Abstract
Four putative apyrase genes were identified from the model legume Medicago truncatula. Two of the genes identified from M. truncatula (Mtapy1 and Mtapy4) are expressed in roots and are inducible within 3 h after inoculation with Sinorhizobium meliloti. The level of mRNA expression of the other two putative apyrases, Mtapy2 and Mtapy3, was unaffected by rhizobial inoculation. Screening of a bacterial artificial chromosome library of M. truncatula genomic DNA showed that Mtapy1, Mtapy3, and Mtapy4 are present on a single bacterial artificial chromosome clone. This apyrase cluster was mapped to linkage group seven. A syntenic region on soybean linkage group J was found to contain at least two apyrase genes. Screening of nodulation deficient mutants of M. truncatula revealed that two such mutants do not express apyrases to any detectable level. The data suggest a role for apyrases early in the nodulation response before the involvement of root cortical cell division leading to the nodule structure.
Apyrases are enzymes that have the ability to hydrolyze nucleotide tri- and diphosphates to nucleotide monophosphates (Komoszynski and Wojtczak, 1996). Apyrases have been identified in almost all organisms investigated and have been proposed to function in very diverse roles, including blood platelet aggregation (Marcus and Safier, 1993), neurotransmission (Sarkis and Salto, 1991; Edwards and Gibb, 1993), protein glycosylation (Abeijon et al., 1993; Gao et al., 1999), and phosphate metabolism (Thomas et al., 1999). Apyrases can be classified as endoapyrases, enzymes that act intracellularly, and as ectoapyrases, enzymes that have their catalytic domain outside the cell. One function proposed for apyrases in animals is the regulation of blood platelet aggregation. Ectoapyrases present in the saliva of many insects that feed on blood can prevent blood platelet aggregation directly by decreasing the ADP concentration in damaged tissues (Marcus and Safier, 1993; Komoszynski and Wojtczak, 1996 and refs. therein). It is normal that the concentration of ADP in the extracellular space increases in response to damage. ADP is then able to bind to P2 purinergic receptors on platelets, causing their concomitant aggregation (Komoszynski, and Wojtczak, 1996). Apyrases found in animal cells have also been proposed to function in neurotransmission by interacting indirectly with P2 purinoreceptors (Edwards and Gibb, 1993). These receptors have a high affinity for ATP and ADP, but a much lower affinity for AMP. Ectoapyrases present in synaptic membranes are thought to degrade ATP in the synaptic space (Sarkis and Salto, 1991). As the pool of ATP is hydrolyzed to AMP, the receptor is released and its stimulation is broken (Komoszynski and Wojtczak, 1996). In addition, apyrase effects on the ATP + ADP/AMP ratio modulate the action of 5′-nucleotidase, an enzyme that is abundant in the synaptic space. This enzyme is inhibited by ATP and ADP, but is activated by AMP.
Two endoapyrases, GDA1 and YND1, identified in Saccharomyces cerevisiae are postulated to regulate protein glycosylation in the Golgi (Abeijon et al., 1993; Gao et al., 1999). These apyrases control the turn over of GDP arising from hydrolysis of GTP sugars. In support of this hypothesis, a yeast strain that is defective in the production of GDA1 and YND1 was found to grow very slowly and exhibited defects in cell wall integrity and protein glycosylation (Gao et al., 1999).
Apyrases identified in plants have been implicated to play functional roles in a variety of different systems. For example, an apyrase purified from pea nuclei was originally reported to be involved in phytochrome responses (Chen and Roux, 1986; Chen, et al., 1987). Enzymatic activity of this protein was stimulated by Ca2+/calmodulin. This pea NTPase was more recently reported to play a role in mediating the uptake of phosphate from the extracellular matrix (Thomas et al., 1999). The pea apyrase has been found not only in nuclei, but also in the plasma membrane. Transgenic Arabidopsis plants expressing the pea apyrase showed enhanced growth compared with the wild type and could utilize exogenous ATP as a source of inorganic phosphate (Thomas et al., 1999). It has been postulated that an apyrase isolated from potato is involved in starch biosynthesis (Handa and Guidotti, 1996).
A recent report indicated that an apyrase purified from the roots of the legume Dolichos biflorus, lectin nucleotide phosphohydrolase (LNP), bound to the lipo-chitin nodulation signals produced by Bradyrhizobium spp. (Etzler et al., 1999). These lipo-chitin signals are essential for nodulation and trigger the de novo organogenesis of the root nodule (for review, see Cohn et al., 1998). The D. biflorus apyrase showed highest affinity for the Nod signals produced by Bradyrhizobium japonicum and Rhizobium spp. NGR234, bacteria that are able to nodulate D. biflorus. LNP was localized by immunofluorescence to the surface of the root hairs, the primary site of rhizobial infection. Binding of Nod signals to LNP stimulated ATPase activity. Two apyrases have recently been characterized in soybean, one of which is transcriptionally activated by rhizobia (Day et al., 2000). The protein product of this gene, GS52, was immunolocalized to Suc gradient fractions enriched in plasma membrane, suggesting that this protein is plasma membrane associated. In addition, pretreatment of roots with antiserum raised against LNP or GS52 inhibited the ability of rhizobia to nodulate D. biflorus or soybean, respectively (Etzler et al., 1999; Day et al., 2000). These data, taken together, suggest that LNP and GS52, an orthologous protein from soybean, are promising candidates for Nod signal receptors involved in the nodulation response.
Due to their postulated role in nodulation and possible importance to plant metabolism we initiated a study to characterize the apyrase genes found in Medicago truncatula. This plant has been proposed as a model for the genetic study of legumes and is currently the subject of intensive investigation (Barker et al., 1990; Cook, 1999). Using primers designed from conserved apyrase motifs, four unique apyrase cDNAs were isolated from M. truncatula. Genetic mapping analysis indicated that at least three of these genes share synteny with apyrase genes characterized in soybean. To our knowledge, this is the first report of synteny between soybean and M. truncatula. Analysis of mRNA expression indicated that M. truncatula apyrases are differentially expressed in various tissues. Two of these genes are rapidly induced upon rhizobial inoculation, similar to the expression pattern of a group of genes that have been postulated to be involved in the nodulation process, i.e. early nodulins (ENODs). Proteins that are specifically expressed during nodulation have been termed nodulins. ENODs are expressed within 48 h after rhizobial inoculation. The rapid accumulation of apyrase mRNA in response to rhizobia classifies these apyrases as ENODs. Consistent with the possibility that these genes play an important role in nodulation, apyrase expression was dramatically reduced in nodulation deficient mutant lines of M. truncatula.
RESULTS
Identification of a cDNA Encoding a Putative Apyrase Gene
An M. truncatula cDNA was identified by touch-down-PCR (TD-PCR) using primers designed from conserved domains found in a variety of apyrases (Fig. 1). The 850-bp fragment obtained by TD-PCR was extended using 3′- and 5′-RACE (Frohman et al., 1988) to yield a full-length cDNA sequence. One of the clones generated was designated plasmid pJRC63, which contains the full-length Mtapy1 cDNA.
Figure 1.
Amino acid sequence comparison of Mtapy1 with other known apyrase proteins. The boxed regions represent the four conserved apyrase domains. See text for details.
The sequence of Mtapy1 (GenBank accession no. AF288132) predicts an open reading frame of 466 amino acids, encoding a protein with a calculated molecular mass of 51,600. Hydropathy analysis of the predicted protein sequence indicated that Mtapy1 has a single strong transmembrane helix of 20 amino acids at the N terminus (data not shown). This transmembrane region was also predicted to be a cleavable N-terminal signal sequence according to the methods of von Heijne et al. (1986; Emanuelsson et al., 2000).
Comparison of the sequence of Mtapy1 with other genes in the nonredundant National Center for Biotechnology Information protein sequence database using a BLASTP search identified several possible orthologs (Altschul et al., 1997). We also identified possible orthologous genes using a dbEST search (Pearson et al., 1997). Alignment of Mtapy1 with the sequence of other known genes in the database revealed that Mtapy1 contained all four of the putative conserved apyrase domains (Fig. 1; Handa and Guidotti, 1996).
Mtapy1 Is Induced Rapidly in Response to Sinorhizobium meliloti Inoculation
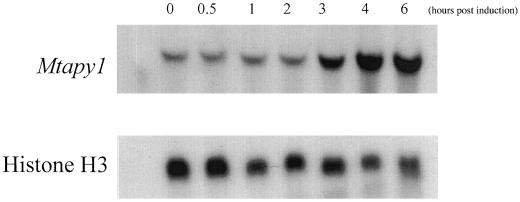
Since the D. biflorus apyrase has been implicated in nodulation (Etzler et al., 1999), northern-blot analysis was used to see if the level of apyrase mRNA significantly increased upon rhizobial inoculation. We performed a time course of expression of Mtapy1 using total RNA isolated from roots of wild-type M. truncatula line A17 inoculated with S. meliloti strain ABS7M. As seen in Figure 2, Mtapy1 mRNA accumulated significantly in response to S. meliloti within 3 h after inoculation.
Figure 2.
