Highlights
-
•
Non-specific chronic low back pain is one of the most disabling conditions in the world, yet the cause remains unknown.
-
•
Eventhough race is a social construct, Non-Hispanic Blacks (African Americans) with chronic low back experience poorer outcomes, greater pain severity, and more disability than Non-Hispanic Whites (White Americans).
-
•
Differentially methylated genes significantly enriched pathways of relevance to pain – Dopamine-DARPP32 Feedback in cAMP signaling, GABA Receptor Signaling, Opioid Signaling, and Endocannabinoid Neuronal Synapse.
-
•
More pathways were enriched in Non-Hispanic Blacks than Non-Hispanic Whites with chronic low back pain.
Abbreviations: NHB, Non-Hispanic Black; NHW, Non-Hispanic White; RRBS, Reduced representation bisulfite sequencing; cLBP, Chronic low back pain; PFC, Pain free control; DML, DIfferentially methylated loci; DNAm, DNA methylation
Keywords: Racial pain disparities, RRBS, Pain mechanisms, Chronic low back pain, Non-specific chronic low back pain, DNA methylation, Epigenetics
Abstract
Compared to Non-Hispanic Whites (NHWs), individuals who self-identify as Non-Hispanic Blacks (NHBs) in the United States experience more severe and disabling chronic low back pain (cLBP). We hypothesized that differences in DNA methylation (DNAm) play a role in racial disparities in cLBP.
Purpose
To determine the relationship between DNAm levels and racial group differences in adults with cLBP. Our study’s secondary purpose was to perform a race-stratified comparison of adults with cLBP and pain-free controls and identify functional genomic pathways enriched by annotated differentially methylated genes.
Patients and Methods
We recruited 49 NHBs and 49 NHWs (49 cLBP and 49 pain-free controls, PFCs), analyzed DNAm from whole blood using reduced representation bisulfite sequencing, and identified enriched genomic pathways.
Results
Among participants with cLBP, we identified 2873 differentially methylated loci (DML; methylation differences of at least 10% and p < 0.0001), many of which were annotated to genes of importance to pain pathology. These DMLs significantly enriched pathways to involved in nociception/pain processing (Dopamine-DARPP32 Feedback in cAMP signaling, GABA Receptor Signaling, Opioid Signaling) and neuronal differentiation (e.g., Calcium Signaling, Axon Guidance Signaling, and Endocannabinoid Neuronal Synapse). Our race stratified analyses of individuals with cLBP versus PFCs revealed 2356 DMLs in NHBs and 772 DMLs in NHWs with p < 0.0001 and > 10% methylation difference. Ingenuity Pathway Analysis revealed that many pathways of significance to pain such as Corticotropin Releasing Hormone Signaling, White Adipose Tissue Browning, and GABA Receptor Signaling pathways, were more significant in NHBs than NHWs.
Conclusion
Even though an individual’s self-identified race is a social construct, not a biological variable, racism associated with that classification affects virtually every aspect of life, including disease risk. DNAm induced alterations in stress signaling pathways may explain worse pain outcomes in NHBs.
Introduction
Chronic low back pain (cLBP) is a significant public health problem in the United States and one of the leading causes of disability worldwide. (Clark and Horton, 2018, GBD, 2017) Despite its high prevalence, the disease process of cLBP is poorly understood. (Hartvigsen et al., 2018) An individual’s race is one predictor of pain sensitivity, severity, and interference with daily activities. (Aroke et al., 2020a, Meints et al., 2018) Compared to non-Hispanic Whites (NHW), Non-Hispanic Blacks (NHB) experience more chronic pain, report more pain-related chronic medical conditions, more chronic pain-related disability, and have a more inferior pain-associated quality of life. (Kim et al., 2017, Edwards et al., 2001) The reasons for these racial differences in cLBP are poorly understood. This may be related to the fact that race is a socially constructed categorization of people, different from biological traits such as ancestry, and not an inherent disease risk. (Aroke et al., 2019)
Genetic, (Battie et al., 1976, Battie et al., 2007) environmental, (Battie et al., 1976, Suri et al., 2017) and psychosocial factors (Moix et al., 2011, Pinheiro et al., 2016, Fernandez et al., 2017) have been implicated in the pathogenesis of cLBP. Studies have shown that psychosocial factors such as depressive symptoms, (Fernandez et al., 2017) catastrophizing, (Moix et al., 2011) and lower socioeconomic status (Hill and Fritz, 2011, Katz, 2006) predict worse cLBP outcomes. NHBs report more severe depressive symptoms and endorse more pain catastrophizing than NHWs. Also, NHBs are more likely to report lower socioeconomic status and lower social support than NHWs. NHBs experience more poverty, residential segregation, aggressive policing, interpersonal racial harassment, and microaggressions. (Sue et al., 2007, Williams, 2021) These psychosocial factors may contribute to racial differences in cLBP, but some studies have suggested that racial differences in pain persist after controlling for psychosocial factors. (Fuentes et al., 2007) NHBs are more likely to experience adverse environmental factors such as violence, racial discrimination, injustice, and adverse childhood experiences. (Nelson et al., 2018, Sack and Murphey, 2018, Clark et al., 1999) Differences in lived experience may thus contribute to racial differences in cLBP. Still, evidence of the mechanism by which psychosocial factors and adverse life exposures contribute to race differences in cLBP remains indeterminate.
Overwhelming evidence indicates that modifications in the human genome (epigenomic modifications) regulate what genes turn-on or turn-off leading to tissue differentiation. (Huang et al., 2014) Thus, epigenetic changes are critical in understanding the mechanisms of human physiologic and pathophysiologic states. (Kanherkar et al., 2014, Syed and Nemeroff, 2017) DNA methylation (DNAm) is a stable, heritable, and well-studied epigenetic modification. (Aroke et al., 2019, Dupont et al., 2009) Generally, DNAm is tissue-specific, however, in rat models with chronic pain, Massart and colleagues found a strong correlation between DNAm in the prefrontal cortex and peripheral blood T-cells; peripheral T-cell methylation predicted chronic pain with 80% accuracy. (Massart et al., 2016) Other researchers have also utilized peripheral blood methylation as a less-invasive sample to study methylation patterns in chronic pain conditions, including fibromyalgia, persistent post-mastectomy pain, generalized musculoskeletal pain, and cLBP. (Tajerian et al., 2011, Kawi et al., 2018, Sukenaga et al., 2016, Burri et al., 2016, Stephens et al., 2017)
We previously suggested that a lifetime of adversity, chronic stress, poor coping, and racial discrimination may induce epigenetic modifications, affecting adaptation and resulting in racial differences in adulthood disorders such as chronic pain. (Aroke et al., 2019) Emerging evidence suggests that DNAm changes that occur throughout the lifetime may play an essential role in the pathology of cLBP. (Gerra et al., 2017) Tajerian and colleagues have previously reported cLBP correlated with DNAm levels in secreted protein acidic and cysteine-rich (SPARC) gene. (Tajerian et al., 2011) Using reduced representation bisulfite sequencing (RRBS) and functional genomic enrichment analysis, we found several differentially methylated regions and important biologic pathways to be differentially enriched in individuals with cLBP compared to pain-free controls. The differentially methylated regions contained important genes such as CELSR1, NAV1, MINK1, and KIF11, which have previously been associated with pain, cell-cell adhesion/migration, and neural differentiation. (Aroke et al., 2020b) However, little is known about whether DNAm and functional genomic pathways vary as functions of an individual’s self-identified race within the context of cLBP.
To gain insight into the role of epigenetic changes in racial differences in cLBP, we determined the relationship between DNAm levels and racial group differences in adults with cLBP. Specifically, we investigated how DNAm levels of NHBs with cLBP compare with those of NHWs with cLBP. Our study’s secondary purpose was to perform a race-stratified comparison of adults with cLBP and pain-free controls. Furthermore, we performed functional genomic pathway analyses to identify biologically relevant pathways enriched by the differentially methylated genes.
Methods
Participants
Participants were enrolled as part of an ongoing study: Examining Racial And SocioEconomic Disparities in cLBP (R01MD010441). All participants were recruited through flyers posted at the Pain Treatment Clinic – University of Alabama at Birmingham (UAB) Department of Anesthesiology and the surrounding UAB community. Details about Enrollment criteria have been described in previous publications. (Aroke et al., 2020b, Penn et al., 2020) Briefly, we enrolled 25 NHWs and 25 NHBs with non-specific cLBP, ages of 19 and 85 years, from June 2018 to September 2019. To qualify as cLBP, participants must have experienced pain for at least half the days, for three or more consecutive months. Illnesses that could confound the interpretation of results were grounds for exclusion, including other pain conditions, malignancy, trauma, ankylosis for spondylitis, infection, poorly controlled diabetes, chronic inflammatory diseases (e.g., systemic lupus erythematosus, fibromyalgia, rheumatoid arthritis), severe psychiatric disorders requiring hospitalization in the last 12 months, and neurological disorders (e.g., epilepsy, multiple sclerosis, Parkinson’s). The diagnosis was confirmed through medical records. The joint clinical practice guidelines for the American College of Physicians and the American Pain Society were used to identify non-specific cLBP in participants. (Chou et al., 2016)
For comparison, we also recruited and enrolled 50 (25 NHWs and 25 NHBs) pain-free controls (PFCs). Inclusion criteria for our pain-free controls included men and women 19 to 85 years of age, without a recent history of pain, not pregnant or breastfeeding, and able to write and read English. PFC participants were excluded using the same cLBP criteria. Participants with cLBP self-reported data about pain severity and pain interference were assessed using the Brief Pain Inventory (BPI) – Short Form, a widely used questionnaire to evaluate pain and its impact on functioning. (Cleeland and Ryan, 1994) The University of Alabama at Birmingham Institutional Review Board (IRB) for Human Subject Research approved this study. All participants had the opportunity to have any questions answered and provided written informed consent.
Measurement of DNA methylation
Peripheral whole blood samples were collected into ethylene-diamine-tetra-acetic acid (EDTA) anticoagulant tubes. Genomic DNA was isolated using the Gentra Puregene DNA Purification Protocol (Qiagen, Valencia, CA, USA) and stored at −80 °C. The NanoDrop 2000 was used to quantify the extracted DNA. Determination of purity via spectrometry indicated an absorbance between 260 and 280 nm ratio of >1.8 for all samples.
