In this review, Burgess et al. discuss strategies and mechanisms that control mRNA decay, modification, and translation in animal virus-infected cells. Besides settling infection outcomes, post-transcriptional gene regulation in virus-infected cells epitomizes fundamental physiological stress responses in health and disease.
Keywords: RNA decay, RNA modification, animal viruses, infection stress, translational control
Abstract
With their categorical requirement for host ribosomes to translate mRNA, viruses provide a wealth of genetically tractable models to investigate how gene expression is remodeled post-transcriptionally by infection-triggered biological stress. By co-opting and subverting cellular pathways that control mRNA decay, modification, and translation, the global landscape of post-transcriptional processes is swiftly reshaped by virus-encoded factors. Concurrent host cell-intrinsic countermeasures likewise conscript post-transcriptional strategies to mobilize critical innate immune defenses. Here we review strategies and mechanisms that control mRNA decay, modification, and translation in animal virus-infected cells. Besides settling infection outcomes, post-transcriptional gene regulation in virus-infected cells epitomizes fundamental physiological stress responses in health and disease.
Responses to environmental and physiological stress demand swift, coordinated remodeling of the genome-wide expression landscape. Such abrupt adaptation provoked by stimuli that upset homeostasis is often controlled post-transcriptionally by mRNA decay and translation. As a powerful, genetically tractable model, virus infection provides a window to interrogate how mammalian cells react to biological stress and identify how virus gene products shape host cell responses by accentuating or subverting them. Indeed, virus-encoded effectors interact with cellular targets to regulate virus and host gene expression post-transcriptionally and determine infection outcomes. Virus mRNAs are absolutely reliant on host ribosomes and compete for them with cellular mRNAs. Similarly, differential mRNA accumulation regulated in part by RNA modification and decay is exploited by viruses to complete their reproductive cycle and by host immune defenses to limit virus replication. Besides revealing fundamental mechanisms regulating gene expression and infection biology, viruses provide insight into human disease. Here, we review molecular interactions between select animal viruses and their hosts that regulate mRNA decay, modification, and translation, highlighting developments pertaining to genome-wide changes and stress responses.
Synopsis of virus genome structure and reproductive strategies
Viruses are obligate intracellular parasites unable to reproduce outside of host cells. Within eukaryotic hosts, viruses replicate in the cell cytoplasm or nucleus. Not all infections cause clinical disease, and outcomes vary depending on host, cell type, and immune status. Although acute infection results in virus reproduction and often host cell destruction, persistent infections exhibiting chronic or episodic virus production may endure over the host's lifetime. Viruses are classified by genome structure and replication strategies, each of which influences how viral mRNAs engage and impact cellular post-transcriptional regulatory pathways.
Composed of single- or double-stranded (ds) DNA or RNA, virus genome structures and sizes are diverse, ranging from ≤10 kb for small RNA or DNA viruses to >200 kb for the largest human DNA viruses. Still bigger megabase genomes exist for DNA viruses that infect Acanthamoeba (Schulz et al. 2017). RNA virus genomes are composed of single or multiple nucleic acid segments. Even large genomes maximize coding capacity by using proteases to generate multiple polypeptides from one single ORF, overlapping ORFs, and frameshifting (Atkins et al. 2016; Jan et al. 2016; Penn et al. 2020). Virus genome structure impacts mRNA biogenesis. Single-strand RNA viruses with genome polarity identical to that of mRNA [(+)-strand RNA viruses], with the exception of retrovirus virion RNA inside incoming virus particles, are translated upon infection, whereas those with opposite polarity [(−)-strand RNA viruses] or dsRNA genomes require a virus-encoded RNA-dependent RNA polymerase (RdRp) to produce mRNA. Following reverse transcription of their (+)-strand RNA genome and chromosomal integration, retrovirus mRNA biogenesis requires the host RNA polymerase II (RNAPII) and associated processing factors. While nuclear-replicating DNA viruses also rely on host mRNA biogenesis and processing factors, poxviruses such as vaccinia virus (VacV) replicate in the cytoplasm and encode viral functions for transcription, capping, and 3′ end processing.
While mRNA biogenesis and genome replication strategies vary, virus reproduction and protein synthesis are contingent on host ribosomes. Viruses may interfere with host protein synthesis by diverse mechanisms, ranging from near-global inhibition called host shutoff, which often involves destabilizing host mRNAs or inactivating translation factors, to triaging which host mRNAs are translated. Besides fostering virus mRNA translation, host shutoff restricts antiviral immune responses including interferon (IFN) production, which antagonizes virus reproduction and spread. Host antiviral effectors, many of which are induced by IFN, similarly target RNA stability and protein synthesis to suppress virus replication. Accordingly, post-transcriptional control of host cell-intrinsic immune defenses and virus reproduction is often achieved by regulating mRNA translation, modification, and decay.
Shaping the infected cell mRNA landscape
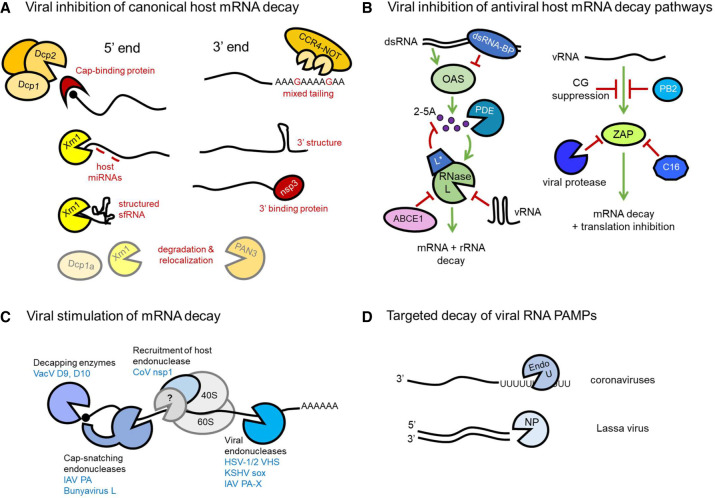
Evading, co-opting, or supplementing the host RNA decay machinery with viral factors remodels infected cell mRNA composition. Deadenylation-dependent decay is the canonical mechanism for bulk mRNA turnover whereby the stabilizing 3′-terminal poly(A) tail of most cellular mRNAs is degraded by deadenylase complexes CCR4–NOT and PAN2/3 (Heck and Wilusz 2018). Once shortened to preclude binding of cytoplasmic poly(A)-binding protein (PABP), an RNA-decapping complex is recruited, comprised of enzyme Dcp2, Dcp1, and decapping enhancer proteins (Fig. 1A). Upon protective m7G cap removal, the cellular 5′–3′ exonuclease Xrn1 degrades mRNAs bearing an exposed 5′ monophosphate (Fig. 1A). Alternatively, the RNA exosome complex can degrade deadenylated RNAs 3′–5′. Deadenylase recruitment by sequence-dependent binding of discrete RNA-binding proteins (RNA-BPs), such as the AU-rich element (ARE) BP tristetraprolin (TTP) or microRNAs, targets specific mRNAs for decay. Conversely, RNA-BPs that impede decay promoting factor binding stabilize specific mRNAs (Fukao and Fujiwara 2017).
Figure 1.
Viral strategies to oppose and promote mRNA decay. (A) Inhibition of cellular proteins that effect 5′–3′ mRNA decay can be achieved by protection from decapping by cap binding proteins (VPg) and obstruction of the exonuclease Xrn1 by miRNAs and RNA structure (sfRNA). 3′ end decay is similarly opposed by recruitment of viral proteins and formation of RNA structure, and “mixed tailing” can also inhibit poly(A) tail deadenylation by CCR4–NOT. Both 5′ and 3′ targeting cellular mRNA decay proteins may be degraded or relocalized during infection. (B) Viruses inhibit OAS/RNase L and ZAP antiviral cellular RNA decay pathways using diverse strategies. Recognition of dsRNA by OAS is blocked by viral dsRNA-BPs. Viral phosphodiesterases (PDEs) degrade second messenger 2-5A. RNase L activation can also be blocked by viral proteins, up-regulated cellular negative regulator ABCE1, or viral RNA structures. ZAP recognition of virus RNA (vRNA) is evaded by CG suppression, viral ZAP-binding proteins, and ZAP cleavage by virus proteases. (C) Viral proteins stimulate mRNA decay via multiple modalities. They can contain direct enzymatic activity such as mRNA decapping and endonucleolytic cleavage, which may be cap-proximal. In contrast, coronavirus nsp1 binds the 40S ribosome and, although no direct nucleolytic activity has been identified, effects mRNA cleavage in infected cells, leading to speculation that it recruits a host decay enzyme. (D) Viral RNA PAMPs are directly controlled by viral dsRNA-specific exonucleases or U-specific endonucleases that, by limiting the potential for 5′ (−)-strand poly(U) sequences to complex with A-rich sequences, prevent dsRNA formation.
Antiviral roles for mRNA decay and viral defensive lines
To evade the host RNA decay machinery, virus RNAs contain structures that impede degradation and sequences that recruit stabilizing RNA-BPs (Fig. 1A). Viruses also manipulate stabilizing and destabilizing host factor availability. Most viral RNAs possess a 5′ m7G cap and poly(A) tail akin to cellular mRNAs or features that protect them from exonucleases. m7G caps are added to RNAPII transcribed viral mRNAs by the cellular capping machinery in the nucleus (most DNA viruses and retroviruses), adjoined by viral capping enzymes (poxviridae and many RNA viruses), or acquired by excising a short 5′-terminal fragment from host mRNAs (“cap snatching”) that in turn primes virus RdRp-directed transcription (orthomyxoviridae [e.g., influenza], arenaviridae, and bunyaviridae [e.g., Lassa fever virus]). Alternatively, a viral protein (VPg) covalently attached to (+)-sense virus genomes (caliciviridae and picornaviridae) can protect the RNA 5′ end (Decroly and Canard 2017). Unusually, two copies of liver-specific cellular microRNA miR-122 bind to a 5′ sequence in the (+)-sense hepatitis C virus (HCV) RNA genome, blocking Xrn1-mediated decay and promoting translation (Henke et al. 2008; Li et al. 2013). HCV replication further requires DEAD-box RNA helicase eIF4A2, which interacts with the virus replicon in a mIR122-dependent manner and is critical for gene regulation by microRNAs (Ahmed et al. 2018; Wilczynska et al. 2019).
Distinct tactics preserve poly(A) 3′ tails, which are added by the nuclear host machinery (most DNA viruses), by viral poly(A) polymerases (poxviridae), or by transcription of a poly(U) template (e.g., paramyxoviridae, rhabdoviridae, and orthomyxoviridae). Viral poly(A) tail stability may be enhanced by non-A nucleotide incorporation to slow deadenylation, so-called “mixed tailing,” promoted by virus-encoded sequences (Fig. 1A; Hyrina et al. 2019; Kim et al. 2020). Following recognition of stem–loops in hepatitis B virus (HBV) RNAs and human cytomegalovirus (HCMV) noncoding (nc) RNAs by the cellular RNA-BP ZCCHC14, recruitment of TENT4 noncanonical poly(A) polymerases allows non-A nucleotide incorporation (often Gs) into viral poly(A) tails (Kim et al. 2020). HCMV also up-regulates cytoplasmic polyadenylation machinery, extending virus and cellular mRNA poly(A) tails (Batra et al. 2016). While flaviviride, bunyaviridae, and arenaviridae encode a 3′-terminal stem–loop to block exonucleases, an RNA 3′-terminal sequence bound by rotavirus protein NSP3 (Fig. 1A) stimulates translation and likely protects from exosome action (Brinton et al. 1986; Meyer and Southern 1993; Deo et al. 2002; Geerts-Dimitriadou et al. 2012; Gratia et al. 2015).
To disrupt RNA decay, viruses obstruct host factor function using structured RNA and/or by perturbing their localization (Fig. 1A). Proteases encoded by porcine reproductive and respiratory syndrome virus (PRRSV), poliovirus (PV), and coronavirus (CoV) cleave decapping activator Dcp1a (Dougherty et al. 2011; Tao et al. 2018; Zhu et al. 2020). PV also induces degradation of deadenylase PAN3 and exonuclease Xrn1 and disrupts processing bodies (P-bodies), cytoplasmic sites enriched for RNA decay proteins (Dougherty et al. 2011). During rotavirus infection, Nsp1 degrades PAN3, both Xrn1 and Dcp1a are relocalized from the cytoplasm to nucleus (Bhowmick et al. 2015), and additional host decay proteins are relocalized to cytoplasmic inclusions (Dhillon and Rao 2018; Dhillon et al. 2018). Similarly, adenovirus (Ad) relocalizes decay proteins including Xrn1 to cytoplasmic aggresomes (Greer et al. 2011). Degrading or sequestering these host mRNA decay effectors is expected to slow viral and cellular RNA turnover in infected cells, but has not been fully examined. In a different approach, a structured 3′ UTR in flavivirus (+)-sense RNA genomes limits complete 5′–3′ digestion by Xrn1 (Pijlman et al. 2008). Short, noncoding subgenomic flavivirus RNAs (sfRNAs) accumulate as a result (Silva et al. 2010; Chapman et al. 2014a,b), functionally sequestering XRN1, as demonstrated by global host mRNA stabilization in HCV-infected cells (Moon et al. 2015). While expected to extend flavivirus RNA genome longevity, how host mRNA dysregulation contributes to infection pathogenesis remains unknown.
Host mRNA quality control pathways that ensure transcripts bearing mutations or processing errors and spurious transcripts are destroyed also dispose of viral RNAs. RNA viruses with sequentially arranged ORFs are subject to nonsense-mediated decay (NMD), which targets transcripts that have a long distance between a stop codon and the poly(A) tail, and can restrict virus replication (Popp et al. 2020; May and Simon 2021). With long 3′ UTR-containing RNAs vulnerable to NMD, retroviral RNA elements in Moloney murine leukemia virus (MMLV) and Rous sarcoma virus prevent NMD machinery detection and cleavage, respectively (Weil and Beemon 2006; Hogg and Goff 2010). The cellular helicase UPF1, which is required for NMD target recognition (Rao et al. 2019), is down-regulated by human immunodeficiency virus 1 (HIV-1) and inhibited by human T-lymphotropic virus type 1 (HTLV-1) protein tax (Mocquet et al. 2012; Nakano et al. 2013). Zika virus (ZIKV), Semliki forest virus (SFV), and CoV capsid proteins also interact with UPF1 and can antagonize NMD (Fontaine et al. 2018; Wada et al. 2018; Gordon et al. 2020; Contu et al. 2021).
Multiple antiviral host RNA decay pathways are encoded by IFN-stimulated genes (ISGs). Oligo adenylate synthetases (OASs) 1, 2, and 3 bind and are activated by dsRNA, a pathogen-associated molecular pattern (PAMP) generated by RNA virus replication or from complementary transcripts of opposing DNA virus genome strands. Activated OAS produces 2′–5′ oligo adenylate (2-5A), which stimulates latent RNase L (Fig. 1B), an endoribonuclease that broadly attacks mRNA and rRNA to inhibit translation and promote apoptosis. Viral countermeasures (Fig. 1B) include shielding dsRNA from OAS detection by deploying dsRNA-BPs such as herpes simplex virus 1 (HSV-1) Us11 (Sànchez and Mohr 2007), HCMV TRS1 and IRS1 (Marshall et al. 2009), VacV E3L (Chang et al. 1992), influenza virus A (IAV) NS1 (Min and Krug 2006), and reovirus σ3 (Imani and Jacobs 1988). Downstream from OAS activation, CoV and rotavirus phosphodiesterases cleave 2-5A to prevent RNase L activation (Zhao et al. 2012; Zhang et al. 2013; Thornbrough et al. 2016; Goldstein et al. 2017), while Theiler's murine encephalomyelitis virus L* protein inhibits RNase L activation by blocking 2-5A binding (Sorgeloos et al. 2013; Drappier et al. 2018), and PV RNA elements competitively inhibit RNase L (Han et al. 2007; Townsend et al. 2008). In contrast, EMCV and HIV up-regulate ABCE1 (Martinand et al. 1998, 1999), a host RNase L BP that prevents activation by 2-5A (Bisbal et al. 1995), turning an endogenous RNase L regulator against the host.
Zinc finger antiviral protein (ZAP) is an ISG-encoded RNA-BP with a preference for GC-rich sequences and thus functions to detect “nonself” RNA (Takata et al. 2017). ZAP activity has been ascribed to translational inhibition and RNA decay in RNA virus infections, the latter linked to RNA exosome recruitment (Guo et al. 2007) and the host endonuclease KHNYN (Ficarelli et al. 2019). CG dinucleotide suppression in HIV-1 and HCMV genomes, mimicking mammalian DNA genomes, is a tactic to resist ZAP (Fig. 1B; Takata et al. 2017; Lin et al. 2020; Gonzalez-Perez et al. 2021). Whereas IAV protein PB2 inhibits RNA binding by ZAP (Tang et al. 2017), ZAP is cleaved by PRRSV and enterovirus A71 (EV-A71) proteases (Xie et al. 2018; Zhao et al. 2020) and sequestered by VacV protein C16 (Peng et al. 2020). ZAP is expressed as long (L) and short (S) isoforms, both of which contain RNA recognition motifs but differ in intracellular localization and RNA targeting (Schwerk et al. 2019; Kmiec et al. 2021). ZAP-S also binds to the SARS-CoV-2 programmed ribosomal frameshifting (PRF) RNA element to inhibit PRF, which is essential for RdRp and nonstructural protein production (Zimmer et al. 2021).
Last, ISG20 possesses 3′–5′ exoribonuclease activity in vitro (Nguyen et al. 2001) and suppresses RNA virus replication (Espert et al. 2003; Zhou et al. 2011). Antiviral action in vivo, however, relies more on ISG20 inhibiting viral mRNA translation and modulating ISG expression than RNA decay (Weiss et al. 2018; Wu et al. 2019).
Proviral roles for mRNA decay—viruses on the offensive
To ensure translational dominance, some virus mRNAs rely on their sheer abundance compared with host mRNAs, rather than specific cis-elements or trans factors, to competitively acquire ribosomes (Bercovich-Kinori et al. 2016; Finkel et al. 2021). By encoding endoribonucleases that accelerate RNA decay (Fig. 1C), Kaposi's sarcoma-associated herpesvirus (KSHV) and HSV reduce infected cell mRNA abundance (Glaunsinger and Ganem 2004; Pheasant et al. 2018; Friedel et al. 2021). Broad targeting of host and viral mRNAs is observed, the latter facilitating sharp temporal transitions in virus gene expression (Pasieka et al. 2008). The HSV-1 virion host shutoff endonuclease (vhs) is targeted to mRNAs by associating with translation initiation factors eIF4H and eIF4A (Doepker et al. 2004; Feng et al. 2005; Page and Read 2010), although spliced mRNAs are transiently protected from vhs cleavage until exon junction complex removal by the first round of translation (Sadek and Read 2016). A loose target consensus sequence was found for the KSHV endoribonuclease sox as well as a RNA element that protects select host immune mRNAs from decay (Gaglia et al. 2015; Muller and Glaunsinger 2017). Besides its shrewd use by segmented, (−)-sense RNA viruses to acquire 5′ ends, cap snatching results in decapitated host transcript decay (Fig. 1C). The endoribonuclease is provided by virus RdRp components (IAV PA protein and bunyavirus large “L” protein). During IAV PA translation, a ribosomal frameshift generates a second endoribonuclease, PA-X (Fig. 1C), to further enforce host shutoff (Jagger et al. 2012; Bavagnoli et al. 2015; Chaimayo et al. 2018). PA-X interacts with host mRNA processing factors and preferentially targets spliced mRNAs (Gaucherand et al. 2019).
