Abstract
Background
The limited knowledge about the PTEN hamartoma tumor syndrome (PHTS) makes its diagnosis a challenging task. We aimed to define the clinical and genetic characteristics of this syndrome in the Spanish population and to identify new genes potentially associated with the disease.
Results
We reviewed the clinical data collected through a specific questionnaire in a series of 145 Spanish patients with a phenotypic features compatible with PHTS and performed molecular characterization through several approaches including next generation sequencing and whole exome sequencing (WES). Macrocephaly, mucocutaneous lesions, gastrointestinal polyposis and obesity are prevalent phenotypic features in PHTS and help predict the presence of a PTEN germline variant in our population. We also find that PHTS patients are at risk to develop cancer in childhood or adolescence. Furthermore, we observe a high frequency of variants in exon 1 of PTEN, which are associated with renal cancer and overexpression of KLLN and PTEN. Moreover, WES revealed variants in genes like NEDD4 that merit further research.
Conclusions
This study expands previously reported findings in other PHTS population studies and makes new contributions regarding clinical and molecular aspects of PHTS, which are useful for translation to the clinic and for new research lines.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13023-021-02079-7.
Keywords: PTEN hamartoma tumor syndrome, Cowden syndrome, PTEN gene, NGS, Exome
Background
The PTEN hamartoma tumor syndrome (PHTS; MIM 158350) encompasses several clinical entities with overlapping phenotypic characteristics, that are associated with germline pathogenic variants in the PTEN gene. Cowden syndrome (CS) and Bannayan-Riley-Ruvalcaba syndrome (BRRS) are the 2 principal entities of PHTS [1]. In addition to these, PHTS also includes some cases of Proteus-like syndromes, autism spectrum disorder (ASD) associated with macrocephaly, and VATER syndrome [2–5]. Macrocephaly, mucocutaneous lesions, Lhermitte-Duclos disease (LDD) and hamartomatous polyposis are generally considered characteristic features of PHTS. Heterozygous germline variants in the tumor suppressor gene PTEN cause PHTS. These variants are inherited in an autosomal dominant pattern and are found in up to 80–85% of CS patients, and in 60–65% of BRRS patients [2]. PTEN encodes a lipid and protein phosphatase that among other functions antagonizes the proliferative PI3K/AKT/mTOR pathway [6].
Besides numerous non-neoplastic multisystemic features, PHTS also entails increased risks for thyroid, breast, endometrial, colon, renal cell, and melanoma cancers [7]. These high cancer risks reinforce the importance of an early diagnosis that allows appropriate surveillance strategies. Reality, however, is quite the opposite: PHTS diagnosis usually takes a long time to be made, given the rareness of the disease, its considerable clinical heterogeneity, and the lack of awareness and the limited knowledge of this entity based on scarce studies. To improve this situation, several diagnostic criteria and recommendations have been proposed in the last decade [7–10]. However, these recommendations are based on a limited number of patients from only a few populations and there is still no consensus regarding their clinical and diagnostic utility [7, 9].
Furthermore, PTEN germline variants are not detected in a considerable number of patients who meet the clinical diagnostic criteria [2], reason why multiple efforts have been made to identify other genes involved in these syndromes. Variants in SDH-B, SDH-D, PIK3CA, AKT1, TTN and SEC23B, together with hypermethylation of the KLLN promoter, have been previously reported in certain CS, CS-like and BRRS patients [11–16].
Here, we describe the clinical and molecular aspects of 145 Spanish patients with a phenotype compatible with PHTS entities, to contribute to the knowledge of this rare disease by defining its characteristics in a new population and searching for other relevant genes.
Results
Spectrum of alterations in PTEN
Almost half of the patients (46% of the total series) had a germline alteration in the PTEN gene: 52 patients (36%) carried pathogenic point variants, 7 individuals (5%) had large deletions and 7 additional patients (5%) carried variants of unknown significance (VUS). The complete list of PTEN alterations is shown in Additional file 1: Tables S1-S3. No pathogenic variants nor VUS were found in the promoter of PTEN. We were able to trace the origin of the PTEN variants in 21 cases through genetic testing of their relatives. Thus, we confirmed the presence of 14 de novo (24% of the PTEN variant carriers) and 7 familial variant cases (12%).
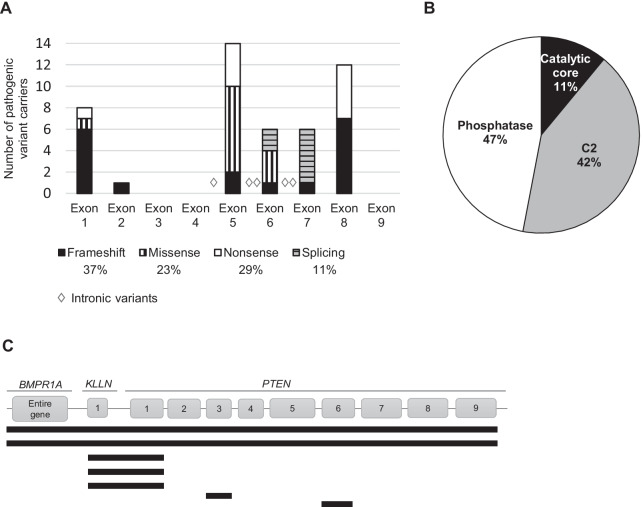
We will refer to carriers of PTEN pathogenic variants and large deletions as PTEN-mut, excluding VUS carriers. The pathogenic variants appeared at higher frequencies in exons 5 to 8 of PTEN, but we also found a considerable number of patients carrying pathogenic variants in exon 1 (Fig. 1A). Several types of variants were observed (Fig. 1A) that affected the different PTEN protein domains, except the C-terminal end (residues 352 to 403) (Fig. 1B). Some pathogenic variants in PTEN appeared recurrently in our series: c.1003C > T, p.(Arg335*) (n = 5 unrelated patients); c.388C > T, p.(Arg130*) (n = 3); c.39_40del, p.(Arg14Glufs*29) (n = 2); c.406T > C, p.(Cys136Arg) (n = 2); and c.697C > T, p.(Arg233*) (n = 2).
Fig. 1.
