Abstract
Initial studies on the inositol phosphates metabolism were enabled by the social amoeba Dictyostelium discoideum. The abundant amount of inositol hexakisphosphate (IP6 also known as Phytic acid) present in the amoeba allowed the discovery of the more polar inositol pyrophosphates, IP7 and IP8, possessing one or two high energy phosphoanhydride bonds, respectively. Considering the contemporary growing interest in inositol pyrophosphates, it is surprising that in recent years D. discoideum, has contributed little to our understanding of their metabolism and function. This work fulfils this lacuna, by analysing the ip6k, ppip5k and ip6k-ppip5K amoeba null strains using PAGE, 13C-NMR and CE-MS analysis. Our study reveals an inositol pyrophosphate metabolism more complex than previously thought. The amoeba Ip6k synthesizes the 4/6-IP7 in contrast to the 5-IP7 isomer synthesized by the mammalian homologue. The amoeba Ppip5k synthesizes the same 1/3-IP7 as the mammalian enzyme. In D. discoideum, the ip6k strain possesses residual amounts of IP7. The residual IP7 is also present in the ip6k-ppip5K strain, while the ppip5k single mutant shows a decrease in both IP7 and IP8 levels. This phenotype is in contrast to the increase in IP7 observable in the yeast vip1Δ strain. The presence of IP8 in ppip5k and the presence of IP7 in ip6k-ppip5K indicate the existence of an additional inositol pyrophosphate synthesizing enzyme. Additionally, we investigated the existence of a metabolic relationship between inositol pyrophosphate synthesis and inorganic polyphosphate (polyP) metabolism as observed in yeast. These studies reveal that contrary to the yeast, Ip6k and Ppip5k do not control polyP cellular level in amoeba.
Keywords: Metabolism, ip6k, ppip5k, Inorganic polyphosphates, Inositol, Phosphate, Amoeba, Development
1. Introduction
The social amoeba Dictyostelium discoideum was one of the primary experimental models to study inositol phosphate metabolism and signalling in the 1980s (Europe-Finner et al., 1991). The lipid independent route to IP6 synthesis was identified (Stephens and Irvine, 1990) in D. discoideum, and inositol species more polar than the fully phosphorylated ring of IP6, the inositol pyrophosphate (see below) were also discovered in this organism (Stephens et al., 1993). By the middle of the 90s, it was discovered that phospholipase C in D. discoideum is not required to produce IP3 nor to control calcium signalling (Van Dijken et al., 1995, 1997). Perhaps for these reasons, the interest of inositol scientists in this organism faded over the years. However, the interest in D. discoideum did not completely disappear. At the start of the new millennium, the social amoeba was used to study the effect on the inositol phosphate metabolism of lithium and other mood stabilizing drugs (King et al., 2010; Williams et al., 1999, 2002). More recently, D. discoideum was used to characterise the roles of inositol polyphosphate in programmed cell death (Al-Anbaky et al., 2018) and to characterise the phytocannabinoid-dependent mTORC1 regulation by the inositol polyphosphate multikinase (Damstra-Oddy et al., 2021). However, the precise description of the inositol phosphate metabolic pathway and the characterization of the different inositol kinase mutants is still missing in the amoeba. To our knowledge, only the IP6K (Ip6K also referred to as I6kA) null strain (ip6k, i6kA) has previously been generated. The previously characterized ip6k amoeba (Luo et al., 2003) possess a biochemical phenotype, the absence of inositol pyrophosphate, similar to the mutant of the homologous yeast Saccharomyces cerevisiae gene Kcs1 (kcs1Δ) (Saiardi et al., 2000).
The IP6K, as the name indicates, phosphorylates IP6 to generate the inositol pyrophosphate IP7 (Saiardi et al., 1999). There is at least one other class of kinases able to synthesize inositol pyrophosphate known as PPIP5K (S. cerevisiae Vip1; Schizosaccharomyces pombe Asp1) that, as the name suggests, primarily phosphorylates IP7, also abbreviated to PP-IP5, IP8 or (PP)2-IP4 (Choi et al., 2007; Fridy et al., 2007; Mulugu et al., 2007).
The yeast and mammalian IP6K and PPIP5K have been extensively characterized. The mammalian IP6K is able to pyro-phosphorylate position five of the inositol ring generating the isomer 5PP-IP5 (hereafter 5-IP7) (Draskovic et al., 2008), while mammalian PPIP5K phosphorylate position one physiologically generating 1,5(PP)2-IP4 (hereafter 1,5-IP8) (Lin et al., 2009; Wang et al., 2012).
D. discoideum possesses one Ip6k gene and a Ppip5k homologous gene (see below). The similar enzymology between human, yeast and amoeba suggests a similar inositol pyrophosphate metabolism. However, this appears not to be the case since 1H,31P-NMR spectroscopy and enzymology studies (Laussmann et al., 1996, 1997) suggest that the amoeba possesses a different form of IP8, the isomer 5,6(PP)2-IP4 (hereafter 5,6-IP8).
Inositol pyrophosphates are attracting a growing interest due to their link to metabolic disorders including obesity and diabetes (Mukherjee et al., 2020), human diseases, such as cancer and Alzheimer's (Crocco et al., 2016), combined with improved tools to facilitate their analysis in vivo (Harmel et al., 2019; Ito et al., 2018; Qiu et al., 2020; Wilson et al., 2015). The picture that is emerging is that inositol pyrophosphates regulate basic energy metabolism (Szijgyarto et al., 2011) through their ability to control phosphate homeostasis (Azevedo and Saiardi, 2017; Saiardi, 2012). The discovery that in S. cerevisiae, the IP7 synthesizing enzyme Kcs1 regulates the metabolism of polymeric linear chains of phosphate groups, also known as inorganic polyphosphate (polyP) (Lonetti et al., 2011), led to the discovery of a novel signalling paradigm involving the SPX protein domain (Wild et al., 2016). In this case, the interaction between IP7 and the SPX domain of the S. cerevisiae polyP-synthesizing enzyme, Vtc4, stimulates polyP synthesis (Gerasimaite et al., 2017). The SPX domain is present in dozens of plant proteins involved in phosphate homeostatic regulation (Azevedo and Saiardi, 2017; Secco et al., 2012). Due to the limited availability and the socioeconomic importance of phosphate as fertiliser, the roles played by inositol pyrophosphate in regulating plant phosphate absorption and metabolism is now one area of intense investigation (Dong et al., 2019; Riemer et al., 2021; Zhu et al., 2019). However, recent work carried out in S. pombe, indicates that it is not Kcs1 but the PPIP5K homologous enzyme, Asp1/Vip1, which regulates Vtc4-driven polyP synthesis in fission yeast (Pascual-Ortiz et al., 2021). Therefore, our understanding of the exact roles played by inositol pyrophosphates in polyP synthesis and phosphate homehostasis is far from complete.
Nevertheless, the absence of polyP in S. cerevisiae kcs1Δ has been highly influential. In fact, D. discoideum ip6k knockout has been utilized as a proxy, using unrefined supporting data, which demonstrates that the amoeba has low polyP level (Suess and Gomer, 2016). Amoeba synthesize polyP through the polyphosphate kinase (Ppk1), a gene acquired from bacteria by horizontal gene transfer (Livermore et al., 2016). We discovered that polyP hugely accumulates during the D. discoideum developmental program (Livermore et al., 2016). The polyP-induced synthesis following the starvation/aggregation signal leads to its secretion (Suess and Gomer, 2016).
Like inositol pyrophosphates, polyP is also attracting a growing interest. PolyP, has been described as an important primordial chaperone able to regulate the aggregation of proteins which form in neurodegenerative disorders (Cremers et al., 2016). However, it is also important to mitochondrial physiology (Solesio et al., 2021) driving a novel protein post translational modification, lysine-polyphosphorylation (Azevedo et al., 2015); and controlling several aspects of the blood coagulation cascade (Morrissey et al., 2012). Since the many important roles attributed to polyP, it is of fundamental importance to understand if the link between inositol pyrophosphates (either IP7 or IP8) and polyP synthesis is an evolutionarily conserved feature.
D. discoideum offers the unique opportunity to address these issues since it is an excellent experimental model for both inositol pyrophosphate and polyP studies (Desfougeres and Saiardi, 2020). Here we characterise the amoeba inositol pyrophosphate metabolic pathway by creating the ip6k, the ppip5k and the double ip6k-ppip6k strain in the AX2 genetic background to verify if these enzymes are regulating polyP synthesis.
2. Materials and methods
2.1. Identification D. discoideum inositol phosphate kinase genes
To identify the amoeba inositol phosphate genes, we performed Protein Basic Local Alignment Search Tool (BLAST) searches using all inositol phosphate kinases found in S. cerevisiae and H. sapiens against the D. discoideum complete genome as previous described (Laha et al., 2021).
2.2. Genetic manipulations
Yeast transformations were performed using the lithium/acetate method (Gietz and Woods, 2002). Yeast knockouts were generated using well-described procedures (Janke et al., 2004). The correct removal of the genes were first verify by PCR and then phenotypically characterising the inositol phosphate profile by 3H-inositol-labeling Sax-HPLC analysis (Azevedo and Saiardi, 2006).
2.3. Cloning D. discoideum Ppip5K in yeast expression vector
Codon-optimisation of D. discoideum Ppip5K sequences for yeast expression was designed through an interface from SciTools® (Integrated DNA Technology). Restriction sites were added at the 5′ SalI and 3’ NotI to cloned Ppip5K in a pADH-GST plasmid (Azevedo et al., 2015).
2.4. Growth of yeast and amoeba
Yeast were grown in rich (YPD: 1% yeast extract, 2% peptone, 2% dextrose) or synthetic complete medium (SC, Formedium) in the absence or presence of uracil to select for auxotrophy. For [3H]-inositol labelling, the cells were grown in inositol-free media (SC-inositol, Formedium). The list of the yeast strains used in this study is given in Table 1. D. discoideum lines were isogenic to the axenic strain AX2. Amoeba were cultivated at 22 °C in HL5 media, either in Petri dishes or in flasks at 120 rpm. Cells were diluted every 1–2 days to avoid confluence of dishes or when cell densities exceeded 5 × 106 cells/ml.
