Abstract
The change in cell state from normal to malignant is driven fundamentally by oncogenic mutations in cooperation with epigenetic alterations of chromatin. These alterations in chromatin can be a consequence of environmental stressors or germline and/or somatic mutations that directly alter the structure of chromatin machinery proteins, their levels, or their regulatory function. These changes can result in an inability of the cell to differentiate along a predefined lineage path, or drive a hyperactive, highly proliferative state with addiction to high levels of transcriptional output. We discuss how these genetic alterations hijack the chromatin machinery for the oncogenic process to reveal unique vulnerabilities and novel targets for cancer therapy.
Cancer is a panoply of diseases caused by a combination of inherited (germline) and newly acquired (somatic) genetic mutations that converge on chromatin, which results in dysregulation of gene expression leading to abnormal cell growth. Oncogenic mutations that directly influence chromatin biology can be divided into two major categories: (1) chromatin machinery errors that comprise mutations, translocations, or copy number alterations effecting chromatin machinery genes, and (2) regulatory element errors that include genomic alterations in noncoding elements that change how chromatin regulates transcription. Similar to thermodynamic entropy, genetic replication errors accumulate with cellular age (Tomasetti and Vogelstein 2015), such that cancer occurs proportionally to the rate and number of cell divisions in the tissue of origin. This principally explains why age is the strongest risk factor for cancer overall (National Cancer Institute, Surveillance, Epidemiology, and End Results Program, www.seer.cancer.gov). Rising disorder in genetic information, both coding and noncoding errors, are entropic to intra- and intercellular communication circuits that mediate through the epigenome (Gryder et al. 2013; Nijman 2020).
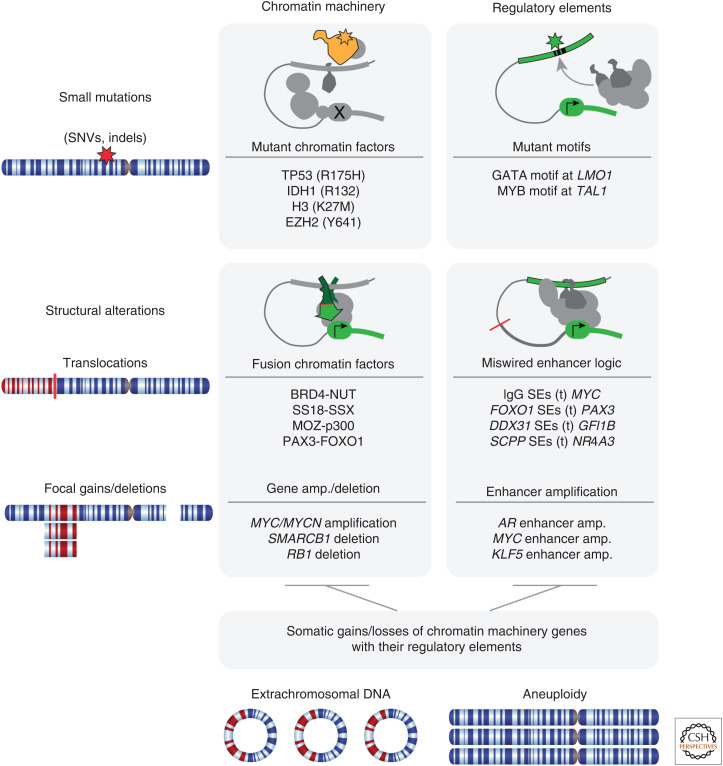
Genetic errors in chromatin-related processes include gain-of-function (GOF), loss-of-function (LOF), or swap-of-function (SOF) with simultaneous loss and gain (Shen and Vakoc 2015). Both small mutations (single-nucleotide variants [SNVs] or insertions or deletions [indels]) and large structural rearrangements effect both the chromatin genes and regulatory elements in cancer; therefore, the combination of small mutation or structural alterations affecting either chromatin machinery or regulatory elements generates four distinct types of major chromatin-based cancer-driving events (Fig. 1). These categories are often co-occurrent, as in the case of extrachromosomal circular DNA amplifications and aneuploidy effecting whole chromosome gains and losses where they can alter both the chromatin machinery gene copy number along with their regulatory elements.
Figure 1.
Categorization of genetic alterations impacting chromatin in cancer. (SNVs) Single-nucleotide variants, (SEs) superenhancers.
In this review, we focus on the mechanisms directly impinging upon chromatin machinery malfunction and gene misregulation as well as their effect on higher-order topological changes that alter the behavior of a cell (Franke et al. 2016; Ochs et al. 2019; Johnstone et al. 2020). We will not address cancer-causing mutations that influence chromatin indirectly (e.g., mutant RAS creating aberrant superenhancers [SEs] in the nucleus via signaling through MEK/ERK hyperphosphorylation) as, arguably, all oncogenic mechanisms must ultimately impinge on chromatin regulation to produce and maintain their malignant state.
THE CHROMATIN MACHINERY
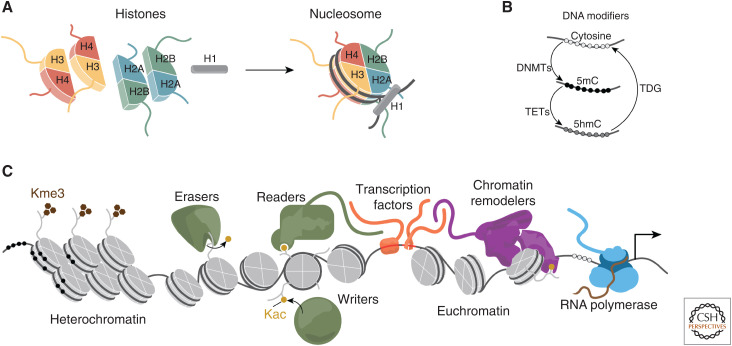
Broadly, any protein that alters the 3D structure of DNA and its function can be considered part of the chromatin machinery. Of central importance to chromatin are the histones that assemble into an octameric bead with a structured core that DNA wraps around twice (147 bp/octamer). This fundamental unit of chromatin is termed the nucleosome. The typical histone octamer contains two each of H2A, H2B, H3, and H4. Serving as a stabilizing linker of the bead is the linker histone, H1, which is not a member of the core octamer (Fig. 2). One function of the nucleosome core particle is to block RNA polymerase from spuriously transcribing (Kornberg and Lorch 2020). Each nucleosome has eight disordered tails that are subject to diverse posttranslational modifications to alter their biochemical properties (Fig. 2A; Strahl and Allis 2000).
Figure 2.
Chromatin machinery including (A) histone assembly around 147 bp of DNA to form the nucleosome, (B) the DNA modification cycle to methylate cytosine nucleotides, and (C) the variety of chromatin machinery including readers, writers, and erasers of posttranslational marks, DNA-binding transcription factors, chromatin remodeling complexes, and RNA polymerases.
Gene expression is also controlled directly by DNA methylation of the nucleotide cytosine to 5-methylcytosine (5mC) or 5-hydroxymethylcytosine (5hmC). Methylated DNA has altered stiffness and an impaired ability to be recognized by transcription factors (TFs), effecting gene regulation significantly (Hörberg and Reymer 2018). DNA methyltransferases convert cytosine to 5mC, TET proteins convert to 5hmC, and TDG proteins convert cytosine back to its unmodified form (Fig. 2B). Hypermethylation of cytosine in CpG islands near gene promoters is a primary gene-silencing mechanism (Naveh-Many and Cedar 1981).
Some histone tail modifications block transcription, for example, by methylation of histone tail lysines in regions of densely packed heterochromatin, while others such as acetylation of histone tail lysines, loosen the histone's grip on DNA to enable nucleosome eviction, DNA accessibility, RNA polymerase movement, and transcription in regions termed euchromatin (Li et al. 2007). Histone modifications are controlled by “writers” and “erasers” (Schreiber and Bernstein 2002). Appropriately modified nucleosomes serve as binding sites for “reader” domains through which many proteins, and some large complexes, can engage the nucleosome (Fig. 2C). Many such complexes such as BAF (also known as mSWI/SNF) possess chromatin remodeling activity through ATPase-driven motors that can slide or evict nucleosomes to enhance access to DNA (Fig. 2C; Wilson and Roberts 2011; Kadoch 2019).
The location of histone modifications in relationship to the underlying DNA sequence is essential to faithful transcription and thus gives importance to sequence-specific DNA-binding proteins (i.e., TFs that direct histone modifiers and chromatin remodelers to specific positions in the epigenome) (Fig. 2C). There are >1500 TFs encoded in the human genome, and roughly 70 of them are ubiquitously expressed and play routine roles (such as CCCTC-binding factor [CTCF]), while the rest are restricted to and define specific cell types (Gerstein et al. 2012). TFs are further distinguished into nonmutually exclusive functional groups based on characteristics such as (1) ability to sense external signaling cascades, (2) ligand-binding capacity, (3) preference for recruitment of coactivator complexes or corepressor complexes, (4) ability to “pioneer” into closed chromatin, (5) downstream gene pathway regulation, (6) preference for promoters or distal regulatory elements, and (7) the structural features they enable (Wingender et al. 2013).
MUTATIONS THAT ALTER CHROMATIN MACHINERY
Remarkably, cancerous aberrations are found for every class of the chromatin machinery, including histones, histone modifiers, chromatin remodelers, TFs, and DNA modifiers. Both small mutations and large-scale genetic alterations have been identified in cancer that perturb these proteins, derailing the healthy checks and balances of the human epigenome and that can be targeted for therapy (Jones et al. 2016).
