Summary
Traditional replication timing (RT) experiments divide S phase into two phases: early and late. However, there is an increasing awareness that variation in RT can occur during the course of S phase and impact our understanding of RT patterns and regulation. Here, we describe a RT protocol in RPE-1 cells for collecting four phases within S and the library preparation that takes advantage of a commercial kit for methyl-DNA. This step allows BrdU-labeled DNA sequencing and assessment of RT genome wide.
For complete details on the use and execution of this protocol, please refer to Van Rechem et al. (2021).
Subject areas: Cell isolation, Flow Cytometry/Mass Cytometry, Genomics, Sequencing, Molecular Biology
Graphical abstract

Highlights
-
•
Collecting four phases of S phase allows to profile replication timing in mid S phase
-
•
The use of a commercial kit for library preparation simplifies the protocol
-
•
This protocol can be adapted to any cycling cells
Traditional replication timing (RT) experiments divide S phase into two phases: early and late. However, there is an increasing awareness that variation in RT can occur during the course of S phase and impact our understanding of RT patterns and regulation. Here, we describe a RT protocol in RPE-1 cells for collecting four phases within S and the library preparation that takes advantage of a commercial kit for methyl-DNA. This step allows BrdU-labeled DNA sequencing and assessment of RT genome wide.
Before you begin
Experimental conditions described in this protocol apply to hTERT RPE-1 cells. However, this protocol should be applicable to any cycling cell type.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-BrdU (1/250 dilution) | BD Pharmingen | Cat# 555627 |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s Modified Eagle’s Medium - High Glucose | Sigma-Aldrich | Cat# D5648 |
| FBS | GIBCO | Cat# 26140-079 |
| Penicillin-Streptomycin | Life Technologies | Cat# 15140122 |
| L-Glutamine | Life Technologies | Cat# 25030-081 |
| Propidium Iodide | Sigma-Aldrich | Cat# P4864 |
| Trypsin-0.25% EDTA | Life Technologies | Cat# 2520056 |
| Ribonuclease A | Sigma-Aldrich | Cat# R4875 |
| Proteinase K | Sigma-Aldrich | Cat# P6556 |
| 5-Bromo-2-deoxyuridine | Sigma-Aldrich | Cat# B5002 |
| AMPure XP | Beckman Coulter | Cat# A63881 |
| Dynabeads Protein G | Thermo Fisher Scientific | Cat# 10003D |
| Phenol:Chloroform:Isoamyl Alcohol 25:24:1 Saturated with 10 mM Tris, pH 8.0, 1 mM EDTA | Sigma-Aldrich | Cat# P2069-400ML |
| Glycogen | Roche (now Sigma) | Cat# 10901393001 |
| Critical commercial assays | ||
| Accel-NGS Methyl-Seq DNA Library Kit | Swift Biosciences | Cat# 30024 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Cat # Q32851 |
| High Sensitivity D1000 ScreenTape | Agilent | Cat # 5067-5584 |
| High Sensitivity D1000 Reagents | Agilent | Cat # 5067-5585 |
| Deposited data | ||
| Raw data | (Van Rechem et al., 2021) | GEO: GSE175752 |
| Experimental models: Cell lines | ||
| hTERT RPE-1 | Nicholas Dyson’s lab | N/A |
| RPE-GFP-CTRL | (Van Rechem et al., 2021) | N/A |
| RPE-GFP-KDM4A | (Van Rechem et al., 2021) | N/A |
| Oligonucleotides | ||
| qPCR primers (optional step): see step 35 | N/A | N/A |
| Other | ||
| FACS sorter | N/A | N/A |
| Magnetic stand for 1.5 mL tubes | N/A | N/A |
| Magnetic stand for 96 wells plate | N/A | N/A |
| Sonicator | QSonica | Q700 |
| Qubit | Thermo Fisher Scientific | N/A |
| TapeStation or Bioanalyser | N/A | N/A |
| Thermocycler | N/A | N/A |
Step-by-step method details
Cell sorting
Timing: 3–4 days
During this step cells are labeled with BrdU and sorted based on DNA content.
Note: It is necessary to use BrdU rather than EdU because it will be immunoprecipitated at a later step.
-
1.
Seed 2 × 106 RPE in four 15 cm2 plates for 48 h.
Note: Cells need to be at ∼70% confluence when harvested.
-
2.
Label the cells with 100 μM BrdU for 2 h at 37°C.
Note: BrdU incubation time depends on the growth rate of the cells and can need to be adjusted down to 1 h for very rapidly proliferating cells or even longer for slow dividing cells.
-
3.
Rinse the cells with 1× PBS and add 2.5 mL of trypsin per 15 cm2 plate.
