In this study, Saito et al. sought to understand how the N6-methyladenosine (m6A) reader and RNA helicase YTHDC2 switches cells from mitotic to meiotic gene expression programs. Their findings provide insight into YTHDC2's mechanism, and they propose a model in which YTHDC2 binds transcripts independent of m6A status and regulates gene expression during multiple stages of meiosis by distinct mechanisms.
Keywords: meiosis, N6-methyladenosine, RNA-binding proteins, RNA helicase, spermatogenesis, Ythdc2
Abstract
Mechanisms regulating meiotic progression in mammals are poorly understood. The N6-methyladenosine (m6A) reader and 3′ → 5′ RNA helicase YTHDC2 switches cells from mitotic to meiotic gene expression programs and is essential for meiotic entry, but how this critical cell fate change is accomplished is unknown. Here, we provide insight into its mechanism and implicate YTHDC2 in having a broad role in gene regulation during multiple meiotic stages. Unexpectedly, mutation of the m6A-binding pocket of YTHDC2 had no detectable effect on gametogenesis and mouse fertility, suggesting that YTHDC2 function is m6A-independent. Supporting this conclusion, CLIP data defined YTHDC2-binding sites on mRNA as U-rich and UG-rich motif-containing regions within 3′ UTRs and coding sequences, distinct from the sites that contain m6A during spermatogenesis. Complete loss of YTHDC2 during meiotic entry did not substantially alter translation of its mRNA binding targets in whole-testis ribosome profiling assays but did modestly affect their steady-state levels. Mutation of the ATPase motif in the helicase domain of YTHDC2 did not affect meiotic entry, but it blocked meiotic prophase I progression, causing sterility. Our findings inform a model in which YTHDC2 binds transcripts independent of m6A status and regulates gene expression during multiple stages of meiosis by distinct mechanisms.
Meiosis occurs in a dedicated germ cell lineage in most well-studied metazoans (Kimble 2011; Spradling et al. 2011; Lesch and Page 2012). Following mitotic expansion, extrinsic signals trigger transcriptional and post-transcriptional regulators to remodel the germ cell transcriptome to transition cells from mitotic to meiotic gene expression programs (Kimble 2011; Spradling et al. 2011; Griswold 2016). The mitosis-to-meiosis switch is a fundamental and intricately controlled event, and its misregulation can lead to tumorous mitotic expansion of the germline, depletion of the germline stem cell pool, or meiotic failure (Kimble 2011; Spradling et al. 2011; Griswold 2016).
While meiotic entry control has unifying themes across organisms (for instance, translational control by RNA-binding proteins is often used), the regulatory pathways appear to be organism-specific and are not broadly conserved (Kimble 2011). In mammals, extrinsic signals including retinoic acid and nutrient restriction coupled with cell-intrinsic factors induce the STRA8 complex to bind and up-regulate transcription of a broad array of genes whose expression triggers meiotic entry (Baltus et al. 2006; Bowles et al. 2006; Koubova et al. 2006; Anderson et al. 2008; Lin et al. 2008; Matson et al. 2010; Kojima et al. 2019; Ishiguro et al. 2020; Zhang et al. 2021). Beyond this transcriptional control, however, meiotic entry remained poorly understood. Furthermore, little is known about how entry into meiosis and the subsequent progression of cells through meiotic prophase to meiotic divisions is coordinated.
We and others previously identified YTHDC2 as a critical regulator of meiotic entry (Abby et al. 2016; Bailey et al. 2017; Hsu et al. 2017; Soh et al. 2017; Wojtas et al. 2017; Jain et al. 2018). YTHDC2, along with its binding partner and functional collaborator MEIOC, constitutes a post-transcriptional control pathway that switches cells from mitotic to meiotic gene expression programs. YTHDC2 harbors multiple RNA interaction domains that may directly engage RNA to modulate gene expression. These include an N-terminal R3H domain, a C-terminal YTH domain, and a helicase core module with sequence motifs characteristic of superfamily 2 DExH-box helicases, an ankyrin repeat pair insertion, and two C-terminal HA2 and OB domain extensions (Bailey et al. 2017; Hsu et al. 2017; Wojtas et al. 2017; Jain et al. 2018). YTHDC2 has 3′ → 5′ RNA helicase activity and similarity within its YTH domain to an N6-methyladenosine (m6A) recognition pocket that has been shown to bind m6A-modified RNAs in vitro (Xu et al. 2015; Bailey et al. 2017; Hsu et al. 2017; Wojtas et al. 2017; Jain et al. 2018).
On the basis of this domain architecture and biochemical activities, the current model is that the YTH domain mediates direct interaction with m6A to select targets and regulate their stability and/or translation during the mitosis-to-meiosis transition (Bailey et al. 2017; Hsu et al. 2017; Wojtas et al. 2017; Jain et al. 2018). Direct tests of this model have been lacking, however. For example, it is unknown whether interaction of the YTH domain with m6A is the sole determinant of target selectivity, and the functional significance of YTHDC2 helicase activity is not understood.
Here we used a structure–function approach to dissect how YTHDC2 shapes the gene expression landscape during spermatogenesis. Surprisingly, interaction of the YTH domain with m6A is dispensable for spermatogenesis, whereas ATP hydrolysis activity of the helicase domain is essential for progression through meiotic prophase I. YTHDC2 binds to regions containing U-rich and UG-rich motifs on mRNA targets and affects their steady-state levels, but does not appear to substantially impact target translation during the mitosis-to-meiosis transition.
Results
Mutations in the m6A-binding pocket of YTHDC2 do not disrupt spermatogenesis
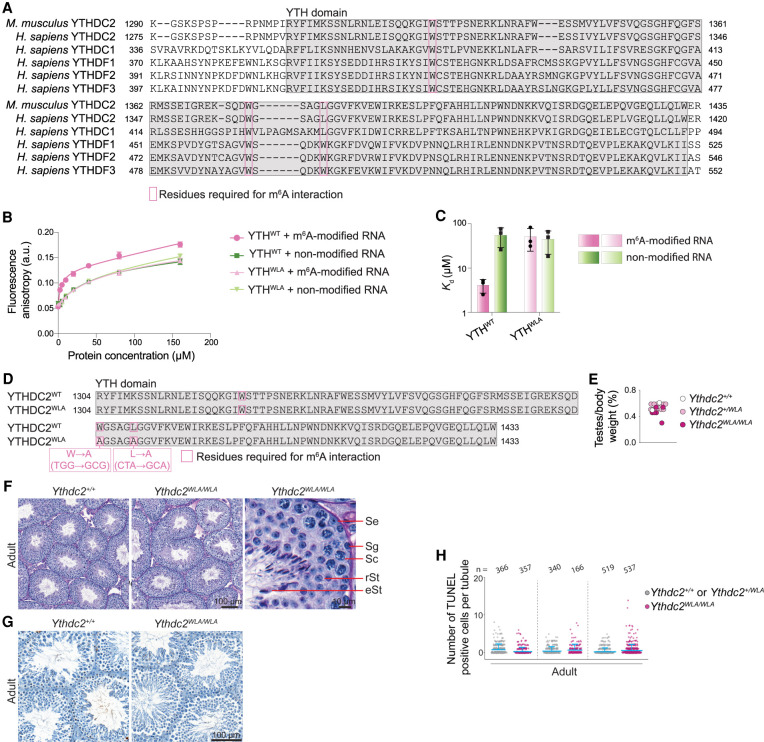
YTH domains in YTHDF1, YTHDF2, YTHDF3, and YTHDC1 recognize m6A residues on RNAs via an aromatic cage composed of two or three tryptophans that encase the methyl group (Luo and Tong 2014; Theler et al. 2014; Xu et al. 2014, 2015; Zhu et al. 2014; Patil et al. 2018). These residues are conserved in human and mouse YTHDC2 (Fig. 1A). To directly test m6A binding, we expressed and purified the YTH domain of mouse YTHDC2 and performed fluorescence anisotropy measurements of RNA interaction with m6A-modified or nonmodified RNAs (Fig. 1B,C). We observed a higher affinity for m6A-containing RNA (Kd = 4.1 µM ± 1.4 µM) compared with nonmodified RNA (Kd = 54 µM ± 26 µM). Mutating two of the three predicted m6A-interacting residues (W1375A and L1380A) eliminated the preferential binding to m6A-modified RNA (Fig. 1B,C). These results are consistent with previous measurements for the YTH domain of human YTHDC2 (Kd = 61.6 µM for unmodified RNA and 8.9 µM for m6A-modified RNA), and mutating the equivalent tryptophan in human YTHDC2 (W1360A) abolished preferential m6A interaction (Xu et al. 2015; Wojtas et al. 2017). This confirms that the YTH domain of mouse YTHDC2 confers m6A binding ability in vitro.
Figure 1.
m6A binding is dispensable for YTHDC2 function. (A) Multiple sequence alignment obtained using Clustal Omega (Sievers et al. 2011) of YTH domains (gray highlight) from mouse YTHDC2 (Uniprot B2RR83) and all five human YTH domain-containing proteins: YTHDC2 (Uniprot Q9H6S0), YTHDC1 (Uniprot Q96MU7), YTHDF1 (Uniprot Q9BYJ9), YTHDF2 (Uniprot Q9Y5A9), and YTHDF3 (Uniprot Q7Z739). Pink boxes indicate positions of m6A-binding residues. (B) Fluorescence anisotropy binding curves for the interaction of the mouse YTHDC2 YTH domain (YTHWT) or the YTH domain carrying m6A-binding pocket mutations W1375A and L1380A (YTHW1375A/L1380A) with m6A-modified or nonmodified RNA. Data points (arbitrary units [a.u.]) represent mean ± SD from three technical replicates and are fitted with a one site total binding equation (GraphPad Prism 9) to obtain the dissociation constant (Kd). (C) Bar graph showing Kd (mean ± SD) obtained from B. (D) The YTH domain (gray highlight) of the CRISPR/Cas9-induced Ythdc2WLA allele with nucleotide and amino acid changes indicated. (E) Ratios of testis weight to body weight for 10- to 24-wk-old mice. (F) Bouin's fixed and periodic acid Schiff (PAS)-stained seminiferous tubule sections from adult (14 wk) testes are shown. (Se) Sertoli cells, (Sg) spermatogonia, (Sc) spermatocytes, (rSt) round spermatids, (eSt) elongated spermatids. (G) Adult (14-wk) testis sections stained with TUNEL and hematoxylin. (H) Quantification of TUNEL staining in three littermate pairs of adult (10- and 14-wk) Ythdc2WLA/WLA and control littermates. The number of seminiferous tubules counted from each animal is indicated (n). Bars indicate mean and SD.