VacV uses a distinct tactic by encoding two mRNA decapping enzymes (D9 and D10) (Fig. 1C) produced at different times during infection, each of which contains a nudix hydrolase motif similar to that found in cellular Dcp2 (Parrish and Moss 2007; Parrish et al. 2007). While Dcp2 is active within a complex of decapping-enhancing proteins needed for mRNA targeting, D9 and D10 require no additional proteins for activity and contribute to global host shutoff (Parrish and Moss 2007; Parrish et al. 2007). Viral transcripts are not immune to D9 and D10 (Liu et al. 2015), and whether these enzymes show any target selectivity remains unknown.
Although discovered as host shutoff effectors that promote virus mRNA ribosome access, viral mRNA decay enzymes unexpectedly control immunogenic viral dsRNA accumulation. In addition to limiting protein synthesis (via OAS/RNase L), dsRNA stimulates IFN production and antiviral ISG expression (Liu and Gack 2020). Attenuated replication and hyperactivation of host defenses associated with dsRNA accumulation occur during infection with virus mutants deficient for VacV decapping enzymes (Liu et al. 2015), IAV PA-X (Jagger et al. 2012; Hayashi et al. 2015; Rigby et al. 2019), and HSV-1 vhs (Strelow and Leib 1995; Pasieka et al. 2008; Burgess and Mohr 2018; Dauber et al. 2019). Instead of destabilizing RNA globally, other viruses focus their RNA decay enzymes on virus RNAs detected as PAMPs (Fig. 1D). CoVs deploy a ribonuclease endoU that degrades (−)-sense viral RNA bearing 5′-U tracts copied from (+)-strand poly(A) tails (Kindler et al. 2017; Hackbart et al. 2020), while Lassa fever virus (LASV) NP has 3′–5′ exoribonuclease activity that degrades dsRNA (Qi et al. 2010; Hastie et al. 2011; Mateer et al. 2020). Viruses lacking these proteins accumulate dsRNA and replicate poorly.
Viruses also disrupt targeted decay pathways. The generally destabilizing AU-rich element (ARE)-BP AUF1 is sequestered by a ncRNA (EBER1) produced by Epstein-Barr virus (EBV), a herpesvirus subfamily member, and this is proposed to extend virus RNA half-life (Lee et al. 2012). AUF1 is degraded by picornaviruses, and while it directly binds picornaviral RNA, its capacity to inhibit infection has been linked to mRNA translation and stability (Rozovics et al. 2012; Cathcart et al. 2013; Wong et al. 2013). Conversely, RNA virus genomes recruit stabilizing ARE-BP HuR (also known as ELAVL1) (Sokoloski et al. 2010; Nadar et al. 2011; Shwetha et al. 2015).
Host decay enzymes can be co-opted to play proviral roles too. Xrn1 is required by VacV to degrade decapped mRNA and suppress accumulation of dsRNA from complementary viral transcripts (Burgess and Mohr 2015; Liu and Moss 2016). A similar role for Xrn1 is likely in Sindbis virus (SINV) and IAV-infected cells as enhanced IFN induction and impaired virus replication are observed in Xrn1 knockout cells (Garcia-Moreno et al. 2019; Liu et al. 2021). The SKI complex, an RNA helicase cofactor of the cytosolic exosome, stimulates CoV and IAV replication (Weston et al. 2020), possibly via interactions with virus dsRNA BPs (IAV NS1 and MERS ORF4a) that suppress IFN signaling (Niemeyer et al. 2013; Ayllon and García-Sastre 2015). Although endonucleolytic activity has not been found in vitro, CoV Nsp1 reportedly promotes host mRNA decay (Fig. 1C), suggesting an unidentified host endoribonuclease may be needed (Kamitani et al. 2009; Nakagawa and Makino 2021). Finally, viruses repurpose host decay factors. RNA (+) strand viruses including HCV conscript the Lsm1–7 complex, which usually stimulates mRNA decapping, to promote virus mRNA translation and RNA replication (Díez et al. 2000; Scheller et al. 2009; Jungfleisch et al. 2015).
Although infection changes host and viral RNA decay dynamics, the secondary impact on RNA-BP availability and activity is poorly understood. Broad RNA destabilization reduces the abundance of RNAs targeted by RNA-BPs, while virus RNA (vRNA) accumulation qualitatively alters the pool of RNA-bound versus available RNA-BPs. RNA decay triggered by viral endonucleases perturbs transcription and nuclear RNA processing, the latter resulting in part from nuclear accumulation of normally cytoplasmic PABP (Kumar and Glaunsinger 2010; Abernathy et al. 2015). HuR sequestration by SINV destabilizes host HuR-regulated mRNAs (Barnhart et al. 2013), and appropriation of RNA-BPs that regulate ISG mRNA translation by dengue virus (DENV) limits the IFN response (Bidet et al. 2014). New RNA–protein interactome approaches highlight the impact of RNA-BPs sequestered by vRNA during RNA virus infection, including SARS-CoV-2, yielding new avenues for investigation (Flynn et al. 2021; Kamel et al. 2021; Lee et al. 2021; Schmidt et al. 2021; Iselin et al. 2022).
Targeting decay and translation by RNA modification
N6-adenosine methylation (m6A) can alter gene expression post-transcriptionally and is the most widespread internal mRNA base modification in eukaryotes (Zaccara et al. 2019; He and He 2021). The m6A epitranscriptomic landscape is shaped by rival activities of a methyltransferase writer core complex (METTL3 catalytic subunit, METTL14, and WTAP) that installs m6A on nascent mRNA cotranscriptionally and “eraser” demethylases FTO and ALKBH5, which remove m6A marks in vitro (Jia et al. 2011; Zheng et al. 2013; Liu et al. 2014; Ke et al. 2017; Rosa-Mercado et al. 2017). Deposition of m6A by METTL3/14 occurs largely at consensus DRACH (D = A,G,T; R = A,G; and H = A,C,U) motifs, only a fraction of which are modified (He and He 2021). While detected throughout transcripts, m6A is enriched within terminal exons near stop codons and poly(A) signals (He and He 2021). Recognition of m6A-modified sites by “reader” RNA-BPs, including YTH domain-containing proteins in the cytoplasm (YTHDF1, YTHDF2, and YTHDF3) or nucleus (YTHDC1), differentially recruits effector proteins that impact RNA nuclear processing and export and mRNA stability and translation (Zaccara et al. 2019; He and He 2021). YTHDF1,2,3 bind to the same RNAs and act redundantly to regulate RNA decay (Lasman et al. 2020; Zaccara and Jaffrey 2020).
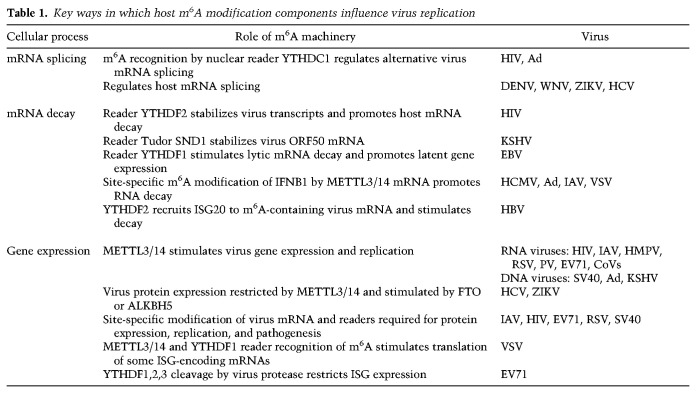
Virus mRNAs are m6A-modified (Williams et al. 2019), and the major ways by which host m6A modification and recognition components post-transcriptionally regulate virus gene expression and replication are summarized in Table 1. Replication of many RNA viruses (HIV, IAV, human metapneumovirus [HMPV], respiratory syncytial virus [RSV], PV, EV71, and CoVs) was stimulated by METTL3/14 and reduced by m6A demethylases. In some but not all cases, specific m6A sites and readers have been identified that are required for protein expression, replication, and pathogenesis (Kennedy et al. 2016; Tirumuru et al. 2016; Lichinchi et al. 2016a; Courtney et al. 2017; Hao et al. 2019; Xue et al. 2019; Han et al. 2020; Yao et al. 2020). The nuclear reader YTHDC1 regulates HIV alternate splicing, while m6A recognition by cytoplasmic YTHDF2, which promotes decay of m6A-containing host mRNAs, stabilized virus transcripts. Thus, sequence context of YTHDF2 m6A recognition influences transcript fate (Tsai et al. 2021). Remarkably, m6A was detected in RNA genomes of viruses that replicate exclusively in the cytoplasm (RSV, HMPV, PV, EV71, ZIKV; HCV, DENV, West Nile virus [WNV], and yellow fever virus [YFV]), including seasonal and pandemic CoVs, whose reproduction was suppressed by depletion of METTL3 or YTHDF1,3 cytoplasmic m6A readers (Burgess et al. 2021; Li et al. 2021). METTL3 catalytic activity was further required for efficient CoV RNA synthesis, protein accumulation, and replication (Burgess et al. 2021). Further work is needed to identify the specific m6A modification sites in virus RNAs and host RNAs that might influence CoV replication. In contrast, METTL3/14 restricted HCV protein expression and virus reproduction without changing RNA replication, whereas FTO stimulated HCV protein expression (Gokhale et al. 2016; Gonzales-van Horn and Sarnow 2017). ZIKV reproduction was also repressed by METTL3/14 and stimulated by FTO and ALKBH5 (Lichinchi et al. 2016b). How nuclear host m6A modification components accumulate in the cytoplasm and how they recognize virus RNA remain outstanding questions. One exciting possibility involves association with METTL3 with a virus RdRp (Hao et al. 2019).
Table 1.
Key ways in which host m6A modification components influence virus replication
Interfering with METTL3 also reduced gene expression of nuclear-replicating DNA viruses SV40, Ad, HBV, and herpesviruses, which produce m6A-containing mRNAs (Lavi and Shatkin 1975; Moss et al. 1977; Hesser et al. 2018; Imam et al. 2018; Tsai et al. 2018). Reader YTHDF2 is needed for SV40 replication and m6A acceptor site ablation reduced virus reproduction (Tsai et al. 2018). By binding to YTHDF2, ISG20 is recruited to m6A-containing HBV transcripts and stimulates their decay (Imam et al. 2020). Nuclear m6A-interacting factors concentrated at sites of nascent Ad RNA synthesis and METTL3 loss reduced late gene expression by deregulating viral RNA processing and reducing splicing efficiency (Price et al. 2020). How specific virus m6A acceptor sites impact the spectrum of discrete alternatively spliced isoforms remains unanswered. The m6A pathway becomes progressively less important for HSV-1 gene expression over time, as the virus ICP27 protein redistributes nuclear methyltransferase components into the cytoplasm, reducing RNA modification on host and virus mRNAs (Srinivas et al. 2021). This could represent another way HSV-1 limits host RNA processing, as most late virus RNAs do not contain introns. EBV transcriptome m6A modification stimulated viral latent gene expression in part by mRNA stabilization and repression of lytic genes via YTHDF1-stimulated RNA decay (Lang et al. 2019; Xia et al. 2021). While most lytic KSHV transcripts were m6A-modified (Tan et al. 2018), the host m6A machinery was shown to have a complex proviral and antiviral impact on viral gene expression depending on cell type (Hesser et al. 2018). By binding to a m6A-modified hairpin within the KSHV ORF50 mRNA, which encodes a potent virus regulatory protein (Baquero-Perez et al. 2019), the host Tudor SND1 protein stabilizes ORF50 mRNA and is essential for KSHV early gene expression.
Host antiviral immune defenses, including IFNB1 transcript accumulation and ISG expression (Table 1), are regulated by m6A (Shulman and Stern-Ginossar 2020; McFadden and Horner 2021). METTL3/14 depletion stimulated IFNB1 accumulation and inhibited DNA (HCMV and Ad) and RNA (VSV and IAV) virus reproduction, whereas ALKBH5 depletion restricted IFNB1 accumulation and stimulated HCMV reproduction (Rubio et al. 2018; Winkler et al. 2019). Cellular m6A modification components also regulate IFNB1 mRNA accumulation in uninfected cells exposed to dsDNA (Rubio et al. 2018). This established that responses to nonmicrobial dsDNA in uninfected cells, which shape host immunity and contribute to autoimmune disease, are regulated by enzymes controlling m6A epitranscriptomic changes. Indeed, IFNB1 mRNA is m6A-modified at specific sites, which regulates IFNB1 mRNA decay in HCMV-infected cells and IFNB1 mRNA biogenesis and decay in uninfected, dsDNA-treated cells (Rubio et al. 2018; Winkler et al. 2019). The m6A reader YTHDF3, together with eIF4G2 and PABPC1, stimulates FOXO3 mRNA translation, the product of which negatively regulates ISG transcription and stimulated VSV replication (Zhang et al. 2019). Many ISG mRNAs are m6A-modified, and this reportedly stimulates translation of a subset (McFadden et al. 2021). Indeed, YTHDF1,2,3 are cleaved by EV71 2a protease, and this has been proposed to antagonize ISG expression in infected cells (Kastan et al. 2021). Reduced m6A on mRNA encoding α-ketoglutarate dehydrogenase promoted RNA decay, limiting metabolite (itaconate) accumulation required for VSV replication (Liu et al. 2019b). Virus-induced alterations to host transcript m6A content influences their splicing or translation and regulates infection by Flaviviridae members DENV, WNV, ZIKV, and HCV (Gokhale et al. 2020).
Cytosine methylation and acetylation at C5 (m5C) and N4 positions (ac4C) regulate retrovirus reproduction (Squires et al. 2012; Li et al. 2017; Arango et al. 2018). MLV genomic RNA modification by m6A and m5C stimulate virus replication (Courtney et al. 2017; Eckwahl et al. 2020). HIV-1 transcripts contain m5C, and interfering with m5C nuclear methyltransferase NSUN2 dysregulated HIV-1 mRNA alternative splicing, correlated with reduced virus mRNA ribosome recruitment, and inhibited virus replication (Courtney et al. 2019). Similarly, ac4C enhanced mRNA translation and stability (Arango et al. 2018) and was detected on genomic HIV-1 virion RNA (McIntyre et al. 2018). Disrupting ac4C HIV mRNA acceptor sites without altering coding content reduced virus gene expression, and reducing expression of N-acetyltransferase 10 (NAT10), which installs ac4C, inhibited HIV-1 replication by increasing RNA decay (Tsai et al. 2020). The extent to which other RNA modifications (McIntyre et al. 2018; Wiener and Schwartz 2021) impact virus reproduction is an exciting research direction, potentially exposing new ways gene expression is shaped post-transcriptionally by physiological stress.
Appropriating host ribosomes in virus-infected cells
Mechanisms and regulation of 40S ribosome loading on virus mRNAs
All viral mRNAs must capture cellular ribosomes. This begins with 40S recruitment, a necessary, regulated translation initiation step. It is coordinated in eukaryotes by initiation factors (eIFs) that assemble a specialized ribonucleoprotein complex on the mRNA 5′ end to engage 40S loaded with methionine-charged initiator tRNA (met-tRNAi). Recognition of m7G-capped virus mRNAs typically relies on the host cap-binding protein eIF4E, which together with the large scaffold eIF4G1 and the RNA helicase eIF4A forms a heterotrimeric complex called eIF4F (Pelletier and Sonenberg 2019). Association of eIF3-bound 40S with eIF4F via binding to eIF4G1 enables 40S loading onto m7G-capped mRNA. Diverse virus strategies preserve cellular eIF4F accessibility and activity and subvert host defenses that curtail eIF4F activity (Fig. 2). To stimulate virus capped mRNA translation, repressive host eIF4E-binding protein family members including 4E-BP1 are inactivated. By interacting with eIF4E, 4E-BP1 stoichiometrically restricts eIF4E binding to eIF4G1. Accordingly, 4E-BP1 represses cap-dependent translation by limiting eIF4E binding to eIF4G and preventing (1) eIF4F assembly and (2) 40S loading onto capped mRNAs (Pelletier and Sonenberg 2019). Phosphorylation of 4E-BP1 by the Ser/Thr kinase mTORC1 liberates eIF4E from the 4E-BP1 repressor and allows tuning of cap-dependent translation to physiological cues that regulate mTORC1 (Fig. 2). The latter can be subverted to constitutively stimulate anabolic programs such as protein synthesis and restrict catabolic outcomes such as autophagy during acute infection and lytic virus growth (Rubio and Mohr 2019). Alternatively, it can be harnessed to balance whether an infection remains latent or lytic reproduction ensues, as interfering with 4E-BP1 inactivation by mTORC1 stimulates latent HSV-1 genomes in neurons to reactivate and commence their reproductive lytic cycle (Kobayashi et al. 2012b; Hu et al. 2019). As transient mTOR inhibition in axons stimulates reactivation (Kobayashi et al. 2012b), differential mRNA translation localized within axons is possibly required. Identifying these mRNAs will further our mechanistic understanding of how HSV sustains and transitions out of latency in neurons.
Figure 2.
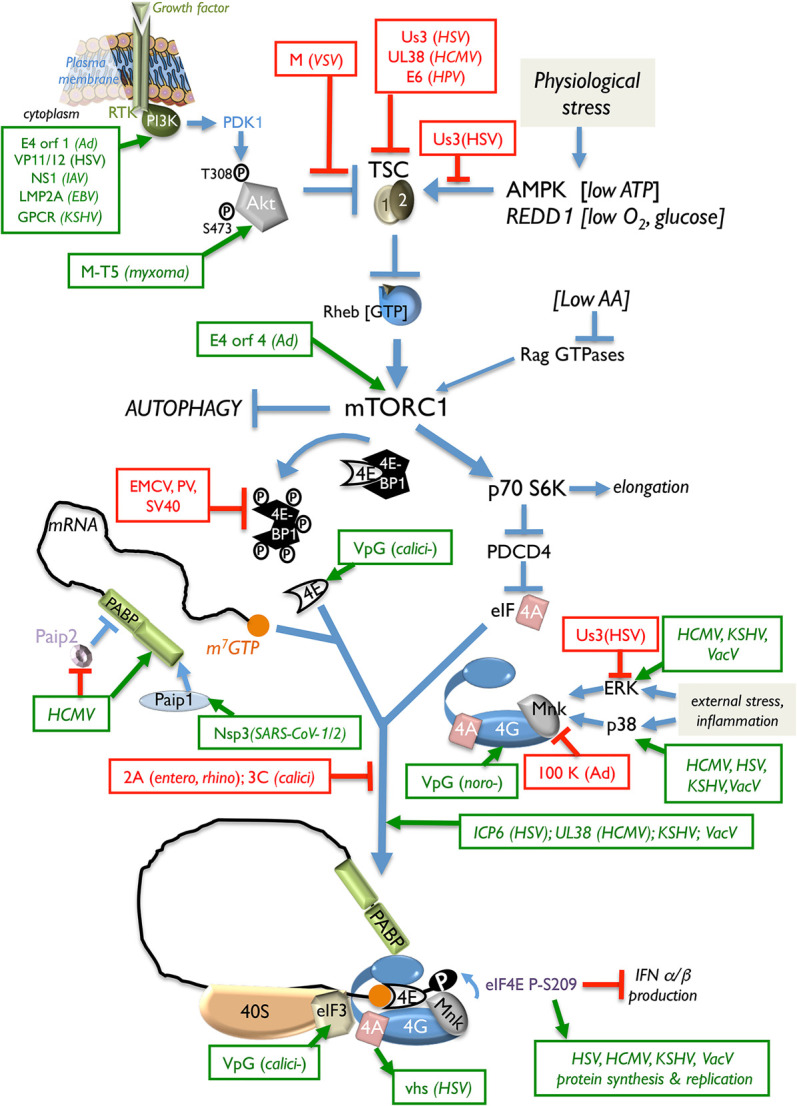
Controlling cap-dependent 40S ribosome loading in virus-infected cells. Sampling extracellular and intracellular cues (growth factor, nutrients, O2, and energy) allows 40S ribosome loading onto capped mRNAs to be responsive to changing environmental and physiological conditions, including virus infection. Receptor tyrosine kinase (RTK) signaling through PI3-kinase (PI3-K) and PDK1 stimulates Akt phosphorylation (T308 and S473). Once activated, Akt represses the tuberous sclerosis complex (TSC1/2), which results in Rheb-GTP accumulation and mTORC1 activation. Physiological stress (energy, O2, or nutrient insufficiency) prevents mTORC1 activation. The translational repressor 4E-BP1 regulates the availability of the cap-binding protein eIF4E. Hyperphosphorylation and inactivation of 4E-BP1 releases eIF4E, which interacts with eIF4G and the DEAD-box helicase eIF4A to assemble the multisubunit eIF4F complex on m7G-capped mRNA. Regulated eIF4F assembly controls eIF3-bound 40S loading onto capped mRNA. Virus-encoded effectors that stimulate (green) or repress (red) the indicated host factors to control 40S loading and translation initiation are shown. (SV40) Simian virus 40, (calici) calicivirus, (noro) norovirus, (entero) enterovirus, (rhino) rhinovirus.
