Abstract
The walls deposited by growing pollen tubes contain two types of β-glucan, the (1,3)-β-glucan callose and the (1,4)-β-glucan cellulose, as well as various α-linked pectic polysaccharides. Pollen tubes of Nicotiana alata Link et Otto, an ornamental tobacco, were therefore used to identify genes potentially encoding catalytic subunits of the callose synthase and cellulose synthase enzymes. Reverse transcriptase-polymerase chain reactions (RT-PCR) with pollen-tube RNA and primers designed to conserved regions of bacterial and plant cellulose synthase (CesA) genes amplified a fragment that corresponded to an abundantly expressed cellulose-synthase-like gene named NaCslD1. A fragment from a true CesA gene (NaCesA1) was also amplified, but corresponding cDNAs could not be identified in a pollen-tube library, consistent with the very low level of expression of the NaCesA1 gene. RT-PCR with pollen-tube RNA and primers designed to regions conserved between the fungal FKS genes [that encode (1,3)-β-glucan synthases] and their presumed plant homologs (the Gsl or glucan-synthase-like genes) amplified a fragment that corresponded to an abundantly expressed gene named NaGsl1. A second Gsl gene detected by RT-PCR (NaGsl2) was expressed at low levels in immature floral organs. The structure of full-length cDNAs of NaCslD1, NaCesA1, and NaGsl1 are presented. Both NaCslD1 and NaGsl1 are predominantly expressed in the male gametophyte (developing and mature pollen and growing pollen tubes), and we propose that they encode the catalytic subunits of two β-glucan synthases involved in pollen-tube wall synthesis. Different β-glucans deposited in one cell type may therefore be synthesized by enzymes from different gene families.
Cell walls are primary determinants of plant growth and an important determinant of resistance to pathogens (Bacic et al., 1988). Polysaccharides are the major components of these walls, but the processes by which wall polysaccharides are synthesized and then assembled into a growing wall are not well known. In recent years, genes that may encode some of the polysaccharide synthase enzymes have been identified in the genomes of Arabidopsis and other plants, often using similarity to bacterial or fungal gene sequences. We are now faced with the task of determining the functions in plant wall synthesis of these candidate genes.
Cellulose synthase (CelS; EC 2.4.1.12) assembles the (1,4)-β-glucan backbone of cellulose. The first genes proposed to encode plant CelS, now named GhCesA1 and GhCesA2, were identified using a plant cell highly enriched in cellulose, the cotton fiber (Pear et al., 1996; Delmer, 1999). Plant and bacterial CesA genes share small, highly conserved regions such as the D,D,D,QXXRW motif that may be important for substrate binding and catalysis in the family 2 β-glycosyl transferases (Saxena et al., 1995; Campbell et al., 1997). The cotton CesA protein is predicted to be a membrane-bound protein of approximately 110 kD with eight transmembrane helices, two at the N terminus and six at the C terminus, plus a central cytoplasmic domain containing the catalytic site. Genetic complementation of cellulose-deficient mutants in Arabidopsis has shown that the CesA genes encode CelS enzymes (Arioli et al., 1998; Taylor et al., 1999).
A relatively large number of Arabidopsis and other plant sequences are related to CesA, and this set of genes has been divided into the “true” CesA family plus six distinct groups of cesA-like (Csl) genes labeled Csl A, B, C, D, E, and G (Holland et al., 2000; Richmond and Somerville, 2000). To date, no definite function has been assigned to any Csl gene, although they all contain the D,D,D,QXXRW motif and thus probably encode β-glycosyl transferases. The CesA and Csl genes might all encode CelS enzymes that form cellulose in the different cell types and developmental stages in plants, or alternatively some may encode other β-glycosyltransferases that synthesize the backbone of polymers such as xyloglucans, mannans, xylans, mixed-linkage glucans, and callose (Carpita and Vergara, 1998; Delmer, 1999; Richmond and Somerville, 2000). It is interesting that, although most bacterial genes related to the plant CesA/Csl gene superfamily encode (1,4)-β-glucan synthases, the Agrobacterium CrdS gene is involved in (1,3)-β-glucan (callose) synthesis (Stasinopoulos et al., 1999). Expression profiles of members of the CesA/Csl superfamily in plants are currently being determined in an effort to gain insight into the functions of these genes (Holland et al., 2000).
Callose synthase (CalS; EC 2.4.1.34) assembles the (1,3)-β-glucan backbone of callose. Callose is deposited at a range of locations during plant development, as well as in response to wounding. A number of unpublished reports describe plant genes with sequence similarity to the fungal (1,3)-β-glucan synthase FKS genes (Douglas et al., 1994) and have proposed that these genes encode CalS in cotton fibers (Cui et al., 1999), suspension-cultured Lolium endosperm cells (Wardak et al., 1999), Hieracium ovules (Paech et al., 1999), Nicotiana pollen tubes (Doblin et al., 2000), Arabidopsis flowers (Østergaard et al., 2000), and cultured tobacco cells (Hong et al., 2000). The encoded enzymes are very large with the cotton CFL cDNA, for example, predicted to encode a 1,899 amino acid polypeptide with a molecular mass of 219 kD (Cui et al., 1999). The Arabidopsis genome contains 12 genes with similarity to FKS, and these are provisionally known as glucan-synthase-like (Gsl) genes (http://cellwall.stanford.edu/).
Pollen tubes, the male gametophyte generation of flowering plants, provide an excellent system in which to study the synthesis of these wall glucans. Pollen tubes are formed when pollen grains germinate, normally on the receptive surface of the female stigma, and then grow extracellularly through the stylar tissue to the embryo sac (Derksen et al., 1995). Sufficient pollen of Nicotiana alata (ornamental tobacco) can be collected to grow relatively large amounts of tubes in liquid culture with a morphology and wall structure similar to those of tubes growing through compatible stylar tissue (Read et al., 1993a, 1993b; Li et al., 1999). Whereas the wall polysaccharides of N. alata pollen grains are typical of those of most somatic plant cells and include cellulose, xyloglucan, and pectins, the major component of the pollen-tube wall is (1,3)-β-glucan (callose). Callose constitutes 86% by weight of pollen-tube wall carbohydrate with the other constituents being (1,4)-β-glucan (cellulose, 5%) and two pectins, a neutral, linear (1,5)-α-l-arabinan (4%) and an acidic (1,4)-α-d-galacturonan (5%) (Li et al., 1999). The tubes grow by tip extension, producing an outer wall layer that is believed to contain solely arabinan and galacturonan. Cellulose is deposited 5 to 15 μm behind the growing tube tip, then this inner secondary wall layer is thickened from approximately 30 μm behind the tip with callose (Ferguson et al., 1998). This spatial separation of cellulose and callose deposition in pollen tubes is consistent with the existence of two different glucan synthase complexes in agreement with the separate CelS and CalS enzyme complexes reported from somatic cells (Kudlicka and Brown, 1997).
Membrane preparations from in vitro-grown pollen tubes of N. alata contain a novel and highly active CalS enzyme that does not require Ca2+ for activity. This enzyme is developmentally regulated with activity appearing 1 to 2 h after grain hydration and displays activation kinetics consistent with initial synthesis as an inactive zymogen (Schlüpmann et al., 1993, 1994; Li et al., 1997, 1999). The pollen-tube CalS is thus different from the CalS activity of wounded somatic cells that has been suggested to be an altered form of CelS (Delmer, 1999). As for most other plant cells, effectively no CelS activity can be detected in disrupted pollen tubes (Schlüpmann et al., 1993, 1994).