Small-Scale Mutations Altering Chromatin Protein Function
Mutations in Histones
A growing number of histone mutations have been cataloged in cancer, and are collectively termed “oncohistones” (Nacev et al. 2019). Histone H3 has the longest disordered tail, is subject both to the highest number of writing/reading/erasing modifications, and harbors mutations that prevent proper posttranslational modification in various cancer contexts. For example, 78% of diffuse intrinsic pontine gliomas (DIPGs) have the oncohistone mutation H3K27M, and H3K27 is a key lysine residue that is acetylated in active euchromatin and methylated in heterochromatin (Schwartzentruber et al. 2012). H3.3 (histone 3 variant 3) is mutated to methionine at lysine 36 (H3.3 K36M) in 95% of chondroblastomas and H3.3 is mutated at glycine 34 (H3.3 G34W/L) in 92% of giant cell tumors of bone. The exact mechanisms by which these mutations in the histone tails impact gene regulation are not completely resolved for each mutant, but studies on H3K27M suggest that this amino acid substitution leads to an inability of PRC2 (Polycomb Repressive Complex 2) to maintain the repressive H3K27me3 status, globally impairing transcriptional silencing in DIPG (Lewis et al. 2013).
Whereas the above mutations occur in the disordered histone tail, many newly reported mutations accumulate in the acidic patch of the nucleosome and are found in all four core histone particles (Ghiraldini et al. 2021). Because the acidic patch at the surface of the nucleosome binds the H-tail binding from neighboring nucleosomes, it is thought that mutations in the acidic patch prevents higher-order chromatin compaction. The histone acidic patch, which is negatively charged, is also a key docking site for positively charged amino acid modules in many chromatin-associated proteins such as the mSWI/SNF complex (BAF), and acidic patch mutants may result in the loss of these interactions (He et al. 2020), leading to gene expression dysregulation. In all, the list of histone variants and their functional roles in cancer is growing and includes H2AFZ, H2AFV, H2AFY, H2AFY2, H3F3A, and H3F3B, across a diverse set of human tumors (Ghiraldini et al. 2021).
Mutations in Transcription Factors
The best-established mutations identified in any TF are those in the “guardian of the genome,” the TP53 gene, encoding the p53 protein (Darnell 2002). TP53 is the most commonly mutated gene in cancer, and although these mutations lead to loss of normal tumor suppressor activity, hotspot nonsynonymous mutations frequently result in GOF through aberrant binding across the genome and increased transcription of MLL1, MLL2 (both histone methyltransferases), and MOZ (a lysine acetyltransferase) through binding to ETS2 (ETS family of TFs). Increased MLL and MOZ expression leads to elevated methylation and acetylation of histones, respectively, which results in alteration of chromatin structure and gene expression, and ultimately to increased proliferation of cancer cells (Zhu et al. 2015). Other TFs that play dominant roles in cancer do so without becoming themselves mutant, such as MYC. One exception is the ultra-rare but lethal spindle cell and sclerosing rhabdomyosarcoma (RMS) (Agaram et al. 2019), where MYOD1 is mutated such that the amino acid sequence of its DNA-binding domain becomes identical to MYC (Kohsaka et al. 2014).
Although rare in other cancers, within the context of prostate cancer, mutations in the TF androgen receptor (AR) do not result in LOF of DNA binding, nor are they tumor-initiating (Taplin et al. 1995; Armenia et al. 2018). AR mutants arise predominantly in the ligand-binding domain, where testosterone binds to activate AR's recruitment to chromatin, and substitute amino acids to alter binding to antiandrogens, such as bicalutamide or enzalutamide, thereby evading the therapeutic pressure of androgen deprivation, and sometimes creating tumors that depend on anti-AR ligands for growth even when AR mutants cause an antagonist-to-agonist functional switch (Dai et al. 2017).
Mutations in Chromatin Remodeling Complexes
It is estimated that >20% of human cancers harbor mutations in one of the many subunits of the mammalian SWI/SNF (also known as BAF) complex (Kadoch et al. 2013). Whereas other chromatin remodelers are mutated in cancer (CHD, ISWI, INO80, others), these are numerically dwarfed by the high frequency of BAF mutants (Valencia and Kadoch 2019). Molecular, epigenomic, and biochemical studies have been used to dissect the mechanisms by which the growing catalog of the BAF alterations in cancer are causing gene misregulation (Kadoch and Crabtree 2013; Michel et al. 2018; Valencia et al. 2019; Centore et al. 2020; Mashtalir et al. 2020; Shi et al. 2020). One important emerging theme is that because BAF plays diverse roles in a lineage-specific fashion, mutational studies are finding that certain cancers are associated with mutations of specific subunits in this multimodal complex. For example, mutations in SMARCB1 disable the BAF complex from properly binding to the acidic patch of the core nucleosome particle, resulting in diminished enhancer DNA accessibility (Valencia et al. 2019). These mutations are likely pathogenic in multiple cancers including meningioma, adenocarcinoma, schwannoma, and others (Kadoch and Crabtree 2015; Collord et al. 2018; Pereira et al. 2019; Schaefer and Hornick 2021). Solid tumors are often defined by BAF mutations with 100% of patients harboring the hallmark genetic lesion, such as fusion of BAF subunits SSX with SS18 in synovial sarcoma (also see below) (Kadoch and Crabtree 2013).
Mutations in Genes Effecting DNA Methylation and 3D Genome Architecture
Methylation of CpGs have been shown to eliminate CTCF-binding sites inhibiting its insulator function, which can lead to profound changes in gene expression by modulating enhancer access to the gene promoter through regulation of an enhancer boundary (Bell and Felsenfeld 2000). The ways human cancers are altering gene regulation through DNA methylation mechanisms is likely more complex than currently appreciated, but some clear examples have been elucidated in tumors harboring deficiencies in DNA methylation-related enzymes. For example, genes encoding for isocitrate dehydrogenases 1 and 2, IDH1 and IDH2, are mutated in several cancers (Parsons et al. 2008; Schnittger et al. 2010; Yang et al. 2012). Mutations in these genes results in a loss of activity of the enzymatic function with lower production of α-ketoglutarate (α-KG), combined with a GOF, causing an increase in 2-hydroxyglutarate (2-HG) levels. 2-HG competes with α-KG and competitively suppress the activity of α-KG-dependent dioxygenases, including both lysine histone demethylases (KDMs) and the ten-eleven translocation (TET) family of DNA hydroxylases (Fig. 2B; Yang et al. 2012). This has been shown to result in hypermethylation of CTCF-binding sites, with loss of insulation around the locus of driver oncogenes such as the PDGFRA gene, resulting in high expression and kinase activity in glioma (Flavahan et al. 2016). Similarly, SDH-deficient gastrointestinal stromal tumors (GISTs) also exhibit the hypermethylation phenotype, resulting in a lineage-specific enhancer acting out-of-bounds, in this case driving up oncogenic levels of different kinases, FGF4 at one locus and KIT at another (Flavahan et al. 2019). The emerging theme here is that lineage-specific TFs driving SEs can become tumorigenic when global hypermethylation removes the topological guardrails on chromatin folding. This is an elegant example of an activation of a known glioma oncogene through loss of a topologically associated domain (TAD) boundary function through aberrant methylation as a result of a single-nucleotide somatic variant.
Mutations in Chromatin Readers, Writers, and Erasers
Another sizable class of cancer-causing mutations arises in proteins that read, write, and erase histone modifications. The result is a mis-reading, mis-writing, mis-erasing, and, ultimately, a mis-interpretation of epigenomic information (Chi et al. 2010).
Mutant Epi-Writers
Histone modifications are generally divided into active marks such as H3K27ac, and gene repressive marks such as H3K27me3. Because of this, generally the enzymes that write marks associated with active chromatin are considered activating epi-writers (i.e., CPB/p300, which catalyze H3K27 → H3K27ac) or repressive epi-writers (i.e., Enhancer of Zeste 2 Polycomb Repressive Complex 2 Subunit [EZH2], which catalyzes H3K27 → H3K27me3). The repressive epi-writer EZH2 is part of the Polycomb Repressive Complex 2 (PRC2) and is recurrently mutated in 21% of diffuse large B-cell lymphomas (Morin et al. 2010). Most commonly, the mutation occurs at Y641 that resides in the SET domain, which is the enzymatic pocket responsible for adding methylation to lysine substrates (McCabe et al. 2012). Biochemical studies revealed that lymphomas with this mutation experience a GOF effect on PRC2 and global increases in H3K27me3 levels, resulting in widespread repression of gene expression, blocking B-cell differentiation (Shen and Vakoc 2015). The path to cancer is not one sided for the PRC2 complex, as LOF can also cause cancer; for example, deletion of EZH2 or another PRC2 subunit, SUZ12, is frequent in malignant peripheral nerve sheath tumors (MPNSTs), resulting in complete loss of H3K27me3 and unchecked proliferation (De Raedt et al. 2014). Thus, PRC2 can contribute to cancer as either a gained mutant oncogene, or a lost tumor suppressor, a duality that no doubt reflects different roles of PRC2 in varied cancer cells of origin (Piunti and Shilatifard 2016).
Mutant Epi-Readers
MLLT1 super elongation complex (SEC) subunit (protein ENL) is a chromatin reader of the SEC that recognizes histone acetylation using its YEATS domain. The SEC suppresses transient pausing of the RNA polymerase II, increasing transcription (Lin et al. 2010). Approximately 5% of patients with Wilms tumors have GOF mutations clustered in the Yaf9, ENL, AF9, Taf14, Sas5 (YEATS) reader domain, which results in increased expression of genes that favor a premalignant cell fate (Wan et al. 2020).