Many DNA and RNA virus effectors co-opt host PI3-kinase–Akt–mTORC1 signaling in part to antagonize 4E-BP1 and stimulate translation of capped virus mRNAs (Fig. 2). Herpesviruses, adenoviruses, poxviruses, paramyxoviruses, and orthomyxoviruses (IAV) constitutively stimulate mTORC1 during their lytic reproductive cycle (O'Shea et al. 2005; Werden et al. 2007; Moorman et al. 2008; Walsh et al. 2008; Arias et al. 2009; Chuluunbaatar et al. 2010; Hale et al. 2010; Kuss-Duerkop et al. 2017; Zhan et al. 2020). By mimicking the cellular kinase Akt, the HSV-1 Ser/Thr kinase Us3 inactivates the host tuberous sclerosis complex (TSC) via directly phosphorylating TSC2 on Akt target sites (Chuluunbaatar et al. 2010). Us3 further disrupts host AMPK-dependent responses to energy insufficiency by preventing TSC2-dependent mTORC1 inhibition by AMPK (Vink et al. 2017). Additionally, Us3 uncouples mTORC1 activation from amino acid sufficiency signals (Vink et al. 2018). Both of these functions enable sustained mTORC1 activation and virus replication during physiological stress. Inactivation of 4E-BP1 is accompanied by virus-induced eIF4F assembly, which proceeds via distinct mechanisms. The HSV-1 ICP6 protein N terminus interacts with the eIF4G N terminus, enhances eIF4E binding to eIF4G, and stimulates virus mRNA translation (Walsh and Mohr 2006). While cytoplasmic PABP availability in HSV-1-infected cells is reduced by its nuclear accumulation, the viral ICP27 RNA-binding protein stimulates 40S recruitment in a PABP- and eIF4G-dependent manner. By recruiting PABP, which in turn interacts with eIF4G, ICP27 stimulates translation of capped mRNAs, imitating a mechanism used by cellular RNA-BP Dazl (deleted in azoospermia-like) (Smith et al. 2017). In contrast, eIF and PABP abundance increases upon infection with HCMV, a related herpesvirus from a distinct subfamily. Unlike HSV-1 infection, host protein synthesis proceeds, and the genome-wide translational landscape is remodeled by HCMV. Differentially translated cellular mRNAs have been identified that stimulate virus growth or host defenses (McKinney et al. 2014; Tirosh et al. 2015). RNA structure remodeling among regulated host genes in part confers HCMV infection responsiveness (Mizrahi et al. 2018) along with mTORC1 activation by the viral UL38 protein, which stimulates cap-dependent translation of mRNAs containing a terminal oligopyrimidine (TOP) sequence element including PABP (McKinney et al. 2012, 2014). Reduced eIF4F assembly and virus growth were observed by interfering with HCMV-induced PABP1 accumulation (McKinney et al. 2012). By raising PABP abundance, HCMV overcomes an unexpected host antiviral response that increases PABP-interacting protein 2 (Paip2) levels, which inhibits PABP binding to eIF4G and poly(A) RNA (McKinney et al. 2013). PABP is also targeted in SARS-CoV-1/2 replicon transfected cells by nsp3, which interacts with PABP-interacting protein-1 (Paip1) and 40S/80S ribosomes to enhance virus but not host protein synthesis (Lei et al. 2021). In lieu of controlling eIF abundance, DNA viruses that replicate in the cytoplasm (VacV and ASFV) sequester eIF4E, eIF4G, and PABP within and around discrete replication compartments (RCs) to increase their effective local concentration (Katsafanas and Moss 2007; Walsh et al. 2008; Castelló et al. 2009; Zaborowska et al. 2012). Cells infected with mammalian orthoreovirus, a dsRNA virus, also accumulate eIFs in cytoplasmic RCs (Desmet et al. 2014). How these factors concentrate within specific cytoplasmic regions is unknown and might inform mechanisms underlying local mRNA translation.
The host eIF4G-associated Ser/Thr kinase Mnk-1, which is activated by ERK and p38, influences infected cell protein synthesis by phosphorylating eIF4E. Binding of eIF4E to eIF4G delivers Mnk-1 to its substrate eIF4E, stimulating eIF4E S209 phosphorylation (Fig. 2). Situated near the eIF4E cap-binding pocket, S209 phosphorylation weakened cap binding affinity, (Scheper et al. 2002; Zuberek et al. 2004; Slepenkov et al. 2006), possibly accelerating cap release during initiation. The mechanism underlying how eIF4E phosphorylation regulates translation of a subset of mRNAs involved in proliferation, circadian rhythms, stress response, inflammation, and memory formation remains elusive (Furic et al. 2010; Herdy et al. 2012; Cao et al. 2015; Bramham et al. 2016; Proud 2019). Many viruses (large DNA viruses, CoV, and flavi, noro, and paramyxo viruses) promote eIF4E phosphorylation by stimulating ERK and/or p38, which activates Mnk1 (Mizutani et al. 2004; Walsh and Mohr 2004, 2006; Walsh et al. 2005, 2008; Royall et al. 2015; Roth et al. 2017; Proud 2019; Zhan et al. 2020), which stimulates translation of virus mRNAs (Walsh and Mohr 2004) and mRNA encoding the NF-κB inhibitor IκB (Herdy et al. 2012). Mnk1 recruitment by eIF4F is regulated by eIF3 subunit e, and eIF4E phosphorylation is eIF3e-dependent, consistent with eIF4F assembly preceding eIF4E phosphorylation. It further illustrates how modifying a cap recognition complex in response to eIF3-bound 40S loading regulates mRNA translation. (Walsh and Mohr 2014). Although how eIF4E phosphorylation influences selective mRNA translation is unknown, RNA binding activities displayed by eIF3 subunits (Hinnebusch 2006) could play a role. Reduced IκB mRNA translation, NF-κB activation, and IFN production result when unphosphorylated eIF4E accumulates in infected cells (Ad and many RNA viruses) (Jan et al. 2016). At late times in Ad-infected cells, binding of Ad 100K to eIF4G displaces Mnk1 and results in unphosphorylated eIF4E accumulation (Cuesta et al. 2004), which correlates with reduced host cell mRNA translation while allowing high-level virus mRNA translation via a cap-dependent, noncanonical ribosome shunting mechanism discussed below. Exiting virus latency is also stimulated by eIF4F assembly and eIF4E phosphorylation, as inhibiting the eIF4E kinase Mnk1 reduced accumulation of the KSHV transactivator RTA needed for lytic replication (Arias et al. 2014). Thus, whether latent infection persists or productive, lytic reproduction is triggered can be determined by critical cell signaling pathways (MAPK and PI3K–Akt–mTOR) that regulate translation.
Once loaded onto capped mRNA, 40S-containing complexes search for the AUG start codon by translocating along the 5′ UTR in an ATP-dependent process termed “scanning” (Merrick and Pavitt 2018). Ribosome shunting cis-elements that mediate nonlinear 40S translocation, whereby a 5′ UTR section is bypassed and scanning resumed downstream, have been identified in virus (Ad, HPV, and HBV) and host (hsp70) mRNAs (Kwan and Thompson 2019). Shunting supports initiation on capped mRNAs, which typically requires 5′ UTR unwinding by eIF4A, when initiation is suppressed by stress such as heat shock or infection that interferes with eIF4F. Ad 100K protein facilitates shunting by binding to the 5′ noncoding region of virus late mRNA (called the tripartite leader) and eIF4G, which in turn enhances PABP and 40S loading (Xi et al. 2004). A different cis-element surrounding the AUG codon mediates translation initiation on leaderless mRNAs (TILM) that are capped and have very short or no 5′ UTRs. HPV E6 oncoprotein expression uses TILM-directed initiation, which requires the cap structure, eIF4E, and eIF4A1 and could drive E6 production by cancer cells (García et al. 2021).
While eIF4E stimulates translation, a related host cap recognition protein, 4EHP, directs transcript-specific repression. Up-regulation of miR34a, which targets IFNB1, in RNA virus-infected cells, results in 4EHP-dependent translational repression of IFNβ (Zhang et al. 2021). Besides demonstrating 4EHP's role in translational silencing and cell-intrinsic immunity, this illustrates how distinct cellular cap-binding proteins manipulate infection outcomes.
In lieu of stimulating eIF4F, certain viruses obstruct eIF4F, which restricts host cap-dependent translation to achieve host shutoff and requires alternative strategies to initiate translation on virus mRNAs. Many RNA viruses that replicate in the cytoplasm rely on cis-elements termed internal ribosome entry sites (IRESs) to load 40S subunits onto viral mRNA in a cap-independent manner (Jaafar and Kieft 2019; Stern-Ginossar et al. 2019). IRES-directed initiation allows virus mRNAs to evade host defenses that repress cap-dependent translation, such as 4E-BP1; prevent synthesis of host antiviral proteins; and ensure selective virus mRNA translation proceeds while host protein synthesis is impaired. Comprised of stable and dynamic RNA structures that form 40S high-affinity ligands, IRESs are classified by structure, initiation factor requirements, and initiation mechanism (Jan et al. 2016; Johnson et al. 2017; Jaafar and Kieft 2019; Stern-Ginossar et al. 2019; Arhab et al. 2020). Type 1 and 2 IRESs are larger and require nearly all eIFs except eIF4E. IRES transacting factors (ITAFs) that remodel RNA structure are also required. Initiation by type 1 or 2 IRESs depends on binding to full-length eIF4G (EMCV) or an eIF4G fragment lacking the N-terminal eIF4E-binding domain (PV) that interacts with eIF4A. Whereas type 1 IRESs recruit 40S upstream of coding regions and scan to locate the start codon, type 2 IRESs secure the initiation complex to the start codon without scanning (Yu et al. 2011a; Sweeney et al. 2014). Structurally similar to type 1 and 2 IRESs, the hepatitis A virus (HAV) type 3 IRES requires eIF4E binding to eIF4G, which increases IRES binding and stimulates eIF4A unwinding (Avanzino et al. 2017). Despite needing all eIF4F subunits unlike other IRESs, HAV RNAs are not capped, precluding a role for cap recognition by eIF4E. Somehow, eIF4E binding to eIF4G stimulates high-affinity binding of eIF4G to the HAV IRES and facilitates IRES structural remodeling (Avanzino et al. 2017). This could involve eIF4E altering eIF4G conformation or impacting the interaction of eIF4F with the HAV IRES (Ali et al. 2001; Borman et al. 2001). Type 4 IRESs (HCV and CSFV) directly bind 40S and eIF3, displacing eIF3 from its normal position on 40S, altering 40S conformation, and positioning the AUG without scanning (Spahn et al. 2001; Siridechadilok et al. 2005; Hashem et al. 2013; Quade et al. 2015). The HCV IRES also associates with the 40S subunit of a translating 80S ribosome without disrupting protein synthesis, and this captured 40S is likely hijacked for subsequent IRES-directed initiation (Yokoyama et al. 2019). Delivery of met-tRNAi either by eIF2 or by eIF2A or eIF2D enables IRES function during physiological stress when canonical eIF2-mediated delivery is impaired (discussed later). Uniquely dependent on DDX29, which likely remodels a stem– loop that sequesters the initiating AUG, the aichivirus (AV) type 5 IRES eIF4G-binding domain is structurally distinct from type 1 and 2 IRESs (Yu et al. 2011b). In contrast, dicistroviruses such as CrPV contain two IRESs. The 5′ UTR IRES requires eIF3 but docks very differently with 40S subunits compared with type 4 IRESs (Neupane et al. 2020). The CrPV intergenic region (IGR) type 6 IRES, however, does not require eIFs for 40S binding and 80S assembly, can initiate translation from a non-AUG codon (Wilson et al. 2000; Muhs et al. 2015; Murray et al. 2016), and repositions some ribosomes to bypass 12 codons and resume +1 frame translation at a non-AUG codon (Kerr et al. 2018). Dicistrovirus IGR IRESs share similar structures consisting of a ribosome recruitment domain and a smaller domain containing pseudoknot I (PK I). Unlike canonical initiation, where the ribosome P-site is occupied by Met-tRNAi and elongator tRNAs load into the A-site, the IGR IRES PK I-containing domain docks into the P-site. While CrPV IRES codon–anticodon mimicking PK initially occupies the ribosome A-site, 40S rotation and eEF2-dependent translocation into the P-site are required to expose the A-site for elongation to commence (Costantino et al. 2008). By imitating an intermediate ribosomal state with hybrid tRNAs, IRES PK I from Israeli acute paralysis virus (IAPV) within the A-site blocks eIF1/eIF1A binding and promotes 60S joining followed by codon translocation to the P-site in a related elongation factor recruitment strategy (Costantino et al. 2008; Acosta-Reyes et al. 2019). A simpler mechanism used by Halastavi árva virus positions the IRES PK into the P-site, averting the need for eEF2-mediated translocation prior to commencing decoding (Abaeva et al. 2020).
Besides stimulating selective virus mRNA translation, IRES-directed translation proceeds when viral functions inhibit cap-dependent 40S loading. Thus, by subverting normal host translation regulatory circuits, viruses impose a potent host shutoff where virus IRES-mediated translation proceeds while host protein synthesis is suppressed (Fig. 2). Hypophosphorylated 4E-BP1 in PV- or EMCV-infected cells limits eIF4E binding to eIF4G, inhibiting eIF4F assembly (Gingras et al. 1996). By cleaving eIF4G to sever the eIF3- and eIF4A-binding segment from the N-terminal eIF4E-binding fragment, PV 2A proteinase selectively disables eIF4E-dependent 40S loading onto host capped transcripts (Gradi et al. 1998). Inhibition of host cap-dependent translation was better correlated with virus proteinase cleavage of eIF4G3 (formerly eIF4GII) rather than eIF4G1 (formerly eIF4GI), although eIF4G3 was less sensitive to 2A cleavage than eIF4G1 (Gradi et al. 1998). Cleavage of eIF4G by PV or group A rhinovirus 2A protease is stimulated by eIF4E (Aumayr et al. 2017; Avanzino et al. 2017). The 3C proteinase produced by PV and EMCV also cleaves host PABP1 (Rivera and Lloyd 2008; Kobayashi et al. 2012a). Finally, EV71 infection stimulates cellular microRNA miR-141 expression to diminish eIF4E levels and curb host protein synthesis (Ho et al. 2011).
Using a termination–reinitiation mechanism (Kronstad et al. 2013, 2014; Royall and Locker 2016), some viruses express multiple proteins from a polycistronic transcript. Following upstream ORF translation, a termination upstream ribosomal binding site (TURBS) in calicivirus RNAs retains 40S post-termination. TURBS promote reinitiation at a nearby AUG or non-AUG codon via 18S rRNA base pairing (Luttermann and Meyers 2014; Royall and Locker 2016). Reinitiation in vitro requires eIF2, eIF1, and eIF1A but not eIF3 (Zinoviev et al. 2015).
Preserving initiator tRNA loading onto 40S subunits and subverting the integrated stress response
By detecting nutrient insufficiency, proteostasis deficiencies, and virus infection, the integrated stress response (ISR) allows mammalian cells to re-establish homeostasis by reprogramming gene expression (Costa-Mattioli and Walter 2020). Once new protein synthesis initiation is arrested by stress, specialized mRNA translation produces proteins required to implement a new transcription program that restores homeostasis. Virus models have illuminated how translation initiation is globally repressed by stress and the fundamental importance of this process to innate immune responses.
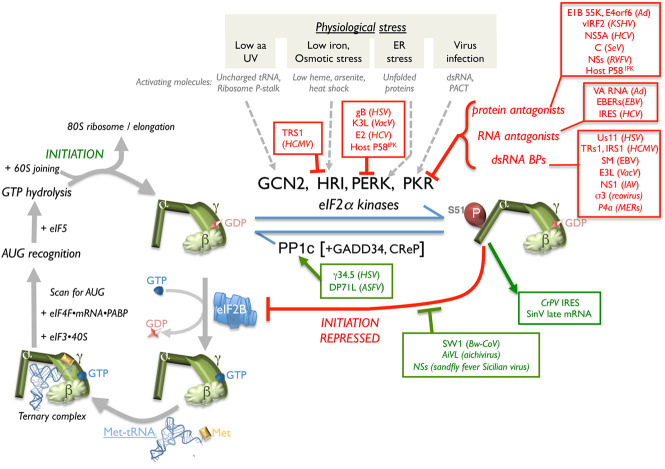
Four mammalian Ser/Thr kinases, each activated by discrete physiological stressors, can globally inhibit protein synthesis by preventing 40S ribosomes from acquiring met-tRNAi required to initiate translation from most mRNAs (Fig. 3). This is achieved by phosphorylating the α subunit of eIF2, a heterotrimeric initiation factor that assembles and delivers a ternary complex (TC) comprised of eIF2•GTP•met-tRNAi to the 40S subunit. Upon AUG start codon recognition by TC-loaded 40S, GTP hydrolysis stimulated by eIF5 promotes 60S joining and translation elongation. Recycling eIF2•GDP into the active, GTP-bound form requires the guanine nucleotide exchange factor (GEF) eIF2B (Fig. 3). Phosphorylated eIF2, however, binds eIF2B with high affinity and inhibits its GEF activity (Fig. 3). Initiation is arrested as eIF2B is present in limiting amounts, allowing small changes in phospho-eIF2 to have large effects on protein synthesis (Jan et al. 2016; Adomavicius et al. 2019; Stern-Ginossar et al. 2019). Phosphorylation of eIF2α also induces stress granule formation—cytoplasmic foci containing mRNA, RNA-BPs, eIFs, and 40S RPs whose roles in infection have been reviewed (McCormick and Khaperskyy 2017; Gaete-Argel et al. 2019).
Figure 3.
Viral strategies to preserve initiator tRNA loading onto 40S subunits. (Left panel) eIF2 is a GTP-binding initiation factor comprised of three subunits (α, β, and γ) that loads eIF3-bound 40S with Met-tRNAi. After recruitment of mRNA containing eIFs and AUG identification, eIF5-dependent GTP hydrolysis is followed by 60S joining and initiation, and subsequent 80S-mediated elongation. The GEF activity of the eIF2B pentamer–dimer recycles inactive eIF2•GDP to the active GTP-bound form. S51 eIF2α phosphorylation precludes initiation by associating with and inhibiting eIF2B, obstructing GDP-GTP exchange. eIF2α phosphorylation is controlled by four eIF2α kinases, each activated by distinct biological stress, and PP1c with its cognate constitutive (CrEP) or inducible (GADD34) regulatory subunit. Virus-encoded functions that antagonize (red) host eIF2α kinases, stimulate PP1, or support translation despite phospho-eIF2 accumulation (green) are indicated. (aa) Amino acid.
