Abstract
We demonstrate here that mitoribosomal protein synthesis, responsible for the synthesis of oxidative phosphorylation (OXPHOS) subunits encoded by the mitochondrial genome, occurs at high levels during glycolysis fermentation and in a manner uncoupled from OXPHOS complex assembly regulation. Furthermore, we provide evidence that the mitospecific domain of Mrp7 (bL27), a mitoribosomal component, is required to maintain mitochondrial protein synthesis during fermentation but is not required under respiration growth conditions. Maintaining mitotranslation under high-glucose-fermentation conditions also involves Mam33 (p32/gC1qR homologue), a binding partner of Mrp7’s mitospecific domain, and together they confer a competitive advantage for a cell’s ability to adapt to respiration-based metabolism when glucose becomes limiting. Furthermore, our findings support that the mitoribosome, and specifically the central protuberance region, may be differentially regulated and/or assembled, under the different metabolic conditions of fermentation and respiration. On the basis of our findings, we propose that the purpose of mitotranslation is not limited to the assembly of OXPHOS complexes, but also plays a role in mitochondrial signaling critical for switching cellular metabolism from a glycolysis- to a respiration-based state.
INTRODUCTION
Eukaryotic cells have the capacity to switch their metabolism from glycolysis to one dependent on mitochondrial oxidative phosphorylation (OXPHOS), also referred to as aerobic respiration. In mammals, a glycolysis-based metabolism, for example, is favored by proliferating embryonic and hematopoietic stem cells for their energy production, and a switch to OXPHOS-based metabolism is often correlated with their loss of pluripotency and a commitment to differentiation pathways (Suda et al., 2011; Ito and Suda, 2014; Snoeck, 2017; Papa et al., 2019; Tsogtbaatar et al., 2020; Zhu et al., 2020). On the other hand, differentiated postmitotic cells generally favor OXPHOS-based metabolism for their energy production. A preference for glycolysis-based metabolism, the so-called Warburg effect, is also characteristic of many cancer cells and considered to be an important determinant of cell proliferation and tumor formation (Park et al., 2020; Schiliro and Firestein, 2021). The yeast Saccharomyces cerevisiae can utilize a wide range of carbon sources for growth but prefers to ferment glucose through glycolysis-based metabolism when available, even if oxygen is present. However, upon transition to limiting glucose levels and/or when cultivated on a nonfermentable carbon source, such as glycerol and/or ethanol, the yeast cell adapts and switches to respiratory-based metabolism, producing ATP aerobically through mitochondrial OXPHOS. Switching from one metabolic state to another involves multiple cellular signaling pathways, and how specifically mitochondria may directly contribute to these signaling pathways remains unclear.
The OXPHOS enzymes are multisubunit complexes located within the mitochondrial inner membrane system and are mosaic in their genetic origin. The majority of OXPHOS protein subunits (and their assembly factors) are encoded by nuclear genes, and a minor subset, all enzymatically key OXPHOS subunits, are encoded by the mitochondrial genome (mtDNA). In S. cerevisiae the mtDNA-encoded OXPHOS proteins include cytochrome b (Cytb) of the cytochrome bc1 complex, Cox1, Cox2, and Cox3 of the cytochrome c oxidase (COX) enzyme, and Atp6, Atp8, and Atp9, F0-sector subunits of the F1F0-ATP synthase. Consequently, the synthesis and assembly of the OXPHOS enzymes require the expression of two genome systems and the translational activity of both cytosolic and mitochondrial ribosomes (mitoribosomes). Mitoribosomes display a number of conserved features from their bacterial ancestors; however, they have also acquired novel protein features (either novel proteins or extensions on conserved proteins), termed “mitospecific” elements, during their evolution. The activity of mitoribosomes has been suggested to require more complex levels of regulation to ensure temporal and physical coordination of both nuclear and mitochondrial genetic systems to coordinate OXPHOS complex assembly, and the novel mitospecific elements of the mitoribosomes may contribute to this regulation (Box et al., 2017).
Mitoribosomes have been proposed to exhibit translational plasticity to regulate OXPHOS complex assembly by ensuring that the synthesis levels of mtDNA-encoded proteins adapt to the availability of their imported nuclear-encoded partner proteins (Couvillion et al., 2016; Richter-Dennerlein et al., 2016; Dennerlein et al., 2017). The coupling of mitoribosomal protein synthesis with OXPHOS complex assembly demands has been proposed to reduce the burden of unnecessary protein synthesis/turnover and/or minimize the accumulation of unproductive and potentially reactive OXPHOS complex assembly intermediates (Khalimonchuk et al., 2010; Mick et al., 2011; McStay et al., 2013; Mayorga et al., 2016; Soto and Barrientos, 2016). Under glucose fermentation conditions, however, yeast cells actively repress respiratory metabolism, a process that involves glucose repression of the nuclear genes encoding OXPHOS protein subunits, resulting in an overall down-regulation of OXPHOS complex assembly and activity levels. Significant differences therefore exist in the content of OXPHOS-related enzymes in mitochondria isolated from yeast cells grown under (glucose) fermentation versus aerobic respiration conditions, and as demonstrated by a recent quantitative analysis of the mitochondrial proteome (Morgenstern et al., 2017). If regulated so as to adapt to the level of nuclear-encoded OXPHOS proteins being imported into the mitochondrion as previously discussed, a prediction would be that the translational activity of the mitoribosomes should be suppressed under the low-OXPHOS-demand conditions of glucose fermentation growth. This study, which represents a comparative analysis of cellular mitotranslation levels under different metabolic conditions, experimentally addresses this prediction.
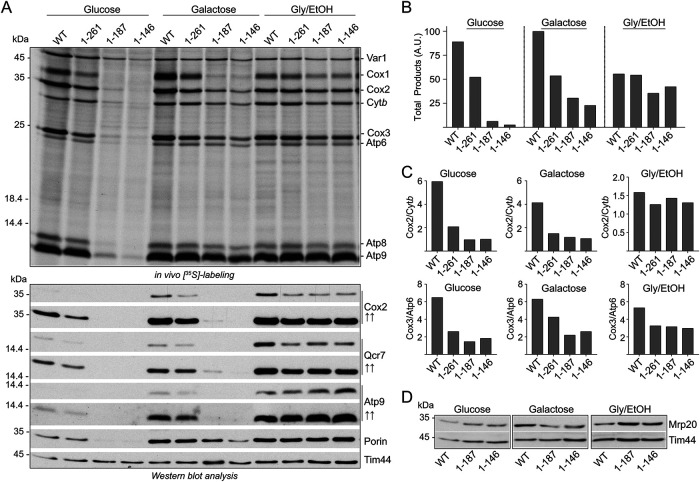
Previous analysis of the large mitoribosomal subunit (mtLSU) proteins MrpL36 (bL31) and MrpL35 (mL38) have pointed to a possible regulatory role of the central protuberance (CP) region of the mtLSU in coordinating mitoribosome translation with downstream OXPHOS assembly processes (Prestele et al., 2009; Box et al., 2017). In this study we have functionally analyzed the mitospecific domain of Mrp7 (bL27), another protein component of the CP and located close to the interface with the small mitoribosomal subunit (mtSSU) (Amunts et al., 2014; Desai et al., 2017). Mrp7 contains a L27-homology domain located at the extreme N-terminus of the mature protein (residues 28–108) (i.e., following its cleavable mitochondrial targeting signal, residues 1–27). The bulk of the Mrp7 protein (residues 109–371) represents its C-terminal mitospecific domain (Figure 1A) and is located toward the exterior surface of the CP region of the mtLSU (Amunts et al., 2014; Desai et al., 2017). The functional relevance of the mitospecific domain of Mrp7 and its possible role in regulating the mitoribosomal translational activity in response to differing demands for OXPHOS complex assembly represents the central focus of this study.
FIGURE 1:
C-terminal truncations in the mitospecific domain of Mrp7 result in a respiratory deficient phenotype. (A) Image of Mrp7 structure generated using PDB ID: 5MRC: blue denotes the L27-homology domain (corresponds to residues 28–108 of Mrp7 precursor protein), yellow denotes mitospecific domain (residues 109–371). Truncation points used for C-terminal deletion mrp7 mutants are indicated. (B) qPCR analysis of COX1 (mtDNA) to ACT1 (nDNA) content ratios compared in Mrp7 (+RNR1), mrp7(1-187) (+RNR1), and mrp7(1-146) (+RNR1) strains grown in glucose, galactose, or glycerol + ethanol synthetic minimal media. Ratios in each media normalized to corresponding wild-type (WT) strain in the same media. A.U., arbitrary units. (C) Tenfold serial dilutions of Δmrp7 strains with pRS413 plasmid encoding WT Mrp7 protein (MRP7) or one of the truncated mrp7 derivates (mrp7(1-261), mrp7(1-187), or mrp7(1-146)), compared with corresponding strains containing the YEplac112 plasmid encoding the Rnr1 protein (+RNR1), as indicated. Strains were taken from 24 h growths on selective SD plates and spotted onto YP plates containing either glucose (YPD) (2d) or glycerol + ethanol (YPGE) (3d and 6d) and incubated at either 30°C or 37°C.
We demonstrate here that under glucose fermentation conditions, expression of the mitochondrial genome, as measured by cellular levels of mitoribosomal translation, is not repressed. Rather, our findings indicate that mitoribosomal protein synthesis continues to occur at high levels under glucose fermentation conditions and uncoupled from regulation by the reduced demands of OXPHOS complex assembly. Moreover, we report that truncation of Mrp7’s mitospecific domain had severe consequences for the assembled mitoribosomes’ ability to perform protein synthesis under fermentation metabolism, but not under respiration growth conditions. Our findings point to the important role of Mrp7 and associated proteins, MrpL36 (bL31) and Mam33, in maintaining mitoribosomal translational capacity under fermentation conditions. Furthermore, we show that the mitospecific domain of Mrp7 interacts with Mam33 and their function to preserve mitoribosomal protein synthesis during fermentation is required for the cells’ ability to effectively adapt from fermentation to respiration growth.
RESULTS
Truncation of the C-terminal mitospecific domain of Mrp7 impacts its ability to support respiration growth
To experimentally analyze the functional relevance of the mitospecific domain of the yeast Mrp7 (bL27) protein, a series of C-terminal truncation mrp7 derivatives representing the successive removal of key structural regions of the Mrp7 protein were constructed and expressed from a centromeric plasmid (under the endogenous MRP7 promoter) in a Δmrp7 null mutant strain (Figure 1A and Supplemental Figure S1A). The mrp7(1-325) derivative represents a truncation of the extreme C-terminal region encompassing a short parallel β-strand structure in contact with a mitospecific extension of the 21S rRNA. Mrp7’s mitospecific domain resembles a “cork-screw and handle” (Amunts et al., 2014), and the mrp7(1-261) and mrp7(1-187) derivatives represent the removal of half or all of the “handle” feature of the Mrp7 protein, respectively. The fourth truncated derivative, mrp7(1-146), represents removal of both the “handle” and the “cork-screw” features, leaving a short α-helical region (residues 135–146) following the universally conserved mitospecific domain (residues 108–134).
