Abstract
Eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1) encodes the 4EBP1 protein, a negative regulator of mRNA translation and a substrate of the mechanistic target of rapamycin (mTOR), whose function and relevance in cancer is still under debate. Here, we analyzed EIF4EBP1 expression in different glioma patient cohorts and investigated its mode of transcriptional regulation in glioblastoma cells. We verified that EIF4EBP1 mRNA is overexpressed in malignant gliomas, including isocitrate dehydrogenase (IDH)-wildtype glioblastomas, relative to non-neoplastic brain tissue in multiple publically available datasets. Our analyses revealed that EIF4EBP1 overexpression in malignant gliomas is neither due to gene amplification nor to altered DNA methylation, but rather results from aberrant transcriptional activation by distinct transcription factors. We found seven transcription factor candidates co-expressed with EIF4EBP1 in gliomas and bound to the EIF4EBP1 promoter, as revealed by chromatin immunoprecipitation (ChIP)-sequencing data. We investigated the ability of these candidates to activate the EIF4EBP1 promoter using luciferase reporter assays, which supported four transcription factors as candidate EIF4EBP1 regulators, namely MYBL2, ETS1, HIF-1A, and E2F6. Finally, by employing transient knock-down experiments to repress either of these transcription factors, we identified MYBL2 and ETS1 as the relevant transcriptional drivers of enhanced EIF4EBP1 expression in malignant glioma cells. Taken together, our findings confirm enhanced expression of EIF4EBP1 in malignant gliomas relative to non-neoplastic brain tissue and characterize the underlying molecular pathomechanisms.
Subject terms: CNS cancer, Transcription
Introduction
Eukaryotic initiation factor 4E binding protein 1 (EIF4EBP1) encodes 4EBP1, a substrate of the nutrient-responsive hub mechanistic target of rapamycin complex 1 (mTORC1). Upon nutrient deprivation, 4EBP1 gets activated [1] and in turn inhibits mRNA translation initiation by binding the mRNA cap-binding protein eIF4E [2]. The role of 4EBP1 in cancer is still being debated, as 4EBP1 exhibits both tumor-suppressive [3–6] and pro-tumorigenic functions [7, 8], depending on the tumor types. Accordingly, the clinical relevance of EIF4EBP1 expression is strongly dependent on the tumor entity. On the one hand, loss of EIF4EBP1 and low 4EBP1 levels have been linked to poor survival of patients with head and neck squamous cell carcinoma [3] or prostate cancer [9]. On the other hand, EIF4EBP1, as part of the 8p11-12 amplicon, is frequently amplified in breast cancer [10, 11]. Furthermore, high EIF4EBP1 levels are associated with poor survival in all The Cancer Genome Atlas (TCGA) cancer entities combined [12], as well as in breast cancer [10, 11], colorectal cancer [13], hepatocellular carcinoma [14] or diffuse large B-cell lymphoma [15]. However, the prognostic relevance of EIF4EBP1 expression in other individual tumor entities is poorly established, and the mechanisms regulating EIF4EBP1 expression in distinct types of cancer warrant further investigations.
To date, only a few transcription factors have been characterized to bind the EIF4EBP1 promoter and stimulate EIF4EBP1 transcription in normal and cancer cells. These include the MYC oncoprotein [15], the androgen receptor [16], the stress response regulators ATF4 [15] and ATF5 [17], as well as HIF-1A [18]. In particular, MYC and ATF4 have been shown to co-regulate EIF4EBP1 transcription in cancer cells [15], providing one potential mechanism underlying EIF4EBP1 overexpression in cancer. The possible involvement of yet other transcription factors in regulating EIF4EBP1 expression in human cancers remains to be investigated.
Glioblastoma is the most common and most malignant primary glial tumor type of the central nervous system (CNS) that according to the World Health Organization (WHO) classification of CNS tumors corresponds to CNS WHO grade 4 [19, 20]. This tumor entity nowadays comprises only isocitrate dehydrogenase (IDH)-wildtype tumors [20], as opposed to the previous WHO classification of CNS tumors which also included IDH-mutant tumors [19]. IDH-wildtype glioblastomas are diffuse astrocytic gliomas that grow invasively in the brain parenchyma, are highly proliferative and angiogenic, and are characterized by the presence of hypoxic and necrotic regions [21]. Median survival time is only around 15 months after diagnosis [19], despite standard of care treatment [22, 23]. The initiation and progression of IDH-wildtype glioblastomas are driven by genetic alterations that inactivate tumor suppressor genes like PTEN, CDKN2A, RB1, NF1, and TP53, or activate cellular oncogenes like EGFR, PDGFRA, CDK4, MDM2, and PIK3CA [24]. In addition, epigenetic changes and alteration of transcription factor-driven gene expression contribute to glioblastoma pathogenesis [25].
Using different publically available malignant glioma datasets and chromatin immunoprecipitation (ChIP)-sequencing data, we confirmed that EIF4EBP1 mRNA expression is elevated in malignant glioma tissues, relative to non-neoplastic brain tissue, and identified seven transcription factor candidates supporting EIF4EBP1 overexpression. We showed with promoter-reporter assays and genetic knockdown experiments that among these factors, ETS1 and MYBL2 regulate EIF4EBP1 transcription in IDH-wildtype glioblastoma cells.
Results
EIF4EBP1 mRNA levels in malignant gliomas are elevated independently of gene amplification or promoter methylation
Based on a glioma dataset from TCGA database, a recent study reported on overexpression of EIF4EBP1 in glioblastoma tissue samples compared to non-neoplastic brain tissues [12]. To further delineate the expression of EIF4EBP1 in malignant gliomas including IDH-wildtype and IDH-mutant tumors, we determined the levels of EIF4EBP1 in additional publically available glioma datasets and investigated its association with common genetic alterations as well as EIF4EBP1 gene copy number alteration and promoter methylation. We confirmed and extended the reported finding [12] in six independent and non-overlapping patient datasets, namely REMBRANDT [26], SUN [27], FRENCH [28], HEGI [29], TUYSUZ [30], and DONSON [31] (a pediatric glioblastoma dataset). Thereby, we confirmed that malignant glioma tissues showed higher levels of EIF4EBP1 mRNA expression compared to non-neoplastic brain tissues in each of the analyzed cohorts (Fig. 1A and Fig. S1A, B). We then asked whether EIF4EBP1 mRNA expression is associated with common genetic and epigenetic alterations found in malignant gliomas. Specifically, we analyzed EIF4EBP1 mRNA expression levels in EGFR-amplified and EGFR-non-amplified as well as in O6-methylguanine DNA methyltransferase (MGMT) promoter-methylated and promoter-unmethylated IDH-wildtype glioblastoma patient samples using publically available datasets [32]. We found that EIF4EBP1 mRNA level is not impacted by either of these alterations (Fig. S1C, D). We also investigated the potential association of EIF4EBP1 expression with the IDH mutation status in primary glioma samples and found that EIF4EBP1 mRNA expression is not dependent on the IDH mutation status in three independent datasets (Fig. S1E–G). Among IDH-mutant gliomas, there was no difference in EIF4EBP1 expression levels in 1p/19q-codeleted oligodendrogliomas versus 1p/19q-intact astrocytomas included in the FRENCH cohort dataset [28] (Fig. S1H) or TCGA dataset [32] (Fig. S1I).
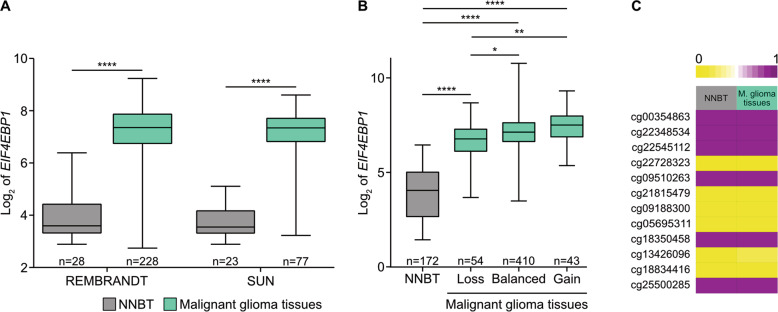
Fig. 1. Increased expression of EIF4EBP1 mRNA in malignant gliomas relative to non-neoplastic brain tissue.