While all eIF2α kinases may impact infection biology, the dsRNA-activated protein kinase PKR is a universal threat to animal viruses and a critical component of cell-intrinsic immune responses. Constitutively present but not activated, PKR is one of a suite of IFN-induced proteins needed to establish an antiviral state refractory to virus reproduction. PKR is activated by dsRNA and also by a protein, PACT (Stern-Ginossar et al. 2019). To neutralize PKR, viruses may produce dsRNA BPs to camouflage dsRNA from cellular sensors, degrade PKR as in Rift Valley fever virus (RVFV)-infected cells (Mudhasani et al. 2016), encode PKR inhibitory proteins, or produce a ncRNA (Ad VA-RNA) that associates with PKR to prevent kinase dimer formation needed for activity (Fig. 3; Jan et al. 2016; Stern-Ginossar et al. 2019). Similarly, dsRNA regions within host circular (circ) RNAs have been proposed to bind PKR and prevent its activation. Significantly, synthetic dsRNA exposure or EMCV infection triggered OAS/RNase L activation and rapid circRNA degradation (Liu et al. 2019a).
Combinatorial tactics incorporating multiple viral effectors shield eIF2 from attack by other eIF2α kinases activated by stresses distinct from dsRNA. HSV-1 (γ34.5) and African swine fever virus (DP71L) encode a protein phosphatase 1α (PP1α) regulatory subunit, which engages the cellular PP1 catalytic subunit (PP1c) and broadly counteracts eIF2α kinases by dephosphorylating phospho-eIF2α (Rojas et al. 2015; Barber et al. 2017). Specific eIF2α kinase antagonists including the HSV-1 Us11 dsRNA BP, which inhibits PKR, and glycoprotein B, which restricts PERK activation, synergize with the γ34.5 PP1α regulatory subunit to limit eIF2α phosphorylation via discrete mechanisms. Using the viral eIF2α pseudosubstrate K3L (Sood et al. 2000; Seo et al. 2008) and the dsRNA BP E3L, which limits PKR activation, poxviruses also restrict eIF2α phosphorylation using independent effectors. Viral eIF2α kinase antagonists also inhibit multiple kinases, like HCMV TRS1, which inhibits PKR and HRI (Vincent et al. 2017). Cellular p58IPK induction by stress also reportedly prevents PKR, PERK, and GCN2 activation (Roobol et al. 2015).
The extent of genome coding capacity and deployment of viral antagonists to preserve eIF2 activity emphasizes its importance in the virus reproductive cycle. Mutant viruses lacking functions to challenge host dsRNA-responsive defenses such as PKR are often hypersensitive to IFN, are attenuated (Mulvey et al. 2004; White and Jacobs 2012; Liu et al. 2015), and display an altered host range (Haller et al. 2014; Carpentier et al. 2016; Peng et al. 2016; Cao et al. 2020; Park et al. 2021). Host eIF2α kinase activation may at times benefit virus replication. By promoting type I IFN receptor degradation, which precludes IFN responses (Liu et al. 2009), activation of unfolded protein response and PERK in infected cells supports HCV and VSV replication. Conversely, RNA viruses CrPV and SINV dispense with any eIF2 requirement by relying on cis-elements that direct eIF2-independent initiation (Wilson et al. 2000; Spahn et al. 2004; Kerr et al. 2016; Sanz et al. 2019). How cellular proteins and RNA structures regulate and contribute to eIF2-independent initiation in CrPV- and SINV-infected cells remains actively investigated.
A distinctive mechanism that attacks eIF2B•phospho-eIF2 interaction dynamics, an integral ISR feature, was identified in RNA virus-infected cells. While unrelated in sequence or predicted structure, the beluga whale CoV (Bw-CoV SW1) multifunctional AcP10 protein and the AiVL protein (Fig. 3) encoded by human aichivirus, a picornavirus that infects the GI tract, prevent phospho-eIF2 from binding to the eIF2B GEF (Rabouw et al. 2020). Both viral factors, however, did not interfere with eIF2B binding to unphosphorylated eIF2 (Rabouw et al. 2020). This enables AiVL and AcP10 to antagonize the ISR. In contrast, nonstructural protein s (NSs), encoded by the arthropod transmitted sandfly fever Sicilian virus, associates with and modifies eIF2B such that the NSs–eIF2B complex resists inhibition by phospho-eIF2 (Wuerth et al. 2020). Future studies are needed to reveal how NSs association with eIF2B subunits achieves this and whether any host cell factor is capable of a related feat.
Ribosomal proteins and quality control processes regulate infected cell protein synthesis
While long considered as invariant mRNA decoding machines, ribosome composition and ribosome protein (RP) abundance and stoichiometry in different cells and tissues may be heterogeneous and can regulate gene expression (Shi et al. 2017). Indeed, ribosome catalytic activity tolerates loss of several RPs, and specific RP requirements have been identified for discrete host mRNAs (Xue et al. 2015). Individual RPs also shape host responses to infection by regulating MHC class I peptide generation (Wei et al. 2019). Post-translational modifications (PTMs) of RPs and rRNAs and association with noncanonical RPs accentuate ribosome heterogeneity. Recent attention has focused on how virus infection impacts ribosomes and their role in regulating, as opposed to executing, protein synthesis (Fig. 4).
Figure 4.
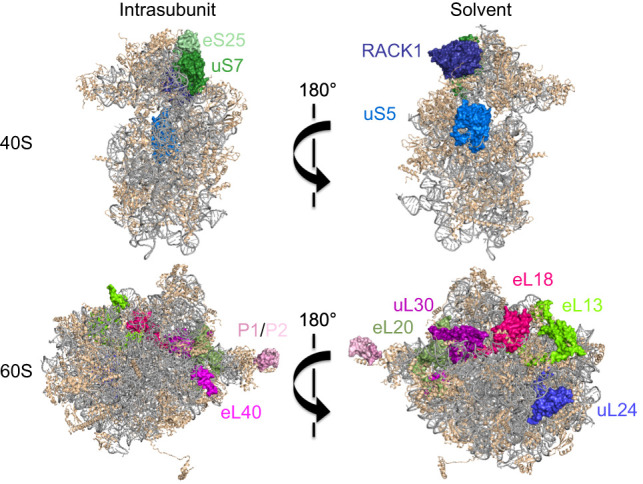
Ribosome proteins (RPs) that regulate translation in virus-infected cells. The figure depicts solvent and intrasubunit surfaces of 40S and 60S subunits with ribosomal RNA (gray) and RPs (tan). Highlighted RPs represent RPs important for virus IRES-mediated translation (green) or proviral functions (pink), or post-translationally modified during viral infection (blue). (Ribosome structure derived from PDB entry 4v88; figure based on data from Ben-Shem et al. [2011]).
To translate their mRNAs, viruses may be more reliant on RPs that are not essential for bulk host protein synthesis. Virus IRESs require a different RP subset from transcripts that rely on cap-dependent initiation (Fig. 4). Structurally and functionally diverse IRESs require eS25 (RPS25) for 40S–IRES complex formation (Landry et al. 2009). Largely dispensable for cap-dependent translation, eS25 sits within the ribosomal E-site with a projection extending toward the P-site. CrPRV IRES binding to 40S requires eS25, as does an ensuing, stabilizing conformation change (Walters et al. 2020). Ribosome shunting also requires eS25 (Hertz et al. 2013). Host 40S RP RACK1 contributes to PV, EMCV, CrPV, and HCV IRES-mediated translation (Majzoub et al. 2014; LaFontaine et al. 2020). Insufficient RACK1 levels reduced PV plaque size and impaired virion release (LaFontaine et al. 2020). The HCV IRES binds to eL20 (RPL18A) and uS7 (RPS5) (Dhar et al. 2006), the latter required for IRES function (Fukushi et al. 2001; Bhat et al. 2015). Tethered to IRESs by binding to DDX3, eL13 (RPL13) stimulates translation from FMDV, SVV, and CSFV IRESs with minimal impact on host translation (Han et al. 2020).
While not essential for cellular and IRES-driven translation, eL40 (RPL40) is needed for cap-dependent translation of VSV, rabies, and measles transcripts (Lee et al. 2013). P1 (RPLP1) and P2 (RPLP2) bind 60S to form the ribosomal stalk, which is anchored by uL10 (RPLP0) and needed for DENV, YFV, and ZIKV replication, but dispensable for host translation (Campos et al. 2017). P1/P2 mitigates ribosome pausing on DENV RNA (Campos et al. 2020), and DENV NS1 protein interacts with and relocalizes eL18 (RPL18), eL20, and uL30 (RPL7), although how this influences translation is unclear (Cervantes-Salazar et al. 2015). While some virus mRNAs require specific RPs dispensable for general host translation, the extent to which diverse viruses might rely on discrete individual RPs or combinatorial RP subsets to differentially impact virus mRNA translation in varied cell types, tissues, or host/vector systems remains unknown and ripe for investigation. In contrast, protein synthesis in HSV1-infected compared with uninfected cells is far less dependent on host RPs. Under conditions where RP insufficiency limits translation and ribosome availability, the HSV-1 late protein VP22 cosediments with initiating and elongating ribosomes, promotes polysome accumulation, and enforces translation (Vink et al. 2021). How VP22 compensates for RP insufficiency, which could support protein synthesis despite cell type and stress-induced RP variations, requires further study.
While it remains conceivable although unproven that virus mRNA translation might rely on a ribosome subpopulation with a specific RP stoichiometry, virus infection can modify ribosomes to impart selectivity for specific transcripts. One way host ribosomes are modified is through virus-encoded, ribosome-associated proteins. IAV NS1 protein associates with ribosomes and stimulates initiation on mRNAs, except those with a dicistrovirus IGR IRES (Panthu et al. 2017). By triaging which transcripts access ribosomes, SARS-CoV-1/2 Nsp1 promote host shutoff and virus protein synthesis (Huang et al. 2011; Lokugamage et al. 2015). SARS-CoV-1 Nsp1 inhibits cap-dependent and IRES-mediated translation by binding 40S via interactions with uS3 (RPS3), uS5 (RPS2), and 18S rRNA helix h18, and preventing 60S joining (Kamitani et al. 2009). Moreover, Nsp1 locks the 40S into a conformation that prevents mRNA loading and preinitiation complex formation by binding eIF1 and eIF3j (Thoms et al. 2020; Yuan et al. 2020; Lapointe et al. 2021). The Nsp1 C terminus occupies the mRNA entry channel, inhibiting translation by preventing most host mRNAs, including those encoding immune defenses, from entering ribosomes (Schubert et al. 2020). The SL1 hairpin within the virus mRNA 5′ UTR interacts with and displaces Nsp1 (Shi et al. 2020; Mendez et al. 2021; Tidu et al. 2021) from the entry channel to selectively enable CoV mRNA translation. Not all host transcripts are excluded from ribosomes by Nsp1, as translation of mRNAs containing 5′ TOP elements is stimulated (Rao et al. 2021). Further study is needed to discern whether this mechanism shares similarity to the one used by CoV mRNAs.
Another way virus infection (VacV, HSV-1, and VSV) modifies ribosomes is through RP and ribosome-associated protein PTM, which could regulate infected cell protein synthesis (DiGiuseppe et al. 2020). The VacV protein kinase B1R phosphorylates uS5, a 40S RP located near the mRNA entry channel (Fig. 4), enabling VacV protein synthesis (Banham et al. 1993; Beaud et al. 1994; DiGiuseppe et al. 2020). By stimulating RACK1 (Fig. 4) phosphorylation, VacV remodels ribosome transcript selectivity. The resulting negative charge on RACK1 Ser/Thr residues within an extended loop increases the swivel motion of the 40S head domain and broadens the translational capacity of the human ribosome, enabling preferential translation of viral mRNAs containing A-rich 5′ UTRs (Jha et al. 2017; Rollins et al. 2021). Finally, ubiquitin fold modifier (UFM1) conjugation to uL24 (RPL26) (Fig. 4) is required for HAV translation (Kulsuptrakul et al. 2021).
Rather than altering core RP stoichiometry, PV, ZIKV, and DENV remodel the host polysome-associated protein landscape. While host factors that trigger RNA sequestration, RNA degradation, antiviral responses, and eIFs were displaced, proteins required for proline hydroxylation were recruited to polysomes in ZIKV- and DENV-infected cells. (Aviner et al. 2021) This enables cotranslational proline hydroxylation of the viral polyprotein, which is essential for transmembrane domain folding and topology and is required for replication (Aviner et al. 2021)
Virus mRNA translation is also regulated by ribosome quality control (RQC) surveillance. Upon identification of aberrant mRNAs, including those lacking in-frame stop codons due to premature poly(A) addition within coding sequences, or ribosome stall-inducing translation events, RQC triggers degradation of mRNA/nascent protein and recycles stalled ribosomes (Meydan and Guydosh 2021). Elevated protein synthesis in virus-infected cells may provoke ribosome stalling or stress responses resolved by RQC. The multifunctional host protein ABCE1, which also inhibits RNase L, is needed for translation termination and ribosome recycling and in part facilitates RQC (Anderson et al. 2019). Virus mRNA translation and replication in cells infected with (−)-strand RNA viruses (measles, mumps, and RSV) was more reliant on ABCE1 than host mRNAs. In response to detecting stall-inducing A-rich sequences on translating mRNA, ribosome-bound ZNF598 E3 ubiquitin ligase triggers RQC and restricts ribosome read-through by ubiquitination of select 40S RPs (Garzia et al. 2017; Juszkiewicz and Hegde 2017; Sundaramoorthy et al. 2017). Unexpectedly, ZNF598 E3 ligase and uS10 (RPS20) site-specific ubiquitination were required for poxvirus replication and translation of virus mRNAs, many of which contain an unusual A-rich 5′ UTR (DiGiuseppe et al. 2018; Sundaramoorthy et al. 2021). Stimulation of translation by ZNF598 is in contrast to its role in 80S stalling and RQC, although both might involve ZNF598-sensing poly(A) or A-rich sequences. Instead, ZNF598 might potentially impact scanning or possibly be repurposed to stimulate translation by poxviruses (DiGiuseppe et al. 2018). Virus infection might also induce ribosome collisions, and ZNF598-mediated RQC may rescue stalled ribosomes and recycle the subunits to stimulate virus mRNA translation (Sundaramoorthy et al. 2021). Further studies are needed to elucidate the underlying mechanism. Finally, disrupting RQC stimulates cyclic GMP–AMP synthase (cGAS), a cytosolic DNA sensor that reportedly binds to collided ribosomes in vitro (Wan et al. 2021). Increased cGAS activity triggers ISG expression, which restricts virus replication. One such IFN-induced protein, IFIT2, is repurposed by IAV to prevent ribosome pausing on host and virus AU-rich mRNAs that could trigger RQC (Tran et al. 2020).
Closing thoughts
By appropriating and subverting cellular RNA decay and modification pathways together with unconditional reliance on host ribosomes, viruses remain enduring models to probe how gene expression is controlled post-transcriptionally and remodeled by physiological stress. Fundamental findings revealed by virus infection have implications for post-transcriptional mechanisms impacting health and disease including roles for eIF2α kinase PKR in tuning protein synthesis to stress responses and new ways to subvert the ISR by phospho-eIF2α–eIF2B, the control of innate immune responses by m6A, how 40S ribosomes are captured by a structured RNA without initiation factors, and how the cellular mRNA landscape is globally remodeled during infection stress. Understanding these processes offers new therapeutic possibilities to treat virus infections. Variants of the dsRNA sensor OAS1 are known to protect against severe COVID19 (Soveg et al. 2021; Wickenhagen et al. 2021). Inhibition of PA endonuclease activity by baloxavir represents a first in class new influenza antiviral (Heo 2018). SARs-CoV-2 Nsp1•40S interactions may also prove amenable to antiviral drug development. Cellular decay factors, including the SKI and TRAMP-like complex, and m6A modification by METTL3 on which viruses depend but whose short-term inhibition is tolerated by the host also present druggable targets less likely to be overcome by virus resistance (Weston et al. 2020; Burgess et al. 2021; Ho et al. 2021; Kulsuptrakul et al. 2021). By tinkering with functions that regulate eIF2α phosphorylation, a safe, attenuated HSV-1 that preferentially replicates in cancer cells with impaired cell-intrinsic immune responses was engineered (Taneja et al. 2001). This unexpected property led to development of an approved immunotherapeutic oncolytic virus to treat melanoma (Ribas et al. 2017). Continued exploration of diverse animal virus models will undoubtedly drive our understanding of how infection and physiological stress regulate gene expression post-transcriptionally and fuel unanticipated future opportunities to treat a spectrum of unmet medical needs.
Acknowledgments
We apologize to colleagues whose work was not cited owing to space limitations. Work in the author's laboratories is supported by grants from the National Institute of Allergy and Infectious Diseases to H.M.B. (AI151436 and AI166638) and I.M. (AI073898 and AI152543), and from the National Institute of General Medical Sciences to I.M. (GM056927).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.349276.121.
Competing interest statement
The authors declare no competing interests.
