Summary
Background
Pancreatic ductal adenocarcinoma (PDAC) is a malignant tumor with an extremely poor prognosis. Effective targets for anticancer therapy confirmed in PDAC are limited. However, the characteristics of genomics have not been fully elucidated in large-scale patients with PDAC from China.
Methods
We collected both blood and tissue samples from 1080 Chinese patients with pancreatic cancer and retrospectively investigated the genomic landscape using next-generation sequencing (NGS).
Findings
We found recurrent somatic mutations in KRAS (83.2%), TP53 (70.6%), CDKN2A (28.8%), SMAD4 (23.0%), ARID1A (12.8%) and CDKN2B (8.9%) in Chinese PDAC patients. Compared with primary pancreatic cancers, more genomic alterations accumulated especially cell cycle regulatory gene variants (45.4% vs 31.6%, P < 0.001) were observed in metastatic tumors. The most common mutation site of KRAS is p.G12D (43.6%) in pancreatic cancer. Patients with KRAS mutations were significantly associated with older age and mutations in the other three driver genes, while KRAS wild-type patients contained more fusion mutations and alternative mechanisms of RTK/Ras/MAPK pathway including a number of clinically targetable mutations. KRAS mutations in Chinese cohort were significantly lower than those in Western cohorts (all P < 0.05). A total of 252 (23.3%) patients with the core DNA damage response (DDR) gene mutations were detected. ATM (n =59, 5.5%) was the most frequent mutant DDR gene in patients with pancreatic cancer from China. Patients with germline DDR gene mutations were younger (P = 0.018), while patients with somatic DDR gene mutations were more likely to accumulate in metastatic lesions (P < 0.001) and had higher TMB levels (P < 0.001). In addition, patients with mutant DDR genes and patients carrying TP53 mutation were observed mutually exclusive (P < 0.001).
Interpretation
We demonstrated the real-world genomic characteristics of large-scale patients with pancreatic cancer from China which may have promising implications for further clinical significance and drug development.
Funding
The funders are listed in the Acknowledgement.
Keywords: Pancreatic ductal adenocarcinoma, genomic mutation, next-generation sequencing, KRAS, DNA damage response
Research in context.
Evidence before this study
Effective targets for anticancer therapy confirmed in pancreatic cancer are limited. Most current investigations about genetic characteristics of pancreatic cancer are based on Caucasian populations. However, the characteristics of genomics have not been fully elucidated in large-scale patients with pancreatic cancer from China.
Added value of this study
We retrospectively analyzed the characteristics of genomics from 1080 Chinese patients with pancreatic cancer detected by NGS and found recurrent somatic mutations in KRAS (83.2%), TP53 (70.6%), CDKN2A (28.8%), SMAD4 (23.0%), ARID1A (12.8%) and CDKN2B (8.9%), among others. Different lesion locations, KRAS status and population cohorts of pancreatic cancers were shown different genomic characteristics. A total of 252 (23.3%) patients with the core DDR gene mutations were detected. ATM (n =59, 5.5%) was the most frequent DDR mutant gene in patients with pancreatic cancer from China. Patients with somatic and germline DDR gene mutations had different clinicopathological and molecular characteristics.
Implications of all the available evidence
This study comprehensively reported the real-world genomic characteristics of large-scale patients with pancreatic cancer by clinical sequencing from China which is conducive to identify potentially targetable and predictive biomarkers, screen exceptional genetic subtypes in respond to specific treatment, and explore the clinical practice and novel agent development for patients with pancreatic cancer.
Alt-text: Unlabelled box
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal malignant tumors with a 5-year overall survival (OS) rate of less than 10%.1 Pancreatic cancer is the sixth leading cause of cancer death in China,2 and its incidence rate has rapidly increased worldwide over the last decade.3 Although nab-paclitaxel plus gemcitabine4 and the FOLFIRINOX5 chemotherapy regimens have been shown a significant survival benefit for pancreatic cancer patients, developing treatments with greater efficacy remains challenging. Genetic heterogeneity is common in patients with pancreatic cancer.6 Therefore, it is a fundamental problem to optimize anticancer treatment selection according to genetic variations.7
In the era of precision medicine, comprehensive genomic profiling of malignant tumor specimens has identified potential targets for therapeutic intervention. As a result, the combination of genetic testing and targeted therapy has enabled significant achievements in the treatment of lung cancer8 and breast cancer.9 However, few effective targets for anticancer therapy have been confirmed in PDAC. Genomic alterations of KRAS, TP53, CDKN2A and SMAD4 have been widely reported as pancreatic cancer driver genes that are associated with PDAC patients’ outcomes; however, few of these genes has been clinically directly targeted through the current therapeutic regimens.10 DNA damage response (DDR) deficiency is a hallmark of PDAC, which contains at least 200 related genes with different functional pathways.11, 12, 13 Prior studies demonstrated that germline DDR alteration carriers with PDAC had a significantly longer overall survival (OS) than noncarriers.14,15 The genes of BRCA family are thought to predict preferentially sensitive to platinum-based chemotherapy or PARP inhibition.16, 17, 18 A comprehensive understanding of the genetic characteristics and molecularly guided therapy have proved to a substantial impact on survival in patients with pancreatic cancer.19, 20, 21 Recently, studies have reported the genomic characteristics of Chinese pancreatic cancer patients, however, lack of blood paired germline or based on small cases, which is difficult to reflect the real-world genomic characteristics of pancreatic cancer from China.22,23
To determine the significant role played by genomics, we investigated 1080 patients with pancreatic cancer from China using panel-based next-generation sequencing (NGS) to explore the real-world genomic alterations and evaluate the potential clinical significance.
Methods
Patients and specimens
All participants provided written informed consent. The study protocol was reviewed and approved by the Ethics Committee of Shanghai Jiao Tong University School of Medicine Affiliated Renji Hospital (protocol No.2016089). Patients with pathologically diagnosed, anti-tumor therapy-naive, primary PDAC who were enrolled between January 2017 to August 2020. Both blood and formalin-fixed, paraffin-embedded (FFPE) tissue samples (n = 1080) were collected from each patient. The genes contained in our NGS panels were listed (381-gene panel and 733-gene panel) in Supplementary Table 1-2. All FFPE samples forwarded for DNA extraction contained at least 20% tumor nuclei. Genomic profiling was performed in a Clinical Laboratory Improvement Amendments–certified and College of American Pathologists–accredited laboratory (3D Medicines, Inc., Shanghai, China).
