Abstract
Background
SARS-CoV-2 is responsible for COVID-19, a clinically heterogeneous disease, ranging from being completely asymptomatic to life-threating manifestations. An unmet clinical need is the identification at disease onset or during its course of reliable biomarkers allowing patients' stratification according to disease severity. In this observational prospective cohort study, patients' immunologic and laboratory signatures were analyzed to identify independent predictors of unfavorable (either death or intensive care unit admission need) or favorable (discharge and/or clinical resolution within the first 14 days of hospitalization) outcome.
Methods
Between January and May 2021 (third wave of the pandemic), we enrolled 139 consecutive SARS-CoV-2 positive patients hospitalized in Northern Italy to study their immunological and laboratory signatures. Multiplex cytokine, chemokine, and growth factor analysis, along with routine laboratory tests, were performed at baseline and after 7 days of hospital stay.
Results
According to their baseline characteristics, the majority of our patients experienced a moderate to severe illness. At multivariate analysis, the only independent predictors of disease evolution were the serum concentrations of IP-10 (at baseline) and of C-reactive protein (CRP) after 7 days of hospitalization. Receiver-operating characteristic (ROC) curve analysis confirmed that baseline IP − 10 > 4271 pg/mL and CRP > 2.3 mg/dL at 7 days predict a worsening in clinical conditions (87% sensitivity, 66% specificity, area under the curve (AUC) 0.772, p < 0.001 and 83% sensitivity, 73% specificity, AUC 0.826, p < 0.001, respectively).
Conclusions
According to our results, baseline IP-10 and CRP after 7 days of hospitalization could be useful in driving clinical decisions tailored to the expected disease trajectory in hospitalized COVID-19 patients.
1. Introduction
In December 2019, many cases of pneumonia of unknown origin were reported in Wuhan (Hubei province, China). Since then, this illness rapidly spread across China and all over the world. The causative agent of this new disease has been identified in SARS-CoV-2, a positive, single-stranded RNA virus with a genome length of less than 30 kb, belonging to the same β-coronavirus genus of SARS-CoV and MERS-CoV [1–4]. SARS-CoV-2 virus enters host's cells by binding the angiotensin-converting enzyme 2 (ACE2) expressed in nasal epithelium, low airways, and lungs [4–7]. This new viral agent is characterized by a high interhuman transmission rate: according to the current evidence, World Health Organization (WHO) reports that respiratory droplets and direct contact represent the most common routes of infection [4–6, 8–10].
Patients affected by COVID-19 show a wide range of clinical manifestations, ranging from a nearly asymptomatic or mild flu-like condition to severe interstitial pneumonia and acute respiratory distress syndrome. It is well-recognized that severe clinical manifestations depend not only on viral infection but also on a heavy inflammatory response [1–4, 11, 12]. Pathogen recognition by antigen presenting cells activates both innate and adaptive immune cells to produce large amounts of proinflammatory mediators, in some cases leading to systemic spread of the aberrant immune response leading—in the most severe cases—to multiple organ failure and death [2, 13–15].
Reliable biomarkers allowing clinicians to identify, at an early stage, those patients whose condition will deteriorate would facilitate patients' stratification into risk groups for optimal resources allocation. To meet this need, many clinical research groups, including ours, analyzed their case series, identifying several demographic (i.e., age, gender, and comorbidities) and laboratory (i.e., CRP (C-reactive protein), IL-6, D-dimer, and creatinine) parameters that could be used as prognostic biomarkers [14, 16–21]. Unfortunately, the retrospective design of these studies has many potential biases.
The present prospective, observational monocentric study is aimed at investigating cytokine, chemokine, and growth factor expression in an Italian cohort of SARS-CoV-2 positive patients, by using a large and unbiased multiplex quantification approach. To this purpose, patients were studied at the time of hospital admission and along their hospital stay, during which they received per protocol corticosteroids and heparin, to identify possible biomarkers able to predict disease severity and evolution.
2. Methods
2.1. Patients
We performed a prospective, observational cohort study during the third wave of COVID-19 epidemic in Italy: 139 consecutive patients were enrolled in non-ICU (intensive care unit) wards (including high-dependency/subintensive units) of “Maggiore della Carità” University Hospital in Novara, Italy, between January and May 2021. This study is part of the large multicenter observational study “BIAS” (Baseline Immunity status effect on sArs-cov2 presentation and evolution: comparison between immunocompetent and immunocompromised patients). The study protocol was approved by the local ethical committee (CE 7/21) and was conducted in strict accordance with the Declaration of Helsinki. Patients were selected according to precise inclusion and exclusion criteria and were asked to give a written consent. To be eligible for the study, patients should be adults (>18 years), hospitalized for confirmed SARS-CoV-2 infection (molecular RT-PCR or quick test positivity), with clinical symptoms not exceeding 12 days. Patients needing an immediate ICU admission or with a very severe clinical presentation (stage V renal failure, severe oncological condition) were excluded.
All patients meeting inclusion criteria for the study received treatment according to the “Maggiore della Carità” Hospital internal protocol. Specifically, all patients received oxygen supplementation, corticosteroids, and low molecular weight heparin (LMWH) unless contraindicated.
2.2. Endpoints Definition
The expected endpoints were the following: (1) identification of biomarkers predicting at baseline and after 7 days of hospitalization an adverse disease evolution (death or ICU admission); (2) identification of clinical or immunological biomarkers predicting at baseline and after 7 days of hospitalization a rapid clinical recovery (discharge from hospital and/or National Early Warning Score 2 (NEWS2) ≤ 2 for at least 24 hours within the first 14 days of hospitalization).
2.3. Blood Sample Collection
Blood samples for routine analysis and for multiplex quantifications were collected by venous puncture using EDTA as anticoagulant at baseline (t0) and after 7 days of hospitalization (t7). Blood fractions were immediately separated by centrifugation and stored at -80°C until the time of analysis.
2.4. Routine Laboratory Evaluation
For each patient, routine laboratory studies included a complete blood cell count, a common biochemistry panel (i.e., creatinine, alanine aminotransferase (ALT), and aspartate aminotransferase (AST)), as well as inflammatory markers (i.e., CRP and ferritin) and markers of coagulation and fibrinolysis (including D-dimer).
2.5. Multiplex Analysis
Twenty-seven plasma cytokines, chemokines, and growth factors were analyzed using the Bio-Plex Pro Human Cytokine 27-plex panel (Bio-Rad Laboratories Inc, Hercules, CA, USA) following the manufacturer instructions. Prior to quantification, plasma samples were diluted 1 : 4 in standard diluent (provided by the manufacturer). By using such panel, serum samples were screened for interleukin (IL)-1β, IL-1RA, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12, IL-13, IL-15, IL-17, eotaxin, basic fibroblast growth factor (FGF), granulocyte-colony stimulating factor (G-CSF), granulocyte-macrophage colony stimulating factor (GM-CSF), interferon (IFN)-γ, IFN-γ induced protein-10 (IP-10), monocyte chemoattractant protein-1 (MCP-1), macrophage inflammatory protein (MIP)-1 α/β (MIP-1α, MIP-1β), platelet-derived growth factor (PDGF), RANTES (regulated on activation normal T-cell expressed and secreted), tumor necrosis factor α (TNF-α), and vascular endothelial growth factor (VEGF) concentrations. Fluorescent signals were recorded using a Bio-Plex 200 System instrument and analyzed using the Bio-Plex Manager Software (Bio-Rad Laboratories Inc, Hercules, CA, USA). The software fitted samples' median fluorescence intensity (MFI) values versus standards' MFI and converted it to concentration (pg/mL) by applying a five-parameter logistic regression (as suggested by the manufacturer).
2.6. Data Collection and Statistical Analysis
A web-based database (RedCap platform) was used to store and manage clinical and laboratory data of each patient (demographics, clinical parameters, therapeutic schedule, and laboratory parameters). Clinical and routine laboratory data were collected by carefully reviewing medical records of each patient, starting from the time of admission (baseline, t0), until discharge (or for a maximum of 28 days) or study exit (death or ICU admission). Data extracted from the RedCap database and multiplex quantifications underwent univariate and multivariate statistical analysis to evaluate their significance toward the expected endpoints. For continuous variables, the measures of central tendency and dispersion chosen were medians and interquartile range (IQR). Categorical variables were presented as frequencies (percentage). Variables were compared with the Mann-Whitney U test (continuous variables) or Pearson χ2 test (categorical variables). Data obtained from univariate analysis were used to build multiple regression models. Receiver-operating characteristic (ROC) curves were drawn to identify the prognostic cut-off for clinical parameters according to the corresponding AUC (area under the curve) score. The threshold chosen to indicate statistical significance was 0.05 (two-tailed). Statistical tests were performed either with the software package Statistica for Windows, release 12 (TIBCO Software Inc., Palo Alto, CA, USA) or the MedCalc® Statistical Software, version 20.014 (MedCalc Software Ltd., Ostend, Belgium).
3. Results
From January until May 2021, 139 SARS-CoV-2-positive patients admitted to non-ICU wards of “Maggiore della Carità” Hospital in Novara meeting the study inclusion criteria were enrolled to the present study: 86 were males (61.9%), and 53 (38.1%) were females.
The most common symptoms at hospital admission (t0) were dyspnea (62.6%) and dry cough (38.9%). Moreover, among the 139 patients, 83 started a COVID-19-related treatment before hospital admission (corticosteroids (52.5%), azithromycin (35.2%), and heparin (30.9%)). The majority of the enrolled patients at the time of hospital admission showed a moderate (74.1%) or severe (6.5%) respiratory failure (we defined the respiratory failure as moderate when 100 ≤ PiO2/FiO2 < 200 and severe when PiO2/FiO2 < 100). Consistently median NEWS2 score recorded at the time of admission in our cohort was 5 (IQR: 4-6), a value indicating a potentially serious clinical deterioration in patient's conditions, requiring a close clinical monitoring [22]. The detailed demographical and baseline (t0) clinical description of the selected population is shown in Table 1.