References
- Abaeva IS, Vicens Q, Bochler A, Soufari H, Simonetti A, Pestova TV, Hashem Y, Hellen CUT. 2020. The Halastavi árva virus intergenic region IRES promotes translation by the simplest possible initiation mechanism. Cell Rep 33: 108476. 10.1016/j.celrep.2020.108476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abernathy E, Gilbertson S, Alla R, Glaunsinger B. 2015. Viral nucleases induce an mRNA degradation-transcription feedback loop in mammalian cells. Cell Host Microbe 18: 243–253. 10.1016/j.chom.2015.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta-Reyes F, Neupane R, Frank J, Fernández IS. 2019. The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs. EMBO J 38: e102226. 10.15252/embj.2019102226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adomavicius T, Guaita M, Zhou Y, Jennings MD, Latif Z, Roseman AM, Pavitt GD. 2019. The structural basis of translational control by eIF2 phosphorylation. Nat commun 10: 2136. 10.1038/s41467-019-10167-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed CS, Winlow PL, Parsons AL, Jopling CL. 2018. Eukaryotic translation initiation factor 4AII contributes to microRNA-122 regulation of hepatitis C virus replication. Nucleic Acids Res 46: 6330–6343. 10.1093/nar/gky262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali IK, McKendrick L, Morley SJ, Jackson RJ. 2001. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J Virol 75: 7854–7863. 10.1128/JVI.75.17.7854-7863.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DE, Pfeffermann K, Kim SY, Sawatsky B, Pearson J, Kovtun M, Corcoran DL, Krebs Y, Sigmundsson K, Jamison SF, et al. 2019. Comparative loss-of-function screens reveal ABCE1 as an essential cellular host factor for efficient translation of Paramyxoviridae and Pneumoviridae. MBio 10: e00826-19. 10.1128/mBio.00826-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, Hosogane M, Sinclair WR, Nanan KK, Mandler MD, et al. 2018. Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175: 1872–1886.e24. 10.1016/j.cell.2018.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhab Y, Bulakhov AG, Pestova TV, Hellen CUT. 2020. Dissemination of internal ribosomal entry sites (IRES) between viruses by horizontal gene transfer. Viruses 12: 612. 10.3390/v12060612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C, Walsh D, Harbell J, Wilson AC, Mohr I. 2009. Activation of host translational control pathways by a viral developmental switch. PLoS Pathog 5: e1000334. 10.1371/journal.ppat.1000334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, Holdorf M, Weissman JS, Ganem D. 2014. KSHV 2.0: a comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog 10: e1003847. 10.1371/journal.ppat.1003847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV. 2016. Ribosomal frameshifting and transcriptional slip-page: from genetic steganography and cryptography to adventitious use. Nucleic Acids Res 44: 7007–7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumayr M, Schrempf A, Üzulmez O, Olek KM, Skern T. 2017. Interaction of 2A proteinase of human rhinovirus genetic group A with eIF4E is required for eIF4G cleavage during infection. Virology 511: 123–134. 10.1016/j.virol.2017.08.020 [DOI] [PubMed] [Google Scholar]
- Avanzino BC, Fuchs G, Fraser CS. 2017. Cellular cap-binding protein, eIF4E, promotes picornavirus genome restructuring and translation. Proc Natl Acad Sci 114: 9611–9616. 10.1073/pnas.1704390114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviner R, Li KH, Frydman J, Andino R. 2021. Cotranslational prolyl hydroxylation is essential for flavivirus biogenesis. Nature 596: 558–564. 10.1038/s41586-021-03851-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayllon J, García-Sastre A. 2015. The NS1 protein: a multitasking virulence factor. Curr Top Microbiol Immunol 386: 73–107. [DOI] [PubMed] [Google Scholar]
- Banham AH, Leader DP, Smith GL. 1993. Phosphorylation of ribosomal proteins by the vaccinia virus B1R protein kinase. FEBS Lett 321: 27–31. 10.1016/0014-5793(93)80614-Z [DOI] [PubMed] [Google Scholar]
- Baquero-Perez B, Antanaviciute A, Yonchev ID, Carr IM, Wilson SA, Whitehouse A. 2019. The Tudor SND1 protein is an m6A RNA reader essential for replication of Kaposi's sarcoma-associated herpesvirus. Elife 8: e47261. 10.7554/eLife.47261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber C, Netherton C, Goatley L, Moon A, Goodbourn S, Dixon L. 2017. Identification of residues within the African swine fever virus DP71L protein required for dephosphorylation of translation initiation factor eIF2α and inhibiting activation of pro-apoptotic CHOP. Virology 504: 107–113. 10.1016/j.virol.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MD, Moon SL, Emch AW, Wilusz CJ, Wilusz J. 2013. Changes in cellular mRNA stability, splicing, and polyadenylation through HuR protein sequestration by a cytoplasmic RNA virus. Cell Rep 5: 909–917. 10.1016/j.celrep.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra R, Stark TJ, Clark E, Belzile JP, Wheeler EC, Yee BA, Huang H, Gelboin-Burkhart C, Huelga SC, Aigner S, et al. 2016. RNA-binding protein CPEB1 remodels host and viral RNA landscapes. Nat Struct Mol Biol 23: 1101–1110. 10.1038/nsmb.3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavagnoli L, Cucuzza S, Campanini G, Rovida F, Paolucci S, Baldanti F, Maga G. 2015. The novel influenza A virus protein PA-X and its naturally deleted variant show different enzymatic properties in comparison to the viral endonuclease PA. Nucleic Acids Res 43: 9405–9417. 10.1093/nar/gkv926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaud G, Sharif A, Topa-Masse A, Leader DP. 1994. Ribosomal protein S2/Sa kinase purified from HeLa cells infected with vaccinia virus corresponds to the B1R protein kinase and phosphorylates in vitro the viral ssDNA-binding protein. J Gen Virol 75: 283–293. 10.1099/0022-1317-75-2-283 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529. 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- Bercovich-Kinori A, Tai J, Gelbart IA, Shitrit A, Ben-Moshe S, Drori Y, Itzkovitz S, Mandelboim M, Stern-Ginossar N. 2016. A systematic view on influenza induced host shutoff. Elife 5: e18311. 10.7554/eLife.18311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat P, Shwetha S, Sharma DK, Joseph AP, Srinivasan N, Das S. 2015. The β hairpin structure within ribosomal protein S5 mediates interplay between domains II and IV and regulates HCV IRES function. Nucleic Acids Res 43: 2888–2901. 10.1093/nar/gkv110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick R, Mukherjee A, Patra U, Chawla-Sarkar M. 2015. Rotavirus disrupts cytoplasmic P bodies during infection. Virus Res 210: 344–354. 10.1016/j.virusres.2015.09.001 [DOI] [PubMed] [Google Scholar]
- Bidet K, Dadlani D, Garcia-Blanco MA. 2014. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog 10: e1004242. 10.1371/journal.ppat.1004242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T. 1995. Cloning and characterization of a RNase L inhibitor. A new component of the interferon-regulated 2–5A pathway. J Biol Chem 270: 13308–13317. 10.1074/jbc.270.22.13308 [DOI] [PubMed] [Google Scholar]
- Borman AM, Michel YM, Kean KM. 2001. Detailed analysis of the requirements of hepatitis A virus internal ribosome entry segment for the eukaryotic initiation factor complex eIF4F. J Virol 75: 7864–7871. 10.1128/JVI.75.17.7864-7871.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramham CR, Jensen KB, Proud CG. 2016. Tuning specific translation in cancer metastasis and synaptic memory: control at the MNK-eIF4E axis. Trends Biochem Sci 41: 847–858. 10.1016/j.tibs.2016.07.008 [DOI] [PubMed] [Google Scholar]
- Brinton MA, Fernandez AV, Dispoto JH. 1986. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 153: 113–121. 10.1016/0042-6822(86)90012-7 [DOI] [PubMed] [Google Scholar]
- Burgess HM, Mohr I. 2015. Cellular 5′-3′ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe 17: 332–344. 10.1016/j.chom.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess HM, Mohr I. 2018. Defining the role of stress granules in innate immune suppression by the herpes simplex virus 1 endoribonuclease vhs. J Virol 92: e00829-18. 10.1128/JVI.00829-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess HM, Depledge DP, Thompson L, Srinivas KP, Grande RC, Vink EI, Abebe JS, Blackaby WP, Hendrick A, Albertella MR, et al. 2021. Targeting the m6A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication. Genes Dev 35: 1005–1019. 10.1101/gad.348320.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos RK, Wong B, Xie X, Lu YF, Shi PY, Pompon J, Garcia-Blanco MA, Bradrick SS. 2017. RPLP1 and RPLP2 are essential flavivirus host factors that promote early viral protein accumulation. J VIrol 91: e01706-16. 10.1128/JVI.01706-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos RK, Wijeratne HRS, Shah P, Garcia-Blanco MA, Bradrick SS. 2020. Ribosomal stalk proteins RPLP1 and RPLP2 promote biogenesis of flaviviral and cellular multi-pass transmembrane proteins. Nucleic Acids Res 48: 9872–9885. 10.1093/nar/gkaa717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Gkogkas CG, de Zavalia N, Blum ID, Yanagiya A, Tsukumo Y, Xu H, Lee C, Storch KF, Liu AC, et al. 2015. Light-regulated translational control of circadian behavior by eIF4E phosphorylation. Nat Neurosci 18: 855–862. 10.1038/nn.4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Varga J, Deschambault Y. 2020. Poxvirus encoded eIF2α homolog, K3 family proteins, is a key determinant of poxvirus host species specificity. Virology 541: 101–112. 10.1016/j.virol.2019.12.008 [DOI] [PubMed] [Google Scholar]
- Carpentier KS, Esparo NM, Child SJ, Geballe AP. 2016. A single amino acid dictates protein kinase R susceptibility to unrelated viral antagonists. PLoS Pathog 12: e1005966. 10.1371/journal.ppat.1005966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelló A, Quintas A, Sánchez EG, Sabina P, Nogal M, Carrasco L, Revilla Y. 2009. Regulation of host translational machinery by African swine fever virus. PLoS Pathog 5: e1000562. 10.1371/journal.ppat.1000562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathcart AL, Rozovics JM, Semler BL. 2013. Cellular mRNA decay protein AUF1 negatively regulates enterovirus and human rhinovirus infections. J Virol 87: 10423–10434. 10.1128/JVI.01049-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Salazar M, Angel-Ambrocio AH, Soto-Acosta R, Bautista-Carbajal P, Hurtado-Monzon AM, Alcaraz-Estrada SL, Ludert JE, Del Angel RM. 2015. Dengue virus NS1 protein interacts with the ribosomal protein RPL18: this interaction is required for viral translation and replication in Huh-7 cells. Virology 484: 113–126. 10.1016/j.virol.2015.05.017 [DOI] [PubMed] [Google Scholar]
- Chaimayo C, Dunagan M, Hayashi T, Santoso N, Takimoto T. 2018. Specificity and functional interplay between influenza virus PA-X and NS1 shutoff activity. PLoS Pathog 14: e1007465. 10.1371/journal.ppat.1007465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HW, Watson JC, Jacobs BL. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci 89: 4825–4829. 10.1073/pnas.89.11.4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EG, Costantino DA, Rabe JL, Moon SL, Wilusz J, Nix JC, Kieft JS. 2014a. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 344: 307–310. 10.1126/science.1250897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EG, Moon SL, Wilusz J, Kieft JS. 2014b. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. Elife 3: e01892. 10.7554/eLife.01892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuluunbaatar U, Roller R, Feldman ME, Brown S, Shokat KM, Mohr I. 2010. Constitutive mTORC1 activation by a herpesvirus Akt surrogate stimulates mRNA translation and viral replication. Genes Dev 24: 2627–2639. 10.1101/gad.1978310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contu L, Balistreri G, Domanski M, Uldry AC, Mühlemann O. 2021. Characterisation of the Semliki forest virus-host cell interactome reveals the viral capsid protein as an inhibitor of nonsense-mediated mRNA decay. PLoS Pathog 17: e1009603. 10.1371/journal.ppat.1009603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Walter P. 2020. The integrated stress response: from mechanism to disease. Science 368: eaat5314. 10.1126/science.aat5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. 2008. tRNA–mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol 15: 57–64. 10.1038/nsmb1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney DG, Kennedy EM, Dumm RE, Bogerd HP, Tsai K, Heaton NS, Cullen BR. 2017. Epitranscriptomic enhancement of influenza a virus gene expression and replication. Cell Host Microbe 22: 377–386.e5. 10.1016/j.chom.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, Swanstrom R, Holley CL, Cullen BR. 2019. Epitranscriptomic addition of m5C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe 26: 217–227.e6. 10.1016/j.chom.2019.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta R, Xi Q, Schneider RJ. 2004. Structural basis for competitive inhibition of eIF4G-Mnk1 interaction by the adenovirus 100-kilodalton protein. J Virol 78: 7707–16. 10.1128/JVI.78.14.7707-7716.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber B, Saffran HA, Smiley JR. 2019. The herpes simplex virus host shutoff (vhs) RNase limits accumulation of double stranded RNA in infected cells: evidence for accelerated decay of duplex RNA. PLoS Pathog 15: e1008111. 10.1371/journal.ppat.1008111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E, Canard B. 2017. Biochemical principles and inhibitors to interfere with viral capping pathways. Curr Opin Virol 24: 87–96. 10.1016/j.coviro.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo RC, Groft CM, Rajashankar KR, Burley SK. 2002. Recognition of the rotavirus mRNA 3′ consensus by an asymmetric NSP3 homodimer. Cell 108: 71–81. 10.1016/S0092-8674(01)00632-8 [DOI] [PubMed] [Google Scholar]
- Desmet EA, Anguish LJ, Parker JSL. 2014. Virus-mediated compartmentalization of the host translational machinery. mBio 5: e01463-14. 10.1128/mBio.01463-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar D, Mapa K, Pudi R, Srinivasan P, Bodhinathan K, Das S. 2006. Human ribosomal protein L18a interacts with hepatitis C virus internal ribosome entry site. Arch Virol 151: 509–524. 10.1007/s00705-005-0642-6 [DOI] [PubMed] [Google Scholar]
- Dhillon P, Rao CD. 2018. Rotavirus induces formation of remodeled stress granules and P bodies and their sequestration in viroplasms to promote progeny virus production. J Virol 92: e01363-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon P, Tandra VN, Chorghade SG, Namsa ND, Sahoo L, Rao CD. 2018. Cytoplasmic relocalization and colocalization with viroplasms of host cell proteins, and their role in rotavirus infection. J Virol 92: e00612-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez J, Ishikawa M, Kaido M, Ahlquist P. 2000. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc Natl Acad Sci 97: 3913–3918. 10.1073/pnas.080072997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiuseppe S, Rollins MG, Bartom ET, Walsh D. 2018. ZNF598 plays distinct roles in interferon-stimulated gene expression and poxvirus protein synthesis. Cell Rep 23: 1249–1258. 10.1016/j.celrep.2018.03.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiuseppe S, Rollins MG, Astar H, Khalatyan N, Savas JN, Walsh D. 2020. Proteomic and mechanistic dissection of the poxvirus-customized ribosome. J Cell Sci 134: jcs246603. 10.1242/jcs.246603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doepker RC, Hsu WL, Saffran HA, Smiley JR. 2004. Herpes simplex virus virion host shutoff protein is stimulated by translation initiation factors eIF4B and eIF4H. J Virol 78: 4684–99. 10.1128/JVI.78.9.4684-4699.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD, White JP, Lloyd RE. 2011. Poliovirus-mediated disruption of cytoplasmic processing bodies. J Virol 85: 64–75. 10.1128/JVI.01657-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drappier M, Jha BK, Stone S, Elliott R, Zhang R, Vertommen D, Weiss SR, Silverman RH, Michiels T. 2018. A novel mechanism of RNase L inhibition: Theiler's virus L* protein prevents 2-5A from binding to RNase L. PLoS Pathog 14: e1006989. 10.1371/journal.ppat.1006989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckwahl M, Xu R, Michalkiewicz J, Zhang W, Patel P, Cai Z, Pan T. 2020. 5-methylcytosine RNA modifications promote retrovirus replication in an ALYREF reader protein-dependent manner. J Virol 94: e00544-20. 10.1128/JVI.00544-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espert L, Degols G, Gongora C, Blondel D, Williams BR, Silverman RH, Mechti N. 2003. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J Biol Chem 278: 16151–16158. 10.1074/jbc.M209628200 [DOI] [PubMed] [Google Scholar]
- Feng P, Everly DN Jr, Read GS. 2005. mRNA decay during herpes simplex virus (HSV) infections: protein–protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A. J Virol 79: 9651–9664. 10.1128/JVI.79.15.9651-9664.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarelli M, Wilson H, Pedro Galão R, Mazzon M, Antzin-Anduetza I, Marsh M, Neil SJ, Swanson CM. 2019. KHNYN is essential for the zinc finger antiviral protein (ZAP) to restrict HIV-1 containing clustered CpG dinucleotides. Elife 8: e46767. 10.7554/eLife.46767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel Y, Gluck A, Nachshon A, Winkler R, Fisher T, Rozman B, Mizrahi O, Lubelsky Y, Zuckerman B, Slobodin B, et al. 2021. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature 594: 240–245. 10.1038/s41586-021-03610-3 [DOI] [PubMed] [Google Scholar]
- Flynn RA, Belk JA, Qi Y, Yasumoto Y, Wei J, Alfajaro MM, Shi Q, Mumbach MR, Limaye A, DeWeirdt PC, et al. 2021. Discovery and functional interrogation of SARS-CoV-2 RNA–host protein interactions. Cell 184: 2394–2411.e16. 10.1016/j.cell.2021.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine KA, Leon KE, Khalid MM, Tomar S, Jimenez-Morales D, Dunlap M, Kaye JA, Shah PS, Finkbeiner S, Krogan NJ, et al. 2018. The cellular NMD pathway restricts zika virus infection and is targeted by the viral capsid protein. MBio 9: e02126-18. 10.1128/mBio.02126-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel CC, Whisnant AW, Djakovic L, Rutkowski AJ, Friedl MS, Kluge M, Williamson JC, Sai S, Vidal RO, Sauer S, et al. 2021. Dissecting herpes simplex virus 1-induced host shutoff at the RNA level. J Virol 95: e01399-20. 10.1128/JVI.01399-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao A, Fujiwara T. 2017. The coupled and uncoupled mechanisms by which trans-acting factors regulate mRNA stability and translation. J Biochem 161: 309–314. [DOI] [PubMed] [Google Scholar]
- Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino FB, Katayama K. 2001. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J Biol Chem 276: 20824–20826. 10.1074/jbc.C100206200 [DOI] [PubMed] [Google Scholar]
- Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, et al. 2010. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci 107: 14134–14139. 10.1073/pnas.1005320107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaete-Argel A, Márquez CL, Barriga GP, Soto-Rifo R, Valiente-Echeverría F. 2019. Strategies for success. Viral infections and membraneless organelles. Front Cell Infect Microbiol 9: 336. 10.3389/fcimb.2019.00336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglia MM, Rycroft CH, Glaunsinger BA. 2015. Transcriptome-wide cleavage site mapping on cellular mRNAs reveals features underlying sequence-specific cleavage by the viral ribonuclease SOX. PLoS Pathog 11: e1005305. 10.1371/journal.ppat.1005305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García A, Maldonado G, González JL, Svitkin Y, Cantú D, García-Carrancá A, Sonenberg N, Hernández G. 2021. High-risk human papillomavirus-18 uses an mRNA sequence to synthesize oncoprotein E6 in tumors. Proc Natl Acad Sci 118: e2108359118. 10.1073/pnas.2108359118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno M, Noerenberg M, Ni S, Järvelin AI, González-Almela E, Lenz CE, Bach-Pages M, Cox V, Avolio R, Davis T, et al. 2019. System-wide profiling of RNA-binding proteins uncovers key regulators of virus infection. Mol Cell 74: 196–211.e11. 10.1016/j.molcel.2019.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia A, Jafarnejad SM, Meyer C, Chapat C, Gogakos T, Morozov P, Amiri M, Shapiro M, Molina H, Tuschl T, et al. 2017. The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat Commun 8: 16056. 10.1038/ncomms16056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucherand L, Porter BK, Levene RE, Price EL, Schmaling SK, Rycroft CH, Kevorkian Y, McCormick C, Khaperskyy DA, Gaglia MM. 2019. The influenza a virus endoribonuclease PA-X usurps host mRNA processing machinery to limit host gene expression. Cell Rep 27: 776–792.e7. 10.1016/j.celrep.2019.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerts-Dimitriadou C, Lu YY, Geertsema C, Goldbach R, Kormelink R. 2012. Analysis of the tomato spotted wilt virus ambisense S RNA-encoded hairpin structure in translation. PLoS One 7: e31013. 10.1371/journal.pone.0031013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Svitkin Y, Belsham GJ, Pause A, Sonenberg N. 1996. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci 93: 5578–5583. 10.1073/pnas.93.11.5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaunsinger B, Ganem D. 2004. Highly selective escape from KSHV-mediated host mRNA shutoff and its implications for viral pathogenesis. J Exp Med 200: 391–398. 10.1084/jem.20031881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, et al. 2016. N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20: 654–665. 10.1016/j.chom.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NS, McIntyre ABR, Mattocks MD, Holley CL, Lazear HM, Mason CE, Horner SM. 2020. Altered m6A modification of specific cellular transcripts affects flaviviridae infection. Mol Cell 77: 542–555.e8. 10.1016/j.molcel.2019.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein SA, Thornbrough JM, Zhang R, Jha BK, Li Y, Elliott R, Quiroz-Figueroa K, Chen AI, Silverman RH, Weiss SR. 2017. Lineage A betacoronavirus NS2 proteins and the homologous torovirus berne pp1a carboxy-terminal domain are phosphodiesterases that antagonize activation of RNase L. J VIrol 91: e02201-16. 10.1128/JVI.02201-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales-van Horn SR, Sarnow P. 2017. Making the mark: the role of adenosine modifications in the life cycle of RNA viruses. Cell Host Microbe 21: 661–669. 10.1016/j.chom.2017.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez AC, Stempel M, Wyler E, Urban C, Piras A, Hennig T, Ganskih S, Wei Y, Heim A, Landthaler M, et al. 2021. The zinc finger antiviral protein ZAP restricts human cytomegalovirus and selectively binds and destabilizes viral UL4/UL5 transcripts. MBio 12: e02683-20. 10.1128/mBio.02683-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O'Meara MJ, Rezelj VV, Guo JZ, Swaney DL, et al. 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583: 459–468. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi A, Svitkin YV, Imataka H, Sonenberg N. 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci 95: 11089–11094. 10.1073/pnas.95.19.11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratia M, Sarot E, Vende P, Charpilienne A, Baron CH, Duarte M, Pyronnet S, Poncet D. 2015. Rotavirus NSP3 is a translational surrogate of the poly(A) binding protein–poly(A) complex. J Virol 89: 8773–8782. 10.1128/JVI.01402-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer AE, Hearing P, Ketner G. 2011. The adenovirus E4 11 k protein binds and relocalizes the cytoplasmic P-body component Ddx6 to aggresomes. Virology 417: 161–168. 10.1016/j.virol.2011.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Ma J, Sun J, Gao G. 2007. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci 104: 151–156. 10.1073/pnas.0607063104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbart M, Deng X, Baker SC. 2020. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci 117: 8094–8103. 10.1073/pnas.1921485117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale BG, Kerry PS, Jackson D, Precious BL, Gray A, Killip MJ, Randall RE, Russell RJ. 2010. Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein. Proc Natl Acad Sci 107: 1954–1959. 10.1073/pnas.0910715107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller SL, Peng C, McFadden G, Rothenburg S. 2014. Poxviruses and the evolution of host range and virulence. Infect Genet Evol 21: 15–40. 10.1016/j.meegid.2013.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JQ, Townsend HL, Jha BK, Paranjape JM, Silverman RH, Barton DJ. 2007. A phylogenetically conserved RNA structure in the poliovirus open reading frame inhibits the antiviral endoribonuclease RNase L. J Virol 81: 5561–5572. 10.1128/JVI.01857-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Sun S, Li P, Liu Q, Zhang Z, Dong H, Sun M, Wu W, Wang X, Guo H. 2020. Ribosomal protein L13 promotes IRES-driven translation of foot-and-mouth disease virus in a helicase DDX3-dependent manner. J VIrol 94: e01679-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Hao S, Chen H, Chen Z, Zhang Y, Wang J, Wang H, Zhang B, Qiu J, Deng F, et al. 2019. N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res 47: 362–374. 10.1093/nar/gky1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. 2013. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 503: 539–543. 10.1038/nature12658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. 2011. Structure of the lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc Natl Acad Sci 108: 2396–2401. 10.1073/pnas.1016404108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, MacDonald LA, Takimoto T. 2015. Influenza a virus protein PA-X contributes to viral growth and suppression of the host antiviral and immune responses. J Virol 89: 6442–6452. 10.1128/JVI.00319-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He PC, He C. 2021. m6A RNA methylation: from mechanisms to therapeutic potential. EMBO J 40: e105977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck AM, Wilusz J. 2018. The interplay between the RNA decay and translation machinery in eukaryotes. Cold Spring Harb Persepct Biol 10: a032839. 10.1101/cshperspect.a032839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JI, Goergen D, Zheng J, Song Y, Schüttler CG, Fehr C, Jünemann C, Niepmann M. 2008. MicroRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 27: 3300–3310. 10.1038/emboj.2008.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo YA. 2018. Baloxavir: first global approval. Drugs 78: 693–697. 10.1007/s40265-018-0899-1 [DOI] [PubMed] [Google Scholar]
- Herdy B, Jaramillo M, Svitkin YV, Rosenfeld AB, Kobayashi M, Walsh D, Alain T, Sean P, Robichaud N, Topisirovic I, et al. 2012. Translational control of the activation of transcription factor NF-κB and production of type I interferon by phosphorylation of the translation factor eIF4E. Nat Immunol 13: 543–550. 10.1038/ni.2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz MI, Landry DM, Willis AE, Luo G, Thompson SR. 2013. Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol Cell Biol 33: 1016–1026. 10.1128/MCB.00879-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesser CR, Karijolich J, Dominissini D, He C, Glaunsinger BA. 2018. N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi's sarcoma-associated herpesvirus infection. PLoS Pathog 14: e1006995. 10.1371/journal.ppat.1006995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. 2006. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci 31: 553–562. 10.1016/j.tibs.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Ho BC, Yu SL, Chen JJ, Chang SY, Yan BS, Hong QS, Singh S, Kao CL, Chen HY, Su KY, et al. 2011. Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 9: 58–69. 10.1016/j.chom.2010.12.001 [DOI] [PubMed] [Google Scholar]
- Ho JSY, Zhu Z, Marazzi I. 2021. Unconventional viral gene expression mechanisms as therapeutic targets. Nature 593: 362–371. 10.1038/s41586-021-03511-5 [DOI] [PubMed] [Google Scholar]
- Hogg JR, Goff SP. 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389. 10.1016/j.cell.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HL, Shiflett LA, Kobayashi M, Chao MV, Wilson AC, Mohr I, Huang TT. 2019. TOP2β-dependent nuclear DNA damage shapes extracellular growth factor responses via dynamic AKT phosphorylation to control virus latency. Mol Cell 74: 466–480.e4. 10.1016/j.molcel.2019.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. 2011. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. J Virol 85: 638–643. 10.1128/JVI.01806-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrina A, Jones C, Chen D, Clarkson S, Cochran N, Feucht P, Hoffman G, Lindeman A, Russ C, Sigoillot F, et al. 2019. A genome-wide CRISPR screen identifies ZCCHC14 as a host factor required for hepatitis B surface antigen production. Cell Rep 29: 2970–2978.e6. 10.1016/j.celrep.2019.10.113 [DOI] [PubMed] [Google Scholar]
- Imam H, Khan M, Gokhale NS, McIntyre ABR, Kim GW, Jang JY, Kim SJ, Mason CE, Horner SM, Siddiqui A. 2018. N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc Natl Acad Sci 115: 8829–8834. 10.1073/pnas.1808319115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam H, Kim GW, Mir SA, Khan M, Siddiqui A. 2020. Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified Hepatitis B virus transcripts. PLoS Pathog 16: e1008338. 10.1371/journal.ppat.1008338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imani F, Jacobs BL. 1988. Inhibitory activity for the interferon-induced protein kinase is associated with the reovirus serotype 1 σ3 protein. Proc Natl Acad Sci 85: 7887–7891. 10.1073/pnas.85.21.7887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iselin L, Palmalux N, Kamel W, Simmonds P, Mohammed S, Castello A. 2022. Uncovering viral RNA-host cell interactions on a proteome-wide scale. Trends Biochem sci 47: 23–38. 10.1016/j.tibs.2021.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafar ZA, Kieft JS. 2019. Viral RNA structure-based strategies to manipulate translation. Nat Rev Microbiol 17: 110–123. 10.1038/s41579-018-0117-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, et al. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337: 199–204. 10.1126/science.1222213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Mohr I, Walsh D. 2016. A cap-to-tail guide to mRNA translation strategies in virus-infected cells. Ann Rev Virol 3: 283–307. 10.1146/annurev-virology-100114-055014 [DOI] [PubMed] [Google Scholar]
- Jha S, Rollins MG, Fuchs G, Procter DJ, Hall EA, Cozzolino K, Sarnow P, Savas JN, Walsh D. 2017. Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature 546: 651–655. 10.1038/nature22814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. 2011. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7: 885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AG, Grosely R, Petrov AN, Puglisi JD. 2017. Dynamics of IRES-mediated translation. Philos Trans R Soc Lond B Biol Sci 372: 20160177. 10.1098/rstb.2016.0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungfleisch J, Chowdhury A, Alves-Rodrigues I, Tharun S, Díez J. 2015. The Lsm1–7–Pat1 complex promotes viral RNA translation and replication by differential mechanisms. RNA 21: 1469–1479. 10.1261/rna.052209.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Hegde RS. 2017. Initiation of quality control during Poly(A) translation requires site-specific ribosome ubiquitination. Mol Cell 65: 743–750.e4. 10.1016/j.molcel.2016.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel W, Noerenberg M, Cerikan B, Chen H, Järvelin AI, Kammoun M, Lee JY, Shuai N, Garcia-Moreno M, Andrejeva A, et al. 2021. Global analysis of protein-RNA interactions in SARS-CoV-2-infected cells reveals key regulators of infection. Mol Cell 81: 2851–2867.e7. 10.1016/j.molcel.2021.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. 2009. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol 16: 1134–1140. 10.1038/nsmb.1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan JP, Tremblay MW, Brown MC, Trimarco JD, Dobrikova EY, Dobrikov MI, Gromeier M. 2021. Enterovirus 2Apro cleavage of the YTHDF m6A readers implicates YTHDF3 as a mediator of type I interferon-driven JAK/STAT signaling. MBio 12: e00116-21. 10.1128/mBio.00116-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsafanas GC, Moss B. 2007. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2: 221–228. 10.1016/j.chom.2007.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE Jr, Darnell RB. 2017. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev 31: 990–1006. 10.1101/gad.301036.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Bogerd HP, Kornepati AV, Kang D, Ghoshal D, Marshall JB, Poling BC, Tsai K, Gokhale NS, Horner SM, et al. 2016. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19: 675–685. 10.1016/j.chom.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr CH, Ma ZW, Jang CJ, Thompson SR, Jan E. 2016. Molecular analysis of the factorless internal ribosome entry site in cricket paralysis virus infection. Sci Rep 6: 37319. 10.1038/srep37319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr CH, Wang QS, Moon KM, Keatings K, Allan DW, Foster LJ, Jan E. 2018. IRES-dependent ribosome repositioning directs translation of a +1 overlapping ORF that enhances viral infection. Nucleic Acids Res 46: 11952–11967. 10.1093/nar/gky1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Lee YS, Jung SJ, Yeo J, Seo JJ, Lee YY, Lim J, Chang H, Song J, Yang J, et al. 2020. Viral hijacking of the TENT4–ZCCHC14 complex protects viral RNAs via mixed tailing. Nat Struct Mol Biol 27: 581–588. 10.1038/s41594-020-0427-3 [DOI] [PubMed] [Google Scholar]
- Kindler E, Gil-Cruz C, Spanier J, Li Y, Wilhelm J, Rabouw HH, Züst R, Hwang M, V'Kovski P, Stalder H, et al. 2017. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog 13: e1006195. 10.1371/journal.ppat.1006195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec D, Lista MJ, Ficarelli M, Swanson CM, Neil SJD. 2021. S-farnesylation is essential for antiviral activity of the long ZAP isoform against RNA viruses with diverse replication strategies. PLoS Pathog 17: e1009726. 10.1371/journal.ppat.1009726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Arias C, Garabedian A, Palmenberg AC, Mohr I. 2012a. Site-specific cleavage of the host poly(A) binding protein by the encephalomyocarditis virus 3C proteinase stimulates viral replication. J Virol 86: 10686–10694. 10.1128/JVI.00896-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Wilson AC, Chao MV, Mohr I. 2012b. Control of viral latency in neurons by axonal mTOR signaling and the 4E-BP translation repressor. Genes Dev 26: 1527–1532. 10.1101/gad.190157.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad LM, Brulois KF, Jung JU, Glaunsinger BA. 2013. Dual short upstream open reading frames control translation of a herpesviral polycistronic mRNA. PLoS Pathog 9: e1003156. 10.1371/journal.ppat.1003156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad LM, Brulois KF, Jung JU, Glaunsinger BA. 2014. Reinitiation after translation of two upstream open reading frames (ORF) governs expression of the ORF35–37 Kaposi's sarcoma-associated herpesvirus polycistronic mRNA. J Virol 88: 6512–6518. 10.1128/JVI.00202-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulsuptrakul J, Wang R, Meyers NL, Ott M, Puschnik AS. 2021. A genome-wide CRISPR screen identifies UFMylation and TRAMP-like complexes as host factors required for hepatitis A virus infection. Cell Rep 34: 108859. 10.1016/j.celrep.2021.108859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar GR, Glaunsinger BA. 2010. Nuclear import of cytoplasmic poly(A) binding protein restricts gene expression via hyperadenylation and nuclear retention of mRNA. Mol Cell Biol 30: 4996–5008. 10.1128/MCB.00600-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss-Duerkop SK, Wang J, Mena I, White K, Metreveli G, Sakthivel R, Mata MA, Muñoz-Moreno R, Chen X, Krammer F, et al. 2017. Influenza virus differentially activates mTORC1 and mTORC2 signaling to maximize late stage replication. PLoS Pathog 13: e1006635. 10.1371/journal.ppat.1006635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T, Thompson SR. 2019. Noncanonical translation initiation in eukaryotes. Cold Spring Harb Perspect Biol 11: a032672. 10.1101/cshperspect.a032672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFontaine E, Miller CM, Permaul N, Martin ET, Fuchs G. 2020. Ribosomal protein RACK1 enhances translation of poliovirus and other viral IRESs. Virology 545: 53–62. 10.1016/j.virol.2020.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry DM, Hertz MI, Thompson SR. 2009. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev 23: 2753–2764. 10.1101/gad.1832209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang F, Singh RK, Pei Y, Zhang S, Sun K, Robertson ES. 2019. EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PLoS Pathog 15: e1007796. 10.1371/journal.ppat.1007796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe CP, Grosely R, Johnson AG, Wang J, Fernández IS, Puglisi JD. 2021. Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. Proc Natl Acad Sci 118: e2017715118. 10.1073/pnas.2017715118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasman L, Krupalnik V, Viukov S, Mor N, Aguilera-Castrejon A, Schneir D, Bayerl J, Mizrahi O, Peles S, Tawil S, et al. 2020. Context-dependent functional compensation between Ythdf m6A reader proteins. Genes Dev 34: 1373–1391. 10.1101/gad.340695.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavi S, Shatkin AJ. 1975. Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc Natl Acad Sci 72: 2012–2016. 10.1073/pnas.72.6.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Pimienta G, Steitz JA. 2012. AUF1/hnRNP D is a novel protein partner of the EBER1 noncoding RNA of Epstein-Barr virus. RNA 18: 2073–2082. 10.1261/rna.034900.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Burdeinick-Kerr R, Whelan SP. 2013. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci 110: 324–329. 10.1073/pnas.1216454109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee YS, Choi Y, Son A, Park Y, Lee KM, Kim J, Kim JS, Kim VN. 2021. The SARS-CoV-2 RNA interactome. Mol cell 81: 2838–2850.e6. 10.1016/j.molcel.2021.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Ma-Lauer Y, Han Y, Thoms M, Buschauer R, Jores J, Thiel V, Beckmann R, Deng W, Leonhardt H, et al. 2021. The SARS-unique domain (SUD) of SARS-CoV and SARS-CoV-2 interacts with human Paip1 to enhance viral RNA translation. EMBO J 40: e102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Masaki T, Yamane D, McGivern DR, Lemon SM. 2013. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc Natl Acad Sci 110: 1881–1886. 10.1073/pnas.1213515110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Li X, Tang H, Jiang B, Dou Y, Gorospe M, Wang W. 2017. NSUN2-mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J Cell Biochem 118: 2587–2598. 10.1002/jcb.25957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Hui H, Bray B, Gonzalez GM, Zeller M, Anderson KG, Knight R, Smith D, Wang Y, Carlin AF, et al. 2021. METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep 35: 109091. 10.1016/j.celrep.2021.109091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM. 2016a. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 1: 16011. 10.1038/nmicrobiol.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, Rana TM. 2016b. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe 20: 666–673. 10.1016/j.chom.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Chiweshe S, McCormick D, Raper A, Wickenhagen A, DeFillipis V, Gaunt E, Simmonds P, Wilson SJ, Grey F. 2020. Human cytomegalovirus evades ZAP detection by suppressing CpG dinucleotides in the major immediate early 1 gene. PLoS Pathog 16: e1008844. 10.1371/journal.ppat.1008844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Gack MU. 2020. Distinct and orchestrated functions of RNA sensors in innate immunity. Immunity 53: 26–42. 10.1016/j.immuni.2020.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Moss B. 2016. Opposing roles of double-stranded RNA effector pathways and viral defense proteins revealed with CRISPR–Cas9 knockout cell lines and vaccinia virus mutants. J Virol 90: 7864–7879. 10.1128/JVI.00869-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, HuangFu WC, Kumar KG, Qian J, Casey JP, Hamanaka RB, Grigoriadou C, Aldabe R, Diehl JA, Fuchs SY. 2009. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe 5: 72–83. 10.1016/j.chom.2008.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. 2014. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10: 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SW, Katsafanas GC, Liu R, Wyatt LS, Moss B. 2015. Poxvirus decapping enzymes enhance virulence by preventing the accumulation of dsRNA and the induction of innate antiviral responses. Cell Host Microbe 17: 320–331. 10.1016/j.chom.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, Xue W, Cui Y, Dong K, Ding H, et al. 2019a. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177: 865–880 e821. 10.1016/j.cell.2019.03.046 [DOI] [PubMed] [Google Scholar]
- Liu Y, You Y, Lu Z, Yang J, Li P, Liu L, Xu H, Niu Y, Cao X. 2019b. N6-methyladenosine RNA modification-mediated cellular metabolism rewiring inhibits viral replication. Science 365: 1171–1176. 10.1126/science.aax4468 [DOI] [PubMed] [Google Scholar]