The RRBS libraries were generated at the Heflin Center for Genomic Sciences at UAB, using the Ovation RRBS Methyl-Seq System (NuGEN, Tecan Genomics, Redwood City, CA, USA), according to the manufacturer’s instructions. High-quality MspI digested genomic DNA fragments were ligated to adaptors and bisulfite converted (using the Qiagen Epitect kit for bisulfite conversion) strands sequenced on Illumina NextSq 500 platform to generate raw RRBS reads. The quantity and quality of the libraries were assessed using a Qubit fluorometer.
Data processing and analysis
Demographic and clinical data were summarized and compared using means (with standard deviation) and frequency (with percentage) depending on the level of measurement. Quality control (QC) of read was performed using FastQC. (Fast, 2019) We used Trim Galore (Trim, 2019) to trim low quality reads, and remove adapters before aligning and mapping to the human reference genome (hg19) using Bismark. (Krueger and Andrews, 2011) DNA methylation level of each locus was further extracted based on the aligned reads for the downstream statistical analysis.
To test the association of DNAm with cLBP status within each racial group, we performed differential methylation analyses comparing cLBP versus PFC within NHBs group and NHWs, respectively. Specifically, we employed linear regression models followed by the empirical Bayes moderated t-statistics test, which were implemented in the R limma package. (Ritchie et al., 2015) In this model, the methylation level of a CpG loci was the outcome variable, the pain status (cLBP or PFC) was the predictor variable, adjusting for age and sex as covariates. Differentially methylated loci (DML) were defined as methylation differences of at least 10% and p < 0.0001. Hypomethylated DMLs were those with significantly lower percent methylation in cLBP than in PFCs; and hypermethylated DMLs with significantly higher percent methylation in cLBP than in PFCs. To correct for multiple testing, we used the Benjamini-Hochberg method to determine q-values, (Wang et al., 2011) which is the equivalence of the false discovery rate (FDR). (Akalin et al., 2012) We annotated the location of DMLs in relation to genomic features (intron, exons, intergenic regions, and promoters regions) or CpG features (CpG island, CpG shores). CpG island features included CG fractions > 0.5, CG length of at least 200 bp, and an observed to expected CpG ratio of > 0.6. CpG shores were identified as positions adjacent to CpG islands with a length of at least 2000 bp. All these annotation analyses were performed using R methylKit package. (Akalin et al., 2012)
To examine the race disparity in relation to methylation level of subjects with cLBP, we performed differential methylation analysis comparing NHWs and NHBs within cLBP participants similarly using linear regression via R limma package. To be specific, the race (NHW or NHB) was the predictor variable, the methylation level of a CpG site was the outcome, adjusting for age and sex as covariates. To correct for multiple testing, we used the Benjamini-Hochberg method to obtain q-values. Hypomethylated DMLs were defined as those with significantly lower percent methylation NHBs and hypermethylated DMLs with significantly higher percent methylation in NHBs. The putative DMLs were annotated to genomic features (intron, exons, intergenic regions and promoter regions) or CpG features (CpG island, CpG shores).
To examine whether the association between pain and methylation were moderated by race, we further evaluated the pain-race interaction. We adopted linear regression models via R limma package. In this model, a methylation level for each CpG site was the outcome variable, race, pain status, and pain by race interaction were predictors, and age and gender were covariates. A significant pain by race interaction would imply the pain effect on methylation level could be different by race. As no DMLs from the interaction analysis were found to be statistically significant after correcting for multiple testing, we defined DMLs as nominal p < 0.0001.
To identify potential pathways driving racial disparities in pain, we used functional enrichment analyses to determine if any biological processes were overrepresented in genes based on DMLs. Following the removal of duplicates, genes annotated to DMLs showing significant differences (p < 0.0001) were imported into Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Redwood City, CA), and genomic pathways over-represented by these genes determined. The analyses were carried out for (i) race stratified analysis in the association of DNAm with cLBP status; (ii) race disparity analysis in relation to methylation level of subjects with cLBP; and (iii) pain-race interaction analysis. Over-represented pathways in IPA with p-values < 0.01 (-log10p-value = 2) were considered statistically significant.
Results
Characteristics of study participants
Of the 100 participants recruited, two participants were excluded from analysis because they selected multiple racial backgrounds. Our final sample included 49 NHBs and 49 NHWs. The participants’ mean age was 44.6 ± 12.8 years; there were slightly more women (53.1%) than men (46.9%). There was no statistically significant racial group difference in terms of age or gender of participants. Table 1 describes demographics and a description of pain outcomes. On average, NHBs reported greater pain severity and interference compared to NHWs with cLBP. The difference in pain severity was statistically significant (p < 0.001).
Table 1.
Characteristics of study participants.
| Non-Hispanic Blacks | Non-Hispanic Whites | p-values | |
|---|---|---|---|
| Chronic Low Back Pain Cases | |||
| (n = 25) | (n = 24) | ||
| Age, mean (SD) | 43.5 (10.6) | 45.8 (14.9) | 0.541 |
| Sex, N (%) | |||
| Men | 9 (36) | 12 (50) | 0.321 |
| Women | 16 (64) | 12 (50) | |
| BPI, mean (SD) | |||
| Severity | 5.9 (2.2) | 3.4 (1.9) | <0.001 |
| Interference | 4.4 (2.8) | 2.8 (2.0) | 0.077 |
| Pain-Free Controls | |||
| (n = 24) | (n = 25) | ||
| Age, mean (SD) | 40.7 (16.5) | 39.3 (12.6) | 0.749 |
| Sex, N (%) | |||
| Men | 13 (54.2) | 12 (48) | 0.778 |
| Women | 11 (45.8) | 13 (52) | |
| BPI, mean (SD) | |||
| Severity | 0.1 (0.5) | 0.1 (0.1) | 0.891 |
| Interference | 0.1 (0.5) | 0 | 0.19 |
Notes: SD = standard deviation, BPI = brief pain inventory.
Quality of the samples and reproducibility of the data
Each of the 103 data files comprised, on average, approximately 45 million trimmed sequence reads, with each file about 7 GB in size. Among these 103 samples, there were 5 technical replicates and 98 unique biological samples. Each file was mapped to the reference human genome (hg19) using bismark to build bisulfite genome libraries. The bisulfite conversion rate was >99% for all study samples. The mapping efficiency ranged from 68% to 75%, consistent with other research using bismark to map bisulfite sequence data. DNAm profiles of the five randomly selected biological replicates matched perfectly. As depicted in Supplemental Fig. 1, a pairwise comparison of the technical samples revealed a correlation coefficient of>0.9 for all comparisons, which indicates high levels of reproducibility. (Bock, 2012)
Race stratified DNA methylation of adults with and without cLBP
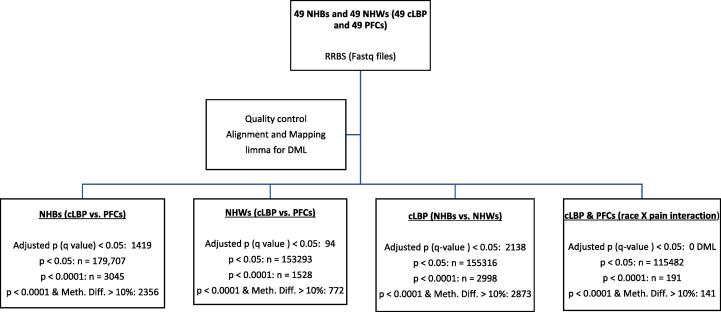
We have previously identified DMLs and differentially enriched biological pathways in a mixed racial group of adults with cLBP versus PFCs. (Aroke et al., 2020b) To further explicate the role of DNAm in racial differences in cLBP, we compared DMLs and over-represented pathways within each racial group: NHBs (cLBP versus PFC) and NHWs (cLBP and PFC). Fig. 1 depicts an overview of the genome-wide differential methylation analysis.
Fig. 1.
Overview of the genome wide differential methylation analysis. Notes: NHBs = Non-Hispanic Blacks; NHWs = Non-Hispanic Whites; cLBP = chronic low back pain; PFCs = pain-free controls; Meth.Diff = absolute methylation difference between two groups; DML = differentially methylated loci.
Differentially methylated loci between cLBP- and PFCs in NHBs
After controlling for age and sex, differential methylation analysis revealed 3045 DMLs exhibiting significant differences at p < 0.0001, and 1419 at q < 0.05 in cLBP vs. PFCs NHBs. Of the 3045 DMLs, 2851 were hypomethylated and 194 were hypermethylated. Also, 2356 DMLs had an absolute methylation difference of >10 percent and p < 0.0001 in cLBP vs. PFC NHBs. Table 2 summarizes the top 20 DMLs that significantly predict cLBP in NHBs sorted by the p-values. Detailed examination of the DMLs revealed that majority mapped to intronic region (33%), followed by intergenic region (31%), promoter regions (25%), and exonic regions (10%) (Supplemental Fig. 2A). Also, 29% of the DMLs belonged to CpG island, 18% of the DMLs belonged to CpG shores (Supplemental Fig. 2B).
Table 2.