Northern blot showing the expression of Mtapy1 in response to inoculation with S. meliloti wild-type strain ABS7M (top). Total RNA was isolated from roots inoculated 0, 0.5, 1, 2, 3, 4, 5, and 6 h after inoculation with bacteria. Blots were also probed with Histone H3 as an RNA loading control (bottom). Each lane was loaded with RNA isolated from approximately 0.2 g of root tissue. See text for details.
Identification of a Second Putative Apyrase Gene from M. truncatula
A search of the dbEST sequence database using the Mtapy1 sequence identified a distinct apyrase from a cDNA library made from M. truncatula root hair tissue (Covitz et al., 1998; GenBank accession no. AA660474). Using the sequence in the database, primers were designed and used in a reverse transcriptase- (RT) PCR reaction to clone the corresponding cDNA. This approach yielded a plasmid JRC201 containing a partial cDNA clone of 430 bp. DNA sequence analysis revealed that this PCR fragment was identical to the sequence found in the dbEST database, but distinct from Mtapy1. Therefore, this apyrase cDNA was designated Mtapy2. Sequence analysis was also confirmed by the identification of a second expressed sequence tag (EST) sequence from a different EST library (dbEST accession no. AI974272). This EST clone, pKV01 M6, was kindly provided by Dr. Kate VandenBosch (Texas A&M University, College Station) and the clone was sequenced. The overlapping regions in this sequence and the original EST sequence (accession no. AA660474) indicated that they represent the same gene, namely, Mtapy2. The additional sequence information was critical for subsequent phylometric analyses.
M. truncatula Linkage Group 7 Contains an Apyrase Gene Cluster
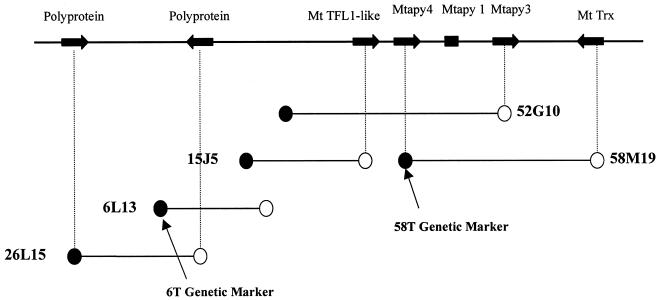
Mtapy1 was used as a probe to screen a M. truncatula bacterial artificial chromosome (BAC) library (Nam et al., 1999). Two hybridizing BAC clones, 52G10 and 58 M19, were isolated and confirmed by Southern-blot hybridization (data not shown). As shown in Figure 3, a larger region of contiguous BAC DNA was identified by PCR amplification of the BAC library multiplex using oligonucleotide primers designed from BAC sequence information. Taken together, DNA sequencing of BAC ends or internal sequencing with Mtapy1 primers identified five putative genes in this region, and two retrotransposon-like sequences (Fig. 3; Table I). Additional sequence information for the five predicted genes was obtained by a primer walking strategy on purified BAC DNA or on subcloned BAC DNA fragments. As shown in Table I, three of these predicted genes have their highest homology to characterized apyrase genes from legumes. One of these genes was verified as Mtapy1, based on sequence identity, whereas the other two were distinct from Mtapy2 and were designated as Mtapy3 and Mtapy4. The advent of numerous M. truncatula EST sequences in the National Center for Biotechnology Information GenBank allowed the immediate verification of Mtapy3 and a thioredoxin-like sequence as transcribed genes. For Mtapy4 a combination of RT-PCR and 3′-RACE was used to isolate a 1,017-bp cDNA PCR product (GenBank accession no. AF288133) that was identical to sequenced regions of the Mtapy4 genomic clone. Thus, the BAC contig shown in Figure 3 contains a minimum of three apyrase genes with high homology to other legume arpyases. Similar, but less extensive analysis was conducted with Mtapy2. In brief, a single BAC clone designated 20K21 was identified by means of hybridization to high-density filter arrays. BAC end sequencing of 20K21 revealed the presence of a putative gene with best protein homology to NADPH quinone reductase (Table I).
Figure 3.
BAC contig map of a region containing a cluster of apyrase genes. The map indicates the location of the Mtapy1, Mtapy3, and Mtapy4. The BAC map indicates at least five genes present in this region, including a putative TFL1 ortholog and thioredoxin. Two retrotransposon-like sequences were also identified in this region. The genetic markers 58T and 6T are also present in the contiguous region surrounding the apyrase cluster, which were useful in mapping the cluster on the M. truncatula genome.
Table I.
Identified and predicted genes
| GenBank Accession No. | Designation | Mt Linkage Group | Strand | BAC Clone | Best Protein Homologa | Identification Methodb | BLAST × E Valuec | Predicted Gene Product |
|---|---|---|---|---|---|---|---|---|
| AZ124289 | Polyprotein | 7 | − | 26L15 | AF105716 | Prediction | 2.00E-20 | Copia-type polyprotein |
| AZ124288 | Polyprotein | 7 | + | 26L15, 6L13 | Y12432 | Prediction | 4.00E-58 | Putative reverse transcriptase |
| AZ142497 | MtTFL-like | 7 | + | 15J5, 52G10 | AC001229 | Prediction | 3.00E-12 | Similar to Arabidopsis terminal flower |
| 4720bp | Mtapy 3 | 7 | + | 52G10, 58M19 | AF156780 | Contains AW257004, AW776612c | 3.00E-53 | Putative apyrase |
| AF288132 | Mtapy 1 | 7 | n.d.d | 52G10, 58M19 | AB027616 | Contains Mtapyl cDNA | 5.00E-52 | Putative apyrase |
| AF288133 | Mtapy 4 | 7 | + | 52G10, 58M19 | AF139807 | Contains Mtapy4 cDNA | 9.00E-06 | Putative apyrase |
| AZ124328 | MtTrx | 7 | − | 58M19 | T00710 | Contains AW774567, AW229187 | 3.20E-35 | Similar to Arabidopsis thioredoxin homolog |
| AZ124253 | MtNQO-like | 4 | n.d. | 20K21 | T10203 | Prediction | 7.00E-16 | Similar to NADPH quinone reductase |
| Mtapy 2 | 4 | n.d. | 20K21 | Hybridization | Putative apyrase |
GenBank accession nos. of the closest protein homolog in the the nr database.
Accession nos. are given for exact EST or rtPCR cDNA matches. In the absence of matches to cDNA sequences, predictions are based on the best nr protein homolog.
For definition of Blast Expect (E) values, see http://www.ncbi.nlm.nih.gov/BLAST.
n.d., No data.
AW776612 is identical to, and completely contained within, the Mtapy3 genomic sequence, except for 2 of 267 base pairs.
Genetic Mapping of Apyrases
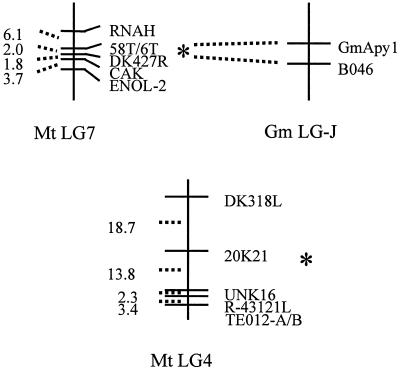
To determine the genetic map position of the apyrase cluster (Mtapy1, Mtapy3, and Mtapy4) and of Mtapy2, codominant PCR-based cleaved amplified polymorphic sequence (CAPS) markers were developed. In brief, BAC end sequence information from BACs 6L13 and 58 M19 (corresponding to the apyrase cluster) and from BAC 20K21 (corresponding to Mtapy2) was used to design oligonucleotide primers. PCR amplification and sequencing of genomic DNA from M. truncatula mapping genotypes A17 and A20 allowed the development of three CAPS markers, as detailed in Table II. As shown in Figure 4, analysis of these CAPS markers on the basic mapping population of 93 F2 individuals indicated that the apyrase cluster maps to M. truncatula linkage group 7, whereas Mtapy2 maps to linkage group 4.
Table II.
Genetic marker information
| Marker Name | Marker Homology | Oligonucleotide Sequence | PCR Conditions | Product Size | Restriction Enzyme | Size of Cleaved Product |
|---|---|---|---|---|---|---|
| 58Ta | Mtapy4 | AAGGGCTTTTAATTTCTGTCTGCCAAACCAATCTAAAATAATAAAC | 90°C, 4 min; 50°C, 30 s; 72°C, 50 s | 691 bp | BsiEI | A17: 233 + 458 bp; A20: 691 bp |
| 6Tb | Mtapy contig | ATTTTAATTGAACGTATCTTTTTGAGGCTCCTAATTTTTGACTGTTG | 90°C, 4 min | 740 bp | Bsp1286I | A17: 332 + 408 bp; A20: 740 bp |
| 20Sc | Mtapy2 | GATGGTCTGGCAACTGTAGGGAGGACTTTTCTTAG | 90°C, 4 min; 50°C, 30 s; 72°C, 50 s | 409 bp | RsaI | A17: 119 + 290 bp; A20: 691 bp |
| DK427 R | Soybean RFLP B046 | CCAAACAAGGAAAAGTGTTGGTGTCAATGAGAAACTTTTGAAATTTAGGATACGATAG | 90°C, 4 min; 50°C, 30 s; 72°C, 50 s | 500 bp | BsmAI | A17: 60 + 440 bp; A20: 500 bp |
85T corresponds to BAC 58M19.