Table 1.
List of the yeast strains used in this study.
| Name | Relevant genotype | Reference |
|---|---|---|
| DDY1810 | MATA leu2 ura3-52 trp1 prb1-1122 pep4-3 pre1-451 | |
| kcs1Δ | DDY1810 KCS1::LEU2 | Onnebo and Saiardi, (2009) |
| vip1Δ | DDY1810 VIP1::KANMX4 | Onnebo and Saiardi, (2009) |
| kcs1Δvip1Δ | DDY1810 KCS1::LEU2 VIP1::KANMX4 | Onnebo and Saiardi, (2009) |
| kcs1Δddp1Δ | DDY1810 DDP1::LEU2 KCS1::KANMX4 | This study |
| EY1109 | MATa leu2::PHO84pr:GFP::LEU2 ADE2 | Thomas and O'Shea, (2005) |
| EY1109 pho81Δ | MATa leu2::PHO84pr:GFP::LEU2 ADE2 PHO81::NatNT2 | Desfougeres et al., (2016) |
| EY1109 vip1Δ | MATa leu2::PHO84pr:GFP::LEU2 ADE2 VIP1::NatNT2 | This study |
2.5. Quantification of the PHO pathway activation by fluorescence-activated cell sorting
Logarithmic growing yeast grown in Sc-Ura media carrying pADH-Ppip5k or empty vector were washed and shifted in media with or without phosphate 10 μM for 3 h. Before FACS measurement, 50 μl of the yeast culture was diluted into 950 μl of TBS and immediately analysed using an LSRII flow cytometer (BD Biosciences).
2.6. 13C-NMR analysis
D. discoideum AX2 and ppip5k were grown for 5–7 days in SIH media (Formedium) supplemented with 13C6-inositol (400 μM) synthesized as described (Harmel et al., 2019). Amoeba extracts were separated by PAGE to purify IP7 and IP8. In brief, whole-cell extracts from 350 ml labelled cultures were extracted with perchloric acid and run on single-lane 33% PAGE gels. Bands corresponding to each inositol pyrophosphate species were cut and elute over 24 h by rotation in alternating solutions of water and 1:1 water/methanol (Loss et al., 2011). The combined solutions were acidified with 0.1M perchloric acid and inositol pyrophosphate recovered by TiO2 purification as described (Wilson et al., 2015). The 13C,1H-NMR analysis was performed as previously described (Harmel et al., 2019) using a Bruker AVANCE III spectrometer.
2.7. Extraction and analysis of inositol polyphosphates form yeast
Logarithmic growing yeast were inoculated at OD600 = 0.01 in 5 ml of SC-Ura-inositol supplemented with 5 μCi/ml of [3H]-inositol. The yeast were grow for 16–20 h at 30 °C with shacking. Radiolabelled inositol phosphates were extracted and analysed by Sax-HPLC as described (Azevedo and Saiardi, 2006).
2.8. Extraction and PAGE analysis of inositol polyphosphates and polyP from amoeba
D. discoideum cells centrifuged (500 g, 4 min, 4 °C) and washed once with KK2 (20 mM potassium phosphate pH 6.8). Pellets were resuspended in 40–100 μl perchloric acid (1M), incubated in ice and vortexed for 10 s every 2 min for a total period of 10 min. The extracts were centrifuged at (18000 g, 5 min, 4 °C) and the supernatants were neutralised with 1M potassium carbonate supplemented with 3 mM EDTA at 4 °C for 2 h and subsequently centrifuged. For polyP analysis cell were extracted using acidic phenol procedure (Livermore et al., 2016). PAGE analysis over 33% acrylamide gel was performed as previously described (Losito et al., 2009). Briefly, gels were pre-run for 30 min before loading samples and running overnight at 700 V and 5 milliamps at 4 °C until the Orange G dye had run through 2/3 of the gel. Gels were stained by toluidine blue solution (20% methanol, 2% glycerol, 0.05% Toluidine Blue) at room temperature for 30 min with gentle agitation. Toluidine gels were destained twice in 20% methanol and scanned with an Epson Perfection 4990 Photo Scanner. Image quantification was carried using Image-J software package.
2.9. Generation of D. discoideum null strains
The ip6k and ppip5k strain were constructed using the TMO1 deletion vector (Muramoto et al., 2012). Regions of DNA flanking the gene of interest were amplified by PCR from AX2 genomic DNA using the oligo listed in Table 2. Knockout plasmids TMO1-IP6K-Bsr, TMO1-PPIP5K-Bsr, were generated by inserting these sequences into the plasmid TMO1, using NotI and EcoRI sites for the 5′ arm and the HindIII and KpnI sites for the 3’ arm. The resultant plasmids were then digested using BssHII and used to transform AX2 cells by electroporation using a Biorad Inc. genepulser and exposed to a single pulse of 0.65 kV at 25 μF. Amoebas were subjected to blastocidin selection before screening the transformants by PCR, southern and northern blot and biochemically by PAGE analysis. To generate the ip6k-ppip5k strain the Blastocidin Resistance gene was excised from the ppip5k strain using the pDex-Cre-NLS plasmid (dictybase stock centre (Faix et al., 2004), before knocking out the Ip6k gene using the strategy described above. Southern blotting and Northern blotting were performed using a standard procedure.
Table 2.
Oligo use to generate the Ip6K and Ppip5k deletion constructs.
| Gene | Primer | Primer sequence | Product length | Restriction site |
|---|---|---|---|---|
| Ip6k | 5′ arm Forward | GCAGCGGCCGCCTCAATCCACCCACACTCAC | 1050 | NotI |
| 5′ arm Reverse | GCAGAATTCGTTGTTGTTGGGCTTCTTGG | EcoRI | ||
| 3′ arm Forward | GCGAAGCTTGCGGTAGTAAACCTCAACCTTC | 835 | HindIII | |
| 3′ arm Reverse | GCAGGTACCCTCTTCACTGGTGACACCTATGC | KpnI | ||
| Ppip5k | 5′ arm Forward | GCAGCGGCCGCGTAGTCAGCAAATTTACCAC | 1069 | NotI |
| 5′ arm Reverse | GCAGAATTCCTATCTACAATTGGCCATTC | EcoRI | ||
| 3′ arm Forward | GCGAAGCTTGATATATTACGTGTTAATGG | 832 | HindIII | |
| 3′ arm Reverse | GCAGGTACCTCAAATGGTCAAATTGCTGG | KpnI |
2.10. D. discoideum development
Amoebas were starved by transferring cells from rapidly dividing vegetative cultures onto KK2 2% agar plates. 1 × 107 cells were plated on a 35 mm plate. Cells were allowed to develop at room temperature. Cells were collected after 1 h, ∼8 h when cells were beginning to aggregate, ∼16 h, when cells had coalesced to form slugs and after ∼24 h when fruiting bodies were fully formed. For development on filters, three Whatman® Grade 3 filter papers were layered and covered by a Whatman® Grade 50 quantitative filter paper (hardened low-ash). The filters were soaked in the specified buffer. Following the removal of excess liquid, cells were resuspended in 500 μl of buffer (KK2, TrisHCl 20 mM pH7.0 and HEPES 20 mM pH7.0) and allowed to flow into the filter by delivery with a pipette in an outward spiral movement. Cells were harvested with a plastic scraper. Fruiting body images were then taken on an Olympus camera mounted on a dissecting microscope.
2.11. D. discoideum CE-MS analysis
D. discoideum extract for CE-MS analysis were prepared from vegetative growing strains in HL5 medium. Cultures (20–30 ml of 1–3 X 106 cell/ml) were spun at 500 g for 5 min; the cell pellet was washed in 1 ml KK2 and the inositol phosphate were extracted with 500 μl of perchloric acid 1M in the presence of 5 mM EDTA. The inositol phosphates in the perchloric acid extract were purified using TiO2 (Wilson et al., 2015) before subjecting them to CE-MS-Q-TOF analysis. The analysis was performed as previously described with internal standards (Qiu et al., 2020) using an Agilent 7100 capillary electrophoresis system coupled to a Q-TOF (6520, Agilent) mass spectrometer. Data were collected with Agilent OpenLAB CDS Chemstation 2.3.53 and Agilent MassHunter Workstation Acquisition for Q-TOF B.04.00.
3. Results and discussion
3.1. Identification of D. discoideum inositol phosphate kinases
The screening of D. discoideum genome revealed the presence of seven inositol phosphate kinases (Table 3). Out of these, only the Ip6k (gene name i6kA) has been characterized through the generation of the ip6k (i6kA) strain (Luo et al., 2003). Two other genes, IpkA and Ipmk, were used in overexpression studies but not biochemically characterized (Damstra-Oddy et al., 2021; King et al., 2010). As opposed to the four kinases found in yeast (Laha et al., 2021; Saiardi et al., 2018; Tsui and York, 2010), the seven D. discoideum kinases pointed towards a higher complexity of inositol phosphate synthesis in the amoeba, more similar to mammalian cells. Indeed, like the human genome, amoeba, possess Itpk1, the enzyme which drives the cytosolic route of IP6 synthesis (Desfougeres et al., 2019). The amoeba also possesses one IP5-2Kinase (yeast Ipk1 and mammalian IPPK) and one PPIP5K gene. The inositol kinase enzymology of amoeba is similar to the mammalian counterpart, therefore it is peculiar, as stated in the introduction, that amoeba and human inositol pyrophosphate species differ in their isomeric nature. We, decided to reinvestigate this issue by performing the new structural studies using the newly developed 1H,13C-NMR approach.
Table 3.
