Abstract
The aggressive nature of pancreatic ductal adenocarcinoma (PDAC) mandates the development of improved therapies. Since KRAS mutations are found in 95% of PDAC and are critical for tumor maintenance, one promising strategy involves exploiting KRAS-dependent metabolic perturbations. The macrometabolic process of autophagy is upregulated in KRAS-mutant PDAC, and PDAC growth is reliant on autophagy. However, inhibition of autophagy as monotherapy using the lysosomal inhibitor hydroxychloroquine (HCQ) has shown limited clinical efficacy. To identify strategies that can improve PDAC sensitivity to HCQ, we applied a CRISPR-Cas9 loss-of-function screen and found that a top sensitizer was the receptor tyrosine kinase (RTK) insulin-like growth factor 1 receptor (IGF1R). Additionally, RPPA pathway activation mapping profiled the signaling pathways altered by CQ treatment. Activating phosphorylation of RTKs, including IGF1R, were common compensatory increases in response to CQ. Inhibition of IGF1R increased autophagic flux and sensitivity to CQ-mediated growth suppression both in vitro and in vivo. Co-targeting both IGF1R and pathways that antagonize autophagy, such as ERK-MAPK axis, was strongly synergistic. IGF1R and ERK inhibition converged on suppression of glycolysis, leading to enhanced dependence on autophagy. Accordingly, concurrent inhibition of IGF1R, ERK, and autophagy induced cytotoxicity in PDAC cell lines, and decreased viability in human PDAC organoids. In conclusion, targeting IGF1R together with ERK enhances the effectiveness of autophagy inhibitors in PDAC.
Significance
Compensatory upregulation of IGF1R and ERK-MAPK signaling limits the efficacy of autophagy inhibitors chloroquine/hydroxychloroquine, and their concurrent inhibition synergistically increases autophagy dependence and chloroquine sensitivity in pancreatic ductal adenocarcinoma.
Introduction
Metabolic reprogramming, wherein cancer cells acquire metabolic adaptations to support sustained proliferation, is a hallmark of cancer (1). Pancreatic ductal adenocarcinoma (PDAC) is driven by mutational activation of the KRAS oncogene and KRAS-mutant PDAC exhibits a spectrum of altered metabolic processes (2). One such metabolic process is elevated macroautophagy, a process of “self-eating” whereby cancer cells degrade and recycle macromolecules and defective organelles to supply amino acids and nucleotides to support their increased metabolic needs. Pharmacologic or genetic suppression of autophagy impairs the tumorigenic growth of PDAC, supporting anti-autophagy therapies as tractable treatment strategies (3-6). Although clinical evaluation of the autophagy inhibitor hydroxychloroquine (HCQ) as monotherapy did exhibit anti-tumor activity, disappointingly, it did not result in increased overall survival in PDAC patients (7,8).
The limited clinical efficacy of the only two FDA approved autophagy inhibitors, HCQ and the related analog chloroquine (CQ), has been speculated to involve their limited potency and indirect mechanism of action in blocking autophagy (9). HCQ/CQ impair lysosomal activities and disrupt the final stage of the autophagy pathway, specifically by impairing autophagosome-lysosome fusion and degradation of cargo for nutrient release. Because they target lysosomal activity, HCQ/CQ additionally disrupt non-autophagy related activities that also terminate in the lysosome. Current strategies to develop more effective autophagy inhibitors include identification of more potent and selective lysosomal inhibitors, as well as inhibitors of other more specific components of the autophagy machinery (e.g., ULK1) (9). However, to date, more specific autophagy inhibitors have not been assessed clinically.
Recently we assessed whether mutant KRAS acts as a driver of elevated autophagy in PDAC, and observed that genetic suppression of KRAS or pharmacological inhibition of the key KRAS effector network, the RAF-MEK-ERK mitogen-activated protein kinase (MAPK) cascade, unexpectedly resulted in enhancement of autophagy (10). We determined that ERK inhibition of other key metabolic processes contributed to a compensatory increase in autophagy in PDAC. Specifically, KRAS/ERK suppression in KRAS-mutant PDAC further elevated autophagy in part by perturbation of glycolysis. We and others showed that KRAS upregulates glycolysis by ERK-driven increased transcription of genes involved in virtually every step of the glycolytic pathway (10,11). Inhibition of glycolysis both activates AMPK and inhibits mTORC1, a negative regulator of the initiating step in the autophagy pathway. Our findings, together with two other independent studies (10,12,13), have led to the initiation of clinical trials investigating the combination of MEK/ERK inhibitors and HCQ in RAS-mutant cancers (NCT03825289, NCT04145297, NCT04132505, NCT04214418, NCT03979651, NCT04566133, NCT04735068, and NCT04386057).
A limitation of essentially all targeted anti-cancer therapies is treatment-induced compensatory activities to offset the deleterious consequences of target inhibition (14). Identification of such compensatory mechanisms may reveal combination treatment strategies capable of eliciting more durable responses. In this study, we hypothesized that treatment-induced compensatory mechanisms may also contribute to the limited clinical efficacy of HCQ. To address this possibility, we first applied a CRISPR-Cas9 loss-of-function screen to identify genes that modulate PDAC sensitivity to the HCQ analog, CQ. Second, we applied reverse phase protein array (RPPA) pathway activation mapping to profile compensatory signaling activation and/or expression changes in response to CQ treatment. Both analyses converged on the RTK, IGF1R (insulin-like growth factor 1 receptor). We found that inhibition of IGF1R increased autophagic flux and sensitized cells to CQ. Additionally, we investigated known pathways that antagonize autophagy, and found that concurrent inhibition of IGF1R and ERK synergistically increased PDAC cell dependence on autophagy. We found that concurrent inhibition of IGF1R and ERK converged on and suppressed glycolysis, leading to AMPK activation and inhibition of mTORC1. We propose that dual targeting of IGF1R and ERK together with autophagy inhibition may be a more durable therapeutic approach for PDAC.
Materials and Methods
Cell culture
Patient-derived xenograft (PDX)-derived human KRAS-mutant cell lines Pa01C (RRID:CVCL_1638), Pa02C (RRID:CVCL_E302), Pa04C (RRID:CVCL_E299), Pa14C (RRID:CVCL_1638), and Pa16C (RRID:CVCL_1639) were provided by A. Maitra (MD Anderson Cancer Center, Houston, TX) (15). The mouse tumor-derived Kras-mutant cell line mT4 [KPC (KrasG12D/+; Tp53R172H/+; Pdx-Cre)] and the KRAS WT hF39 and hF43 cell lines were provided by D. Tuveson (Cold Spring Harbor Laboratory, NY). The KRAS WT PATC153 cell line was provided by C. Taniguchi (MD Anderson Cancer Center, Houston, TX). HEK 293T (RRID:CVCL_0063), HPAC (RRID:CVCL_3517), MIA PaCa-2 (RRID:CVCL_HA89), PANC-1 (RRID:CVCL_0480), and BxPC-3 (RRID:CVCL_0186) KRAS-mutant cell lines were obtained from the American Type Culture Collection (ATCC). hTERT-HPNE (HPNE-DT, RRID:CVCL_C466), hTERT-HPNE E6/E7 (HPNE, RRID:CVCL_C467), and hTERT-HPNE E6/E7/KRASG12D (HPNE-KRASG12D, RRID:CVCL_C469) cell lines were described previously (16). HPDE E6/E7 (HPDE, RRID:CVCL_S975) and KRAS-expressing HPDE cells (HPDE-KRASG12V) cell lines were provided by M. Tsao (Ontario Cancer Institute at Princess Margaret Hospital, Ontario, Canada). All cell lines were cultured in DMEM or RPMI (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin-streptomycin solution and passaged for 1 month or 10 passages before a new aliquot was thawed. Cells were maintained in a humidified chamber at 37°C in 5% CO2. All human cell lines were most recently authenticated by short-tandem-repeat (STR) profiling in April 2017. All cell lines were tested monthly for mycoplasma using MycoAlert (Lonza).
