Abstract
Two 51-kD aluminum (Al)-induced proteins (RMP51, root membrane proteins of 51 kD) were recently discovered in an aluminum-resistant cultivar of wheat (Triticum aestivum) cv PT741 (Basu et al., 1994a). These proteins segregate with the aluminum resistance phenotype in a segregating population arising from a cross between Al-resistant cv PT741 and Al-sensitive cv Katepwa (Taylor et al., 1997). The proteins have been purified by continuous elution electrophoresis and analyzed by peptide microsequencing. Sequence analysis of the purified peptides revealed that they are homologous to the B subunit of the vacuolar H+-ATPase (V-ATPase) and the α- and β-subunits of the mitochondrial ATP synthase (F1F0-ATPase). To confirm that these ATPases are induced by Al, ATPase activity and transcript levels were analyzed under Al stress. Both V-ATPase and F1F0-ATPase activities were induced by Al and responded in a dose-dependent manner to 0 to 150 μm Al. In contrast, plasma membrane H+-ATPase (P-ATPase) activity decreased to 0.5× control levels, even when plants were exposed to 25 μm Al. Northern analysis showed that the transcript encoding the B subunit of V-ATPase increased by 2.2× in a dose-dependent manner, whereas levels of the transcript encoding the α-subunit of F1F0-ATPase remained constant. The effect of Al on ATPase activity in other cultivars was also examined. The Al-resistant cultivar, cv PT741, was the only cultivar to show induction of V- and F1F0-ATPases. These results suggest that the V-ATPase in cv PT741 is responding specifically to Al stress with the ATP required for its activity supplied by ATP synthase to maintain energy balance within the cell.
A wide range of proteins are induced by Al stress in wheat and other plant species. Al-induced proteins include membrane-bound (Basu et al., 1994a; Cruz-Ortega et al., 1997; Taylor et al., 1997), cytosolic (Richards et al., 1998), cytoskeletal (Cruz-Ortega et al., 1997), and exudate (Basu et al., 1994b, 1997) proteins. Many of these have been implicated as general stress response proteins (Snowden et al., 1995; Hamel et al., 1998). Others have been associated with oxidative and other stresses (Hamel et al., 1998; Richards et al., 1998). The potential roles of others are still unclear. However, few have been functionally characterized as to their possible roles in Al tolerance.
The majority of Al-induced gene products have been identified at the RNA level. These include the wali (wheat Al induced) and war (wheat Al regulated) gene products (Snowden and Gardner, 1993; Richards et al., 1994; Hamel et al., 1998), β-1,3-glucanase (Cruz-Ortega et al., 1997), glutathione S-transferase (Richards et al., 1998), and a fimbrin-like protein (Cruz-Ortega et al., 1997) from wheat. In Arabidopsis, transcripts encoding aldolase, peroxidase, glutathione S-transferase, blue-copper-binding protein, superoxide dismutase, and a reticuline:oxygen oxidoreductase homolog are all induced by Al (Richards et al., 1998). These gene products appear to be involved in general stress response, since they are also induced by oxidative stress (Richards et al., 1998), other toxic metals, low calcium levels, and wounding (Snowden et al., 1995; Hamel et al., 1998). Expression of genes encoding the Arabidopsis blue copper-binding protein and tobacco glutathione S-transferase, peroxidase, and GDP-dissociation inhibitor recently have been shown to confer resistance to Al in transgenic Arabidopsis (Ezaki et al., 2000).
Several gene products have been shown to be Al-induced by analysis of their activity. These include Glc-6-P dehydrogenase, 6-phosphogluconate dehydrogenase (Slaski, 1996), and vacuolar H+-ATPase (Kasai et al., 1992, 1993). In contrast, plasma membrane H+-ATPase (Matsumoto, 1988; Widell et al., 1994; Sasaki et al., 1995) is inhibited by Al. Glucan synthase II is inhibited by Al in vitro (Widell et al., 1994), but synthesis of its end-product, callose, is stimulated by Al stress in vivo (Zhang et al., 1994).
Another group of gene products have been shown to be Al-induced by comparing protein profiles of Al-sensitive and Al-resistant cultivars during Al stress (Delhaize et al., 1991; Ownby and Hruschka, 1991; Picton et al., 1991; Cruz-Ortega and Ownby, 1993; Somers et al., 1996). Only three Al-induced proteins have been shown to co-segregate with the Al-resistance phenotype. One of these is a 23-kD root exudate protein (Basu et al., 1997), whereas the other two are 51-kD, tonoplast associated proteins (RMP51), which are the focus of this study. These 51-kD proteins are specifically induced by Al in root tips of an Al-resistant cultivar of wheat (cv PT741). They are newly synthesized once Al stress begins, accumulate in a dose- and time-dependent manner, and then decline to control levels with the removal of Al stress (Basu et al., 1994). The identity of these proteins remained to be determined.
The aim of the present study was to identify and characterize the RMP51 proteins to better understand their role in the Al stress response. We have purified these proteins from an endomembrane-enriched membrane fraction isolated from an Al-resistant cultivar of wheat (Triticum aestivum) cv PT741, grown in the presence of Al and determined their identity using peptide microsequence data. Purified peptides were homologous to the B subunit of the V-ATPase and to the α- and β-subunits of F1F0-ATPase (mitochondrial ATP synthase). Both V-ATPase and F1F0-ATPase activities were induced by Al treatment in cv PT741 and not in other cultivars tested. These results suggest that up-regulation of ATPase activity in cv PT741 may be an adaptive response involved in Al resistance.
RESULTS
Purification of RMP51
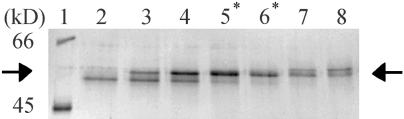
The first step in the purification of RMP51 was preparation of endomembrane-enriched membranes from Al-treated seedlings of wheat cv PT741. Membrane enrichment was confirmed using marker enzyme analysis (Briskin et al., 1987). Positive markers for the tonoplast (bafilomycin A1- and nitrate-sensitive ATPase activities) were enriched by 1.43× and 3.35×, respectively, whereas negative markers (vanadate-sensitive ATPase, glucan synthase II, and cytochrome c oxidase activities) were reduced to 0.45×, 0.69×, and 0.35× (Table I). Other endomembrane-associated proteins were then separated from RMP51 by continuous elution electrophoresis using the Miniprep Cell (Bio-Rad Laboratories, Hercules, CA). Separating conditions were optimized to purify the RMP51 band in a single step (Fig. 1). The optimized procedure was repeated five times with the RMP51 proteins isolated as a single band each time.
Table I.