The mrp7 mutants were tested for their ability to support OXPHOS metabolism, as evidenced by their ability to grow under nonfermentable carbon (glycerol/ethanol) conditions. Strains were initially cultivated under fermentation carbon (glucose) conditions before their respiratory-growth phenotypes were tested. The Mrp7 null mutant (Δmrp7) was unable to grow on the glycerol/ethanol medium, showing that the presence of Mrp7 is required to support respiratory-based growth (Supplemental Figure S1A). Expression of the mrp7(1-325) derivative was observed to restore growth of the Δmrp7 mutant to the same extent as the full-length (wild-type) Mrp7 protein, at both optimal (30°C) and elevated (37°C) stress temperatures, indicating that the extreme C-terminal region of Mrp7 (residues 326–371) was not required to support respiratory-based growth (Supplemental Figure S1A). (Note that owing to the lack of a detectable phenotype, the mrp7(1-325) mutant was not further analyzed in this study.) We observed a decreased capacity for respiratory-based growth with the mrp7(1-261) (at 37°C) and the mrp7(1-187) and mrp7(1-146) strains (both at 30°C and 37°C) (Supplemental Figure S1A). Subsequent analysis revealed that the mrp7(1-187) and mrp7(1-146) mutants displayed a tendency to lose their mtDNA when maintained on fermentable carbon sources (Supplemental Figure S2A), and we therefore constructed the Mrp7 wild-type and the mrp7 mutant strains in the presence of the RNR1 suppressor. RNR1, which encodes a subunit of the dNTP checkpoint enzyme ribonucleotide reductase, has previously been shown to suppress the mtDNA instability commonly observed in yeast mitoribosomal mutant strains, including the mrp7 null mutant strain (Zeng et al., 2018). To assess the retention of mtDNA in the mrp7(+RNR1) mutants, quantitative PCR (qPCR) was employed to compare the content of the COX1 gene (a measure for mtDNA content) with that of ACT1 (a nuclear DNA [nDNA] marker) (Goke et al., 2020). The mtDNA:nDNA content ratios in the mrp7(1-187) (+RNR1) and mrp7(1-146) (+RNR1) mutants (Figure 1B and Supplemental Figure S2B) were found to be similar to those of the wild-type Mrp7 (+RNR1) control strain, irrespective of the cultivation conditions, that is, fermentation (glucose or galactose) or respiration (glycerol/ethanol) growth conditions. We conclude therefore that in the presence of the RNR1 suppressor the mrp7 mutants display mtDNA stability under fermentation and respiration conditions.
Analysis of the mrp7 (+RNR1) strains revealed that the initially observed respiration growth phenotypes persisted (Figure 1C). The ability of the mrp7(1-261) (+RNR1), mrp7(1-187) (+RNR1), and mrp7(1-146) (+RNR1) strains to support respiratory-based growth at 37°C was significantly compromised. Both the mrp7(1-187) (+RNR1) and mrp7(1-146) (+RNR1) strains also had compromised respiration growth phenotypes at 30°C. The respiration growth behaviors of the mrp7(1-261), mrp7(1-187), and mrp7(1-146) strains in the presence of the RNR1 suppressor were indistinguishable from those without the RNR1 suppressor analyzed in parallel, confirming that the presence of the RNR1 suppressor did not influence the observed mrp7 growth phenotypes (Figure 1C). Note that all further analyses of the mrp7 mutants in this study were performed with the +RNR1 strains to exclude any possible mtDNA instability side effects, especially under fermentation growth conditions.
Although severely compromised, the mrp7(1-187) and mrp7(1-146) strains did retain a capacity for respiratory-based growth at 30°C, as their growth was noticeable at later time points (Figure 1C, compare 3-d with 6-d results at 30°C). The observed growth after the extended incubation period suggests that these mrp7 mutant strains either exhibit a pronounced adaptation lag coming from glucose to glycerol media and/or an overall slower respiration growth rate relative to wild-type control. Preadaptation of the mrp7(1-146) strain (and to a lesser extent the mrp7(1-187) strain) to nonfermentable carbon sources before phenotype testing was observed to improve its subsequent respiration growth at 30°C (Supplemental Figure S1B). As this growth advantage occurred in an RNR1-independent manner, we conclude that it is unlikely to be related to retention of mtDNA content, but rather may indicate that the C-terminal region of Mrp7 may play a role in the cell’s ability to adapt from fermentation to respiration metabolic states.
Taken together these results indicate that the C-terminal mitospecific domain of Mrp7 plays a role in Mrp7’s ability to support normal respiratory-based growth and in a manner that may also involve the ability of the yeast to adapt from fermentation to respiration metabolic conditions under normal growth temperature conditions.
Loss of the C-terminal mitospecific domain of Mrp7 results in a general OXPHOS defect
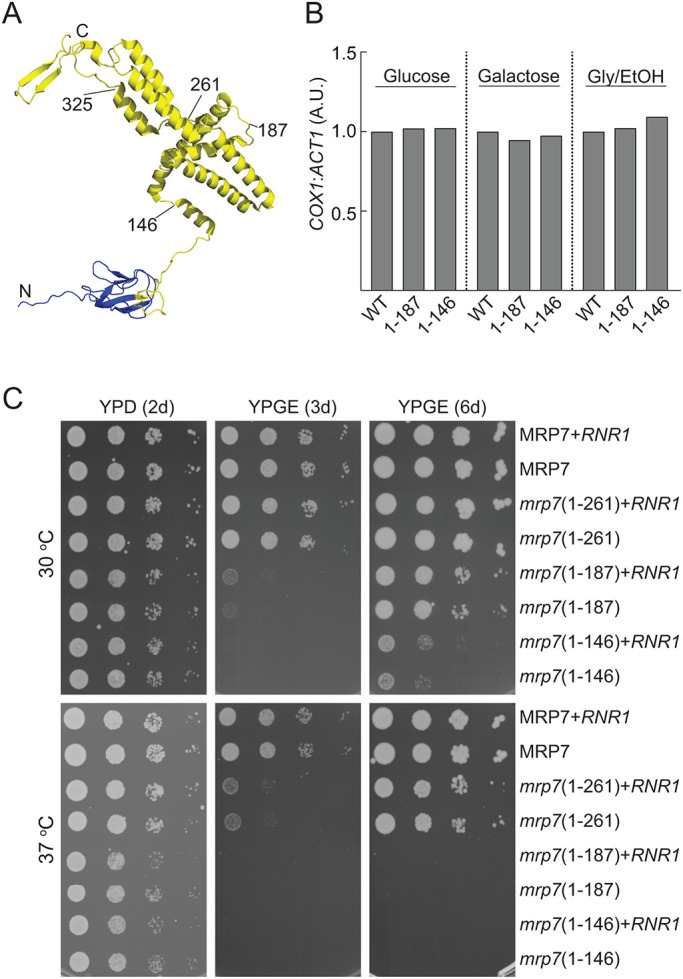
As the mrp7(1-146) and mrp7(1-187) strains retained some ability to grow on nonfermentable carbon sources at 30°C, albeit at reduced capacity, we can conclude that they are not completely deficient in the ability to assemble OXPHOS complexes. To further explore the impact of Mrp7 truncation on mitoribosomal translation and OXPHOS assembly during respiratory-based metabolism, mitochondria were isolated from each of the mrp7 (+RNR1) strains that had been adapted to and grown in glycerol/ethanol at the permissive temperature (Figure 2). In organello protein translation indicated that the overall translational capacity of the mitoribosomes was not adversely impacted in the mrp7 mutant mitochondria isolated from these respiratory grown cells. An alteration in the profile of proteins synthesized was, however, observed in the mrp7 mutant mitochondria, where an increased ratio in Cytb:Cox2 and Atp6:Cox3 synthesis relative to the wild-type control was recorded (Figure 2A). The steady state levels of the mitoribosomes (as indicated by analysis of Mrp20 [uL23], MrpL36 [bL31], and Mrp2 [uS14] levels) in the mrp7 mitochondria isolated from respiratory grown cells were similar to those of the wild-type control (Figure 2B), a finding consistent with their normal activity levels of mitochondrial translation. Efforts to directly assess the amount of truncated mrp7 derivatives in the isolated mitochondria were limited in success because the epitope of the available Mrp7 monoclonal antibody appears not to be present in the shorter two of the mrp7 derivatives (unpublished data). The Mrp7 monoclonal antibody did, however, recognize the mrp7(1-261) and mrp7(1-325) derivatives, and their levels were similar to those of the full-length wild-type Mrp7 protein. However, given the observed levels of mitotranslation activity in the glycerol/ethanol-isolated mrp7(1-187) and mrp7(1-146) mitochondria (Figure 2A) (and the steady state and assembly levels of mrp7(1-146) mitoribosomes; see later in Figure 4), we can indirectly conclude that the mrp7(1-187) and mrp7(1-146) mutant derivatives were expressed and present.
FIGURE 2:
mrp7 mutants maintain the ability to translate in respiration growth conditions yet exhibit defects in COX and cytochrome bc1 assembly. Mitochondria were isolated from Mrp7 wild-type (WT) (all +RNR1) and indicated mrp7 strains grown in SGE (Gly/EtOH) media at 30°C and analyzed in panels A–D. (A) In organello labeling in the presence of [35S]methionine was performed at 30°C for 15 min, followed by the addition of puromycin and cold methionine for an additional 5 min. [35S]methionine-labeled proteins were analyzed by SDS–PAGE, followed by autoradiography. (B) The steady state levels of the indicated proteins in the SGE isolated mitochondria (50 μg, 100 μg protein) were analyzed following SDS–PAGE and Western blotting with the indicated antibodies. Tim44 was used as a loading control. (C) Specific activities of the COX (left panel) and cytochrome bc1 (right panel) enzyme complexes were measured in the indicated SGE mitochondria as detailed in Materials and Methods. Quadruplicate activity measurements are presented as percentage of wild-type Mrp7 (WT) control, with means and SDs indicated. The significance between measurements was determined by one-way ANOVA using GraphPad Prism 9. Comparison of each mutant to the WT control resulted in p < 0.0001. (D) Mitochondria (30 μg protein) isolated from the indicated strains were solubilized in digitonin (1%) and subjected to BN-PAGE analysis, Western blotting, and immunedecoration with antibodies against an F1F0-ATP synthase (V) subunit (α-F1α/β). Dimeric and monomeric forms (Vdim and Vmon, respectively) are indicated.
