Abstract
Background
We studied a young woman with atypical diabetes associated with mild intellectual disability, lymphedema distichiasis syndrome (LDS) and polymalformative syndrome including distichiasis. We used different genetic tools to identify causative pathogenic mutations and/or copy number variations.
Results
Although proband’s, diabetes mellitus occurred during childhood, type 1 diabetes was unlikely due to the absence of detectable autoimmunity. DNA microarray analysis first identified a de novo, heterozygous deletion at the chr16q24.2 locus. Previously, thirty-three pathogenic or likely pathogenic deletions encompassing this locus have been reported in patients presenting with intellectual deficiency, obesity and/or lymphedema but not with diabetes. Of note, the deletion encompassed two topological association domains, whose one included FOXC2 that is known to be linked with LDS. Via whole-exome sequencing, we found a heterozygous, likely pathogenic variant in WFS1 (encoding wolframin endoplasmic reticulum [ER] transmembrane glycoprotein) which was inherited from her father who also had diabetes. WFS1 is known to be involved in monogenic diabetes. We also found a likely pathogenic variant in USP9X (encoding ubiquitin specific peptidase 9 X-linked) that is involved in X-linked intellectual disability, which was inherited from her mother who had dyscalculia and dyspraxia.
Conclusions
Our comprehensive genetic analysis suggested that the peculiar phenotypes of our patient were possibly due to the combination of multiple genetic causes including chr16q24.2 deletion, and two likely pathogenic variants in WFS1 and USP9X.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13023-022-02248-2.
Keywords: Childhood onset diabetes, Genotype–phenotype relations, Genetic disorders, Genetic analysis, Intellectual disability, Wolfram syndrome
Background
Type 2 diabetes is a non-autoimmune, multifactorial metabolic disorder characterized by chronic hyperglycemia resulting from impaired insulin secretion and altered action of insulin [1]. Type 2 diabetes depends on various environmental components while having a high heritability ranging from 40 to 70% [2]. Apart from common type 2 diabetes that affects most cases with non-autoimmune diabetes, there are atypical, monogenic forms of diabetes which are usually rare and severe, with an early onset. Pathogenic mutations in more than 40 genes have now been described in patients with monogenic diabetes. They cause various conditions including neonatal diabetes, maturity-onset diabetes of the young (MODY), and diabetes-associated syndromes (e.g. Wolfram syndrome due to variants in WFS1, Wolcott–Rallison syndrome) [3]. In Wolfram syndrome (OMIM #222300), juvenile-onset- diabetes mellitus can be isolated or associated with more complex phenotypes including optic atrophy, diabetes insipidus, and deafness. Several genes linked with monogenic diabetes are actionable (such as KCNJ11 [5], ABCC8 [5], GCK [5], HNF1A [5], HNF1B [5, 6] or HNF4A [5]), implying a substantial change in care for the carriers of a pathogenic variant (such as specific therapeutic management and monitoring) [4, 5]. Therefore, the genetic diagnosis of patients with a suspicion of monogenic diabetes has become crucial.
Here, we performed karyotyping, DNA microarray, and whole-exome sequencing in order to detect point mutations and/or copy number variations (CNVs) putatively causing a complex syndrome (including an atypical diabetes and polymalformative syndrome) in a young woman.
Results
Phenome of the case report
A 17-year-old girl was referred after a pediatric follow-up for a non-autoimmune diabetes and a mild intellectual disability. She was from a non-consanguineous family and had two healthy siblings.
On the maternal side, her mother is an only child, and presented with mild dyscalculia and dyspraxia. None other members of this part of the family had an intellectual disability.
On the paternal side, her father has had type 2 diabetes since age 50. There was no history of diabetes in any of the three patient’s uncles. Her paternal grandmother presented with a late isolated cataract (from age ~ 80).
She was born at 41 weeks gestation after an uneventful pregnancy. Her birth weight was 3.620 kg (67th percentile), her size was 51 cm (67th percentile), and had a normal head circumference (85th percentile). Diabetes was diagnosed at age 7 while the young girl suffered from sudden polydipsia, polyuria, and unexplained weight loss (− 1.50 kg). At hospital admission, venous glycaemia level was 5.34 g/L, with 2 + in urinary ketone testing, glycated hemoglobin A1c (HbA1c) was very high (12.8%; Normal range: < 6.5%) and C-peptide was low (0.27 ng/mL; Normal range: 1.2–4.5 ng/mL). The venous pH, blood sodium and potassium levels were normal, with alkaline reserve at 26 mmol/L. Neither islet autoantibodies, nor anti-insulin antibodies were present. Anti-GAD, anti-IA2, anti-ZNT8 antibodies were not initially analyzed. Insulin therapy was initiated and the patient was regularly followed up. HbA1c had remained between 7.5% and 8.5% overtime without episodes of ketosis or severe hypoglycemia. At the last clinical investigation at age 24, no chronic complications of diabetes were reported. Anti-GAD, anti-ZnT8 and anti-IA2 antibodies were still negative. Pancreas imaging did not show any pancreatic atrophy.