Tissue processing, DNA extractions, targeted sequencing and data processing
DNA extraction and sequencing were performed as previously described with some modifications.24 Genomic DNA was isolated from tissue samples using the ReliaPrep™ FFPE gDNA Miniprep System (Promega) and quantified using the Qubit™ dsDNA HS Assay Kit (Thermo Fisher Scientific). DNA extracts (30-200ng) were sheared into 250-bp fragments using an S220 focused-ultrasonicator (Covaris). Libraries were prepared using the KAPA Hyper Prep Kit (KAPA Biosystems).
For targeted capture of genomic regions, indexed libraries were subjected to probe-based hybridization with a customized NGS panel targeting 381 or 733 cancer-related genes, where the probe baits were individually synthesized, 5’biotinylated, 120-bp DNA oligonucleotides (IDT), and repetitive elements were filtered out from intronic baits according to the annotation provided by University of California Santa Cruz (UCSC) Genome RepeatMasker.
The captured libraries were loaded onto a NovaSeq 6000 platform (Illumina) for 100-bp paired-end sequencing with a mean sequencing depth of 500×. Raw data of tissue samples were mapped to the reference human genome hg19 using the Burrows-Wheeler Aligner (v0.7.12). Variant calling was performed only in the targeted regions. Somatic single nucleotide variants (SNVs) were detected using MuTect (v1.1.7) (https://github.com/broadinstitute/mutect), and somatic insertions and deletions (indels) were detected using Pindel (v0.2.5a8) (http://gmt.genome.wustl.edu/packages/pindel) with default parameters. Copy number variations (CNVs) were called by an in-house developed script with a cutoff of 6 copies. Gene rearrangements were identified by analyzing the clipped reads that could be extracted by the tag information of bam files mapped by bwa software. Single-nucleotide polymorphisms (SNPs) and indels were annotated by ANNOVAR against the following databases: dbSNP (v138) (https://www.ncbi.nlm.nih.gov/snp/), 1000 Genomes (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/), and ESP6500 (population frequency > 0.015) (https://evs.gs.washington.edu/EVS/). Only missense, stopgain, frameshift, nonframeshift, and splicing were retained. The human sequence data generated in this study were not publicly available due to China human genetic resources law and patient privacy requirements, but were available upon reasonable request from the corresponding authors.
Categorization of genomic alterations
Genomic alterations were categorized based on published literature.21,25 Aberrant signaling pathways and genes with alterations were listed in Supplementary Table 3. In keeping with recent references, the core DDR genes (n = 21) including ATM, ATR, ATRX, BARD1, BLM, BRCA1, BRCA2, CHEK2, ERCC1, FANCA, FANCD2, MLH1, MSH2, MSH6, MUTYH, PALB2, POLE, PMS2, RAD50, RAD51, and SMARCA4 involved in this study, which were contained in our NGS panels.11, 12, 13
The public pancreatic cancer dataset
The Cancer Genome Atlas (TCGA) 186 pancreatic cancer somatic mutations were downloaded from https://cbioportal-datahub.s3.amazonaws.com/paad_tcga.tar.gz. 150 patients with primary pancreatic cancer were included in the study. International Cancer Genome Consortium (ICGC) 99 pancreatic cancer somatic mutations were downloaded from https://cbioportal-datahub.s3.amazonaws.com/paad_icgc.tar.gz. University of Texas Southwestern Medical Center (UTSW) 109 pancreatic cancer somatic mutations were downloaded from https://cbioportal-datahub.s3.amazonaws.com/paad_utsw_2015.tar.gz. Queensland Centre for Medical Genomics (QCMG) 456 pancreatic cancer somatic mutations were downloaded from https://cbioportal-datahub.s3.amazonaws.com/paad_qcmg_uq_2016.tar.gz. 283 patients with tumor histologic subtype PDAC and sample type primary were included in the study. CNV in all the dataset was excluded in the comparative studies between our cohort and the public cohort.
Tumor mutational burden (TMB) analysis
TMB was defined as somatic mutation counts in coding regions per megabase (muts/Mb) of genome examined. Although this study was based on panels with different cancer gene numbers, the method used to calculate TMB was the same. SNVs included both synonymous and nonsynonymous mutations, as well as stopgain, stoploss, and splicing variants. Indel variants included both frameshift or nonframeshift insertions and deletions. Noncoding alterations were not counted.
Microsatellite instability (MSI) analysis
The MSI phenotype detection method is a read count distribution-based approach. An in-house developed R script was employed to evaluate the distribution of read counts among various repeat lengths for each microsatellite locus of each sample.26 An MSI score was defined as the percentage of unstable loci. Any sample with an MSI score of ≥ 0.4 was classified as MSI-high, and samples were otherwise classified as microsatellite stability (MSS).
Statistical analyses
For normally distributed continuous variables, Student's t-test was utilized to determine the differences between two groups; otherwise, the Mann-Whitney U test was utilized. Fisher's exact test or the chi-square test was employed to identify the association of two categorical variables. All reported P values were two-tailed, and P < 0.05 was considered to be significant unless otherwise specified. All the analyses were performed by using GraphPad Prism 8.0 (GraphPad Software, USA) and R software (version 3.6.0, https://cran.r-project.org).
Role of funders
The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Results
Characteristics of the study population
In this study, tumor tissues and matched blood samples were obtained from a total of 1080 patients with pathologically diagnosed PDAC (Table 1). Briefly, 58.0% (626/1080) of the patients were male, and the median age was 60 (range, 20–87). The median TMB level was 4.03 (range, 0–1124) in 1051 patients. Only 6 of 1051 (0.6%) patients were confirmed to have MSI-high status. A total of 754 (69.8%) samples originated from the primary pancreas. The remaining 326 (30.2%) samples represented distant metastases obtained from the liver (n = 227), peritoneum (n = 28), lymph nodes (n = 16), lung (n = 4) and other sites (n = 50).
Table 1.