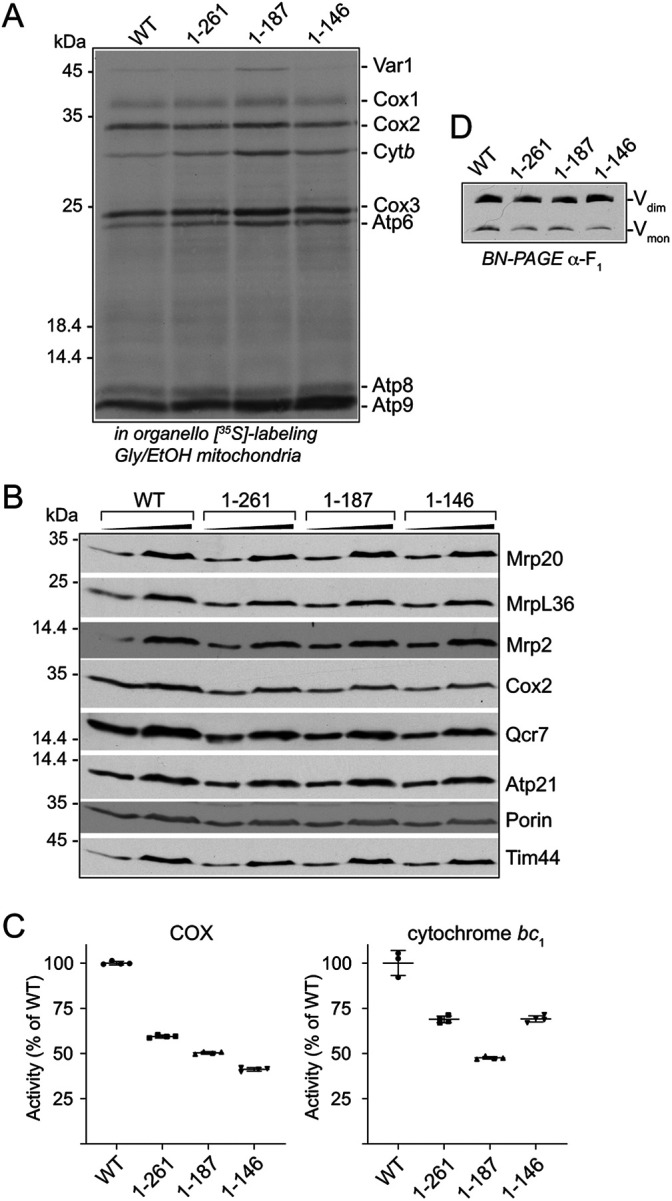
FIGURE 4:
mrp7 truncations result in altered CP assembly profile of glucose mitoribosomes. Mitochondria were isolated from the indicated Mrp7 wild-type (WT) and mrp7(1-146) strains (all +RNR1) grown in SD media at 30°C and used in panels A, B, and C, whereas those isolated from respiratory grown (SGE) cultures were used in panel D. (A) In organello labeling in the presence of [35S]methionine was performed in the indicated glucose mitochondria at 30°C for 15 min, followed by the addition of puromycin and cold methionine for an additional 5 min. The [35S]methionine-labeled proteins were analyzed by SDS–PAGE followed by autoradiography. (B) The steady state amounts of the indicated mitoribosomal proteins were analyzed in isolated glucose mitochondria (50 μg, 100 μg protein), as described in Figure 2B. Tim44 was used as a loading control. (C) Isolated WT and mrp7(1-146) glucose mitochondria (450 μg protein) were solubilized in an octyl-glucopyranoside (1.5%)-containing buffer and fractionated on a linear sucrose gradient (25%–40%), as described in Materials and Methods. Equal-volume fractions were collected (1 to 14, bottom to top of the gradient) and TCA precipitated and analyzed with SDS–PAGE. Western blotting with antibodies against the large mitoribosomal subunit (Mrp20, MrpL36) and the small mitoribosomal subunit (Mrp2) was performed, as indicated. (D) WT and mrp7(1-146) mitochondria (450 μg protein) isolated from strains grown in respiratory (SGE, Gly/EtOH) media at 30°C were lysed and fractionated and analyzed as detailed in C.
Western blotting of the mitochondrial extracts indicated that the steady state levels of subunits of the COX (Cox2) and cytochrome bc1 (Qcr7) complexes were partially reduced in all the mrp7 mutant mitochondria, including in the mrp7(1-261) mitochondria (Figure 2B). Consistently, COX enzyme activity levels were determined to be reduced in the mrp7(1-261) mitochondria and to a greater extent in the mrp7(1-187) and mrp7(1-146) mitochondria (Figure 2C). The cytochrome bc1 enzyme activity levels were also lower in the mrp7 mutant mitochondria, with levels lower in the mrp7(1-187) mitochondria than in the mrp7(1-146) mutant (Figure 2C), consistent with the previous observation that the mrp7(1-146) grew better than the mrp7(1-187) strain following prior adaptation to glycerol media (Supplemental Figure S1, A and B). The levels of F1F0-ATP synthase complex, the third OXPHOS complex containing mtDNA-encoded subunits, did not appear to be adversely affected in the mrp7 mutants, as indicated by both blue native (BN)-PAGE analysis of the complex (Figure 2D) and steady state analysis of the Atp21 (Su e) subunit (Figure 2B), a component of the F1F0-ATP synthase that is susceptible to turnover if assembly of the complex is perturbed.
Taken together, we conclude from these data that the truncation of the mitospecific domain of Mrp7 does not reduce the capacity of the mitoribosomes to support translation in mitochondria isolated from respiratory grown cells. Rather, our findings indicate that the observed respiration growth phenotypes of the mrp7 mutants may be attributed to impacts downstream of translation, that is, the stable assembly of the proteins synthesized by mitoribosomes into functional COX and cytochrome bc1 complexes.
Under fermentation conditions, mitochondrial translational levels are not coordinated with OXPHOS assembly and require the integrity of the Mrp7 mitospecific domain
Prior adaptation of the mrp7(1-146) strain to a nonfermentable carbon source partially improved its subsequent respiration growth (Supplemental Figure S1, A and B), suggesting that truncation of the mitospecific C-terminal region of Mrp7 may compromise a cell’s ability to transition from glycolysis-based fermentation to aerobic respiration–based growth conditions. Using an in vivo radiolabeling approach, we next explored the impact of truncation of Mrp7’s mitospecific domain on the ability of the mitoribosomes to perform protein synthesis under fermentation and respiration (glycerol/ethanol) growth conditions (Figure 3). For fermentation we compared two conditions, glucose, which represses mitochondrial OXPHOS complex assembly and thus respiration, and galactose, a nonrepressing fermentable sugar. Focusing first on the wild-type Mrp7 (+RNR1) control results, we observed that both the overall level and profile pattern of cellular mitoribosomal translation were not consistent between cells grown on the three different carbon sources (Figure 3, A, top panel, and B). In these control wild-type cells, the cellular levels of mitochondrial translation were unexpectedly lowest in respiratory-growth conditions (Figure 3, A, top panel, and B), that is, where demand for the assembly levels of OXPHOS complexes was the highest, as confirmed by parallel Western blotting of the cellular extracts with antibodies against OXPHOS enzyme subunits (Cox2/COX complex, Qcr7/cytochrome bc1 complex, and Atp9/F1F0-ATP synthase) (Figure 3A, bottom panel). Furthermore, mitochondrial translation was not repressed under glucose fermentation, despite that these metabolic conditions repress nuclear gene expression and assembly of high levels of OXPHOS complexes, and as confirmed by steady state levels of the Cox2, Qcr7, and Atp9 proteins. Indeed, kinetic analysis indicated that in vivo radiolabeling of mitochondrial translation products (in particular Var1, Cox1, Cox2, Cox3, Atp8, and Atp9) occurred faster under glucose fermentation conditions relative to respiratory-based (glycerol/ethanol) labeling conditions (Supplemental Figure S3). Additionally, the turnover rate of glucose-synthesized mitochondrial translation products was found to be faster than those synthesized under respiratory conditions (with the exception of Atp6 and Atp9) (Supplemental Figure S4), highlighting the increased vulnerability of the mtDNA-encoded proteins synthesized under conditions that do not promote their efficient assembly into productive OXPHOS enzyme complexes. Under galactose (nonrepressing fermentation) conditions, the level of mitochondrial translation output was the highest although an intermediate level of OXPHOS complexes was observed, in comparison to glucose- and glycerol/ethanol- grown cells. From these data it would appear that while OXPHOS complex assembly is repressed in glucose growth conditions, mitoribosomal translation activity is not, indicating that a coordinated expression of mtDNA-encoded and nuclear-encoded proteins in wild-type cells does not occur under fermentation conditions.
FIGURE 3:
mrp7 mutants are unable to maintain mitochondrial translation under fermentation conditions. (A) Indicated mrp7 strains (all +RNR1) adapted to either glucose, galactose, or glycerol + ethanol (Gly/EtOH) selective synthetic media (as described in Materials and Methods) were grown at 30°C to mid–log phase. The mitochondrial translation capacity was analyzed in vivo with the equivalent of OD600 0.6 absorbance units of whole cells for 10 min at 30°C in the presence of cycloheximide (0.3 mg/ml) and [35S]methionine. Cells were isolated and solubilized, and newly synthesized proteins were analyzed by SDS–PAGE followed by autoradiography (top panel). The equivalent of OD600 0.2 of cells were loaded on each gel. Steady state levels of mitochondrial proteins from these cell extracts were also analyzed by Western blotting and immunedecoration with the indicated antibodies (bottom panel). Additional darker exposures (indicated by “↑↑”) are also shown for Cox2, Qcr7, and Atp9 decorations so that their levels in glucose-grown cells can be seen. (B) The total of newly synthesized proteins (i.e., the sum of Var1, Cox1, Cox2, Cox3, Cytb, Atp6, Atp8, and Atp9 synthesis signals) from the top panel in A were quantified by phosphorimaging, with the total products expressed as a percentage of wild-type Mrp7 (WT) in galactose conditions. A.U., arbitrary units. (C) Newly synthesized proteins in the top panel in A were quantified by phosphorimaging, and the levels of Cox2 and Cox3 were expressed as a ratio to the levels of Cytb or Atp6, respectively, synthesized in the same 10 min time period. (D) WT, mrp7(1-187), and mrp7(1-146) strains were adapted to and grown in the indicated synthetic media, and the equivalent of OD600 0.2 absorbance units of whole cells were isolated and solubilized. Steady state levels of Mrp20 in these cell extracts were determined following SDS–PAGE and Western blotting with the indicated antibodies. Tim44 was used as a loading control within each growth medium. Adaptation was performed for each strain as indicated in A.
Consistent with the in organello translation results reported earlier (Figure 2A), the mrp7 mutant strains (all +RNR1) grown under respiratory conditions largely retained their capacity for mitochondrial translation (Figure 3, A, top panel, and B). The translational capacity of the mrp7(1-261) mutant was reduced in both glucose and galactose relative to the wild-type control, but not in glycerol/ethanol-grown cells, indicating that the performance of the mitoribosome was partially compromised by this truncation in the Mrp7 protein under fermentation conditions only. A more pronounced deficit in cellular mitoribosomal translation under fermentation conditions, especially under glucose-repressing conditions, was observed in the mrp7(1-187) and mrp7(1-146) cells, where overall mitochondrial translation, with the possible exception of Var1 synthesis, was strongly reduced (Figure 3, A, top panel, and B). Parallel Western blotting of Cox2, Qcr7, and Atp9 proteins indicated a strong reduction in OXPHOS complexes in the glucose- and galactose-grown mrp7 mutant cells, relative to their wild-type controls, particularly in the mrp7(1-187) and mrp7(1-146) mutants, where it correlated with the observed defect in mitotranslation under fermentation conditions (Figure 3A, bottom panel).