A Expression levels of EIF4EBP1 in non-neoplastic brain tissue (NNBT) and glioblastoma tissues from the REMBRANDT [26] and SUN [27] cohorts. B Expression levels of EIF4EBP1 in 172 NNBT samples (BERCHTOLD [67]) and according to EIF4EBP1 copy number variation in 507 malignant gliomas of CNS WHO grade 4 of TCGA cohort [32] categorized as EIF4EBP1 copy number loss (hemizygous deletion [loss]), EIF4EBP1 balanced copy number (balanced), or EIF4EBP1 low-level copy number gain (gain). C DNA methylation levels of 12 CpG sites located within the EIF4EBP1 promoter region (hg19; Chr8: 37,886,520–37,889,020) using the datasets GSE112179 and GSE156374 for NNBT (n = 13) and GSE119774 for malignant glioma (M. glioma) tissues (n = 40) with 0 representing unmethylated and 1 representing fully methylated CpG sites. Note identical methylation patterns in normal brain tissue and the glioblastoma samples. Significance in A, B was calculated using an unpaired and two-tailed parametric t test (*p < 0.05, **p < 0.01, ****p < 0.0001).
Next, we asked whether EIF4EBP1 overexpression in malignant gliomas might be caused by EIF4EBP1 gene amplification. Analyzing the copy number status of EIF4EBP1 in 507 malignant glioma samples did not reveal any amplification of EIF4EBP1 (Fig. 1B). This observation stands in contrast to a previous report stating that EIF4EBP1 is amplified in approximately 13% of breast cancers [11]. While approximately 8.5% of TCGA malignant glioma cases analyzed here exhibited a low-level gain of EIF4EBP1 [33, 34], there was no association with higher EIF4EBP1 mRNA expression as compared to tumors without EIF4EBP1 copy number gain (Fig. 1B and Table S1). We then assessed whether EIF4EBP1 mRNA overexpression is due to differential promoter methylation in non-neoplastic brain versus malignant glioma tissues. We analyzed the DNA methylation level of 12 CpG sites within the EIF4EBP1 promoter region (hg19; Chr8: 37,886,520–37,889,020), which showed that non-neoplastic brain tissues and malignant glioma tissues exhibited a very similar methylation profile (Fig. 1C). This goes along with a previous study reporting no difference of EIF4EBP1 promoter methylation in glioma compared to control samples [35]. Based on these analyses, we can exclude EIF4EBP1 gene amplification or altered EIF4EBP1 promoter methylation as possible mechanisms driving EIF4EBP1 overexpression in malignant gliomas.
Identification of potential transcription factors driving enhanced transcription of EIF4EBP1 in malignant gliomas
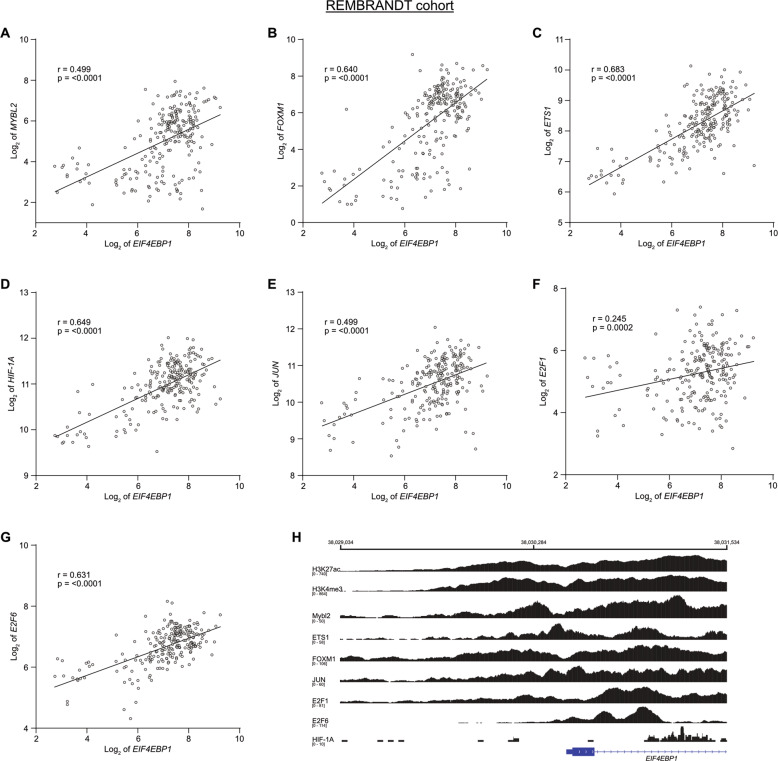
We next reasoned that the increased EIF4EBP1 mRNA expression in malignant gliomas might be driven by specific transcription factors. To identify potential transcription factor candidates, we searched for transcription factors that are positively co-expressed with EIF4EBP1 in malignant gliomas, overexpressed in these tumors as compared to non-neoplastic brain tissues, and known to bind the endogenous EIF4EBP1 promoter by ChIP. This allowed us to uncover seven transcription factors that fulfilled these criteria. We searched for transcription factors that are positively co-expressed with EIF4EBP1 in gliomas and found EIF4EBP1 mRNA expression to be significantly and positively associated with the mRNA expression levels of MYBL2, FOXM1, ETS1, HIF-1A, JUN, E2F1, and E2F6 in the REMBRANDT dataset [26] (Fig. 2A–G). These associations were validated for each of these transcription factors, excluding E2F1, in at least three additional glioma cohorts, including the SUN [27] (Fig. S2A–G), KAWAGUCHI [36], FRENCH [28], or FREIJE [37] datasets (Table S2). In support of the co-expression data, we analyzed the expression of these transcription factors in malignant glioma tissues using TCGA [32, 38] and the REMBRANDT [26] datasets, as well as non-neoplastic brain tissues [39]. This demonstrated a significant overexpression of MYBL2, FOXM1, ETS1, HIF-1A, and JUN in both glioma cohorts compared to non-neoplastic brain tissues (Fig. S3A, B). Expression of E2F1 and E2F6 was previously reported to be higher in glioblastomas (using TCGA dataset) compared to non-neoplastic brain tissues [40], which we validated in the REMBRANDT dataset [26] (Fig. S3B). Of note, the expression of these transcription factors was independent of the IDH mutation status in malignant gliomas, except for ETS1 (Fig S3C). Finally, we analyzed existing ChIP-sequencing (seq) data from the Encode consortium [41, 42], which demonstrated direct binding of FOXM1, ETS1, E2F1, and E2F6 to the EIF4EBP1 promoter region, exon 1 and intron 1 (−1500 to +1000) in various normal and cancer cells, however not including glioblastoma cells (Fig. 2H). The transcriptional regulatory region for EIF4EBP1 is not restricted to its promoter but also encompasses exon 1 and the 5’ region of intron 1, as indicated by histone H3K27 acetylation and H3K4 trimethylation signals (Fig. 2H). In addition, by using other ChIP-seq datasets [43, 44] we found signals for MYBL2 and HIF-1A binding to the EIF4EBP1 promoter (Fig. 2H). In accordance, ChIP analyses demonstrating HIF-1A binding to its putative responsive element within the EIF4EBP1 promoter segment −278 to +64 have been published [18]. Taken together, these data indicate that seven transcription factors could contribute to driving increased expression of EIF4EBP1 in malignant gliomas.
Fig. 2. Co-expression of EIF4EBP1 and EIF4EBP1 promoter binding transcription factor genes in glioblastoma tissue samples.
A–G Expression levels of EIF4EBP1 mRNA in glioblastoma patient samples plotted against the mRNA expression levels of (A) MYBL2, (B) FOXM1, (C) ETS1, (D) HIF-1A, (E) JUN, (F) E2F1 or (G) E2F6 in the REMBRANDT cohort (n = 228 patients) [26]. Co-expression levels were quantified by calculating the Pearson correlation coefficient. H ChIP peak locations within the human EIF4EBP1 promoter, exon 1 and part of intron 1 (−1500 to +1000; hg38; Chr8: 38,029,034–38,031,534) from ChIP-sequencing data for histone H3K27 acetylation (H3K27ac) and H3K4 trimethylation (H3K4me3), ETS1, FOXM1, JUN, E2F1, and E2F6 (Encode consortium, Encyclopedia of DNA Elements at UCSC; [41, 42]), HIF-1A (accession code GSE39089; name GSM955978; run SRR518265 [43]) and MYBL2 (accession code GSE119972; name GSM3389599 [44]).