The human PDAC patient-derived organoid cultures were provided by Dr. David Tuveson (Cold Spring Harbor Laboratory; hM1A, hT105, and hT106) and Dr. Calvin Kuo (Stanford University; PT3, PT6, and PT8). The latter cultures were initially established in an air-liquid interface as described previously (17) and subsequently grown in the Matrigel dome method. Organoids were cultured at 37°C in 5% CO2 in growth factor reduced Matrigel (Corning) domes in complete human feeding medium: Advanced DMEM/F12 (Thermo Fisher Scientific) based WRN conditioned medium (L-WRN (ATCC CRL-3276)), 1x B27 supplement, 10 mM HEPES, 0.01 μM GlutaMAX (all from Thermo Fisher Scientific), 10 mM nicotinamide (Sigma-Aldrich), 50 ng/mL hEGF (Peprotech), 100 ng/mL hFGF10 (Peprotech), 0.01 μM hGastrin I (TOCRIS), 500 nM A83-01 (TOCRIS), and 1.25 mM (hM1A, hT105, hT106) or 1 mM (PT3, PT6, PT8) N-acetylcysteine (Sigma-Aldrich). The media for PT3, PT6, and PT8 additionally contained 10 μM SB202190 (Sigma). Y27632 (Selleckchem, 10.5 μM) was added for the first two days after reseeding single cells.
CRISPR-Cas9 screen
The CRISPR-Cas9 barcoded druggable genome library targeting 2,240 cancer signaling genes has been described previously (18,19). PDAC cells were infected with the library at a MOI of 0.2 and placed under puromycin selection. Genomic DNA was harvested immediately after selection (2 days post infection, day 0) to determine library coverage, and after sufficient time to generate stable knockouts and to achieve ~1,000x coverage of the library (9 days, day 7) to determine gene essentiality. Cells were then split into DMSO or CQ (3.125 μM) treatment groups. After two weeks of treatment, DNA was extracted with the DNeasy Blood & Tissue Kit (QIAGEN) and analyzed by next generation sequencing (NGS). sgRNA abundances were quantified for each gene using MAGeCK analysis (20).
Sample preparation and Reverse Phase Protein Array (RPPA) pathway activation mapping
Cells were seeded in 6-well plates and treated with CQ (10 μM, 72 hours). Cell lysis, protein quantification, and array analysis was performed as described previously (21). Principal component analysis was used to assess the consistency among replicates. Based on sample distribution, a two-sided parametric t-test was used to compare the population means of treated and untreated (DMSO control) cells and p-values were corrected for multiple comparisons using the Benjamini-Hochberg procedure. Statistical analysis was performed using the R software package.
Syngeneic PDAC mouse model
All in vivo experiments were overseen and approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill (protocol 18-162). Eight- to ten-week-old male C57Bl/6 mice were acclimated for two weeks on an AIN-93G diet, at three mice per cage. Mice were injected subcutaneously with 2.5 x 105 mT4 cells in the right flank. Tumors were allowed to develop until palpable, whereupon mice were blocked and randomized to ensure uniform initial tumor size and random assignment to treatment groups. All mice were then injected daily with either vehicle control, BMS-754807 (12.5 mg/kg), HCQ (SelleckChem, 60 mg/kg), or both in a vehicle of 2% DMSO and 30% PEG300 in phosphate-buffered saline. Tumors were monitored bi-weekly by palpation and caliper measurements taken. All mice were sacrificed when tumor sizes in any group reached 15 mm in any direction, and tumors were excised and weighed. Statistical significance of tumor weight was evaluated using ANOVA and Dunnett’s test for multiple comparisons.
Chemical reagents, antibodies, reagents
All reagents and antibodies are listed in the Supplementary Materials and Methods.
Generation of stable cell lines
Generation of stable cell lines was performed as described in Supplementary Materials and Methods.
Immunoblotting
Details are provided in Supplementary Materials and Methods.
Immunofluorescence
Details are provided in Supplementary Materials and Methods
Drug response testing
All methods are described in Supplementary Materials and Methods.
Flow cytometry
Analyses were performed as described in Supplementary Details and Methods.
1H NMR for identification of extracellular metabolites
Analyses were performed as described in Supplementary Materials and Methods.
Microarray gene expression profiling
Analyses were performed as described in Supplemental Materials and Methods.
Data Availability
Microarray data is available at GEO (accession number GSE163348). All other datasets generated for the current study are available from the corresponding author upon request.
Code Availability
No unique code was generated for this study.
Results
CRISPR-Cas9 loss-of-function screen and signaling pathway activation profiling identify IGF-1-axis and mTORC1 as sensitizers to autophagy inhibition
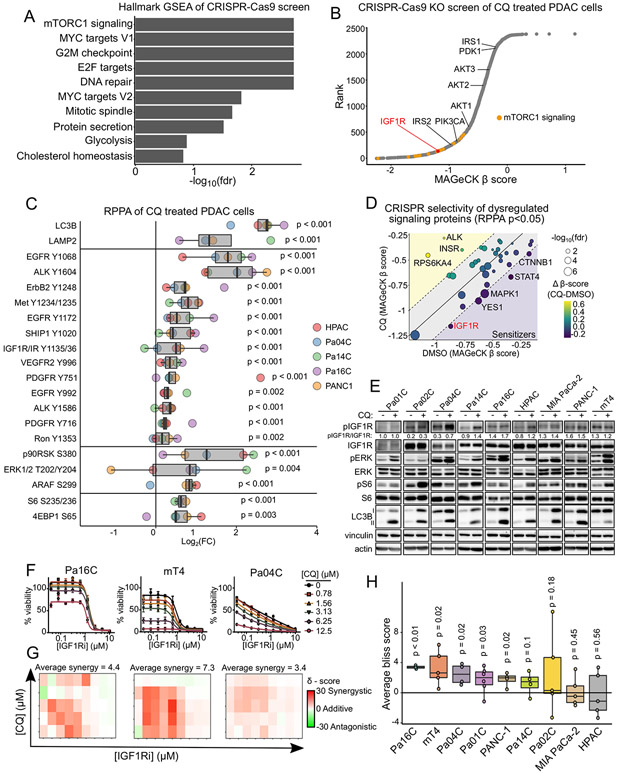
To identify potential therapeutic targets capable of enhancing the growth inhibitory activity of HCQ/CQ, we applied a lentiviral CRISPR-Cas9 loss-of-function screen targeting 2,240 genes representing major cancer signaling networks (18,19). We performed deep sequencing to analyze sgRNA abundance in four KRAS-mutant PDAC cell lines treated for two weeks with either vehicle or a sublethal dose (3.125 μM) of the HCQ analog chloroquine (CQ). We used principal component and Spearman correlation analyses to confirm sample quality (Supplementary Fig S1A and S1B). Changes in sgRNA abundances were quantified for each gene using MAGeCK analysis. To validate the screen, we compared MAGeCK β-scores of essential and non-essential genes in vehicle control cells to the median CERES scores across all pancreatic cancer cell lines deposited in DepMap ([EH5358], 21Q1). We found that essential genes predicted to be lost by DepMap (Supplementary Fig S1C) were consistently lost in our screen compared to non-essential genes (Supplementary Fig S1D) (20). We then performed Gene Set Enrichment Analysis (GSEA) of genes ranked by their MAGeCK β score in CQ treated cells and identified the Hallmark mTORC1 gene set among the most significantly depleted (Fig. 1A). Interestingly, E2F, MYC and G2M gene targets, the major transcriptional outputs of the ERK MAPK pathway (18) scored as the three next most depleted Hallmark gene sets. This finding is consistent with our recent demonstration that concurrent ERK inhibition synergistically enhanced CQ growth inhibitory activity (10).
Figure 1.