Clinicopathologic characteristics in PDACs.
| Features | Total (%) |
|---|---|
| No. of Patients | 1080 |
| Age | |
| Median (Range) | 60 (20-87) |
| <=60 | 546 (50.6%) |
| >60 | 534 (49.4%) |
| Gender | |
| Male | 626 (58.0%) |
| Female | 454 (42.0%) |
| TMB Level | |
| Median (Range) | 4.03 (0-1124) |
| No information | 29 (2.7%) |
| MSI Status | |
| MSI-H | 6 (0.6%) |
| MSS | 1045 (96.8%) |
| No information | 29 (2.7%) |
| Sample Location | |
| Pancreas | 754 (69.8%) |
| Distant metastasis | 326 (30.2%) |
| Liver | 228 (21.1%) |
| Peritoneum | 28 (2.6%) |
| Lymph node | 16 (1.5%) |
| Lung | 4 (0.4%) |
| Others | 50 (4.6%) |
Abbreviations: PDAC, pancreatic ductal adenocarcinoma
Molecular landscape of somatic genomic alterations in PDAC
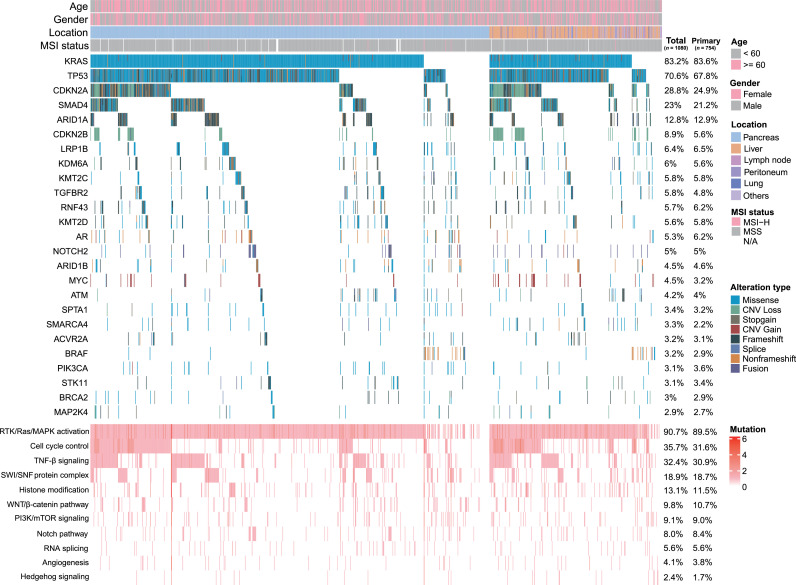
Among the 1080 pancreatic cancer patients, 8329 genomic alterations were identified in 652 genes. The most common mutational type was missense (62.3%) followed by frameshift (9.3%) and truncation (9.0%). A total of 598 (55.4%) patients were evaluated by 381-gene NGS panel, and 3432 genomic alterations were detected in 279 genes. Similarly, 482 (44.6%) patients were tested by 733-gene NGS panel, and 4897 genomic alterations were detected in 638 genes. KRAS was clearly the most frequently occurring somatic mutation in PDAC patients and the prevalence of this mutation was 83.2% (n = 899, Figure 1). Other genomic changes involved in decreasing order, TP53 (70.6%), CDKN2A (28.8%), SMAD4 (23.0%), ARID1A (12.8%) and CDKN2B (8.9%), among others. The key signaling pathways altered in PDAC were also demonstrated, 90.7% (n = 980) of PDAC patients had genomic alterations in the RTK/Ras/MAPK signaling pathway followed by cell cycle control (35.7%) and TGF-β signaling pathway (32.4%).
Figure 1.
Summary of the common prevalent genomic alterations and signaling pathways in 1080 Chinese PDAC patients. Genomic alterations include missense, nonframeshift, fusion, stopgain, splice, and copy number variations (CNV) gain or loss. The majority of pathways altered in PDAC contain RTK/Ras/MAPK activation, cell cycle control, TGF-β signaling, histone modification, SWI/SNF complex, PI3K/mTOR signaling, WNT/β-catenin pathway, RNA splicing, Notch pathway, angiogenesis, and Hedgehog signaling. The mutational frequencies of total samples and primary lesion samples were listed. Germline mutations are not performed.
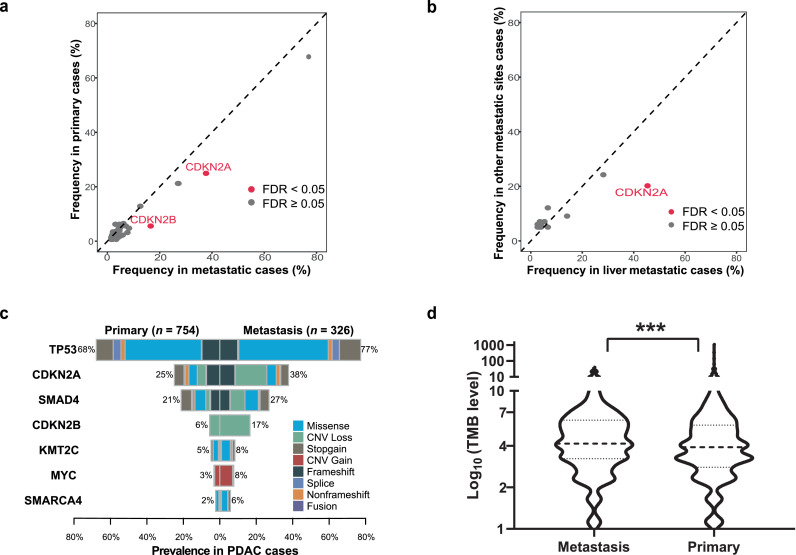
The genomic changes associated with available clinicopathological data for 1080 samples were summarized in Table 2. Older patients (> 60 years old) with PDAC were significantly more likely to exhibit KRAS mutations (P = 0.003 [two-sided Fisher's exact test]). Female patients with PDAC more often exhibited genomic alterations in CDKN2B and AR. A comparison of primary and metastatic PDAC showed that TP53, CDKN2A, SMAD4, MYC and CDKN2B gene alterations were likely to be observed in metastases (all P < 0.05 [two-sided Fisher's exact test]). There were no significant differences in KRAS mutations (83.6% vs 82.5%, P = 0.741 [two-sided Fisher's exact test]) between primary and metastatic tumors. A subgroup analysis by metastatic location demonstrated that liver metastases more frequently harbored alterations in KRAS, TP53, CDKN2A and CDKN2B (all P < 0.05 [two-sided Fisher's exact test]). GNAS gene mutations were likely to be observed in other metastatic sites. To further confirm the difference of molecular characteristics between primary and metastatic tumors, we found CDKN2A and CDKN2B harbored higher mutation frequencies in metastases than in primary after false discovery rate (FDR) correction (FDR < 0.05 [two-sided Fisher's exact test], Figure 2a). In the metastatic subgroup, liver metastasis with CDKN2A mutations were more frequent than those of the other metastatic lesions (FDR < 0.05 [two-sided Fisher's exact test], Figure 2b). Then, compared with primary tumors, we illustrated mutational types of representative differential genes and found CDKN2A, SMAD4 and CDKN2B contained much more CNV loss, while TP53, KMT2C and SMARCA4 accumulated much more missense in metastases (Figure 2c). Metastatic PDAC patients were also observed significantly harbored cell signaling control pathway (45.4% vs 31.6%, P < 0.001 [two-sided Fisher's exact test], Figure 1) and carried significantly higher TMB level (P < 0.001 [Mann-Whitney U test], Figure 2d) than those in patients with primary tumors. We also analyzed the prevalence of CNV alterations in our cohort and found MYC (n=44, 4.1%) was most common gene with CNV gain, following by FGFR1 (n=23, 2.1%), AKT2 (n=20, 1.9%) and KRAS (n=20, 1.9%), among others. However, the genes with the most frequent CNV loss were CDKN2B (n=95, 8.8%), CDKN2A (n=91, 8.4%) and SMAD4 (n=42, 3.9%), respectively (Supplementary Figure).
