Abstract
EUS-guided tissue acquisition carries certain risks from unnecessary needle puncture in the low-likelihood lesions. Artificial intelligence (AI) system may enable us to resolve these limitations. We aimed to assess the performance of AI-assisted diagnosis of pancreatic ductal adenocarcinoma (PDAC) by off-line evaluating the EUS images from different modes. The databases PubMed, EMBASE, SCOPUS, ISI, IEEE, and Association for Computing Machinery were systematically searched for relevant studies. The pooled sensitivity, specificity, diagnostic odds ratio (DOR), and summary receiver operating characteristic curve were estimated using R software. Of 369 publications, 8 studies with a total of 870 PDAC patients were included. The pooled sensitivity and specificity of AI-assisted EUS were 0.91 (95% confidence interval [CI], 0.87–0.93) and 0.90 (95% CI, 0.79–0.96), respectively, with DOR of 81.6 (95% CI, 32.2–207.3), for diagnosis of PDAC. The area under the curve was 0.923. AI-assisted B-mode EUS had pooled sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of 0.91, 0.90, 0.94, and 0.84, respectively; while AI-assisted contrast-enhanced EUS and AI-assisted EUS elastography had sensitivity, specificity, PPV, and NPV of 0.95, 0.95, 0.97, and 0.90; and 0.88, 0.83, 0.96 and 0.57, respectively. AI-assisted EUS has a high accuracy rate and may potentially enhance the performance of EUS by aiding the endosonographers to distinguish PDAC from other solid lesions. Validation of these findings in other independent cohorts and improvement of AI function as a real-time diagnosis to guide for tissue acquisition are warranted.
Keywords: artificial intelligence, computer-assisted diagnosis, computer-assisted image analysis, EUS, machine learning, pancreatic cancer
INTRODUCTION
Pancreatic adenocarcinoma (PDAC) is the seventh leading cause of cancer-related deaths worldwide, with an average 5-year survival rate of only 5%–9%.[1,2] Despite advancements in the detection and treatment modalities, most patients are diagnosed at an unresectable stage at the time of diagnosis. To date, a number of diagnostic tools are available, including tri-phasic pancreatic protocol computerized tomography, magnetic resonance imaging (MRI), and EUS-guided fine-needle aspiration (FNA) and/or biopsy (FNB) for cytological and histological diagnosis.[3] Among these, EUS is only the test enabling tissue acquisition, hence its popularity and satisfactory diagnostic yield for PDAC. At present, the joint American Society for Gastrointestinal Endoscopy/American College of Gastroenterology recommends the use of EUS-FNA for the diagnosis of pancreatic lesions, particularly for solid lesions.[4] EUS-guided tissue sampling is very accurate with prior meta-analyses reporting pooled sensitivities and specificities as high as 85%–89% and 96%–99%, respectively.[5,6,7,8] However, EUS-guided tissue sampling is an invasive procedure and does carry a small but real risk of pancreatitis, infection, pancreatic duct leak, malignant seeding, hemorrhage, and even death. EUS is particularly useful for the detection of small pancreatic lesions not identified on other modalities, particularly, tumor <2 cm in size, with higher sensitivity.[9,10] Since EUS is an operator-dependent procedure and the diagnostic yield may drop in the beginners or less-experienced operators.[11,12] They may miss to detect a small lesion, some types of lesions, e.g. diffusely infiltrating PDAC, and fail to make the correct diagnosis in the less typical PDAC EUS-images.[13,14] Moreover, interobserver variability creates subjective operator dependency in the diagnostic yield of EUS.[10]
Artificial intelligence (AI) has increasingly been applied to various areas of medicine, including gastroenterology.[15] The use of AI or computer-aided diagnosis system may therefore enable us to resolve aforementioned limitations. The role of AI in assisting EUS for PDAC diagnosis has gained a lot of attention recently. However, most studies had a limited number of sample size. We therefore proposed to perform a systematic review and meta-analysis to have a more precise estimate of the performance of AI-assisted EUS for PDAC diagnosis.
In this study, we focused on a specific area of AI called machine learning (ML). To train ML systems, pairs of input and answer are fed into the ML systems, then the systems learn underlying relationship between set of inputs and answers by themselves.[16] The trained ML system can then be used to predict answers from unseen inputs. Our main objective is to evaluate the effectiveness of AI for the diagnosis of PDAC in offline EUS images. We focused on the performance of the ML in the characterization of pancreatic lesions, i.e. malignant versus benign pancreatic diseases.
MATERIALS AND METHODS
The study was conducted in accordance with the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) guidelines.[17] The protocol was registered with PROSPERO (CRD42021232014).
Data sources and searches
Literature search was conducted in Ovid MEDLINE, EMBASE, SCOPUS, International Scientific Indexing databases. The Computer Sciences and Engineering databases including Institute of Electrical and Electronics Engineers and Association for Computing Machinery were also used for the search. The search was conducted from the inception of databases through April 05, 2020. We searched for studies on AI in PDAC and EUS. The search was limited to human studies and studies published in the English language. The following keywords were used for the search: (artificial intelligence OR machine learning OR neural network OR deep learning OR support vector machine OR SVM OR digital image processing OR digital image analysis OR parameter analysis) AND (pancreas OR pancreatic) AND (malignancy OR malignant OR tumor OR mass OR neoplasm OR cancer OR adenocarcinoma) AND (endoscopic ultrasound OR endoscopic ultrasonography OR EUS). We also used PubMed Automatic Term Mapping, which automatically mapped the search keywords to MeSH terms. The full search strategies for each database are described in Supplemental Table 1.
Supplemental Table 1.