Enrichment of marker enzymes in endomembrane-enriched membrane fractions isolated from root tips of wheat cv PT741
| Marker Enzyme | Cellular Location | Enrichment in
Endomembrane Fractiona
|
|
|---|---|---|---|
| Mean | se | ||
| Bafilomycin A1-sensitive ATPase | Tonoplast | 1.43 | 0.24 |
| Nitrate-sensitive ATPase | Tonoplast | 3.35 | 1.15 |
| Vanadate-sensitive ATPase | Plasma membrane | 0.45 | 0.19 |
| Glucan synthase II | Plasma membrane | 0.69 | 0.21 |
| Cytochrome c oxidase | Mitochondria | 0.35 | 0.02 |
Endomembrane-enriched membranes were isolated from microsomal membranes using a two-step gradient of Dextran T70 (2% and 10%), from which the interface was collected. Data are representative of three independent membrane preparations. Values shown are means ± se.
Enrichment in endomembrane fraction is calculated as endomembrane specific activity/microsomal specific activity.
Figure 1.
Fractionation of endomembrane-associated proteins by continuous elution electrophoresis. Total endomembrane proteins were isolated from root tips of wheat cv PT741 after 48 h of treatment with 100 μm AlCl3. Selected fractions (lanes 2–8) were analyzed for the presence of RMP51 (arrow) by SDS-PAGE and silver staining. Apparent molecular mass of protein standards are given in kilodaltons. Fractions pooled for further analysis are indicated by asterisks.
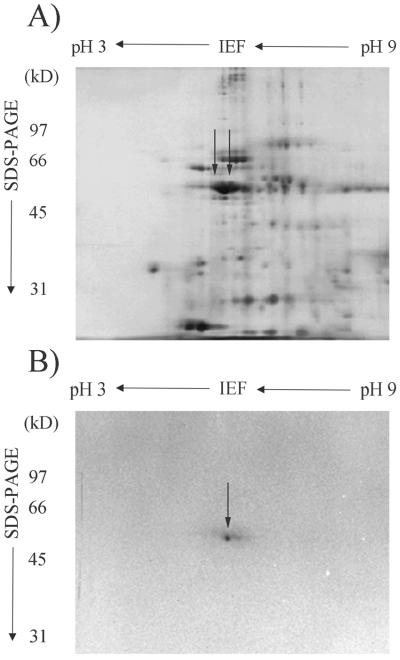
To determine whether or not both RMP51 proteins were present in the purified 51-kD band, aliquots were analyzed by two-dimensional electrophoresis. Although two RMP51 spots were visible in the crude endomembrane fraction (Fig. 2A), only one spot was visible in the purified sample (Fig. 2B). This may have been caused by the presence of residual SDS (even after detergent removal and addition of nonionic CHAPS {3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid} or by loss of one of the RMP51 proteins during purification. Only one protein spot was observed from every preparation. Total protein yield from five independent preparations was approximately 15 μg, sufficient for subsequent analysis by tryptic digestion, and peptide microsequencing.
Figure 2.
Two-dimensional gel analysis of crude endomembrane proteins (A) and purified RMP51 (B). Apparent molecular mass of protein standards (kD) are indicated on the left and the pH scale is shown on the top. RMP51 is indicated by solid arrows. Data shown are representative of five independent trials.
Peptide Microsequence Analysis
After tryptic digestion and fragment purification by HPLC, four peptides were sequenced and subsequently identified using the BLAST sequence algorithm (Altschul et al., 1997). Peptide 1 (QIYPPINVLPSLSR) was identical to amino acids 365 to 378 of the V-ATPase B subunit from barley (Hordeum vulgare, accession no. Q40078). Peptide 2 (FVAQGAYDTR) also showed 100% identity to the barley V-ATPase B subunit (accession no. Q40078, amino acids 440–449). Peptide 3 (FTAQANSEVSALLGR) showed 100% identity to amino acids 338 to 349 of the F1F0-ATPase β-subunit from wheat (accession no. P20858) but no significant homology to the V-ATPase B subunit. Peptide 4 (TGSIVDVPAGK) showed 100% identity to amino acids 93 to 103 of the F1F0-ATPase α-subunit from wheat (accession no. P12862).
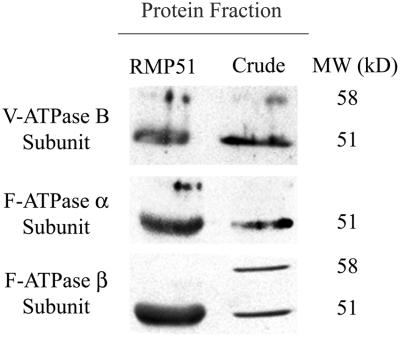
Results from the BLAST sequence comparison suggest that the four tryptic fragments were derived from the ATP-binding and catalytic subunits of related ATPases (V-ATPase and F1F0-ATPase). The possibility that they are all derived from a single protein was checked by aligning the four RMP51 peptide sequences with the B subunit of the V-ATPase from barley and the α- and β-subunits of the F1F0-ATPase from wheat. The three ATPase subunits share only approximately 25% identity, and the peptide sequences are not derived from regions with significant sequence homology between them. Therefore, it is unlikely that any single protein could contain all four peptides. The fact that more than one protein is present in the RMP51 band was confirmed by western analysis (Fig. 3). Antibodies raised against the V-ATPase B subunit from mung bean (Vigna radiata) and the F1F0-ATPase α- and β-subunits from Brewer's yeast (Saccharomyces cerevisiae) all cross-react with RMP51, suggesting that all three proteins are present. It is interesting that this contrasts with previous observations (Taylor et al., 1997) that antibodies raised against the B subunit of V-ATPase did not cross-react with RMP51. This inconsistency is most likely due to the more stringent binding conditions (37°C) used here or misalignment of the blots with protein gels in previous experiments, where alignment of cross-reacting species with a single band in a complex mixture had to be determined. The use of purified protein in the present experiments eliminated this complexity. Another possibility is that the cross-reaction reported here was due to lack of specificity in our antibodies. To rule out this possibility, our antibodies were tested against yeast strains with null mutations in the respective ATPase subunits (data not shown). Non-specific binding was not observed under the conditions used.
Figure 3.
Identification of RMP51 by immunoblotting. RMP51 and crude microsomal proteins isolated from Al-treated root tips of wheat cv PT741 were separated by SDS-PAGE, immunoblotted, and probed with polyclonal antibodies specific to the V-ATPase B subunit (from mung bean) and the F1F0-ATPase α- and β-subunits (from S. cerevisiae). Antibody labeling was detected using horseradish peroxidase-conjugated secondary antibodies and chemiluminescent detection. Apparent molecular mass of the cross-reacting bands indicated on the right are estimates based on migration of prestained molecular mass markers.