Note: A single cell suspension is critical for the sorting.
-
4.
Wash the cells with 1× PBS with 1% FBS.
-
5.
Centrifuge for 2 min at 100 g and resuspend the cells in ice cold 2.5 mL 1× PBS with 1% FBS.
-
6.
Add 7.5 mL EtOH slowly and dropwise while vortexing.
Pause point: Cells can be stored in the dark (foil wrap) at −20°C for several months.
-
7.
Centrifuge the cells 5 min at 100 g at 4°C.
-
8.
Remove the supernatant and resuspend the cells in 3 mL 1× PBS with 1% FBS.
-
9.
Add 1/100 Propidium Iodide and RNAse A to 250 μg/mL. Incubate 1 h at 18°C–25°C in the dark.
-
10.
Keep the cells on ice until sorting.
-
11.
FACS Sort the cells with a BD FACS Fusion Tex Red A laser. A typical sort is represented in Figure 1. To place the gates: place P5 (G1) and P6 (G2) first, then divide the remaining four phases across S phase in equal fractions. The P5 fraction goes from the base of the large peak (1N) to 50% of the slope on the right. The P6 fraction goes from the top of the second peak (2N) to the base on the right. Collect P1 (S1) to P4 (S4) in 500 μL of PBS (P5 and P6 are optional), at least 250,000 cells per fraction. Troubleshooting 1.
Note: Any sorter with a laser line of 488 to excite Propidium Iodide can be used.
Note: There is no need to correct for a potential overestimation of numbers of events from the sorter compared to the actual number of collected cells.
Figure 1.
Gating for cell sorting
DNA extraction
Timing: 2 days
During this step DNA is purified and sonicated.
-
12.
Add SDS to 0.8% and proteinase K to 20 μg/mL to each fraction, incubate 2 h at 55°C .
-
13.
Add one volume of phenol-chloroform, vortex, centrifuge for 1 min at maximum speed. Collect as much as the upper phase possible and transfer in 15 mL conical tubes.
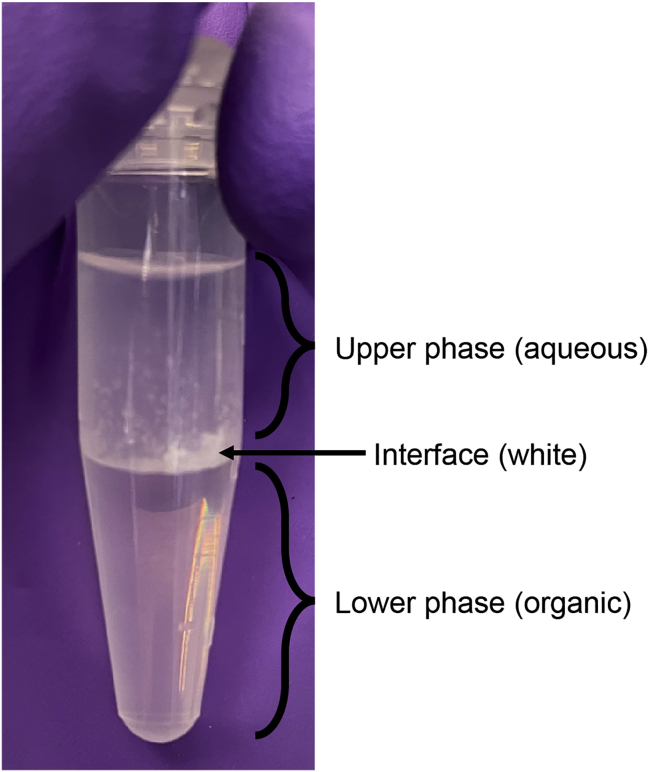
Note: Be careful not to transfer the interface (see Figure 2).
-
14.
Add 1/9 volume of 3 M sodium acetate pH 5.2, 2.5 volumes of ice cold 100% EtOH and 1 μL (20 μg) of glycogen. Vortex and incubate 12–20 h at −80°C.
Note: volume refers to the volume of upper phase collected in the previous step.
Pause point: DNA can be precipitated at −80°C for a few days if needed.
-
15.
Transfer 2 mL of each sample to 2 mL low-bind DNA tubes, centrifuge 17 min at maximum speed at 4°C. Remove the supernatant without disturbing the pellet.
-
16.
Repeat step 15 until all the material has been precipitated in the same tube.
-
17.
Wash the pellet with 500 μL of 70% EtOH, centrifuge 2 min at maximum speed at 4°C.
-
18.
Remove the supernatant and air dry the pellet.
-
19.
Resuspend the DNA in 100 μL of 10 mM Tris pH 8 and in 0.5 mL thin wall PCR tubes.