Top 20 Differentially Methylated Loci Between cLBP Vs. PFC in NHBs.
| Chr | Position | Beta | SE | 95% CI |
P-value | q-value | Genes | Gene name/description | Genomic Features | |
|---|---|---|---|---|---|---|---|---|---|---|
| LL | UL | |||||||||
| 3 | 186,629,673 | −0.19 | 0.02 | −0.23 | −0.14 | 6.39E-11 | 1.66E-04 | intergenic | ||
| 2 | 121,106,655 | −0.21 | 0.03 | −0.26 | −0.16 | 1.96E-10 | 2.48E-04 | INHBB | Inhibin Subunit Beta B | intergenic |
| 6 | 111,591,940 | −0.16 | 0.02 | −0.21 | −0.12 | 2.86E-10 | 2.48E-04 | KIAA1919 | Sodium-Dependent Glucose Transporter | exons |
| 15 | 102,255,559 | −0.11 | 0.01 | −0.14 | −0.08 | 3.94E-10 | 2.56E-04 | TARSL2 | Threonyl-tRNA Synthetase-Like 2 | intergenic |
| 12 | 20,922,311 | 0.18 | 0.02 | 0.13 | 0.22 | 7.14E-10 | 3.71E-04 | intergenic | ||
| 10 | 9,595,069 | −0.19 | 0.02 | −0.24 | −0.14 | 8.59E-10 | 3.72E-04 | intergenic | ||
| 8 | 144,408,266 | 0.35 | 0.05 | 0.26 | 0.44 | 1.24E-09 | 4.14E-04 | TOP1MT | DNA Topoisomerase I Mitochondrial | introns |
| 2 | 242,428,016 | −0.16 | 0.02 | −0.20 | −0.12 | 1.28E-09 | 4.14E-04 | FARP2; STK25 | Serine/Threonine Kinase 25 | introns |
| 21 | 10,597,920 | −0.16 | 0.02 | −0.21 | −0.12 | 2.37E-09 | 6.84E-04 | intergenic | ||
| 1 | 10,670,667 | −0.10 | 0.01 | −0.13 | −0.07 | 2.92E-09 | 7.59E-04 | PEX14 | Peroxisomal Biogenesis Factor 14 | introns |
| 19 | 40,177,900 | −0.17 | 0.02 | −0.22 | −0.12 | 6.87E-09 | 1.62E-03 | LGALS17A | Galectin 14 pseudogene | intergenic |
| 11 | 8,285,031 | −0.24 | 0.03 | −0.31 | −0.17 | 9.18E-09 | 1.89E-03 | LMO1 | LIM Domain only 1; Rhombotin 1 | intergenic |
| 9 | 100,683,907 | −0.27 | 0.04 | −0.35 | −0.19 | 9.46E-09 | 1.89E-03 | C9orf156 | Chromosome 9 open reading frame 156 | intergenic |
| 10 | 35,465,538 | −0.10 | 0.02 | −0.14 | −0.07 | 1.08E-08 | 2.01E-03 | CREM | cAMP Responsive Element Modulator | introns |
| 14 | 24,951,201 | 0.35 | 0.05 | 0.25 | 0.46 | 1.70E-08 | 2.82E-03 | intergenic | ||
| 12 | 28,343,050 | −0.31 | 0.05 | −0.41 | −0.22 | 1.84E-08 | 2.82E-03 | CCDC91 | Coiled-Coil Domain Containing 91 | promoters |
| 17 | 8,926,988 | −0.26 | 0.04 | −0.33 | −0.18 | 1.85E-08 | 2.82E-03 | NTN1 | Netrin 1 | introns |
| 5 | 176,166,533 | −0.25 | 0.04 | −0.32 | −0.17 | 2.24E-08 | 2.99E-03 | RP11-375B1.2 | Long noncoding RNA | intergenic |
| 15 | 86,118,741 | −0.17 | 0.02 | −0.22 | −0.12 | 2.40E-08 | 2.99E-03 | AKAP13 | A-Kinase Anchor Protein 13 | intergenic |
| 19 | 10,297,587 | 0.25 | 0.04 | 0.17 | 0.32 | 2.44E-08 | 2.99E-03 | DNMT1 | DNA Methyltransferase 1 | introns |
Notes: Chr = chromosome, CI = confidence interval; LL = lower limit of the confidence interval; UL = upper limit of the confidence interval; SE = standard error; cLBP = chronic low back pain; PFC = pain-free control; NHBs = non-Hispanic Blacks.
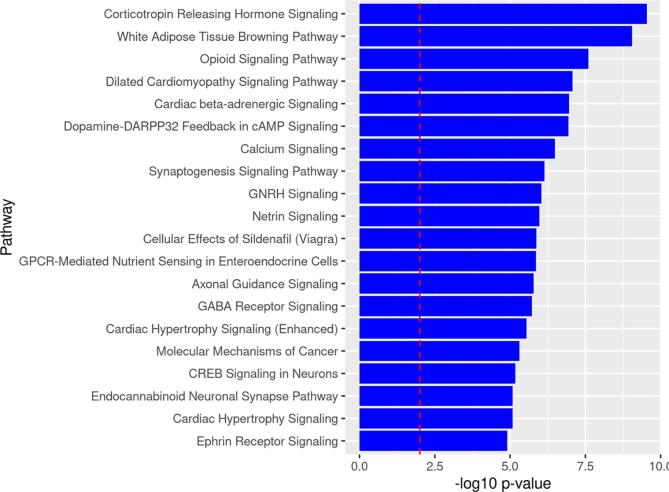
Pathway analysis of DMLs in NHBs
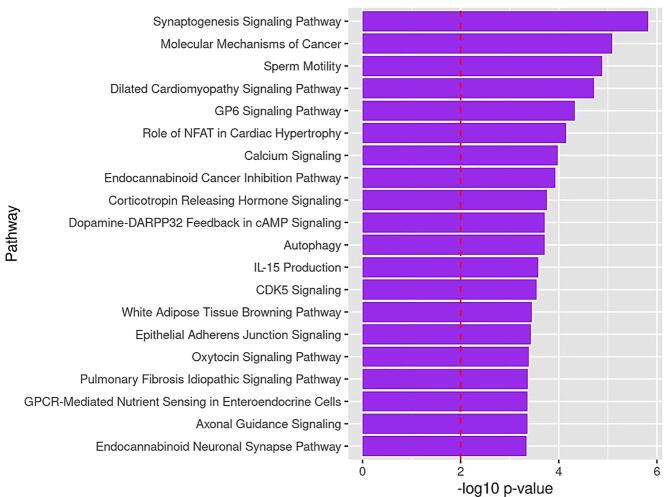
To infer the functional role of the identified DMLs, we performed a pathway enrichment analysis of genes annotated to DMLs with p-values <0.0001. From the list of 2110 differentially methylated genes, IPA mapped 110 canonical pathways (p < 0.01) over-represented in the NHB (cLBP versus PFC) data set. Many of the overrepresented pathways are of relevance to pain (e.g., opioid signaling), chronic stress (e.g., corticotropin releasing hormone signaling, white adipose tissue browning, and dopamine-DARPP32 Feedback in cAMP signaling) and neuronal differentiation (e.g., synaptogenesis signaling, calcium signaling, and CREB signaling in neurons). Fig. 2 depicts the top 20 over-represented canonical pathways from differentially methylated genes between NHBs with CLBP and NHB PFCs. Supplemental Table 1 summarizes the differentially methylated genes associated with the over-represented pathways.
Fig. 2.
Top 20 Over-represented pathways by identified from genes containing DMLs between cLBP and PFCs in NHBs. Note: The dotted line depicts the statistically significant level (p < 0.01).
Differentially methylated loci between cLBP- and PFCs in NHWs
After controlling for age and sex, we identified 1528 (1451hypomethylatedand77hypermethylated) DMLs with p values < 0.0001, and 94 DMLs with q values < 0.05 in models predicting cLBP in NHWs. Of the 1528 DMLs, 772 had a methylation difference of at least 10 percent in NHWs with cLBP compared to NHW PFCs. Table 3 summarizes the annotated genes of the top 20 DMLs based on smallest p-values. Detailed examination of the DMLs revealed that majority mapped to intronic (32%), followed by intergenic regions (30%), promoter regions (28%), and exon regions (10%) (Supplemental Fig. 3A). In addition, 33% of the DMLs belonged to CpG island, 17% of the DMLs belonged to CpG shores (Supplemental Fig. 3B).
Table 3.
Top 20 Differentially Methylated Loci Between cLBP vs. PFCs in NHWs.
| Chr | Position | Beta | SE | 95% CI |
P-value | q-value | Genes | Gene Name/Description | Genomic Features | |
|---|---|---|---|---|---|---|---|---|---|---|
| LL | UL | |||||||||
| 15 | 102,255,559 | −0.13 | 0.02 | −0.17 | −0.10 | 2.24E-10 | 5.81E-04 | TARSL2 | Threonyl-tRNA Synthetase-Like 2 | intergenic |
| 1 | 148,556,709 | −0.10 | 0.01 | −0.13 | −0.07 | 5.02E-09 | 6.52E-03 | NBPF15 | Neuroblastoma breakpoint family, member 15 | intergenic |
| 17 | 47,633,779 | −0.14 | 0.02 | −0.18 | −0.10 | 3.06E-08 | 1.87E-02 | RP5-1029 K10.2 | Long non-coding RNA | intergenic |
| 12 | 40,499,027 | −0.08 | 0.01 | −0.11 | −0.06 | 3.48E-08 | 1.87E-02 | SLC2A13 | Solute Carrier Family 2 Member 13 | promoters |
| 21 | 46,975,705 | −0.10 | 0.02 | −0.13 | −0.07 | 3.83E-08 | 1.87E-02 | – | introns | |
| 11 | 2,009,641 | −0.22 | 0.03 | −0.29 | −0.15 | 4.31E-08 | 1.87E-02 | MRPL23 | Mitochondrial Ribosomal Protein L23 | introns |
| 10 | 119,304,081 | −0.24 | 0.04 | −0.32 | −0.16 | 6.71E-08 | 2.25E-02 | EMX2 | Empty Spiracles Homeobox 2 | promoters |
| 17 | 40,718,967 | −0.21 | 0.03 | −0.27 | −0.14 | 7.70E-08 | 2.25E-02 | COASY; MLX | Coenzyme A Synthase; Max-Like Protein X | promoters |
| 20 | 8,942,669 | −0.07 | 0.01 | −0.10 | −0.05 | 7.81E-08 | 2.25E-02 | PLCB1 | Phospholipase C Beta 1 | intergenic |
| 19 | 36,736,335 | −0.09 | 0.01 | −0.12 | −0.06 | 1.02E-07 | 2.66E-02 | ZNF565 | Zinc Finger Protein 565 | intergenic |
| 16 | 1,382,020 | −0.14 | 0.02 | −0.18 | −0.09 | 1.19E-07 | 2.79E-02 | UBE2I; BAIAP3 | Ubiquitin Conjugating Enzyme E2 I | intergenic |
| 1 | 222,638,844 | −0.06 | 0.01 | −0.09 | −0.04 | 1.29E-07 | 2.79E-02 | CICP13 | capicua transcriptional repressor pseudogene | intergenic |
| 1 | 167,424,858 | −0.15 | 0.02 | −0.20 | −0.10 | 1.53E-07 | 2.85E-02 | CD247 | T-cell receptor coding | intergenic |
| 2 | 44,223,044 | −0.07 | 0.01 | −0.10 | −0.05 | 1.64E-07 | 2.85E-02 | LRPPRC | Leucine Rich Pentatricopeptide Repeat Containing Protein | intergenic |
| 2 | 111,877,924 | −0.10 | 0.02 | −0.13 | −0.07 | 1.67E-07 | 2.85E-02 | ACOXL; BCL2L11 | Acyl-CoA Oxidase Like | intergenic |
| 18 | 40,656,876 | −0.14 | 0.02 | −0.19 | −0.09 | 1.97E-07 | 2.85E-02 | RIT2 | Ras Like Without CAAX 2 | introns |
| 16 | 88,497,586 | −0.21 | 0.03 | −0.27 | −0.14 | 2.03E-07 | 2.85E-02 | ZNF469 | Zing-finger protein 469 | intergenic |
| 15 | 96,885,264 | −0.11 | 0.02 | −0.15 | −0.07 | 2.10E-07 | 2.85E-02 | NR2F2 | Nuclear Receptor Subfamily 2 Group F Member 2 | intergenic |
| 13 | 107,186,569 | −0.18 | 0.03 | −0.24 | −0.12 | 2.30E-07 | 2.85E-02 | EFNB2 | Ephrin B2 | intergenic |
| 18 | 19,746,392 | −0.13 | 0.02 | −0.18 | −0.09 | 2.32E-07 | 2.85E-02 | GATA6-AS1; GATA6 | GATA Binding Protein 6 | intergenic |
Notes: Chr = chromosome, CI = confidence interval; LL = lower limit of the confidence interval; UL = upper limit of the confidence interval; SE = standard error; cLBP = chronic low back pain; NHW = non-Hispanic White.
