Abstract
The metabolism of polyamines (putrescine, spermidine, and spermine) has become the target of genetic manipulation because of their significance in plant development and possibly stress tolerance. We studied the polyamine metabolism in non-transgenic (NT) and transgenic cells of poplar (Populus nigra × maximowiczii) expressing a mouse Orn decarboxylase (odc) cDNA. The transgenic cells showed elevated levels of mouse ODC enzyme activity, severalfold higher amounts of putrescine, a small increase in spermidine, and a small reduction in spermine as compared with NT cells. The conversion of labeled ornithine (Orn) into putrescine was significantly higher in the transgenic than the NT cells. Whereas exogenously supplied Orn caused an increase in cellular putrescine in both cell lines, arginine at high concentrations was inhibitory to putrescine accumulation. The addition of urea and glutamine had no effect on polyamines in either of the cell lines. Inhibition of glutamine synthetase by methionine sulfoximine led to a substantial reduction in putrescine and spermidine in both cell lines. The results show that: (a) Transgenic expression of a heterologous odc gene can be used to modulate putrescine metabolism in plant cells, (b) accumulation of putrescine in high amounts does not affect the native arginine decarboxylase activity, (c) Orn biosynthesis occurs primarily from glutamine/glutamate and not from catabolic breakdown of arginine, (d) Orn biosynthesis may become a limiting factor for putrescine production in the odc transgenic cells, and (e) assimilation of nitrogen into glutamine keeps pace with an increased demand for its use for putrescine production.
Polyamines (putrescine, spermidine, and spermine) are low Mr polycations found in all living organisms. At the cellular level, polyamines are involved in DNA and protein synthesis, stabilization of membranes, scavenging of free radicals, and modulation of enzyme activities (Minocha and Minocha, 1995; Watson and Malmberg, 1996, Walden et al., 1997; Kumar and Minocha, 1998). It has often been suggested that their biosynthesis may compete with the biosynthesis of ethylene (Kushad and Dumbroff, 1991; Minocha and Minocha, 1995; Turano et al., 1997), which has a major developmental role in plants (Kende, 1993; Kieber, 1997). Due to their richness in amine groups, and their presence in millimolar quantities in plant cells, polyamines could also play a role in the modulation of reduced nitrogen and in the sequestration of free ammonia produced inside the cells (Lovatt, 1990; Slocum and Weinstein, 1990).
Despite their importance in cellular and developmental processes in plants, little experimental evidence for the regulation of polyamine metabolism has been forthcoming. Most studies reported thus far have emphasized the correlative changes in cellular polyamines and a developmental and/or a physiological response of the plant (Evans and Malmberg, 1989; Walden et al., 1997; Cohen, 1998, and references therein). This is in contrast to an abundance of literature on polyamine metabolism in animals where significant progress has been made in biochemical and molecular characterization of the polyamine biosynthetic enzymes and their genes (Cohen, 1998). Until recently, the most common approach to modulate cellular polyamines in plants has been to use chemical inhibitors. Some limitations of this approach include the issues related to differential rates of uptake of the inhibitors, their metabolic conversions, the lack of their specificity, and their deleterious effects on membrane properties. The inhibitors additionally often do not allow an up-regulation of the cellular polyamines. The transgenic gene expression, on the other hand, provides a means of both up- and down-regulating specific metabolic steps in a pathway (Kinney, 1998; Lindsey, 1998; Nuccio et al., 1999). The latter approach can reveal mechanisms of metabolic regulation that may not be seen simply by mutant analysis or inhibitor studies. As we move toward modulating specific aspects of cellular metabolism in plants through genetic engineering, it would be prudent to analyze the impact of manipulating single reactions in a pathway on the regulation of the entire pathway and also on other related pathways that use the same precursors and intermediates. The present report deals with the results of such a study with respect to the metabolism of polyamines.
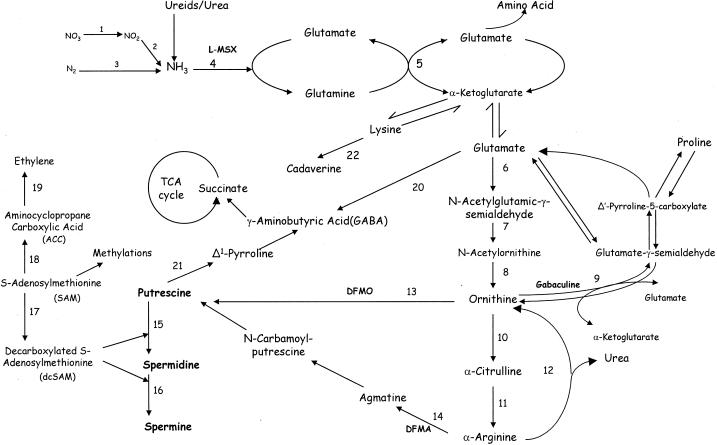
In plants, biosynthesis of putrescine occurs from Orn and/or Arg (Fig. 1) and is regulated by the enzymes Orn decarboxylase (ODC) and Arg decarboxylase (ADC) (Slocum, 1991; Cohen, 1998). Spermidine and spermine are synthesized from putrescine by sequential additions of aminopropyl groups derived from decarboxylated S-adenosyl-Met (SAM), the reactions being catalyzed by spermidine and spermine synthases. Decarboxylated SAM is produced from SAM by SAM decarboxylase (SAMDC). The three decarboxylases share a common property of having relatively short half-lives (≤1 h) in the cells (Cohen, 1998). While genes for both odc and adc are believed to be present in most plants, their contribution to putrescine production is often tissue specific and/or developmentally regulated (Minocha and Minocha, 1995; Kumar et al., 1997; Walden et al., 1997).
Figure 1.
Polyamine biosynthesis and related nitrogen metabolism. The enzymes are: 1, nitrate reductase; 2, nitrite reductase; 3, nitrogenase; 4, Gln synthetase (GS); 5, Glu synthase (GOGAT); 6, Glu reductase; 7, acetylglutamic-γ-semialdehyde transaminase; 8, acetylornithinase; 9, Orn aminotransferase (OAT); 10, Orn transcarbamylase; 11, Arg synthase; 12, arginase; 13, Orn decarboxylase (ODC); 14, Arg decarboxylase (ADC); 15, spermidine synthase; 16, spermine synthase; 17, SAM decarboxylase (SAMDC); 18, ACC synthase; 19, ACC oxidase; 20, Glu decarboxylase (GAD); 21, diamine oxidase; and 22, Lys decarboxylase (LDC).
In recent years, polyamine metabolism has become the target of genetic manipulation both in animals and in plants (for review, see Kumar and Minocha, 1998). Hamill et al. (1990) demonstrated the use of a yeast odc cDNA in tobacco plants to modulate the metabolism of putrescine and nicotine, an alkaloid derived from putrescine. The cellular contents of spermidine and spermine were not affected. In earlier studies, we reported increased production of putrescine in tobacco plants (DeScenzo and Minocha, 1993) and carrot cells (Bastola and Minocha, 1995) overexpressing a mouse odc cDNA. Whereas most of the transgenic tobacco plants were phenotypically normal, carrot cell cultures, however, exhibited an increased frequency of somatic embryogenesis. It was subsequently demonstrated that in the transgenic carrot cells not only were the rates of putrescine biosynthesis higher the catabolism of putrescine was also enhanced as compared with the non-transgenic (NT) cells (Andersen et al., 1998). Noh and Minocha (1994) reported that the leaves of transgenic tobacco plants over-expressing a human samdc cDNA contained significantly higher levels of spermidine and reduced levels of putrescine. Kumar et al. (1996, 1997), using an antisense construct of potato samdc cDNA, observed a reduction in spermidine production and accompanying abnormal phenotypes in the transgenic tubers.
Two different groups (Masgrau et al., 1996; Burtin and Michael, 1997) have published results on transformation of tobacco and one group on transformation of rice (Capell et al., 1998) with an oat adc cDNA. Whereas their results differ somewhat from each other, in all cases increased putrescine accumulation was observed in transgenic plants with relatively small change in spermidine and spermine. No detailed analysis of the metabolism of polyamines in the transgenic cells was reported in any of these studies.
The presence of two alternative pathways (ODC and ADC) for putrescine production in many tissues complicates the situation regarding their metabolic regulation, particularly when the substrates of the two pathways (Orn and Arg) are also interconvertible (Fig. 1, steps 10–12; also see Ireland, 1997). It is thus conceivable that the metabolic effects of overexpression of the odc or the adc gene may be limited by substrate availability. Furthermore, ADC and ODC activities may be subject to feedback regulation by polyamine concentrations in the cells (Primikirios and Roubelakis-Angelakis, 1999). The present report provides an insight into the regulation of polyamine metabolism and its relationship to the metabolism of Arg, Orn, and Gln in transgenic and NT cells of an angiospermic woody plant poplar (Populus nigra × maximowiczii). Some of the specific questions addressed in this study are: (a) What are the effects of overexpression of an odc gene on cellular levels and biosynthetic rates of putrescine, spermidine and spermine? (b) Does Orn become limiting in the transgenic cells due to its excessive use by ODC, and does it affect the availability of Arg to ADC? (c) What is the primary source of Orn in the cells—is it Glu or Arg? An additional objective was to test the hypothesis that ADC activity in plants is subject to feedback regulation by either cellular putrescine or total polyamines.