We observed a severe gain of weight at age 8, concomitant to insulin treatment, reaching the 97th percentile of body mass index (BMI) (Fig. 1). Weight gain persisted throughout puberty. No endocrine causes of obesity were found, but low limbs swelling were regularly reported by the patient suggesting lymphedema. At age 16, the patients suddenly needed more insulin (1 UI/kg). Furthermore, metformin (1 g per day) was added to insulin injections (Fig. 1). This additional treatment led to an initial and persistent weight loss and to a 40% reduction in insulin doses. Then, we have tested the action of the incretin pathway agonists on diabetes control and insulin requirement. First, a dipeptidyl peptidase 4 (DPP4) inhibitor was added to her insulin injections, and we observed a decrease in HbA1c after 3 months (8.9 to 7.9%) and 1 kg weight loss (Fig. 1). Then, we have switched from prandial insulin to glucagon-like peptide-1 receptor agonist (GLP1-RA) (Liraglutide 1.8 mg per day) associated with basal insulin only. The patient has further lost 3.6 kg (Fig. 1).
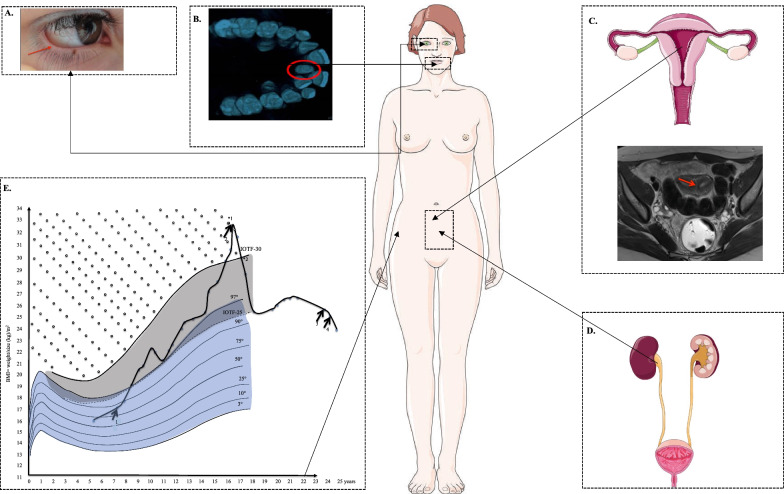
Fig. 1.
Clinical features of the patient—Polymalformative syndrome including distichiasis (red arrow in a), palatine tooth (circled in red in b), uterine septum (red arrow in c), vesicoureteral reflux (d) and overweight (e). In part E we reported the increase in insulin requirement during childhood (arrow #1: 1.1 ui/kg) followed by the decrease in insulin requirement after the addition of metformin (arrow #2: 0.7 UI/kg). We show the addition effects of DPP4 inhibitors (arrow #3), and GLP1-RA (arrow #4). The dark area shows the values corresponding to overweight, while the area with points shows the values corresponding to obesity
In addition, after ruling out deafness, we assessed psychomotor retardation of the patient during her childhood. She used sentences at age 6. At age 8, the intelligence quotient (IQ) test showed a mild intellectual disability (WISC III test: total IQ 68). Moreover, at age 5, the patient was diagnosed with a bilateral distichiasis in the lower and superior eyelids, complicated with keratitis, which required surgery followed by laser and recurrent epilation. She also had dental malposition and palatine tooth requiring a surgery during her childhood. At age 8, she had diurnal enuresis. Urodynamic examinations showed that the patient also had vesicoureteral reflux with retro-urethral meatal stenosis and overactive bladder. Kidney ultrasound showed renal calyceal, hypotonia and duplex kidney. Despite anticholinergic drugs, the injection of botulinum toxin was required to manage bladder instability. After puberty, at age 13, the patient suffered from painful dysmenorrhea. Pelvic ultrasound and magnetic resonance imaging showed that the patient presented with uterine septum, which required surgery.