Full search strategies for each database
| Database | Search fields | Search query | Search hits |
|---|---|---|---|
| PubMed | PubMed automatic term mapping* searched by MeSH terms and all fields | ((artificial intelligence) OR (machine learning) OR (neural network) OR (deep learning) OR (support vector machine) OR (“SVM”) OR (digital image processing) OR (digital image analysis) OR (parameter analysis)) AND ((pancreas) OR (pancreatic)) AND ((malignancy) OR (malignant) OR (tumor) OR (mass) OR (neoplasm) OR (cancer) OR (adenocarcinoma)) AND ((EUS) OR (endoscopic ultrasonography) OR (“EUS”)) | 42 |
| SCOPUS | Title, abstract, and keyword fields | TITLE-ABS-KEY (((artificial intelligence) OR (machine learning) OR (neural network) OR (deep learning) OR (support vector machine) OR (“SVM”) OR (digital image processing) OR (digital image analysis) OR (parameter analysis)) AND ((pancreas) OR (pancreatic)) AND ((malignancy) OR (malignant) OR (tumor) OR (mass) OR (neoplasm) OR (cancer) OR (adenocarcinoma)) AND ((EUS) OR (endoscopic ultrasonography) OR (“EUS”))) | 138 |
| ISI | Topic and title fields | TS = ((artificial intelligence) OR (machine learning) OR (neural network) OR (deep learning) OR (support vector machine) OR (“SVM”) OR (digital image processing) OR (digital image analysis) OR (parameter analysis)) AND ((pancreas) OR (pancreatic)) AND ((malignancy) OR (malignant) OR (tumor) OR (mass) OR (neoplasm) OR (cancer) OR (adenocarcinoma)) AND ((EUS) OR (endoscopic ultrasonography) OR (“EUS”)) OR TI = ((artificial intelligence) OR (machine learning) OR (neural network) OR (deep learning) OR (support vector machine) OR (“SVM”) OR (digital image processing) OR (digital image analysis) OR (parameter analysis)) AND ((pancreas) OR (pancreatic)) AND ((malignancy) OR (malignant) OR (tumor) OR (mass) OR (neoplasm) OR (cancer) OR (adenocarcinoma)) AND ((EUS) OR (endoscopic ultrasonography) OR (“EUS”)) | 103 |
| EMBASE | “Multi-purpose” (mp) field, which encompasses abstract, candidate term word, device manufacturer, device trade name, drug manufacturer, drug trade name, floating subheading word, heading word, keyword, original title and title | #1 artificial intelligence.mp. or exp artificial intelligence/ #2 machine learning.mp. or exp machine learning/ #3 exp artificial neural network/ #4 deep learning.mp. #5 support vector machine.mp. or exp support vector machine/ #6 digital image processing.mp. #7 digital image analysis.mp. #8 parameter analysis.mp. #9 pancreas.mp. or exp pancreas/ #10 pancreatic.mp. #11 malignancy.mp. #12 exp solid malignant neoplasm/or exp malignant neoplasm/or malignant.mp. #13 tumor.mp. or exp neoplasm/ #14 mass.mp. or exp mass/ #15 neoplasm.mp. #16 cancer.mp. #17 exp pancreas adenocarcinoma/or adenocarcinoma.mp. or exp adenocarcinoma/ #18 exp pancreas tumor/or pancreatic mass.mp. or pancreatic adenocarcinoma.mp. or exp pancreas adenocarcinoma/or pancreatic cancer.mp. or exp pancreas cancer/ #19 EUS.mp. or exp endoscopic ultrasonography/ #20 EUS.mp. Then, the following Boolean operators were used to pool the search results: (#1 OR #2 OR #3 OR #4 OR #5 OR #6 OR #7 OR #8) AND ((#9 OR #10) AND (#11 OR #12 OR #13 OR #14 OR #15 OR #16 OR #17)) OR 18) AND (#19 OR #20) |
64 |
| IEEE | All metadata | ((artificial intelligence) OR (machine learning) OR (neural network) OR (deep learning) OR (support vector machine) OR (“SVM”) OR (digital image processing) OR (digital image analysis) OR (parameter analysis)) AND ((pancreas) OR (pancreatic)) AND ((malignancy) OR (malignant) OR (tumor) OR (mass) OR (neoplasm) OR (cancer) OR (adenocarcinoma)) AND ((EUS) OR (endoscopic ultrasonography) OR (“EUS”)) | 2 |
| ACM | All fields (limit search to the ACM full-text collection) | [[All: artificial intelligence] OR [All: machine learning] OR [All: neural network] OR [All: deep learning] OR [All: support vector machine] OR [All: “svm”] OR [All: digital image processing] OR [All: digital image analysis] OR [All: parameter analysis]] AND [[All: pancreas] OR [All: pancreatic]] AND [[All: malignancy] OR [All: malignant] OR [All: tumor] OR [All: mass] OR [All: neoplasm] OR [All: cancer] OR [All: adenocarcinoma]] AND [[All: EUS] OR [All: endoscopic ultrasonography] OR [All: “eus”]] | 20 |
*PubMed automatic term mapping yielded the following search queries
Artificial intelligence: "Artificial intelligence" [MeSH Terms] OR ("artificial" [All Fields] AND "intelligence"[All Fields]) OR "artificial intelligence"[All Fields]
Machine learning: "Machine learning" [MeSH Terms] OR ("machine" [All Fields] AND "learning" [All Fields]) OR "machine learning" [All Fields]
Neural network: "Neural networks, computer" [MeSH Terms] OR ("neural" [All Fields] AND "networks" [All Fields] AND "computer" [All Fields]) OR "computer
neural networks" [All Fields] OR ("neural" [All Fields] AND "network" [All Fields]) OR "neural network" [All Fields]
Deep learning: "Deep learning" [MeSH Terms] OR ("deep" [All Fields] AND "learning" [All Fields]) OR "deep learning"[All Fields]
Support vector machine: "Support vector machine" [MeSH Terms] OR ("support" [All Fields] AND "vector" [All Fields] AND "machine" [All Fields]) OR "support vector machine" [All Fields]
Digital image processing: "Image processing, computer-assisted" [MeSH Terms] OR ("image" [All Fields] AND "processing" [All Fields] AND "computer-assisted" [All Fields]) OR "computer-assisted image processing" [All Fields] OR ("digital" [All Fields] AND "image" [All Fields] AND "processing" [All Fields]) OR "digital image processing" [All Fields]
Digital: "Digital" [All Fields] OR "digitalization" [All Fields] OR "digitalized" [All Fields] OR "digitalization" [All Fields] OR "digitalize" [All Fields] OR "digitalized" [All Fields] OR "digitalizer" [All Fields] OR "digitalizing"[All Fields] OR "digitally" [All Fields] OR "digitals" [All Fields] OR "digitization" [All Fields] OR "digitizations" [All Fields] OR "digitize" [All Fields] OR "digitized" [All Fields] OR "digitizer" [All Fields] OR "digitizers" [All Fields] OR "digitizes" [All Fields] OR "digitizing" [All Fields]
Image: "Image" [All Fields] OR "image's" [All Fields] OR "imaged" [All Fields] OR "imager" [All Fields] OR "imager's: [All Fields] OR "imagers" [All Fields] OR "images" [All Fields] OR "imaging" [All Fields] OR "imaging's" [All Fields] OR "imagings" [All Fields]
Analysis: "Analysis" [Subheading] OR "analysis" [All Fields]
Parameter: "Parameter" [All Fields] OR "parameter's" [All Fields] OR "parameters" [All Fields]
Analysis: "analysis" [Subheading] OR "analysis" [All Fields]
Pancreas: "Pancrea" [All Fields] OR "pancreas" [MeSH Terms] OR "pancreas" [All Fields]
