Abstract
Type 2 diabetes mellitus (T2DM) is one of the common metabolic diseases in the world. Due to the rise in morbidity and mortality, it has become a global health problem. To date, T2DM still cannot be cured, and its intervention measures mainly focus on glucose control as well as the prevention and treatment of related complications. Interestingly, the gut microbiota plays an important role in the development of metabolic diseases, especially T2DM. In this review, we introduce the characteristics of the gut microbiota in T2DM population, T2DM animal models, and diabetic complications. In addition, we describe the molecular mechanisms linking host and the gut microbiota in T2DM, including the host molecules that induce gut microbiota dysbiosis, immune and inflammatory responses, and gut microbial metabolites involved in pathogenesis. These findings suggest that we can treat T2DM and its complications by remodeling the gut microbiota through interventions such as drugs, probiotics, prebiotics, fecal microbiota transplantation (FMT) and diets.
Keywords: gut microbiota, glucose metabolism, insulin resistance, type 2 diabetes mellitus, pathogenesis
Introduction
Type 2 diabetes mellitus (T2DM) is a progressive metabolic disease characterized by pancreatic β-cell dysfunction and peripheral insulin resistance, leading to defects in glucose metabolism and chronic low-grade inflammation. The development of this chronic disease is closely related to both genetic and environmental factors, and certain environmental factors, such as caloric intake, nutrient composition, ambient air pollution and physical inactivity, are important reasons for the continuous increase in its prevalence (Kahn et al., 2014; Kang et al., 2022). It was reported that more than 460 million adults worldwide had diabetes in 2019, and this number would rise to 700 million by 2045 (Saeedi et al., 2019). Since T2DM contributes to the occurrence of harmful macrovascular and microvascular outcomes (Sehgal et al., 2021; Zeng et al., 2021; Zhang X. et al., 2021; Zhao et al., 2021), it not only brings physical and mental pain to patients, but also leads to a major medical economic burden. Therefore, it is of great significance to explore the novel pathogenesis and treatment of T2DM.
The gut microbiota is considered to be a complex ecosystem in the gastrointestinal tract environment, which is composed of bacteria, archaea, fungi, viruses and protozoa (Wang et al., 2021). Growing evidence indicates that the gut microbiota plays a vital role in human health, and its dysbiosis is associated with a variety of pathological processes (Crittenden and Goepp, 2021; Han et al., 2021; Mayneris-Perxachs et al., 2021). Notably, many studies have linked the gut microbiota to T2DM (Kashtanova et al., 2018; Takagi and Naito, 2020). For example, researchers found that 43 bacterial taxa were significantly different between Chinese obese individuals with T2DM and healthy people by LEfSe analysis, and Acidaminococcales, Bacteroides plebeius and Phascolarctobacterium sp.CAG207 might be potential biomarkers for T2DM (Wang T.Y. et al., 2020). Here, we summarize the characteristics of the gut microbiota in T2DM population, T2DM animal models, and diabetic complications, as well as the molecular mechanisms linking host and the gut microbiota in T2DM, which may provide new ideas for the treatment of T2DM.
Gut Microbiota and T2DM
The gut microbiota is involved in obesity, non-alcoholic fatty liver (NAFL), insulin resistance and chronic inflammation, which are related to the development of T2DM (Saad et al., 2016; Mouries et al., 2019; Lee J.Y. et al., 2021). Obese individuals generally have a low abundance of intestinal bacteria (Le Chatelier et al., 2013). Comprehensive analysis of targeted metagenomics and metabolomics found that Italian NAFL patients showed lower levels of Oscillospira and higher levels of 2-butanone and 1-pentanol compared to healthy controls, indicating that the gut microbiota might play an important role in liver steatosis (Del Chierico et al., 2017). Moreover, Horne et al. discovered that a high-fat and high-fructose diet could change the composition of the gut microbiota in Syrian hamsters, leading to dyslipidemia and hepatomegaly (Horne et al., 2020). Among them, Ruminiclostridium 9 and Tyzzerella were positively correlated with fasting cholesterol levels, while Tyzzerella and Ruminococceace NK4A214 group were positively correlated with fasting triglyceride levels. These findings suggest that the gut microbiota is closely linked to a cluster of metabolic disorders. Of note, the gut microbiota has been demonstrated to be altered in diabetic populations and animal models, implying that it may be an important participant in the pathogenesis of T2DM (Figure 1).
Figure 1.
The association between the gut microbiota and T2DM. Dysbiosis of the gut microbiota has been demonstrated not only in T2DM populations, but also in certain animal models, including mice, cats and zebrafish. Furthermore, the gut microbiota is closely associated with various diabetic complications, such as diabetic nephropathy, diabetes-induced cognitive impairment, diabetic retinopathy, and diabetic peripheral neuropathy.
Gut Microbiota Dysbiosis in T2DM Population
To date, substantial evidence of gut microbiota dysbiosis has been found in T2DM individuals (Table 1). Previous research found that the gut microbiota was moderately dysregulated in Chinese T2DM patients (Qin et al., 2012). Specifically, T2DM patients showed an increase in multiple pathogenic bacteria, such as Clostridium hathewayi, Clostridium symbiosum and Escherichia coli, while healthy controls had a high abundance of butyrate-producing bacteria. Another study confirmed that European women with T2DM had a higher abundance of four Lactobacillus species and a lower abundance of five Clostridium species compared with the normal glucose tolerance individuals (Karlsson et al., 2013). Importantly, Lactobacillus species were positively correlated with fasting glucose and glycosylated hemoglobin (HbA1c), while Clostridium species were negatively correlated with fasting glucose, HbA1c and plasma triglycerides, suggesting that these bacterial taxa might be related to the development of T2DM. Similarly, the levels of Lactobacillus were significantly increased, while the levels of Clostridium coccoides and Clostridium leptum were significantly decreased in newly diagnosed T2DM patients (Chen et al., 2019). Notably, Shih et al. studied the characteristics of the intestinal microbiota of patients with refractory T2DM (RT2D), whose HbA1c still increased by at least 8% under treatment (Shih et al., 2020). Compared with T2DM controls, RT2D patients showed higher abundance of Bacteroides vulgatus and Veillonella denticariosi, and lower abundance of Akkermansia muciniphila and Fusobacterium. Of these, the relative levels of A. muciniphila were negatively correlated with HbA1c.
Table 1.
Major findings from the studies of patients with T2DM.
| Patient cohort | Strategy | Characteristics | Main observations | References |
|---|---|---|---|---|
| T2DM | Whole-genome metagenomics shotgun | Chinese, mean age 47.0±4.5 years | Acidaminococcales, Bacteroides plebeius and Phascolarctobacterium sp. CAG207 might be potential biomarkers for T2DM | Wang T.Y. et al., 2020 |
| T2DM | A two-stage metagenome-wide association study based on deep shotgun sequencing | Chinese, aged 13–86 years | T2DM patients showed an increase in multiple pathogenic bacteria, while healthy controls had a high abundance of butyrate-producing bacteria | Qin et al., 2012 |
| T2DM | Whole-genome metagenomics shotgun | 70-year-old European women | European women with T2DM had a higher abundance of four Lactobacillus species and a lower abundance of five Clostridium species | Karlsson et al., 2013 |
| Newly diagnosed T2DM | 16S rRNA sequencing | Taiwanese individuals, mean age 51±12 years | The levels of Lactobacillus were increased, while the levels of Clostridium coccoides and Clostridium leptum were decreased in newly diagnosed T2DM patients | Chen et al., 2019 |
| RT2D | 16S rRNA sequencing | Taiwanese individuals, mean age 64.37±2.194 years | RT2D patients showed higher abundance of Bacteroides vulgatus and Veillonella denticariosi, and lower abundance of Akkermansia muciniphila and Fusobacterium | Shih et al., 2020 |
| Prediabetes | 16S rRNA sequencing | Danish, aged 57–68 years | The abundance of Clostridium genus and A. muciniphila was decreased in Danish patients with prediabetes | Allin et al., 2018 |
| Prediabetes | Whole-genome metagenomics shotgun | Chinese, mean age 61.75 ± 9.09 years | The abundance of MLGs from the Clostridia class and Faecalibacterium prausnitzii was lower, while that from E. coli, Streptococcus salivarius, and Eggerthella sp. was higher in prediabetics | Zhong et al., 2019 |
| DN | 16S rRNA sequencing | Chinese, mean age 52.93 ± 9.98 years | Escherichia-Shigella and Prevotella_9 genera could distinguish DN patients from T2DM without renal disease patients | Tao et al., 2019 |
| DN | 16S rRNA sequencing | Chinese, mean age 60.86 ± 5.69 years | Patients with stage IV DN had a higher abundance of Haemophilus and Lachnospiraceae_UCG-004 compared with patients with stage III DN | Du et al., 2021 |
| DCI | 16S rRNA sequencing | Chinese, mean age 69.41 ± 4.16 years | DCI patients showed a decreased abundance of Bifidobacterium, Tenericutes and unrank-RF39, as well as an increased abundance of Peptococcus and unrank-Leuconostocaceae | Zhang et al., 2021a |
| DR | 16S rRNA sequencing | Chinese, mean age 60.28 ± 10.5 years | Pasteurellaceae might be used as a non-invasive biomarker for the diagnosis of DR | Huang et al., 2021 |
Prediabetes is a pathological condition in which blood glucose levels are higher than normal but lower than diabetes thresholds, and individuals with prediabetes are at increased risk of T2DM (Tabák et al., 2012). Intriguingly, prediabetics also have aberrant gut microbiota (Zhou et al., 2019a; Chávez-Carbajal and Pizano-Zárate, 2020). Compared with individuals with normal glucose regulation, the most significant feature of the gut microbiota in Danish patients with prediabetes was the decreased abundance of Clostridium genus and A. muciniphila (Allin et al., 2018). Zhong et al. observed that there was no significant difference in microbial gene-based richness between treatment-naïve T2DM, prediabetes and normal glucose tolerance Chinese individuals (Zhong et al., 2019). However, compared to normal glucose tolerance individuals, the abundance of metagenomic linkage groups (MLGs) from the Clostridia class and Faecalibacterium prausnitzii was lower, while that from E. coli, Streptococcus salivarius, and Eggerthella sp. was higher in prediabetics (Zhong et al., 2019). Meanwhile, MLGs annotated to E. coli were also enriched in prediabetics compared to treatment-naïve T2DM individuals. These findings indicate that different stages of T2DM show diverse characteristics of the gut microbiota, and there may be complex interactions between host and the gut microbiota.
Gut Microbiota in T2DM Animal Models
Consistently, the association between the gut microbiota and T2DM has also been confirmed in several animal models, including mice, cats and zebrafish (Kieler et al., 2019; Okazaki et al., 2019; Wang Y. et al., 2020). Compared with normal Wistar rats, Goto-Kakizaki (GK) rats, a genetic T2DM animal model produced by repeated inbreeding of Wistar rats, showed significant enrichment of the genera Prevotella_9, Blautia, Roseburia, Allobaculum and Prevotella_1 (Peng et al., 2019). db/db mouse is an animal model of spontaneous diabetes caused by leptin receptor deficiency (Suriano et al., 2021). Similarly, db/db mice had abnormal gut microbiota composition, and the fecal microbiota transplantation (FMT) from db/db mice induced increased body weight and fasting blood glucose, as well as changes in microbiota composition in pseudo-germ-free mice (Yu et al., 2019a). Moreover, Beli et al. found that certain bacterial taxa showed loss of diurnal oscillatory rhythms or phase shifts in their peak levels in db/db mice (Beli et al., 2019). Zucker diabetic fatty (ZDF) rat with a mutant leptin receptor is another common T2DM model, characterized by insulin resistance and metabolic disorders (Griffen et al., 2001). Of note, the relative abundance of Ruminococcus and Allobaculum was positively correlated with the levels of random blood glucose, while the relative abundance of Lactobacillus and Turicibacter was negatively correlated with the levels of random blood glucose in ZDF rats, indicating that these bacterial taxa might be implicated in the pathogenesis of T2DM (Zhou et al., 2019b). In high-fat diet (HFD) and streptozotocin (STZ) treated mice, genistein, an active isoflavone, alleviated insulin resistance and inflammation response by regulating the abundance of genera Bacteroides, Prevotella, Helicobacter and Ruminococcus, suggesting that the gut microbiota could be a potential target for the treatment of T2DM (Yang and Jia, 2021).