To examine the contribution of m6A interaction to substrate selectivity and YTHDC2 function in vivo, we used CRISPR–Cas9-based genome editing to generate a Ythdc2 allele with both W1375A and L1380A mutations that abolish preferential m6A interaction (referred to here as Ythdc2WLA) (Fig. 1B–D). Heterozygotes (Ythdc2+/WLA) had normal fertility and Mendelian transmission of the mutations (25.3% Ythdc2+/+, 53.5% Ythdc2+/WLA, and 21.2% Ythdc2WLA/WLA from heterozygote × heterozygote crosses; average litter size = 5; n = 269 mice). Surprisingly, however, Ythdc2WLA/WLA homozygotes also displayed normal fertility: Ythdc2WLA/WLA males and females bred to wild type had litter sizes comparable with heterozygotes (average litter size = 6 and n = 32 mice for Ythdc2WLA/WLA females; average litter size = 5 and n = 31 mice for Ythdc2WLA/WLA males). Testes from adult Ythdc2WLA/WLA males were sized similarly to those from heterozygous and wild-type littermates (Fig. 1E) and contained the full array of spermatogenic cells, including spermatocytes and round and elongated spermatids (Fig. 1F). TUNEL staining detected no evidence of an increased frequency of apoptotic cells (Fig. 1G,H), unlike Ythdc2-null animals (Bailey et al. 2017; Hsu et al. 2017; Wojtas et al. 2017; Jain et al. 2018). These results indicate that specific interaction with m6A is dispensable for YTHDC2 function in the germline.
An additional, truncating allele that removes part of the YTH domain resulted in a phenotype similar to a Ythdc2 null, but also yielded undetectable protein levels in immunoblots of testis extracts from juvenile mice (Supplemental Fig. S1). Because of the apparent instability of the truncated protein, we were unable to draw conclusions about whether the YTH domain contributes an important RNA-binding function separate from m6A recognition.
YTHDC2 binds regions containing U-rich and UG-rich motifs in testis mRNAs
Because m6A binding appears dispensable, we focused on identifying the determinants of YTHDC2 RNA substrate selectivity. To this end, we performed YTHDC2 cross-linking and immunoprecipitation (CLIP) in mouse testes. YTHDC2 is expressed throughout meiotic prophase I during spermatogenesis (Abby et al. 2016; Bailey et al. 2017; Jain et al. 2018), so we performed CLIP on testes from adult mice that contain all spermatogenic stages. In addition, because YTHDC2 has critical functions early, when cells transition from mitotic to meiotic divisions (Bailey et al. 2017; Hsu et al. 2017; Wojtas et al. 2017; Jain et al. 2018), we also performed CLIP in testes from animals at 8 and 10 d postpartum (dpp). At these ages, testes are enriched for cells that are about to transition or have just transitioned into meiotic prophase I, respectively (Bellve et al. 1977).
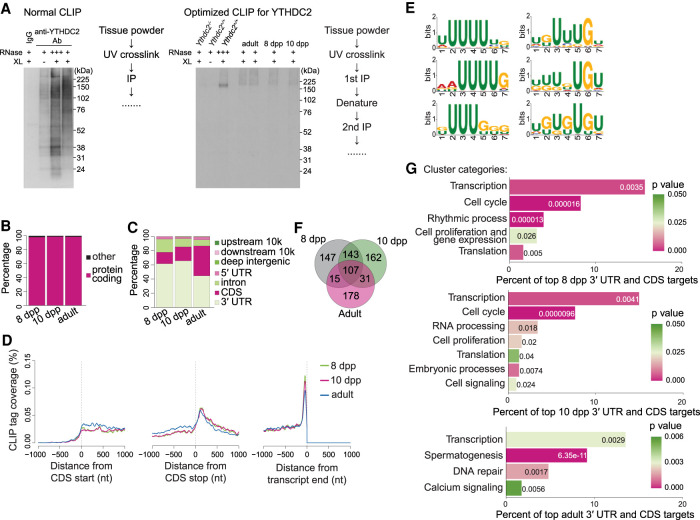
First, we optimized YTHDC2 immunoprecipitation (IP). We identified two commercially available YTHDC2 antibodies that have high specificity and IP efficiency in testis extracts when examined by IP and immunoblotting. Unexpectedly, however, we observed high nonspecific autoradiographic signals in YTHDC2 CLIP samples labeled with T4 polynucleotide kinase and γ-32P ATP (Fig. 2A, left). We therefore optimized the CLIP methodology (Darnell et al. 2018), including steps involving cell lysis, washes, DNase and RNase treatments, and denaturation. This established a new, tailored YTHDC2 CLIP protocol involving two sequential IPs with the two specific antibodies identified plus an intermediate denaturation step (see the Supplemental Material).
Figure 2.
Optimized YTHDC2 CLIP in juvenile and adult testes. (A) Autoradiograph images of YTHDC2 CLIP samples labeled with T4 polynucleotide kinase and γ-32P ATP using normal and optimized conditions. Frozen tissue powder from 8-dpp, 10-dpp, and adult (8.5-wk) testes was subjected to UV cross-linking (XL) and either high RNase (+++) or low RNase (+) digestion conditions prior to IP of YTHDC2. RNA fragments were subsequently used for cDNA library preparation and analysis. (B) Biotype of RNA with YTHDC2 CLIP peaks. (C) Distribution of YTHDC2 CLIP peak locations across genic regions. (D) Metatranscript coverage profiles of YTHDC2 CLIP tags on protein-coding transcripts. The Y-axis indicates the percentage of YTHDC2 CLIP tags mapped at the indicated transcript positions (in nucleotides [nt]) when anchored to translation start (left), translation stop (middle), or transcript end (right). Unique CLIP tags obtained after collapsing PCR duplicates were used for analysis. (E) YTHDC2-binding motifs enriched in cross-linking-induced truncation sites (CITSs) by MEME-ChIP motif analysis (Machanick and Bailey 2011). Fifty-nucleotide-long sequences flanking CITSs (P < 0.001) on both 5′ and 3′ ends were used for motif analysis. Significant motifs (E-value ≤ 0.05) were clustered by similarity and ordered by E-value using MEME-ChIP. Shown are the three most significant motifs from the most significant cluster (left panel; E-value ≤ 7.2 × 10−1799) and the next most significant cluster (right panel; E-value ≤ 1.2 × 10−466). (F) Overlap of top CLIP transcripts between mice of different ages. (G) Functional clusters of GO terms (P < 0.05) enriched in the top YTHDC2 CLIP targets. P-values are indicated by bar colors and are listed in or adjacent to bars.
Our optimized CLIP protocol yielded a single autoradiographic band at the expected size of YTHDC2 (161 kDa) in high RNase A-treated and UV cross-linked testis extracts from wild type (Fig. 2A, right, third lane). This signal was absent in Ythdc2-null and no UV cross-linked negative controls (Fig. 2A, right, first and second lanes). The expected smeared signals were detected in low RNase A-treated and UV cross-linked testis extracts from adult, 8-dpp and 10-dpp animals (Fig. 2A, right, fourth through seventh lanes).
We performed three or four CLIP biological replicates at each mouse age examined to generate totals of 345,785, 404,997, and 1,636,477 unique YTHDC2 CLIP tags at 8 dpp and 10 dpp and in adults, respectively. This analysis yielded 2478, 3764, and 21,313 YTHDC2 CLIP peaks (with filters for P < 0.05, peak height [PH] ≥ 5, biological complexity [BC] ≥ 2, and CLIP tag in Ythdc2 null = 0) at 8 dpp and 10 dpp and in adults, respectively.
Over 98.4% of total YTHDC2 CLIP peaks were within protein-coding transcripts at all ages examined (Fig. 2B). We also observed YTHDC2 peaks within piRNA precursor transcripts (Supplemental Fig. S2), consistent with a previous report that investigated YTHDC2 testis targets by formaldehyde fixation followed by YTHDC2 IP (Bailey et al. 2017). Moreover, 57.9% (8 dpp) and 65.4% (10 dpp) of YTHDC2 peaks were located within the 3′ UTRs of transcripts, and 12.4% and 17.2% were within protein-coding regions (Fig. 2C). This binding pattern shifted modestly toward the coding region in adults, with 48.8% of peaks in 3′ UTRs and 36.9% in protein-coding regions (Fig. 2C). Metaplot analysis revealed a sharp enrichment of YTHDC2 CLIP tags near the ends of 3′ UTRs (Fig. 2D) at all ages examined.
Motif analysis in cross-linking-induced truncation sites (CITSs) (Zhang and Darnell 2011; Moore et al. 2014; Ule et al. 2018) identified enrichment for a series of U-rich and UG-rich motifs (Fig. 2E). Most CITSs (64.9%) contained U-rich or UG-rich sequences within a 10-nt window encompassing the CITS (53.2% contained UUU and 27.6% contained UGU within a 10-nt window).
Comparison of the top CLIP targets (PH ≥ 8, PH ≥ 10, and PH ≥ 40 for 8 dpp, 10 dpp, and adult, respectively) between different ages indicates that YTHDC2 targets vary across germ cell developmental stages. Testes at 8 dpp contain premeiotic spermatogonia and cells in early meiotic prophase I (Bellve et al. 1977). Testes at 10 dpp additionally contain germ cells that have progressed further into meiotic prophase I. Consistent with this, 60.7% of 8-dpp CLIP targets overlapped with those at 10 dpp (Fig. 2F).
Testes from adult mice contain a mixture of postmeiotic cells and cells in late stages of meiotic prophase I, as well as cells in early meiotic prophase I, with the latter category comprising a small proportion of total cell types. Furthermore, the germ cell transcriptomes of cells in early and late substages of meiotic prophase I stages are vastly different (Ball et al. 2016), so we expect adult CLIP targets to reflect YTHDC2-binding preferences during late meiotic prophase I stages. Of the 8-dpp CLIP targets, 29.6% were also top targets in adults, but more than half of adult targets (53.8%) were unique to adults (not shared with 8 or 10 dpp) (Fig. 2F).