E2F6, ETS1, HIF-1A, and MYBL2 induce EIF4EBP1 promoter activity
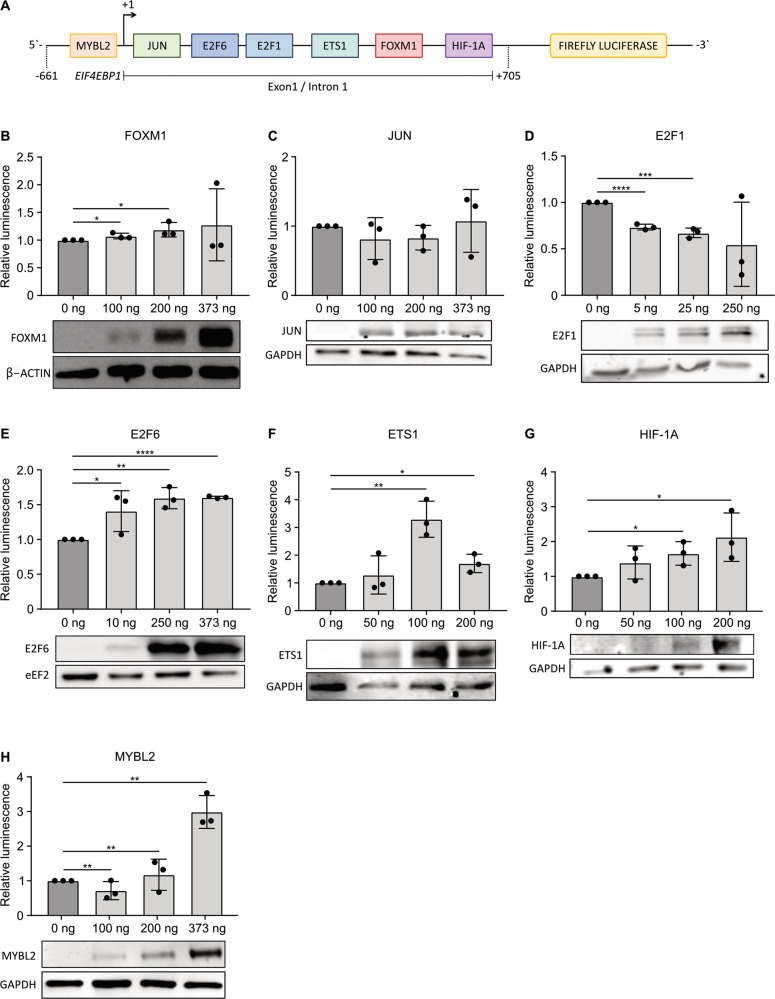
We next investigated the ability of the seven transcription factor candidates to induce EIF4EBP1 promoter activity, which was only reported for HIF-1A [18]. To assess promoter activity, we used a luciferase reporter containing the −661 to +705 EIF4EBP1 promoter region, exon 1, and part of intron 1 (Fig. 3A), as this region is predicted to be bound by the seven transcription factor candidates based on the ChIP-seq data (Fig. 2H). Overexpression of FOXM1 (Fig. 3B) or JUN (Fig. 3C) did not unequivocally induce EIF4EBP1 promoter activity. While we noticed a significant increase of luciferase activity with low (100 ng) and medium (200 ng) amounts of FOXM1, this was below 1.5-fold and therefore was not considered as biologically relevant. Unexpectedly, overexpression of E2F1, a well-characterized transcriptional activator, led to a decrease of EIF4EBP1 promoter activity in a dose-dependent manner (Fig. 3D). On the contrary, forced expression of E2F6, a known transcriptional repressor, caused induction of EIF4EBP1 promoter activity even with low E2F6 expression level (Fig. 3E). Additionally, we showed that ectopic expression of either ETS1 (Fig. 3F), HIF-1A (Fig. 3G), or MYBL2 (Fig. 3H) was able to increase EIF4EBP1 promoter activity in a dose-dependent manner. The overexpression of each transcription factor was validated by immunoblot analyses (Fig. 3B–H). These experiments proved that among the seven transcription factor candidates, E2F6, ETS1, HIF-1A, and MYBL2 were able to induce EIF4EBP1 promoter activity. Given that HIF-1A has been previously reported to stimulate EIF4EBP1 promoter activity [18], we focused on the three other transcription factor candidates for further investigation.
Fig. 3. Induction of EIF4EBP1 promoter activity by E2F6, ETS1, HIF-1A, and MYBL2.
A Scheme of the luciferase reporter construct containing the EIF4EBP1 promoter, exon 1, and part of intron 1 (−661; +705), coupled to Firefly luciferase, with the indicated binding sites of transcription factor candidates. B–H HEK293-T cells were transfected with the −661; +705 EIF4EBP1 promoter reporter construct, together with increasing amounts of plasmids expressing either one of the indicated transcription factors and a vector expressing Renilla luciferase. Luciferase activities were detected using the Dual-Luciferase Reporter Assay. Firefly luciferase activity was normalized to Renilla luciferase activity and the ratio was normalized to the corresponding 0 ng condition. Data represent the mean of three independent replicates ± standard deviation (SD). Significance was calculated using an unpaired and one-tailed parametric t-test (*p < 0.05, **p < 0.01, ***p < 0.001 ****p < 0.0001). Below each diagram, a representative immunoblot analyzing overexpression of each of the indicated transcription factors is presented.
ETS1 and MYBL2 regulate 4EBP1 mRNA and protein expression
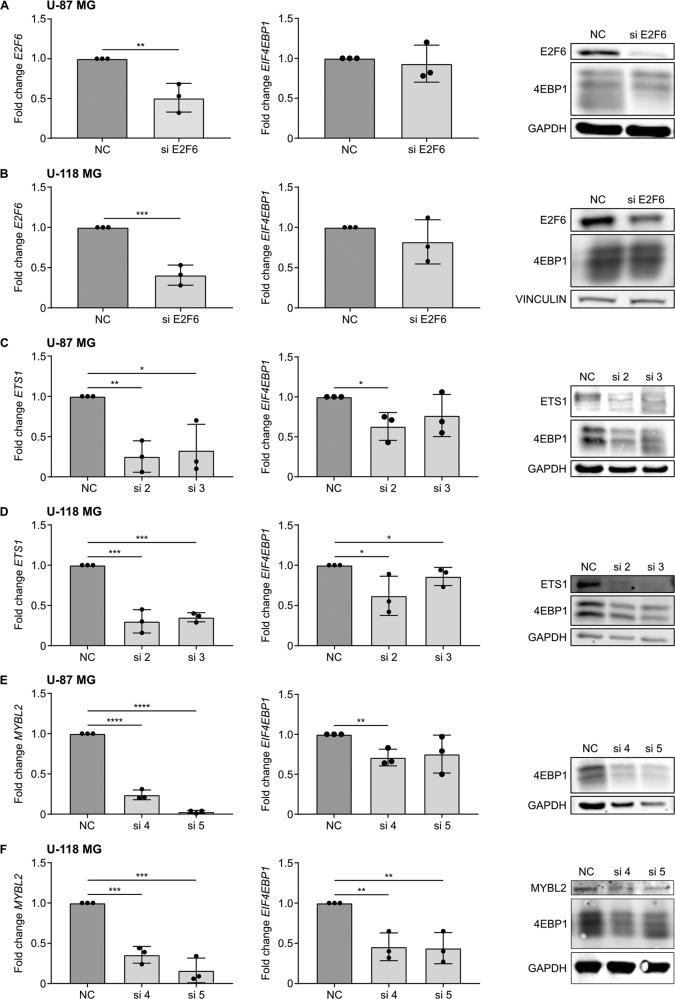
To determine whether ETS1, E2F6, and MYBL2 activate the transcription of endogenous EIF4EBP1 in glioblastoma cells, each transcription factor was transiently knocked down in U-87 MG and U-118 MG glioblastoma cell lines. At the mRNA level, we achieved at least 50% knock-down for MYBL2, ETS1, and E2F6 in both cell lines (Fig. 4A–F). This was confirmed at the protein level, as we observed a decrease of ETS1 and E2F6 in U-118 MG and U-87 MG, and of MYBL2 in U-118 MG upon knock-down. However, while the knock-down of MYBL2 in U-87 MG was strong at the mRNA level, we could not detect it at the protein level due to low endogenous MYBL2 levels in this cell line (Fig. 4E, F). We then assessed the effect of the respective transcription factor knock-downs on 4EBP1 transcript and protein levels. With the half-life of 4EBP1 being longer than 48 h [18], we transfected cells twice with siRNA over a period of 192 h to ensure that 4EBP1 protein is degraded and thus allow for observing potential changes of 4EBP1 protein levels. We observed that E2F6 knock-down in U-87 MG (Fig. 4A) and U-118 MG (Fig. 4B) had no impact on 4EBP1 mRNA and protein levels, eliminating E2F6 as a transcriptional regulator of EIF4EBP1 in these glioblastoma cell lines. In contrast, transient knock-down of either ETS1 or MYBL2 resulted in a significant decrease of 4EBP1 mRNA and protein levels in both glioblastoma cell lines (Fig. 4C–F). These results were confirmed at the protein levels, i.e., MYBL2 or ETS1 knock-down each resulted in lower 4EBP1 protein levels in both cell lines (Fig. 4C–F). Based on these results, we identified two transcription factors, ETS1 and MYBL2, that regulate EIF4EBP1 expression in glioblastoma cells.