IGF1R inhibition sensitizes cells to autophagy inhibition. A, Gene set enrichment analysis (GSEA) of Hallmark gene sets (MSigDB v 6.2) was performed on MAGeCK β scores from a CRISPR-Cas9 loss-of-function screen in Pa01C, Pa02C, Pa14C, and PANC-1 human PDAC cell lines treated with chloroquine (CQ, 3.125 μM, 14 days). B, Ranked MAGeCK β scores, with the mTORC1 gene set highlighted (orange), and IGF1R and IGF1-axis related genes annotated. C, Fold-change values from RPPA analysis of Pa04C, Pa14C, Pa16C, HPAC, and PANC-1 cell lines treated with either DMSO control or CQ (10 μM, 72 hours). Boxplots and significance values were calculated across all cell lines from at least three independent replicates per condition per cell line using a two-sample t-test, and p-values were corrected for multiple comparisons using the Benjamini-Hochberg procedure. D, MAGeCK β scores for genes corresponding to proteins significantly dysregulated (p<0.05) in the RPPA analyses from C. Significance represents false discovery rate for CQ-treated β score. E, Immunoblot analysis of IGF1R (Y1131/Y1135/Y1136), ERK1/2 (Y202/Y204), and ribosomal S6 (S235/S236) phosphoproteins, and LC3B-I/II proteoforms following treatment with CQ (10 μM, 72 hours) in a panel of PDAC cell lines. F, Cell viability assays following treatment with BMS-754807 (IGF1Ri, 72 hours) and increasing concentrations of the autophagy inhibitor CQ. Values represent the average +/− SD of three independent experiments. G, Excess over Bliss synergy values were calculated from data represented in F using SynergyFinder and represented as a 2D-contour. Plots are representative of at least four independent experiments. H, Average synergy values were calculated by taking the mean synergy score across the drug matrix represented in G from at least four independent experiments.
Previous studies demonstrated that inhibition of mTORC1 enhances metabolic reliance on autophagy (22), consistent with our finding that loss of mTORC1 signaling components enhanced CQ sensitivity and growth inhibition. We found that multiple genes within the mTORC1 pathway were depleted in CQ-treated cells, with the receptor tyrosine kinase (RTK) IGF1R among the strongest hits (Fig. 1B). IGF1R signals through the SHC adaptor to activate the RAS-ERK signaling network, and through the IRS1/2 adaptors to the PI3K-AKT-mTORC1 signaling network (23). Multiple proteins within the IRS-mediated component of IGFR1 signaling were also identified (e.g., IRS1/2, AKT1-3), supporting the role of PI3K signaling in IGF1R-mediated resistance to CQ.
We next sought to characterize signaling changes that accompany CQ treatment by using reverse phase protein array (RPPA) analyses (24) (Fig. 1C). Consistent with its mechanism of impairing autophagosome degradation, CQ dramatically increased the autophagosome-associated protein LC3B and the lysosomal protein LAMP2 (Fig. 1C, Supplementary Fig S1E). CQ treatment also markedly increased phosphorylation and activation of RTKs, including IGF1R (Fig. 1C). Because RTKs can activate signaling that antagonizes autophagy, we speculated that RTK activation may be a compensatory response to offset CQ treatment-associated growth suppression. We then interrogated our CRISPR screening data to determine if the loss of RTKs or other CQ-dysregulated signaling proteins sensitized PDAC cells to CQ and found IGF1R to be a strong sensitizer (Fig. 1D). Considering our independent identification of IGF1R compensatory activation in response to CQ and as a CQ sensitizer through two distinct but complementary screens, we focused on this RTK for further evaluation.
We first verified the RPPA results in a panel of KRAS-mutant human PDAC cell lines and a Kras+/LSL-G12D/p53+/LSL-R172H;Pdx1-Cre (KPC) mouse tumor-derived cell line (mT4) (25). We found that CQ treatment led to the accumulation of the LC3B-II proteoform, consistent with an inhibition of autophagosome processing (Fig 1E). Additionally, we found CQ induced IGF1R phosphorylation and phosphorylation of downstream effectors ERK1/2 and ribosomal protein S6 in the majority of cell lines (Fig. 1E). Lastly, we determined that a clinically evaluated IGF1R tyrosine kinase inhibitor (IGF1Ri), BMS-754807 (26), sensitized PDAC cells to CQ (Fig. 1F and G; Supplementary Fig S1F-I). Concurrent treatment with an IGF1Ri and CQ resulted in a synergistic or additive decrease in cell viability in over half of the KRAS MT cell lines tested (Fig. 1H) and KRAS WT PDAC cell lines harboring alternative oncogenic driver mutations in PIK3CA (Supplementary Fig S1H-I).
IGF1R inhibition increases autophagic flux in PDAC cells
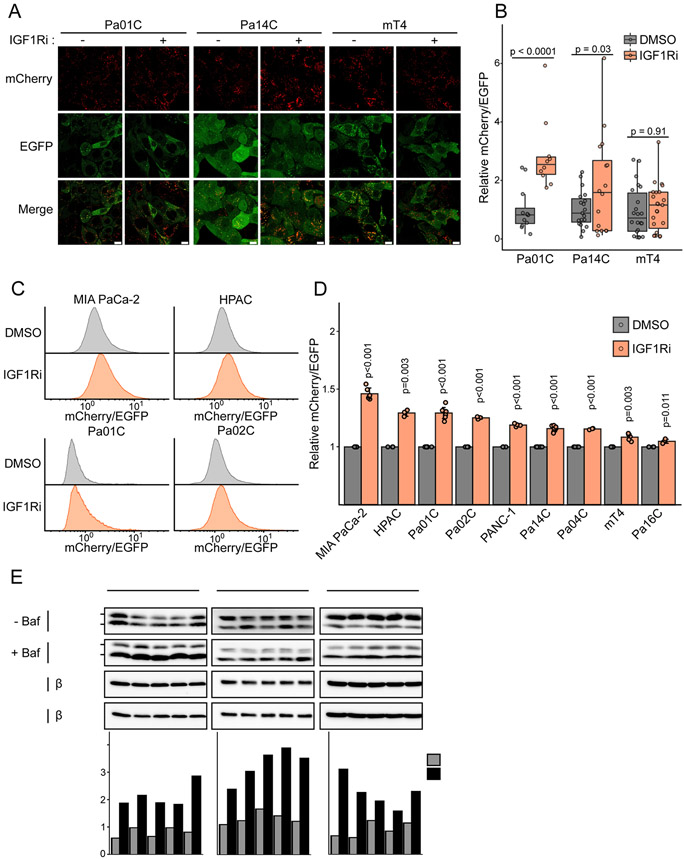
Having validated that IGF1Ri sensitized cells to CQ treatment, and that CQ induced IGF1R phosphorylation, we reasoned that IGF1R activity would impact autophagic flux. To address this possibility, we utilized PDAC cells stably expressing the autophagy flux reporter, mCherry-EGFP-LC3B, where the ratio of mCherry/EGFP fluorescence is a direct readout of autophagic flux (10). We found that IGF1Ri increased autophagic flux in KRAS-mutant human (Pa01C and Pa14C) and mouse (mT4) PDAC cell lines (Fig. 2A and B; Supplementary Fig. S1A). Expanding our analyses to six additional human KRAS-mutant PDAC cell lines, we found that IGF1Ri treatment increased autophagic flux in all nine cell lines analyzed (Fig. 2C and D; Supplementary Fig. S2).
Figure 2.