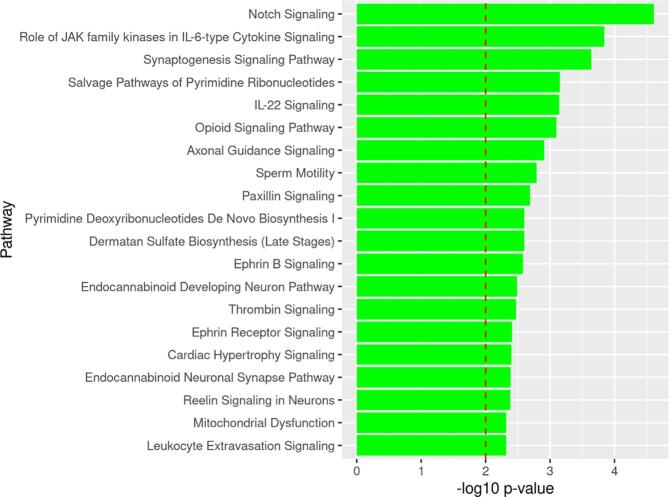
Pathway analysis of DMLs in NHWs
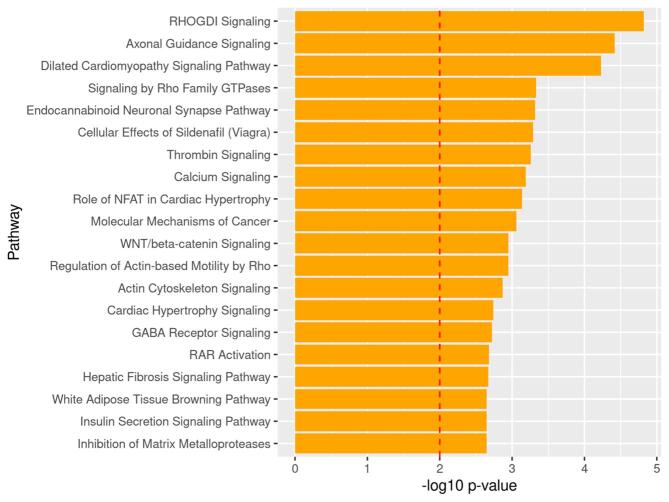
From the list of 1330 differentially methylated genes annotated to DMLs with p < 0.0001, IPA mapped over-represented pathways. IPA revealed 31 canonical pathways over-represented (p < 0.01) from the list of differentially methylated genes. Top over-represented pathways were related to inflammation and immunity (notch signaling, role of JAK family kinases in IL-6-type Cytokine signaling and IL-22 signaling), and neuronal differentiation (synaptogenesis signaling and axon guidance signaling) pathways (Fig. 3). Supplemental Table 2 summarizes the list of over-represented pathways from differentially methylated genes between NHWs with cLBP and PFCs.
Fig. 3.
Top 20 Over-represented pathways by identified from genes containing DMLs between cLBP and PFCs in NHWs. Note: The dotted line depicts the statistically significant level (p < 0.01).
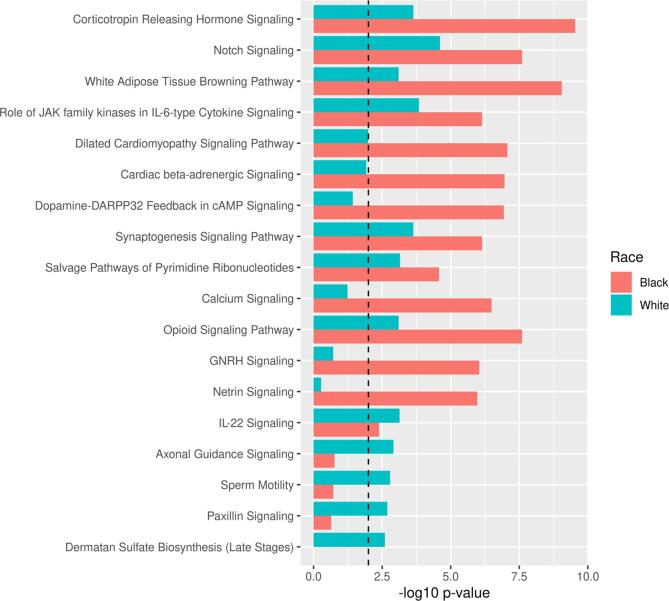
Comparing over-represented pathways in race stratified analysis
As previously mentioned, IPA revealed 110 and 31 overrepresented pathways among genes containing significant DMLs between cLBP cases and PFCs in NHBs and NHWs, respectively. For comparison, we classified the top 20 over-represented pathways according to calculated p-values. As depicted in Fig. 4, most of the calculated p-values were more significant in NHBs than NHW data set, suggesting that these pathways have a larger effect size in NHBs than NHWs.
Fig. 4.
Comparison of top the 20 over-represented pathways in race stratified analysis. Note: The dotted line depicts the statistically significant level (p < 0.01).
Differentially methylated loci in associated with cLBP in NHBs versus NHWs
To investigate differential methylation between NHBs and NHWs related to cLBP, we used linear models to predict DML using race, controlling for sex, and age. At p < 0.0001 level, we identified 2998 (1458hypomethylatedand1540hypermethylated) DMLs. Of these, 2873 DMLs had at least a 10 percent methylation difference between NHBs and NHWs with cLBP. After controlling for multiple testing, 2138 DMLs remained statistically significant (q < 0.05). The top 20 DMLs identified according to the order of p-values from low to high are shown in Table 4. The data indicated that majority of the mapped to intronic region (38%), followed by intergenic regions (34%), promoter regions (18%) and exon regions (10%) (Supplemental Fig. 4A). 61% of the DMLs belonged to CpG island, 22% of the DMLs belonged to CpG shores (Supplemental Fig. 4B).
Table 4.
Top 20 Differentially Methylated Loci in Associated with cLBP in NHBs versus NHWs.
| Chr | Position | Beta | SE | 95% CI |
P-value | q-value | Genes | Gene Name/Description | Genomic Features | |
|---|---|---|---|---|---|---|---|---|---|---|
| LL | UL | |||||||||
| 1 | 1,419,278 | 0.76 | 0.05 | 0.66 | 0.87 | 1.20E-18 | 3.12E-12 | ATAD3B | ATPase Family AAA Domain Containing 3B | intergenic |
| 17 | 36,590,735 | 0.71 | 0.05 | 0.60 | 0.81 | 8.82E-18 | 1.14E-11 | ARHGAP23 | Rho GTPase Activating Protein 23 | intergenic |
| 18 | 11,550,683 | 0.69 | 0.06 | 0.58 | 0.81 | 3.69E-16 | 3.19E-10 | RP11-712C7.2 | Long-noncoding RNA | intergenic |
| 5 | 176,190,238 | −0.67 | 0.06 | −0.78 | −0.56 | 7.66E-16 | 4.97E-10 | intergenic | ||
| 1 | 53,970,911 | 0.72 | 0.06 | 0.60 | 0.85 | 1.68E-15 | 8.74E-10 | GLIS1 | GLIS Family Zinc Finger 1 | intergenic |
| 19 | 4,028,691 | 0.70 | 0.06 | 0.57 | 0.83 | 1.93E-14 | 8.37E-09 | PIAS4 | Protein Inhibitor Of Activated STAT 4 | intergenic |
| 6 | 95,534,202 | 0.73 | 0.07 | 0.59 | 0.86 | 3.08E-14 | 1.14E-08 | intergenic | ||
| 1 | 160,347,421 | 0.68 | 0.06 | 0.55 | 0.80 | 4.60E-14 | 1.39E-08 | NHLH1 | Nescient Helix-Loop-Helix 1 | intergenic |
| 12 | 50,862,234 | −0.73 | 0.07 | −0.87 | −0.59 | 4.80E-14 | 1.39E-08 | LARP4 | La Ribonucleoprotein 4 | intergenic |
| 20 | 62,149,177 | 0.64 | 0.06 | 0.52 | 0.76 | 6.01E-14 | 1.56E-08 | PPDPF | Pancreatic Progenitor Cell Differentiation And Proliferation Factor | intergenic |
| 18 | 71,841,430 | −0.61 | 0.06 | −0.73 | −0.49 | 1.47E-13 | 3.48E-08 | intergenic | ||
| 10 | 127,291,526 | 0.61 | 0.06 | 0.49 | 0.73 | 2.82E-13 | 6.10E-08 | TEX36 | Testis Expressed 36 | introns |
| 4 | 177,637,599 | −0.54 | 0.06 | −0.66 | −0.43 | 6.64E-13 | 1.33E-07 | VEGFC | Vascular Endothelial Growth Factor C | intergenic |
| 10 | 128,944,081 | −0.64 | 0.07 | −0.77 | −0.51 | 7.26E-13 | 1.35E-07 | DOCK1 | Dedicator Of Cytokinesis 1 | introns |
| 15 | 74,592,862 | −0.60 | 0.06 | −0.73 | −0.48 | 1.10E-12 | 1.82E-07 | CCDC33 | Coiled-Coil Domain Containing 33 | intergenic |
| 2 | 195,597,960 | 0.38 | 0.04 | 0.30 | 0.46 | 1.12E-12 | 1.82E-07 | AC006196.1 | Long non-coding RNA | intergenic |
| 19 | 51,801,429 | 0.67 | 0.07 | 0.53 | 0.81 | 1.51E-12 | 2.30E-07 | intergenic | ||
| 4 | 3,578,876 | 0.36 | 0.04 | 0.28 | 0.44 | 3.67E-12 | 5.28E-07 | LINC00955 | Long non-coding RNA | promoters |
| 1 | 11,395,415 | 0.67 | 0.07 | 0.52 | 0.81 | 3.86E-12 | 5.28E-07 | intergenic | ||
| 1 | 16,455,052 | −0.64 | 0.07 | −0.78 | −0.50 | 5.51E-12 | 7.16E-07 | EPHA2 | Ephrin type A receptor 2 | intergenic |
Notes: Chr = chromosome, CI = confidence interval; LL = lower limit of the confidence interval; UL = upper limit of the confidence interval; SE = standard error; cLBP = chronic low back pain; NHW = non-Hispanic White; NHBs = non-Hispanic Blacks.