Fig. 4. Regulation of EIF4EBP1 mRNA and protein expression by MYBL2 and ETS1 in glioblastoma cells.
A–F U-118 MG and U-87 MG glioblastoma cells were transiently transfected with negative control siRNAs (NC), and an siRNA pool targeting (A, B) E2F6 (si E2F6) or two different siRNAs each targeting either (C, D) ETS1 (si 2 and si 3) or (E, F) MYBL2 (si 4 and si 5). Cells were re-transfected after 96 h with their corresponding siRNA and incubated for a total of 192 h. MRNA and protein were harvested to determine the expression levels of EIF4EBP1/4EBP1 and (A, B) E2F6, (C, D) ETS1 or (E, F) MYBL2 by qRT-PCR and immunoblots. Data obtained by qRT-PCR represent the mean of three independent replicates ±SD and the fold change in expression was normalized to the negative control. Results of representative immunoblot are depicted on the right-hand side of the diagrams representing the qRT-PCR results. Significance was calculated using an unpaired and one-tailed parametric t-test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
EIF4EBP1 is co-expressed with MYBL2, but not with ETS1, in other non-CNS cancer types
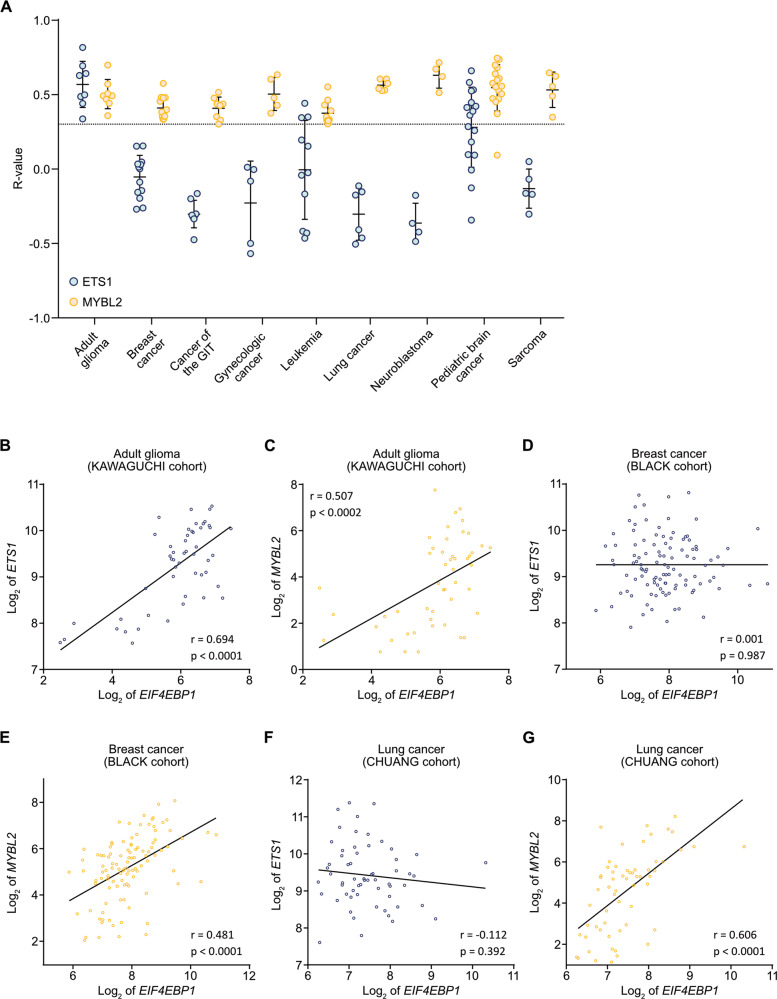
We further analyzed the potential co-expression of EIF4EBP1 and either ETS1 or MYBL2 at the mRNA level in multiple different cancer types using datasets available in R2 AMC (Table S3). These studies indicated that EIF4EBP1 expression correlates positively with MYBL2 expression in each of the analyzed tumor entities, whereas co-expression of EIF4EBP1 with ETS1 was restricted to CNS tumors (adult-type gliomas and certain pediatric brain cancers) (Fig. 5A). In particular, while we observed that both MYBL2 and ETS1 are co-expressed with EIF4EBP1 in adult-type glioma, as exemplified by the KAWAGUCHI cohort [36] (Fig. 5B, C), only MYBL2 mRNA levels, but not ETS1 mRNA levels, showed a positive correlation with EIF4EBP1 mRNA levels in non-CNS tumor entities, such as breast and lung cancers, as exemplified by the BLACK and CHUANG cohorts [45, 46], respectively (Fig. 5D–G). These analyses indicate that the co-expression between MYBL2 and EIF4EBP1 is not restricted to glioblastomas, suggesting that MYBL2 might represent a more general regulatory mechanism driving EIF4EBP1 expression in different cancer entities.
Fig. 5. Co-expression of EIF4EBP1 and MYBL2 or ETS1 in different cancer entities.
A Correlation between the mRNA expression levels of EIF4EBP1 and ETS1 (light blue dots) or MYBL2 (yellow dots) in the indicated human cancer types (Table S3). Co-expression levels were quantified by calculating the Pearson correlation coefficient. Each dot represents the R-value for one cohort. The dotted line corresponds to an R-value of 0.3, chosen as the cut-off for positive correlation. B–H Representative co-expression analysis between EIF4EBP1 mRNA and B, D, F ETS1 (light blue dots) or C, E, G MYBL2 (yellow dots) mRNA levels in the indicated tumor type and cohort. The represented cohorts are (B, C) glioma (KAWAGUCHI cohort; n = 50) [36], D, E breast cancer (BLACK cohort; n = 107) [45], and F, G lung cancer (CHUANG cohort; n = 60) cohort [46]. Co-expression levels were quantified by calculating the Pearson correlation coefficient. GIT, gastrointestinal tract.
Discussion
EIF4EBP1 gene expression and its clinical relevance in cancer are highly tumor-type specific [47]. We found that EIF4EBP1 is overexpressed in glioblastoma tissue samples in different patient cohorts as compared to non-neoplastic brain tissues, thus extending previous observations made in the TCGA cohort [12]. Elevated mRNA expression may lead to increased active 4EBP1 protein levels in glioblastoma, as it was reported that mTOR activity is reduced regionally in this tumor entity, thus leading to 4EBP1 activation in poorly vascularized areas [48]. We searched for the underlying causes of increased EIF4EBP1 mRNA expression in malignant gliomas and observed that the EIF4EBP1 gene is not amplified in glioblastomas although amplification of 8p11.23, which encompasses EIF4EBP1, has been reported in other cancer entities, such as lung squamous cell carcinoma, bladder cancer, and breast cancer, and correlated with higher EIF4EBP1 expression [49]. By bioinformatic analysis, we identified seven transcription factors that may potentially drive overexpression of EIF4EBP1 in gliomas. Each of these transcription factors harbors oncogenic or tumor-promoting functions and some of them were reported to be overexpressed in cancer, including overexpression of E2F1, E2F6 [40], FOXM1, and MYBL2 [50] in glioblastomas. Among the seven transcription factor candidates, we found that HIF-1A, E2F6, ETS1, and MYBL2 activated the EIF4EBP1 promoter in vitro while E2F1, JUN, and FOXM1 did not. Surprisingly, E2F1 a transcriptional activator repressed EIF4EBP1 promoter activity while E2F6, which is a transcriptional repressor, induced EIF4EBP1 promoter activity. Of note, E2F1 has been shown to repress transcription of YAP1 by binding to the transcription factor TEAD [51], so we cannot exclude that E2F1 may repress the endogenous EIF4EBP1 promoter. While JUN was not validated as a transcriptional regulator of EIF4EBP1 promoter with our assays, this may be explained by the absence of a consensus binding motif (5’-TGAC/GTCA-3’) [52] within the −661; +705 EIF4EBP1 promoter construct we used. Of note, the endogenous EIF4EBP1 promoter contains two JUN consensus binding motifs, which are located further upstream and downstream of the −661; +705 promoter region, suggesting that JUN is still a possible candidate that might regulate the EIF4EBP1 promoter.