- Liu YC, Mok BW, Wang P, Kuo RL, Chen H, Shih SR. 2021. Cellular 5′–3′ mRNA exoribonuclease XRN1 inhibits interferon β activation and facilitates influenza a virus replication. MBio 12: e0094521. 10.1128/mBio.00945-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage KG, Narayanan K, Nakagawa K, Terasaki K, Ramirez SI, Tseng CT, Makino S. 2015. Middle east respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J VIrol 89: 10970–10981. 10.1128/JVI.01352-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttermann C, Meyers G. 2014. Two alternative ways of start site selection in human norovirus reinitiation of translation. J Biol Chem 289: 11739–11754. 10.1074/jbc.M114.554030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majzoub K, Hafirassou ML, Meignin C, Goto A, Marzi S, Fedorova A, Verdier Y, Vinh J, Hoffmann JA, Martin F, et al. 2014. RACK1 controls IRES-mediated translation of viruses. Cell 159: 1086–1095. 10.1016/j.cell.2014.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall EE, Bierle CJ, Brune W, Geballe AP. 2009. Essential role for either TRS1 or IRS1 in human cytomegalovirus replication. J Virol 83: 4112–4120. 10.1128/JVI.02489-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinand C, Salehzada T, Silhol M, Lebleu B, Bisbal C. 1998. RNase L inhibitor (RLI) antisense constructions block partially the down regulation of the 2-5A/RNase L pathway in encephalomyocarditis-virus-(EMCV)-infected cells. Eur J Biochem 254: 248–255. 10.1046/j.1432-1327.1998.2540248.x [DOI] [PubMed] [Google Scholar]
- Martinand C, Montavon C, Salehzada T, Silhol M, Lebleu B, Bisbal C. 1999. RNase L inhibitor is induced during human immunodeficiency virus type 1 infection and down regulates the 2-5A/RNase L pathway in human T cells. J Virol 73: 290–296. 10.1128/JVI.73.1.290-296.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateer EJ, Maruyama J, Card GE, Paessler S, Huang C. 2020. Lassa virus, but not highly pathogenic new world arenaviruses, restricts immuno-stimulatory double-stranded RNA accumulation during infection. J Virol 94: e02006-19. 10.1128/JVI.02006-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JP, Simon AE. 2021. Targeting of viral RNAs by Upf1-mediated RNA decay pathways. Curr Opin VIrol 47: 1–8. 10.1016/j.coviro.2020.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick C, Khaperskyy DA. 2017. Translation inhibition and stress granules in the antiviral immune response. Nat Rev Immunol 17: 647–660. 10.1038/nri.2017.63 [DOI] [PubMed] [Google Scholar]
- McFadden MJ, Horner SM. 2021. N6-methyladenosine regulates host responses to viral infection. Trends Biochem Sci 46: 366–377. 10.1016/j.tibs.2020.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden MJ, McIntyre ABR, Mourelatos H, Abell NS, Gokhale NS, Ipas H, Xhemalce B, Mason CE, Horner SM. 2021. Post-transcriptional regulation of antiviral gene expression by N6-methyladenosine. Cell Res 34: 108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre W, Netzband R, Bonenfant G, Biegel JM, Miller C, Fuchs G, Henderson E, Arra M, Canki M, Fabris D, et al. 2018. Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res 46: 5776–5791. 10.1093/nar/gky029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Perez C, Mohr I. 2012. Poly(A) binding protein abundance regulates eukaryotic translation initiation factor 4F assembly in human cytomegalovirus-infected cells. Proc Natl Acad Sci 109: 5627–5632. 10.1073/pnas.1202829109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Yu D, Mohr I. 2013. A new role for the cellular PABP repressor Paip2 as an innate restriction factor capable of limiting productive cytomegalovirus replication. Genes Dev 27: 1809–1820. 10.1101/gad.221341.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Zavadil J, Bianco C, Shiflett L, Brown S, Mohr I. 2014. Global reprogramming of the cellular translational landscape facilitates cytomegalovirus replication. Cell Rep 6: 9–17. 10.1016/j.celrep.2013.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez AS, Ly M, Gonzalez-Sanchez AM, Hartenian E, Ingolia NT, Cte JC, Glaunsinger BA. 2021. The N-terminal domain of SARS-CoV-2 nsp1 plays key roles in suppression of cellular gene expression and preservation of viral gene expression. Cell Rep 37: 109841. 10.1016/j.celrep.2021.109841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC, Pavitt GD. 2018. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb Persect Biol 10: a033092. 10.1101/cshperspect.a033092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meydan S, Guydosh NR. 2021. A cellular handbook for collided ribosomes: surveillance pathways and collision types. Curr Genet 67: 19–26. 10.1007/s00294-020-01111-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ, Southern PJ. 1993. Concurrent sequence analysis of 5′ and 3′ RNA termini by intramolecular circularization reveals 5′ nontemplated bases and 3′ terminal heterogeneity for lymphocytic choriomeningitis virus mRNAs. J Virol 67: 2621–2627. 10.1128/jvi.67.5.2621-2627.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JY, Krug RM. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′–5′ oligo (A) synthetase/RNase L pathway. Proc Natl Acad Sci 103: 7100–7105. 10.1073/pnas.0602184103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi O, Nachshon A, Shitrit A, Gelbart IA, Dobesova M, Brenner S, Kahana C, Stern-Ginossar N. 2018. Virus-induced changes in mRNA secondary structure uncover cis-regulatory elements that directly control gene expression. Mol Cell 72: 862–874.e5. 10.1016/j.molcel.2018.09.003 [DOI] [PubMed] [Google Scholar]
- Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. 2004. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem Biophys Res Commun 319: 1228–1234. 10.1016/j.bbrc.2004.05.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocquet V, Neusiedler J, Rende F, Cluet D, Robin JP, Terme JM, Duc Dodon M, Wittmann J, Morris C, Le Hir H, et al. 2012. The human T-lymphotropic virus type 1 tax protein inhibits nonsense-mediated mRNA decay by interacting with INT6/EIF3E and UPF1. J Virol 86: 7530–7543. 10.1128/JVI.07021-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SL, Blackinton JG, Anderson JR, Dozier MK, Dodd BJ, Keene JD, Wilusz CJ, Bradrick SS, Wilusz J. 2015. XRN1 stalling in the 5′ UTR of hepatitis C virus and bovine viral diarrhea virus is associated with dysregulated host mRNA stability. PLoS Pathog 11: e1004708. 10.1371/journal.ppat.1004708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, Shenk T. 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3: 253–262. 10.1016/j.chom.2008.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B, Gershowitz A, Stringer JR, Holland LE, Wagner EK. 1977. 5′-terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. J Virol 23: 234–239. 10.1128/jvi.23.2.234-239.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudhasani R, Tran JP, Retterer C, Kota KP, Whitehouse CA, Bavari S. 2016. Protein kinase R degradation is essential for rift valley fever virus infection and is regulated by SKP1–CUL1–F-box (SCF)FBXW11–NSs E3 ligase. PLoS Pathog 12: e1005437. 10.1371/journal.ppat.1005437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhs M, Hilal T, Mielke T, Skabkin MA, Sanbonmatsu KY, Pestova TV, Spahn CM. 2015. Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES. Mol Cell 57: 422–432. 10.1016/j.molcel.2014.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Glaunsinger BA. 2017. Nuclease escape elements protect messenger RNA against cleavage by multiple viral endonucleases. PLoS Pathog 13: e1006593. 10.1371/journal.ppat.1006593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M, Camarena V, Mohr I. 2004. Full resistance of herpes simplex virus type 1-infected primary human cells to α interferon requires both the Us11 and γ134.5 gene products. J Virol 78: 10193–10196. 10.1128/JVI.78.18.10193-10196.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J, Savva CG, Shin BS, Dever TE, Ramakrishnan V, Fernández IS. 2016. Structural characterization of ribosome recruitment and translocation by type IV IRES. Elife 5: e13567. 10.7554/eLife.13567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadar M, Chan MY, Huang SW, Huang CC, Tseng JT, Tsai CH. 2011. HuR binding to AU-rich elements present in the 3′ untranslated region of classical swine fever virus. Virol J 8: 340. 10.1186/1743-422X-8-340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K, Makino S. 2021. Mechanisms of coronavirus Nsp1-mediated control of host and viral gene expression. Cells 10: 300. 10.3390/cells10020300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Ando T, Yamagishi M, Yokoyama K, Ishida T, Ohsugi T, Tanaka Y, Brighty DW, Watanabe T. 2013. Viral interference with host mRNA surveillance, the nonsense-mediated mRNA decay (NMD) pathway, through a new function of HTLV-1 Rex: implications for retroviral replication. Microbes Infect 15: 491–505. 10.1016/j.micinf.2013.03.006 [DOI] [PubMed] [Google Scholar]
- Neupane R, Pisareva VP, Rodriguez CF, Pisarev AV, Fernández IS. 2020. A complex IRES at the 5′-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate. Elife 9: e54575. 10.7554/eLife.54575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LH, Espert L, Mechti N, Wilson DM III. 2001. The human interferon- and estrogen-regulated ISG20/HEM45 gene product degrades single-stranded RNA and DNA in vitro. Biochemistry 40: 7174–7179. 10.1021/bi010141t [DOI] [PubMed] [Google Scholar]
- Niemeyer D, Zillinger T, Muth D, Zielecki F, Horvath G, Suliman T, Barchet W, Weber F, Drosten C, Müller MA. 2013. Middle east respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol 87: 12489–12495. 10.1128/JVI.01845-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea C, Klupsch K, Choi S, Bagus B, Soria C, Shen J, McCormick F, Stokoe D. 2005. Adenoviral proteins mimic nutrient/growth signals to activate the mTOR pathway for viral replication. EMBO J 24: 1211–1221. 10.1038/sj.emboj.7600597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page HG, Read GS. 2010. The virion host shutoff endonuclease (UL41) of herpes simplex virus interacts with the cellular cap-binding complex eIF4F. J Virol 84: 6886–6890. 10.1128/JVI.00166-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panthu B, Terrier O, Carron C, Traversier A, Corbin A, Balvay L, Lina B, Rosa-Calatrava M, Ohlmann T. 2017. The NS1 protein from influenza virus stimulates translation initiation by enhancing ribosome recruitment to mRNAs. J Mol Biol 429: 3334–3352. 10.1016/j.jmb.2017.04.007 [DOI] [PubMed] [Google Scholar]
- Park C, Peng C, Rahman MJ, Haller SL, Tazi L, Brennan G, Rothenburg S. 2021. Orthopoxvirus K3 orthologs show virus- and host-specific inhibition of the antiviral protein kinase PKR. PLoS Pathog 17: e1009183. 10.1371/journal.ppat.1009183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S, Moss B. 2007. Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses. J Virol 81: 12973–12978. 10.1128/JVI.01668-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S, Resch W, Moss B. 2007. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc Natl Acad Sci 104: 2139–2144. 10.1073/pnas.0611685104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasieka TJ, Lu B, Crosby SD, Wylie KM, Morrison LA, Alexander DE, Menachery VD, Leib DA. 2008. Herpes simplex virus virion host shutoff attenuates establishment of the antiviral state. J Virol 82: 5527–5535. 10.1128/JVI.02047-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. 2019. The organizing principles of eukaryotic ribosome recruitment. Ann Rev Biochem 88: 307–335. 10.1146/annurev-biochem-013118-111042 [DOI] [PubMed] [Google Scholar]
- Peng C, Haller SL, Rahman MM, McFadden G, Rothenburg S. 2016. Myxoma virus M156 is a specific inhibitor of rabbit PKR but contains a loss-of-function mutation in Australian virus isolates. Proc Natl Acad Sci 113: 3855–3860. 10.1073/pnas.1515613113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Wyatt LS, Glushakow-Smith SG, Lal-Nag M, Weisberg AS, Moss B. 2020. Zinc-finger antiviral protein (ZAP) is a restriction factor for replication of modified vaccinia virus Ankara (MVA) in human cells. PLoS Pathog 16: e1008845. 10.1371/journal.ppat.1008845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn WD, Harrington HR, Schlebach JP, Mukhopadhyay S. 2020. Regulators of viral frameshifting: more than RNA influences translation events. Ann Rev Virol 7: 219–238. 10.1146/annurev-virology-012120-101548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pheasant K, Möller-Levet CS, Jones J, Depledge D, Breuer J, Elliott G. 2018. Nuclear-cytoplasmic compartmentalization of the herpes simplex virus 1 infected cell transcriptome is co-ordinated by the viral endoribonuclease vhs and cofactors to facilitate the translation of late proteins. PLoS Pathog 14: e1007331. 10.1371/journal.ppat.1007331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, et al. 2008. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4: 579–591. 10.1016/j.chom.2008.10.007 [DOI] [PubMed] [Google Scholar]
- Popp MW-L, Cho H, Maquat LE. 2020. Viral subversion of nonsense-mediated mRNA decay. RNA 26: 1509–1518. 10.1261/rna.076687.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AM, Hayer KE, McIntyre ABR, Gokhale NS, Abebe JS, Della Fera AN, Mason CE, Horner SM, Wilson AC, Depledge DP, et al. 2020. Direct RNA sequencing reveals m6A modifications on adenovirus RNA are necessary for efficient splicing. Nat Commun 11: 6016. 10.1038/s41467-020-19787-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG. 2019. Phosphorylation and signal transduction pathways in translational control. Cold Spring Harb Prespect Biol 11: a033050. 10.1101/cshperspect.a033050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Lan S, Wang W, Schelde LM, Dong H, Wallat GD, Ly H, Liang Y, Dong C. 2010. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468: 779–783. 10.1038/nature09605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quade N, Boehringer D, Leibundgut M, van den Heuvel J, Ban N. 2015. Cryo-EM structure of hepatitis C virus IRES bound to the human ribosome at 3.9-Å resolution. Nat Commun 6: 7646. 10.1038/ncomms8646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouw HH, Visser LJ, Passchier TC, Langereis MA, Liu F, Giansanti P, van Vliet ALW, Dekker JG, van der Grein SG, Saucedo JG, et al. 2020. Inhibition of the integrated stress response by viral proteins that block p-eIF2–eIF2B association. Nat Microbiol 5: 1361–1373. 10.1038/s41564-020-0759-0 [DOI] [PubMed] [Google Scholar]
- Rao S, Amorim R, Niu M, Breton Y, Tremblay MJ, Mouland AJ. 2019. Host mRNA decay proteins influence HIV-1 replication and viral gene expression in primary monocyte-derived macrophages. Retrovirolgy 16: 3. 10.1186/s12977-019-0465-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Hoskins I, Tonn T, Garcia PD, Ozadam H, Sarinay Cenik E, Cenik C. 2021. Genes with 5′ terminal oligopyrimidine tracts preferentially escape global suppression of translation by the SARS-CoV-2 Nsp1 protein. RNA 1025–1045. 10.1261/rna.078661.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas A, Dummer R, Puzanov I, VanderWalde A, Andtbacka RHI, Michielin O, Olszanski AJ, Malvehy J, Cebon J, Fernandez E, et al. 2017. Oncolytic virotherapy promotes intratumoral T cell infiltration and improves anti-PD-1 immunotherapy. Cell 170: 1109–1119.e10. 10.1016/j.cell.2017.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby RE, Wise HM, Smith N, Digard P, Rehwinkel J. 2019. PA-X antagonises MAVS-dependent accumulation of early type I interferon messenger RNAs during influenza A virus infection. Sci Rep 9: 7216. 10.1038/s41598-019-43632-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera CI, Lloyd RE. 2008. Modulation of enteroviral proteinase cleavage of poly(A)-binding protein (PABP) by conformation and PABP-associated factors. Virology 375: 59–72. 10.1016/j.virol.2008.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M, Vasconcelos G, Dever TE. 2015. An eIF2α-binding motif in protein phosphatase 1 subunit GADD34 and its viral orthologs is required to promote dephosphorylation of eIF2α. Proc Natl Acad Sci 112: E3466–E3475. 10.1073/pnas.1501557112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins MG, Shasnai M, Meade N, Astar H, Shen PS, Walsh D. 2021. Negative charge in the RACK1 loop broadens the translational capacity of the human ribosome. Cell Rep 36: 109663. 10.1016/j.celrep.2021.109663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roobol A, Roobol J, Bastide A, Knight JR, Willis AE, Smales CM. 2015. p58IPK is an inhibitor of the eIF2α kinase GCN2 and its localization and expression underpin protein synthesis and ER processing capacity. Biochem J 465: 213–225. 10.1042/BJ20140852 [DOI] [PubMed] [Google Scholar]
- Rosa-Mercado NA, Withers JB, Steitz JA. 2017. Settling the m6A debate: methylation of mature mRNA is not dynamic but accelerates turnover. Genes Dev 31: 957–958. 10.1101/gad.302695.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth H, Magg V, Uch F, Mutz P, Klein P, Haneke K, Lohmann V, Bartenschlager R, Fackler OT, Locker N, et al. 2017. Flavivirus infection uncouples translation suppression from cellular stress responses. MBio 8: e02150-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royall E, Locker N. 2016. Translational control during calicivirus infection. Viruses 8: 104. 10.3390/v8040104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royall E, Doyle N, Abdul-Wahab A, Emmott E, Morley SJ, Goodfellow I, Roberts LO, Locker N. 2015. Murine norovirus 1 (MNV1) replication induces translational control of the host by regulating eIF4E activity during infection. J Biol Chem 290: 4748–4758. 10.1074/jbc.M114.602649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozovics JM, Chase AJ, Cathcart AL, Chou W, Gershon PD, Palusa S, Wilusz J, Semler BL. 2012. Picornavirus modification of a host mRNA decay protein. MBio 3: e00431-00412. 10.1128/mBio.00431-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio RM, Mohr I. 2019. Inhibition of ULK1 and Beclin1 by an α-herpesvirus Akt-like Ser/Thr kinase limits autophagy to stimulate virus replication. Proc Natl Acad Sci 116: 26941–26950. 10.1073/pnas.1915139116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio RM, Depledge DP, Bianco C, Thompson L, Mohr I. 2018. RNA m6A modification enzymes shape innate responses to DNA by regulating interferon β. Genes Dev 32: 1472–1484. 10.1101/gad.319475.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadek J, Read GS. 2016. The splicing history of an mRNA affects its level of translation and sensitivity to cleavage by the virion host shutoff endonuclease during herpes simplex virus infections. J Virol 90: 10844–10856. 10.1128/JVI.01302-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sànchez R, Mohr I. 2007. Inhibition of cellular 2′–5′ oligoadenylate synthetase by the herpes simplex virus type 1 Us11 protein. J Virol 81: 3455–3464. 10.1128/JVI.02520-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz MA, Almela EG, García-Moreno M, Marina AI, Carrasco L. 2019. A viral RNA motif involved in signaling the initiation of translation on non-AUG codons. RNA 25: 431–452. 10.1261/rna.068858.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller N, Mina LB, Galão RP, Chari A, Giménez-Barcons M, Noueiry A, Fischer U, Meyerhans A, Díez J. 2009. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci 106: 13517–13522. 10.1073/pnas.0906413106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper GC, van Kollenburg B, Hu J, Luo Y, Goss DJ, Proud CG. 2002. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J Biol Chem 277: 3303–3309. 10.1074/jbc.M103607200 [DOI] [PubMed] [Google Scholar]
- Schmidt N, Lareau CA, Keshishian H, Ganskih S, Schneider C, Hennig T, Melanson R, Werner S, Wei Y, Zimmer M, et al. 2021. The SARS-CoV-2 RNA–protein interactome in infected human cells. Nat Microbiol 6: 339–353. 10.1038/s41564-020-00846-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut M, Thiel V, Muhlemann O, Ban N. 2020. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 27: 959–966. 10.1038/s41594-020-0511-8 [DOI] [PubMed] [Google Scholar]
- Schulz F, Yutin N, Ivanova NN, Ortega DR, Lee TK, Vierheilig J, Daims H, Horn M, Wagner M, Jensen GJ, et al. 2017. Giant viruses with an expanded complement of translation system components. Science 356: 82–85. 10.1126/science.aal4657 [DOI] [PubMed] [Google Scholar]
- Schwerk J, Soveg FW, Ryan AP, Thomas KR, Hatfield LD, Ozarkar S, Forero A, Kell AM, Roby JA, So L, et al. 2019. RNA-binding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions. Nat Immunol 20: 1610–1620. 10.1038/s41590-019-0527-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo EJ, Liu F, Kawagishi-Kobayashi M, Ung TL, Cao C, Dar AC, Sicheri F, Dever TE. 2008. Protein kinase PKR mutants resistant to the poxvirus pseudosubstrate K3L protein. Proc Natl Acad Sci 105: 16894–16899. 10.1073/pnas.0805524105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Fujii K, Kovary KM, Genuth NR, Rost HL, Teruel MN, Barna M. 2017. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol Cell 67: 71–83.e7. 10.1016/j.molcel.2017.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Wang L, Fontana P, Vora S, Zhang Y, Fu TM, Lieberman J, Wu H. 2020. SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism. bioRxiv. 10.1101/2020.09.18.302901 [DOI] [Google Scholar]