Inhibition of IGF1R increases autophagic flux in murine and human PDAC cells. A, Confocal microscopy of indicated cell lines stably expressing mCherry-EGFP-LC3B treated with DMSO or BMS-754807 (IGF1Ri, 1 μM, 24 hours). Scale bar = 10 μm. Enlarged images in Supplementary Fig. S2A to visualize puncta. B, Quantification of the mCherry/EGFP ratio of LC3B puncta in indicated cell lines treated as in A. Data points represent mCherry/EGFP ratio from each field (> 10 fields per condition). Plot is representative of at least three independent experiments and statistics calculated by two-sided t-test. C, Flow cytometry analysis of indicated cell lines expressing mCherry-EGFP-LC3B treated with DMSO or BMS-754807 (IGF1Ri, 1 μM, 24 hours). Histograms represent the ratio of mCherry to EGFP fluorescence. D, Median values from flow cytometry analysis of mCherry-EGFP-LC3B expressing PDAC cells following treatment with DMSO or BMS-754807 (IGF1Ri, 1 μM, 24 hours). Data represents average +/− SD from at least four independent experiments and statistics calculated by a two-sided t-test. E, Immunoblot analyses of cell lysates from indicated cell lines for LC3B-I/LC3B-II (autophagic flux) and β-tubulin (loading control) treated with DMSO or BMS-754807 (IGF1Ri, 1 μM, 24 hours) followed by an incubation with DMSO (control) or bafilomycin A1 (Baf, 200 nM, 2 hours). LC3B-I/LC3B-II bands were quantified by densitometry and the ratio is plotted (bottom). Data are representative of three independent experiments.
To complement these methods, we also assayed endogenous autophagic flux by treating cells with IGF1Ri for 24 hours before acute (2-hour) treatment with the vacuolar ATPase inhibitor, bafilomycin A1 (Baf). Baf treatment causes accumulation of autophagic vesicles by preventing acidification of autophagolysosomes and results in an increased ratio of the autophagosome marker LC3B-II to cytosolic LC3B-I (27). We determined that LC3B-II levels increased in IGF1Ri treated Pa14C, Pa01C, and mT4 cells (Fig. 2E). We conclude that IGF1R inhibition increased autophagic flux in PDAC cells.
Concurrent IGF1R and autophagy inhibition blocks PDAC growth in vitro and in vivo
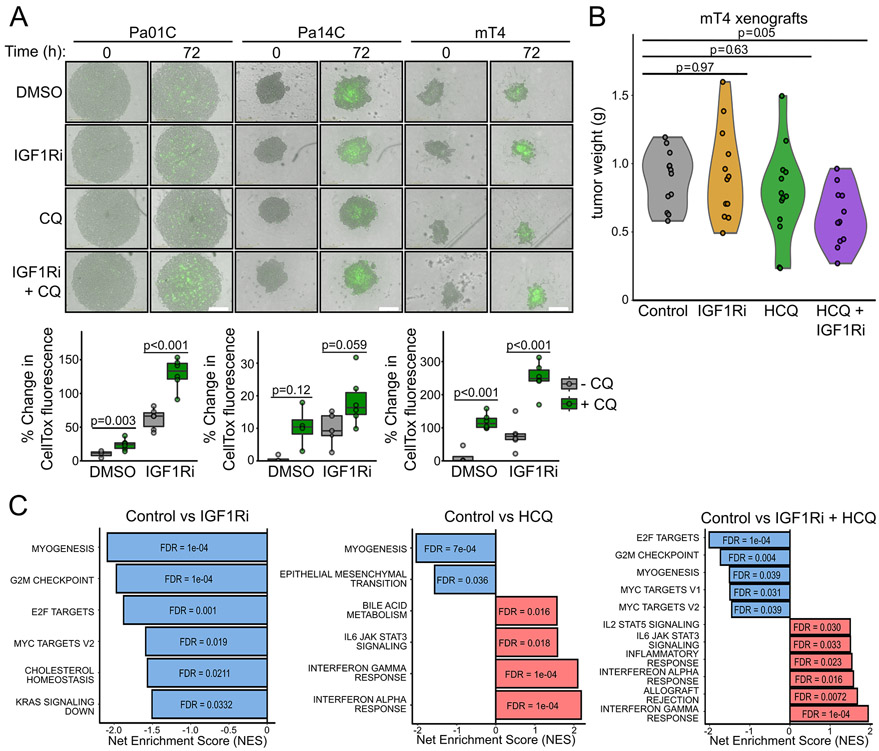
We next evaluated the efficacy of concurrent inhibition of IGF1R and autophagy in more physiologically relevant preclinical cell models of PDAC. First, we propagated Pa01C, Pa14C, and mT4 cell lines as three-dimensional (3D) non-adherent spheroid cultures (28,29). We found that cytotoxicity was increased, to variable levels, by CQ treatment alone using the DNA-binding dye CellTox™ green that is excluded from viable cells but not cells that have impaired membrane permeability upon cell death (Fig. 3A). Notably, we found that combined CQ and IGF1Ri further enhanced toxicity in all cell lines, particularly in Pa01C and mT4.
Figure 3.
Dual inhibition of IGF1R and autophagy impairs PDAC proliferation in 3D and in vivo models. A, Human (Pa01C, Pa14C) and murine (mT4) PDAC cell lines were grown as spheroids and treated with or without BMS-754807 (IGF1Ri, 200 nM) and/or CQ (6 μM) for 72 hours and cytotoxicity was assayed via the addition of the apoptotic dye CellTox Green (500 nM). Representative images at 0 and 72 hours of drug treatment are shown (top). Scale bar = 400 μm. Total integrated intensity of CellTox fluorescence at 72 hours after treatment is plotted (bottom). Integrated CellTox fluorescence intensity is representative of at least four independent experiments. B, Ex vivo tumor weight of mT4 mouse xenografts following treatment with vehicle, BMS-754807 (IGF1Ri, 12.5 mg/kg), HCQ (60 mg/kg), and the combination. C, GSEA of Hallmark gene sets following Clariom S Assay HT mouse microarray of mRNA prepared from six representative mT4 tumors per condition from B.
We then evaluated the efficacy of concurrent IGF1Ri and HCQ treatment in vivo using mT4 cells grown as xenograft tumors in syngeneic BL6 mice. Similar to previous studies (7,30), neither IGF1Ri nor HCQ treatment alone significantly reduced tumor weight compared with vehicle control (Fig. 3B). However, consistent with our results in adherent and spheroid cultures, the combination of HCQ and IGF1Ri resulted in a 40% reduction in final tumor burden in vivo (Fig. 3B). The combination treatment was well-tolerated, with only minor changes in mouse body weight and non-fasted blood glucose levels (Supplementary Fig. S3A and S3B).
To determine a mechanistic basis for the potent anti-tumor activity of the combination, we performed mRNA microarray analysis to investigate the effects of each treatment, alone or in combination, on gene expression. GSEA of Hallmark gene sets demonstrated that IGF1Ri treatment alone induced downregulation of genes involved in regulating cell cycle progression, in particular, G2/M checkpoints, and of E2F- and MYC-regulated targets. In contrast, HCQ treatment alone stimulated transcription of genes associated with pro-inflammatory responses (Fig. 3C). The combination treatment induced gene expression changes that corresponded essentially to the sum of the gene expression changes seen with each inhibitor alone.
