Abstract
Although the mode of action of the ribosomes, the multi-component universal effective protein-synthesis organelles, has been thoroughly explored, their mere appearance remained elusive. Our earlier comparative structural studies suggested that a universal internal small RNA pocket-like segment called by us the protoribosome, which is still embedded in the contemporary ribosome, is a vestige of the primordial ribosome. Herein, after constructing such pockets, we show using the "fragment reaction" and its analyses by MALDI-TOF and LC–MS mass spectrometry techniques, that several protoribosome constructs are indeed capable of mediating peptide-bond formation. These findings present strong evidence supporting our hypothesis on origin of life and on ribosome's construction, thus suggesting that the protoribosome may be the missing link between the RNA dominated world and the contemporary nucleic acids/proteins life.
INTRODUCTION
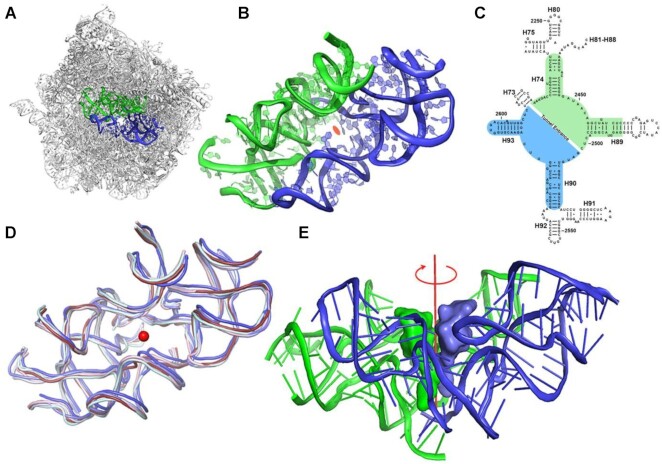
The vital role of the ribosomes and their universality stimulated profound hypotheses regarding their prebiotic origin and activity (1–4). The advent of ribosome's structures (5,6) revealed their various active sites and allowed the identification of their mode of function in genetic code decoding and protein synthesis. All ribosomes are built of two unequal subunits that associate to form the active ribosome. The small subunit provides the decoding site while the large one comprises the site for peptide bond formation, which is composed almost solely of ribosomal RNA (rRNA) chains and is named the peptidyl transferase center (PTC) (7). The PTC, situated at the heart of the contemporary ribosome, is built of a semi-symmetrical pocket-like RNA (Figure 1A, B) (8), a unique feature within the otherwise asymmetric ribosome. Analyses of structures of ribosomes from organisms throughout all kingdoms of life as well as those from organelles showed that despite the ribosomal size differences, in all cases, the fold of this pocket region and the sequence of its rRNA chains are highly conserved (Figure 1C, D) (8). In addition, structural analyses of ribosome complexes with transfer RNA molecules (tRNAs), as well as with their shorter analogs, which mimic their amino acylated 3’ termini that bind to both PTC sides (called A and P-sites, according to their accommodation of the A- and P-tRNAs) indicated that the CCA-3’-ends of the PTC bound tRNAs (9–11) are also related by a semi 2-fold symmetry (Figure 1E) (12).
Figure 1.
The protoribosome concept. (A) The symmetrical region, marked in blue (A-reg) and green (P-reg), within the rRNA scaffold of the large ribosomal subunit of D. radiodurans (PDBID 1NKW). (B) A close-up of the protoribosome where the 2-fold semi-symmetrical parts are shown. The view is along the pseudo-symmetry 2-fold axis. The center of the PTC is marked by an orange ellipse. (C) A two-dimensional structure diagram of the rRNA surrounding the PTC depicting the symmetry. The A- and P-reg nucleotides are marked using blue and green backgrounds, respectively. 23S rRNA helices numbers are marked in black labels. Nucleotides numbering according to E. coli is shown. (D) Overlay of the symmetrical region of ribosome structures from various organisms representative of various phylogenetic classes: bacterial (D. radiodurans and E. coli in slate and light blue, respectively), Yeast (S. cerevisiae in pale cyan), parasite (L. donovani in blue) and Human ribosomes (cytosolic and mitochondrial in ruby and light pink, respectively) (PDBID used are: 1NKW, 4V4Q, 4V7R, 3JCS, 4U60 and 3J7Y, respectively). The central red dot represents the position of the putative symmetry axis, which is perpendicular to the plane. (E) CCA-3’ end of A-site and P-site tRNAs were superimposed on the symmetrical region of the bacterial ribosome (PDBID 1NKW). The view is perpendicular to the semi-symmetrical 2-fold axis, shown in red.
This semi-symmetrical pocket-like region, which accounts for only ∼6% of the entire large subunit rRNA in prokaryotes, provides the scaffold for peptide bond formation by anchoring the 3’ CCA ends of the ribosomal substrates, namely the aminoacyl and the peptidyl tRNA molecules (9–11). We named the two sides of these pocket-forming RNA regions, according to their interactions with the tRNA substrates (8), A- and P-region (A-reg and P-reg), respectively. Both regions include a stem–elbow–stem (SES) structural motif, which is an ancient RNA fold found in tRNAs as well as ribozymes (13), thus leading to postulations regarding their common prebiotic origin (14,15). The elaborate architecture of this pocket provides the required framework for the catalytic activity of the contemporary ribosomes, by positioning the substrates in a spatial orientation optimal for peptide bond formation (16,17), allowing for their A-site to P-site motion (18), for substrate-mediated catalysis (19,20) and for navigating the nascent chains into the protein exit tunnel (6,12). Additionally, by confining the peptide bond formation site, this pocket limits the void required for the translocation of the tRNA 3’-end, which is imperative for successive peptide bond formation. Based on the above observations, we proposed that this pocket, which is still implanted in the modern ribosome, is actually the protoribosome, namely the remnant of an ancient machinery that presumably was initially useful in various primordial RNA-mediated reactions analogous to current biochemical enzymatic RNA processes such as replication, cleavage, ligation, splicing, phosphorylation etc. (1,21). This protoribosome hypothesized to be made exclusively of RNA molecules that self-assemble and could self-replicate to create an active pocket, was probably further snatched by various activated forms of amino acids, to form peptide bonds. In addition, the findings of self-replicating RNA molecules that possess catalytic activities (22–24), led us to hypothesize that the protoribosome is the origin of the modern ribosome. This proposal was further supported by quantum mechanical calculations (25). An additional support to our hypothesis arose from the detailed analysis of the structure of modern ribosomes, based on the abundance of rRNA A-minor motif (26). This study, which presents a hierarchal evolution of the assembly of the 23S rRNA, suggested the PTC as the origin of the ribosome. Additional studies described scenarios in which the rRNA gradually expanded from a rather small entity to its current dimension through the addition of RNA chains and ribosomal proteins, which served as structural and functional elements that are needed for the accurate and efficient activity of the contemporary ribosomes. (27,28). However, so far none of the above proposals was directly supported by experimental evidence.
Ever since the discovery that ribosomes mediate the formation of peptide bond even between minimal substrates and inhibitors, i.e., fMet-tRNAfMet (29) or CAACAA-formyl-methionine (30) and puromycin, in a reaction termed the ‘fragment reaction’, efforts were made to identify the smallest set of ribosomal components that are competent of implementing it. Thus, initially it was shown that the large ribosomal subunit provides the stereochemistry for the fragment reaction even without the presence of the small ribosomal subunit. Additional studies illustrated that identical RNA structures can be stabilized by unrelated proteins (31) suggesting that the ribosomal proteins are probably needed for the ribosomal subunit stabilization rather than for peptide bond formation catalysis. Furthermore, it has been demonstrated that transcripts of 23S rRNA could be reconstituted in vitro into catalytically active 50S ribosomal subunits demonstrating that posttranslational rRNA modifications are less needed for the principal activity of the ribosome (32,33). As the combination of several intact ribosomal components preserved partial ribosomal functionality, efforts were made to obtain such activity with minimal constructs derived from the 23S rRNA chains adjacent to the PTC (34). Interestingly, RNA ribozymes, irrespective of their origin, were shown to exhibit peptidyl transferase activity in restricted space (35). Wet-dry reaction cycles of the appropriate substrates could have also been a complementary approach to peptide bond formation in the absence of RNA (36).
In order to examine our protoribosome hypothesis empirically, RNA chains derived from the conserved pocket region were designed, prepared and tested for their ability to dimerize and catalyze the formation of a peptide bond. The latter activity was examined using the above-mentioned fragment reaction and the substrates CCA–phenyl alanine–caproic acid–biotin (CCA-pcb) and C-Puromycin (C-Pmn) (11). Gratifyingly, several of the protoribosome constructs led to the formation of a peptide bond between these substrates. To the best of our knowledge, this is the only case in which the protoribosome hypothesis is directly supported through the activity of PTC-derived RNA constructs. Herein, we discuss these findings, which present strong evidence supporting the hypothesis that the modern ribosome evolved from an ancestral protoribosome that was capable of forming peptide bonds. Thus, we claim that the protoribosome may be the missing link between the RNA dominated world and the contemporary nucleic acids and proteins world.