In addition, the abundance of several butyrate-producing bacterial genera, such as Dialister, Anaerotruncus and unknown Ruminococcaceae, was reduced in diabetic cats compared to healthy lean cats (Kieler et al., 2019). At the same time, the gut microbiota was related to specific clinical parameters in diabetic cats (Kieler et al., 2019). Specifically, the relative abundance of Enterobacteriaceae was positively correlated with serum fructosamine levels, which reflected the long-term blood glucose levels of cats, while the relative abundance of Ruminococcaceae was negatively correlated with the levels of serum amyloid A. Okazaki et al. established a T2DM zebrafish model by overfeeding for 4 weeks and found lower bacterial diversity in the T2DM zebrafish intestine (Okazaki et al., 2019). Intriguingly, certain members of the gut microbiota were necessary for early pancreatic β-cell development in zebrafish (Hill et al., 2016). Further studies found that some Aeromonas strains could secrete β-cell expansion factor A (BefA), which promoted β-cell expansion by inducing proliferation. More importantly, the specific bacterial species in humans could also secrete BefA-like proteins, and these proteins had similar functions, which provided new ideas for T2DM treatment (Hill et al., 2016). Interestingly, several bacterial taxa, such as Bacteroides (Zhang et al., 2013; Zhang et al., 2021b), A. muciniphila (Yan et al., 2016; Zhang J. et al., 2021), and Lactobacillus (Sato et al., 2014; Gu et al., 2016), showed consistent trends in T2DM patients and animal models. Therefore, it is of great interest to explore the molecular mechanisms by which these bacterial taxa participate in the development of diabetes.
Gut Microbiota and Diabetic Complications
Notably, the gut microbiota is closely associated with a variety of diabetic complications, such as diabetic nephropathy (DN), diabetes-induced cognitive impairment (DCI), diabetic retinopathy (DR), and diabetic peripheral neuropathy (DPN) (Salguero et al., 2019; Jayasudha and Das, 2020; Xie et al., 2020b; Zhang et al., 2021a). DN is one of the common microvascular complications of diabetes mellitus, which eventually develops into end-stage renal failure. Previous studies have found significant differences in the composition of the intestinal microbiota between DN, T2DM without renal disease and healthy individuals (Tao et al., 2019). Specifically, the Escherichia-Shigella and Prevotella_9 genera could accurately distinguish DN patients from T2DM without renal disease patients, while the Prevotella_9 genus could accurately distinguish T2DM without renal disease patients from healthy controls. Moreover, patients with stage IV DN had a higher abundance of Haemophilus and Lachnospiraceae_UCG-004 compared with patients with stage III DN, indicating that the gut microbiota was involved in the progression of DN (Du et al., 2021). Further research demonstrated that the dysregulated gut microbiota inhibited adenosine 5’-monophosphate-activated protein kinase α (AMPKα) by activating G protein-coupled receptor 43 (GPR43), which in turn led to insulin resistance-mediated podocyte and kidney damage (Lu et al., 2021). Importantly, the gut microbiota depletion induced by broad-spectrum antibiotics or the gut microbiota improvement by FMT could reduce glomerular damage in diabetic rats, indicating that the gut microbiota played a key role in the development of DN. Of note, the gut microbiota-derived metabolites, including short chain fatty acids (SCFAs), trimethylamine N-oxide (TMAO) and phenyl sulfate, are also related to the development of DN (Al-Obaide et al., 2017; Kikuchi et al., 2019; Zhao et al., 2019). For example, phenyl sulfate could predict early proteinuria in patients with DN (Kikuchi et al., 2019). In diabetic mice, phenyl sulfate induced proteinuria by promoting podocyte damage, indicating that this metabolite might be a potential target for the treatment of DN.
Diabetes predisposes to cognitive impairment and may be related to the increased incidence of dementia in elderly diabetic patients. However, the research on the pathogenesis of DCI is still in the preliminary stage. Compared to T2DM patients with normal cognition, patients with cognitive impairment showed a unique composition of intestinal microbiota, which was characterized by decreased abundance of Bifidobacterium, Tenericutes and unrank-RF39, as well as increased abundance of Peptococcus and unrank-Leuconostocaceae (Zhang et al., 2021a). Meanwhile, gut microbiota dysbiosis was also found in DCI animal models (Wang et al., 2018; Yu et al., 2019b). Zheng et al. discovered that vancomycin, a broad-spectrum antibiotic, reduced the levels of synaptophysin (SYP) by inhibiting acetate-producing bacteria, thereby accelerating cognitive impairment in diabetic mice (Zheng et al., 2021). Interestingly, this cognitive impairment could be reversed by FMT or exogenous acetate supplementation, indicating that acetate-producing bacteria were necessary to maintain normal cognitive functions. It is worth noting that several interventions, including intermittent fasting, synbiotics and traditional Chinese medicine (TCM), have shown certain efficacy in treating DCI (Gu et al., 2017; Liu and Dai, 2020; Morshedi et al., 2020). Intermittent fasting improved the cognitive dysfunction in db/db mice by reconstructing the gut microbiota and changing microbial metabolites, and the mechanism might be related to the regulation of mitochondrial biogenesis and energy metabolism in the hippocampus (Liu and Dai, 2020). These findings indicate that the gut microbiota and microbial metabolites may be ideal targets for the treatment of DCI.
The latest research pointed out that the gut microbial signatures composed of 25 bacterial families could distinguish DR patients from diabetic patients without retinopathy and healthy controls (Huang et al., 2021). Among them, the relative abundance of Pasteurellaceae was the lowest in DR patients, which alone could be used as a non-invasive biomarker for the diagnosis of DR. Significantly, plasma TMAO levels in DR individuals were significantly higher than those in non-retinopathy T2DM individuals, and its levels were positively correlated with the incidence of DR, indicating that TMAO might be involved in the development of DR (Liu W. et al., 2021). Xie et al. discovered that STZ-induced DPN rats showed gut microbiota dysbiosis, which was characterized by enrichment of Klebsiella, Coprococcus, Prevotella and other bacterial taxa (Xie et al., 2020a). Importantly, The TCM compound Jinmaitong improved the phenotype of peripheral neuropathy by regulating the gut microbiota composition and increasing the levels of neuregulin 1 (NRG1) in DPN rats. In addition, the gut microbiota was also associated with diabetic macrovascular complications and diabetes-induced reproductive system damages (Sanchez-Alcoholado et al., 2017; Tian et al., 2021; Zhu et al., 2021). Together, these data suggest that the gut microbiota and microbial metabolites play a vital role in the complications of diabetes. However, the molecular mechanisms underlying them still need to be studied in depth.
Molecular Mechanisms Linking Host and the Gut Microbiota in T2DM
As mentioned above, there is a complex relationship between the gut microbiota and T2DM. Therefore, it is significant to understand the interaction between host and microbiota in the context of abnormal glucose metabolism. In pathological conditions, the dysregulation of host molecules can lead to changes in the composition of the gut microbiota. In turn, the microbiota plays a regulatory role to participate in the development of T2DM.
Host Molecules That Induce Gut Microbiota Dysbiosis
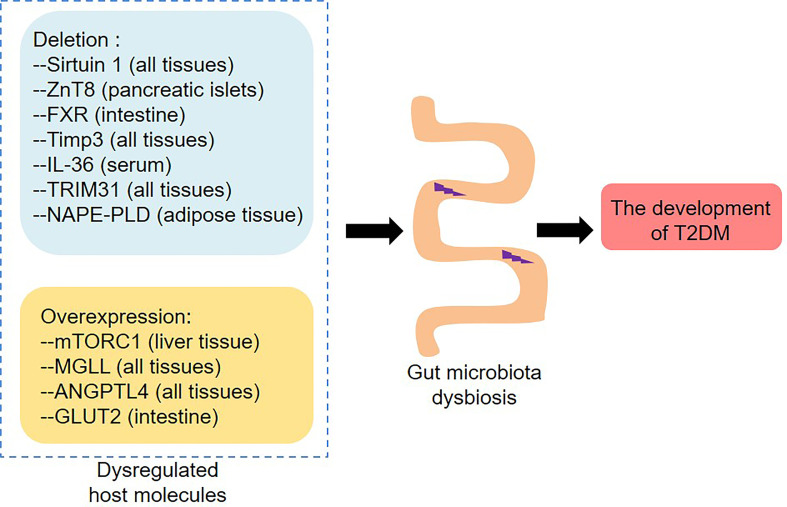
Although T2DM is related to the dysbiosis of the gut microbiota, the reason for such dysbiosis is still unclear. Of note, gut microbial homeostasis is subject to regulation by several host molecules (Figure 2). Previous studies have shown that the protein deacetylase sirtuin 1 protects HFD-fed mice from metabolic disorders mainly by regulating the abundance of Firmicutes and Bacteroidetes (Caron et al., 2014). HFD-fed mice lacking sirtuin 1 exhibited adipose tissue hypertrophy, fatty liver and insulin resistance. Zinc transporter 8 (ZnT8) is a transmembrane protein enriched in pancreatic β-cells, which is involved in the development of diabetes mellitus (Lee et al., 2008; Wan et al., 2017). ZnT8 deficiency induced fat accumulation and glucose intolerance in part by regulating the intestinal morphology and composition of the gut microbiota, which might increase the risk of T2DM and obesity (Mao et al., 2019). It is worth noting that db/db mice treated with farnesoid X receptor (FXR) agonist fexaramine showed decreased serum cholesterol and free fatty acid levels, increased glucagon-like peptide-1 (GLP-1) secretion, as well as improved insulin sensitivity (Pathak et al., 2018). In-depth research found that fexaramine activated the Takeda G protein-coupled receptor 5 (TGR5)/GLP-1 signaling pathway by increasing the abundance of Acetatifactor and Bacteroides that produced lithocholic acid, thereby improving glucose metabolism. Mavilio et al. observed that knockout of tissue inhibitor of metalloproteinase 3 (Timp3) induced liver steatosis and glucose intolerance in HFD-fed mice (Mavilio et al., 2016). Mechanistically, loss of Timp3 led to gut microbiota dysbiosis, which further triggered systemic inflammation through interleukin 6 (IL-6) signaling. Antibiotic treatment improved metabolism and reduced inflammation, indicating that the gut microbiota mediated the role of Timp3 in regulating metabolism (Mavilio et al., 2016). Furthermore, the anti-diabetic effects of host molecules such as IL-36, tripartite motif-containing protein 31 (TRIM31) and N-acylphosphatidylethanolamine phospholipase D (NAPE-PLD) are also related to the gut microbiota (Geurts et al., 2015; Cheng et al., 2018; Giannoudaki and Hernandez-Santana, 2019). Overall, these host proteins can improve metabolism to a certain extent by reshaping the gut microbiota, thereby preventing obesity and T2DM.
Figure 2.
Dysregulated host molecules change the composition of the gut microbiota and thus contribute to the development of T2DM. Deletion of Sirtuin 1, ZnT8, FXR, Timp3, IL-36, TRIM31 and NABE-PLD, as well as overexpression of mTORC1, MGLL, ANGPTL4 and GLUT2, induce gut microbiota dysbiosis. ZnT8, Zinc transporter 8; FXR, Farnesoid X receptor; Timp3, Tissue inhibitor of metalloproteinase 3; IL-36, Interleukin 36; TRIM31, Tripartite motif-containing protein 31; NAPE-PLD, N-acylphosphatidylethanolamine phospholipase D; mTORC1, Mechanistic target of rapamycin complex 1; MGLL, Monoglyceride lipase; ANGPTL4, Angiopoietin-like 4; GLUT2, Glucose transporter 2.
On the contrary, certain host proteins can promote the development of T2DM and related metabolic perturbations by inducing gut microbiota dysbiosis (Jung et al., 2016; Dione et al., 2020; Patil and Arvindekar, 2021). Resveratrol was a specific inhibitor of mechanistic target of rapamycin complex 1 (mTORC1), which could improve glucose intolerance and insulin resistance in HFD-fed mice, indicating that mTORC1 played an important role in glucose homeostasis (Jung et al., 2016). Importantly, resveratrol reduced the relative abundance of Oscillibacter, Clostridium XI, Lactococcus, Flavonifractor and Hydrogenoanaerobacterium in HFD-fed mice, and these bacterial taxa were positively correlated with fasting blood glucose levels and HOMA2-IR index, suggesting that changes in the gut microbiota induced by mTORC1 dysregulation might be related to diabetic phenotypes. Another study found that HFD-fed monoglyceride lipase (MGLL) knockout mice showed improved glucose tolerance and decreased adiposity, and that the gut microbiota of MGLL knockout mice and wild-type mice responded differently to HFD feeding (Dione et al., 2020). For example, HFD increased the abundance of Lactobacillus in wild-type mice, but did not change its abundance in MGLL knockout mice. These data indicate that MGLL may be a potential target for the treatment of metabolic diseases. Moreover, loss of angiopoietin-like 4 (ANGPTL4) improved glucose tolerance and increased insulin levels in mice that were fed diets rich in unsaturated fatty acids, cholesterol and fructose (Janssen et al., 2018). Angptl4−/− mice had a higher abundance of Adlercreutzia, Lactobacillus and SMB53, and a lower abundance of Allobaculum, and the gut microbiota inhibition by antibiotics reduced the glucose tolerance in Angptl4−/− mice, indicating that the effect of ANGPTL4 on glucose metabolism might depend on the regulation of the gut microbiota. Analogously, the gut-specific deletion of glucose transporter 2 (GLUT2) might improve the glucose homeostasis by changing the composition of the gut microbiota in mice (Schmitt et al., 2017). Together, blocking mTORC1, MGLL, ANGPTL4 or GLUT2 by drugs may be a therapeutic strategy for T2DM.