Table 1.
Demographic and baseline characteristics of the studied population. § refers to data obtained with oxygen supplementation.
| Demographics, parameters, and clinical scores | Median [IQR] |
|---|---|
| Age (years) | 63.8 [56.2-71.9] |
| Heart rate (beats/min) | 85 [75-95] |
| Respiratory rate (breaths/min)§ | 21 [18-26] |
| SpO2 (%)§ | 96 [94-98] |
| Temperature (°C) | 36.5 [36.1-36.8] |
| Systolic pressure (mmHg) | 125 [115-140] |
| Diastolic pressure (mmHg) | 75 [70-85] |
| NEWS2 | 5 [4-6] |
| Days from illness onset to hospital admission | 6 [5-8] |
| Laboratory findings | |
| Hemoglobin (g/dL) | 14.2 [12.6-15.0] |
| RDW-CV (%) | 13.3 [12.8-14.0] |
| White blood cells (cell count × 103/μL) | 7.03 [5.05-9.52] |
| Neutrophils (cell count × 103/μL) | 5.67 [4.20-8.56] |
| Eosinophils (cell count × 103/μL) | 0.00 [0.00-0.00] |
| Lymphocytes (cell count × 103/μl) | 0.71 [0.54-0.95] |
| Platelets (cell count × 103/μL) | 205 [161-263] |
| ALT (U/L) | 37 [28-55] |
| AST (U/L) | 41 [32-57] |
| Bilirubin (mg/dL) | 0.6 [0.5-0.8] |
| Creatinine (mg/dL) | 0.8 [0.64-0.96] |
| Glomerular filtration rate (mL/min) | 90 [71-103] |
| CRP (mg/dL) | 8.2 [4.4-13.0] |
| LDH (U/L) | 718 [554-871] |
| Erythrocyte sedimentation rate (mm/h) | 40 [27-52] |
| Troponin I (ng/mL) | 7 [3-15] |
| Ferritin (ng/mL) | 826 [401-1348] |
| D-dimer (μg/L) | 697 [517-1275] |
| Albumin (g/dL) | 4.0 [3.7-4.2] |
| IL-6 (pg/mL) | 11.6 [5.00-31.30] |
| Arterial blood gas test§ | |
| pO2 (mmHg) | 70.0 [59.6-79.6] |
| pH | 7.46 [7.44-7.49] |
| pCO2 (mmHg) | 36 [33-39] |
| PiO2/FiO2 | 146 [120-180] |
Among the 139 patients initially enrolled to the study, 29 died during hospital stay or were transferred to ICU, while of the remaining 110 patients, 91 were discharged or reached a NEWS2 ≤ 2 for at least 24 hours within the first 14 days of hospitalization.
3.1. Outcomes
We compared data obtained at baseline and at 7 days from the 29 patients who died or were transferred to ICU during hospital stay to those obtained from all the other patients
Univariate statistical analysis is shown in Tables 2 and 3. At baseline, patients evolving toward a more severe form of the disease showed lower platelet and lymphocyte counts and glomerular filtration rates, in addition to higher RDW-CV (red cell distribution width–coefficient of variation), creatinine, LDH (lactic dehydrogenase), ferritin, D-dimer, IL-6, IL-8, IP-10, and MCP-1 values (Table 2). After 7 days of hospital stay, patients with a worse disease evolution showed a statistically significant lower level in lymphocyte and platelet counts as well as in TNF-α levels and an increment in neutrophil to lymphocyte ratio, CRP, LDH, erythrocyte sedimentation rate, troponin I, ferritin, D-dimer, G-CSF, IFN-γ, IL-6, IL-8, IP-10, MCP-1, and MIP-1α values (Table 3).
Table 2.
Baseline routine laboratory findings and multiplex quantifications in patients with an adverse disease evolution (death/ICU admitted) vs. all the other patients. Data are expressed as medians (IQR). Bold text highlights the statistically significant results.
| Laboratory findings at baseline (t0) | Adverse disease evolution (n = 29) | All other patients (n = 110) | Z | p value |
|---|---|---|---|---|
| Hemoglobin (g/dL) | 14.3 [11.9-15.1] | 14.2 [12.9-15.0] | 0.3708 | 0.7108 |
| RDW-CV (%) | 13.6 [13.0-14.4] | 13.3 [12.8-13.8] | 2.0285 | 0.0425 |
| White blood cells (cell count × 103/μL) | 8.12 [5.12-9.87] | 6.99 [5.05-9.21] | 0.2385 | 0.8115 |
| Neutrophils (cell count × 103/μL) | 5.70 [4.20-9.15] | 5.67 [4.20-7.93] | 0.3940 | 0.6936 |
| Eosinophils (cell count × 103/μL) | 0.00 [0.00-0.00] | 0.00 [0.00-0.00] | 0.2413 | 0.8093 |
| Lymphocytes (cell count × 103/μL) | 0.62 [0.39-0.77] | 0.74 [5.70-0.99] | -2.2837 | 0.0224 |
| Neutrophil/lymphocyte ratio | 11.90 [4.97-17.60] | 7.88 [4.70-10.55] | 1.7909 | 0.0733 |
| Platelets (cell count × 103/μL) | 162 [128-200] | 219 [176-273] | -4.1859 | 0.0001 |
| ALT (U/L) | 36 [27-57] | 37 [28-55] | -0.1949 | 0.8455 |
| AST (U/L) | 43 [36-70] | 41 [31-56] | 1.2971 | 0.1946 |
| Bilirubin (mg/dL) | 0.7 [0.5-0.8] | 0.6 [0.5-0.8] | 1.1019 | 0.2705 |
| Creatinine (mg/dL) | 0.91 [0.77-1.24] | 0.78 [0.64-0.90] | 2.6236 | 0.0087 |
| Glomerular filtration rate (mL/min) | 75 [61-90] | 93 [74-103] | -2.8489 | 0.0044 |
| CRP (mg/dL) | 10.2 [5.6-15.5] | 7.7 [4.1-12.0] | 1.8531 | 0.0639 |
| LDH (U/L) | 784 [694-1010] | 690 [535-864] | 2.1414 | 0.0322 |
| Erythrocyte sedimentation rate (mm/h) | 44 [25-56] | 40 [31-50] | 0.6988 | 0.4847 |
| Troponin I (ng/mL) | 13 [4-17] | 7 [2-14] | 1.7752 | 0.0759 |
| Ferritin (ng/mL) | 1155 [608-1921] | 793 [371-1245] | 2.2487 | 0.0245 |
| D-dimer (μg/L) | 1175 [593-1773] | 667 [481-1056] | 2.2821 | 0.0225 |
| Albumin (g/dL) | 4.0 [3.7-4.3] | 4.0 [3.7-4.2] | 0.6087 | 0.5427 |
| Eotaxin (pg/mL) | 3.45 [2.97-4.13] | 3.68 [2.62-4.78] | -0.0467 | 0.9628 |
| FGF (pg/mL) | 12.68 [0.00-17.49] | 15.72 [5.03-22.55] | -1.6272 | 0.1037 |
| G-CSF (pg/mL) | 61.47 [43.47-111.57] | 64.27 [43.47-91.38] | -0.6610 | 0.5086 |
| GM-CSF (pg/mL) | 0.00 [0.00-0.00] | 0.00 [0.00-0.00] | 1.8897 | 0.0588 |
| IFN-γ (pg/mL) | 7.64 [5.56-9.70] | 7.37 [5.20-9.98] | 0.1919 | 0.8478 |
| IL-1β (pg/mL) | 0.16 [0.00-1.03] | 0.53 [0.00-1.43] | -0.5697 | 0.5689 |
| IL1-RA (pg/mL) | 0.00 [0.00-168.65] | 0.00 [0.00-168.65] | -0.2656 | 0.7906 |
| IL-2 (pg/mL) | 0.07 [0.00-1.79] | 1.50 [0.00-4.34] | -1.8431 | 0.0653 |
| IL-4 (pg/mL) | 0.58 [0.00-0.90] | 0.32 [0.00-0.73] | 1.1959 | 0.2317 |
| IL-5 (pg/mL) | 0.00 [0.00-48.48] | 0.00 [0.00-107.56] | -1.4608 | 0.1441 |
| IL-6 (pg/mL) | 20.12 [11.51-42.41] | 8.17 [3.8-22.45] | 2.7546 | 0.0059 |
| IL-7 (pg/mL) | 7.44 [1.93-19.94] | 8.57 [1.93-22.56] | -0.7296 | 0.4656 |
| IL-8 (pg/mL) | 22.60 [16.77-44.72] | 15.95 [11.18-25.20] | 2.7034 | 0.0069 |
| IL-9 (pg/mL) | 456.18 [401.78-493.75] | 456.86 [403.68-553.68] | -1.0004 | 0.3171 |
| IL-10 (pg/mL) | 2.05 [0.13-3.11] | 1.27 [0.00-4.42] | 0.4075 | 0.6837 |
| IL-12 (pg/mL) | 0.95 [0.00-3.23] | 2.21 [0.00-4.68] | -1.8208 | 0.0686 |
| IL-13 (pg/mL) | 0.00 [0.00-0.49] | 0.00 [0.00-0.79] | -0.1276 | 0.8985 |
| IL-15 (pg/mL) | 0.00 [0.00-84.08] | 0.00 [0.00-273.20] | -1.0831 | 0.2788 |
| IL-17 (pg/mL) | 4.37 [3.65-5.95] | 4.37 [2.91-6.33] | 0.5474 | 0.5841 |
| IP-10 (pg/mL) | 7520.77 [4953.57-12322.33] | 3451.44 [2102.05-5850.57] | 4.8026 | 0.0001 |
| MCP-1 (pg/mL) | 121.31 [67.56-203.32] | 75.11 [40.74-103.27] | 2.6877 | 0.0072 |
| MIP-1α (pg/mL) | 1.89 [1.29-2.95] | 1.69 [1.04-2.66] | 0.6768 | 0.4985 |
| MIP-1β (pg/mL) | 222.66 [198.75-241.60] | 228.00 [194.67-257.58] | -0.8683 | 0.3853 |
| PDGF (pg/mL) | 1040.46 [614.45-1910.19] | 1370.46 [723.44-2622.26] | -1.4074 | 0.1593 |
| RANTES (pg/mL) | 5525.30 [2952.02-8016.74] | 5300.72 [2982.24-10540.92] | -0.9823 | 0.3260 |
| TNF-α (pg/mL) | 19.12 [15.42-20.63] | 21.91 [16.17-27.47] | -1.9106 | 0.0561 |
| VEGF (pg/mL) | 178.91 [19.79-230.35] | 198.68 [48.82-291.68] | -1.1787 | 0.2385 |
Table 3.