In the work reported here, we use the unique characteristics of the pollen-tube system as a means to allocate functions to the different families of β-glucan synthase genes. A reverse transcriptase (RT)-PCR approach using known bacterial, yeast, and plant sequences was used to target (1,4)-β- and (1,3)-β-glucan synthase genes expressed in Nicotiana pollen tubes, and these were then related to the various genes described in Arabidopsis. We report that Nicotiana pollen tubes do not express any “true” CesA genes and instead express a member of the related CslD family (NaCslD1). We also report that a member of the Gsl gene family (NaGsl1) is abundantly expressed in pollen tubes.
RESULTS
Isolation of a CslD Gene Expressed in Pollen Tubes
An RT-PCR approach was used to clone the CesA/Csl genes expressed in N. alata pollen tubes. A consensus sequence surrounding the conserved residues of the D,D,D,QXXRW motif was identified by aligning the CesA genes of Acetobacter xylinum and Agrobacterium tumefaciens (known to encode (1,4)-β-glucan synthases) with several rice and Arabidopsis CesA expressed sequence tags (EST). The degenerate primers UGF and DOMB1R were then designed to recognize the sequence context of the first two D residues. Using these primers, an approximately 700-bp fragment was amplified by RT-PCR from pollen-tube RNA (Fig. 1, lane 2). This fragment was approximately 150-bp larger than the bands amplified using these primers on D47622, a rice CesA EST (Fig. 1, lane 1), or on N. alata leaf cDNA (Fig. 1, lane 3), indicating additional sequences in the pollen-tube-expressed gene. The 700-bp fragment was isolated and cloned, and the inserts of 21 independent clones were sequenced. Six of these had identical sequences, including matches to the back-translated amino acid sequence to which the primers were designed; other clones were not derived from genes encoding target sequences. Comparison of the six identical sequences to databases and to sequences collated and categorized by the Somerville laboratory (http://cellwall.stanford.edu; Richmond and Somerville, 2000) indicated that the gene they represent was most similar to the cellulose-synthase-like CslD gene family rather than to the CesA gene family. The UGF and DOMB1R primers were therefore suitable for amplification of this region in the CslD genes as well as the CesA genes to which they were designed.
Figure 1.
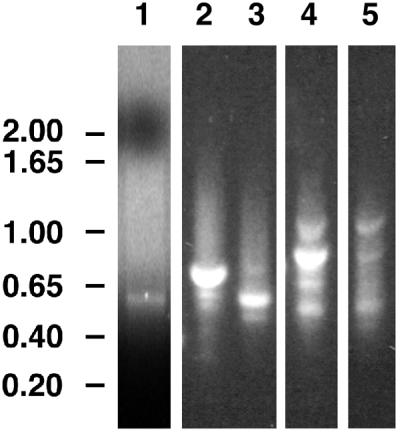
RT-PCR amplification of fragments encoding putative β-glucan synthases. Lanes 1 through 3 show products of reactions containing primers UGF and DOMB1R for (1,4)-β-glucan synthases. Lanes 4 and 5 show products of reactions containing primers Fks3F and Fks7R for (1,3)-β-glucan synthases. Lane 1 (control) used the rice CesA EST D47622 as template; lanes 2 and 4 used cDNA from N. alata pollen tubes grown in vitro for 12 h as template; and lanes 3 and 5 used cDNA from young expanding N. alata leaves as template. Numbers to the left show fragment sizes in kilobase pairs.
The entire CslD cDNA isolated from a pollen-tube library is 3,621 bp in length, has an open reading frame (ORF) of 3,381 bp, and is predicted to encode a 1,127 amino acid polypeptide with a molecular mass of 125 kD and a pI of 6.9. Based on its similarity with the CslD genes, this cDNA was named NaCslD1 and has been deposited in GenBank with accession no. AF304375.
Identification of Other CesA/Csl Family Members Expressed in Pollen Tubes
In addition to the major 700-bp band, the products of RT-PCR amplification with the UGF and DOMB1R primers included minor products of approximately 450 bp and approximately 550 bp (Fig. 1, lane 2). These smaller fragments were pooled, cloned, and inserts of 23 clones were sequenced. Six had identical sequences and included matches to the back-translated amino acid sequence to which the primers were designed. Database comparisons indicated that they represented a member of the CesA gene family, and thus the gene was named NaCesA1. However, extensive screening of the pollen-tube library failed to identify any cDNAs that hybridized to the Na-CesA1 fragment. A full-length cDNA was isolated from a N. alata pistil library, and comparison of this sequence with other sequences confirmed that this cDNA encoded a CesA protein (Fig. 2). The sequence has been deposited in GenBank with accession no. AF304374.
Figure 2.
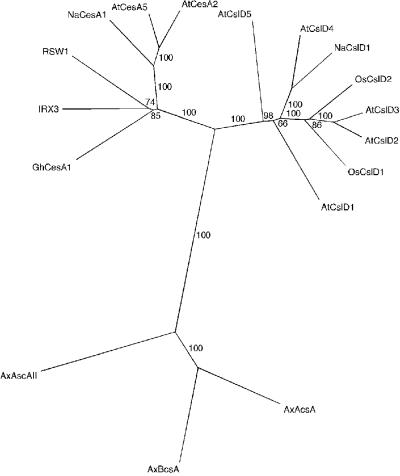
Distance cladogram for polypeptide sequences deduced from CesA and CslD genes. Three A. xylinum CesA sequences (AcsAB, BcsA, and AcsAII; Wong et al., 1990; Saxena and Brown, 1995) were included as an outgroup. Bootstrap supports are indicated along the relevant branches.
Isolation of a Gsl Gene Expressed in Pollen Tubes
Regions conserved between the fungal FKS genes that encode (1,3)-β-glucan synthases, the recently cloned CFL1 gene from cotton (Cui et al., 1999) and 12 similar Arabidopsis genes (named Gsl genes; http://cellwall.stanford.edu/), were identified from an alignment of the encoded polypeptides and used to design three forward and three reverse “Fks” primers.
When pairs of these Fks primers were used to amplify sequences from pollen-tube RNA, the fragments observed were similar in size to those predicted from the sequences of the CFL1 and the Arabidopsis Gsl genes (data not shown). Primers Fks3F and Fks7R were expected to amplify an approximately 820-bp fragment, and a fragment of this size was the most abundant product in the reaction (Fig. 1, lane 4). A fragment of the expected size was also obtained using leaf RNA but was far less abundant (Fig. 1, lane 5). The approximately 820-bp pollen-tube fragment was cloned, and inserts of 19 clones were sequenced. Two different fragments, both with significant sequence similarity (>65% at the amino acid level) to the plant Gsl sequences, were identified. The two sequences had only 66% identity with each other and so it is likely that these fragments are the amplification products of distinct Gsl genes. These genes were therefore named NaGsl1 and NaGsl2, respectively.
Based on the expression profile of NaGsl1 and NaGsl2 (see below), the RT-PCR fragment of NaGsl1 was chosen to screen the pollen-tube cDNA library and isolate a full-length cDNA. The entire cDNA sequence of NaGsl1 is 6,266 bp in length and contains an ORF of 5,793 bp that is predicted to encode a 1,931 amino acid polypeptide with a molecular mass of 221 kD and a pI of 9.6. The full sequence of NaGsl1 and the partial sequence of NaGsl2 have been deposited in GenBank with accession nos. AF304372 and AF304373.
Sequence Analysis of NaCslD1
Figure 2 shows a cladogram of selected polypeptides deduced for the plant CesA/Csl family. NaCslD1 groups with the other CslD sequences with a bootstrap value of 100%. This grouping is robust and was seen when trees were built using other algorithms available in PAUP (Swafford, 1999), when amino acids and gaps were included in the alignment, and when only the “informative” regions (defined as those regions in which all aligned polypeptides have blocks of overlapping amino acid sequence) were included. The deduced NaCesA1 polypeptide grouped with the other CesA proteins (Fig. 2).