MATERIALS AND METHODS
RNA constructs design
In order to identify a minimal rRNA component that mimics the pocket-like structure of the contemporary ribosome and forms peptide bonds, a series of RNA constructs was designed and prepared, based on the sequences of segments of the ribosome's pocket. Particularly, the initial proto-ribosome constructs were planned on the basis of nucleotide sequences derived from the 23S rRNA of T. thermophilus (tt), which are identical to those of D. radiodurans in these regions. These minimal constructs, which consist of two helices, namely H90 and H93 (named tt_A1) or H74 and H89 (named tt_P1) (see Figure S1 for the corresponding regions within the 2D representation of the 23S rRNA and Table S1 for their sequences), form the minimal part of the 2-fold symmetrical region, in which H90 and H93 are symmetrically related to H74 and H89 (min-A and -P). A few modifications were made in the actual construct's synthesis. For example, the first nucleotide in the tt_P1 region, adenosine 2058 which is conserved in eubacteria was replaced by guanosine in our designed construct (37) to meet the requirement of T7 RNAP transcript. Since this G is conserved in eukaryotes and archaea in that location, this change was not expected to result in adverse effects. Furthermore, short loops of up to six nucleotides were inserted (marked with green lines in Figure S1 and in red fonts in Table S1) within positions where the original rRNA helices were truncated such that the minimal constructs include only the semi-symmetrical region (9). The sequence of the stable hexamer hairpin 5’-CUUCGG-3’, which is found in the center of palindrome-like patterns of various mRNA sequences and is predicted to form stem loops (38), was selected to serve that purpose. Interestingly, this hairpin is present in the 16S rRNA (starting in positions 207, 419, 1028, 1449) (38) as well as in the 23S rRNA (starting at nucleotide 1691), of available ribosome structures where it folds as loops at the tips of well-defined rRNA helices.
H92 and H80 of the contemporary ribosome hold the A and P-loops, respectively. These highly conserved loops, which reside within the semi-symmetrical region form Watson-Crick base-pairs with the 3’ CCA end of the tRNA substrates (11). Appreciating their significance, these loops, and the rRNA helices in which they are embedded, were added into the sequence of tt_A1 and tt_P1 to form extended constructs, namely tt_A2 and tt_P2, each bearing the entire nucleotide sequence of either the extended A- or P-reg within the contemporary ribosome (Figures S1, S2A, S2B and Tables 1 and S1). Further extended proto-ribosome constructs, termed tt_A1P1 and tt_A2P2 were designed based on the sequence of the entire semi-symmetrical region, either lacking or holding H92 and H80, respectively (Figures S1, S2C and Tables 1 and S1).
Within the contemporary ribosome, the interactions between the two parts of the semi-symmetrical region involve an A-minor motif (39) between a GNRA tetraloop (Where N can be any nucleotide, and R is purine) in H93 (GUGA) at the A-reg and a receptor located at H74 in the P-reg (Figure S3). As H74 is part of both tt_P1 and tt_P2 constructs, we envisaged, based on previous studies (40), that incorporation of the GUGA sequence into these constructs, at the location corresponding to H89, instead of the hyper stable hairpin CUUCGG would increase the affinity between the monomeric constructs such that they would form tighter homodimers. These GUGA holding proto-ribosome constructs were termed tt_P1c and tt_P2c, respectively (Tables 1 and S1).
In addition to T. thermophilus based constructs, additional ones were designed on the basis of sequences from Escherichia coli, Enterococcus facium and Staphylococcus aureus, and were termed ec_P1c, ef_P1c and sa_P1c, respectively. However, as the PTC is highly conserved, the sequences varied only in few bases (Figure S2D). Additional minimal P-reg constructs named tt_P1n and tt_P1m, were designed with different closing loops aimed to mimic the tt_P1 H74 natural fold, namely, without the addition of the synthetic loop CUUCGG (Tables 1 and S1, Figures S3 and S2D). In addition to the rational design described above, prior to their actual preparation, the 2D structures of the above mentioned 13 RNA constructs were predicted using free energy minimization calculations. It was found that they arrange in 2D structures that resemble their fold within the 23S rRNA contemporary structure (Figures S4 and S5), thus strengthening our hypothesis that they would fold, and function as independent entities. In addition, the propensity of the constructs tt_A1 and tt_P1 to form tight homodimers using direct complementarity criteria, rather than active pockets was examined and excluded. Alignment of sequences derived from the symmetrical region of different organisms was performed using Clustal omega (41) or MAFFT (42) servers. The aligned sequences were imaged using Jalview (43). Coot was used to superimpose structures of the symmetrical region from different organisms (44). Free energy minimization (45) was used for RNA secondary structure predictions using mfold (46) and RNAstructures (47). OligoCalc (48) was used to predict the propensity of the constructs tt_A1 and tt_P1 to form tight homodimers. Images of 3D structures were prepared using Pymol (49).
Preparation of the plasmids for in vitro transcription of RNA
The RNA constructs: ec_P1c, tt_P1m, tt_P1n, tt_A1, tt_A2 were purchased from Dharmacon, Inc. (Colorado, USA). The DNA primers for the constructs tt_P1c, tt_P1, tt_P2, tt_P2c, tt_A1P1, tt_A2P2 were obtained from Sigma-Aldrich™ and inserted into pUC57 plasmid using restriction enzymes. Since the sequences of sa_P1c and ef_P1c had high similarity to tt_P1c, site-directed mutagenesis of the tt_P1c-pUC57 plasmid was performed to achieve sa_P1c and ef_P1c plasmids. A transfer PCR was set up using Phusion Polymerase. The newly prepared plasmids (called sa_P1c-pUC57 and ef_P1c-pUC57 hereafter) were then transformed to DH5α cells and subsequently purified using Qiagen Mini-prep kit and sent for sequencing. Plasmids were stored at –20°C for future use. The plasmids were then transformed into chemically competent E. coli DH5α cells using heat shock, multiplied, and purified according to Qiagen Midi prep kit instructions.
In vitro transcription—preparation of the RNA constructs
Plasmid was linearized in batches of 1mg using HgaI restriction enzymes (New England Biolabs. Inc.) according to the instructions provided by the manufacturer. 1.4–1.6 mg of linearized plasmid was mixed with 16 mM NTP mix, 10 mM DTT, 20 mM MgCl2, 40 mM Tris-pH 8.1, 1 mM spermidine, 0.01% triton100 and 0.4 mg/ml T7 RNA polymerase purified in the lab (50), or using Thermo Scientific T7 in vitro transcription kit according to manufacturer's protocol. Promega RNAsin plus was added in transcription mix to get rid of any RNase present. Precipitated PPI was removed by centrifugation for 5 min at 2570 RCF. EDTA (pH 8.0) was added to the supernatant to a final concentration of 50 mM to cease the reaction.
Purification of RNA using preparative PAGE
The transcription mixture was extracted with phenol/chloroform/isoamyl alcohol (Sigma P3803) thrice to remove the proteins. Excess phenol was removed with ethanol precipitation, by adding 0.3 M NaAc and 0.7 V ethanol, and incubation for 1 h at –20°C or on ice. Centrifugation at 2570 RCF yielded an RNA pellet which was purified by preparative PAGE (Poly Acrylamide gel Electrophoresis). The RNA samples were mixed at a 2:1 ratio with 2×BCD dye (2× bromophenol cyanol dye loading dye consisted of 90% (v/v) formamide, 2 mM EDTA pH 8.0, 100 mM Tris˙HCl, pH 7.8, 0.25% (w/v) bromophenol blue and 0.25% (w/v) xylene cyanol), loaded on a 10% preparative gel (1:19 acrylamide: Bis acrylamide, 7 M urea and 1×TBE) and ran at 150 V in a gel of 17.5 cm × 20 cm × 1 mm dimension for 7–14 h. The RNA was extracted from the gel using 0.5 M NaAc pH 5.0 at RT and precipitated with EtOH at –20°C. The concentration of the RNA was further checked using Implen Nanophotometer® NP80 and its purity was assured from the 260/280 and 260/230 values. The purity of all the extracted RNA oligomers was further evaluated on a denaturing PAGE (Figure S6). In addition, denaturing gels after peptide formation assay is shown in (Figure S6D) to demonstrate that RNA constructs are not degraded during the assay duration.
The capability of RNA constructs to form peptide bonds
The capability of all 13 RNA constructs to allow peptide bond formation was probed using the fragment reaction (11). Each such experiment involved incubation of the relevant construct with CCA-pcb (Dharmacon, Inc. Colorado, USA) as a P-site substrate and C-Pmn (Dharmacon, Inc. Colorado, USA) as an A-site substrate. The reactions were performed as follows: RNA sample to the final concentration of 2.8–100 μM in milli-Q water was heated for 1 min at 90°C and cooled down for 2–5 min at 4°C. The RNA was then mixed with 49 μM of CCA-pcb and 19.6 μM of C-Pmn, in 50 mM Sodium phosphate pH 7.5, 20 mM KCl and 10 mM MgCl2 to a total volume of 10.2 μl. The reaction mixture was then incubated for 24 h at either 15, 25, 37 or 50°C. Large ribosomal subunit from E. coli (E50S), purified according to a reported protocol (51) was used as a positive control. However, no melting and annealing cycle was followed for E50S. Negative controls did not contain RNA or E50S.
