Abstract
Class III pistil-specific extensin-like proteins (PELPIII) are specifically localized in the intercellular matrix of tobacco (Nicotiana tabacum) styles. After pollination the majority of PELPIII are translocated into the callosic layer and the callose plugs of the pollen tubes, which could suggest a function of PELPIII in pollen tube growth. PELPIII may represent one of the chemical and/or physical factors from the female sporophytic tissue that contributes to the difference between in vivo and in vitro pollen tube growth. PELPIII glycoproteins were purified and biochemically characterized. Because of their high proline (Pro) and hydroxy-Pro (Hyp) content, PELPIII proteins belong to the class of Pro/Hyp-rich glycoproteins. The carbohydrate moiety of PELPIII is attached through O-glycosidic linkages and comprises more than one-half the total glycoprotein. Deglycosylation of PELPIII revealed two backbones, both reacting with PELPIII-specific antibodies. N-terminal amino acid sequencing of these backbones showed that PELPIII is encoded by the MG14 and MG15 genes. Two heterogeneous N-terminal sequences of MG14 and MG15, both starting downstream of the predicted signal peptide cleavage site, seem to be present, which indicates a novel N-terminal processing. Monosaccharide analysis showed that the carbohydrate moiety of PELPIII almost completely consists of arabinose and galactose in an equal molar ratio. Carbohydrate linkage analysis showed terminal and 2-linked arabinofuranosyl residues, as well as terminal and 6-, 3-, and 3,6-linked galactopyranosyl residues to be present, indicating the presence of both extensin-like and Type II arabinogalactan oligosaccharide units. The ability of β-glucosyl Yariv reagent to bind with PELPIII confirmed the arabinogalactan protein-like characteristics of these proteins.
The ultimate goal of pollination in angiosperms is the delivery of the male gametes of the pollen to the female gametophyte in the ovule. In many angiosperms, such as tobacco (Nicotiana tabacum), a pistil separates the ovule from the landing site (stigma) of the pollen. Upon germination, pollen extrudes a tube that grows toward the ovary through the intercellular matrix (IM) of the stylar transmitting tissue of the pistil. Apart from providing a physical pathway for the pollen tubes, transmitting tissue cells are also involved in providing appropriate physiological conditions for the growth of compatible pollen by secreting molecules such as free sugars, amino acids, fatty acids, lipids, polysaccharides, and glycolipids into the IM (Knox, 1984). It has been proposed that these IM components might function in a variety of processes related to pollen tube growth such as nutrition, protection/defense, guidance, and signaling (Sanders and Lord, 1992; Cheung et al., 1995). Biochemical and molecular analyses of secreted proteins found in the IM and their genes or cDNAs led to the identification of several hydroxy-Pro (Hyp)-rich glycoproteins (HRGPs), including arabinogalactan proteins (AGPs), extensin-like glycoproteins, and Pro/HRGPs (P/HRGPs; Chen et al., 1992; Goldman et al., 1992; Cheung et al., 1993; Wu et al., 1993; Lind et al., 1994; Du et al., 1996; Sommer-Knudsen et al., 1996). The biological function of most of these pistil-specific glycoproteins has not yet been established, although some involvement in pollen tube growth has been reported for some of these proteins (Cheung et al., 1995; Lind et al., 1996; Sommer-Knudsen et al., 1998b).
A class of genes encoding for the class III pistil-specific extensin-like proteins (PELPIII) was isolated from tobacco and subsequently characterized at the mRNA level (Goldman et al., 1992). The PELPIII mRNA comprises at least two members (MG14 and MG15) localized specifically in the transmitting tissue cells of the tobacco style. Immunoblot and immunolocalization experiments demonstrated that the accumulation of PELPIII in the pistil transmitting tissue begins during the early stages of pistil maturation. At flower anthesis the proteins are located in the stylar IM of non-pollinated pistils. After pollination the majority of the PELPIII are translocated into the callosic layer and the callose plugs of the pollen tubes (de Graaf, 2000). These observations suggest that the biological function of PELPIII directly or indirectly relates to pollen tube growth. In this paper we describe the purification and the biochemical characterization of PELPIII and show that these proteins have both extensin- and AGP-like properties. Our results provide a better insight into the nature of this class of proteins and are important in elucidating the function of these proteins in the stylar transmitting tissue.
