Abstract
On November 24, 2021, the SARS-CoV-2 Omicron variant (B.1.1.529) was first identified in South Africa. The World Health Organization (WHO) declared the Omicron as a variant of concern (VoC) because of the unexpected and large numbers of mutations occurred in the genome, higher viral transmission and immune evasions. The present study was performed to explore the interactions of SARS-CoV-2 spike glycoprotein receptor-binding domain (SGp RBD) of the three variants (Omicron, Delta, and WT) with the receptor hACE2. The structural changes occurred in Omicron due to the mutations at key positions improved the ability to mediate SARS-CoV-2 viral infection compared to other VoCs. The phytochemicals limonin and glycyrrhizic acid were docked with the SGp RBD of the variants WT, Delta and Omicron. The computed dock score revealed that limonin and glycyrrhizic acid binds effectively at the SGp RBD of all three variants, and showed almost similar binding affinity at the binding interface of ACE2. Therefore, despite the multiple mutations occurred in Omicron and its viral transmission is comparatively high, the computed binding affinity of the phytochemicals limonin and glycyrrhizic acid supported that the traditional medicines can be useful in formulating adjuvant therapies to fight against the SARS-CoV-2 Omicron.
Keywords: SARS-CoV-2 Omicron, Molecular docking, Dynamics simulations, Limonin, Glycyrrhizic acid
Graphical abstract
1. Introduction
SARS-CoV-2 is the viral strain behind the ongoing global pandemic and periodically the concern on the pandemic is enhancing due to the large number of unexpected mutations occurring in its genome. The recently (November 24, 2021) evolved SARS-CoV-2 Omicron variant (B.1.1.529) in South Africa was declared as a variant of concern (VoC) by the World Health Organization (WHO) [1]. Out of the five VoCs (alpha, beta, gamma, Delta and Omicron), the other highly transmitted and antibody evasion variant is Delta: B.1.617.2 that played an enormous role in the global pandemic [2,3]. The different variants of this respiratory virus SARS-CoV-2 affects mainly the lungs, but also capable to infect other vital organs, such as heart, kidney and brain [4]. Virus invasion requires receptor binding of the host system. SARS-CoV-2 recognises the human host receptor angiotensin converting enzyme 2 (ACE2). The C-terminal domain (CTD), also known as the receptor-binding domain (RBD) of the SARS-CoV-2 spike glycoprotein (SGp) interacted first with the human receptor ACE2 [5,6]. Among the various SARS-CoV-2 variants, the large number of mutations occurred in the Omicron RBD is expected to favour the interaction with ACE2 that resulted immune evasions and the higher viral transmission [7].
The interactions of ACE2 acidic amino acids GLU and ASP was enhanced by the mutated SGp RBD amino acids ARG and LYS by ion-ion interaction [8]. The surface electrostatic interactions study revealed that the binding interface changes to a highly positive patch in Omicron RBD, which favours ACE2 binding due to the presence of negatively charged GLU and ASP residues [9]. It is also important to mention here that there are more than thirty-four mutations occurred in the Omicron SGp and distributed across all domains of the trimer protein compared to wild-type (WT) Wuhan virus B.1.1.7 variant [10]. Omicron RBD function is impaired by TYR505 mutation resulting in the increase in the risk of disease establishment [11]. Omicron may be ten times more infectious than the original SARS-CoV-2, and twice as infectious as the Delta variant due to its RBD mutations ASN440LYS, THR478LYS, and ASN501TYR. Based on 185 antibody RBD complex 3D structures, it was found that Omicron might be able to evade current vaccines by 88% [12,13]. Therefore, there is still bourgeoning interest on anti-SARS-CoV-2 research to search suitable drugs and phytochemicals from special databases by employing the computational approaches, like protein-protein docking, protein-ligand docking and molecular dynamics simulations [[14], [15], [16], [17], [18], [19], [20]].
Based on the recent reports on SARS-CoV-2 Omicron and our interest to search potential lead compounds from phytochemicals for formulating adjuvant therapies to combat the ongoing viral pandemic, we performed in silico computational studies like sequence alignment and molecular docking to examine the interactions of the SARS-CoV-2 SGp RBD of the variants WT, Delta and Omicron with the human host receptor ACE2. Then, the molecular docking of two important phytochemicals limonin and glycyrrhizic acid was performed with the SGp RBD of the variants WT, Delta and Omicron. The literatures supported the importance of phytochemicals in formulating adjuvant therapeutic approaches to fight against SARS-CoV-2 [21,22]. The therapeutic nature of the limonoids and triterpenoids based phytochemicals like limonin, glycyrrhizic acid, maslinic acid, obacunone,7-deacetyl-7-benzoylgedunin, corosolic acid and ursolic acid was well-known against HIV-1, SARS-CoV, cancers, and other viral and fungal infections [[23], [24], [25], [26]]. Also, our recent results supported the role of limonoids and triterpenoids against therapeutic protein targets of SARS-CoV-2, such as spike glycoprotein, RdRp, PLpro, 3CLpro, NSP13, NSP14 and NSP15 as well as host receptors ACE2, furin and TMPRSS2 involved in the host-pathogen interactions [27,28]. The dock score and mode of interactions in the limonin-RBD and glycyrrhizic acid-RBD complexes were discussed. The limonin and glycyrrhizic acid showed almost equal binding affinity at the RBD of the variants WT, Delta and Omicron, which supported that the reported research on WT/Delta variants for formulating traditional medicines as adjuvant therapies may also be useful to combat Omicron infection and transmission.