The CslD clade includes polypeptides predicted from all five full-length Arabidopsis CslD genes (AtCslD1–AtCslD5) and both full-length rice CslD genes (OsCslD1 and OsCslD2) (Fig. 2). The overall amino acid identity between NaCslD1 and the other CslD proteins ranges from 64% (AtCslD5, 78% similarity) to 82% (AtCslD4, 91% similarity). The NaCslD1 gene may therefore be the N. alata homolog of AtCslD4.
NaCslD1 had lower identity with the deduced Arabidopsis CesA polypeptides (46% with RSW1/AtCesA1 and 48% with IRX3/AtCesA7). However, the CslD and CesA proteins still share many features and are more related to each other than either is to members of other Csl clades. Figure 3A illustrates some of these common features. Thus, NaCslD1 contains all the regions expected in a plant CesA protein (Pear et al., 1996), including the three homology domains (H-1, H-2, and H-3), the plant-conserved region (P-CR) and a hypervariable region (HVR) (Fig. 3A). Within the homology domains lie more highly conserved regions containing the conserved D residues and the QXXRW sequence. In NaCslD1, as in the CesA and other CslD polypeptides, the two amino acids after the Q residue are V and L, forming a D,D,D,QVLRW motif for this clade. The N-terminal domains of the CesA and CslD proteins contain a Cys-rich region (the LIM domain) that is not present in other Csl proteins (Fig. 3A).
Figure 3.
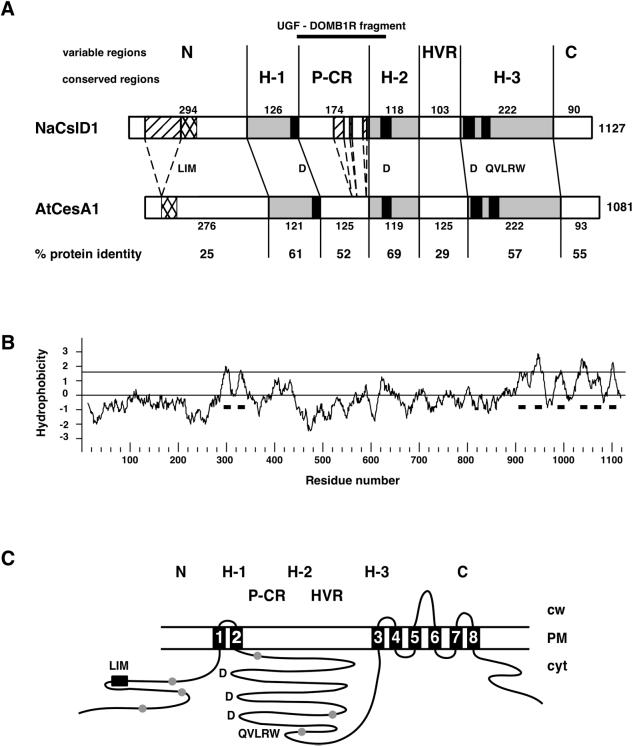
Domain structure and topology of the predicted NaCslD1 protein. A, Predicted NaCslD1 protein with predicted AtCesA1/RSW1 protein (Arioli et al., 1998) as comparison, drawn to scale as boxes. Length of total amino acid sequence is shown at the end of each box. Proteins are divided into the seven domains defined by Pear et al. (1996). H-1, H-2, and H-3 (gray boxes) are homology domains; P-CR, plant conserved region; HVR, hypervariable region; N and C refer to the N- and C-terminal domains, respectively. Numbers above the NaCslD1 box and below the AtCesA1 box show the number of amino acids within each domain. Amino acid identity (%) within a domain is given below the figure. Black boxes define the regions containing motifs of the conserved D,D,D,QXXRW domain as defined by Pear et al. (1996). A hatched box indicates the Cys-rich LIM domain. Dashed boxes in the NaCslD1 sequence indicate the insertion in the N-terminal domain, and the three insertions in the P-CR region. The position corresponding to the 700-bp UGF-DOMB1R fragment produced by RT-PCR is also shown. B, Hydropathy plot of the predicted NaCslD1 protein. Horizontal black bars indicate the eight predicted TMHs. C, Topology model of NaCslD1 based on the GhCesA1 model proposed by Pear et al. (1996). Cw, Cell wall; PM, plasma-membrane; cyt, cytoplasm; ●, location of potential N-glycosylation sites; 1 through 8, the TMHs.
A hydropathy plot of NaCslD1 indicated the presence of eight transmembrane helices (TMHs), two at the N terminus and six at the C terminus (Fig. 3B), with the N- and C-terminal sets of helices separated by a region of approximately 60 kD. Like CesA polypeptides, NaCslD1 lacks obvious signal sequences, and the first TMH is presumed to direct insertion into the endomembrane system. NaCslD1 can be folded in a similar way to GhCesA1 (Fig. 3C) and has six potential N-glycosylation sites (Fig. 3C; Pear et al., 1996), two of which (Asn 217 and 860) are at similar places to sites in GhCesA1, but none of these sites are predicted to be extracellular and thus NaCslD1 may not be N-glycosylated.
Despite the high degree of similarity, several features distinguish NaCslD1 from the CesA proteins. Within their central region, the proteins encoded by NaCslD1 and the Arabidopsis and rice CslD genes have three short insertions that are not found in the proteins encoded by CesA or other Csl family members (Fig. 3A). An additional insertion is also found in the N-terminal domain. This domain in NaCslD1 contains 294 amino acids and is comparable in length with corresponding regions from Arabidopsis and rice CslD proteins, which range in length from 268 to 319 amino acids. (An exception to this is AtCslD1 in which the N-terminal domain is 184 amino acids in length.) The length of the N-terminal domains of the CesA proteins also varies, but at 174 to 283 amino acids is generally shorter than those of the CslD proteins. The LIM domain (see below) is at least 100 amino acids from the N-terminal end of NaCslD1 and four of the five Arabidopsis CslD proteins, but is no more than 40 amino acids from the N terminus of CesA proteins.
The Cys-rich regions of the CesA and CslD proteins most closely resemble the double-zinc-finger consensus sequence of a LIM domain (Sánchez-García and Rabbitts, 1994). LIM domains may play a role in protein-protein or protein-lipid interactions. The 8 Cys residues in the CesA LIM domain are spaced symmetrically, whereas in the CslD proteins the spacing of Cys residues is variable and not symmetrical.
The HVR is reasonably well conserved within the CslD proteins. The NaCslD1 HVR is 103 amino acids in length and has 56% amino acid identity with the HVR of AtCslD5 and 81% identity with the HVR of AtCslD4. However, the NaCslD1 HVR has only 25% to 48% identity with the HVRs of the Arabidopsis CesA proteins and does not contain the stretches of basic amino acids (KKK, RKK) or the Cys residues interspersed by acidic (D, E) residues that are typical of the HVR of the CesA sequences (Pear et al., 1996; Carpita and Vergara, 1998; Fig. 3; data not shown).