RESULTS
Purification of PELPIII
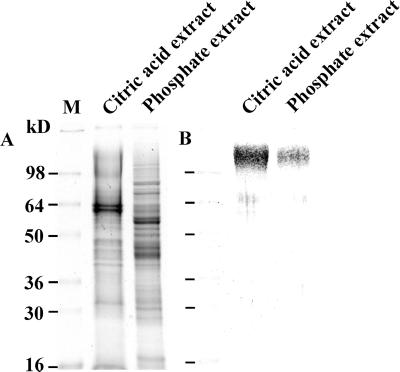
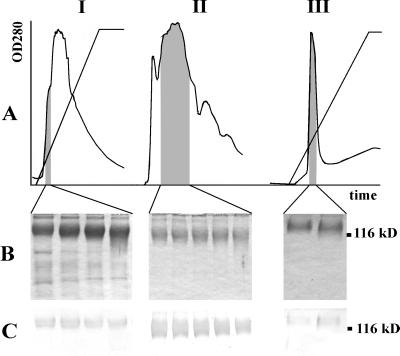
Total stylar protein extracts were prepared using buffers of various pH and salt concentrations, and extracts were tested for the presence of PELPIII. Phosphate buffers supplemented with increasing concentrations of NaCl did not result in an increase in the amount of extracted PELPIII (data not shown). A comparison of the stylar proteins that can be extracted with a citric acid buffer (84 mm citric acid, 2 mm Na2S2O4, and 15 mm β-mercaptoethanol [pH 3]; Cheung et al., 1995) and a phosphate buffer (50 mm sodium phosphate and 15 mm β-mercaptoethanol [pH 6]) is shown in Figure 1. The use of the citric acid buffer had a 2-fold advantage because it released absolute and relatively higher quantities of PELPIII. Densitometric measurements of equal amounts of protein extract showed at least four times more PELPIII after extraction with citric acid buffer than with the phosphate buffer. This property of the citric acid buffer has also been described for other Pro-rich glycoproteins (Cheung et al., 1995; Sommer-Knudsen et al., 1998b). The alkaline character (pI 9) and the high Mr of these proteins (Fig. 1) led us to apply a procedure by which PELPIII was purified in three sequential chromatographic steps: cation exchange chromatography on sulfopropyl Sepaharose, gel filtration chromatography on superose 12, and a cation exchange chromatography on methyl-sulfonate (Fig. 2). SDS-PAGE and immunoblot detection analysis of PELPIII showed that the glycoproteins migrate as a smear from 110 to 140 kD (Fig. 2, B and C).
Figure 1.
Extraction of PELPIII from the stylar tissue of tobacco. Tobacco styles were extracted with 84 mm citric acid and 2 mm Na2S2O4 (pH 3; Cheung et al., 1995) and 50 mm sodium phosphate (pH 6). Both buffers contained 15 mm β-mercaptoethanol. Each extract (5 μg) was fractionated on a 10% (w/v) polyacrylamide gel and stained with Coomassie (A) or electroblotted onto nitrocellulose and immunostained with the antibodies raised against PELPIII (B).
Figure 2.
Purification of PELPIII from tobacco styles. A, Chromatographs of the three purification steps. The fractions in the gray area were pooled for the next purification step. I, Cation exchange chromatography on SP Sepharose after buffer exchange of the citric acid protein extract into 50 mm sodium phosphate (pH 6). Eluent A, 50 mm sodium phosphate (pH 6); eluent B, 50 mm sodium phosphate (pH 6) + 1 m NaCl. Gradient: 0% to 100% (v/v) eluent B. II, Superose 12 gel filtration chromatography using 50 mm sodium phosphate (pH 6) as eluent. III, Methyl sulfonate cation exchange chromatography. Same conditions as in I. B, 10% (w/v) SDS-polyacrylamide gel of the pooled fractions. Proteins were detected by Coomassie staining. C, Immunostaining of the corresponding western blot using the PELPIII antibody.