To explore the biological pathways regulated by YTHDC2, we performed gene ontology (GO) analysis followed by functional annotation clustering of top CLIP targets (Fig. 2G). YTHDC2 target mRNAs in juvenile testes were enriched for transcripts involved in cell cycle regulation, including Ccna2. This is consistent with YTHDC2 function in regulating the mitosis-to-meiosis transition, with previously reported GO analysis of YTHDC2 testis targets identified by formaldehyde fixation followed by YTHDC2 IP (Bailey et al. 2017), and with misregulation of CCNA2 protein expression during meiotic entry in Ythdc2 mutants (Bailey et al. 2017; Jain et al. 2018). CLIP targets at 8 and 10 dpp were also enriched for transcripts involved in regulation of transcription (e.g., Sox9, Suz12, Tgif1, Cbx3, Cbx4, Smad7, Hdac2, Hira, E2f6, and Mga), suggesting that YTHDC2 may indirectly regulate transcription as germ cells enter meiosis.
The adult target set is expected to be enriched for YTHDC2 targets in late meiotic prophase I substages. Consistent with this, the most significant GO term was spermatogenesis (Fig. 2G). This category contained several transcripts involved in spermatid development (e.g., Prm1, Prm2, Spink2, Odf2, Meig1, Tnp1, and Agfg1). It also contained Meioc, a binding partner and functional collaborator of YTHDC2 (Abby et al. 2016; Soh et al. 2017) that was identified as a potential YTHDC2 target previously (Bailey et al. 2017). Other targets in this cluster included genes implicated in meiotic prophase I progression (Hspa2 and Rpl10l) and the piRNA pathway (Mael and Pld6). Additionally, transcripts involved in DNA repair (e.g., H2ax, Rad23b, Setx, Yy1, Cul4a, Mum1, and Polh) were enriched among adult targets, implicating YTHDC2 in potentially modulating meiotic recombination and/or meiotic sex chromosome inactivation processes during later prophase I stages. As in juveniles, the top adult targets were also enriched for transcription factors, including Mybl1. MYBL1 is a master regulator of meiotic genes required for cell cycle progression through late meiotic prophase (Bolcun-Filas et al. 2011). Thus, YTHDC2 may shape diverse meiotic processes to regulate meiotic progression.
YTHDC2-binding sites do not overlap with m6A-modified sites
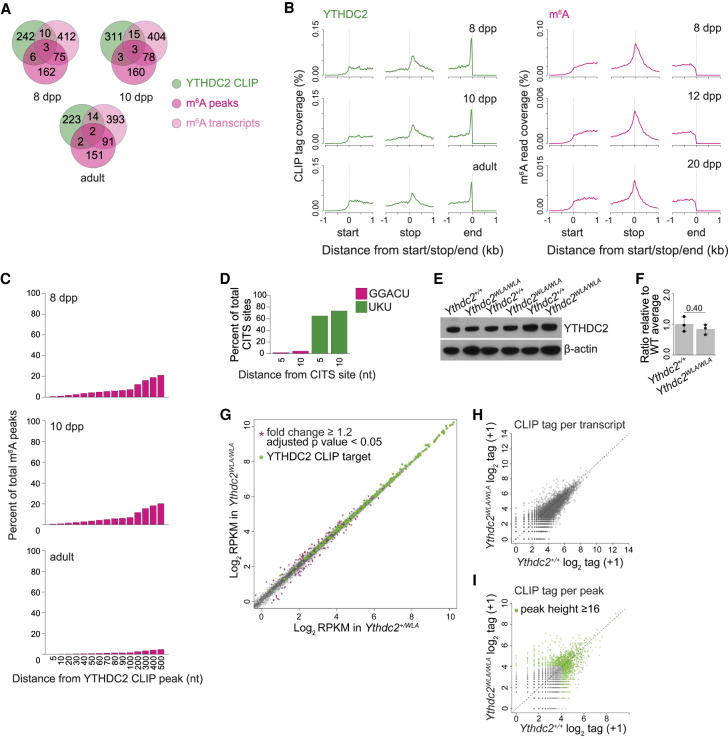
To determine whether YTHDC2 binds m6A in vivo, we compared our YTHDC2 CLIP maps with existing testis m6A maps (Wojtas et al. 2017). Because the m6A landscape changes as cells progress through spermatogenesis (Wojtas et al. 2017), we compared 8- and 10-dpp CLIP targets with 8- and 12-dpp m6A maps, respectively, and compared adult CLIP targets with m6A maps generated from 20-dpp mice.
There was remarkably little congruence between the YTHDC2 CLIP and m6A maps. m6A-enriched transcripts (which we defined as transcripts containing the top 250 m6A peaks) mapped onto 246, 244, and 246 genes at 8 dpp and 10 dpp and in adults, respectively. Very few of these transcripts overlapped with the top YTHDC2 targets at any age (Fig. 3A). For example, at 8 dpp, only nine of 246 m6A-enriched transcripts overlapped with the 261 top YTHDC2 targets. Similarly, <4% of the top 500 m6A genes ranked by Wojtas et al. (2017) were among the top YTHDC2 CLIP targets (Fig. 3A) at any of the ages examined.
Figure 3.
YTHDC2 binds distinct transcripts at distinct positions relative to the m6A mark. (A) Venn diagrams showing overlap between the top YTHDC2 CLIP targets and transcripts containing m6A in testes. Transcripts containing the top 250 m6A peaks identified by reanalysis of raw data from Wojtas et al. (2017) are shown in dark pink, and the list of the top 500 m6A-containing transcripts reported by Wojtas et al. (2017) are shown in light pink. CLIP data from 8 dpp, 10 dpp, and adults were compared with m6A data from 8-, 12-, and 20-dpp testes, respectively. (B) Metatranscript coverage profiles of YTHDC2 CLIP unique tags and reads from m6A IPs on protein-coding transcripts. The Y-axis indicates the percentage of total CLIP tags or IP reads mapped within 1 kb of the translation start sites (left), translation stop sites (middle), or the ends of transcripts (right). (C) Bar graph showing the distribution of distances between m6A peaks and YTHDC2 CLIP peaks when present on the same transcript. The Y-axis shows the percentage of total m6A peaks that are within a defined distance from a YTHDC2 CLIP peak (X-axis). (nt) Nucleotides. (D) Proportion of YTHDC2 CITSs containing either the m6A consensus motif GGACU or UKU (where K is U or G) within 5 or 10 nt. (E) Immunoblot analysis of YTHDC2 protein levels in testis extracts from adult (20- and 24-wk) Ythdc2WLA/WLA and wild-type animals. Immunoblot for β-actin served as a loading control. (F) Quantification of YTHDC2 protein levels in E. Mean values from three mice per genotype are plotted. SD are indicated, and P-value is listed (Student's t-test). (G) Gene expression (reads per thousand mapped [RPKM]) in testes from adult (20- and 24-wk) Ythdc2WLA/WLA and Ythdc2+/WLA littermates. Three mice per genotype were analyzed. Gray dots represent transcripts with comparable values between Ythdc2+/WLA and Ythdc2WLA/WLA. Purple stars denote transcripts with significantly altered expression in Ythdc2WLA/WLA compared with Ythdc2+/WLA (fold change ≥ 1.2 and adjusted P-value < 0.05). Green dots show the top YTHDC2 CLIP targets in adults. (H,I) Comparison of YTHDC2 CLIP between Ythdc2+/+ and Ythdc2WLA/WLA testes from adult animals (20 and 24 wk). Scatter plots show the correlation of total YTHDC2 CLIP unique tag numbers (obtained by independent CLIP assays in three animals per genotype) on individual transcripts (H) or CLIP peaks (I). A pseudocount of one was added for calculating log2 scores used in the X- and Y-axes. Each black dot represents a transcript (H) or a YTHDC2 CLIP peak (I). Each green dot in I represents a top ranked CLIP peak (peak height ≥ 16) in both Ythdc2+/+ and Ythdc2WLA/WLA.
Metaplot analysis of m6A read coverage showed the most enrichment over the end of the coding region (stop codon) (Wojtas et al. 2017), which is in contrast to the dramatic enrichment of YTHDC2 CLIP tags near the transcript end (Figs. 2D, 3B). We also observed a modest enrichment of YTHDC2 near the start of 3′ UTRs, but this enrichment was shifted into the 3′ UTRs as compared with the m6A enrichment over the stop codons. When individual transcripts were compared for YTHDC2 binding and m6A presence, <0.67% and <7.4% of m6A peaks were detected in 5- and 100-nt windows, respectively, located upstream of or downstream from YTHDC2 CLIP peaks (Fig. 3C). Consistent with this, the m6A consensus motif (GGACU) was present in <4.3% of 5- and 10-nt windows located upstream of and downstream from YTHDC2 CITSs (Fig. 3D). In contrast, 64.9% and 73.5% of sites contained the U/UG-rich motifs in these 5- and 10-nt window ranges, respectively (Fig. 3D). We conclude that YTHDC2 recognizes distinct transcripts and distinct positions compared with the reported distribution of m6A in testes.
To examine the effect of abrogating the m6A-binding ability of YTHDC2 on its RNA interaction landscape, we performed YTHDC2 CLIP in testes from Ythdc2WLA/WLA adults. The YTHDC2 protein level was comparable between Ythdc2+/+ and Ythdc2WLA/WLA (Fig. 3E,F). RNA sequencing of Ythdc2WLA/WLA adults showed no significant changes in the abundance of YTHDC2 targets in Ythdc2WLA/WLA compared with heterozygous littermate controls (Fig. 3G). The levels of just a few transcripts were significantly altered (72 transcripts up and 95 transcripts down), but none of these were top CLIP targets. Three CLIP biological replicates in Ythdc2+/+ and Ythdc2WLA/WLA generated totals of 788,570 and 662,315 unique YTHDC2 CLIP tags, respectively. We observed strong positive correlations in YTHDC2 CLIP tag numbers per transcript (R = 0.99) (Fig. 3H), as well as per CLIP peak (R = 0.98) (Fig. 3I) between Ythdc2+/+ and Ythdc2WLA/WLA. These findings reveal that loss of the ability of YTHDC2 to engage specifically with m6A does not substantially alter its RNA-binding preferences in vivo.