Co-targeting the ERK-MAPK cascade and IGF1R upregulates autophagy through inhibition of glucose metabolism and activation of AMPK
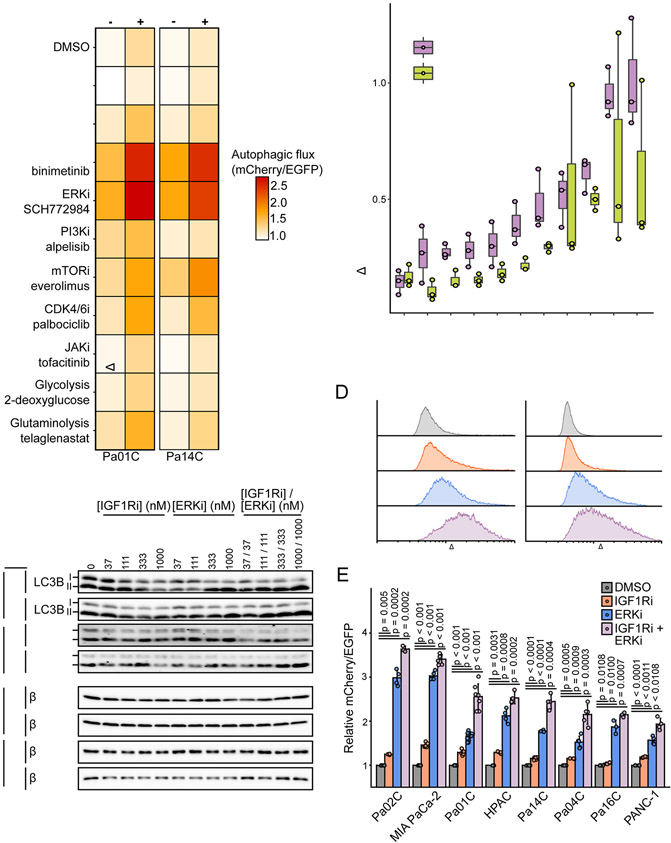
While the combination of IGF1Ri and autophagy inhibition was effective in a panel of human PDAC cell lines and a syngeneic murine in vivo tumor model, we were interested in determining if compensatory pathway activation would limit IGF1Ri-induced autophagy. We consulted our RPPA (Fig. 1C) and CRISPR screen results (Fig. 1D) to select a panel of 11 clinically relevant inhibitors for further evaluation and included additional inhibitors that were previously established to modulate autophagy. We then established a fluorescence-based assay that utilized PDAC cells stably expressing the autophagic flux reporter and performed flow cytometry following treatment with each inhibitor alone or in combination with IGF1Ri (Fig. 4A). Except the SHP2 and JAK inhibitors, which either reduced or did not affect basal autophagy levels, we found that all other inhibitors increased autophagic flux. As we determined previously, MEKi and ERKi potently enhanced autophagy (10,12,13), with no other inhibitor stimulating autophagy to the same degree.
Figure 4.
ERK-MAPK blockade potentiates IGF1Ri induced autophagic flux. A, Median EGFP/mCherry ratios of mCherry-EGFP-LC3B expressing cells analyzed by flow cytometry. Cells were treated for 24 hours with the following inhibitors: SHP2i SHP-099 (10 μM), pan-RAFi lifirafenib/BGB823 (1 μM), MEKi binimetinib (1 μM), ERKi SCH772984 (1 μM), PI3Kαi alpelisib (1 μM), mTORC1i everolimus (100 nM), CDK4/6i palbociclib (500 nM), JAKi tofacitinib (1 μM), phosphoglucose isomerase inhibitor 2-deoxyglucose (5 mM in glucose-free DMEM), glutaminase inhibitor telaglenastat (1 μM). Cells were treated with or without co-treatment with BMS-754807 (IGF1Ri, 500 nM). Data represents the average of three independent experiments. B, Change in median mCherry/EGFP following addition of BMS-754807 (IGFRi, 500 nM) in A is plotted. Boxplots represent three independent experiments. C, Immunoblot analyses of cell lysates from indicated cell lines for LC3B-I/LC3B-II (autophagic flux) and β-tubulin (loading control) treated with BMS-754807 (IGF1Ri), SCH772984 (ERKi), or the combination (24 hours) and then treated with or without bafilomycin A1 (Baf, 200 nM, 2 hours) before lysis. Data are representative of two independent experiments. D, Representative histograms of the ratiometric determination of mCherry/EGFP fluorescence by flow cytometry in mCherry-EGFP-LC3B expressing PDAC cells following treatment with BMS-7754807 (IGF1Ri, 1 μM), SCH-772984 (ERKi, 1 μM), or the combination (24 hours). E, Quantification of median mCherry/EGFP fluorescence as shown in D from four independent experiments.
We also profiled the ability of each inhibitor to induce autophagic flux in the presence of IGF1Ri. We found that combining IGF1Ri with MEKi or ERKi induced the highest autophagic flux when used together, where the combination induced a synergistic increase in flux greater than would be predicted by their effects alone (Fig. 4B). To further assess the ability of the ERKi-IGF1Ri combination to increase autophagy, we performed Baf-clamp experiments in Pa01C and Pa14C cells with the combination. Consistent with our findings here and as reported previously (10), IGF1Ri or ERKi alone increased autophagy, with the combination inducing the greatest level of LC3B-II (Fig. 4C, Supplementary Fig. S4A). Finally, to establish the generality of these findings, we extended the evaluation to additional KRAS-mutant PDAC cell lines. We found that the combination consistently upregulated autophagy more than either inhibitor alone, in all cell lines evaluated (Fig. 4D and 4E, Supplementary Fig. S4B and S4C).
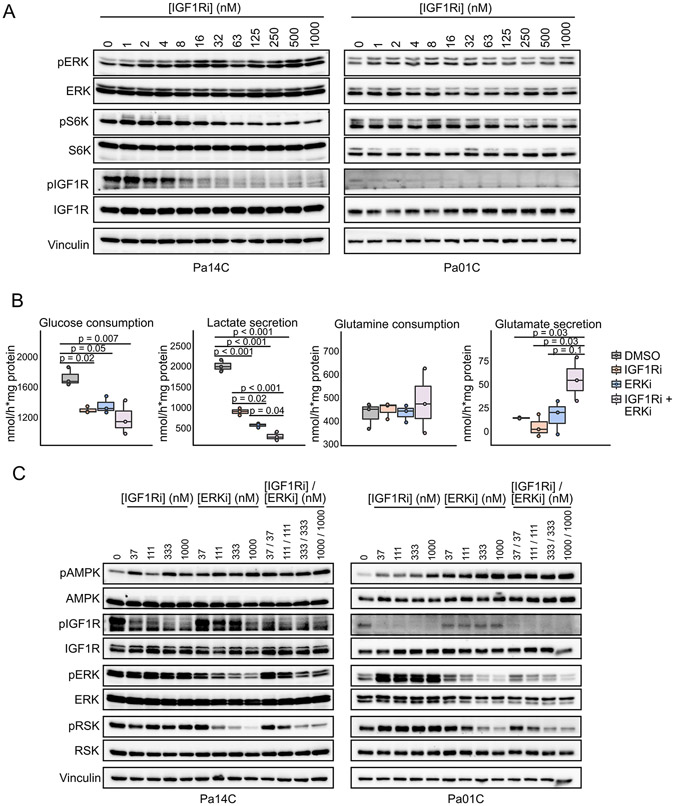
We next evaluated a signaling basis for the potent autophagy induction observed upon concurrent ERKi and IGR1Ri treatment. We speculated that the potency of this combination may involve two complementary mechanisms. First, we observed recently that ERKi treatment is associated with compensatory activation of IGF1R, contributing to downstream ERK reactivation (10). Thus, concurrent IGF1Ri treatment negates an ERKi-induced compensatory activity. Second, IGF1R signals through both PI3K-mTOR and RAS-ERK effector pathways. However, we found that IGF1Ri treatment impaired mTOR (pS6K) but not ERK (pERK) activity (Fig. 5A, Supplementary Fig. S5A), most likely because of the presence of mutant KRAS, a dominant driver of ERK activation. Notably, IGF1Ri treatment upregulated pERK. Therefore, we speculated that concurrent ERKi treatment is needed to effectively block ERK signaling in KRAS-mutant PDAC. Consistent with this possibility, when we applied a 525-oncology drug screen to identify combinations that enhanced RAF/MEK/ERK or PI3K/AKT/mTOR inhibitor efficacy in PDAC cell lines, IGF1Ri was among the strongest sensitizers to RAF/MEK/ERK inhibition, but not PI3K/AKT/mTOR (Supplementary Fig. S5B). In summary, the potent stimulation of autophagy observed with the MEKi/ERKi and IGF1Ri combination is mediated, in part, by each inhibitor disrupting signaling activities that limit the efficacy of the other inhibitor when used alone.
Figure 5.