V-ATPase and F1F0-ATPase Are Al Induced
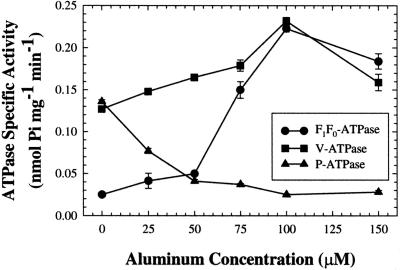
Because RMP51 protein levels increase in a dose-dependent manner with Al treatment (Taylor et al., 1997), levels of V-ATPase and F1F0-ATPase activity in Al-treated and control seedlings were compared. Activities were measured in total microsomal membrane fractions isolated from 5-d-old seedlings of wheat cv PT741 exposed to 0 to 150 μm AlCl3 for 2 d (Fig. 4). V-ATPase and F1F0-ATPase activities were both induced during Al stress, although to different extents. Vacuolar ATPase activity increased by 1.6× as concentrations of AlCl3 increased from 0 to 100 μm, followed by a decline to 1.1× the control level at 150 μm. Mitochondrial F1F0-ATPase activity increased by 7.3× as concentrations of AlCl3 increased from 0 to 75 μm, followed by a decline to 5.6× the control level. Plasma membrane ATPase activity decreased to 0.5× of the control level at 25 μm AlCl3 and to 0.2× control at 100 μm. Because both F1F0-ATPase and V-ATPase were induced by Al in a dose-dependent manner, it remains possible that the Al-induced RMP51 band initially identified by Basu et al. (1994a) consisted of both vacuolar and mitochondrial components. This is supported by western analysis of Al-treated microsomal fractions, which showed that protein levels of the V-ATPase B subunit and F1F0-ATPase α- and β-subunits all increased in response to Al (data not shown).
Figure 4.
The effect of Al on activity of the V-ATPase, F1F0-ATPase, and P-ATPase. ATPase specific activity was measured in total microsomal membranes prepared from 1-cm root tips of the Al-resistant cv PT741. Five-day-old seedlings were treated with different concentrations of Al (0, 25 50, 75, 100, and 150 μm) for 48 h. Values are means ± se of three biological replicates and are representative of three independent trials.
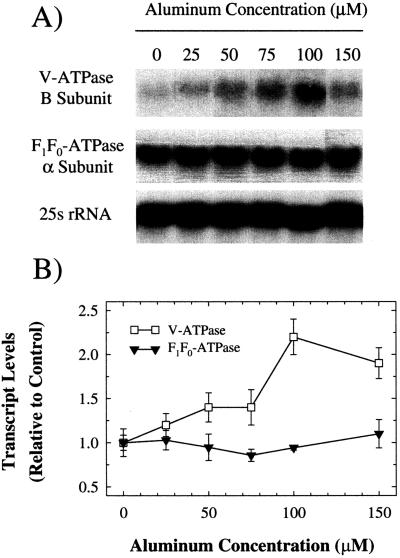
To determine whether or not there is a transcriptional component to the induction of these ATPases, northern analysis was performed on RNA isolated from the same tissues used for ATPase assays (Fig. 5). Levels of the transcript encoding the B subunit of the V-ATPase showed an increase to 2.2× control from 0 to 100 μm AlCl3. This increase, although relatively small, was consistent in three independent experiments and showed a pattern similar to that observed for V-ATPase activity (Fig. 4). This suggests that induction of V-ATPase activity by Al may be transcriptionally mediated, although participation of translational and post-translational mechanisms cannot be excluded. In contrast, levels of the transcript encoding the α-subunit of the F1F0-ATPase remained constant over the entire range of Al concentrations tested. Since F1F0-ATPase activity levels increased 7.3× over this range, it appears that this increase may be due to a translational or post-translational mechanism.
Figure 5.
The effect of Al on transcript levels of V-ATPase and F1F0-ATPase subunits. RNA was isolated from the same tissues used for ATPase activity measurements in Figure 4 and northern blotted. A, Autoradiographs of northern blots probed with 32-P labeled cDNAs encoding the V-ATPase B subunit from barley and the F1F0-ATPase α-subunit from N. plumbaginafolia. To ensure equal RNA loading and transfer, membranes were also probed with a cDNA clone encoding the 25s rRNA from Glycine max. Results shown are representative of three independent trials. B, Quantification of transcript levels shown in A. Transcript levels are expressed as the density of each band relative to 25S rRNA on the autoradiograph. Values are means ± se of three independent replicates.
Cultivar Screen for ATPase Induction
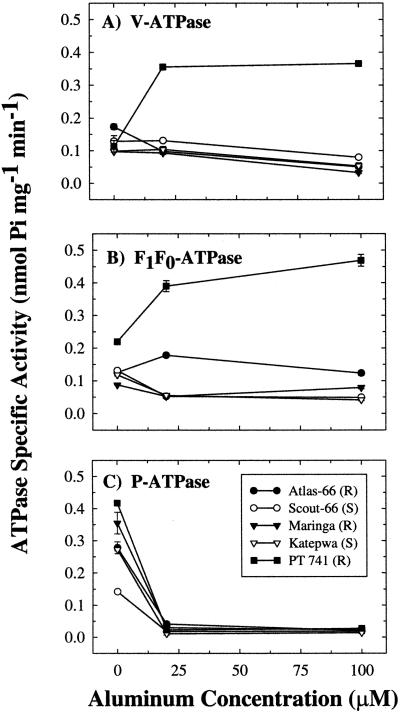
If induction of V-ATPase and F1F0-ATPase activities are general responses to Al stress, then induction should be observed in Al-resistant and Al-sensitive cultivars of wheat under stress conditions. To test this hypothesis, V-ATPase, F1F0-ATPase, and P-ATPase activities were measured in three Al-resistant and two Al-sensitive cultivars of wheat in control (0 μm AlCl3) conditions, conditions stressful to sensitive cultivars (20 μm AlCl3), and conditions stressful to resistant cultivars (100 μm AlCl3; Taylor et al., 1997). In cv PT741, V-ATPase activity increased by 2.1×, even at 20 μm AlCl3 (Fig. 6A). Vacuolar ATPase activities in cv Atlas-66 and cv Maringa (Al-resistant) were unaffected at 20 μm AlCl3 but reduced to 0.7× control at 100 μm AlCl3. In cv Scout-66 and cv Katepwa (Al-sensitive), a similar pattern was observed with reductions to 0.7× and 0.6× of control observed at 100 μm AlCl3.
Figure 6.
The effect of Al on ATPase specific activity in total microsomal membranes prepared from 1-cm root tips of cv PT741, cv Atlas-66, and cv Maringa (Al resistant), and from cv Scout-66 and cv Katepwa (Al sensitive). Five-day-old seedlings of each cultivar were exposed to 0, 20, or 100 μm Al for 48 h. Values represent means ± se of three independent replicates.