Following the reactions, the samples were either directly analyzed by Matrix-Assisted Laser Desorption-Time of Flight (MALDI-TOF) spectrometry, Bruker® MALDI TOF, or Waters SYNAPT-XS Q-Tof High Resolution mass spectrometer (Manchester, UK) with an electrospray ionization (ESI) interface. If not analyzed immediately, the samples were stored at -80°C for maximum 24 hours prior to the MALDI or LC–MS determination.
Analysis of the fragment reaction using MALDI-TOF
The reaction mixtures were analyzed using α-cyano-4-hydroxycinnamic acid (CHCA) as the matrix. For analyses, 1 μl of the CHCA matrix prepared in the lab was combined with 1 μl of the reaction mixture and the combined solution was left to air dry on the MALDI plate. After drying, the samples were examined using the Reflector Negative Mode. All experiments were repeated at least thrice to ensure consistency.
To determine the role of the dimers in product formation, the above described peptide activity assay was performed also in the absence of MgCl2, but in the presence of KCl, or in the absence of both salts for tt_P1c, tt_P1 and tt_A1P1. The remaining protocol was similar to that described above.
Preparation of α-cyano-4-hydroxycinnamic acid (CHCA) as a matrix for MALDI TOF
5–10 mg of CHCA were dissolved in a 500 μl solution of 50% milli-Q water, 47% acetonitrile and 3% trifluoroacetic acid to provide a saturated solution, which was centrifuged. The supernatant was used as matrix for MALDI-TOF.
LC−MS analysis for further validation of peptide bond formation
To further assure the product formation, LC–MS experiments were performed. Peptide activity assays were performed with all the five constructs (tt_P1c, tt_P1, tt_A1P1, sa_P1c, ef_P1c) that were able to show peptide bond formation with MALDI-TOF. The reactions were performed as follows: RNA sample with a final concentration of 2.8 μM in milli-Q water was heated for 1 min at 90°C and cooled down for 2–5 min at 4°C. The RNA was then mixed with 49 μM of CCA-pcb and 49 μM of C-Pmn, in 50 mM Sodium phosphate pH 7.5, 20 mM KCl and 10 mM MgCl2 to a total volume of 50 μl. The reaction mixture was then incubated for 24 h at 37°C. Large ribosomal subunit from E. coli (E50S), was used as a positive control. However, no melting and annealing cycle was followed for E50S. Negative controls did not contain RNA or E50S.
The reaction mixtures for tt_P1c, tt_P1, sa_P1c, ef_P1c, tt_A1P1 were then concentrated to a volume 10 μl and analyzed by UPLC–MS without further dilution. However, for E50S concentration of the sample was not necessary. The spectra were recorded in the negative ion mode within a mass range from 400 to 1400 m/z. The parameters were set as follows: capillary voltage at 2.2 kV, cone gas flow at 30 l/h, source temperature at 140 °C, and cone voltage at 30 V(−). The desolvation temperature was set at 600°C, and the desolvation gas (N2) flow rate was set at 800 l/h. Lock spray was acquired with Leucine Encephalin (m/z 554.2615 in negative mode) at a concentration of 200 ng/ml and a flow rate of 10 μl/min once every 10 s for 1 s period. Waters MassLynx v4.2 software was used for data acquisition and data processing.
The analytes were separated using Waters Premier Acquity UPLC system. The aqueous mobile phase A consisted of 50 mM 1,1,1,3,3,3-hexafluoro-2-propanol (Sigma-Aldrich, 105228) and 2.5 mM hexylamine (Sigma-Aldrich, 219703). The organic mobile phase B consisted of methanol with 50 mM 1, 1,1,3,3,3-hexafluoro-2-propanol and 2.5 mM hexylamine. The separation was achieved with Waters Acquity Premier BEH C18 Column, 1.7 μm, 2.1 × 50 mm at 0.3 mlL/min flow rate, 60°C. After a one min hold at 10% B, the gradient was ramped to 50% B in 6.5 min, then the column was maintained at 95% B for 2.5 min and re-equilibrated at initial conditions.
To ensure that a peptide bond was indeed formed, the stability of the product in basic conditions was examined. Following the reaction termination, the reaction mixture of tt_P1c was divided into two parts. One part was set aside, while 0.5 M NaOH was added to the second part till pH 10.0 was achieved. This mixture was then incubated at 25ºC for 60 min. After 60 min, the pH of the reaction, monitored using pH strips, was brought to pH 7.0 using 0.5 M acetic acid. Both samples were then examined by MALDI.
Dimerization assay of RNA constructs (EMSA native gel)
RNA samples were diluted with milli-Q water to a final concentration of 2 μM, annealed for 1 min at 90°C and chilled on ice. The samples were then mixed with a dimerization buffer DB comprising of 89 mM Tris-borate (TB) pH 7.5 and 15 mM MgCl2, incubated for 30–45 min at 37°C and chilled on ice for 15 min to 18 h. Alternatively, to assess the role of MgCl2 in folding and dimer formation, samples were mixed with Tris–boric acid EDTA (TBE) pH 7.5 instead of DB buffer. The samples were then run on a native gel of 12% PAGE and 15 mM MgCl2 at 4°C in the DB buffer. The constructs tt_P1c, ec_P1c, tt_P1, tt_P2, tt_P2c, tt_P1m, tt_P1n, tt_A1, tt_A2 and tt_A1P1 were run at 40 V for 14 h on a 12.5 cm × 6.5 cm × 0.75 mm gel while for the longer construct, i.e. tt_A2P2 the gel was run at 80V for 10 h on a gel of similar dimension. The gels were stained with Ethidium Bromide (EtBr) and imaged using ChemiDoc™ XRS + Imager.
Dimerization of the RNA constructs sa_P1c and ef_P1c was achieved using the slow annealing method. Samples of sa_P1c and ef_P1c were mixed with milli-Q water and DB to a final concentration of 1.8 μM. RNA samples were placed in a heat block at 90ºC for 30 s to denature them. They were then gradually denatured stepwise at the rate of 1.5ºC per minute to room temperature (25ºC) gradually over a period of 30–40 min. The RNA was then brought to 4ºC by keeping it on ice for a minimum of 2 h. They were stored at 4ºC and used within 24 h. The samples were incubated at 37ºC for 30 min and then on ice for 15 min. To the tubes, 2× RNA loading dye was added. An electrophoretic mobility shift assay (EMSA) was performed on a 12% native PAGE gel. The gels were run at 80V for 5–8 h on a 12.5 cm × 6.5 cm × 0.75 mm gel. They were then stained with EtBr and imaged using ChemiDoc™ XRS + Imager.
Dimerization at higher concentrations for tt_A1, tt_A2 and ec_P1c was done at 20 μM final RNA concentration. The procedures were followed exactly as done previously for the corresponding samples at 2 μM RNA concentration.
RNA 3’-end fluorescent labelling and purification
In order to determine the dimers dissociation constant (Kd) the RNA 3’-end of the constructs were fluorescently labelled with Fluorecein-5-thiosemicarbazide (FTSC) using a published protocol (Figure S7) (52). In brief, 250 pmol of RNA were oxidized by incubating it with NaIO4 (0.535 μg, 2500 pmol) and 50.5 mM NaAc pH 5.6, to a total volume of 40 μl for 90 min at 25°C in the dark. Excess of oxidant was then quenched by the addition of Na2SO3 (0.63 μg, 5000 pmole) and further 15 min incubation at 25°C in the dark. Fluorecein-5-thiosemicarbazide (FTSC), (3.16 μg, 7500 pmole) dissolved in DMF was then added to the oxidized RNA solution and the mixture incubated for 3–12 h at 37°C in the dark. Following that manipulation, the excess of FTSC was removed with illustra MicoSpin G-25 Columns (GE life sciences). The size and purity of the labelled RNAs were assessed using denaturing gels.
Determination of the dimers dissociation constant (Kd)
In order to estimate the stability of the laboratory constructed dimers, RNA dimerization was approximated using a native EMSA gel. A constant amount of labelled RNA, namely 0.15μM for tt_P1, tt_P1c, tt_P1m, tt_P1n, sa_P1c, ef_P1c and 1.5 μM of tt_A1P1 were titrated with increasing 0–42 μM concentration of the unlabelled RNA. Seemingly, due to the longer length of tt_A1P1, the labelling with a fluorophore was less efficient than that achieved with the shorter constructs. The samples were then treated as described above for the dimerization assay of corresponding samples. Next, the gels were imaged with Typhoon FLA9500 under fluorescent reader. For tt_P1, tt_P1c, tt_A1P1, tt_P1m, tt_P1n, sa_P1c and ef_P1c fluorescent bands of monomer and dimer were quantified by the software ImageJ. The percentage of dimers was then calculated using Eq. (2) of SI page 45.. The calculated percentage of the dimers were plotted vs. the concentration of the unlabelled monomers in the following equation using Mathematica 11.3 in order to calculate the dissociation constant (Kd) (Derivation of this equation has been presented in SI Page 45):
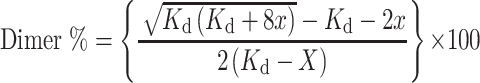 |
(1) |
where X = M0 or concentration of the unlabelled oligomers, Kd = dissociation constant of RNA constructs, Y = dimer %.