PTEN pathogenic variants found in our series. A PTEN point variants: numbers, distribution and types. B PTEN protein domains affected by the exonic variants. C Schematic representation of the gene regions affected by each of the 7 large deletions involving PTEN
Array comparative genomic hybridization (aCGH) allowed exhaustive characterization of 5 large deletions which also encompassed the proximal gene KLLN. Two of these rearrangements involved regions of 8 and 10 Mb, affecting other genes such as BMPR1A (Fig. 1C, Additional file 1: Table S3).
The remaining 79 probands (54%) were negative for pathogenic variants, large rearrangements or VUS in the PTEN gene. We will refer to these patients as PTEN-wt.
Clinical characterization and criteria for PTEN study
The series of probands consisted mostly in adults (84%) but also included 25 patients (16%) under the age of 18 years old. 59% of the adults were women and 25% men, plus 12% young men and 4% young women. In the series of PTEN-mut+—excluding VUS—there were 32 women and 27 men. Mean age was 30 years in the mutation carriers (PTEN-mut+) and 46 in the PTEN-wt.
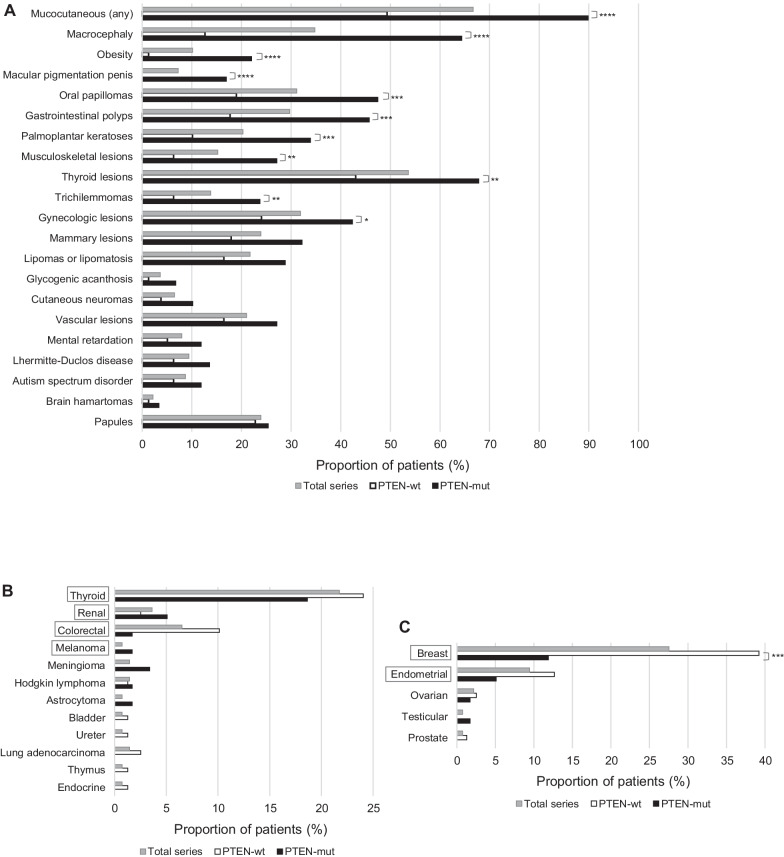
Excluding the PTEN VUS carriers (described in Additional file 1: Table S2), we compared the phenotype between the PTEN-mut and the PTEN-wt groups, and found several clinical features that were significantly more common among PTEN-mut individuals, such as macrocephaly, mucocutaneous lesions and obesity (body mass index ≥ 30), suggesting their usefulness as criteria to identify PTEN pathogenic variant carriers (Fig. 2A). On the other hand, other classical clinical features of PHTS, such as LDD, ASD and vascular lesions, did not significantly discriminate between PTEN-mut and PTEN-wt individuals (Fig. 2A), suggesting they are poorer indicators of an individual carrying a PTEN pathogenic variant. This was supported by regression analyses (Additional file 1: Table S4).
Fig. 2.
Proportion of individuals showing the indicated clinical manifestations in our series. A Occurrence of benign clinical features. B Occurrence of the different cancer types (not linked to sex). C Occurrence of sex-linked cancer types. PHTS-associated cancer types are boxed. Chi-square or Fisher test significance (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001) is shown for comparisons of absolute numbers of the PTEN-wt and PTEN-mut groups
The incidence of cancer was higher in the PTEN-wt compared to the PTEN-mut patients (58% (n = 46) versus 39% (n = 23); p = 0.0017). The 3 most frequent cancer types in the series (excluding the VUS carriers) were thyroid, colorectal and renal cancer, while breast, endometrial and ovarian cancers were the most frequent sex-linked cancers (Fig. 2B,C). Of the 69 cancer cases, 64 individuals had suffered some cancer within the spectrum of PHTS (PHTS-associated cancer): 20 PTEN-mut (31% of the PHTS-associated cancer patients) and 44 PTEN-wt (69%). Nevertheless, we also found other cancer types not so frequently associated with PHTS, like Hodgkin lymphoma, meningioma and astrocytoma in PTEN-mut patients (Fig. 2B, C).
We noticed that 18 patients of our series were referred for presenting only PHTS-associated cancers, with apparently no other feature of the disease. None of these individuals were carriers of PTEN pathogenic variants and they accounted for 23% of the PTEN-wt patients, suggesting that only the presentation of certain types of cancer might not be sufficient criterion to perform PTEN genetic testing. This is also supported by regression analyses, which shows a poor contribution of the presence of cancer alone to the probability of finding a PTEN pathogenic variant in the patient (Additional file 1: Table S5).
Interestingly, we found that a considerable proportion of our patients (14% of the PTEN-mut and only 3% of the PTEN-wt individuals; p = 0.013) developed cancer in childhood or adolescence (Table 1). Some of the cancer types we encountered are very rare in this age range, such as endometrial carcinoma or clear cell renal cell carcinoma. This suggests a risk to develop cancer at a very early age for carriers of germline pathogenic variants in PTEN.
Table 1.