Amino Acid Analysis
The amino acid composition of the purified PELPIII is shown in Table I. The values obtained from the amino acid analysis were almost identical to those deduced from the MG14 and MG15 cDNA clones (Goldman et al., 1992). Hyp (18.9 mol%) and Pro (13.5 mol%) were the most abundant amino acids, followed by Leu (9.0 mol%), Lys (8.8 mol%), and Ser (7.6 mol%). Of the original Pro residues (32.4 mol%), 58% was posttranslationally modified to Hyp.
Table I.
Comparison of the amino acid composition of purified PELPIII with the amino acid composition derived from the MG14 and MG15 cDNA clones (described by Goldman et al., 1992)
| Amino Acid | PELPIII | MG14 | MG15 |
|---|---|---|---|
| mol% | |||
| Ala | 3.9 | 3.5 | 3.6 |
| Arg | 1.5 | 2.3 | 1.5 |
| Asp/Asn | 6.6 | 5.9 | 6.0 |
| Cys | nd | 1.8 | 1.7 |
| Glu/Gln | 5.6 | 4.8 | 5.5 |
| Gly | 3.1 | 3.0 | 3.0 |
| His | 0.4 | 0.0 | 0.3 |
| Hyp | 18.9 | – | – |
| Pro | 13.6 | 31.6 | 30.7 |
| Ile | 3.6 | 4.2 | 3.8 |
| Leu | 9.0 | 9.8 | 9.5 |
| Lys | 8.8 | 9.2 | 8.9 |
| Met | 0.9 | 1.0 | 1.6 |
| Phe | 3.5 | 4.0 | 3.9 |
| Ser | 7.6 | 8.2 | 7.3 |
| Thr | 3.8 | 3.5 | 3.4 |
| Trp | nd | 0.0 | 1.3 |
| Tyr | 3.0 | 2.4 | 2.3 |
| Val | 5.7 | 4.9 | 5.7 |
The deduced amino acid compositions of MG14 and MG15 did not change significantly when the downstream N-terminal processing sites, as revealed by N-terminal amino acid sequencing (Table II; Fig. 3), were taken into account for the calculations.
Table II.
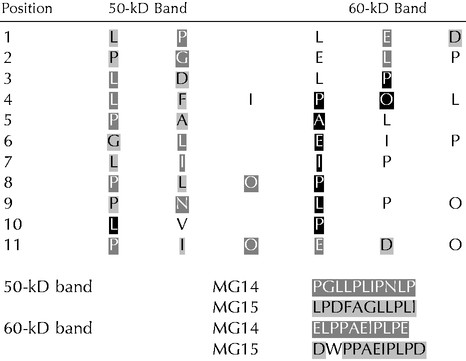
N-Terminal sequencing results of the two backbone bands after deglycosylation
 |
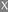 , Matching with MG14;
, Matching with MG14;
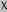 , matching with MG15;
, matching with MG15;
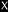 , matching both MG14 and MG15.
, matching both MG14 and MG15.
Figure 3.
Deduced amino acid sequences of the PELPIIIcDNA
clones MG14 and MG15. The predicted signal
peptide cleavage site is denoted by ↓. The N-terminal sequences
obtained from amino acid sequencing are shown in the gray boxes
 . ▾ Shows the corresponding
cleavage sites that would generate these N-terminal sequences.
. ▾ Shows the corresponding
cleavage sites that would generate these N-terminal sequences.