YTHDC2 loss alters levels of direct targets with little or no effect on their ribosome occupancy
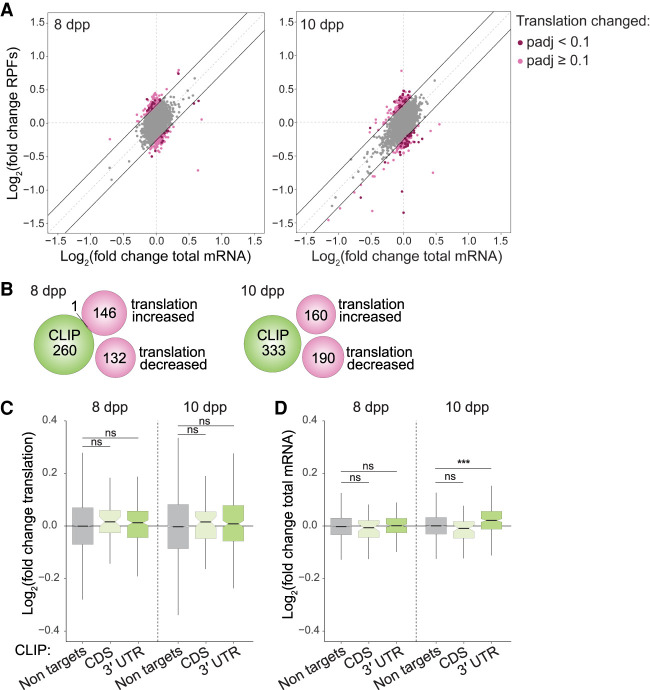
Depletion of YTHDC2 has been reported to affect translation of m6A-containing transcripts and of luciferase reporter constructs in HEK293 cells (Mao et al. 2019), so we asked whether YTHDC2 controls translation of its targets during spermatogenesis. We examined ribosome occupancy as a readout for translation by performing ribosome profiling in Ythdc2−/− testes. Because Ythdc2−/− testis cells begin to exhibit cell death at ∼10 dpp as they transition into meiosis during the first wave of spermatogenesis (Bellve et al. 1977; Abby et al. 2016; Bailey et al. 2017; Jain et al. 2018), we examined testes from 8- and 10-dpp wild-type and Ythdc2 knockout mice. Ribosome profiling showed a mild change in Ythdc2−/− translation efficiency (TE) compared with control littermates for 25 and 53 transcripts (TE fold change ≥ 1.2 and adjusted P-value < 0.1) at 8 and 10 dpp, respectively (Fig. 4A). None of the changed transcripts were among the top YTHDC2 targets at the corresponding ages.
Figure 4.
Molecular effects of complete YTHDC2 loss on direct RNA targets during meiotic entry. (A) Scatter plots showing log2 fold changes for the mRNA input and ribosome protected fragments (RPFs) as analyzed by anota2seq (Oertlin et al. 2019) in whole testes from Ythdc2 knockouts compared with wild-type or heterozygous littermates at 8 and 10 dpp. Three pairs of knockout and wild-type or heterozygous littermates were examined for each age. The default anota2seq parameter of translational efficiency (TE; i.e. RPF/input) fold change ≥ 1.2 (threshold denoted by solid black lines) was applied. Transcripts meeting this criterion are shown in light pink. Transcripts that met this criterion and also had an adjusted P-value < 0.1 were considered to have significantly different TEs and are shown in dark pink. (B) Venn diagrams illustrating the almost complete lack of overlap of top YTHDC2 CLIP targets with transcripts that show TE fold change ≥ 1.2 (irrespective of P-value; light-pink transcripts in A) at 8 and 10 dpp. (C) Comparison of the change in TE in Ythdc2 knockout at the indicated ages for nontargets and the top 250 (based on CLIP peak height) YTHDC2 targets, categorized according to the location of CLIP peaks. (D) Change in steady-state mRNA levels of the top 250 (based on CLIP peak height) YTHDC2 targets in Ythdc2 knockout compared with wild type at 8 and 10 dpp. The results of Mann–Whitney tests are shown in C and D. (ns) Not significant (P > 0.05), (***) P < 0.001.
To search for evidence of more subtle effects on translation, we reduced the stringency of the cutoff to expand the set of potentially affected transcripts (namely, including those that had a TE fold change ≥ 1.2 and adjusted P ≥ 0.1). The additional transcripts defined this way still showed no overlap with the top YTHDC2 target set at 10 dpp and yielded only one overlapping transcript (encoding Rad9a) at 8 dpp (Fig. 4B). Furthermore, the top YTHDC2 CLIP targets at both ages examined did not show a significant change in translation efficiency in Ythdc2−/− compared with wild type (P = 0.178 and P = 0.201 by Mann–Whitney test at 8 and 10 dpp, respectively) (Fig. 4C), irrespective of whether they were bound by YTHDC2 in the CDS or 3′ UTR.
Instead, transcript levels of the top YTHDC2 3′ UTR targets at 10 dpp were modestly but significantly increased in Ythdc2−/− (P = 3.07 × 10−10 by Mann–Whitney test for 3′ UTR targets compared with nontargets) (Fig. 4D), consistent with previous reports (Bailey et al. 2017; Hsu et al. 2017; Wojtas et al. 2017; Jain et al. 2018). The increased abundance of targets upon loss of YTHDC2 indicates that the normal function of YTHDC2 may be to enhance their degradation when bound in the 3′ UTR. Taken together, these findings suggest that YTHDC2 may directly modulate RNA levels, but control of RNA translation contributes more modestly, if at all, to the mechanism of YTHDC2 action during the mitosis-to-meiosis transition.
Mutating the helicase ATPase motif confers dominant male sterility
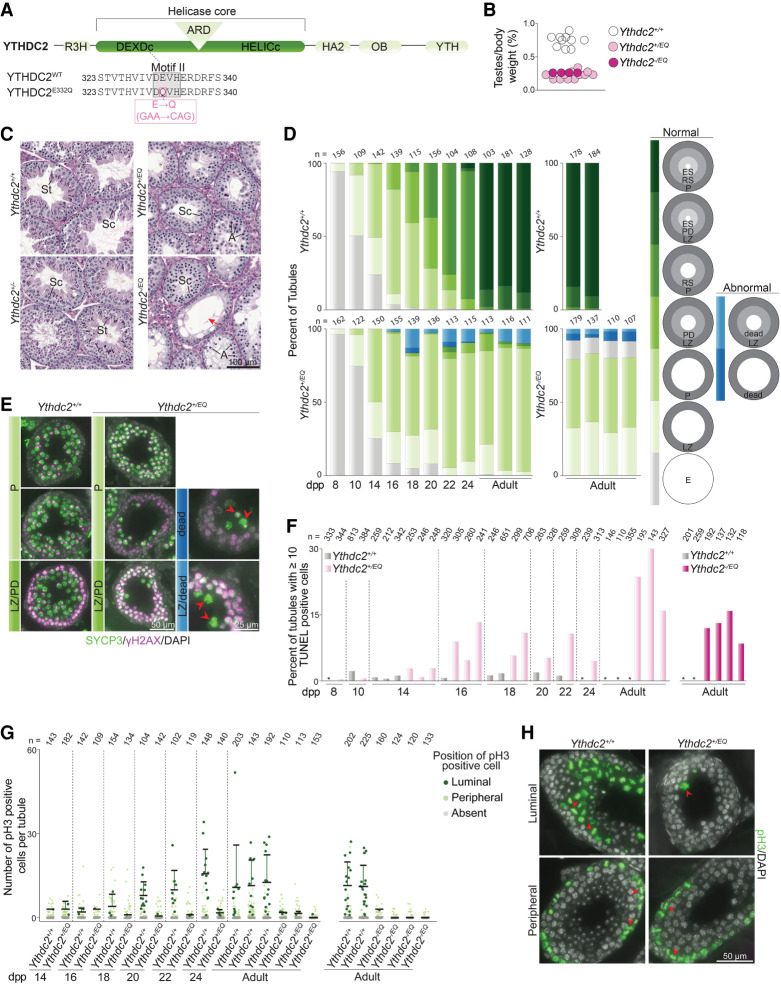
We asked whether the helicase activity of YTHDC2 is important. To answer this question, we generated mice harboring a single point mutation (E332Q) within motif II in the helicase DEXDc module of YTHDC2 using TALENs (Fig. 5A). This mutation is expected to maintain ATP binding while disrupting hydrolysis, and the equivalent mutation in human YTHDC2 has been shown to abolish ATP hydrolysis and helicase activity in vitro (Wojtas et al. 2017). Heterozygous females carrying the E332Q allele (referred to here as Ythdc2EQ) were fertile and showed normal Mendelian transmission of the mutation (51.2% Ythdc2+/+ and 48.5% Ythdc2+/EQ from heterozygote × wild type crosses; n = 344 mice). However, Ythdc2+/EQ heterozygous males were sterile; none of the seven animals tested sired offspring when bred with wild-type females for >14 wk, and testes were 33.5% of the size of those in littermate controls (Fig. 5B). Histopathology turned up no somatic defects in male or female heterozygotes, suggesting that the E332Q mutation does not impact nonmeiotic cells when heterozygous (see the Supplemental Material).
Figure 5.
YTHDC2 ATPase mutation disrupts meiotic progression. (A) Schematic of mouse YTHDC2 domain structure (top) and sequences around motif II (gray highlight) within the helicase core domain (bottom) are shown. The amino acid change (pink box) and corresponding nucleotide changes in motif II of the TALEN-induced Ythdc2EQ allele are indicated. (B) Testis weight to body weight ratios for 5- to 36-wk-old mice. (C) Bouin's fixed and PAS-stained seminiferous tubule sections from 9-wk-old animals harboring Ythdc2EQ and Ythdc2 knockout (Ythdc2−) alleles and from wild-type littermates. (Sc) Spermatocytes, (St) spermatids, (A) cells with compacted apoptotic-like nuclei, (red arrow) tubules with only a single layer of cells (spermatogonia plus Sertoli cells). (D) Quantification of the proportion of seminiferous tubule types in testes from Ythdc2+/EQ, Ythdc2−/EQ, and wild-type littermates of the indicated ages. Normal tubule types are categorized based on the meiotic prophase I stage of cell layers present within them as judged by SYCP3 and γH2AX staining. Cell layers are as follows: empty with no SYCP3 staining (E), leptonema or zygonema (LZ), pachynema (P), pachynema or diplonema (PD), round spermatids (RS), and elongating spermatids (ES). Abnormal tubule types in Ythdc2+/EQ and Ythdc2−/EQ that contained cells with fragmented and intense SYCP3 staining indicative of dying cells were also quantified. (E) Example images of tubule categories quantified in D taken from 24-dpp mice. Arrowheads indicate presumptive dying cells (based on SYCP3 staining). (F) Quantification of TUNEL staining (images in Supplemental Fig. S3A) in Ythdc2+/EQ, Ythdc2−/EQ, and control littermates. Wild-type animals with 0% tubules containing ≥10 TUNEL-positive cells are indicated with an asterisk. Each lane represents one animal, and the number of seminiferous tubules counted is indicated (n). (G) Quantification of pH3 staining in seminiferous tubules from Ythdc2+/EQ, Ythdc2−/EQ, and control littermates of the indicated ages. pH3-positive cells were categorized based on their location within the tubule (luminal or peripheral). The number of seminiferous tubules counted is indicated (n), and bars show mean and SD for the distribution of pH3-positive cells that are luminally located. (H) Example images from 24-dpp mice of tubules quantified in G. Arrowheads point to pH3-positive cells that were categorized as being luminally located (top) or located near the periphery (bottom). (Top right) A rare tubule type containing one luminally-located pH3-positive cell is shown for Ythdc2+/EQ. In D, F, and G, adult Ythdc2+/EQ animals were 5 and 14 wk old, and Ythdc2−/EQ animals were 8–9 wk old.