Immune and Inflammatory Responses
It is reported that the crosstalk between the gut microbiota and host regulates local or systemic immunity and inflammation, which in turn contributes to the development of T2DM (Tilg and Moschen, 2014). In this process, the intestinal barrier function plays an important role (Figure 3A). The intestinal barrier protects the body from intestinal contents, and its dysfunction increases the leakage of bacteria or bacterial products and thus leads to chronic inflammation and metabolic diseases (Scheithauer et al., 2020). In vivo and in vitro studies have confirmed that hyperglycemia increases intestinal barrier permeability by changing the integrity of tight and adherence junctions, resulting in systemic influx of microbial products (Thaiss et al., 2018). Importantly, the lack of GLUT2 in intestinal epithelial cells maintained barrier integrity by retaining zonula occludens-1 (ZO-1) and E-cadherin, indicating that GLUT2 might be a target for improving intestinal-related inflammation and infection. In addition, the gut microbiota is also involved in the regulation of intestinal permeability in obesity and diabetes (Everard et al., 2013; Xu et al., 2020). One of the mechanisms is that the gut microbiota affects the intestinal barrier function by regulating the secretion of GLP-2 during diabetes (Cani et al., 2009; Zhang et al., 2020). Moreover, microbial anti-inflammatory molecule (MAM), a metabolite produced by F. prausnitzii, restored the damaged intestinal barrier by increasing ZO-1 expression in db/db mice (Xu et al., 2020). The abundance of A. muciniphila was reduced in the T2DM mouse model, and A. muciniphila treatment could improve metabolic endotoxemia and glucose metabolism by restoring mucus layer thickness (Everard et al., 2013). Interestingly, A. muciniphila-derived extracellular vesicles (EVs) reduced intestinal permeability by promoting the expression of tight junction proteins and thus improved glucose homeostasis in diabetic mice (Chelakkot et al., 2018). Another research found that the protein Amuc_1100 derived from the outer membrane of A. muciniphila maintained the integrity of the intestinal barrier by interacting with toll-like receptor 2 (TLR2), thereby alleviating obesity and insulin resistance in HFD-fed mice (Plovier et al., 2017). These data indicate that A. muciniphila is a beneficial gut bacteria and plays an important role in controlling intestinal permeability.
Figure 3.
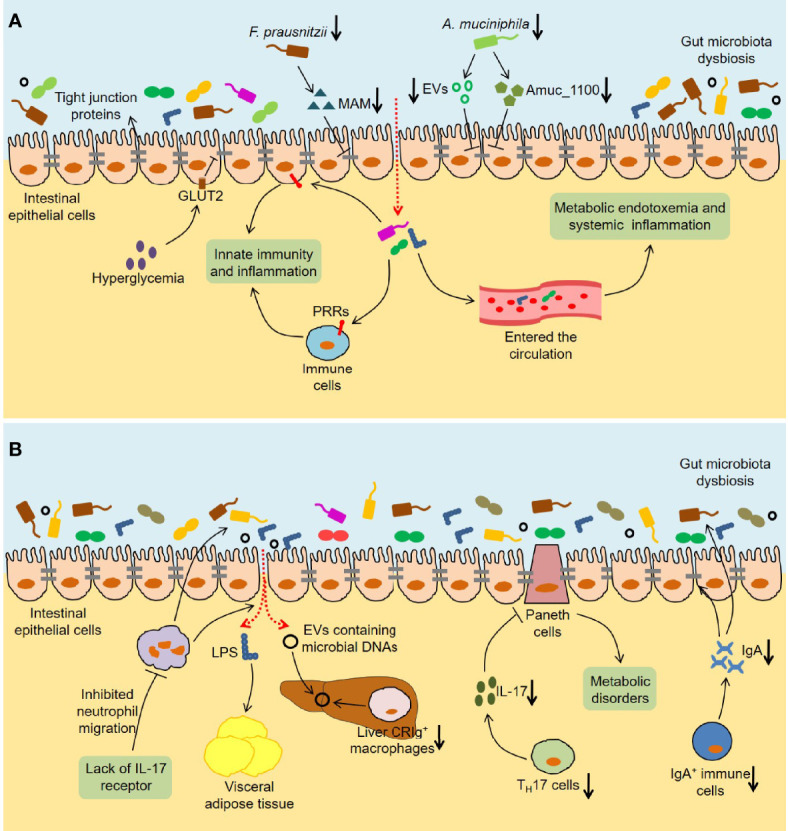
Immune and inflammatory responses induced by the gut microbiota in T2DM. (A) The intestinal barrier is disturbed in T2DM. Hyperglycemia disrupts the integrity of tight and adherent junctions by GLUT2. F. prausnitzii abundance and its metabolite MAM levels are downregulated in db/db mice, which results in damaged intestinal barrier by reducing ZO-1 expression. Analogously, A. muciniphila abundance and the levels of A. muciniphila-derived EVs and Amuc_1100 are reduced in diabetic mice, which also disrupts intestinal barrier integrity. The increased intestinal permeability leads to the translocation of bacteria and bacterial products, which not only activates the innate immunity and inflammation in the gut, but also causes metabolic endotoxemia and systemic inflammation. (B) Multiple immune cells are involved in the influence of the gut microbiota on host metabolism. Lack of IL-17 receptor promotes intestinal dysbiosis and LPS translocation into visceral adipose tissue by inhibiting the migration of neutrophils in the intestinal mucosa. Reduced liver CRIg+ macrophages leads to an accumulation of EVs containing gut microbial DNAs in obesity, thereby promoting tissue inflammation and insulin resistance. The number of TH17 cells is reduced in the intestinal tissues of obese mice, which leads to metabolic disorders by regulating the functions of Paneth cells. Furthermore, HFD-fed mice show a decrease in IgA+ immune cell percentage and secretory IgA concentrations in colon tissues, which impairs glucose metabolism by increasing intestinal permeability and changing microbial composition. GLUT2, Glucose transporter 2; MAM, Microbial anti-inflammatory molecule; ZO-1, Zonula occludens-1; EVs, Extracellular vesicles; PRRs, Pattern recognition receptors; IL-17, Interleukin 17; LPS, Lipopolysaccharide; CRIg, Complement receptor of the immunoglobulin superfamily.
It is well known that innate immunity exists in the organism as a rapid anti-infection effect, and the interaction of pattern recognition receptors (PRRs) and microbe-associated molecular patterns (MAMPs) initiates this process. Invasive bacteria and bacterial products could induce inflammation in the intestinal tract by triggering innate immunity. Notably, some PRRs, namely TLRs and nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), are highly related to metabolic diseases (Winer et al., 2016). Previous studies found that TLR5-deficient mice exhibited insulin resistance and obesity, which was related to changes in the gut microbiota composition (Vijay-Kumar et al., 2010). Importantly, transplantation of the gut microbiota from TLR5-deficient mice caused metabolic syndrome in wild-type germ-free mice, suggesting that the crosstalk between TLR5 and the gut microbiota might play an important role in metabolic disease. Similarly, TLR2 knockout mice had differences in the gut microbiota compared with wild-type mice, and exhibited glucose intolerance and insulin resistance (Guadagnini et al., 2019). Another study discovered that myeloid differentiation primary-response gene 88 (MyD88), a TLR adapter, could regulate glucose and lipid metabolism in part by altering the composition of the gut microbiota (Duparc et al., 2017). The activation of NOD1 contributed to the inflammation and insulin intolerance caused by HFD (Schertzer et al., 2011). Conversely, bacterial cell wall-derived muramyl dipeptide promoted the expression of interferon regulatory factor 4 (IRF4) through NOD2, thereby improving inflammation and insulin resistance in obese mice (Cavallari et al., 2017). Additionally, increased intestinal permeability causes more bacteria and bacterial products to cross the barrier and enter the systemic circulation. Cani et al. observed that HFD-fed mice showed an increase in the proportion of lipopolysaccharide (LPS)-containing bacteria in the intestine and plasma LPS levels (Cani et al., 2007). Meanwhile, they simulated the metabolic impairment and inflammation induced by a HFD in mice through continuous subcutaneous infusion of LPS, indicating that LPS in the systemic circulation might contribute to the development of T2DM. Consistently, antibiotic treatment improved endotoxemia, inflammation, and metabolic parameters in both HFD-fed and ob/ob mice, indicating that the gut microbiota dysbiosis regulated intestinal permeability and further led to metabolic endotoxemia (Cani et al., 2008).
It is worth noting that various immune cells are involved in the role of the gut microbiota in host metabolism (Figure 3B). The lack of IL-17 receptor might promote intestinal dysbiosis and LPS translocation into visceral adipose tissue by inhibiting the migration of neutrophils in the intestinal mucosa, leading to glucose intolerance and insulin resistance in HFD-fed mice (Pérez et al., 2019). Complement receptor of the immunoglobulin superfamily (CRIg) is expressed on human macrophages and plays an important role in immune responses (Duan et al., 2021). Luo et al. observed an accumulation of EVs containing gut microbial DNAs and a decrease in the number of liver CRIg+ macrophages in obesity (Luo et al., 2021). Significantly, CRIg+ macrophages could clear these EVs in a complement C-dependent manner, thereby improving tissue inflammation and insulin resistance. Furthermore, the crosstalk between CD4+ T cells and the gut microbiota is closely related to the development of T2DM. On the one hand, the gut microbiota dysbiosis induced by HFD led to the decrease of TH17 cells in the lamina propria of the small intestine and thus promoted the occurrence of T2DM (Garidou et al., 2015). On the other hand, the number of TH17 cells was significantly reduced in the intestinal tissues of obese mice, and the further reduction of TH17 cells by a HFD lacking vitamin A aggravated glucose intolerance and insulin resistance (Hong et al., 2017). In-depth research found that TH17 cells might regulate the functions of Paneth cells and metabolism-related intestinal microbiota through IL-17, thereby maintaining metabolic homeostasis (Hong et al., 2017). Luck et al. discovered that HFD-fed mice showed a decrease in IgA+ immune cell percentage and secretory IgA concentrations in colon tissues, and that glucose metabolism disorders were more serious in HFD-fed IgA-deficient mice (Luck et al., 2019). Importantly, IgA could improve glucose metabolism by regulating intestinal permeability and microbial compositions, indicating that the treatments targeting IgA+ immune cells might have therapeutic value for T2DM.
Gut Microbial Metabolites
Conspicuously, various gut microbial metabolites, such as SCFAs, TMAO and tryptophan-derived metabolites, have been reported to be closely related to the pathogenesis of T2DM (Jia et al., 2019; Mercer et al., 2020; Noureldein et al., 2020) (Figure 4). Previous studies found that SCFAs inhibited insulin signaling by activating G protein-coupled receptor 43 (GPR43) in adipocytes, thereby suppressing fat accumulation in adipose tissue and promoting energy metabolism in other tissues (Kimura et al., 2013). In addition, SCFAs inhibited the expression of Neurogenin 3 through forkhead box O1 (FOXO1) O-GlcNAcylation and thus suppressed L cell development (Zhao M. et al., 2020). The protein O-GlcNAcylation deficiency in intestinal epithelial cells promoted L cell hyperplasia and GLP-1 secretion in mice, thereby improving glucose metabolism. These findings indicate that SCFAs may be involved in the development of T2DM. Among them, butyrate is one of the well-studied SCFAs produced by the gut microbiota. It could ameliorate the progression of T2DM via a variety of mechanisms, including maintaining the integrity of the intestinal epithelial barrier (Xu et al., 2018), promoting liver glycogen metabolism (Zhang et al., 2019), as well as regulating the function of mitochondria (Zhao T. et al., 2020). Another SCFA propionate promoted the release of GLP-1 and peptide YY (PYY) in rats and mice by activating GPR43 (Psichas et al., 2015). Likewise, acute supplementation of propionate increased postprandial plasma levels of these two gut hormones in humans, and its long-term supplementation prevented weight gain and worsening insulin sensitivity in overweight adults (Chambers et al., 2015). Moreover, propionate activated intestinal gluconeogenesis by the GPR41-dependent gut-brain neural circuit, thereby maintaining glucose and energy homeostasis (De Vadder et al., 2014). It is worth noting that acetate is related to diabetes-associated reproductive abnormalities. Acetate not only improved the hypothalamic-pituitary-ovarian function of female patients with T2DM by inhibiting histone deacetylase-5 (HDAC5) (Olaniyi et al., 2021a), but also alleviated the testicular dysfunction in male patients with diabetes by inhibiting proprotein convertase subtilisin/kexin type 9 (PCSK9) (Olaniyi et al., 2021b). Collectively, SCFAs, such as acetate, propionate and butyrate, have played an important role in the development of T2DM.