2. Results and discussion
2.1. Mutations in SGp RBD of Omicron
The pairwise sequence alignment of the SGp RBD of the Omicron was first performed with the variants WT and Delta to predict the structural and functional similarity. The FASTA sequences of SGp RBD of the variants WT, Delta and Omicron were retrieved from NCBI database (https://www.ncbi.nlm.nih.gov/protein) of GI: 2043688783, GI: 2106681814 and GI: 2171220934 (Fig. S1). These sequences were compared by global sequence alignment (www.ebi.ac.uk/Tools/psa/) “for two sequences” using Needleman-Wunsch algorithm [29]. The two amino sequences of 201 residues of SGp RBD of Omicron and WT aligned for pairwise sequence alignment scoring gap penalty of 10.0 with similarity (180/201) and identity (175/201) is 89.6% and 87.1%, respectively (Fig. 1 ) [30]. The gap score of the two sequences alignment is 957.0. However, the two amino sequences of 201 residues of SGp RBD of Omicron and Delta aligned for pairwise sequence alignment scoring gap penalty 10.0 with similarity (190/201) and identity (186/201) is 94.5% and 92.5%. respectively (Fig. 2 ). The gap score of the two sequences alignment is 1014.0. The pairwise sequence alignment study clearly supported the multiple mutations occurred in the SGp RBD of Omicron in compared to the SGp RBD of Delta and WT variants.
Fig. 1.
Pairwise sequence alignment of SGp RBD of Omicron and WT variants.
Fig. 2.
Pairwise sequence alignment of SGp RBD of Omicron and Delta variants.
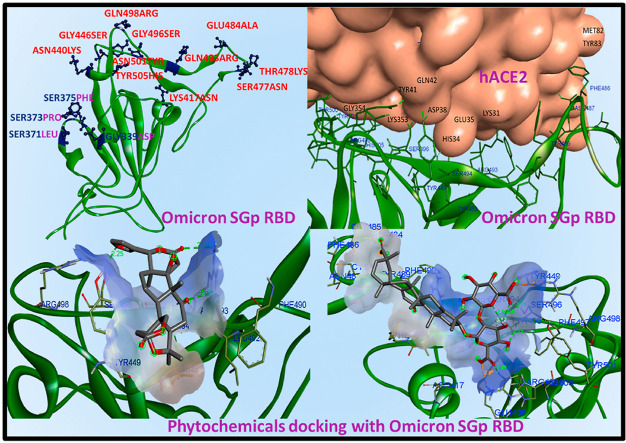
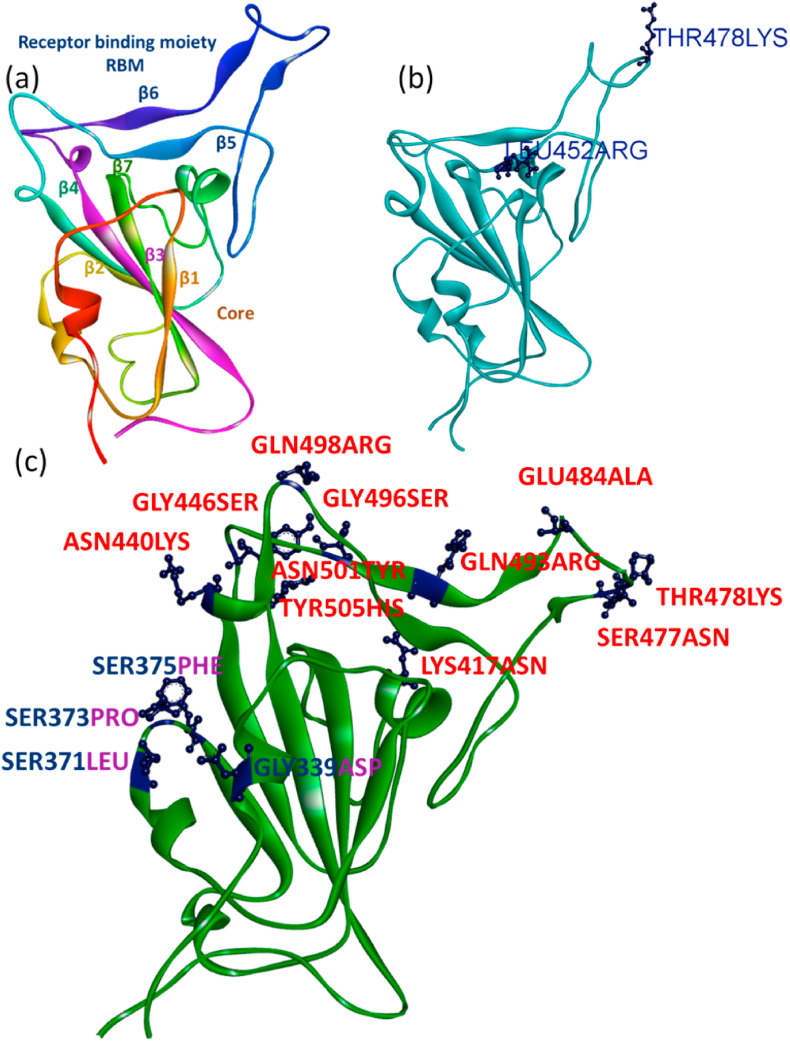
The 3D ribbon structures of SGp RBD of Omicron, Delta and WT was drawn to compare the structural changes occurred due to the mutations (Fig. 3 ). There are thirty-four mutations occurred in the SGp Omicron in which fifteen mutations occurred only at the RBD. In compared to WT (Fig. 3a), the SGp RBD of Delta was reported to be mutated with amino acid residues as LEU452ARG and THR478LYS (Fig. 3b). Surprisingly, the Omicron SGp RBD showed significant conformational and surface modifications compared to WT ribbon structure due to the multiple mutations occurred with the residues, such as THR478LYS, GLU484ALA, GLN493ARG, GLY496SER, GLN498ARG, SER477ASN, GLY446SER, LYS417ASN, TYR505HIS, ASN501TYR, ASN440LYS, GLY339ASP, SER373PHE, SER375PHE, SER371LEU [31,32] (Fig. 3c), where the residue THR478LYS was commonly mutated in both Omicron and Delta RBD. The residues LYS417ASN, ASN440LYS, GLY446SER, SER477ASN, THR478LYS, GLU484ALA, GLN493ARG, GLY496SER, GLN498ARG, ASN501TYR and TYR505HIS are mutated in the β5 and β6 sheets of WT, whereas the residues SER371LEU, SER373PRO and SER375PHE are mutated in the WT-β2 sheet. As the SGp RBD initiated the host pathogen interaction, the multiple mutations at the SGp RBD Omicron is expected to show better affinity towards the human host receptor ACE2 than the WT and Delta variants due to the variations occurred in the hydrophobic and/or electrostatic surfaces [33].