Genetic investigations
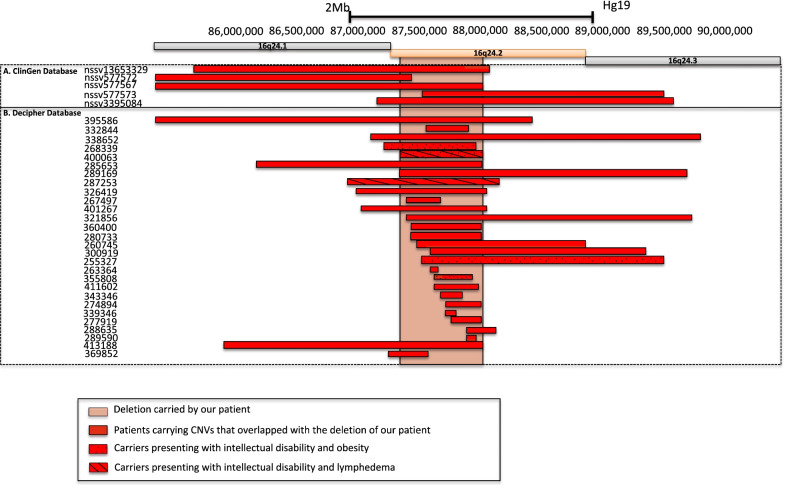
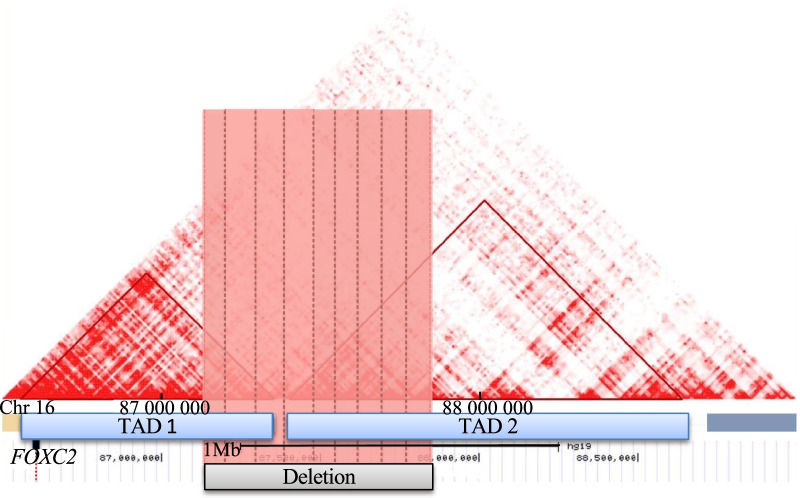
The karyotype was normal. Through array comparative genomic hybridization (CGH) performed in the trio (proband versus both parents), we found a de novo heterozygous deletion of 693 kb (chr16q24.2; 16: 87,152,792–87,845,741 [hg19]) in the proband. The deleted region includes six protein-coding genes (Table 1). This result was confirmed by real-time PCR. According to the Human Gene Mutation Database (HGMD), none of these genes were consistently found to be linked with monogenic forms of diabetes, obesity, kidney disorders, intellectual deficiency, lymphedema and/or distichiasis. However, other deletions encompassing chr16q24.2 have already been reported in 33 patients according to ClinGen and Decipher databases (Fig. 2 and Table 2). Among them, 25 patients presented with intellectual disability (Fig. 2 and Table 2). It is noteworthy that four patients shared intellectual disability, obesity or lymphedema as observed in our patient (Fig. 2 and Table 2). Furthermore, we observed that FOXC2 which is linked with lymphedema distichiasis syndrome (LDS) (Additional file 1: Table S1) was located 550 kb downstream to the chr16q24.2 deletion. This gene encodes a transcription factor belonging to the forkhead family. Using HiC data obtained from human fetal brain, we found that the chr16q24.2 deletion encompassed two subsequent topologically association domains (TADs) (Fig. 3). FOXC2 was located in the first TAD (Fig. 3). Therefore, we have hypothesized that the deletion might disrupt the transcriptional network of FOXC2, possibly leading to lymphedema in the carriers of chr16q24.2 deletion [7].
Table 1.
List of genes and their transcripts included in the chr16q24 deletion carried by the proband (UCSC Genomic Institute. UCSC Genomic Institute (University of California Santa Cruz)
| Gene | Transcript | Chr | Tx-Start [hg19] | Tx-End [hg19] | Exon Count | Strand | Function | Disease |
|---|---|---|---|---|---|---|---|---|
| C16orf95 | NM_001195124 | 16 | 87,336,420 | 87,350,998 | 7 | − | Unknown | Unknown |
| FBXO31 | NM_024735 | 16 | 87,360,592 | 87,417,382 | 9 | − | Degradation via Skp1-Cul1- Fbox protein complex; cell cycle regulator [33] | Intellectual disability autosomal recessive |
| MAP1LC3B | NM_022818 | 16 | 87,425,941 | 87,438,380 | 4 | + | Autophagy pathway [33] | Unknown |
| ZCCHC14 | NM_015144 | 16 | 87,439,853 | 87,526,630 | 13 | − | Regulates tumor progression [34] | Unknown |
| JPH3 | NM_001271604 | 16 | 87,636,113 | 87,638,254 | 2 | + | cross talk between cell surface and intracellular ion channels by junctional complexes (OMIM* 60,526) | Huntington’s Disease Like 2 |
| KLHDC4 | NM_017566 | 16 | 87,741,417 | 87,799,592 | 12 | − | Unknown | Unknown |
Chr Chromosome, Tx transcription
Fig. 2.
Pathogenic or likely pathogenic deletions encompassing chr16q24.2, which were found in 33 patients from Decipher and ClinGen databases
Table 2.