Mutant Epi-Erasers
Among the epigenetic erasers, it is a curious fact that, very rarely, are the erasers of histone acetylation (HDACs) mutated, whereas mutations of erasers of lysine methylation (KDM proteins) are found much more commonly (Dawson 2017; Donaldson-Collier et al. 2019; Han et al. 2019; Wang and Shilatifard 2019). KDM mutants are thought to cause methylation imbalances that provide a more promiscuous epigenome with increased access to genetic programs (Flavahan et al. 2017), and our perspective on the negative data suggesting a lack of commonly observed HDAC mutants is that these enzymes are under more acute and purifying negative selection pressure, preventing their accumulation in cancer, because they are essential to global gene transcription (Gryder et al. 2012, 2019c).
Large-Scale Alterations that Alter Chromatin Machinery
Some cancers, for example sarcomas and pediatric cancers, have a low somatic mutational burden of <0.5 per megabase despite their aggressive nature (Pugh et al. 2013; Brohl et al. 2014; Shern et al. 2014). Many of these cancers with simple genomes are driven by translocation events with the production of chimeric or fusion oncogenes (Aplan et al. 2021). Chromosome translocation within the cancer genome can lead to the juxtaposition of two regions that would ordinarily not be collocated leading to oncogenic transformation through profound alterations of the chromatin landscape (Kadoch and Crabtree 2013; Boulay et al. 2017; Gryder et al. 2017, 2019a). This could be through the generation of novel in-frame oncogenic chimeric protein and/or the relocation of an intact gene to cis sites near active promoter/enhancer elements of a partner gene leading to increased or aberrant expression of the intact gene (Battey et al. 1983; Kadoch and Crabtree 2013; Boulay et al. 2017; Gryder et al. 2017, 2019a; Lancho and Herranz 2018). Although fusion events can activate kinase genes such as BCR-ABL fusions in CML or ALL or ALK fusions in solid tumors, which ultimately lead to transcriptional dysregulation through their downstream effects on chromatin content and architecture, it will not be discussed further (Salesse and Verfaillie 2002; Nambiar et al. 2008).
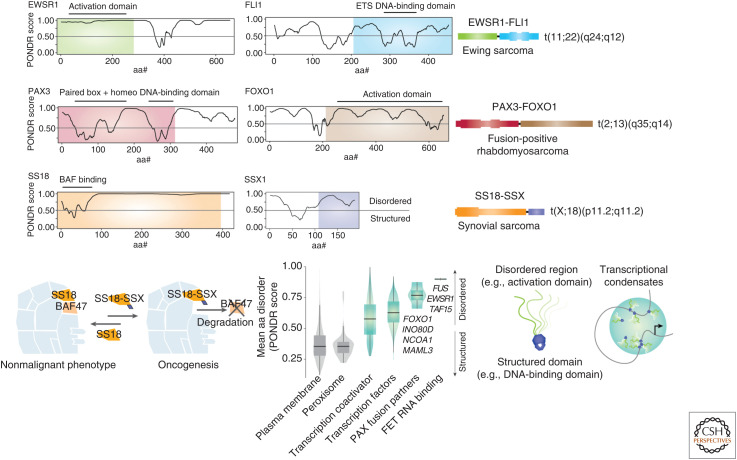
Chimeric TFs often have an ordered DNA-binding domain of a protein that anchors it to specific regions of the genomes, with a partner protein that is intrinsically disordered leading to multimerization and physiological liquid–liquid phase separation. This results in the coalescence of the transcription machinery around the target region, which has been described to lead to subsequent opening of chromatin and alteration in the expression of target genes in disease states (Couthouis et al. 2011,2012; Kwon et al. 2013; Schwartz et al. 2013). Examples of ordered DNA-binding domains of TFs with the disordered domains of proteins include the t(2;13) translocation leading to the generation of the PAX3-FOXO1 in fusion-positive RMS and the t(11;22) translocation leading to the generation of EWSR1-FLI1 in EWS (Fig. 3A,B).
Figure 3.
Fusion oncogenes involving intrinsically disordered chromatin proteins. (A) PONDR score of disorder by amino acid number for EWSR1 and FLI1, with the portions found in Ewing sarcoma fusion oncogene t(11;22)(q24;q12) highlighted in green and blue. (B) PONDR score of disorder by amino acid number for PAX3 and FOXO1, with the portions found in fusion oncogene-positive rhabdomyosarcoma t(2;13)(q35;q14) highlighted in red and brown. (C) PONDR score of disorder by amino acid number for SS18 and SSX1, with the portions fused in t(X;18)(p11.2;q11.2)-positive synovial sarcoma highlighted in orange and purple. (D) Model of BAF dysfunction in synovial sarcoma, where fusion oncogene SS18-SSX disrupts BAF47 incorporation and hinders proper assembly of the complete BAF complex. (Panel D adapted from data in Kadoch and Crabtree 2013.) (E) Violin box plot of mean PONDR scores among plasma membrane proteins, peroxisome proteins, transcription coactivators, transcription factors, PAX fusion partners, and FET RNA-binding proteins. (Panel E adapted from data in Gryder et al. 2020.) (F) Illustration of the cooperation between structured and disordered protein partners commonly found in chromatin-bound oncogenes, and their potential role as contributors to aberrant transcriptional condensates.
In RMS, the t(2;13) fusion results in the expression of the PAX3-FOXO1 fusion gene that acts as a pioneering factor and activates a core regulatory TF circuitry locking the cells in a myoblastic state (Gryder et al. 2017, 2019a). In EWS, gene expression is altered through the recruitment of the BAF complex by the EWS-FLI1 fusion protein to tumor-specific enhancers through phase transitions by the prion-like regions in the intrinsically disordered domain of EWSR1 (Boulay et al. 2017). In synovial sarcoma, which is associated with t(X;18) translocations, the SS18 gene on chromosome 18 joins on one of three synovial sarcoma X (SSX) genes (SSX1, SSX2, or rarely SSX4) on the X chromosome. In the SS18-SSX chimeric protein, almost the entire SS18 protein is retained except the last eight amino acids, which are replaced by the 78 amino acids from the carboxy terminal of SSX, a very disordered region (Fig. 3C). Recent work has shown that the oncogenic mechanism of this fusion is displacement of a tumor-suppressing subunit from the SWI/SNF chromatin-remodeling complex with subsequent altered expression of pluripotency genes (Kadoch and Crabtree 2013) through changes in the BAF complex (Fig. 3D).
In general, TFs appear to select for highly disordered fusion partner proteins (Fig. 3E), such that the combination of a highly structured domain (i.e., DNA-binding domain of a TF) and a disordered domain creates an enhanced capacity for transcriptional condensate formation, thus accelerating transcription in an uncontrolled manner (Fig. 3F). It is hypothesized that the resultant changes of the chromatin composition at the target regions alter the epigenetic landscape of the cell that lock the cells in a blast- or stem-like state (Gryder et al. 2017, 2019a). These fusion TFs cause higher-order topological alteration, such as in PAX3-FOXO1-driven RMS where the fusion oncogene creates disease-specific TADs and H3K27ac-mediated promoter/enhancer loops in compartments ranging from 200 kb to 1.5 Mb in size (Gryder et al. 2019a).
Copy Number Gains and Losses of Chromatin Machinery Genes in Cancer
MYC/MYCN Gene Amplifications
The MYC family of TFs are a basic helix-loop-helix transcription-leucine zipper (bHLH-Zip) factor family whose members dimerize with MAX and bind to specific DNA sequences called E-boxes and has been referred to as the master regulator of the cancer epigenome and transcriptome. It is activated in up to 70% of human cancers by posttranslational modification or overexpressed through gain or amplification of DNA (Dang 2012). MYC recruits histone acetyltransferases (HATs) including lysine acetyltransferase 2A (KAT2A) and two related coactivators CBP/p300 that acetylate lysine residue on histone tails leading to a more open chromatin, promoting recruiting of the transcriptional machinery and increased expression. More broadly, MYC–MAX complexes interact directly or indirectly with chromatin-modifying cofactors including chromatin writers, readers, and erasers (Poole and van Riggelen 2017).
SMARCB1 Deletions
The SWI/SNF subunits have been found to be mutated in 20% of cancers. For example, the SMARCB1 subunit consisting of INI1/SNF5/BAF47 is inactivated in the large majority of rhabdoid tumors (Kim and Roberts 2014). Although SMARCB1, components of the BAF and PBAF complex, may contribute to alterations in canonical cancer pathways, these complexes bind to the promoters and enhancers of about one-third of all genes, and thus it is likely that LOF mutations such as deletions will lead to context-specific changes in pathways specific to that lineage (Nakayama et al. 2017). Loss of SMARCB1 leads to oncogenesis through enhanced SWI/SNF occupancy at SEs while disrupting SWI/SNF binding at typical enhancers that are needed for differentiation (Wang et al. 2017).
RB1 Deletions
The RB Transcriptional Corepressor 1 gene (RB1) is mutated or deleted in ∼7% and the RB pathway disrupted in >30% of all cancers (Knudsen et al. 2020). RB blocks cell-cycle progression by repressing the E2F target gene transcription through the recruitment of transcriptional corepressors and/or chromatin remodeling protein factors, including HDAC and SWI/SNF, at promoter regions (Talluri and Dick 2012). RB also recruits Brm or BRG1 through their LXCXE domains, repressing gene expression and inducing cell-cycle arrest. RB also binds to histone methyltransferase and demethylases, and DNA methyltransferase 1 (DNMT1), which promotes the RB-dependent regulation of gene expression by changing the chromatin structure to a repressive form near the pRB-E2F target promoters (Talluri and Dick 2012). Thus, RB pathway disruption leads to loss of tumor-suppressive activity through epigenetic mechanisms.