-
20.
Sonicate at amplitude 70% for 20 s ON, 40 s OFF for 12 min of total time on, using a QSonica Q700 containing cold water (ideally in a cold room).
Note: We found this sonicator optimal for DNA sonication, however equivalent bath sonicators could be used and will need adapting for the sonication time and amplitude.
Note: The bath should never get hot, change the water if necessary.
-
21.
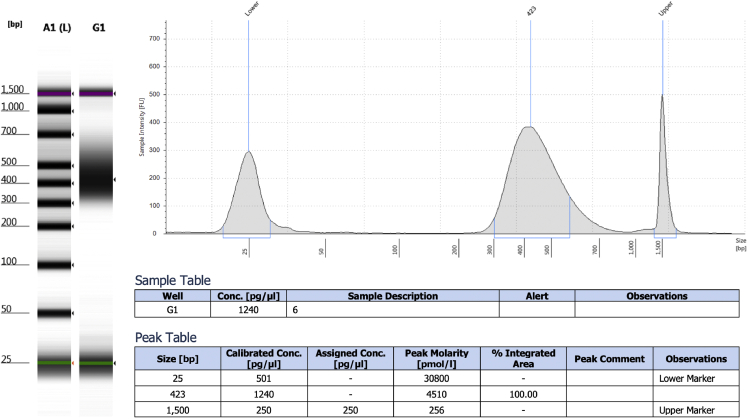
Evaluate the size of the DNA using a TapeStation D1000 or a Bioanalyzer. The DNA should be < 300 bp fragments. Troubleshooting 2 and 3.
Note: A typical TapeStation result is presented in Figure 3.
Note: Refer to Figure 3 for a typical yield of DNA given by Tape Station, please note that one can proceed with at least twice as less DNA than presented in the figure.
Pause point: DNA can be stored at −80°C for several months.
Figure 2.
Picture of a typical phenol-chloroform extraction
Figure 3.
TapeStation analyses for a typical DNA extraction performed with this protocol
Immunoprecipitation
Timing: 2 days
During this step the DNA labeled with BrdU is immunoprecipitated and purified.
-
22.
Heat denature the DNA 2 min at 95°C. Move immediately on ice and spin down the contents when cold.
Note: The BrdU antibody only works on single-stranded DNA (see antibody’s technical data sheet).
-
23.
Take 75 μL of DNA, add 45 μL of 10× IP buffer (10× PBS, 0.5% Triton-X100) and 330 μL of RNAse free H2O.
-
24.
Wash 50 μL of Protein G magnetic beads with 1× IP buffer and resuspend in 1× IP buffer.
-
25.
Add 50 μL of washed beads and 2 μL of anti-BrdU antibody to the DNA prepared in step 23. Incubate 2.5 h under rotation at 18°C–25°C in the dark.
-
26.
Place the tubes on a magnetic rack and remove the supernatant. Wash the beads with 1 mL 1× IP buffer, vortex thoroughly, place the tubes on the magnetic stand and remove the supernatant.
-
27.
Repeat the previous step for 4 more washes.
-
28.
Elute the samples by resuspending the beads in 200 μL 1× IP buffer containing 250 μg/mL proteinase K. Incubate 2 h at 55°C in the dark.
-
29.
Take the eluates off the beads, add 200 μL phenol/chloroform, vortex, centrifuge 1 min at maximum speed. Collect as much as the upper phase possible.
Note: Be careful not to transfer the interface.
-
30.
Add 1/9 volume of 3 M sodium acetate pH 5.2, 2.5 volumes of ice cold 100% EtOH and 1 μL of glycogen. Vortex and incubate o/n at −80°C.
Note: Volume refers to the volume of upper phase collected in the previous step.
Pause point: DNA can be precipitated at −80°C indefinitely.
-
31.
Centrifuge 17 min at maximum speed at 4°C. Remove the supernatant without disturbing the pellet.
-
32.
Wash the pellet with 500 μL of 70% EtOH, centrifuge 2 min at maximum speed at 4°C.
-
33.
Remove the supernatant and air dry the pellet.
-
34.
Resuspend the DNA in 75 μL of 10 mM Tris pH 8.
Note: If the pellet is difficult to resuspend the tubes can be placed 20 min at 55°C.
-
35.
Optional step: qPCR verification of IP. Set up 10 μL qPCR reactions using 0.5 μL of IP and 1% input (1% input: 0.75 μL denaturated DNA (non-IPed) diluted to 75 μL) and actin and satellite 2 primers. Troubleshooting 4 and 5.
Note: Actin replicates early so the qPCR should be prominent in early S phases while satellite 2 replicates late so should be enriched in late S phases (Black et al., 2010).