RESULTS
Transformation, ODC Activity, and Polyamine Content of Cells
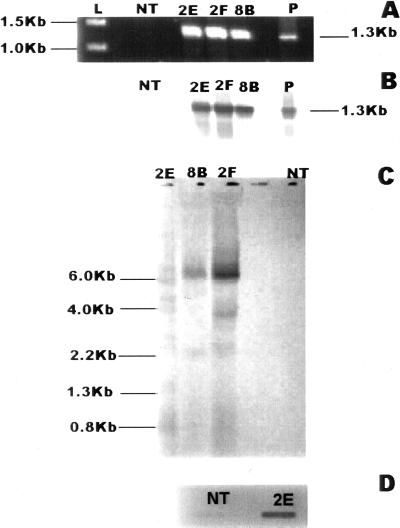
Over the period of 2 months, a total of three transgenic cell lines transformed with the gus gene and 15 transgenic cell lines transformed with the mouse odc gene were selected on kanamycin. Following several rounds of subculture on solid medium, suspension cultures were initiated for most cell lines and maintained on a weekly subculture routine. Each cell line was first characterized with respect to the presence of the respective transgene by PCR. For cell lines transformed with the plasmid pCW122-odc, genomic DNA only from the putative transgenic cells yielded the expected PCR product of 1.3 kb (Fig. 2A), which hybridized with the labeled probe for mouse odc cDNA (Fig. 2B). Cells transformed with the plasmid pCW122 tested positive for the presence of nptII gene and the gus gene, yielding the expected PCR products (data not shown). Genomic Southern analysis revealed that the transgenic line 2E, used in the study here, had more than two copies of the transgene (Fig. 2C). There was no hybridization signal observed in the DNA from NT cells using a labeled probe of mouse odc DNA. The transcription product (mRNA) of the mouse odc transgene was detectable by northern slot-blot analysis of total RNA only in the transgenic cells (Fig. 2D). Again, no signal was observed in RNA from the NT cells. Several of the transgenic cell lines were tested for the presence of NPT protein using ELISA kit (5′ → 3′, Inc., Westchester, PA) and all were found to be positive (data not shown).
Figure 2.
Molecular analysis of transgenic cells. A, Amplification product of PCR using genomic DNA from different cell lines and mouse odc-specific primers: NT, non-transgenic cells; 2E, 2F, 8B, different transgenic lines; and P, control plasmid. B, Southern hybridization of PCR amplification products from Figure 2A above using DIG-labeled probe made from mouse odc-cDNA. C, Southern hybridization of HindIII-restricted genomic DNA from different cell lines using DIG-labeled probe made from mouse odc-cDNA. D, Slot-blot northern hybridization of total RNA from and NT and 2E cells using DIG-labeled probe made from mouse odc-cDNA.
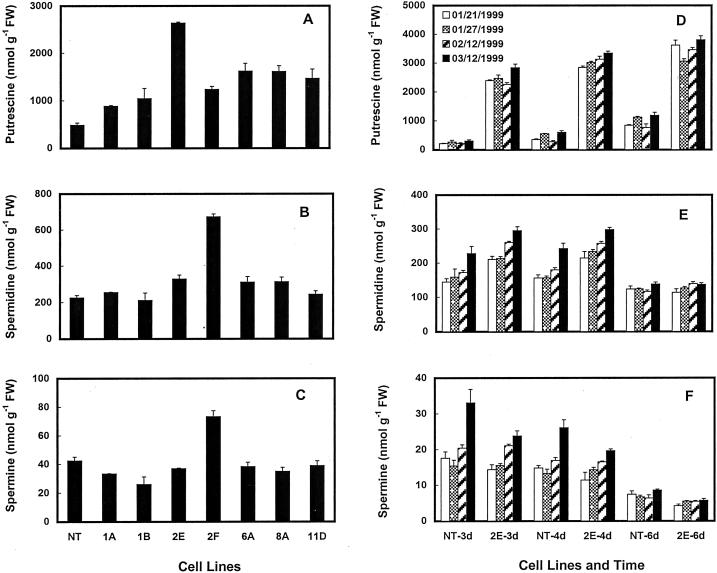
Ten of the 15 odc-transgenic cell lines, three gus cell lines, and two NT cell lines were analyzed several times for their polyamine contents. Putrescine contents were generally 2- to 10-fold higher in most of the odc-transgenic cell lines on any given day as compared with the NT and the gus-transgenic cells. Typical data of putrescine content in several cell lines are shown in Figure 3A. However, it must be pointed out that the content of putrescine in any given cell line varied on different days of analysis (Fig. 3D). Putrescine contents in the NT and the gus-transgenic cells were quite comparable on any given day (data not shown). Putrescine concentration in some of the transgenic cells was as high as 6.5 μmol/g fresh weight. Spermidine contents of transgenic cells were either similar to those in the NT cells or were slightly higher in the former on some days but not on others (Fig. 3, B and E). Spermine in most transgenic cell lines was often lower than the NT cells (Fig. 3, C and F). One cell line (2F) that showed a small increase in spermidine as well as spermine on some days did not consistently show a major increase in putrescine and was not followed up for further experimentation. The presence of kanamycin in the medium did not affect cellular polyamine content of the transgenic cells (data not shown).
Figure 3.
A through C, Amounts of polyamines in NT and several transgenic cell lines of poplar grown for 7 d on solid medium. D through F, Amounts of free polyamines in NT and a transgenic (2E) cell line grown in liquid medium for 3, 4, and 6 d. Data for D through F are from four different experiments conducted over a period of 2 months. Each bar represents mean ± se of four replicates.
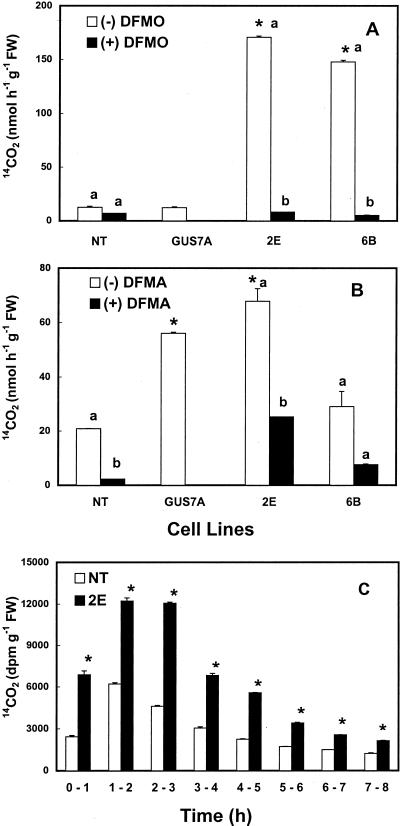
Several cell lines were analyzed for enzyme activity of mouse ODC, native (plant) ODC, and native ADC. The mouse ODC activity can be distinguished from the plant ODC by its sensitivity to dl-α-difluoromethyl-Orn (DFMO) and its pH optimum at 6.8 versus 8.2 for the plant ODC (DeScenzo and Minocha, 1993). The NT and the gus-transgenic cells showed very little of either mouse-type or native ODC activity in both the cell extracts as well as intact cells (data for native ODC not shown). Substantial activity of mouse ODC measured either in cell extracts (data not shown) or intact cells (Fig. 4A) was observed in most of the odc-transgenic cell lines. This activity was almost completely inhibited by 2 mm DFMO. Little or no native ODC (at pH 8.2) activity was observed in the transgenic cells (data not shown). As with the putrescine content, the actual amounts of enzyme activity varied in different cell lines and also in the same cell line on different days of analysis. The activity of ADC was found to be either comparable in all cell lines (transgenic or NT) or was somewhat higher in the transgenic cells on some days (Fig. 4B). The activity of ADC was inhibited by dl-α-difluoromethyl-Arg (DFMA) by as much as 60% to 100%.
Figure 4.
The rate of 14CO2 production from l-[1-14C]Orn (A), dl-[1-14C]Arg (B), and l-[U-14C]Orn (C) by non-transgenic (NT) and transgenic (2E) cells of poplar. Data in Figure 3, A and B, are from standard enzyme assays using intact cells (Minocha et al., 1999a) in the absence or presence of DFMO or DFMA. Bars represent mean ± se of two replicates. An asterisk indicates that values for transformed cell lines are significantly different (P ≤ 0.05) from NT cells; different letters indicate that values are significantly different (P ≤ 0.05) for the presence (+) and absence (−) of the inhibitor within the same cell line. Cell line GUS7A was transformed with plasmid pCW122, and 2E and 6B with pCW122-odc. C, Intact cells were incubated with l-[U-14C]Orn for 8 h and the production of 14CO2 analyzed at 1-h intervals. Bars represent mean ± se of three replicates. An asterisk indicates that values for 2E are significantly different (P ≤ 0.05) from NT cells at a given time.