6T corresponds to BAC 6L13.
20T corresponds to BAC 20K21.
Figure 4.
Genetic maps of apyrases from M. truncatula. The maps indicate that the apyrase cluster, including at least Mtapy1, Mtapy3, and Mtapy4 is located on linkage group (MtLG) 7 based on analysis of the map positions of the 58T marker. Mtapy2 was determined to be located on linkage group 4 based on 6T genetic CAPS marker developed from sequence analysis of the BAC clone 20K21. An apyrase cluster was also found on soybean linkage group J (Gm LG-J), based on analysis of the map positions of two apyrases identified in soybean (i.e. GS50 and GS52; Day et al., 2000). The apyrase cluster in soybean is closely linked to the B046 RFLP marker, which is orthologous to the 58T/6T marker in M. truncatula. Thus, the B046 homology is tightly linked to an apyrase cluster in M. truncatula and soybean. These data are suggestive of conserved synteny between the two genomes and are consistent with close ancestry of these two apyrase gene clusters.
The position of the M. truncatula apyrase markers is shown relative to selected core markers on these linkage groups (D.-J. Kim and D.R. Cook, personal communication). Because several of the M. truncatula core markers were developed based on homology to mapped restriction fragment-length polymorphism (RFLP) clones from soybean genetic map (http://ars-genome.cornell.edu/cgi-bin/WebAce/webace?db=3Dsoybase; D.-J. Kim and D.R. Cook, unpublished data), it is possible to identify putative syntenic relationships between the M. truncatula and soybean genomes. One such marker designated DK427-R maps in close proximity to the apyrase cluster marker 58T (Fig. 4). MtDK427 has homology to soybean RFLP clone B046, the sequence of which (GenBank accession no. AQ841833) is similar to an EST from cotton (GenBank accession no. AI727823). In soybean, B046 maps to two loci, including marker B046b on soybean linkage group J. To ascertain if the M. truncatula apyrase cluster might share a conserved genome context with previously identified soybean apyrase genes (Day et al., 2000) we conducted a similar analysis with soybean apyrase genes GS50 and GS52. In brief, a total of nine soybean BAC clones in three soybean contigs were identified. Two of these contigs were positive for GS50 and GS52. One of these contigs could be assigned to a previously described locus on soybean LG-J (Day et al., 2000) based on its restriction fragment pattern. The map locations of the other two apyrase-associated contigs in soybean were not determined. It is significant that the apyrase locus on soybean LG-J lies in close proximity to RFLP marker B046, just as in M. truncatula. Moreover, this contig contains at least two hybridizing fragments, one showing homology to GS50 only and a second with homology to GS50 and GS52, indicating a possible apyrase cluster. Thus, the B046 homology is tightly linked to an apyrase cluster in M. truncatula and soybean. These data are suggestive of conserved synteny between the two genomes, and are consistent with close ancestry of these two apyrase gene clusters.
Phylogenetic Analysis of Apyrases
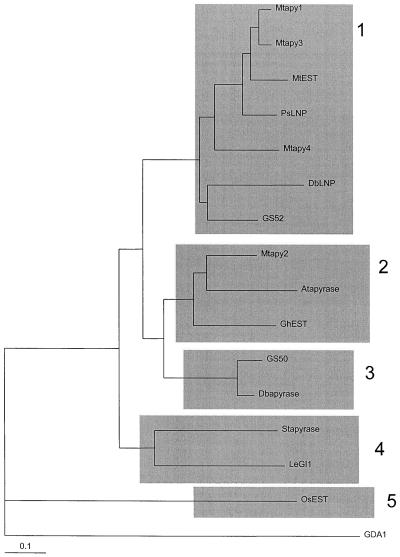
A recent report indicated that plant apyrases are separated into two distinct families based on phylogenetic studies (Roberts et al., 1999). It was suggested that LNP-like genes from a number of different sources were members of a distinct class of apyrases that may be unique to legumes. We performed a similar phylogenetic analysis with the apyrase sequences from M. truncatula by aligning 16 partial sequences of putative plant apyrases, and a yeast protein, GDA1. One additional EST sequence of a putative apyrase from M. truncatula (MtEST, accession no. AJ388942) that is distinct from Mtapy1, Mtapy2, Mtapy3, and Mtapy4 was included in the phylogenetic analysis. A neighbor-joining tree generated from this alignment shows that there appear to be at least five subfamilies of apyrases found in plants (Fig. 5). Subfamily 1 contains Mtapy1, Mtapy3, Mtapy4, and the newly identified MtEST. This subfamily also includes the pea NTPase, LNP from D. biflorus, and GS52, a recently identified apyrase from soybean (Day et al., 2000). Subfamily 2 consists of Mtapy2, an apyrase from Arabidopsis, and an EST identified in cotton (GhEST). Subfamily 3 consists of a second apyrase isolated from soybean, GS50 (Day et al., 2000), and a second apyrase from D. biflorus that was identified subsequently to DbLNP (Roberts et al., 1999). Subfamily 4 consists of the potato apyrase (Handa and Guidotti, 1996) and a tomato EST (LeGl1). Subfamily 5 consists of a single gene represented by an EST identified in rice (OsEST) (GenBank accession no. AU065811). These results support the earlier results of Roberts et al. (1999), indicating that LNP from D. biflorus is a member of distinct family of apyrases that are present only in legumes. It is quite interesting that Mtapy1, Mtapy3, and Mtapy4, which are all clustered in the M. truncatula genome, are all members of this apyrase family. It is possible that these genes arose from duplication events during the evolution of M. truncatula.
Figure 5.
Neighbor-joining tree representing alignment of apyrases from selected sources. Sixteen partial sequences of plant apyrases and the yeast protein GDA1 were aligned for analysis. The apyrases fall into five distinct subfamilies. Mtapy1, Mtapy3, Mtapy4, a newly identified EST from M. truncatula, MtEST (GenBank accession no. AJ388942), LNP (accession no. AF139807), and GS52 (accession no. AF207688; Day et al., 2000) fall into the first class. Mtapy2 was found to fall into subfamily 2, along with an Arabidopsis apyrase (accession no. AF156783; the second Arabidopsis apyrase [accession no. AF141671] falls in a similar position, not shown), and an EST from cotton, GhEST (accession no. AI729322). Additional sequences include G. soja GS50 (accession no. AF207687; Day et al., 2000), D. biflorus Dbapyrase (accession no. AF156781), potato apyrase (accession no. U58597), tomato EST cLED13N21 (accession no. AI488720), rice OsEST, (accession no. AU065811), and the S. cerevisiae GDA1 (accession no. 6320793).
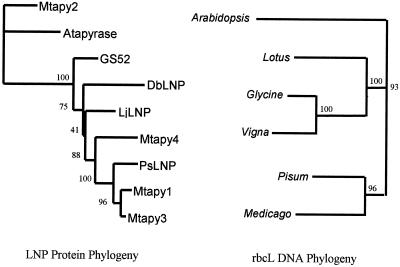
A further phylogenetic analysis was performed with the apyrases from M. truncatula to study their evolutionary relationships. To infer the phylogeny of the Mtapy genes, amino acid sequences of deduced LNP proteins were aligned to the amino acid sequences of Mtapy1-4 using CLUSTALW (Thompson et al., 1994). An apyrase from Arabidopsis was also included in the alignment as an out-group. The results of this analysis showed that Mtapy1, Mtapy3, and Mtapy4 all group together with the LNP sequences, whereas Mtapy2 is more closely related to the out-group apyrase from Arabidopsis (Fig. 6). The appearance of multiple copies of M. truncatula apyrase sequences in the LNP clade strongly implies that gene duplication may be an ongoing phenomenon in this apyrase subfamily. To confirm this, a neighbor-joining phylogeny from rbcL sequences was constructed of closely related species (Fig. 6). The results showed that the topology of the two trees is not the same. Whereas the rbcL tree shows a substantial divergence between Lotus/Glycine/Vigna and Pisum/Medicago (bootstrap support 93%), the LNP clearly shows the Glycine soja sequence GS52 as the only outgroup (bootstrap support 100%). In the rbcL tree Lotus is a sister taxon to the Glycine/Vigna clade, but in the LNP phylogeny, the Lotus sequence LjLNP is a sister taxon to a clade formed of three Medicago sequences (Mtapy1, Mtapy3, and Mtapy4) and the Pisum sequence PsLNP. The conflicts between the rbcL and LNP trees can be explained as the result of gene transfer or multiple LNP paralogues that have not yet been sequenced. Because the LNP tree clearly shows three Medicago LNP paralogues (Mtapy1, Mtapy3, and Mtapy4), undiscovered paralogues in other legume species would be the preferred explanation over gene transfer.
Figure 6.