Figure 4.
Molecular mechanisms of gut microbial metabolites involved in T2DM. Various gut microbial metabolites, including SCFAs, bile acids, BCAAs, tryptophan-derived metabolites, TMAO, succinate, imidazole propionate, N-formyl peptide and isovanillic acid 3-O-sulfate, contribute to the development of T2DM through complex molecular mechanisms. SCFAs, Short chain fatty acids; BCAAs, Branched-chain amino acids; TMAO, Trimethylamine N-oxide; GPR43, G protein-coupled receptor 43; FOXO1, Forkhead box O1; GLP-1, Glucagon-like peptide-1; PYY, Peptide YY; HDAC5, Histone deacetylase-5; PCSK9, Proprotein convertase subtilisin/kexin type 9; TGR5, Takeda G-protein receptor 5; cAMP, Cyclic AMP; D2, Type 2 iodothyronine deiodinase; FGF21, Fibroblast growth factor 21; UCP1, Uncoupling protein 1; BCKAs, Branched-chain α-keto acids; mTORC1, Mechanistic target of rapamycin complex 1; GLUT4, Glucose transporter 4; PI3K, Phosphoinositide 3-kinase.
The primary bile acids are products of cholesterol metabolism in the liver, which are involved in the absorption and transport of lipids and fat-soluble vitamins. When primary bile acids are secreted into the intestine, they are metabolized by the gut microbiota into secondary bile acids through deconjugation and 7-dehydroxylation (Yesair and Himmelfarb, 1970; Devlin and Fischbach, 2015; Wahlström et al., 2016). It is well known that most secondary bile acids promote the secretion of GLP-1 from intestinal L cells mainly by activating Takeda G-protein receptor 5 (TGR5) (Katsuma et al., 2005; Thomas et al., 2009; Chaudhari et al., 2021). Moreover, bile acids could improve metabolic control by inducing energy expenditure in adipose tissue (Watanabe et al., 2006). Mechanistically, bile acids increased cyclic AMP (cAMP) production and type 2 iodothyronine deiodinase (D2) activity by interacting with TGR5, thereby promoting the conversion of inactive thyroxine (T4) into active thyroid hormone (T3). Interestingly, bile acid-induced activation of TGR5 ameliorated insulin resistance and chronic inflammation in obese mice, which was related to macrophage migration and chemokine expression in macrophages (Perino et al., 2014). These data indicate that bile acids participate in the pathogenesis of T2DM by regulating energy metabolism and inflammation response.
Growing evidence suggests that individuals with diabetes or insulin resistance exhibit increased levels of branched-chain amino acids (BCAAs) (Pedersen et al., 2016; Vanweert et al., 2021). In HFD-fed mice, BCAAs aggravated liver insulin resistance by inhibiting Akt2 signaling, that is, increasing liver gluconeogenesis and inhibiting liver adipogenesis (Zhao H. et al., 2020). Of note, a low-isoleucine diet increased liver insulin sensitivity and energy expenditure by activating the fibroblast growth factor 21 (FGF21)-uncoupling protein 1 (UCP1) axis in mice, while a low-leucine diet had no such effects, indicating that each BCAA might have different contributions to host metabolism (Yu et al., 2021). Normally, BCAAs are metabolized into branched-chain α-keto acids (BCKAs) by branched-chain aminotransferases (BCATs), which are further irreversibly decarboxylated by branched-chain α-ketoacid dehydrogenase (BCKDH) complex (Lynch and Adams, 2014). db/db mice showed decreased activity of BCKDH complex and increased levels of BCKAs (Liu S. et al., 2021). In-depth studies found that BCKAs could induce mitochondrial oxidative stress and cytokine production in macrophages, thereby exacerbating inflammation and organ damage in T2DM (Liu S. et al., 2021).
Virtue et al. observed that microbial metabolites of tryptophan, such as indole and indoxyl sulfate, slowed down the progress of inflammation and insulin resistance by inhibiting the expression of miR-181 in white adipose tissue (Virtue et al., 2019). Intriguingly, indole could regulate GLP-1 secretion from L cells through a complex mechanism (Chimerel et al., 2014). On the one hand, indole acutely promoted GLP-1 secretion by increasing the intracellular Ca2+ concentration. On the other hand, long-term indole stimulation reduced GLP-1 secretion by inhibiting the production of ATP. These results suggest that tryptophan-derived metabolites may serve as important signal molecules that affect host metabolism. Nutrients containing trimethylamine are metabolized by gut microbiota to form trimethylamine, which is then converted into TMAO in the liver. Higher plasma TMAO concentrations were associated with an increased risk of T2DM (Shan et al., 2017). TMAO could block insulin signaling in the liver and induce inflammation in adipose tissues, thus aggravating insulin resistance in HFD-fed mice (Gao et al., 2014). Additionally, other microbiota-produced metabolites, such as succinate, imidazole propionate, N-formyl peptide and isovanillic acid 3-O-sulfate, are also related to host glucose metabolism, which may provide new targets for T2DM treatment strategies (De Vadder et al., 2016; Koh et al., 2018; Houghton et al., 2019; Wollam et al., 2019).
Other Mechanisms
Notably, the gut microbiota composition in elderly T2DM and periodontitis patients has changed compared with healthy controls and elderly patients with T2DM alone, especially the increase in the abundance of Prevotella copri and the decrease in the abundance of F. prausnitzii, indicating that the gut microbiota may mediated the influence of periodontitis on diabetes (Li J. et al., 2020). In C57BL/6J mice, periodontitis induced alterations in the gut microbiota and high levels of fasting blood glucose and glucose intolerance, while the glucose metabolism was improved after antibiotic treatment (Li et al., 2021). Further research found that Porphyromonas gingivalis exacerbated fasting and postprandial hyperglycemia by inducing changes in the gut microbiota in db/db mice, which might be related to excessive hepatic gluconeogenesis (Kashiwagi et al., 2021). Grasset et al. observed that GLP-1-induced insulin secretion and gastric emptying were disrupted in HFD-fed mice (Grasset et al., 2017). Mechanistically, the dysregulated gut microbiota damaged the GLP-1-activated gut-brain axis through reducing the nitric oxide production by enteric neurons and thus led to GLP-1 resistance. Furthermore, the relative abundance of Bacteriodes uniformis and Phascolarctobacterium faecium was decreased in T2DM patients and was negatively correlated with insulin and fasting blood glucose (Li L. et al., 2020). Importantly, their abundance was negatively correlated with serum miR-122-5p levels, indicating that the interaction between the gut microbiota and miRNAs might be involved in the development of T2DM.
Conclusion and Future Perspective
As mentioned above, the gut microbiota plays an important role in the development of T2DM, which greatly expands our understanding of the pathogenesis of this metabolic disease. Meanwhile, accumulating evidence links changes in the gut microbiota with T1DM, gestational diabetes and latent autoimmune diabetes in adults (LADA), indicating that the gut microbiota may be a key regulator of host glucose metabolism and immune response (Zheng et al., 2020; Fang et al., 2021; Qin et al., 2021). However, most of the research focuses on gut bacteria, and little attention is paid to viruses and fungi in the intestinal tract. Recent studies found that the diversity of gut viruses in obese T2DM patients was reduced compared with lean controls (Yang et al., 2021). Moreover, the fungal communities of T2DM patients were different from those of healthy individuals, showing high abundance of Malessezia firfur and unclassified Davidiella as well as low abundance of unclassified Basidiomycota (Al Bataineh et al., 2020). These findings indicate that the gut microbiota is a complex ecosystem and is inextricably linked to T2DM. Nevertheless, further studies are needed to explore which specific microbes are responsible for diabetes physiopathology and the molecular mechanisms at work.
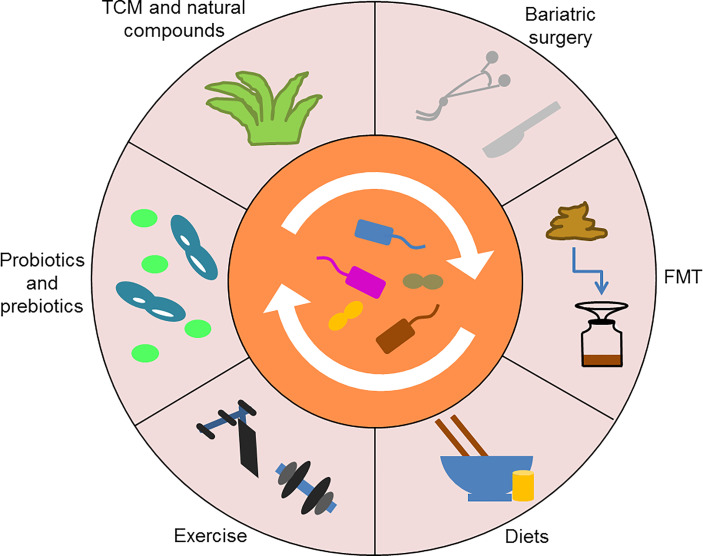
Significantly, various antidiabetic interventions targeting the gut microbiota are being explored (Figure 5). Among them, the most promising ones for clinical application are probiotics and prebiotics (Lee Y. et al., 2021; Watanabe et al., 2021). Lactobacillus rhamnosus LRa05 reduced fasting blood glucose and insulin resistance by regulating the composition of the gut microbiota in T2DM mice (Wu et al., 2021). Chang et al. demonstrated that the polysaccharides derived from Ganoderma lucidum improved insulin resistance and inflammation in HFD-fed mice by reversing the gut microbiota dysbiosis and maintaining intestinal barrier integrity, indicating that the polysaccharides might be an effective prebiotic for diabetes treatment (Chang et al., 2015). In addition, TCM and natural compounds show great potential for restoring homeostasis in the intestinal microenvironment. For example, ginsenoside Rg5 could modulate the abundance of multiple bacterial taxa, such as Firmicutes, Verrucomicrobia and Bacteroidetes, in db/db mice and thus exert anti-diabetic effects (Wei et al., 2020). Of note, certain non-drug therapies, such as bariatric surgery, FMT, diets and exercise, can also slow the progression of T2DM by reshaping the gut microbiota (Wei et al., 2018; Liu et al., 2020; Wang C. et al., 2020; Ng et al., 2021). In conclusion, the gut microbiota is closely related to glucose metabolism, which may provide novel ideas for the treatment of T2DM.
Figure 5.
Various antidiabetic interventions targeting the gut microbiota. Probiotics and prebiotics, TCM and natural compounds, as well as certain non-drug therapies, such as bariatric surgery, FMT, diets, and exercise, can reshape the gut microbiota to treat T2DM. TCM, Traditional Chinese medicine; FMT, Fecal microbiota transplantation.
Author Contributions
ZZ and BS contributed to drafting and revising the article. DY and CZ contributed to the conception and design. All authors approved the final version.