Cultivar PT741 also showed induction of F1F0-ATPase activity in this experiment (Fig. 6B). Control levels of F1F0-ATPase activity were slightly greater in cv PT741 than in the other cultivars tested, and activity increased by 1.8× at 20 μm AlCl3 and by 2.1× at 100 μm AlCl3. In cv Atlas-66, F1F0-ATPase activity increased to 1.4× at 20 μm AlCl3, but activity declined to control levels at 100 μm AlCl3. All other cultivars tested showed reduced F1F0-ATPase activities (cv Maringa, cv Scout-66, and cv Katepwa reduced to 0.1×, 0.6×, and 0.7× of control, respectively) at 20 μm AlCl3.
In contrast to the above observations, decreased activity of the plasma membrane ATPase was a general effect of Al treatment in all cultivars tested (Fig. 6C). Activities decreased to near zero in all cultivars even at 20 μm AlCl3. In summary, V-ATPase and F1F0-ATPase activities were specifically induced in cv PT741, whereas P-ATPase activity declined in all cultivars tested.
DISCUSSION
Differential screening of mRNA and protein profiles between Al-treated and -untreated tissues or between Al-resistant and Al-sensitive cultivars has revealed that expression of many genes is induced during Al stress (Snowden and Gardner, 1993; Richards et al., 1994; Snowden et al., 1995; Cruz-Ortega et al., 1997; Hamel et al., 1998). However, only three Al-induced proteins (Basu et al., 1994b; Taylor et al., 1997) have been shown to cosegregate with the Al-resistance phenotype. To better understand the role of these proteins, their identities must be determined.
Al stress causes a general decline in protein synthesis (35S-Met incorporation) in Al-sensitive cultivars of wheat, whereas there is little to no effect in Al-resistant cultivars (Ownby and Hruschka, 1991; Rincon and Gonzales, 1991; Basu et al., 1994a). Similar results have been observed in Medicago sativa (Campbell et al., 1994). Because pretreatment of Al-resistant cultivars of wheat (cv Atlas-66 and cv Grana) improves the resistance of plants to subsequent exposures, Aniol (1984) hypothesized that induction of new protein synthesis was involved in Al resistance. This hypothesis was supported by the subsequent observation that Al pretreatment induced Al tolerance in an Al-resistant cultivar of Phaseolus vulgaris L. (Cumming et al., 1992). We previously reported that two tonoplast-associated proteins are induced by Al stress in root tips of an Al-resistant cultivar of wheat, cv PT741 (Basu et al., 1994a) and that these proteins segregate with the Al-resistance phenotype in a cross between cv PT741 and cv Katepwa (Taylor et al., 1997). In this work, we used continuous elution electrophoresis to purify the 51-kD band containing these proteins from Al-treated seedlings of wheat cv PT741 (Fig. 1). Sequence analysis of purified peptides was then used to identify RMP51. Of four peptides sequenced, two were identical to the V-ATPase B subunit from barley, one was identical to the F1F0-ATPase α-subunit from wheat and one was identical to the F1F0-ATPase β-subunit from wheat. Although RMP51 was originally identified as two proteins, it was nonetheless surprising that both mitochondrial and vacuolar proteins were present in the 51-kD band, which had been purified from an endomembrane-enriched fraction (Table I). However, antibodies specific to the B subunit of the V-ATPase and the α- and β-subunits of the F1F0-ATPase all cross-reacted with the RMP51 band (Fig. 3), suggesting that there were in fact at least three proteins present in the purified band.
To determine whether the V-ATPase and the F1F0-ATPase are Al induced, ATPase activities were measured at various Al concentrations. Basu et al. (1994a) showed previously that RMP51 protein levels increased in an Al dose-dependent manner. If RMP51 is in fact a subunit of the V-ATPase or F1F0-ATPase, then activities of these enzymes should show a similar response to Al. Our data showed that V-ATPase and F1F0-ATPase activities increased by 1.6× and 7.3×, respectively, with Al treatment, whereas P-ATPase activity decreased to 0.2× control levels (Fig. 4). This parallels increases in protein levels of the V-ATPase B subunit and the F1F0-ATPase α- and β-subunits (data not shown). Induction of V-ATPase (and H+-PPiase) by Al has been previously demonstrated in tonoplast-enriched membrane vesicles from barley roots by Kasai et al. (1992, 1993). These authors observed a 40% to 53% increase in V-ATPase activity upon Al exposure, depending upon the external Ca2+ concentration. These results were confirmed by Zhang et al. (1998). Inhibition of P-ATPase activity has also been previously reported. Matsumoto (1988) observed a 50% decrease in P-ATPase activity in barley roots treated with 100 μm AlCl3 at pH 6.5, and a 45% decrease after treatment with 1 mm AlCl3 at pH 5.5 (Matsumoto et al., 1992). In subsequent experiments, Sasaki et al. (1995) showed that P-ATPase activity in wheat declined by 13% to 19% after treatment with 50 μm AlCl3 at pH 4.5. Widell et al. (1994) observed a similar effect in Picea abies and wheat.
The effect of Al treatment on F1F0-ATPase activity has not previously been investigated. We were initially surprised by the induction of F1F0-ATPase activity since localization experiments showed no enrichment of the RMP51 band in mitochondrial fractions (Taylor et al., 1997). However, western analysis of Al-treated wheat root microsomal fractions showed that band intensities corresponding to the α- and β-subunits do in fact increase (data not shown). It is possible that F1F0-ATPase activity is being modulated at the post-translational level (Stevens and Forgac, 1997). This is consistent with our observation that steady-state transcript levels remained constant, whereas activity increased by 7.3× (Figs. 4 and 5). Since Al induces F1F0-ATPase activity, increased ATP synthase activity may be required to support V-ATPase induction and other energy-dependent processes involved in Al resistance.
If induction of V-ATPase and F1F0-ATPase activities is a general effect of Al stress, then a similar response should be observed in all cultivars, whether Al-resistant or Al-sensitive. If induction is involved in mediating a general Al-resistance mechanism, induction should occur only in Al-resistant cultivars. Alternatively, induction may be a cultivar-specific resistance mechanism in cv PT741. To differentiate between these hypotheses, ATPase activities were measured in two additional Al-resistant cultivars (cv Atlas-66 and cv Maringa) and two Al-sensitive cultivars (cv Scout-66 and cv Katepwa). Our data show that Al-induction of V-ATPase and F1F0-ATPase is unique to cv PT741 (Fig. 6). In all other cultivars, V-ATPase and F1F0-ATPase activities remained constant or declined with Al treatment.
It is interesting that this phenomenon was observed only in cv PT741 and not in the other two Al-resistant cultivars tested, neither of which showed increased levels of RMP51 protein (data not shown). The lack of an effect in cv Maringa is perhaps not surprising since research on this cultivar has correlated exudation of a 23 kD, Al-binding polypeptide with Al resistance, and Al-resistance is controlled by a single dominant gene (Basu et al., 1997). In cv Atlas 66, Al resistance has also been correlated with enhanced exudation of malate during Al stress (Basu et al., 1994c).