Metabolism of l-[U-14C]Orn and l-[U-14C]Arg
One of the odc-transgenic cell line (2E) and one NT cell line were chosen for further analysis of the metabolism of l-[U-14C]Orn and l-[U-14C]Arg. Three-day-old cells were incubated with l-[U-14C]Orn or l-[U-14C]Arg for varying lengths of time, and data were collected on the release of 14CO2 and the conversion of labeled precursor into labeled polyamines over intervals of 1 to 8 h (in some cases up to 72 h). The rates of 14CO2 production from l-[U-14C]Orn were typically 2- to 3-fold higher in the odc-transgenic cells as compared with the NT cells during the entire 8-h period of incubation (Fig. 4C). The NT and the gus-transgenic cells showed similar rates of 14CO2 production from [U-14C]Orn (data not shown).
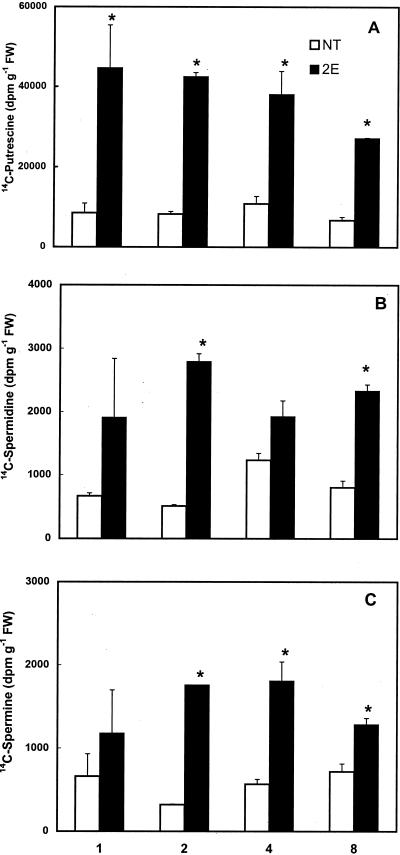
Figure 5 shows the amounts of radioactivity recovered in the three major polyamines in NT and 2E cells at different times of incubation with [U-14C]Orn. The incorporation of label from [U-14C]Orn into putrescine was significantly higher in the odc-transgenic cells than the NT cells (Fig. 5A). This was true at all times of analysis. In the 2E cells, the amount of label in the putrescine fraction was seen to decline slightly after the first 4 h of incubation, whereas in the NT cells, the amount of [14C]putrescine did not change much with the time of incubation. The radioactivity in the spermidine and the spermine fractions was also generally higher in the 2E cells as compared with the NT cells (Fig. 5, B and C). However, the total amount of label recovered in putrescine was severalfold higher than that in the other two polyamines. Both cell lines showed similar rates of 14CO2 production from [U-14C]Arg as well as its incorporation into the three polyamines (data not shown).
Figure 5.
Incorporation of radioactivity from l-[U-14C]Orn into free putrescine (A), spermidine (B), and spermine (C) in a non-transgenic (NT) and a transgenic (2E) cell line of poplar at different times of incubation. Three-day-old cells (approximately 1 g in 10 mL) were incubated with 0.2 μCi of l-[U-14C]Orn for various time periods. Each bar represents mean ± se of two replicates. An asterisk indicates that values for 2E are significantly different (P ≤ 0.05) from NT cells at a given time.
To determine whether the uptake of Orn and Arg in the transgenic cells was different from the NT cells, radioactivity in the aqueous fraction of the dansylation reaction mix (after partitioning of polyamines into toluene) was counted. This fraction contains all of the dansylated Orn, Arg, and other amino acids but no polyamines. The amount of radioactivity in this fraction was comparable in the two cell lines at different times for both Arg and Orn, showing that similar amounts of Orn and Arg were taken up by the two cell types (data not shown).
The Effect of Exogenous Supply of Arg, Orn, Urea, Gln, and Inhibitors
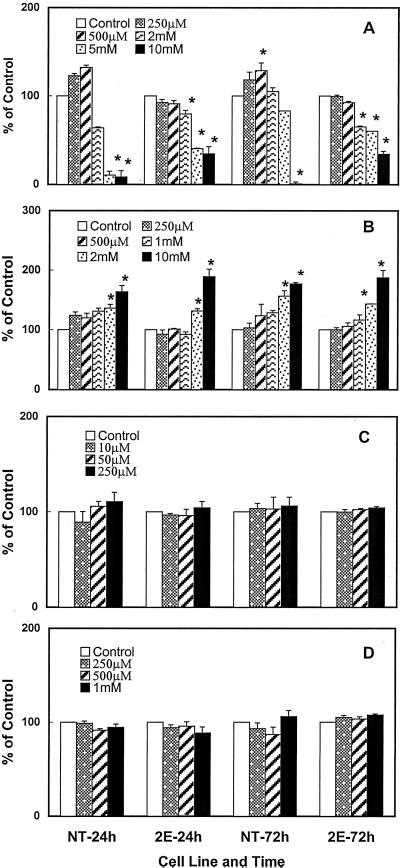
Both the transgenic cells (2E) and the NT cells were incubated with varying concentrations of Arg, Orn, urea, or Gln, and the polyamine content of treated cells were determined at 24 and 72 h. Data presented in Figure 6 A show that 0.25 to 0.5 mm Arg caused a small increase in the putrescine content of NT cells but not the 2E cells. At higher concentrations (2–10 mm), however, there was a significant decrease in putrescine content in both cell lines, the effect being more pronounced in NT cells. Addition of Orn, particularly at 10 mm, resulted in a significant increase in putrescine at both 24 and 72 h in both cell lines (Fig. 6B). Lower concentrations of Orn had smaller effect. Figure 6, C and D, shows that no further increase in putrescine content was seen in either of the cell line at 24 or 72 h in response to the addition of up to 250 μm urea or 1.0 mm of the amino acid Gln. The contents of spermidine and spermine were not affected in either the NT or 2E cell line with any of the treatments mentioned above (data not shown).
Figure 6.
The effects of different concentrations of Arg (A), Orn (B), urea (C), and Gln (D) on cellular putrescine levels in 3-d-old non-transgenic (NT) and transgenic (2E) cells of poplar at 24 and 72 h. Data are expressed as percentage of control for the respective cell lines. Each bar represents mean ± se of three (2 and 10 mm Orn and Arg) or four (all other treatments) replicates. The control values ranged as follows: NT = 380 to 580 nmol g−1 fresh weight putrescine at 24 h; 2E = 2,350 to 4,750 nmol g−1 fresh weight putrescine at 24 h; NT = 627 to 1,000 nmol g−1 fresh weight putrescine at 72 h; and 2E = 2,550 to 4,270 nmol g−1 fresh weight putrescine at 72 h. An asterisk indicates that values for treated cells are significantly different (P ≤ 0.05) from the untreated cells within the same cell line at a given time.
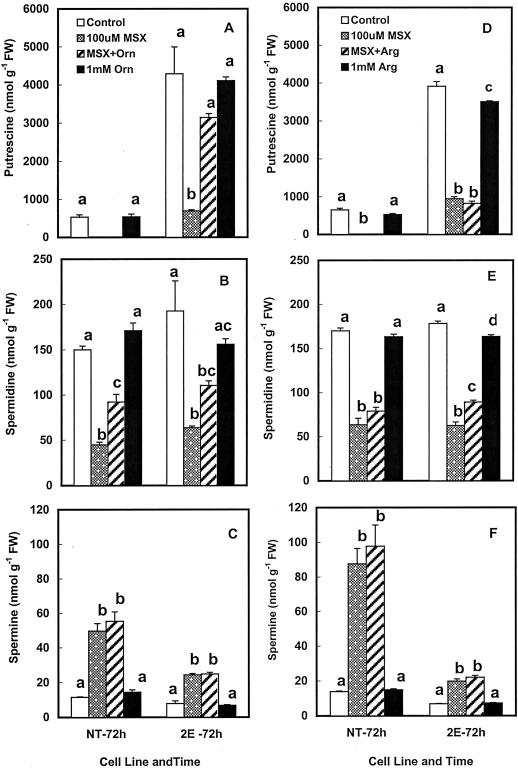
Both Orn and Arg are produced from Gln via Glu (Fig. 1). Met sulfoximine (MSX) is a strong inhibitor of Gln synthetase (Fig. 1, step 4) and effectively causes nitrogen limitation in the cells (Leason et al., 1982; Florencio and Vega, 1983). It consequently would be expected to rapidly reduce the biosynthesis of Orn and Arg in the cells, thus affecting the rates of putrescine biosynthesis via both ODC and ADC. The addition of MSX to the medium resulted in a significant reduction in cellular levels of putrescine within 24 h in both the NT and 2E cells (data not shown); a more dramatic reduction being observed at 72 h (Fig. 7, A and D). Whereas putrescine levels in the NT cells were below the detection limits, the 2E cells showed a 5- to 8-fold reduction in putrescine. Cellular spermidine content was also significantly lower in the MSX-treated cells, both the cell lines showing similar levels of reduction (Fig. 7, B and E). Spermine, on the other hand, was not affected by MSX at 24 h but showed a severalfold increase in MSX-treated cells at 72 h (Fig. 7, C and F). In the presence of MSX in the medium, transgenic cells contained significant amounts of another polyamine, namely cadaverine, which was never seen in the NT cells (data not shown). Cadaverine is produced via the decarboxylation of Lys by the mouse ODC, which is capable of using this amino acid as an alternate substrate to Orn, albeit at a low efficiency (Pegg and McGill, 1979; Persson, 1981). The addition of 1.0 mm Orn in the presence of MSX caused a substantial (but never complete) reversal of the effect of this inhibitor on cellular putrescine in the transgenic cells and a partial reversal of spermidine in both the cell lines. However, MSX effects on spermine were not reversed (Fig. 7, A–C). Arg was largely ineffective in reversing the MSX effects on polyamines in either of the cell lines (Fig. 7, D–F).