Table 2.
Clinicopathologic characteristics related to genomic alterations.
| Clinicopathologic features | n (%) vs. n (%) | p |
|---|---|---|
| Age <= 60 vs age > 60 | ||
| KRAS | 436 (80) vs 463 (87) | 0.003 |
| Female vs male | ||
| CDKN2B | 53 (12) vs 43 (7) | 0.009 |
| AR | 40 (9) vs 17 (3) | <0.001 |
| Primary vs metastasis | ||
| TP53 | 511 (68) vs 252 (77) | 0.002 |
| CDKN2A | 188 (25) vs 123 (38) | <0.001 |
| SMAD4 | 160 (21) vs 88(27) | 0.046 |
| MYC | 24 (3) vs 25 (8) | 0.002 |
| CDKN2B | 42 (6) vs 54 (17) | <0.001 |
| SMARCA4 | 17 (2) vs 19 (6) | 0.005 |
| FGFR1 | 14 (2) vs 16 (5) | 0.009 |
| KMT2C | 36 (5) vs 27 (8) | 0.034 |
| Liver vs other metastatic sites | ||
| KRAS | 196 (86) vs 73 (74) | 0.009 |
| TP53 | 186 (82) vs 66 (67) | 0.004 |
| CDKN2A | 103 (45) vs 20 (20) | <0.001 |
| CDKN2B | 50 (22) vs 4 (4) | <0.001 |
| GNAS | 4 (2) vs 7 (7) | 0.021 |
Figure 2.
Molecular characteristics depended on tumor location in Chinese PDAC patients. a. Molecular differences between primary and metastatic PDAC tumors. False-discovery-rate (FDR) correction was used to adjust for multiple comparisons. Each dot represents one gene and red dots represent FDR < 0.05, two-sided Fisher's exact test. b. Molecular differences between liver metastasis and other site metastatic PDAC tumors. c. Comparison of representative differential genes’ mutational types between primary and metastatic tumors. d. Violin plots of TMB in the corresponding clinical subgroups of primary and metastatic PDAC tumors (n =1051). The outline of the violins shows the mirrored kernel density, with the black dotted line indicating the median and quartile. Statistical comparisons were performed using Mann-Whitney U test. ***P < 0.001. Germline mutations are not included.
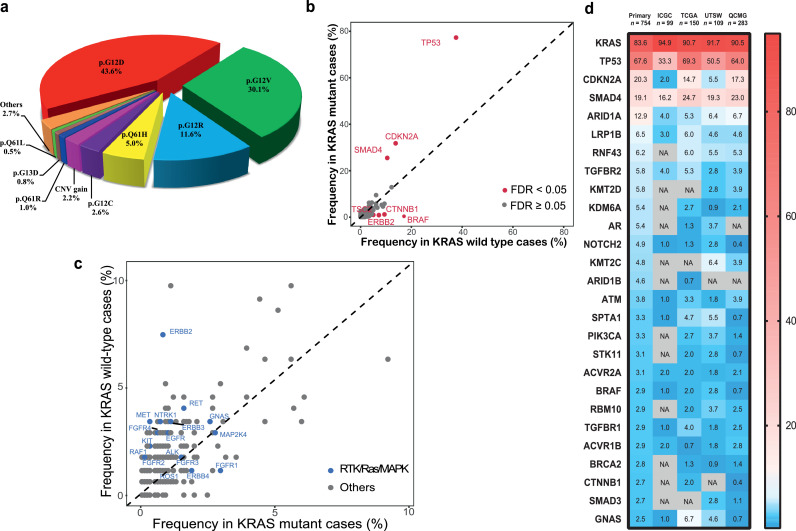
In the analysis of KRAS mutation sites, the most frequently observed alterations occurred on codon 12 of exon 2. These mutations included p.G12D (43.6%), p.G12V (30.1%), p.G12R (11.6%), p.Q61H (5.0%), p.G12C (2.6%) and CNV gain (2.2%). Other rare (<1%) point mutation sites could be also detected (Figure 3a). In Chinese population of PDAC patients, KRAS alterations were absent in 181 of 1080 (16.8%) cases. KRAS wild-type patients were significantly younger than those of KRAS mutant status (56.7 vs 60.6 years old, P < 0.001 [Student's t-test]). We found that patients with KRAS mutations were significantly associated with mutations in the other three driver genes, namely, TP53 (77.3% versus 37.6%), CDKN2A (31.8% versus 13.8%) and SMAD4 (25.5% versus 10.5%) (all FDR< 0.05 [two-sided Fisher's exact test], Figure 3b). However, KRAS wild-type patients were significantly associated with BRAF (0.4% versus 17.1%), CTNNB1 (1.2% versus 9.4%), ERBB2 (0.9% versus 7.2%) and TSC2 (1.0% versus 5.0%) mutations (all FDR< 0.05 [two-sided Fisher's exact test]). While BRAF mutations were the most prevalent mutations in the KRAS wild-type cohort, merely 4 of 899 (0.4%) KRAS-mutant samples had mutations in BRAF. Similarly, as alternative mechanisms of RTK/Ras/MAPK pathway, ERBB2, MET, KIT, RET, NTRK1, ERBB3, FGFR4, EGFR, FGFR2, RAF1, ROS1, GNAS, and ALK alterations were more likely to be observed in the KRAS wild-type cohort (Figure 3c). We also detected 124 genomic fusion alterations from 110 (10.2%) patients. Various fusion genes involving NOTCH2, BRAF, RET, ERBB2, KMT2C, RAF1, ALK, FGFR1 and EGFR were identified in 26 (14.4%) KRAS wild-type pancreatic cancer patients, significantly higher than those in the patients with KRAS mutations (9.3%, P = 0.042 [Two-sided Chi-square]).
Figure 3.