Pathogenic or likely pathogenic deletions encompassing chr16q24.2, which were found in 33 patients from Decipher and ClinGen databases
| Sample's ID | Position (Hg19) | Size (Mb) | Pathogenicity | Phenotypes |
|---|---|---|---|---|
| nssv577572 | chr16: 83,912,597–87,257,444 | 3.3 | Pathogenic | Developmental delay and/or other significant developmental or morphological phenotypes |
| nssv13653329 | chr16: 85,491,404–87,883,528 | 2.4 | Pathogenic | Premature birth, respiratory distress, obsolete malformation of the heart and great vessels |
| nssv577567 | chr16: 78,738,172–87,852,948 | 9.1 | Pathogenic | Low-set ears |
| nssv577573 | chr16: 87,340,135–89,335,487 | 2 | Pathogenic | Developmental delay and/or other significant developmental or morphological phenotypes |
| nssv3395084 | chr16: 86,983,712–89,402,222 | 2.4 | Pathogenic | Autistic behavior, cleft palate, delayed fine motor development, delayed gross motor development, delayed speech and language development, failure to thrive, hydronephrosis, omphalocele, premature birth, seizures, ventricular septal defect |
| 395,586 | chr16: 66,542,499–88,242,499 | 22 | Likely pathogenic | Aplasia/Hypoplasia of the hallux, broad forehead, broad hallux, delayed closure of the anterior fontanelle, epicanthus, flat occiput, frontal bossing, high anterior hairline, intellectual disability, microcephaly, muscular hypotonia, proportionate short stature, supernumerary nipple |
| 413,188 | chr16: 85,817,324–87,841,184 | 2 | Likely pathogenic | Unknown |
| 285,653 | chr16: 86,011,033–87,846,920 | 1.8 | Pathogenic | Unknown |
| 287,253 | chr16: 86,761,351–87,972,139 | 1.2 | Likely pathogenic | Intellectual disability, lymphedema |
| 326,419 | chr16: 86,804,086–87,887,066 | 1.1 | Likely pathogenic | Abnormality of movement, microcephaly, seizure |
| 401,267 | chr16: 86,877,615–87,868,052 | 0.99 | Likely pathogenic | Abnormality of prenatal development or birth, broad nasal tip, hyperextensibility of the finger joints, hypertelorism, intellectual disability, umbilical hernia |
| 338,652 | chr16: 86,910,262–89,623,832 | 2.7 | Unknown | Acute lymphoblastic leukemia |
| 268,339 | chr16: 87,044,630–87,779,387 | 0.73 | Unknown | Barrel-shaped chest, delayed speech and language development, feeding difficulties in infancy, intellectual disability, obesity |
| 369,852 | chr16: 87,054,643–87,410,554 | 0.36 | Uncertain | Bilateral tonic–clonic seizure with focal onset, low-set ears, macrocephaly, neonatal hypotonia |
| 400,063 | chr16: 87,152,792–87,845,741 | 0.69 | Likely pathogenic | Abnormality of the uterus, borneal ulceration, diabetes, distichiasis, intellectual disability, misalignment of teeth, predominantly lower limb lymphedema, ureter duplex, vesicoureteral reflux |
| 289,169 | chr16: 87,183,661–89,520,803 | 2.3 | Uncertain | Abnormal facial shape, abnormality of the kidney, congenital thrombocytopenia, developmental delay, increased susceptibility to fractures, mild intrauterine growth retardation, multiple skeletal anomalies, ptosis |
| 267,497 | chr16: 87,213,968–87,504,677 | 0.29 | Unknown | Unknown |
| 321,856 | chr16: 87,219,866–89,561,087 | 2.3 | Likely pathogenic | Depressed nasal bridge, failure to thrive in infancy, moderate global developmental delay, prominent metopic ridge, rocker bottom foot, thick eyebrow, wide nose |
| 280,733 | chr16: 87,257,185–87,845,882 | 0.59 | Unknown | 2–3 toe syndactyly, behavioral abnormality, developmental delay, synophrys |
| 360,400 | chr16: 87,257,185–87,845,882 | 0.59 | Likely pathogenic | Unknown |
| 260,745 | chr16: 87,319,450–88,669,353 | 1.4 | Unknown | Depressed nasal bridge, hypotonia, hypertelorism, infra-orbital crease, prominent forehead, thoracic hypoplasia |
| 255,327 | chr16: 87,340,135–89,335,428 | 2 | Unknown | Coarse facial features, intellectual disability, macrocephaly, obesity, proportionate short stature, short foot, short palm, synophrys |
| 332,844 | chr16: 87,381,358–87,713,863 | 0.33 | Uncertain | Developmental delay |
| 300,919 | chr16: 87,432,206–89,199,829 | 1.8 | Uncertain | Congenital horizontal nystagmus, high myopia, intellectual disability, joint hypermobility |
| 263,364 | chr16: 87,433,214–87,485,161 | 0.052 | Unknown | Unknown |
| 355,808 | chr16: 87,439,917–87,753,353 | 0.31 | Uncertain | Intellectual disability, obesity |
| 411,602 | chr16: 87,485,102–87,814,273 | 0.33 | Uncertain | Intellectual disability |
| 343,346 | chr16: 87,504,276–87,670,757 | 0.17 | Uncertain | Intellectual disability |
| 339,346 | chr16: 87,539,375–87,637,824 | 0.098 | Unknown | Unknown |
| 274,894 | chr16: 87,539,375-87,845,741 | 0.31 | Unknown | Developmental delay |
| 277,919 | chr16: 87,609,569–87,848,970 | 0.24 | Unknown | Behavioral abnormality |
| 289,590 | chr16: 87,709,267–87,802,845 | 0.094 | Uncertain | Behavioral abnormality, intellectual disability |
| 288,635 | chr16: 87,709,267–87,957,415 | 0.25 | Uncertain | Developmental delay, tetralogy of Fallot |
Fig. 3.