K d was not calculated for the RNA constructs tt_P2, tt_P2c and tt_A2P2, since along with the increase in concentration of the monomer, the amount of multimer formed also increased precluding the possibility of reliable Kd calculations.
RESULTS
RNA constructs mediate peptide bond formation
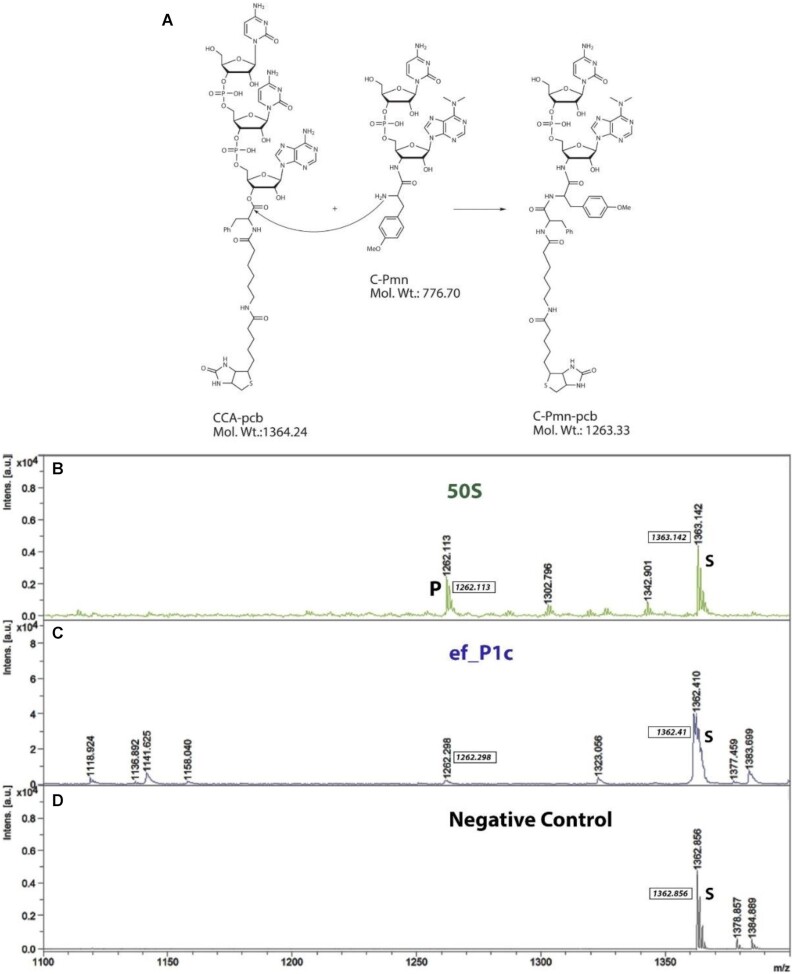
A series of 13 rRNA constructs was designed based on the structure and sequence conservation of the PTC according to various natural sequences (see Methods section for details) (Tables 1 and S1 and Figures S1–S3). The capability of all our RNA constructs to lead to peptide bond formation was probed by applying the fragment reaction followed by analyses of the reaction mixtures using Matrix-Assisted Laser Desorption-Time of Flight (MALDI-TOF) mass spectrometry analysis (53) which was further confirmed by LC–MS. For this reaction, CCA-pcb and C-Pmn were chosen as the P and A-site analogs, respectively (Figure 2A). A fruitful condensation between these counterparts provides the product C-puromycin–phenylalanine–caproic acid–biotin (C-Pmn-pcb molecular weight of 1263.33), which could be detected using MALDI-TOF and LC–MS, in a negative mode, as evidenced from the analysis of the reaction mixture of the E. coli 50S subunit with these substrates (Figures 2B and 3C).
Figure 2.
The fragment reaction and analyses of peptide bond formation using MALDI. (A) Schematic representation of a fruitful condensation between the substrates CCA-phenylalanine-biotin (CCA-pcb) and C-puromycin (C-Pmn) to afford the product C-Puromycin-phenylalanine-caproic acid-biotin (C-Pmn-pcb). (B–D) Representative MALDI-TOF spectra of fragment reaction mixtures recorded in negative ion reflector mode. The peak of CCA-pcb (starting material) and the C-Pmn-pcb (product peak) are marked with S and P, respectively. (B) E. coli large ribosomal subunit (50S) (positive control). (C) RNA construct ef_P1c. (D) Substrates without RNA or E. coli 50S (negative control).
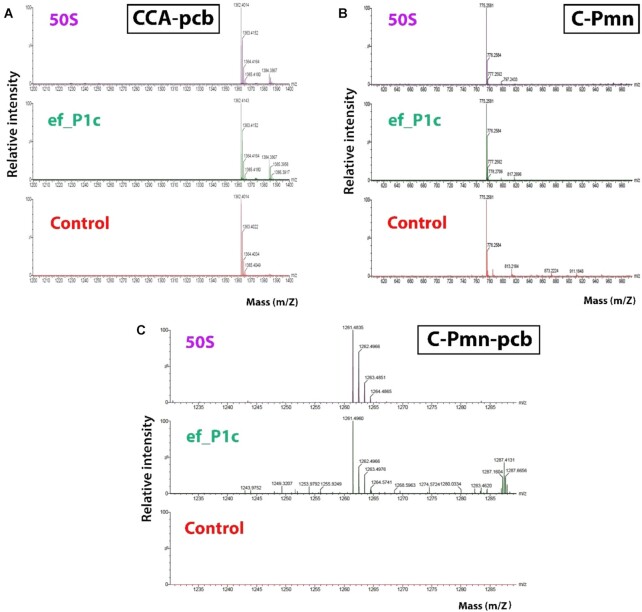
Figure 3.
Analyses of peptide bond formation using LC–MS. Representative LC–MS spectra of fragment reaction mixtures of 50S (positive control), ef_P1c and Negative control (No ribosome or RNA). The peaks of the starting materials CCA-pcb and C-Pmn and the product C-Pmn-pcb (A–C).
To test the ability of our protoribosome RNA constructs to promote this reaction and provide the above-mentioned product, they were incubated with the same substrates for 24 h at 37°C (Figures 2C, 3C and Figure S8). The reaction of the E. coli 50S was used as a positive control, and with milli-Q water as a replacement of the RNA constructs, as a negative control (Figure 2B and D, respectively). In the case of the construct tt_P1c, its complementary sequence com_tt_P1c (Table S1), which was constructed according to base-paring rules, and did not catalyze the reaction, served as an additional negative control (Figure S8H). Analyses of the MALDI data clearly showed that 5 of our designed constructs, namely, ef_P1c, tt_P1c, sa_P1c, tt_P1 and tt_A1P1, mediated peptide bond formation at 37°C, as evident from the presence of the desired product molecular ion peak in their respective spectra (Figure 2C and Figure S8). However, the remaining 8 RNA constructs did not show any activity, as indicated from the absence of the product molecular ion peak in the MALDI spectra of their reaction mixtures. The reactions in the presence of the protoribosome constructs were significantly less effective with much lower product peak intensities as compared to those obtained in the presence of the E. coli 50S.
To further authenticate the identity of the product peak, the reactions of the working constructs, i.e. tt_P1c, tt_P1, tt_A1P1, sa_P1c, ef_P1c, E50S (positive control), as well as the negative control (no E50S or RNA construct) were repeated and their mixtures were analyzed using LC–MS. The LC–MS experiment showed a peak corresponding to the product C-Pmn-pcb in all cases except negative control (Figure 3 and Figure S9). Comparison of the UPLC chromatograms of the constructs mentioned above with that of the E50S ribosomal subunit clearly indicate that the product's intensities (ion count) are much lower than the latter. That is, while in the case of the E50S the product intensity is of 2.06e5, ef_P1c, tt_P1, sa_P1c, tt_A1P1, tt_P1c shows product intensities of 472, 435, 522, 1800 and 1160 respectively (Figure S10) despite of the construct samples being approximately 5 times more concentrated than that of the E50S.
To validate that the product presenting the desired mass indeed holds a peptide bond, the reaction mixture of tt_P1c was treated with 0.5 M NaOH followed by neutralization with acetic acid, prior to its MALDI analysis. As expected, a peak corresponding to a product of phosphodiester bond hydrolysis, i.e. Pmn-pcb, was observed (Figure S11). The fact that after this base treatment a product having the required mass was obtained, suggests that a peptide bond, which is stable under these conditions, is indeed present in the product.
Our results further demonstrate that constructs which include A- and P-loops, i.e. tt_A2, tt_P2, tt_P2c and tt_A2P2, are not capable of catalyzing the peptidyl transfer reaction. In the case of tt_P2, tt_P2c and tt_A2P2 the addition of the A- and P-loops impedes the activity of the shorter analogous constructs tt_P1, tt_P1c and tt_A1P1. A plausible explanation for this observation is that these extended constructs may fold to different conformations and accordingly form various dimers, which hinder the binding of the substrates, as seen in Figure 4B. In the case of the construct tt_A2, however, the addition of the A-loop did not change the disability of its shorter analog tt_A1 to catalyse the reaction.