ALTERATIONS OF CIS-REGULATORY ELEMENTS IN CANCER
The majority of the human genome is noncoding, and what was once largely considered to be “junk DNA” with little function has become the focus of intense study. The ENCODE project “wrote the eulogy” for the concept of junk DNA in 2018, proposing that as much as 80% of the noncoding space has the potential for biological function (Gerstein et al. 2012). Functions of noncoding DNA include (1) cis regulation of gene expression, driven primarily by transcription factor-binding sequences, (2) noncoding RNA transcripts, and (3) structural, architectural, and mechanical features of certain DNA sequences. In cancer, the two emerging mechanisms that strictly involve noncoding variation are small mutations that cause gain or loss of TF binding and large structural alterations that allow proto-oncogenes to gather a strong boost in transcription by enhancer hijacking.
Small Mutations Altering Transcription Factor Binding
It is striking that the majority of disease-associated mutations identified in genome-wide association studies (GWAS) are in noncoding regions (Edwards et al. 2013). While full functional validation of causality is sparse for the catalog of GWAS hits, there are a few examples where a direct link to cancer has been made. Yet, an early clue resides in the observation that 93% of single-nucleotide polymorphisms (SNPs) are found in a noncoding space, and these SNPs are found most frequently in enhancers, especially in clusters of interconnected enhancers with unusually high occupancy of chromatin machinery known as SEs (Whyte et al. 2013).
A prominent example of this mode of action was discovered in childhood neuroblastoma (NB) in which the expression of GATA3, which is highly expressed and a core regulatory TF (Durbin et al. 2018b), is altered by a SNP rs2168101 (G > T). This alteration disrupts an SE by removing a GATA-binding site, thereby reducing LMO1 protein levels and conferring reduced risk of disease (Oldridge et al. 2015).
Whereas GWAS is designed largely to detect germline mutations that predispose to a disease state, a few examples of somatically acquired mutations have been described that recurrently alter TF binding to drive cancer. The first such example was found in melanoma, first as germline/familial mutation, then as a somatic mutation in sporadic melanoma, where a single SNP in the promoter of the TERT gene creates a binding site for ETS family TFs to increase TERT expression and drive the disease (Horn et al. 2013). A longer-range polymorphism was soon thereafter reported in T-cell acute lymphoblastic leukemias (T-ALLs) where a small insertion, 2–18 bp in length, was somatically acquired in some tumors, seeding a de novo SE 7500 bp distal to the TAL1 gene (Mansour et al. 2014). The recurrent theme among insertions at this site is the addition of a sequence recognizable by the MYB TF, now recognized as a critical oncogenic TF driving this T-ALL via complex formation with TAL1 across that cancer's epigenome (Mansour et al. 2014).
How widespread are small variants that alter the cis regulome? Each cancer epigenome has upward of 20,000 enhancers, many of which are specific to the disease state (Akhtar-Zaidi et al. 2012; Morrow et al. 2018; Zhang et al. 2020), but most of these have not yet been linked to an obvious and recurrent causal SNP or INDEL. Yet, some recent studies indicate these events are in fact widespread, and often effect TFs that are involved in the etiology and lineage/origin of the cancer type. Studies in ovarian cancer have uncovered a diversity of somatic mutations in several distinct enhancers, all of which converge on the PAX8 pathway (Corona et al. 2020). A catalog of INDELS found in H3K27ac data revealed that, in a diverse collection of cancer types, many important oncogenes have enhancers with a somatically altered sequence, and those that have been investigated directly alter oncogene expression (Abraham et al. 2017). These kinds of alterations are invisible to exome or targeted sequencing efforts, including copy number assessment. Thus, in cancers with no obvious protein coding mutation, perhaps especially some childhood cancers, explanatory driver events could be sought by diligent investigation of noncoding variants.
Large Structural Alterations Causing Enhancer Hijacking
Structural alterations can not only result in the production of chimeric proteins, as discussed earlier, but also result in the juxtaposition of whole genes that are normally not expressed to regions near enhancers that are naturally active in the cell of origin of that cancer, a term known as enhancer hijacking (Weischenfeldt et al. 2017). This is particularly noted in B- or T-cell lymphoid malignancies where MYC translocate to IG or TCR regions that are highly active in the respective cell lineage or origin resulting in the hijacking of these enhancers and activation of MYC expression (Ryan et al. 2015). Another example is salivary gland acinic cell carcinoma (AciCC), which has a recurrent t(4;9)(q13;q31) translocation that brings the active enhancer regions from the SCPP gene cluster to the region upstream of Nuclear Receptor Subfamily 4 Group A Member 3 (NR4A3) at 9q31 (PMID: 30664630). Overexpression of NR4A3 target genes increases cell proliferation and is thought to be part of the oncogenic mechanism (Haller et al. 2019). A further example is in medulloblastoma where disparate genomic structural variants lead to activation of the growth factor independent 1 family proto-oncogenes, GFI1 and GFI1B (Northcott et al. 2014). In RMS, the t(2;13) fusion event, which results in the expression of PAX3-FOXO1, is driven by enhancer hijacking of the translocated FOXO1 SE to the PAX3 region resulting in continual expression of the fusion gene (Gryder et al. 2020).
Large Focal Structural Alterations of Enhancer Amplification
Noncoding amplification of regions containing enhancers or SEs have been shown to increase transcription of oncogenes including the control of MYC expression reported in lung adenocarcinoma and endometrial carcinoma (Zhang et al. 2016) and in castrate-resistant prostate cancer an amplification of an AR enhancer located 650 kb centromeric of AR leads to its overexpression (Takeda et al. 2018). Recently, somatic SE duplications and hotspot mutations have been shown to lead to oncogenic activation of the oncogenic KLF5 TF in squamous cell carcinomas (Zhang et al. 2018).
Extrachromosomal Focal Alterations Involving Oncogenes and Their Regulatory Elements
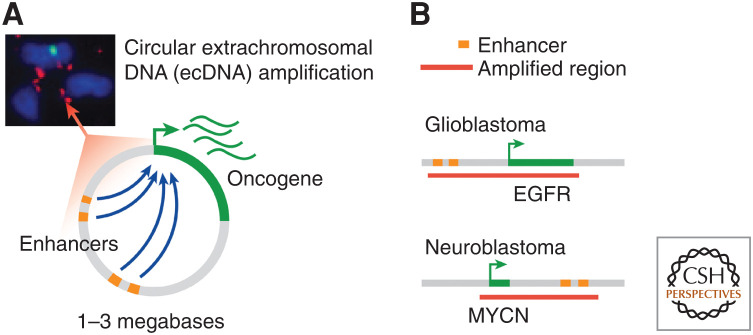
Genomic fragmentation events can circularize via non- or micro-homologous end joining and be propagated as extrachromosomal DNA (ecDNA) at massive copy number (10–200 copies per cell), which are typically 1–3 Mb in size (Verhaak et al. 2019). They autonomously replicate and are subject to non-Mendelian segregation during cell division. Also known as “double minutes,” ecDNA was first observed in cancer cells in the 1960s (Mark 1967) and ecDNA amplifications are now estimated to occur in 14% of all human cancers (Kim et al. 2020). They are particularly prevalent in aggressive cancers notoriously difficult to treat, including glioblastoma, NB, and medulloblastoma (Kim et al. 2020). Numerous active enhancer elements undergo selection on ecDNA and engage the oncogene in a novel 3D conformation, mediating high expression of the oncogene (Fig. 4).
Figure 4.
Extrachromosomal DNA includes (A) circularized megabase scale units that include enhancers with abnormal topology and a key oncogene. (B) Examples include extrachromosomal DNAs (ecDNAs) amplified with enhancers for oncogene EGFR in glioblastoma and MYCN in neuroblastoma.
Functional studies using CRISPRi revealed that nearly every enhancer incorporated on circular ecDNA functionally contributes to proliferative fitness (Morton et al. 2019). Although double minutes have been observed for decades, it has not been appreciated that the regulatory DNA sequences in addition to oncogenes on double minutes are important. These findings, corroborated by multiple laboratories (Morton et al. 2019; Wu et al. 2019; Helmsauer et al. 2020), indicate that circular DNAs in tumor cells are not simply carriers of oncogenes. Rather, the circular DNA is a wholly functional entity with a unique topological configuration, free from highest-order chromosome-level 3D constraints, that drives tumor growth. Additionally, this finding shifts the 40-yr research paradigm that oncogenes on DNA amplifications are the sole drivers of cancer and points to a repertoire of enhancers as additional crucial contributors.
Aneuploidy, Gene Expression, and Nuclear Structure in Cancer
Aneuploidy is defined as a state in which a cell contains abnormal numbers of chromosomes. Germline aneuploidy is a cause of diverse genetic syndromes, most commonly three copies, such as chromosome 21 trisomy in Down syndrome and miscarriages, while somatic aneuploidy is found in nearly 70% of cancers. Interestingly, somatic aneuploidy is unevenly observed across the spectrum of malignancies. At the low end of this aneuploid spectrum are the hematological malignancies, where most genetic alterations occur as chromosomal translocations. For instance, in chronic myeloid leukemia, we can invariably observe a translocation between chromosomes 9 and 22. The consequence of this translocation is the generation of a fusion protein that results in increased tyrosine kinase activity (Koretzky 2007). As discussed above, in Burkitt lymphoma one can frequently detect a translocation between chromosome 8 and 14. Here, the MYC oncogene is brought under the control of the promotor for the immunoglobulin locus, and is therefore transcribed more highly than under physiological conditions (Wiegant et al. 1991; Atallah-Yunes et al. 2020). Notably, both examples deregulate the expression of a single gene, which, in hematological cells, is sufficient for malignant transformation.