Routine laboratory findings and multiplex quantifications after 7 days hospitalization in patients with an adverse disease evolution (death/ICU admitted) vs. all the other patients. Data are expressed as medians (IQR). Bold text highlights the statistically significant results.
| Laboratory findings after 7 days (t7) | Adverse disease evolution (n = 18) | All other patients (n = 95) | Z | p value |
|---|---|---|---|---|
| Hemoglobin (g/dL) | 13.5 [11.8-13.9] | 13.5 [12.6-14.8] | -1.3108 | 0.1899 |
| RDW-CV (%) | 13.4 [12.9-14.0] | 13.0 [12.6-13.8] | 1.3590 | 0.1742 |
| White blood cells (cell count × 103/μL) | 10.42 [6.96-13.04] | 9.49 [7.52-11.02] | 0.6708 | 0.5023 |
| Neutrophils (cell count × 103/μL) | 9.21 [6.18-11.81] | 7.24 [5.70-8.99] | 1.9144 | 0.0556 |
| Eosinophils (cell count × 103/μL) | 0.01 [0.01-0.02] | 0.02 [0.01-0.08] | -1.2358 | 0.2165 |
| Lymphocytes (cell count × 103/μL) | 0.46 [0.28-0.64] | 1.38 [0.76-1.94] | -4.2958 | 0.0001 |
| Neutrophil/lymphocyte ratio | 19.64 [12.98-33.23] | 6.23 [3.19-10.63] | 4.8488 | 0.0001 |
| Platelets (cell count × 103/μL) | 295 [214-323] | 347 [289-415] | -3.1070 | 0.0019 |
| ALT (U/L) | 48 [31-130] | 58 [37-98] | -0.3288 | 0.7423 |
| AST (U/L) | 33 [25-60] | 29 [23-44] | 0.9947 | 0.3199 |
| Bilirubin (mg/dL) | 0.8 [0.6-0.9] | 0.6 [0.5-0.9] | 1.0531 | 0.2923 |
| Creatinine (mg/dL) | 0.78 [0.60-1.09] | 0.71 [0.61-0.84] | 0.8046 | 0.4210 |
| Glomerular filtration rate (mL/min) | 93 [68-101] | 97 [88-105] | -1.4266 | 0.1537 |
| CRP (mg/dL) | 3.7 [2.7-9.8] | 1.0 [0.3-2.3] | 4.7086 | 0.0001 |
| LDH (U/L) | 687 [540-892] | 575 [469-666] | 2.7176 | 0.0066 |
| Erythrocyte sedimentation rate (mm/h) | 58 [41-66] | 29 [16-46] | 2.8219 | 0.0048 |
| Troponin I (ng/mL) | 6 [3-34] | 4 [2-8] | 2.1054 | 0.0353 |
| Ferritin (ng/mL) | 933 [734-1912] | 719 [379-1017] | 2.2958 | 0.0217 |
| D-dimer (μg/L) | 1718 [1117-6381] | 1001 [594-1811] | 2.5272 | 0.0115 |
| Albumin (g/dL) | 3.4 [3.2-3.5] | 3.6 [3.4-3.8] | -1.4590 | 0.1446 |
| Eotaxin (pg/mL) | 4.15 [3.04-5.79] | 5.07 [3.62-7.40] | -1.1539 | 0.2485 |
| FGF (pg/mL) | 18.09 [12.68-25.30] | 21.36 [10.82-28.33] | -0.2850 | 0.7756 |
| G-CSF (pg/mL) | 121.15 [89.00-192.46] | 79.31 [61.47-103.07] | 3.1603 | 0.0016 |
| GM-CSF (pg/mL) | 0.00 [0.00-0.00] | 0.00 [0.00-0.00] | -0.6118 | 0.5407 |
| IFN-γ (pg/mL) | 9.70 [6.61-38.83] | 6.83 [4.27-10.50] | 2.2561 | 0.0241 |
| IL-1β (pg/mL) | 0.16 [0.00-1.80] | 0.38 [0.00-1.43] | -0.7482 | 0.4544 |
| IL1-RA (pg/mL) | 0.00 [0.00-135.38] | 0.00 [0.00-135.38] | 0.3655 | 0.7147 |
| IL-2 (pg/mL) | 2.01 [0.00-3.74] | 0.79 [0.00-3.42] | -0.1423 | 0.8869 |
| IL-4 (pg/mL) | 0.32 [0.00-0.84] | 0.68 [0.14-1.10] | -0.6687 | 0.5037 |
| IL-5 (pg/mL) | 0.00 [0.00-72.05] | 0.00 [0.00-72.05] | -0.1147 | 0.9087 |
| IL-6 (pg/mL) | 11.51 [9.12-28.52] | 3.34 [0.00-8.50] | 3.8190 | 0.0001 |
| IL-7 (pg/mL) | 5.55 [0.00-9.00] | 8.57 [0.00-22.26] | -0.7029 | 0.4821 |
| IL-8 (pg/mL) | 28.59 [15.89-54.90] | 11.02 [6.49-19.72] | 3.6798 | 0.0002 |
| IL-9 (pg/mL) | 460.16 [316.26-488.06] | 460.82 [404.15-524.46] | -0.8905 | 0.3732 |
| IL-10 (pg/mL) | 1.25 [0.00-3.70] | 1.25 [0.00-4.42] | -0.0189 | 0.9849 |
| IL-12 (pg/mL) | 0.95 [0.00-1.73] | 1.58 [0.00-4.68] | -1.1909 | 0.2337 |
| IL-13 (pg/mL) | 0.00 [0.00-0.49] | 0.00 [0.00-0.78] | -0.1755 | 0.8607 |
| IL-15 (pg/mL) | 0.00 [0.00-0.00] | 0.00 [0.00-203.24] | -1.2231 | 0.2213 |
| IL-17 (pg/mL) | 3.93 [3.42-5.43] | 4.87 [3.65-6.49] | -0.6190 | 0.5360 |
| IP-10 (pg/mL) | 2893.33 [2672.14-6710.26] | 737.64 [396.95-1440.44] | 4.6502 | 0.0001 |
| MCP-1 (pg/mL) | 335.38 [106.19-712.15] | 60.87 [42.11-103.27] | 3.8958 | 0.0001 |
| MIP-1α (pg/mL) | 3.55 [2.56-5.64] | 2.26 [1.51-3.02] | 3.0059 | 0.0027 |
| MIP-1β (pg/mL) | 230.67 [180.96-234.94] | 231.62 [202.97-256.98] | -1.1378 | 0.2552 |
| PDGF (pg/mL) | 1622.08 [622.51-2464.00] | 2270.91 [1399.66-3625.82] | -1.6820 | 0.0926 |
| RANTES (pg/mL) | 5444.86 [2013.72-10073.76] | 7544.65 [4221.08-12495.32] | -1.2120 | 0.2256 |
| TNF-α (pg/mL) | 18.41 [13.91-19.89] | 24.04 [17.03-28.63] | -2.4685 | 0.0136 |
| VEGF (pg/mL) | 75.81 [0.00-264.14] | 119.46 [0.00-210.59] | 0.0444 | 0.9646 |
Parameters being significant (p < 0.05) at univariate analysis were used to build multivariate analysis models to identify independent predictors of outcome (Tables 4 and 5). Higher RDW-CV, IP-10, and D-dimer values and lower platelet count at baseline predicted a worsening in clinical conditions (i.e., death or need to ICU admission) (Table 4) also after correction for demographic and COVID-19 severity variables (Table 5), while after 7 days of hospitalization, the only biomarker with prognostic significance was the increase in CRP levels (Table 6) again confirmed after correction for demographic and COVID-19 severity variables (multivariate analysis: CRP β 0.3372, p = 0.0049; sex β -0.1960, p = 0.0779; PiO2/FiO2β -0.1776, p = 0.1664; NEWS2 β 0.1302, p = 0.2975; age β 0.0216, p = 0.8701).
(2) We evaluated which biomarkers were useful to early identify patients who had a faster clinical recovery (hospital discharge or NEWS2 ≤ 2 for at least 24 hours within 14 days of hospitalization) with respect to all the other patients
Table 4.
Multivariate analysis of baseline statistically significant routine laboratory findings and multiplex quantifications predicting an adverse disease evolution (death/ICU admission). Bold text highlights the statistically significant results.
| Prognostic biomarkers (t0) | β ∗ | p value |
|---|---|---|
| IP-10 | 0.2614 | 0.0059 |
| Platelets | -0.1983 | 0.0245 |
| D-dimer | 0.1744 | 0.0353 |
| RDW-CV | 0.1710 | 0.0477 |
| Ferritin | 0.1152 | 0.2143 |
| IL-6 | 0.1165 | 0.2213 |
| MCP-1 | 0.1240 | 0.2223 |
| IL-8 | -0.0921 | 0.3135 |
| Glomerular filtration rate | -0.0910 | 0.4844 |
| Lymphocytes | -0.0379 | 0.6523 |
| Creatinine | -0.0494 | 0.6850 |
| LDH | -0.0158 | 0.8647 |
Table 5.