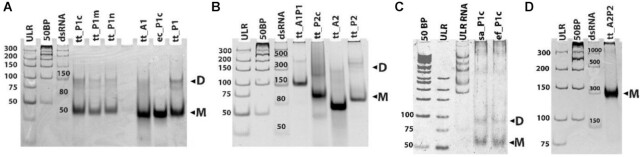
Figure 4.
Dimerization capability of the RNA constructs. EMSA Native gels of the RNA constructs. (A) Monomer constructs (M) tt_P1c, tt_P1m, tt_p1n and tt_P1 form dimers (D) (lanes 4–6, 10). Constructs tt_A1 and ec_P1c do not dimerize (lanes 8–9). (B) Monomer constructs (M) tt_A1P1, tt_P2c and tt_P2 form dimers (D) (lanes 4–5,7). Construct tt_A2 does not dimerize (lane 6). (C) Monomer constructs (M) sa_p1c and ef_p1c form dimers (D), small amounts of higher order oligomers can also be seen at the upper part of the gel (lanes 4–5). (D) Monomer constructs (M) tt_A2P2 forms multimer (lanes 4). (A–D) Two DNA ladders and one RNA ladder were used as reference (lanes 1–3).
Exploring reaction conditions
In an attempt to understand the effect various parameters have on the reaction outcome a set of additional experiments were performed. We first studied the effect of the experiment duration on the reaction. Accordingly, the reaction with tt_P1c was performed for variable time durations ranging from 30 min to 10 h in addition to almost all other experiments that were run for 24 h. As evident from the MALDI spectra (Figure S12A), the desired product as well as substrates were observed in all cases ranging from 2 to 10 h. However, no product was obtained in reactions preformed for 30 min or 1 h. This observation suggests that in the presence of our construct a minimum of 2 h reaction is required to obtain significant amount of product such that it would be detectable using MALDI. The fact that no significant difference was observed between the reactions ran for 10 and 24 h suggests that, extended time is not beneficial in respect to the reaction completion. In case of the E50S subunit, however, 1 h was sufficient to catalyse the product formation such that it would be detectable using MALDI. When the reaction duration was reduced to 30 min no product was observed even in the presence of the 50S ribosomal subunits (Figure S12B). In order to examine whether the reactions in presence of RNA constructs do not run to completion due to their instability under the reaction conditions, i.e. prolonged time, the 24 h reaction mixture was run on a denaturing polyacrylamide gel. Single bands of RNA constructs corresponding to their expected size were observed, suggesting that degradation of the RNA constructs is not the reason for the incomplete reactions (Figure S6D).
Next, we examined the effect of the concentrations of the reaction components. Accordingly, variable concentrations of the construct tt_P1c, i.e. 10, 20, 30, 40, 50 and 100 μM, were used instead of the typical concentration of 2.8 μM, while all other parameters were left unchanged (i.e. CCA-pcb and C-Pmn were 49.9 and 19.9 μM, respectively). As evident from the MALDI spectra which offers qualitative information rather than quantitative (54) (Figure S13), the increase in the construct concentration did not significantly change the reaction outcome, which led in all cases to the desired product. Thus, changing the stoichiometry of the reaction from ∼1:20:8 of RNA construct/CCA-pcb/C-Pmn up to ∼35:20:8 namely increasing the RNA amount by a factor of 35 did not affect the reaction outcome. Similarly, varying the concentrations of the substrates CCA-pcb and C-Pmn, i.e. from 49.9 and 19.9 μM, respectively (ratio of 20:8) to 19 or 49 μM of both CCA-pcb and C-Pmn (ratio of 1:1), in presence of the construct tt_P1c for 24 h reaction time, did not show any major effect on the reaction, which was successful in all cases (Figure S14).
Finally, in order to evaluate the effect of the reaction temperature on its outcome, in addition to 37°C, the reaction was performed at 15, 25 and 50°C. Interestingly, according to the MALDI data, the constructs tt_P1c, tt_P1 and tt_A1P1 as well as the 50S ribosomal subunit mediated peptide bond formation at these temperatures. The constructs sa_P1c and ef_P1c, on the other hand, were found inactive under these conditions (Figures S15–S17).
Dimerization capability of the RNA constructs
Aiming to understand the activity pattern displayed by our RNA constructs, we examined their state under conditions similar to that of the activity assay reaction. This was sought, as we envisaged that dimerized constructs might imitate the pocket-like structure surrounding the PTC and provide activity. The dimerization capability of our 13 RNA constructs was tested using the Electrophoretic Mobility Shift Assay (EMSA) (Fig. 4A–D) (55). This assay showed that constructs mimicking the minimal P-reg (min-P), namely, tt_P1, tt_P1c, tt_P1n and tt_P1m tend to form dimers (Figure 4A and C), whereas those derived from the minimal A-reg (min-A); tt_A1 and tt_A2, do not (Figure 4A and B). The latter constructs even failed to form dimers under higher concentrations (Figure S18F). The ability of the P-reg constructs to dimerize was further observed with the tt_P1c analogs, sa_P1c and ef_P1c derived from the PTC sequences of S. aureus and E. facium ribosomes, respectively (Figure 4C). The construct ec_P1c originating from the E. coli sequence, did not form a dimer, even at higher concentrations (Figure S18F). Evidently, in some cases introducing slight changes in the RNA sequence had a significant effect on its dimerization capabilities.
Extended P-reg constructs, namely, tt_P2 and tt_P2c formed few multimeric conformations with no clear dominance of a single band (Figure 4B) in contrast to the minimal P-reg constructs, tt_P1, tt_P1c, tt_P1n and tt_P1m which showed two distinct bands (Figure 4A). Thus, it seems that the P-loop addition to the constructs gives rise to the observed conformational variability. The constructs mimicking the entire symmetrical region gave results that further exhibit this pattern. Whereas tt_A1P1 showed distinct two bands (Figure 4B), tt_A2P2, gave a mixture of higher order multimers (Fig. 4D). Since the A-reg constructs did not form dimers (Figure 4A and B), it appears reasonable to believe that the minimal P-reg is actually the driving force for dimerization of the entire symmetrical region constructs. The preference of P-reg constructs to form homodimers might indicate on the earlier prebiotic origin of that region, a time when no difference between the two sides of the pocket existed, in contrast to its contemporary form, in which the A site attracts the aa-tRNA and the P-site pushes out the free tRNA.
Interestingly, and in support to our above hypothesis, our results point out that only the constructs that dimerized, thus potentially mimicking a pocket structure, mediated peptide bond formation (Table 1). The constructs which did not dimerize, failed to exhibit such activity. However, although required, it appears that dimerization by itself does not assure peptide bond formation, as evidenced from the lack of such activity, within the conditions tested, by the constructs tt_P1m, tt_P1n and com_tt_P1c, albeit their dimerization.
Table 1.
RNA constructs used, their dimerization capability and activity in peptide bond formation
| RNA Construct | Organism origin | 2D PTC location | Length | Dimer Kd (μM) | Peptide bond | Remarks |
|---|---|---|---|---|---|---|
| tt_A1 | T. thermophilus |
|
64 | ✗ | ✗ | min-A |
| tt_P1 | T. thermophilus |
|
71 | ✓ 47.43 ± 7.10 | ✓ | min-P |
| tt_A1P1 | T. thermophilus |
|
135 | ✓ 8.55 ± 2.53 | ✓ | min-A + min-P |
| tt_A2 | T. thermophilus |
|
91 | ✗ | ✗ | A-reg (includes A loop) |
| tt_P2 | T. thermophilus |
|
102 | ✓ | ✗ | P-reg (includes P loop) |
| tt_A2P2 | T. thermophilus |
|
193 | ✓ | ✗ | A-reg + P-reg |
| tt_P2c | T. thermophilus |
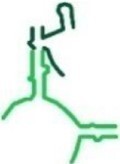
|
98 | ✓ | ✗ | P-reg and GNRA tetraloop |
| tt_P1c | T. thermophilus |
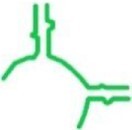
|
67 | ✓ 10.57 ± 1.16 | ✓ | min-P + GNRA tetraloop |
| ec_P1c | E. coli |
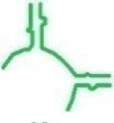
|
67 | ✗ | ✗ | min-P + GNRA tetraloop |
| sa_P1c | S. aureus |
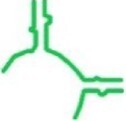
|
67 | ✓ 0.73 ± 0.19 | ✓ | min-P + GNRA tetraloop |
| ef_P1c | E. faecium |
|
67 | ✓ 1.00 ± 0.20 | ✓ | min-P + GNRA tetraloop |
| tt_P1m | T. thermophilus |
|
64 | ✓ 6.57 ± 1.28 | ✗ | min-P + UAA loop |
| tt_P1n | T. thermophilus |
|
65 | ✓ 32.11 ± 4.19 | ✗ | min-P + UUAA loop |
To further substantiate the significance of construct dimerization in peptide bond formation, the reactions with tt_P1, tt_A1P1 and tt_P1c were performed either in the absence of all salts, i.e. potassium chloride and magnesium chloride, or in the presence of the former and absence of the latter. It was interesting to evaluate these conditions since magnesium chloride, specifically, was found essential for the construct's dimerization, as dimers were not observed in its absence (Figure S18 A–C). Potassium chloride, on the other hand did not promote dimerization (Figure S18D) although it is known to bind to ribosomal RNA and in some cases was shown to replace the hydrated Mg2+ ion (56). Evidently, reactions under both conditions were not fruitful as no product was visible on the MALDI-TOF spectra of any of the three construct mixtures (Figure S19), suggesting that indeed dimer formation might be a necessary condition for peptide bond formation. To rule out any contribution of the substrates CCA-pcb and C-Pmn to the dimer formation even in the absence of any salt, i.e. MgCl2 and KCl, EMSA of the reaction mixtures, without salts, were also performed. As evident from the gels obtained (Figure S18E) the presence of the substrates did not promote dimer formation. Thus, the above experiments suggest that the presence of Mg2+ ions is crucial for the formation of dimers which in-turn seems necessary for the constructs activity. The observations further imply that both the substrates and K+ ions cannot replace Mg2+ in affecting dimerization or peptide bond formation.