Fig. 3.
Ribbon structures of SGp RBD of Omicron, Delta and WT: (a) Alfa RBD structure represented by the beta sheets and labelled with respective colour of the ribbon, (b) Delta SGp cyan ribbon structure with labelled mutations and (c) Omicron SGp RBD green ribbon structure with labelled mutations (residues labelled in red are present at the binding interface to ACE2).
2.2. Protein-protein interactions
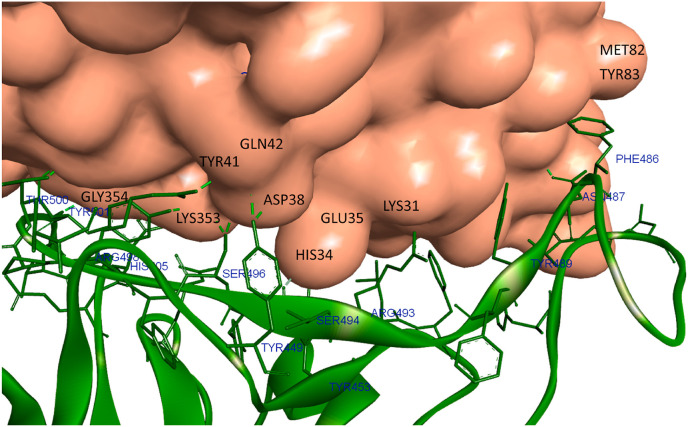
The protein-protein complexes between SGp-RBD of different variants (Omicron, Delta and WT) with hACE2 receptor were analysed to understand the key residues involved during the pathogen-host interaction. The viral infection is so effective to human because of the high binding affinity of the ACE2 receptor towards SARS-CoV-2 SGp. At the binding interface of SGp RBD-ACE2, the residues 19–42, 82–83, 330 and 352–357 of ACE2 make close contacts with the residues present in the receptor-binding motif (RBM) of SGp-RBD with residues from 438 to 506. During the infection, the WT SGp RBD mediating with ACE2 by various residual contacts [34]. For example, the residues THR500, GLY502, TYR505, GLY496, TYR449, GLN493, ASN487, ARG417 were formed conventional hydrogen bonds respectively with ASN330, ASP355, LYS353, GLU37, LYS353, GLN42, GLN24, TYR83, ASP30. The WT charge attractions was mediated by ARG403 with GLU37. The Pi-sigma, Pi-alkyl and Pi-Pi interactions were observed between the LEU455-HIS34, PHE486-MET82 and TYR505-GLY354, respectively (Fig. S2). The key residues involved in the mediation of Omicron RBD and ACE2 interaction are PHE488 forming Pi-sulphur with MET82, and the residues ASN487, TYR453, TYR449, TYR501, THR500 and GLY502 are forming conventional hydrogen bonds with TYR83, HIS34, ASP38, GLN42, LYS353, TYR41 and GLY354 (Fig. 4 ) [7]. In Delta RBD-ACE2 complex (Fig. S3), the residues PHE486, TYR453, GLY496, SER494 of Delta variant interacted with MET82, HIS34, ASP38, HIS34 of ACE2 by carbon bond. In addition, the residues TYR449, THR500, ASN501 formed hydrogen bonds with ASP38, TYR41. Some of the Delta variant residues, i.e., LYS417, PHE486, TYR489 and TYR505 mediated by charged and Pi-alkyl interactions with ASP80, TYR83, LYS31 and LYS353. The close analysis of the crystal structure of the SGp-RBD of different variants (Omicron, Delta and WT) with hACE2 receptor revealed that the key interactions present in WT SGp-RBD-ACE2 complex was also observed with the Delta and Omicron variants and the mode of binding conformation is almost similar, however, the mutations occurred at the RBM greatly affects the nature of bonding at the interface of Omicron SGp RBD-ACE2. The mutations greatly enhanced the electrostatic attraction at the binding interface of Omicron SGp RBD-ACE2 due to the enhancement of positive charge region in RBD. The mutated residues GLN498ARG, GLN493ARG, THR478LYS and GLU484ALA creates a region of positive patch in the Omicron RBD, which expected to favour the binding with ACE2 because of the presence of negatively charged residues GLU and ASP.