Funding
This work was supported by funding from the National Natural Science Foundation of China (No. 82104307) and the Natural Science Foundation of Hunan Province (No. 2021JJ40865).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Al Bataineh M., Dash N., Bel Lassen P., Banimfreg B., Nada A., Belda E., et al. (2020). Revealing Links Between Gut Microbiome and Its Fungal Community in Type 2 Diabetes Mellitus Among Emirati Subjects: A Pilot Study. Sci. Rep. 10 (1), 9624. doi: 10.1038/s41598-020-66598-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allin K. H., Tremaroli V., Caesar R., Jensen B. A. H., Damgaard M. T. F., Bahl M. I., et al. (2018). Aberrant Intestinal Microbiota in Individuals With Prediabetes. Diabetologia 61 (4), 810–820. doi: 10.1007/s00125-018-4550-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Obaide M. A. I., Singh R., Datta P., Rewers-Felkins K. A., Salguero M. V., Al-Obaidi I., et al. (2017). Gut Microbiota-Dependent Trimethylamine-N-Oxide and Serum Biomarkers in Patients With T2DM and Advanced CKD. J. Clin. Med. 6 (9), 86. doi: 10.3390/jcm6090086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beli E., Prabakaran S., Krishnan P., Evans-Molina C., Grant M. B. (2019). Loss of Diurnal Oscillatory Rhythms in Gut Microbiota Correlates With Changes in Circulating Metabolites in Type 2 Diabetic Db/Db Mice. Nutrients 11 (10), 2310. doi: 10.3390/nu11102310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani P. D., Amar J., Iglesias M. A., Poggi M., Knauf C., Bastelica D., et al. (2007). Metabolic Endotoxemia Initiates Obesity and Insulin Resistance. Diabetes 56 (7), 1761–1772. doi: 10.2337/db06-1491 [DOI] [PubMed] [Google Scholar]
- Cani P., Bibiloni R., Knauf C., Waget A., Neyrinck A., Delzenne N., et al. (2008). Changes in Gut Microbiota Control Metabolic Endotoxemia-Induced Inflammation in High-Fat Diet-Induced Obesity and Diabetes in Mice. Diabetes 57 (6), 1470–1481. doi: 10.2337/db07-1403 [DOI] [PubMed] [Google Scholar]
- Cani P. D., Possemiers S., Van de Wiele T., Guiot Y., Everard A., Rottier O., et al. (2009). Changes in Gut Microbiota Control Inflammation in Obese Mice Through a Mechanism Involving GLP-2-Driven Improvement of Gut Permeability. Gut 58 (8), 1091–1103. doi: 10.1136/gut.2008.165886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron A. Z., He X., Mottawea W., Seifert E. L., Jardine K., Dewar-Darch D., et al. (2014). The SIRT1 Deacetylase Protects Mice Against the Symptoms of Metabolic Syndrome. FASEB J. 28 (3), 1306–1316. doi: 10.1096/fj.13-243568 [DOI] [PubMed] [Google Scholar]
- Cavallari J., Fullerton M., Duggan B., Foley K., Denou E., Smith B., et al. (2017). Muramyl Dipeptide-Based Postbiotics Mitigate Obesity-Induced Insulin Resistance via IRF4. Cell Metab. 25 (5), 1063–1074.e1063. doi: 10.1016/j.cmet.2017.03.021 [DOI] [PubMed] [Google Scholar]
- Chambers E., Viardot A., Psichas A., Morrison D., Murphy K., Zac-Varghese S., et al. (2015). Effects of Targeted Delivery of Propionate to the Human Colon on Appetite Regulation, Body Weight Maintenance and Adiposity in Overweight Adults. Gut 64 (11), 1744–1754. doi: 10.1136/gutjnl-2014-307913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Lin C., Lu C., Martel J., Ko Y., Ojcius D., et al. (2015). Ganoderma Lucidum Reduces Obesity in Mice by Modulating the Composition of the Gut Microbiota. Nat. Commun. 6, 7489. doi: 10.1038/ncomms8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari S., Harris D., Aliakbarian H., Luo J., Henke M., Subramaniam R., et al. (2021). Bariatric Surgery Reveals a Gut-Restricted TGR5 Agonist With Anti-Diabetic Effects. Nat. Chem. Biol. 17 (1), 20–29. doi: 10.1038/s41589-020-0604-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez-Carbajal A., Pizano-Zárate M. L. (2020). Characterization of the Gut Microbiota of Individuals at Different T2D Stages Reveals a Complex Relationship With the Host. Microorganisms 8 (1), 94. doi: 10.3390/microorganisms8010094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelakkot C., Choi Y., Kim D. K., Park H. T., Ghim J., Kwon Y., et al. (2018). Akkermansia Muciniphila-Derived Extracellular Vesicles Influence Gut Permeability Through the Regulation of Tight Junctions. Exp. Mol. Med. 50 (2), e450. doi: 10.1038/emm.2017.282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. C., Chien Y. W., Yang S. C. (2019). The Alteration of Gut Microbiota in Newly Diagnosed Type 2 Diabetic Patients. Nutrition 63-64, 51–56. doi: 10.1016/j.nut.2018.11.019 [DOI] [PubMed] [Google Scholar]
- Cheng J., Xue F., Zhang M., Cheng C., Qiao L., Ma J., et al. (2018). TRIM31 Deficiency Is Associated With Impaired Glucose Metabolism and Disrupted Gut Microbiota in Mice. Front. Physiol. 9, 24. doi: 10.3389/fphys.2018.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimerel C., Emery E., Summers D., Keyser U., Gribble F., Reimann F. (2014). Bacterial Metabolite Indole Modulates Incretin Secretion From Intestinal Enteroendocrine L Cells. Cell Rep. 9 (4), 1202–1208. doi: 10.1016/j.celrep.2014.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden S., Goepp M. (2021). Prostaglandin E(2) Promotes Intestinal Inflammation via Inhibiting Microbiota-Dependent Regulatory T Cells. Sci. Adv. 7 (7), eabd7954. doi: 10.1126/sciadv.abd7954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Chierico F., Nobili V., Vernocchi P., Russo A., De Stefanis C., Gnani D., et al. (2017). Gut Microbiota Profiling of Pediatric Nonalcoholic Fatty Liver Disease and Obese Patients Unveiled by an Integrated Meta-Omics-Based Approach. Hepatology 65 (2), 451–464. doi: 10.1002/hep.28572 [DOI] [PubMed] [Google Scholar]
- De Vadder F., Kovatcheva-Datchary P., Goncalves D., Vinera J., Zitoun C., Duchampt A., et al. (2014). Microbiota-Generated Metabolites Promote Metabolic Benefits via Gut-Brain Neural Circuits. Cell 156, 84–96. doi: 10.1016/j.cell.2013.12.016 [DOI] [PubMed] [Google Scholar]
- De Vadder F., Kovatcheva-Datchary P., Zitoun C., Duchampt A., Bäckhed F., Mithieux G. (2016). Microbiota-Produced Succinate Improves Glucose Homeostasis via Intestinal Gluconeogenesis. Cell Metab. 24 (1), 151–157. doi: 10.1016/j.cmet.2016.06.013 [DOI] [PubMed] [Google Scholar]
- Devlin A., Fischbach M. (2015). A Biosynthetic Pathway for a Prominent Class of Microbiota-Derived Bile Acids. Nat. Chem. Biol. 11 (9), 685–690. doi: 10.1038/nchembio.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dione N., Lacroix S., Taschler U. (2020). Mgll Knockout Mouse Resistance to Diet-Induced Dysmetabolism Is Associated With Altered Gut Microbiota. Cells 9 (12), 2705. doi: 10.3390/cells9122705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y., Chu H., Brandl K., Jiang L., Zeng S., Meshgin N., et al. (2021). CRIg on Liver Macrophages Clears Pathobionts and Protects Against Alcoholic Liver Disease. Nat. Commun. 12 (1), 7172. doi: 10.1038/s41467-021-27385-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Liu J., Xue Y., Kong X., Lv C., Li Z., et al. (2021). Alteration of Gut Microbial Profile in Patients With Diabetic Nephropathy. Endocrine 73 (1), 71–84. doi: 10.1007/s12020-021-02721-1 [DOI] [PubMed] [Google Scholar]
- Duparc T., Plovier H., Marrachelli V., Van Hul M., Essaghir A., Ståhlman M., et al. (2017). Hepatocyte MyD88 Affects Bile Acids, Gut Microbiota and Metabolome Contributing to Regulate Glucose and Lipid Metabolism. Gut 66 (4), 620–632. doi: 10.1136/gutjnl-2015-310904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A., Belzer C., Geurts L., Ouwerkerk J. P., Druart C., Bindels L. B., et al. (2013). Cross-Talk Between Akkermansia Muciniphila and Intestinal Epithelium Controls Diet-Induced Obesity. Proc. Natl. Acad. Sci. U. S. A. 110 (22), 9066–9071. doi: 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Zhang C., Shi H., Wei W., Shang J., Zheng R., et al. (2021). Characteristics of the Gut Microbiota and Metabolism in Patients With Latent Autoimmune Diabetes in Adults: A Case-Control Study. Diabetes Care 44 (12), 2738–2746. doi: 10.2337/dc20-2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Liu X., Xu J., Xue C., Xue Y., Wang Y. (2014). Dietary Trimethylamine N-Oxide Exacerbates Impaired Glucose Tolerance in Mice Fed a High Fat Diet. J. Biosci. Bioeng. 118 (4), 476–481. doi: 10.1016/j.jbiosc.2014.03.001 [DOI] [PubMed] [Google Scholar]
- Garidou L., Pomié C., Klopp P., Waget A., Charpentier J., Aloulou M., et al. (2015). The Gut Microbiota Regulates Intestinal CD4 T Cells Expressing Rorγt and Controls Metabolic Disease. Cell Metab. 22 (1), 100–112. doi: 10.1016/j.cmet.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Geurts L., Everard A., Van Hul M., Essaghir A., Duparc T., Matamoros S., et al. (2015). Adipose Tissue NAPE-PLD Controls Fat Mass Development by Altering the Browning Process and Gut Microbiota. Nat. Commun. 6, 6495. doi: 10.1038/ncomms7495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoudaki E., Hernandez-Santana Y. E. (2019). Interleukin-36 Cytokines Alter the Intestinal Microbiome and Can Protect Against Obesity and Metabolic Dysfunction. Nat. Commun. 10 (1), 4003. doi: 10.1038/s41467-019-11944-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasset E., Puel A., Charpentier J., Collet X., Christensen J., Tercé F., et al. (2017). A Specific Gut Microbiota Dysbiosis of Type 2 Diabetic Mice Induces GLP-1 Resistance Through an Enteric NO-Dependent and Gut-Brain Axis Mechanism. Cell Metab. 25 (5), 1075–1090.e1075. doi: 10.1016/j.cmet.2017.04.013 [DOI] [PubMed] [Google Scholar]
- Griffen S. C., Wang J., German M. S. (2001). A Genetic Defect in Beta-Cell Gene Expression Segregates Independently From the Fa Locus in the ZDF Rat. Diabetes 50 (1), 63–68. doi: 10.2337/diabetes.50.1.63 [DOI] [PubMed] [Google Scholar]
- Guadagnini D., Rocha G., Santos A., Assalin H., Hirabara S., Curi R., et al. (2019). Microbiota Determines Insulin Sensitivity in TLR2-KO Mice. Life Sci. 234, 116793. doi: 10.1016/j.lfs.2019.116793 [DOI] [PubMed] [Google Scholar]
- Gu C., Yang Y., Xiang H., Li S., Liang L., Sui H., et al. (2016). Deciphering Bacterial Community Changes in Zucker Diabetic Fatty Rats Based on 16S rRNA Gene Sequences Analysis. Oncotarget 7 (31), 48941–48952. doi: 10.18632/oncotarget.10597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C., Zhou W., Wang W., Xiang H., Xu H., Liang L., et al. (2017). ZiBuPiYin Recipe Improves Cognitive Decline by Regulating Gut Microbiota in Zucker Diabetic Fatty Rats. Oncotarget 8 (17), 27693–27703. doi: 10.18632/oncotarget.14611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q., Wang J., Li W., Chen Z. J., Du Y. (2021). Androgen-Induced Gut Dysbiosis Disrupts Glucolipid Metabolism and Endocrinal Functions in Polycystic Ovary Syndrome. Microbiome 9 (1), 101. doi: 10.1186/s40168-021-01046-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. H., Franzosa E. A., Huttenhower C., Guillemin K. (2016). A Conserved Bacterial Protein Induces Pancreatic Beta Cell Expansion During Zebrafish Development. Elife 5, e20145. doi: 10.7554/eLife.20145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C., Park A., Yang B., Yun C., Kwak M., Lee G., et al. (2017). Gut-Specific Delivery of T-Helper 17 Cells Reduces Obesity and Insulin Resistance in Mice. Gastroenterology 152 (8), 1998–2010. doi: 10.1053/j.gastro.2017.02.016 [DOI] [PubMed] [Google Scholar]