The situation in solid tumors, specifically in those of epithelial origin (i.e., carcinomas) is very different. These carcinomas are defined by chromosomal aneuploidies, which result in an imbalanced number of chromosomes (Heim and Mitelman 2009; Ried 2009). Other chromosomal aberrations observed commonly in solid tumors are unbalanced chromosomal translocations or so-called isochromosomes consisting of an aberrant chromosome that contains two identical arms of the parental chromosome, both mechanisms resulting in transcriptional deregulation of hundreds to thousands of genes.
These aneuploidies and the ensuing genomic imbalances are cancer-type specific; for instance, cervical carcinomas invariably carry extra copies of the long arm of chromosomes 1 and 3 (Heselmeyer et al. 1996). Colorectal carcinomas, on the other hand, frequently carry extra copies of chromosomes 7 and 13, the long arm of chromosome 8 and 20, along with losses of chromosome 18 (Ried et al. 1996). These changes occur in premalignant lesions; for instance, cervical intraepithelial dysplastic lesions that are ultimately prone to progress already have extra copies of chromosome 3q (Heselmeyer et al. 1996). In the colorectum, those polyps that show progression to invasive cancer carry extra copies of chromosome 7 (Fiedler et al. 2019). The reason for this tissue specificity is not entirely known; however, recent evidence suggests that the aneuploidies observed in the respective cancers hard-wire gene expression levels that are found in the cognate normal tissues (Patkar et al. 2021).
A fundamental question in cancer biology is how aneuploidies affect the expression levels of resident genes. In other words, do aneuploidies target only a few genes or do they affect gene expression levels of all genes on the aneuploid chromosomes. This question can now be answered with the advent of global gene expression analyses based on array-based technologies or by RNA sequencing (Upender et al. 2004; Wangsa et al. 2018). Results from extensive global sequencing of solid tumors have shown that aneuploidies result in gene expression changes of most resident genes (i.e., function follows form) (Ben-David and Amon 2020). Therefore, aneuploidies result in a massive deregulation of the transcriptional equilibrium. This is in strong contrast to hematological malignancies. These results have been confirmed in experimental models, in which single chromosomes were introduced into karyotypically normal cells (Wangsa et al. 2018). The results were also confirmed in a model system, in which normal colon epithelium was cultured under hypoxic conditions (Braun et al. 2019). These normal colon cells spontaneously transform upon the acquisition of an extra copy of chromosome 7, mimicking the karyotype of colonic polyps. It is therefore conceivable that therapeutically targeting single genes, as it is possible with CML using imatinib (Gleevec), is not sufficient to kill cancer cells that are aneuploid. Rather, the aneuploid state needs to be targeted, which is a formidable task.
Another central question in cancer biology is how aneuploidy affects nuclear structure. This question can now be addressed with the advent of techniques that monitor chromosome conformation on a global level, most notably chromosome conformation capture (i.e., Hi-C) (Ried and Rajapakse 2017). Here, contacts between chromatin are determined by global sequence analysis after fixing interactions with formalin. The vicinity of chromatin regions is then determined by the frequency of sequence contacts (Lieberman-Aiden et al. 2009).
The application of Hi-C to cancer cell lines (i.e., the colorectal cancer cell line HT-29) revealed a strict structure–function relationship (Seaman et al. 2017). In fact, the karyotype of this cell line could be reconstructed from the Hi-C maps (i.e., copy number changes and translocation directly affected nuclear organization). This was also observed in the above-described model system of colorectal cancer, in which the cells acquired an extra copy of chromosome 7 (Braun et al. 2019).
TADs are megabase-long genomic regions along the contiguous genome that interact with each other more frequently than with sequences outside the TAD and are enriched for the insulator-binding protein CTCF (Dixon et al. 2012) and can be defined by chromosome conformation capture assays such as Hi-C. Although TADs are generally stable from cell type to cell type, genomic events such as duplications of noncoding regions can result in the formation of new chromatin domains (neo-TADs) that result in misexpression of genes leading to profound phenotypic effects (Franke et al. 2016). Of interest, it has been reported that evolutionarily conserved chromosome loop anchors are frequently mutated in cancers (Katainen et al. 2015) and those bound by CTCF and cohesin are vulnerable to DNA double-strand breaks (DSBs) mediated by topoisomerase 2B (TOP2B) (Canela et al. 2017). In addition, the frequency of recurrent DSBs at these sites positively correlates with transcriptional output and directionality, and may contribute to the occurrence of genomics rearrangements such as oncogenic fusion events at these transcriptionally active sites (Gothe et al. 2019). Thus, loop anchors are fragile sites where DSBs lead to chromosomal rearrangements, aneuploidy, and alterations of the 3D genome.
CHROMATIN MACHINERY ESSENTIAL TO CANCER IN THEIR NONMUTANT FORM
Large-scale RNAi screens in cancer revealed critical cancer-specific dependencies (Zuber et al. 2011). In more recent years, this has been greatly expanded by CRISPR-Cas9-based genome editing (Wang et al. 2014; Meyers et al. 2017). Because CRISPR mutagenesis can be guided to any exon of a gene, special attention has been given to domain-focused screening of chromatin regulators and TFs, revealing not only which chromatin proteins are essential but which functional domains are contributing to that essentiality (Shi et al. 2015). This domain-focused “biochemistry via genetics” is critical because most chromatin-associated proteins wear many hats, such as EP300 that is endowed with both a bromodomain, which is an acetylation reader domain, and a HAT domain to write the same mark it can read.
The results of these efforts have revealed three predominant themes: First, a handful of broadly expressed chromatin coactivator proteins are pan-cancer lethal such as BRD4 (Shi et al. 2015), CHD4 (Marques et al. 2020), and SMARCA4 (Dempster et al. 2019). Second, the selectively essential factors of each cancer subtype are predominantly the lineage-specific master TFs (Reddy et al. 2019), also called core regulatory TFs, such as PAX5 in chronic lymphocytic leukemia (CLL) (Ott et al. 2018), HAND2 in NB (Durbin et al. 2018a), and MYOD in RMS (Gryder et al. 2019a). Third, select cancers with specific mutations have synthetic lethal dependence on nonmutant chromatin factors, such as noncanonical SWI/SNF complex members in BAF-mutant tumors (Michel et al. 2018).
The first two themes are unified at the chemical–genomic level, as therapeutics that target these broadly required coactivators (BRD4 and CHD4/NuRD) selectively impair the function of core regulatory TFs specific to that cancer. For example, in RMS, MYOD is the top essential TF in the disease, and co-binds with BRD4 at virtually every SE, and BET inhibitors selectively shut down the genes that are SE driven (Gryder et al. 2017, 2019b). A straightforward means to identify the cell-type-specific TFs that are recruiting BRD4 to enact their oncogenic enhancer network can be by motif analysis of BRD4 peaks, which report the TFs that direct its positioning at enhancers and SEs. BRD4, HDACs, CPB/p300, EZH2, and many other chromatin-associated factors are traditionally more amenable to drug discovery than TFs, and represent a key strategy to drugging chromatin for anticancer benefit (Filippakopoulos et al. 2010; Delmore et al. 2011; Roe et al. 2015; Lasko et al. 2017; Chen et al. 2018; Gryder et al. 2019b).
OUTLOOK
Chromatin machinery controls gene expression and its deregulation has a central role during carcinogenesis. The manifestation of epigenetic errors leading to cancer can be thought of as either an inability to differentiate along a predefined lineage path, or a hyperactive, highly proliferative state with addiction to high levels of transcriptional output (Bradner et al. 2017). These are not mutually exclusive, as poorly differentiated (or stem-cell-like) cells in normal human biology often have a proliferative advantage (Burdon et al. 2002). Maintenance of these abnormal states can be attributed to a lack of transcriptional control, or an inability of chromatin to properly respond to external cues, to leading to a loss of cellular homeostasis (Flavahan et al. 2017). Many of the mutations summarized above are selected for in cancer because they lead to these same phenotypic outcomes. Future research should be directed toward an understanding of structure–function relationships in cancer cells. Knowing how cancer genome-specific aberrations affect chromatin and gene expression is essential for a fundamental, systematic understanding of cancer biology, and for developing novel potent precision therapies for cancer.