Fig. 4.
Omicron SGp-RBD binding with human ACE2 receptor.
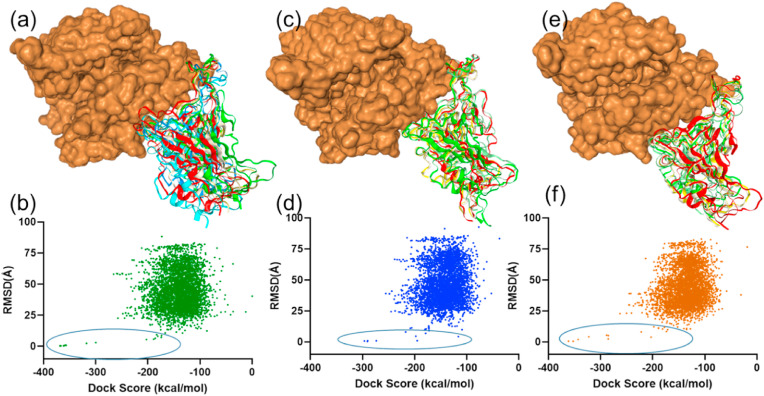
In order to get fuller insight on the SGp RBD-ACE2 complex formation, the protein-protein docking experiments were performed in HDOCK server [35] and the binding affinity of the SGp RBD of the three variants (Omicron, Delta and WT) with ACE2 was compared. The server computed maximum 4329 binding conformations, and their dock scores are summarized in Table S1. Also, the dock scores of the 4329 binding conformations with least RMSD are shown in Fig. 5 along with the best dock scores are highlighted. In addition, the pose between the SGp RBD of the variants (Omicron, Delta and WT) with ACE2 for the best dock score are also shown in Fig. 5. The Omicron SGp RBD showed best docking score of −368.44 kcal/mol with an average RMSD 0.35 Å. The presence of multiple mutated residues on the surface of the RBD enhanced the receptor binding affinity with the human receptor ACE2. The Delta SGp RBD showed best dock score of −296.34 kcal/mol with an interaction RMSD of 0.39 Å, whereas the WT SGp RBD showed the best dock score of −361.13 kcal/mol with ligand RMSD 0.56 Å. The β-sheet receptor binding motif (RBM) residues of SGp RBD interact with the ACE2 α-helices residues extended from SER19 to TYR83 during the host-pathogen mediation. The different interaction pairs observed in the best pose of the SGp RBD (Omicron, Delta and WT)-ACE2 complexes with bond distance are tabulated in Table S2. The protein-protein docking studies revealed that the RBM of Omicron bind with ACE2 receptor more efficiently than the WT and Delta variants. The mutation enhanced the hydrophobic surface of the Omicron RBM in comparison to WT and Delta variants. In addition, the increase in nonpolar amino acids in Omicron RBD favoured the interaction with ACE2 containing polar amino acids like ASP and GLU. In addition, the mutations like LYS417ASN, ASN440LYS, GLN493ARG, and GLN498ARG in Omicron RBD is expected to reduce the affinity of vaccine due to change in the electrostatic influences on the protein surface [13]. Delta variant showed comparatively lower binding affinity, and also its mutated residues LEU452ARG and THR487LYS do not participate in the bond formation with ACE2. However, the mutation induced conformational changes in ACE2 enhanced the electrostatic interactions by forming salt-bridge with SARS-CoV-2 SGp RBD [36]. Also, the TMPRSS2 play an important role in the viral fusion of Delta variant compared to the Omicron variant of SARS-CoV-2 is expected to make it a VoC [37].
Fig. 5.
Protein-protein docking and respective dock scores of 4329 conformations: (a) Omicron SGp RBD best pose with ACE2 and (b) the dock scores plot with average RMSD (Å) of 4329 poses of Omicron SGp RBD with ACE2; (c) Delta SGp RBD best pose with ACE2 and (d) the dock scores plot with average RMSD (Å) of 4329 poses of Delta SGp RBD with ACE2; (e) WT SGp RBD pose with ACE2 and (f) the dock scores plot with average RMSD (Å) of 4329 poses of WT SGp RBD with ACE2.
2.3. Molecular docking with phytochemicals
The in silico computational docking and simulations were performed extensively with large number of phytochemicals with the various protein targets of SARS-CoV-2 from the early 2020 to propose the lead compounds against COVID-19. Limonin found mainly in citrus fruits whereas glycyrrhizic acid in licorice root was predicted to be important lead compounds to fight against COVID-19 [23,27,28]. The molecular docking of limonin and glycyrrhizic acid was performed at the SGp RBD site of the variants WT, Omicron and Delta, followed by the computed binding conformation and dock score was compared. The dock score of pose with SGp RBD at the binding interface of ACE2 of the three variants with limonin and glycyrrhizic acid, and the residues interacting with the phytochemicals are summarized in Table 1 .
Table 1.