Inositol phosphate kinases in Dictyostelium discoideum.
| Gene ID | Dicty base name | Gene name | Inositol Kinase Family | Chr. | Yeast homol. | Mammalian homol. | Ref. |
|---|---|---|---|---|---|---|---|
| DDB_G0281737 | - | Ipmk | IPK superfamily Pfam 03770 |
3 | Arg82 | IPMK | Damstra-Oddy et al., (2021) |
| DDB_G0278739 | I6kA | Ip6k | IPK superfamily Pfam 03770 |
3 | Kcs1 | IP6K 1-3 | Luo et al., (2003) |
| DDB_G0271760 | ipkA1 | Ipka | IPK superfamily Pfam 03770 |
2 | – | – | King et al., (2010) |
| DDB_G0283863 | - | Ipkb | IPK superfamily Pfam 03770 |
4 | – | – | |
| DDB_G0269746 | Itpk1 | Itpk1 | ITPK1 family Pfam 17927 |
1 | – | ITPK1 | |
| DDB_G0288351 | - | Ipk1 | IP5-2K family Pfam 06090 |
5 | Ipk1 | IPPK | |
| DDB_G0284617 | - | Ppip5k | PPIP5K family Pfam 18086 |
4 | Vip1 | PPIP5K 1-2 |
3.2. 13C-NMR characterization of D. discoideum IP7 and IP8
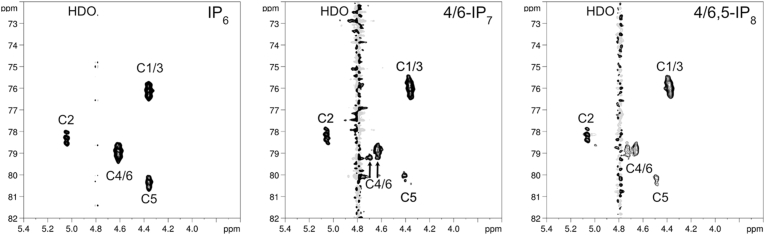
Previous NMR studies of IP7 and IP8 purified from amoeba were performed using two-dimensional 1H,31P-NMR (Laussmann et al., 1996). This approach has limited sensitivity. Conversely, the newly developed 1H,13C-NMR offers higher sensitivity since the chemical shift dispersion of 13C is superior to 31P, and the magnetization transfer via 1J(1H,13C) one-bond couplings is more efficient (Harmel et al., 2019). We fed wild type AX2 amoeba with 13C6-inositol and after extracting and purifying the inositol pyrophosphates using TiO2, we analysed them using 1H,13C-NMR spectroscopy. The 2-dimensional inverse H,C correlation spectra (Fig. 1) confirm previous studies. The IP7 spectra reveal the carbon 4/6 split signal, while the IP8 spectra additionally reveal the shift towards the left of the signal of carbon 5. These are the typical signatures of pyrophosphate moieties at these two carbons (Harmel et al., 2019). Therefore, the inositol pyrophosphate isomers present in the social amoeba are indeed the 4/6-IP7 and the 4/6,5-IP8 forms. Of note, neither the myo-inositol 4/6 carbon positions nor the 1/3 carbon positions can be distinguished by NMR as they are enantiotopic. The different inositol pyrophosphate isomers present in amoeba and mammals, despite similar enzymology, suggests that the IP6K or the PPIP5K enzyme could pyro-phosphorylate different inositol ring positions depending on the species analysed.
Fig. 1.
13C-NMR spectra of IP6, IP7 and IP8 extracted from D. discoideum AX2. Inositol pyrophosphates (extracted and TiO2-purified from D. discoideum grown in presence of 13C6-inositol) were analysed using a Bruker AVANCE III spectrometer operating at 600 MHz 600 MHz for proton, and at 151 MHz for carbon. Two-dimensional 1H,13C NMR spectrum for IP6 (left), IP7 (centre) and InsP8 (right) reveals the presence of 4/6-IP7 and 4/6,5-IP8 in wild type amoeba. The arrows indicate the spitted 4/6 carbon signal in 4/6-IP7. The positions of the carbon atoms and the solvent signal of deuterium water (HDO) are indicated.
3.3. D. discoideum Ppip5K rescues yeast vip1Δ phenotypes
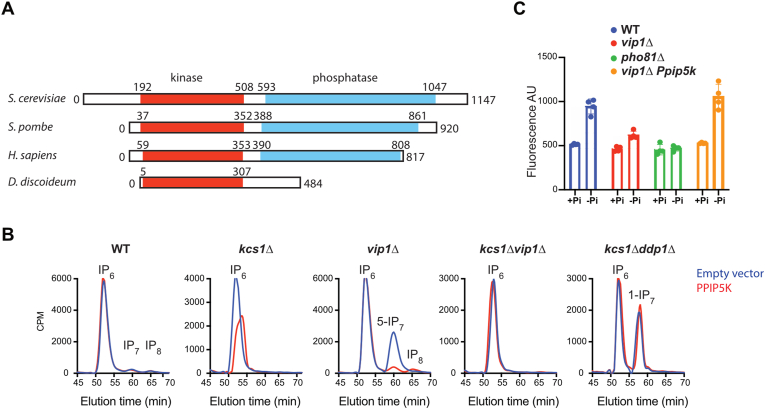
To gain further insight into D. discoideum inositol pyrophosphate metabolism, we focused our attention on Ppip5k. The Ppip5k (DDB_G0284617) homologue in D. discoideum encodes a 56 kDa protein, compared to the 130 kDa yeast protein and ∼150 kDa in mammalian cells (Fig. 2A). In both yeast and mammals, the PPIP5K encodes a protein containing both a kinase domain and a phosphatase domain (Dollins et al., 2020; Pascual-Ortiz et al., 2018). Interestingly, the D. discoideum gene encodes a much smaller enzyme, which completely lacks the phosphatase domain. The absence of this phosphatase domain in the amoeba might abolish the futile cycle proposed for this type of kinase (Randall et al., 2020).
Fig. 2.
D. discoideum Ppip5k structure and its ability to rescue yeast vip1Δ phenotypes. The schematic representation of Ppip5k structural organization (A) from different organisms illustrates the absence of the phosphatase domain in D. discoideum protein. Sax-HPLC analysis of 3H-inositol labelled yeast expressing amoeba Ppip5K (B) reveals the ability of this enzyme to revert the biochemical phenotype of the vip1Δ strain, i.e. an increase in 5-IP7. The activation of the PHO pathway under phosphate starvation was monitored by FACS analysis (C). In the indicated strains, GFP is under the control of the promoter of the high affinity phosphate transporter Pho84 and is thus a readout for the PHO pathway activation. The different yeast carrying empty vector or pADH-Ppip5k were washed and shifted in media with (+Pi) or without (-Pi) phosphate for 3 h. The defect in the response observed in the vip1Δ is fully rescued upon expression of Ppip5k. The pho81Δ strain, which constitutively represses the PHO pathway, is used as a control. The results are from four independent experiments.
The D. discoideum proteome has evolved to encode peptides with long poly-glutamine or poly-asparagine tracts (Santarriaga et al., 2015). The Ppip5k coding sequence possesses two long poly-asparagine stretches that have prevented us obtaining recombinant Ppip5k from bacterial expression systems. Similarly, D. discoideum Ip6k (DDB_G0278739) contains six poly-asparagine and one poly-glutamine repeats, which have also prevented us obtaining recombinant protein. Therefore, to test the amoeba Ppip5k activity, we cloned the gene into a yeast expression vector and transformed it into an array of S. cerevisiae mutants (Fig. 2B). The amoeba Ppip5k is able to completely reverse the increase in 5-IP7, observable in vip1Δ yeast. Ppip5k do not appear to use as substrate the 1-IP7 that accumulate in kcs1Δddp1Δ or the IP6 present in kcs1Δ or kcs1Δvip1Δ strains.
To verify if the biochemical phenotype generates a product functionally equivalent to the Vip1 generated IP8, we investigated if amoeba Ppip5k rescues the vip1Δ PHO response defect (Choi et al., 2017). In low phosphate conditions, a set of genes, named the PHO genes, are up-regulated. The expression of these genes is repressed in phosphate-rich conditions. One such gene is PHO84 that encodes a high affinity phosphate transporter. The induction of the PHO genes expression can be monitored by recording the expression of a reporter-protein cloned behind the PHO gene promoter. We took advantage of the strain (EY1109) developed by Thomas and O'Shea that expresses GFP under the control of the PHO84 promoter (Thomas and O'Shea, 2005). In this background, the deletion of Vip1 leads to a repression of the PHO genes expression, as previously demonstrated (Choi et al., 2017). This is similar to what is observed when PHO81 is deleted (Desfougeres et al., 2016). Expression of the amoeba Ppip5k in the vip1Δ strain fully rescues the expression of the reporter (Fig. 2C) indicating that, in vivo, the product of the enzymatic reaction catalysed by Ppip5k is functionally equivalent to the Vip1 product. The rescue of both biochemical and physiological vip1Δ phenotypes demonstrates that D. discoideum Ppip5K is a genuine PPIP5K enzyme able to phosphorylate 5-IP7 to, likely, 1,5-IP8.