Several observations suggest that induction of V-ATPase and F1F0-ATPase could be involved in mediating Al resistance in wheat cv PT741. First, subunits of these enzymes are newly synthesized upon Al treatment and RMP51 protein accumulates in an Al dose-dependent manner (Basu et al., 1994a). Second, accumulation of V-ATPase and F1F0-ATPase subunits (RMP51) segregates with the Al-resistance phenotype (Taylor et al., 1997). Third, V-ATPase and F1F0-ATPase activities increase in an Al dose-dependent manner only in the Al-resistant cv PT741.
Induction of V-ATPase activity has been linked to salt tolerance in several species (Ballesteros et al., 1996; Kirsch et al., 1996). Induction of V-ATPase activity is thought to be a homeostatic mechanism required to provide energy for Na+/H+ antiport, which delivers Na+ to the vacuole (Matsumoto and Chung, 1988; Reuveni et al., 1990; Nakamura et al., 1992; Colombo and Cerana, 1993). Kasai et al. (1992) suggested that a similar mechanism could be responsible for Al resistance in wheat with an Al+/H+ antiport system driving sequestration of Al in the vacuole. This suggestion should be taken with caution, however, since there is no direct evidence for an Al+/H+ co-transporter. Increased V-ATPase activity could also be required as a homeostatic mechanism to maintain the cytoplasmic pH near neutrality. We have shown that Al exposure decreases plasma membrane ATPase activity in agreement with results reported previously (Matsumoto, 1988; Matsumoto et al., 1992; Widell et al., 1994; Sasaki et al., 1995). This could cause a decrease in cytoplasmic pH with adverse physiological effects. Increased activity of V-ATPase, with energy balance maintained by increased ATP synthase activity, could counteract these changes by transporting protons into the vacuole. A similar phenomenon has been observed in Lactobacillus acidophilus, which expresses a pH-inducible F1F0-ATPase to extrude protons and maintain cytoplasmic pH (Kullen and Klaenhammer, 1999).
In summary, we have purified the Al-induced RMP51 proteins from the Al-resistant cultivar of wheat, cv PT741 and shown that the RMP51 band consists of the B subunit of the V-ATPase and the α- and β-subunits of the F1F0-ATPase. Vacuolar ATPase activity increased by 1.6 to 2.1×, while levels of the transcript encoding the B subunit increased by 2.2×, from 0 to 100 μm AlCl3. Mitochondrial F1F0-ATPase activity increased by 2.1 to 7.3× with a constant level of the transcript encoding the α-subunit over the same range of Al concentrations. Increased V-ATPase and F1F0-ATPase activities are observed only in cv PT741, suggesting that induction of V-ATPase and F1F0-ATPase activities is not simply symptomatic of stress. These results allow us to put forward the hypothesis that induction of the V-ATPase and the F1F0-ATPase plays a role in Al resistance. We are now testing this hypothesis using transgenic Arabidopsis expressing the V-ATPase B subunit gene in the antisense orientation to determine whether lack of these activities causes hypersensitivity to Al. Moreover, the demonstration that tolerance to Al in cv PT741 is mediated by a different mechanism than is observed in other varieties of wheat (e.g. cv Maringa) suggests the possibility of combining these traits genetically to produce a cultivar with enhanced Al tolerance.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of Al-resistant (cv PT741, cv Atlas-66, and cv Maringa) and Al-sensitive (cv Scout-66 and cv Katepwa) cultivars of wheat (Triticum aestivum) were surface sterilized in 10% (v/v) sodium hypochlorite for 15 to 20 min and germinated overnight in a 0.005 g L−1 solution of the antifungal agent Vitavax (Uniroyal Chemical Ltd, Calgary, AB, Canada) in double distilled water to limit fungal growth. Seedlings were grown for 5 d on nylon mesh floating over 15 L of an aerated mineral nutrient solution (pH 4.30) containing: 2,900 μm NO3, 300 μm NH4, 100 μm PO4, 800 μm K, 1,000 μm Ca, 300 μm Mg, 101 μm SO4, 34 μm Cl, 60 μm Na, 10 μm Fe, 6 μm B, 2 μm Mn, 0.15 μm Cu, 0.5 μm Zn, 0.1 μm Mo, and 10 μm EDTA. For Al exposure, 5-d-old seedlings were transferred to solutions containing: 1,000 μm Ca, 300 μm Mg, 300 μm NH4, 2,900 μm NO3, and 0 to 150 μm AlCl3 at pH 4.30 (Taylor et al., 1997). The seedlings were grown in a growth chamber (16-h light, 20°C, 68% relative humidity and 8-h darkness, 16°C, 85% relative humidity) for 2 d. Taylor et al. (1997) demonstrated that 100 μm AlCl3 is optimal for induction of the proteins of interest in cv PT741. After 2 d of Al exposure, 1-cm root tips were harvested for isolation of endomembranes and subsequent purification of the 51-kD proteins (RMP51).
Isolation of Endomembrane-Enriched Membranes
All steps involved in endomembrane preparation were carried out at 4°C. Root tips (1 cm) were finely chopped and immediately homogenized in a Proctor-Silex blender in homogenization buffer (0.25 m Suc, 50 mm MOPS [3-(N-morpholino)propanesulfonic acid]-Tris, pH 7.5, 5 mm EDTA, and 5 mm ascorbic acid), 1 mL g−1 root tissue. The homogenate was then filtered through miracloth (Calbiochem, San Diego) and centrifuged at 20,000g for 15 min. The supernatant was collected and centrifuged at 100,000g for 1 h. The microsomal membrane pellet was resuspended in gradient buffer (0.25 m sorbitol, 5 mm HEPES-BTP, pH 7.0) and loaded onto a two-step gradient of 5 mL each of 2% and 10% (w/w) Dextran T-70 prepared in gradient buffer (Kasai et al., 1992). The gradient was centrifuged at 70,000g for 2 h, and the interface containing endomembranes was collected, diluted with gradient buffer, and centrifuged at 120,000g for 1 h. The endomembrane-enriched membrane pellet was resuspended in 10 mm Tris-acetate, pH 7.9, 10% (v/v) glycerol and either frozen at −80°C for marker enzyme analysis, or used immediately for further purification.