The evolution of the LNP proteins has been markedly different from the evolution of the rbcL sequences from closely related species. LNP protein phylogeny (left) was inferred by the neighbor-joining method using a 188-amino acid alignment of LNP proteins out-grouped using the apyrase from Arabidopsis (accession no. AF156783; identical results were obtained using a second Arabidopsis apyrase [accession no. AF141671], not shown). Sequences shown are Mtapy2 (this study), GS52 (Day et al., 2000), D. biflorus LNP (accession no. AF139807), Lotus japonicus LNP (accession no. AF156780), Mtapy4 (this study), pea LNP (accession no. Z32743), and Mtapy1 and Mtapy3 (this study). RbcL DNA phylogeny (right) was also inferred by the neighbor-joining method using a 1,310-nucleotide alignment of rbcL DNA sequences out-grouped using the rbcL of Arabidopsis (accession no. U91966). Other sequences shown are from Lotus corniculatus (accession no. U74213), soybean (accession no. Z95552), cowpea (accession no. Z95543), pea (accession no. X03853), and alfalfa (accession no. X04975). Bootstrap values out of 100 bootstrap replicates are shown at nodes of both trees.
MtApy1 and Mtapy4 Are Transiently Induced after Inoculation with S. meliloti
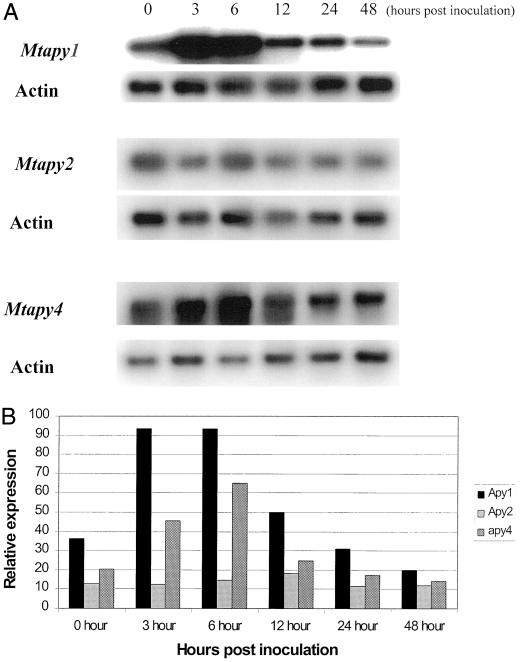
Due to the high degree of sequence similarity shared between the apyrases identified in M. truncatula, we wanted to identify which of the genes were induced upon rhizobial inoculation. As shown in Figure 7, RT-PCR results using gene-specific primers showed that the level of Mtapy1 transcript significantly increased within 3 h after inoculation. These data are consistent with our previous northern results using a radiolabeled Mtapy1 probe (Fig. 2). Quantification of the data shown in Figure 7A showed that the level of Mtapy1 increased approximately 3-fold within 6 h postinoculation (Fig. 7B). Likewise, similar results were obtained using gene specific primers for Mtapy4 (Fig. 7A). Quantification of these data indicated that Mtapy4 induction by inoculation is only slightly less than Mtapy1 (Fig. 7B). In the cases of Mtapy1 and Mtapy4, induction after inoculation is transient. The highest levels of expression were seen 3 to 6 h postinoculation with expression declining to basal levels by 12 to 24 h after inoculation.
Figure 7.
A, RT-PCR analysis of the level of mRNA expression of apyrase genes in response to wild-type S. meliloti strain ABS7M. RNA was collected from roots 0, 3, 6, 12, 24, and 48 h post-rhizobial inoculation. Actin primers were included in each reaction as a loading control. See text for details. B, Quantification of the data shown in A. The relative level of expression was determined by the following calculation: (total counts apyrase/total counts actin) × 100 = relative expression. The sd between relative expression values for two separate experiments was less than 10%.
In contrast to these results, the level of Mtapy2 mRNA was not significantly enhanced upon rhizobial inoculation (Fig. 7A). In these experiments Mtapy3 mRNA was not detected, consistent with the lack of expression of this gene in roots (see below).
Apyrase Genes Are Differentially Expressed in M. truncatula Tissues
To analyze the tissue-specific expression of apyrase genes in M. truncatula we used gene-specific primers and RT-PCR to analyze mRNA levels in various tissues. We isolated total RNA from the roots, hypocotyls, and cotyledons of 5-d-old seedlings, and from the stems, leaves, and flowers of 5-week-old plants. These plants were vernalized at 4°C for 14 d to shorten the flowering time. In addition, we isolated total RNA from 10-d-old nodules that were harvested from plants inoculated with wild-type S. meliloti strain 1021.
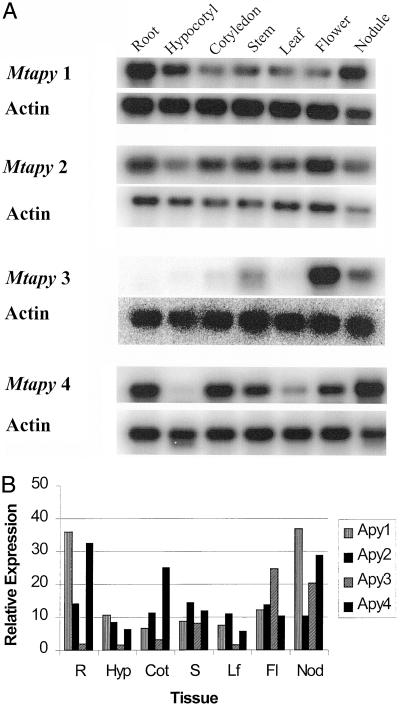
As shown in Figure 8, Mtapy1 was expressed predominantly in roots, hypocotyls, and nodule tissue. Mtapy4 was also expressed in root and nodule, but also showed significant expression in cotyledons, stem, and flower. No expression was found in hypocotyl tissue. Mtapy2 mRNA was detected in roughly equivalent amounts in all tissues. Mtapy3 showed the highest level of tissue specificity, showing appreciable expression only in flower tissue, with lesser amounts found in stem and nodule tissue. Consistent with previous RT-PCR results (see above), Mtapy3 mRNA was not detected in root tissue.
Figure 8.
A, RT-PCR analysis of the expression of apyrase genes from M. truncatula in various tissues. R, Five-day-old roots; Hyp, 5-d-old hypocotyls; Cot, 5-d-old cotyledon; S, 6-week-old stems; Lf, 6-week-old leaves; Fl, 6-week-old flowers; Nod, nodules isolated 10 d post-inoculation with wild-type S. meliloti strain 1021. Actin primers were included in each reaction as a loading control. See text for details. B, Quantification of the data shown in A. The relative level of expression was determined by the following calculation: (total counts apyrase/total counts actin) × 100 = relative expression. The sd between relative expression values for two separate experiments was less than 10%.
Apyrase Expression Is an Early Nodulation Event
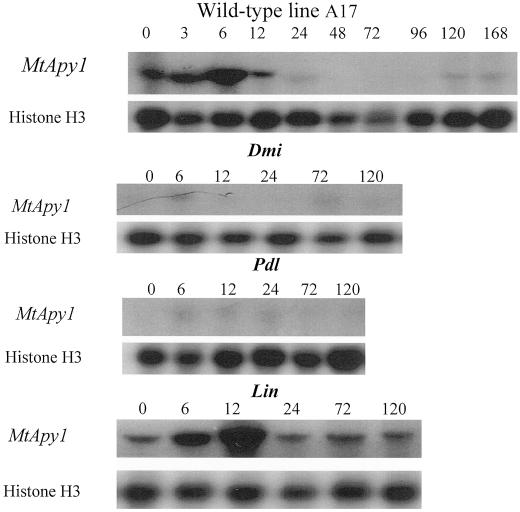
If apyrases are important to the nodulation response, then plant mutants defective in nodulation may also show reduced or altered apyrase expression. Moreover, such nodulation mutants can be used to determine when, during nodule ontogeny, a gene is expressed. Therefore, we analyzed Mtapy1 expression in three genetically distinct non-nodulation mutants of M. truncatula (Prabhu, 1998). As described in Table III, use of these mutants allowed for the analysis of apyrase expression at distinct, development time points during early nodulation. For example, these mutants were either nonresponsive to the bacterial symbiont (i.e. very early, mutant dmi1), blocked for formation of nodule primordia (i.e. a later step, mutant pdl1), or in the formation and/or maintenance of epidermal cell infection (i.e. a step subsequent to primordium formation, mutant lin). As shown in Figure 9, little or no Mtapy1 expression was detected in root tissue isolated from dmi1 or pdl, as measured by RT-PCR with Mtapy1-specific primers. As a positive control, Mtapy1 mRNA levels increased upon inoculation in root tissue isolated from the wild-type M. truncatula line A17 (Fig. 9). Consistent with previous results, the level of this expression peaked at approximately 6 h and was undetectable by 12 h postinoculation. The basal level of Mtapy1 expression was also significantly lower than that found in the wild type. The level of Mtapy1 expression in root tissue from the lin mutant showed a similar time course to that of the wild type, although maximal expression was slightly delayed, peaking at 12 h and reaching basal levels by 24 h postinoculation. This delay in Mtapy1 expression may reflect a slight delay in nodule development in the lin mutant. Analysis of Mtapy4 expression gave similar results (data not shown).
Table III.
Phenotypes of Medicago truncatula nodulation mutants
| Mutant Name | Gene Name | Infection Phenotype | Nodule Primordia Phenotype | Reference |
|---|---|---|---|---|
| Poodle | pdl | Wild-type nos. of infections; rare progression of infections past epidermis | None | R.V. Penmetsa and D.R. Cook, unpublished data |
| Lumpy infections | lin | Reduced nos. of infections; infections arrest soon after infection thread formation | Nodule primordia evident; no infection threads in nodule primordia | R.V. Penmetsa and D.R. Cook, unpublished data |
| Doesn't make infections | dmil | None | None | Penmetsa and Cook (1997); Catoira et al. (2000) |
Figure 9.