Pathway analysis of DMLs between NHBs and NHWs with cLBP
To investigate the potential physiological processes that influence racial differences in epigenetic changes associated with cLBP, 1695 genes containing significant DMLs with p < 0.0001 were further analyzed using the IPA software. At p-value < 0.01, IPA revealed 79 overrepresented pathways from the list of differentially methylated genes. Many of the top overrepresented pathways are of relevance to nociception/pain processing (corticotropin releasing hormone signaling, dopamine-DARPP32 feedback in cAMP signaling, GABA receptor signaling, opioid signaling) and neuronal differentiation (e.g., synaptogenesis signaling pathway, calcium signaling, axon guidance signaling, and endocannabinoid neuronal synapse). Fig. 5 shows the list of top 20 over-represented pathways from genes containing DMLs between NHB and NHW individuals with cLBP. The differentially methylated genes overrepresented in the pathways are shown in Supplemental Table 3.
Fig. 5.
Top 20 over-represented pathways by genes containing DMLs between NHBs and NHWs with cLBP. Note: The dotted line depicts the statistically significant level (p < 0.01).
Differentially methylated loci by race-pain interaction
Analysis of the race by pain interaction effects identified 191 statistically significant DMLs at p < 0.0001. These DMLs annotated to important regulatory genes and those of relevance to pain pathology, including ELL2, CTD-3179p9.2, ZNF808, SYN3, SOX7, and LRFN5. Table 5 summarizes the top 20 DMLs by race-pain interaction. However, at 5% false discovery rate (FDR) there is no DML, implying no interaction effect between race and pain. Future large studies are needed to replicate these results.
Table 5.
Top 20 Differentially Methylated Loci Based on Race-Pain Interaction Effects.
| Chr | Position | Beta | SE | 95% CI |
P-values | q-value | Genes | Gene Name/Description | Genomic Features | |
|---|---|---|---|---|---|---|---|---|---|---|
| LL | UL | |||||||||
| 12 | 103,311,112 | −0.09 | 0.02 | −0.12 | −0.05 | 1.84E-06 | 0.96 | PAH | Phenylalanine Hydroxylase | promoters |
| 5 | 95,296,026 | 0.22 | 0.04 | 0.13 | 0.3 | 2.14E-06 | 0.96 | ELL2 | Elongation Factor For RNA Polymerase II 2 | introns |
| 20 | 61,185,059 | 0.29 | 0.06 | 0.17 | 0.41 | 4.36E-06 | 0.96 | – | intergenic | |
| 5 | 117,618,581 | −0.16 | 0.03 | −0.22 | −0.09 | 5.76E-06 | 0.96 | CTD-3179P9.2 | Long Intergenic Non-Protein Coding RNA | intergenic |
| 19 | 53,031,512 | −0.16 | 0.03 | −0.23 | −0.1 | 5.93E-06 | 0.96 | ZNF808 | Zinc Finger Protein 808 | intergenic |
| 1 | 195,667,563 | 0.15 | 0.03 | 0.09 | 0.21 | 5.94E-06 | 0.96 | – | intergenic | |
| 16 | 31,342,748 | −0.22 | 0.05 | −0.32 | −0.13 | 6.24E-06 | 0.96 | ITGAM | Integrin Subunit Alpha M | introns |
| 3 | 53,301,594 | −0.06 | 0.01 | −0.09 | −0.04 | 8.13E-06 | 0.96 | – | intergenic | |
| 12 | 8,046,968 | 0.11 | 0.02 | 0.06 | 0.16 | 8.98E-06 | 0.96 | SLC2A14 | Solute Carrier Family 2 Member 14 | intergenic |
| 22 | 32,955,596 | −0.15 | 0.03 | −0.21 | −0.08 | 9.20E-06 | 0.96 | SYN3 | Synapsin III | intergenic |
| 1 | 228,318,664 | 0.11 | 0.02 | 0.06 | 0.16 | 1.00E-05 | 0.96 | RP11-520H14.1 | Pseudogene | intergenic |
| 11 | 117,665,622 | −0.33 | 0.07 | −0.47 | −0.19 | 1.01E-05 | 0.96 | DSCAML1 | Down Syndrome Cell Adhesion Molecule Like 1 | intergenic |
| 8 | 10,589,079 | −0.12 | 0.02 | −0.17 | −0.07 | 1.01E-05 | 0.96 | SOX7 | SRY-related HMG-Box Transcription Factor 7 | introns |
| 17 | 3,790,089 | 0.3 | 0.07 | 0.17 | 0.44 | 1.08E-05 | 0.96 | CAMKK1 | Calcium/Calmodulin Dependent Protein Kinase Kinase 1 | intergenic |
| 1 | 205,023,204 | 0.23 | 0.05 | 0.13 | 0.33 | 1.16E-05 | 0.96 | CNTN2 | Contactin 2 | introns |
| 14 | 42,074,670 | 0.06 | 0.01 | 0.04 | 0.09 | 1.18E-05 | 0.96 | LRFN5 | Leucine Rich Repeat and Fibronectin Type III domain containing 5 | intergenic |
| 7 | 1,867,522 | −0.09 | 0.02 | −0.13 | −0.05 | 1.32E-05 | 0.96 | MAD1L1 | Mitotic Arrest Deficient 1 like 1 | intergenic |
| 3 | 179,390,332 | −0.05 | 0.01 | −0.08 | −0.03 | 1.37E-05 | 0.96 | USP13 | Ubiquitin Specific Peptidase 13 | intergenic |
| 1 | 2,392,971 | 0.16 | 0.03 | 0.09 | 0.23 | 1.39E-05 | 0.96 | PLCH2 | Phospolipase C eta 2 | intergenic |
| 20 | 44,552,516 | −0.05 | 0.01 | −0.08 | −0.03 | 1.47E-05 | 0.96 | – | intergenic | |
Notes: Chr = chromosome, CI = confidence interval; LL = lower limit of the confidence interval; UL = upper limit of the confidence interval; SE = standard error; cLBP = chronic low back pain.
Pathways analysis of genes containing DMLs in race-pain interactions
After removing duplicate, 160 genes containing DMLs with p < 0.0001 were subjected to IPA to determine functional canonical pathways enriched by the interactions of race, pain and DML in NHBs and NHW with cLBP. We identified 34 pathways that were significantly enriched by the genes (p < 0.01). Fig. 6 depicts the top 20 enriched pathways, including those relevant to pain pathologies such as axon guidance signaling, endocannabinoids neuronal synapse, calcium signaling, and Wnt/beta-catenin signaling pathways. Supplemental Table 4 summarizes the list of over-represented pathways and associated differentially methylated genes containing DMLs with race-by-pain interactions.
Fig. 6.
Top 20 over-represented pathways by genes containing DMLs from the race by pain interaction among adults with cLBP. Note: The dotted line depicts the statistically significant level (p < 0.01).
Discussion
Epigenetic modifications, particularly DNAm, induced by environmental exposures may be responsible for heritable changes in gene expression and account for a substantial fraction of variability in physiologic and disease processes. (van Dongen et al., 2016) Multiple studies have documented an elevated risk for many chronic diseases, including cLBP among NHBs. (Aroke et al., 2020a, Meints et al., 2018) While an individual’s race is not a biological trait or an inherent disease risk, societal racism, environmental exposures, and chronic stress may contribute to racial disparities in pain through epigenetic markings on the DNA, thus linking lived experience and cLBP risk. (Aroke et al., 2019) Other investigators have supported the potential mechanistic role of epigenetic modifications as “biological memories of experiences acquired earlier in our own lives” and environment (p. 801). (Thayer and Kuzawa, 2011) Specifically, epigenetic changes can link environmental exposures to biological processes that affect disease risk. Using RRBS analysis followed by pathway analysis, we identified DMLs between NHBs and NHWs with cLBP and over-represented pathways from genes containing the DMLs, thus revealing potential molecular pathways implicated in racial disparities in cLBP.
To our knowledge, this is the first study to apply RRBS to investigate associations between DNAm and racial disparities in cLBP and use integrative computational analysis to identify functional pathways over-represented by the genes with DMLs. Our results demonstrate well-defined racial category-associated differential methylation patterns among individuals with cLBP. As expected, our genome-wide methylation analysis revealed DML in various regions, including intronic, intergenic, promoters, and exons, as well as CpG island and shores. Generally, methylation of CpGs in the promoter region of genes blocks transcription factors from binding and inhibits gene expressions. Typically CpG islands span 200–1000 base pairs and can overlap the first intron with gene promoter regions, regulating transcription. (Moore et al., 2013) However, the effect of methylation in intergenic or non-promoter gene regions on gene expression is less straightforward. Thus, given the high number of annotated differentially methylated genes and varied distribution of the DMLs, we will focus our discussion on the over-represented pathways.