Deglycosylation
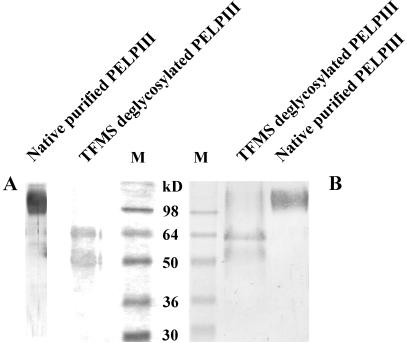
The high content of Hyp and Ser, both of which can be O-glycosylated, the running pattern on SDS-PAGE (110–140 kD) versus the predicted backbone size of 42 kD, and the ability of periodic acid to stain the purified protein (data not shown) indicate that these proteins are extensively glycosylated. Removal of the O-linked glycans by chemical deglycosylation with trifluoromethanesulphonic acid (TFMS) resulted in two backbones that migrated at about 50 and 60 kD on SDS-PAGE (Fig. 4A). Deglycosylation with anhydrous hydrogen fluoride (HF) gave the same results. No decrease in Mr could be seen when both the native and the chemically deglycosylated PELPIII were treated with N-glycosidase F (data not shown).
Figure 4.
Chemical deglycosylation of PELPIII. Native PELPIII proteins were chemically deglycosylated with TFMS and fractionated on a 10% (w/v) polyacrylamide gel and stained with silver (A) or electroblotted to nitrocellulose and immunostained with the PELPIII antibody (B). Both silver staining and immunodetection of the deglycosylated proteins show the presence of two backbones (Mr of 50 and 60). Lanes were loaded with 0.5 to 3 μg of protein. M, Mr marker.
Both backbones reacted with the PELPIII antibody (Fig. 4B). The observation that neither of the two backbones migrated at the predicted 42 kD may be explained by the fact that SDS does not bind as well to proteins with a high content of Hyp (Desai et al., 1983) and therefore has a slower electrophoretic mobility than expected for its size (Gleeson et al., 1989).
Amino Acid Sequencing
To address the nature of the two backbones present after deglycosylation of PELPIII, we subjected both bands to N-terminal amino acid sequencing. The N-terminal amino acids obtained are listed in Table II. The fact that each band (Fig. 4) turned out to be a mixture of two backbones made unambiguous assignments difficult. However, by comparison with the amino acid sequences deduced from the cDNA clones (MG14 and MG15), we were able to establish that the 50-kD and 60-kD bands resulting from chemical deglycosylation both consist of MG14 and MG15 gene products (Table II). Figure 3 shows the position of the sequences obtained in these two members of PELPIII. The N-terminal sequence of the 60-kD band started with Glu-4 of MG14 and Asp-39 of MG15. This is in contrast with the predicted signal peptide cleavage site, which would result in an N-terminal sequence starting with Lys-24 of MG15 (5′ sequence of the MG14 cDNA clone is missing). The 50-kD band started with Pro-76 of MG14 and Leu-109 of MG15. The difference of 73 and 70 amino acids, respectively, between the N termini of MG14 and MG15 in both protein backbones matches the Mr difference of about 7 kD between these two bands. Two independent deglycosylation steps and subsequent amino acid sequencing gave the same results. The reproducibility and specificity of the two backbones on SDS gel, together with the fact that deglycosylation with either TFMS or anhydrous HF resulted in the same protein backbones, suggest that acid hydrolysis of the proteins did not occur.
Carbohydrate Analysis
To determine the nature of the carbohydrate moiety of PELPIII, we performed both monosaccharide analysis and carbohydrate linkage analysis. The monosaccharide composition of PELPIII is shown in Table III. The carbohydrate part consisted predominantly of Ara (48 mol%) and Gal (50 mol%) in almost equal amounts. Glc (2 mol%) was also present, but in such a small amount that its presence was probably due to contamination. The absence of GlcNAc is in agreement with the presence of only O-linked glycans. The linkage composition of the monosaccharide residues showed terminal and 2-linked arabinofuranosyl residues as well as terminal and 6-, 3-, and 3,6-linked galactopyranosyl residues (Table IV). The ratio of branching 3,6-galactosyl residues, linear (non-branching) residues and terminal residues was approximately 1:2:2.5. The presence of low levels of 1,2,3,5-linked Araf and 1,2,3,4,6-linked Galp is probably due to slight undermethylation of the PELPIII glycoprotein.