Figure 7.
The effects of 100 μm MSX on cellular putrescine (A and D), spermidine (B and E), and spermine (C and F) levels in 3-d-old non-transgenic (NT) and transgenic (2E) cells of poplar and its reversal by Orn (A–C) or Arg (D–F). Treatments were given for 72 h. Each bar represents mean ± se of four replicates. Different letters above the bars indicate that values are significantly different (P ≤ 0.05) from each other for the same cell line.
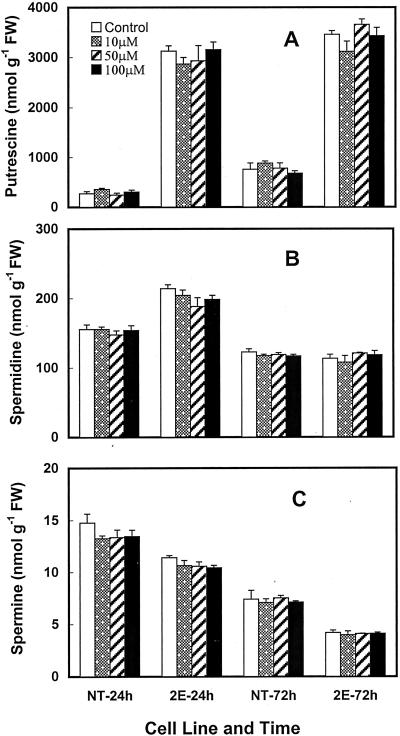
In plants, Pro is generally synthesized from Glu (Fig. 1; also see Ireland, 1997). It can also be synthesized from Orn. Gabaculine is an inhibitor of Orn aminotransferase (OAT), an enzyme that regulates the conversion of Orn into Pro via Glu-γ-semi-aldehyde (Fig. 1, step 9). If there was a competition between the two pathways (i.e. putrescine and Pro biosynthesis) for Orn use (steps 9 and 13 in Fig. 1), less Orn may become available to ODC for putrescine production when Pro production is high. Therefore, the inhibition of Pro biosynthesis from Orn would be expected to increase the availability of Orn for mouse ODC. The data in Figure 8 show that treatment with 10 to 100 μm gabaculine for up to 72 h had little effect on the cellular content of any of the three polyamines in either the NT or the transgenic cells.
Figure 8.
The effects of different concentrations of gabaculine on cellular putrescine (A), spermidine (B), and spermine (C) levels in 3-d-old non-transgenic (NT) and transgenic (2E) cells of poplar at 24 and 72 h. Each bar represents mean ± se of three replicates. None of the values for treatments were significantly different from the untreated cells of the same cell line at a given time.
DISCUSSION
The roles of the 5′- and the 3′-untranslated region (UTR) in the regulation of mammalian odc (which has some of the longest UTRs seen in animal mRNAs) have been variously discussed (Grens and Scheffler, 1990; Wallstrom and Persson, 1999). Previous studies from our laboratory (DeScenzo and Minocha, 1993; Bastola and Minocha, 1995) showed that the presence of only 59 bp at the 3′ end of the 737-bp-long 5′-UTR was sufficient for expression of the mammalian odc cDNA in plants. The results presented here clearly demonstrate that the expression of mouse odc in plants does not require any part of either the 5′- or the 3′-UTRs (see also Wallstrom and Persson, 1999). Likewise, the PEST amino acid domain (Li and Coffino, 1993) at the C terminus of the ODC protein is not essential for the activity of this enzyme. In fact, the expression of mammalian cDNA containing sequence for the 3′-PEST region of 37 amino acids caused a much smaller increase in ODC activity in transgenic tobacco than the truncated version without this sequence (DeScenzo and Minocha, 1993). This could be attributed to a rapid turnover of the full-length mouse enzyme protein in the transgenic cells. These results are consistent with the reported properties of mammalian ODC and with studies on the transgenic expression of mammalian odc in animal cells (Halmekytö et al., 1991; Kauppinen and Alhonen, 1995).
Polyamines in Transgenic Cells
A number of studies on transgenic expression of homologous and heterologous sequences of odc in animals and odc as well as adc in plants have been reported (for review, see Kumar and Minocha, 1998). In transgenic mice overexpressing human odc gene, most tissues (except brain and testes) showed a homeostatic response to increased ODC activity, i.e. normal levels of putrescine were maintained in these tissues (Halmekytö et al., 1991). However, the brain and the testes did accumulate putrescine in higher amounts without significant changes in either spermidine or spermine. The maintenance of near normal levels of putrescine in most transgenic tissues was later shown to be the result of increased export/excretion of putrescine from these tissues (Halmekytö et al., 1993; Seiler et al., 1996). In several studies with plants, including the present one, significant increases in the content of putrescine in transgenic cells have been observed with the expression of either odc or adc cDNAs (for review, see Kumar and Minocha, 1998). In most cases, however, little or no increase in spermidine or spermine was observed. Also, in studies where significant changes in putrescine content were observed during normal development or in response to a variety of abiotic stress factors, only a little or no concomitant change in spermidine or spermine content was observed (Minocha et al., 1993, 1995, 1999b; Zhou et al., 1995; Bouchereau et al., 1999). The situation with transgenic poplar cells overexpressing the mouse odc cDNA seems to be consistent with these reports. Davis et al. (1992) have proposed that animal cells, although able to tolerate high concentrations of putrescine, are unable to tolerate high concentrations of spermidine and spermine. If true, this would require a tight metabolic regulation of spermidine and spermine content within the cells. This probably is achieved by the regulation of key enzymes needed for their biosynthesis, namely SAMDC and spermidine/spermine synthases, and/or increased turnover or secretion of putrescine from the cells as mentioned above. Studies to date reveal that increased spermidine and spermine biosynthesis in animal cells is often accompanied by increased catabolic breakdown of these compounds through induction of spermidine/spermine acetyltransferase and polyamine oxidase activities (Cohen, 1998). Whether or not in plants also the levels of spermidine and spermine are regulated by similar compensatory mechanisms involving increase in their catabolic turnover by polyamine oxidases is presently unknown.
In plants, in addition to serving as a precursor for spermidine biosynthesis, putrescine can be conjugated with a variety of phenolic acids and in some cases converted to secondary metabolites, e.g. alkaloids (Walden et al., 1997). Genetic manipulation of putrescine biosynthesis to modulate nicotine content in tobacco has been attempted by using a yeast odc cDNA (Hamill et al., 1990). Because an immediate catabolic product of putrescine in plants is γ-aminobutyric acid (GABA), which plays a variety of important roles in plant development (Shelp et al., 1999), increased putrescine production in transgenic plant cells may also have far-reaching implications for their physiological responses involving this compound. Neither the conjugation of putrescine with phenolic compounds nor the cellular content of GABA has yet been analyzed in transgenic plant cells overproducing putrescine. A small decrease in spermine content often seen in the odc transgenic cells has no obvious explanation at present and may be physiologically insignificant, since spermine constitutes only a small proportion (less than 5%) of the total polyamines.
Are the Substrates for ODC and ADC Limiting?