3.4. Generation of the D. discoideum ppip5k strain
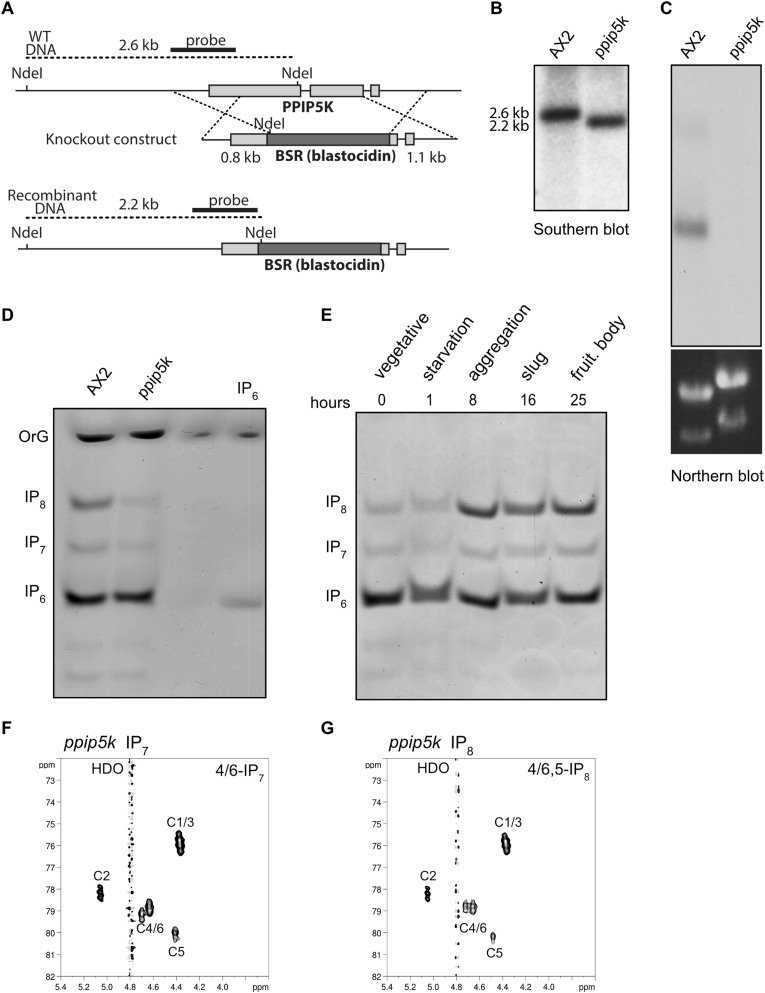
Since NMR studies indicates that D. discoideum do not possess 1,5-IP8 ((Laussmann et al., 1996) Fig. 1), we decided to knockout Ppip5k to characterise the effect of the absence of this kinase on the amoeba inositol pyrophosphate metabolism. A homologous recombination approach was used to generate amoeba knockout. This approach involved cloning two regions flanking the target gene and inserting them on either side of a Blastocidin resistance marker gene. The strategy for deletion of the Ppip5k involved cloning 1.1 kb of genomic sequence overlapping slightly with the 5′ region of the gene and 0.8 kb 3’ of the gene (Fig. 3A). The generated ppip5k strains were confirmed by Southern blot (Fig. 3B); while Northern blot analysis (Fig. 3C) confirms the loss of the Ppip5K transcript in the ppip5k strain.
Fig. 3.
Generation of D. discoideum ppip5k strain. The homologous recombination strategy to generate ppip5k amoeba (A) highlights the screening approach and probe location. Southern blot analysis (B) of AX2 and ppip5k amoeba is consistent with the strategy design prediction. Norther blot analysis (C) reveals the absence of any Ppip5k transcript in the ppip5k amoeba. Ethidium bromide staining (bottom panel) of the ribosomal RNA confirms equal loading. Neutralised acidic extracts from AX2 and ppip5k- (5 × 106 cells) were resolved on 33% PAGE and the inositol phosphates were visualised by toluidine blue staining (D). This analysis reveals a substantial decrease in the level of IP7 and especially IP8, in the ppip5k strain. The PAGE is a representative result of an experiment repeated 4 or more times. The recorded accumulation of IP8 during development (Pisani et al., 2014) is not altered in the ppip5k amoeba (E) as revealed by PAGE analysis of neutralised cell extracts collected at different developmental stages. The experiment was repeated twice giving identical results. Two-dimensional 1H,13C NMR spectrum of ppip5k extracted IP7 (F) and IP8 (G) reveals the presence of 4/6-IP7 and 4/6,5-IP8 like in wild type amoeba. The positions of the carbon atoms and the solvent signal of deuterium water (HDO) are indicated.
The analysis by PAGE of the ppip5k inositol pyrophosphates profile reveals a 49.6 ± 7.8% (n = 4) reduction in IP8 levels while a 14.3 ± 6.7% (n = 4) reduction in the level of IP7 is recorded (Fig. 3D). This biochemical defect is remarkably different from the one reported for yeast vip1Δ in which a substantial increase in IP7 is observed (Onnebo and Saiardi, 2009). The decrease of both IP7 and IP8 observed in ppip5k amoeba grown in rich HL5 medium, prompted us to verify if the reported increase on IP8 level during D. discoideum development (Laussmann et al., 2000; Pisani et al., 2014) is under Ppip5k control. The developmental analysis of inositol pyrophosphate profile in ppip5k amoeba reveals a consistent accumulation of IP8 during the late stage of development. In conclusion, ppip5K regulate inositol pyrophosphate metabolism in the vegetative stage but not its modulation during amoeba development (Fig. 3E).
We next assessed the isomeric nature of IP7 and IP8 in the ppip5k strain using 13C-NMR. After feeding ppip5k amoeba with 13C6-inositol IP7 and IP8 were extracted and subjected to 13C-NMR analysis (Fig. 3F and G). The ppip5k-purified IP7 and IP8 spectra show the characteristic signature of pyrophosphate moiety at position 4/6 and 5 carbons. Like the wild type AX2 amoeba, the ppip5k strain possesses the 4/6-IP7 and 4/6,5-IP8 isomers. This is not surprising since PPIP5K are kinases thought to phosphorylate position one of the inositol ring (Lin et al., 2009; Wang et al., 2012); furthermore our vip1Δ rescue experiments (Fig. 2B and C) are also indicative of this specificity. While 13C-NMR sensitivity might fail to detect minor species of IP7 and of IP8 species, our theoretical consideration and our analysis suggest that amoeba Ppip5k, while not participating directly in the synthesis of the abundant 4/6,5-IP8, is nevertheless able to regulate its cellular levels.
3.5. Generation of the D. discoideum ip6k and ip6k-ppip5k strain
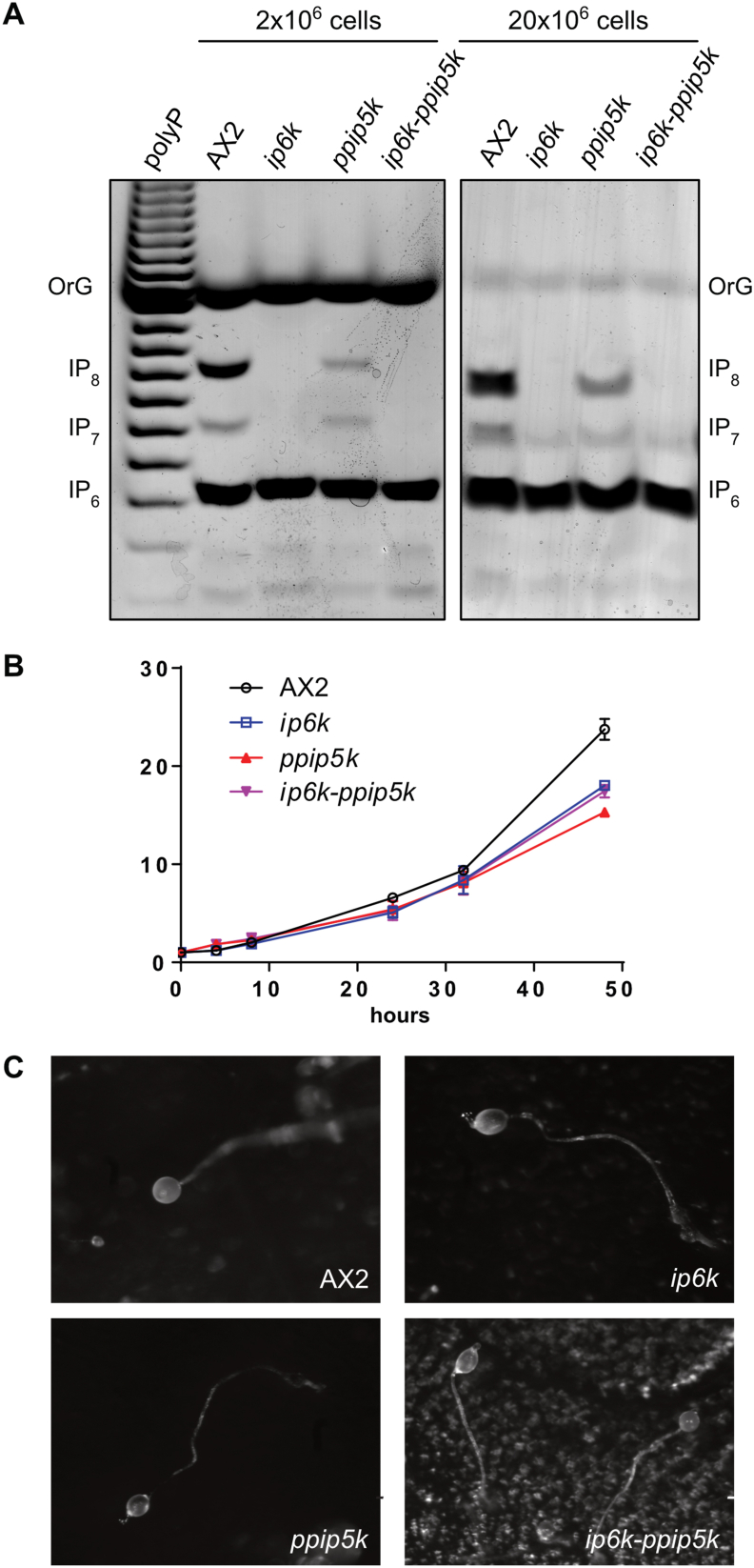
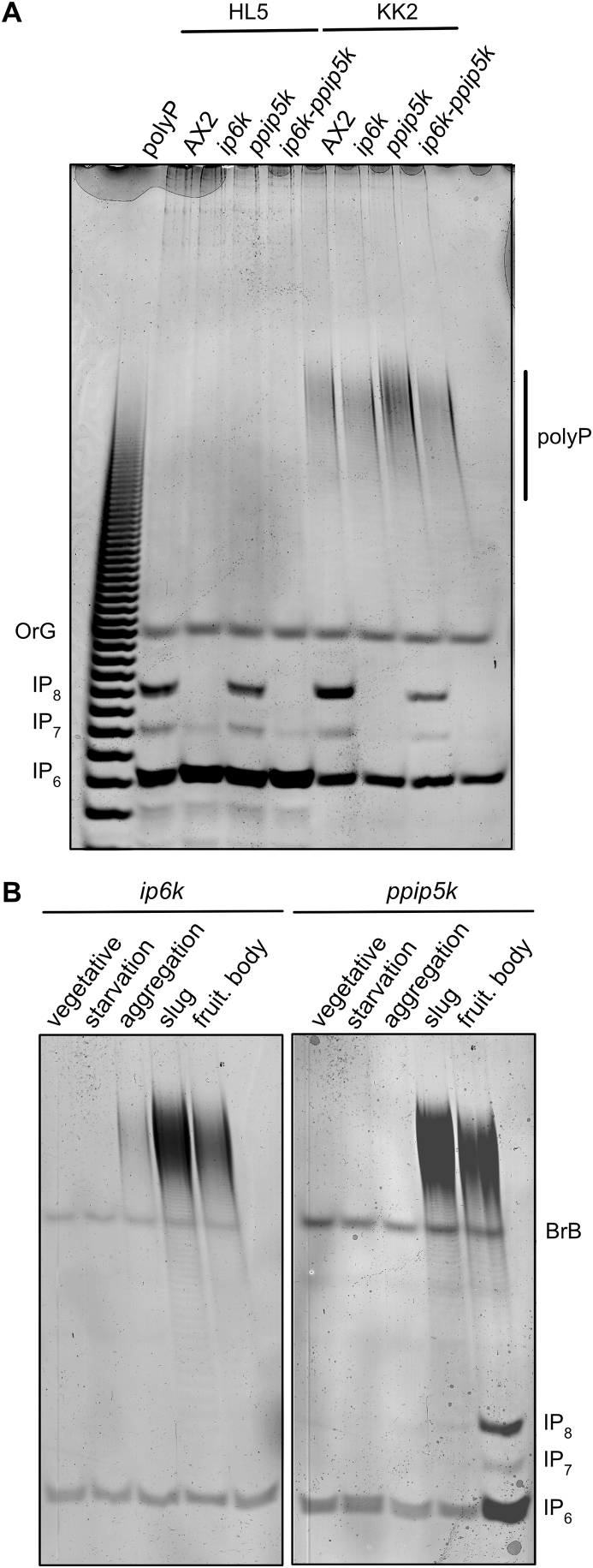