By functional knockdown experiments, we uncovered that ETS1 and MYBL2 regulate the transcription of endogenous EIF4EBP1 in glioblastoma cells, highlighting novel regulators of EIF4EBP1 transcription that complement the transcription factors previously reported, including MYC [15], the androgen receptor [16], ATF4 [15], ATF5 [17], and HIF-1A [18]. Since ETS1 and MYBL2 as well as EIF4EBP1 are overexpressed in other cancer entities, for instance in colorectal cancer [12, 13, 53, 54] or breast cancer [12, 53, 55], these transcription factors might also regulate EIF4EBP1 expression in cancers outside the CNS. In support of this assumption, we found that MYBL2, but not ETS1, is co-expressed with EIF4EBP1 at the mRNA level in a variety of non-CNS cancer entities, suggesting that MYBL2 might represent a general transcriptional driver of EIF4EBP1 overexpression in human cancers while ETS1-dependent regulation of EIF4EBP1 may be more restricted to CNS tumors. The molecular mechanisms underlying MYBL2 and ETS1 overexpression in malignant gliomas are to date unknown. In the case of MYBL2, this may be due to EGFR signaling, which is frequently amplified and overexpressed in IDH-wildtype glioblastomas [56] and was reported to activate the MYBL2 promoter in association with E2F1 [57]. ETS1 activity is directly induced by the RAS/RAF/MEK/ERK pathway [53], which is overactive in a large number of IDH-wildtype glioblastomas [58] and leads to ETS1 promoter activation [53].
Given that we found EIF4EBP1 to be a target gene of the ETS1 and MYBL2 oncoproteins in malignant gliomas, 4EBP1 may possibly contribute to ETS1 and MYBL2 tumorigenic functions in these tumors. Functions of both transcription factors as well as 4EBP1 have been linked to support angiogenesis. Indeed, ETS1 is known to regulate the VEGF promoter and its transcription [59], and ETS1 expression is associated with a higher density of microvessels in tumors [60]. MYBL2 expression was reported to be induced under ischemic conditions in rat brains [61], stabilized by HIF-2α [62], and to protect cells toward hypoxia-induced apoptosis [63]. Additionally, 4EBP1 has been shown to promote the selective translation of VEGF or HIF-1A mRNAs in response to hypoxia [7]. Taken together, this raises the possibility that the induction of EIF4EBP1 expression by ETS1 and MYBL2 in glioblastoma cells may be a previously unrecognized mechanism mediating angiogenesis in this tumor type. Independently of ETS1 or MYBL2, 4EBP1 may exhibit other functions in glioblastomas. It has been reported that 4EBP1 is required for oncogenic RAS transformation of mouse embryonic fibroblasts in vitro and in vivo [64], pointing to a tumor-supporting role of 4EBP1. Thus, it is possible that 4EBP1 may also contribute to glioma tumorigenesis by supporting oncogenicity.
In summary, we elucidated molecular mechanisms of enhanced EIF4EBP1 levels in glioblastoma cells, revealing the oncogenic transcription factors ETS1 and MYBL2 as responsible transcriptional regulators.
Materials and methods
Data availability and bioinformatics analysis
We used publically available cancer datasets (Table S3) as well as glioma and non-neoplastic brain tissue datasets derived from various cohorts for correlative analyses of RNA expression data. Table S4 provides an overview of the glioma datasets that were used including accession numbers, patient numbers, original diagnoses, and information on IDH mutation status, if available. As these datasets were generated before the current WHO classification, the provided diagnoses are mostly based on histological classification only. RNA expression data were analyzed with the Gepia website [38] using the publicly available GTEx non-neoplastic brain tissue and TCGA [32] (tumor tissues) datasets or obtained from the R2 Genomic Analysis Visualization Platform (R2 AMC; http://r2.amc.nl) using the REMBRANDT [26] datasets to analyze the expression levels of EIF4EBP1, MYBL2, FOXM1, ETS1, HIF-1A, JUN, E2F1, and E2F6 in non-neoplastic brain tissue versus malignant glioma patient samples. Additionally, the expression levels of EIF4EBP1 were analyzed with R2 AMC using the SUN [27], FRENCH [28], HEGI [29], DONSON [31] (microarray platforms u133p2) and TUYSUZ [30] (microarray platform hugene21t) datasets. For co-expression analyses, the above-mentioned cohorts as well as the KAWAGUCHI [36], FREIJE [37], and PAUGH [65] cohorts were used. Expression data of IDH-wildtype glioblastoma patient samples according to the MGMT promoter methylation status were retrieved from cBioportal [33, 34] (TCGA [32]) and data related to the EGFR amplification status in IDH-wildtype glioblastomas were retrieved with R2 AMC using the FRENCH [28] cohort. Expression data according to 1p/19q codeletion were obtained for IDH-mutant CNS WHO grade 2, 3, and 4 gliomas from R2 AMC using the FRENCH [28] cohort or from https://portal.gdc.cancer.gov using TCGA datasets for lower-grade glioma and glioblastoma [32]. MRNA expression data according to IDH mutation status were analyzed using the CGGA [66], FRENCH [28], and TCGA [32] datasets for EIF4EBP1 expression and TCGA dataset [32] for the expression of the transcription factors. TCGA data were accessed using cBioportal [33, 34]. Copy number variations for EIF4EBP1 and corresponding EIF4EBP1 expression in glioma patient samples were acquired from cBioportal and R2 AMC, respectively [33, 34] (TCGA [32]) and compared to expression data of EIF4EBP1 in non-neoplastic brain tissue [67] from R2 AMC. DNA methylation data were downloaded from R2 AMC (GSE112179 [68] and GSE156374 [69] for non-neoplastic brain tissue and GSE119774 [70] for tumor tissues). CpG sites included within the −1500 to +1000 of EIF4EBP1 (human genome GRCh 38/hg38; Chr8: 38,029,034–38,031,534) were selected for analysis and the mean was determined for each group and CpG site. ChIP-seq data for H3K27ac (UCSC Accession: wgEncodeEH000030, wgEncodeEH000997, wgEncodeEH000111, wgEncodeEH000055, wgEncodeEH000043, wgEncodeEH000064, wgEncodeEH000097), H3K4me3 (wgEncodeEH000913, wgEncodeEH000909, wgEncodeEH002876, wgEncodeEH001882), ETS1 (wgEncodeEH002290; wgEncodeEH001580), FOXM1 (wgEncodeEH002529), JUN (wgEncodeEH000746, wgEncodeEH000719, wgEncodeEH002805, wgEncodeEH000620), E2F1 (wgEncodeEH000699, wgEncodeEH000688, wgEncodeEH000693) and E2F6 (wgEncodeEH000692 wgEncodeEH000676; wgEncodeEH001598) were downloaded from ENCODE (Encyclopedia of DNA Elements at UCSC; [41, 42]) using the human genome GRCh 38/hg 38, whereas NCBI Geo datasets were used to access ChIP-seq data for HIF-1A (human genome GRCh 38/hg 38; accession code GSE39089; name GSM955978; run SRR518265; [43]) and MYBL2 (human genome GRCh 37/hg 19; accession code GSE119972; name GSM3389599; [44]). Fastq files for HIF-1A were aligned to human reference genome hg38 using STAR v2.4.1d, whereas MYBL2 data were re-aligned from hg19 to hg38. ChIP seq data from ENCODE [41, 42] included data from seven cell lines. These files were combined into a single BAM file. BAM files were then visualized using IGV version 2.9.1 (https://igv.org; [71]).
Statistical analyses
Unpaired t-tests were performed when comparing gene expression in gliomas versus non-neoplastic brain tissues samples, as well as between IDH-mutant glioma groups stratified according to 1p/19q co-deletion, or IDH-wildtype glioblastoma groups stratified according to EGFR amplification and MGMT promoter methylation status. ANOVA analysis was used to determine the significance of copy number status between glioma and non-neoplastic brain tissue samples. Correlation analyses were performed by calculating Pearson correlation. GraphPad Prism version 7.04 (GraphPad Software, San Diego, CA, USA) was used for the statistical analysis.
Cell culture
HEK293-T embryonic kidney cells as well as the human glioblastoma cell lines U-118 MG and U-87 MG were originally obtained from American Type Culture Collections (ATCC). Cells were maintained in Dulbecco’s modified Eagle Medium (10569010, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (10270-106, Thermo Fisher Scientific) and 1% penicillin/streptomycin (10270-106, Sigma Aldrich, St Louis, USA) and cultured in a humidified incubator at 37 °C with 5% CO2. The cell lines were confirmed to be mycoplasma-free by Venor GeM Classic (11-1050, Minerva Biolabs, Berlin, Germany) kit and validated by STR-profiling (Genomics & Transcriptomics Labor (GTL), Heinrich Heine University, Düsseldorf, Germany).
siRNA transfection
Cells were transfected in 6-well plates at 70% confluency with 25 nM control siRNA (D-001206-14-50, Dharmacon, Cambridge, UK) or negative control siPool (siTools Biotech, Planegg, Germany) or siRNAs targeting ETS1 (D-003887-02-0010 & D-003887-03-0010, Dharmacon), MYBL2 (D-010444-04-0005 and D-010444-05-0005, Dharmacon) or E2F6 (siTools Biotech) using siLentFect transfection reagent (1703362, Biorad, Hercules, CA, USA) (see Table S5 for siRNA sequences). Briefly, a master mix containing 125 µl Opti-MEM (31985-070, Thermo Fisher Scientific) and 3 µl siLentFect was prepared and incubated for 5 min at room temperature (RT). Meanwhile, 125 µl Opti-MEM were mixed with 25 nM of siRNA for each well. The siRNA mix was mixed 1:1 with the master mix, incubated for 20 min at RT, and added dropwise onto the cells. The medium was changed the day after transfection. Cells were re-transfected after 96 h. At 192 h following the first transfection, RNA and protein were harvested for further analysis.