Sequence Analysis of NaGsl1
Analyses of the deduced NaGsl1 polypeptide, the full-length plant Gsl polypeptide sequences, and representative fungal FKS polypeptide sequences showed that, as a group, the plant Gsl sequences were more closely related to each other than to any of the fungal FKS sequences (Fig. 4). The plant Gsl sequences fell into three clades with NaGsl1 grouping to the largest clade containing eight of the Arabidopsis Gsl sequences. CFL1 (= GhGsl1), a rice Gsl, and two Arabidopsis Gsl sequences (AtGsl8 and AtGsl10) formed a second, smaller clade, and the RT-PCR fragment of NaGsl2 grouped within this clade. Two other Arabidopsis Gsl sequences (AtGsl1 and AtGsl5) formed the third clade. The AtGsl1 and AtGsl5 polypeptides are clearly distinct from other Arabidopsis Gsl members, being approximately 100 amino acids shorter and being encoded by genes that have two and three exons, respectively, compared with the approximately 40 exons of the other Arabidopsis Gsl family members (http://cellwall.stanford.edu/cellwall/index.html). The overall amino acid identity of NaGsl1 with the 12 Arabidopsis Gsl genes ranged from approximately 48% (AtGsl1, AtGsl5, AtGsl8, and AtGsl10, approximately 68% similarity) to 81% (AtGsl2, 90% similarity), and NaGsl1 may therefore be the N. alata homolog of AtGsl2.
Figure 4.
Distance cladogram for polypeptide sequences deduced from plant Gsl genes. Five fungal FKS sequences (ScFks1, AfFks1, FnFKS1, PbFKS1, and CaGSC1; Douglas et al., 1994; Kelly et al., 1996) were included as an outgroup. Bootstrap supports are indicated along the relevant branches.
Figure 5A shows the structure of the NaGsl1 gene and a comparison with the fully sequenced cotton cDNA. A hydropathy plot of the deduced NaGsl1 protein predicted six TMHs near the N terminus and 10 THMs at the C terminus (Fig. 5B). The yeast FKS1 polypeptide (ScFKS1) is predicted to have a similar hydrophobicity profile, with the N terminus, the C terminus, and the large, central, hydrophilic region all on the cytoplasmic side of the membrane (Douglas et al., 1994); NaGsl1 may have the same topology (Fig. 5C). Comparison of NaGsl1 with CFL1 and the Arabidopsis and rice Gsl polypeptides indicated that these regions are generally of a similar length, whereas the fungal polypeptides all have a shorter central domain and longer C-terminal domain. Like the CesA and CslD polypeptides, NaGsl1 and the other plant Gsl polypeptides lack obvious signal sequences. NaGsl1 has eight potential N-glycosylation sites (Fig. 5C), one of which (Asn 530) is predicted to be extracellular.
Figure 5.
Domain structure and topology of the predicted NaGsl1 protein. A, Predicted NaGsl1 protein, with predicted CFL1 (Cui et al., 1999) as comparison, drawn to scale as boxes. Length of total amino acid sequence is shown at the end of each box. Proteins are divided into the four domains defined by Douglas et al. (1994). N, N-terminal domain; TM1 and TM2, first and second transmembrane domains; cyt, cytoplasmic domain. Numbers above the NaGsl1 box and below the CFL1 box show the number of amino acids within each domain. Amino acid identity (%) within a domain is given below the figure. The position corresponding to the 820-bp Fks3F-7R fragment produced by RT-PCR is also shown. B, Hydropathy plot of the predicted NaGsl1 protein. Horizontal black bars indicate the 16 TMHs. C, Topology model of NaCslD1 based on the yeast FKS1 model (Douglas et al., 1994). 1 through 16, The tMHs. Abbreviations and locations of potential N-glycosylation sites are as for Figure 3.
Overall, NaGsl1 shares 48% identity and 68% similarity at the amino acid level with CFL1, with highest levels of sequence identity being in the central region (56%) and C-terminal domain (55%) (Fig. 5A). Both NaGsl1 and CFL1 lack the D,D,D,QXXRW motif found in the family 2 inverting β-glycosyl transferases such as those encoded by the CesA and Csl genes (Saxena et al., 1995; Campbell et al., 1997), as well as the proposed UDP-Glc-binding motif present in family 1 inverting β-glycosyl transferases such as the glucosyl- and glucuronosyl-transferases that transfer single glycosyl residues (Mackenzie et al., 1997). The Gsl polypeptides also do not contain sequences that resemble the UDP-Glc-binding consensus (R/K) XGG found in glycogen synthase (Farkas et al., 1990). Last, the products of the fungal FKS and plant Gsl genes share no significant sequence similarity to the catalytic subunit polypeptide of the bacterial (1,3)-β-glucan synthase, CrdS (Stasinopoulos et al., 1999). Thus, novel sequence motifs within the Gsl polypeptides specify the amino acids critical for substrate binding and catalysis.
Expression Analysis of NaCslD1
The RT-PCR fragment of NaCslD1 was used to probe blots of RNA isolated from various N. alata tissues, including pollen tubes grown in vitro. An abundant transcript of 4.0 kb was detected in mature pollen and in pollen tubes growing actively in culture (Fig. 6A). This transcript was also present in anthers containing binucleate pollen grains (5–7-cm flower buds; Dodds et al., 1993) but was not found in anthers before pollen mitosis (2- to 3-cm flower buds), in other floral organs, in leaves, or stems, or in rapidly growing suspension-cultured cells (of N. plumbaginifolia). The transcript was present, however, at a low level in roots. NaCslD1 is therefore expressed abundantly in male gametophyte tissues after pollen mitosis.
Figure 6.
RNA-blot analysis of NaCslD1, NaCesA1, NaGsl1, and NaGsl2 expression. N. alata tissues: SE, sepal; PE, petal; AN, anther; ST, style; OV, ovary; MP, mature pollen; 4 through 24, pollen tubes grown for various times, given as h; L, leaf; R, root; S, stem; LS, light-grown seedling; and DS, dark-grown seedling. N. plumbaginifolia tissues: CC, suspension-cultured cells. Transcript sizes are indicated on the right of the figure. A, Blots probed with DIG-labeled UGF-DOMB1R fragment of NaCslD1. Flower buds and vegetative tissues: hybridized and washed at 35°C, exposed for 2 min. Pollen and pollen tubes: hybridized and washed at 42°C, exposed for 2 min. B, Flower buds, pollen, and pollen tubes: blots probed with 32P-labeled UGF-DOMB1R fragment of NaCesA1, hybridized, and washed at 42°C, exposed for 21 d. Vegetative tissues: blots probed with DIG-labeled UGF-DOMB1R fragment of NaCesA1, hybridized, and washed at 37°C, exposed for 2 h. C, Blots probed with DIG-labeled Fks3F-Fks7R fragment of NaGsl1. Flower buds, pollen, pollen tubes, and vegetative tissues: hybridized at 42°C and washed at 65°C, exposed for 1.5 h. D, Blots probed with DIG-labeled Fks3F-Fks7R fragment of NaGsl2. Flower buds, pollen, pollen tubes, and vegetative tissues: hybridized at 42°C, washed at 65°C, and exposed for 1.5 h.
Expression Analysis of NaCesA1
Using the NaCesA1 RT-PCR fragment as a probe, transcripts of 3.8 kb were detected at a low level in a range of floral and vegetative tissues including leaf, stem, root, sepals, and petals but were not detected in the male gametophyte (pollen or anthers) at any stage of development (Fig. 6B). NaCesA1 transcripts were very much less abundant than those of NaCslD1 (compare Fig. 6, A and B). Weak cross-hybridization to NaCslD1 transcripts of 4.0 kb was observed in mature pollen and anther samples (Fig. 6B).
NaCesA1 transcripts are thus undetectable in pollen tubes by RNA-blot hybridization but are detectable after RT-PCR. No other Csl products were detected by RT-PCR using the UGF and DOMB1R primers on pollen-tube RNA (although CslA and CslC transcripts would give products smaller than the 450- to 700-bp range analyzed). Pollen tubes thus appear to express one CslD gene abundantly and one CesA gene at a very low level.