Cancer occurrence in individuals ≤ 18 years old from our series. Each case corresponds to a unique proband
| Cancer type | Number of probands | Age of onset | PTEN germline status |
|---|---|---|---|
| Papillary thyroid cancer | 1 | 16 | p.(Lys6Argfs*4) |
| 1 | 16 | p.(Arg335*) | |
| Follicular thyroid cancer | 1 | 14 | p.(Arg335*) |
| Endometrial adenocarcinoma | 1 | 15 | p.(Cys136Arg) |
| Ovarian endodermal sinus tumor | 1 | 6 | p.(Cys136Arg) |
| Clear cell renal cell carcinoma | 1 | 18 | p.0? |
| Hodgkin lymphoma | 1 | 18 | p.(Gln17*) |
| Testicular mixed germ cell tumor | 1 | 18 | p.(Thr277Asnfs*21) |
| Hodgkin lymphoma | 1 | 6 | WT |
| Thyroid cancer (unspecified type) | 1 | 14 | WT |
Genotype–phenotype correlations
In order to search for genotype–phenotype correlations, we evaluated the clinical features as a function of the location of the variants along the PTEN sequence. We found several associations, most notably an association between renal cancer and pathogenic variants in PTEN exon 1 (p = 0.045; Additional file 1: Table S6).
Search for other genetic factors
Given that not all patients with a clinical phenotype compatible with the PHTS entities are found to carry PTEN alterations, we searched for other genetic factors that could be involved as causal elements in these PTEN-wildtype patients. Considering the large variability of phenotypes found along the PHTS spectrum, we also searched for genes that could be involved as phenotype modifiers.
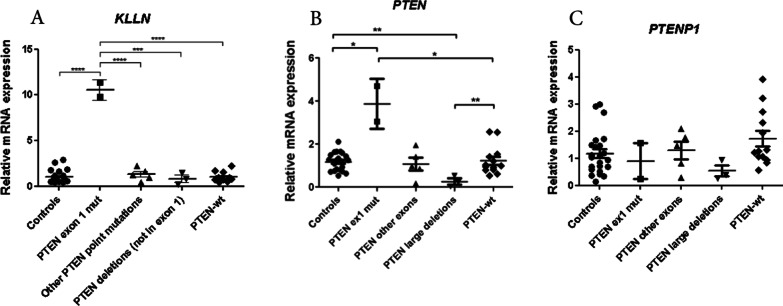
We first studied the mRNA expression of KLLN in our patient series. Strikingly, we found that the expression of KLLN was similar in patients and controls, except for patients carrying PTEN variants in exon 1, who showed overexpression of KLLN (Fig. 3A). These individuals also showed an unexpected high expression of PTEN (Fig. 3B), and we could rule out an upregulation of the pseudogene PTENP1 (Fig. 3C). Therefore, only variants in exon 1 of PTEN seem to alter the expression levels of PTEN and its neighbor gene KLLN, although it is unclear how this contributes to PHTS etiology or pathogenesis. The only distinctive clinical feature of the patients with KLLN and PTEN overexpression was an increased presence of renal cancer, as noted above.
Fig. 3.
mRNA expression analyses. Relative mRNA expression of KLLN (A), PTEN (B) and PTENP1 (C), determined by qPCR using 36B4 as a reference gene. Control individuals were compared to subgroups of PHTS patients according to the variant status of PTEN. Only significant differences are indicated (two tailed t-test; *p < 0.05; **p < 0.01; ***p < 0.001; **** p < 0.0001). Each dot represents the mean value of the expression of the gene under study for each patient (assessed in triplicate). The mean value of each group ± SEM is indicated
We also searched for alterations in mTOR pathway genes that could be involved either as phenotype modifiers in the PTEN-mut patients or as causal factors in the PTEN-wt patients. For this purpose, we screened almost our entire series (127 patients, including PTEN-mut and PTEN-wt) by using a multigene panel (see the full list of genes in Additional file 1: Table S7). None of the variants found (Additional file 1: Table S8) has been described to trigger the mTOR pathway and they were classified as VUS until additional experiments are performed. Moreover, the number of variants in mTOR pathway genes was similar in the groups of PTEN-mut and PTEN-wt patients, and we did not find any clear correlation between these variants and a specific phenotype in the carriers. The gene panel also targeted genes associated with other cancer predisposition syndromes (such as FLCN) and we found some PTEN-wt patients with rare variants in these genes (Additional file 1: Table S8). This prompted us to consider a clinical reevaluation of these patients.
In order to expand the search for genes involved in PHTS phenotypes other than PTEN and mTOR pathway genes, we performed WES in 11 unrelated PTEN-wt patients selected for their clinical features (see Methods) and we found several potentially relevant germline variants (Additional file 1: Fig. S1). On the one hand, we found variants in known cancer genes (such as MUTYH, associated with colorectal cancer risk) that might explain a certain feature of the patient’s phenotype, but not the development of a syndrome that resembles PHTS. On the other hand, we found several candidate variants to be further assessed in following studies (e.g. in RNF135, NEDD4 and HERC1).
Additionally, available tumor DNAs were studied using the mentioned gene panel and the somatic variants found were in agreement with each cancer type, including a splicing variant in PTEN in the lung adenocarcinoma sample, and BRAF p.(V600E) and PIK3CA p.(G1049R) in thyroid cancer samples (Additional file 1: Table S9).
Discussion
Our study describes clinical and molecular findings in a series of 145 patients with clinical features of PHTS entities, the largest one studied in the Spanish population thus far [17]. In agreement with previous studies in other populations [7, 8, 10], the pathogenic variants found in our series were located along the PTEN sequence, with hotspots in exons 5 and 8. In contrast to those other studies, however, we found a relatively high number of individuals with pathogenic variants in exon 1 [7, 8, 10]. All variants in exon 1 differed from each other, ruling out a founder effect. Our sample size does not allow us to support the conclusion that this is a specific characteristic of Spanish PHTS patients. The study of more individuals in other populations and in ours is required to better address this hypothesis.
When analyzing large rearrangements, we found several cases in which the deletion also affected other genes, such as KLLN, a neighbor gene of PTEN, but also BMPR1A, a further upstream gene associated with juvenile polyposis syndrome (JPS; MIM 174,900) and colorectal cancer risk. Hence, the 2 patients with a deletion affecting both PTEN and BMPR1A showed clinical features of both PHTS and JPS. This suggests an additive effect of the two genes and a possibly increased risk of colorectal cancer in these individuals. This finding is not uncommon in PHTS patients, as it has been previously reported [18–20]. Therefore, we recommend to extend the study of large deletion carriers by using methods such as aCGH or gene panels that allow copy number variant identification, in order to identify other genes that might also be deleted and cause additional clinical risks for the patient.