Limonin and glycyrrhizic acid interactions with SGp RBD of WT, Delta and Omicron variants at the binding interface of ACE2.
| Phytochemicals | Variants | Interaction with RBD at the binding interface of ACE2 | Binding energy, kcal/mol |
|---|---|---|---|
| Limonin | WT | Conventional hydrogen bond: GLN493, GLY496, Van der Waal: TYR505, HIS498, ASN501, TYR 495, TYR449, TYR 453, LEU455, ARG403, and ARG417. | −8.2 [13] |
| Delta | Conventional hydrogen bond: GLY496, ASN501, Pi Alkyl: TYR505, Carbon bond: TYR495, ARG403, Van de Waals: PHE497, GLN493, TYR453, SER494 and GLN498 | −8.3 | |
| Omicron | Conventional hydrogen bond: SER494, ARG493, SER496, Van der Waals: LEU452, LEU492, TYR449, and ARG498. | −8.3 | |
| Glycyrrhizic acid | WT | Conventional hydrogen bond: SER494, GLN493, LEU492 Van der Waal: TYR489, PHE456, LEU455, TYR449, GLU484, THR470, ILE472, GLY482, ASN481. Unfavorable Acceptor: PHE490, GLU471, Pi-Sigma: PHE490. | −8.5 |
| Delta | Conventional hydrogen bond: ASN501, GLY496, SER494, ARG452, Van de Waals: PHE497, GLN498, TYR505, ARG403, TYR495, TYR449, PHE490, GLN493. Attractive charge: ARG452. | −8.7 | |
| Omicron | Conventional hydrogen bond: SER496, Van der Waals: HIS505, TYR495, TYR453, ARG493, LEU455, TYR489, ALA484, GLY485, CYS488, PHE456. Carbon bond: SER494, Unfavorable bond: TYR449, Attractive charge: ARG498, ARG403. | −8.4 |
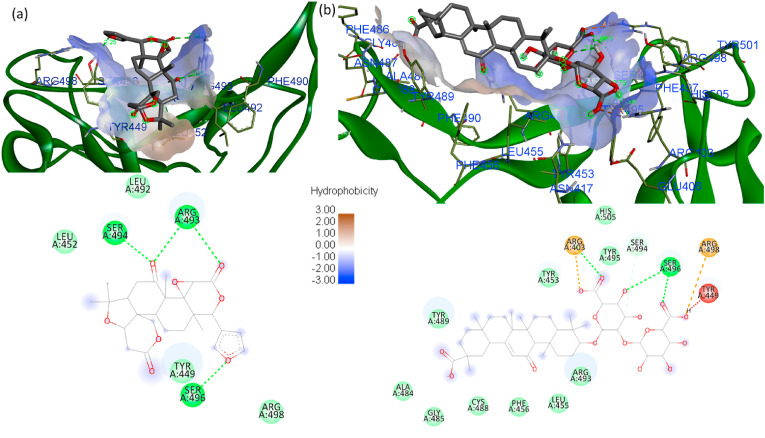
Limonin obeyed the ADMET limitations and drug-likeness, and showed a dock score of −8.2 kcal/mol with SGp RBD at the binding interface of ACE2 [23]. The binding of limonin with WT SGp RBD hydrophobic surface was shown in Fig. S4. It is interacting with the residues GLN493 and GLY496 with the conventional hydrogen bond. The ligand oxygen is forming a close interaction with the receptor H with closest bond distance of 2.20 Å. The surface of the receptor is exhibiting van der Waals interactions with TYR505, HIS498, ASN501, TYR495, TYR449, TYR453, LEU455, ARG403 and ARG417 residues. Similar to WT, limonin is also interacting with Delta RBD GLY496 residue. It is also forming conventional hydrogen bond with ASN501 (2.67 Å) and Pi-alkyl bond with TYR505 (Fig. S5), the key residues involved in the host receptor ACE2 binding. In addition, limonin is forming conventional carbon bond with TYR495 and ARG403 residues. In Omicron SGp RBD, limonin is effectively binding with the receptor-binding site forming four conventional hydrogen bonds with minimum binding energy of −8.3 kcal/mol and low bond distance with the residues SER494 (2.25 Å), ARG493 (2.94 Å, 2.88 Å) and SER496 (2.25 Å) (Fig. 6 a). In addition, limonin poses van der Waals interactions with the LEU452, LEU492, TYR449 and ARG498 residues. From the docking studies, it was observed that the limonin interacting at the SGp RBD of the variants Omicron, Delta and WT with almost similar binding affinity. Also, the limonin is interacting with the key residues at the SGp RBD that played essential role during the interaction with human receptor ACE2 for the establishment of SARS-COV-2 infection.
Fig. 6.
(a) Limonin and (b) glycyrrhizic acid pose at hydrophobic cavity of SGp RBD Omicron of SARS CoV-2 at the binding interface of ACE2.
The glycyrrhizic acid showed best dock score of −9.3, −8.9 and −8.7 kcal/mol at the SGp RBD site of the variants WT, Omicron and Delta. However, the pose of the best dock score was docked slightly away from the mutated region of RBM. Therefore, the next pose of glycyrrhizic acid was examined that docked at the mutated region of RBD and also complexed at the binding interface of ACE2. As tabulated in Table 1, the glycyrrhizic acid also showed nearly similar binding affinity like limonin at the SGp RBD of the three variants of WT, Delta and Omicron with dock score of −8.5, −8.7 and −8.4 kcal/mol, respectively. The binding of glycyrrhizic acid with WT and Delta SGp RBD hydrophobic surface was shown in Fig. S6 and Fig. S7, respectively. The glycyrrhizic acid is forming conventional hydrogen bonds with the residues SER494, GLN493 and LEU492. The ligand oxygen is forming a close contact with the receptor SER494 with the bond distance of 2.24 Å and 2.75 Å. The surface of the receptor is exhibiting van der Waals interactions with the residues TYR489, PHE456, LEU455, TYR449, GLU484, THR470, ILE472, GLY482 and ASN481. Similar to WT, the glycyrrhizic acid is also interacting with Delta RBD residues ASN501, GLY496, SER494 and ARG452 by conventional hydrogen bonds with bond distance of ∼2.87, 2.72, 2.83, 2.87 and 2.51 Å respectively and charge attraction with ARG452. In addition, the residues PHE497, GLN498, TYR505, ARG403, TYR495, TYR449, PHE490, and GLN493 were formingVan der Waals attraction with glycyrrhizic acid. In Omicron SGp RBD, the glycyrrhizic acid is effectively binding with the receptor-binding site forming four conventional hydrogen bonds with SER496 with bond distance of 2.44 and 2.15 Å (Fig. 6b). In addition, glycyrrhizic acid poses Van der Waal interactions with the residues HIS505, TYR495, TYR453, ARG493, LEU455, TYR489, ALA484, GLY485, CYS488 and PHE456. The docking results revealed that the phytochemicals are binding effective at the SGp RBD of the three variants (Omicron, Delta and WT) with almost similar binding affinity. There is no significant consequence on the binding affinity of the phytochemicals at the SGp RBD because of the large number of unexpected mutations occurred in Omicron.