The inability to produce recombinant Ip6k and Ppip5k proteins to assess their biochemistry in vitro, prompted us to develop the full array of knockout strains to perform in vivo analyses. We re-generate the ip6k strain (see material and methods) isogenic to our AX2 background. Identically to the previously generated ip6k mutant, the new strain has no detectable level of IP8 and an almost completely depleted level of IP7 (Fig. 4A). Double mutants, in which both ppip5k and ip6k genes were disrupted, were generated starting from the ppip5k strain in which the blastocidin resistance gene (BSR) was excised by overexpressing a recombinant Cre (Faix et al., 2004). The Ip6k gene was then disrupted to generate the ip6k-ppip5k strain (Fig. 4A). PAGE analysis of the ip6k-ppip5k reveals an inositol pyrophosphate profile similar to ip6k amoeba. Both the ip6k and the ip6k-ppip5k strains possess residual amounts of IP7 detectable by PAGE when extracts from 20 million cells were loaded on gel (Fig. 4A right panel), indicating the presence of an additional enzyme able to synthesize inositol pyrophosphates.
Fig. 4.
Comparative biochemical, growth and developmental analysis of D. discoideum wild type and knockout strains. Inositol pyrophosphates extracted from 1 × 106 (left) or 20 × 106 (right) AX2 and mutants ip6k, ppip5k and ip6k-ppip5- amoeba were resolved on 33% PAGE and visualised by staining with toluidine blue (A). Inorganic polyphosphate polyP is used to orientate the gel, OrangeG (OrG) is used as migrating dye. A reduction but not ablation of both IP8 and IP7 is noticeable in ppip5k strain. Neither IP7 nor IP8 is detectable in extracts from two million cells of either ip6k or ip6k-ppip5k (left panel). However, residual levels of IP7 are detectable in extracts from 20 millions of cells of both ip6k and ip6k-ppip5k (right panel). The gel is representative of at least three independent experiments. To investigate the effect of the altered inositol pyrophosphate metabolism on general fitness, the growth of AX2 was compared to ip6k, ppip5k and ip6k-ppip5k mutants (B). WT, ip6k, ppip5k and ip6k-ppip5k were grown for 48 h in HL5 media starting at a density of 1 × 105 cells per ml The figure shows averages ± SD from three independent experiments. All three mutants strain displayed very slight growth defects not reaching statistical significance. Developmental analysis (C) performed under standard KK2-agar conditions shows no obvious developmental phenotype as revealed by the photos of the fruiting body. The result is representative of an experiment repeated at least three times.
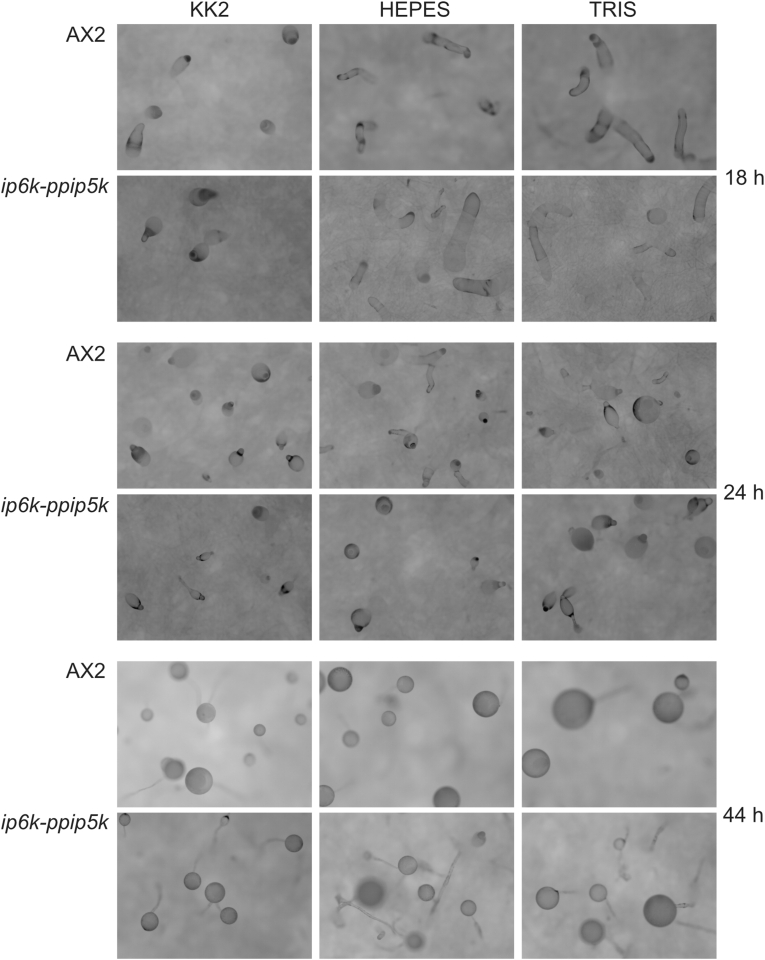
To verify the effect on the amoeba general fitness of the deletion of the known inositol pyrophosphate synthesizing enzymes, we characterized the growth rate of AX2, ip6k, ppip5k, and ip6k-ppip5k amoebas (Fig. 4B). We did not observe major growth defects when the null strains were grown in rich HL5 synthetic media. Although we could observe the tendency for the mutant strains to grow slowly, this difference does not reach statistical significance. We next assessed the ability of the mutants to undergo starvation-induced development. All strains succeeded to follow development under standard KK2 agar conditions and form fruiting bodies (Fig. 4C). To detect any developmental phenotype that may have gone unnoticed in a phosphate rich buffer as KK2 (20 mM potassium phosphate buffer pH 6.8), the process of development of the double mutant ip6k-ppip5k was examined under complete phosphate starvation using TrisHCl or HEPES as buffer on cellulose filters. Despite variable differences in timing, not attributable to differing buffer conditions, ip6k-ppip5k completed development, culminating in the formation of fruiting bodies slightly smaller than the AX2 strain (Fig. 5).
Fig. 5.
Developmental progression of wild type AX2 and the double knockout ip6k-ppip5k strain. To assess if phosphate affects the developmental process of the ip6k-ppip5k strain, the AX2 and ip6k-ppip5k were developed on buffered cellulose filters. The buffers used were KK2 (potassium phosphate buffer), the phosphate-free HEPES buffer, and TRIS buffer. While developmental timings were inconstant between experiments, due to the variable amount of liquid in the imbedded filter, no consistent phenotype was detected.
3.6. CE-MS analysis of D. discoideum inositol pyrophosphate metabolism
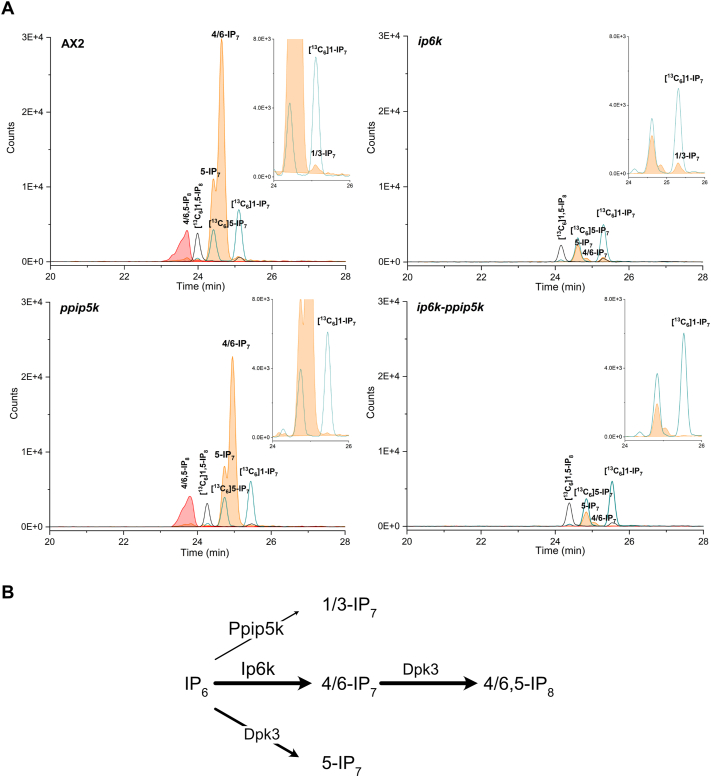
To better elucidate the inositol pyrophosphate metabolism in the mutant strains, we next performed Capillary Electrophoresis Mass Spectrometry (CE-MS) analysis (Qiu et al., 2020). This sensitive analytical technique resolves with unprecedented resolution the different isomers of IP7 and IP8. CE-MS studies complement 13C-NMR analysis, which offers unique structural information, but lacks the degree of sensitivity of mass spectrometry detection.