It is presently unknown as to how the cellular levels of putrescine are regulated in plant cells and what factors determine the upper limits of putrescine accumulation in the transgenic cells over-expressing the mouse odc cDNA. However, we do know that poplar cells can actually tolerate and maintain much higher levels of putrescine than they normally do, as shown by the levels of putrescine in transgenic cells compared with the NT cells. For NT cells, some possibilities for regulation of cellular putrescine content include: (a) limitation of the substrate Arg since these cells primarily use ADC and do not possess much ODC activity; (b) limitation of the enzyme ADC; (c) feedback regulation of ADC activity by putrescine; and (d) increased putrescine catabolism. For transgenic cells, it can be hypothesized that a constitutive overexpression of the mouse odc cDNA could lead to a depletion of their Orn pools since this amino acid is being used at a high rate by the mouse ODC. The depletion of Orn could in turn reduce the availability of Arg (for ADC) since it is also the precursor of Arg (Fig. 1, steps 10 and 11). This would then limit the amount of putrescine that can be synthesized in these cells via ADC. To test this hypothesis, the cells were exogenously supplied with Orn or Arg and analyzed for their polyamine contents. Based upon the data presented here, it can be argued that: (a) Commensurate with increased use of Orn, its biosynthesis is also enhanced in the transgenic cells without affecting its cellular pools, (b) this enhancement is still insufficient to saturate the available ODC enzyme in these cells, and (c) exogenous Orn can probably be converted to Arg in NT cells providing additional substrate for ADC and causing increased putrescine production. This indicates the existence of a homeostatic regulatory mechanism, which induces increased Orn production concomitant with its increased use. The observed inhibition of putrescine accumulation by high concentrations of Arg in both cell lines is difficult to explain at present. Although Arg metabolism has been extensively studied (Wu and Morris, 1998), relatively little is known about homeostatic regulation of Orn pools in plant cells.
Mammalian ODC is known to be regulated by feedback mechanisms that operate both at transcriptional and translational levels (Kanamoto et al., 1986, 1993; Glass and Gerner, 1986; Nilsson et al., 1997; Cohen, 1998). Also, the turnover of ODC in animals is promoted by excess polyamines via the induction of an antizyme protein (Nishiyama et al., 1989; Hayashi and Murakami, 1995). The existence of similar controls for ODC and ADC in plants have not been demonstrated. Primikirios and Roubelakis-Angelakis (1999) have hinted at the existence of a feedback regulation of the amounts of ADC enzyme by exogenous putrescine in Vitis vinifera. The data presented here on transgenic Populus cells, and also the results published earlier from our laboratory with transgenic tobacco (DeScenzo and Minocha, 1993) and carrot cells (Andersen et al., 1998), clearly show that at least in these species there is no evidence of a feedback regulation of ADC either by putrescine or by total polyamine levels in the cells. The transgenic cells exhibits as much (or more) ADC activity as the NT cells even though the former contain severalfold higher amounts of putrescine. Although the subcellular location of ADC in poplar cells is not known, it is conceivable that cellular ADC may be compartmentalized away from the increased putrescine produced by mouse ODC, which is presumably present in the cytoplasm.
What Is the Source of Orn?
Orn biosynthesis in plants occurs largely from Gln/Glu using several enzymes (Fig. 1; also see Davis, 1986; Ireland, 1997). Orn alternatively can be produced from Arg by arginase as a part of the urea cycle (Fig. 1, step 12). Assuming that Orn levels in transgenic cells were limiting (see argument above) and the urea cycle pathway was an important source of Orn in the transgenic cells one would expect a depletion of Arg in these cells. This would in turn make it a limiting factor for putrescine production via ADC also. Exogenous supply of Arg consequently should promote both the ADC-produced putrescine and the amount of Orn available to ODC, resulting in an increase in putrescine levels in both the NT and the transgenic cells. However, exogenous Arg supplied to transgenic cells did not cause increased putrescine production nor was the conversion of [14C]Arg into putrescine altered in the transgenic cells (for similar results with carrot cells, see also Andersen et al., 1998 ). Therefore, it can be argued that most of the Orn in plant cells comes directly from Glu and not from Arg. This explanation is consistent with the mitochondrial location of plant arginase and its overall low activity in plant cells (Jenkinson et al., 1996). The above conclusion is further supported by the results of MSX treatment, which inhibits ammonia assimilation into Gln and Glu (Leason et al., 1982; Florencio and Vega, 1983), thus limiting the amounts of Glu available for Orn production. The effects of MSX were partially reversed by the addition of exogenous Orn but not Arg.
The apparent lack of an effect of exogenous Gln on polyamines in the transgenic cells leads us to postulate that the production of Gln/Glu from nitrate and ammonium in the medium is keeping pace with its increased use for Orn production and that nitrogen in the medium is not a limiting factor for this pathway. This argument is further supported by the results from urea addition to the medium, which also had no effect on polyamine levels in either the NT or the transgenic cells. It can thus be concluded that as long as a source of inorganic nitrogen is available to the cells, its conversion into Gln/Glu and, subsequently into Orn, is not a limiting factor for polyamine biosynthesis. In other words, the primary regulation of putrescine biosynthesis is achieved by ODC or ADC activities and not by substrate availability.
Both putrescine and Pro accumulate in plants under conditions of abiotic stress (Bouchereau et al., 1999). Gabaculine is a strong inhibitor of OAT, an enzyme that channels Orn toward Pro biosynthesis (Davis, 1986; Ireland, 1997). This inhibitor had no significant effect on cellular putrescine in either the NT or the transgenic cells, indicating that there probably is little competition between ODC and OAT for the use of Orn as a substrate by these two enzymes. This argument is compatible with the conclusion stated above that the rates of Orn biosynthesis are regulated by its overall consumption in the polyamine biosynthetic pathway. Thus, a stimulation of Gln/Glu biosynthesis, Orn biosynthesis, and its consumption in putrescine production, and Pro biosynthesis must all be part of a coordinated response to stress in plants. An enhancement of this pathway may also be important for the regulation of free ammonia in the cells, as well as for inhibition of ethylene production, since the latter uses the same substrate (SAM) as the higher polyamines and the two pathways presumably compete with each other. In addition, increased catabolism of putrescine via diamine oxidase could result in increased GABA production, thus making polyamines important players in stress response of plants in more than one way, i.e. through effects on Pro as well as GABA production (Bouchereau et al., 1999).
From the data presented here, it can be concluded that: (a) transgenic expression of a heterologous odc gene can be used to modulate putrescine metabolism in poplar cells; (b) overproduction of putrescine and its accumulation in high amounts does not affect the native ADC activity and its contribution to putrescine production; (c) Orn biosynthesis occurs primarily from Gln/Glu and not from a catabolic breakdown of Arg; (d) Orn biosynthesis may become a limiting factor for putrescine production in the odc transgenic cells; and (e) assimilation of nitrogen into Gln keeps pace with an increased demand for its use for putrescine production and possibly also for Pro production. It is also clear from the data presented here, and from the results published earlier from several laboratories including ours (for references, see Kumar and Minocha, 1998 ) that: (a) Although cellular putrescine levels in plant cells can fluctuate widely, the levels of spermidine and spermine are regulated tightly and are not limited by the rates of putrescine biosynthesis and (b) the ODC and the ADC pathways work independently in the transgenic cells. Whether or not a similar situation exists in those wild-type plant cells, which contain both ADC and ODC activities, is not yet clear.
MATERIALS AND METHODS
Plasmid Construction
The plasmid pucODC-1 (DeScenzo and Minocha, 1993) was used to amplify a PCR product containing the coding sequence of the mouse odc gene. A Kozak consensus sequence (Kozak, 1991) was added as part of the forward primer (5′GAACCATGGGCAGCTTTAC3′) and a translation termination codon was added as part of the reverse primer (5′CTACTACATGGCTCTGGATCTGTTTCA3′) at a site 111 bp upstream of the original translation termination site of the mouse odc cDNA (Kahana and Nathans, 1985). This resulted in a cDNA sequence that lacked the 737-bp 5′-UTR, the 342-bp 3′-UTR, and also the coding sequence of 37 C terminus amino acids, which constitute a PEST region supposedly responsible for rapid turnover of the enzyme (Ghoda et al., 1989, 1992). The PCR product was gel purified and ligated into the pCW122 expression vector (Walter et al., 1998) from which the gus gene had been removed by restriction with HindIII and BamHI. Blunt-end ligation was performed following a filling-in reaction (Klenow polymerase) and dephosphorylation of the vector. Electroporated Escherichia coli (DH10B) containing the reconstituted plasmid were selected on ampicillin and tested for correct orientation of the mouse cDNA by restriction analysis and by sequencing of the junction between the promoter and the coding sequence (data not shown). The reconstituted plasmid called pCW122-odc contains the truncated mouse odc cDNA regulated by a 2× 35S cauliflower mosaic virus (CaMV) promoter and a CaMV 3′-termination sequence. The plasmid also contains a nptII gene under the control of a single 35S CaMV promoter for selection of transgenic plant cells on kanamycin. Plasmid DNA prepared by the Promega Megaprep kit (Promega, Madison, WI) was used in the transformation of poplar (Populus nigra × maximowiczii) cells by biolistic bombardment.
