo Alto, CA: Annual Reviews, Inc.; 1974. pp. 423–458. [Google Scholar]
- Trebst A. Inhibitors in electron flow: tools for the functional and structural localization of carriers and energy conservation sites. Methods Enzymol. 1980;69:675–715. [Google Scholar]
- Tsinoremas N, Schaefer M, Golden S. Blue and red light reversibly control psbA expression in the cyanobacterium Synechococcus sp. strain PCC 7942. J Biol Chem. 1994;269:16143–16147. [PubMed] [Google Scholar]
- Vassiliev I, Jung Y-S, Yang F, Golbeck J. PsaC subunit of photosystem I is oriented with iron-sulsur cluster FB as the immediate electron donor to ferredoxin and flavodoxin. Biophys J. 1998;74:2029–2035. doi: 10.1016/S0006-3495(98)77909-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vioque A. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acid Res. 1992;20:6333–6337. doi: 10.1093/nar/20.23.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A, Churin Y, Schubert H, Borner T. Disruption of a Synechocystis sp. PCC 6803 gene with partial similarity to phytochrome genes alters growth under changing light qualities [published erratum appears in FEBS Lett 1997 Jul 28;412(2):404] FEBS Lett. 1997;406:89–92. doi: 10.1016/s0014-5793(97)00248-2. [DOI] [PubMed] [Google Scholar]
- Williams WP, Allen JF. State 1/state 2 changes in higher plants and algae. Photosynth Res. 1987;13:19–45. doi: 10.1007/BF00032263. [DOI] [PubMed] [Google Scholar]
- Yeh KC, Wu SH, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]