- Shulman Z, Stern-Ginossar N. 2020. The RNA modification N6-methyladenosine as a novel regulator of the immune system. Nat Immunol 21: 501–512. 10.1038/s41590-020-0650-4 [DOI] [PubMed] [Google Scholar]
- Shwetha S, Kumar A, Mullick R, Vasudevan D, Mukherjee N, Das S. 2015. HuR displaces polypyrimidine tract binding protein to facilitate la binding to the 3′ untranslated region and enhances hepatitis C virus replication. J Virol 89: 11356–11371. 10.1128/JVI.01714-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva PA, Pereira CF, Dalebout TJ, Spaan WJ, Bredenbeek PJ. 2010. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J Virol 84: 11395–11406. 10.1128/JVI.01047-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. 2005. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310: 1513–1515. 10.1126/science.1118977 [DOI] [PubMed] [Google Scholar]
- Slepenkov SV, Darzynkiewicz E, Rhoads RE. 2006. Stopped-flow kinetic analysis of eIF4E and phosphorylated eIF4E binding to cap analogs and capped oligoribonucleotides: evidence for a one-step binding mechanism. J Biol Chem 281: 14927–14938. 10.1074/jbc.M601653200 [DOI] [PubMed] [Google Scholar]
- Smith RWP, Anderson RC, Larralde O, Smith JWS, Gorgoni B, Richardson WA, Malik P, Graham SV, Gray NK. 2017. Viral and cellular mRNA-specific activators harness PABP and eIF4G to promote translation initiation downstream of cap binding. Proc Natl Acad Sci 114: 6310–6315. 10.1073/pnas.1610417114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloski KJ, Dickson AM, Chaskey EL, Garneau NL, Wilusz CJ, Wilusz J. 2010. Sindbis virus usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells. Cell Host Microbe 8: 196–207. 10.1016/j.chom.2010.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R, Porter AC, Ma K, Quilliam LA, Wek RC. 2000. Pancreatic eukaryotic initiation factor-2α kinase (PEK) homologues in humans, Drosophila melanogaster and Caenorhabditis elegans that mediate translational control in response to endoplasmic reticulum stress. Biochem J 346 Pt 2: 281–293. 10.1042/bj3460281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgeloos F, Jha BK, Silverman RH, Michiels T. 2013. Evasion of antiviral innate immunity by Theiler's virus L* protein through direct inhibition of RNase L. PLoS Pathog 9: e1003474. 10.1371/journal.ppat.1003474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soveg FW, Schwerk J, Gokhale NS, Cerosaletti K, Smith JR, Pairo-Castineira E, Kell AM, Forero A, Zaver SA, Esser-Nobis K, et al. 2021. Endomembrane targeting of human OAS1 p46 augments antiviral activity. Elife 10: e71047. 10.7554/eLife.71047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science 291: 1959–1962. 10.1126/science.1058409 [DOI] [PubMed] [Google Scholar]
- Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. 2004. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell 118: 465–475. 10.1016/j.cell.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. 2012. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 40: 5023–5033. 10.1093/nar/gks144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas KP, Depledge DP, Abebe JS, Rice SA, Mohr I, Wilson AC. 2021. Widespread remodeling of the m6A RNA-modification landscape by a viral regulator of RNA processing and export. Proc Natl Acad Sci 118: e2104805118. 10.1073/pnas.2104805118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Thompson SR, Mathews MB, Mohr I. 2019. Translational control in virus-infected cells. Cold Spring Harb Perspect Biol 11: a033001. 10.1101/cshperspect.a033001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelow LI, Leib DA. 1995. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J Virol 69: 6779–6786. 10.1128/jvi.69.11.6779-6786.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, Bennett EJ. 2017. ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol Cell 65: 751–760.e4. 10.1016/j.molcel.2016.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy E, Ryan AP, Fulzele A, Leonard M, Daugherty MD, Bennett EJ. 2021. Ribosome quality control activity potentiates vaccinia virus protein synthesis during infection. J Cell Sci 134: jcs257188. 10.1242/jcs.257188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney TR, Abaeva IS, Pestova TV, Hellen CU. 2014. The mechanism of translation initiation on type 1 picornavirus IRESs. EMBO J 33: 76–92. 10.1002/embj.201386124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata MA, Gonçalves-Carneiro D, Zang TM, Soll SJ, York A, Blanco-Melo D, Bieniasz PD. 2017. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 550: 124–127. 10.1038/nature24039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B, Liu H, Zhang S, da Silva SR, Zhang L, Meng J, Cui X, Yuan H, Sorel O, Zhang SW, et al. 2018. Viral and cellular N6-methyladenosine and N6,2′-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat Microbiol 3: 108–120. 10.1038/s41564-017-0056-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja S, MacGregor J, Markus S, Ha S, Mohr I. 2001. Enhanced antitumor efficacy of a herpes simplex virus mutant isolated by genetic selection in cancer cells. Proc Natl Acad Sci 98: 8804–8808. 10.1073/pnas.161011798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Wang X, Gao G. 2017. The short form of the zinc finger antiviral protein inhibits influenza a virus protein expression and is antagonized by the virus-encoded NS1. J Virol 91: e01909-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Fang L, Bai D, Ke W, Zhou Y, Wang D, Xiao S. 2018. Porcine reproductive and respiratory syndrome virus nonstructural protein 4 cleaves porcine DCP1a to attenuate its antiviral activity. J Immunol 201: 2345–2353. 10.4049/jimmunol.1701773 [DOI] [PubMed] [Google Scholar]
- Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, Kratzat H, Hayn M, Mackens-Kiani T, Cheng J, et al. 2020. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369: 1249–1255. 10.1126/science.abc8665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornbrough JM, Jha BK, Yount B, Goldstein SA, Li Y, Elliott R, Sims AC, Baric RS, Silverman RH, Weiss SR. 2016. Middle east respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. MBio 7: e00258. 10.1128/mBio.00258-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidu A, Janvier A, Schaeffer L, Sosnowski P, Kuhn L, Hammann P, Westhof E, Eriani G, Martin F. 2021. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA 27: 253–264. 10.1261/rna.078121.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh O, Cohen Y, Shitrit A, Shani O, Le-Trilling VT, Trilling M, Friedlander G, Tanenbaum M, Stern-Ginossar N. 2015. The transcription and translation landscapes during human cytomegalovirus infection reveal novel host-pathogen interactions. PLoS Pathog 11: e1005288. 10.1371/journal.ppat.1005288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. 2016. N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife 5: e15528. 10.7554/eLife.15528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend HL, Jha BK, Han JQ, Maluf NK, Silverman RH, Barton DJ. 2008. A viral RNA competitively inhibits the antiviral endoribonuclease domain of RNase L. RNA 14: 1026–1036. 10.1261/rna.958908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran V, Ledwith MP, Thamamongood T, Higgins CA, Tripathi S, Chang MW, Benner C, Garcia-Sastre A, Schwemmle M, Boon ACM, et al. 2020. Influenza virus repurposes the antiviral protein IFIT2 to promote translation of viral mRNAs. Nat Microbiol 5: 1490–1503. 10.1038/s41564-020-0778-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Courtney DG, Cullen BR. 2018. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathog 14: e1006919. 10.1371/journal.ppat.1006919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Jaguva Vasudevan AA, Martinez Campos C, Emery A, Swanstrom R, Cullen BR. 2020. Acetylation of cytidine residues boosts HIV-1 gene expression by increasing viral RNA stability. Cell Host Microbe 28: 306–312.e6. 10.1016/j.chom.2020.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Bogerd HP, Kennedy EM, Emery A, Swanstrom R, Cullen BR. 2021. Epitranscriptomic addition of m6A regulates HIV-1 RNA stability and alternative splicing. Genes Dev 35: 992–1004. 10.1101/gad.348508.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent HA, Ziehr B, Moorman NJ. 2017. Mechanism of protein kinase R inhibition by human cytomegalovirus pTRS1. J Virol 91: e01574-16. 10.1128/JVI.01574-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink EI, Smiley JR, Mohr I. 2017. Subversion of host responses to energy insufficiency by Us3 supports herpes simplex virus 1 replication during stress. J Virol 91: e00295-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink EI, Lee S, Smiley JR, Mohr I. 2018. Remodeling mTORC1 responsiveness to amino acids by the herpes simplex virus UL46 and Us3 gene products supports replication during nutrient insufficiency. J Virol 92: e01377-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink EI, Andrews J, Duffy C, Mohr I. 2021. Preventing translational inhibition from ribosomal protein insufficiency by a herpes simplex virus-encoded ribosome-associated protein. Proc Natl Acad Sci 118: e2025546118. 10.1073/pnas.2025546118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M, Lokugamage KG, Nakagawa K, Narayanan K, Makino S. 2018. Interplay between coronavirus, a cytoplasmic RNA virus, and nonsense-mediated mRNA decay pathway. Proc Natl Acad Sci 115: E10157–E10166. 10.1073/pnas.1811675115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I. 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev 18: 660–672. 10.1101/gad.1185304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I. 2006. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev 20: 461–472. 10.1101/gad.1375006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I. 2014. Coupling 40S ribosome recruitment to modification of a cap-binding initiation factor by eIF3 subunit e. Genes Dev 28: 835–840. 10.1101/gad.236752.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Perez C, Notary J, Mohr I. 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J Virol 79: 8057–8064. 10.1128/JVI.79.13.8057-8064.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Arias C, Perez C, Halladin D, Escandon M, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Mohr I. 2008. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol Cell Biol 28: 2648–2658. 10.1128/MCB.01631-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters B, Axhemi A, Jankowsky E, Thompson SR. 2020. Binding of a viral IRES to the 40S subunit occurs in two successive steps mediated by eS25. Nuc Acids Res 48: 8063–8073. 10.1093/nar/gkaa547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Juszkiewicz S, Blears D, Bajpe PK, Han Z, Faull P, Mitter R, Stewart A, Snijders AP, Hegde RS, et al. 2021. Translation stress and collided ribosomes are co-activators of cGAS. Mol Cell 81: 2808–2822.e10. 10.1016/j.molcel.2021.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Kishton RJ, Angel M, Conn CS, Dalla-Venezia N, Marcel V, Vincent A, Catez F, Ferré S, Ayadi L, et al. 2019. Ribosomal proteins regulate MHC class I peptide generation for immunosurveillance. Mol Cell 73: 1162–1173.e5. 10.1016/j.molcel.2018.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil JE, Beemon KL. 2006. A 3′ UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA 12: 102–110. 10.1261/rna.2129806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss CM, Trobaugh DW, Sun C, Lucas TM, Diamond MS, Ryman KD, Klimstra WB. 2018. The interferon-induced exonuclease ISG20 exerts antiviral activity through upregulation of type I interferon response proteins. mSphere 3: e00209-18. 10.1128/mSphere.00209-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werden SJ, Barrett JW, Wang G, Stanford MM, McFadden G. 2007. M-T5, the ankyrin repeat, host range protein of myxoma virus, activates Akt and can be functionally replaced by cellular PIKE-A. J Virol 81: 2340–2348. 10.1128/JVI.01310-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston S, Baracco L, Keller C, Matthews K, McGrath ME, Logue J, Liang J, Dyall J, Holbrook MR, Hensley LE, et al. 2020. The SKI complex is a broad-spectrum, host-directed antiviral drug target for coronaviruses, influenza, and filoviruses. Proc Natl Acad Sci 117: 30687–30698. 10.1073/pnas.2012939117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SD, Jacobs BL. 2012. The amino terminus of the vaccinia virus E3 protein is necessary to inhibit the interferon response. J VIrol 86: 5895–5904. 10.1128/JVI.06889-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickenhagen A, Sugrue E, Lytras S, Kuchi S, Noerenberg M, Turnbull ML, Loney C, Herder V, Allan J, Jarmson I, et al. 2021. A prenylated dsRNA sensor protects against severe COVID-19. Science 374: eabj3624. 10.1126/science.abj3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener D, Schwartz S. 2021. The epitranscriptome beyond m6A. Nat Rev Genet 22: 119–131. 10.1038/s41576-020-00295-8 [DOI] [PubMed] [Google Scholar]
- Wilczynska A, Gillen SL, Schmidt T, Meijer HA, Jukes-Jones R, Langlais C, Kopra K, Lu WT, Godfrey JD, Hawley BR, et al. 2019. eIF4A2 drives repression of translation at initiation by Ccr4-not through purine-rich motifs in the 5′UTR. Genome biol 20: 262. 10.1186/s13059-019-1857-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GD, Gokhale NS, Horner SM. 2019. Regulation of viral infection by the RNA modification N6-methyladenosine. Ann Rev VIrol 6: 235–253. 10.1146/annurev-virology-092818-015559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Pestova TV, Hellen CU, Sarnow P. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102: 511–520. 10.1016/S0092-8674(00)00055-6 [DOI] [PubMed] [Google Scholar]
- Winkler R, Gillis E, Lasman L, Safra M, Geula S, Soyris C, Nachshon A, Tai-Schmiedel J, Friedman N, Le-Trilling VTK, et al. 2019. m6A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol 20: 173–182. 10.1038/s41590-018-0275-z [DOI] [PubMed] [Google Scholar]
- Wong J, Si X, Angeles A, Zhang J, Shi J, Fung G, Jagdeo J, Wang T, Zhong Z, Jan E, et al. 2013. Cytoplasmic redistribution and cleavage of AUF1 during coxsackievirus infection enhance the stability of its viral genome. FASEB J 27: 2777–2787. 10.1096/fj.12-226498 [DOI] [PubMed] [Google Scholar]
- Wu N, Nguyen XN, Wang L, Appourchaux R, Zhang C, Panthu B, Gruffat H, Journo C, Alais S, Qin J, et al. 2019. The interferon stimulated gene 20 protein (ISG20) is an innate defense antiviral factor that discriminates self versus non-self translation. PLoS Pathog 15: e1008093. 10.1371/journal.ppat.1008093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerth JD, Habjan M, Kainulainen M, Berisha B, Bertheloot D, Superti-Furga G, Pichlmair A, Weber F. 2020. eIF2B as a target for viral evasion of PKR-mediated translation inhibition. MBio 11: e00976-20. 10.1128/mBio.00976-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q, Cuesta R, Schneider RJ. 2004. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev 18: 1997–2009. 10.1101/gad.1212504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia TL, Li X, Wang X, Zhu YJ, Zhang H, Cheng W, Chen ML, Ye Y, Li Y, Zhang A, et al. 2021. N(6)-methyladenosine-binding protein YTHDF1 suppresses EBV replication and promotes EBV RNA decay. EMBO Rep 22: e50128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Lu B, Zheng Z, Miao Y, Liu Y, Zhang Y, Zheng C, Ke X, Hu Q, Wang H. 2018. The 3C protease of enterovirus A71 counteracts the activity of host zinc-finger antiviral protein (ZAP). J Gen Virol 99: 73–85. 10.1099/jgv.0.000982 [DOI] [PubMed] [Google Scholar]
- Xue S, Tian S, Fujii K, Kladwang W, Das R, Barna M. 2015. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 517: 33–38. 10.1038/nature14010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Zhao BS, Zhang Z, Lu M, Harder O, Chen P, Lu Z, Li A, Ma Y, Xu Y, et al. 2019. Viral N6-methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus. Nat Commun 10: 4595. 10.1038/s41467-019-12504-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Dong Y, Wang Y, Liu H, Ma H, Zhang H, Zhang L, Cheng L, Lv X, Xu Z, et al. 2020. N6-methyladenosine modifications enhance enterovirus 71 ORF translation through METTL3 cytoplasmic distribution. Biochem Biophys Res Commun 527: 297–304. 10.1016/j.bbrc.2020.04.088 [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Machida K, Iwasaki W, Shigeta T, Nishimoto M, Takahashi M, Sakamoto A, Yonemochi M, Harada Y, Shigematsu H, et al. 2019. HCV IRES captures an actively translating 80S ribosome. Mol Cell 74: 1205–1214.e8. 10.1016/j.molcel.2019.04.022 [DOI] [PubMed] [Google Scholar]
- Yu Y, Abaeva IS, Marintchev A, Pestova TV, Hellen CU. 2011a. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res 39: 4851–4865. 10.1093/nar/gkr045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Sweeney TR, Kafasla P, Jackson RJ, Pestova TV, Hellen CU. 2011b. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J 30: 4423–4436. 10.1038/emboj.2011.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Peng L, Park JJ, Hu Y, Devarkar SC, Dong MB, Shen Q, Wu S, Chen S, Lomakin IB, et al. 2020. Nonstructural protein 1 of SARS-CoV-2 is a potent pathogenicity factor redirecting host protein synthesis machinery toward viral RNA. Mol Cell 80: 1055–1066.e6. 10.1016/j.molcel.2020.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborowska I, Kellner K, Henry M, Meleady P, Walsh D. 2012. Recruitment of host translation initiation factor eIF4G by the vaccinia virus ssDNA-binding protein I3. Virology 425: 11–22. 10.1016/j.virol.2011.12.022 [DOI] [PubMed] [Google Scholar]
- Zaccara S, Jaffrey SR. 2020. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell 181: 1582–1595.e18. 10.1016/j.cell.2020.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S, Ries RJ, Jaffrey SR. 2019. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 20: 608–624. 10.1038/s41580-019-0168-5 [DOI] [PubMed] [Google Scholar]
- Zhan Y, Yu S, Yang S, Qiu X, Meng C, Tan L, Song C, Liao Y, Liu W, Sun Y, et al. 2020. Newcastle disease virus infection activates PI3K/Akt/mTOR and p38 MAPK/Mnk1 pathways to benefit viral mRNA translation via interaction of the viral NP protein and host eIF4E. PLoS Pathog 16: e1008610. 10.1371/journal.ppat.1008610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Jha BK, Ogden KM, Dong B, Zhao L, Elliott R, Patton JT, Silverman RH, Weiss SR. 2013. Homologous 2′,5′-phosphodiesterases from disparate RNA viruses antagonize antiviral innate immunity. Proc Natl Acad Sci 110: 13114–13119. 10.1073/pnas.1306917110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Zhang X, Wang J, Ma Y, Zhang L, Cao X. 2019. RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc Natl Acad Sci 116: 976–981. 10.1073/pnas.1812536116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chapat C, Wang P, Choi JH, Li Q, Luo J, Wiebe S, Kim SH, Robichaud N, Karam IF, et al. 2021. MicroRNA-induced translational control of antiviral immunity by the cap-binding protein 4EHP. Mol Cell 81: 1187–1199.e5. 10.1016/j.molcel.2021.01.030 [DOI] [PubMed] [Google Scholar]
- Zhao L, Jha BK, Wu A, Elliott R, Ziebuhr J, Gorbalenya AE, Silverman RH, Weiss SR. 2012. Antagonism of the interferon-induced OAS–RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11: 607–616. 10.1016/j.chom.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Song Z, Bai J, Liu X, Nauwynck H, Jiang P. 2020. Porcine reproductive and respiratory syndrome virus Nsp4 cleaves ZAP to antagonize its antiviral activity. Vet Microbiol 250: 108863. 10.1016/j.vetmic.2020.108863 [DOI] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. 2013. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49: 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Wang N, Woodson SE, Dong Q, Wang J, Liang Y, Rijnbrand R, Wei L, Nichols JE, Guo JT, et al. 2011. Antiviral activities of ISG20 in positive-strand RNA virus infections. Virology 409: 175–188. 10.1016/j.virol.2010.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Chen J, Tian L, Zhou Y, Xu S, Long S, Wang D, Fang L, Xiao S. 2020. Porcine deltacoronavirus nsp5 cleaves DCP1A to decrease its antiviral activity. J VIrol 94: e02162-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer MM, Kibe A, Rand U, Pekarek L, Ye L, Buck S, Smyth RP, Cicin-Sain L, Caliskan N. 2021. The short isoform of the host antiviral protein ZAP acts as an inhibitor of SARS-CoV-2 programmed ribosomal frameshifting. Nat Commun 12: 7193. 10.1038/s41467-021-27431-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoviev A, Hellen CUT, Pestova TV. 2015. Multiple mechanisms of reinitiation on bicistronic calicivirus mRNAs. Mol Cell 57: 1059–1073. 10.1016/j.molcel.2015.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberek J, Jemielity J, Jablonowska A, Stepinski J, Dadlez M, Stolarski R, Darzynkiewicz E. 2004. Influence of electric charge variation at residues 209 and 159 on the interaction of eIF4E with the mRNA 5′ terminus. Biochemistry 43: 5370–5379. 10.1021/bi030266t [DOI] [PubMed] [Google Scholar]