Under the glycerol/ethanol (respiratory) growth conditions, cellular contents of Cox2 and Qcr7, but not Atp9, were somewhat reduced in all of the mrp7 mutants (relative to the wild-type control), indicating that although the mutants largely retained their capacity to synthesize the mtDNA-encoded OXPHOS proteins, their subsequent efficient assembly into stable COX and cytochrome bc1 complexes was perturbed. This analysis of the cellular OXPHOS complex levels in the respiratory grown mrp7 mutant cells was consistent with analysis performed with the mrp7 mitochondria isolated from the glycerol/ethanol-grown cells, described above (Figure 2, B–D).
The comparative in vivo radiolabeling analysis also enabled us to observe that the profile of newly synthesized proteins in the wild-type control cells differed between the fermentation and respiratory conditions. In glucose- (and galactose)-cultivated wild-type cells, an increased ratio of Cox2:Cytb (and to a lesser extent Cox3:Atp6), relative to their respiratory grown counterparts, was observed (Figure 3, A, top panel, and C). The mrp7 mutant mitoribosomes, however, adopted a similar synthesis profile preference irrespective of their metabolic environment. Indeed, the profile of translation, that is, the decreased Cox2:Cytb and Cox3:Atp6 synthesis ratios observed in the mrp7 mutants under fermentation conditions, resembled that of the wild-type control (and of the mrp7 mutants) achieved under respiration conditions, where a decrease in Cox2:Cytb synthesis was recorded (relative to glucose translation).
Finally, the observed decreased translation in the mrp7(1-187) and mrp7(1-146) mutants under glucose fermentation conditions was not attributed to the lack of mitoribosomes in these cells, as the Mrp20/uL23 levels in these mrp7 mutants were found to be similar (or even elevated) relative to those of the wild-type control (Figure 3D); the stability of Mrp20 is secured by its assembly into the mtLSU (Kaur and Stuart, 2011). Furthermore, the inhibition of mitotranslation in the glucose mrp7(1-187) and mrp7(1-146) mutants was also not due to a possible reduction in Var1 synthesis. Var1 is a mtDNA-encoded component of the mtSSU, and thus a decrease in its synthesis in turn potentially may compromise mitoribosomal function. Allotopic expression of a universal Var1 (Var1u) protein (i.e., as a functional chimera with a mitochondrial targeting signal) (Seshadri et al., 2020) did not restore the translational capacity of glucose-grown mrp7(1-187) and mrp7(1-146) cells, indicating that the mtVar1 levels were not the limiting factor for mitotranslation in these mutants under fermentation conditions (Supplemental Figure S5).
In summary, we conclude that mitoribosomes display differences in their level of translational output and the profile of proteins synthesized under fermentation and respiration conditions. Furthermore, our findings indicate that the mitospecific C-terminal domain of Mrp7 is required to support the translational capacity and synthesis profile of the mitoribosomes under fermentation but not respiration growth conditions. Together these findings suggest that mitoribosomes may be differentially regulated under different metabolic conditions and the mitospecific C-terminal region of the Mrp7 protein may play a role in this process.
Mitoribosomes assemble in the glucose-grown mrp7(1-146) mutant, albeit with an altered profile
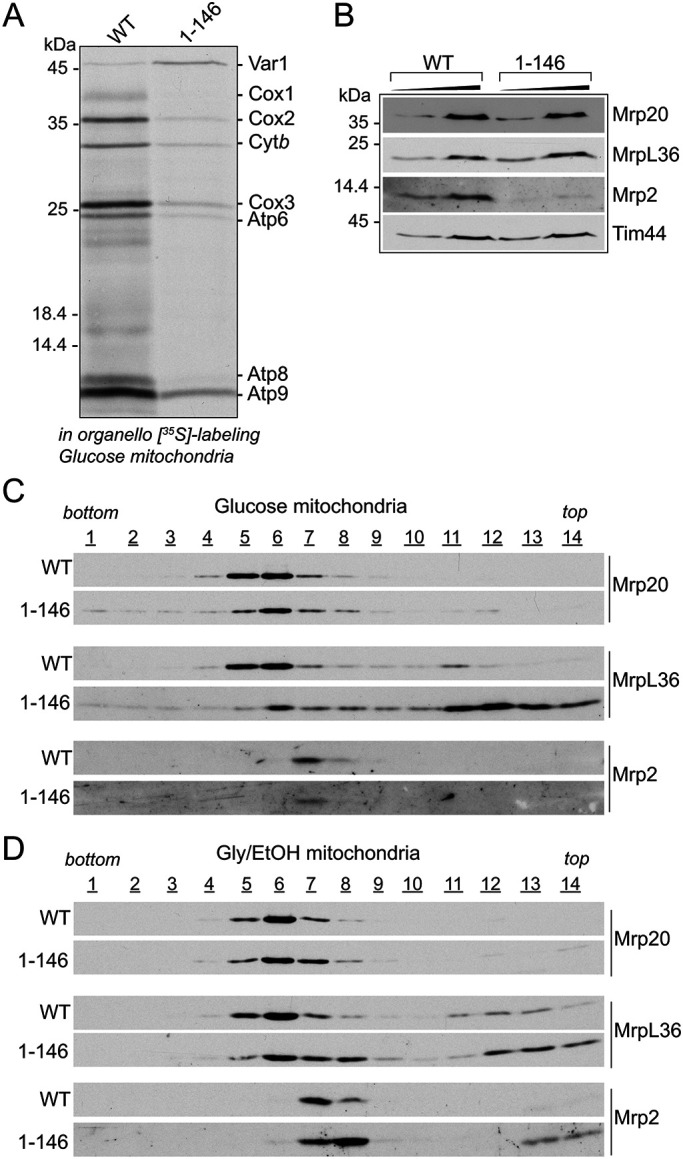
The normal levels of the Mrp20 protein in the glucose-grown mrp7(1-146) (and mrp7(1-187)) mutant cells (Figure 3D) would suggest that mitoribosomes are assembled and stable in these mutants when grown under fermentation conditions, but that their translational activity is compromised. To directly probe the mrp7(1-146) fermentation mitoribosomes, their activity, content, and assembly state, mitochondria were isolated from glucose-grown mrp7(1-146) and the control wild-type Mrp7 cells. In organello translation confirmed that mitochondrial translational capacity (with the exception of mtVar1 synthesis) was greatly inhibited in the isolated glucose mrp7(1-146) mitochondria (Figure 4A), consistent with the in vivo glucose mitotranslational analysis performed in Figure 3A. The steady state analysis of Mrp20 (uL23 and located at the polypeptide exit pore of the mtLSU) and MrpL36 (bL31, located in the CP region of the mtLSU) in the isolated glucose-grown mrp7(1-146) mitochondria indicated that the mtLSU levels were similar to those of the parallel Mrp7 wild-type control; however, a decrease in the level of the mtSSU component Mrp2 (uS14) was observed in the glucose mrp7(1-146) mitochondria (Figure 4B). Sucrose gradient centrifugation of mitochondrial extracts confirmed that the mtLSU was assembled in the glucose mrp7(1-146) mitochondria; however, the fractionation behavior of the Mrp20 and MrpL36 proteins indicated some differences from the mtLSU of the glucose wild-type Mrp7 control analyzed in parallel (Figure 4C). First, Mrp20 largely peaked in fractions 5 and 6 in the wild-type control, whereas in the mrp7(1-146) sample it was divided between fractions 5 and 8. This minor shift in the fractionation profile of Mrp20 suggests that a population of mtLSU may exist in in the mrp7(1-146) mitochondria that was slightly smaller than that of the wild-type control. Second, in the wild-type control the fractionation behavior of MrpL36 mirrored that of Mrp20; however, in the mrp7(1-146) glucose mitoribosomes, only a minor subpopulation of the MrpL36 protein peaked along with Mrp20/mtLSU fractions. Instead, the vast majority of the MrpL36 protein was recovered toward the top of the gradient in fractions 11–14 (i.e., in smaller complexes) on the mrp7(1-146) gradient, indicating that the stable assembly of MrpL36 into the CP region of the mtLSU was perturbed in the glucose mrp7(1-146) mitoribosomes (Figure 4C). Taking both the Mrp20 and MrpL36 data together, we conclude that the gross inhibition of mitochondrial translation observed in the glucose-grown mrp7(1-146) cells is not attributed to an absence of mitoribosome assembly. Rather, the altered profile of MrpL36 assembly indicates that the integrity and function of the CP region of the mtLSU may be impacted in the mrp7(1-146) mitochondria from glucose-grown cells, a perturbation that may contribute to the observed translation defect under fermentation conditions.
The assembly states of mitoribosomes isolated from respiratory grown (glycerol/ethanol) Mrp7 wild-type and mrp7(1-146) cells was next compared with their glucose-grown counterparts to assess whether the Mrp7 truncation had a differential impact on the MrpL36 assembly in the CP region of respiratory mitoribosomes, where mitotranslation ability is maintained. The mtLSU from respiratory grown wild-type control cells (as judged by Mrp20 and MrpL36 fractionation behaviors) peaked more robustly in fraction 6 (Figure 4D), displaying slight profile differences from its glucose counterpart, which was distributed more between fractions 5 and 6 (Figure 4C). Similar to that observed for the glucose mrp7(1-146) mitoribosomes, a subtle shift in the mtLSU, as evidenced from the Mrp20 profile (i.e., more was recovered in fractions 7 and 8), was observed for the respiratory mrp7 mutant mitoribosomes. However, in contrast to that observed with the glucose mrp7(1-146) mitoribosomes, a significantly larger fraction of the total MrpL36 remained assembled with the mtLSU from the respiratory mrp7(1-146) mitochondria, with a correspondingly smaller MrpL36 subpopulation recovered toward the top of the gradient (fractions 11–14) (Figure 4D). We also noticed a shift in the mtSSU profile (as indicated by profiling Mrp2) in the respiratory mitoribosomes from the mrp7(1-146) mutant, showing that truncation of the mitospecific domain of Mrp7 in this manner also impacted the mtSSU assembly.
Taken together, these data indicate that the mitoribosomes are assembled in the mrp7 mutants, even in glucose-grown mrp7(1-146) cells where the mitochondrial translation activity was pronouncedly compromised. On the basis of the observed behaviors of MrpL36, however, we conclude that truncation of the C-terminal mitospecific domain of Mrp7 has more deleterious consequences for the integrity of the CP region of mitoribosomes assembled under fermentation than under respiration conditions. These differences, although molecularly unclear at this stage, may contribute to the inhibition of mitotranslation observed in the mrp7 mutants under fermentation, but not under respiration, conditions and point to possible differences in the composition and/or organization of mitoribosomes performing under different metabolic conditions.