Northern-blot analysis of the level of expression of Mtapy1 in wild-type and EMS-mutagenized lines of M. truncatula. Total RNA was purified from roots of the A17 wild-type line at 0, 3, 6, 12, 24, 48, 72, 96, 120, and 168 h postinoculation with wild-type S. meliloti strain ABS7M. Total RNA was purified from the roots of dmi, pdl, and lin plants at 0, 6, 12, 24, 72, and 120 h postinoculation with wild-type S. meliloti strain ABS7M. Blots were first probed with an Mtapy1-specific probe and then reprobed with a histone probe as a loading control.
DISCUSSION
Many of the very early nodulation responses generated in legume roots by rhizobial inoculation (including the induction of a nodule structure) can also be elicited by treatment with the appropriate, purified lipo-chitin nodulation signal (for review, see Dénarié et al., 1996; Long, 1996; Minami et al., 1996; Cohn et al., 1998). Nodulation signals act with high specificity at very low concentrations and, therefore, they are thought to mediate their effects through interaction with a specific, protein receptor. The search for such a receptor is an area of intensive research.
A promising candidate for a Nod signal receptor is the apyrase protein isolated from the roots of the legume D. biflorus, LNP. This protein was shown to be localized on the root hair surface and to bind Nod signals from rhizobia capable of nodulating D. biflorus (Etzler et al., 1999). Moreover, binding of the Nod signal to the apyrase stimulated ATPase activity, suggesting a possible mode for signal transduction. In addition, an orthologue of LNP characterized from soybean, GS52, was localized to the plasma membrane of soybean roots (Day et al., 2000). One striking result from work with LNP and the soybean orthologue was the demonstration that antiserum raised against either protein inhibited the ability of rhizobia to nodulate their respective hosts, D. biflorus and soybean. These data suggest that apyrases play a critical role in the early events of nodulation. It is unfortunate that D. biflorus is a little-studied legume with limited information available on its nodulation properties, genetics, preferred symbiont, etc. Although soybean is well characterized, the large genome size and the fact that it is an ancestral tetraploid make genetic analysis of this plant difficult.
M. truncatula has recently been touted as a promising model plant for genetic studies of legumes (Barker et al., 1990; Cook, 1999). A number of laboratories worldwide are now working to develop this plant as a model system. The accumulating information on M. truncatula should be an aid to studies of nodulation. Therefore, we undertook a study to examine the apyrase genes of this plant as an initial step in defining the role of these proteins in the nodulation response.
Four apyrase genes were isolated. Due to the close sequence similarity among these genes we utilized gene-specific primers and RT-PCR to examine mRNA expression levels. Consistent with a possible role in nodulation, mRNA levels of Mtapy1 and Mtapy4 were rapidly elevated upon inoculation of roots with S. meliloti. The expression pattern of these two genes classifies them as ENODs. There are only two other examples of legume genes that respond this quickly to rhizobia (Pichon et al., 1992; Journet et al., 1994; Cook et al., 1995). The gene encoding ENOD12 has been reported to be inducible by rhizobia within 1 h after rhizobial inoculation (Pichon et al., 1992; Horvath et al., 1993; Journet et al., 1994). At present, there is no known biochemical function for the Pro-rich ENOD12 protein. In fact, a M. truncatula subspecies that does not express ENOD12 is not affected in its ability to nodulate (Csanádi et al., 1994). One ENOD that does have a proposed function is rip1 (Rhizobium-induced peroxidase). Rip1 mRNA was transiently induced in M. truncatula roots within 3 h after S. meliloti inoculation (Cook et al., 1995). This expression pattern is similar to what was seen for Mtapy1 and Mtapy4. Rip1 has been postulated to play a role in oxidative processes that occur very early in the nodulation response (Cook et al., 1995)
RT-PCR analysis of gene expression shows that induction of Mtapy1 and Mtapy4 after S. meliloti inoculation is transient, reaching a peak of expression 3 to -6 h postinoculation, but returning to basal levels in 12 to 24 h. These results are similar to what has been recently found for the expression pattern of a soybean apyrase, GS52 (Day et al., 2000). The expression of this gene was transiently increased in response to inoculation with the soybean symbiont, B. japonicum within 6 h after inoculation. If these apyrases are indeed Nod signal receptors, it seems at first counterintuitive to see the level of expression of these genes increase in response to their agonist (i.e. rhizobia). However, these results are consistent with several reports of the activation of receptor genes in response to agonist addition in other systems (Eriksson et al., 1991; Ng et al., 1997; Helmrath et al., 1998; Kisselgof and Oettgen, 1998; Perera et al., 1999).
Analysis of the level of expression of Mtapy1 and Mtapy4 in response to inoculation with rhizobial mutants indicated that S. meliloti strains that are not able to produce Nod signals did not significantly affect the level of transcription of Mtapy1 or Mtapy4 (data not shown). These data suggested that only rhizobia that are able to produce Nod signals are able to induce the transcription of apyrases in M. truncatula. However, further experiments are necessary to make any conclusions about specific bacterial factors that might affect apyrase mRNA expression.
The data indicate that Mtapy2 and Mtapy3 are not induced in response to rhizobial inoculation. However, the Mtapy2 gene was identified as an EST sequence from a cDNA library generated from mRNA isolated from M. truncatula root hair tissue. The root hairs are the site of rhizobial infection and, therefore, this location would be consistent with a role in nodulation. Although Mtapy3 mRNA expression was not detected in roots, there was appreciable expression in nodule tissue. In fact, the tissue expression pattern of Mtapy3 found in nodules and flowers is similar to the expression pattern found for the ENOD gene, ENOD40 (Crespi et al., 1994). Since the focus of this work was on early nodulation events, Mtapy3 expression was not extensively examined.
Sequencing and genetic mapping placed Mtapy1, Mtapy3, and Mtapy4 close together on linkage group 7 of the M. truncatula genome. These three genes are found on the same BAC contig. Southern hybridization data of this BAC contig is consistent with the presence of three apyrase genes (data not shown), although we cannot rule out the possibility of additional apyrase genes with reduced nucleotide homology in this cluster. PCR analysis indicated that Mtapy2 was not present on this BAC contig, but instead Mtapy2 hybridized strongly to BAC 20K21 that mapped to M. truncatula linkage group 4.
Apyrase genes have also been characterized from soybean (Day et al., 2000). Hybridization of these apyrases to high-density BAC filters indicates that, as we have determined for M. truncatula, the legume-specific apyrase genes are also clustered in the soybean genome (N.D. Young, unpublished data). Moreover, genetic mapping data demonstrated that in M. truncatula and soybean, these homologous apyrase clusters are tightly linked to a conserved genetic marker with homology to cotton EST AI727823. The finding of a conserved genetic locus that contains a cluster of apparently legume-specific apyrase genes provides the first indication of conserved genome structure between crop and model legume systems, and it should provide the basis for more detailed analysis of the conservation of this gene family within the legume subfamily Papilionoideae.
If apyrases play an important role in the nodulation response, then plant mutants defective in nodulation may show an altered profile of apyrase expression. Use of such mutants also allowed us to examine apyrase expression in relation to distinct developmental events during early nodulation. Northern analysis of ethyl methane sulfatonate- (EMS) generated, nodulation-defective lines of M. truncatula identified mutants defective in apyrase expression. Little or no Mtapy1 mRNA expression was seen in root tissue from the pdl or dmi mutants. dmi1 is nearly devoid of symbiotic responses, lacking bacterial infection and formation of nodule primordia, and has been proposed to function in transduction of the Nod signal (Catoira et al., 2000), whereas the pdl mutant develops numbers of infections comparable with wild-type plants, but fails to develop nodule primordia. In contrast to dmi1 and pdl, the lin mutant develops visible nodule primordia, but has a reduced infection phenotype. The numbers of infection events in lin are reduced relative to wild type, and those infections that do form are arrested soon after infection thread initiation. Thus, the observation that symbiotic expression of Mtapy1 is absent in dmi1 and pdl, but present in lin suggests that Mtapy1 expression is correlated with the formation of nodule primordia, but is apparently independent of rhizobial infection. The low expression of Mtapy1 in the absence of inoculation in the dmi1 and pdl mutant lines indicates that these mutants are not only defective in the induction of apyrase expression, but also in basal expression. Therefore, it would appear that the same signaling circuitry involved in inducible Mtapy1 expression are also involved in maintaining a basal level of expression. It is interesting that genetic mapping data indicates that the apyrase cluster shown in Figure 3 is located within 2 cM of the pdl locus (data not shown). Therefore, it is possible that the apyrase genes are clustered in an area of the M. truncatula genome that contains other symbiotically relevant loci.