Membrane Marker Assays
To ensure that the isolated membrane fraction was enriched for endomembranes, marker enzyme analysis was carried out according to Briskin et al. (1987). Activities of the NO3−- and bafilomycin-sensitive ATPases, vanadate-sensitive ATPase and glucan synthase II (GSII), and cytochrome c oxidase were used as markers for tonoplast, plasma membrane, and mitochondria, respectively. Adenosine triphosphatase activity was assayed in a reaction mixture containing 30 mm Tris-MES (pH 8.0), 3 mm MgSO4, 0.2% (v/v) Triton X-100, 50 mm KCl, and 3 mm ATP-Tris, in the presence or absence of 250 μm Na3VO4 (P-Type ATPase activity), 1 mm NaN3 (F1F0-type ATPase activity), 100 nm bafilomycin A1 or 50 mm KNO3 (V-type ATPase activity). Phosphatase activity was measured as described by Ames (1966).
Purification of RMP51
Separation of the 51-kD proteins (RMP51) from other endomembrane-associated proteins was achieved by continuous elution electrophoresis (Miniprep Cell, Bio-Rad Laboratories). Total endomembrane protein (up to 500 μg per run), was prepared for electrophoresis by adding an equivalent volume of SDS-PAGE loading buffer (0.125 m Tris-HCl, pH 6.8, 4% [w/v] SDS, 20% [v/v] glycerol, 10% [v/v] β-mercaptoethanol, 0.002% [v/v] bromphenol blue) and heating at 95°C for 5 min. Separating conditions for SDS-PAGE were as follows: 1.5 cm (4%) stacking gel, 10 cm (10%) separating gel. Electrophoresis/elution was carried out at 4°C at a constant current of 5 mA. Fractions (200 μL) were collected after the dye front had run off the gel and analyzed for the presence of RMP51 by SDS-PAGE (Mini Protean II, Bio-Rad Laboratories). Fractions containing RMP51 were pooled and desalted using Sephadex G-50 gel filtration chromatography (Nick Column, Amersham-Pharmacia Biotech, Uppsala). Desalted samples were then analyzed for purity using two-dimensional electrophoresis (Mini Protean II 2D Cell, Bio-Rad Laboratories) according to the manufacturer's directions.
SDS-PAGE
Samples were prepared for SDS-PAGE by adding an equivalent volume of SDS-PAGE loading buffer and heating at 95°C for 5 min. Running conditions for SDS-PAGE (Laemmli, 1970) were: 4% (w/v) stacking gel (at 12.5 mA) and 10% (w/v) resolving gel (at 25 mA).
Two-Dimensional Electrophoresis
First-dimension isoelectric focusing gels (4% [w/v] acrylamide, 9.2 m urea, 2.0% [w/v] CHAPS, 1.6% [v/v] 5/7 ampholyte [Bio-Rad Laboratories], and 0.4% [v/v] 3/10 ampholyte [Bio-Rad Laboratories]) were prefocused at 200 V/10 min, 300 V/15 min, and 400 V/15 min. Microsomal membrane samples were extracted and precipitated as described by Hurkman and Tanaka (1986). Approximately 250 ng of purified protein or 20 μg of crude protein was then mixed with isoelectric focusing sample buffer (9.5 m urea, 2.0% [w/v] CHAPS, 5% [v/v] β-mercaptoethanol, 1.6% [v/v] 5/7 ampholyte [Bio-Rad Laboratories], 0.4% [v/v] 3/10 ampholyte [Bio-Rad Laboratories]) and heated at 70°C for 20 min. After changing electrolytes and sample loading, isoelectric focusing was run at 500 V for 10 min and 750 V for 3.5 h using the Mini Protean II 2D Cell (Bio-Rad Laboratories). Second dimension electrophoresis was carried out as described above.
Protein Quantitation and Visualization
Quantitation of protein present in membrane samples was performed using either the Bradford assay (Bradford, 1976) or comparing the band density of samples and known quantities of a standard protein (chicken egg ovalbumin: model A5503, Sigma, St. Louis) of similar molecular mass (45 kD) on denaturing polyacrylamide gels. Proteins present in polyacrylamide gels were visualized using either a modified Morrissey silver stain procedure (Merril et al., 1981; Morrissey, 1981) or staining with Coomassie Brilliant Blue R-250 (CBB R-250 Staining Kit, Bio-Rad Laboratories).
Immunoblotting
Microsomal proteins and RMP51 were separated by SDS-PAGE and electroblotted onto nitrocellulose (0.45 μm, Bio-Rad Laboratories) membranes using the Mini Trans-Blot Cell (Bio-Rad Laboratories) with a transfer buffer of 25 mm Tris, 192 mm Gly (pH 8.3), and 20% (v/v) methanol. Transfer was performed at 100 V at 4°C for 1 h. Membranes were blocked overnight in Tris-buffered saline plus Tween 20 (TBST; 20 mm Tris, pH 7.5, 140 mm NaCl, 0.1% [v/v] Tween 20) and 5% (w/v) skim milk powder. Membranes were incubated with primary antibodies diluted in TBST at 37°C for 1 h, followed by three 15-min washes in PBST (80 mm Na2HPO4, 20 mm NaH2PO4, 100 mm NaCl, 0.1% [v/v] Tween 20). Membranes labeled with primary antibody were incubated with horseradish peroxidase-conjugated secondary antibodies (A6154, Sigma) diluted 1:15, 000 in PBST at 4°C for 1 h, followed by three 15-min washes. Chemiluminescent detection (Kirkegaard and Perry Laboratories, 54–61–00) was carried out according to the manufacturer's directions.
Peptide Microsequence Analysis
A 10-μg sample of purified RMP51 was excised from a denaturing polyacrylamide gel and sent to the Harvard Microchemistry Facility (Cambridge, MA) for tryptic digestion, peptide separation, and microsequence analysis. Sequence analysis of the isolated peptides was performed using the BLAST algorithm (Altschul et al., 1997).
RNA Isolation, Northern Hybridization, and Analysis
RNA was isolated from 1-cm root tips using the RNeasy Plant Mini Kit (Qiagen USA, Valencia, CA) according to the manufacturer's directions. Each 100 mg of tissue sample yielded approximately 50 μg of RNA. RNA concentration was estimated by measuring A260 and checked by running aliquots on non-denaturing 1% (w/v) agarose gels. Size standards ( RNA ladder, Gibco-BRL, Cleveland) were included on all gels. Northern transfers were carried out using GeneScreen Plus (DuPont-Dow Elastomers L.L.C., Wilmington, DE) membranes according to the manufacturer's directions.