Table III.
Monosaccharide composition of PELPIII
| Monosaccharide | Mol% |
|---|---|
| Ara | 48 |
| Gal | 50 |
| Glc | 2 |
| GlcNAc | 0 |
Table IV.
Carbohydrate linkage analysis of PELPIII
| Monosaccharide | Linkage Type | Mol% |
|---|---|---|
| Araf | Terminal | 23 |
| 1,2-Linked | 17 | |
| 1,2,3,5-Linked | 3 | |
| Galp | Terminal | 16 |
| 1,3-Linked | 6 | |
| 1,6-Linked | 12 | |
| 1,3,6-Linked | 16 | |
| 1,2,3,4,6-Linked | 4 | |
| Glcp | 1,4-Linked | 3 |
Binding to β-Glucosyl Yariv Reagent
Classical AGPs have been defined as being rich in Ara and Gal and containing high levels of Ala, Ser, and Hyp. In addition, an often-used criterion for defining whether a protein can be classified as an AGP is its ability to bind β-glucosyl Yariv reagent (Yariv et al., 1967; Fincher et al., 1983; Baldwin et al., 1993). Figure 5 shows that the purified PELPIII reacted strongly with β-glucosyl Yariv reagent, which indicates that these proteins have at least some AGP-like properties.
Figure 5.
Protein blot stained with β-glucosyl Yariv reagent. Lane A, Purified PELPIII (3 μg); lane B, stylar proteins (5 μg) extracted with the citric acid buffer; lane C, Gum Arabic AGP (2 μg); M, Prestained Mr marker.
DISCUSSION
Based on the amino acid sequence deduced from the MG14 and MG15 cDNA clones PELPIII were described as chimeric proteins containing a Pro-rich extensin-like domain and a non-Pro-rich C-terminal domain (Fig. 3; Goldman et al., 1992). Amino acid analysis, after a purification of PELPIII based on solubility, charge properties, and Mr, showed that these proteins are rich in both Pro (13.6 mol%) and Hyp (18.9 mol%). As such they belong to the HRGPs. This family includes extensins, P/HRGPs, AGPs, and solanaceous lectins (Showalter, 1993). Many proteins, however, cannot be assigned to one class only, as they may share motifs and properties usually attributed to one or more different classes (Kieliszewski and Lamport, 1994; Sommer-Knudsen et al., 1998a). The low Tyr content (3 mol%) and the low number (four–five) of repeating Ser-Pro4 motifs distinguishes PELPIII from the extensins (6–15 mol% Tyr, high no. of Ser-Pro4 motifs; Showalter and Varner, 1989). The observation that the synthetic chromophore β-glucosyl Yariv phenylglycoside (β-d-Glc)3 (Yariv et al., 1962) binds to the purified PELPIII (Fig. 5) showed that these proteins have AGP-like characteristics. AGPs are implicated in many aspects of plant growth and development, including cell fate, cell proliferation, and cell expansion, but other functions like protection and nutrition have also been proposed (Knox, 1995; Du et al., 1996; Ding and Zhu, 1997; Nothnagel, 1997; Cheung and Wu, 1999; Majewska-Sawka and Nothnagel, 2000). The AGP family can be divided into two classes designated as “classical” AGPs and “nonclassical” AGPs (Du et al., 1996). “Classical” AGPs contain a hydrophobic transmembrane domain at their C terminus, which in the mature AGPs is replaced by a glycosylphosphatidylinositol lipid anchor (Schultz et al., 1998; Youl et al., 1998; Oxley and Bacic; 1999; Majewska-Sawka and Nothnagel, 2000). They also tend to have a neutral to acidic protein backbone and a protein content typically lower than 10% by weight and rich in Hyp/Pro, Ala, and Ser/Thr (Fincher et al., 1983; Showalter, 1993; Du et al., 1996). PELPIII has a rather hydrophilic C terminus, a basic protein backbone, a protein content between 35% and 50%, and a low Ala content. As such these glycoproteins can be classified as P/HRGPs with characteristics of “nonclassical” AGPs.