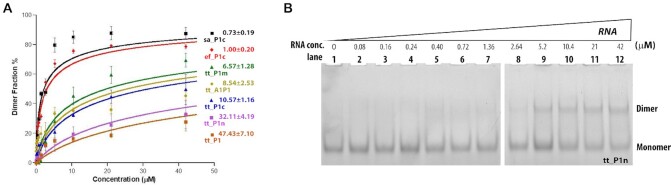
To assess the stability of the RNA dimers we set to quantify the dissociation constants (Kd) of tt_P1, tt_P1c, tt_P1m, tt_P1n, tt_A1P1, sa_P1c and ef_p1c using EMSA (55). Accordingly, RNA constructs of increasing concentrations were incubated with constant concentration of fluorescently labelled RNA of each specific construct and the mixtures were loaded on a native polyacrylamide gel. The gels were then quantitatively analyzed using a fluorescence reader (Figure 4, Figure S20) and qualitatively analyzed post ethidium bromide staining (Figure S21). In all cases similar to our qualitative observations, two bands, corresponding to the monomer and dimer, appeared predominantly even upon RNA concentration increase.
The thus determined dimer dissociation constants, from equation (1) in ‘Material and Methods’ section (Figure 5 and Table 1) present variability in the tendency of the various RNA constructs to homodimerize. Whereas ef_P1c and sa_P1c form strong dimers as evident from their low Kd values, 1.00 and 0.73 μM respectively, a Kd value of 32.11 μM was obtained for tt_P1n and 47.43 μM for tt_P1 indicating the formation of less stable dimers. The constructs tt_A1P1, tt_P1c and tt_P1m, on the other hand, were found to have Kd values of 8.54, 10.57 and 6.57 μM, respectively, indicating they form dimers of intermediate strength. The dimerization assay also indicated on variability in the maximal dimer percentage each construct reached. While sa_P1c and ef_P1c displayed the highest percentage of dimers, i.e., 88% and 79% respectively, tt_P1m and tt_P1c achieved lower, yet still, a considerably high percentage of dimers, i.e. 69% and 50%, respectively, while tt_A1P1, tt_P1n and tt_P1 attained only 45%, 32% and 28% dimerization, respectively. The observed constructs' Kd values completely correlate with the maximal dimer percentage observed for the constructs. While the constructs which show a lower dimer percentage show a higher Kd, the ones with a higher dimer percentage show a considerably lower Kd.
Figure 5.
Determination of the Kd values of the RNA construct dimers. (A) The average mean fluorescence intensity of varying concentration of dimer vs. monomer obtained was plotted to determine the dissociation constant Kd. Each experiment was repeated three times. An error bar represents the standard deviation of mean. (B) A representative gel (of RNA construct tt_P1n) used to determine the Kd value. Increasing concentration of RNA solutions, from 0 μM (left) to 42 μM (right) were mixed with a constant concentration of labelled RNA (0.15 μM). Gels were imaged fluorescently.
DISCUSSION
The studies reported herein represent attempts to explore our working hypothesis of ribosomal origin. Our results highlight the P-reg as an effective dimer forming RNA chain, in comparison to the A-reg. Accordingly, we suggest that the P-reg dimers may have served as the ancestral sequence which formed an RNA machine capable to mediate RNA needed reactions as well as peptide bond formation. This molecular machine, however, did not possess the ability to support substrate motion from one site of the symmetrical pocket to the other, as required in the contemporary mode of protein biosynthesis. According to this hypothesis, during the evolution of this peptide forming molecular machine, its two parts were independently mutated such to adapt to the polypeptides formation requirement, including the entrance and exit of the substrates and products, as well as their motion from side to side. This resulted in sequence variation between the A and P-reg, which exists in contemporary ribosomes. Consequently, in all current ribosome structures, the fold of the PTC exhibits semi-symmetry but hardly any sequence identity as observed between the constructs tt_A1 and tt_P1 and between tt_A2 and tt_P2. Hence, the detected complementarity between the nucleotides that constitute the two halves of the PTC cavity may hint on the evolutionary path from P-reg homodimers to a heterodimeric AP region (57). In this respect, preliminary studies have shown that in some cases, constructs derived from the A and P regions formed heterodimers that promoted peptide bond formation. A thorough and systematic study in this direction is underway.
In relation to the actual mechanism by which our RNA constructs promote peptide bond formation, we hypothesize that the substrates CCA-pcb and C-Pmn bind to our constructed pockets, which mimic the A- and P-regions, in a similar manner to which the 3’-end of tRNAs interact with the PTC of the contemporary ribosome. Namely, in proximity and proper spatial orientation, which permits C-Pmn nucleophilic attack on CCA-pcb to provide the peptide bond. Aiming to shed light on the suggested mechanism, we have examined the fragment reaction using various conditions. Specifically, altering the reaction components stoichiometry by increasing the concentrations of the RNA and the substrates, as well as varying the reaction duration. However, these variations did not affect the reaction outcome. As the expected products obtained in all cases with the starting materials, it appears that the reaction reaches its maximal possible level rather rapidly.
Among the protoribosome constructs that form peptide bonds, tt_A1P1 is an exception, as in principal, in its monomeric form, prior to dimerization, it includes the entire protoribosome entity. Interestingly, under the dimerization conditions tt_A1P1 indeed mediated peptide bond formation, while it was not active in this reaction in the absence of salts, where dimerization was not obtained. As we cannot rule out the possibility that the salts are also needed for maintaining an optimal folding of an active monomeric RNA construct, the significance of tt_A1P1dimerization for its catalytic activity is only hypothesized. On the other hand, since the construct tt_P1 forms dimers and exhibits activity, it is possible that a dimer of tt_A1P1, in which tt_P1 is embedded, is actually formed through the P1 part which mediates the peptide bond formation. The findings that the addition of A- and P-loops to the various constructs did not promote peptide bond formation by these extended constructs suggests that these loops might have gained necessity only once the ribosome and its substrates evolved to larger entities.
In addition, we observed that typically the PTC SES motif may promote dimerization/pocket formation but not in all cases (i.e. constructs tt_A1, tt_A2, ec_P1c). Thus, although required, dimerization by itself, apparently is not sufficient to promote peptide bond activity as exemplified by constructs, tt_P2, tt_A2P2, tt_P2c, tt_P1m and tt_P1n.
Earlier studies by Zhang and Cech (35) demonstrated in vitro selection of a ribozyme that perform peptidyl transferase reaction as the ribosome. This ribozyme lack specificity with respect to the amino acid, a feature necessary for a generalized protein-synthesizing enzyme. Nevertheless, the generated sequences are not related to the contemporary ribosome conserved active site. Later studies by Anderson et. al. (34) generated a minimal construct that comprises much of the 23S rRNA peptidyl transferase center, including the central loop and the A- and P-loops but their minimal rRNA domain was inactive for peptide bond formation under all tested conditions. In addition, while working on this manuscript a study (58), which is based on our suggestions (12,21,25,59–66), indicating that a 9 lysines oligomer could be produced by a larger RNA construct encapsulating the ribosomal PTC, was published. However, their constructs include additional extensions to the minimal protoribosome and are 2x110-mer long. Such length of RNA sequences conserving the structure and function of the PTC was shown to have infinitesimal probability to emerge by chance (67), hence it seems to relate to an evolutionary step following the protoribosome, i.e. towards the formation of the actual ribosome.