A role for apyrases in early nodulation has been suggested previously by Etzler et al. (1999) and Day et al. (2000). In particular, a subclass of apyrase genes with uniquely high homology among legumes is implicated in the perception of the Nod signal. Here we have determined that M. truncatula contains multiple apyrase genes. It is interesting that several members of the legume-specific clade of apyrases are clustered in the M. truncatula genome, and transcripts for each of these apyrase genes can be detected in nodules by northern-blot analysis or by surveys of nodule derived ESTs in M. truncatula. Two of these apyrases, designated Mtapy1 and Mtapy4, are rapidly induced upon rhizobial inoculation of roots, thus classifying these genes as ENODs. The differential expression of Mtapy1 in dmi/pdl in relation to lin establishes a correlation between expression of Mtapy1 and nodule morphogenesis. Future studies of this important gene family should benefit from the advent of an intensively studied model legume system such as M. truncatula, and the opportunities provided by an increasingly well-characterized genome (Cook, 1999) and facile transformation technologies (Trieu et al., 2000).
MATERIAL AND METHODS
Culture Media and Growth Conditions
Escherichia coli strains were grown and maintained on Luria-Bertani medium (Sambrook et al., 1989) at 37°C. Sinorhizobium meliloti strains were grown and maintained on tryptone-yeast extract (TY) medium (Somasegaren and Hoben, 1994) at 30°C. Antibiotics used in this study for selection and plasmid maintenance were 100 μg mL−1 ampicillin, 50 μg mL−1 kanamycin, or 10 μg mL−1 tetracycline.
Plant Material and Growth Conditions
Prior to germination Medicago truncatula seeds were scarified by treatment with concentrated H2SO4 for 8 min, rinsed thoroughly with sterile distilled water, and treated with 5% (v/v) hypochlorite for 3 min. Following thorough rinsing, the seeds were imbibed in sterile distilled water for 3 to 4 h at room temperature. Treated seeds were placed into sealed Petri plates on moist filter paper at 4°C in the dark for at least 48 h and were then transferred to room temperature in the dark for 24 h. Germinated seedlings were transferred to growth pouches (Mega International, Minneapolis) or to aeroponic chambers (Lullien et al., 1987; Gallusci et al., 1991). Seedlings were grown in inorganic nutrient medium (Lullien et al., 1987) under a 16-h day/8-h night cycle (22°C light/18°C dark).
Bacterial Inoculation of Plants
Prior to plant inoculation, bacteria were grown to late log phase in TY medium (Somasegaran and Hoben, 1994) supplemented with appropriate antibiotics. Bacteria were then centrifuged, washed, and resuspended in sterile distilled water. Bacteria were added directly to nitrogen-deficient plant nutrient solution (Lullien et al., 1987) in the aeroponic chambers. Nodulation was rapid and uniform under these conditions. Plants were grown under a 16-h-light (22°C)/8-h-dark (18°C) cycle.
For inoculation of plants grown in plastic growth pouches, bacteria were grown to log phase in TY medium and an aliquot was transferred to fresh medium to monitor growth. Bacterial cultures were grown to an OD600 of 1.0, centrifuged, washed, and then resuspended in inorganic nutrient medium (Lullien et al., 1987) to yield 107 cells mL−1. Each plant was inoculated with 1 mL of the bacterial suspension. Nodulation was also rapid and uniform under these conditions.
RNA Isolation
Plant tissue samples were frozen in liquid nitrogen and stored at −80°C until use. RNA was isolated using a protocol modified (Cook et al., 1995) from the guanidine-HCl extraction protocol (described in Sambrook et al., 1989). The purified RNA pellet was resuspended in RNase-free, diethyl pyrocarbonate- (Sigma, St. Louis) treated sterile distilled water and stored at −80°C until use.
TD-PCR
For the initial cloning of the M. truncatula apyrase cDNAs using TD-PCR (Don et al., 1991), degenerate primers were designed from conserved apyrase motifs present in the pea (Hseih et al., 1996), potato (Handa and Guidotti, 1996), D. biflorus (Etzler et al., 1999), and soybean apyrases (Day et al., 2000). Primers were used in a final concentration of 0.2 μm in all reactions. The primers used for TD-PCR were as follows: TD-apyrase forward, 5′-ATTGATGGAACCCAAGAAGG -3′; and TD-apyrase reverse, 5′-GGYAAAGMTGATATGGCTTC-3′ (Y = C/T; M = A/C). The conditions were as follows: 94°C, 1 min; X°C, 2 min; and 72°C, 3 min, where X represents changing annealing temperatures. The annealing temperature was steadily decreased by 2°C every 3 cycles starting at 60°C and ending at 42°C. PCR products were visualized by agarose gel electrophoresis and ethidium bromide staining (Sambrook et al., 1989).
Cloning of PCR Products
PCR products of the expected size were cloned into the TA-cloning vector pCR2.1 using a TA Cloning Kit (Invitrogen, Carlsbad, CA), or into the TOPO cloning vector (Invitrogen) following the manufacturer's instructions. In certain cases ligation products were cloned using the P-GEM-T-EASY kit (Promega, Madison, WI) and transformed into E. coli strain JM109 according to Sambrook et al. (1989). Plasmid DNA was isolated using the alkaline lysis method (Sambrook et al., 1989) or using the Wizard Plus Mini Prep Kit (Promega). Plasmid DNA was digested with EcoRI restriction endonuclease (Promega) and was analyzed by gel electrophoresis (Sambrook et al., 1989). DNA sequencing of cloned PCR products was performed at the University of Tennessee Molecular Biology Resource Facility using an ABI 373 DNA sequencer (Perkin-Elmer, Foster City, CA) with the ABI Prism Dye Terminator Cycle Sequencing reaction kit. In all cases, both strands of at least two independent clones were sequenced.
Isolation of a Full-Length cDNA Encoding Mtapy1
Extension of the cDNA ends of the 850-bp Mtapy1 product was performed using the 5′- and 3′-RACE kit (Gibco-BRL, Gaithersburg, MD), according to the manufacturer's protocol. For extension of the 3′ end, the primer 3′apy1 GSP1 was employed: 3′-RACEGSP1, 5′-TGCTCGTTGATGGATTTGGC-3′. For extension of the 5′ end, the following primers were employed: 5′apy1GSP1, 5′-ACCTTCATAATCTCAGCTCG-3′; and 5′apy1GSP2, 5′-CTCCAAGATCCATTACTCCC-3′. Cloning and sequencing of the PCR generated products was performed as described above.
After sequence analysis of the 5′- and 3′-RACE products, primers were designed to amplify the predicted coding region from the start (first in frame ATG) to the stop codon. Primers used for amplification of the full-length cDNA were 5′-Mt46fullORFE and 3′-Mt46fullORFE, respectively. The primers were used in an RT-PCR reaction as described above using 0.2 μm of each primer and 2 units of Pfu DNA polymerase (Stratagene, La Jolla, CA). The reaction was carried out as described above, except a single annealing temperature of 52°C was used. Cloning and sequencing were performed as described above.
Cloning of Mtapy2
To compare the full sequence of Mtapy1 with known sequences in the database, a BLAST search was performed (Altschul et al., 1997). When compared with the EST database dbEST (Pearson et al., 1997), it was discovered that a very similar gene had been described from the roots of M. truncatula (Covitz et al., 1998; accession no. AA660474). The available sequence was used to generate primers for use in an RT-PCR reaction. The following primers were used: 5′-Mt46–2, 5′- GGGGCAACTGCAGGTTTAAGGGC-3′; and 3′-Mt46–2, 5′-GAGCCAGATAAGATACACGGG-3′. The product of PCR was a 450-bp fragment that showed 85% sequence similarity to Mtapy1, and was designated Mtapy2.
BAC Clone Isolation and Manipulation
BAC clones of M. truncatula genotype A17 were identified by means of hybridization to high-density filter arrays obtained from the Clemson University Genomics Institute (http://www.genome.clemson.edu), or by PCR screening of a multiplexed DNA copy of the BAC library as described by Nam et al. (1999). BAC end sequencing was performed on whole BAC clones using the ABI PRISM BigDye Primer Cycle Sequencing reaction kit and oligonucleotide primers complementary to the pBeloBAC11 vector (“left primer,” AACGCCAGGGTTTTCCCAGTCACGACG; “right primer,” ACACAGGAAACAGCTATGACCATGATTACG). Internal sequencing of selected regions of BAC clones was performed by a primer walking strategy on whole BAC clones or on DNA fragments subcloned into pBluescript (Stratagene). The BAC contig shown in Figure 3 was extended by multiplex PCR using primers designed to amplify sequences corresponding to the outermost BAC ends.
Genetic markers were developed based on BAC end sequence information obtained from BAC clones with homology to characterized M. truncatula apyrase cDNA products or to the soybean RFLP clone B046. In brief, oligonucleotide primers (Table II) were designed based on the corresponding BAC end sequence information and were used to PCR amplify and sequence genomic DNA from M. truncatula genotypes A17 and A20. Comparing differences in restriction enzyme patterns between the parental genotypes identified restriction enzyme polymorphisms. The resulting CAPS markers were mapped on a population of 93 F2 progeny from a genotype A17 × A20 cross (Penmetsa and Cook, 2000). Polymorphic DNAs were resolved on a 1.5% (w/v) agarose gel and were visualized by ethidium bromide staining. Primers, PCR conditions, and restriction enzyme information is given in Table II. DNA was extracted from the mapping population by means of the Nucleon Phytopure kit (Amersham Life Sciences, Buckinghamshire, UK), according to manufacturer's instructions.