Footnotes
Editors: Ana Pombo, Martin W. Hetzer, and Tom Misteli
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Abraham BJ, Hnisz D, Weintraub AS, Kwiatkowski N, Li CH, Li Z, Weichert-Leahey N, Rahman S, Liu Y, Etchin J, et al. 2017. Small genomic insertions form enhancers that misregulate oncogenes. Nat Commun 8: 14385. 10.1038/ncomms14385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaram NP, LaQuaglia MP, Alaggio R, Zhang L, Fujisawa Y, Ladanyi M, Wexler LH, Antonescu CR. 2019. MYOD1-mutant spindle cell and sclerosing rhabdomyosarcoma: an aggressive subtype irrespective of age. A reappraisal for molecular classification and risk stratification. Mod Pathol 32: 27–36. 10.1038/s41379-018-0120-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, et al. 2012. Epigenomic enhancer profiling defines a signature of colon cancer. Science 336: 736–739. 10.1126/science.1217277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplan PD, Shern JF, Khan J. 2021. Molecular and genetic basis of childhood cancer. Kluwer, Amsterdam. [Google Scholar]
- Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, Chatila WK, Chakravarty D, Han GC, Coleman I, et al. 2018. The long tail of oncogenic drivers in prostate cancer. Nat Genet 50: 645–651. 10.1038/s41588-018-0078-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atallah-Yunes SA, Murphy DJ, Noy A. 2020. HIV-associated Burkitt lymphoma. Lancet Haematol 7: e594–e600. 10.1016/S2352-3026(20)30126-5 [DOI] [PubMed] [Google Scholar]
- Battey J, Moulding C, Taub R, Murphy W, Stewart T, Potter H, Lenoir G, Leder P. 1983. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell 34: 779–787. 10.1016/0092-8674(83)90534-2 [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485. 10.1038/35013100 [DOI] [PubMed] [Google Scholar]
- Ben-David U, Amon A. 2020. Context is everything: aneuploidy in cancer. Nat Rev Genet 21: 44–62. 10.1038/s41576-019-0171-x [DOI] [PubMed] [Google Scholar]
- Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, Awad ME, Rengarajan S, Volorio A, McBride MJ, et al. 2017. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 171: 163–178.e19. 10.1016/j.cell.2017.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner JE, Hnisz D, Young RA. 2017. Transcriptional addiction in cancer. Cell 168: 629–643. 10.1016/j.cell.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun R, Ronquist S, Wangsa D, Chen H, Anthuber L, Gemoll T, Wangsa D, Koparde V, Hunn C, Habermann JK, et al. 2019. Single chromosome aneuploidy induces genome-wide perturbation of nuclear organization and gene expression. Neoplasia 21: 401–412. 10.1016/j.neo.2019.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, Patidar R, Hurd L, Chen L, Shern JF, et al. 2014. The genomic landscape of the Ewing sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet 10: e1004475. 10.1371/journal.pgen.1004475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T, Smith A, Savatier P. 2002. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol 12: 432–438. 10.1016/S0962-8924(02)02352-8 [DOI] [PubMed] [Google Scholar]
- Canela A, Maman Y, Jung S, Wong N, Callen E, Day A, Kieffer-Kwon KR, Pekowska A, Zhang H, Rao SSP, et al. 2017. Genome organization drives chromosome fragility. Cell 170: 507–521.e18. 10.1016/j.cell.2017.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centore RC, Sandoval GJ, Soares LMM, Kadoch C, Chan HM. 2020. Mammalian SWI/SNF chromatin remodeling complexes: emerging mechanisms and therapeutic strategies. Trends Genet 36: 936–950. 10.1016/j.tig.2020.07.011 [DOI] [PubMed] [Google Scholar]
- Chen L, Alexe G, Dharia NV, Ross L, Iniguez AB, Conway AS, Wang EJ, Veschi V, Lam N, Qi J, et al. 2018. CRISPR-Cas9 screen reveals a MYCN-amplified neuroblastoma dependency on EZH2. J Clin Invest 128: 446–462. 10.1172/JCI90793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. 2010. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10: 457–469. 10.1038/nrc2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collord G, Tarpey P, Kurbatova N, Martincorena I, Moran S, Castro M, Nagy T, Bignell G, Maura F, Young MD, et al. 2018. An integrated genomic analysis of anaplastic meningioma identifies prognostic molecular signatures. Sci Rep 8: 13537. 10.1038/s41598-018-31659-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona RI, Seo JH, Lin X, Hazelett DJ, Reddy J, Fonseca MAS, Abassi F, Lin YG, Mhawech-Fauceglia PY, Shah SP, et al. 2020. Non-coding somatic mutations converge on the PAX8 pathway in ovarian cancer. Nat Commun 11: 2020. 10.1038/s41467-020-15951-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, et al. 2011. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci 108: 20881–20890. 10.1073/pnas.1109434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Mojsilovic-Petrovic J, Panossian S, et al. 2012. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet 21: 2899–2911. 10.1093/hmg/dds116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Heemers H, Sharifi N. 2017. Androgen signaling in prostate cancer. Cold Spring Harb Perspect Med 7: a030452. 10.1101/cshperspect.a030452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. 2012. MYC on the path to cancer. Cell 149: 22–35. 10.1016/j.cell.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE. 2002. Transcription factors as targets for cancer therapy. Nat Rev Cancer 2: 740–749. 10.1038/nrc906 [DOI] [PubMed] [Google Scholar]
- Dawson MA. 2017. The cancer epigenome: concepts, challenges, and therapeutic opportunities. Science 355: 1147–1152. 10.1126/science.aam7304 [DOI] [PubMed] [Google Scholar]
- Delmore JE, Issa Ghayas C, Lemieux Madeleine E, Rahl Peter B, Shi J, Jacobs Hannah M, Kastritis E, Gilpatrick T, Paranal Ronald M, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917. 10.1016/j.cell.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster JM, Pacini C, Pantel S, Behan FM, Green T, Krill-Burger J, Beaver CM, Younger ST, Zhivich V, Najgebauer H, et al. 2019. Agreement between two large pan-cancer CRISPR-Cas9 gene dependency data sets. Nat Commun 10: 5817. 10.1038/s41467-019-13805-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N, Helin K, Hornick JL, Mautner V, Kehrer-Sawatzki H, et al. 2014. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 514: 247–251. 10.1038/nature13561 [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson-Collier MC, Sungalee S, Zufferey M, Tavernari D, Katanayeva N, Battistello E, Mina M, Douglass KM, Rey T, Raynaud F, et al. 2019. EZH2 oncogenic mutations drive epigenetic, transcriptional, and structural changes within chromatin domains. Nat Genet 51: 517–528. 10.1038/s41588-018-0338-y [DOI] [PubMed] [Google Scholar]
- Durbin AD, Zimmerman MW, Dharia NV, Abraham BJ, Iniguez AB, Weichert-Leahey N, He S, Krill-Burger JM, Root DE, Vazquez F, et al. 2018a. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat Genet 50: 1240–1246. 10.1038/s41588-018-0191-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin AD, Zimmerman MW, Dharia NV, Abraham BJ, Iniguez AB, Weichert-Leahey N, He S, Krill-Burger JM, Root DE, Vazquez F, et al. 2018b. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat Genet 50: 1240–1246. 10.1038/s41588-018-0191-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Beesley J, French JD, Dunning AM. 2013. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet 93: 779–797. 10.1016/j.ajhg.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler D, Heselmeyer-Haddad K, Hirsch D, Hernandez LS, Torres I, Wangsa D, Hu Y, Zapata L, Rueschoff J, Belle S, et al. 2019. Single-cell genetic analysis of clonal dynamics in colorectal adenomas indicates CDX2 gain as a predictor of recurrence. Int J Cancer 144: 1561–1573. 10.1002/ijc.31869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073. 10.1038/nature09504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. 2016. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529: 110–114. 10.1038/nature16490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Gaskell E, Bernstein BE. 2017. Epigenetic plasticity and the hallmarks of cancer. Science 357: eaal2380. 10.1126/science.aal2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Johnstone SE, Hemming ML, Tarjan DR, Hegazi E, Shareef SJ, Javed NM, Raut CP, Eschle BK, et al. 2019. Altered chromosomal topology drives oncogenic programs in SDH-deficient GISTs. Nature 575: 229–233. 10.1038/s41586-019-1668-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, Kraft K, Kempfer R, Jerković I, Chan WL, et al. 2016. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538: 265–269. 10.1038/nature19800 [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature 489: 91–100. 10.1038/nature11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiraldini FG, Filipescu D, Bernstein E. 2021. Solid tumours hijack the histone variant network. Nat Rev Cancer 21: 257–275. 10.1038/s41568-020-00330-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothe HJ, Bouwman BAM, Gusmao EG, Piccinno R, Petrosino G, Sayols S, Drechsel O, Minneker V, Josipovic N, Mizi A, et al. 2019. Spatial chromosome folding and active transcription drive DNA fragility and formation of oncogenic MLL translocations. Mol Cell 75: 267–283.e12. 10.1016/j.molcel.2019.05.015 [DOI] [PubMed] [Google Scholar]
- Gryder BE, Sodji QH, Oyelere AK. 2012. Targeted cancer therapy: giving histone deacetylase inhibitors all they need to succeed. Future Med Chem 4: 505–524. 10.4155/fmc.12.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryder B, Nelson C, Shepard S. 2013. Biosemiotic entropy of the genome: mutations and epigenetic imbalances resulting in cancer. Entropy 15: 234–261. 10.3390/e15010234 [DOI] [Google Scholar]