Mrp7 and Mam33 proteins interact with each other, and both serve to support the adaptation of cells from glucose fermentation to respiration conditions
The observation that preconditioning the mrp7 mutants to glycerol/ethanol medium improved their subsequent respiration growth suggested that the mutants may struggle with adaptation from glucose fermentation to respiration growth conditions (Supplemental Figure S1B). The time required to adapt from glucose (fermentation growth) to glycerol/ethanol (respiratory) media was therefore determined for each of the mrp7 mutants (all +RNR1). Glucose-grown mrp7(1-187) and mrp7(1-146) strains were found to require a longer time to adapt to the glycerol/ethanol media than the wild-type control or mrp7(1-261) cells, with the mrp7(1-146) strain being the slowest (Table 1). Once adapted to glycerol/ethanol, the mrp7 mutants displayed respiration growth capacities albeit at different efficiencies, as illustrated by the differences in their measured doubling times (Table 1) and consistent with the phenotype testing (Supplemental Figure S1B). The prolonged adaptation times of the mrp7(1-187) and mrp7(1-146) mutants suggest that the ability to perform mitochondrial translation under fermentation conditions may aid in supporting the events required for effective glucose derepression and respiratory adaptation.
TABLE 1:
mrp7 mutant strains display delayed adaptation times to nonfermentive carbon source, similar to those of Δmam33 strains.
| Strain | Time to initial doubling (h) | SGE log-phase doubling time (h) |
|---|---|---|
| Wild type | 14.56 ± 0.77 | 3.18 ± 0.24 |
| mrp7(1-261) | 16.38 ± 0.43 | 4.98 ± 0.72 |
| mrp7(1-187) | 95.75 ± 0.94 | 6.36 ± 0.34 |
| mrp7(1-146) | 202.44 ± 2.38 | 10.06 ± 0.86 |
| WT;Δmam33 | 33.63 ± 2.26 | 9.18 ± 0.52 |
| mrp7(1-261);Δmam33 | 39.56 ± 3.17 | 10.36 ± 0.60 |
| mrp7(1-187);Δmam33 | 85.25 ± 2.28 | 10.92 ± 1.18 |
| mrp7(1-146);Δmam33 | 203.19 ± 5.13 | 10.11 ± 0.46 |
The indicated strains were grown to mid–log phase in selective SD (glucose) media. Cells were harvested, washed, and resuspended in SGE (glycerol/ethanol) media and incubated with continuous shaking at 30°C, as described in Materials and Methods. The times (hours) required by each strain to double its following initial transfer from SD to SGE media, and their doubling times in the SGE media once they had achieved logarithmic respiration growth, are given. Data shown are an average of quadruplicate readings, ±SD. Comparisons in time to initial doubling were compared with unpaired Student’s t test for all strains relative to the WT control (p <0.01) and again with all Δmam33 mutant strains relative to the WT; Δmam33 control (p < 0.05).
The expression of many nuclear-encoded proteins destined for mitochondria is repressed under high-glucose-growth conditions and includes proteins such as the abundant isoforms of porin and citrate synthase. Consistently, decreased cellular porin levels were observed in the glucose/fermentation-grown wild-type control cells and relative to respiratory cells grown in parallel (Figure 3A, bottom panel). The cellular porin content in the mrp7(1-187) and mrp7(1-146) strains grown in glucose (and galactose) fermentation conditions was. however, even further decreased relative to their parallel mrp7(1-261) mutant and Mrp7 wild-type controls, suggesting that the glucose repression of porin was more extensive in the mrp7 mutants that displayed defective mitochondrial translation under fermentation conditions (Figure 3A, bottom panel). The kinetics of induction of porin upon transition from high-glucose (2%) to glucose-limiting (0.05%) conditions (as measured by porin protein steady state levels) was observed to be significantly compromised in the mrp7(1-146) mutant relative to the wild-type Mrp7 control (Figure 5A). In the wild-type Mrp7 cells, the levels of porin were closer to those in respiratory grown cells 2 h after the transition to low glucose. Little increase in porin levels was observed in the mrp7(1-146) mutant over the same time period (Figure 5A). However, the mrp7(1-187) and mrp7(1-146) mutants displayed normal cellular porin content levels once they had been adapted to and propagated on glycerol/ethanol media before harvesting (Figures 2B and 3A, bottom panel). The mitochondrial content of another glucose-repressed mitochondrial enzyme, citrate synthase, was also significantly reduced (by about 50%) in the mitochondria isolated from glucose-grown mrp7(1-146) cells relative to their glucose wild-type control, but displayed levels closer to those of the corresponding wild-type control in mrp7(1-146) mitochondria isolated from cultures preadapted to respiratory (glycerol/ethanol) conditions (Figure 5B). In contrast to porin and citrate synthase, the levels of the Tim44 in the mrp7(1-146) mutant were similar to those of the wild-type Mrp7 control (Figure 5A). The expression of Tim44 (and of mitoribosomal proteins) is not repressed under high-glucose concentrations (Morgenstern et al., 2017), so the observed normal levels of Tim44 (and of mitoribosomal proteins assayed in Figure 4A) are indicative that no gross mitochondrial import defect occurs in the mrp7(1-146) mutant under these fermentation conditions.
FIGURE 5:
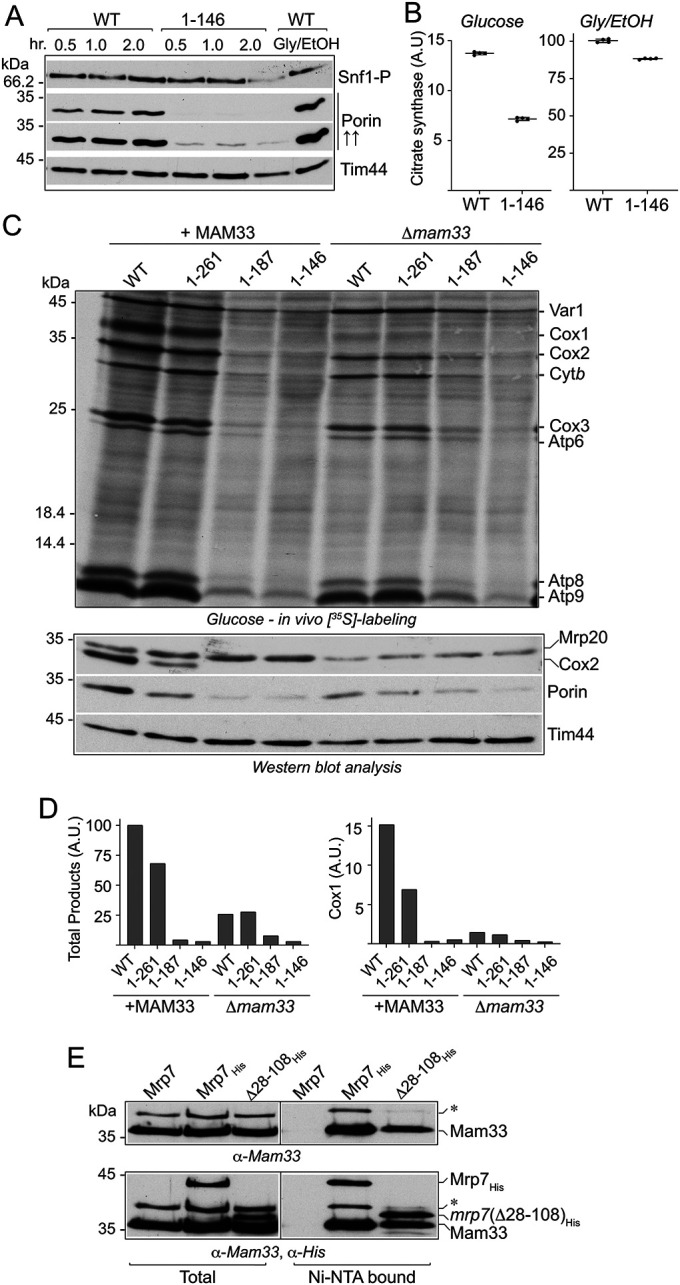
Mam33 physically interacts with the mitospecific domain of Mrp7. (A) Wild-type Mrp7 (WT) and mrp7(1-146) strains (all +RNR1) were grown in selective SD media (2% glucose) to mid–log phase, and equivalent amounts of cells (50 ml at OD600 0.5) were pelleted, washed twice with dH2O, and resuspended in synthetic media containing 0.05% glucose and incubated further at 30°C. Samples equivalent to OD600 0.6 of cells were taken at the indicated time points and subjected to TCA precipitation and subsequent analysis by SDS–PAGE, Western blotting, and immunedecoration with porin and Snf1-P(T172 phosphorylation) antibodies. An additional darker exposure (indicated by “↑↑”) of porin decoration is also provided so that the levels of porin in the mrp7(1-146) mutant can be seen. Tim44 was used as loading control. (B) Citrate synthase activity was measured in mitochondria isolated from glucose (left panel)- and glycerol/ethanol (Gly/EtOH, right panel)-grown WT and mrp7(1-146) strains, as described in Materials and Methods. Quadruplicate activity measurements are presented and expressed as a percentage of the WT glycerol + ethanol control, with means and SDs indicated. The significance between measurements was determined by an unpaired Student’s t test using GraphPad Prism 9. Comparison of each mutant to its respective control resulted in p < 0.0001. A.U., arbitrary units. (C) In vivo labeling of mitotranslation was performed in the indicated glucose-grown strains (each equivalent to OD600 of 0.6 absorbance units) that contained (+MAM33) or were deficient (Δmam33) in the Mam33 protein. Radiolabeling and subsequent analysis of proteins were performed as described in Figure 3A, top and bottom panels. (D) The newly synthesized proteins from the top panel in A were quantified by phosphorimaging. The sum of total products (as described in Figure 3B) (left) and Cox1 (right) amounts were determined and each expressed as a percentage of its corresponding Mrp7 wild-type (WT)+MAM33 control. A.U., arbitrary units. (E) Isolated mitochondria harboring His-tagged Mrp7 WT (Mrp7His) or His-tagged mrp7(Δ28-108) were solubilized in a buffer containing DDM (1%) and subjected to affinity purification with Ni-NTA beads as described in Materials and Methods. The purified material was analyzed with SDS–PAGE, Western blotting, and immunedecoration initially with antibodies against Mam33 (top panel), followed by decoration with an α-His antibody (bottom panel). A 38 kDa protein that cross-reacts with the Mam33 antisera is indicated by *. His-tagged mrp7 derivatives are indicated. Total, 5% of solubilized material; Ni-NTA bound, 100% of affinity-purified material recovered from the Ni-NTA beads.