In summary, our findings that five of our designed protoribosome RNA constructs, which vary in their sequences, mediated peptide bond formation, strongly support our hypothesis that self-dimerized protoribosome entities that form peptide bonds could have existed in a prebiotic world. Our results demonstrate the necessity of dimerization for peptide bond formation. So far, functional RNA oligomers have rarely been observed in nature, although in vitro studies show that RNA has a high potential to multimerize through stable tertiary contact motifs, such as the GNRA tetraloop-receptors (68). In addition, our results suggest that a homodimer, formed from P-reg based protoribosome, could have served as an ancestor of the evolving heterodimers. A scenario where a homodimeric P-reg active RNA entity that was generated by fusion, and later underwent mutagenesis or replication of one of the two P-reg copies, to create the A-reg of the contemporary heterodimeric ribosome, seems plausible. This fashion may be reflected in the contemporary ribosome activity as the initiator tRNA substrate binds the P-site of the PTC in the large ribosomal subunit (69). In addition, our results suggest a rather limited catalytic activity of the protoribosome, compared to the current contemporary ribosome efficiency. Hence, we speculate that the superb catalytic abilities of the contemporary ribosome evolved later, upon the stabilization of the ancestral translation machinery by the addition of longer rRNA chains and ribosomal proteins, which could even adopt parts of RNA bases (e.g. the imidazole ring of purines by histidine (70)). Mutually stabilizing interactions between proto-peptides and RNA (71) could have also occurred. Thus, althoug doutfall until recently (72), the survival of the protoribosome association was shown. Furthermore, the ability of some of the constructs to form peptide bonds at diverse temperatures may imply that the protoribosome could maintain its activity in the prebiotic world where temperature and environmental conditions were suggested to be erratic (73).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ella Zimmerman, Donna Matzov, Matthew Belousoff and Hagen Hofmann for useful discussions and comments; Shoshana Tel-Or, Miriam Lachever and Maggie Kessler for their interest and experimental support; Arie Tishbee for sharing with us his MS expertise, Zvi Hayouka and Heli Bochnic-Tamir from The Robert H Smith Faculty of Agriculture, Food and Environment for their indispensable assistance with the MALDI spectrometry. We also thank Dr Vaijayanti A. Kumar from CSIR-National Chemical Laboratory, India for her useful discussion and comments on MALDI spectrometry.
Notes
Present address: Chen Davidovich, Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Faculty of Medicine, Nursing and Health Sciences, Monash University, Clayton, Victoria, Australia.
Present address: Miri Krupkin, Department of Structural Biology, Stanford University School of Medicine, Stanford, CA, USA.
Present address: Nikita Dinger, Department of Chemical, Materials and Production Engineering, University of Napoli Federico II, Naples, Italy.
Contributor Information
Tanaya Bose, Department of Chemical and Structural Biology, Weizmann Institute of Science 7610001 Rehovot, Israel.
Gil Fridkin, Department of Chemical and Structural Biology, Weizmann Institute of Science 7610001 Rehovot, Israel; Department of Organic Chemistry, Israel Institute for Biological Research, P.O. Box 19, Ness Ziona 7410001, Israel.
Chen Davidovich, Department of Chemical and Structural Biology, Weizmann Institute of Science 7610001 Rehovot, Israel.
Miri Krupkin, Department of Chemical and Structural Biology, Weizmann Institute of Science 7610001 Rehovot, Israel.
Nikita Dinger, Department of Chemical and Structural Biology, Weizmann Institute of Science 7610001 Rehovot, Israel.
Alla H Falkovich, Department of Chemical and Structural Biology, Weizmann Institute of Science 7610001 Rehovot, Israel.
Yoav Peleg, Department of Life Sciences Core Facilities (LSCF), Weizmann Institute of Science, Rehovot, Israel.
Ilana Agmon, Institute for Advanced Studies in Theoretical Chemistry, Schulich Faculty of Chemistry-Technion-Israel Institute of Technology, Haifa 3200003, Israel; Fritz Haber Research Center for Molecular Dynamics, Hebrew University, Jerusalem 9190401, Israel.
Anat Bashan, Department of Chemical and Structural Biology, Weizmann Institute of Science 7610001 Rehovot, Israel.
Ada Yonath, Department of Chemical and Structural Biology, Weizmann Institute of Science 7610001 Rehovot, Israel.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council, Horizon 2020 Framework Programmes [322581 (NOVRIB) to A.Y.]; National Institutes of Health [GM34360]; Adams Foundation (to C.D. and M.K.); the Kimmelman Center for Macromolecular Assemblies; A.Y. holds the Martin S. and Helen Kimmel Professorial Chair at the Weizmann Institute of Science. Funding for open access charge: Horizon 2020 Framework Programmes grant.
Conflict of interest statement. None declared
REFERENCES
- 1. Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P.. Molecular Biology of the Cell. 2002; 4th ednGarland Science. [Google Scholar]
- 2. Gilbert W. Origin of life: the RNA world. Nature. 1986; 319:618–618. [Google Scholar]
- 3. Bowman J.C., Petrov A.S., Frenkel-Pinter M., Penev P.I., Williams L.D.. Root of the tree: the significance, evolution, and origins of the ribosome. Chem. Rev. 2020; 120:4848–4878. [DOI] [PubMed] [Google Scholar]
- 4. Fox G.E. Origin and evolution of the ribosome. Cold Spring Harbor Perspect. Biol. 2010; 2:a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A.. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000; 289:905–920. [DOI] [PubMed] [Google Scholar]
- 6. Harms J., Schluenzen F., Zarivach R., Bashan A., Gat S., Agmon I., Bartels H., Franceschi F., Yonath A.. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001; 107:679–688. [DOI] [PubMed] [Google Scholar]
- 7. Moore P.B., Steitz T.A.. The roles of RNA in the synthesis of protein. Cold Spring Harbor Perspect. Biol. 2011; 3:a003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Agmon I., Bashan A., Zarivach R., Yonath A.. Symmetry at the active site of the ribosome: structural and functional implications. Biol. Chem. 2005; 386:833–844. [DOI] [PubMed] [Google Scholar]
- 9. Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J., Noller H.F.. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001; 292:883–896. [DOI] [PubMed] [Google Scholar]
- 10. Hansen J.L., Schmeing T.M., Moore P.B., Steitz T.A.. Structural insights into peptide bond formation. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11670–11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmeing T.M., Seila A.C., Hansen J.L., Freeborn B., Soukup J.K., Scaringe S.A., Strobel S.A., Moore P.B., Steitz T.A.. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 2002; 9:225–230. [DOI] [PubMed] [Google Scholar]
- 12. Bashan A., Agmon I., Zarivach R., Schluenzen F., Harms J., Berisio R., Bartels H., Franceschi F., Auerbach T., Hansen H.A.. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol Cell. 2003; 11:91–102. [DOI] [PubMed] [Google Scholar]
- 13. de Farias S.T., Rêgo T.G., José M.V.. Origin of the 16S ribosomal molecule from ancestor tRNAs. Sci. 2020; 2:69. [DOI] [PubMed] [Google Scholar]
- 14. Caetano-Anollés D., Caetano-Anollés G. Piecemeal buildup of the genetic code, ribosomes, and genomes from primordial tRNA building blocks. Life. 2016; 6:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Farias S.T., Rêgo T.G., José M.V.. Origin and evolution of the peptidyl transferase center from proto-tRNAs. FEBS Open Biol. 2014; 4:175–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gregory S.T., Dahlberg A.E.. Peptide bond formation is all about proximity. Nat. Struct. Mol. Biol. 2004; 11:586–587. [DOI] [PubMed] [Google Scholar]
- 17. Rodnina M.V., Wintermeyer W.. Peptide bond formation on the ribosome: structure and mechanism. Curr. Opin. Struct. Biol. 2003; 13:334–340. [DOI] [PubMed] [Google Scholar]
- 18. Sato N.S., Hirabayashi N., Agmon I., Yonath A., Suzuki T.. Comprehensive genetic selection revealed essential bases in the peptidyl-transferase center. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:15386–15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weinger J.S., Parnell K.M., Dorner S., Green R., Strobel S.A.. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat. Struct. Mol. Biol. 2004; 11:1101–1106. [DOI] [PubMed] [Google Scholar]
- 20. Bashan A., Yonath A.. Correlating ribosome function with high-resolution structures. Trends Microbiol. 2008; 16:326–335. [DOI] [PubMed] [Google Scholar]
- 21. Agmon I., Davidovich C., Bashan A., Yonath A.. Identification of the prebiotic translation apparatus within the contemporary ribosome. Nat. Precedings. 2009; 10.1038/npre.2009.2921.1. [DOI] [Google Scholar]
- 22. Kruger K., Grabowski P.J., Zaug A.J., Sands J., Gottschling D.E., Cech T.R.. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell. 1982; 31:147–157. [DOI] [PubMed] [Google Scholar]
- 23. Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S.. The RNA moiety of ribonuclease p is the catalytic subunit of the enzyme. Cell. 1983; 35:849–857. [DOI] [PubMed] [Google Scholar]