Multivariate analysis of baseline statistically significant routine laboratory findings and multiplex quantifications predicting an adverse disease evolution (death/ICU admission) which were significant at multivariate analysis including demographic and COVID-19 severity-related variables. Bold text highlights the statistically significant results.
| Predictors (t0) | β ∗ | p value |
|---|---|---|
| IP-10 | 0.2930 | 0.0002 |
| D-dimer | 0.2331 | 0.0017 |
| Platelets | -0.1799 | 0.0212 |
| RDW-CV | 0.1508 | 0.0462 |
| PiO2/FiO2 | -0.1269 | 0.0888 |
| Age | 0.1293 | 0.0989 |
| Sex | -0.1031 | 0.1870 |
| NEWS2 | 0.0795 | 0.2729 |
Table 6.
Multivariate analysis of statistically significant routine laboratory findings and multiplex quantifications after 7 days of hospitalization. Bold text highlights the statistically significant results.
| Prognostic biomarkers (t7) | β ∗ | p value |
|---|---|---|
| CRP | 0.3772 | 0.0049 |
| IP-10 | 0.3356 | 0.1261 |
| IL-6 | 0.4479 | 0.2018 |
| LDH | -0.2065 | 0.2128 |
| MCP-1 | -0.5202 | 0.2777 |
| IL-8 | 1.8435 | 0.2950 |
| TNF-α | -1.4239 | 0.3563 |
| Platelets | -0.1067 | 0.3914 |
| D-dimer | -0.2360 | 0.4291 |
| INF-γ | 0.7433 | 0.4583 |
| Ferritin | 0.1041 | 0.4744 |
| Erythrocyte sedimentation rate | 0.0657 | 0.5932 |
| Neutrophil/lymphocyte ratio | 0.0590 | 0.7066 |
| G-CSF | -0.6803 | 0.7070 |
| MIP-1α | -0.2429 | 0.8606 |
| Troponin I | -0.0182 | 0.8626 |
| Lymphocytes | -0.0159 | 0.9039 |
Univariate statistical analysis (Tables 7 and 8) highlighted a significant alteration in some parameters between the two considered populations. In particular, it has been observed that, at baseline, patients with a faster clinical resolution showed a reduction in RDW-CV, creatinine, CRP, neutrophil to lymphocyte ratio, troponin I, D-dimer, IL-4, IL-6, IL-8, IL-10, IP-10, MCP-1, and MIP-1α in addition to an increase in lymphocytes and platelets count and glomerular filtration rate (Table 7). At t7, indeed, it has been observed that patients with a more favorable prognosis showed an increase in platelets and lymphocytes counts, glomerular filtration rate, ALT, and albumin, in addition to a decrease in neutrophil to lymphocyte ratio, RDW-CV, neutrophil count, CRP, LDH, erythrocyte sedimentation rate, troponin I, D-dimer, G-CSF, IFN-γ, IL-6, IL-8, IP-10, MCP-1, and MIP-1α (Table 8).
Table 7.
Baseline routine laboratory findings and multiplex quantifications in patients who had a faster clinical recovery (discharged or reaching NEWS2 ≤ 2 for at least 24 hours within 14 days) vs. all the other patients. Data are expressed as medians (IQR). Bold text highlights the statistically significant results.
| Laboratory findings at baseline (t0) | Faster clinical recovery (n = 91) | All other patients (n = 48) | Z | p value |
|---|---|---|---|---|
| Hemoglobin (g/dL) | 14.3 [12.9-15.1] | 13.9 [12.4-15.0] | 1.0547 | 0.2916 |
| RDW-CV (%) | 13.2 [12.7-13.8] | 13.7 [13.0-14.4] | -2.9173 | 0.0035 |
| White blood cells (cell count × 103/μL) | 6.80 [4.91-9.09] | 7.80 [5.24-10.06] | -1.2448 | 0.2132 |
| Neutrophils (cell count × 103/μL) | 5.39 [4.09-7.73] | 6.50 [4.25-9.59] | -1.3910 | 0.1642 |
| Eosinophils (cell count × 103/μL) | 0.00 [0.00-0.00] | 0.00 [0.00-0.00] | -0.1312 | 0.8956 |
| Lymphocytes (cell count × 103/μL) | 0.74 [0.57-0.99] | 0.65 [0.44-0.89] | 1.9738 | 0.0484 |
| Neutrophil/lymphocyte ratio | 7.62 [4.62-10.26] | 10.16 [5.63-16.94] | -2.4407 | 0.0147 |
| Platelets (cell count × 103/μL) | 219 [175-284] | 186 [155-223] | 2.7953 | 0.0052 |
| ALT (U/L) | 41 [29-56] | 33 [23-52] | 1.7894 | 0.0735 |
| AST (U/L) | 42 [31-54] | 41 [33-63] | -0.6809 | 0.4959 |
| Bilirubin (mg/dL) | 0.6 [0.5-0.8] | 0.6 [0.5-0.8] | 0.2432 | 0.8078 |
| Creatinine (mg/dL) | 0.78 [0.62-0.88] | 0.86 [0.67-1.13] | -2.5478 | 0.0108 |
| Glomerular filtration rate (mL/min) | 95 [78-105] | 73 [62-91] | 4.1183 | <0.0001 |
| CRP (mg/dL) | 7.3 [4.1-11.9] | 9.7 [5.3-15.4] | -2.1905 | 0.0285 |
| LDH (U/L) | 688 [533-870] | 780 [629-904] | -1.4145 | 0.1572 |
| Erythrocyte sedimentation rate (mm/h) | 40 [29-50] | 42 [26-56] | -0.8566 | 0.3916 |
| Troponin I (ng/mL) | 7 [2-13] | 12 [4-30] | -2.3845 | 0.0171 |
| Ferritin (ng/mL) | 826 [371-1278] | 1032 [465-1650] | -1.1114 | 0.2664 |
| D-dimer (μg/L) | 660 [449-1088] | 873 [577-1377] | -2.0546 | 0.0399 |
| Albumin (g/dL) | 4.0 [3.6-4.2] | 4.0 [3.8-4.2] | -0.2944 | 0.7685 |
| Eotaxin (pg/mL) | 3.59 [2.48-4.62] | 3.57 [2.99-5.71] | -1.1962 | 0.2316 |
| FGF (pg/mL) | 12.68 [4.55-21.63] | 16.61 [4.55-23.26] | -0.6986 | 0.4848 |
| G-CSF (pg/mL) | 59.83 [40.36-86.73] | 71.35 [52.62-109.62] | -1.8984 | 0.0577 |
| GM-CSF (pg/mL) | 0.00 [0.00-0.00] | 0.00 [0.00-0.00] | 1.4044 | 0.1602 |
| IFN-γ (pg/mL) | 6.61 [4.73-9.59] | 7.69 [5.61-11.05] | -1.6863 | 0.0917 |
| IL-1β (pg/mL) | 0.35 [0.00-1.43] | 0.53 [0.14-1.84] | -0.8948 | 0.3709 |
| IL1-RA (pg/mL) | 0.00 [0.00-135.38] | 0.00 [0.00-226.94] | -1.2088 | 0.2267 |
| IL-2 (pg/mL) | 1.06 [0.00-3.74] | 1.06 [0.00-4.36] | -0.5243 | 0.6001 |
| IL-4 (pg/mL) | 0.32 [0.00-0.68] | 0.63 [0.00-0.99] | -2.1481 | 0.0317 |
| IL-5 (pg/mL) | 0.00 [0.00-102.42] | 0.00 [0.00-114.12] | -0.1084 | 0.9136 |
| IL-6 (pg/mL) | 7.66 [3.13-19.51] | 17.87 [8.17-39.87] | -3.3250 | 0.0009 |
| IL-7 (pg/mL) | 7.44 [1.93-18.09] | 8.57 [1.93-25.07] | -1.1357 | 0.2561 |
| IL-8 (pg/mL) | 15.26 [10.84-25.05] | 22.04 [16.50-35.79] | -3.6703 | 0.0002 |
| IL-9 (pg/mL) | 446.49 [391.10-529.21] | 469.28 [405.64-539.06] | -0.3455 | 0.7297 |
| IL-10 (pg/mL) | 1.25 [0.00-3.91] | 2.28 [0.52-5.67] | -2.1681 | 0.0302 |
| IL-12 (pg/mL) | 2.21 [0.00-4.68] | 1.65 [0.00-3.96] | 0.7296 | 0.4657 |
| IL-13 (pg/mL) | 0.00 [0.00-0.49] | 0.24 [0.00-0.68] | -1.4539 | 0.1460 |
| IL-15 (pg/mL) | 0.00 [0.00-229.24] | 0.00 [0.00-271.68] | -0.6564 | 0.5116 |
| IL-17 (pg/mL) | 4.37 [2.91-5.95] | 4.41 [3.33-6.20] | -0.6518 | 0.5145 |
| IP-10 (pg/mL) | 3117.30 [1993.37-5384.47] | 6964.02 [4616.95-10457.34] | -5.2205 | <0.0001 |
| MCP-1 (pg/mL) | 73.46 [40.74-99.83] | 110.84 [52.14-203.81] | -2.8661 | 0.0042 |
| MIP-1α (pg/mL) | 1.59 [0.95-2.40] | 1.95 [1.26-3.25] | -2.0564 | 0.0397 |
| MIP-1β (pg/mL) | 226.33 [194.67-253.07] | 224.25 [193.76-263.00] | -0.2658 | 0.7904 |
| PDGF (pg/mL) | 1294.37 [719.67-2587.88] | 1399.54 [653.99-2487.55] | 0.0775 | 0.9382 |
| RANTES (pg/mL) | 5290.36 [3133.84-10501.29] | 5729.65 [2483.56-9480.44] | 0.1617 | 0.8716 |
| TNF-α (pg/mL) | 21.28 [15.42-25.97] | 20.10 [15.42-25.97] | 0.0709 | 0.9435 |
| VEGF (pg/mL) | 168.66 [0.00-273.00] | 211.99 [58.45-287.78] | -0.8201 | 0.4121 |
Table 8.