Our findings suggest that differentially methylated genes between NHBs and NHWs with cLBP were over-represented in pathways involved in chronic pain, chronic stress, and the brain reward system, notably, Corticotropin-Releasing Hormone Signaling, Dopamine-DARPP32 Feedback in cAMP Signaling, and White Adipose Tissue Browning Pathway. At the physiological level, chronic stress and chronic pain share several overlapping processes. (Abdallah and Geha, 2017) Enrichment of Corticotropin-Releasing Hormone Signaling and Dopamine-DARPP32 Signaling pathways have previously been linked with chronic postsurgical pain in adolescents undergoing spinal surgery. (Chidambaran et al., 2019) Others have reported that long-term exposure to stress and early-life adversity correlate with epigenetically induced alterations in related stress adaptation pathways. (Silberman et al., 2016, Géranton, 2019) Also, chronic pain and stress can induce depressive symptoms, potentially through epigenetically induced long-term neuroplasticity in the central nervous system. For instance, epigenetic alterations in pathways involved in dopamine metabolism have been associated with depressive symptoms and chronic pain conditions. (Géranton, 2019, Kerr and Burri, 2017) Similarly, Sidossis and colleagues reported that prolonged stress epigenetically induces browning of subcutaneous white adipose tissue in children recovering from burns. (Sidossis et al., 2015) Consistent with our previous hypothesis, (Aroke et al., 2019) these findings suggest the potential role of chronic stress, such as the stress of racial discrimination on racial disparities in cLBP.
Our sample observed that Synaptogenesis Signaling, Wnt/β-catenin Signaling, Axon Guidance Signaling, Netrin Signaling, and Calcium Signaling pathways were over-represented in genes differentially methylated between NHBs and NHWs with cLBP. This observation aligns with previous publications indicating that disruptions in neuronal proliferation and sensitizing could play a role in chronic pain. (Babcock et al., 2011, Yang et al., 2020, Tang, 2014, Zhao and Yang, 2018, Patil and Andry, 2017, Shi et al., 2016) Netrin and Wnt signaling play an essential role in neuronal development (axon guidance) through a multifunctional protein complex that interacts with transcriptional factors and affects target gene expression. (Nusse, 2005, Gao et al., 2020) Wnt signaling ligands and receptors (e.g., Frizzled family of receptors) are expressed along neural pain pathways in the dorsal root ganglion (DRG) (Tang, 2014) and play a role in the etiology of neuropathic and inflammatory pain. (Zhang et al., 2013, Wu et al., 2020) Using animal models, Zheng and colleagues reported that nerve injury and bone cancer increase Wnt expression and β-catenin levels in the sensory neurons in the DRG. At the same time, spinal blockade of Wnt signaling suppressed the induction of pain and delayed pain processing for up to 14 days. (Zhang et al., 2013) Other investigators have also found that increased expression of WNT5B plays an essential role in the transition from acute to chronic pain and the development of opioid withdrawal symptoms. (Wu et al., 2020) Similarly, increased expression of Netrin signaling pathways appears to be neuroprotective after spinal code injury. (Gao et al., 2020) Chronic stress affects genes’ expression in Netrin and Wnt signaling pathways and has been associated with anxiety-like behavior, attention deficit hyperactivity disorder, insulin resistance, depressive symptoms, and increased blood pressure. (Odaka et al., 2017, Yde Ohki et al., 2020, Torres-Berrío et al., 2020) Thus, it is plausible that higher levels of perceived stress (e.g., the stress of racial discrimination) induce alterations in Netrin and Wnt signaling pathways and result in higher cLBP outcomes in NHBs.
Glutamate, γ-amino-butyric acid (GABA) Receptor Signaling, Opioid Signaling, Endocannabinoid Neuronal Synapse, and G-Protein Couple Receptor-Mediated Nutrient Sensing in Enteroendocrine cells pathways were over-represented. It is well known that neurotransmitters such as GABA, substance P, endorphins, catecholamines, inflammatory cytokines, and endothelins affect nociception and pain perception via G-protein coupled signaling, which is a primary target for many analgesics. (Geppetti et al., 2015, Nourbakhsh et al., 2018) There is evidence from prior studies that some of the identified pathways are involved in chronic pain conditions and pain management. (Chidambaran et al., 2019, Pedersen et al., 2015, Duan et al., 2021, Wang and Burrell, 2018, Hossain et al., 2020, Siuda et al., 2015, Isensee et al., 2017, Gomes et al., 2020, Louwies et al., 2019, Montesino-Goicolea et al., 2020) For instance, Montesino-Goicolea et al. reported that enrichment of GABA Receptor Signaling pathways by differentially methylated genes between adults chronic pain and healthy controls. (Montesino-Goicolea et al., 2020) Also, enrichment of GABA Receptor Signaling pathways have previously been associated with chronic postoperative pain, (Chidambaran et al., 2019) while Opioid Signaling and Endocannabinoid Neuronal Synapse Pathways have previously been associated with addiction and pain management. (Gomes et al., 2020, Ramesh et al., 2018, Dawley et al., 2017, Regan et al., 2012) Thus, epigenetically induced changes in multiple pathways may play an essential role in cLBP and more significant pain severity in NHBs. These support our findings that racial disparities in non-specific cLBP may be related to epigenetic modification of genes in different pathways signaling pathways. These observations align with prior works suggesting the need to consider the interaction of immune and neurological systems as potential novel targets for pain. (Montesino-Goicolea et al., 2020)
Finally, our stratified race analysis comparing individuals with cLBP against racially similar PFCs suggests the need for race-specific epigenetic studies. One hundred and ten (110) pathways were significantly over-represented in the NHBs compared to 31 pathways in the NHWs group. Nineteen (Fuentes et al., 2007) of the pathways over-represented in NHWs were also over-represented in NHBs, but the p-values were much smaller in NHBs than NHWs. While the p-value is not a perfect predictor of effect size, a smaller p-value between data of equal sample size suggests a more robust effect size. The effect size of the over-represented pathways may be greater in NHBs than NHWs. Also, more genes may be differentially methylated in the NHB than NHW groups. Thus, future epigenetic studies of cLBP should recruit cases and controls from the same racial background.
Strengths and limitations
Our study has several strengths and limitations that must be considered when generalizing the findings. First, to our knowledge, this study provides the first evidence of a potential mechanistic important role for DNAm in racial disparities in cLBP. By using an epigenetic approach, our findings lend themselves to future therapeutic targets to reduce the disparities in cLBP. Also, using racial group-stratified comparisons is a major strength because epigenetic changes are influenced by environmental exposure. Thus, uncovering mechanistic roles requires a within-group comparison to account for differences in distinct lived experiences. This approach indirectly controls for the social nature of the racial group construct and avoids using one racial group as the reference group. Finally, using an epigenomic approach gives novel insight into potential pathways without the limitations of a priori knowledge.
There are some limitations to our study. First, despite being the first and largest study to date, our sample size was relatively small. Second, the epigenomic approach gives insight into novel pathways, but the precise mechanism by which DNAm changes affect the signaling pathways and cLBP phenotype need to be confirmed in future studies through the measure of gene expression. It would be highly relevant to investigate the influence of each gene marker on gene expression and cLBP phenotype using targeted sequencing epigenetic approach. Also, some of the racial differences may be due to genetics because genetic polymorphisms can lead to changes in methylation. Third, racial categories in our study were self-identified by the participants. Given that race is a social construct, we believed that using self-identified race would most appropriately capture the potential effects of lived experiences on the epigenome, which may explain the racial disparities in cLBP. Our study did not include a secondary validation of the differentially methylated genes. However, it provides a solid basis for further investigations with larger cohorts and deeper explorations of specific genes or signaling pathways. Additionally, it provides proof-of-concept data addressing the larger issue of how race relates to the risk of chronic diseases in the United States. Finally, the use of blood samples rather than nervous tissues is a limitation. Epigenetic changes are tissue-specific, but DNAm changes in the nervous system have been shown to correlate with changes in blood samples. The use of blood samples provides a readily available marker that can be used clinically. With the use of blood samples, DNAm differences could represent variations in the proportion of cell subtype populations. We will address this in future studies by either directly determining cell population sizes via flow cytometry or single-cell RNAseq or statistically using methods described previously. (Houseman et al., 2012, Jaffe and Irizarry, 2014, Karmaus and Chen, 2017) Finally, we limited this study to individuals with nonspecific cLBP, eliminating many individuals with cLBP (e.g., trauma, rheumatoid arthritis, other chronic pain conditions, etc.). Similarly, while we did an epigenetic race by pain interaction analysis, there was a strong correlation between race and pain severity. Thus, the observed racial differences may be related to pain severity.
Conclusion
In the United States, individuals who self-identify as NHBs report more severe and disabling cLBP compared to NHWs. Even though an individual’s self-identified race is not a biological variable, that system of classifying people and associated racism affects virtually every aspect of life, including disease risk. Our findings suggest a potential mechanistic role of epigenetic modifications in racial disparities in cLBP. Specifically, our results indicate that DNAm alterations in pathways involving G-protein coupled receptors, neuronal transmission, and chronic stress may explain some of the variances in racial disparities in cLBP. While future studies are required in larger prospective cohorts with a longitudinal evaluation of DNAm and gene expression with replication of our findings, these results are promising and open novel avenues of epigenetic-based pain disparities research. Given that epigenetic modifications are dynamic and reversible, future targeted epigenetics interventions may help reduce cLBP disparities and improve cLBP management.
Ethics approval and consent of participants
The Institutional Review Board (IRB) at the University of Alabama at Birmingham approved all research included in this study (IRB-170119003). Written informed consent was obtained from all participants under the approval from the IRB.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors would like to extend sincere appreciation to all the participants, all the recruiting and participating centers and technicians, and other members of Biobehavioral Pain labs for support and discussion.