Pancreatic: "Pancreas" [MeSH Terms] OR "pancreas" [All Fields] OR "pancreatic" [All Fields] OR "pancreatitides" [All Fields] OR "pancreatitis"[MeSH Terms] OR "pancreatitis" [All Fields]
Malignancy: "Malign" [All Fields] OR "malignance" [All Fields] OR "malignances" [All Fields] OR "malignant" [All Fields] OR "malignants" [All Fields] OR "malignities" [All Fields] OR "malignity" [All Fields] OR "malignization" [All Fields] OR "malignized" [All Fields] OR "maligns" [All Fields] OR "neoplasms" [MeSH Terms] OR "neoplasms" [All Fields] OR "malignancies" [All Fields] OR "malignancy" [All Fields]
Malignant: "Malign" [All Fields] OR "malignance" [All Fields] OR "malignances" [All Fields] OR "malignant" [All Fields] OR "malignants" [All Fields] OR "malignities" [All Fields] OR "malignity" [All Fields] OR "malignization" [All Fields] OR "malignized" [All Fields] OR "maligns" [All Fields] OR "neoplasms" [MeSH Terms] OR "neoplasms" [All Fields] OR "malignancies" [All Fields] OR "malignancy" [All Fields]
Tumor: "Cysts" [MeSH Terms] OR "cysts" [All Fields] OR "cyst" [All Fields] OR "neurofibroma" [MeSH Terms] OR "neurofibroma" [All Fields] OR "neurofibromas" [All Fields] OR "tumor's"[All Fields] OR "tumoral" [All Fields] OR "tumorous" [All Fields] OR "tumour" [All Fields] OR "neoplasms" [MeSH Terms] OR "neoplasms" [All Fields] OR "tumor" [All Fields] OR "tumour's"[All Fields] OR "tumoural" [All Fields] OR "tumourous" [All Fields] OR "tumours" [All Fields] OR "tumors" [All Fields]
Mass: "Molecular weight" [MeSH Terms] OR ("molecular" [All Fields] AND "weight" [All Fields]) OR "molecular weight" [All Fields] OR "mass" [All Fields]
Neoplasm: "Neoplasm's" [All Fields] OR "neoplasms" [MeSH Terms] OR "neoplasms" [All Fields] OR "neoplasm" [All Fields]
Cancer: "Cancer's"[All Fields] OR "cancerated" [All Fields] OR "canceration" [All Fields] OR "cancerization" [All Fields] OR "cancerized" [All Fields] OR "cancerous" [All Fields] OR "neoplasms" [MeSH Terms] OR "neoplasms" [All Fields] OR "cancer" [All Fields] OR "cancers" [All Fields]
Adenocarcinoma: "Adenocarcinoma" [MeSH Terms] OR "adenocarcinoma" [All Fields] OR "adenocarcinomas" [All Fields] OR "adenocarcinoma's" [All Fields]
EUS: "Endosonography" [MeSH Terms] OR "endosonography" [All Fields] OR ("endoscopic" [All Fields] AND "ultrasound" [All Fields]) OR "EUS" [All Fields]
Endoscopic ultrasonography: "Endosonography" [MeSH Terms] OR "endosonography" [All Fields] OR ("endoscopic" [All Fields] AND "ultrasonography" [All Fields]) OR "endoscopic ultrasonography" [All Fields].
IEEE: Institute of electrical and electronics engineers, ACM: Association for computing machinery, SVM: Support vector machine
A Population, Intervention, Comparison, Outcome framework was used to formulate a review question and identify relevant studies [Table 1]. We included studies focusing on the utilization of AI in any aspects of diagnostic yield and lesion classification. The studies that did not reported our desired outcomes, i.e. sensitivity and specificity, were not included. Review articles, editorial review and commentaries, letters to the Editors, and abstracts published in conference proceedings and scientific meetings were not included. Full papers published in the conference books were included if the met the selection criteria.
Table 1.
Population, intervention, comparison, outcomes framework for study selection
| PICO | Description of detail |
|---|---|
| Population | Pancreatic ductal adenocarcinoma, patients, males and females, datasets, worldwide |
| Intervention | Use of computer-assisted diagnosis, AI, machine learning |
| Comparison | Benign pancreatic diseases |
| Outcome | Lesion classification |
AI: Artificial intelligence
Study selection and data extraction
Two authors (TT and TP) independently screened titles and abstracts of all studies identified by the search and reached for the full text of potential relevant studies. Disagreements were identified and discussed with the third author (RC). From full-text review, the following criteria were used to select the studies: (i) cohort or case–control study; (ii) PDAC diagnosed by histopathology; (iii) ML model; (iv) diagnostic performance as outcome of interest; and (v) study that provided adequate information to calculate true positive (TP), false positive (FP), true negative (TN), and false negative (FN). Reviewers were not blinded to authors’ names and affiliations. Regarding publications of overlapping cohort from the same investigator group with similar research outcome and ML model, only the publication with a larger cohort were included.[18,19]
Data were independently extracted from the full-text articles by two authors (TT and TP). Disagreements were discussed with the third author (RC). The following information were extracted: (1) first author's name; (2) year of publication; (3) country where a study was conducted; (4) EUS mode; (5) diagnoses of cases and controls; (6) number of patients and ultrasound images and (6) type of AI model; (7) number of patients in training and test cohort; (8) validation method (e.g. k-fold cross validation, independent test set, and external clinical validation cohort); (9) sensitivity and specificity; and (10) crude number of TP, FP, TN, and FN.
Quality assessment
Quality assessment of diagnostic accuracy studies (QUADAS-2) was used to evaluate methodological quality, risk of bias, and applicability concerns of the studies.[20] Four domains, including patient selection, index test, reference standard, and flow and timing, with 12 questions were used to assess quality of the included studies. In the index test domain, we modified the questions to reasonably assess AI systems [Supplemental Methods]. Since all included studies are in a pre-clinical phase, i.e. the results of the AI systems’ predictions were not used to make decisions in a real clinical setting, we did not consider whether the index tests or reference standards were conducted with knowledge of the results of each other. Furthermore, a question regarding pre-specified threshold was not directly considered because these thresholds are often used with tests of which outputs are numerical scales (e.g. laboratory values). The purpose of pre-specification of thresholds is to ensure that the threshold has been set before initiation of the study and is not post hoc adjusted by researchers to get optimum results. In the context of AI systems, this idea is similar to asking whether the performance of the developed AI systems is validated in another independent cohort. We therefore answer this question by instead assessing whether there is any validation cohort, e.g. external clinical validation cohorts, internal independent cohort – “test set,” cross validation.