Expression Analysis of NaGsl1 and NaGsl2
The RT-PCR fragments of NaGsl1 and NaGsl2 were also used to probe blots of RNA isolated from various N. alata tissues. The NaGsl1 fragment detected an abundant transcript of approximately 6.3 kb that was expressed in late-stage anthers, mature pollen, and pollen tubes grown in culture (Fig. 6C). Transcript levels peaked in mature pollen and remained at a high level over 24 h of pollen-tube growth. The NaGsl1 fragment detected no transcripts in other floral tissues during their development or in any vegetative tissue tested (Fig. 6C).
The RT-PCR fragment of NaGsl2 also detected an approximately 6.3-kb transcript, but this was much less abundant than the NaGsl1 transcript (Fig. 6D). The NaGsl2 transcript was present at low levels in a number of immature floral organs including ovaries from immature flower buds, as well as in mature pollen and pollen tubes grown in culture for 4 h but not in older pollen tubes.
DISCUSSION
Three groups of plant wall polysaccharide synthase genes have been identified by the sequence homology approach to date. The CesA gene family includes two genes that have been deduced to encode CelS enzymes through mutant complementation (Arioli et al., 1998; Taylor et al., 1999), although no direct proof of enzyme activity is available and corresponding polypeptides have not been identified (Delmer, 1999). The products of different CesA genes are believed to be responsible for synthesis of cellulose in primary and secondary walls (Delmer, 1999; Holland et al., 2000). The functions of the various groups of cellulose-synthase-like (Csl) genes are unknown (Richmond and Somerville, 2000), although probably all encode β-glycosyl transferases. Last, the glucan-synthase-like (Gsl) genes resemble fungal (1,3)-β-glucan synthase (FKS) genes (Douglas et al., 1994). Gene families not yet identified include those encoding the Golgi-localized enzymes that synthesize the backbones and branches of pectin and other non-cellulosic polysaccharides (hemicelluloses), including the mixed-linkage β-glucan of graminaceous monocots. However, individual genes encoding a galactosyltransferase involved in galactomannan biosynthesis in fenugreek seed endosperm (Edwards et al., 1999) and a xyloglucan fucosyltransferase from Arabidopsis (Perrin et al., 1999) have been cloned using information from the relevant purified plant proteins; both are type II membrane proteins with a single transmembrane sequence.
Pollen-tube walls are simpler than the walls of somatic cells, containing only the two β-glucans callose and cellulose, plus α-linked pectic polysaccharides (Li et al., 1999). They are the only plant cells in which the (1,3)-β-glucan callose forms the major wall structural component and is not subject to rapid turnover. The Nicotiana pollen-tube CalS enzyme is highly active and is distinct from the wound-activated CalS enzyme of other cell types (Schlüpmann et al., 1993; Li et al., 1997, 1999). We report here that two β-glucan synthase genes from different families (NaCslD1 and NaGsl1) are abundantly and specifically expressed in the male gametophyte (developing and mature pollen grains, and growing pollen tubes), and present full-length cDNA sequences for these genes. Further, we propose that the best explanation for this result and the associated sequence data is that the NaCslD1 gene encodes the pollen-tube (1,4)-β-glucan synthase (CelS) enzyme, and the NaGsl1 gene encodes the pollen-tube (1,3)-β-glucan synthase (CalS) enzyme.
A “true” CesA gene (NaCesA1) was a minor amplification product from pollen-tube mRNA, but the corresponding gene could not be detected in a pollen-tube cDNA library, consistent with the very low NaCesA1 transcript levels detected on RNA blots. The expression data suggest that NaCesA1 encodes a CelS enzyme that is active in immature anthers and mature pollen grains before dehydration, and thus responsible for deposition of the small amount of cellulose present in the intine of the maturing pollen grain; residual levels of the transcript remain in the growing tube. Instead, the major amplification product using the CesA primers was a CslD gene (NaCslD1) that was highly expressed in pollen grains and growing tubes. Several pieces of corroborative evidence support the hypothesis that NaCslD1 encodes the CelS catalytic subunit of pollen tubes. First, NaCslD1 is likely to be involved in the synthesis of β-linked polysaccharides, since the predicted polypeptide sequence contains the D,D,D,QXXRW motif. Second, the CslD family has the most sequence identity (45%–50% at the protein level) with the CesA genes that encode CelS enzymes (Richmond and Somerville, 2000). Third, the number and position of predicted membrane-spanning helices in the NaCslD1 protein is similar to CesA proteins, indicating that these probably have a similar plasma-membrane topology.
The hypothesis that NaCslD1 encodes the catalytic subunit of the pollen-tube CelS enzyme, rather than any CesA gene, implies that, over the whole plant body, both the CesA genes and the CslD genes encode CelS enzyme components. This is consistent with the finding that the CesA genes are more closely related to the CslD genes than they are to any of the other Csl gene families. There are estimated to be 12 CesA and five CslD genes in Arabidopsis, implying that there could be a total of approximately 17 different genes encoding CelS catalytic subunits. Pollen tubes only express one CslD gene, and minimal levels of one CesA gene, so expression of multiple CesA genes in other plant systems may partly reflect a diversity of cell types being present (Holland et al., 2000). The detection of NaCslD1 transcripts in male gametophyte tissues and (weakly) in roots indicates that CslD genes may be specifically involved in cellulose synthesis in cell types that undergo tip growth, such as the vegetative cell of pollen tubes, and root hairs. This is consistent with CslD ESTs being identified in root cDNA libraries of Medicago truncatula and Glycine max (http://cellwall.stanford.edu/cellwall/index.html). In addition, the rhizoids of mosses and liverworts and fern prothalli all elongate via tip growth, and to date only a CslD EST, and not a CesA EST, has been identified in the moss Ceratodon purpureus.
Pollen tubes also express a gene (NaGsl1) similar to the FKS genes of fungi. Several pieces of evidence support the hypothesis that NaGsl1 encodes the pollen-tube CalS. First, NaGsl1 shares low but significant levels of sequence identity with the fungal FKS genes, which encode the membrane-bound catalytic subunit of the (1,3)-β-glucan synthase of fungi (Douglas et al., 1994). Highest sequence identity between NaGsl1 and the fungal polypeptides lies in a C-terminal portion of the central cytoplasmic region that is proposed to contain the catalytic domain, as would be expected for proteins that are functional homologs. Second, NaGsl1 is predicted to encode a polypeptide of similar size to the fungal FKS proteins (approximately 220 kD) with a similar transmembrane topology (Douglas et al., 1994). Furthermore, the polypeptide encoded by the NaGsl1 cDNA is of a similar size to the 190-kD polypeptide that is enriched in the product-entrapment pellet containing the most highly enriched pollen-tube CalS (Turner et al., 1998). The final size of the active pollen-tube CalS may be slightly less than that predicted from the ORF due to possible proteolytic activation of an initially synthesized zymogen (Li et al., 1999). Third, the NaGsl1 gene is expressed abundantly and specifically in pollen grains and tubes, consistent with the deposition of large amounts of callose in pollen-tube walls.
One implication of NaCslD1 and NaGsl1 being expressed together is that the pollen-tube plasma-membrane contains two unrelated β-glucan synthases. Fungi also express both types of gene with the CHS genes encoding enzymes with a D,D,D,QXXRW motif that synthesize chitin (Bulawa et al., 1986) and the FKS genes encoding enzymes without this motif that synthesize (1,3)-β-glucan (Douglas et al., 1994). In contrast, bacteria appear to use β-glucan synthases with a D,D,D,QXXRW motif to synthesize both (1,4)-β-linked glucan (cellulose: Matthysse et al., 1995; Saxena et al., 1995) and (1,3)-β-linked glucan (curdlan: Stasinopoulos et al., 1999).