KRAS status and genetic comparison with Western populations in Chinese PDAC patients. a. The frequency of KRAS mutational type in PDAC patients. b. Comparison of alternative driver genes depended on PDAC patients with KRAS status. FDR correction was used to adjust for multiple comparisons. Each dot represents one gene and red dots represent FDR < 0.05, two-sided Fisher's exact test. c. Comparison of RTK/Ras/MAPK pathway related genes depended on PDAC patients with KRAS status. Each dot represents one gene and blue dots represent targetable genes. d. Heatmap showing the frequency comparison of somatic mutant genes without copy number variation (CNV) among Chinese primary patients (n = 754) and the Western cohorts from International Cancer Genome Consortium (ICGC, n = 99), The Cancer Genome Atlas (TCGA, n = 150), University of Texas Southwestern Medical Center (UTSW, n =109), and Queensland Centre for Medical Genomics (QCMG, n = 283). Germline mutations are not performed. The genes are listed in the order of mutation frequency in our cohort. The gradient color from red to blue represents the mutation frequency from high to low. NA. denotes not detected.
To further explore the differences in the mutational features between Chinese patients and Western patients, we compared our cohort (only primary without CNV alterations, n = 754) with public available database from the International Cancer Genome Consortium (ICGC, n = 99), the Cancer Genome Atlas (TCGA, n = 150), the University of Texas Southwestern Medical Center (UTSW, n = 109) and the Queensland Centre for Medical Genomics (QCMG, n = 283). We observed KRAS status as the most frequently occurring genomic mutation in our primary PDAC patients (83.6%) had significantly lower mutation frequency than that in all western cohorts (ICGC, 94.9%; TCGA, 90.7%; UTSW, 91.7%; QCMG, 90.5%; all P < 0.05 [two-sided Fisher's exact test], Figure 3d). While ARID1A, KDM6A, BRCA2 and ARID1B in our Chinese cohort were observed higher mutation rate than those in all western cohorts, although not all comparison reached statistical significance.
Characteristics of somatic and germline DDR gene mutations in Chinese PDAC patients
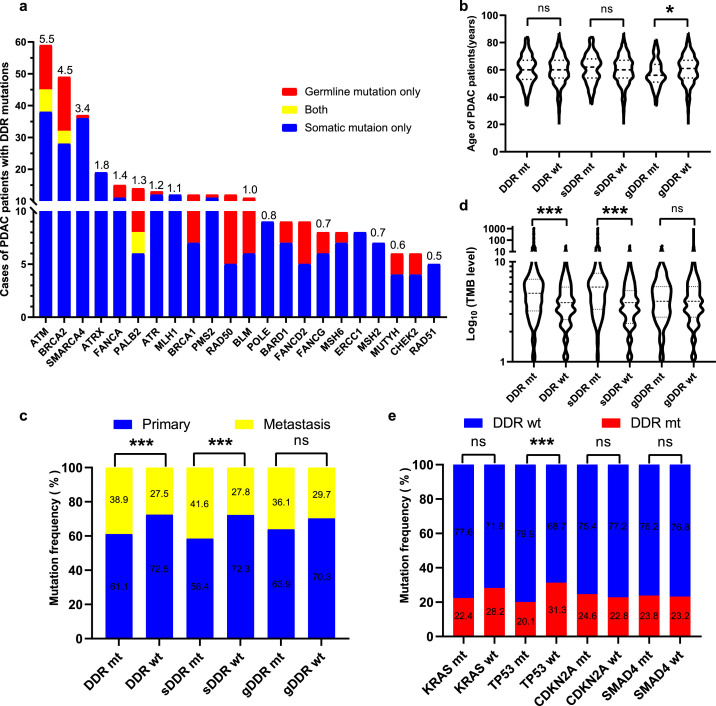
Our previous work comprehensively revealed the characteristics of DDR deficiency in patients with pancreatic cancer.27 Here, we evaluated somatic and germline mutations in 21 core DDR genes covered by our NGS panels. We found 252 (23.3%) PDAC patients were accompanied by the core DDR gene mutations. All core DDR mutant genes were displayed in Figure 4a. The most frequently mutated DDR gene in Chinese PDAC patients was ATM (n =59, 5.5%). The additional DDR deficiency were commonly seen in BRCA2 (4.5%), SMARCA4 (3.4%), ATRX (1.8%), FANCA (1.4%), PALB2 (1.3%), and ATR (1.2%), among others. Interestingly, 7 patients with ATM, 4 patients with BRCA2 and 2 patients with PALB2, respectively had both somatic and germline mutations.
Figure 4.
The core DDR genes with somatic and germline mutations in Chinese PDAC patients. a. The number of patients with the core DDR gene mutations in our cohort. Red bar represents germline DDR mutation only, blue bar represents somatic DDR mutation only, and yellow bar represents both. The mutational frequency of each gene is above bar. b. Violin plots compared the relationship between DDR status, somatic DDR status and germline DDR status and the age of the corresponding patients. The outline of the violins shows the mirrored kernel density, with the black dotted line indicating the median and quartile. Statistical comparisons were performed using Student's t-test. NS, not significant; *P < 0.05. c. Comparison of DDR status, somatic DDR status and germline DDR status with primary and metastatic PDAC tumors. Mutational frequencies of primary and metastasis were listed. Statistical comparisons were performed using two-sided Fisher's exact test. NS, not significant; ***P < 0.001. d. Violin plots compared the relationship between DDR status, somatic DDR status and germline DDR status and TMB level of the corresponding patients (n =1051). The outline of the violins shows the mirrored kernel density, with the black dotted line indicating the median and quartile. Statistical comparisons were performed using Mann-Whitney U test. NS, not significant; ***P < 0.001. e. Comparison of DDR status with 4 driver genes in Chinese PDAC patients. Mutational frequencies of primary and metastasis were listed. Statistical comparisons were performed using two-sided Fisher's exact test. NS, not significant; ***P < 0.001.