Our series of patients comes from very different medical specialists and not in all cases we have an exhaustive description of their clinical picture. Understanding that the percentages of each clinical trait should be interpreted as a minimum estimate, we can draw some valuable conclusions by comparing those patients with and without mutation in the PTEN gene.
As expected, several clinical features such as macrocephaly and mucocutaneous lesions were significantly more frequent among the PTEN pathogenic variant carriers, suggesting their usefulness as clinical diagnostic criteria. Together with these features, another good indicator of the presence of a PTEN pathogenic variant was obesity, present in 22% of our PTEN-mut individuals. Obesity rates in the Spanish population were: men 15.1%, women 13.1%, boys 10.6%, girls 11.8% (Global Obesity Observatory, https://data.worldobesity.org/country/spain-199/). This feature was hardly noted in the other patient series described in the literature [7, 8, 10]. Obesity has a very heterogeneous origin, and high rates observed in our series could be related with some features of the syndrome (e.g. thyroid disorders), but the association of PTEN and obesity could also be related with the involvement of PTEN in the insulin pathway [21, 22]. Despite of the limitations of our sample size it is important to highlight this finding since obesity is a known risk factor for cancer, with a strong causal link for breast, uterine, colon and renal cancers [23], and we observed that half (6 out of 13) of the obese PTEN-mut patients suffered cancer with a median onset age of 22 years old (versus 33 for non-obese patients). We therefore suggest that attention should be given to obesity in the surveillance of PHTS patients. In fact, it might be advisable to consider PI3K inhibitors in the treatment of PHTS patients not only to reverse classical PHTS lesions (such as skin hamartomas) [24], but also to treat obesity [25, 26].
On the other hand, some characteristics classically associated with CS, such as LDD (OR = 3.78, p = 0.11) and the presence of cancer (OR = 0.40, p = 0.05) alone, were poor predictors of a PTEN pathogenic variant. In the present study, we noticed that many clinicians find the presence of 2 or more CS-associated cancers a sufficient diagnostic criterion to refer a patient for PTEN testing. However, we found that all patients with CS-associated cancers but without any other reported clinical feature of CS (such as macrocephaly or mucocutaneous lesions) were PTEN-wt, i.e., none of them carried an alteration in PTEN. Therefore, the presence of CS-associated cancers should not be criterion to refer a patient for PTEN testing, unless this is accompanied by other features of the disease.
Cancer has been usually described to develop during adulthood in PTEN pathogenic variant carriers [7, 8, 10], and only one study highlighted the risk of early cancer development, especially thyroid cancer [27]. In fact, the guidelines of the U.S. National Comprehensive Cancer Network already suggest a yearly thyroid ultrasound for PHTS patients under 18 [28]. Based on our findings, 8 patients developed cancer under 18 among 59 patients PTEN-mut+, (Table 1), if a cancer is diagnosed at young age, it should be consider also genetic testing for PTEN variants. A large prospective study in young PHTS patients would be useful to establish the appropriate age at which screening should begin for each cancer type in these individuals.
We explored if the different clinical diagnostic criteria proposed in the literature [7, 9] could retrospectively identify the patients from our series (Additional file 1: Fig. S2). The International Cowden Consortium (ICC) and the Cleveland Clinic (CC) score had a good performance by identifying around 90% of our PTEN-mut patients (as expected, given the similarity of our criteria to these), together with 40% of the PTEN-wt patients. By contrast, when using the revised criteria proposed by Pilarski et al. [9] we failed to identify a considerable amount of the patients carrying PTEN pathogenic variants (more than 50% of these individuals).
To date, there are no strong genotype–phenotype correlations in PHTS, mainly due to the small sample sizes of the studies. Bearing in mind the limitations due to the size of our cohort, we did find some significant associations between the location of the PTEN pathogenic variant and the phenotype. Interestingly, we observed an association between renal cancer and PTEN exon 1 variants (p = 0.045). As KLLN was described as a possible phenotype modifier [15], we explored its role in our PHTS patients and found an overexpression of KLLN that correlated with an overexpression of PTEN (which may indicate a co-regulation of these genes due to their shared bidirectional promoter). However, no allelic–specific expression studies have been done, so we can´t determine if the upregulation is limited to the mutated allele. In consequence, the mechanism underlying this observation and its hypothetical association with an increase in risk for renal cancer in the individuals that harbored variants in PTEN exon 1, is currently unclear.
Since a considerable proportion of CS, CS-like, BRRS and ASD-macrocephaly patients (from 20 to 90%, depending on the clinical entity) do not carry a PTEN germline pathogenic variant [2–5], it is relevant to continue searching for new genetic factors involved in PHTS development, to improve counseling, risk assessment and therapeutic measures for each patient. Even though we did not find any clear candidate, several variants in known cancer predisposition genes (such as FLCN, MUTYH and BAP1) were observed, which made us reconsider the clinical diagnosis of these patients. As an example, one patient harbored a probably pathogenic variant in FLCN, a gene that is associated with the Birt-Hogg-Dubé (BHD) syndrome which includes cutaneous lesions that can resemble CS lesions. This example suggests the need for performing a differential diagnosis considering syndromes that have overlapping clinical features with the PHTS entities. We also performed WES and found some interesting variants that could account for etiological factors in PTEN-wt patients: variants in RNF135, associated with overgrowth, macrocephaly and facial dysmorphism [29]; variants in UBN2, associated with autism [30]; and variants in NEDD4 and HERC1, which encode two ubiquitin ligases involved in PTEN and TSC2 degradation, respectively [31, 32]. We did not find candidate variants in SDH-B, SDH-D, PIK3CA, AKT1, TTN or SEC23B; these genes were suggested in the literature to be involved in CS and BRRS [11–14].
Several LDD patients do not present PTEN alterations and recently EGFR was proposed as a novel candidate for LDD susceptibility [33, 34]. Thus, we also sought for other LDD genes through WES in 4 PTEN-wt patients with this cerebellar tumor. We did not find any relevant alterations in EGFR, but of note, we found an as yet unreported variant in FGFR1, which encodes a receptor involved in PI3K signaling [35, 36].