2.4. Molecular dynamics simulations
The molecular dynamics simulation of the best pose of limonin and glycyrrhizic acid with Omicron SGp RBD was performed for 100 ns to examine the time-dependent conformational and binding stability. The spatial orientation of the limonin (Fig. 7 ) and glycyrrhizic acid (Fig. 8 ) within the RBD surface pocket provides a clear insight of the possible conformational stability and binding affinity. The strong binding affinity and stability are of utmost importance for the limonin and glycyrrhizic acid to mediate the suppression of SGp RBD activity during the SARS-CoV-2 disease establishment. Also, the dynamics analysis by GROMACS package provides a comprehensive examination of the conformational landscape of protein-ligand complex near to physiological conditions. The resulting MD trajectory was utilised to analyse the dynamics behaviour of limonin and glycyrrhizic acid within the binding pocket of Omicron SGp RBD from the computed root mean square deviations (RMSD), root mean square fluctuations (RMSF), number of hydrogen bond contacts, radius of gyration (Rg) and solvent-accessible surface area of the complexes.
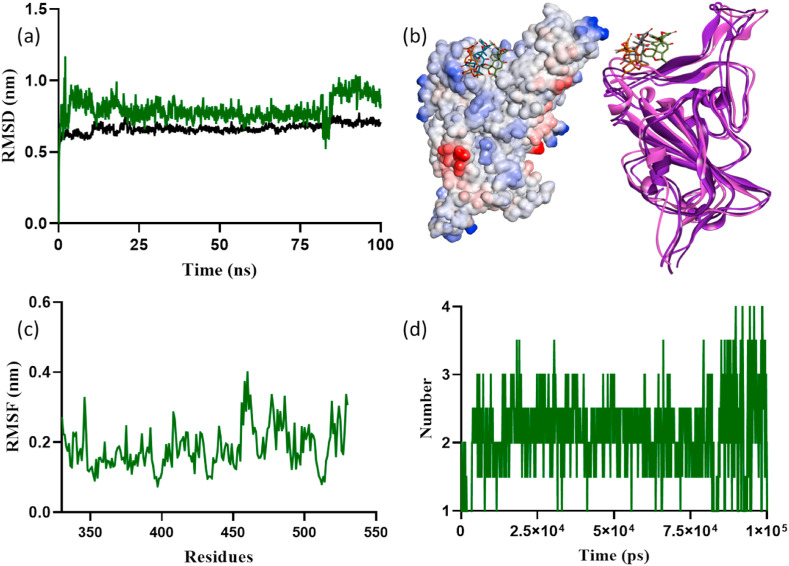
Fig. 7.
Molecular dynamics simulations result of limonin with Omicron SGp RBD: (a) RMSD plot, (b) surface and ribbon structures showing the confirmations of limonin with RBD, (c) RMSF plot, and (d) hydrogen bond contacts of limonin in the active pocket of RBD.
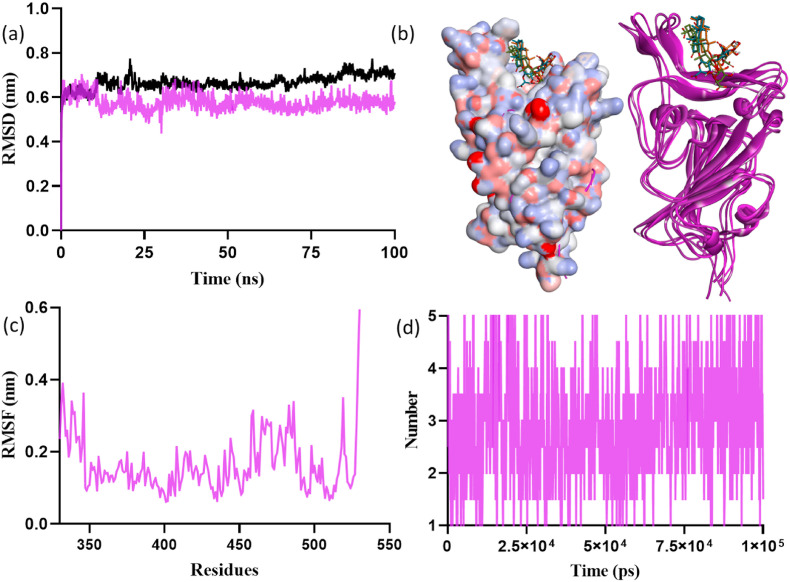
Fig. 8.