Routine laboratory findings and multiplex quantifications after 7 days of hospitalization in patients who had a faster clinical recovery (discharged or reaching NEWS2 ≤ 2 for at least 24 hours within 14 days) vs. all the other patients. Data are expressed as median (IQR). Bold text highlights the statistically significant results.
| Laboratory findings after 7 days (t7) | Faster clinical recovery (n = 76) | All other patients (n = 37) | Z | p value |
|---|---|---|---|---|
| Hemoglobin (g/dL) | 13.5 [12.5-15.0] | 13.6 [12.3-14.2] | 0.9946 | 0.3199 |
| RDW-CV (%) | 13.0 [12.5-13.6] | 13.4 [12.9-13.9] | -2.1164 | 0.0343 |
| White blood cells (cell count × 103/μL) | 9.40 [7.22-10.91] | 10.36 [8.32-12.13] | -1.7866 | 0.0740 |
| Neutrophils (cell count × 103/μL) | 6.79 [5.59-8.37] | 8.99 [7.22-10.55] | -3.3804 | 0.0007 |
| Eosinophils (cell count × 103/μL) | 0.02 [0.01-0.08] | 0.02 [0.00-0.04] | 1.3076 | 0.1910 |
| Lymphocytes (cell count × 103/μL) | 1.50 [0.85-1.95] | 0.62 [0.39-1.03] | 4.4788 | 0.0001 |
| Neutrophil/lymphocyte ratio | 4.82 [3.16-8.98] | 13.19 [7.74-32.29] | -4.8490 | 0.0001 |
| Platelets (cell count × 103/μL) | 348 [303-417] | 302 [260-371] | 2.2241 | 0.0261 |
| ALT (U/L) | 63 [38-112] | 41 [27-69] | 2.4130 | 0.0158 |
| AST (U/L) | 30 [24-45] | 30 [23-40] | 0.5323 | 0.5945 |
| Bilirubin (mg/dL) | 0.6 [0.5-0.9] | 0.7 [0.6-0.9] | -0.3411 | 0.7330 |
| Creatinine (mg/dL) | 0.72 [0.62-0.85] | 0.73 [0.55-0.83] | 0.5298 | 0.5963 |
| Glomerular filtration rate (mL/min) | 98 [88-107] | 91 [69-102] | 1.9951 | 0.0460 |
| CRP (mg/dL) | 0.8 [0.3-1.9] | 3.7 [2.3-7.8] | -6.1875 | <0.0001 |
| LDH (U/L) | 551 [449-650] | 645 [561-819] | -3.3632 | 0.0008 |
| Erythrocyte sedimentation rate (mm/h) | 25 [16-43] | 50 [32-63] | -3.1074 | 0.0019 |
| Troponin I (ng/mL) | 4 [2-7] | 6 [3-22] | -2.2200 | 0.0264 |
| Ferritin (ng/mL) | 751 [452-998] | 793 [520-1346] | -1.2909 | 0.1968 |
| D-dimer (μg/L) | 903 [567-1438] | 1755 [1132-2639] | -3.5792 | 0.0003 |
| Albumin (g/dL) | 3.6 [3.4-3.8] | 3.4 [3.2-3.7] | 1.9968 | 0.0458 |
| Eotaxin (pg/mL) | 5.06 [3.59-7.92] | 4.26 [3.68-6.09] | 0.8166 | 0.4142 |
| FGF (pg/mL) | 19.63 [10.82-28.33] | 20.40 [12.68-28.66] | -0.3791 | 0.7046 |
| G-CSF (pg/mL) | 78.28 [59.83-98.47] | 93.75 [77.86-156.42] | -2.5911 | 0.0096 |
| GM-CSF (pg/mL) | 0.00 [0.00-0.00] | 0.00 [0.00-0.00] | 1.7318 | 0.0833 |
| IFN-γ (pg/mL) | 6.14 [3.82-9.72] | 9.57 [6.61-13.09] | -2.8483 | 0.0044 |
| IL-1β (pg/mL) | 0.38 [0.00-1.09] | 0.59 [0.00-1.43] | -0.3944 | 0.6933 |
| IL1-RA (pg/mL) | 0.00 [0.00-117.10] | 0.00 [0.00-168.65] | -1.1670 | 0.2432 |
| IL-2 (pg/mL) | 0.79 [0.00-2.77] | 2.01 [0.00-4.22] | -0.5259 | 0.5989 |
| IL-4 (pg/mL) | 0.58 [0.14-0.92] | 0.68 [0.00-1.10] | -0.2517 | 0.8013 |
| IL-5 (pg/mL) | 0.00 [0.00-72.05] | 0.00 [0.00-72.05] | 0.2073 | 0.8357 |
| IL-6 (pg/mL) | 2.13 [0.00-6.63] | 9.60 [5.39-17.71] | -4.3785 | 0.0001 |
| IL-7 (pg/mL) | 1.93 [0.00-18.09] | 9.00 [0.91-22.56] | -1.4369 | 0.1507 |
| IL-8 (pg/mL) | 9.42 [6.49-14.56] | 21.19 [11.85-28.97] | -4.0087 | 0.0001 |
| IL-9 (pg/mL) | 460.38 [403.05-520.05] | 460.82 [368.17-516.36] | 0.1341 | 0.8933 |
| IL-10 (pg/mL) | 0.97 [0.00-3.63] | 2.05 [0.10-4.88] | -1.3278 | 0.1842 |
| IL-12 (pg/mL) | 1.34 [0.00-3.96] | 1.56 [0.00-3.96] | 0.0000 | 1.0000 |
| IL-13 (pg/mL) | 0.00 [0.00-0.79] | 0.00 [0.00-0.35] | 0.6089 | 0.5426 |
| IL-15 (pg/mL) | 0.00 [0.00-172.77] | 0.00 [0.00-84.08] | 0.6610 | 0.5029 |
| IL-17 (pg/mL) | 4.73 [3.65-6.49] | 4.94 [3.42-6.37] | -0.1294 | 0.8970 |
| IP-10 (pg/mL) | 668.26 [377.20-1164.56] | 1897.80 [1022.41-4465.42] | -4.4249 | 0.0001 |
| MCP-1 (pg/mL) | 59.34 [38.81-100.05] | 106.19 [60.87-359.83] | -3.2373 | 0.0012 |
| MIP-1α (pg/mL) | 2.23 [1.51-2.69] | 3.25 [2.40-4.36] | -3.5158 | 0.0004 |
| MIP-1β (pg/mL) | 234.90 [207.93-256.98] | 230.67 [181.26-246.32] | 0.9721 | 0.3310 |
| PDGF (pg/mL) | 2270.91 [1463.58-3558.49] | 1918.40 [929.93-3491.39] | 0.9099 | 0.3629 |
| RANTES (pg/mL) | 7255.70 [4279.12-12430.58] | 7635.33 [2738.21-10391.71] | 0.5172 | 0.6050 |
| TNF-α (pg/mL) | 23.77 [16.92-28.63] | 19.89 [15.42-25.97] | 1.1116 | 0.2663 |
| VEGF (pg/mL) | 105.40 [0.00-188.97] | 119.46 [0.00-264.76] | -1.0418 | 0.2975 |
Again, multivariate models were built to identify the laboratory findings better identifying patients with a faster recovery (hospital discharge or NEWS ≤ 2 within 14 days of hospitalization) (Tables 9 and 10). Such analysis highlighted that patients with a more favorable prognosis showed lower IP-10 and neutrophil to lymphocyte ratio values and higher glomerular filtration rate at the time of hospital admission (t0) (Table 9). Interestingly, after correction for age, gender, and severity of clinical presentation (NEWS2 score and PiO2/FiO2 at baseline), only low IP-10 values appeared to be related to a favorable prognosis (Table 10). After 7 days of hospitalization, indeed, the only laboratory finding with prognostic significance for a more positive outcome was a low CRP value (Table 11) even after correction for age, gender, and severity of the disease (NEWS2 and PiO2/FiO2 at 7 days) (multivariate analysis: CRP β 0.4786, p = 0.0001; NEWS2 β -0.2143, p = 0.0602; PiO2/FiO2β 0.2175, p = 0.0622; age β -0.1319, p = 0.2723; sex β -0.0430, p = 0.6647).
Table 9.
Multivariate analysis for baseline routine laboratory findings and multiplex quantifications predicting a faster clinical recovery (hospital discharge or NEWS ≤ 2 within 14 days of hospitalization). Bold text highlights the statistically significant results.
| Prognostic biomarkers (t0) | β ∗ | p value |
|---|---|---|
| Glomerular filtration rate | 0.3828 | 0.0046 |
| IP-10 | -0.2388 | 0.0226 |
| Neutrophil/lymphocyte ratio | -0.2033 | 0.0138 |
| Creatinine | 0.2719 | 0.1363 |
| RDW-CV | -0.1229 | 0.1524 |
| D-dimer | -0.1115 | 0.2090 |
| IL-6 | -0.1222 | 0.1952 |
| MIP-1α | 0.2804 | 0.1768 |
| MCP-1 | -0.0815 | 0.4383 |
| Troponin I | -0.0861 | 0.5313 |
| IL-8 | -0.1307 | 0.3177 |
| Platelets | 0.0731 | 0.3932 |
| IL-10 | -0.1754 | 0.5810 |
| CRP | -0.0061 | 0.9450 |
| Lymphocytes | -0.0011 | 0.9882 |
| IL-4 | 0.0668 | 0.8293 |
Table 10.
Multivariate analysis for baseline routine laboratory findings and multiplex quantifications predicting a faster clinical recovery (hospital discharge or NEWS ≤ 2 within 14 days of hospitalization) including demographic and COVID-19 severity-related variables. Bold text highlights the statistically significant results.
| Predictors (t0) | β ∗ | p value |
|---|---|---|
| IP-10 | -0.2999 | 0.0004 |
| Age | -0.2653 | 0.0076 |
| PiO2/FiO2 | 0.1990 | 0.0085 |
| Neutrophil/lymphocyte ratio | -0.1490 | 0.4120 |
| NEWS2 | -0.0590 | 0.0602 |
| Sex | -0.0293 | 0.7031 |
| Glomerular filtration rate | -0.0168 | 0.8723 |
Table 11.