ASD has a complex etiology, with at least 1,000 susceptibility genes reported [30] and variants in PTEN account for a relevant amount of individuals with ASD and macrocephaly [37]. Five PTEN-wt patients of our series showed this phenotype. Through WES in one of these individuals, we found a missense variant in the ATR gene (malfunction of its protein can impair fragile site stability, which can be a risk for autism), a stop gain variant in UBN2 and two different variants in EP400; variants in the latter two genes have been suggested to be associated with autism [30].
We found several variants in other genes besides PTEN that could account for a subset of the patients who tested negative for alterations in PTEN, but their importance remains to be determined before a translation to the clinical setting can be considered. Moreover, we did not find a common gene altered in several patients, similar to the results of other authors [38, 39], and only certain gene variants could explain specific individual cases. Therefore, it is possible that PTEN is the only high susceptibility gene of CS, CS-like, BRRS or ASD-macrocephaly, and other yet to be discovered factors might explain the disease in individuals with no variants in PTEN. Other approaches such as RNA-sequencing, genome sequencing or methylation assays might shed light on this issue.
The fact that the somatic variants found in our work in the cancer tissues were to be expected for each cancer type could point towards a similar evolution pattern of tumors in PHTS and their sporadic counterparts, although we were only able to study a small sample set.
Conclusions
Our findings suggest that to improve diagnosis, focus should be put on macrocephaly, mucocutaneous lesions, obesity and gastrointestinal polyposis when performing the clinical evaluation, as these were features that best suggested the presence of a PTEN pathogenic variant. Once the PTEN germline status of the patient is known, it is relevant to perform a differential diagnosis in case no pathogenic variants were found. In this last scenario or when finding large rearrangements, it is important to expand the search to other genes that might be altered causing additional or unexpected clinical risks. Finally, regarding the management and follow-up recommendations for PHTS, we suggest regularly monitoring weight and considering cancer screenings at an earlier age in young individuals. Prospective studies of PHTS patients will aid in the determination of their clinical risks.
Materials and methods
Patients and clinical evaluation
One hundred and forty-five probands (unrelated individuals from unique families) meeting relaxed clinical criteria from the International Cowden Consortium (ICC) [7], including patients who a) met the pathognomonic criteria, b) met 1 major criterion and 2 minor criteria, and c) suffered any 2 of the following cancer types within the PHTS spectrum: breast, thyroid or endometrial cancer, were included in our series. All patients were Caucasians of Spanish origin (age range from 1 to 76 years old; 84% adults and 16% ≤ 18 years old). 26 patients were seen in the consultancy of our group at the University Hospital of Fuenlabrada (UHF, Madrid, Spain). The remainder 119 patients were referred to our laboratory through collaboration with medical specialists of 35 different hospitals from Spain (PHTS Working Group). Biological samples, clinical information and signed informed consent were referred to our laboratory at the Familial Cancer Clinical Unit (CNIO, Madrid, Spain). Written formal consent was obtained from the parents or guardians of individuals under 18 years old. The patients’ phenotype information was collected through a clinical questionnaire specifically designed for this project (Additional file 4: Methods S1). We sent 119 questionnaires to medical professionals from 9 different specialties. 51 questionnaires were returned to us completed. The project has the approval of the Ethics Committee of the UHF (approval number: 20/28).
PTEN genetic analyses
DNA was extracted from samples of peripheral blood leukocytes of each proband. The presence of germline variants was evaluated in all 9 exons of PTEN (together with the intron–exon boundaries) by PCR and Sanger sequencing. Variants were named in relation to NM_000314.4. Positive results were confirmed in a second blood sample using the MyTaqBlood PCR (Bioline) reagent. Large rearrangement analysis was performed through multiplex ligation-dependent probe amplification (MLPA) with SALSA P225D1 (MRC Holland). The SurePrint G3 Unrestricted CGH 4x180K microarray (Agilent) was used for cases with large deletions involving the 5’ end of PTEN to interrogate the extent of the deletion and the location of the breakpoints. The PTEN promoter was analyzed in 31 patients in whom no potentially relevant changes in the PTEN gene were detected. Primers are mentioned in Additional file 5: Methods S2.
mRNA expression analyses
RNA was extracted from peripheral blood leukocytes with TRIzol reagent (Thermo Fisher). Concentration and integrity of the RNA were checked using NanoDrop (ND-1000 V3.7.1; Thermo Fisher). The High Capacity cDNA reverse transcriptase kit (Applied Biosystems) was used to synthesize cDNAs. cDNAs from unaffected donors were used as controls for expression levels in the qPCR reaction. Samples (n = 23) and controls (n = 23) were analyzed in triplicate. Primers for PTEN, KLLN and PTENP1 are listed in Additional file 4: Methods S1. The GoTaq® qPCR Master Mix (Promega) was used for the reaction and the qPCR was performed in an ABI QuantStudio S6 Flex System (Applied Biosystems). 36B4 was used as the reference gene to calculate relative mRNA expression using the 2−∆∆Ct method for qPCR analysis.
Next generation sequencing (NGS) panel
A total of 131 DNA samples (127 from blood and 4 from paraffin-embedded tumor tissue samples) were included in a custom NGS panel from NimbleGen (Roche) to look for other possible genetic factors involved as phenotype modifiers or with a causal role. Variant filtering consisted of maintaining only the variants in canonical transcripts (APPRIS) with high or moderate functional effect—refer to the Variant Effect Predictor (VEP, Ensembl) calculated impacts, with moderate impact corresponding to coding non-synonymous variants (e.g. missense, in-frame indels) and high to loss-of-function variants (e.g. nonsense, those disrupting canonical splice sites, frameshift indels)—, a variant allele frequency (VAF) between 0.3 and 0.6 for heterozygotes and > 0.9 for homozygotes, and a minor allele frequency (MAF) < 0.1% (gnomAD). Somatic variants were called with Mutect2 (GATK4, ref) and variants were filtered out following the following criteria: “LOW” and “MODIFIER” categories according to VEP; variants present in the matched-paired blood sample (germline variants); variants with a VAF < 15% “ were excluded. Further details are described in the Additional file 4: Methods S1 and Additional file 1: Table S7.