The qualitative analysis of AX2 amoeba reveals 4/6-IP7 and 4/6,5-IP8 to be the major inositol pyrophosphates species, confirming the 13C-NMR studies (Fig. 6). However, two additional, IP7 isomers could be identified; 5-IP7 constituting about 20% of the entire IP7 pools and 1-IP7 representing roughly <5% of the entire IP7 pools. As expected, the analysis of ppip5k strain reveals the absence of the minor 1/3-IP7 species demonstrating that the D. discoideum Ppip5k likely phosphorylates the 1 position similarly to the mammalian counterpart (Wang et al., 2012) and as our yeast rescues experiment suggested (Fig. 2B and C). The Ppip5k synthesized 1/3-IP7, while not participating directly in the synthesis of the 4/6,5-IP8, still regulates its cellular level (Fig. 3, Fig. 4A).
Fig. 6.
CE-MS analysis of D. discoideum of AX2 and ip6k and ip6-ppip5k strains extracts. Qualitative CE-MS separation of TiO2-purified D. discoideum extracts (A). Empty peak area indicates the migration of the indicated 13C6-inositol standard while the filled peak area represents the elution of the amoeba extracted IP7s (orange) and IP8 (red). Enlarged inserts for the IP7 region are presented to highlight the minor species of 1-IP7 and 5-IP7. This analysis reveals the absence of 1-IP7 in ppip5- and of 4/6-IP7 in ip6k stains and of both in ip6k-ppip5k-. From this, we could deduce the inositol pyrophosphate pathway presented in (B) where Dpk3 stand for Diphosphate kinase number 3. This analysis was repeated three times giving identical results.
Surprisingly, the analysis of ip6k mutant reveals the disappearance of the major IP7 isomer the 4/6-IP7 species (Fig. 6A). Thus, to the contrary of the mammalian IP6Ks D. discoideum homologous enzyme pyrophosphorylate position 4/6 of the inositol ring generating 4/6-IP7. In light of these observations, in the ip6k-ppip5k amoeba, only the 5-IP7 isomer could be detected. Therefore, D. discoideum must possess an additional kinase that we named Diphospho kinase 3 (Dpk3) responsible for 5-IP7 synthesis and that together with the Ip6k would generate 4/6,5-IP8. Fig. 6B summarises D. discoideum inositol pyrophosphate metabolism revealed by these analyses where Dpk3 represents an as-yet uncharacterised kinase capable of producing inositol pyrophosphates. The presence of three IP7 isomers suggests the possibility for amoeba to synthesize three IP8 species however only 4/6,5-IP8 is detectable in our current CE-MS-qTOF experimental setup.
3.7. polyP metabolism is not affected by D. discoideum Ip6k or Ppip5k
Based on our understanding of the link between inositol pyrophosphates and phosphate metabolism in yeast, the altered inositol pyrophosphates present in ip6k, ppip5k, and ip6k-ppip5k could influences directly or indirectly phosphate homeostasis in D. discoidem. The social amoeba possess sub-millimolar concentration of the phosphate-rich IP6, IP7, and IP8 thus changing their concentration influences a large pool of cellular phosphate. Alternatively, inositol pyrophosphate could influence phosphate availability by regulating polyP metabolism, particularly as the primary function of polyP is to buffer cellular free phosphate concentration. We previously demonstrated that in S. cerevisiae polyP synthesis is under Kcs1 (the IP6K) control (Lonetti et al., 2011), while in S. pombe it is Asp1/Vip1 (the PPIP5K) that regulates polyP metabolism (Pascual-Ortiz et al., 2021). Therefore, in yeast, there is a clear link between inositol pyrophosphate and polyP cellular level even if the precise inositol phosphate kinase regulating polyP metabolism differs between yeast species.
The D. discoideum ip6k, ppip5k, and ip6k-ppip5k strains offer the opportunity to verify in an organism belonging to a different taxon if the synthesis of polyP is under control of Ip6K, of Ppip5k, of both or neither of the two enzymes. We extracted polyP using acidic phenol procedure from fast-dividing AX2, ip6k, ppip5k, and ip6k-ppip5k amoeba grown on rich HL5 medium and from KK2 agar plates for 16 h, a condition we previously demonstrated to induce polyP synthesis (Livermore et al., 2016). PAGE analysis of the extracted polyP revealed that while polyP is undetectable in this experimental setup from amoeba grown in HL5 medium, polyP under starvation conditions is detected in all four D. discoideum strains (Fig. 7A). We next followed the developmental synthesis and accumulation of polyP (Livermore et al., 2016). The ip6k and ppip5k amoebae were transferred on KK2 agar and cells were collected at different time points corresponding to the diverse developmental stages. PAGE analysis of phenol extract from ip6k and ppip5k amoebae revealed the dramatic accumulation of polyP during development as reported previously (Livermore et al., 2016). Therefore, in the social amoeba nether, the Ip6K or the Ppip5k are able to control polyP metabolism.
Fig. 7.
D. discoideum polyP analysis of vegetative and development states of the inositol phosphate kinase mutants. Wild type AX2 and ip6k and ip6k-ppip5- strains growing exponentially in rich medium HL5 were collected (20 × 106 cells) or plated on KK2 agar plates for 16 h to stimulate developmentally induced polyP synthesis. The plates were scraped to recover D. discoideum and a neutralised acidic extract of the samples in vegetative stage (HL5) or after starvation (KK2) was prepared before loading on a 33% PAGE gel. The gels were stained with Toluidine blue to visualise inositol phosphate and polyP. While in this experimental set up polyP could not be detected in vegetative amoeba, polyP induction during development is clearly evident in both the wild type or mutant stains. OrangeG (OrG) is used as migrating dye. Developmental accumulation of polyP (B) is present in both ip6k (left panel) and in ppip5k (right panel) strains. Bromophenol Blue (BrB) is used as migrating dye. The figure is representative of experiments repeated at least three times.
4. Conclusion
Studying the inositol pyrophosphate metabolism in ip6k, ppip6k and ip6k-ppip5k amoeba revealed interesting features. The amoeba Ip6k synthesizes 4/6-IP7 instead of the 5-IP7 isomer synthesized by its mammalian counterpart. Therefore, the definition of the inositol pyrophosphate species present in one specific organism cannot be extrapolated by sequence homology, but must be tested experimentally.
Conversely, the amoeba Ppip5k similarly to its mammalian counterpart, does pyro-phosphorylate position 1/3 of the inositol ring producing 1/3-IP7, and therefore could not participate directly to the synthesis of the abundant 4/6,5-IP8 isomer. The 1/3-IP7 is by far the minor species of the three IP7 isomer found in amoeba, but it does indirectly regulate IP8 synthesis since pppip5k possess a 50% decrease in 4/6,5-IP8 level. These results indicate the existence of a third kinase, likely regulated by Ppip5k or its product, able to pyro-phosphorylate position five, synthesizing 5-IP7 and 4/6,5-IP8. The recent discovery that Arabidopsis thaliana possesses three isomers of IP7 and like in D. discoideum the most abundant is the 4/6-IP7 species (Riemer et al., 2021), suggests the amoeba inositol pyrophosphates metabolism is conserved across many species.
Our work also reveals that in amoeba neither the Ip6k or the Ppip6k are involved in regulating polyP metabolism. This should not come as a surprise, since the synthesis of polyP in amoeba and yeast occurs using different enzymology. While yeast Vtc4 possesses an SPX domain that could be regulated by inositol pyrophosphates, the amoeba Ppk1 does not. Our work highlights how incorrect it is to extrapolate polyP yeast discoveries to other species when the mechanism of polyP synthesis is different as in amoeba or unknown as in mammals.
Surprisingly, ip6k, ppip6k and ip6k-ppip5k amoebas do not show major growth or developmental defects. We could not exclude that the minor species of IP7 present in ip6k and the 5-IP7 present in ip6k-ppip5k is sufficient to play signalling roles preventing the manifestation of inositol pyrophosphate-specific phenotypes. For this reason, it is imperative to identify the D. discoideum enzyme responsible to pyro-phosphorylate position five of the inositol ring Dpk3 (Fig. 6B) and thus responsible for the synthesis of the 5-IP7 present in ip6k-ppip5k and for the synthesis of the abundant 4/6,5-IP8 present in wild type amoeba. After identifying this additional kinase, the generation of the triple mutant strain might reveal the amoeba phenotypes associated with the absence of inositol pyrophosphates.
Credit author statement
Yann Desfougères: Investigation, Visualization, Writing- Reviewing and Editing. Paloma Portela-Torres: Investigation, Visualization, Writing – review & editing. Danye Qiu: Investigation, Visualization, Writing – review & editing. Thomas M Livermore: Investigation, Writing – review & editing. Robert K Harmel: Investigation, Visualization, Writing – review & editing. Filipy Borghi: Writing – review & editing. Henning J. Jessen: Supervision, Formal analysis, Funding acquisition, Dorothea Fiedler: Supervision, Formal analysis, Funding acquisition, Adolfo Saiardi; Conceptualization, Methodology, Supervision, Formal analysis, Funding acquisition, Writing – original draft, Writing – review & editing.