Data analysis
All statistical analyses were performed using R statistical software, version 3.6.3, Vienna, Austria.[21] Pooled sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and diagnostic odds ratio (DOR) along with 95% confidence interval (95% CI) of AI-assisted EUS for PDAC diagnosis were estimated from the crude number of TP, FP, TN, and FN of each study using a random-effects model. The summary receiver operator characteristic (SROC) curve was generated; the area under the curve (AUC) was calculated to determine the diagnostic accuracy of the AI models. The AUC values of 0.5–0.7, 0.7–0.9, and 0.9–1 indicate low, moderate, and high accuracy, respectively.[22] Heterogeneity among studies was assessed using I2 and Cochran's Q and P value. An I2 value of >50% suggests substantial heterogeneity. Publication bias was assessed using the Deeks’ plot and P value.
RESULTS
Literature search
Figure 1 summarizes the search results and process of article selection. There were 369 potentially relevant articles identified from the 6 databases. Subsequently, 141 duplicates were removed, and 183 articles were excluded as they were clearly irrelevant articles (n = 63); not studies on EUS (n = 56); reviews, comment, editorial, abstract, or meta-analysis (n = 45); not studies on PDAC (n = 10); and not studies on AI (n = 9). A total of 45 articles remained for reading abstract and the entire text. Of these, 37 were excluded due to not met the inclusion criteria (n = 34), duplicate patient cohort (n = 2), and non-English language (n = 1). Finally, the remaining 8 articles met the selection criteria and were included for analysis.[19,23,24,25,26,27,28,29]
Figure 1.
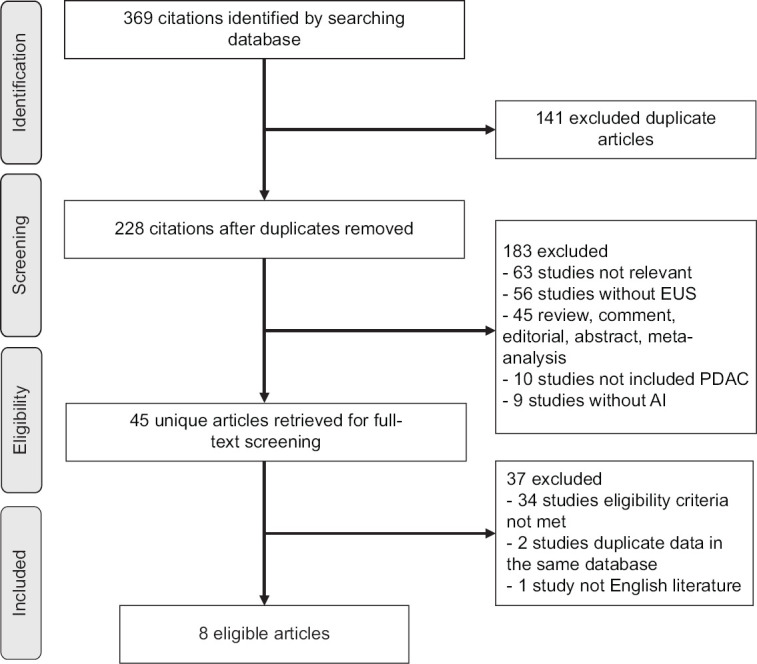
Flow chart
Study characteristics
Eight studies were included in our meta-analysis, with a total of 870 PDAC patients [Table 2].[19,23,24,25,26,27,28,29] Three studies were conducted in the US, whereas two studies were from Europe, two were from Asia, and the other one was from Turkey. Six studies were conducted in a single center while two were multicenter studies. Six publications determined the performance of AI model on B-mode EUS images; one publication applied an AI model on contrast-enhanced EUS images, and the last one used an AI model on EUS elastography. In terms of AI classifier, neural network (NN) was used as an AI model in 4 studies and non-NN was used in the other 4 studies, i.e. support vector machine (SVM) in 2 studies and linear discriminant analysis (LDA) in 2 studies. Regarding validation methods, 3 used cross-validation method, 3 used training and testing set, and the remaining 1 used leave-one-out method. One study did not have a validation method.
Table 2.
Characteristics of included studies
| Reference/year | Country | Study design | AI classifier | EUS mode | Development cohort | Validation cohort | Validation Methods | Gold standard diagnosis | Sensitivity (%) | Specificity (%) | TP | FP | FN | TN | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||||||||
| Case | Control | Case | Control | Malignancy | Benign | ||||||||||||
| Nortonet al., 2001[23] | USA | Retrospective case-control | ANN | B-mode | 21 PC | 14 FC | None | None | None | Pathology | Pathology | 100 | 50 | 21 | 0 | 7 | 7 |
| Zhanget al., 2010[24] | China | Retrospective case-control | SVM | B-mode | 76 PC | 32 noncancer (CP, NP) | 77 PC | 31 noncancer (CP, NP) | Independent test set | Cytology/pathology/clinical 12 months | Cytology/pathology/clinical 12 months | 94.3 | 99.4 | 73 | 4 | 0 | 31 |
| Kumonet al., 2010[25] | USA | Prospective cohort | LDA | B-mode | 13 PC | 7 CP | Cross validation* | Pathology | Clinical | 84.6 | 71.4 | 11 | 2 | 2 | 5 | ||
| Kumonet al., 2012[26] | USA | Prospective cohort | LDA | B-mode | 15 PC | 15 CP | Cross validation* | Cytology/pathology | Diagnostic criteria | 80 | 87 | 12 | 3 | 2 | 13 | ||
| Saftoiuet al., 2012[27] | Europe$ | Prospective cohort | ANN | Elastography | 211 PC (645 videos)** | 47 CP (129 videos)** | Cross validation* | Cytology/pathology/clinical ≥6 months | Cytology/pathology/clinical ≥6 months | 87.5 | 82.9 | 565 | 80 | 22 | 107 | ||
| Zhuet al., 2013[28] | China | Retrospective case-control | SVM | B-mode | 262 PC | 126 CP | Cross validation* (leave-one-out) | Cytology | Diagnostic criteria | 91.6 | 95.1 | 240 | 22 | 6 | 120 | ||
| Saftoiuet al., 2015[19] | Europe$ | Prospective cohort | ANN | Contrast- enhanced | 112 PC | 55 CP | 70% training, 15% validation, 15% testing | Independent test set | Cytology/pathology | Diagnostic criteria | 94.6 | 94.4 | 106 | 6 | 3 | 52 | |
| Ozkanet al., 2016[29] | Turkey | Prospective cohort | ANN | B-mode | 160 PC | 100 NP | 42 PC | 30 NP | Independent test set | NA | NA | 83.3 | 93.3 | 35 | 7 | 2 | 28 |
$ European EUS Elastography Multicentric Study Group (Romania, Denmark, Germany, Spain, Italy, France, Norway, UK); *In cross- validation method, the performance results are averaged over entire development dataset; **The unit of analysis was number of video. AI: Artificial intelligence; PC: Pancreatic cancer; FC: Focal pancreatitis; CP: Chronic pancreatitis; NP: Normal pancreas; SVM: Support vector machine; ANN: Artificial neural network; MLR: Multilinear regression; TP: True positive; FP: False positive; FN: False negative; TN: True negative; LDA: Linear discriminant analysis; NA: Not available
Qualities of the eligible studies were assessed using the QUADAS-2 criteria. The percentage of low risk of bias in the sections of patient selection, index test, reference standard, and flow and timing varied from 62.5%–100%, 75%–100%, 100%, and 100%, respectively [Table 3].