Probes for northern blots were prepared from cDNA fragments isolated from pHTB1 (Berkelman et al., 1994) and pAH (Chaumont et al., 1988) plasmids (V-ATPase B subunit from barley (Hordeum vulgare) and F1F0-ATPase α-subunit from Nicotiana plumbaginafolia, respectively). Plasmids were digested with appropriate restriction enzymes (Amersham-Pharmacia Biotech) and the released cDNA inserts were isolated from agarose gels using the QiaQuick gel extraction kit (Qiagen, USA). Probes were then prepared by random priming (Sambrook et al., 1989) using 100 ng of each DNA template. Unincorporated nucleotides (including [32P]dCTP) were removed using Sephadex G-50 size exclusion chromatography (Nick Column, Amersham-Pharmacia Biotech). Membranes were prehybridized at 42°C with 100 μg/mL denatured, sheared, herring sperm DNA (Sigma) for 4 h. Hybridization was carried out overnight at 42°C and washed twice with 2× sodium chloride/sodium phosphate/EDTA (1× SSPE: 0.15 m NaCl, 0.01 m NaH2PO4-H2O, 0.001 m EDTA-Na2, pH 7.4) for 15 min at room temperature, twice with 2× SSPE, 2% (w/v) SDS for 45 min at 65°C, and twice with 0.1× SSPE for 15 min at room temperature. Membranes were exposed to X-OMAT x-ray film (Kodak, Rochester, NY) at −80°C. Transcript levels were measured by densitometry of autoradiographs using an Alphaimager 2000 Documentation and Analysis System (Version 5.1, Alpha Innotech, San Leandro, CA). For figure preparation, representative lanes of triplicate samples were selected from the same exposure of the same blot and compiled using Adobe PhotoshopR version 5.5.
ACKNOWLEDGMENTS
We wish to thank Dr Francois Chaumont (University of Louvain) for providing the plasmid encoding the mitochondrial F1-ATPase α-subunit from N. plumbaginafolia and Lance Larka (Genetic Resources Conservation Program, University of California) for providing the cDNA clone of the V-ATPase B subunit from barley. Polyclonal antibodies raised against the α- and β-subunits of the yeast F1-ATPase were generously provided by A. Lewin (University of Florida). Antibodies raised against the B subunit of the Vigna radiata V-ATPase were provided by M. Maeshima (Nagoya University).
Footnotes
This work was supported by the Research Grants Program of the Natural Sciences and Engineering Research Council of Canada, by the Canadian Wheat Board Graduate Fellowship Program, and by the University of Alberta, Department of Biological Sciences (to C.A.H.).
LITERATURE CITED
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN. Assay of inorganic phosphate, total phosphate and phosphatase. Methods Enzymol. 1966;8:115–118. [Google Scholar]
- Aniol A. Induction of aluminum tolerance in wheat seedlings by low doses of aluminum in the nutrient solution. Plant Physiol. 1984;75:551–555. doi: 10.1104/pp.76.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros E, Donaire JP, Belver A. Effects of salt stress on H+-ATPase and H+-PPiase activities of tonoplast-enriched vesicles isolated from sunflower roots. Physiol Plant. 1996;97:259–268. [Google Scholar]
- Basu A, Basu U, Taylor GJ. Induction of microsomal membrane proteins in roots of an aluminum-resistant cultivar of Triticum aestivum L. under conditions of aluminum stress. Plant Physiol. 1994a;104:1007–1013. doi: 10.1104/pp.104.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu U, Basu A, Taylor GJ. Differential exudation of polypeptides by roots of aluminum-resistant and aluminum-sensitive cultivars of Triticum aestivum L. in response to aluminum stress. Plant Physiol. 1994b;106:151–158. doi: 10.1104/pp.106.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu U, Godbold D, Taylor GJ. Aluminum resistance in Triticum aestivum associated with enhanced exudation of malate. J Plant Physiol. 1994c;144:747–753. [Google Scholar]
- Basu U, McDonald-Stephens JL, Archambault DJ, Good AG, Briggs KG, Taing-Aung, Taylor GJ. Genetic and physiological analysis of doubled-haploid, aluminum resistant lines of wheat provide evidence for the involvement of a 23 kD, root exudate polypeptide in mediating resistance. Plant Soil. 1997;196:283–288. [Google Scholar]
- Berkelman T, Houtchens KA, Dupont FM. Two cDNA clones encoding isoforms of the B subunit of the vacuolar ATPase from barley roots. Plant Physiol. 1994;104:287–288. doi: 10.1104/pp.104.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Briskin DP, Leonard RT, Hodges TK. Isolation of the plasma membrane: membrane markers and general principals. In: Packer L, Douce R, editors. Methods in Enzymology. Vol. 148. New York: Academic Press; 1987. pp. 542–558. [Google Scholar]
- Campbell TA, Jackson PR, Xia ZL. Effects of aluminum stress on alfalfa root proteins. J Plant Nutr. 1994;17:461–471. [Google Scholar]
- Chaumont F, Boutry M, Briquet M, Vassarotti A. Sequence of the gene encoding the mitochondrial F1-ATPase α subunit from Nicotiana plumbaginafolia. Nucleic Acids Res. 1988;16:6247. doi: 10.1093/nar/16.13.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo R, Cerana R. Enhanced activity of tonoplast pyrophosphatase in NaCl-grown cells of Daucus carota. J Plant Physiol. 1993;142:226–229. [Google Scholar]
- Cruz-Ortega R, Cushman JC, Ownby JD. cDNA clones encoding 1,3-β-glucanase and a fimbrin-like cytoskeletal protein are induced by Al toxicity in wheat roots. Plant Physiol. 1997;114:1453–1460. doi: 10.1104/pp.114.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ortega R, Ownby JD. A protein similar to PR (pathogenesis-related) proteins is elicited by metal toxicity in wheat roots. Physiol Plant. 1993;89:211–219. [Google Scholar]
- Cumming JR, Buckelew Cumming A, Taylor GJ. Patterns of root respiration associated with the induction of aluminum tolerance in Phaseolus vulgaris L. J Exp Bot. 1992;43:1075–1081. [Google Scholar]
- Delhaize E, Higgins TJV, Randall PJ. Aluminum tolerance in wheat: analysis of polypeptides in the root apices of tolerant and sensitive genotypes. In: Wright RJ, Baligar VC, Murrman RP, editors. Plant-Soil Interactions at Low pH: Proceedings of the Second International Symposium on Plant-Soil Interactions at Low pH, 24–29 June 1990, Beckley, WV. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 1071–1079. [Google Scholar]
- Ezaki B, Gardner RC, Ezaki Y, Matsumoto H. Expression of aluminum-induced genes in transgenic Arabidopsis plants can ameliorate aluminum stress and/or oxidative stress. Plant Physiol. 2000;122:657–665. doi: 10.1104/pp.122.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel F, Breton C, Houde M. Isolation and characterization of wheat aluminum-regulated genes: possible involvement of aluminum as a pathogenesis response regulator. Planta. 1998;205:531–538. doi: 10.1007/s004250050352. [DOI] [PubMed] [Google Scholar]