Identification of two TADs (from fetal brain) encompassing the deleted region at the 16q24.2 locus. Deletion found in the patient is represented by a grey rectangle. TAD 1 and TAD 2 are represented by triangles. The deleted region, containing part of the TADs, is highlighted in red
Based on the WES analyses of the trio (proband versus both parents), we did not find in the proband any de novo pathogenic or likely pathogenic variant located in the 1321 genes linked with monogenic forms of diabetes, obesity, kidney disorders, lymphedema, intellectual deficiency and/or distichiasis (Additional file 1: Table S1). However, we found one new heterozygous, likely pathogenic, missense variant in WFS1 (NM_006005.3: c.424G > C/p.V142L). The heterozygous WFS1 variants have previously been found to cause autosomal dominant diabetes [8]. This mutation that has never been reported before and that was not listed in the Genome Aggregation Database (gnomAD; v2.1.1), was inherited from her father who presented with type 2 diabetes. Furthermore, we found a likely pathogenic missense variant in USP9X (NM_001039590.2: c.3803A > G/p.Y1268C) involved in X-linked intellectual disability [9], which was inherited from her mother who had academic difficulties, related to dyscalculia and dyspraxia. One of the two healthy brothers of the patient did not carry neither WFS1 p.V142L variant or USP9X p.Y1268C variant. In the proband, we did not identify any rare coding mutations in FOXC2.
Discussion
We describe the case of a young female whose diabetes was difficult to associate with intellectual disability, polymalformative syndrome and lymphedema distichiasis syndrome (LDS).
On the one hand, initially, the young age of the patient, the presence of ketone bodies and the low C-peptide level at diagnosis, argued in favor of type 1 diabetes [10]. However, the absence of acidosis and of islets antibodies at diagnosis, the absence of anti-GAD, anti-IA2 and anti-ZNT8 antibodies at age 24 [11, 12], as well as the normal size of pancreas [13], suggested instead an atypical non-autoimmune diabetes. Furthermore, as the introduction of metformin in addition to insulin led to weight loss and a decrease in insulin needs, we hypothesized that insulin resistance was also involved in the patient’s metabolic phenotype. More recently, the unexpected effect of Liraglutide on the discontinuation of prandial insulin has been an additional argument in favor of atypical non-autoimmune diabetes [14–16].
On the other hand, we performed genetic investigations in front of a polymalformative syndrome and an intellectual disability.
Through our comprehensive genetic analyses, we identified a de novo 693-kb deletion in chr16q24.2. Intellectual disability, obesity, lymphedema and distichiasis have already been described in individuals carrying a pathogenic deletion at the same locus chr16q24.2 [17–19]. Interestingly, FOXC2 that is a major gene linked with LDS (OMIM# 153400 [20]) was located close to the chr16q24.2 deletion (550 kb downstream). The specific function of FOXC2 has not yet been fully explored; however, the encoded transcription factor plays an important role in the development and maintenance of venous and lymphatic valves and of mesenchymal tissues [21]. Considering that the activity of a transcription factor depends on its expression, we hypothesized that LDS might be the result of altered expression of FOXC2, triggered by the deletion of a long-range regulatory element in chr16q24.2 [7, 17, 22]. Indeed, a growing body of research suggests that deletions can exert a pathogenic effect through disruption of DNA structural elements such as TADs, and disruption of boundaries as a major mechanism [23]. TAD can regulate gene expression notably by the activation of an enhancer-promoter loop [24]. As the chr16q24.2 deletion encompassed two TADs, it might cause TAD disruption, and altered FOXC2 expression leading to LDS [7, 23]. However, further studies are needed to demonstrate this assumption.
Furthermore, we identified a novel heterozygous, likely pathogenic, missense mutation in WFS1 (p.V142L) inherited from the father with diabetes. Interestingly, we observed different presentations of diabetes in the proband and her father. Contrary to the proband, diabetes in the father was polygenic-like type 2 diabetes, as it was diagnosed in adulthood and was treated with metformin and DPP4 inhibitor without any insulin. Wolfram syndrome 1 caused by homozygous or compound heterozygous mutations in WFS1 (encoding wolframin, a 100-kDa transmembrane glycoprotein localized in the secretory granules and endoplasmic reticulum [ER]). Heterozygous mutations in WFS1 have been reported to be involved in less severe phenotypes including isolated adult-onset diabetes (OMIM #614296) [8]. A recent study has shown that GLP1-RA could be efficient for patients with diabetes and heterozygous, pathogenic WFS1 variants [16]. The authors proposed that GLP1-RA might improve glucose regulation due to decreased ER stress leading to conservation of β-cell function and insulin secretion [16]. Here, we hypothesized that the likely pathogenic heterozygous variant in WFS1 might possibly explain the patient's early-onset diabetes and the efficacy of GLP1-RA on prandial insulin interruption.