- Horne R. G., Yu Y., Zhang R., Abdalqadir N., Rossi L., Surette M., et al. (2020). High Fat-High Fructose Diet-Induced Changes in the Gut Microbiota Associated With Dyslipidemia in Syrian Hamsters. Nutrients 12 (11), 3557. doi: 10.3390/nu12113557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton M., Kerimi A., Mouly V., Tumova S., Williamson G. (2019). Gut Microbiome Catabolites as Novel Modulators of Muscle Cell Glucose Metabolism. FASEB J. 33 (2), 1887–1898. doi: 10.1096/fj.201801209R [DOI] [PubMed] [Google Scholar]
- Huang Y., Wang Z., Ma H., Ji S., Chen Z., Cui Z., et al. (2021). Dysbiosis and Implication of the Gut Microbiota in Diabetic Retinopathy. Front. Cell Infect. Microbiol. 11, 646348. doi: 10.3389/fcimb.2021.646348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A. W. F., Katiraei S., Bartosinska B., Eberhard D., Willems van Dijk K., Kersten S. (2018). Loss of Angiopoietin-Like 4 (ANGPTL4) in Mice With Diet-Induced Obesity Uncouples Visceral Obesity From Glucose Intolerance Partly via the Gut Microbiota. Diabetologia 61 (6), 1447–1458. doi: 10.1007/s00125-018-4583-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasudha R., Das T. (2020). Gut Mycobiomes are Altered in People With Type 2 Diabetes Mellitus and Diabetic Retinopathy. PLoS One 15 (12), e0243077. doi: 10.1371/journal.pone.0243077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J., Dou P., Gao M., Kong X., Li C., Liu Z., et al. (2019). Assessment of Causal Direction Between Gut Microbiota-Dependent Metabolites and Cardiometabolic Health: A Bidirectional Mendelian Randomization Analysis. Diabetes 68 (9), 1747–1755. doi: 10.2337/db19-0153 [DOI] [PubMed] [Google Scholar]
- Jung M. J., Lee J., Shin N. R., Kim M. S., Hyun D. W., Yun J. H., et al. (2016). Chronic Repression of mTOR Complex 2 Induces Changes in the Gut Microbiota of Diet-Induced Obese Mice. Sci. Rep. 6, 30887. doi: 10.1038/srep30887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn S. E., Cooper M. E., Del Prato S. (2014). Pathophysiology and Treatment of Type 2 Diabetes: Perspectives on the Past, Present, and Future. Lancet 383 (9922), 1068–1083. doi: 10.1016/s0140-6736(13)62154-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang N., Chen G., Tu R., Liao W., Liu X., Dong X., et al. (2022). Adverse Associations of Different Obesity Measures and the Interactions With Long-Term Exposure to Air Pollutants With Prevalent Type 2 Diabetes Mellitus: The Henan Rural Cohort Study. Environ. Res. 207, 112640. doi: 10.1016/j.envres.2021.112640 [DOI] [PubMed] [Google Scholar]
- Karlsson F. H., Tremaroli V., Nookaew I., Bergström G., Behre C. J., Fagerberg B., et al. (2013). Gut Metagenome in European Women With Normal, Impaired and Diabetic Glucose Control. Nature 498 (7452), 99–103. doi: 10.1038/nature12198 [DOI] [PubMed] [Google Scholar]
- Kashiwagi Y., Aburaya S., Sugiyama N., Narukawa Y., Sakamoto Y., Takahashi M., et al. (2021). Porphyromonas Gingivalis Induces Entero-Hepatic Metabolic Derangements With Alteration of Gut Microbiota in a Type 2 Diabetes Mouse Model. Sci. Rep. 11 (1), 18398. doi: 10.1038/s41598-021-97868-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashtanova D. A., Tkacheva O. N., Doudinskaya E. N., Strazhesko I. D., Kotovskaya Y. V., Popenko A. S., et al. (2018). Gut Microbiota in Patients With Different Metabolic Statuses: Moscow Study. Microorganisms 6 (4), 98. doi: 10.3390/microorganisms6040098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuma S., Hirasawa A., Tsujimoto G. (2005). Bile Acids Promote Glucagon-Like Peptide-1 Secretion Through TGR5 in a Murine Enteroendocrine Cell Line STC-1. Biochem. Biophys. Res. Commun. 329 (1), 386–390. doi: 10.1016/j.bbrc.2005.01.139 [DOI] [PubMed] [Google Scholar]
- Kieler I. N., Osto M., Hugentobler L., Puetz L. (2019). Diabetic Cats Have Decreased Gut Microbial Diversity and a Lack of Butyrate Producing Bacteria. Sci. Rep. 9 (1), 4822. doi: 10.1038/s41598-019-41195-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Saigusa D., Kanemitsu Y., Matsumoto Y., Thanai P., Suzuki N., et al. (2019). Gut Microbiome-Derived Phenyl Sulfate Contributes to Albuminuria in Diabetic Kidney Disease. Nat. Commun. 10 (1), 1835. doi: 10.1038/s41467-019-09735-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura I., Ozawa K., Inoue D., Imamura T., Kimura K., Maeda T., et al. (2013). The Gut Microbiota Suppresses Insulin-Mediated Fat Accumulation via the Short-Chain Fatty Acid Receptor GPR43. Nat. Commun. 4, 1829. doi: 10.1038/ncomms2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A., Molinaro A., Ståhlman M., Khan M., Schmidt C., Mannerås-Holm L., et al. (2018). Microbially Produced Imidazole Propionate Impairs Insulin Signaling Through Mtorc1. Cell 175 (4), 947–961.e917. doi: 10.1016/j.cell.2018.09.055 [DOI] [PubMed] [Google Scholar]
- Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., et al. (2013). Richness of Human Gut Microbiome Correlates With Metabolic Markers. Nature 500 (7464), 541–546. doi: 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Bae E., Kim H. Y., Lee K. M., Yoon S. S., Lee D. C. (2021). High-Fat-Diet-Induced Oxidative Stress Linked to the Increased Colonization of Lactobacillus Sakei in an Obese Population. Microbiol. Spectr. 9 (1), e0007421. doi: 10.1128/Spectrum.00074-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. H., Kang E. S., Kim S. H., Han S. J., Kim C. H., Kim H. J., et al. (2008). Association Between Polymorphisms in SLC30A8, HHEX, CDKN2A/B, IGF2BP2, FTO, WFS1, CDKAL1, KCNQ1 and Type 2 Diabetes in the Korean Population. J. Hum. Genet. 53 (11-12), 991–998. doi: 10.1007/s10038-008-0341-8 [DOI] [PubMed] [Google Scholar]
- Lee Y., Lee D., Park G., Ko S., Park J., Lee Y., et al. (2021). Lactobacillus Plantarum HAC01 Ameliorates Type 2 Diabetes in High-Fat Diet and Streptozotocin-Induced Diabetic Mice in Association With Modulating the Gut Microbiota. Food Funct. 12 (14), 6363–6373. doi: 10.1039/d1fo00698c [DOI] [PubMed] [Google Scholar]
- Li L., Bao J., Chang Y., Wang M., Chen B., Yan F. (2021). Gut Microbiota May Mediate the Influence of Periodontitis on Prediabetes. J. Dental Res. 100 (12), 1387–1396. doi: 10.1177/00220345211009449 [DOI] [PubMed] [Google Scholar]
- Li L., Li C., Lv M., Hu Q., Guo L., Xiong D. (2020). Correlation Between Alterations of Gut Microbiota and miR-122-5p Expression in Patients With Type 2 Diabetes Mellitus. Ann. Trans. Med. 8 (22), 1481. doi: 10.21037/atm-20-6717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lu H., Wu H., Huang S., Chen L., Gui Q., et al. (2020). Periodontitis in Elderly Patients With Type 2 Diabetes Mellitus: Impact on Gut Microbiota and Systemic Inflammation. Aging 12 (24), 25956–25980. doi: 10.18632/aging.202174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Dai X. (2020). Gut Microbiota Mediates Intermittent-Fasting Alleviation of Diabetes-Induced Cognitive Impairment. Nat. Commun. 11 (1), 855. doi: 10.1038/s41467-020-14676-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Li L., Lou P., Zhao M., Wang Y., Tang M., et al. (2021). Elevated Branched-Chain α-Keto Acids Exacerbate Macrophage Oxidative Stress and Chronic Inflammatory Damage in Type 2 Diabetes Mellitus. Free Radical Biol. Med. 175, 141–154. doi: 10.1016/j.freeradbiomed.2021.08.240 [DOI] [PubMed] [Google Scholar]
- Liu Y., Wang Y., Ni Y., Cheung C., Lam K., Wang Y., et al. (2020). Gut Microbiome Fermentation Determines the Efficacy of Exercise for Diabetes Prevention. Cell Metab. 31 (1), 77–91.e75. doi: 10.1016/j.cmet.2019.11.001 [DOI] [PubMed] [Google Scholar]
- Liu W., Wang C., Xia Y., Xia W., Liu G., Ren C., et al. (2021). Elevated Plasma Trimethylamine-N-Oxide Levels Are Associated With Diabetic Retinopathy. Acta Diabetol. 58 (2), 221–229. doi: 10.1007/s00592-020-01610-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Chen P. P., Zhang J. X., Li X. Q., Wang G. H., Yuan B. Y., et al. (2021). GPR43 Deficiency Protects Against Podocyte Insulin Resistance in Diabetic Nephropathy Through the Restoration of Ampkα Activity. Theranostics 11 (10), 4728–4742. doi: 10.7150/thno.56598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck H., Khan S., Kim J., Copeland J., Revelo X., Tsai S., et al. (2019). Gut-Associated IgA Immune Cells Regulate Obesity-Related Insulin Resistance. Nat. Commun. 10 (1), 3650. doi: 10.1038/s41467-019-11370-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Ji Y., Gao H., Gomes Dos Reis F., Bandyopadhyay G., Jin Z., et al. (2021). CRIg Macrophages Prevent Gut Microbial DNA-Containing Extracellular Vesicle-Induced Tissue Inflammation and Insulin Resistance. Gastroenterology 160 (3), 863–874. doi: 10.1053/j.gastro.2020.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch C., Adams S. (2014). Branched-Chain Amino Acids in Metabolic Signalling and Insulin Resistance. Nat. Rev. Endocrinol. 10 (12), 723–736. doi: 10.1038/nrendo.2014.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z., Lin H., Su W., Li J., Zhou M., Li Z., et al. (2019). Deficiency of ZnT8 Promotes Adiposity and Metabolic Dysfunction by Increasing Peripheral Serotonin Production. Diabetes 68 (6), 1197–1209. doi: 10.2337/db18-1321 [DOI] [PubMed] [Google Scholar]
- Mavilio M., Marchetti V., Fabrizi M., Stöhr R., Marino A., Casagrande V., et al. (2016). A Role for Timp3 in Microbiota-Driven Hepatic Steatosis and Metabolic Dysfunction. Cell Rep. 16 (3), 731–743. doi: 10.1016/j.celrep.2016.06.027 [DOI] [PubMed] [Google Scholar]
- Mayneris-Perxachs J., Cardellini M., Hoyles L., Latorre J., Davato F., Moreno-Navarrete J. M., et al. (2021). Iron Status Influences Non-Alcoholic Fatty Liver Disease in Obesity Through the Gut Microbiome. Microbiome 9 (1), 104. doi: 10.1186/s40168-021-01052-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer K., Yeruva L., Pack L., Graham J., Stanhope K., Chintapalli S., et al. (2020). Xenometabolite Signatures in the UC Davis Type 2 Diabetes Mellitus Rat Model Revealed Using a Metabolomics Platform Enriched With Microbe-Derived Metabolites. American Journal of Physiology. Gastrointest. Liver. Physiol. 319 (2), G157–G169. doi: 10.1152/ajpgi.00105.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morshedi M., Saghafi-Asl M., Hosseinifard E. S. (2020). The Potential Therapeutic Effects of the Gut Microbiome Manipulation by Synbiotic Containing-Lactobacillus Plantarum on Neuropsychological Performance of Diabetic Rats. J. Transl. Med. 18 (1), 18. doi: 10.1186/s12967-019-02169-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouries J., Brescia P., Silvestri A., Spadoni I., Sorribas M., Wiest R., et al. (2019). Microbiota-Driven Gut Vascular Barrier Disruption Is a Prerequisite for Non-Alcoholic Steatohepatitis Development. J. Hepatol. 71 (6), 1216–1228. doi: 10.1016/j.jhep.2019.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., Xu Z., Mak J., Yang K., Liu Q., Zuo T., et al. (2021). Microbiota Engraftment After Faecal Microbiota Transplantation in Obese Subjects With Type 2 Diabetes: A 24-Week, Double-Blind, Randomised Controlled Trial. Gut, gutjnl-2020-323617. doi: 10.1136/gutjnl-2020-323617 [DOI] [PubMed]
- Noureldein M., Bitar S., Youssef N., Azar S., Eid A. (2020). Butyrate Modulates Diabetes-Linked Gut Dysbiosis: Epigenetic and Mechanistic Modifications. J. Mol. Endocrinol. 64 (1), 29–42. doi: 10.1530/jme-19-0132 [DOI] [PubMed] [Google Scholar]