Cell Cultures
Liquid and solid cultures of hybrid poplar cells were maintained on 50 mL of Murashige and Skoog medium (Murashige and Skoog, 1962) containing vitamins of B-5 medium (Gamborg et al., 1968), 2% (w/v) Suc, and 0.5 mg/L 2,4-d. The pH of the medium was adjusted to 5.7 before autoclaving. Suspension cultures were maintained by transferring 7 mL of the 7-d-old cell suspensions to 50 mL of fresh medium in a 125 mL of Erlenmeyer flask, and kept on a gyratory shaker at 160 rpm. Callus on solid medium was subcultured at 3- to 4-week intervals. All cultures were maintained at 25°C ± 1°C under 12-h photoperiod (80 ± 10 μE m−2 s−1). The medium for maintenance of transgenic cell lines contained 100 mg/L kanamycin; however, the antibiotic was not present during the experimental treatments.
Transformation
The biolistic bombardment technique was modified from Walter et al. (1998) for transformation of suspension cultures. Gold particles (1.0 μm, Bio-Rad Laboratories, Hercules, CA) were coated with either the plasmid pCW122 (gus + nptII gene) or pCW122-odc (odc + nptII gene) DNA (2 μg of DNA/μg of gold particles) in the presence of 1.0 m CaCl2 and 16.7 mm spermidine. Rupture discs of 1,350 psi were used for bombardment. For preparation of tissue, 1 mL of 3-d-old cell suspension (containing about 100 mg fresh weight of cells) was vacuum-filtered onto a sterilized 60-mm-diameter #1 filter paper (Whatman, Clifton, NJ). The filter paper was placed in the center of a Petri dish containing Murashige and Skoog medium with 0.2 m sorbitol for 16 to 20 h prior to bombardment. Following bombardment, the cells were kept for 3 d on the same medium and then the filter papers were transferred to the selection medium containing100 mg/L kanamycin but no sorbitol. When the cells had grown to 5-mm clumps on the filter paper, they were transferred directly onto solid medium containing kanamycin. Following several subcultures, suspension cultures were initiated by transferring cell masses from solid medium to liquid medium and placing them on the shaker.
The transgenic cell lines were characterized with respect to the presence of the mouse odc or the gus DNA by PCR and Southern hybridization of the PCR-amplified product, as well as by Southern hybridization of the HindIII-restricted genomic DNA. Genomic DNA was isolated by minor modifications of the method of Webb and Knapp (1990) or by using the Phytopure Plant DNA Isolation Kit (Nucleon Biosciences, Coatbridge, UK). The PCR reaction was carried out using “Ready-to-go” PCR beads (Amersham-Pharmacia, Piscataway, NJ). The odc primers were the same as described earlier. The gus primers were 5′TTATGCGGGCAACGTCGTGTATCA3′ and 5′TGTTCGGCGTGGTGTAGAGCAT3′. The reaction conditions for both amplifications were: 35 cycles at 94°C for 30 s, 62°C for 30 s, and 72°C for 30 s, followed by 72°C for 5 min. The PCR products, separated by electrophoresis on 1% (w/v) Sea-Kem GTG agarose (FMC, Rockport, ME), were transferred to a nylon membrane (0.2 μm of Nytran, S & S, Keene, NH) and confirmed to represent odc or gus by Southern hybridization (65°C) with DIG-labeled probes (Boehringer Mannheim, Indianapolis) followed by washes at 68°C (0.1× SSC). The genomic DNA (15 μg) was digested with HindIII overnight and separated on 1.0% (w/v) Sea-Kem GTG agarose. The transfer, prehybridization, and hybridization conditions were the same as for PCR products. Total RNA was extracted from 2 g of 4-d-old cells by the method of Chomczynski and Sacchi (1987). For slot-blot analysis, 10 μg of total RNA was collected on a nylon membrane (0.45 μm of Nytran, S & S) by S & S Minifold I, using vacuum according to manufacturer's instructions. The membranes were treated the same way as for DNA hybridization.
Polyamine Analysis
Several times during the period of this study, cell samples were collected from NT, gus-transgenic, and odc-transgenic cell lines growing in suspension cultures with or without kanamycin. This was done by vacuum filtering 5 to 10 mL of 3- to 5-d-old suspensions onto Miracloth and transferring 200 mg fresh weight of cells to 800 μL of 5% (v/v) perchloric acid (PCA) in a microfuge tube. These samples were then frozen and thawed three times before dansylation (Minocha et al., 1994). Following centrifugation (13,000g, 15 min), 50 or 100 μL of the PCA extract was dansylated, the dansyl-polyamines extracted with toluene, dried in Speed-Vac, redissolved in methanol, and analyzed by HPLC using a gradient of acetonitrile (40%–100%) and 10 mm heptanesulfonic acid, pH 3.4, on a reversed-phase Pecosphere C18 column (4.6 × 33 mm, 3 μm) using a HPLC system (Perkin-Elmer Applied Biosystems, Foster City, CA) (Minocha et al., 1990). Polyamines were quantified by a fluorescence detector set at excitation and emission wavelengths of 340 and 515 nm, respectively.
Enzyme Analysis
The activities of ODC and ADC were measured in cell homogenates (Robie and Minocha, 1989) as well as in intact cells (Minocha et al., 1999a) using radiolabeled substrates. The reaction mix contained 200 μL cell extract or 100 mg fresh weight of intact cells in 0.1 m Tris-EDTA buffer (pH 6.8 for mouse ODC and pH 8.4 for plant ADC and ODC) containing 5.0 mm pyridoxal phosphate, 1.0 mm dithiothreitol, the labeled substrate (0.05 or 0.1 μCi of l-[1-14C]Orn, specific activity 56 mCi/mmol, Moravek Biochemicals, Brea, CA; or 0.1 μCi dl-[1-14C]Arg, specific activity 61 mCi/mmol, Amersham-Pharmacia Biotech), and the unlabeled substrate (2 mm l-Orn or l-Arg) in a total volume of 300 μL. The reaction tubes were incubated at 37°C for 60 min. The 14CO2 was adsorbed during the reaction on to a 2 cm2 3MM filter paper (Whatman, Clifton, NJ) soaked with 50 μL of Scintigest (Fisher Scientific, Fair Lawn, NJ). The reaction was stopped by injecting 0.5 mL of 0.5 n sulfuric acid into each tube through the stopper and the tubes incubated for an additional 30 min to adsorb all of the released 14CO2. The filter paper was removed and counted for radioactivity in 10 mL of ScintiLene (Fisher Scientific) in a liquid-scillintation counter (model 7,000, Beckman Instruments, Fullerton, CA). The rate of decarboxylation was linear for at least 90 min. For inhibitor effects, 50 μL of either DFMO or DFMA stocks were used to achieve the desired concentration. The enzyme was pre-incubated with the inhibitor for 15 min prior to the addition of substrate.
Incorporation of Labeled Precursors
For incorporation of the labeled precursors, cell suspensions from several flasks grown in kanamycin-free medium for 3 d were pooled and subdivided into 10 mL fractions in 25-mL Erlenmeyer flasks to achieve a cell density of approximately 1.0 g per flask. To each flask, either 0.2 or 0.5 μCi of l-[U-14C]Orn (specific activity 257 mCi/mmol, Amersham-Pharmacia Biotech) or l-[U-14C]Arg (specific activity 272 mCi/mmol, Moravek Biochemicals) was added and the flask was fitted with a polypropylene well containing 2 cm2 3MM filter paper soaked with 50 μL Scintigest (Robie and Minocha, 1989). The flasks were incubated for various lengths of time at 25°C at 100 rpm on a gyratory shaker. At the end of incubation, the cap was removed and the filter paper transferred to a scintillation vial for counting of radioactivity to determine the rate of 14CO2 production from l-[U-14C]Orn and l-[U-14C]Arg. To each flask containing cell suspension, cold Orn or Arg was added to a final concentration of 2 mm, the cells collected onto Miracloth by vacuum filtration, washed with 2 mm ice-cold Orn or Arg, weighed, and stored frozen in double volume of 7.5% (v/v) PCA at −20°C. Following three cycles of freezing, thawing, and centrifugation (13,000g, 5 min), the PCA extracts were dansylated as described in Andersen et al. (1998). A parallel set of 0.4 mm standards of polyamines (Sigma, St. Louis) was also prepared in the same way. The dansyl-polyamines were extracted in 1.0 mL of toluene. A 20-μL aliquot of toluene and 20 μL of the aqueous phase were counted for radioactivity. The latter fraction mostly contained the unused 14C-Orn and 14C- Arg taken up by the cells and provides data on the uptake of labeled substrates. Eight hundred microliters of the toluene fraction was dried in a Speed-vac, redissolved in 50 μL of methanol, of which 45 μL were spotted on 5 × 25 cm thin-layer chromatography plates (Whatman LK6D silica gel 60). The thin-layer chromatography plates were developed in a solvent mixture of chloroform:triethylamine (5:1 v/v). When the solvent front had moved 15 cm from origin, the plates were air-dried and viewed under UV light to mark the spots of the three polyamines. The bands corresponding to the three polyamines were scraped and counted for radioactivity in ScintiLene.