Multivariate statistical analysis for routine laboratory findings and multiplex quantifications predicting a faster clinical recovery (hospital discharge or NEWS ≤ 2 within 14 days of hospitalization) after 7 days of hospitalization. Bold text highlights the statistically significant results.
| Prognostic biomarkers (t7) | β ∗ | p value |
|---|---|---|
| CRP | -0.4455 | 0.0024 |
| Glomerular filtration rate | 0.2647 | 0.1159 |
| G-CSF | 3.8136 | 0.0612 |
| Troponin I | 0.1827 | 0.1591 |
| MIP-1α | -2.5994 | 0.1009 |
| LDH | 0.2445 | 0.1679 |
| IL-8 | -1.2701 | 0.2966 |
| IP-10 | -0.2288 | 0.2800 |
| Neutrophils | -0.1053 | 0.5295 |
| D-dimer | -0.3139 | 0.3026 |
| ALT | -0.0981 | 0.4299 |
| MCP-1 | 0.3322 | 0.3968 |
| IL-6 | 0.2213 | 0.5519 |
| Platelets | 0.0784 | 0.5607 |
| Lymphocytes | 0.0239 | 0.8613 |
| Neutrophil/lymphocyte ratio | -0.1745 | 0.4233 |
| Albumin | 0.0452 | 0.7511 |
| Erythrocyte sedimentation rate | 0.0770 | 0.5592 |
| IFN-γ | 0.0536 | 0.9560 |
| RDW-CV | -0.0024 | 0.8355 |
Finally, to calculate the accuracy of the identified prognostic biomarkers, ROC curves were drawn, and cut-off values were identified according to the corresponding AUC (Figures 1–3). For IP-10 measured at baseline, the best cut-off value (0.7 < AUC ≤ 0.9) was 4271 pg/mL: IP-10 higher than 4271 pg/mL predicted a worsening in clinical conditions (87% sensitivity, 66% specificity), while IP-10 lower than 4271 pg/mL predicted a more favorable disease evolution (71% sensitivity, 78% specificity) (Figures 1(a) and 1(b)).
Figure 1.
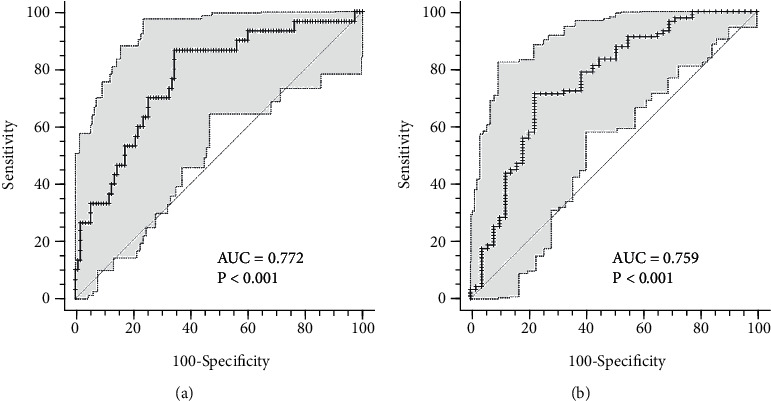
ROC curves for IP-10 at the time of hospital admission. (a) ROC curve predicting severe disease evolution; (b) ROC curve predicting shorter hospital stay. AUC : area under the curve.
Figure 2.
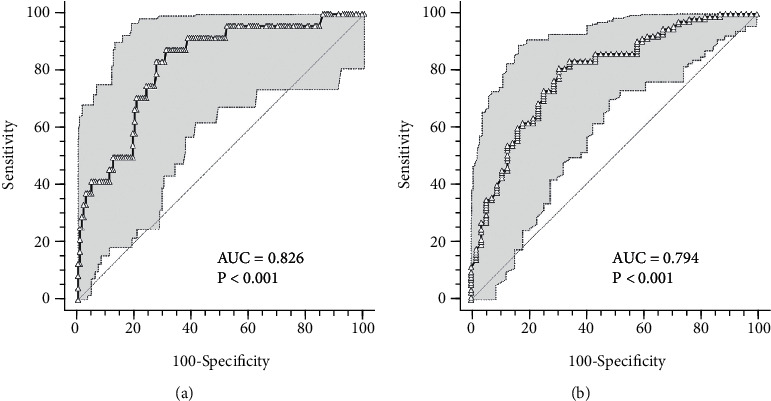
ROC curves for CRP after 7 days hospitalization. (a) ROC curve predicting severe disease evolution; (b) ROC curve predicting shorter hospital stay. AUC: area under the curve.
Figure 3.
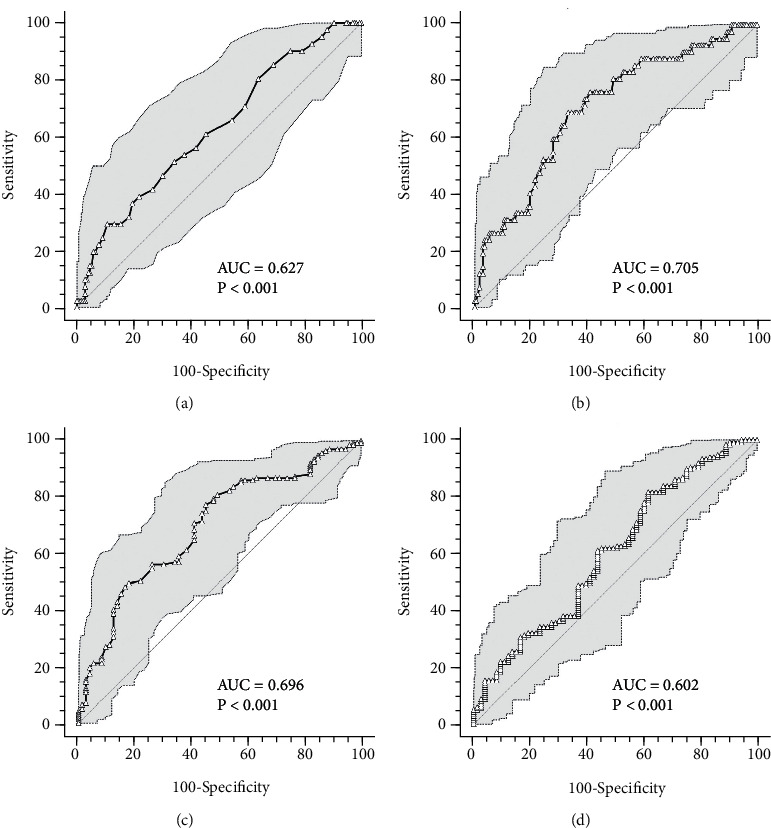
ROC curves for RDW-CV, platelet count, glomerular filtration rate, and neutrophil to lymphocyte ratio at the time of hospital admission. (a) RDW-CV ROC curve predicting severe disease evolution; (b) platelet count ROC curve predicting severe disease evolution; (c) glomerular filtration rate ROC curve predicting a shorter hospital stay; (d) neutrophil to lymphocyte ratio ROC curve predicting a shorter hospital stay. AUC: area under the curve.
With regard to CRP, ROC curve analysis highlighted that after 7 days of hospitalization, the cut-off allowing the most accurate patients' stratification (0.79 < AUC < 0.82) was 2.3 mg/mL: CRP higher than 2.3 mg/mL predicted a worsening in clinical conditions (83% sensitivity, 73% specificity), while CRP lower than 2.3 mg/mL predicted a shorter hospitalization (81% sensitivity, 69% specificity) (Figures 2(a) and 2(b)).
ROC curves were built also for baseline values of RDW-CV, platelet count, glomerular filtration rate, and neutrophil to lymphocyte ratio (Figure 3). RDW-CV values higher than 14.3% (AUC = 0.63, 29.3% sensitivity, and 89.4% specificity) (Figure 3(a)) and platelet count lower than 192 × 103/μL (AUC = 0.70, 69.1% sensitivity, and 66.9% specificity) (Figure 3(b)) predicted a more severe outcome, while glomerular filtration rate higher than 75 mL/min (AUC = 0.7, 77.4% sensitivity, and 54.8% specificity) (Figure 3(c)) and neutrophil to lymphocyte ratio lower than 11.78 × 103 (AUC = 0.60, 81.8% sensitivity, and 38.4% specificity) (Figure 3(d)) predicted a more favorable disease evolution.
4. Discussion
COVID-19 severe manifestations affect 10-20% of SARS-CoV-2-positive patients, due to a severe pneumonia depending on host's aberrant immune response [13, 15, 23]. To date, it is known that severe COVID-19 features are hypercytokinemia, hyperferritinemia, hemodynamic instability, and multiorgan failure [2, 4, 12, 13, 15, 23, 24]. Moreover, several specific abnormalities of metabolic parameters and extracellular vesicles have been observed correlated with disease refractoriness or evolution [25–28]. However, specific disease-related or severity-related accurate predictors are still lacking.
Therefore, in this observational prospective cohort, we screened a wide range of cytokines, chemokines, and routine laboratory markers to identify the best biomarkers predicting disease evolution and prognosis in hospitalized patients.
Since selected patients were hospitalized due to a moderate or severe respiratory failure, all were treated with a standardized protocol including steroids as dexamethasone or methylprednisolone and LMWH, aimed to manage the hyperinflammatory state and prothrombotic and hypercoagulable state observed in these patients [29].
By comparing the cytokine signature at hospital admission of the most severe patients to that of those experiencing milder forms of the disease, it appears that many bioactive molecules are involved (IL-4, IL-6, IL-8, IL-10, IP-10, MCP-1, MIP-1α, TNF-α, G-CSF, and IFN-γ); however, after multivariate statistical analysis, the only chemokine showing a clear predictive value at hospital admission is IP-10, showing both a positive association with greater disease severity and adverse prognosis, and an inverse association with faster recovery [30, 31]. Additionally, among laboratory biomarkers tested at 7 days, only CRP was predictive either of a worse or a good prognosis. IP-10 and CRP were the only laboratory parameters that retained a prognostic relevance even after correction for demographics (age and gender) and variables linked to disease severity (PiO2/FiO2 and NEWS2 score either at baseline or at 7 days) [14, 32–35].