Our indication that a Gsl gene encodes the developmentally regulated pollen-tube CalS does not specifically address the issue of the wound-activated enzyme, as we cannot detect any wound-activated, Ca2+-dependent CalS activity in pollen tubes (Schlüpmann et al., 1993). There is a long-standing proposal that the wound-activated CalS enzyme may be a deregulated form of the CelS enzyme (Jacob and Northcote, 1985; Delmer, 1999). There is already precedent for a bacterial member of the CesA/Csl family synthesizing (1,3)-β-glucan (CrdS; Stasinopoulos et al., 1999), and there is direct genetic evidence that the Dictyostelium CesA gene product that makes (1,4)-β-glucan in vivo also makes (1,3)-β-glucan in vitro (Blanton and Northcote, 1990; Blanton et al., 2000). A similar phenomenon is observed with the mixed-linkage glucan synthase of corn or barley as, when isolated Golgi membranes are assayed in the presence of UDP-Glc, a (1,3)-β-glucan synthase activity is detected although no callose is deposited in vivo (Gibeaut and Carpita, 1993; Becker et al., 1995). Oomycetes, filamentous organisms that deposit a wall containing both cellulose and callose, have (1,3)-β-glucan synthases that are biochemically distinct from those of plants and fungi (Billongrand et al., 1997; Antelo et al., 1998) and that may be a third type of enzyme. Our data also do not bear on the function of the Csl A, B, C, E, and G gene families in plants, and these may encode components of other polysaccharide synthases, such as those responsible for synthesis of the (1,4)-β-linked backbones of xyloglucan, xylan, or glucomannan.
Transcripts of NaGsl1 and NaCslD1 accumulate after the mitotic division in developing anthers, reach a maximum level in mature pollen, and remain high throughout pollen-tube growth. This male gametophyte expression profile indicates that both are “late” pollen-expressed genes proposed to be involved in pollen maturation, germination, and pollen-tube growth (Mascarenhas, 1993), with the relative levels of the NaCslD1 and NaGsl1 transcripts in pollen and pollen-tube RNA reflecting the relative levels of their glucan products in the pollen-tube wall. However, the differential timing of cellulose and callose deposition indicates that the relevant genes or enzymes are likely to undergo differential regulation. There is some evidence for specific post-transcriptional regulation of expression of NaGsl1, as no CalS activity or callose is detectable in pollen grains (Li et al., 1999) despite the presence of large amounts of the NaGsl1 transcript before grain dehydration. Therefore, it appears that NaGsl1 transcripts are stored in the grain before desiccation and then translated upon germination like other late pollen-expressed genes (Mascarenhas, 1993). There is also evidence for specific post-translational control of CalS activity in pollen tubes through activation of a zymogen (Li et al., 1999). The sequences of NaCslD1 and NaGsl1 also indicate the potential for translational control of gene expression, as both contain in-frame ATG codons and stop codons at their 5′ end. In any case, the identification of genes proposed to encode the major β-glucan polysaccharide synthases of pollen tubes now allows a more detailed analysis of the mechanisms that regulate the timing and location of deposition of these wall polymers.
Proof of gene identities will require either a genetic/transgenic approach, or a proteomics approach. A recent comparative analysis of CesA family members (Holland et al., 2000) concluded that orthologs (functional homologs across species) are often more similar than paralogs (other homologs within species), which suggests that AtCslD4 and AtGsl2 could be the Arabidopsis pollen-tube β-glucan synthases and thus the preferred targets for genetic manipulation to determine gene function. In contrast, Nicotiana remains the better system for biochemical and proteomic studies of pollen tubes, and the ability to enrich an active and stable pollen-tube CalS (Turner et al., 1998) will allow the complementary proteomics approach to gene identification.
MATERIALS AND METHODS
Plant Material and Pollen-Tube Culture
Plants of Nicotiana alata Link et Otto (ornamental tobacco, self-incompatibility genotypes S2S2) were grown and maintained in a pollinator-proof glasshouse under standard conditions. Suspension-cultured cells of N. plumbaginifolia were maintained as in Sims and Bacic (1995). Pollen from open flowers was collected for in vitro growth experiments. Pollen-tube growth medium and culturing conditions were as described by Schlüpmann et al. (1993) with the modifications of Li et al. (1997). Pollen tubes were harvested by filtration on a GF/A filter disc (Whatman) and frozen in liquid nitrogen for RNA extraction.
Primer Design and RT-PCR
Degenerate primers UGF (5′ GGGAATTCTGYTAYRTNTCHGAYGAYG 3′) and DOMB1R (5′ CCGGATCCTGRTCRCARTCVASRTTVA 3′) were designed based on the sequence of conserved motifs within homology domains H-1 and H-2 of the bacterial CesA genes (Wong et al., 1990; Matthysse et al., 1995; Saxena et al., 1995; Saxena and Brown, 1995) and rice and Arabidopsis CesA ESTs (accession nos. D41261, D47622, D41986, and T20782; Pear et al., 1996). The rice EST D47622 was obtained from the Japanese Ministry of Agriculture, Forestry, and Fisheries stock center. The sequences of degenerate primers Fks3F (5′ CARACWYTKKMHAGRACWRT 3′) and Fks7R (5′ AWWCCWGCRWARATRTCYTC 3′) were based on back-translations of two short peptides conserved in the fungal FKS and the cotton and Arabidopsis FKS-like proteins (Douglas et al., 1994; sequences now assembled at http://cellwall.stanford.edu/). The level of degeneracy was reduced using the Nicotiana codon bias table (http://www. kazusa.or.jp/codon/).
Total RNA was isolated essentially as described by McClure et al. (1990). First-strand cDNA was synthesized in a 25-μL reaction that contained 1 μg of total RNA, 1× PCR buffer (Life Technologies/Gibco-BRL, Cleveland), 2.5 mm MgCl2, 400 μm dNTPs, 10 pmol oligo d(T) Ad1Ad2 primer (5′ CTGAGAGAACTAGTCTCGAGCTCTAGAACAAGCTTTTTTTTTTTTTTTTT 3′), 10 mm dithiothreitol, 40 units of RNase inhibitor (Promega, Madison, WI) and 200 units of Superscript II reverse transcriptase (Life Technologies/Gibco-BRL). Reactions were heated to 70°C for 10 min and template RNA removed by adding 1.5 units of RNase H (Promega).
cDNA (2.0 μL) was amplified by PCR in 50-μL reactions that contained standard reagents and enzymes and either the UGF and DOMB1R primers or the Fks3F and Fks7R primers (150–350 ng each primer). DNA was heated to 96°C for 2 min, followed by a program of 94°C for 20 s, 45°C for 45 s, and 72°C for 1.5 min, for 55 cycles. PCR products were analyzed, cloned, and sequenced using standard recombinant DNA methods (Sambrook et al., 1989) and commercially available reagents.
Construction and Screening of cDNA Libraries
RNA was isolated from N. alata pollen tubes grown in culture for 12 h, and poly(A+) RNA prepared using the PolyATtract system (Promega). The ZAP-cDNA kit (Stratagene, La Jolla, CA) was used for synthesis of cDNA from approximately 5 μg poly(A+) RNA. The estimated size of the unamplified pollen-tube library was 2.4 × 106 plaque-forming units. The library was amplified before screening. The amplified N. alata S2S6 pistil cDNA library is described in Royo et al. (1996).
DNA probes for library screening were randomly labeled with [α-32P]dCTP (3,000 Ci/mmol; Amersham, Buckinghamshire, UK) using the Prime-A-Gene labeling kit (Promega). Plaque lifts on nylon membranes (Hybond N+, Amersham or Magna, MSI) were hybridized with radioactive probes at 37°C overnight. Filters were then washed twice with 2× SSC, 0.1% (w/v) SDS (5 min, room temperature), then twice with 1× SSC, 0.5% (w/v) SDS (15 min, 37°C), and were exposed to film (RX [Fuji Photo Film, Tokyo] or XAR [Kodak, Rochester, NY]) with an intensifying screen at −70°C for 1 to 4 d. Positive plaques were purified to homogeneity and converted into plasmids using ExAssist (Stratagene).