- Gryder BE, Yohe ME, Chou HC, Zhang X, Marques J, Wachtel M, Schaefer B, Sen N, Song Y, Gualtieri A, et al. 2017. PAX3-FOXO1 establishes myogenic super enhancers and confers BET bromodomain vulnerability. Cancer Discov 7: 884–899. 10.1158/2159-8290.CD-16-1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryder BE, Pomella S, Sayers C, Wu XS, Song Y, Chiarella AM, Bagchi S, Chou HC, Sinniah RS, Walton A, et al. 2019a. Histone hyperacetylation disrupts core gene regulatory architecture in rhabdomyosarcoma. Nat Genet 51: 1714–1722. 10.1038/s41588-019-0534-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryder BE, Wu L, Woldemichael GM, Pomella S, Quinn TR, Park PMC, Cleveland A, Stanton BZ, Song Y, Rota R, et al. 2019b. Chemical genomics reveals histone deacetylases are required for core regulatory transcription. Nat Commun 10: 3004. 10.1038/s41467-019-11046-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryder BE, Wu L, Woldemichael GM, Pomella S, Quinn TR, Park PMC, Cleveland A, Stanton BZ, Song Y, Rota R, et al. 2019c. Chemical genomics reveals histone deacetylases are required for core regulatory transcription. Nat Commun 10: 3004. 10.1038/s41467-019-11046-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryder BE, Wachtel M, Chang K, El Demerdash O, Aboreden NG, Mohammed W, Ewert W, Pomella S, Rota R, Wei JS, et al. 2020. Miswired enhancer logic drives a cancer of the muscle lineage. iScience 23: 101103. 10.1016/j.isci.2020.101103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller F, Bieg M, Will R, Körner C, Weichenhan D, Bott A, Ishaque N, Lutsik P, Moskalev EA, Mueller SK, et al. 2019. Enhancer hijacking activates oncogenic transcription factor NR4A3 in acinic cell carcinomas of the salivary glands. Nat Commun 10: 368. 10.1038/s41467-018-08069-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Jia L, Lv W, Wang L, Cui W. 2019. Epigenetic enzyme mutations: role in tumorigenesis and molecular inhibitors. Front Oncol 9: 194. 10.3389/fonc.2019.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Wu Z, Tian Y, Yu Z, Yu J, Wang X, Li J, Liu B, Xu Y. 2020. Structure of nucleosome-bound human BAF complex. Science 367: 875–881. 10.1126/science.aaz9761 [DOI] [PubMed] [Google Scholar]
- Heim S, Mitelman F. 2009. Cancer cytogenetics. Wiley, Hoboken, NJ. [Google Scholar]
- Helmsauer K, Valieva ME, Ali S, Chamorro González R, Schöpflin R, Röefzaad C, Bei Y, Dorado Garcia H, Rodriguez-Fos E, Puiggros M, et al. 2020. Enhancer hijacking determines extrachromosomal circular MYCN amplicon architecture in neuroblastoma. Nat Commun 11: 5823. 10.1038/s41467-020-19452-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heselmeyer K, Schrock E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T. 1996. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci 93: 479–484. 10.1073/pnas.93.1.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörberg J, Reymer A. 2018. A sequence environment modulates the impact of methylation on the torsional rigidity of DNA. Chem Commun (Camb) 54: 11885–11888. 10.1039/C8CC06550K [DOI] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. 2013. TERT promoter mutations in familial and sporadic melanoma. Science 339: 959–961. 10.1126/science.1230062 [DOI] [PubMed] [Google Scholar]
- Johnstone SE, Reyes A, Qi Y, Adriaens C, Hegazi E, Pelka K, Chen JH, Zou LS, Drier Y, Hecht V, et al. 2020. Large-scale topological changes restrain malignant progression in colorectal cancer. Cell 182: 1474–1489.e23. 10.1016/j.cell.2020.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Issa JP, Baylin S. 2016. Targeting the cancer epigenome for therapy. Nat Rev Genet 17: 630–641. 10.1038/nrg.2016.93 [DOI] [PubMed] [Google Scholar]
- Kadoch C. 2019. Diverse compositions and functions of chromatin remodeling machines in cancer. Sci Transl Med 11: eaay1018. 10.1126/scitranslmed.aay1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Crabtree GR. 2013. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 153: 71–85. 10.1016/j.cell.2013.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Crabtree GR. 2015. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci Adv 1: e1500447. 10.1126/sciadv.1500447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. 2013. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 45: 592–601. 10.1038/ng.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katainen R, Dave K, Pitkänen E, Palin K, Kivioja T, Välimäki N, Gylfe AE, Ristolainen H, Hänninen UA, Cajuso T, et al. 2015. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet 47: 818–821. 10.1038/ng.3335 [DOI] [PubMed] [Google Scholar]
- Kim KH, Roberts CW. 2014. Mechanisms by which SMARCB1 loss drives rhabdoid tumor growth. Cancer Genet 207: 365–372. 10.1016/j.cancergen.2014.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Nguyen NP, Turner K, Wu S, Gujar AD, Luebeck J, Liu J, Deshpande V, Rajkumar U, Namburi S, et al. 2020. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat Genet 52: 891–897. 10.1038/s41588-020-0678-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen ES, Nambiar R, Rosario SR, Smiraglia DJ, Goodrich DW, Witkiewicz AK. 2020. Pan-cancer molecular analysis of the RB tumor suppressor pathway. Commun Biol 3: 158. 10.1038/s42003-020-0873-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka S, Shukla N, Ameur N, Ito T, Ng CKY, Wang L, Lim D, Marchetti A, Viale A, Pirun M, et al. 2014. A recurrent neomorphic mutation in MYOD1 defines a clinically aggressive subset of embryonal rhabdomyosarcoma associated with PI3K-AKT pathway mutations. Nat Genet 46: 595–600. 10.1038/ng.2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koretzky GA. 2007. The legacy of the Philadelphia chromosome. J Clin Invest 117: 2030–2032. 10.1172/JCI33032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. 2020. Primary role of the nucleosome. Mol Cell 79: 371–375. 10.1016/j.molcel.2020.07.020 [DOI] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. 2013. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155: 1049–1060. 10.1016/j.cell.2013.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancho O, Herranz D. 2018. The MYC enhancer-ome: long-range transcriptional regulation of MYC in cancer. Trends Cancer 4: 810–822. 10.1016/j.trecan.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, Hansen TM, Risi RM, Frey R, Manaves V, et al. 2017. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550: 128–132. 10.1038/nature24028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. 2013. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857. 10.1126/science.1232245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. 2007. The role of chromatin during transcription. Cell 128: 707–719. 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. 2010. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 37: 429–437. 10.1016/j.molcel.2010.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan SE, Silverman LB, et al. 2014. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346: 1373–1377. 10.1126/science.1259037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark J. 1967. Double-minutes—a chromosomal aberration in Rous sarcomas in mice. Hereditas 57: 1–22. 10.1111/j.1601-5223.1967.tb02091.x [DOI] [PubMed] [Google Scholar]
- Marques JG, Gryder BE, Pavlovic B, Chung Y, Ngo QA, Frommelt F, Gstaiger M, Song Y, Benischke K, Laubscher D, et al. 2020. NuRD subunit CHD4 regulates super-enhancer accessibility in rhabdomyosarcoma and represents a general tumor dependency. eLife 9: e54993. 10.7554/eLife.54993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashtalir N, Suzuki H, Farrell DP, Sankar A, Luo J, Filipovski M, D'Avino AR, St Pierre R, Valencia AM, Onikubo T, et al. 2020. A structural model of the endogenous human BAF complex informs disease mechanisms. Cell 183: 802–817.e24. 10.1016/j.cell.2020.09.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A III, Diaz E, et al. 2012. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492: 108–112. 10.1038/nature11606 [DOI] [PubMed] [Google Scholar]
- Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, et al. 2017. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat Genet 49: 1779–1784. 10.1038/ng.3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel BC, D'Avino AR, Cassel SH, Mashtalir N, McKenzie ZM, McBride MJ, Valencia AM, Zhou Q, Bocker M, Soares LMM, et al. 2018. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat Cell Biol 20: 1410–1420. 10.1038/s41556-018-0221-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. 2010. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42: 181–185. 10.1038/ng.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow JJ, Bayles I, Funnell APW, Miller TE, Saiakhova A, Lizardo MM, Bartels CF, Kapteijn MY, Hung S, Mendoza A, et al. 2018. Positively selected enhancer elements endow osteosarcoma cells with metastatic competence. Nat Med 24: 176–185. 10.1038/nm.4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton AR, Dogan-Artun N, Faber ZJ, MacLeod G, Bartels CF, Piazza MS, Allan KC, Mack SC, Wang X, Gimple RC, et al. 2019. Functional enhancers shape extrachromosomal oncogene amplifications. Cell 179: 1330–1341.e13. 10.1016/j.cell.2019.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacev BA, Feng L, Bagert JD, Lemiesz AE, Gao J, Soshnev AA, Kundra R, Schultz N, Muir TW, Allis CD. 2019. The expanding landscape of “oncohistone” mutations in human cancers. Nature 567: 473–478. 10.1038/s41586-019-1038-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama RT, Pulice JL, Valencia AM, McBride MJ, McKenzie ZM, Gillespie MA, Ku WL, Teng M, Cui K, Williams RT, et al. 2017. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat Genet 49: 1613–1623. 10.1038/ng.3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar M, Kari V, Raghavan SC. 2008. Chromosomal translocations in cancer. Biochim Biophys Acta 1786: 139–152. [DOI] [PubMed] [Google Scholar]
- Naveh-Many T, Cedar H. 1981. Active gene sequences are undermethylated. Proc Natl Acad Sci 78: 4246–4250. 10.1073/pnas.78.7.4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SMB. 2020. Perturbation-driven entropy as a source of cancer cell heterogeneity. Trends Cancer 6: 454–461. 10.1016/j.trecan.2020.02.016 [DOI] [PubMed] [Google Scholar]
- Northcott PA, Lee C, Zichner T, Stütz AM, Erkek S, Kawauchi D, Shih DJ, Hovestadt V, Zapatka M, Sturm D, et al. 2014. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511: 428–434. 10.1038/nature13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs F, Karemore G, Miron E, Brown J, Sedlackova H, Rask MB, Lampe M, Buckle V, Schermelleh L, Lukas J, et al. 2019. Stabilization of chromatin topology safeguards genome integrity. Nature 574: 571–574. 10.1038/s41586-019-1659-4 [DOI] [PubMed] [Google Scholar]