Table 3.
Quality assessment of included studies using quality assessment of diagnostic accuracy studies
| Reference/year | Risk of bias | Applicability concerns | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Patient selection[1] | Index test[2] | Reference standard[3] | Flow and timing[4] | Patient selection[5] | Index test[6] | Reference standard[7] | |
| Nortonet al., 2001[23] | HR | HR | LR | LR | LR | LR | LR |
| Zhanget al., 2010[24] | LR | LR | LR | LR | LR | LR | LR |
| Kumonet al., 2010[25] | LR | LR | LR | LR | LR | LR | LR |
| Kumonet al., 2012[26] | UR | LR | LR | LR | LR | LR | LR |
| Saftoiuet al., 2012[27] | LR | LR | LR | LR | LR | LR | LR |
| Zhuet al., 2013[28] | LR | LR | LR | LR | LR | LR | LR |
| Saftoiuet al., 2015[19] | LR | UR | LR | LR | LR | LR | LR |
| Ozkanet al., 2016[29] | UR | LR | UR | UR | LR | LR | LR |
Low risk, Unclear risk, High risk
Meta-analysis
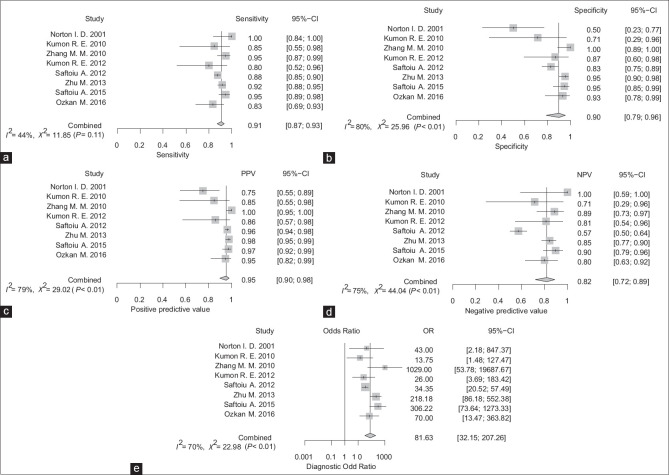
AI-assisted EUS for the diagnosis of PDAC had a pooled sensitivity and specificity of 0.91 (95% CI, 0.87–0.93) and 0.90 (95% CI, 0.79–0.96), respectively [Figures 2a and b]. PPV and NPV were 0.95 (95% CI, 0.90–0.98) and 0.82 (95% CI, 0.72–0.89), respectively, with DOR of 81.6 (95% CI, 32.2–207.3) [Figure 2c-e]. The pooled PLR and NLR were 6.635 (95% CI, 3.453–12.751) and 0.122 (0.085–0.175), respectively. Figure 3 shows the SROC curves of AI-assisted EUS, with AUC of 0.923, indicating the high accuracy of AI-assisted EUS.
Figure 2.
Sensitivity (a), specificity (b), positive predictive value (c), negative predictive value (d), and diagnostic odds ratio (e) of artificial intelligence-assisted EUS for diagnosis of pancreatic cancer
Figure 3.
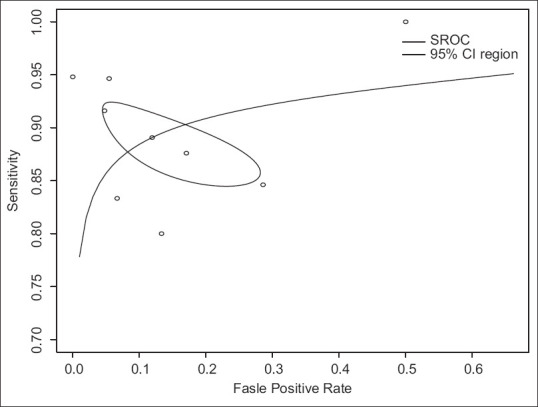
Summary receiver operator characteristics curves demonstrating performance of artificial intelligence-assisted EUS
A subgroup analysis of the 6 studies on the B-mode EUS alone, without image-enhanced technique, the AI system had a pooled sensitivity, specificity, PPV, and NPV of 0.91 (95% CI, 0.88–0.94), 0.90 (95% CI, 0.73–0.97), 0.94 (95% CI, 0.84–0.98), and 0.84 (95% CI, 0.79–0.88), respectively, for differentiating between PDAC and other non-cancerous lesions [Supplemental Figure 1a (829.9KB, tif) -e (829.9KB, tif) ]. The I2 was 0% for the pooled sensitivity and NPV but 79% and 78% for the pooled specificity and PPV, respectively, suggestive of substantial heterogeneity of the pooled specificity and PPV. When subgrouped by type of AI classifiers, the studies using LDA, SVM, and artificial NN (ANN) achieved pooled sensitivities of 0.82 (95% CI, 0.64–0.92), 0.92 (95% CI, 0.89–0.95), and 0.94 (95% CI, 0.49–1.00); pooled specificities of 0.82 (95% CI, 0.60–0.93), 0.96 (95% CI, 0.92–0.98), and 0.79 (95% CI, 0.36–0.96); PPVs of 0.85 (95% CI, 0.67–0.94), 0.98 (95% CI, 0.96–0.99), and 0.87 (95% CI, 0.66–0.96); and NPVs of 0.78 (95% CI, 0.57–0.91), 0.85 (95% CI, 0.79–0.90), and 0.83 (95% CI, 0.69–0.92), respectively [Supplemental Figure 2a (1.2MB, tif) -e (1.2MB, tif) ]. The within-subgroup heterogeneity of the latter analysis was markedly decreased, i.e. all I2 values were 0 for the LDA and SVM subgroups and 0%–79% for the ANN subgroup, emphasizing that the performance of AI systems was in part determined by the types of AI classifiers. The remaining high heterogeneity in the ANN subgroup was likely driven by the study of which the AI performance was estimated from the development cohort but not from the validation/test cohort.[23]
For implementing the AI-assisted EUS into practice, it is uttermost importance to validate the performance of the developed AI system on an independent dataset. Among studies with validation set, the pooled sensitivity and specificity were 0.90 (95% CI, 0.87–0.93) and 0.92 (95% CI, 0.86–0.96, respectively [Supplemental Figure 3a (270.2KB, tif) and b (270.2KB, tif) ]. In addition, when categorized by risk of bias, the group of studies with low risk of bias had a pooled sensitivity and specificity of 0.90 (95% CI, 0.86-0.93) and 0.92 (95% CI, 0.78-0.97), respectively, while studies with high risk of bias had a pooled sensitivity and specificity of 0.91 (95%CI, 0.82-0.96) and 0.87 (95%CI, 0.66-0.96), respectively [Supplemental Figure 4a (443.6KB, tif) and b (443.6KB, tif) ].