YTHDC2 ATPase mutation causes spermatocyte death after meiotic entry
Complete loss of YTHDC2 function in Ythdc2−/− causes sterility and hypogonadism because of spermatocyte apoptosis during the mitosis-to-meiosis transition (Bailey et al. 2017; Wojtas et al. 2017; Jain et al. 2018). In contrast, the E332Q mutation does not appear to compromise the meiotic entry function of YTHDC2: Testis sections from Ythdc2+/EQ adults contained meiotic spermatocytes but lacked postmeiotic round and elongated spermatids that were present in wild-type littermate controls (Fig. 5C). This suggested that the sterility in Ythdc2+/EQ animals is caused by a defect during meiotic prophase I, so we evaluated this phenotype more closely.
We tracked meiotic progression during the first wave of spermatogenesis when cells enter meiosis semisynchronously (Bellve et al. 1977) by immunostaining testis sections for the diagnostic γH2AX and SYCP3 markers that display distinctive patterns as cells progress through meiotic prophase (Fig. 5D). In normal meiosis, chromosomes acquire DSBs early in prophase I (leptonema and zygonema), resulting in florid staining of the DSB marker γH2AX (Mahadevaiah et al. 2001); this staining is lost with γH2AX restricted to the sex chromosomes in late prophase (pachynema and diplonema) when the bulk of DSBs are repaired. Concurrently, chromosomes start to form SYCP3-positive protein-rich backbones called axial elements during leptonema (Lammers et al. 1994; Zickler and Kleckner 2015); these elongate during zygonema as homologous chromosomes pair to form a proteinaceous scaffold called the synaptonemal complex (SC). The SC connects homologs along their lengths in pachynema, and then the SC disassembles at the end of prophase in diplonema. These meiotic prophase stages (leptonema, zygonema, pachynema, and diplonema) are present as distinguishable cell layers within individual seminiferous tubules.
In wild type, tubules containing a layer of cells in leptonema were present at 8 dpp, and by 14 dpp, 25% of tubules contained leptonema/zygonema and 50% contained cells that had progressed to pachynema (Fig. 5D). At 16 dpp, most tubules contained cells in pachynema, and 18% of tubules contained cells in late pachynema or diplonema along with a second, new layer of cells entering meiosis (which were in leptonema or zygonema). By 18 dpp, the latter tubules had progressed to contain postmeiotic round spermatids and cells in pachynema. Tubules containing elongating spermatids appeared at 24 dpp and were the predominant tubule type in adults.
Tubule patterns in Ythdc2+/EQ animals were like wild type at 8, 10, and 14 dpp (25% and 24% of tubules contained leptonema/zygonema and 50% and 50% contained pachynema at 14 dpp in wild type and heterozygotes, respectively) (Fig. 5D). However, abnormal patterns emerged at later ages. From 16 dpp onward, we observed fewer tubules in the mutant containing one layer of cells in late pachynema/diplonema plus a layer in leptonema/zygonema (1% at 16 dpp for heterozygotes vs. 18% in wild type) (Fig. 5D). Moreover, tubules in this category, when present, had markedly fewer cells in the late pachytene/diplotene layer (Fig. 5E). We also observed a depletion in pachytene cells in some tubules that contained a single pachytene layer (Fig. 5E). Heterozygote testes also had abnormal tubules containing cells that appeared to be dying 16 dpp onward (Fig. 5D,E). Consistent with this, TUNEL staining detected apoptotic tubules in heterozygotes at 16 dpp and later ages, while tubules from younger animals were similar to wild-type littermates (Fig. 5F; Supplemental Fig. S3A,B). Tubules containing round or elongating spermatids were absent in Ythdc2+/EQ (Fig. 5D). These results indicate that meiotic prophase I progression is blocked in Ythdc2+/EQ animals during late pachynema and/or diplonema.
The meiotic prophase I arrest observed in Ythdc2+/EQ is in contrast to the phenotype observed upon complete loss of YTHDC2. Ythdc2−/− cells carry out an abortive attempt at meiosis: They express hallmark meiotic proteins and initiate meiotic recombination but proceed prematurely to an aberrant metaphase-like state (Bailey et al. 2017; Wojtas et al. 2017; Jain et al. 2018). As a result, early prophase I stages (leptonema/zygonema) are depleted in Ythdc2−/−juveniles and adults, whereas they were present in Ythdc2+/EQ (Fig. 5D). We also did not detect evidence of rampant apoptosis during early ages in Ythdc2+/EQ (8, 10, and 14 dpp in Fig. 5F; Supplemental Fig. S3A,B), unlike in Ythdc2−/−. Moreover, we could not find cells that contained abnormally condensed metaphase-like chromosomes in Ythdc2+/EQ adults (Fig. 5C); such cells are present within the nearly empty tubules from Ythdc2−/− adults.
To closely assess metaphase, we stained Ythdc2+/EQ testis sections for the metaphase marker histone H3 phosphorylated on serine 10 (pH3). pH3 staining in both wild type and Ythdc2−/− detects mitotic spermatogonia located near the periphery of tubules (Bailey et al. 2017; Jain et al. 2018). Ythdc2−/− tubules additionally contain pH3-positive cells near the tubule lumen at 14 dpp due to premature entry of abnormal spermatocytes into metaphase, whereas luminally located pH3-positive cells are expected to appear later in wild type, at 18 dpp, when normal spermatocytes reach the first meiotic division (Bellve et al. 1977). pH3 staining detected the appearance of luminally located metaphase cells in wild type at 18 dpp and in older animals, as expected (Fig. 5G,H). These were absent or extremely rare in Ythdc2+/EQ littermates (Fig. 5G,H), consistent with the observed depletion of cells in late meiotic prophase (Fig. 5D). We also did not observe luminally located pH3-positive cells at younger ages in Ythdc2+/EQ (Fig. 5G), confirming that Ythdc2+/EQ mutants do not have the premature metaphase entry phenotype characteristic of complete loss of YTHDC2 function.
Collectively, these findings suggest that the E332Q mutation does not disrupt the meiotic entry function of YTHDC2, but instead causes defects in progression through late meiotic prophase I. To assess whether the meiotic entry function of YTHDC2 may be conferred by the presence of wild-type protein in Ythdc2+/EQ, we analyzed testis sections from Ythdc2−/EQ hemizygotes (mice containing the E332Q mutation along with a knockout allele). Ythdc2−/EQ adult tubules were devoid of postmeiotic cells, as expected (Fig. 5C). Largely empty tubules containing early spermatogenic cells and cells with apoptotic morphology were present alongside tubules containing meiotic spermatocytes, suggesting that cells can progress into prophase I in the absence of YTHDC2 ATPase activity, but likely arrest after meiotic entry, resulting in the empty tubules. Compared with wild-type littermates, Ythdc2−/EQ had fewer tubules containing a single pachytene layer (47%–51% for mutants vs. 86%–92% in wild type with additional spermatid layers) and no tubules containing one layer of late pachynema/diplonema plus a layer in leptonema/zygonema (0% for mutants vs. 8%–14% in wild type with an additional spermatid layer) (Fig. 5D), consistent with an arrest during late pachynema. This was accompanied by the presence of apoptotic tubules in Ythdc2−/EQ adults (Fig. 5F; Supplemental Fig. S3A,B), and testes were 33.4% of the size in littermate controls (Fig. 5B). Additionally, we observed none of the abnormal metaphase-like cells characteristic of Ythdc2-null mutants (no luminally located cells in 416 tubules counted from three adult Ythdc2−/EQ mice) (Fig. 5C), and pH3-positive luminally located metaphase cells were absent or extremely rare (Fig. 5G). We observed infertility in both Ythdc2−/EQ females and males: None of the four females or three males paired with wild type for 13 and 5 wk, respectively, led to pregnancies or birth of offspring. We conclude that YTHDC2 helicase activity is dispensable for meiotic entry, but is required for meiotic progression.
Discussion
We previously identified YTHDC2 as a meiotic regulator in a forward genetics screen (Jain et al. 2018), and here we lay down principles underlying its molecular function in shaping the meiotic gene expression landscape. YTHDC2 is an m6A reader in vitro, but this activity is dispensable in vivo during gametogenesis. YTHDC2 helicase activity is not essential for meiotic entry but is required for normal progression through prophase I. Disruption of helicase activity causes catastrophic failure of spermatogenesis and leads to both male and female sterility.
There are five YTH domain-containing proteins in mammalian genomes, and structural studies have demonstrated that an aromatic cage composed mainly of tryptophans in the YTH domain encases the methyl group and confers selectivity for m6A binding (Zaccara et al. 2019). On the basis of the conservation of the m6A-interacting residues, the prevailing model has been that YTHDC2 exerts its effects by recognizing m6A. However, our findings unexpectedly show that m6A is not a significant determinant of target selectivity during spermatogenesis. YTHDC2 binds distinct transcripts at distinct positions compared with those reported to contain m6A. Furthermore, disruption of the aromatic cage causes no detectable changes during gametogenesis and does not substantially alter the identity of transcripts bound or the extent of transcript binding in vivo. Of note, the YTH domain of YTHDC2 binds m6A with ∼20-fold lower affinity than that of other YTH domain-containing proteins in vitro (Xu et al. 2015) and, in contrast to the other four mammalian YTH-containing proteins, YTHDC2 has additional nucleic acid-interacting modules, including the helicase domain with HA2 and OB domain extensions and an R3H domain (Patil et al. 2018). Target selectivity may be conferred principally by these other domains and/or may be modulated by its protein partners, such as MEIOC.