Media Supplementation with Precursors and Inhibitors
Cell cultures were grown for 3 d in 10 mL of kanamycin-free medium in 50-mL Erlenmeyer flasks. Appropriate amounts of l-Arg, l-Orn, l-Gln, urea, MSX, or gabaculine were added to achieve the desired concentrations in each case (for details of concentrations, see “Results”). Following 24- and 72-h incubation on a gyratory shaker, cells were collected by filtration and processed for polyamine analysis as described above. At least three concentrations were tested for each compound.
Statistical Analysis
For all experiments involving quantitative analysis, three or four replicate flasks were used for each treatment. Each experiment was repeated at least twice, most being repeated three to four times. Data from a single representative experiment are presented here for each treatment. The data were subjected to analysis of variance using SYSTAT version 7.0 Student's t test was used to determine significance at P ≤ 0.05.
ACKNOWLEDGMENTS
The authors would like to express their gratitude to Dr. Dale Smith for arranging a visit of S.C.M. and R.M. to New Zealand Forest Research Institute. The help of Heather Hinton, Simone Donaldson, Armin Wagner, Cathy Hargreaves, Cathy Reeves, and Lynette Grace in construction of the plasmid and the help of Mr. Benjamin Mayer with polyamine analysis are duly acknowledged. The authors are also thankful to Dr. Chris Neefus for help with statistical analysis and to Drs. John Wallace and Curtis Givan for valuable suggestions in improvement of the manuscript.
Footnotes
This work was supported by the University of New Hampshire Undergraduate Research Opportunity Program, by the U.S. Department of Agriculture Forest Service, and by the New Zealand Forest Research Institute (Rotorua). This is New Hampshire Agricultural Experiment Station contribution no. 2052.
LITERATURE CITED
- Andersen SE, Bastola DR, Minocha SC. Metabolism of polyamines in transgenic cells of carrot expressing a mouse ornithine decarboxylase cDNA. Plant Physiol. 1998;116:629–638. doi: 10.1104/pp.116.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastola DR, Minocha SC. Increased putrescine biosynthesis through transfer of mouse ornithine decarboxylase cDNA in carrot promotes somatic embryogenesis. Plant Physiol. 1995;109:63–71. doi: 10.1104/pp.109.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchereau A, Aziz A, Larher F, Martin-Tanguy J. Polyamines and environmental challenges: recent developments. Plant Sci. 1999;140:103–125. [Google Scholar]
- Burtin D, Michael AJ. Over-expression of arginine decarboxylase in transgenic plants. Biochem J. 1997;325:331–337. doi: 10.1042/bj3250331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell T, Escobar C, Lui H, Burtin D, Lepri O, Christou P. Over-expression of the oat arginine decarboxylase cDNA in transgenic rice (Oryza sativa L.) affects normal development patterns in vitro and results in putrescine accumulation in transgenic plants. Theor Appl Genet. 1998;97:246–254. [Google Scholar]
- Chomczynski P, Sacchi N. Single step method for RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cohen SS. A Guide to the Polyamines. New York: Oxford University Press; 1998. pp. 1–595. [Google Scholar]
- Davis RH. Compartmental and regulatory mechanisms in the arginine pathways of Neurospora crassa and Saccharomyces cerevisiae. Microbiol Rev. 1986;50:280–313. doi: 10.1128/mr.50.3.280-313.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RH, Morris DR, Coffino P. Sequestered end products and enzyme regulation: the case of ornithine decarboxylase. Microbiol Rev. 1992;56:280–290. doi: 10.1128/mr.56.2.280-290.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeScenzo RA, Minocha SC. Modulation of cellular polyamines in tobacco by transfer and expression of mouse ornithine decarboxylase cDNA. Plant Mol Biol. 1993;22:113–127. doi: 10.1007/BF00039000. [DOI] [PubMed] [Google Scholar]
- Evans PT, Malmberg RL. Do polyamines have roles in plant development? Annu Rev Plant Physiol. 1989;40:235–269. [Google Scholar]
- Florencio FJ, Vega JM. Utilization of nitrate, nitrite and ammonium by Chlamydomonas reinhardtii. Planta. 1983;158:288–293. doi: 10.1007/BF00397329. [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Ghoda L, Sidney D, Macrae M, Coffino P. Structural elements of ornithine decarboxylase required for intracellular degradation and polyamine-dependent regulation. Mol Cell Biol. 1992;12:2178–2185. doi: 10.1128/mcb.12.5.2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoda L, van Daalen Wetters T, Macrae M, Ascherman D, Coffino P. Prevention of rapid intracellular degradation of ODC by a carboxyl-terminal truncation. Science. 1989;243:1493–1495. doi: 10.1126/science.2928784. [DOI] [PubMed] [Google Scholar]
- Glass JR, Gerner EW. Polyamine-mediated turnover of ornithine decarboxylase in Chinese-hamster ovary cells. Biochem J. 1986;236:351–357. doi: 10.1042/bj2360351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grens A, Scheffler IE. The 5′- and 3′-untranslated regions of ornithine decarboxylase mRNA affect the translational efficiency. J Biol Chem. 1990;265:11810–11816. [PubMed] [Google Scholar]
- Halmekytö M, Alhonen L, Alakuijala L, Jänne J. Transgenic mice over-producing putrescine in their tissues do not convert the diamine into higher polyamines. Biochem J. 1993;291:505–508. doi: 10.1042/bj2910505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halmekytö M, Hyttinen J-M, Sinervirta R, Utriainen M, Myöhänen S, Voipio H-M, Wahlforst J, Syrjänen S, Syrjänen K, Alhonen L. Transgenic mice aberrantly expressing human ornithine decarboxylase gene. J Biol Chem. 1991;266:19746–19751. [PubMed] [Google Scholar]
- Hamill JD, Robins RJ, Parr AJ, Evans DM, Furze JM, Rhodes MJC. Over-expressing a yeast ornithine decarboxylase gene in transgenic roots of Nicotiana rustica can lead to enhanced nicotine accumulation. Plant Mol Biol. 1990;15:27–38. doi: 10.1007/BF00017721. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Murakami Y. Rapid and regulated degradation of ornithine decarboxylase. Biochem J. 1995;306:1–10. doi: 10.1042/bj3060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland R. Amino acid and ureide biosynthesis. In: Dennis DT, Turpin DH, Lefebvre DD, Layzell DB, editors. Plant Metabolism. Ed 2. Singapore: Longman; 1997. pp. 478–494. [Google Scholar]
- Jenkinson CP, Grody WW, Cederbaum SD. Comparative properties of arginases. Comp Biochem Physiol. 1996;114B:107–132. doi: 10.1016/0305-0491(95)02138-8. [DOI] [PubMed] [Google Scholar]
- Kahana C, Nathans D. Nucleotide sequence of murine ornithine decarboxylase. Proc Natl Acad Sci USA. 1985;82:1673–1677. doi: 10.1073/pnas.82.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamoto R, Kameji T, Iwashita S, Igarashi K, Hayashi S. Spermidine-induced destabilization of ornithine decarboxylase (ODC) is mediated by accumulation of antizyme in ODC-overproducing variant cells. J Biol Chem. 1993;268:9393–9399. [PubMed] [Google Scholar]
- Kanamoto R, Utsunomiya K, Kameji T, Hayashi S. Effects of putrescine on synthesis and degradation of ornithine decarboxylase in primary cultured hepatocytes. Eur J Biochem. 1986;154:539–544. doi: 10.1111/j.1432-1033.1986.tb09432.x. [DOI] [PubMed] [Google Scholar]
- Kauppinen RA, Alhonen LI. Transgenic animals as models in the study of the neurobiological role of polyamines. Prog Neurobiol. 1995;47:545–563. doi: 10.1016/0301-0082(95)00037-2. [DOI] [PubMed] [Google Scholar]
- Kende H. Ethylene biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:283–307. [Google Scholar]
- Kieber JJ. The ethylene response pathway in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:277–296. doi: 10.1146/annurev.arplant.48.1.277. [DOI] [PubMed] [Google Scholar]