The delayed growth adaptation phenotype of the mrp7(1-187) and mrp7(1-146) mutants parallels that of the Δmam33 null mutant, which displays prolonged adaptation times transitioning from glucose repression to respiratory-inducing growth conditions (Roloff and Henry, 2015). Mam33, an acidic matrix protein of the mitochondrial matrix and homologue of the mammalian p32/gC1qR protein, has recently been shown to physically interact with a number of mitoribosomal proteins, including Mrp7 (Box et al., 2017; Hillman and Henry, 2019). Deletion of the MAM33 gene in Mrp7 wild-type and mrp7(1-261) cells caused an extension of their adaptation times, requiring almost twice as long before respiratory-based growth was observed (Table 1), consistent with the results previously published for the Δmam33 null mutant (Roloff and Henry, 2015). The absence of Mam33, however, did not further impact the already extended adaptation times of the mrp7(1-187) and mrp7(1-146) mutants, indicating that the process required for adaptation of yeast cells from glucose fermentation to respiration growth that is impacted in the absence of Mam33 may already be affected by the C-terminal truncation of Mrp7 (Table 1).
The presence of Mam33 has been reported to support Cox1 synthesis under fermentation, but not respiration conditions (Hillman and Henry, 2019). In vivo labeling experiments performed with the mrp7 and mrp7;Δmam33 strains indicated that the perturbation in mitotranslation in the absence of Mam33 included, but was not limited to, Cox1 production (Figure 5, C and D). Rather, analysis of both the wild-type and mrp7(1-261) strains indicated that the overall ability to perform mitochondrial translation was adversely impacted under glucose fermentation conditions when Mam33 was absent (Figure 5, C, top panel, and D). Furthermore, an altered ratio of Cox2:Cytb synthesis was observed when Mam33 was absent in the wild-type and mrp7(1-261) cells (Figure 5C, top panel), which resembled that observed in the respiratory grown wild-type cells and the mrp7 mutants (under both glucose fermentation and respiratory conditions) (Figure 3A). The cellular levels of porin were also reduced in the glucose-grown wild-type and mrp7(1-261) strains when Mam33 was absent (Figure 5C, bottom panel), consistent with our earlier observation in Figure 3A that the porin levels are reduced when cellular mitotranslation is perturbed.
Although already strongly inhibited under fermentation conditions, mitochondrial translation in the glucose-grown mrp7(1-187) and mrp7(1-146) mutants was not further inhibited through the additional absence of Mam33 and in the case of the mrp7(1-187);Δmam33 mutant may even have been partially improved (Figure 5, C, top panel, and D). The previously observed reduction in cellular porin levels in the glucose-grown mrp7(1-187) and mrp7(1-146) mutants was similar whether Mam33 was present or absent (Figure 5C, bottom panel). Furthermore, a strong overexpression of Mam33 (achieved through additional expression of an epitope-tagged Mam33 derivative under the galactose-inducible promoter GAL10) did not improve the mitotranslation defect of the mrp7(1-146) mutant under fermentation nor did it improve the ability of this strain to grow under respiration growth conditions (Supplemental Figure S6, A and B).
In contrast to the observed reduction in mitotranslation levels during glucose growth, the deletion of the MAM33 gene did not reduce the mitochondrial translation efficiencies (or alter the Cytb:Cox2 synthesis ratios) of the wild-type control or mrp7 mutants (with the exception of the mrp7(1-146) mutant) under respiratory-based growth conditions (Supplemental Figure S7, top panel), indicating that the absence of Mam33 negatively impacts mitochondrial translation only under fermentation conditions. Consistent with the observed normal levels of mitotranslation, the cellular content of porin appeared to be unaffected by the absence of Mam33 (wild-type control or mrp7 mutants) when cells had been adapted to and grown under respiratory metabolism conditions (Supplemental Figure S7, bottom panel).
Taken together the data indicate that the phenotypes resulting from the absence of Mam33, namely delayed fermentation to respiration adaptation times, decreased cellular porin levels, an overall suppression of mitochondrial translation, and an alteration in the profile of proteins synthesized under glucose fermentation conditions, closely parallel the phenotypes that result from the truncation of the mitospecific domain of Mrp7.
Finally, Mam33 has been reported to have a direct physical interaction with Mrp7 in the mitochondrial matrix, with the postulated interaction site being the unstructured, positively charged, N-terminal L27-homology domain (residues 28–108) of Mrp7 (Figure 1A) (Hillman and Henry, 2019). Given the parallels observed in the Δmam33 mutant and the mrp7(1-187) and mrp7(1-146) mutant phenotypes, we explored whether the C-terminal mitospecific region of Mrp7 could be an interaction site for recruiting the Mam33 protein (Figure 5E). C-terminal, His-tagged, full-length Mrp7 and a Mrp7 derivative that lacked the bL27 N-terminal domain, mrp7(Δ28-108)His, were constructed and expressed in yeast. Mitochondria were isolated from the resulting yeast strains, and Ni-NTA purification of the His-tagged Mrp7 derivatives and their associated proteins was performed. The specific recovery of Mam33 with both full-length Mrp7His and the mrp7(Δ28-108)His derivative indicated that Mam33 binding site(s) reside within Mrp7’s mitospecific domain (i.e., residues 109–371) and that the presence of the N-terminal L27-homology domain (residues 28–108) is not required for the Mam33 and Mrp7 interaction (Figure 5E).
DISCUSSION
In this study we have evaluated the functional significance of the C-terminal mitospecific element of the Mrp7 (bL27) protein and the role it plays in supporting mitotranslation under different metabolic conditions, namely glycolysis and aerobic respiration. Our data here indicate that truncation of the mitospecific region of Mrp7 did not impair the ability of the mitoribosome to perform the synthesis of mtDNA-encoded OXPHOS components under respiration growth conditions but resulted in a gross defect in mitochondrial translation under fermentation growth conditions. Although mitotranslation appeared normal in the mrp7(1-187) and mrp7(1-146) mutants under respiration growth conditions, the subsequent assembly of the newly synthesized proteins into functional OXPHOS complexes, in particular the subunits of the cytochrome bc1 and COX complexes, was found to be partially affected. A similar uncoupling of mitotranslation and OXPHOS assembly was also observed in mutants of the mitospecific MrpL35 (mL38) protein, a component of the CP region and physical partner of Mrp7 (Box et al., 2017). Together these results highlight a potential role of the CP region as a regulatory hub of the mitoribosome, acting under respiration growth conditions (i.e., OXPHOS assembly-promoting conditions) to coordinate protein synthesis with downstream OXPHOS assembly events to regulate bioenergetic capacity of the mitochondria.
The levels of many mitochondrial metabolic enzymes, including the OXPHOS enzymes such as the COX and cytochrome bc1 complexes, are strongly reduced under glucose fermentation conditions as the yeast cell relies largely on aerobic glycolysis for energy production (Morgenstern et al., 2017). Despite the reduced demand for the assembly of OXPHOS enzymes under fermentation conditions, our findings indicate that the process of mitochondrial translation, that is, the synthesis of mtDNA-encoded OXPHOS subunits, is not repressed under glucose growth conditions. Indeed, we observed an increase in in vivo radiolabeling of mitoribosomal translational products (overall levels and rates) under fermentation conditions relative to respiration growth conditions, the latter representing the metabolic conditions that promote a significant up-regulation of OXPHOS complex assembly. Importantly, our findings illustrate that cellular mitochondrial translation activity is not always tightly coordinated to the productive assembly of OXPHOS enzymes, in contrast to previous proposals (Fontanesi et al., 2011; Mick et al., 2011; Dennerlein et al., 2017). We conclude that the reduction of OXPHOS complex content observed under fermentation conditions can likely be attributed to the glucose repression of nuclear-encoded OXPHOS subunits rather than a suppression of mtDNA gene expression. The functional availability of translation initiation factors of mtDNA-encoded proteins within the mitochondrial matrix (such as Mss51 for Cox1 [ Siep et al., 2000]) must be differentially regulated under glucose fermentation conditions, such that they maintain a high level of mitochondrial protein synthesis during repression of OXPHOS complex assembly. It is possible that the various proteases within the mitochondria (e.g., Oma1, Yta10/12, Yta11, Pim1) whose mitochondrial contents are similar between fermentation and respiration growth conditions (Morgenstern et al., 2017) play a critical role in this respect during fermentation. Furthermore an increased proteolytic vulnerability of mitochondrially synthesized proteins under glucose growth conditions was observed here. Efficient turnover of the newly synthesized proteins by the mitochondrial protease system under fermentation conditions would ensure the availability of translational activators, such as Mss51, to promote the continued synthesis of mtDNA-encoded proteins under glucose conditions where their subsequent assembly into OXPHOS complexes is repressed.
We report here that the ability of the mitoribosome to maintain translational capacity under fermentation conditions involves the integrity of the CP region of the mtLSU. Truncation of the Mrp7’s mitospecific domain, that is, removal of residues 188–371 (or of residues 147–371), resulted in a strongly impaired capacity to support mitochondrial translation under fermentation but not under respiration growth conditions. Failure to support glucose mitotranslation in the mrp7 mutants was not due to an inability to assemble or maintain normal mtLSU levels under these fermentation conditions. However, our findings indicate that truncation of Mrp7’s mitospecific domain adversely impacted the organization of the CP region of the glucose mitoribosome, as judged by MrpL36’s (bL31m) altered association with the mtLSU in the mrp7(1-146) mutant. The CP region of the mtLSU is adjacent to the mtSSU interface, and a number of physical contacts/bridges between both ribosomal subunits involve proteins of the CP region, including the MrpL36 (bL31), MrpL17 (mL46), and MrpL7 (uL5m) proteins (Amunts et al., 2014; Desai et al., 2017). The altered assembly of MrpL36 in the glucose mrp7(1-146) mitoribosomes thus may have consequences for the arrangement of the mtLSU with the mtSSU and have contributed to the observed decrease in Mrp2 (uS14m) levels. Mrp2’s physical partners within the mtSSU, namely Sws2 (uS13m), Rsm19 (uS19m), and Rsm10 (uS10m) are all directly involved in bridges between mtSSU and the CP region of the mtLSU subunit, with Rsm19 (uS19m) directly contacting MrpL36 (Desai et al., 2017). Importantly, we observed that the impact of the truncation of Mrp7 on MrpL36’s association with the mtLSU (or on the steady state levels of Mrp2) was not as pronounced in the mitochondria isolated from the respiratory grown cells. We suggest that the composition and/or regulatory modification of the mitoribosome, in particular of the CP region, may differ between fermentation and respiration grown cells, such that the truncation of Mrp7 exerts differential effects.