Molecular dynamics simulations result of glycyrrhizic acid with Omicron SGp RBD: (a) RMSD plot, (b) surface and ribbon structures showing the confirmations of glycyrrhizic acid with RBD, (c) RMSF plot, and (d) hydrogen bond contacts of glycyrrhizic acid in the active pocket of RBD.
The protein-ligand RMSD plots for limonin-SGp RBD complex indicated that the docked complex attained stability after 21 ns (Fig. 7). The RMSD variation was observed as ∼0.25 nm for overall simulation time with respect to residual protein fluctuations. The limonin maintained a stable binding affinity and stability for 100 ns, but a shift in binding position of limonin was observed after ∼80 ns at the binding pocket of Omicron SGp RBD (Fig. S8). The Omicron SGp RBD residues ASN450, SER446 and LYS444 formed hydrogen bonds with limonin. The residue TYR449 formed Pi-alkyl, whereas the residues ASN448, GLY447 and ARG498 formed van der Waals interactions with limonin. The RMSF plot of protein residual peak variations noticed at 330–335, 360–395, 410–435, 455–506 and 515–530 (Fig. 7c), when compared with the RMSD plot. Some of the amino acid residues in this window were pocket residues that are promoting ligand binding. The mutated residues, such as LYS478, ALA484, ARG493, ASN477, LYS440 and PHE375 are promoting high fluctuation at 0.30, 0.28, 0.25, 0.33, 0.24 and 0.24 nm, respectively. The number of hydrogen bond contacts with protein observed during the overall simulation time was shown in Fig. 7d. In addition, the intramolecular hydrogen bonds plotted for average number of hydrogens involved in protein dynamics, the compactness calculated by the radius of gyration (Rg) parameter, the solvent-accessible surface area of the complex over the simulation time and area per residue over the trajectory computed data on average 0.65 nm2 in most of the frames were shown in Fig. S9.
The computed MD trajectory of glycyrrhizic acid-SGp RBD complex for 100 ns supported better conformational and binding stability (Fig. 8). The RMSD variations was maintained as 0.58 nm during whole simulation time (Fig. 8a). The RMSF plot of protein residual peak variations was noticed at 315–530, 360–395, 405–450 and 455–495 (Fig. 8c). The residues ASP339, ASN477, ALA484 and LYS478 showed high fluctuations at 0.30, 0.27, 0.25 and 0.24 nm, respectively. The residues maintained four to five hydrogen bond interactions with glycyrrhizic acid during the 100 ns simulation (Fig. 8d). In addition, the other computed MD parameters (Fig. S10), i.e., average number of intramolecular hydrogen bonds plotted for protein dynamics, the radius of gyration (Rg) parameter compactness, the solvent-accessible surface area of the complex over the simulation time and the average area per residue over the trajectory computed data clearly delineated the conformational and binding stability of glycyrrhizic acid-SGp RBD complex. Overall, the MD simulation of docked complexes of limonin and glycyrrhizic acid with Omicron SGp RBD showed satisfactory time-dependent conformational stability, which supported the potential of phytochemicals to obstruct the viral infection due to the Omicron.
3. Conclusions
The in silico studies of pairwise sequence alignment, molecular docking and molecular dynamics simulation were performed to examine the binding affinity between the SGp RBD of the variants Omicron, Delta and WT with the human receptor ACE2. Large alterations occurred in the Omicron SGp RBD may increase the binding specificity and affinity with hACE2 resulting in a faster transmission rate and a significant influence on pathogenesis when compared to the WT and Delta variants. The in silico results revealed that the Omicron SGp RBD showed higher binding affinity towards ACE2 compared to WT and Delta variants. The higher binding affinity between Omicron SGp RBD and ACE2 is due to the changes occurred in the surface electrostatics upon mutations that altered the nature of bonding. However, the binding conformation was found to be similar in SGp RBD of the three variants with hACE2. The next important part of the present study was to compare the binding affinity of two important phytochemicals (limonin and glycyrrhizic acid) at the SGp RBD of Omicron, Delta and WT variants. Both the phytochemicals are predicted to be potential compounds against the WT variant of SARS-CoV-2. Interestingly, both the phytochemicals bind effectively at the SGp RBD of the variants Omicron, Delta and WT with almost equal binding affinity. Also, the mutations had no significant effects to the binding of phytochemicals at the SGp RBD site of mutated variants Omicron and Delta. Further, the efficient binding and conformational stability of limonin and glycyrrhizic acid at the binding pocket of Omicron SGp RBD was complemented by employing molecular dynamics simulation for 100 ns. Overall, despite the multiple mutations occurred in Omicron and its viral transmission is comparatively high, the present study supported that the traditional medicines can be useful in formulating adjuvant therapies to fight against the SARS-CoV-2 Omicron.