In recent years more evidence has appeared that indicates that oligosaccharide units of glycoconjugates, such as glycoproteins, proteoglycans, and glycolipids, play an important role in a broad range of biological processes, such as providing signals for cell surface recognition as well as providing structural, protective, and stabilizing features (Kobata, 1992; Varki, 1993). As a consequence, we further analyzed the carbohydrate moiety of PELPIII. The carbohydrate moiety of PELPIII consists predominantly of Ara and Gal in a 1:1 molar ratio (Table III). The carbohydrate linkage characteristics shown in Table IV suggest that the PELPIII proteins have AGP-like carbohydrate chains, typically composed of mainly β-(1–3)-galactan chains with β-(1–6)-galactosyl side chains terminated primarily with arabinosyl residues (Fincher et al., 1983), as well as extensin-like carbohydrate chains due to the presence of 1,2-linked Ara residues (Fong et al., 1992). The presence of eight 1,3,6-linked Gal branching residues for every three 1,3-linked Gal residues suggests that the galactosyl residues are arranged in highly branched chains, as was also suggested for the style-specific 120-kD glycoprotein characterized in Nicotiana alata (Lind et al., 1994).
Amino acid sequencing showed the presence of two distinct peptides, which indicates that both the MG14 and MG15 genes are transcriptionally active and that both transcripts are translated. This corresponds with the Southern analysis data showing that PELPIII is encoded by a small gene family composed of two or three members (de Graaf, 2000). The MG14 and MG15 gene products each seem to have two heterogeneous N-terminal sequences, both starting downstream of the site predicted from signal sequence cleavage analysis (Fig. 3). This could derive from nonspecific cleavage of peptide bonds during chemical deglycosylation, as described previously by Vogeler et al. (1990). Proteolytic cleavage has also been suggested for negatively charged (rich in Glu and Asp) N-terminal regions that may be highly exposed to the solvent (Van Beeumen et al., 1993). The absence of low-Mr products after deglycosylation argues against nonspecific hydrolysis of the protein backbone. In addition, the lack of a negatively charged N terminus, together with the reproducible appearance of the two backbones after chemical deglycosylation using TFMS and anhydrous HF, also indicate that nonspecific hydrolysis did not occur. Together, these observations could imply a novel type of N-terminal processing of PELPIII proteins. Unusual posttranslational processing of backbones has also been proposed to occur at the N and C terminus for other HRGPs (e.g. Mau et al.; 1995, Schultz et al., 1997). These findings suggest that such processing may be more common than previously thought and could be important for their function.
There are several P/HRGPs in the styles of tobacco and N. alata with an architecture comparable to that of PELPIII (for references, see Cheung et al., 1993; Lind et al., 1994; Sommer-Knudsen et al., 1996; Schultz et al., 1997). The protein that shows most resemblance to PELPIII is the N. alata 120-kD glycoprotein. Comparison of the predicted backbone encoded by MG15 with the predicted backbone of the 120-kD glycoprotein shows that the C-terminal domain is 70% identical. However, the identity between the Pro-rich domains is much lower (39%). Like PELPIII, the carbohydrate moiety of the 120-kD glycoprotein contains linkages characteristic for both AGPs and extensins (Lind et al., 1994). However, the amount of 1,2-linked Ara (17 mol% and 36 mol% in PELPIII and the 120-kD glycoprotein, respectively) and of 1,3,6-linked Gal (16 mol% and 8.5 mol%, respectively) differs significantly between these two glycoproteins. Moreover, in contrast to PELPIII, the 120-kD glycoprotein does not bind to β-glucosyl Yariv reagent (Lind et al., 1994). Like PELPIII, this glycoprotein is located in the IM of the transmitting tract and after pollination can also be detected in the pollen tube cell walls. However, unlike PELPIII, it is also found in the pollen tube cytoplasm (Lind et al., 1996). The function of this glycoprotein is still unknown.