- 24. Joyce G.F., Szostak J.W.. Protocells and RNA self-replication. Cold Spring Harbor Perspect. Biol. 2018; 10:a034801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang L., Krupkin M., Bashan A., Yonath A., Massa L.. Protoribosome by quantum kernel energy method. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:14900–14905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bokov K., Steinberg S.V.. A hierarchical model for evolution of 23S ribosomal RNA. Nature. 2009; 457:977–980. [DOI] [PubMed] [Google Scholar]
- 27. Hsiao C., Mohan S., Kalahar B.K., Williams L.D.. Peeling the onion: ribosomes are ancient molecular fossils. Mol. Biol. Evol. 2009; 26:2415–2425. [DOI] [PubMed] [Google Scholar]
- 28. Noller H.F. The driving force for molecular evolution of translation. Rna. 2004; 10:1833–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bretscher M., Marcker K.. Polypeptidyl-sigma-ribonucleic acid and amino-acyl-sigma-ribonucleic acid binding sites on ribosomes. Nature. 1966; 211:380–384. [DOI] [PubMed] [Google Scholar]
- 30. Monro R., Marcker K.. Ribosome-catalysed reaction of puromycin with a formylmethionine-containing oligonucleotide. J Mol Biol. 1967; 25:347–350. [DOI] [PubMed] [Google Scholar]
- 31. Klein D., Moore P., Steitz T.. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol. 2004; 340:141–177. [DOI] [PubMed] [Google Scholar]
- 32. Monro R.E. Catalysis of peptide bond formation by 50 s ribosomal subunits from Escherichia coli. J Mol Biol. 1967; 26:147–151. [DOI] [PubMed] [Google Scholar]
- 33. Khaitovich P., Tenson T., Kloss P., Mankin A.S.. Reconstitution of functionally active thermus aquaticus large ribosomal subunits with in vitro-transcribed rRNA. Biochemistry. 1999; 38:1780–1788. [DOI] [PubMed] [Google Scholar]
- 34. Anderson R.M., Kwon M., Strobel S.A.. Toward ribosomal RNA catalytic activity in the absence of protein. J. Mol. Evol. 2007; 64:472–483. [DOI] [PubMed] [Google Scholar]
- 35. Zhang B., Cech T.R.. Peptide bond formation by in vitro selected ribozymes. Nature. 1997; 390:96–100. [DOI] [PubMed] [Google Scholar]
- 36. Forsythe J.G., Yu S.S., Mamajanov I., Grover M.A., Krishnamurthy R., Fernández F.M., Hud N.V.. Ester-mediated amide bond formation driven by wet–dry cycles: a possible path to polypeptides on the prebiotic earth. Angew. Chem. Int. Ed. 2015; 54:9871–9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smith C.W. RNA-Protein Interactions: A Practical Approach: A Practical Approach. 1998; UK: Oxford University Press. [Google Scholar]
- 38. Tuerk C., Gauss P., Thermes C., Groebe D.R., Gayle M., Guild N., Stormo G., d’Aubenton-Carafa Y., Uhlenbeck O.C., Tinoco I.. CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc. Natl. Acad. Sci. U.S.A>. 1988; 85:1364–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nissen P., Ippolito J.A., Ban N., Moore P.B., Steitz T.A.. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jaeger L., Leontis N.B.. Tecto-RNA: one-dimensional self-assembly through tertiary interactions. Angew. Chem. Int. Ed. 2000; 39:2521–2524. [DOI] [PubMed] [Google Scholar]
- 41. Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J.. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 2011; 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Katoh K., Standley D.M.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J.. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009; 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of coot. Acta Crystallogr. D: Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Doshi K.J., Cannone J.J., Cobaugh C.W., Gutell R.R.. Evaluation of the suitability of free-energy minimization using nearest-neighbor energy parameters for RNA secondary structure prediction. BMC bioinformatics. 2004; 5:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bellaousov S., Reuter J.S., Seetin M.G., Mathews D.H.. RNAstructure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013; 41:W471–W474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kibbe W.A. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007; 35:W43–W46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schrödinger L. The PyMOL molecular graphics system, version 1.8. 2015; [Google Scholar]
- 50. Grodberg J., Dunn J.J.. ompT encodes the escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J. Bacteriol. 1988; 170:1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Simons S.P., McLellan T.J., Aeed P.A., Zaniewski R.P., Desbonnet C.R., Wondrack L.M., Marr E.S., Subashi T.A., Dougherty T.J., Xu Z.. Purification of the large ribosomal subunit via its association with the small subunit. Analyt. Biochem. 2009; 395:77–85. [DOI] [PubMed] [Google Scholar]
- 52. Wu T.-p., Ruan K.-c., Liu W.-y.. A fluorescence-labeling method for sequencing small RNA on polyacrylamide gel. Nucleic Acids Res. 1996; 24:3472–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hillenkamp F., Karas M., Beavis R.C., Chait B.T.. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Analyt. Chem. 1991; 63:1193A–1203A. [DOI] [PubMed] [Google Scholar]
- 54. Bucknall M., Fung K.Y., Duncan M.W.. Practical quantitative biomedical applications of MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 2002; 13:1015–1027. [DOI] [PubMed] [Google Scholar]
- 55. Heffler M.A., Walters R.D., Kugel J.F.. Using electrophoretic mobility shift assays to measure equilibrium dissociation constants: GAL4-p53 binding DNA as a model system. Biochem. Mol. Biol. Educ. 2012; 40:383–387. [DOI] [PubMed] [Google Scholar]
- 56. Klein D.J., Moore P.B., Steitz T.A.. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA. 2004; 10:1366–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Agmon I. Sequence complementarity at the ribosomal peptidyl transferase centre implies self-replicating origin. FEBS Lett. 2017; 591:3252–3258. [DOI] [PubMed] [Google Scholar]
- 58. Xu D., Wang Y.. Protein-free ribosomal RNA scaffolds can assemble poly-lysine oligos from charged tRNA fragments. Biochem. Biophys. Res. Commun. 2021; 544:81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Agmon I., Auerbach T., Baram D., Bartels H., Bashan A., Berisio R., Fucini P., Hansen H.A., Harms J., Kessler M.. On peptide bond formation, translocation, nascent protein progression and the regulatory properties of ribosomes: delivered on 20 october 2002 at the 28th FEBS meeting in istanbul. Eur. J. Biochem. 2003; 270:2543–2556. [DOI] [PubMed] [Google Scholar]
- 60. Agmon I., Amit M., Auerbach T., Bashan A., Baram D., Bartels H., Berisio R., Greenberg I., Harms J., Hansen H.A.. Ribosomal crystallography: a flexible nucleotide anchoring tRNA translocation, facilitates peptide-bond formation, chirality discrimination and antibiotics synergism. FEBS Lett. 2004; 567:20–26. [DOI] [PubMed] [Google Scholar]
- 61. Bashan A., Zarivach R., Schluenzen F., Agmon I., Harms J., Auerbach T., Baram D., Berisio R., Bartels H., Hansen H.A.. Ribosomal crystallography: peptide bond formation and its inhibition. Biopolymers: Orig. Res. Biomol. 2003; 70:19–41. [DOI] [PubMed] [Google Scholar]
- 62. Yonath A., Bashan A.. Ribosomal crystallography: initiation, peptide bond formation, and amino acid polymerization are hampered by antibiotics. Annu. Rev. Microbiol. 2004; 58:233–251. [DOI] [PubMed] [Google Scholar]
- 63. Zarivach R., Bashan A., Berisio R., Harms J., Auerbach T., Schluenzen F., Bartels H., Baram D., Pyetan E., Sittner A.. Functional aspects of ribosomal architecture: symmetry, chirality and regulation. J. Phys. Org. Chem. 2004; 17:901–912. [Google Scholar]
- 64. Yonath A. Antibiotics targeting ribosomes: resistance, selectivity, synergism, and cellular regulation. Annu. Rev. Biochem. 2005; 74:649–679. [DOI] [PubMed] [Google Scholar]
- 65. Krupkin M., Matzov D., Tang H., Metz M., Kalaora R., Belousoff M.J., Zimmerman E., Bashan A., Yonath A.. A vestige of a prebiotic bonding machine is functioning within the contemporary ribosome. Philos. Trans. R. Soc. B: Biol. Sci. 2011; 366:2972–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fox G.E., Tran Q., Yonath A.. An exit cavity was crucial to the polymerase activity of the early ribosome. Astrobiology. 2012; 12:57–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Agmon I.C. Could a proto-ribosome emerge spontaneously in the prebiotic world?. Molecules. 2016; 21:1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bou-Nader C., Zhang J.. Structural insights into RNA dimerization: motifs, interfaces and functions. Molecules. 2020; 25:2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Söll D., Soll D., Rajbhandary U., Rajbhandary T.. tRNA: Structure, Biosynthesis, and Function. 1995; ASM Press. [Google Scholar]
- 70. Vázquez-Salazar A., Becerra A., Lazcano A.. Evolutionary convergence in the biosyntheses of the imidazole moieties of histidine and purines. PloS one. 2018; 13:e0196349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Frenkel-Pinter M., Haynes J.W., Mohyeldin A.M., Martin C., Sargon A.B., Petrov A.S., Krishnamurthy R., Hud N.V., Williams L.D., Leman L.J.. Mutually stabilizing interactions between proto-peptides and RNA. Nat.Commun. 2020; 11:3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tirumalai M.R., Rivas M., Tran Q., Fox G.E.. The Peptidyl Transferase Center: a Window to the Past. Microbiol. Mol. Biol. Rev. 2021; 15:e0010421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Deamer D.W. Assembling Life: How Can Life Begin on Earth and Other Habitable Planets?. 2018; Oxford University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.