Funding
This work was supported by the Medical Research Council (MRC) grant MR/T028904/1, and by the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie Grant agreement PHEMDD 752903. This study was supported by the Deutsche Forschungsgemeinschaft (DFG) under Germany's Excellence Strategy (CIBBS, EXC-2189, Project ID 390939984 to HJJ). D.Q. gratefully acknowledges financial support from the Brigitte-Schlieben-Lange-Programm. R.K.H. gratefully acknowledges funding from the Leibniz-Gemeinschaft (SAW-2017-FMP-1).
Declaration of competing interest
The authors declare none conflict of interest. The funding bodies do not have any role in the study design, and in data collection and analysis.
Acknowledgements
We thank Antonella Riccio for suggestions and helpful comments. We thank Erin O'Shea for providing EY1109 yeast. We would also like to thank the core staff members at the LMCB for facilitating our research. The research data are available upon request.
Abbreviations
- IP6
inositol hexakisphosphate, phytic acid
- IP7
diphosphinositol pentakisphosphate
- IP8
bis-diphosphinositol tetrakisphosphate
- IP6K
inositol hexakisphosphate kinase
- PPIP5K
diphosphoinositol pentakisphosphate kinase
- Vtc4
vacuolar transporter chaperone 4
- PPK1
polyphosphate Kinase 1
- polyP
inorganic polyphosphate
- SPX
protein domain named after SYG1/Pho81/XPR1
- AX2
axenic strains 2
- NMR
nuclear magnetic resonance
- CE-MS
capillary electrophoresis mass spectrometry
- qTOF
quadrupole time of flight
- BSR
blastocidin resistance gene
- PAGE
poly acrylamide gel electrophoresis
- OrG
Orange G
- BrB
bromophenol blue
- DpK3
diphospho kinase 3
References
- Al-Anbaky Q., Al-Karakooly Z., Connor R., Williams L., Yarbrough A., Bush J., Ali N. Role of inositol polyphosphates in programed cell death in Dictyostelium discoideum and its developmental life cycle. Mol. Cell. Biochem. 2018;449(1–2):237–250. doi: 10.1007/s11010-018-3360-6. [DOI] [PubMed] [Google Scholar]
- Azevedo C., Livermore T., Saiardi A. Protein polyphosphorylation of lysine residues by inorganic polyphosphate. Mol. Cell. 2015;58(1):71–82. doi: 10.1016/j.molcel.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Azevedo C., Saiardi A. Extraction and analysis of soluble inositol polyphosphates from yeast. Nat. Protoc. 2006;1(5):2416–2422. doi: 10.1038/nprot.2006.337. [DOI] [PubMed] [Google Scholar]
- Azevedo C., Saiardi A. Eukaryotic phosphate homeostasis: the inositol pyrophosphate perspective. Trends Biochem. Sci. 2017;42(3):219–231. doi: 10.1016/j.tibs.2016.10.008. [DOI] [PubMed] [Google Scholar]
- Choi J., Rajagopal A., Xu Y.F., Rabinowitz J.D., O'Shea E.K. A systematic genetic screen for genes involved in sensing inorganic phosphate availability in Saccharomyces cerevisiae. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0176085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.H., Williams J., Cho J., Falck J.R., Shears S.B. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J. Biol. Chem. 2007;282(42):30763–30775. doi: 10.1074/jbc.M704655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremers C.M., Knoefler D., Gates S., Martin N., Dahl J.U., Lempart J., Xie L., Chapman M.R., Galvan V., Southworth D.R., Jakob U. Polyphosphate: a conserved modifier of amyloidogenic processes. Mol. Cell. 2016;63(5):768–780. doi: 10.1016/j.molcel.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocco P., Saiardi A., Wilson M.S., Maletta R., Bruni A.C., Passarino G., Rose G. Contribution of polymorphic variation of inositol hexakisphosphate kinase 3 (IP6K3) gene promoter to the susceptibility to late onset Alzheimer's disease. Biochim. Biophys. Acta. 2016;1862(9):1766–1773. doi: 10.1016/j.bbadis.2016.06.014. [DOI] [PubMed] [Google Scholar]
- Damstra-Oddy J.L., Warren E.C., Perry C.J., Desfougeres Y., Fitzpatrick J.K., Schaf J., Costelloe L., Hind W., Downer E.J., Saiardi A., Williams R.S.B. Phytocannabinoid-dependent mTORC1 regulation is dependent upon inositol polyphosphate multikinase activity. Br. J. Pharmacol. 2021;178(5):1149–1163. doi: 10.1111/bph.15351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desfougeres Y., Gerasimaite R., Jessen H.J., Mayer A. Vtc5, a novel subunit of the vacuolar transporter chaperone complex, regulates polyphosphate synthesis and phosphate homeostasis in yeast. J. Biol. Chem. 2016;2016(42):291. doi: 10.1074/jbc.M116.746784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desfougeres Y., Saiardi A. Dictyostelium discoideum as a model to study inositol polyphosphates and inorganic polyphosphate. Methods Mol. Biol. 2020;2091:59–71. doi: 10.1007/978-1-0716-0167-9_5. [DOI] [PubMed] [Google Scholar]
- Desfougeres Y., Wilson M.S.C., Laha D., Miller G.J., Saiardi A. ITPK1 mediates the lipid-independent synthesis of inositol phosphates controlled by metabolism. Proc. Natl. Acad. Sci. U. S. A. 2019;116(49):24551–24561. doi: 10.1073/pnas.1911431116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollins D.E., Bai W., Fridy P.C., Otto J.C., Neubauer J.L., Gattis S.G., Mehta K.P.M., York J.D. Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling. Proc. Natl. Acad. Sci. U. S. A. 2020;117(17):9356–9364. doi: 10.1073/pnas.1908875117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Ma G., Sui L., Wei M., Satheesh V., Zhang R., Ge S., Li J., Zhang T.E., Wittwer C., Jessen H.J., Zhang H., An G.Y., Chao D.Y., Liu D., Lei M. Inositol pyrophosphate InsP8 acts as an intracellular phosphate signal in Arabidopsis. Mol. Plant. 2019;12(11):1463–1473. doi: 10.1016/j.molp.2019.08.002. [DOI] [PubMed] [Google Scholar]
- Draskovic P., Saiardi A., Bhandari R., Burton A., Ilc G., Kovacevic M., Snyder S.H., Podobnik M. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem. Biol. 2008;15(3):274–286. doi: 10.1016/j.chembiol.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Europe-Finner G.N., Gammon B., Newell P.C. Accumulation of [3H]-inositol into inositol polyphosphates during development of Dictyostelium. Biochem. Biophys. Res. Commun. 1991;181(1):191–196. doi: 10.1016/s0006-291x(05)81400-7. [DOI] [PubMed] [Google Scholar]
- Faix J., Kreppel L., Shaulsky G., Schleicher M., Kimmel A.R. A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res. 2004;32(19) doi: 10.1093/nar/gnh136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridy P.C., Otto J.C., Dollins D.E., York J.D. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J. Biol. Chem. 2007;282(42):30754–30762. doi: 10.1074/jbc.M704656200. [DOI] [PubMed] [Google Scholar]
- Gerasimaite R., Pavlovic I., Capolicchio S., Hofer A., Schmidt A., Jessen H.J., Mayer A. Inositol pyrophosphate specificity of the SPX-dependent polyphosphate polymerase VTC. ACS Chem. Biol. 2017;12(3):648–653. doi: 10.1021/acschembio.7b00026. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Woods R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Harmel R.K., Puschmann R., Nguyen Trung M., Saiardi A., Schmieder P., Fiedler D. Harnessing (13)C-labeled myo-inositol to interrogate inositol phosphate messengers by NMR. Chem. Sci. 2019;10(20):5267–5274. doi: 10.1039/c9sc00151d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Fujii N., Wittwer C., Sasaki A., Tanaka M., Bittner T., Jessen H.J., Saiardi A., Takizawa S., Nagata E. Hydrophilic interaction liquid chromatography-tandem mass spectrometry for the quantitative analysis of mammalian-derived inositol poly/pyrophosphates. J. Chromatogr. A. 2018;1573:87–97. doi: 10.1016/j.chroma.2018.08.061. [DOI] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21(11):947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- King J., Keim M., Teo R., Weening K.E., Kapur M., McQuillan K., Ryves J., Rogers B., Dalton E., Williams R.S., Harwood A.J. Genetic control of lithium sensitivity and regulation of inositol biosynthetic genes. PLoS One. 2010;5(6) doi: 10.1371/journal.pone.0011151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha D., Portela-Torres P., Desfougeres Y., Saiardi A. Inositol phosphate kinases in the eukaryote landscape. Adv. Biol. Regul. 2021;79 doi: 10.1016/j.jbior.2020.100782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laussmann T., Eujen R., Weisshuhn C.M., Thiel U., Vogel G. Structures of diphospho-myo-inositol pentakisphosphate and bisdiphospho-myo-inositol tetrakisphosphate from Dictyostelium resolved by NMR analysis. Biochem. J. 1996;315(Pt 3):715–720. doi: 10.1042/bj3150715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laussmann T., Pikzack C., Thiel U., Mayr G.W., Vogel G. Diphospho-myo-inositol phosphates during the life cycle of Dictyostelium and Polysphondylium. Eur. J. Biochem. 2000;267(8):2447–2451. doi: 10.1046/j.1432-1327.2000.01264.x. [DOI] [PubMed] [Google Scholar]