Whole exome sequencing (WES)
Eleven individuals with no pathogenic variants or variants of unknown significance (VUS) identified in PTEN (7 meeting Pilarski’s clinical diagnostic criteria [9], 3 with LDD, and 1 pediatric case with macrocephaly, autism and overgrowth) were selected for whole exome sequencing (WES). Germline genomic DNA samples from these individuals were quantified using Quant-iT PicoGreen dsDNA reagent (Thermo Fisher), their quality was checked using the NanoDrop spectrophotometer (ND-1000 V3.7.1; Thermo Fisher) and degradation was assessed by agarose gel electrophoresis. WES (12 Gb, 100X coverage) and primary bioinformatics analysis were performed at the Novogene Bioinformatics Institute (Beijing, China). Further details are described in the Additional file 4: Methods S1.
Interpretation and validation of variants
Variants were considered deleterious if they were described as such in public databases (ClinVar, HGMD and LOVD), or when the specific study of cDNA sequence supported a deleterious consequence. The presence of variants was confirmed in a second sample using a different method: MyTaq Blood-PCR Kit (Bioline).
Statistical analyses
Chi-square or Fisher’s exact tests were used to evaluate differences between our cohort and other previously published cohorts [7, 8, 10] using R. Logistic regression was used to evaluate associations and risks using SPSS. qPCR analyses (t-test, Mann–Whitney) were done using GraphPad Prism (GraphPad Software, Inc.). Bilateral p values < 0.05 were considered statistically significant.
Supplementary Information
Additional file 1: Fig. S1. Clinical manifestations and selected gene variants for the 11 PTEN-wt patients studied through WES. Group A: patients who meet strict Pilarski* diagnostic criteria; group B: patients who developed LDD but do not meet strict Pilarski* diagnostic criteria; group C: pediatric case with macrocephaly together with neurological alterations and overgrowth. *Pilarski et al. 2013. Fig. S2. Proportion of individuals in each group of our series identified using the indicated diagnostic criteria from the literature. (See references in Additional file 3: Appendix S2). Table S1. PTEN pathogenic point variants found in our series. Variants are named in relation to NM_000314.4. Table S2. PTEN variants of unknown significance (VUS) found in our series. Variants are named in relation to NM_000314.4. Table S3. Deleted chromosomal regions found in our series through MLPA and aCGH. Table S4. Risk of carrying a PTEN germline pathogenic variant according to different clinical features, based on analysis of our patient series. Odds ratios obtained with logistic regression. Table S5. Risk of carrying a PTEN germline pathogenic variant when the patient presents only a certain clinical feature. Odds ratios obtained with logistic regression. Table S6. Genotype–phenotype associations found in our series. Chi-square or t-test significance is shown. Table S7. List of genes included in the multigene panel. Table S8. Germline VUS found in mTOR-related genes by NGS. Table S9. Germline PTEN status and NGS findings in the indicated tumor samples (each one from a different proband).
Additional file 2: Appendix S1. List of authors and affiliations of the PHTS working group.
Additional file 3: Appendix S2. References related to supporting information.
Additional file 4: Methods S1. Clinical questionnaire.
Additional file 5: Methods S2. Primers and further information regarding the NGS and WES.
Acknowledgements
We thank the families and their caregivers for providing consent to participate in this study.
Abbreviations
- aCGH
Array comparative genomic hybridization
- ASD
Autism spectrum disorder
- BHD
Birt-Hogg-Dubé
- BRRS
Bannayan-Riley-Ruvalcaba syndrome
- CC
Cleveland Clinic
- CS
Cowden syndrome
- HGMD
Human gene mutation database
- ICC
International Cowden Consortium
- LDD
Lhermitte-Duclos disease
- JPS
Juvenile polyposis syndrome
- LOVD
Leiden open variation database
- MAF
Minor allele frequency
- MLPA
Multiplex ligation-dependent probe amplification
- PHTS
PTEN hamartoma tumor syndrome
- NGS
Next generation sequencing
- VAF
Variant allele frequency
- VATER
Vertebral, anal, tracheal, esophageal and renal abnormalities
- VUS
Variant of unknown significance
- WES
Whole exome sequencing
Authors' contributions
LP-C collected, evaluated and interpreted clinical and molecular data. ME: recruited patients and revised the manuscript. FM, PC and RL: performed and interpreted genetic studies. DG-S: analyzed results of WES. LI-P: performed statistical analysis. MS, JL and CRA: evaluated and interpreted results of NGS panel. DG-A, AH and PHTS Working Group: cared and recruited patients. MR and JP: conceptualized the study, interpreted clinical and genetic data, reviewed and revised the manuscript. MU: cared and recruited patients, designed and supervised the study and reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Spanish Ministry of Economy and Competitiveness (MINECO) and the European Regional Development Fund in the framework of project PI14/00459, and by a grant from the Federación Española de Enfermedades Raras (FEDER). FPI fellowship BES‐2015‐071383 to LP-C, funded by MINECO and the European Social Fund (ESF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The dataset of current study are not publicly available but are available from the corresponding author on reasonable request.
Declarations
Ethical approval and consent for participation and publication
This study was approved by the research ethics committee of the University Hospital of Fuenlabrada (approval number 20/28). Parents or guardians of individuals under 18 years old signed written informed consents forms for the investigation and publication of articles.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
A list of the group authors and their affiliations is provided in the Additional file 2: Appendix S1.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Miguel Urioste, Email: murioste@cnio.es.