Previous efforts to identify YTHDC2 targets relied on RNA immunoprecipitation (RIP) from mouse testes (Abby et al. 2016; Bailey et al. 2017; Hsu et al. 2017; Soh et al. 2017), but the lists of putative targets differed between studies. We found that YTHDC2 immunoprecipitation during conventional CLIP yields substantial amounts of RNA that is not cross-linked directly to YTHDC2 and is removed in our optimized protocol, consistent with prior findings of indirect and/or high nonspecific background results with RIP (Mili and Steitz 2004; Darnell 2010). These indirect RNA interactions may be biologically meaningful (for instance, if YTHDC2 is part of a larger complex that contains other RNA-interacting proteins), but they do not appear to represent direct YTHDC2-binding targets. Thus, limited stringency of anti-YTHDC2 RNA IP may explain at least some of the disparities in previous reports. Differences in the ages of the mice examined may also contribute. YTHDC2–RNA interactions have also been profiled in cell lines (Patil et al. 2016; Hsu et al. 2017; Mao et al. 2019), but these may not reflect function in germ cells with their unique transcriptome.
A previous study using YTHDC2 CLIP in adult mouse testes (Hsu et al. 2017) reported enrichment for the m6A consensus GGACU motif and clustering of binding sites around stop codons where m6A is typically enriched, including during spermatogenesis (Bailey et al. 2017; Hsu et al. 2017; Wojtas et al. 2017). Our CLIP data disagree: We observed poor enrichment of GGACU near cross-linked YTHDC2–RNA sites. While it is formally possible that YTH domain binding to m6A-containing sites occurs but was not detected, we consider this unlikely because the UV cross-linking step used in our CLIP protocol is standard for tissues, the YTH domain of YTHDC2 (specifically the presumptive RNA-interacting tryptophan cage) is highly similar to that of other YTH domain-containing proteins that have been previously analyzed by CLIP, and it is unlikely that the YTH domain of YTHDC2 would uniquely be a poor cross-linker. Our findings are consistent with the previously reported lower affinity of YTHDC2 for m6A compared with other YTH domain-containing proteins in vitro (Xu et al. 2015).
While we did observe a small enrichment of YTHDC2 binding downstream from translation stop sites, this was offset from where m6A is enriched and is retained in the YTH domain mutant, and thus is unlikely to be related to m6A recognition. Additionally, the proportion of YTHDC2 cross-linking sites that harbor the m6A consensus sequence (GGACU) in their vicinity (<4%) does not change in the Ythdc2WLA/WLA mutant. We infer that our optimized CLIP assay, which involves purification to a thus far unprecedented degree of stringency and has single-nucleotide resolution, is more likely to have captured true in vivo interactions of YTHDC2 with RNA during spermatogenesis. In addition, the lack of a meiotic phenotype and the lack of significant alterations in the YTHDC2–RNA interaction landscape in YTH domain mutants reinforce that YTHDC2 does not selectively engage m6A during spermatogenesis. Instead, our CLIP data suggest that YTHDC2 binds at or adjacent to U-rich and UG-rich motifs in the 3′ UTR and coding sequence of transcripts, consistent with our previous finding that YTHDC2 is more efficient at unwinding RNA duplexes with a 3′ poly(U) overhang than a 3′ poly(A) overhang in vitro (Jain et al. 2018).
We previously reported that the YTH domain of YTHDC2 orthologs has been lost independently in multiple metazoan lineages, suggesting that this domain is an evolutionarily and biochemically modular contributor to YTHDC2 function (Jain et al. 2018). Why has mammalian YTHDC2 retained this domain when it does not mediate functionally significant m6A interactions during gametogenesis? Truncation leads to protein loss, so this domain might contribute to normal protein folding/stability, or the YTH domain may contribute to RNA interactions that do not involve the W1375 and L1380 residues. The YTH domain of Schizosaccharomyces pombe Mmi1 binds a specific RNA sequence using a protein surface that is distinct from that used by other m6A-binding YTH domains (Harigaya et al. 2006; Yamashita et al. 2012; Chatterjee et al. 2016; Wang et al. 2016). An additional possibility is that the YTH domain is important for nonmeiotic functions of YTHDC2. We did not observe gross pathological defects in Ythdc2WLA/WLA (see the Supplemental Material) or Ythdc2−/− mice (Jain et al. 2018), but we cannot rule out the possibility that YTHDC2 interaction with m6A is important under pathological conditions, such as during cell stress or tumorigenesis.
What is the molecular outcome of YTHDC2 binding to target RNAs? Two opposing models have been proposed in which the principal function of YTHDC2 is to stabilize transcripts (Abby et al. 2016) or to destabilize them (Hsu et al. 2017; Soh et al. 2017; Wojtas et al. 2017). Juvenile mice that lack YTHDC2 showed a modest but significant increase in the levels of transcripts to which YTHDC2 binds in the 3′ UTR. Therefore, we favor a version of the latter model in which YTHDC2 binding to the 3′ UTR leads to mRNA destabilization during meiotic entry. YTHDC2 binding may directly modulate RNA stability, consistent with identification of the 5′ → 3′ RNA exonuclease XRN1 as a potential partner of YTHDC2 (Abby et al. 2016; Hsu et al. 2017; Wojtas et al. 2017). However, we note that changes in RNA levels in Ythdc2 mutants is quantitatively small, so YTHDC2 binding may primarily affect other processes, with RNA destabilization as a secondary consequence.
The functional significance of YTHDC2 binding within coding regions remains to be determined. This binding increases modestly in adult mice compared with juveniles, which may indicate that this binding pattern predominates later during meiosis. Recently, a study in HEK293 cells suggested that YTHDC2 binding to coding regions promotes translation (Mao et al. 2019), and another reported that YTHDC2 directly interacts with the small ribosomal subunit (Kretschmer et al. 2018). YTHDC2 has been proposed to promote translation also during spermatogenesis, based on measures of apparent translation efficiency of two putative targets (Hsu et al. 2017). Additionally, IP/MS experiments in testes identified potential YTHDC2 interactors with roles in translation (Abby et al. 2016; Hsu et al. 2017; Wojtas et al. 2017). Paradoxically, however, YTHDC2 has also been proposed to repress translation based on analogy with its fly homolog Bgcn, which has well-documented translational inhibition functions when germ cells transition into meiosis in Drosophila melanogaster (Bailey et al. 2017; Jain et al. 2018). We cannot exclude the possibility that whole-testis ribosome profiling may not detect YTHDC2 effects on translation; for instance, if effects in meiotic cells are masked by unchanged transcripts in somatic cells in the input ribosome profiling samples. However, any such underlying effects on translation are likely to be quantitatively subtle. Therefore, while ribosome profiling in testes from mice lacking YTHDC2 cannot definitively rule out effects on translation, it does not support a role in controlling translation of direct targets during the mitosis-to-meiosis transition. Moreover, control of translation remains a possible role during later meiotic stages.
An additional nonexclusive possibility is that YTHDC2 regulates subcellular localization of RNAs. For example, YTHDC2 may target mRNAs to germ cell RNA granules or other cellular compartments (Bailey et al. 2017). Our CLIP data show that YTHDC2 binds directly to piRNA precursors, also suggested by previous RIP approaches (Bailey et al. 2017). This implicates YTHDC2 as part of the piRNA regulatory machinery, components of which are typically localized within cytoplasmic germ granules (Aravin et al. 2009; Meikar et al. 2014). Consistent with this, YTHDC2 forms perinuclear puncta in spermatocytes (Bailey et al. 2017) and was identified as a component of granules called chromatoid bodies that begin to assemble in pachytene spermatocytes (Meikar et al. 2011, 2014). We found that mRNAs encoding factors involved in transcription are enriched among YTHDC2 targets, so indirect regulation of transcription may be yet another way in which YTHDC2 shapes the germ cell gene expression landscape.
We unexpectedly observed that germ cells lacking YTHDC2 helicase activity arrested during meiotic prophase, days after meiotic entry. This result is likely attributable to loss of a later function of YTHDC2 during meiosis, supported by a recent study that conditionally deleted Ythdc2 after meiotic entry during spermatogenesis and indicated a role for YTHDC2 during mid-meiotic prophase (Liu et al. 2021). Our helicase-dead allele showed a meiotic arrest phenotype similar to that described for the conditional Ythdc2 allele. Additionally, our helicase mutant revealed that the mechanism of action of YTHDC2, and possibly the molecular outcome of its action, is context-dependent: Helicase activity is dispensable for the meiotic entry function of YTHDC2 but is required for its function during mid-meiotic prophase. The gene expression signature of meiotic cells is vastly different between early and late meiotic stages (Ball et al. 2016), and we found that YTHDC2 engages a distinct set of RNAs in juvenile mice compared with adults. Other RNA-binding proteins decorating the different YTHDC2-bound transcripts or distinct sets of YTHDC2-interacting partners could dictate differences in how YTHDC2 regulates target RNAs.
Evolutionary analyses revealed a Ythdc2 gene duplication event in schizophoran flies, which likely diversified to create the Bgcn subfamily (Jain et al. 2018). Bgcn is essential for controlling the mitosis-to-meiosis transition in Drosophila (Kimble 2011; Insco et al. 2012). Bgcn contains alterations in the ATPase motif (motif II) that are expected to preclude helicase activity (Ohlstein et al. 2000); thus, Bgcn function during the mitosis-to-meiosis transition in flies must not involve helicase activity. The mouse YTHDC2 helicase mutant provides an intriguing parallel and shows that helicase activity is also not essential during the mammalian mitosis-to-meiosis transition but plays a later function that perhaps is lacking in flies. Thus, our findings provide insight into how gene expression regulation may have diversified during evolution to support organism-specific aspects of meiosis in mammals.
Materials and methods
Mice
Mice were maintained and euthanized under United States federal regulatory standards, and experiments were approved by the Institutional Animal Care and Use Committees of Memorial Sloan Kettering Cancer Center (MSK) and Rutgers University. Construction and genotyping of the Ythdc2WLA, Ythdc2trunc, and Ythdc2EQ alleles are detailed in the Supplemental Material. The Ythdc2− allele used here was the previously described Ythdc2em1Sky allele (Jain et al. 2018). Wild-type mice used for CLIP were C57BL/6J, purchased from Jackson Laboratory.