Note: Actin genomic primers forward TCCAAAGGAGACTCAGGTCAG reverse CGCCCTTTCTCACTGGTTC, satellite 2 primers forward CATCGAATGGAAATGAAAGGAGTC reverse ACCATTGGATGATTGCAGTCAA.
Note: Expected qPCR results should be 1.5%–10% input in the fractions the regions are expected to replicate.
Libraries preparation for sequencing
Timing: 1 day
During this step the DNA will be prepped to be sequenced with Illumina Nextseq 550 or related Illumina sequencer.
We used the Accel-NGS Methyl-Seq DNA Library Kit (Swift Biosciences).
Swift Biosciences- https://swiftbiosci.com/wp-content/uploads/2020/02/PRT-019-Methyl-Seq-Protocol-Rev-3.pdf
Note: This is the only library prep kit that we optimized successfully with BrdU labeled DNA.
We are annotating the main steps below as an overview, please refer to the guide for details.
-
36.
Start with 15 μL of immunoprecipitated DNA.
-
37.
Denaturation (page 9).
-
38.
Adaptase (page 9).
-
39.
Extension (page 10).
Note: To cleanup follow the instructions for < 10 ng gDNA.
-
40.
Ligation (page 10).
Note: To cleanup follow the instructions for gDNA.
-
41.
Indexing PCR (page 11).
Note: We performed 7 PCR cycles. The numbers of PCR cycles could be increased slightly if very little DNA was obtained at step 21 (∼2 cycles); however too many cycles will create PCR duplicates that will need to be removed from the sequencing results.
Note: To cleanup follow the instructions for gDNA.
-
42.
Validate the libraries using a TapeStation or a Bioanalyzer and proceed to sequencing.
Note: A typical TapeStation result is presented in Figure 4.
Note: Libraries should yield at a minimum 800 ng/mL (Qubit concentration).
Figure 4.
TapeStation analyses for a typical library performed with this protocol
Expected outcomes
Expected outcomes were annotated in the protocol. The final libraries should yield at a minimum 800 ng/mL (Qubit concentration).
Limitations
N/A.
Troubleshooting
Problem 1
Step 1. Cells are clumpy during sorting.
Potential solution
Make sure to resuspend the cells in a single cell suspension and dilute more the cells before sort (sort in a higher volume).
Problem 2
Step 21. DNA fragments above 300 bp after sonication.
Potential solution
Sonicate more.
Problem 3
Step 21. No or very little DNA observed on TapeStation or Bioanalyzer after DNA extraction.
Potential solution
Collect more cells per fraction during the sort.
Problem 4
Step 35. qPCR test of replication timing did not work: no amplification in the expected fractions.
Potential solution
Increase the labeling time with BrdU.
Increase the incubation time with the antibody.
Problem 5
Step 35. qPCR test of replication timing did not work: amplification everywhere.
Potential solution
Increase the washes after IP: vortex more thoroughly and add more washes.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Capucine Van Rechem (cvrechem@stanford.edu).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This study is supported by R01GM097360 and R35GM144131 (J.R.W.), NIH/NCI Cancer Center Support grant P30 CA006927 (J.R.W.), and the American Lung Association Lung Cancer Discovery Award. Graphical Abstract created with BioRender.com.
Author contributions
C.V.R. and J.R.W. worked together to optimize the conditions that are described in this methods article. C.V.R. conducted the methods.
Declaration of interests
In the past year, J.R.W. has consulted for Qsonica (manufacturer of Q800R system). J.R.W. has served as a consultant or advisor for Salarius Pharmaceuticals, Daiichi Sankyo, and VYNE Therapeutics. J.R.W. has also received sponsored research from Salarius Pharamceuticals. C.V.R. declares no competing interests.
Contributor Information
Johnathan R. Whetstine, Email: johnathan.whetstine@fccc.edu.
Capucine Van Rechem, Email: cvrechem@stanford.edu.
Data and code availability
The published article includes all datasets and codes generated during this study.
References
- Black J.C., Allen A., Van Rechem C., Forbes E., Longworth M., Tschop K., Rinehart C., Quiton J., Walsh R., Smallwood A., et al. Conserved antagonism between JMJD2A/KDM4A and HP1gamma during cell cycle progression. Mol. Cell. 2010;40:736–748. doi: 10.1016/j.molcel.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Van Rechem C., Ji F., Chakraborty D., Black J.C., Sadreyev R.I., Whetstine J.R. Collective regulation of chromatin modifications predicts replication timing during cell cycle. Cell Rep. 2021;37:109799. doi: 10.1016/j.celrep.2021.109799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The published article includes all datasets and codes generated during this study.