- Okazaki F., Zang L., Nakayama H., Chen Z., Gao Z., Chiba H., et al. (2019). Microbiome Alteration in Type 2 Diabetes Mellitus Model of Zebrafish. Sci. Rep. 9 (1), 867. doi: 10.1038/s41598-018-37242-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaniyi K., Amusa O., Ajadi I., Alabi B., Agunbiade T., Ajadi M. (2021. a). Repression of HDAC5 by Acetate Restores Hypothalamic-Pituitary-Ovarian Function in Type 2 Diabetes Mellitus. Reprod. Toxicol. (Elmsford N.Y.) 106, 69–81. doi: 10.1016/j.reprotox.2021.10.008 [DOI] [PubMed] [Google Scholar]
- Olaniyi K., Badejogbin O., Saliu S., Olatunji L. (2021. b). Rescue Effect of Sodium Acetate in Diabetes Mellitus-Associated Testicular Dysfunction Is Accompanied by PCSK9 Modulation. Biochimie 184, 52–62. doi: 10.1016/j.biochi.2021.02.004 [DOI] [PubMed] [Google Scholar]
- Pathak P., Xie C., Nichols R. G., Ferrell J. M., Boehme S., Krausz K. W., et al. (2018). Intestine Farnesoid X Receptor Agonist and the Gut Microbiota Activate G-Protein Bile Acid Receptor-1 Signaling to Improve Metabolism. Hepatology 68 (4), 1574–1588. doi: 10.1002/hep.29857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil R., Arvindekar A. (2021). Glycation of Gut Proteins Initiates Microbial Dysbiosis and can Promote Establishment of Diabetes in Experimental Animals. Microb. Pathog. 152, 104589. doi: 10.1016/j.micpath.2020.104589 [DOI] [PubMed] [Google Scholar]
- Pedersen H., Gudmundsdottir V., Nielsen H., Hyotylainen T., Nielsen T., Jensen B., et al. (2016). Human Gut Microbes Impact Host Serum Metabolome and Insulin Sensitivity. Nature 535 (7612), 376–381. doi: 10.1038/nature18646 [DOI] [PubMed] [Google Scholar]
- Peng W., Huang J., Yang J., Zhang Z., Yu R., Fayyaz S., et al. (2019). Integrated 16s rRNA Sequencing, Metagenomics, and Metabolomics to Characterize Gut Microbial Composition, Function, and Fecal Metabolic Phenotype in Non-Obese Type 2 Diabetic Goto-Kakizaki Rats. Front. Microbiol. 10, 3141. doi: 10.3389/fmicb.2019.03141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez M., Martins L., Dias M., Pereira C., Leite J., Gonçalves E., et al. (2019). Interleukin-17/Interleukin-17 Receptor Axis Elicits Intestinal Neutrophil Migration, Restrains Gut Dysbiosis and Lipopolysaccharide Translocation in High-Fat Diet-Induced Metabolic Syndrome Model. Immunology 156 (4), 339–355. doi: 10.1111/imm.13028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perino A., Pols T., Nomura M., Stein S., Pellicciari R., Schoonjans K. (2014). TGR5 Reduces Macrophage Migration Through mTOR-Induced C/Ebpβ Differential Translation. J. Clin. Invest. 124 (12), 5424–5436. doi: 10.1172/jci76289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plovier H., Everard A., Druart C., Depommier C., Van Hul M., Geurts L., et al. (2017). A Purified Membrane Protein From Akkermansia Muciniphila or the Pasteurized Bacterium Improves Metabolism in Obese and Diabetic Mice. Nat. Med. 23 (1), 107–113. doi: 10.1038/nm.4236 [DOI] [PubMed] [Google Scholar]
- Psichas A., Sleeth M., Murphy K., Brooks L., Bewick G., Hanyaloglu A., et al. (2015). The Short Chain Fatty Acid Propionate Stimulates GLP-1 and PYY Secretion via Free Fatty Acid Receptor 2 in Rodents. Int. J. Obes. (2005) 39 (3), 424–429. doi: 10.1038/ijo.2014.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q., Chen Y., Li Y., Wei J., Zhou X., Le F., et al. (2021). Intestinal Microbiota Play an Important Role in the Treatment of Type I Diabetes in Mice With BefA Protein. Front. Cell. Infect Microbiol. 11, 719542. doi: 10.3389/fcimb.2021.719542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., et al. (2012). A Metagenome-Wide Association Study of Gut Microbiota in Type 2 Diabetes. Nature 490 (7418), 55–60. doi: 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- Saad M. J., Santos A., Prada P. O. (2016). Linking Gut Microbiota and Inflammation to Obesity and Insulin Resistance. Physiol. (Bethesda) 31 (4), 283–293. doi: 10.1152/physiol.00041.2015 [DOI] [PubMed] [Google Scholar]
- Saeedi P., Petersohn I., Salpea P., Malanda B., Karuranga S., Unwin N., et al. (2019). Global and Regional Diabetes Prevalence Estimates for 2019 and Projections for 2030 and 2045: Results From the International Diabetes Federation Diabetes Atlas, 9(Th) Edition. Diabetes Res. Clin. Pract. 157, 107843. doi: 10.1016/j.diabres.2019.107843 [DOI] [PubMed] [Google Scholar]
- Salguero M. V., Al-Obaide M. A. I., Singh R., Siepmann T., Vasylyeva T. L. (2019). Dysbiosis of Gram-Negative Gut Microbiota and the Associated Serum Lipopolysaccharide Exacerbates Inflammation in Type 2 Diabetic Patients With Chronic Kidney Disease. Exp. Ther. Med. 18 (5), 3461–3469. doi: 10.3892/etm.2019.7943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Alcoholado L., Castellano-Castillo D., Jordán-Martínez L., Moreno-Indias I., Cardila-Cruz P., Elena D., et al. (2017). Role of Gut Microbiota on Cardio-Metabolic Parameters and Immunity in Coronary Artery Disease Patients With and Without Type-2 Diabetes Mellitus. Front. Microbiol. 8, 1936. doi: 10.3389/fmicb.2017.01936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato J., Kanazawa A., Ikeda F., Yoshihara T., Goto H., Abe H., et al. (2014). Gut Dysbiosis and Detection of “Live Gut Bacteria” in Blood of Japanese Patients With Type 2 Diabetes. Diabetes Care 37 (8), 2343–2350. doi: 10.2337/dc13-2817 [DOI] [PubMed] [Google Scholar]
- Scheithauer T. P. M., Rampanelli E., Nieuwdorp M., Vallance B. A., Verchere C. B., van Raalte D. H., et al. (2020). Gut Microbiota as a Trigger for Metabolic Inflammation in Obesity and Type 2 Diabetes. Front. Immunol. 11, 571731. doi: 10.3389/fimmu.2020.571731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertzer J., Tamrakar A., Magalhães J., Pereira S., Bilan P., Fullerton M., et al. (2011). NOD1 Activators Link Innate Immunity to Insulin Resistance. Diabetes 60 (9), 2206–2215. doi: 10.2337/db11-0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C. C., Aranias T., Viel T., Chateau D., Le Gall M., Waligora-Dupriet A. J., et al. (2017). Intestinal Invalidation of the Glucose Transporter GLUT2 Delays Tissue Distribution of Glucose and Reveals an Unexpected Role in Gut Homeostasis. Mol. Metab. 6 (1), 61–72. doi: 10.1016/j.molmet.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal P., Mathew S., Sivadas A., Ray A., Tanwar J. (2021). LncRNA VEAL2 Regulates PRKCB2 to Modulate Endothelial Permeability in Diabetic Retinopathy. EMBO J. 40 (15), e107134. doi: 10.15252/embj.2020107134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Z., Sun T., Huang H., Chen S., Chen L., Luo C., et al. (2017). Association Between Microbiota-Dependent Metabolite Trimethylamine-N-Oxide and Type 2 Diabetes. Am. J. Clin. Nutr. 106 (3), 888–894. doi: 10.3945/ajcn.117.157107 [DOI] [PubMed] [Google Scholar]
- Shih C. T., Yeh Y. T., Lin C. C., Yang L. Y., Chiang C. P. (2020). Akkermansia Muciniphila is Negatively Correlated With Hemoglobin A1c in Refractory Diabetes. Microorganisms 8 (9), 1360. doi: 10.3390/microorganisms8091360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suriano F., Vieira-Silva S., Falony G., Roumain M., Paquot A., Pelicaen R., et al. (2021). Novel Insights Into the Genetically Obese (Ob/Ob) and Diabetic (Db/Db) Mice: Two Sides of the Same Coin. Microbiome 9 (1), 147. doi: 10.1186/s40168-021-01097-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabák A. G., Herder C., Rathmann W., Brunner E. J., Kivimäki M. (2012). Prediabetes: A High-Risk State for Diabetes Development. Lancet 379 (9833), 2279–2290. doi: 10.1016/s0140-6736(12)60283-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi T., Naito Y. (2020). Changes in the Gut Microbiota Are Associated With Hypertension, Hyperlipidemia, and Type 2 Diabetes Mellitus in Japanese Subjects. Nutrients 12 (10), 2996. doi: 10.3390/nu12102996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao S., Li L., Li L., Liu Y., Ren Q., Shi M., et al. (2019). Understanding the Gut-Kidney Axis Among Biopsy-Proven Diabetic Nephropathy, Type 2 Diabetes Mellitus and Healthy Controls: An Analysis of the Gut Microbiota Composition. Acta Diabetol. 56 (5), 581–592. doi: 10.1007/s00592-019-01316-7 [DOI] [PubMed] [Google Scholar]
- Thaiss C. A., Levy M., Grosheva I., Zheng D., Soffer E., Blacher E., et al. (2018). Hyperglycemia Drives Intestinal Barrier Dysfunction and Risk for Enteric Infection. Science 359 (6382), 1376–1383. doi: 10.1126/science.aar3318 [DOI] [PubMed] [Google Scholar]
- Thomas C., Gioiello A., Noriega L., Strehle A., Oury J., Rizzo G., et al. (2009). TGR5-Mediated Bile Acid Sensing Controls Glucose Homeostasis. Cell Metab. 10 (3), 167–177. doi: 10.1016/j.cmet.2009.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R., Liu H., Feng S., Wang H., Wang Y., Wang Y., et al. (2021). Gut Microbiota Dysbiosis in Stable Coronary Artery Disease Combined With Type 2 Diabetes Mellitus Influences Cardiovascular Prognosis. Nutr. Metab. Cardiovasc. Dis. 31 (5), 1454–1466. doi: 10.1016/j.numecd.2021.01.007 [DOI] [PubMed] [Google Scholar]
- Tilg H., Moschen A. R. (2014). Microbiota and Diabetes: An Evolving Relationship. Gut 63 (9), 1513–1521. doi: 10.1136/gutjnl-2014-306928 [DOI] [PubMed] [Google Scholar]
- Vanweert F., de Ligt M., Hoeks J., Hesselink M., Schrauwen P., Phielix E. (2021). Elevated Plasma Branched-Chain Amino Acid Levels Correlate With Type 2 Diabetes-Related Metabolic Disturbances. J. Clin. Endocrinol. Metab. 106 (4), e1827–e1836. doi: 10.1210/clinem/dgaa751 [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M., Aitken J., Carvalho F., Cullender T., Mwangi S., Srinivasan S., et al. (2010). Metabolic Syndrome and Altered Gut Microbiota in Mice Lacking Toll-Like Receptor 5. Science (N.Y. N.Y.) 328 (5975), 228–231. doi: 10.1126/science.1179721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtue A., McCright S., Wright J., Jimenez M., Mowel W., Kotzin J., et al. (2019). The Gut Microbiota Regulates White Adipose Tissue Inflammation and Obesity via a Family of microRNAs. Sci. Trans. Med. 11 (496), eaav1892. doi: 10.1126/scitranslmed.aav1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlström A., Sayin S., Marschall H., Bäckhed F. (2016). Intestinal Crosstalk Between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 24 (1), 41–50. doi: 10.1016/j.cmet.2016.05.005 [DOI] [PubMed] [Google Scholar]
- Wang Y., Ouyang M., Gao X., Wang S., Fu C., Zeng J., et al. (2020). Phocea, Pseudoflavonifractor and Lactobacillus Intestinalis: Three Potential Biomarkers of Gut Microbiota That Affect Progression and Complications of Obesity-Induced Type 2 Diabetes Mellitus. Diabetes Metab. Syndr. Obes. 13, 835–850. doi: 10.2147/dmso.s240728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Tang R., Li B., Ma X., Schnabl B., Tilg H. (2021). Gut Microbiome, Liver Immunology, and Liver Diseases. Cell. Mol. Immunol.. 18 (1), 4–17. doi: 10.1038/s41423-020-00592-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang F., Zhang Y., Xiong H., Zhang Y., Zhuang P., et al. (2018). Diabetic Cognitive Dysfunction Is Associated With Increased Bile Acids in Liver and Activation of Bile Acid Signaling in Intestine. Toxicol. Lett. 287, 10–22. doi: 10.1016/j.toxlet.2018.01.006 [DOI] [PubMed] [Google Scholar]