- Laussmann T., Reddy K.M., Reddy K.K., Falck J.R., Vogel G. Diphospho-myo-inositol phosphates from Dictyostelium identified as D-6-diphospho-myo-inositol pentakisphosphate and D-5,6-bisdiphospho-myo-inositol tetrakisphosphate. Biochem. J. 1997;322(Pt 1):31–33. doi: 10.1042/bj3220031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Fridy P.C., Ribeiro A.A., Choi J.H., Barma D.K., Vogel G., Falck J.R., Shears S.B., York J.D., Mayr G.W. Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J. Biol. Chem. 2009;284(3):1863–1872. doi: 10.1074/jbc.M805686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore T.M., Chubb J.R., Saiardi A. Developmental accumulation of inorganic polyphosphate affects germination and energetic metabolism in Dictyostelium discoideum. Proc. Natl. Acad. Sci. U. S. A. 2016;113(4):996–1001. doi: 10.1073/pnas.1519440113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetti A., Szijgyarto Z., Bosch D., Loss O., Azevedo C., Saiardi A. Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 2011;286(37):31966–31974. doi: 10.1074/jbc.M111.266320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losito O., Szijgyarto Z., Resnick A.C., Saiardi A. Inositol pyrophosphates and their unique metabolic complexity: analysis by gel electrophoresis. PLoS One. 2009;4(5) doi: 10.1371/journal.pone.0005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loss O., Azevedo C., Szijgyarto Z., Bosch D., Saiardi A. Preparation of quality inositol pyrophosphates. JoVE. 2011;55 doi: 10.3791/3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H.R., Huang Y.E., Chen J.C., Saiardi A., Iijima M., Ye K., Huang Y., Nagata E., Devreotes P., Snyder S.H. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell. 2003;114(5):559–572. doi: 10.1016/s0092-8674(03)00640-8. [DOI] [PubMed] [Google Scholar]
- Morrissey J.H., Choi S.H., Smith S.A. Polyphosphate: an ancient molecule that links platelets, coagulation, and inflammation. Blood. 2012;119(25):5972–5979. doi: 10.1182/blood-2012-03-306605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Haubner J., Chakraborty A. Targeting the inositol pyrophosphate biosynthetic enzymes in metabolic diseases. Molecules. 2020;25(6) doi: 10.3390/molecules25061403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulugu S., Bai W., Fridy P.C., Bastidas R.J., Otto J.C., Dollins D.E., Haystead T.A., Ribeiro A.A., York J.D. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science. 2007;316(5821):106–109. doi: 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- Muramoto T., Cannon D., Gierlinski M., Corrigan A., Barton G.J., Chubb J.R. Live imaging of nascent RNA dynamics reveals distinct types of transcriptional pulse regulation. Proc. Natl. Acad. Sci. U. S. A. 2012;109(19):7350–7355. doi: 10.1073/pnas.1117603109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onnebo S.M., Saiardi A. Inositol pyrophosphates modulate hydrogen peroxide signaling. Biochem. J. 2009;423(1):109–118. doi: 10.1042/BJ20090241. [DOI] [PubMed] [Google Scholar]
- Pascual-Ortiz M., Saiardi A., Walla E., Jakopec V., Kunzel N.A., Span I., Vangala A., Fleig U. Asp1 bifunctional activity modulates spindle function via controlling cellular inositol pyrophosphate levels in Schizosaccharomyces pombe. Mol. Cell Biol. 2018;38(9) doi: 10.1128/MCB.00047-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Ortiz M., Walla E., Fleig U., Saiardi A. The PPIP5K family member Asp1 controls inorganic polyphosphate metabolism in S. pombe. J. Fungi (Basel) 2021;7(8) doi: 10.3390/jof7080626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani F., Livermore T., Rose G., Chubb J.R., Gaspari M., Saiardi A. Analysis of Dictyostelium discoideum inositol pyrophosphate metabolism by gel electrophoresis. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0085533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D., Wilson M.S., Eisenbeis V.B., Harmel R.K., Riemer E., Haas T.M., Wittwer C., Jork N., Gu C., Shears S.B., Schaaf G., Kammerer B., Fiedler D., Saiardi A., Jessen H.J. Analysis of inositol phosphate metabolism by capillary electrophoresis electrospray ionization mass spectrometry. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-19928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall T.A., Gu C., Li X., Wang H., Shears S.B. A two-way switch for inositol pyrophosphate signaling: evolutionary history and biological significance of a unique, bifunctional kinase/phosphatase. Adv. Biol. Regul. 2020;75 doi: 10.1016/j.jbior.2019.100674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer E., Qiu D., Laha D., Harmel R.K., Gaugler P., Gaugler V., Frei M., Hajirezaei M.R., Laha N.P., Krusenbaum L., Schneider R., Saiardi A., Fiedler D., Jessen H.J., Schaaf G., Giehl R.F.H. ITPK1 is an InsP6/ADP phosphotransferase that controls phosphate signaling in Arabidopsis. Mol. Plant. 2021 doi: 10.1016/j.molp.2021.07.011. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A. How inositol pyrophosphates control cellular phosphate homeostasis? Adv. Biol. Regul. 2012;52(2):351–359. doi: 10.1016/j.jbior.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Saiardi A., Azevedo C., Desfougeres Y., Portela-Torres P., Wilson M.S.C. Microbial inositol polyphosphate metabolic pathway as drug development target. Adv. Biol. Regul. 2018;67:74–83. doi: 10.1016/j.jbior.2017.09.007. [DOI] [PubMed] [Google Scholar]
- Saiardi A., Caffrey J.J., Snyder S.H., Shears S.B. The inositol hexakisphosphate kinase family. Catalytic flexibility and function in yeast vacuole biogenesis. J. Biol. Chem. 2000;275(32):24686–24692. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- Saiardi A., Erdjument-Bromage H., Snowman A.M., Tempst P., Snyder S.H. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 1999;9(22):1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- Santarriaga S., Petersen A., Ndukwe K., Brandt A., Gerges N., Bruns Scaglione J., Scaglione K.M. The social amoeba Dictyostelium discoideum is highly resistant to polyglutamine aggregation. J. Biol. Chem. 2015;290(42):25571–25578. doi: 10.1074/jbc.M115.676247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D., Wang C., Shou H., Whelan J. Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 2012;586(4):289–295. doi: 10.1016/j.febslet.2012.01.036. [DOI] [PubMed] [Google Scholar]
- Solesio M.E., Xie L., McIntyre B., Ellenberger M., Mitaishvili E., Bhadra-Lobo S., Bettcher L.F., Bazil J.N., Raftery D., Jakob U., Pavlov E.V. Depletion of mitochondrial inorganic polyphosphate (polyP) in mammalian cells causes metabolic shift from oxidative phosphorylation to glycolysis. Biochem. J. 2021;478(8):1631–1646. doi: 10.1042/BCJ20200975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens L., Radenberg T., Thiel U., Vogel G., Khoo K.H., Dell A., Jackson T.R., Hawkins P.T., Mayr G.W. The detection, purification, structural characterization, and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s) J. Biol. Chem. 1993;268(6):4009–4015. [PubMed] [Google Scholar]
- Stephens L.R., Irvine R.F. Stepwise phosphorylation of myo-inositol leading to myo-inositol hexakisphosphate in Dictyostelium. Nature. 1990;346(6284):580–583. doi: 10.1038/346580a0. [DOI] [PubMed] [Google Scholar]
- Suess P.M., Gomer R.H. Extracellular polyphosphate inhibits proliferation in an autocrine negative feedback loop in Dictyostelium discoideum. J. Biol. Chem. 2016;291(38):20260–20269. doi: 10.1074/jbc.M116.737825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szijgyarto Z., Garedew A., Azevedo C., Saiardi A. Influence of inositol pyrophosphates on cellular energy dynamics. Science. 2011;334(6057):802–805. doi: 10.1126/science.1211908. [DOI] [PubMed] [Google Scholar]
- Thomas M.R., O'Shea E.K. An intracellular phosphate buffer filters transient fluctuations in extracellular phosphate levels. Proc. Natl. Acad. Sci. U. S. A. 2005;102(27):9565–9570. doi: 10.1073/pnas.0501122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui M.M., York J.D. Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv. Enzym. Regul. 2010;50(1):324–337. doi: 10.1016/j.advenzreg.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijken P., Bergsma J.C., Van Haastert P.J. Phospholipase-C-independent inositol 1,4,5-trisphosphate formation in Dictyostelium cells. Activation of a plasma-membrane-bound phosphatase by receptor-stimulated Ca2+ influx. Eur. J. Biochem. 1997;244(1):113–119. doi: 10.1111/j.1432-1033.1997.00113.x. [DOI] [PubMed] [Google Scholar]
- Van Dijken P., de Haas J.R., Craxton A., Erneux C., Shears S.B., Van Haastert P.J. A novel, phospholipase C-independent pathway of inositol 1,4,5-trisphosphate formation in Dictyostelium and rat liver. J. Biol. Chem. 1995;270(50):29724–29731. doi: 10.1074/jbc.270.50.29724. [DOI] [PubMed] [Google Scholar]
- Wang H., Falck J.R., Hall T.M., Shears S.B. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat. Chem. Biol. 2012;8(1):111–116. doi: 10.1038/nchembio.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild R., Gerasimaite R., Jung J.Y., Truffault V., Pavlovic I., Schmidt A., Saiardi A., Jessen H.J., Poirier Y., Hothorn M., Mayer A. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science. 2016;352(6288):986–990. doi: 10.1126/science.aad9858. [DOI] [PubMed] [Google Scholar]
- Williams R.S., Cheng L., Mudge A.W., Harwood A.J. A common mechanism of action for three mood-stabilizing drugs. Nature. 2002;417(6886):292–295. doi: 10.1038/417292a. [DOI] [PubMed] [Google Scholar]
- Williams R.S., Eames M., Ryves W.J., Viggars J., Harwood A.J. Loss of a prolyl oligopeptidase confers resistance to lithium by elevation of inositol (1,4,5) trisphosphate. EMBO J. 1999;18(10):2734–2745. doi: 10.1093/emboj/18.10.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.S., Bulley S.J., Pisani F., Irvine R.F., Saiardi A. A novel method for the purification of inositol phosphates from biological samples reveals that no phytate is present in human plasma or urine. Open Biol. 2015;5(3) doi: 10.1098/rsob.150014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Lau K., Puschmann R., Harmel R.K., Zhang Y., Pries V., Gaugler P., Broger L., Dutta A.K., Jessen H.J., Schaaf G., Fernie A.R., Hothorn L.A., Fiedler D., Hothorn M. Two bifunctional inositol pyrophosphate kinases/phosphatases control plant phosphate homeostasis. Elife. 2019;8 doi: 10.7554/eLife.43582. [DOI] [PMC free article] [PubMed] [Google Scholar]