Soybean BAC Library Screening with GS50 and GS52
An 8× redundant soybean BAC library was spotted onto high-density nylon membranes at the Clemson University Genomics Center (Clemson, SC). 32P-labeled probes of GS50 and GS52 were generated using the Random Primers DNA Labeling System (Gibco-BRL) and the BAC filters were hybridized as described in Danesh et al. (1998).
Placing BAC Contigs onto the Soybean Genetic Map
GS50 and GS52 hybridizations potentially revealed more than one location in the soybean genome. For this reason positive BACs were sorted into contigs and those corresponding to previously mapped apyrase loci were determined. BAC DNA was purified using Polyfiltronic 96-well Uni-Filter 800s (Whatman, Clifton, NJ), as recommended by the manufacturer. All clones that were positive with GS50 or GS52 along with genomic DNA from the parents of an recombinant inbred mapping population described previously (Concibido et al., 1996) were digested with the restriction enzyme originally used to map GS50 (Day et al., 2000). Southern blots were then hybridized with radiolabeled GS50 and GS52 to visualize corresponding restriction fragments. BACs were grouped into contigs based on similar banding patterns and those BACs corresponding to the originally mapped locus were identified by comparing the digestion patterns.
Phylogenetic Analysis of M. truncatula Apyrase Genes
For the tree generated in Figure 5, protein sequences were aligned using CLUSTALW (Thompson et al., 1994) under default parameters. End gaps were trimmed, leaving a 214-amino acid multiple sequence alignment for phylogenetic inference. Translations of EST sequences were generated for TFASTX3 (Pearson et al., 1997; alignments with Db-LNP, AF139807).
LNP protein sequences and rbcL DNA sequences were aligned using CLUSTALW (Thompson et al., 1994), running under default parameters. For both alignments, end gaps were trimmed. The final alignment for the LNP protein sequences contained 188 amino acid positions; the final alignment for the rbcL sequences contained 1,310 nucleotide positions. DNA and protein distances were inferred using the DNADIST and PROTDIST programs of the PHYLIP package (version 3.5c, J. Felsenstein, Department of Genetics, University of Washington, Seattle), respectively. Bootstrap and neighbor-joining analysis were performed using the SEQBOOT and NEIGHBOR programs, also from the PHYLIP package. Both trees in Figure 6 were rooted by the out-group Arabidopsis sequences.
Northern-Blot Analysis
Total root RNA was isolated from roots at various times between 0 and 6 h after inoculation with S. meliloti strain ABS7. Total RNA was electrophoresed on denaturing 1% (w/v) agarose formaldehyde gels and blotted as described by Sambrook et al. (1989). Each lane was loaded with RNA equivalent to 0.2 g fresh weight of root tissue. The blot was probed with a 32P-labeled full-length Mtapy1 cDNA probe, stripped, and then reprobed with Histone H3 from M. truncatula. In all cases probes were radiolabeled with 32P using random primer labeling (Promega). Hybridizations were performed at 60°C followed by successive washings as previously described (Day et al., 2000).
RT-PCR Analysis of Gene Expression
Total RNA was collected from 5-d-old roots, hypocotyls, and cotyledons, from 6-week-old stems, leaves, and flowers, and 10-d-old nodules using the guanidine-HCl method described above. Total root RNA was also isolated from M. truncatula A17 plants inoculated with S. meliloti ABS7M at 0, 3, 6, 12, 24, and 48 h postinoculation.
Due to the high degree of sequence similarity between all of the putative apyrases from M. truncatula, a PCR-based approach was used to distinguish the expression of the different genes. Gene-specific primers were designed from the available sequence of all of the genes from regions of sequence divergence and from the 3′-untranslated regions. The primers used in these reactions were as follows: 5′Mt46fullORFE, 5′-GGAATTCATGGTCTTACTTTGGCAAAACACC-3′; 3′Mt46fullORFE, 5′-GGCTTAAGTTAAACAAAATACATCATTCG-3′; Mtapy1 GS forward, 5′-CTGGGGCTAATTTTAATGGATGC-3′; Mtapy1 GS reverse, 5′-GTGGTACCCTCAATAGAAAAAACATGTCGG-3′; Mtapy2 GS forward, 5′-GATGCTGATGCGGTTACAGTGTTG-3′; Mapy2 GS reverse, 5′-CCATGGCAGCCTCAGACTCA-3′; Mtapy3 GS forward, 5′-GGTCACACATAATTCTCCCAACCC-3′; Mtapy3 GS reverse, 5′-GCAGCGTTAAACTTGAGCG-3′; Mtapy4 GS forward, 5′-GTTCAGTAACAGAAGTACCCTCAACG-3′; and Mtapy4 GS reverse, 5′-CACCTACTGTAATCTCTTTTCGTGGAC-3′.
These primers were first used in PCR reactions against reverse transcribed total RNA isolated from pooled M. truncatula line A17 tissues. The PCR products generated were cloned, sequenced to confirm gene identity, and subsequently used as hybridization probes for Southern analysis. RT-PCR reactions were performed as previously described (Day et al., 2000). In all of the reactions, primers designed to amplify actin were included as internal controls in each tube. The sequence of the actin primers were as follows: 5′Mt ACTIN, 5′-GCAGATGCTGAGGATATTAACCCC-3′; 3′ Mt ACTIN, 5′-CGACCACTTGCATAGAGGGAGAGG-3′.
The RT-PCR products were separated by gel electrophoresis, blotted, and hybridized with the appropriate 32P-labeled cDNA probe (Sambrook et al., 1989). Blots were first probed with the respective apyrase genes, stripped, and reprobed with actin as an internal control. All blots were counted for total radioactivity using an Instant Imager (Packard Instrument Co., Meriden, CT). The total counts of apyrase probe hybridization were divided by total counts of actin probe hybridization to yield a ratio of the relative intensity of hybridization of apyrase. The value obtained was multiplied by 100 to yield a value referred to as relative expression.
Screening of Nodulation-Deficient Mutant Lines of M. truncatula
Penmetsa and Cook (2000) described an efficient EMS mutagenesis of M. truncatula. Based on screening of the resulting progeny numerous nodulation mutants were identified, including several lines affected for symbiotic development (Penmetsa and Cook, 1997; Catoira et al., 2000; R.V. Penmetsa and D.R. Cook, unpublished data). Three genetically separable mutants with distinct non-nodulation phenotypes (Table III) were used for analysis of apyrase gene expression by RT-PCR as described above. The wild-type M. truncatula A17 and nodulation defective lines pdl, dmi1, and lin were grown aeroponically and inoculated with wild-type S. meliloti strain ABS7. At each time point, a 5-cm section of root was sampled surrounding the zone of nodulation. RNA isolation, northern blotting, and hybridizations were performed as described previously.
ACKNOWLEDGMENTS
The authors would like to thank Marilynn Etzler for providing the sequence of the D. biflorus apyrase prior to publication. We would also like to thank Kate VandenBosch for kindly providing us with the EST clone pVK0–1 M6.
Footnotes
This work was supported by the U.S. Department of Energy (grant no. DE–FG02–97ER20260 to G.S.) and by the National Science Foundation (grant no. 9872664 to D.R.C. and N.D.Y.).