- Wang T. Y., Zhang X. Q., Chen A. L., Zhang J., Lv B. H., Ma M. H., et al. (2020). A Comparative Study of Microbial Community and Functions of Type 2 Diabetes Mellitus Patients With Obesity and Healthy People. Appl. Microbiol. Biotechnol. 104 (16), 7143–7153. doi: 10.1007/s00253-020-10689-7 [DOI] [PubMed] [Google Scholar]
- Wang C., Zhang H., Liu H., Zhang H., Bao Y., Di J., et al. (2020). The Genus Sutterella Is a Potential Contributor to Glucose Metabolism Improvement After Roux-En-Y Gastric Bypass Surgery in T2D. Diabetes Res. Clin. Pract. 162, 108116. doi: 10.1016/j.diabres.2020.108116 [DOI] [PubMed] [Google Scholar]
- Wan H., Merriman C., Atkinson M. A., Wasserfall C. H., McGrail K. M., Liang Y., et al. (2017). Proteoliposome-Based Full-Length ZnT8 Self-Antigen for Type 1 Diabetes Diagnosis on a Plasmonic Platform. Proc. Natl. Acad. Sci. U. S. A. 114 (38), 10196–10201. doi: 10.1073/pnas.1711169114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Houten S., Mataki C., Christoffolete M., Kim B., Sato H., et al. (2006). Bile Acids Induce Energy Expenditure by Promoting Intracellular Thyroid Hormone Activation. Nature 439 (7075), 484–489. doi: 10.1038/nature04330 [DOI] [PubMed] [Google Scholar]
- Watanabe A., Tochio T., Kadota Y., Takahashi M., Kitaura Y., Ishikawa H., et al. (2021). Supplementation of 1-Kestose Modulates the Gut Microbiota Composition to Ameliorate Glucose Metabolism in Obesity-Prone Hosts. Nutrients 13 (9), 2983. doi: 10.3390/nu13092983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S., Han R., Zhao J., Wang S., Huang M., Wang Y., et al. (2018). Intermittent Administration of a Fasting-Mimicking Diet Intervenes in Diabetes Progression, Restores β Cells and Reconstructs Gut Microbiota in Mice. Nutr. Metab. 15, 80. doi: 10.1186/s12986-018-0318-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Yang H., Zhu C., Deng J., Fan D. (2020). Hypoglycemic Effect of Ginsenoside Rg5 Mediated Partly by Modulating Gut Microbiota Dysbiosis in Diabetic Db/Db Mice. J. Agric. Food Chem. 68 (18), 5107–5117. doi: 10.1021/acs.jafc.0c00605 [DOI] [PubMed] [Google Scholar]
- Winer D., Luck H., Tsai S., Winer S. (2016). The Intestinal Immune System in Obesity and Insulin Resistance. Cell Metab. 23 (3), 413–426. doi: 10.1016/j.cmet.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Wollam J., Riopel M., Xu Y., Johnson A., Ofrecio J., Ying W., et al. (2019). Microbiota-Produced N-Formyl Peptide fMLF Promotes Obesity-Induced Glucose Intolerance. Diabetes 68 (7), 1415–1426. doi: 10.2337/db18-1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Zhang Y., Li W., Zhao Y., Long H., Muhindo E., et al. (2021). Lactobacillus Rhamnosus LRa05 Ameliorate Hyperglycemia Through a Regulating Glucagon-Mediated Signaling Pathway and Gut Microbiota in Type 2 Diabetic Mice. J. Agric. Food. Chem. 69 (31), 8797–8806. doi: 10.1021/acs.jafc.1c02925 [DOI] [PubMed] [Google Scholar]
- Xie J., Song W., Liang X., Zhang Q., Shi Y., Liu W., et al. (2020. a). Jinmaitong Ameliorates Diabetic Peripheral Neuropathy in Streptozotocin-Induced Diabetic Rats by Modulating Gut Microbiota and Neuregulin 1. Aging (Albany N. Y.) 12 (17), 17436–17458. doi: 10.18632/aging.103750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Song W., Liang X., Zhang Q., Shi Y., Liu W., et al. (2020. b). Protective Effect of Quercetin on Streptozotocin-Induced Diabetic Peripheral Neuropathy Rats Through Modulating Gut Microbiota and Reactive Oxygen Species Level. BioMed. Pharmacother. 127, 110147. doi: 10.1016/j.biopha.2020.110147 [DOI] [PubMed] [Google Scholar]
- Xu Y., Gao C., Guo H., Zhang W., Huang W., Tang S., et al. (2018). Sodium Butyrate Supplementation Ameliorates Diabetic Inflammation in Db/Db Mice. J. Endocrinol. 238 (3), 231–244. doi: 10.1530/joe-18-0137 [DOI] [PubMed] [Google Scholar]
- Xu J., Liang R., Zhang W., Tian K., Li J., Chen X., et al. (2020). Faecalibacterium Prausnitzii-Derived Microbial Anti-Inflammatory Molecule Regulates Intestinal Integrity in Diabetes Mellitus Mice via Modulating Tight Junction Protein Expression. J. Diabetes. 12 (3), 224–236. doi: 10.1111/1753-0407.12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Jia Q. (2021). Genistein Ameliorates Inflammation and Insulin Resistance Through Mediation of Gut Microbiota Composition in Type 2 Diabetic Mice. Eur. J. Nutr. 60 (4), 2155–2168. doi: 10.1007/s00394-020-02403-0 [DOI] [PubMed] [Google Scholar]
- Yang K., Niu J., Zuo T., Sun Y., Xu Z., Tang W., et al. (2021). Alterations in the Gut Virome in Obesity and Type 2 Diabetes Mellitus. Gastroenterology 161 (4), 1257–1269.e13. doi: 10.1053/j.gastro.2021.06.056 [DOI] [PubMed] [Google Scholar]
- Yan M., Song M. M., Bai R. X., Cheng S., Yan W. M. (2016). Effect of Roux-En-Y Gastric Bypass Surgery on Intestinal Akkermansia Muciniphila. World J. Gastrointest. Surg. 8 (4), 301–307. doi: 10.4240/wjgs.v8.i4.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesair D., Himmelfarb P. (1970). Hydrolysis of Conjugated Bile Acids by Cell-Free Extracts From Aerobic Bacteria. Appl. Microbiol. 19 (2), 295–300. doi: 10.1128/am.19.2.295-300.1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Han W., Zhan G., Li S., Jiang X., Wang L., et al. (2019. a). Abnormal Gut Microbiota Composition Contributes to the Development of Type 2 Diabetes Mellitus in Db/Db Mice. Aging (Albany N. Y.) 11 (22), 10454–10467. doi: 10.18632/aging.102469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Han W., Zhan G., Li S., Xiang S., Zhu B., et al. (2019. b). Abnormal Gut Microbiota Composition Contributes to Cognitive Dysfunction in Streptozotocin-Induced Diabetic Mice. Aging (Albany N. Y.) 11 (10), 3262–3279. doi: 10.18632/aging.101978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Richardson N., Green C., Spicer A., Murphy M., Flores V., et al. (2021). The Adverse Metabolic Effects of Branched-Chain Amino Acids are Mediated by Isoleucine and Valine. Cell Metab. 33 (5), 905–922.e906. doi: 10.1016/j.cmet.2021.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M., Wen J., Ma Z., Xiao L. (2021). FOXO1-Mediated Downregulation of RAB27B Leads to Decreased Exosome Secretion in Diabetic Kidneys. Diabetes 70 (7), 1536–1548. doi: 10.2337/db20-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y., Chen J., Yi K., Peng L., Xie J., Gou X., et al. (2020). Phlorizin Ameliorates Obesity-Associated Endotoxemia and Insulin Resistance in High-Fat Diet-Fed Mice by Targeting the Gut Microbiota and Intestinal Barrier Integrity. Gut Microbes 12 (1), 1–18. doi: 10.1080/19490976.2020.1842990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lu S., Yang Y., Wang Z., Wang B., Zhang B., et al. (2021. a). The Diversity of Gut Microbiota in Type 2 Diabetes With or Without Cognitive Impairment. Aging Clin. Exp. Res. 33 (3), 589–601. doi: 10.1007/s40520-020-01553-9 [DOI] [PubMed] [Google Scholar]
- Zhang J., Ni Y., Qian L., Fang Q., Zheng T., Zhang M., et al. (2021). Decreased Abundance of Akkermansia Muciniphila Leads to the Impairment of Insulin Secretion and Glucose Homeostasis in Lean Type 2 Diabetes. Adv. Sci. (Weinheim Baden-Wurttemberg Germany) 8 (16), e2100536. doi: 10.1002/advs.202100536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shen D., Fang Z., Jie Z., Qiu X., Zhang C., et al. (2013). Human Gut Microbiota Changes Reveal the Progression of Glucose Intolerance. PLoS One 8 (8), e71108. doi: 10.1371/journal.pone.0071108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wu T., Li W., Zhao Y., Long H., Liu R., et al. (2021. b). Lactobacillus Casei LC89 Exerts Antidiabetic Effects Through Regulating Hepatic Glucagon Response and Gut Microbiota in Type 2 Diabetic Mice. Food Funct. 12 (18), 8288–8299. doi: 10.1039/d1fo00882j [DOI] [PubMed] [Google Scholar]
- Zhang W., Zhao T., Gui D., Gao C., Gu J., Gan W., et al. (2019). Sodium Butyrate Improves Liver Glycogen Metabolism in Type 2 Diabetes Mellitus. J. Agric. Food Chem. 67 (27), 7694–7705. doi: 10.1021/acs.jafc.9b02083 [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhao S., Yuan Q., Zhu L., Li F., Wang H., et al. (2021). TXNIP, a Novel Key Factor to Cause Schwann Cell Dysfunction in Diabetic Peripheral Neuropathy, Under the Regulation of PI3K/Akt Pathway Inhibition-Induced DNMT1 and DNMT3a Overexpression. Cell Death Dis. 12 (7), 642. doi: 10.1038/s41419-021-03930-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Gu J., Zhang H., Wang Z., Zhang W., Zhao Y., et al. (2020). Sodium Butyrate-Modulated Mitochondrial Function in High-Insulin Induced HepG2 Cell Dysfunction. Oxid. Med. Cell. Longev. 2020, 1904609. doi: 10.1155/2020/1904609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Lou H., Peng Y., Chen S., Zhang Y., Li X. (2019). Comprehensive Relationships Between Gut Microbiome and Faecal Metabolome in Individuals With Type 2 Diabetes and Its Complications. Endocrine 66 (3), 526–537. doi: 10.1007/s12020-019-02103-8 [DOI] [PubMed] [Google Scholar]
- Zhao G., Lu H., Liu Y., Zhao Y., Zhu T., Garcia-Barrio M. T., et al. (2021). Single-Cell Transcriptomics Reveals Endothelial Plasticity During Diabetic Atherogenesis. Front. Cell Dev. Biol. 9, 689469. doi: 10.3389/fcell.2021.689469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Ren K., Xiong X., Cheng M., Zhang Z., Huang Z., et al. (2020). Protein O-GlcNAc Modification Links Dietary and Gut Microbial Cues to the Differentiation of Enteroendocrine L Cells. Cell Rep. 32 (6), 108013. doi: 10.1016/j.celrep.2020.108013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Zhang F., Sun D., Wang X., Zhang X., Zhang J., et al. (2020). Branched-Chain Amino Acids Exacerbate Obesity-Related Hepatic Glucose and Lipid Metabolic Disorders via Attenuating Akt2 Signaling. Diabetes 69 (6), 1164–1177. doi: 10.2337/db19-0920 [DOI] [PubMed] [Google Scholar]
- Zheng W., Xu Q., Huang W., Yan Q., Chen Y., Zhang L., et al. (2020). Gestational Diabetes Mellitus Is Associated With Reduced Dynamics of Gut Microbiota During the First Half of Pregnancy. mSystems 5 (2), e00109–20. doi: 10.1128/mSystems.00109-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Xu P., Jiang Q., Xu Q., Zheng Y., Yan J., et al. (2021). Depletion of Acetate-Producing Bacteria From the Gut Microbiota Facilitates Cognitive Impairment Through the Gut-Brain Neural Mechanism in Diabetic Mice. Microbiome 9 (1), 145. doi: 10.1186/s40168-021-01088-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H., Ren H., Lu Y., Fang C., Hou G., Yang Z., et al. (2019). Distinct Gut Metagenomics and Metaproteomics Signatures in Prediabetics and Treatment-Naïve Type 2 Diabetics. EBioMedicine 47, 373–383. doi: 10.1016/j.ebiom.2019.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Sailani M. R., Contrepois K., Zhou Y., Ahadi S., Leopold S. R., et al. (2019. a). Longitudinal Multi-Omics of Host-Microbe Dynamics in Prediabetes. Nature 569 (7758), 663–671. doi: 10.1038/s41586-019-1236-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Xu H., Zhan L., Lu X., Zhang L. (2019. b). Dynamic Development of Fecal Microbiome During the Progression of Diabetes Mellitus in Zucker Diabetic Fatty Rats. Front. Microbiol. 10, 232. doi: 10.3389/fmicb.2019.00232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Du Q., Jiao N., Shu A., Gao Y., Chen J., et al. (2021). Catalpol Ameliorates Diabetes-Induced Testicular Injury and Modulates Gut Microbiota. Life Sci. 267, 118881. doi: 10.1016/j.lfs.2020.118881 [DOI] [PubMed] [Google Scholar]