In addition, we found a likely pathogenic missense variant in USP9X (p.Y1268C) which was inherited from her mother who had dyscalculia and dyspraxia during her school career. USP9X encodes an enzyme that plays a key role in human neural development and the p.Y1268C mutation has been shown to be involved in X-linked intellectual disability [9]. Furthermore, USP9X is an X-chromosome gene that escapes X-inactivation [25]. Female patients with de novo pathogenic mutations present with mild to moderate intellectual disability (motor and language delay), and several other clinical features (including notably urogenital abnormality, hearing impairment, cleft palate or bifid uvula) [26]. The intellectual disability could therefore be due to the likely pathogenic missense variant in USP9X associated with the impact of the deletion.
The major limitation of our study is the fact that we did not analyze the functional effect of the deletion on FOXC2 expression, and of the two variants in WFS1 and USP9X on the activity of respective encoded proteins.
Conclusions
In conclusion, we suggest that the patient’s complex syndrome might be due to several genetic events including a new likely pathogenic heterozygous WFS1 variant, a likely pathogenic missense USP9X variant and a chr16q24.2 deletion possibly causing the dysregulation of FOXC2 via TADs disruption (in addition to possibly other abnormalities due to the multi-gene deletion). This study demonstrates the relevance and usefulness of comprehensive genetic analyses in cases presenting with very complex phenotypes.
Methods
Patient
We collected clinical data of a female patient from pediatric and adult diabetology medical records. Family medical data have been also collected. In accordance with French laws, patient’s and her parents’ consent were obtained.
DNA extraction
Genomic DNA samples from the proband and her two parents were extracted from peripheral blood using the QIAamp DNA Blood Midi kit (Qiagen, Valencia, CA, USA).
Karyotyping
Conventional cytogenetic analysis was performed on peripheral blood lymphocytes from the patient using the 550-band including GTG (G-bands after trypsin and Giemsa) and RHG (R-bands by heating using Giemsa) banding for family members.
DNA microarray
Array CGH was performed in the proband, using Agilent 180 k oligoarrays (SurePrint G3 Human CGH Microarray 4 × 180 k). Random primer labelling and hybridization were carried out with sex-matched reference DNA according to the manufacturer's recommendations and results were analyzed using cytogenomics (v3.0.6.6) software (Agilent Technologies) using ADM2 algorithm and a three-point filter.
CNV presence was confirmed in the proband and both parents by real-time PCR using primers targeting JPH3 (that was included in the CNV). Primers for the JPH3 was designed and tested using standard procedures (forward primer 5′-CCTGTGCGTCTGATCTGCT-3′, reverse primer 5′-GTCCTGCTGCTCCTTCTGAC-3′) on a LightCycler 480 Real-Time PCR System (Roche Diagnostics, Basel, Switzerland).
Whole-exome sequencing (WES)
WES was performed in the proband and both parents. For this purpose, we used SeqCap EZ MedExome + Probes (Roche) or Human Core Exome Kit (Twist Bioscience), for Illumina sequencing (on the NovaSeq6000 system). Alignment of sequence reads to the human genome (GRCh38/hg38), variant calling and variant annotation were done as previously described [27]. For the proband and both parents, more than 98% of the target was covered with more than eight reads. Based on the clinical data of the patient (Fig. 1), we analyzed 1321 genes linked with monogenic diabetes, monogenic obesity, monogenic kidney disorders, lymphedema and/or intellectual deficiency (Additional file 1: Table S1). To assess the pathogenicity of the variants we used the standards and guidelines of the American College of Medical Genetics and Genomics (ACMG) [28]. For the moderate pathogenic criterion PM2 and the supporting pathogenic criterion PP2, we used gnomAD browser (v2.1.1). For the supporting pathogenic criterion PP3, we used PolyPhen-2 (HumDiv), and Mutation Taster [29, 30]. The variants were written according to the nomenclature of the Human Genome Variation Society (HGVS).
TAD prediction
To integrate recent high-throughput technologies based on chromatin interaction data at the chr16q24.2 locus, we used the 3D Genome Browser (http://3dgenome.org), provided by the Bioinformatics and Genomics Program, Pennsylvania State University [31]. TADs were predicted from Hi-C analysis in Human fetal brain GZ [32].
Supplementary Information
Additional file 1: Supplementary Table 1. Characteristics of the 1,321 genes that have been specifically analyzed in the WES data, according to the phenotypes of the patient.
Acknowledgements
The authors would like to thank the patient and her family for their cooperation. We are specifically thankful to Genetic laboratory, American Hospital, University hospital Robert Debre (Reims, France) for their cooperation and genetic investigation. We are also thankful to Hematology laboratory (University hospital, Reims France) and Pediatric diabetology Department, American Hospital (University hospital, Reims, France) for his participation.