PHTS Working Group:
Miguel Ángel Alonso, Raquel Andrés, Sara Arévalo, María del Mar Arias, Judith Balmaña, Elena Beristain, Ignacio Blanco, Mauro Boronat, Joan Brunet, María Victoria Cózar, Miguel del Campo, Arantza Díaz, Elisabeth Gabau, María Jesús Barcina, Margarita González, Miriam Guitart, Imma Hernán, Héctor Salvador Hernández, Susana Hernando, Carmen Lacambra, Adriana Lasa, Enrique Lastra, Gemma Llort, María del Rosario Marín, David Marrupe, Francisco Martínez, Víctor Martínez, Loreto Martorell, María Orera, Susana Pedrinaci, Pedro Pérez, Marta Pineda, Ana María Plasencia, Teresa Ramón y Cajal, Luis Robles, Diana Rodà, Nuria Rodríguez, Jordi Rosell, Raquel Sáez, Mónica Salvat, Antonio Sánchez, Alfredo Santana, José Luis Soto, Agustín Toll, Anna Tuneu, and Carlos Vázquez
References
- 1.Marsh DJ, Kum JB, Lunetta KL, Bennett MJ, Gorlin RJ, Ahmed SF, et al. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan–Riley–Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet. 1999;8:1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- 2.Eng C. PTEN: one gene, many syndromes. Hum Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 3.Zbuk KM, Eng C. Hamartomatous polyposis syndromes. Nat Clin Pract Gastroenterol Hepatol. 2007;4:492–502. doi: 10.1038/ncpgasthep0902. [DOI] [PubMed] [Google Scholar]
- 4.Orloff MS, Eng C. Genetic and phenotypic heterogeneity in the PTEN hamartoma tumour syndrome. Oncogene. 2008;27:5387–5397. doi: 10.1038/onc.2008.237. [DOI] [PubMed] [Google Scholar]
- 5.Spinelli L, Black FM, Berg JN, Eickholt BJ, Leslie NR. Functionally distinct groups of inherited PTEN mutations in autism and tumour syndromes. J Med Genet. 2015;52:128–134. doi: 10.1136/jmedgenet-2014-102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 7.Tan M-H, Mester J, Peterson C, Yang Y, Chen J-L, Rybicki LA, et al. A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. Am J Hum Genet. 2011;88:42–56. doi: 10.1016/j.ajhg.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nieuwenhuis MH, Kets CM, Murphy-Ryan M, Yntema HG, Evans DG, Colas C, et al. Cancer risk and genotype–phenotype correlations in PTEN hamartoma tumor syndrome. Fam Cancer. 2014;13:57–63. doi: 10.1007/s10689-013-9674-3. [DOI] [PubMed] [Google Scholar]
- 9.Pilarski R, Burt R, Kohlman W, Pho L, Shannon KM, Swisher E. Cowden syndrome and the PTEN hamartoma tumor syndrome: systematic review and revised diagnostic criteria. J Natl Cancer Inst. 2013;105:1607–1616. doi: 10.1093/jnci/djt277. [DOI] [PubMed] [Google Scholar]
- 10.Bubien V, Bonnet F, Brouste V, Hoppe S, Barouk-Simonet E, David A, et al. High cumulative risks of cancer in patients with PTEN hamartoma tumour syndrome. J Med Genet. 2013;50:255–263. doi: 10.1136/jmedgenet-2012-101339. [DOI] [PubMed] [Google Scholar]
- 11.Mahdi H, Mester JL, Nizialek EA, Ngeow J, Michener C, Eng C. Germline PTEN, SDHB-D, and KLLN alterations in endometrial cancer patients with Cowden and Cowden-like syndromes: an international, multicenter, prospective study. Cancer. 2015;121:688–696. doi: 10.1002/cncr.29106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orloff MS, He X, Peterson C, Chen F, Chen JL, Mester JL, et al. Germline PIK3CA and AKT1 mutations in Cowden and Cowden-like syndromes. Am J Hum Genet. 2013;92:76–80. doi: 10.1016/j.ajhg.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yehia L, Ni Y, Eng C. Germline TTN variants are enriched in PTEN-wildtype Bannayan–Riley–Ruvalcaba syndrome. npj Genom Med. 2017;2:37. doi: 10.1038/s41525-017-0039-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yehia L, Niazi F, Ni Y, Ngeow J, Sankunny M, Liu Z, et al. Germline heterozygous variants in SEC23B are associated with Cowden Syndrome and enriched in apparently sporadic thyroid cancer. Am J Hum Genet. 2015;97:661–676. doi: 10.1016/j.ajhg.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nizialek EA, Mester JL, Dhiman VK, Smiraglia DJ, Eng C. KLLN epigenotype–phenotype associations in Cowden syndrome. Eur J Hum Genet. 2015;23:1538–1543. doi: 10.1038/ejhg.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mester J, Eng C. When overgrowth bumps into cancer: the PTEN-opathies. Am J Med Genet C Semin Med Genet. 2013;163C:114–121. doi: 10.1002/ajmg.c.31364. [DOI] [PubMed] [Google Scholar]
- 17.Bussaglia E, Pujol RM, Jesus Gil M, Martí RM, Tuneu A, Febrer MI, et al. PTEN mutations in eight Spanish families and one Brazilian family with Cowden syndrome. J Invest Dermatol. 2002;118:639–644. doi: 10.1046/j.1523-1747.2002.01728.x. [DOI] [PubMed] [Google Scholar]
- 18.Delnatte C, Sanlaville D, Mougenot J-F, Vermeesch J-R, Houdayer C, de Blois M-C, et al. Contiguous gene deletion within chromosome arm 10q is associated with juvenile polyposis of infancy, reflecting cooperation between the BMPR1A and PTEN tumor-suppressor genes. Am J Hum Genet. 2006;78:1066–1074. doi: 10.1086/504301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Hattem WA, Brosens LAA, De Leng WWJ, Morsink FH, Lens S, Carvalho R, et al. Large genomic deletions of SMAD4, BMPR1A and PTEN in juvenile polyposis. Gut. 2008;57:623–627. doi: 10.1136/gut.2007.142927. [DOI] [PubMed] [Google Scholar]
- 20.Waisbourd-Zinman O, Mamula P, Piccoli DA. Chromosome 10q23 deletion syndrome: an overlap of Bannayan–Riley–Ruvalcaba syndrome and juvenile polyposis syndrome. J Paediatr Child Health. 2016;52:852. doi: 10.1111/jpc.13289. [DOI] [PubMed] [Google Scholar]
- 21.Pal A, Barber TM, Van de Bunt M, Rudge SA, Zhang Q, Lachlan KL, et al. PTEN mutations as a cause of constitutive insulin sensitivity and obesity. N Engl J Med. 2012;367:1002–1011. doi: 10.1056/NEJMoa1113966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakashima N, Sharma PM, Imamura T, Bookstein R, Olefsky JM. The tumor suppressor PTEN negatively regulates insulin signaling in 3T3-L1 adipocytes. J Biol Chem. 2000;275:12889–12895. doi: 10.1074/jbc.275.17.12889. [DOI] [PubMed] [Google Scholar]
- 23.Arnold M, Lam F, Ervik M, Soerjomataram I. Cancer and obesity: global burden of cancer attributable to excess weight. Lyon, Fr. Int Agency Res Cancer. 2015. http://gco.iarc.fr/obesity. Accessed 2 Mar 2020.