Sequencing and Sequence Manipulation
The NaCslD1 and NaGsl1 cDNAs were initially sequenced using vector-specific primers. Primers based on these sequences were used to sequence further into the clones, a procedure that was repeated until both strands had been completely sequenced. The 5′ end of NaGsl1 was obtained by primer extension using 12-h pollen tubes as the RNA source.
The NaCesA1 cDNA could not be sequenced using this procedure. Instead, the clone was amplified by PCR using the UGF and DOMB1R primers and an aliquot of lysed phage stock as template. Amplification products were purified and sequenced. The 5′ and 3′ ends of the clone were obtained by PCR using the T3 and T7 primers in combination with UGR or DOMB1F, respectively, and the resulting fragments purified and sequenced. Additional primers were used to amplify the remainder of the clone.
Sequences were compared with databases using software available through the Australian National Genome Information Service (http://mel1.angis.org.au). Sequences were aligned using PileUp (Version 8, Genetics Computer Group, Madison, WI) and edited using SeqApp 1.9 (http://iubio.bio.indiana.edu/molbiol/). Phylogenetic trees were constructed from these alignments using PAUP Version 4.0.b2a (Swafford, 1999). Robustness of branches was estimated using 100 boot-strapped replicates with the heuristic search option.
RNA-Blot Analyses
Total RNA (10–20 μg) was fractionated on a 1% (w/v) agarose-formaldehyde gel in 1× MOPS (3-(N-morpholino)propanesulfonic acid) running buffer (20 mm MOPS, 5 mm NaOAc, 1 mm EDTA, pH 7.0) and transferred to nylon membranes (Hybond N+, Amersham or Magna) by a standard protocol (Sambrook et al., 1989). After transfer, membranes were briefly rinsed in 2× SSC, air-dried, and the RNA fixed to the membrane using UV light. The RNA blots were then hybridized in 50% (v/v) formamide, 5× SSPE, 2% (w/v) blocking agent (Boehringer Mannheim/Roche, Basel) in 0.1 m maleic acid/NaOH, pH 7.5, 0.15 m NaCl, 0.1% (w/v) lauroyl sarcosine, 7% (w/v) SDS, with either a radioactive DNA probe as for cDNA library screening or a digoxygenin (DIG)-labeled DNA probe (Roche), then washed in 1× SSC, 0.5% (w/v) SDS at 35°C to 42°C (NaCslD1 and NaCesA1 probes) or 0.5× SSC, 0.5% (w/v) SDS at 65°C (NaGsl1 and NaGsl2 probes), as described in figure legends. RNA blots probed with a radioactive probe were exposed to film at −70°C with an intensifying screen. Blots probed with DIG-labeled DNA were developed using a chemiluminescent detection method (Boehringer Mannheim/Roche). Transcript sizes were estimated using an RNA marker (Promega). rRNA detected with ethidium bromide or with a DIG-labeled fragment of the N. alata 18S rRNA gene was used for loading comparisons.
ACKNOWLEDGMENTS
The authors thank Professor D.P. Delmer (University of California, Davis) and Professor B.A. Stone (La Trobe University, Australia) for sharing before publication the identity of a rice CesA EST and the sequence of a bacterial CrdS gene, respectively. We are grateful to Professor C.M. Douglas (Merck Research Laboratories, NJ) for the yeast FKS clone and to the rice and Arabidopsis stock centers for providing clones. We thank the Somerville laboratory (Stanford University), particularly Dr T. Richmond, for maintaining excellent web pages on the biosynthesis of plant cell walls, and Bruce McGinness (Plant Cell Biology Research Centre, University of Melbourne) for his assistance in the glasshouse.
Footnotes
This work was supported by the Sir John and Lady Higgins postgraduate scholarship from the University of Melbourne (to M.S.D.), by a Special Research Centre grant from the Australian Research Council (to A.B.), and by a Functional Genomics grant from the Grains Research and Development Corporation (to A.B. and E.N.).
LITERATURE CITED
- Antelo L, Cosio EG, Hertkorn N, Ebel J. Partial purification of a GTP-insensitive (1,3)-β-glucan synthase from Phytophthora sojae. FEBS Lett. 1998;433:191–195. doi: 10.1016/s0014-5793(98)00904-1. [DOI] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- Bacic A, Harris PJ, Stone BA. Structure and function of plant cell walls. In: Priess J, editor. The Biochemistry of Plants. New York: Academic Press; 1988. pp. 297–371. [Google Scholar]
- Becker M, Vincent C, Reid JSG. Biosynthesis of (1,3)(1,4)-β-glucan and (1,3)-β-glucan in barley (Hordeum vulgare L): properties of the membrane-bound glucan synthases. Planta. 1995;195:331–338. doi: 10.1007/BF00202589. [DOI] [PubMed] [Google Scholar]
- Billongrand C, Marais MF, Joseleau JP, Girard V, Gay L, Fevre M. A novel 1,3-β-glucan synthase from the oomycete Saprolegnia monoica. Microbiology. 1997;143:3175–3187. doi: 10.1099/00221287-143-10-3175. [DOI] [PubMed] [Google Scholar]
- Blanton RL, Fuller D, Iranfar N, Grimson MJ, Loomis WF. The cellulose synthase gene of Dictyostelium. Proc Natl Acad Sci USA. 2000;97:2391–2396. doi: 10.1073/pnas.040565697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton RL, Northcote DH. A 1,4-β-d-glucan-synthase system from Dictyostelium discoideum. Planta. 1990;180:324–332. doi: 10.1007/BF00198783. [DOI] [PubMed] [Google Scholar]
- Bulawa CE, Slater M, Cabib E, Au-Young J, Sburlati A, Adair WL, Jr, Robbins PW. The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986;46:213–225. doi: 10.1016/0092-8674(86)90738-5. [DOI] [PubMed] [Google Scholar]
- Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarity. Biochem J. 1997;326:929–942. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita N, Vergara C. A recipe for cellulose. Science. 1998;279:672–673. doi: 10.1126/science.279.5351.672. [DOI] [PubMed] [Google Scholar]
- Cui X, Shin H, Brown RM. A novel gene from cotton shows homology to the yeast β-1,3-glucan synthase subunit FKS1 (abstract no. 184). Baltimore, MD: Proceedings of the American Society of Plant Physiologists Plant Biology Annual Meeting; 1999. [Google Scholar]
- Delmer DP. Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- Derksen J, Rutten T, van Amstel T, de Win A, Doris F, Steer M. Regulation of pollen tube growth. Acta Bot Neerl. 1995;44:93–119. [Google Scholar]
- Doblin MS, de Melis L, Read S, Newbigin EJ, Bacic A (2000) The cloning of two putative β-glucan synthase genes from pollen tubes of Nicotiana alata (abstract no. 310). Plant Biology 2000: Proceedings of the American Society of Plant Physiologists Annual Meeting, San Diego
- Dodds P, Bönig I, Du H, Rödin J, Anderson MA, Newbigin E, Clarke AE. S-RNase gene of Nicotiana alata is expressed in developing pollen. Plant Cell. 1993;5:1771–1782. doi: 10.1105/tpc.5.12.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas CM, Foor F, Marrinan JA, Morin N, Nielsen JB, Dahl AM, Mazur P, Baginsky W, Li WL, Elsherbeini M. The Saccharomyces cerevisiae Fks1 (Etg1) gene encodes an integral membrane protein which is a subunit of 1: 3-β-d-glucan synthase. Proc Natl Acad Sci USA. 1994;91:12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards ME, Dickson CA, Chengappa S, Sidebottom C, Gidley MJ, Reid JSG. Molecular characterization of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J. 1999;19:691–697. doi: 10.1046/j.1365-313x.1999.00566.x. [DOI] [PubMed] [Google Scholar]