Another architecturally related protein characterized in tobacco, called transmitting tissue-specific protein (TTS), contains a C-terminal domain that shares 54% identity with that of PELPIII. TTS is translocated into the pollen tube wall after pollination but not into the pollen tube cytoplasm. This protein, as well as the N. alata counterpart N. alata TTS (NaTTS), has been shown to promote pollen tube elongation and to attract pollen tubes grown in a semi-in vivo pollen tube culture system (Cheung et al., 1995; Wu et al., 2000). Moreover, TTS is deglycosylated by pollen tube-bound deglycosylating enzymes (Wu et al., 1995). A very similar glycoprotein, galactose-rich style glycoprotein (GaRSGP), isolated from the styles of N. alata did not promote pollen tube growth and attract pollen tubes in culture assays or get deglycosylated by pollen tube enzymes in vivo (Sommer-Knudsen et al., 1998b). So far there is no indication that growing tobacco pollen tubes in vivo significantly modify PELPIII (de Graaf, 2000), which makes a role in nutrition unlikely and suggests a more structural function of these proteins. Although the structural similarities of several abundant P/HRGPs in the stylar transmitting tissue initially suggest functional redundancy to be present, biochemical characterization and immunolocalization experiments have shown significant differences between these proteins, which necessitates their further functional analyses.
The differences between in vivo and in vitro pollen tube growth indicate a major contribution of chemical and/or physical factors from the female sporophytic tissue to pollen tube growth in vivo (Heslop-Harrison et al., 1985; Lush et al., 1997; Cheung and Wu, 1999). The accumulation of PELPIII in the transmitting tract and the translocation of these proteins from the IM into the pollen tube walls after pollination (de Graaf, 2000) suggest that this class of proteins could directly or indirectly serve such a role. The availability of purified PELPIII together with bioassays should enable a biochemical approach toward studying their function. The availability of clones for PELPIII alternatively allows a molecular approach to understanding their function.
MATERIALS AND METHODS
Plant Material
Tobacco (Nicotiana tabacum cv Petit Havana SR1) plants were grown under standard greenhouse conditions. Styles at stage 10 through 11 of flower development (Goldberg, 1988) were collected, immediately frozen in liquid nitrogen, and stored at −80°C.
Protein Purification
Protein Extraction
Stylar tissue was ground in liquid nitrogen to a fine powder with a mortar and pestle. To obtain the optimal extraction buffer we added to 0.15-g aliquots of stylar powder several buffers (1 mL, 4°C) with various pH and salt concentrations. Results are shown for two buffers: 84 mm citric acid and 2 mm Na2S2O4 (pH 3; Cheung et al., 1995) and 50 mm sodium phosphate (pH 6). All buffers contained 15 mm β-mercaptoethanol. Proteins were extracted by repeated vortexing (30 s) and incubation on ice (15 min). The extract was centrifuged (14,000g, 15 min, 4°C), and the supernatant was collected. After estimation of the protein concentration according to the method of Bradford (Bradford, 1976; Bio-Rad, Hercules, CA), equal amounts of total protein extracts were analyzed by SDS-PAGE and immunoblots. Densitometric analysis of the polyacrylamide gels and immunoblots were performed with the GS-700 Imaging Densitometer (Bio-Rad) using the Molecular Analist 1.5 software (Bio-Rad).