Funding
This work was supported by the American Association of Nurse Anesthetists Foundation Post-Doctoral Fellowship Award (2018-FS-4) to ENA, The University of Alabama at Birmingham Dean’s Scholar Award to ENA, and the National Institutes of Health grant (R01MD010441) to BRG and (1R01AR079178) to ENA.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynpai.2022.100086.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Clark S., Horton R. Low back pain: a major global challenge. The Lancet. 2018;391(10137):2302. doi: 10.1016/S0140-6736(18)30725-6. [DOI] [PubMed] [Google Scholar]
- GBD 2017 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1789-858.doi: 10.1016/s0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed]
- Hartvigsen J., Hancock M.J., Kongsted A., Louw Q., Ferreira M.L., Genevay S., et al. What low back pain is and why we need to pay attention. The Lancet. 2018;391(10137):2356–2367. doi: 10.1016/S0140-6736(18)30480-X. [DOI] [PubMed] [Google Scholar]
- Aroke E.N., Jackson P., Overstreet D.S., Penn T.M., Rumble D.D., Kehrer C.V., et al. Race, Social Status, and Depressive Symptoms: A Moderated Mediation Analysis of Chronic Low Back Pain Interference and Severity. Clin J Pain. 2020 doi: 10.1097/ajp.0000000000000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meints S.M., Wang V., Edwards R.R. Sex and Race Differences in Pain Sensitization among Patients with Chronic Low Back Pain. Journal of Pain. 2018 doi: 10.1016/j.jpain.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Yang G.S., Greenspan J.D., Downton K.D., Griffith K.A., Renn C.L., et al. Racial and ethnic differences in experimental pain sensitivity: systematic review and meta-analysis. Pain. 2017;158(2):194–211. doi: 10.1097/j.pain.0000000000000731. [DOI] [PubMed] [Google Scholar]
- Edwards C.L., Fillingim R.B., Keefe F. Race, ethnicity and pain. PAIN. 2001;94(2):133–137. doi: 10.1016/S0304-3959(01)00408-0. [DOI] [PubMed] [Google Scholar]
- Aroke E.N., Joseph P.V., Roy A., Overstreet D.S., Tollefsbol T.O., Vance D.E., et al. Could epigenetics help explain racial disparities in chronic pain? Journal of Pain Research. 2019;12:701–710. doi: 10.2147/JPR.S191848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battie MC, Videman T, Levalahti E, Gill K, Kaprio J. Genetic and environmental effects on disc degeneration by phenotype and spinal level: a multivariate twin study. Spine (Phila Pa 1976). 2008;33(25):2801-8.doi: 10.1097/BRS.0b013e31818043b7. [DOI] [PubMed]
- Battie M.C., Videman T., Levalahti E., Gill K., Kaprio J. Heritability of low back pain and the role of disc degeneration. Pain. 2007;131(3):272–280. doi: 10.1016/j.pain.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Suri P., Boyko E.J., Smith N.L., Jarvik J.G., Williams F.M., Jarvik G.P., et al. Modifiable risk factors for chronic back pain: insights using the co-twin control design. Spine J. 2017;17(1):4–14. doi: 10.1016/j.spinee.2016.07.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moix J., Kovacs F.M., Martin A., Plana M.N., Catastrophizing R.A. State Anxiety, Anger, and Depressive Symptoms Do Not Correlate with Disability when Variations of Trait Anxiety Are Taken into Account. A Study of Chronic Low Back Pain Patients Treated in Spanish Pain Units. Pain Medicine. 2011;12(7):1008–1017. doi: 10.1111/j.1526-4637.2011.01155.x. [DOI] [PubMed] [Google Scholar]
- Pinheiro M.B., Ferreira M.L., Refshauge K., Maher C.G., Ordonana J.R., Andrade T.B., et al. Symptoms of depression as a prognostic factor for low back pain: a systematic review. The spine journal. 2016;16(1):105–116. doi: 10.1016/j.spinee.2015.10.037. [DOI] [PubMed] [Google Scholar]
- Fernandez M., Colodro-Conde L., Hartvigsen J., Ferreira M.L., Refshauge K.M., Pinheiro M.B., et al. Chronic low back pain and the risk of depression or anxiety symptoms: insights from a longitudinal twin study. The Spine Journal. 2017;17(7):905–912. doi: 10.1016/j.spinee.2017.02.009. [DOI] [PubMed] [Google Scholar]
- Hill J.C., Fritz J.M. Psychosocial influences on low back pain, disability, and response to treatment. Phys Ther. 2011;91(5):712–721. doi: 10.2522/ptj.20100280. [DOI] [PubMed] [Google Scholar]
- Katz J.N. Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. The Journal of bone and joint surgery. 2006;88(Suppl 2):21–24. doi: 10.2106/jbjs.e.01273. [DOI] [PubMed] [Google Scholar]
- Sue D.W., Capodilupo C.M., Torino G.C., Bucceri J.M., Holder A., Nadal K.L., et al. Racial microaggressions in everyday life: implications for clinical practice. American psychologist. 2007 doi: 10.1037/0003-066X.62.4.271. [DOI] [PubMed] [Google Scholar]
- Williams M.T. Racial Microaggressions: Critical Questions, State of the Science, and New Directions. Perspectives on Psychological Science. 2021;16(5):880–885. doi: 10.1177/17456916211039209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes M., Hart-Johnson T., Green C.R. The association among neighborhood socioeconomic status, race and chronic pain in black and white older adults. Journal of the National Medical Association. 2007;99(10):1160–1169. [PMC free article] [PubMed] [Google Scholar]
- Nelson S., Simons L.E., Logan D. The Incidence of Adverse Childhood Experiences (ACEs) and Their Association With Pain-related and Psychosocial Impairment in Youth With Chronic Pain. Clinical journal of pain. 2018;34(5):402–408. doi: 10.1097/ajp.0000000000000549. [DOI] [PubMed] [Google Scholar]
- Sack V, Murphey D. The prevalence of adverse childhood experiences, nationally, by state, and by race or ethnicity. Child Trends [Internet]. 2018 September 17, 2018; (03). Available from: https://www.childtrends.org/wp-content/uploads/2018/02/ACESBriefUpdatedFinal_ChildTrends_February2018.pdf.doi:.
- Clark R., Anderson N.B., Clark V.R., Williams D.R. Racism as a stressor for African Americans: A biopsychosocial model. American psychologist. 1999 doi: 10.1037//0003-066x.54.10.805. [DOI] [PubMed] [Google Scholar]
- Huang B., Jiang C., Zhang R. Epigenetics: the language of the cell? Epigenomics. 2014;6(1):73–88. doi: 10.2217/epi.13.72. [DOI] [PubMed] [Google Scholar]
- Kanherkar RR, Bhatia-Dey N, Csoka AB. Epigenetics across the human lifespan. Frontiers in cell and developmental biology. 2014;2:49-.doi: 10.3389/fcell.2014.00049. [DOI] [PMC free article] [PubMed]
- Syed S.A., Nemeroff C.B. Early Life Stress, Mood, and Anxiety Disorders. Chronic Stress (Thousand Oaks) 2017 doi: 10.1177/2470547017694461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C., Armant D.R., Brenner C.A. Epigenetics: definition, mechanisms and clinical perspective. Seminars in reproductive medicine. 2009;27(5):351–357. doi: 10.1055/s-0029-1237423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massart R., Dymov S., Millecamps M., Suderman M., Gregoire S., Koenigs K., et al. Overlapping signatures of chronic pain in the DNA methylation landscape of prefrontal cortex and peripheral T cells. Scientific Reports. 2016;6:19615. doi: 10.1038/srep19615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajerian M., Alvarado S., Millecamps M., Dashwood T., Anderson K.M., Haglund L., et al. DNA methylation of SPARC and chronic low back pain. Molecular pain. 2011;7:65. doi: 10.1186/1744-8069-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawi J., Meneses J., Sood K., Tesfagiorgis E. Association between global DNA methylation and exercise in chronic low back pain. The Journal of Pain. 2018;19(3):S103–S104. doi: 10.1016/j.jpain.2017.12.240. [DOI] [Google Scholar]
- Sukenaga N., Ikeda-Miyagawa Y., Tanada D., Tunetoh T., Nakano S., Inui T., et al. Correlation Between DNA Methylation of TRPA1 and Chronic Pain States in Human Whole Blood Cells. Pain Medicine. 2016;17(10):1906–1910. doi: 10.1093/pm/pnv088. [DOI] [PubMed] [Google Scholar]
- Burri A., Marinova Z., Robinson M.D., Kuhnel B., Waldenberger M., Wahl S., et al. Are Epigenetic Factors Implicated in Chronic Widespread Pain? PLoS One. 2016;11(11) doi: 10.1371/journal.pone.0165548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens K.E., Levine J.D., Aouizerat B.E., Paul S.M., Abrams G., Conley Y.P., et al. Associations between genetic and epigenetic variations in cytokine genes and mild persistent breast pain in women following breast cancer surgery. Cytokine. 2017;99:203–213. doi: 10.1016/j.cyto.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerra M.C., Dagostino C., D'Agnelli S., Boggiani L., Rizza V., Marchesini M., et al. Omics as a potential tool to identify biomarkers and to clarify the mechanism of chronic pain development. Scandinavian Journal of Pain. 2017;16(1):187. doi: 10.1016/j.sjpain.2017.04.064. [DOI] [Google Scholar]
- Aroke E.N., Overstreet D.S., Penn T.M., Crossman D.K., Jackson P., Tollefsbol T.O., et al. Identification of DNA methylation associated enrichment pathways in adults with non-specific chronic low back pain. Molecular Pain. 2020;16 doi: 10.1177/1744806920972889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn T.M., Overstreet D.S., Aroke E.N., Rumble D.D., Sims A.M., Kehrer C.V., et al. Perceived Injustice Helps Explain the Association Between Chronic Pain Stigma and Movement-Evoked Pain in Adults with Nonspecific Chronic Low Back Pain. Pain Medicine. 2020 doi: 10.1093/pm/pnaa095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou R., Gordon D.B., de Leon-Casasola O.A., Rosenberg J.M., Bickler S., Brennan T., et al. Management of Postoperative Pain: A Clinical Practice Guideline From the American Pain Society, the American Society of Regional Anesthesia and Pain Medicine, and the American Society of Anesthesiologists' Committee on Regional Anesthesia, Executive Committee, and Administrative Council. The Journal of Pain. 2016;17(2):131–157. doi: 10.1016/j.jpain.2015.12.008. [DOI] [PubMed] [Google Scholar]
- Cleeland C.S., Ryan K.M. Pain assessment: global use of the Brief Pain Inventory. Ann Acad Med Singapore. 1994;23(2):129–138. [PubMed] [Google Scholar]
- Andrew S. Fast QC: a quality control tool for high throughput sequence data 2019 [Available from: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Krueger F. Trim Galore [updated Nov 19, 2019. v0.6.5:[Available from: https://www.bioinformatics.babraham.ac.uk/publications.html.
- Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.Q., Tuominen L.K., Tsai C.J. SLIM: a sliding linear model for estimating the proportion of true null hypotheses in datasets with dependence structures. Bioinformatics. 2011;27(2):225–231. doi: 10.1093/bioinformatics/btq650. [DOI] [PubMed] [Google Scholar]
- Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13(10):R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C. Analysing and interpreting DNA methylation data. Nature Reviews Genetics. 2012;13(10):705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- van Dongen J., Nivard M.G., Willemsen G., Hottenga J.J., Helmer Q., Dolan C.V., et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat Commun. 2016;7:11115. doi: 10.1038/ncomms11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer Z.M., Kuzawa C.W. Biological memories of past environments: Epigenetic pathways to health disparities. Epigenetics. 2011;6(7):798–803. doi: 10.4161/epi.6.7.16222. [DOI] [PubMed] [Google Scholar]
- Moore L.D., Le T., Fan G. DNA Methylation and Its Basic Function. Neuropsychopharmacology. 2013;38(1):23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah CG, Geha P. Chronic Pain and Chronic Stress: Two Sides of the Same Coin? Chronic stress (Thousand Oaks, Calif). 2017;1:10.1177/2470547017704763.doi:. [DOI] [PMC free article] [PubMed]
- Chidambaran V., Zhang X., Geisler K., Stubbeman B.L., Chen X., Weirauch M.T., et al. Enrichment of genomic pathways based on differential DNA methylation associated with chronic postsurgical pain and anxiety in children: a prospective, pilot study. The Journal of Pain. 2019;20(7):771–785. doi: 10.1016/j.jpain.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberman D.M., Acosta G.B., Zubilete M.A.Z. Long-term effects of early life stress exposure: Role of epigenetic mechanisms. Pharmacological Research. 2016 doi: 10.1016/j.phrs.2015.12.033. [DOI] [PubMed] [Google Scholar]
- Géranton S. Does epigenetic ‘memory’of early-life stress predispose to chronic pain in later life? A potential role for the stress regulator FKBP5. Philosophical Transactions of the Royal Society B. 2019;374(1785):20190283.doi:. [DOI] [PMC free article] [PubMed]
- Kerr J.I., Burri A. Genetic and epigenetic epidemiology of chronic widespread pain. J Pain Res. 2017;10:2021–2029. doi: 10.2147/JPR.S143869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidossis L.S., Porter C., Saraf M.K., Børsheim E., Radhakrishnan R.S., Chao T., et al. Browning of subcutaneous white adipose tissue in humans after severe adrenergic stress. Cell metabolism. 2015;22(2):219–227. doi: 10.1016/j.cmet.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock D.T., Shi S., Jo J., Shaw M., Gutstein H.B., Galko M.J. Hedgehog signaling regulates nociceptive sensitization. Current biology : CB. 2011;21(18):1525–1533. doi: 10.1016/j.cub.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Wang X., Zhang X., You S., Feng L., Zhang Y., et al. Sonic Hedgehog Signaling Contributes to Chronic Post-Thoracotomy Pain via Activating BDNF/TrkB Pathway in Rats. Journal of Pain. Research. 2020 doi: 10.2147/JPR.S245515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S.-J. Synaptic activity-regulated Wnt signaling in synaptic plasticity, glial function and chronic pain. CNS & neurological disorders drug targets. 2014;13(5):737–744. doi: 10.2174/1871527312666131223114457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yang Z. Effect of Wnt signaling pathway on pathogenesis and intervention of neuropathic pain. Experimental and therapeutic medicine. 2018;16(4):3082–3088. doi: 10.3892/etm.2018.6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil D., Andry T. Molding young minds: The importance of residency training in shaping residents’ attitudes toward substance use disorders. American Journal on Addictions. 2017;26(1):80–82. doi: 10.1111/ajad.12484. [DOI] [PubMed] [Google Scholar]
- Shi J., Chi S., Xue J., Yang J., Li F., Liu X. Emerging Role and Therapeutic Implication of Wnt Signaling Pathways in Autoimmune Diseases. Journal of Immunology Research. 2016;2016:9392132. doi: 10.1155/2016/9392132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R. Wnt signaling in disease and in development. Cell Research. 2005;15(1):28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- Gao K., Niu J., Dang X. Neuroprotection of netrin-1 on neurological recovery via Wnt/β-catenin signaling pathway after spinal cord injury. Neuroreport. 2020 doi: 10.1097/WNR.0000000000001441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-K., Huang Z.-J., Liu S., Liu Y.-P., Song A.A., Song X.-J. WNT signaling underlies the pathogenesis of neuropathic pain in rodents. The Journal of clinical investigation. 2013;123(5):2268–2286. doi: 10.1172/JCI65364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Li Z., Liang L., Ma P., Cui D., Chen P., et al. Wnt signaling contributes to withdrawal symptoms from opioid receptor activation induced by morphine exposure or chronic inflammation. Pain. 2020;161(3):532–544. doi: 10.1097/j.pain.0000000000001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odaka H., Adachi N., Numakawa T. Impact of glucocorticoid on neurogenesis. Neural Regeneration Research. 2017 doi: 10.4103/1673-5374.211174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yde Ohki C.M., Grossmann L., Alber E., Dwivedi T., Berger G., Werling A.M., et al. The stress–Wnt-signaling axis: a hypothesis for attention-deficit hyperactivity disorder and therapy approaches. Translational Psychiatry. 2020;10(1):315. doi: 10.1038/s41398-020-00999-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Berrío A., Hernandez G., Nestler E.J., Flores C. The netrin-1/DCC guidance cue pathway as a molecular target in depression: translational evidence. Biological Psychiatry. 2020;88(8):611–624. doi: 10.1016/j.biopsych.2020.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geppetti P., Veldhuis Nicholas A., Lieu T., Bunnett N.W. G Protein-Coupled Receptors: Dynamic Machines for Signaling Pain and Itch. Neuron. 2015;88(4):635–649. doi: 10.1016/j.neuron.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Nourbakhsh F., Atabaki R., Roohbakhsh A. The role of orphan G protein-coupled receptors in the modulation of pain: A review. Life sciences. 2018 doi: 10.1016/j.lfs.2018.09.028. [DOI] [PubMed] [Google Scholar]
- Pedersen L.M., Schistad E., Jacobsen L.M., Røe C., Gjerstad J. Serum levels of the pro-inflammatory interleukins 6 (IL-6) and-8 (IL-8) in patients with lumbar radicular pain due to disc herniation: a 12-month prospective study. Brain, behavior, and immunity. 2015:132–136. doi: 10.1016/j.bbi.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Duan H., Shen F., Li L., Tu Z., Chen P., Chen P., et al. Activation of the Notch signaling pathway in the anterior cingulate cortex is involved in the pathological process of neuropathic pain. Pain. 2021 doi: 10.1097/j.pain.0000000000002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Burrell B.D. Endocannabinoid-mediated potentiation of nonnociceptive synapses contributes to behavioral sensitization. Journal of neurophysiology. 2018;119(2):641–651. doi: 10.1152/jn.00092.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MZ, Ando H, Unno S, Kitagawa J. Targeting peripherally restricted cannabinoid receptor 1, cannabinoid receptor 2, and endocannabinoid-degrading enzymes for the treatment of neuropathic pain including neuropathic orofacial pain. International journal of molecular sciences. 2020;21(4):1423.doi:. [DOI] [PMC free article] [PubMed]
- Siuda E.R., Copits B.A., Schmidt M.J., Baird M.A., Al-Hasani R., Planer W.J., et al. Spatiotemporal control of opioid signaling and behavior. Neuron. 2015;86(4):923–935. doi: 10.1016/j.neuron.2015.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee J., Krahé L., Moeller K., Pereira V., Sexton J.E., Sun X., et al. Synergistic regulation of serotonin and opioid signaling contributes to pain insensitivity in Nav1. 7 knockout mice. Science signaling. 2017 doi: 10.1126/scisignal.aah4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes T.M., da Silva D.D., Carmo H., Carvalho F., Silva J.P. Epigenetics and the endocannabinoid system signaling: An intricate interplay modulating neurodevelopment. Pharmacological Research. 2020 doi: 10.1016/j.phrs.2020.105237. [DOI] [PubMed] [Google Scholar]
- Louwies T., Ligon C.O., Johnson A.C., Greenwood-Van M.B. Targeting epigenetic mechanisms for chronic visceral pain: a valid approach for the development of novel therapeutics. Neurogastroenterology & Motility. 2019 doi: 10.1111/nmo.13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesino-Goicolea S, Sinha P, Huo Z, Rani A, Foster TC, Cruz-Almeida Y. Enrichment of genomic pathways based on differential DNA methylation profiles associated with chronic musculoskeletal pain in older adults: An exploratory study. Molecular pain. 2020;16:1744806920966902-.doi: 10.1177/1744806920966902. [DOI] [PMC free article] [PubMed]
- Ramesh D., D'Agata A., Starkweather A.R., Young E.E. Contribution of Endocannabinoid Gene Expression and Genotype on Low Back Pain Susceptibility and Chronicity. Clin J Pain. 2018;34(1):8–14. doi: 10.1097/ajp.0000000000000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawley D., Kessler J., Crow D., Pena A.M., Garmany R., Egleton R., et al. Opioid abuse effects potentially mediated by epigenetic modifications. Biochemistry and Cell Biology. 2017;95(2):180–181. doi: 10.1139/bcb-2017-0043. [DOI] [Google Scholar]
- Regan P.M., Dave R.S., Datta P.K., Khalili K. Epigenetics of μ-opioid receptors: Intersection with HIV-1 infection of the central nervous system. Journal of Cellular Physiology. 2012;227(7):2832–2841. doi: 10.1002/jcp.24004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics. 2012;13(1):1–16. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A.E., Irizarry R.A. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome biology. 2014;15(2):1–9. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmaus W., Chen S. Changes in blood DNA methylation and incomplete adjustment for blood cell composition. International journal of epidemiology. 2017;46(5):1714–1717. doi: 10.1093/ije/dyx082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.