- Farkas IF, Hardy TA, DePaoli-Roach AA, Roach PJ. Isolation of the GSY1 gene encoding yeast glycogen synthase and evidence for the existence of a second gene. J Biol Chem. 1990;265:20879–20886. [PubMed] [Google Scholar]
- Ferguson C, Teeri TT, Siika-aho M, Read SM, Bacic A. Location of cellulose and callose in pollen tubes and grains of Nicotiana tabacum. Planta. 1998;206:452–460. [Google Scholar]
- Gibeaut DM, Carpita NC. Synthesis of (1→3),(1→4)-β-d-glucan in the Golgi apparatus of maize coleoptiles. Proc Natl Acad Sci USA. 1993;90:3850–3854. doi: 10.1073/pnas.90.9.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland N, Holland D, Helentjaris T, Dhugga KS, Xoconostle-Cazares B, Delmer DP. A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol. 2000;123:1313–1323. doi: 10.1104/pp.123.4.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Delauney AJ, Olson JM, Verma DPS (2000) Multiple forms of callose synthases in plants: callose synthase-1 interacts with UDP-glucosyltransferase and phragmoplastin on forming cell plate (abstract no. 324). Plant Biology 2000: Proceedings of the American Society of Plant Physiologists Annual Meeting, San Diego
- Jacob SR, Northcote DH. In vitro glucan synthesis by membranes of celery petioles: the role of the membrane in determining the type of linkage formed. J Cell Sci Suppl. 1985;2:1–11. doi: 10.1242/jcs.1985.supplement_2.1. [DOI] [PubMed] [Google Scholar]
- Kelly R, Register E, Hsu MJ, Kurtz M, Nielsen J. Isolation of a gene involved in 1,3 β-glucan synthesis in Aspergillus nidulans and purification of the corresponding protein. J Bacteriol. 1996;178:4381–4391. doi: 10.1128/jb.178.15.4381-4391.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlicka K, Brown RM., Jr Cellulose and callose biosynthesis in higher plants. Plant Physiol. 1997;115:643–656. doi: 10.1104/pp.115.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Bacic A, Read SM. Activation of pollen tube callose synthase by detergents: evidence for different mechanisms of action. Plant Physiol. 1997;114:1255–1265. doi: 10.1104/pp.114.4.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Bacic A, Read SM. Role of a callose synthase zymogen in regulating wall deposition in pollen tubes of Nicotiana alata. Planta. 1999;208:528–538. [Google Scholar]
- Mackenzie PI, Owens IS, Burchell B, Bock KW, Bairoch A, Belanger A, Fournel-Gigleux S, Green M, Hum DW, Iyanagi T. The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenet. 1997;7:255–269. doi: 10.1097/00008571-199708000-00001. [DOI] [PubMed] [Google Scholar]
- Mascarenhas JP. Molecular mechanisms of pollen tube growth and differentiation. Plant Cell. 1993;5:1303–1314. doi: 10.1105/tpc.5.10.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse AG, White S, Lightfoot R. Genes required for cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1995;177:1069–1075. doi: 10.1128/jb.177.4.1069-1075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Gray JE, Anderson MA, Clarke AE. Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature. 1990;347:757–760. [Google Scholar]
- Østergaard L, Petersen M, Nielsen O, Mundy J. Abstracts of the 6th International Congress of Plant Molecular Biology, Québec, Canada. 2000. A plant β-1,3-glucan synthase (abstract no. S04–24) [Google Scholar]
- Paech NA, Fincher GB, Koltunow AM. Proceedings of the Tenth International Conference on Arabidopsis Research. Parkville, Victoria, Australia: University of Melbourne; 1999. Functional characterization of an Arabidopsis (1,3)-β-glucan synthase homologue (abstract no. 10–18) [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K. Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science. 1999;284:1976–1979. doi: 10.1126/science.284.5422.1976. [DOI] [PubMed] [Google Scholar]
- Read SM, Clarke AE, Bacic A. Requirements for division of the generative nucleus in cultured pollen tubes of Nicotiana. Protoplasma. 1993a;174:101–115. [Google Scholar]
- Read SM, Clarke AE, Bacic A. Stimulation of growth of cultured Nicotiana tabacum W38 pollen tubes by poly(ethylene glycol) and Cu(II) salts. Protoplasma. 1993b;177:1–14. [Google Scholar]
- Richmond TA, Somerville CR. The cellulose synthase superfamily. Plant Physiol. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo J, Nass N, Matton D, Okamoto S, Clarke AE, Newbigin E. A retrotransposon-like sequence linked to the S-locus of Nicotiana alata is expressed in styles in response to touch. Mol Gen Genet. 1996;250:180–188. doi: 10.1007/BF02174177. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis R. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sánchez-García I, Rabbitts TH. The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet. 1994;10:315–320. doi: 10.1016/0168-9525(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Saxena IM, Brown RM., Jr Identification of a second cellulose synthase gene (acsAII) in Acetobacter xylinum. J Bacteriol. 1995;177:5276–5283. doi: 10.1128/jb.177.18.5276-5283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Brown RM, Jr, Fevre M, Geremia RA, Henrissat B. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüpmann H, Bacic A, Read SM. A novel callose synthase from pollen tubes of Nicotiana. Planta. 1993;191:470–481. [Google Scholar]
- Schlüpmann H, Bacic A, Read SM. Uridine diphosphate glucose metabolism and callose synthesis in cultured pollen tubes of Nicotiana alata Link et Otto. Plant Physiol. 1994;105:659–670. doi: 10.1104/pp.105.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims IM, Bacic A. Extracellular polysaccharides from suspension cultures of Nicotiana plumbaginifolia. Phytochemistry. 1995;38:1397–1405. [Google Scholar]
- Stasinopoulos SJ, Fisher PR, Stone BA, Stanisich VA. Detection of two loci involved in (1,3)-β-glucan (curdlan) biosynthesis by Agrobacterium sp. ATCC31749, and comparative sequence analysis of the putative curdlan synthase gene. Glycobiology. 1999;9:31–41. doi: 10.1093/glycob/9.1.31. [DOI] [PubMed] [Google Scholar]
- Swafford DL. PAUP* V 4.0b2a: Phylogenetic Analysis Using Parsimony (*and other methods). Sunderland, MA: Sinauer Associates; 1999. [Google Scholar]
- Taylor NG, Scheible W-R, Cutler S, Somerville CR, Turner SR. The irregular xylem 3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell. 1999;11:769–779. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Bacic A, Harris PJ, Read SM. Membrane fractionation and enrichment of callose synthase from pollen tubes of Nicotiana alata. Planta. 1998;205:380–388. doi: 10.1007/s004250050334. [DOI] [PubMed] [Google Scholar]
- Wardak AZ, Jacobs AK, Anderson MA, Fincher GB, Stone BA. Proceedings of the Australian Society for Biochemistry and Molecular Biology Annual Conference. Gold Coast, Queensland, Australia. 1999. The (1,3)-β-glucan synthase from suspension-cultured Lolium multiflorum (rye grass) endosperm (abstract Mon-04) [Google Scholar]
- Wong HC, Fear AL, Calhoon RD, Eichinger GH, Mayer R, Amikam D, Benziman M, Gelfand DH, Meade JH, Emerick AW. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc Natl Acad Sci USA. 1990;87:8130–8134. doi: 10.1073/pnas.87.20.8130. [DOI] [PMC free article] [PubMed] [Google Scholar]