Publication bias
The slope coefficient of the Deeks’ funnel plot was relatively symmetry (P = 0.9426) [Supplemental Figure 5 (226.4KB, tif) ], suggesting that the publication bias was not present.
DISCUSSION
Conventional brightness mode (B-mode) of EUS utilizes a spectrum of sound wave transmissibility of pancreatic tissue in depicting real-time gray-scale endosonographic images to differentiate tumor from normal pancreatic parenchyma. However, certain pancreatic pathology such as chronic pancreatitis and focal autoimmune pancreatitis may have similar echogenicity as pancreatic tumor, thus mimicking PDAC. Elastography and contrast-enhanced EUS are advanced imaging modalities that enable real-time evaluation of tumor hemodynamics and its elasticity to improve diagnostic yield of EUS. Despite such advanced techniques, EUS is operator-dependent and requires subjective interpretation of endosonographic images for diagnosis and tissue acquisition.
B-mode EUS had a high sensitivity of 95%, however, the specificity was only 53% for distinguishing malignant from benign pancreatic lesions.[30] We found that the specificity of B-mode EUS increased to 90%, while the sensitivity remained high at 91%, when the AI-assisted image analysis system is integrated. For contrast-enhanced EUS, pooled sensitivity and specificity were 91%–93% and 84%–86%, respectively, for diagnosis of malignant versus benign pancreatic lesions.[31,32] In line with the B-mode EUS images, with the AI system, the specificity improved to 94%, with a comparable sensitivity of 95%. EUS elastography had a pooled sensitivity and specificity of 95%–98% and 63%–76% for the diagnosis of malignant pancreatic lesions.[33,34,35,36,37,38,39] Unlike the other 2 modes, the specificity of AI-assisted system increased to 83% but the sensitivity slightly decreased to 88%. It is important to note that the studies on AI-assisted EUS with image-enhanced technique remain sparse. There was only 1 study investigating diagnostic yield of ML system on contrast-enhanced EUS images and 2 studies assessed the accuracy of ML in EUS elastography images.[18,19,27] All the 3 studies were from the same investigator group. However, given the diagnostic performance of the subgroup of conventional B-mode, EUS was not different from that of the main analysis that included studies on conventional B-mode and studies on image-enhanced techniques. This might therefore imply that AI system can potentially be applied to EUS images without image-enhanced technology or with certain image-enhanced technique such as contrast-enhanced technique.
To determine the optimal AI model for EUS, we classified the studies based on the AI techniques used in each study. The AI technique can be broadly categorized into 2 groups, i.e. has and non-NN. Although there is no proven superiority between the two techniques, NN have advantages in their ability to approximate any non-linear relationship between inputs and outputs, while non-NN techniques rely on prior assumptions regarding data distribution. NN has some advantages because they can approximate “any” non-linear relationship between inputs and outputs, while non-NN techniques depend on some assumptions about data distribution. In this meta-analysis, non-NN had a similar pooled sensitivity to NN, i.e. 92% versus 91%, respectively, but had a higher specificity, i.e. 94% versus 86%, respectively. This can be due to the techniques of non-NN applied in these studies. Different non-NN techniques are suitable for modeling different data characteristics. Moreover, all the included studies used features extracted from images as inputs for the AI systems, instead of using the whole images as inputs, performance of the AI systems therefore also depends on choices of these features as well as how well they were extracted.
Because FNA carries some risks of complications,[40] it should be performed only by experienced endoscopists. However, FNA may not be technically feasible in a few circumstances such as in masses with large intertwining vessels; masses in pancreatic tail where transgastric needle approach would significantly increase the risk of malignant seeding; lesions further away from intestinal wall; or patients with large-volume ascites, severe thrombocytopenia, or coagulopathy.[41] Therefore, improvement in diagnostic yield of EUS images will not only increase the efficacy of the procedure but also reduce the complications that may develop from unnecessary puncture on the low-likelihood lesion. Moreover, the knowledge, experience, and skills of individual endoscopist may affect the results obtained from EUS. Therefore, using a computer-aided supporting system that can provide a real-time objective interpretation of EUS images will significantly contribute to more accurate, safer, and easier diagnosis. Even under the experienced endosonographers hands, AI could potentially facilitate the more precise puncture site and perhaps this might reduce the number of punctures.
This is one of the very first meta-analyses of the AI-assisted system in the EUS studies. Although publication bias was not detected, our findings should be interpreted with cautions due to limitation of included studied. First, although the included studies were conducted in various countries, the number of studies was quite small (n = 8), given that AI has just been increasingly involved in EUS field in recent years. In addition, because PDAC is a relatively uncommon cancer, the number of patients in most studies was therefore limited, particularly, the number of controls was very small in 2 studies.[23,25] The small number of controls in these 2 studies can potentially introduce sampling error, leading to a substantial heterogeneity of the pooled specificity, although no significant heterogeneity of the pooled sensitivity. For implementation of the AI system into practice, it is crucial to develop AI systems that can be generalized well to real clinical settings. One way to evaluate generalizability is to validate the AI systems using independent datasets that the systems have never seen before. In this meta-analyses, only 3 studies had an independent dataset used as a validation cohort, however, the validation cohort had similar characteristics to the development cohort.[19,24,29] To ensure the generalizability in different clinical settings, the AI systems should be evaluated on “external” clinical validation cohorts. For the most sophisticated level, AI systems should be evaluated whether they can improve clinical performance, i.e. conducting a randomized controlled trial comparing diagnostic performance between physicians alone and physicians with assistance from AI systems. Finally, all included studies developed AI systems for classifying the type of pancreatic lesions in the offline mode. None had a system for real-time detection and characterization of pancreatic lesion. Real-time operations of the system would increase the chance of clinical use as they will provide physicians guidance, especially for tissue acquisition of the high-yield lesion during procedures.
CONCLUSIONS
AI-assisted EUS image analysis is an effective tool to diagnose PDAC and distinguishing it from other pancreatic condition, with high accuracy. The improvement in AI technology that enables a real-time evaluation can play an important role in tissue-acquisition guidance and avoid unnecessary puncture on the low-likelihood lesion.
Supplementary materials
Supplementary information is linked to the online version of the paper on the Endoscopic Ultrasound website.