IP-10 is an IFN-γ induced protein released by a large number of cells, many of which involved in the immune response, such as T cells, neutrophils, monocytes, and endothelial cells [36, 37]. IP-10 is a proinflammatory mediator involved in leukocyte homing to inflamed tissues and in the perpetuation of the inflammatory response, thus playing a pivotal role in inflammatory tissue damage [36–38]. Due to its known chemotactic action toward T cells, NK cells, monocyte/macrophages, and dendritic cells, this chemokine has been investigated as a potential biomarker for many pathological conditions (i.e., autoimmune diseases and viral infections) [36, 37]. During the 2002 SARS outbreak, many studies highlighted that higher IP-10 blood levels predicted an adverse outcome, an immunological signature also found in SARS-CoV-2 patients [31, 38–42]. In the present study, IP-10 showed a strong predictive power, with a ROC curve-based cut-off of 4271 pg/mL that, if confirmed in other studies, could be used as biomarker to identify patients needing strict clinical and therapeutic monitoring or even to drive the decision to start anti cytokine treatment.
Among the routine laboratory parameters, CRP, an acute phase protein, is known to be a strong indicator of COVID-19 severity [14, 32, 35]. The majority of the available studies confirms CRP as prognostic marker in the early stages of COVID-19 infection [43–46], even before interstitial pneumonia signs become evident at computerized tomography [47, 48]. In this study, CRP values after 7 days of hospitalization showed to have prognostic potential, with CRP higher than 2.3 mg/dL identifying patients undergoing clinical conditions worsening. Therefore, CRP value at 7 days seems to be the most reliable marker of treatment response in our population: we may speculate that the persistence of elevated CRP value at 7 days may be related to several critical conditions like the lack of corticosteroid response or eventually the development of secondary infections [44, 46, 47, 49]. Since biomarkers of treatment response in COVID-19 are lacking, to our knowledge, this result is novel, supporting the use of CRP measurement during the disease course to guide patient management.
As observed in many other studies, also for our study population, other baseline laboratory findings routinely assessed exist that contribute to a more accurate prognosis evaluation, such as RDW-CV, D-dimer levels, platelet count, and neutrophil to lymphocyte ratio at the time of hospital admission. RDW-CV is known to be a prognostic biomarker in several diseases [50–52] and also in COVID-19 patients [19, 53–55]; however, at ROC analysis, we obtained a very low sensitivity (RDW-CV values higher than 14.3% displayed 29.3% sensitivity and 89.4% specificity), therefore limiting its usefulness in clinical practice. Increased D-dimer levels and decreased platelet count were initially linked to the onset of disseminated intravascular coagulation (DIC), but today, thanks to the endlessly increase in scientific knowledge about COVID-19 disease, it is gaining attention the hypothesis of a coagulopathy with specific features different from the classical sepsis-related diffuse intravascular coagulopathy [56–58]. In this study, we observed that a baseline platelet count lower than 192 × 103/μL predicted a worsening in clinical conditions, while D-dimer ROC curve analysis did not allow the identification of a sufficiently accurate cut-off.
Both neutrophilia and lymphopenia are associated to COVID-19 evolution [5, 14, 21, 59, 60]. Higher neutrophil count is generally a nonspecific marker of severity, as it is related to both thromboembolic complications and systemic inflammatory responses [21], while lymphopenia is associated with a dysregulation of immune response [14]. The neutrophil to lymphocyte ratio thus magnifies the prognostic role of both events, with the ability to predict disease severity [19, 61, 62]. We confirmed a role for this biomarker since a low neutrophil to lymphocyte ratio (ROC curve-based cut − off = 11.78 × 103) was associated with a shorter hospital stay.
Our study has several limitations: first, it was based on a single-center enrollment, so that a multicenter validation of our results is needed to make clinical practice recommendations. Moreover, the study was conducted in clinical practice so that slight differences in patients' treatment may have occurred; however, all patients were followed and treated according to a standardized treatment protocol issued at our center, guiding, in particular, but not limited to, steroids and heparin duration and dose, limiting the bias of different clinical approaches.
5. Conclusions
The increasing number of COVID-19 patients requiring hospitalization stressed the national health systems all over the world, thus highlighting the need of early predictors of disease evolution, to assist the medical staff in the patient management as well as in monitoring patients' conditions during hospitalization. In this prospective observational cohort study, we showed that, after a wide screening of different biomarkers and correction for demographic and disease severity variables, baseline IP-10 values and CRP values after 7 days hospitalization are independent predictors of patients' prognosis and in-hospital disease course and may help the physicians to stratify patients treatments.
Acknowledgments
This research is founded by the Italian Ministero della Salute – Ricerca Finalizzata grant (BIAS study grant: COVID-2020-12371760).
Data Availability
Data are available upon reasonable request to be addressed to the corresponding author.
Conflicts of Interest
The authors have no conflict of interest to declare.
Authors' Contributions
Manuela Rizzi and Martina Costanzo contributed equally to this work.
References
- 1.Gomez-Mesa J. E., Galindo-Coral S., Montes M. C., Muñoz Martin A. J. Thrombosis and coagulopathy in COVID-19. Current Problems in Cardiology . 2021;46(3):p. 100742. doi: 10.1016/j.cpcardiol.2020.100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu B., Huang S., Yin L. The cytokine storm and COVID-19. Journal of Medical Virology . 2021;93(1):250–256. doi: 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellan M., Gavelli F., Hayden E., et al. Pattern of emergency department referral during the Covid-19 outbreak in Italy. Panminerva Medica . 2021;63(4) doi: 10.23736/S0031-0808.20.04000-8. [DOI] [PubMed] [Google Scholar]
- 4.Hu B., Guo H., Zhou P., Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nature Reviews Microbiology . 2021;19(3):141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison A. G., Lin T., Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Tends in Immunology . 2020;41(12):1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyerowitz E. A., Richterman A., Gandhi R. T., Sax P. E. Transmission of SARS-CoV-2: a review of viral, host, and environmental factors. Annals of Internal Medicine . 2021;174(1):69–79. doi: 10.7326/M20-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanyal S. How SARS-CoV-2 (COVID-19) spreads within infected hosts – what we know so far. Emerging Topics in Life Sciences . 2020;4(4):383–390. doi: 10.1042/ETLS20200165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rutter H., Parker S., Stahl-Timmins W., et al. Visualising SARS-CoV-2 transmission routes and mitigations. BMJ . 2021;375, article e065312 doi: 10.1136/bmj-2021-065312. [DOI] [PubMed] [Google Scholar]
- 9.Buitrago-Garcia D., Egli-Gany D., Counotte M. J., et al. Occurrence and transmission potential of asymptomatic and presymptomatic SARS-CoV-2 infections: a living systematic review and meta-analysis. PLoS Medicine . 2020;17(9, article e1003346) doi: 10.1371/journal.pmed.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oran D. P., Topol E. J. Prevalence of asymptomatic SARS-CoV-2 infection. A narrative review. Annals of Internal Medicine . 2020;173(5):362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellan M., Sainaghi P. P., Gavelli F., et al. Lessons from the Italian COVID-19 frontline. Minerva Medica . 2020;111(4):303–305. doi: 10.23736/S0026-4806.20.06664-1. [DOI] [PubMed] [Google Scholar]
- 12.Bhaskar S., Sinha A., Banach M., et al. Cytokine storm in COVID-19 – immunopathological mechanisms, clinical considerations and therapeutic approaches: the REPROGRAM consortium position paper. Frontiers in Immunology . 2020;11:p. 1648. doi: 10.3389/fimmu.2020.01648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine and Growth Factor Reviews . 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding L., Zhang W., Zhang F., et al. Prognostic role and diagnostic power of seven indicators in COVID-19 patients. Frontiers in Medicine . 2021;8:p. 733274. doi: 10.3389/fmed.2021.733274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cabaro S., D’Esposito V., Di Matola T., et al. Cytokine signature and COVID-19 prediction models in the two waves of pandemics. Scientific Reports . 2021;11(1):p. 20793. doi: 10.1038/s41598-021-00190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polverino F., Stern D. A., Ruocco G., et al. Comorbidities, cardiovascular therapies, and COVID-19 mortality: a nationwide, Italian observational study (ItaliCO) Frontiers in cardiovascular medicine . 2020;7, article 585866 doi: 10.3389/fcvm.2020.585866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellan M., Patti G., Hayden E., et al. Fatality rate and predictors of mortality in an Italian cohort of hospitalized COVID-19 patients. Scientific reports . 2020;10(1):p. 20731. doi: 10.1038/s41598-020-77698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinato D. J., AJX L., Biello F., et al. Presenting features and early mortality from SARS-CoV-2 infection in cancer patients during the initial stage of the COVID-19 pandemic in Europe. Cancers . 2020;12(7):p. 1841. doi: 10.3390/cancers12071841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellan M., Azzolina D., Hayden E., et al. Simple parameters from complete blood count predict in-hospital mortality in COVID-19. Disease Markers . 2021;2021:7. doi: 10.1155/2021/8863053.8863053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corradini E., Ventura P., Ageno W., et al. Clinical factors associated with death in 3044 COVID-19 patients managed in internal medicine wards in Italy: results from the SIMI-COVID-19 study of the Italian Society of Internal Medicine (SIMI) Internal and emergency medicine . 2021;16(4):1005–1015. doi: 10.1007/s11739-021-02742-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J., He X., Yuan Y., et al. Meta-analysis investigating the relationship between clinical features, outcomes, and severity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pneumonia. American Journal of Infection Control . 2021;49(1):82–89. doi: 10.1016/j.ajic.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gidari A., De Socio G. V., Sabbatini S., Francisci D. Predictive value of National Early Warning Score 2 (NEWS2) for intensive care unit admission in patients with SARS-CoV-2 infection. Infectious Diseases . 2020;52(10):698–704. doi: 10.1080/23744235.2020.1784457. [DOI] [PubMed] [Google Scholar]