Dual IGF1R and ERK inhibition decreases glycolysis that leads to activation of AMPK. A, Cell lines were treated with BMS-754807 (IGF1Ri, 24 hours) and immunoblotted for phosphorylation of ERK (T202/Y204), p70S6K (T389), and IGF1R (Y1131/Y1135/Y1136). Total levels of ERK, S6K, IGF1R, and vinculin (loading) were assessed for normalization. Blots are representative of two independent experiments. B, Extracellular media was analyzed by 1H-NMR to quantify metabolite levels following treatment with BMS-754807 (IGF1Ri, 500 nM), SCH772984 (ERKi, 500 nM) or the combination. Following 18 hours of treatment, fresh medium was added and incubated for six hours. Metabolite levels were normalized to protein content of cell lysates (wet weight). Data represent three independent experiments. C, Cells were treated as in Fig. 4C and cell lysates were immunoblotted for phosphorylation of AMPK (T172, autophagy regulation), IGF1R (Y1131/Y1135/Y1136), ERK (T202/Y204), and RSK (S380). Total AMPK, IGF1R, ERK, RSK, and vinculin were assessed for normalization. Data represent two independent experiments.
Since IGF1R and ERK each stimulate glucose metabolism through distinct mechanisms (10,11,31), we reasoned that dual inhibition would converge on and potently inhibit glycolysis. Impairment of glycolysis is established to activate AMPK, a key inhibitor of mTORC1 activity, thereby stimulating autophagy (32). To address this possibility, we profiled the metabolic inputs and outputs of cells treated with IGF1Ri or ERKi alone or in combination, by analyzing metabolites in the extracellular media using 1H-NMR. ERKi or IGF1Ri treatment alone each significantly decreased glucose consumption and lactate secretion, to comparable degrees (Fig. 5B). These changes were further enhanced in cells treated with the combination, consistent with a more dramatic inhibition of glycolytic activity. Interestingly, we did not find a significant change in glutamine consumption, even though reprogramming of glutamine metabolism has been identified as a KRAS-driven process in PDAC (33). However, we did observe an increase in glutamate secretion by cells treated concurrently with ERKi and IGF1Ri, consistent with decreased utilization of intracellular glutamine. Among the other nutrients, methionine was the only other metabolite altered in cells treated with the combination (Supplementary Fig. S5C). From these data we reasoned that since CQ treatment alone does not lead to a significant depression of glycolysis (Supplementary Fig. S5D) that would lead to pAMPK activation, ERKi and IGF1Ri sensitize cells to CQ by converging on AMPK activation. Finally, we found concurrent treatment with IGF1Ri and ERKi dramatically increased pAMPK levels, establishing a signaling basis for increased autophagy (Fig. 5C, Supplementary Fig. S5E).
Inhibition of ERK, IGF1R, and autophagy induces cell death in PDAC cell lines and sensitizes human PDAC organoids that are less sensitive to ERK inhibition alone
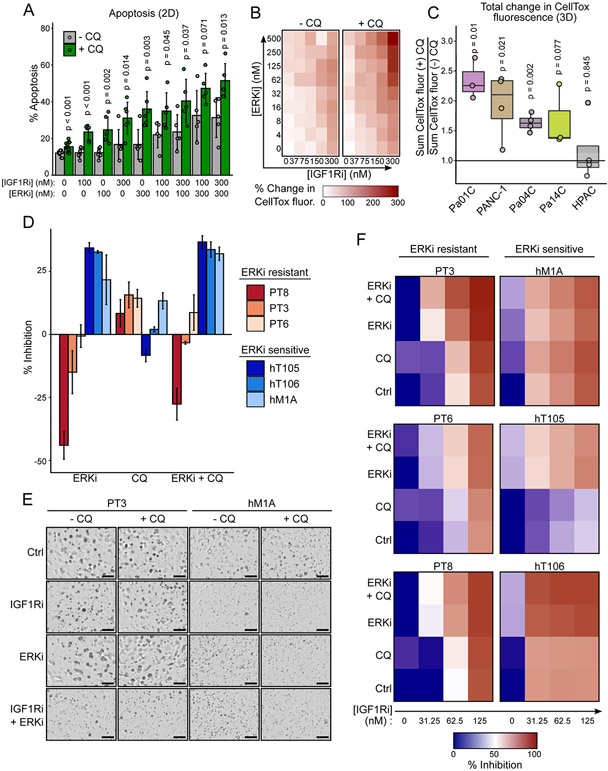
To determine the therapeutic potential of targeting ERK, IGF1R, and autophagy together we evaluated their effects on inducing apoptosis in 2D adherent and 3D spheroid cell cultures of Pa14C cells. Each inhibitor alone stimulated apoptosis to a limited degree, as measured by annexin V staining (2D). Although combined ERKi and IGF1Ri treatment increased apoptosis more than either inhibitor alone (Fig. 6A, Supplementary Fig. S6A), the greatest increase was observed when cells were treated concurrently with ERKi and IGF1Ri together with CQ.
Figure 6.
Simultaneous inhibition of ERK, IGF1R and autophagy is cytotoxic in 2D and 3D culture models and inhibits human PDAC organoid growth. A, Flow cytometric determination of apoptosis measured by FITC-annexin V and propidium iodide staining of Pa14C cells treated with CQ (6 μM), BMS-754807 (IGF1Ri), SCH772984 (ERKi), or in multiple combinations (72 hours) as denoted in the figure. Data represent four independent experiments. B, Integrated CellTox fluorescence of Pa14C spheroids was measured following treatment with a matrix of BMS-754807 (IGF1Ri) and SCH772984 (ERKi) in the presence and absence of CQ (6 μM) and CellTox (500 nM, 72 hours). Percent change from DMSO control was calculated for each condition. Data represent average of at least three independent experiments. C, Percent change of CellTox fluorescence was summed across the entire ERKi and IGF1Ri matrix as shown in B for each cell line and normalized by the sum for the (−) CQ control condition. D, Human pancreatic cancer organoids were treated with SCH772984 (500 nM), CQ (10 μM), or the combination (7 days). Data represent the percent inhibition following CellTiter-Glo 3D assay of three independent experiments. E, Representative images of human PT3 and hM1A organoids following treatment with BMS-754807 (IGF1Ri, 31.25 nM), SCH772984 (ERKi, 500 nM), and the combination, with or without CQ (10 μM, 7 days). F, Percent of growth inhibition based on CellTiter-Glo 3D assay of human PDAC organoids treated with BMS-754807 (IGFRi), alone or in combination with CQ (10 μM), SCH772984 (ERKi, 500 nM), or both CQ and SCH772984. Data represent the average of three independent experiments.
Cytotoxicity was further enhanced when the combined treatment of IGF1Ri, CQ, and ERKi was assessed in spheroid cultures (Fig. 6B, Supplementary Fig. S6B). For example, whereas we observed only a modest (20%) increase in cytotoxicity in Pa14C cells treated with IGF1Ri and CQ (CellTox fluorescence; Fig. 3A), we observed more than a 200% increase when ERKi was added (Fig. 6B). Comparison of the total CellTox fluorescence for a dose-response matrix of ERKi and IGF1Ri to total CellTox fluorescence for the same matrix with the addition of CQ revealed significantly increased cytotoxicity due to the triple combination in spheroids generated from multiple PDAC cell lines (Fig. 6C). This trend was also observed in 2D proliferation and toxicity assays (Supplementary Fig. S6C and S6D) in KRAS mutant PDAC cell lines, but not in the non-transformed immortalized human pancreatic duct derived cell lines (HPNE and HPDE). That HPNE and HPDE cell lines are less sensitive to the triple combination of IGF1Ri, ERKi, and CQ suggests it is selectively toxic to KRAS mutant PDAC cells.