In our cohort, patients with germline DDR gene mutations were significantly younger than those with germline DDR wild-type (57.3 vs 60.2 years old, P =0.018 [Student's t-test]), but no such phenomenon was observed in patients with total DDR gene mutations and somatic DDR gene mutations (Figure 4b). Next, we investigated the correlations between DDR gene mutations and tumor location. We found patients with total DDR gene mutations (38.9% vs 27.5%, P < 0.001 [two-sided Fisher's exact test]) and somatic DDR gene mutations (41.6% vs 27.8%, P < 0.001 [two-sided Fisher's exact test]) were significantly higher than their wild-type patients in metastatic lesions, while there was no significant difference between patients with germline DDR gene mutations (P = 0.22 [two-sided Fisher's exact test]) and lesion location (Figure 4c). We further examined the underlying relationship between DDR gene mutations and TMB level. Patients with total DDR gene mutations (P < 0.001 [Mann-Whitney U test]), especially with somatic DDR mutations (P < 0.001 [Mann-Whitney U test]) had a significantly higher TMB level than did patients with wild-type DDR genes, respectively (Figure 4d). Furthermore, we analyzed the association between the core DDR gene mutations and 4 PDAC driver genes. We observed patients with DDR genes were only significantly associated with TP53 (P < 0.001 [two-sided Fisher's exact test]) and were mutually exclusive (Figure 4e).
Discussion
This study was demonstrated the real-world genomic characteristics in pancreatic cancer based on large-scale patients from China. Every clinical participant provided both tumor specimens and matched blood samples as controls, which were collected and tested for NGS profiles containing exons of 381 or 733 cancer-related genes. In addition, adequate neoplastic cellularity, high-quality DNA and reasonable sequencing depth were strictly implemented for each sample and reliable results could be obtained.
Oncogenic KRAS alteration is the major molecular event in PDAC patients, leading to permanent activation of the KRAS protein, which functions as a genetic switch to activate various cellular signaling pathways and transcription factors, thereby inducing proliferation, invasion, migration and survival.28 In our study, we demonstrated that KRAS mutations were more likely to occur in older patients which was consistent with previous reports.25,29 No difference in KRAS mutation between primary and metastatic lesions was seen; however, this mutation was detected more frequently in liver metastases than in other metastatic sites. Meanwhile, KRAS mutation was significantly closely linked to three tumor suppressor genes, TP53, CDKN2A and SMAD4. Mutational accumulation of these four main driver genes not only represent the specific molecular characteristics of PDAC, but also associate with poor prognosis.10 Furthermore, various KRAS mutational subtypes were demonstrated in our Chinese cohort. p.G12D, as the most prevalent KRAS-activating mutational subtype, accounted for 43.6%, which was consistent with the mutation frequency reported in previous study.28 However, it was demonstrated that p.G12D mutant patients with PDAC had a significantly shorter overall survival compared to p.G12V, p.G12R or wild-type patients.29 KRAS has been considered an undruggable molecule and exhibits escape mechanisms when KRAS is inactivated. Sotorasib (AMG510), a specific and irreversible small-molecule inhibitor targeting KRAS p.G12C mutation, showed encouraging anticancer activity, not only in non-small-cell lung cancer (NSCLC)30 but also in other solid tumors, including pancreatic tumors harboring the KRAS p.G12C alteration.31 Another promising study showed that PDAC patients with the KRAS p.G12R mutation had distinct drug sensitivity.32 In our cohort, 2.6% and 11.9% PDAC cases presented KRAS p.G12C and p.G12R mutations, respectively, and seemed to benefit more from these novel specific therapeutic strategies. However, the majority of KRAS alterations in PDACs are KRAS p.G12D, p.G12V, and rare mutational sites have not been determined to have a definite therapeutic response. Blocking KRAS downstream pathway seems to be a feasible treatment strategy. MEK inhibitors have been identified as a potential option for targeting KRAS to inhibit the RAF–MEK–ERK signaling pathway located downstream of KRAS. A recent study showed that the combination of MEK or ERK inhibitor and autophagy inhibitor led to tumor regression in PDAC patients.33,34 We believe that the proper population of KRAS mutant patients who benefit from various novel therapeutic strategies may be selected, and new anti-KRAS agents may also be designed and developed in the future.
In our Chinese PDAC patients, the proportion of KRAS wild-type patients accounted for 16.8%, which is considerably higher than that of Western population-based reports. KRAS wild-type patients were previously recognized as having a good prognosis, exhibiting a 9-month survival without treatment.28 Interestingly, KRAS wild-type patients were significantly accompanied by alterations in BRAF, CTNNB1, ERBB2, MET, KIT, NTRK1 and FGFR4, which may be primarily related to the activation of the RTK/Ras/MAPK pathway. It was reported that oncogenic BRAF deletions served as potential driver alterations in KRAS wild-type PDACs that responded to the MEK inhibitor.35 Further observation has shown that there is a tendency for other potentially targetable molecular alterations, such as ERBB2, MET, KIT, RET, NTRK1, ERBB3, FGFR4, EGFR, FGFR2, RAF1, ROS1, GNAS, and ALK, in KRAS wild-type PDAC patients to be involved in the other actionable alterations. Nimotuzumab, an EGFR inhibitor, in combination with gemcitabine for the first-line regimen showed a significant survival improvement, increasing survival by 6 months in the KRAS wild-type subgroup with advanced PDAC.36 According to the characteristics of high wild-type KRAS in Chinese pancreatic cancer patients, it appears that a considerable number of patients may benefit from the regimen containing anti-EGFR therapy. Meanwhile, PDAC patients with wild-type KRAS are closely related to other potentially actionable genomic alterations associated with the RTK/RAS/MAPK pathway and specific targetable inhibitors. A number of potential targetable therapeutic strategies need to be carefully designed and evaluated in clinical practice.
In this study, we integrated and analyzed somatic and germline mutations in 21 core DDR genes which were suitable for patients with pancreatic cancer from China and were more convenient and economical for clinical large-scale detection. We observed that 23.3% patients had DDR gene mutations and ATM was the most common DDR mutant gene in Chinese PDAC patients. Patients with germline DDR gene mutation were younger, while patients with somatic DDR mutation were more likely to accumulate in metastatic lesions and had higher TMB levels which was consistent with previous reports.25,37,38 Our core DDR genes could screen part of PDAC population with genetic instability. Detecting more DDR genes can indeed screen a higher proportion of patients with DDR gene mutations,39 but its clinical value needs to be further evaluated. Integrating genomic scars and signature assessment by whole genome sequencing (WGS) and DDR deficiency seems to be more helpful to screen the proper population who benefit from PARP inhibitor or other DNA damaging agents.39, 40, 41 In addition, patients with TP53 mutation accounted for 70.6% of the total population, while TP53 mutation and DDR gene mutations are mutually exclusive. TP53 is one of the important effector genes of the DDR pathway which should be paid more attention to further study.11,12 WEE1 inhibitor, a specific DDR inhibitor was reported to trigger synthetic lethality in the absence of TP53 and enhance chemotherapy cytotoxicity which combined with other targeted agents or immunotherapy may have a promising future in patients with pancreatic cancer.42,43 Furthermore, patients with DDR gene mutations, especially somatic mutations, are associated with higher TMB level which may predict the efficacy of immunotherapy. This is reported in various tumors,44, 45, 46, 47 however, further clinical exploration for DDR mutant subtype is needed in pancreatic cancer.