Plasmid construction
The promoter region of the human EIF4EBP1 gene, spanning from nucleotide −661 to +705 (human genome GRCh 38/hg38; Chr8: 38,029,873–38,031,239), was inserted into the SacI and BglII restriction sites of the Firefly Luciferase expressing pGL4.22 plasmid (E6771, Promega, Madison, WI, USA). Cloning was performed by GENEWIZ Germany GmbH (Leipzig, Germany).
Luciferase reporter assays
HEK 293-T cells were seeded in 12-well plates to reach 50% confluency on the day of transfection. Cells were transfected with 125 ng of the EIF4EBP1 promoter Firefly luciferase plasmid, 2 ng of Renilla luciferase-expressing pRL SV40 plasmid (E2231, Promega), as internal control, and 5–373 ng of either of the transcription factor expressing plasmids, completed to 500 ng total DNA with pCMV-Neo-Bam (16440, Addgene) or pcDNA3.1 (V79020, Thermo Fisher Scientific) plasmids using CalFectinTM Cell Transfection Reagent (SL100478, SignaGen Laboratories; Frederick, MD; USA) according to the manufacturer’s guidelines. The used transcription factor expressing plasmids were pcDNA3 E2F1 (kind gift from Dr. Tony Kouzarides, University of Cambridge, UK), pSG3.1 ETS1 (kindly provided by Dr. Lawrence McIntosh, University of British Columbia, Vancouver, Canada), pcDNA3 FoxM1 (kindly provided by Dr. Pradip Raychaudhuri, University of Illinois Cancer Center, Chicago, IL, USA), pcDNA3 HA-HIF-1A (gift from Dr. William Kaelin [Addgene plasmid # 18949; http://n2t.net/addgene:18949; RRID:Addgene_18949; [72]]), pcDNA3 MYBL2 (gift from Dr. Rob Lewis [Addgene plasmid # 25965; http://n2t.net/addgene:25965; RRID:Addgene_25965; [73]]), pCMV6 JUN (kind gift of Dr. Marguerite Buzza, University of Maryland, College Park, MD, USA). Cells were harvested 48 h post-transfection and the activity of Firefly and Renilla luciferases were sequentially determined using the Dual-Luciferase Reporter Assay System (E1980, Promega) and analyzed with Beckman Coulter microtiter plate reader (Beckman Coulter, Krefeld, Germany). All samples were performed in triplicate and the final luciferase quantification was formulated as the ratio of Firefly luciferase to Renilla luciferase luminescence.
RNA extraction, cDNA synthesis, and qRT-PCR
RNA was extracted using the RNeasy Plus Mini Kit (74136, QIAgen, Hilden, Germany). The extraction was performed according to the protocol provided by the manufacturer. Isolated RNA was retro-transcribed to cDNA using 1 μg of RNA per reaction with either the QuantiTect Reverse Transcription Kit (205311, QIAgen) or the High-Capacity cDNA Reverse Transcription Kit (4368813, Applied Biosystems, Waltham, MA, USA) according to the manufacturer’s protocol. Real-time PCR was performed in triplicates using 1 µl cDNA and 9 µl master mix consisting of 5 µl SYBR Green PCR Mix (4309155, Applied Biosystems), 3 µl H2O and 1 µl of forward and reverse primers (0.5 µM final concentration). PPIA, GusB, and β-actin were used as housekeepers. For primer sequences, see Table S6.
Protein extraction and immunoblot analysis
Cells were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8, 1% Triton X100, 0.5% Sodium deoxycholate, and 0.1% SDS) supplemented with proteinase inhibitor cocktail (11873580001, Roche, Basel, Switzerland) and phosphatase inhibitor (04906837001, Roche). Cell lysates were centrifuged at 14,000 × g for 15 min at 4 °C and supernatants were collected. Protein concentration was quantified using the PierceTM BCA Protein Assay Kit (23225, Thermo Fisher Scientific) according to the manufacturer’s protocol. Twenty micrograms of total protein were loaded either on a 12% polyacrylamide-SDS gel or on a NativePAGE™ 4–12%, Bis-Tris Gels (NP0336BOX, Thermo Fisher Scientific) and transferred to a 0.2 µm nitrocellulose membrane (No10600001, GE Healthcare; Chicago, IL, USA). Membranes were blocked with 5% bovine serum albumin (BSA) (8076.3, Carl Roth, Karlsruhe, Germany) TBS-Tween (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Tween 20) and probed with primary antibodies (as detailed in table S7) diluted 1:1000 in 5% BSA TBS overnight at 4 °C if not stated otherwise. Membranes were then incubated with a corresponding anti-mouse (926-32210, Li-Cor, Bad Homburg, Germany) or anti-rabbit (926-32211, Li-Cor) fluorescent secondary antibody diluted 1:10,000. The fluorescent signal was visualized with the LI-COR Odyssey® CLx system (Li-Cor).
Statistical analysis of experimental data
All experiments were carried out in three biological replicates. Data are represented as mean +/− standard deviation (SD). A one- or two-sided Student’s t-test was used to compare differences between control and experimental groups. Results were considered as being statistically significant at p < 0.05. Statistical tests were calculated with GraphPad Prism version 7.04.
Supplementary information
Acknowledgements
The results published here are in part based on data generated by TCGA Research Network (https://www.cancer.gov/tcga). We would like to acknowledge the ENCODE Consortium and the Bernstein, Farnham, Myers, and Snyder laboratories for generating the respective datasets.
Author contributions
Conception and design: LH, GL. Experimental procedures: LH. Data analysis and interpretation: LH, DP, JM, GL. Critical review and discussion: BR, MR, TG, GR. Manuscript writing: LH, GR, GL. All authors read and approved the final manuscript.
Funding
GL was supported by grants from the Elterninitiative Düsseldorf e.V. (grant no. 701910003), the Research Commission of the Medical Faculty, Heinrich Heine University Düsseldorf (grant no. 2016-056), the Deutsche Forschungsgemeinschaft (grant no. LE 3751/2-1), the German Cancer Aid (grant no. 70112624), and the Dr. Rolf M. Schwiete Stiftung (grant no. 2020-018). LH was funded by the Dr. Rolf M. Schwiete Stiftung (grant no. 2020-018). GR and MR were supported by a joint grant from the Deutsche Forschungsgemeinschaft (grant nos. RE 938 4-1 and RE 2857/2-1). The laboratory of TGPG was supported by the German Cancer Aid (Max-Eder program, 70112257), the Matthias-Lackas foundation, the Dr. Leopold and Carmen Ellinger foundation, and the Barbara and Wilfried Mohr foundation. JM was supported by a scholarship of the Kind-Philipp foundation and the Rudolf und Brigitte Zenner foundation. BR was supported by the Israel Science Foundation (grant no. 1436/19). Open Access funding enabled and organized by Projekt DEAL.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41420-022-00883-z.