Author’s contributions
ABo, MV, PF, ED, EV, ABa, MD performed the sequencing studies and bioinformatics analysis. LLC, BD, MDF, MS, TAM, GJ, PJ, PFS, PCL contributed clinical samples and information. LLC, ABo, BD, PF and MDF wrote and edited the manuscript. All authors read and approved the final manuscript.
Funding
The next-generation sequencing work was supported by grants from the French National Research Agency (ANR-10-LABX-46 [European Genomics Institute for Diabetes] and ANR-10-EQPX-07-01 [LIGAN-PM]), and from the National Center for Precision Diabetic Medicine—PreciDIAB, which is jointly supported by the French National Agency for Research (ANR-18-IBHU-0001), by the European Union (FEDER), by the Hauts-de-France Regional Council and by the European Metropolis of Lille (MEL). A.Bo is supported by the European Research Council (ERC Reg-Seq—715575).
Availability of data and materials
Please contact author for data requests.
Declarations
Ethics approval and consent to participate
All procedures performed were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki declaration and its later amendments. Written informed consent from patients/participants and their parents/legal guardians has been obtained.
Consent for publication
Informed consent from the parents/legal guardians of the patients/participants was obtained for the publication of their data.
Competing interests
The authors declare no competing financial interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Amélie Bonnefond and Martine Doco Fenzy have contributed equally to this work
Contributor Information
Lauriane Le Collen, Email: lle-collen@chu-reims.fr.
Brigitte Delemer, Email: bdelemer@chu-reims.fr.
Philippe Froguel, Email: p.froguel@imperial.ac.uk.
Amélie Bonnefond, Email: amelie.bonnefond@cnrs.fr.
Martine Doco Fenzy, Email: mdocofenzy@chu-reims.fr.
References
- 1.Mahler RJ, Adler ML. Clinical review 102: type 2 diabetes mellitus: update on diagnosis, pathophysiology, and treatment. J Clin Endocrinol Metab. 1999;84:1165–1171. doi: 10.1210/jcem.84.4.5612. [DOI] [PubMed] [Google Scholar]
- 2.Bonnefond A, Froguel P. Rare and common genetic events in type 2 diabetes: what should biologists know? Cell Metab. 2015;21:357–368. doi: 10.1016/j.cmet.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 3.Bonnefond A, Boissel M, Bolze A, Durand E, Toussaint B, Vaillant E, et al. Pathogenic variants in actionable MODY genes are associated with type 2 diabetes. Nat Metab. 2020;2:1126–1134. doi: 10.1038/s42255-020-00294-3. [DOI] [PubMed] [Google Scholar]
- 4.Gloyn AL, Drucker DJ. Precision medicine in the management of type 2 diabetes. Lancet Diabetes Endocrinol. 2018;6:891–900. doi: 10.1016/S2213-8587(18)30052-4. [DOI] [PubMed] [Google Scholar]
- 5.Vaxillaire M, Froguel P. Monogenic diabetes: Implementation of translational genomic research towards precision medicine. J Diabetes. 2016;8:782–795. doi: 10.1111/1753-0407.12446. [DOI] [PubMed] [Google Scholar]
- 6.Bockenhauer D, Jaureguiberry G. HNF1B-associated clinical phenotypes: the kidney and beyond. Pediatr Nephrol. 2016;31:707–714. doi: 10.1007/s00467-015-3142-2. [DOI] [PubMed] [Google Scholar]
- 7.Wallis M, Pope-Couston R, Mansour J, Amor DJ, Tang P, Stock-Myer S. Lymphedema distichiasis syndrome may be caused by FOXC2 promoter-enhancer dissociation and disruption of a topological associated domain. Am J Med Genet A. 2021;185:150–156. doi: 10.1002/ajmg.a.61935. [DOI] [PubMed] [Google Scholar]
- 8.Bonnycastle LL, Chines PS, Hara T, Huyghe JR, Swift AJ, Heikinheimo P, et al. Autosomal dominant diabetes arising from a Wolfram syndrome 1 mutation. Diabetes. 2013;62:3943–3950. doi: 10.2337/db13-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homan CC, Kumar R, Nguyen LS, Haan E, Raymond FL, Abidi F, et al. Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am J Hum Genet. 2014;94:470–478. doi: 10.1016/j.ajhg.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atkinson MA, Eisenbarth GS, Michels AW. Type 1 diabetes. Lancet. 2014;383:69–82. doi: 10.1016/S0140-6736(13)60591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borg H, Marcus C, Sjöblad S, Fernlund P, Sundkvist G. Islet cell antibody frequency differs from that of glutamic acid decarboxylase antibodies/IA2 antibodies after diagnosis of diabetes. Acta Paediatr. 2000;89:46–51. doi: 10.1111/j.1651-2227.2000.tb01186.x. [DOI] [PubMed] [Google Scholar]
- 12.Ongagna JC, Levy-Marchal C. Sensitivity at diagnosis of combined beta-cell autoantibodies in insulin-dependent diabetic children. French Registry of IDDM in Children Study Group. Diabetes Metab. 1997;23:155–160. [PubMed] [Google Scholar]