We then evaluated this treatment strategy utilizing patient-derived KRAS-mutant PDAC organoid cultures, ex-vivo models of PDAC that may more accurately reflect treatment response in the patient (34). We have shown previously that ERK inhibition sensitized human PDAC organoids to CQ treatment (10). Patient-derived organoids better model patient heterogeneity and thus it is not surprising that we observed highly variable growth responses to ERKi alone, or in combination with CQ, when evaluated in six different KRAS-mutant PDAC organoid cultures (Fig. 6D). This variable response to autophagy inhibition is similar to limited clinical observations in PDAC patients treated with MEKi and HCQ with no clear biomarkers to predict responders (35). However, when organoids were stratified by their sensitivity to ERKi, we noted that addition of IGF1Ri dramatically increased growth inhibition only in organoids that were not sensitive to ERKi and CQ treatment (e.g., PT3, PT6, and PT8). Collectively, we observed potent inhibition of organoid viability across all organoid models treated with combined IGF1Ri, ERKi, and CQ (Fig. 6E and 6F).
DISCUSSION
HCQ/CQ, the only clinically approved autophagy inhibitors, have not shown significant clinical benefit as single agent treatment in pancreatic cancer. In the present study, we speculated that de novo or treatment-induced acquired compensatory mechanisms may contribute to the limited efficacy of HCQ. To evaluate this possibility, we applied two distinct and complementary experimental strategies. First, we applied a genetic loss-of-function screen using a cancer signaling network-focused CRISPR-Cas9 library to identify genes that modulate PDAC cell line sensitivity to CQ. Second, we completed RPPA signaling profiling to identify compensatory activities stimulated by CQ treatment. Both strategies converged on IGF1R. We determined that, similar to our recent observations with MEK/ERK inhibitors (10), pharmacologic inhibition of IGF1R enhanced PDAC dependence on autophagy and thereby increased sensitivity to CQ treatment. However, we also found that IGF1R inhibition resulted in compensatory ERK activity, establishing a rationale for co-targeting ERK and IGF1R together to further increase PDAC reliance on autophagy. Finally, we determined that a triple combination concurrently targeting IGF1R, ERK and autophagy may provide a potent therapeutic strategy for the treatment of PDAC.
We have previously reported that genetic suppression of KRAS or inhibition of MEK/ERK in PDAC led to distinct metabolic alterations resulting in increased reliance on autophagy (10). In the present study, we again identified loss of MAPK1 (ERK2) among the top sensitizers to autophagy inhibition. Furthermore, GSEA showed that the CRISPR hits that enhanced CQ sensitivity corresponded to the top transcriptional gene sets that we identified recently as lost upon inhibition of ERK MAPK signaling in PDAC (18). Our RPPA analyses also revealed that CQ induced compensatory activation of RTKs. The activated RTKs identified in this study stimulate metabolic signaling pathways that directly antagonize autophagy, in particular the RAS-RAF-MEK-ERK or PI3K-AKT-mTOR signaling cascades (10,12,13,32). We next leveraged our CRISPR-Cas9 screening results to identify RTKs whose loss would sensitize cells to CQ. This analysis led us to focus on IGF1R. We found that IGF1R inhibition increased autophagy in PDAC cells and synergistically decreased cell viability and in vivo tumor growth with concurrent CQ/HCQ treatment. Tumors treated with IGF1R inhibitors presented depleted gene sets associated with KRAS activity, namely G2/M checkpoint, E2F, and MYC targets (18). Importantly, this finding supports the tumor promoting role of IGF1R in activating the PI3K-AKT-mTOR and RAF-MEK-ERK signaling cascades even in the presence of mutationally activated KRAS (36).
While our in vivo results of reduced tumor burden following combined HCQ and IGF1Ri treatment were encouraging, the induction of autophagy or anti-tumor response due to IGF1R inhibition was not as robust as was observed upon MEK/ERK inhibition (10,12). This observation led us to consider compensatory pathways that would antagonize autophagy, where we identified ERK MAPK activity to be the major limiting factor. This result supports independent findings that IGF1R activation is a compensatory response to KRASG12C/MEK/ERK inhibition (37-39), and conversely, that ERK MAPK activation limits IGF1R inhibitor efficacy (40-42). We have shown here and previously that both IGF1R and ERK MAPK signaling can antagonize autophagy, supporting the hypothesis that concurrent inhibition would synergistically antagonize metabolic activities leading to further reliance on autophagy. Here, we found that dual inhibition of IGF1R and ERK resulted in decreased glycolytic activity, which led to AMPK activation. Taken together, we showed that concurrent targeting of both the ERK MAPK pathway and IGF1R results in potent sensitization to CQ treatment in both cell line and patient-derived organoid models of PDAC.
It is now well-established that the long-term effectiveness of protein kinase inhibitors targeting key cancer signaling drivers is limited by compensatory signaling mechanisms. We now extend this concept to lysosomal inhibitors, where we observed significant RTK activation in response to CQ treatment. The limited clinical efficacy of HCQ is generally ascribed to low potency and to its non-selectivity for autophagy. We suggest that HCQ may be further limited by compensatory signaling, specifically by activation of IGF1R. Additional compensatory mechanisms identified in our study include IGF1Ri reactivation of ERK; conversely, ERKi efficacy is limited by IGF1R activation. Thus, the effectiveness of the triple combination of IGF1Ri, ERKi and CQ is based in part on inhibition of the distinct compensatory activities seen with each inhibitor when used as monotherapy.
Clinical trials investigating MEK/ERK inhibitors together with HCQ are ongoing in RAS-mutant cancers (NCT03825289, NCT04145297, NCT04132505, NCT04214418, NCT03979651, NCT04566133, NCT04735068, and NCT04386057). While these investigations are promising for improving the therapeutic potential of both MEK/ERK inhibitors and HCQ, it is likely that both will be limited by treatment-induced compensatory activities. We have found that activation of IGF1R is a shared compensatory activity, and that addition of an IGF1R inhibitor enhanced the efficacy of concurrent ERK inhibitor and CQ treatment. IGF1R is an attractive target as higher IGF1R protein expression is associated with worse patient outcomes in PDAC, and consequently has been suggested as a biomarker to identify more aggressive cancers that will require multidrug regimens (43). However, it should be noted that it is currently unknown how concurrent treatment with IGF1Ri, ERKi and HCQ will be tolerated in patients or how it will impact cachexia, which is a major concern for patients with advanced disease. Therefore, this combination warrants further investigation to address these issues. In this study we highlight a path to enhance the efficacy of targeted therapies that abrogate IGF1R, MEK/ERK, and autophagy activity by leveraging compensatory pathway activation of each therapeutic approach.
Supplementary Material
Acknowledgments
We thank K. Wood (Duke) for the CRISPR-Cas9 druggable genome library, A. Maitra (MD Anderson Cancer Center) for PDAC cell lines, D. Tuveson (Cold Spring Harbor Laboratory) for cell lines and organoids, and C. Kuo (Stanford) for organoids.
Financial Support:
K.L. Bryant was supported by grants from the Pancreatic Cancer Action Network/AACR (15-70-25-BRYA), the Sky Foundation and from the NCI (P50CA196510 and R37CA251877). C.A. Stalnecker was supported by NCI T32CA009156 and F32CA232529. A.C. Edwards was supported by NIGMS T32GM119999. B. Papke was supported by the Deutsche Forschungsgemeinschaft (DFG PA 3051/1-1). C.M. Goodwin was supported by NCI T32CA009156 and F32CA221005. A.D. Cox and/or C.J. Der were supported by grants from the NCI (R01CA42978, R01CA175747, R01CA223775, P50CA196510, U01CA199235 and P01CA203657 and R35CA232113), Pancreatic Cancer Action Network/AACR (15-90-25-DER), Department of Defense (W81XWH-15-1-0611), and the Lustgarten Foundation (388222). M.F. Coleman and S.D. Hursting were supported by NCI R35CA197627. M. Pierobon and E.F. Petricoin were supported by NCI P01CA203657. The Microscopy Services Laboratory and UNC Biomolecular NMR laboratory Core Facility were supported by P30 CA016086 Cancer Center Core Support grant to the UNC Lineberger Comprehensive Cancer Center.