- Kinney AJ. Manipulating flux through plant metabolic pathways. Curr Opin Plant Biol. 1998;1:173–178. doi: 10.1016/s1369-5266(98)80021-6. [DOI] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- Kumar A, Altabella T, Taylor MR, Tiburcio AF. Recent advances in polyamine research. Trends Plant Sci. 1997;2:124–130. [Google Scholar]
- Kumar A, Minocha SC. Transgenic manipulation of polyamine metabolism. In: Lindsey K, editor. Transgenic Research in Plants. London: Harwood Academic Publishing; 1998. pp. 189–199. [Google Scholar]
- Kumar A, Taylor MR, Mad-Arif SA, Davies H. Potato plants expressing antisense and sense S-adenosylmethionine decarboxylase (SAMDC) transgene show altered levels of polyamines and ethylene: antisense plants display abnormal phenotypes. Plant J. 1996;9:147–158. [Google Scholar]
- Kushad MM, Dumbroff EB. Metabolic and physiological relationship between the polyamine and ethylene biosynthetic pathways. In: Slocum RD, Flores HE, editors. Biochemistry and Physiology of Polyamines in Plants. Boca Raton, FL: CRC Press; 1991. pp. 77–92. [Google Scholar]
- Leason M, Cunliffe D, Parkin D, Lea PJ, Miflin B. Inhibition of pea leaf glutamine synthetase by methionine sulfoximine, phosphoinothricin, and other glutamine analogs. Phytochemistry. 1982;21:855–857. [Google Scholar]
- Li X, Coffino P. Degradation of ornithine decarboxylase: exposure of the C-terminal target by a polyamine-inducible inhibitory protein. Mol Cell Biol. 1993;13:2377–2383. doi: 10.1128/mcb.13.4.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey K, editor. Transgenic Research in Plants, Harwood Academic Publishing, London. 1998. pp. 1–286. [Google Scholar]
- Lovatt CJ. Stress alters ammonia and arginine metabolism. In: Flores HE, Arteca RN, Shannon JC, editors. Polyamines and Ethylene: Biochemistry, Physiology, and Interaction. Boca Raton, FL: CRC Press; 1990. pp. 166–179. [Google Scholar]
- Masgrau C, Altabella T, Farras R, Flores RD, Thompson T, Besford RT, Tiburcio AF. Inducible overexpression of oat ADC in transgenic tobacco. Plant J. 1996;11:465–473. doi: 10.1046/j.1365-313x.1997.11030465.x. [DOI] [PubMed] [Google Scholar]
- Minocha R, Kvaalen H, Minocha SC, Long S. Polyamines in embryogenic cultures of Norway spruce (Picea abies) and red spruce (Picea rubens) Tree Physiol. 1993;13:365–377. doi: 10.1093/treephys/13.4.365. [DOI] [PubMed] [Google Scholar]
- Minocha R, Long S, Maki H, Minocha SC. Assays for the activities of polyamine biosynthetic enzymes using intact tissues. Plant Physiol Biochem. 1999a;37:597–603. [Google Scholar]
- Minocha R, Minocha SC, Simola LK. Somatic embryogenesis and polyamines in woody plants. In: Jain SM, Gupta PK, Newton RJ, editors. Somatic Embryogenesis in Woody Plants. I. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 337–359. [Google Scholar]
- Minocha R, Shortle WC, Long SL, Minocha SC. A rapid and reliable procedure for extraction of cellular polyamines and inorganic ions from plant tissues. J Plant Growth Regul. 1994;13:187–193. [Google Scholar]
- Minocha R, Smith DR, Stewart C, Steele KD, Minocha SC. Polyamine levels during the development of zygotic and somatic embryos of Pinus radiata D Don. Physiol Plant. 1999b;105:155–164. [Google Scholar]
- Minocha SC, Minocha R. Role of polyamines in somatic embryogenesis. In: Bajaj YPS, editor. Biotechnology in Agriculture and Forestry: Somatic Embryogenesis and Synthetic Seed 1. Vol. 30. Berlin: Springer-Verlag; 1995. pp. 53–70. [Google Scholar]
- Minocha SC, Minocha R, Robie CA. High performance liquid chromatographic method for the determination of dansyl-polyamines. J Chromatogr. 1990;511:177–183. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nilsson J, Koskiniemi S, Persson K, Grahn B, Holm I. Polyamines regulate both transcription and translation of the gene encoding ornithine decarboxylase antizyme in mouse. Eur J Biochem. 1997;250:223–231. doi: 10.1111/j.1432-1033.1997.0223a.x. [DOI] [PubMed] [Google Scholar]
- Nishiyama M, Matsufuji S, Kanamoto R, Murakami Y, Hyashi S. Sandwich enzyme immunoassay for ornithine decarboxylase. J Immunoass. 1989;10:19–35. doi: 10.1080/01971528908053225. [DOI] [PubMed] [Google Scholar]
- Noh EW, Minocha SC. Expression of a human S-adenosylmethionine decarboxylase in transgenic tobacco and its effects on polyamine biosynthesis. Transgen Res. 1994;3:26–35. doi: 10.1007/BF01976024. [DOI] [PubMed] [Google Scholar]
- Nuccio ML, Rhodes D, McNeil SD, Hanson AD. Metabolic engineering of plants for osmotic stress resistance. Curr Opin Plant Biol. 1999;2:128–134. doi: 10.1016/s1369-5266(99)80026-0. [DOI] [PubMed] [Google Scholar]
- Pegg AE, McGill S. Decarboxylation of ornithine and lysine in rat tissues. Biochim Biophys Acta. 1979;568:416–421. doi: 10.1016/0005-2744(79)90310-3. [DOI] [PubMed] [Google Scholar]
- Persson L. Decarboxylation of ornithine and lysine by ornithine decarboxylase from kidneys of testosterone treated mice. Acta Chem Scand B. 1981;35:451–455. doi: 10.3891/acta.chem.scand.35b-0451. [DOI] [PubMed] [Google Scholar]
- Primikirios NI, Roubelakis-Angelakis KA. Cloning and expression of an arginine decarboxylase cDNA from Vitis vinifera L. cell-suspension cultures. Planta. 1999;208:574–582. doi: 10.1007/s004250050595. [DOI] [PubMed] [Google Scholar]
- Robie CA, Minocha SC. Polyamines and somatic embryogenesis in carrot: I. The effects of difluoromethylornithine and difluoromethylarginine. Plant Sci. 1989;65:45–54. [Google Scholar]
- Seiler N, Delcros JG, Moulinoux JP. Polyamine transport in mammalian cells: an update. Int J Biochem Cell Biol. 1996;28:843–61. doi: 10.1016/1357-2725(96)00021-0. [DOI] [PubMed] [Google Scholar]
- Shelp BJ, Bown AW, McLean MD. Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 1999;4:446–452. doi: 10.1016/s1360-1385(99)01486-7. [DOI] [PubMed] [Google Scholar]
- Slocum RD. Polyamine biosynthesis in plants. In: Slocum RD, Flores HE, editors. The Biochemistry and Physiology of Polyamines in Plants. Boca Raton, FL: CRC Press; 1991. pp. 23–40. [Google Scholar]
- Slocum RD, Weinstein LH. Stress-induced putrescine accumulation as a mechanism of ammonia detoxification in cereal leaves. In: Flores HE, Arteca RN, Shannon JC, editors. Polyamines and Ethylene: Biochemistry, Physiology, and Interaction. Boca Raton, FL: CRC Press; 1990. pp. 157–165. [Google Scholar]
- Turano FJ, Kramer GF, Wang CY. The effect of methionine, ethylene and polyamine catabolic intermediates on polyamine accumulation in detached soybean leaves. Physiol Plant. 1997;101:510–518. [Google Scholar]
- Walden R, Cordeiro A, Tiburcio AF. Polyamines: small molecules triggering pathways in plant growth and development. Plant Physiol. 1997;113:1009–1013. doi: 10.1104/pp.113.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallstrom EL, Persson L. No role of the 5′-untranslated region of ornithine decarboxylase mRNA in the feedback control of the enzyme. Mol Cell Biochem. 1999;197:71–78. doi: 10.1023/a:1006989808263. [DOI] [PubMed] [Google Scholar]
- Walter C, Grace LJ, Wagner A, White DWR, Walden AR, Donaldson SS, Hinton H, Gardner RC, Smith DR. Stable transformation and regeneration of transgenic plants of Pinus radiata D. Don. Plant Cell Rep. 1998;17:460–468. doi: 10.1007/s002990050426. [DOI] [PubMed] [Google Scholar]
- Watson MB, Malmberg RL. Regulation of Arabidopsis thaliana (L.) Heynh arginine decarboxylase by potassium deficiency stress. Plant Physiol. 1996;111:1077–1083. doi: 10.1104/pp.111.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb DM, Knapp SG. DNA extraction from a previously recalcitrant plant genus. Plant Mol Biol Rep. 1990;8:180–185. [Google Scholar]
- Wu G, Morris SM., Jr Arginine metabolism: nitric oxide and beyond. Biochem J. 1998;336:1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Minocha R, Minocha SC. Physiological responses of suspension cultures of Catharanthus roseus to aluminum: changes in polyamines and inorganic ions. J Plant Physiol. 1995;145:277–284. [Google Scholar]