Consistent with the proposal here that respiratory mitoribosomes may be differentially assembled or modified from their glucose counterparts, our findings also support a differential regulation of the mitoribosomes, depending on their metabolic conditions. The rate and levels of mitotranslation under glucose fermentation conditions were higher than those observed under respiration growth conditions, and this occurs despite the mitoribosomal content being equal or even slightly lower in the glucose-grown cells, relative to their glycerol/ethanol-grown counterparts. These findings suggest that the translational activity of the glucose mitoribosomes may not be as controlled as that of mitoribosomes operating under conditions that promote OXPHOS assembly, that is, where mitotranslation needs to be tightly coordinated with the rates of import and assembly of nuclear-encoded partner proteins to ensure their efficient assembly into the OXPHOS complexes. The translational profile of the mitoribosomes was also observed to be different between the fermentation and respiratory grown cells, with a higher Cytb:Cox2 ratio observed in the latter, suggesting that mitoribosomes may be alternatively “primed” in different metabolic environments and/or quantitative differences in mitotranscript levels may exist under fermentation versus respiration growth conditions. The mrp7 mutants adopted a more “respiratory” translational profile (i.e., a decreased Cox2:Cytb and Cox3:Atp6 synthesis ratio) under glucose fermentation conditions, suggesting that this mitoribosomal synthesis profile alteration may be molecularly determined by the CP region and its organization. A similar shift in the fermentation mitotranslational profile toward a “respiratory output” profile was also observed here in the Δmam33 mutants. As our findings here indicate that Mam33 binds to elements of the C-terminal mitospecific domain of Mrp7, it is possible that the similarities between the mrp7 and Δmam33 mitotranslational profiles reflect a cooperative function between Mrp7 and Mam33 proteins. Taking these observations together, we therefore postulate that the mitoribosome, more specifically the CP region, may be differentially assembled, modified, and/or regulated in fermentation versus respiration conditions. A structural comparative analysis of the mitoribosome isolated from different metabolically grown environments may be warranted for further investigation of these predictions.
Why does the yeast cell invest so much energy under fermentation conditions to maintain a high level of mitochondrial translation, only to degrade the synthesized proteins when their subsequent assembly into OXPHOS complexes is impaired? Analysis of both the mrp7 and Δmam33 mutants indicates that the ability to effectively perform mitochondrial translation under glucose fermentation conditions confers an advantage for the cell when required to shift metabolism and adapt to respiration conditions and consistent with earlier observations made in rho0 yeast cells (Strogolova et al., 2012). While the reduced levels of OXPHOS enzymes in the mrp7 mutant cells under glucose conditions could be attributed solely to impaired mitotranslation, our results here indicate that the levels of other glucose-repressed proteins, that is, those not partnered with mtDNA-encoded proteins such as porin and citrate synthase, were also lower in the cells harboring truncations of Mrp7 and/or deficient in Mam33 under fermentation conditions. In turn we propose that a more robust glucose repression of nuclear genes may be a/the factor contributing to the prolonged adaptation times observed for the mrp7 and Δmam33 mutants when the cells were forced to shift from high-glucose to respiration growth conditions. It is possible therefore that the ability to maintain mitochondrial translation during glycolysis/fermentation may be critical for a cell’s ability to navigate metabolic switching from glucose repression of gene expression to the induction required to support respiratory metabolism. In mammalian cells, a deficiency in the homologue of Mam33, p32 (also known as gC1qR/C1QBP/HABP1), is correlated with decreased mitochondrial translation activity and increased a dependency on glycolysis (Yagi et al., 2012, 2021). Furthermore, dysfunction in p32 has also been correlated with defects in cell differentiation, processes that involve cells switching their metabolism from a glycolysis- to an aerobic respiratory–based one, and evidence suggests that in mammalian cells p32 contributes to sustaining mitochondrial protein synthesis over the course of cell differentiation (Gotoh et al., 2018, 2020; Yagi et al., 2021). It is tempting therefore to speculate that active mitochondrial translation and/or subsequent turnover of the newly synthesized mtDNA-encoded proteins may signal the integrity of mitochondria to the dividing cell and/or play a signaling role in promoting a metabolic shift from glycolysis to aerobic respiration.
In summary, our findings here challenge the current dogma that mitochondrial translation is tightly coordinated with the cellular demand for OXPHOS complex assembly; under glucose fermentation conditions, cellular mitoribosomal activity remains unexpectedly high despite the strong glucose repression of OXPHOS complex assembly. Furthermore, our study demonstrates that the CP region of the mtLSU, more specifically the integrity of the mitospecific region Mrp7 (bL27m) protein, is directly involved in maintaining the translational capacity of the mitoribosome under fermentation but not under respiration growth conditions. Our findings suggest that the composition of the CP-mtSSU interface from the fermentation mitoribosome may differ from that of its respiratory counterpart and this may in turn allow for a differential regulation of the mitoribosome, depending on its metabolic environment. The ability to maintain mitochondrial translation under fermentation conditions appears to confer an advantage to the yeast cell and its ability to effectively transition from glycolysis-based, glucose fermentation to aerobic respiration–promoting growth conditions.
MATERIALS AND METHODS
Yeast strains and growth conditions
All strains used in this study are derivatives of the S. cerevisiae haploid W303-1A genetic background (W303-1A, mat a, leu2, trp1, his3, ade2). They include the Δmrp7 (MRP7::KAN) mutant, with a pRS413 plasmid–borne wild-type MRP7 or mrp7 mutant derivative, along with plasmid-borne RNR1 (YEplac112-RNR1) (Zeng et al., 2018), as indicated. All strains were cultured using standard protocols on minimal synthetic medium (S) supplemented with (when appropriate) uracil, tryptophan, leucine, adenine, or if indicated, on full YP (yeast extract, peptone) media. In vivo labelings and mitochondrial isolations were isolated from cultures grown at 30°C in selective synthetic media with either 2% glucose (SD), 2% galactose (SGal), or 3% glycerol + 2% ethanol (SGE, glycerol/ethanol, Gly/EtOH) as carbon sources, as indicated.
Generation of mrp7 mutants
The yeast chromosomal MRP7 open reading frame (ORF) was replaced with the KAN cassette in a haploid W303-1A yeast strain harboring a wild-type MRP7 genomic insert (corresponding to MRP7 ORF plus 806 base pairs 5′ and 793 base pairs 3′, corresponding to upstream and downstream regulatory regions, respectively) cloned into the centromeric pRS316 (URA3) plasmid (pRS316-MRP7). In the presence of RNR1 (YEplac112-RNR1) (when indicated), a plasmid shuffling approach with 5′-fluoroorotic acid (Box et al., 2017) was employed to replace the pRS316-MRP7 plasmid with the centromeric pRS413 (HIS3) plasmid harboring a similar genomic fragment (i.e., wild-type MRP7 or a truncated mrp7 derivative). The chosen genetic approach (i.e., use of a centromeric plasmid [pRS413] and employment of the endogenous MRP7 5′ and 3′ regulatory regions to drive expression of Mrp7/mrp7 derivatives) was designed to mimic the expression of Mrp7/mrp7 derivatives at levels similar to that of the endogenous Mrp7 protein. Deletions in the C-terminal ORF (while maintaining the integrity of cloned 3′ noncoding region) were generated using a PCR-mediated deletion of pRS413-MRP7 plasmid DNA (Hansson et al., 2008). A mitochondrially targeted universal Var1 derivative (Var1u) was expressed in the indicated strains using the pRS316-VAR1u plasmid (Sanchirico et al., 1995; Seshadri et al., 2020). Mrp7 wild-type and mrp7 mutant strains, which also were deficient in MAM33 (Δmam33; MAM33::URA3), were created through homologous recombination of the URA3 marker to delete the entire MAM33 ORF in the Mrp7/mrp7 mutant strains. Overexpression of the C-terminal epitope-tagged Mam33 derivative was achieved by cloning the ORF encoding Mam33 plus eight His residues into the Yip352 plasmid with a Gal10 promoter and integrating into the ura3 locus of the Mrp7 wild-type and mrp7(1-146) (+RNR1) yeast strains. Induction of the resulting Mam33His was achieved through growing the strains on minimal media containing galactose or glycerol/ethanol medium supplemented with 0.1% galactose.
Generation of His-tagged Mrp7 derivatives
The Yip352-URA3 vector containing the ORF encoding either wild-type Mrp7 or the Mrp7(Δ28-108) derivative as C-terminal His8-tagged proteins (Mrp7His and mrp7(Δ28-108)His, respectively) and downstream of the galactose-inducible GAL10 promoter was created. Plasmids were then linearized with NcoI and integrated into the ura3 loci of the indicated yeast strains, followed by selection of URA+ transformants. Western blotting with a His-specific antibody was employed to confirm the expression of His-tagged proteins before mitochondrial isolation.
Affinity purification of His-tagged proteins
Mitochondria were isolated from His-tagged Mrp7-harboring strains in galactose-based cultures in selective synthetic media at 30°C. Isolated mitochondria (200 μg of protein) were solubilized in lysis buffer (10 mM Tris-HCl, 100 mM KOAc, 20 mM Mg(OAc)2, 0.1 mM spermidine, 5% glycerol, 1% n-dodecyl β-d-maltoside [DDM], 1 mM phenylmethylsulfonyl fluoride [PMSF], 1× cOmplete Protease Inhibitor Cocktail [Roche], pH 7.4) for 30 min on ice. Following a clarifying spin, Ni-NTA purification of His-tagged proteins was performed as previously described (Jia et al., 2007).
In vivo radiolabeling of mitochondrial translation
The adaptation of strains to the indicated growth medium before in vivo labeling was performed by initially growing strains on the respective selective synthetic media plates, followed by growth and passaging in liquid selective synthetic medium (glucose, galactose, and/or glycerol/ethanol, as indicated) for at least 4 d to ensure complete adaptation. Cells were then grown to mid–log phase in the corresponding liquid synthetic medium with the indicated carbon sources, and equivalent amounts of cells (0.6 OD600 absorbance units) were harvested, washed, and resuspended in 40 mM phosphate buffer containing the indicated carbon source. In vivo radiolabeling of mitotranslation with [35S]methionine was performed in the presence of cycloheximide (0.3 mg/ml) for 10 min (unless otherwise indicated) at 30°C, essentially as previously described (Barrientos, 2002; Barrientos et al., 2002). Total cellular proteins were extracted and precipitated by trichloroacetic acid (TCA) and analyzed by SDS–PAGE and autoradiography followed by Western blotting (Barrientos, 2002). For the pulse-chase experiment, in vivo labeling was performed for 10 min, after which translation was stopped by the addition of cold methionine (35 mM) and puromycin (73 µg/ml), cells were isolated by centrifugation, washed twice, and resuspended in minimal media containing glucose or glycerol/ethanol, as indicated, and incubated further in the presence of puromycin/methionine at 30 °C for the indicated time periods.
Sucrose gradient analysis of mitoribosomes
Isolated mitochondria (450 μg) were solubilized in 600 μl of lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM MgCl2, 1 mM PMSF, 1.5% octyl β-d-glycopyranoside, pH 7.2) for 30 min on ice. Following a clarifying spin, the supernatant was layered onto a 10.5 ml linear sucrose gradient (25%–40%) and centrifuged at 26,000 rpm for 18 h at 4°C in a Beckman SW-41 Ti rotor. Fractions (790 μl) were collected, TCA precipitated, and analyzed with SDS–PAGE and Western blotting.