Histology
Fixation and testis tissue processing were performed as described (Jain et al. 2018). Periodic acid Schiff (PAS) staining, immunohistochemical TUNEL assay, and immunofluorescent staining were performed by the MSK Molecular Cytology Core Facility. PAS staining was done using the Autostainer XL (Leica Microsystems) automated stainer for PAS with hematoxylin counterstain. TUNEL assay was done using the Discovery XT processor (Ventana Medical Systems) as described previously (Jain et al. 2017). Immunofluorescent staining was done with the Discovery Ultra processor (Ventana Medical Systems) and is detailed in the Supplemental Material. Stained slides were digitized using Pannoramic Flash 250 (3DHistech) with a 40× objective. Images were produced and analyzed using Pannoramic Viewer or CaseViewer software (3DHistech). Histological examination of somatic tissues is described in the Supplemental Material.
YTHDC2 CLIP
For each independent replicate of CLIP in wild-type adults, testis tissue from one mouse aged 8.5 wk was used. For each replicate in juvenile wild type, testis tissues from 14 and 15 male mice aged 10 dpp and 8 dpp, respectively, were pooled. Four, three, and three replicates were performed in adults, at 10 dpp, and at 8 dpp, respectively. Two replicates of CLIP were performed in Ythdc2−/− (negative control for CLIP tags) using testis tissues from two mice aged 8.5 wk. Three replicates of CLIP were performed in Ythdc2WLA/WLA and three replicates were done in age-matched wild-type controls using testis tissues from one mouse per replicate, aged 20 or 24 wk.
The complete YTHDC2 CLIP protocol applied is detailed in the Supplemental Material. CLIP was performed as described previously (Weyn-Vanhentenryck et al. 2014) with the following critical modifications: Briefly, UV cross-linked YTHDC2–RNA complexes extracted using ice-cold PXL buffer were first immunoprecipitated (first IP) using an anti-YTHDC2 antibody (Abcam ab220160 ) with Dynabeads Protein G magnetic beads. Beads were washed with a series of wash buffers (see the Supplemental Material) and heated for 1 min at 98°C in denature buffer 1, then neutralized with denature buffer 2, and subjected to a second IP using another anti-YTHDC2 antibody (Proteintech 27779-1-AP). The purity of YTHDC2–RNA complexes after standard and optimized YTHDC2 CLIP was compared by visualizing YTHDC2–RNA complexes labeled with 32P-γ-ATP on autoradiography films. Individual YTHDC2 CLIP libraries were multiplexed and sequenced by NextSeq or MiSeq (Illumina) to obtain 100-nt (NextSeq) or 75-nt (MiSeq) single-end reads. CLIP data analyses were done as described previously using the CLIP Tool Kit (Shah et al. 2017; Saito et al. 2019) and are detailed in the Supplemental Material. Top CLIP targets (filters for PH ≥ 8, PH ≥ 10, and PH ≥ 40 for 8 dpp, 10 dpp, and adults, respectively, were chosen according to sequencing depth of CLIP libraries and applied to obtain top targets) are listed in Supplemental Tables S1–S3.
Ribosome profiling and RNA sequencing analysis in Ythdc2−/−
Ribosome profiling was performed on whole testes dissected from three Ythdc2−/− mice and three wild-type littermates aged 8 dpp, as well as on three Ythdc2−/− mice and one wild-type and two heterozygous littermates aged 10 dpp. To generate ribosome profiling libraries, a standard ribosome profiling protocol (McGlincy and Ingolia 2017) was optimized for mouse testis and is detailed along with data analysis steps in the Supplemental Material. Libraries were sequenced on a HiSeq 2500 platform (Illumina) at the Cornell Genomics Core Facility to obtain 50-nt paired-end reads. Differential translation analysis was done using anota2seq (Oertlin et al. 2019). Genes with an adjusted P-value of < 0.1 and a minimum absolute log2 effect size of log2(1.2) for the translation analysis were considered to have significantly differential translation efficiencies between wild type/heterozygotes and Ythdc2−/−. For differential expression analyses of total mRNA between controls and Ythdc2 knockout in juveniles, the log2 fold changes for input mRNA from ribosome profiling were extracted from the same anota2seq analysis pipeline and joined by genes to the respective 8- and 10-dpp top 250 YTHDC2 CLIP targets.
RNA sequencing analyses in Ythdc2WLA/WLA
For gene expression analysis of Ythdc2WLA/WLA, three homozygous mutant animals (same as those used for CLIP) and three heterozygous littermates aged 20 and 24 wk were used. Libraries were prepared and sequenced at the Integrated Genomics Operation of MSK. RNA extracted from whole testes using RNeasy mini kit (Qiagen) underwent polyA capture and library preparation using TruSeq Stranded Total RNA library preparation chemistry (Illumina) and was sequenced on a HiSeq platform (Illumina) to obtain 100-nt paired-end reads. Reads were aligned to the mm10 build (Gencode GRCm38.p6) using STAR (Dobin et al. 2013) and analyzed by differential analysis of raw sequencing counts using DESeq2 (Bioconductor, https://www.bioconductor.org/packages/release/bioc/html/DESeq2.html) (Love et al. 2014).
Antibodies
Primary antibodies used for CLIP were rabbit anti-YTHDC2 (Abcam ab220160) and rabbit anti-YTHDC2 (Proteintech 27779-1-AP). We also tested the following antibodies and did not include them in our CLIP protocol due to poor performance: rabbit anti-YTHDC2 (Bethyl Laboratories A303-026A), rabbit anti-YTHDC2 (Bethyl Laboratories A303-025A), and rabbit anti-YTHDC2 (Invitrogen PA5-57920). The following antibodies were used for immunoblots: rabbit anti-YTHDC2 (1:5000; Abcam ab220160), rabbit anti-YTHDC2 (1:1000; Abcam ab176846), mouse anti-β-Actin (1:20,000; Abcam ab6276), and mouse anti-Vinculin (1:500; Santa Cruz Biotechnology sc-73614). Primary antibodies used for histology were 1 µg/mL mouse anti-SYCP3 (Santa Cruz Biotechnology sc-74569), 0.2 µg/mL rabbit anti-γH2AX (Abcam ab11174), and 1 µg/mL rabbit anti-pH3 (Millipore 06-570).
Fluorescence anisotropy
Recombinant N-terminal hexahistidine (His6)–maltose-binding protein (MBP)–Smt3-tagged mouse YTHDC2 YTH domain (amino acid residues 1353–1436) or YTHDC2 YTH domain with W1375A and L1380A mutations was purified from Escherichia coli as described in the Supplemental Material. Fluorescence anisotropy detailed in the Supplemental Material was performed with 20 nM fluorescein (fl)-conjugated m6A-modified RNA probe [5′-fl-CCGG(m6A)CUGU-3′; Dharmacon] or unmodified RNA (5′-fl-CCGGACUGU-3′; Dharmacon). After 1 h of incubation at 25°C, fluorescence anisotropy was measured using a SpectraMax M5 microplate reader (Molecular Devices) with excitation/emission wavelengths of 485/525 nm. Binding curves did not appear to reach saturation, perhaps due to limitations in protein solubility, but dissociation constants (Kd) were successfully extracted from the data using a one site total binding equation in GraphPad Prism 9.
Data availability
The Gene Expression Omnibus accession numbers for sequencing data are GSE190979, GSE190980, and GSE190981.
Supplementary Material
Acknowledgments
We thank Keeney laboratory member Diana Eng for help with genotyping, and David Ontoso, Meret Arter, Tao Li, Laurent Acquaviva, Marina Marcet, and Shintaro Yamada for help with experiments. We thank Julian Dillon (Memorial Sloan Kettering Cancer Center [MSK] Colony Management Group) for assistance with mouse husbandry, and MSK Molecular Cytology Core members Ning Fan, Afsar Barlas, and Eric Rosiek for help with histology and imaging. We thank Joeseph Giacalone (MSK Mouse Genetics Core) for designing the materials and assays used during mouse construction, Nathalie Lailler (MSK Integrated Genomics Operations) for variant annotation in founder mice, and Sebastian Carrasco and Sebastien Monette (MSK Laboratory of Comparative Pathology Core Facility) for anatomic pathology. MSK core facilities are supported by National Cancer Institute Cancer Center support grant P30 CA08748. This work was supported by Human Genetics Institute of New Jersey laboratory startup funds (to D.J.), the Howard Hughes Medical Institute (to S.K., R.B.D., and C.D.L.), Cell Starr Cancer Consortium grant I14-0031 (to R.B.D.), and National Institutes of Health grants R35 GM118080 (to C.D.L.), R35 NS097404 (to R.B.D.), and P50 HD104454 (to S.R.J.).