LITERATURE CITED
- Abeijon D, Yanagisawa K, Mandon EC, Hausler A, Moreman K, Hirshchberg DB, Robbins PW. Guanosine diphosphatase is required for protein and sphingolipid glycosylation in the Golgi lumen of Saccraromyces cerevisiae. J Biol Chem. 1993;122:307–323. doi: 10.1083/jcb.122.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D, Bianchi S, Blondon F, Dattee Y, Duc G, Essad S, Flament T, Gallusci T, Genier G, Guy P. Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol Biol Rep. 1990;8:40–49. [Google Scholar]
- Catoira R, Galera C, de Billy F, Journet EP, Maillet F, Penmetsa V, Rosenberg C, Gough C, Cook D, Dénarié J. Identification of four genes of Medicago truncatula controlling steps in Nod factor transduction. Plant Cell. 2000;12:1647–1665. doi: 10.1105/tpc.12.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-R, Datta N, Roux SJ. Purification and partial characterization of a calmodulin-stimulated nucleoside triphosphatase from pea nuclei. J Biol Chem. 1987;262:10689–10694. [PubMed] [Google Scholar]
- Chen Y-R, Roux SJ. Characterization of nucleoside triphosphatase activity in isolated pea nuclei and its photoreversible regulation by light. Plant Physiol. 1986;81:609–613. doi: 10.1104/pp.81.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn J, Day RB, Stacey G. Legume nodule organogenesis. Trends Plant Sci. 1998;3:105–110. [Google Scholar]
- Concibido VC, Young ND, Lange D, Denny RL, Danesh D, Orf JH. Targeted comparative genome analysis and qualitative mapping of a major partial-resistance gene to soybean cyst nematode. Theor Appl Genet. 1996;93:234–241. doi: 10.1007/BF00225751. [DOI] [PubMed] [Google Scholar]
- Cook D. Medicago truncatula: a model in the making! Curr Opin Plant Biol. 1999;2:301–304. doi: 10.1016/s1369-5266(99)80053-3. [DOI] [PubMed] [Google Scholar]
- Cook D, Dreyer D, Bonnet D, Howell M, Nony E, VandenBosch K. Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell. 1995;7:43–55. doi: 10.1105/tpc.7.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covitz PA, Smith LS, Long SR. Expressed sequence tags from a root-hair-enriched Medicago truncatula cDNA library. Plant Physiol. 1998;117:1325–1332. doi: 10.1104/pp.117.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi MD, Jurkevitch E, Poiret M, d'Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A. enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994;13:5099–5112. doi: 10.1002/j.1460-2075.1994.tb06839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csanádi G, Szécsi J, Kalo P, Kiss P, Endre G, Kondorosi A, Kondorosi E, Kiss GB. ENOD12, an early nodulin gene is not required for nodule formation and efficient nitrogen fixation in alfalfa. Plant Cell. 1994;6:201–213. doi: 10.1105/tpc.6.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh D, Pe Fluela S, Mudge J, Denny RL, Nordstrom H, Martinez JP, Young ND. A bacterial artificial chromosome library for soybean and identification of clones near a major cyst nematode resistance gene. Theor Appl Genet. 1998;96:196–206. [Google Scholar]
- Day RB, McAlvin CB, Loh JT, Denny RL, Wood TC, Young ND, Stacey G. Differential expression of two soybean apyrases: one of which is an early nodulin. Mol Plant-Microbe Interact. 2000;13:1053–1070. doi: 10.1094/MPMI.2000.13.10.1053. [DOI] [PubMed] [Google Scholar]
- Dénarié JF, Debellé F, Promé JC. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. Touchdown PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards FA, Gibb AJ. ATP: a fast neurotransmitter. FEBS Lett. 1993;325:86–89. doi: 10.1016/0014-5793(93)81419-z. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Eriksson A, Nister M, Leveen P, Westermark B, Heldin CH, Claesson-Welsh L. Induction of platelet-derived growth factor α- and β-receptor mRNA and protein by platelet-derived growth factor BB. J Biol Chem. 1991;266:21138–21144. [PubMed] [Google Scholar]
- Etzler ME, Kalsi G, Eqing NN, Roberts NJ, Day RB, Murphy JB. A nod factor binding lectin with apyrase activity from legume roots. Proc Natl Acad Sci USA. 1999;96:5856–5861. doi: 10.1073/pnas.96.10.5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman MA, Duch MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallusci P, Dedieu A, Journet EP, Huguet T, Barker DG. Synchronous expression of leghaemoglobin genes in Medicago truncatula during nitrogen-fixing root nodule development and response to exogenously supplied nitrate. Plant Mol Biol. 1991;17:335–349. doi: 10.1007/BF00040629. [DOI] [PubMed] [Google Scholar]
- Gao X-D, Kaigorodov V, Jigami Y. YND1, a homologue of GDA1, encodes membrane-bound apyrase required for Golgi N- and O-glycosylation in Saccharomyces cerevisiae. J Biol Chem. 1999;74:21450–21456. doi: 10.1074/jbc.274.30.21450. [DOI] [PubMed] [Google Scholar]
- Handa M, Guidotti G. Purification and cloning of a soluble ATP-diphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum) Biochem Biophys Res Commun. 1996;218:916–923. doi: 10.1006/bbrc.1996.0162. [DOI] [PubMed] [Google Scholar]
- Helmrath MA, Shin CE, Erwin CR, Warner BW. Epidermal growth factor up-regulates the expression of its own intestinal receptor after small bowel resection. J Pediatr Surg. 1998;33:229–234. doi: 10.1016/s0022-3468(98)90437-7. [DOI] [PubMed] [Google Scholar]
- Horvath B, Heidstra R, Lados M, Moerman M, Spaink HP, Promé JC, Van Kammen A, Bisseling T. Lipooligosaccharides of Rhizobium induce infection-related early nodulin gene expression in pea root hairs. Plant J. 1993;4:727–733. doi: 10.1046/j.1365-313x.1993.04040727.x. [DOI] [PubMed] [Google Scholar]
- Hseih HS, Tong DG, Thomas C, Roux SJ. Light-modulated abundance of an mRNA encoding a calmodulin-regulated, chromatin-associated NTPases in pea. Plant Mol Biol. 1996;30:135–147. doi: 10.1007/BF00017808. [DOI] [PubMed] [Google Scholar]
- Journet EP, Pichon M, Dedieu A, de Billy F, Truchet G, Barker DG. Rhizobium maliloti Nod factors elicit cell-specific transcription of the ENOD12 gene in transgenic Alfalfa. Plant J. 1994;6:241–249. doi: 10.1046/j.1365-313x.1994.6020241.x. [DOI] [PubMed] [Google Scholar]
- Kisselgof AB, Oettgen HC. The expression of murine B cell CD23, in vivo, is regulated by its ligand, IgE. Int Immunol. 1998;10:1377–1384. doi: 10.1093/intimm/10.9.1377. [DOI] [PubMed] [Google Scholar]
- Komoszynski M, Wojtczak A. Apyrases (ATP diphosphohydrolases, EC 3.6.1.5): function and relationship to ATPases. Biochem Biophys Acta. 1996;1310:233–241. doi: 10.1016/0167-4889(95)00135-2. [DOI] [PubMed] [Google Scholar]
- Lullien V, Barker DG, de Lajudie P, Huguet T. Plant gene expression in effective and ineffective root nodules of alfalfa (Medicago sativa) Plant Mol Biol. 1987;9:43–48. doi: 10.1007/BF00015878. [DOI] [PubMed] [Google Scholar]
- Long SR. Rhizobium symbiosis: Nod factors in perspective. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus AJ, Safier LB. Thromboregulation: multicellular modulation of platelet reactivity in hemostasis and thrombosis. FASEB J. 1993;7:516–522. doi: 10.1096/fasebj.7.6.8472890. [DOI] [PubMed] [Google Scholar]
- Minami E, Kouchi H, Cohn JR, Ogawa T, Stacey G. Expression of the early nodulin, ENOD40, in soybean roots in response to various lipo-chitin signal molecules. Plant J. 1996;10:23–32. doi: 10.1046/j.1365-313x.1996.10010023.x. [DOI] [PubMed] [Google Scholar]
- Nam Y-W, Penmetsa RV, Endre G, Kim D, Cook DR. Construction of a bacterial artificial chromosome library of Medicago truncatula and identification of clones containing ethylene response genes. Theor Appl Genet. 1999;98:638–646. [Google Scholar]
- Ng GY, Varghese G, Chung HT, Trogadis J, Seman P, O'Dowd BF, George SR. Resistance of the dopamine D2L receptor to desensitization accompanies the up-regulation of receptors on to the surface of Sf9 cells. Endocrinology. 1997;138:4199–4206. doi: 10.1210/endo.138.10.5433. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Wood TC, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24–36. doi: 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Cook D. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science. 1997;275:527–530. doi: 10.1126/science.275.5299.527. [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR. Production and characterization of diverse developmental mutants of Medicago truncatula. Plant Physiol. 2000;123:1387–1398. doi: 10.1104/pp.123.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera SC, Ladd TR, Dhadialla TS, Kress PJ, Sohi SS, Retnakaran A, Palli SR. Studies on two ecdysome receptor isoforms of the spruce budworm, Choristoneura fumiferana. Mol Cell Endocrinol. 1999;152:73–84. doi: 10.1016/s0303-7207(99)00058-1. [DOI] [PubMed] [Google Scholar]
- Pichon M, Journet EP, Dedieu A, de-Billy F, Truchet G, Barker DG. Rhizobium meliloti elicits transient expression of the early nodulin gene ENOD12 in the differentiating root epidermis of transgenic alfalfa. Plant Cell. 1992;4:1199–1211. doi: 10.1105/tpc.4.10.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu M. Characterization of Medicago truncatula mutants defective in infection persistence and defense response during Rhizobium-legume symbiosis. MS thesis. College Station: Texas A&M University; 1998. [Google Scholar]
- Roberts NJ, Brigham J, Wu B, Murphy JB, Volpin H, Philips DA, Etxler ME. A Nod factor-binding lectin is a member of a distinct class of apyrases that may be unique to the legumes. Mol Gen Genet. 1999;262:261–267. doi: 10.1007/s004380051082. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis TA. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sarkis JJ, Salto C. Characterization of a syntaptosomal ATP diphosphohydrolases from the electric organ of Torpedo marmorata. Brain Res Bull. 1991;26:871–876. doi: 10.1016/0361-9230(91)90251-e. [DOI] [PubMed] [Google Scholar]
- Somasegaran P, Hoben JH. Handbook for Rhizobia. New York: Springer-Verlag; 1994. [Google Scholar]
- Thomas C, Sun Y, Naus K, Lloyd A, Roux SJ. Apyrase functions in plant phosphate nutrition and mobilizes phosphate from extracellular ATP. Plant Physiol. 1999;119:543–551. doi: 10.1104/pp.119.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W:improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu AT, Burleigh SH, Kardailsky IV, Maldonado-Mendoza IE, Versaw WK, Blaylock LA, Shin H, Chiou T-J, Katagi H, Dewbre GR. Transformation of Medicago truncatula via infiltration of seedlings or flowering plants with Agrobacterium. Plant J. 2000;22:531–542. doi: 10.1046/j.1365-313x.2000.00757.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleave sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]