There are also several limitations in this study. First, this study is a retrospective study that may suffer from selection bias (e.g., different gene-panels, TMB testing, MSI testing); however, the collection and detection of all samples were in real-time and quality control satisfied. Moreover, although our panel-based NGS analysis involved most of the vital genes in PDAC, some potentially valuable genes may have been overlooked. Combined and comprehensive whole-genome, transcriptome, and proteome multi-omic analyses need to be undertaken. Furthermore, therapeutic profiling and prognosis information matching genomics are limited, and well-designed clinical trials should be conducted in the future.
In summary, this study revealed the characteristics of genomic profiling by clinical sequencing in real-world large-scale patients with PDAC from China. We hope that our findings are conducive to identify potentially targetable and predictive biomarkers, screen exceptional genetic subtypes in respond to specific treatment, and explore the clinical practice and novel agent development for PDAC patients.
Declarations
Contributors
LX and LWW designed and supervised the study. SQC, YZB, LX and LWW had verified the underlying data and had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. XFZ, TBM, BZ, SQC, LX, and LWW created the study methodology. YBL, DLF, YWS, XBZ, QL, XFZ, TBM, HYX, JJC, FJ, DQC, YW, JH, QX, WYG, SML, MY, JYM, JYY, YCW, YLW, DYST and XZ acquired samples and collected clinical data. BZ, SQC, and YZB processed samples and generated experimental data. XFZ, TBM, BZ, SQC, YZB, LX, and LWW designed and conducted the statistical analysis plan. XFZ, TBM, BZ, SQC, YXW, LX, and LWW analyzed and interpreted data. XFZ wrote the manuscript. XFZ and LWW obtained funding. All authors reviewed and approved the manuscript.
Declaration of interests
BZ, SQC, YZB and LX are employees of 3D Medicines Inc. The other authors declare that they have no conflict of interests.
Acknowledgments
Acknowledgements
We owe thanks to the patients in our study and their family members. This work was supported by a senior investigator LWW's fundings from Innovation Group Project of Shanghai Municipal Health Commission (2019CXJQ03), National Natural Science Foundation of China (81874048), Shanghai Municipal Commission of Health and Family Planning (2018ZHYL0223), Fostering Fund of Renji Hospital affiliated to Shanghai Jiao Tong University School of Medicine (PYIV-17-001), Shanghai Municipal Commission of Health and Family Planning Grant (2018ZHYL0223), Clinical Research Plan of SHDC (No. SHDC2020CR1035B), Shanghai Key Clinical Speciality (Oncology), Shanghai leading talents project, Innovative research team of high-level local universities in Shanghai. Also supported by XFZ's grant from Clinical plus Excellence Project (2020ZYA003) from Shanghai Nucleic Acid Chemistry and Nanomedicine Key Laboratory. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data sharing statement
The Cancer Genome Atlas (TCGA), International Cancer Genome Consortium (ICGC), University of Texas Southwestern Medical Center (UTSW), and Queensland Centre for Medical Genomics (QCMG) studies are available at www.cbioportal.org. Data and materials can be provided upon reasonable request to the corresponding author.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103897.
Contributor Information
Lei Xiong, Email: simon.t@3dmedcare.com.
Liwei Wang, Email: liweiwang@shsmu.edu.cn.
Appendix. Supplementary materials
References
- 1.Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395(10242):2008–2020. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 4.Von Hoff DD, Ervin T, Arena FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conroy T, Hammel P, Hebbar M, et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N Engl J Med. 2018;379(25):2395–2406. doi: 10.1056/NEJMoa1809775. [DOI] [PubMed] [Google Scholar]
- 6.Makohon-Moore A, Iacobuzio-Donahue CA. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat Rev Cancer. 2016;16(9):553–565. doi: 10.1038/nrc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grossberg AJ, Chu LC, Deig CR, et al. Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma. CA Cancer J Clin. 2020;70(5):375–403. doi: 10.3322/caac.21626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirsch FR, Suda K, Wiens J, Bunn PA., Jr. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet. 2016;388(10048):1012–1024. doi: 10.1016/S0140-6736(16)31473-8. [DOI] [PubMed] [Google Scholar]
- 9.Network Cancer Genome Atlas. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian ZR, Rubinson DA, Nowak JA, et al. Association of Alterations in Main Driver Genes With Outcomes of Patients With Resected Pancreatic Ductal Adenocarcinoma. JAMA Oncol. 2018;4(3) doi: 10.1001/jamaoncol.2017.3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearl LH, Schierz AC, Ward SE, Al-Lazikani B, Pearl FM. Therapeutic opportunities within the DNA damage response. Nat Rev Cancer. 2015;15(3):166–180. doi: 10.1038/nrc3891. [DOI] [PubMed] [Google Scholar]
- 12.Knijnenburg TA, Wang L, Zimmermann MT, et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018;23(1):239–254. doi: 10.1016/j.celrep.2018.03.076. .e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32(2):185–203. doi: 10.1016/j.ccell.2017.07.007. .e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yadav S, Kasi PM, Bamlet WR, et al. Effect of Germline Mutations in Homologous Recombination Repair Genes on Overall Survival of Patients with Pancreatic Adenocarcinoma. Clin Cancer Res. 2020;26(24):6505–6512. doi: 10.1158/1078-0432.CCR-20-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu H, Zhu Y, Pu N, et al. Association of Germline Variants in Human DNA Damage Repair Genes and Response to Adjuvant Chemotherapy in Resected Pancreatic Ductal Adenocarcinoma. J Am Coll Surg. 2020;231(5):527–535. doi: 10.1016/j.jamcollsurg.2020.06.019. e14. [DOI] [PubMed] [Google Scholar]
- 16.Golan T, Hammel P, Reni M, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381(4):317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rebelatto TF, Falavigna M, Pozzari M, et al. Should platinum-based chemotherapy be preferred for germline BReast CAncer genes (BRCA) 1 and 2-mutated pancreatic ductal adenocarcinoma (PDAC) patients? A systematic review and meta-analysis. Cancer Treat Rev. 2019;80 doi: 10.1016/j.ctrv.2019.101895. [DOI] [PubMed] [Google Scholar]