Author contributions: Y.S. performed CLIP experiments and RNA sequencing analysis in Ythdc2WLA/WLA. B.R.H. performed ribosome profiling and RNA sequencing analysis in Ythdc2−/−. M.R.P. performed biochemistry experiments. D.J. and S.N.S. performed mouse phenotyping. C.D.L., S.R.J., and R.B.D. supervised M.R.P., B.R.H., and Y.S., respectively. D.J., Y.S., and S.K. designed the study with contributions from all authors. D.J. and S.K. wrote the manuscript with contributions from all authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349190.121.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Abby E, Tourpin S, Ribeiro J, Daniel K, Messiaen S, Moison D, Guerquin J, Gaillard JC, Armengaud J, Langa F, et al. 2016. Implementation of meiosis prophase I programme requires a conserved retinoid-independent stabilizer of meiotic transcripts. Nat Commun 7: 10324. 10.1038/ncomms10324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AM, Page DC. 2008. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci 105: 14976–14980. 10.1073/pnas.0807297105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castañeda J, Vagin VV, Hannon GJ, Bortvin A. 2009. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 5: e1000764. 10.1371/journal.pgen.1000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AS, Batista PJ, Gold RS, Chen YG, de Rooij DG, Chang HY, Fuller MT. 2017. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. Elife 6: e26116. 10.7554/eLife.26116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball RL, Fujiwara Y, Sun F, Hu J, Hibbs MA, Handel MA, Carter GW. 2016. Regulatory complexity revealed by integrated cytological and RNA-seq analyses of meiotic substages in mouse spermatocytes. BMC Genomics 17: 628. 10.1186/s12864-016-2865-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltus AE, Menke DB, Hu YC, Goodheart ML, Carpenter AE, de Rooij DG, Page DC. 2006. In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet 38: 1430–1434. 10.1038/ng1919 [DOI] [PubMed] [Google Scholar]
- Bellve AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M. 1977. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol 74: 68–85. 10.1083/jcb.74.1.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E, Bannister LA, Barash A, Schimenti KJ, Hartford SA, Eppig JJ, Handel MA, Shen L, Schimenti JC. 2011. A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development 138: 3319–3330. 10.1242/dev.067645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, Yashiro K, Chawengsaksophak K, Wilson MJ, Rossant J, et al. 2006. Retinoid signaling determines germ cell fate in mice. Science 312: 596–600. 10.1126/science.1125691 [DOI] [PubMed] [Google Scholar]
- Chatterjee D, Sanchez AM, Goldgur Y, Shuman S, Schwer B. 2016. Transcription of lncRNA prt, clustered prt RNA sites for Mmi1 binding, and RNA polymerase II CTD phospho-sites govern the repression of pho1 gene expression under phosphate-replete conditions in fission yeast. RNA 22: 1011–1025. 10.1261/rna.056515.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. 2010. HITS-CLIP: panoramic views of protein–RNA regulation in living cells. Wiley Interdiscip Rev RNA 1: 266–286. 10.1002/wrna.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Mele A, Hung KYS, Darnell RB. 2018. Mapping of In vivo RNA-binding sites by ultraviolet (UV)-cross-linking immunoprecipitation (CLIP). Cold Spring Harb Protoc 2018. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold MD. 2016. Spermatogenesis: the commitment to meiosis. Physiol Rev 96: 1–17. 10.1152/physrev.00013.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. 2006. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442: 45–50. 10.1038/nature04881 [DOI] [PubMed] [Google Scholar]
- Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J, et al. 2017. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res 27: 1115–1127. 10.1038/cr.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insco ML, Bailey AS, Kim J, Olivares GH, Wapinski OL, Tam CH, Fuller MT. 2012. A self-limiting switch based on translational control regulates the transition from proliferation to differentiation in an adult stem cell lineage. Cell Stem Cell 11: 689–700. 10.1016/j.stem.2012.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro KI, Matsuura K, Tani N, Takeda N, Usuki S, Yamane M, Sugimoto M, Fujimura S, Hosokawa M, Chuma S, et al. 2020. MEIOSIN directs the switch from mitosis to meiosis in mammalian germ cells. Dev Cell 52: 429–445.e10. 10.1016/j.devcel.2020.01.010 [DOI] [PubMed] [Google Scholar]
- Jain D, Meydan C, Lange J, Claeys Bouuaert C, Lailler N, Mason CE, Anderson KV, Keeney S. 2017. Rahu is a mutant allele of Dnmt3c, encoding a DNA methyltransferase homolog required for meiosis and transposon repression in the mouse male germline. PLoS Genet 13: e1006964. 10.1371/journal.pgen.1006964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Puno MR, Meydan C, Lailler N, Mason CE, Lima CD, Anderson KV, Keeney S. 2018. Ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. Elife 7: e30919. 10.7554/eLife.30919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J. 2011. Molecular regulation of the mitosis/meiosis decision in multicellular organisms. Cold Spring Harb Perspect Biol 3: a002683. 10.1101/cshperspect.a002683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima ML, de Rooij DG, Page DC. 2019. Amplification of a broad transcriptional program by a common factor triggers the meiotic cell cycle in mice. Elife 8: e43738. 10.7554/eLife.43738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC. 2006. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci 103: 2474–2479. 10.1073/pnas.0510813103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer J, Rao H, Hackert P, Sloan KE, Höbartner C, Bohnsack MT. 2018. The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′–3′ exoribonuclease XRN1. RNA 24: 1339–1350. 10.1261/rna.064238.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers JH, Offenberg HH, van Aalderen M, Vink AC, Dietrich AJ, Heyting C. 1994. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol Cell Biol 14: 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch BJ, Page DC. 2012. Genetics of germ cell development. Nat Rev Genet 13: 781–794. 10.1038/nrg3294 [DOI] [PubMed] [Google Scholar]
- Lin Y, Gill ME, Koubova J, Page DC. 2008. Germ cell-intrinsic and -extrinsic factors govern meiotic initiation in mouse embryos. Science 322: 1685–1687. 10.1126/science.1166340 [DOI] [PubMed] [Google Scholar]
- Liu R, Kasowitz SD, Homolka D, Leu NA, Shaked JT, Ruthel G, Jain D, Lin H, Keeney S, Luo M, et al. 2021. YTHDC2 is essential for pachytene progression and prevents aberrant microtubule-driven telomere clustering in male meiosis. Cell Rep 37: 110110. 10.1016/j.celrep.2021.110110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Tong L. 2014. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc Natl Acad Sci 111: 13834–13839. 10.1073/pnas.1412742111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. 10.1093/bioinformatics/btr189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodríguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS. 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27: 271–276. 10.1038/85830 [DOI] [PubMed] [Google Scholar]
- Mao Y, Dong L, Liu XM, Guo J, Ma H, Shen B, Qian SB. 2019. m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat Commun 10: 5332. 10.1038/s41467-019-13317-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson CK, Murphy MW, Griswold MD, Yoshida S, Bardwell VJ, Zarkower D. 2010. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev Cell 19: 612–624. 10.1016/j.devcel.2010.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy NJ, Ingolia NT. 2017. Transcriptome-wide measurement of translation by ribosome profiling. Methods 126: 112–129. 10.1016/j.ymeth.2017.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikar O, Da Ros M, Korhonen H, Kotaja N. 2011. Chromatoid body and small RNAs in male germ cells. Reproduction 142: 195–209. 10.1530/REP-11-0057 [DOI] [PubMed] [Google Scholar]
- Meikar O, Vagin VV, Chalmel F, Sõstar K, Lardenois A, Hammell M, Jin Y, Da Ros M, Wasik KA, Toppari J, et al. 2014. An atlas of chromatoid body components. RNA 20: 483–495. 10.1261/rna.043729.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Steitz JA. 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 10: 1692–1694. 10.1261/rna.7151404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB. 2014. Mapping argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc 9: 263–293. 10.1038/nprot.2014.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertlin C, Lorent J, Murie C, Furic L, Topisirovic I, Larsson O. 2019. Generally applicable transcriptome-wide analysis of translation using anota2seq. Nucleic Acids Res 47: e70. 10.1093/nar/gkz223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Lavoie CA, Vef O, Gateff E, McKearin DM. 2000. The Drosophila cystoblast differentiation factor, benign gonial cell neoplasm, is related to DExH-box proteins and interacts genetically with bag-of-marbles. Genetics 155: 1809–1819. 10.1093/genetics/155.4.1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. 2016. M6a RNA methylation promotes XIST-mediated transcriptional repression. Nature 537: 369–373. 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil DP, Pickering BF, Jaffrey SR. 2018. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol 28: 113–127. 10.1016/j.tcb.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Yuan Y, Zucker-Scharff I, Fak JJ, Jereb S, Tajima Y, Licatalosi DD, Darnell RB. 2019. Differential NOVA2-mediated splicing in excitatory and inhibitory neurons regulates cortical development and cerebellar function. Neuron 101: 707–720.e5. 10.1016/j.neuron.2018.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A, Qian Y, Weyn-Vanhentenryck SM, Zhang C. 2017. CLIP tool kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics 33: 566–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol Syst Biol 7: 539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh YQS, Mikedis MM, Kojima M, Godfrey AK, de Rooij DG, Page DC. 2017. Meioc maintains an extended meiotic prophase I in mice. PLoS Genet 13: e1006704. 10.1371/journal.pgen.1006704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A, Fuller MT, Braun RE, Yoshida S. 2011. Germline stem cells. Cold Spring Harb Perspect Biol 3: a002642. 10.1101/cshperspect.a002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theler D, Dominguez C, Blatter M, Boudet J, Allain FH. 2014. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res 42: 13911–13919. 10.1093/nar/gku1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Hwang HW, Darnell RB. 2018. The future of cross-linking and immunoprecipitation (CLIP). Cold Spring Harb Perspect Biol 10: a032243. 10.1101/cshperspect.a032243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhu Y, Bao H, Jiang Y, Xu C, Wu J, Shi Y. 2016. A novel RNA-binding mode of the YTH domain reveals the mechanism for recognition of determinant of selective removal by Mmi1. Nucleic Acids Res 44: 969–982. 10.1093/nar/gkv1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, et al. 2014. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep 6: 1139–1152. 10.1016/j.celrep.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. 2017. Regulation of m6A transcripts by the 3′ → 5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol Cell 68: 374–387.e12. 10.1016/j.molcel.2017.09.021 [DOI] [PubMed] [Google Scholar]
- Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. 2014. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol 10: 927–929. 10.1038/nchembio.1654 [DOI] [PubMed] [Google Scholar]
- Xu C, Liu K, Ahmed H, Loppnau P, Schapira M, Min J. 2015. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J Biol Chem 290: 24902–24913. 10.1074/jbc.M115.680389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Shichino Y, Tanaka H, Hiriart E, Touat-Todeschini L, Vavasseur A, Ding DQ, Hiraoka Y, Verdel A, Yamamoto M. 2012. Hexanucleotide motifs mediate recruitment of the RNA elimination machinery to silent meiotic genes. Open Biol 2: 120014. 10.1098/rsob.120014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S, Ries RJ, Jaffrey SR. 2019. Reading, writing and erasing mRNA methylation. Nature reviews Molecular cell biology 20: 608–624. 10.1038/s41580-019-0168-5 [DOI] [PubMed] [Google Scholar]
- Zhang C, Darnell RB. 2011. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol 29: 607–614. 10.1038/nbt.1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gunewardena S, Wang N. 2021. Nutrient restriction synergizes with retinoic acid to induce mammalian meiotic initiation in vitro. Nat Commun 12: 1758. 10.1038/s41467-021-22021-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Roundtree IA, Wang P, Wang X, Wang L, Sun C, Tian Y, Li J, He C, Xu Y. 2014. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res 24: 1493–1496. 10.1038/cr.2014.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. 2015. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 7: a016626. 10.1101/cshperspect.a016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Gene Expression Omnibus accession numbers for sequencing data are GSE190979, GSE190980, and GSE190981.