References
- 1.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–76.. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–9. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z, Feng X, Molinolo AA, Martin D, Vitale-Cross L, Nohata N, et al. 4E-BP1 is a tumor suppressor protein reactivated by mTOR inhibition in head and neck cancer. Cancer Res. 2019;79:1438–50.. doi: 10.1158/0008-5472.CAN-18-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding M, Van der Kwast TH, Vellanki RN, Foltz WD, McKee TD, Sonenberg N, et al. The mTOR targets 4E-BP1/2 restrain tumor growth and promote hypoxia tolerance in PTEN-driven prostate cancer. Mol Cancer Res. 2018;16:682–95.. doi: 10.1158/1541-7786.MCR-17-0696. [DOI] [PubMed] [Google Scholar]
- 5.Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–6. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, et al. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol Cell. 2007;28:501–12. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 8.Dubois L, Magagnin MG, Cleven AH, Weppler SA, Grenacher B, Landuyt W, et al. Inhibition of 4E-BP1 sensitizes U87 glioblastoma xenograft tumors to irradiation by decreasing hypoxia tolerance. Int J Radiat Oncol Biol Phys. 2009;73:1219–27. doi: 10.1016/j.ijrobp.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Graff JR, Konicek BW, Lynch RL, Dumstorf CA, Dowless MS, McNulty AM, et al. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer Res. 2009;69:3866–73. doi: 10.1158/0008-5472.CAN-08-3472. [DOI] [PubMed] [Google Scholar]
- 10.Karlsson E, Waltersson MA, Bostner J, Perez-Tenorio G, Olsson B, Hallbeck AL, et al. High-resolution genomic analysis of the 11q13 amplicon in breast cancers identifies synergy with 8p12 amplification, involving the mTOR targets S6K2 and 4EBP1. Genes Chromosomes Cancer. 2011;50:775–87. doi: 10.1002/gcc.20900. [DOI] [PubMed] [Google Scholar]
- 11.Rutkovsky AC, Yeh ES, Guest ST, Findlay VJ, Muise-Helmericks RC, Armeson K, et al. Eukaryotic initiation factor 4E-binding protein as an oncogene in breast cancer. BMC Cancer. 2019;19:491. doi: 10.1186/s12885-019-5667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu S, Wagner G. Deep computational analysis details dysregulation of eukaryotic translation initiation complex eIF4F in human cancers. Cell Syst. 2021;12:907.e6–23.e6. doi: 10.1016/j.cels.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao MW, Wang LT, Lai CY, Yang XM, Cheng YW, Lee KH, et al. eIF4E binding protein 1 expression is associated with clinical survival outcomes in colorectal cancer. Oncotarget. 2015;6:24092–104. doi: 10.18632/oncotarget.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cha YL, Li PD, Yuan LJ, Zhang MY, Zhang YJ, Rao HL, et al. EIF4EBP1 overexpression is associated with poor survival and disease progression in patients with hepatocellular carcinoma. PLoS ONE. 2015;10:e0117493. doi: 10.1371/journal.pone.0117493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tameire F, Verginadis II, Leli NM, Polte C, Conn CS, Ojha R, et al. ATF4 couples MYC-dependent translational activity to bioenergetic demands during tumour progression. Nat Cell Biol. 2019;21:889–99.. doi: 10.1038/s41556-019-0347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Horn JL, Banda K, Goodman AZ, Lim Y, Jana S, et al. The androgen receptor regulates a druggable translational regulon in advanced prostate cancer. Sci Transl Med. 2019;11:eaaw4993.. doi: 10.1126/scitranslmed.aaw4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juliana CA, Yang J, Rozo AV, Good A, Groff DN, Wang SZ, et al. ATF5 regulates beta-cell survival during stress. Proc Natl Acad Sci USA. 2017;114:1341–6. doi: 10.1073/pnas.1620705114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azar R, Lasfargues C, Bousquet C, Pyronnet S. Contribution of HIF-1alpha in 4E-BP1 gene expression. Mol Cancer Res. 2013;11:54–61. doi: 10.1158/1541-7786.MCR-12-0095. [DOI] [PubMed] [Google Scholar]
- 19.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–20. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 20.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23:1231–51. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 22.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 23.Weller M, van den Bent M, Preusser M, Le Rhun E, Tonn JC, Minniti G, et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol. 2021;18:170–86.. doi: 10.1038/s41571-020-00447-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agnihotri S, Burrell KE, Wolf A, Jalali S, Hawkins C, Rutka JT, et al. Glioblastoma, a brief review of history, molecular genetics, animal models and novel therapeutic strategies. Arch Immunol Ther Exp. 2013;61:25–41. doi: 10.1007/s00005-012-0203-0. [DOI] [PubMed] [Google Scholar]
- 25.Bai H, Harmanci AS, Erson-Omay EZ, Li J, Coskun S, Simon M, et al. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet. 2016;48:59–66. doi: 10.1038/ng.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gusev Y, Bhuvaneshwar K, Song L, Zenklusen JC, Fine H, Madhavan S. The REMBRANDT study, a large collection of genomic data from brain cancer patients. Sci Data. 2018;5:180158. doi: 10.1038/sdata.2018.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Gravendeel LA, Kouwenhoven MC, Gevaert O, de Rooi JJ, Stubbs AP, Duijm JE, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–72. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 29.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–24. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 30.Gulluoglu S, Tuysuz EC, Sahin M, Kuskucu A, Kaan Yaltirik C, Ture U, et al. Simultaneous miRNA and mRNA transcriptome profiling of glioblastoma samples reveals a novel set of OncomiR candidates and their target genes. Brain Res. 2018;1700:199–210. doi: 10.1016/j.brainres.2018.08.035. [DOI] [PubMed] [Google Scholar]
- 31.Griesinger AM, Birks DK, Donson AM, Amani V, Hoffman LM, Waziri A, et al. Characterization of distinct immunophenotypes across pediatric brain tumor types. J Immunol. 2013;191:4880–8. doi: 10.4049/jimmunol.1301966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Court F, Le Boiteux E, Fogli A, Muller-Barthelemy M, Vaurs-Barriere C, Chautard E, et al. Transcriptional alterations in glioma result primarily from DNA methylation-independent mechanisms. Genome Res. 2019;29:1605–21.. doi: 10.1101/gr.249219.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawaguchi A, Yajima N, Tsuchiya N, Homma J, Sano M, Natsumeda M, et al. Gene expression signature-based prognostic risk score in patients with glioblastoma. Cancer Sci. 2013;104:1205–10. doi: 10.1111/cas.12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–10. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 38.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.GT Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao P, Han S, Qu H. Expression, prognosis, and immune infiltrates analyses of E2Fs in human brain and CNS cancer. Biomed Res Int. 2020;2020:6281635. doi: 10.1155/2020/6281635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:D794–801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mimura I, Nangaku M, Kanki Y, Tsutsumi S, Inoue T, Kohro T, et al. Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol Cell Biol. 2012;32:3018–32. doi: 10.1128/MCB.06643-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Musa J, Cidre-Aranaz F, Aynaud MM, Orth MF, Knott MML, Mirabeau O, et al. Cooperation of cancer drivers with regulatory germline variants shapes clinical outcomes. Nat Commun. 2019;10:4128. doi: 10.1038/s41467-019-12071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caldon CE, Sergio CM, Kang J, Muthukaruppan A, Boersma MN, Stone A, et al. Cyclin E2 overexpression is associated with endocrine resistance but not insensitivity to CDK2 inhibition in human breast cancer cells. Mol Cancer Ther. 2012;11:1488–99. doi: 10.1158/1535-7163.MCT-11-0963. [DOI] [PubMed] [Google Scholar]
- 46.Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, Lin CW, et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomark Prev. 2010;19:2590–7. doi: 10.1158/1055-9965.EPI-10-0332. [DOI] [PubMed] [Google Scholar]
- 47.Musa J, Orth MF, Dallmayer M, Baldauf M, Pardo C, Rotblat B, et al. Eukaryotic initiation factor 4E-binding protein 1 (4E-BP1): a master regulator of mRNA translation involved in tumorigenesis. Oncogene. 2016;35:4675–88. doi: 10.1038/onc.2015.515. [DOI] [PubMed] [Google Scholar]
- 48.Kumar S, Sharife H, Kreisel T, Mogilevsky M, Bar-Lev L, Grunewald M, et al. Intra-tumoral metabolic zonation and resultant phenotypic diversification are dictated by blood vessel proximity. Cell Metab. 2019;30:201.e6–11.e6. doi: 10.1016/j.cmet.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 49.Voutsadakis IA. 8p11.23 Amplification in breast cancer: molecular characteristics, prognosis and targeted therapy. J Clin Med. 2020;9:3079. [DOI] [PMC free article] [PubMed]