- 13.Virostko J, Williams J, Hilmes M, Bowman C, Wright JJ, Du L, et al. Pancreas volume declines during the first year after diagnosis of type 1 diabetes and exhibits altered diffusion at disease onset. Diabetes Care. 2019;42:248–257. doi: 10.2337/dc18-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toots M, Seppa K, Jagomäe T, Koppel T, Pallase M, Heinla I, et al. Preventive treatment with liraglutide protects against development of glucose intolerance in a rat model of Wolfram syndrome. Sci Rep. 2018;8. [DOI] [PMC free article] [PubMed]
- 15.Kondo M, Tanabe K, Amo-Shiinoki K, Hatanaka M, Morii T, Takahashi H, et al. Activation of GLP-1 receptor signalling alleviates cellular stresses and improves beta cell function in a mouse model of Wolfram syndrome. Diabetologia. 2018;61:2189–2201. doi: 10.1007/s00125-018-4679-y. [DOI] [PubMed] [Google Scholar]
- 16.Scully KJ, Wolfsdorf JI. Efficacy of GLP-1 agonist therapy in autosomal dominant WFS1-related disorder: a case report. Horm Res Paediatr. 2020;1–6. [DOI] [PubMed]
- 17.Butler MG, Dagenais SL, Garcia-Perez JL, Brouillard P, Vikkula M, Strouse P, et al. Microcephaly, intellectual impairment, bilateral vesicoureteral reflux, distichiasis, and glomuvenous malformations associated with a 16q24.3 contiguous gene deletion and a Glomulin mutation. Am J Med Genet A. 2012;158A:839–849. doi: 10.1002/ajmg.a.35229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Handrigan GR, Chitayat D, Lionel AC, Pinsk M, Vaags AK, Marshall CR, et al. Deletions in 16q24.2 are associated with autism spectrum disorder, intellectual disability and congenital renal malformation. J Med Genet. 2013;50:163–173. doi: 10.1136/jmedgenet-2012-101288. [DOI] [PubMed] [Google Scholar]
- 19.UCSC Genomic Institute. UCSC Genomic Institute (University of California Santa Cruz). UCSC Genome Browser on Human Feb.2009 (GR37/hg 19) Assembly-chr16:87152792-87845741. https://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr16%3A87152792%2D87845741&hgsid=948807615_BA0atwvAXAUEOzrYkusmP8ZsgpOA.
- 20.Mansour S, Brice GW, Jeffery S, Mortimer P. GeneReviews®—Lymphedema-Distichiasis Syndrome. http://www.ncbi.nlm.nih.gov/books/NBK1457/.
- 21.Tissue expression of FOXC2—Summary—The Human Protein Atlas. https://www.proteinatlas.org/ENSG00000176692-FOXC2/tissue.
- 22.Wong JH, Shigemizu D, Yoshii Y, Akiyama S, Tanaka A, Nakagawa H, et al. Identification of intermediate-sized deletions and inference of their impact on gene expression in a human population. Genome Med. 2019;11:44. doi: 10.1186/s13073-019-0656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lupiáñez DG, Spielmann M, Mundlos S. Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet. 2016;32:225–237. doi: 10.1016/j.tig.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Hansen AS, Cattoglio C, Darzacq X, Tjian R. Recent evidence that TADs and chromatin loops are dynamic structures. Nucleus. 2018;9:20–32. doi: 10.1080/19491034.2017.1389365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jolly LA, Parnell E, Gardner AE, Corbett MA, Pérez-Jurado LA, Shaw M, et al. Missense variant contribution to USP9X-female syndrome. NPJ Genom Med. 2020;5:1–11. doi: 10.1038/s41525-020-00162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reijnders MRF, Zachariadis V, Latour B, Jolly L, Mancini GM, Pfundt R, et al. De novo loss-of-function mutations in USP9X cause a female-specific recognizable syndrome with developmental delay and congenital malformations. Am J Hum Genet. 2016;98:373–381. doi: 10.1016/j.ajhg.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaxillaire M, Bonnefond A, Liatis S, Ben Salem Hachmi L, Jotic A, Boissel M, et al. Monogenic diabetes characteristics in a transnational multicenter study from Mediterranean countries. Diabetes Res Clin Pract. 2020;108553. [DOI] [PubMed]
- 28.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Song F, Zhang B, Zhang L, Xu J, Kuang D, et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19. [DOI] [PMC free article] [PubMed]
- 32.Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seeley AH, Durham MA, Micale MA, Wesolowski J, Foerster BR, Martin DM. Macrocerebellum, epilepsy, intellectual disability, and gut malrotation in a child with a 16q24.1–q24.2 contiguous gene deletion. Am J Med Genet A. 2014;164:2062–2068. doi: 10.1002/ajmg.a.36569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi X, Han X, Cao Y, Li C, Cao Y. ZCCHC14 regulates proliferation and invasion of non–small cell lung cancer through the MAPK-P38 signalling pathway. J Cell Mol Med. 2021;25:1406–1414. doi: 10.1111/jcmm.16223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1. Characteristics of the 1,321 genes that have been specifically analyzed in the WES data, according to the phenotypes of the patient.
Data Availability Statement
Please contact author for data requests.