- Oldridge DA, Wood AC, Weichert-Leahey N, Crimmins I, Sussman R, Winter C, McDaniel LD, Diamond M, Hart LS, Zhu S, et al. 2015. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature 528: 418–421. 10.1038/nature15540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott CJ, Federation AJ, Schwartz LS, Kasar S, Klitgaard JL, Lenci R, Li Q, Lawlor M, Fernandes SM, Souza A, et al. 2018. Enhancer architecture and essential core regulatory circuitry of chronic lymphocytic leukemia. Cancer Cell 34: 982–995.e987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patkar S, Heselmeyer-Haddad K, Auslander N, Hirsch D, Camps J, Bronder D, Brown M, Wei-Dong C, Lokanga R, Wangsa D, et al. 2021. Hard-wiring of normal tissue-specific chromosome-wide gene expression levels is a potential factor driving cancer-type-specific aneuploidies. Genome Med 13: 93. 10.1186/s13073-021-00905-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira BJA, Oba-Shinjo SM, de Almeida AN, Marie SKN. 2019. Molecular alterations in meningiomas: literature review. Clin Neurol Neurosurg 176: 89–96. 10.1016/j.clineuro.2018.12.004 [DOI] [PubMed] [Google Scholar]
- Piunti A, Shilatifard A. 2016. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 352: aad9780. 10.1126/science.aad9780 [DOI] [PubMed] [Google Scholar]
- Poole CJ, van Riggelen J. 2017. MYC-master regulator of the cancer epigenome and transcriptome. Genes (Basel) 8: 142. 10.3390/genes8050142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, et al. 2013. The genetic landscape of high-risk neuroblastoma. Nat Genet 45: 279–284. 10.1038/ng.2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy J, Fonseca MAS, Corona RI, Nameki R, Segato Dezem F, Klein IA, Chang H, Chaves-Moreira D, Afeyan L, Malta TM, et al. 2019. Predicting master transcription factors from pan-cancer expression data. bioRxiv 10.1101/839142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried T. 2009. Homage to Theodor Boveri (1862–1915): Boveri's theory of cancer as a disease of the chromosomes, and the landscape of genomic imbalances in human carcinomas. Environ Mol Mutagen 50: 593–601. 10.1002/em.20526 [DOI] [PubMed] [Google Scholar]
- Ried T, Rajapakse I. 2017. The 4D nucleome. Methods 123: 1–2. 10.1016/j.ymeth.2017.06.031 [DOI] [PubMed] [Google Scholar]
- Ried T, Knutzen R, Steinbeck R, Blegen H, Schröck E, Heselmeyer K, du Manoir S, Auer G. 1996. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer 15: 234–245. [DOI] [PubMed] [Google Scholar]
- Roe JS, Mercan F, Rivera K, Pappin Darryl J, Vakoc CR. 2015. BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol Cell 58: 1028–1039. 10.1016/j.molcel.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RJ, Drier Y, Whitton H, Cotton MJ, Kaur J, Issner R, Gillespie S, Epstein CB, Nardi V, Sohani AR, et al. 2015. Detection of enhancer-associated rearrangements reveals mechanisms of oncogene dysregulation in B-cell lymphoma. Cancer Discov 5: 1058–1071. 10.1158/2159-8290.CD-15-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salesse S, Verfaillie CM. 2002. BCR/ABL: from molecular mechanisms of leukemia induction to treatment of chronic myelogenous leukemia. Oncogene 21: 8547–8559. 10.1038/sj.onc.1206082 [DOI] [PubMed] [Google Scholar]
- Schaefer IM, Hornick JL. 2021. SWI/SNF complex-deficient soft tissue neoplasms: an update. Semin Diagn Pathol 38: 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger S, Haferlach C, Ulke M, Alpermann T, Kern W, Haferlach T. 2010. IDH1 mutations are detected in 6.6% of 1414 AML patients and are associated with intermediate risk karyotype and unfavorable prognosis in adults younger than 60 years and unmutated NPM1 status. Blood 116: 5486–5496. 10.1182/blood-2010-02-267955 [DOI] [PubMed] [Google Scholar]
- Schreiber SL, Bernstein BE. 2002. Signaling network model of chromatin. Cell 111: 771–778. 10.1016/S0092-8674(02)01196-0 [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Wang X, Podell ER, Cech TR. 2013. RNA seeds higher-order assembly of FUS protein. Cell Rep 5: 918–925. 10.1016/j.celrep.2013.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DTW, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DAK, Tönjes M, et al. 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231. 10.1038/nature10833 [DOI] [PubMed] [Google Scholar]
- Seaman L, Chen H, Brown M, Wangsa D, Patterson G, Camps J, Omenn GS, Ried T, Rajapakse I. 2017. Nucleome analysis reveals structure–function relationships for colon cancer. Mol Cancer Res 15: 821–830. 10.1158/1541-7786.MCR-16-0374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Vakoc CR. 2015. Gain-of-function mutation of chromatin regulators as a tumorigenic mechanism and an opportunity for therapeutic intervention. Curr Opin Oncol 27: 57–63. 10.1097/CCO.0000000000000151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, Ambrogio L, Auclair D, Wang J, Song YK, et al. 2014. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 4: 216–231. 10.1158/2159-8290.CD-13-0639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. 2015. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 33: 661–667. 10.1038/nbt.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Tao T, Abraham BJ, Durbin AD, Zimmerman MW, Kadoch C, Look AT. 2020. ARID1A loss in neuroblastoma promotes the adrenergic-to-mesenchymal transition by regulating enhancer-mediated gene expression. Sci Adv 6: eaaz3440. 10.1126/sciadv.aaz3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403: 41–45. 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- Takeda DY, Spisák S, Seo JH, Bell C, O'Connor E, Korthauer K, Ribli D, Csabai I, Solymosi N, Szállási Z, et al. 2018. A somatically acquired enhancer of the androgen receptor is a noncoding driver in advanced prostate cancer. Cell 174: 422–432.e13. 10.1016/j.cell.2018.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talluri S, Dick FA. 2012. Regulation of transcription and chromatin structure by pRB: here, there and everywhere. Cell Cycle 11: 3189–3198. 10.4161/cc.21263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taplin ME, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, Keer HN, Balk SP. 1995. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med 332: 1393–1398. 10.1056/NEJM199505253322101 [DOI] [PubMed] [Google Scholar]
- Tomasetti C, Vogelstein B. 2015. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347: 78–81. 10.1126/science.1260825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T. 2004. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res 64: 6941–6949. 10.1158/0008-5472.CAN-04-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia AM, Kadoch C. 2019. Chromatin regulatory mechanisms and therapeutic opportunities in cancer. Nat Cell Biol 21: 152–161. 10.1038/s41556-018-0258-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia AM, Collings CK, Dao HT, St Pierre R, Cheng YC, Huang J, Sun ZY, Seo HS, Mashtalir N, Comstock DE, et al. 2019. Recurrent SMARCB1 mutations reveal a nucleosome acidic patch interaction site that potentiates mSWI/SNF complex chromatin remodeling. Cell 179: 1342–1356.e23. 10.1016/j.cell.2019.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RGW, Bafna V, Mischel PS. 2019. Extrachromosomal oncogene amplification in tumour pathogenesis and evolution. Nat Rev Cancer 19: 283–288. 10.1038/s41568-019-0128-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Chong S, Xuan F, Liang A, Cui X, Gates L, Carroll TS, Li Y, Feng L, Chen G, et al. 2020. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 577: 121–126. 10.1038/s41586-019-1842-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Shilatifard A. 2019. UTX mutations in human cancer. Cancer Cell 35: 168–176. 10.1016/j.ccell.2019.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343: 80–84. 10.1126/science.1246981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lee RS, Alver BH, Haswell JR, Wang S, Mieczkowski J, Drier Y, Gillespie SM, Archer TC, Wu JN, et al. 2017. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet 49: 289–295. 10.1038/ng.3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangsa D, Quintanilla I, Torabi K, Vila-Casadesus M, Ercilla A, Klus G, Yuce Z, Galofre C, Cuatrecasas M, Lozano JJ, et al. 2018. Near-tetraploid cancer cells show chromosome instability triggered by replication stress and exhibit enhanced invasiveness. FASEB J 32: 3502–3517. 10.1096/fj.201700247RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Dubash T, Drainas AP, Mardin BR, Chen Y, Stütz AM, Waszak SM, Bosco G, Halvorsen AR, Raeder B, et al. 2017. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat Genet 49: 65–74. 10.1038/ng.3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegant J, Ried T, Nederlof PM, van der Ploeg M, Tanke HJ, Raap AK. 1991. In situ hybridization with fluoresceinated DNA. Nucleic Acids Res 19: 3237–3241. 10.1093/nar/19.12.3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. 2011. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11: 481–492. 10.1038/nrc3068 [DOI] [PubMed] [Google Scholar]
- Wingender E, Schoeps T, Dönitz J. 2013. TFClass: an expandable hierarchical classification of human transcription factors. Nucleic Acids Res 41: D165–D170. 10.1093/nar/gks1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Turner KM, Nguyen N, Raviram R, Erb M, Santini J, Luebeck J, Rajkumar U, Diao Y, Li B, et al. 2019. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 575: 699–703. 10.1038/s41586-019-1763-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ye D, Guan KL, Xiong Y. 2012. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin Cancer Res 18: 5562–5571. 10.1158/1078-0432.CCR-12-1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD, Meyerson M. 2016. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet 48: 176–182. 10.1038/ng.3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Choi PS, Francis JM, Gao GF, Campbell JD, Ramachandran A, Mitsuishi Y, Ha G, Shih J, Vazquez F, et al. 2018. Somatic superenhancer duplications and hotspot mutations lead to oncogenic activation of the KLF5 transcription factor. Cancer Discov 8: 108–125. 10.1158/2159-8290.CD-17-0532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lee D, Dhiman V, Jiang P, Xu J, McGillivray P, Yang H, Liu J, Meyerson W, Clarke D, et al. 2020. An integrative ENCODE resource for cancer genomics. Nat Commun 11: 3696. 10.1038/s41467-020-14743-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Sammons MA, Donahue G, Dou Z, Vedadi M, Getlik M, Barsyte-Lovejoy D, Al-awar R, Katona BW, Shilatifard A, et al. 2015. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 525: 206–211. 10.1038/nature15251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi JW, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528. 10.1038/nature10334 [DOI] [PMC free article] [PubMed] [Google Scholar]