- 24.Wang Q, Von T, Bronson R, Ruan M, Mu W, Huang A, et al. Spatially distinct roles of class Ia PI3K isoforms in the development and maintenance of PTEN hamartoma tumor syndrome. Genes Dev. 2013;27:1568–1580. doi: 10.1101/gad.216069.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez-Guadamillas E, Muñoz-Martin M, Martinez S, Pastor J, Fernandez-Marcos PJ, Serrano M. PI3Kα inhibition reduces obesity in mice. Aging (Albany NY) 2016;8:2747–2753. doi: 10.18632/aging.101075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortega-Molina A, Lopez-Guadamillas E, Mattison JA, Mitchell SJ, Muñoz-Martin M, Iglesias G, et al. Pharmacological inhibition of PI3K reduces adiposity and metabolic syndrome in obese mice and rhesus monkeys. Cell Metab. 2015;21:558–570. doi: 10.1016/j.cmet.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smpokou P, Fox VL, Tan WH. PTEN hamartoma tumour syndrome: early tumour development in children. Arch Dis Child. 2015;100:34–37. doi: 10.1136/archdischild-2014-305997. [DOI] [PubMed] [Google Scholar]
- 28.Daly MB, Pilarski R, Berry M, Buys SS, Farmer M, Friedman S, et al. NCCN guidelines insights: genetic/familial high-risk assessment: breast and ovarian, version 2.2017. J Natl Compr Cancer Netw. 2017;15:9–20. doi: 10.6004/jnccn.2017.0003. [DOI] [PubMed] [Google Scholar]
- 29.Douglas J, Cilliers D, Coleman K, Tatton-Brown K, Barker K, Bernhard B, et al. Mutations in RNF135, a gene within the NF1 microdeletion region, cause phenotypic abnormalities including overgrowth. Nat Genet. 2007;39:963–965. doi: 10.1038/ng2083. [DOI] [PubMed] [Google Scholar]
- 30.Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aggarwal S, Bhowmik AD, Ramprasad VL, Murugan S, Dalal A. A splice site mutation in HERC1 leads to syndromic intellectual disability with macrocephaly and facial dysmorphism: further delineation of the phenotypic spectrum. Am J Med Genet Part A. 2016;170:1868–1873. doi: 10.1002/ajmg.a.37654. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X-P, Marsh DJ, Morrison CD, Chaudhury AR, Maxwell M, Reifenberger G, et al. Germline inactivation of PTEN and dysregulation of the phosphoinositol-3-kinase/Akt pathway cause human Lhermitte-Duclos disease in adults. Am J Hum Genet. 2003;73:1191–1198. doi: 10.1086/379382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colby S, Yehia L, Niazi F, Chen JL, Ni Y, Mester JL, et al. Exome sequencing reveals germline gain-of-function EGFR mutation in an adult with Lhermitte-Duclos disease. Cold Spring Harb Mol Case Stud. 2016;2:a001230. doi: 10.1101/mcs.a001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starska K, Forma E, Lewy-Trenda I, Stasikowska-Kanicka O, Skóra M, Bryś M. Fibroblast growth factor receptor 1 and 3 expression is associated with regulatory PI3K/AKT kinase activity, as well as invasion and prognosis, in human laryngeal cancer. Cell Oncol. 2018;41:253–268. doi: 10.1007/s13402-017-0367-z. [DOI] [PubMed] [Google Scholar]
- 36.Dey JH, Bianchi F, Voshol J, Bonenfant D, Oakeley EJ, Hynes NE. Targeting fibroblast growth factor receptors blocks PI3K/AKT signaling, induces apoptosis, and impairs mammary tumor outgrowth and metastasis. Cancer Res. 2010;70:4151–4162. doi: 10.1158/0008-5472.CAN-09-4479. [DOI] [PubMed] [Google Scholar]
- 37.Varga EA, Pastore M, Prior T, Herman GE, McBride KL. The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly. Genet Med. 2009;11:111–117. doi: 10.1097/GIM.0b013e31818fd762. [DOI] [PubMed] [Google Scholar]
- 38.Yehia L, Ni Y, Sesock K, Niazi F, Fletcher B, Chen HJL, et al. Unexpected cancer-predisposition gene variants in Cowden syndrome and Bannayan–Riley–Ruvalcaba syndrome patients without underlying germline PTEN mutations. PLoS Genet. 2018;14:e1007352. doi: 10.1371/journal.pgen.1007352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crivelli L, Bubien V, Jones N, Chiron J, Bonnet F, Barouk-Simonet E, et al. Insertion of Alu elements at a PTEN hotspot in Cowden syndrome. Eur J Hum Genet. 2017;25:1087–1091. doi: 10.1038/ejhg.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Clinical manifestations and selected gene variants for the 11 PTEN-wt patients studied through WES. Group A: patients who meet strict Pilarski* diagnostic criteria; group B: patients who developed LDD but do not meet strict Pilarski* diagnostic criteria; group C: pediatric case with macrocephaly together with neurological alterations and overgrowth. *Pilarski et al. 2013. Fig. S2. Proportion of individuals in each group of our series identified using the indicated diagnostic criteria from the literature. (See references in Additional file 3: Appendix S2). Table S1. PTEN pathogenic point variants found in our series. Variants are named in relation to NM_000314.4. Table S2. PTEN variants of unknown significance (VUS) found in our series. Variants are named in relation to NM_000314.4. Table S3. Deleted chromosomal regions found in our series through MLPA and aCGH. Table S4. Risk of carrying a PTEN germline pathogenic variant according to different clinical features, based on analysis of our patient series. Odds ratios obtained with logistic regression. Table S5. Risk of carrying a PTEN germline pathogenic variant when the patient presents only a certain clinical feature. Odds ratios obtained with logistic regression. Table S6. Genotype–phenotype associations found in our series. Chi-square or t-test significance is shown. Table S7. List of genes included in the multigene panel. Table S8. Germline VUS found in mTOR-related genes by NGS. Table S9. Germline PTEN status and NGS findings in the indicated tumor samples (each one from a different proband).
Additional file 2: Appendix S1. List of authors and affiliations of the PHTS working group.
Additional file 3: Appendix S2. References related to supporting information.
Additional file 4: Methods S1. Clinical questionnaire.
Additional file 5: Methods S2. Primers and further information regarding the NGS and WES.
Data Availability Statement
The dataset of current study are not publicly available but are available from the corresponding author on reasonable request.