- 23.Shcherbak S. G., Anisenkova A. Y., Mosenko S. V., et al. Basic predictive risk factors for cytokine storms in COVID-19 patients. Frontiers in Immunology . 2021;12:p. 745515. doi: 10.3389/fimmu.2021.745515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castelli V., Cimini A., Ferri C. Cytokine storm in COVID-19: “when you come out of the storm, you won’t be the same person who walked in”. Frontiers in Immunology . 2020;11:p. 2132. doi: 10.3389/fimmu.2020.02132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barberis E., Timo S., Amede E., et al. Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to SARS-CoV-2. International Journal of Molecular Sciences . 2020;21(22):p. 8623. doi: 10.3390/ijms21228623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barberis E., Amede E., Tavecchia M., et al. Understanding protection from SARS-CoV-2 using metabolomics. Scientific Reports . 2021;11(1):p. 13796. doi: 10.1038/s41598-021-93260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barberis E., Vanella V. V., Falasca M., et al. Circulating exosomes are strongly involved in SARS-CoV-2 infection. Frontiers in Molecular Biosciences . 2021;8:p. 632290. doi: 10.3389/fmolb.2021.632290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cappellano G., Raineri D., Rolla R., et al. Circulating platelet-derived extracellular vesicles are a hallmark of Sars-Cov-2 infection. Cell . 2021;10(1):p. 85. doi: 10.3390/cells10010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braz-de-Melo H. A., Faria S. S., Pasquarelli-do-Nascimento G., Santos I. O., Kobinger G. P., Magalhes K. G. The use of the anticoagulant heparin and corticosteroid dexamethasone as prominent treatments for COVID-19. Frontiers in Medicine . 2021;8:p. 615333. doi: 10.3389/fmed.2021.615333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lev S., Fottesman T., Levin G. A., et al. Observational cohort study of IP-10’s potential as a biomarker to aid in inflammation regulation within a clinical decision support protocol for patients with severe COVID-19. PLoS One . 2021;16(1, article e0245296) doi: 10.1371/journal.pone.0245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y., Shen C., Li J., et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. The Journal of Allergy and Clinical Immunology . 2020;146(1):119–127.e4. doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosquera-Sulbaran J. A., Pedreañez A., Carrero Y., Callejas D. C-reactive protein as an effector molecule in Covid-19 pathogenesis. Reviews in Medical Virology . 2021;31(6, article e2221) doi: 10.1002/rmv.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howe H. S., Ling L. M., Elangovan E., et al. Plasma IP-10 could identify early lung disease in severe COVID-19 patients. Annals of the Academy of Medicine, Singapore . 2021;50(11):856–858. doi: 10.47102/annals-acadmedsg.2021154. [DOI] [PubMed] [Google Scholar]
- 34.Ali A. M., Rostam H. M., Fatah M. H., Noori C. M., Ali K. M., Tawfeeq H. M. Serum troponin, D-dimer, and CRP level in severe coronavirus (COVID-19) patients. Immunity, Inflammation and Disease . 2021 doi: 10.1002/iid3.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yitbarek G. Y., Ayehu G. W., Asnakew S., et al. The role of C-reactive protein in predicting the severity of COVID-19 disease: a systematic review. SAGE Open Medicine . 2021;9:205031212110507–205031212110508. doi: 10.1177/20503121211050755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antonelli A., Ferrari S. M., Giuggioli D., Ferrannini E., Ferri C., Fallahi P. Chemokine (C-X-C motif) ligand (CXCL)10 in autoimmune diseases. Autoimmunity Reviews . 2014;13(3):272–280. doi: 10.1016/j.autrev.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Lee E. Y., Lee Z. H., Song Y. W. The interaction between CXCL10 and cytokines in chronic inflammatory arthritis. Autoimmunity Reviews . 2013;12(5):554–557. doi: 10.1016/j.autrev.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y., Wang J., Liu C., et al. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19. Molecular Medicine . 2020;26(1):p. 97. doi: 10.1186/s10020-020-00230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tripathy A. S., Vishwakarma S., Trimbake D., et al. Pro-inflammatory CXCL-10, TNF-α, IL-1β, and IL-6: biomarkers of SARS-CoV-2 infection. Archives of Virology . 2021;166(12):3301–3310. doi: 10.1007/s00705-021-05247-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y., Zhang C., Huang F., et al. Elevated plasma levels of selective cytokines in COVID-19 patients reflect viral load and lung injury. National Science Review . 2020;7(6):1003–1011. doi: 10.1093/nsr/nwaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Velavan T. P., Meyer C. G. Mild versus severe COVID-19: laboratory markers. International Journal of Infectious Diseases . 2020;95:304–307. doi: 10.1016/j.ijid.2020.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J., Subbarao K. The immunobiology of SARS. Annual Review of Immunology . 2007;25(1):443–472. doi: 10.1146/annurev.immunol.25.022106.141706. [DOI] [PubMed] [Google Scholar]
- 43.Stringer D., Braude P., Myint P., et al. The role of C-reactive protein as a prognostic marker in COVID-19. International Journal of Epidemiology . 2021;50(2):420–429. doi: 10.1093/ije/dyab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L. C-reactive protein levels in the early stage of COVID-19. Médecine et Maladies Infectieuses . 2020;50(4):332–334. doi: 10.1016/j.medmal.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo X., Zhou W., Yan X., et al. Prognostic value of C-reactive protein in patients with coronavirus 2019. Clinical Infectious Diseases . 2020;71(16):2174–2179. doi: 10.1093/cid/ciaa641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen W., Zheng K. I., Liu Z., Yan Z., Xu C., Qiao Z. Plasma CRP level is positively associated with the severity of COVID-19. Annals of Clinical Microbiology and Antimicrobials . 2020;19(1):p. 18. doi: 10.1186/s12941-020-00362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luan Y.-Y., Yin C.-H., Yao Y. M., Yao Y.-M. Update advances on C-reactive protein in COVID-19 and other viral infections. Frontiers in Immunology . 2021;12:p. 720363. doi: 10.3389/fimmu.2021.720363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan C., Huang Y., Shi F., et al. C-reactive protein correlates with computed tomographic findings and predicts severe COVID-19 early. Journal of Medical Virology . 2020;92(7):856–862. doi: 10.1002/jmv.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sahu B. R., Kampa R. K., Padhi A., Panda A. K. C-reactive protein: a promising biomarker for poor prognosis in COVID-19 infection. Clinica Chimica Acta . 2020;509:91–94. doi: 10.1016/j.cca.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bellan M., Soddu D., Zecca E., et al. Association between red cell distribution width and response to methotrexate in rheumatoid arthritis. Reumatismo . 2020;72(1):16–20. doi: 10.4081/reumatismo.2020.1243. [DOI] [PubMed] [Google Scholar]
- 51.Bellan M., Giubertoni A., Piccinino C., et al. Red cell distribution width and platelet count as biomarkers of pulmonary arterial hypertension in patients with connective tissue disorders. Disease Markers . 2019;2019:7. doi: 10.1155/2019/4981982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soddu D., Sola D., Bellan M., et al. Red cell distribution width is a potential predictor of early relapse in polymyalgia rheumatica. Reumatismo . 2021;73(2):117–121. doi: 10.4081/reumatismo.2021.1395. [DOI] [PubMed] [Google Scholar]
- 53.Soni M., Gopalakrishnan R. Significance of RDW in predicting mortality in COVID‐19—an analysis of 622 cases. International Journal of Laboratory Hematology . 2021;43(4):1–3. doi: 10.1111/ijlh.13526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee J. J., Montazerin S. M., Jamil A., et al. Association between red blood cell distribution width and mortality and severity among patients with COVID-19: a systematic review and meta-analysis. Journal of Medical Virology . 2021;93(4):2513–2522. doi: 10.1002/jmv.26797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Banon T., Wortsman J., Moshe S. B., et al. Evaluating red blood cell distribution width from community blood tests as a predictor of hospitalization and mortality in adults with SARS-CoV-2: a cohort study. Annals of Medicine . 2021;53(1):1410–1418. doi: 10.1080/07853890.2021.1968484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Émile C. Risque thrombotique de la Covid-19. OptionBio . 2021;32(629-630):20–21. doi: 10.1016/S0992-5945(21)00048-9. [DOI] [Google Scholar]
- 57.Guglielmetti G., Quaglia M., Sainaghi P. P., et al. "War to the knife" against thromboinflammation to protect endothelial function of COVID-19 patients. Critical Care . 2020;24(1):p. 365. doi: 10.1186/s13054-020-03060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helms J., Tacquard C., Severac F., et al. High risk of thrombosis in patients with severe SARS-CoV-2 infection: a multicenter prospective cohort study. Intensive care medicine . 2020;46:1089–1098. doi: 10.1007/s00134-020-06062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li S., Jiang L., Li X., et al. Clinical and pathological investigation of patients with severe COVID-19. JCI Insight . 2020;5(12, article e138070) doi: 10.1172/jci.insight.138070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J., Li S., Liu J., et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine . 2020;55, article 102763 doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng Z.-Y., Feng S. D., Chen G.-P., Wu J.-N. Predictive value of the neutrophil to lymphocyte ratio for disease deterioration and serious adverse outcomes in patients with COVID-19: a prospective cohort study. BMC Infectious Diseases . 2021;21(1):p. 80. doi: 10.1186/s12879-021-05796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simadibrata D. M., Calvin J., Wijaya A. D., Ibrahim N. A. A. Neutrophil-to-lymphocyte ratio on admission to predict the severity and mortality of COVID-19 patients: a meta-analysis. American Journal of Emergency Medicine . 2021;42:60–69. doi: 10.1016/j.ajem.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available upon reasonable request to be addressed to the corresponding author.