- Hurkman WJ, Tanaka CK. Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 1986;81:802–806. doi: 10.1104/pp.81.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai M, Sasaki M, Tanakamaru S, Yamamoto Y, Matsumoto H. Possible involvement of abscisic acid in increases in activities of two vacuolar H+-pumps in barley roots under aluminum stress. Plant Cell Physiol. 1993;34:1335–1338. [Google Scholar]
- Kasai M, Sasaki M, Yamamoto Y, Matsumoto H. Aluminum stress increases K+ efflux and activities of ATP- and PPi-dependent H+ pumps of tonoplast-enriched membrane vesicles from barley roots. Plant Cell Physiol. 1992;33:1035–1039. [Google Scholar]
- Kirsch M, Zhigang A, Viereck R, Low R, Rausch T. Salt stress induces an increased expression of V-type H+-ATPase in mature sugar beet leaves. Plant Mol Biol. 1996;32:543–547. doi: 10.1007/BF00019107. [DOI] [PubMed] [Google Scholar]
- Kullen MJ, Klaenhammer TR. Identification of the pH-inducible, proton-translocating F1F0-ATPase (atpBEFHAGDC) operon of Lactobacillus acidophilus by differential display: gene structure, cloning and characterization. Mol Microbiol. 1999;33:1152–1161. doi: 10.1046/j.1365-2958.1999.01557.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of the structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Matsumoto H. Inhibition of proton transport activity of microsomal membrane vesicles of barley roots by aluminum. Soil Sci Plant Nutr. 1988;34:499–526. [Google Scholar]
- Matsumoto H, Chung GC. Increase in proton transport activity of tonoplast vesicles as an adaptive response of barley roots to NaCl stress. Plant Cell Physiol. 1988;29:1133–1140. [Google Scholar]
- Matsumoto H, Yamamoto Y, Kasai M. Changes of some properties of the plasma membrane-enriched fraction of barley roots related to aluminum stress: membrane-associated ATPase, aluminum and calcium. Soil Sci Plant Nutr. 1992;38:411–419. [Google Scholar]
- Merril CR, Goldman D, Sedman SA, Ebert MH. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981;211:1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- Morrissey JH. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kasamo K, Shimosato N, Sakata M, Ohta E. Stimulation of the extrusion of protons and H+-ATPase activities with the decline of pyrophosphatase activity of the tonoplast in intact mung bean roots under high NaCl stress and its relation to external levels of Ca2+ ions. Plant Cell Physiol. 1992;33:139–149. [Google Scholar]
- Ownby JD, Hruschka WR. Quantitative changes in cytoplasmic and microsomal proteins associated with aluminum toxicity in two cultivars of winter wheat. Plant Cell Environ. 1991;14:303–309. [Google Scholar]
- Picton SJ, Richards KD, Gardner RC. Protein profiles in root-tips of two wheat (Triticum aestivum L.) cultivars with differential tolerance to aluminum. In: Wright RJ, Baligar VC, Murrmann RP, editors. Plant-Soil Interactions at Low pH. Proceedings of the Second International Symposium on Plant-Soil Interactions at Low pH, 24–29 June 1990, Beckley, WV. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 1063–1070. [Google Scholar]
- Reuveni M, Bennett AB, Bressan RA, Hasegawa PM. Enhanced H+ transport capacity and ATP hydrolysis activity of the tonoplast H+-ATPase after NaCl adaptation. Plant Physiol. 1990;94:524–530. doi: 10.1104/pp.94.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KD, Schott EJ, Sharma YK, Davis KR, Gardner RC. Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiol. 1998;116:409–418. doi: 10.1104/pp.116.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KD, Snowden KC, Gardner RC. Wali6 and wali7: genes induced by aluminum in wheat (Triticum aestivum L.) roots. Plant Physiol. 1994;105:1455–1456. doi: 10.1104/pp.105.4.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon M, Gonzales RA. Induction of protein synthesis by aluminum in wheat (Triticum aestivum L.) root tips. In: Wright RJ, Baligar VC, Murrmann RP, editors. Plant-Soil Interactions at Low pH. Proceedings of the Second International Symposium on Plant-Soil Interactions at Low pH, 24–29 June 1990, Beckley, WV. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 851–858. [Google Scholar]
- Sambrook J, Fritsch T, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sasaki M, Kasai M, Yamamoto Y, Matsumoto H. Involvement of plasma membrane potential in the tolerance mechanism of plant roots to aluminum toxicity. In: Date RA, Grunden NJ, Rayment GE, Probert ME, editors. Plant-Soil Interactions at Low pH. Proceedings of the Third International Symposium on Plant-Soil Interactions at Low pH, 12-16 September 1993, Brisbane, Queensland, Australia. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 285–290. [Google Scholar]
- Slaski JJ. Aluminum resistance in wheat (Triticum aestivum) is associated with rapid, Al-induced changes in activities of glucose-6-phosphate dehydrogenase and 6-phosphoglucanate dehydrogenase in root apices. Physiol Plant. 1996;98:477–484. [Google Scholar]
- Snowden KC, Gardner RC. Five genes induced by aluminum in wheat (Triticum aestivum L.) roots. Plant Physiol. 1993;103:855–861. doi: 10.1104/pp.103.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden KC, Richards KD, Gardner RC. Aluminum-induced genes: induction by toxic metals, low calcium, and wounding and pattern of expression in root tips. Plant Physiol. 1995;107:341–348. doi: 10.1104/pp.107.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DJ, Briggs KG, Gustafson JP. Aluminum stress and protein synthesis in near isogenic lines of Triticum aestivum differing in aluminum tolerance. Physiol Plant. 1996;97:694–700. [Google Scholar]
- Stevens TH, Forgac M. Structure, function and regulation of the vacuolar (H+)-ATPase. Annu Rev Cell Dev Biol. 1997;13:779–808. doi: 10.1146/annurev.cellbio.13.1.779. [DOI] [PubMed] [Google Scholar]
- Taylor GJ, Basu A, Basu U, Slaski JJ, Zhang G, Good A. Al-induced, 51 kilodalton, membrane-bound proteins are associated with resistance to Al in a segregating population of wheat. Plant Physiol. 1997;114:363–372. doi: 10.1104/pp.114.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widell S, Asp H, Jensen P. Activities of plasma membrane-bound enzymes isolated from roots of spruce (Picea abies) grown in the presence of aluminum. Physiol Plant. 1994;92:459–466. [Google Scholar]
- Zhang G, Hoddinott J, Taylor GJ. Characterization of 1,3-β-glucan (callose) synthesis in roots of Triticum aestivum in response to aluminum toxicity. J Plant Physiol. 1994;144:229–234. [Google Scholar]
- Zhang W, Zhang F, Shen Z, Liu Y. Changes of H+ pumps of tonoplast vesicle from wheat roots in vivo and in vitro under aluminum treatment and effect of calcium. J Plant Nutr. 1998;21:2515–2526. [Google Scholar]