Declaration of competing of interest
The authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105367.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. 2022. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern Who.int. [Google Scholar]
- 2.Mlcochova P., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volz E., et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 4.Anifandis G., et al. COVID-19 and human reproduction: a pandemic that packs a serious punch. Syst. Biol. Reprod. Med. 2021;67:3–23. doi: 10.1080/19396368.2020.1855271. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lan J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 7.Han P., et al. Receptor binding and complex structures of human ACE2 to spike RBD from Omicron and Delta SARS-CoV-2. Cell. 2022;185:630–640. doi: 10.1016/j.cell.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanai T., et al. Quantitative in silico analysis of SARS-CoV-2 S-RBD omicron mutant transmissibility. Talanta. 2022;240:123206. doi: 10.1016/j.talanta.2022.123206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rath S., et al. Scanning the RBD-ACE2 molecular interactions in Omicron variant. Biochem. Biophys. Res. Commun. 2022;592:18–23. doi: 10.1016/j.bbrc.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koley T., et al. Structural modeling of Omicron spike protein and its complex with human ACE-2 receptor: molecular basis for high transmissibility of the virus. Biochem. Biophys. Res. Commun. 2022;592:51–53. doi: 10.1016/j.bbrc.2021.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar S., et al. Omicron and Delta variant of SARS‐CoV‐2: a comparative computational study of spike protein. J. Med. Virol. 2022;94:1641–1649. doi: 10.1002/jmv.27526. [DOI] [PubMed] [Google Scholar]
- 12.Chen J., et al. Omicron variant (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J. Chem. Inf. Model. 2022 doi: 10.1021/acs.jcim.1c01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Q., et al. Structural analysis of the SARS-CoV-2 omicron variant proteins. Research. 2021;2021:1–4. doi: 10.34133/2021/9769586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu F., et al. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012;40:D1128–D1136. doi: 10.1093/nar/gkr797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H., et al. Therapeutic target database update 2016: enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res. 2016;44:D1069–D1074. doi: 10.1093/nar/gkv1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y.H., et al. Clinical trials, progression-speed differentiating features and swiftness rule of the innovative targets of first-in-class drugs. Briefings Bioinf. 2020;21:649–662. doi: 10.1093/bib/bby130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J., et al. Structure-based discovery of novel nonpeptide inhibitors targeting SARS-CoV-2 Mpro. J. Chem. Inf. Model. 2021;61:3917–3926. doi: 10.1021/acs.jcim.1c00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., et al. Computational design and modeling of nanobodies toward SARS-CoV-2 receptor binding domain. Chem. Biol. Drug Des. 2021;98:1–18. doi: 10.1111/cbdd.13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., et al. SYNBIP: synthetic binding proteins for research, diagnosis and therapy. Nucleic Acids Res. 2022;50:D560–D570. doi: 10.1093/nar/gkab926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S., et al. RNA-RNA interactions between SARS-CoV-2 and host benefit viral development and evolution during COVID-19 infection. Briefings Bioinf. 2022;23 doi: 10.1093/bib/bbab397. bbab397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuzimoto A.D., et al. The antiviral and coronavirus-host protein pathways inhibiting properties of herbs and natural compounds - additional weapons in the fight against the COVID-19 pandemic? J. Tradit. Complement. Med. 2020;10:405–419. doi: 10.1016/j.jtcme.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vidoni C., et al. Targeting autophagy with natural products to prevent SARS-CoV-2 infection. J. Tradit. Complement. Med. 2022;12:55–68. doi: 10.1016/j.jtcme.2021.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vardhan S., et al. In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19, Comput. Biol. Med. 2020;124:103936. doi: 10.1016/j.compbiomed.2020.103936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae J.R., et al. Role of limonin in anticancer effects of Evodia rutaecarpa on ovarian cancer cells, BMC Complement. Méd. Thérapeutique. 2020;20:94. doi: 10.1186/s12906-020-02890-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan S., et al. Limonin: a review of its pharmacology, toxicity, and pharmacokinetics. Molecules. 2019;24:3679. doi: 10.3390/molecules24203679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang R., et al. Limonin attenuates LPS-induced hepatotoxicity by inhibiting pyroptosis via NLRP3/Gasdermin D signaling pathway. J. Agric. Food Chem. 2021;69:982–991. doi: 10.1021/acs.jafc.0c06775. [DOI] [PubMed] [Google Scholar]
- 27.Vardhan S., et al. Virtual screening by targeting proteolytic sites of furin and TMPRSS2 to propose potential compounds obstructing the entry of SARS-CoV-2 virus into human host cells. J. Tradit. Complement. Med. 2022;12:6–15. doi: 10.1016/j.jtcme.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vardhan S., et al. Exploring the therapeutic nature of limonoids and triterpenoids against SARS-CoV-2 by targeting nsp13, nsp14, and nsp15 through molecular docking and dynamic simulations. J. Tradit. Complement. Med. 2022;12:44–54. doi: 10.1016/j.jtcme.2021.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isa Irawan M., et al. Application of Needleman-Wunch Algorithm to identify mutation in DNA sequences of Corona virus. J. Phys. Conf. Ser. 2019;1218 doi: 10.1088/1742-6596/1218/1/012031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sierk M.L., et al. Improving pairwise sequence alignment accuracy using near-optimal protein sequence alignments. BMC Bioinf. 2010;11:146. doi: 10.1186/1471-2105-11-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C., et al. The impact of receptor-binding domain natural mutations on antibody recognition of SARS-CoV-2. Signal Transduct. Target Ther. 2021;6:132. doi: 10.1038/s41392-021-00536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J., et al. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432:5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pascarella S., et al. The electrostatic potential of the Omicron variant spike is higher than in Delta and Delta-plus variants: a hint to higher transmissibility? J. Med. Virol. 2022;94:1277–1280. doi: 10.1002/jmv.27528. [DOI] [PubMed] [Google Scholar]
- 34.Ortega J.T., et al. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J. 2020;19:410–417. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan Y., et al. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020;15:1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 36.Goher S.S., et al. The Delta variant mutations in the receptor binding domain of SARS-CoV-2 show enhanced electrostatic interactions with the ACE2. Med. Drug Dis. 2022;13:100114. doi: 10.1016/j.medidd.2021.100114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao H., et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells, Emerg. Microb. Infect. 2022;11:277–283. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.