Financial support and sponsorship
This research was funded by the Ratchadapisek Sompoch Endowment Fund (2021) under Telehealth Cluster, Chulalongkorn University, Bangkok, Thailand and the Second Century Fund (C2F), Chulalongkorn University, Bangkok, Thailand.
Conflicts of interest
Pradermchai Kongkam is an Editorial Board Member of the journal. The article was subject to the journal's standard procedures, with peer review handled independently of this editor and his research groups.
Sensitivity (a), specificity (b), positive predictive value (c), negative predictive value (d), and diagnostic odds ratio (e) of a subgroup analysis of the 6 studies on the B-mode EUS alone, without image-enhanced technique
Sensitivity (a), specificity (b), positive predictive value (c), negative predictive value (d), and diagnostic odds ratio (e) of subgroup analysis by type of artificial intelligence classifiers
Pooled sensitivity (a) and specificity (b) of artificial intelligence models with validation set
Pooled sensitivity (a) and specificity (b) of artificial intelligence models classified by risk of bias
Deeks funnel plot
SUPPLEMENTAL METHOD
Twelve questions of QUADAS 2 criteria
-
(1)
Could the selection of patients have introduced bias?
Was a consecutive or random sample of patients enrolled?
Was a case-control design avoided?
Did the study avoid inappropriate exclusions?
-
(2)
Could the conduct or interpretation of the index test have introduced bias?
Is the AI system validated by any means? (e.g. independent test set, k fold cross validation, external clinical validation set)*
-
(3)
Could the reference standard, its conduct, or its interpretation have introduced bias?
Is the reference standard likely to correctly classify the target condition?
-
(4)
Could the patient flow have introduced bias?
Is there an appropriate interval between index test and reference standard?
Did all patients receive a reference standard?
Did patients receive the same reference standard?
Were all patients included in the analysis?
-
(5)
Is there concern that the included patients do not match the review question?
-
(6)
Is there concern that the index test, its conduct, or interpretation differ from the review question?
-
(7)
Is there concern that the target condition as defined by the reference standard does not match the review question?
*This item was modified from the original QUADAS 2 criteria.
**Two items were excluded from the criteria:
Were the index test results interpreted without knowledge of the results of the reference standard?
Were the reference standard results interpreted without the knowledge of the results of the index test?
***Risk of bias in each main question is assessed as “low,” “high,” or “unclear.” If all the subquestions are answered “yes,” the main question will be assessed as low risk of bias. If at least one subquestion is answered “no,” the main question will be assessed as high risk of bias. If there is insufficient information to assess the risk, it will be marked as “unclear”[1]
REFERENCE
- 1.Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
REFERENCES
- 1.Rawla P, Sunkara T, Gaduputi V. Epidemiology of pancreatic cancer: Global trends, etiology and risk factors. World J Oncol. 2019;10:10–27. doi: 10.14740/wjon1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Corral JE, Mareth KF, Riegert-Johnson DL, et al. Diagnostic yield from screening asymptomatic individuals at high risk for pancreatic cancer: A meta-analysis of cohort studies. Clin Gastroenterol Hepatol. 2019;17:41–53. doi: 10.1016/j.cgh.2018.04.065. [DOI] [PubMed] [Google Scholar]
- 4.Best LM, Rawji V, Pereira SP, et al. Imaging modalities for characterising focal pancreatic lesions. Cochrane Database Syst Rev. 2017;4:CD010213. doi: 10.1002/14651858.CD010213.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hewitt MJ, McPhail MJ, Possamai L, et al. EUS-guided FNA for diagnosis of solid pancreatic neoplasms: A meta-analysis. Gastrointest Endosc. 2012;75:319–31. doi: 10.1016/j.gie.2011.08.049. [DOI] [PubMed] [Google Scholar]
- 6.Chen G, Liu S, Zhao Y, et al. Diagnostic accuracy of endoscopic ultrasound-guided fine-needle aspiration for pancreatic cancer: A meta-analysis. Pancreatology. 2013;13:298–304. doi: 10.1016/j.pan.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Hebert-Magee S, Bae S, Varadarajulu S, et al. The presence of a cytopathologist increases the diagnostic accuracy of endoscopic ultrasound-guided fine needle aspiration cytology for pancreatic adenocarcinoma: A meta-analysis. Cytopathology. 2013;24:159–71. doi: 10.1111/cyt.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puli SR, Matthew L, Buxbaum JL, et al. How good is endoscopic ultrasound-guided fine-needle aspiration in diagnosing the correct etiology for a solid pancreatic mass? Pancreas. 2013;42:20–6. doi: 10.1097/MPA.0b013e3182546e79. [DOI] [PubMed] [Google Scholar]
- 9.Costache MI, Cornelia A, Dumitrescu CI, et al. Which is the best imaging method in pancreatic adenocarcinoma diagnosis and staging - CT, MRI or EUS? Curr Health Sci J. 2017;43:132–6. doi: 10.12865/CHSJ.43.02.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitano M, Yoshida T, Itonaga M, et al. Impact of endoscopic ultrasonography on diagnosis of pancreatic cancer. J Gastroenterol. 2019;54:19–32. doi: 10.1007/s00535-018-1519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wani S, Coté GA, Keswani R, et al. Learning curves for EUS by using cumulative sum analysis: Implications for American Society for Gastrointestinal Endoscopy recommendations for training. Gastrointest Endosc. 2013;77:558–65. doi: 10.1016/j.gie.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Wani S, Muthusamy VR, McGrath CM, et al. AGA white paper: Optimizing endoscopic ultrasound-guided tissue acquisition and future directions. Clin Gastroenterol Hepatol. 2018;16:318–27. doi: 10.1016/j.cgh.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 13.Bhutani MS, Gress FG, Giovannini M, et al. The no endosonographic detection of tumor (NEST) study: A case series of pancreatic cancers missed on endoscopic ultrasonography. Endoscopy. 2004;36:385–9. doi: 10.1055/s-2004-814320. [DOI] [PubMed] [Google Scholar]
- 14.Niimi K, Goto O, Kawakubo K, et al. Endoscopic ultrasound-guided fine-needle aspiration skill acquisition of gastrointestinal submucosal tumor by trainee endoscopists: A pilot study. Endosc Ultrasound. 2016;5:157–64. doi: 10.4103/2303-9027.183970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Berre C, Sandborn WJ, Aridhi S, et al. Application of artificial intelligence to gastroenterology and hepatology. 2020;158:76-94. Gastroenterology. 2020;158:76–94e2. doi: 10.1053/j.gastro.2019.08.058. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell TM. Machine Learning. Machine Learning New York: McGraw-Hill, Inc; 1997. [Google Scholar]