- 50.Zhang X, Lv QL, Huang YT, Zhang LH, Zhou HH. Akt/FoxM1 signaling pathway-mediated upregulation of MYBL2 promotes progression of human glioma. J Exp Clin Cancer Res. 2017;36:105. doi: 10.1186/s13046-017-0573-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang P, Pei C, Wang X, Xiang J, Sun BF, Cheng Y, et al. A balance of Yki/Sd activator and E2F1/Sd repressor complexes controls cell survival and affects organ size. Dev Cell. 2017;43:603–17. doi: 10.1016/j.devcel.2017.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li M, Ge Q, Wang W, Wang J, Lu Z. c-Jun binding site identification in K562 cells. J Genet Genomics. 2011;38:235–42. doi: 10.1016/j.jgg.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 53.Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol. 2015;35:20–38. doi: 10.1016/j.semcancer.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 54.Ren F, Wang L, Shen X, Xiao X, Liu Z, Wei P, et al. MYBL2 is an independent prognostic marker that has tumor-promoting functions in colorectal cancer. Am J Cancer Res. 2015;5:1542–52. [PMC free article] [PubMed] [Google Scholar]
- 55.Thorner AR, Hoadley KA, Parker JS, Winkel S, Millikan RC, Perou CM. In vitro and in vivo analysis of B-Myb in basal-like breast cancer. Oncogene. 2009;28:742–51. doi: 10.1038/onc.2008.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanada N, Lo HW, Day CP, Pan Y, Nakajima Y, Hung MC. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol Carcinog. 2006;45:10–7. doi: 10.1002/mc.20147. [DOI] [PubMed] [Google Scholar]
- 58.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–63. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hashiya N, Jo N, Aoki M, Matsumoto K, Nakamura T, Sato Y, et al. In vivo evidence of angiogenesis induced by transcription factor Ets-1: Ets-1 is located upstream of angiogenesis cascade. Circulation. 2004;109:3035–41. doi: 10.1161/01.CIR.0000130643.41587.DB. [DOI] [PubMed] [Google Scholar]
- 60.Arderiu G, Pena E, Aledo R, Espinosa S, Badimon L. Ets-1 transcription is required in tissue factor driven microvessel formation and stabilization. Angiogenesis. 2012;15:657–69. doi: 10.1007/s10456-012-9293-x. [DOI] [PubMed] [Google Scholar]
- 61.Iyirhiaro GO, Zhang Y, Estey C, O’Hare MJ, Safarpour F, Parsanejad M, et al. Regulation of ischemic neuronal death by E2F4-p130 protein complexes. J Biol Chem. 2014;289:18202–13.. doi: 10.1074/jbc.M114.574145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okumura F, Joo-Okumura A, Nakatsukasa K, Kamura T. Hypoxia-inducible factor-2alpha stabilizes the von Hippel-Lindau (VHL) disease suppressor, Myb-related protein 2. PLoS ONE. 2017;12:e0175593. doi: 10.1371/journal.pone.0175593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shao M, Ren Z, Zhang R. MYBL2 protects against H9c2 injury induced by hypoxia via AKT and NFkappaB pathways. Mol Med Rep. 2018;17:4832–8. doi: 10.3892/mmr.2018.8387. [DOI] [PubMed] [Google Scholar]
- 64.Petroulakis E, Parsyan A, Dowling RJ, LeBacquer O, Martineau Y, Bidinosti M, et al. p53-dependent translational control of senescence and transformation via 4E-BPs. Cancer Cell. 2009;16:439–46. doi: 10.1016/j.ccr.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 65.Paugh BS, Broniscer A, Qu C, Miller CP, Zhang J, Tatevossian RG, et al. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol. 2011;29:3999–4006. doi: 10.1200/JCO.2011.35.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Z, Zhang KN, Wang Q, Li G, Zeng F, Zhang Y, et al. Chinese Glioma Genome Atlas (CGGA): a comprehensive resource with functional genomic data from Chinese glioma patients. Genomics Proteomics Bioinformatics. 2021;19:1–12. doi: 10.1016/j.gpb.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci USA. 2008;105:15605–10. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pai S, Li P, Killinger B, Marshall L, Jia P, Liao J, et al. Differential methylation of enhancer at IGF2 is associated with abnormal dopamine synthesis in major psychosis. Nat Commun. 2019;10:2046. doi: 10.1038/s41467-019-09786-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kobow K, Jabari S, Pieper T, Kudernatsch M, Polster T, Woermann FG, et al. Mosaic trisomy of chromosome 1q in human brain tissue associates with unilateral polymicrogyria, very early-onset focal epilepsy, and severe developmental delay. Acta Neuropathol. 2020;140:881–91.. doi: 10.1007/s00401-020-02228-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mack SC, Singh I, Wang X, Hirsch R, Wu Q, Villagomez R, et al. Chromatin landscapes reveal developmentally encoded transcriptional states that define human glioblastoma. J Exp Med. 2019;216:1071–90.. doi: 10.1084/jem.20190196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–46. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 73.Johnson TK, Schweppe RE, Septer J, Lewis RE. Phosphorylation of B-Myb regulates its transactivation potential and DNA binding. J Biol Chem. 1999;274:36741–9. doi: 10.1074/jbc.274.51.36741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We used publically available cancer datasets (Table S3) as well as glioma and non-neoplastic brain tissue datasets derived from various cohorts for correlative analyses of RNA expression data. Table S4 provides an overview of the glioma datasets that were used including accession numbers, patient numbers, original diagnoses, and information on IDH mutation status, if available. As these datasets were generated before the current WHO classification, the provided diagnoses are mostly based on histological classification only. RNA expression data were analyzed with the Gepia website [38] using the publicly available GTEx non-neoplastic brain tissue and TCGA [32] (tumor tissues) datasets or obtained from the R2 Genomic Analysis Visualization Platform (R2 AMC; http://r2.amc.nl) using the REMBRANDT [26] datasets to analyze the expression levels of EIF4EBP1, MYBL2, FOXM1, ETS1, HIF-1A, JUN, E2F1, and E2F6 in non-neoplastic brain tissue versus malignant glioma patient samples. Additionally, the expression levels of EIF4EBP1 were analyzed with R2 AMC using the SUN [27], FRENCH [28], HEGI [29], DONSON [31] (microarray platforms u133p2) and TUYSUZ [30] (microarray platform hugene21t) datasets. For co-expression analyses, the above-mentioned cohorts as well as the KAWAGUCHI [36], FREIJE [37], and PAUGH [65] cohorts were used. Expression data of IDH-wildtype glioblastoma patient samples according to the MGMT promoter methylation status were retrieved from cBioportal [33, 34] (TCGA [32]) and data related to the EGFR amplification status in IDH-wildtype glioblastomas were retrieved with R2 AMC using the FRENCH [28] cohort. Expression data according to 1p/19q codeletion were obtained for IDH-mutant CNS WHO grade 2, 3, and 4 gliomas from R2 AMC using the FRENCH [28] cohort or from https://portal.gdc.cancer.gov using TCGA datasets for lower-grade glioma and glioblastoma [32]. MRNA expression data according to IDH mutation status were analyzed using the CGGA [66], FRENCH [28], and TCGA [32] datasets for EIF4EBP1 expression and TCGA dataset [32] for the expression of the transcription factors. TCGA data were accessed using cBioportal [33, 34]. Copy number variations for EIF4EBP1 and corresponding EIF4EBP1 expression in glioma patient samples were acquired from cBioportal and R2 AMC, respectively [33, 34] (TCGA [32]) and compared to expression data of EIF4EBP1 in non-neoplastic brain tissue [67] from R2 AMC. DNA methylation data were downloaded from R2 AMC (GSE112179 [68] and GSE156374 [69] for non-neoplastic brain tissue and GSE119774 [70] for tumor tissues). CpG sites included within the −1500 to +1000 of EIF4EBP1 (human genome GRCh 38/hg38; Chr8: 38,029,034–38,031,534) were selected for analysis and the mean was determined for each group and CpG site. ChIP-seq data for H3K27ac (UCSC Accession: wgEncodeEH000030, wgEncodeEH000997, wgEncodeEH000111, wgEncodeEH000055, wgEncodeEH000043, wgEncodeEH000064, wgEncodeEH000097), H3K4me3 (wgEncodeEH000913, wgEncodeEH000909, wgEncodeEH002876, wgEncodeEH001882), ETS1 (wgEncodeEH002290; wgEncodeEH001580), FOXM1 (wgEncodeEH002529), JUN (wgEncodeEH000746, wgEncodeEH000719, wgEncodeEH002805, wgEncodeEH000620), E2F1 (wgEncodeEH000699, wgEncodeEH000688, wgEncodeEH000693) and E2F6 (wgEncodeEH000692 wgEncodeEH000676; wgEncodeEH001598) were downloaded from ENCODE (Encyclopedia of DNA Elements at UCSC; [41, 42]) using the human genome GRCh 38/hg 38, whereas NCBI Geo datasets were used to access ChIP-seq data for HIF-1A (human genome GRCh 38/hg 38; accession code GSE39089; name GSM955978; run SRR518265; [43]) and MYBL2 (human genome GRCh 37/hg 19; accession code GSE119972; name GSM3389599; [44]). Fastq files for HIF-1A were aligned to human reference genome hg38 using STAR v2.4.1d, whereas MYBL2 data were re-aligned from hg19 to hg38. ChIP seq data from ENCODE [41, 42] included data from seven cell lines. These files were combined into a single BAM file. BAM files were then visualized using IGV version 2.9.1 (https://igv.org; [71]).
The data that support the findings of this study are available from the corresponding author upon reasonable request.