Footnotes
Conflicts of Interest: C.J. Der is a consultant/advisory board member for Anchiano Therapeutics, Deciphera Pharmaceuticals, Mirati Therapeutics and Revolution Medicines. C.J. Der has received research funding support from SpringWorks Therapeutics, Mirati Therapeutics and Deciphera Pharmaceuticals, and has consulted for Axon Advisors LLC, Sanofi, SmartAnalyst, SVB Leerink, Third Bridge, Ribometrix, Jazz Therapeutics, Turning Point Therapeutics and Eli Lilly. A.D. Cox has consulted for Eli Lilly, Mirati Therapeutics, and SpringWorks Therapeutics. E.F. Petricoin and M. Pierobon receive royalties from Avant Diagnostics. E.F. Petricoin is a consultant to and shareholder of Avant Diagnostics, Inc and Perthera, Inc., and received funding support from Mirati Therapeutics, Genentech, Inc., and Abbvie, Inc. K.L. Bryant has received research funding from Deciphera Pharmaceuticals and SpringWorks Therapeutics.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 2.Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci 2014;39:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seo J-W, Choi J, Lee S-Y, Sung S, Yoo HJ, Kang M-J, et al. Autophagy is required for PDAC glutamine metabolism. Sci Rep 2016;6:37594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev 2011;25:717–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo JY, Chen H-Y, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 2011;25:460–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov 2014;4:905–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boone BA, Bahary N, Zureikat A, Moser AJ, Normolle D, Wu W-C, et al. Safety and Biologic Response of Pre-operative Autophagy Inhibition with Gemcitabine in Patients with Pancreatic Adenocarcinoma. Ann Surg Oncol 2015;22:4402–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolpin BM, Rubinson DA, Wang X, Chan JA, Cleary JM, Enzinger PC, et al. Phase II and Pharmacodynamic Study of Autophagy Inhibition Using Hydroxychloroquine in Patients With Metastatic Pancreatic Adenocarcinoma. The Oncologist 2014;19:637–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onorati A, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer 2018;124:3307–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med 2019;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras Maintains Pancreatic Tumors through Regulation of Anabolic Glucose Metabolism. Cell 2012;149:656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinsey CG, Camolotto SA, Boespflug AM, Guillen KP, Foth M, Truong A, et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med 2019;25:620–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C-S, Lee LC, Yuan TL, Chakka S, Fellmann C, Lowe SW, et al. MAP kinase and autophagy pathways cooperate to maintain RAS mutant cancer cell survival. Proc Natl Acad Sci U S A 2019;116:4508–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trusolino L, Bertotti A. Compensatory pathways in oncogenic kinase signaling and resistance to targeted therapies: six degrees of separation. Cancer Discov 2012;2:876–80. [DOI] [PubMed] [Google Scholar]
- 15.Jones S, Zhang X, Parsons DW, Lin JC-H, Leary RJ, Angenendt P, et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science 2008;321:1801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell PM, Groehler AL, Lee KM, Ouellette MM, Khazak V, Der CJ. K-Ras Promotes Growth Transformation and Invasion of Immortalized Human Pancreatic Cells by Raf and Phosphatidylinositol 3-Kinase Signaling. Cancer Res 2007;67:2098–106. [DOI] [PubMed] [Google Scholar]
- 17.Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018;175:1972–1988.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozkan-Dagliyan I, Diehl JN, George SD, Schaefer A, Papke B, Klotz-Noack K, et al. Low-Dose Vertical Inhibition of the RAF-MEK-ERK Cascade Causes Apoptotic Death of KRAS Mutant Cancers. Cell Rep 2020;31:107764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin KH, Rutter JC, Xie A, Pardieu B, Winn ET, Bello RD, et al. Using antagonistic pleiotropy to design a chemotherapy-induced evolutionary trap to target drug resistance in cancer. Nat Genet 2020;52:408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang B, Wang M, Zhang W, Xiao T, Chen C-H, Wu A, et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat Protoc 2019;14:756–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes TK, Neel NF, Hu C, Gautam P, Chenard M, Long B, et al. Long-Term ERK Inhibition in KRAS-Mutant Pancreatic Cancer Is Associated with MYC Degradation and Senescence-like Growth Suppression. Cancer Cell 2016;29:75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer 2017;17:528–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.B Lw, R El, O Ch, deGraffenried La, H Sd. The Role of the Insulin/IGF System in Cancer: Lessons Learned from Clinical Trials and the Energy Balance-Cancer Link. Front Endocrinol 2015;6:77–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baldelli E, Calvert V, Hodge A, VanMeter A, Petricoin EF, Pierobon M. Reverse Phase Protein Microarrays. Methods Mol Bio 2017;149–69 [DOI] [PubMed] [Google Scholar]
- 25.Boj SF, Hwang C-I, Baker LA, Chio IIC, Engle DD, Corbo V, et al. Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell 2015;160:324–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carboni JM, Wittman M, Yang Z, Lee F, Greer A, Hurlburt W, et al. BMS-754807, a small molecule inhibitor of insulin-like growth factor-1R/IR. Mol Cancer Ther 2009;8:3341–9. [DOI] [PubMed] [Google Scholar]
- 27.Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021;1–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab 2016;23:517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 2016;532:255–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guha M Anticancer IGF1R classes take more knocks. Nat Rev Drug Discov 2013;12:250–250. [DOI] [PubMed] [Google Scholar]
- 31.Waldhart AN, Dykstra H, Peck AS, Boguslawski EA, Madaj ZB, Wen J, et al. Phosphorylation of TXNIP by AKT Mediates Acute Influx of Glucose in Response to Insulin. Cell Rep 2017;19:2005–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 2013;496:12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiriac H, Belleau P, Engle DD, Plenker D, Deschênes A, Somerville TDD, et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov 2018;8:1112–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahib L, Shapiro M, Shrager J, Federowics B, Kinsey C. Abstract 07: Use of a real-world data registry to rapidly generate outcomes data following a case study of a novel treatment combination in pancreatic adenocarcinoma. Clin Cancer Res 2020;26:07–07. [Google Scholar]
- 36.Subramani R, Lopez-Valdez R, Arumugam A, Nandy S, Boopalan T, Lakshmanaswamy R. Targeting Insulin-Like Growth Factor 1 Receptor Inhibits Pancreatic Cancer Growth and Metastasis. PLoS ONE 2014;9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molina-Arcas M, Moore C, Rana S, van Maldegem F, Mugarza E, Romero-Clavijo P, et al. Development of combination therapies to maximize the impact of G12C KRAS inhibitors in lung cancer. Sci Transl Med 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lou K, Steri V, Ge AY, Hwang YC, Yogodzinski CH, Shkedi AR, et al. KRASG12C inhibition produces a driver-limited state revealing collateral dependencies. Sci Signal 2019;12:eaaw9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by co-targeting MEK and IGF-1R/PI3K. Cancer Cell 2010;18:683–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci 2011;36:320–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Q, Zhang Y, Zhu J, Zheng H, Chen S, Chen L, et al. IGF-1R inhibition induces MEK phosphorylation to promote survival in colon carcinomas. Signal Transduct Target Ther 2020;5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flanigan SA, Pitts TM, Newton TP, Kulikowski GN, Tan AC, McManus MC, et al. Overcoming IGF1R/IR Resistance through Inhibition of MEK Signaling in Colorectal Cancer Models. Clin Cancer Res 2013;19:6219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du C, da Silva A, Morales-Oyarvide V, Costa AD, Kozak MM, Dunne RF, et al. Insulin-like growth factor-1 receptor expression and disease recurrence and survival in patients with resected pancreatic ductal adenocarcinoma. Cancer Epidemiol Biomark 2020;29:1586–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data is available at GEO (accession number GSE163348). All other datasets generated for the current study are available from the corresponding author upon request.