- 18.Wattenberg MM, Asch D, Yu S, et al. Platinum response characteristics of patients with pancreatic ductal adenocarcinoma and a germline BRCA1, BRCA2 or PALB2 mutation. Br J Cancer. 2020;122(3):333–339. doi: 10.1038/s41416-019-0582-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 22.Guo S, Shi X, Gao S, et al. The Landscape of Genetic Alterations Stratified Prognosis in Oriental Pancreatic Cancer Patients. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.717989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shui L, Li X, Peng Y, et al. The germline/somatic DNA damage repair gene mutations modulate the therapeutic response in Chinese patients with advanced pancreatic ductal adenocarcinoma. J Transl Med. 2021;19(1):301. doi: 10.1186/s12967-021-02972-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su D, Zhang D, Chen K, et al. High performance of targeted next generation sequencing on variance detection in clinical tumor specimens in comparison with current conventional methods. J Exp Clin Cancer Res. 2017;36(1):121. doi: 10.1186/s13046-017-0591-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singhi AD, George B, Greenbowe JR, et al. Real-Time Targeted Genome Profile Analysis of Pancreatic Ductal Adenocarcinomas Identifies Genetic Alterations That Might Be Targeted With Existing Drugs or Used as Biomarkers. Gastroenterology. 2019;156(8):2242–2253. doi: 10.1053/j.gastro.2019.02.037. .e4. [DOI] [PubMed] [Google Scholar]
- 26.Xiao J, Li W, Huang Y, et al. A next-generation sequencing-based strategy combining microsatellite instability and tumor mutation burden for comprehensive molecular diagnosis of advanced colorectal cancer. BMC Cancer. 2021;21(1):282. doi: 10.1186/s12885-021-07942-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Mao T, Zhang B, et al. Characterization of DNA damage response deficiency in pancreatic cancer patients from China. Cancer Commun (Lond) 2022;42(1):70–74. doi: 10.1002/cac2.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2020;17(3):153–168. doi: 10.1038/s41575-019-0245-4. [DOI] [PubMed] [Google Scholar]
- 29.Bournet B, Muscari F, Buscail C, et al. KRAS G12D Mutation Subtype Is A Prognostic Factor for Advanced Pancreatic Adenocarcinoma. Clin Transl Gastroenterol. 2016;7(3):e157. doi: 10.1038/ctg.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong DS, Fakih MG, Strickler JH, et al. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N Engl J Med. 2020;383(13):1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D, Xue JY, Lito P. Targeting KRAS(G12C): From Inhibitory Mechanism to Modulation of Antitumor Effects in Patients. Cell. 2020;183(4):850–859. doi: 10.1016/j.cell.2020.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hobbs GA, Baker NM, Miermont AM, et al. Atypical KRAS G12R Mutant Is Impaired in PI3K Signaling and Macropinocytosis in Pancreatic Cancer. Cancer Discov. 2020;10(1):104–123. doi: 10.1158/2159-8290.CD-19-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinsey CG, Camolotto SA, Boespflug AM, et al. Protective autophagy elicited by RAF-MEK-ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med. 2019;25(4):620–627. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bryant KL, Stalnecker CA, Zeitouni D, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med. 2019;25(4):628–640. doi: 10.1038/s41591-019-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguirre AJ, Nowak JA, Camarda ND, et al. Real-time Genomic Characterization of Advanced Pancreatic Cancer to Enable Precision Medicine. Cancer Discov. 2018;8(9):1096–1111. doi: 10.1158/2159-8290.CD-18-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultheis B, Reuter D, Ebert MP, et al. Gemcitabine combined with the monoclonal antibody nimotuzumab is an active first-line regimen in KRAS wildtype patients with locally advanced or metastatic pancreatic cancer: a multicenter, randomized phase IIb study. Ann Oncol. 2017;28(10):2429–2435. doi: 10.1093/annonc/mdx343. [DOI] [PubMed] [Google Scholar]
- 37.Hu C, Hart SN, Polley EC, et al. Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer. JAMA. 2018;319(23):2401–2409. doi: 10.1001/jama.2018.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parikh AR, He Y, Hong TS, et al. Analysis of DNA Damage Response Gene Alterations and Tumor Mutational Burden Across 17,486 Tubular Gastrointestinal Carcinomas: Implications for Therapy. Oncologist. 2019;24(10):1340–1347. doi: 10.1634/theoncologist.2019-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casolino R, Paiella S, Azzolina D, et al. Homologous Recombination Deficiency in Pancreatic Cancer: A Systematic Review and Prevalence Meta-Analysis. J Clin Oncol. 2021;39(23):2617–2631. doi: 10.1200/JCO.20.03238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park W, Chen J, Chou JF, et al. Genomic Methods Identify Homologous Recombination Deficiency in Pancreas Adenocarcinoma and Optimize Treatment Selection. Clin Cancer Res. 2020;26(13):3239–3247. doi: 10.1158/1078-0432.CCR-20-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golan T, O'Kane GM, Denroche RE, et al. Genomic Features and Classification of Homologous Recombination Deficient Pancreatic Ductal Adenocarcinoma. Gastroenterology. 2021;160(6):2119–2132. doi: 10.1053/j.gastro.2021.01.220. e9. [DOI] [PubMed] [Google Scholar]
- 42.Cuneo KC, Morgan MA, Sahai V, et al. Dose Escalation Trial of the Wee1 Inhibitor Adavosertib (AZD1775) in Combination With Gemcitabine and Radiation for Patients With Locally Advanced Pancreatic Cancer. J Clin Oncol. 2019;37(29):2643–2650. doi: 10.1200/JCO.19.00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong A, Mehanna H. WEE1 Inhibitor: Clinical Development. Curr Oncol Rep. 2021;23(9):107. doi: 10.1007/s11912-021-01098-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teo MY, Seier K, Ostrovnaya I, et al. Alterations in DNA Damage Response and Repair Genes as Potential Marker of Clinical Benefit From PD-1/PD-L1 Blockade in Advanced Urothelial Cancers. J Clin Oncol. 2018;36(17):1685–1694. doi: 10.1200/JCO.2017.75.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sen T, Rodriguez BL, Chen L, et al. Targeting DNA Damage Response Promotes Antitumor Immunity through STING-Mediated T-cell Activation in Small Cell Lung Cancer. Cancer Discov. 2019;9(5):646–661. doi: 10.1158/2159-8290.CD-18-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362(6411):eaar3593. doi: 10.1126/science.aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mouw KW, Goldberg MS, Konstantinopoulos PA, D'Andrea AD. DNA damage and repair biomarkers of immunotherapy response. Cancer discovery. 2017;7(7):675–693. doi: 10.1158/2159-8290.CD-17-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.