Adaptation growth curve experiments
For glucose to glycerol + ethanol growth curve adaptations, the indicated strains were grown to mid–log phase in SD (2% glucose) media, washed twice with dH2O, and resuspended in selective SGE (3% glycerol + 2% ethanol) media to an OD600 of 0.3 in a 48-well plate. The plate was incubated at 30°C with continuous orbital shaking in a SpectraMax i3x Multi-Mode Microplate Reader. OD600 readings were taken every 15 min.
Statistics and reproducibility
All in vivo labeling experiments, Western blot/immunedecorations, and serial dilution phenotype assays shown are representatives of at least three independent replicates. Enzyme activities and growth curve adaptation measurements were performed with quadruplicate samples, and statistical analysis (unpaired Student’s t test or one-way analysis of variance [ANOVA], as indicated) was performed, and the means and standard deviations (SD) are presented.
Miscellaneous
Mitochondrial isolation, protein determination, and SDS–PAGE were performed as previously described (Dienhart and Stuart, 2008). Spectral measurements of specific enzyme activities of the cytochrome bc1 and COX complexes were performed as previously described (Tzagoloff et al., 1975), and citrate synthase was measured according to the Sigma-Aldrich protocol (MAK057) with the exception that isolated mitochondria were solubilized in 0.1M Tris-HCl with 1 mM DDM, pH 8.1, at a concentration of 1.6 μg/μl before enzyme determinations. BN-PAGE analysis was performed essentially as described previously (Box et al., 2017). In organello monitoring of mitotranslation in the presence of [35S]methionine was performed as previously described (Herrmann et al., 1994; Stuart and Koehler, 2007). The phospho-Snf1(Thr172) antibody used was purchased from Cell Signaling Technology (40H9). All other antibodies used were against the respective purified yeast proteins and generated either in the Stuart lab or received as gifts (see Acknowledgments). Antigen–antibody complexes on Western blots were detected by horseradish peroxidase–coupled secondary antibodies and chemiluminescence detection on x-ray films.
Supplementary Material
Acknowledgments
We acknowledge and thank Emma Schmierer for her preliminary work on the mrp7(1-187) and Δ mam33 analysis and Hannah Buchholz for her qPCR analysis. We are grateful to T. Mason and M. Ott for the valuable gifts of the ribosomal antisera, D. Rapaport for the porin antisera, B. Westermann for providing the Mam33 antisera, V. Zara for the Qcr7 antibody, A. Barrientos and F. Fontanesi for the kind gifts of the pRS316- VAR1 u and YEplac112- RNR1 plasmids, and C. Osman for sharing his qPCR protocol. The research was supported by National Science Foundation (NSF) grant MCB 1817682 to R.A.S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NSF.
Abbreviations used:
- A.U.
arbitrary units
- BN-PAGE
blue native PAGE
- COX
cytochrome c oxidase
- CP
central protuberance
- mtLSU
mitochondrial large ribosomal subunit
- mtSSU
mitochondrial small ribosomal subunit
- OXPHOS
oxidative phosphorylation.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-07-0370) on November 3, 2021.
REFERENCES
- Amunts A, Brown A, Bai XC, Llacer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SHW, Ramakrishnan V (2014). Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A (2002). In vivo and in organello assessment of OXPHOS activities. Methods 26, 307–316. [DOI] [PubMed] [Google Scholar]
- Barrientos A, Korr D, Tzagoloff A (2002). Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh’s syndrome. EMBO J 21, 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box JM, Kaur J, Stuart RA (2017). MrpL35, a mitospecific component of mitoribosomes, plays a key role in cytochrome c oxidase assembly. Mol Biol Cell 28, 3489–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Soto IC, Shipkovenska G, Churchman LS (2016). Synchronized mitochondrial and cytosolic translation programs. Nature 533, 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennerlein S, Wang C, Rehling P (2017). Plasticity of mitochondrial translation. Trends Cell Biol 27, 712–721. [DOI] [PubMed] [Google Scholar]
- Desai N, Brown A, Amunts A, Ramakrishnan V (2017). The structure of the yeast mitochondrial ribosome. Science 355, 528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienhart MK, Stuart RA (2008). The yeast Aac2 protein exists in physical association with the cytochrome bc1-COX supercomplex and the TIM23 machinery. Mol Biol Cell 19, 3934–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi F, Clemente P, Barrientos A (2011). Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae . J Biol Chem 286, 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goke A, Schrott S, Mizrak A, Belyy V, Osman C, Walter P (2020). Mrx6 regulates mitochondrial DNA copy number in Saccharomyces cerevisiae by engaging the evolutionarily conserved Lon protease Pim1. Mol Biol Cell 31, 527–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh K, Kunisaki Y, Mizuguchi S, Setoyama D, Hosokawa K, Yao H, Nakashima Y, Yagi M, Uchiumi T, Semba Y, et al. (2020). Mitochondrial protein synthesis is essential for terminal differentiation of CD45(-) TER119(-)erythroid and lymphoid progenitors. iScience 23, 101654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh K, Morisaki T, Setoyama D, Sasaki K, Yagi M, Igami K, Mizuguchi S, Uchiumi T, Fukui Y, Kang D (2018). Mitochondrial p32/C1qbp is a critical regulator of dendritic cell metabolism and maturation. Cell Rep 25, 1800–1815.e1804. [DOI] [PubMed] [Google Scholar]
- Hansson MD, Rzeznicka K, Rosenback M, Hansson M, Sirijovski N (2008). PCR-mediated deletion of plasmid DNA. Anal Biochem 375, 373–375. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Stuart RA, Craig EA, Neupert W (1994). Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J Cell Biol 127, 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman GA, Henry MF (2019). The yeast protein Mam33 functions in the assembly of the mitochondrial ribosome. J Biol Chem 294, 9813–9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Suda T (2014). Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol 15, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Dienhart MK, Stuart RA (2007). Oxa1 directly interacts with Atp9 and mediates its assembly into the mitochondrial F1F0-ATP synthase complex. Mol Biol Cell 18, 1897–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J, Stuart RA (2011). Truncation of the Mrp20 protein reveals new ribosome-assembly subcomplex in mitochondria. EMBO Rep 12, 950–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O, Bestwick M, Meunier B, Watts TC, Winge DR (2010). Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol Cell Biol 30, 1004–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayorga JP, Camacho-Villasana Y, Shingu-Vazquez M, Garcia-Villegas R, Zamudio-Ochoa A, Garcia-Guerrero AE, Hernandez G, Perez-Martinez X (2016). A novel function of Pet54 in regulation of Cox1 synthesis in Saccharomyces cerevisiae mitochondria. J Biol Chem 291, 9343–9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay GP, Su CH, Tzagoloff A (2013). Stabilization of Cox1p intermediates by the Cox14p-Coa3p complex. FEBS Lett 587, 943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Fox TD, Rehling P (2011). Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat Rev Mol Cell Biol 12, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern M, Stiller SB, Lubbert P, Peikert CD, Dannenmaier S, Drepper F, Weill U, Hoss P, Feuerstein R, Gebert M, et al. (2017). Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep 19, 2836–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa L, Djedaini M, Hoffman R (2019). Mitochondrial role in stemness and differentiation of hematopoietic stem cells. Stem Cells Int 2019, 4067162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Pyun WY, Park HW (2020). Cancermetabolism: phenotype, signaling and therapeutic targets. Cells 9, 2308.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestele M, Vogel F, Reichert AS, Herrmann JM, Ott M (2009). Mrpl36 is important for generation of assembly competent proteins during mitochondrial translation. Mol Biol Cell 20, 2615–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-Dennerlein R, Oeljeklaus S, Lorenzi I, Ronsor C, Bareth B, Schendzielorz AB, Wang C, Warscheid B, Rehling P, Dennerlein S (2016). Mitochondrial protein synthesis adapts to influx of nuclear-encoded protein. Cell 167, 471–483.e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roloff GA, Henry MF (2015). Mam33 promotes cytochrome c oxidase subunit I translation in Saccharomyces cerevisiae mitochondria. Mol Biol Cell 26, 2885–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchirico M, Tzellas A, Fox TD, Conrad-Webb H, Periman PS, Mason TL (1995). Relocation of the unusual VAR1 gene from the mitochondrion to the nucleus. Biochem Cell Biol 73, 987–995. [DOI] [PubMed] [Google Scholar]
- Schiliro C, Firestein BL (2021). Mechanisms of metabolic reprogramming in cancer cells supporting enhanced growth and proliferation. Cells 10, 1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri SR, Banarjee C, Barros MH, Fontanesi F (2020). The translational activator Sov1 coordinates mitochondrial gene expression with mitoribosome biogenesis. Nucleic Acids Res 48, 6759–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siep M, van Oosterum K, Neufeglise H, van der Spek H, Grivell LA (2000). Mss51p, a putative translational activator of cytochrome c oxidase subunit-1 (COX1) mRNA, is required for synthesis of Cox1p in Saccharomyces cerevisiae. Curr Genet 37, 213–220. [DOI] [PubMed] [Google Scholar]
- Snoeck HW (2017). Mitochondrial regulation of hematopoietic stem cells. Curr Opin Cell Biol 49, 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto IC, Barrientos A (2016). Mitochondrial cytochrome c oxidase biogenesis is regulated by the redox state of a heme-binding translational activator. Antioxid Redox Signal 24, 281–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strogolova V, Orlova M, Shevade A, Kuchin S (2012). Mitochondrial porin Por1 and its homolog Por2 contribute to the positive control of Snf1 protein kinase in Saccharomyces cerevisiae . Eukaryot Cell 11, 1568–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart RA, Koehler CM (2007). In vitro analysis of yeast mitochondrial protein import. Curr Protoc Cell Biol Chapter 11, Unit 11 19. [DOI] [PubMed] [Google Scholar]
- Suda T, Takubo K, Semenza GL (2011). Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 9, 298–310. [DOI] [PubMed] [Google Scholar]
- Tsogtbaatar E, Landin C, Minter-Dykhouse K, Folmes CDL (2020). Energy metabolism regulates stem cell pluripotency. Front Cell Dev Biol 8, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Akai A, Needleman RB (1975). Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J Biol Chem 250, 8228–8235. [PubMed] [Google Scholar]
- Yagi M, Toshima T, Amamoto R, Do Y, Hirai H, Setoyama D, Kang D, Uchiumi T (2021). Mitochondrial translation deficiency impairs NAD(+)-mediated lysosomal acidification. EMBO J 40, e105268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi M, Uchiumi T, Takazaki S, Okuno B, Nomura M, Yoshida S, Kanki T, Kang D (2012). p32/gC1qR is indispensable for fetal development and mitochondrial translation: importance of its RNA-binding ability. Nucleic Acids Res 40, 9717–9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R, Smith E, Barrientos A (2018). Yeast mitoribosome large subunit assembly proceeds by hierarchical incorporation of protein clusters and modules on the inner membrane. Cell Metab 27, 645–656.e647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Ying Y, Ji K, Duan X, Mai T, Kim J, Li Q, Yu L, Xu Y (2020). p53 coordinates glucose and choline metabolism during the mesendoderm differentiation of human embryonic stem cells. Stem Cell Res 49, 102067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.