- 17.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Săftoiu A, Vilmann P, Gorunescu F, et al. Neural network analysis of dynamic sequences of EUS elastography used for the differential diagnosis of chronic pancreatitis and pancreatic cancer. Gastrointest Endosc. 2008;68:1086–94. doi: 10.1016/j.gie.2008.04.031. [DOI] [PubMed] [Google Scholar]
- 19.Săftoiu A, Vilmann P, Dietrich CF, et al. Quantitative contrast-enhanced harmonic EUS in differential diagnosis of focal pancreatic masses (with videos) Gastrointest Endosc. 2015;82:59–69. doi: 10.1016/j.gie.2014.11.040. [DOI] [PubMed] [Google Scholar]
- 20.Whiting PF, AW, Westwood ME, et al. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 21.Cohen J, Desilets DJ, Hwang JH, et al. Gastrointestinal endoscopy editorial board top 10 topics: Advances in GI endoscopy in 2018. Gastrointest Endosc. 2019;90:35–43. doi: 10.1016/j.gie.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 22.Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240:1285–93. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- 23.Norton ID, Zheng Y, Wiersema MS, et al. Neural network analysis of EUS images to differentiate between pancreatic malignancy and pancreatitis. Gastrointest Endosc. 2001;54:625–9. doi: 10.1067/mge.2001.118644. [DOI] [PubMed] [Google Scholar]
- 24.Zhang MM, Yang H, Jin ZD, et al. Differential diagnosis of pancreatic cancer from normal tissue with digital imaging processing and pattern recognition based on a support vector machine of EUS images. Gastrointest Endosc. 2010;72:978–85. doi: 10.1016/j.gie.2010.06.042. [DOI] [PubMed] [Google Scholar]
- 25.Kumon RE, Pollack MJ, Faulx AL, et al. In vivo characterization of pancreatic and lymph node tissue by using EUS spectrum analysis: A validation study. Gastrointest Endosc. 2010;71:53–63. doi: 10.1016/j.gie.2009.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumon RE, Repaka A, Atkinson M, et al. Characterization of the pancreas in vivo using EUS spectrum analysis with electronic array echoendoscopes. Gastrointest Endosc. 2012;75:1175–83. doi: 10.1016/j.gie.2012.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Săftoiu A, Vilmann P, Gorunescu F, et al. Efficacy of an artificial neural network-based approach to endoscopic ultrasound elastography in diagnosis of focal pancreatic masses. Clin Gastroenterol Hepatol. 2012;10:84–90e1. doi: 10.1016/j.cgh.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Zhu M, Xu C, Yu J, et al. Differentiation of pancreatic cancer and chronic pancreatitis using computer-aided diagnosis of endoscopic ultrasound (EUS) images: A diagnostic test. PLoS One. 2013;8:e63820. doi: 10.1371/journal.pone.0063820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozkan M, Cakiroglu M, Kocaman O, et al. Age-based computer-aided diagnosis approach for pancreatic cancer on endoscopic ultrasound images. Endosc Ultrasound. 2016;5:101–7. doi: 10.4103/2303-9027.180473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brand B, Pfaff T, Binmoeller KF, et al. Endoscopic ultrasound for differential diagnosis of focal pancreatic lesions, confirmed by surgery. Scand J Gastroenterol. 2000;35:1221–8. doi: 10.1080/003655200750056736. [DOI] [PubMed] [Google Scholar]
- 31.Mei S, Wang M, Sun L. Contrast-enhanced EUS for differential diagnosis of pancreatic masses: A meta-analysis. Gastroenterol Res Pract. 2019;2019:1670183. doi: 10.1155/2019/1670183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Jin H, Liao D, et al. Contrast-enhanced harmonic endoscopic ultrasonography for the differential diagnosis of pancreatic masses: A systematic review and meta-analysis. Mol Clin Oncol. 2019;11:425–33. doi: 10.3892/mco.2019.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pei Q, Zou X, Zhang X, et al. Diagnostic value of EUS elastography in differentiation of benign and malignant solid pancreatic masses: A meta-analysis. Pancreatology. 2012;12:402–8. doi: 10.1016/j.pan.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 34.Hu DM, Gong TT, Zhu Q. Endoscopic ultrasound elastography for differential diagnosis of pancreatic masses: A meta-analysis. Dig Dis Sci. 2013;58:1125–31. doi: 10.1007/s10620-012-2428-5. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Xu W, Shi J, et al. Endoscopic ultrasound elastography for differentiating between pancreatic adenocarcinoma and inflammatory masses: A meta-analysis. World J Gastroenterol. 2013;19:6284–91. doi: 10.3748/wjg.v19.i37.6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mei M, Ni J, Liu D, et al. EUS elastography for diagnosis of solid pancreatic masses: A meta-analysis. Gastrointest Endosc. 2013;77:578–89. doi: 10.1016/j.gie.2012.09.035. [DOI] [PubMed] [Google Scholar]
- 37.Xu W, Shi J, Li X, et al. Endoscopic ultrasound elastography for differentiation of benign and malignant pancreatic masses: A systemic review and meta-analysis. Eur J Gastroenterol Hepatol. 2013;25:218–24. doi: 10.1097/MEG.0b013e32835a7f7c. [DOI] [PubMed] [Google Scholar]
- 38.Ying L, Lin X, Xie ZL, et al. Clinical utility of endoscopic ultrasound elastography for identification of malignant pancreatic masses: A meta-analysis. J Gastroenterol Hepatol. 2013;28:1434–43. doi: 10.1111/jgh.12292. [DOI] [PubMed] [Google Scholar]
- 39.Zhang B, Zhu F, Li P, et al. Endoscopic ultrasound elastography in the diagnosis of pancreatic masses: A meta-analysis. Pancreatology. 2018;18:833–40. doi: 10.1016/j.pan.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 40.ASGE Standards of Practice Committee, Early DS, Acosta RD, et al. Adverse events associated with EUS and EUS with FNA. Gastrointest Endosc. 2013;77:839–43. doi: 10.1016/j.gie.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 41.Wang KX, Ben QW, Jin ZD, et al. Assessment of morbidity and mortality associated with EUS-guided FNA: A systematic review. 2011;73:283- Gastrointest Endosc. 2011;73:283-. doi: 10.1016/j.gie.2010.10.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sensitivity (a), specificity (b), positive predictive value (c), negative predictive value (d), and diagnostic odds ratio (e) of a subgroup analysis of the 6 studies on the B-mode EUS alone, without image-enhanced technique
Sensitivity (a), specificity (b), positive predictive value (c), negative predictive value (d), and diagnostic odds ratio (e) of subgroup analysis by type of artificial intelligence classifiers
Pooled sensitivity (a) and specificity (b) of artificial intelligence models with validation set
Pooled sensitivity (a) and specificity (b) of artificial intelligence models classified by risk of bias
Deeks funnel plot