For the batch purification of PELPIII, the 84 mm citric acid, 2 mm Na2S2O4 (pH 3) buffer was used as the extraction buffer. One thousand frozen styles were ground to a fine powder, and 100 mL of extraction buffer was added. Proteins were extracted as described above. After an initial centrifugation (2,400 rpm, 10 min, 4°C) any particulate material was removed from the soluble protein fraction by filtration through Miracloth; this was followed by a second centrifugation step (14,000g, 15 min, 4°C). The final supernatant was used for further protein purification.
Chromatographic Separation/Purification
After buffer exchange into 50 mm sodium phosphate (pH 6) using a Sephadex G-25 column (PD-10 column, Amersham Pharmacia Biotech, Uppsala), the protein fraction was applied to a sulfopropyl Sepharose cation exchange column (HiTrap SP, Amersham Pharmacia Biotech). Proteins were eluted by a linear gradient of 0 to 1 m NaCl. Fractions containing PELPIII were pooled and concentrated with a Centricon Concentrator (Mr cut-off 30, Millipore-Amicon, Bedford, MA) and separated on a Superose 12 gel filtration column (Amersham Pharmacia Biotech) removing residual high- and low-Mr proteins from the PELPIII-containing fraction. Lingering impurities were removed by running the PELPIII-containing fractions through a methyl-sulfonate cation exchange column (Mono S, Amersham Pharmacia Biotech) using a linear gradient of 0 to 1 m NaCl. Purified PELPIII proteins were stored at −80°C.
Deglycosylation
Chemical Deglycosylation
The glycoprotein, either dried in a speedvac or lyophilized (20–200 μg), was deglycosylated for 3 to 4 h on ice using TFMS as described by Edge et al. (1981). The deglycosylated protein fraction was precipitated by adding diethyl ether and n-hexane (9:1, v:v; Lind et al., 1994) and subsequently incubating the solution for 1.5 h at −80°C. After centrifugation (15 min, 500g, 4°C), the transparent pellet was washed with 95% (v/v) ethanol and dried under vacuum before being solubilized in water. As an alternative to the chemical deglycosylation by TFMS, anhydrous HF was used as described by Du et al. (1994), based on the method of Mort and Lamport (1977).
Enzymatic Deglycosylation
Cleavage of N-linked oligosaccharides from native (10 μg) and TFMS-deglycosylated (5 μg) PELPIII was performed with peptide-N-glycosidase F (recombinant N-glycanase, Oxford Glycosciences, Abingdon Oxon, UK) according to the manufacturer's recommended procedure.
Amino Acid Analysis
Amino acid analysis was carried out by the amino acid analysis laboratory (AAA Laboratory, Mercer Island, WA). A Beckman 7300 Amino Acid Analyzer coupled with System Gold software was used. Analysis after hydrolysis (20 h in 6 n HCl, 0.05% [v/v] β-mercaptoethanol, and 0.02% [w/v] phenol at 115°C) was performed by post-column derivitization with ninhydrin using the ion-exchange chromatographic methods developed by Stein and Moore (1951).
Electrophoretic Separation of Proteins
Protein samples were separated with SDS-PAGE according to the discontinuous buffer system of Laemmli (1970) using 10% or 12% (w/v) SDS-polyacrylamide gels (Mini-Protean II apparatus, Bio-Rad). Proteins were visualized by either Coomassie Brilliant Blue R-250 or by silver staining.
Western-Blot Analysis
The proteins separated by SDS-PAGE were electroblotted onto nitrocellulose in a buffer consisting of 39 mm Gly, 48 mm Tris base, 0.037% (w/v) SDS, and 20% (v/v) methanol (pH 8.3; mini-gel transfer apparatus, Bio-Rad). The nonspecific binding sites for immunoglobulins on the nitrocellulose membrane were blocked overnight with 5% (w/v) nonfat dried milk in phosphate-buffered saline. After blocking, the membrane was incubated for 2 h with immune serum containing antibodies against PELPIII (I-C3P), diluted 1:000 in blocking buffer. Bound antibody was detected with alkaline phosphatase-conjugated goat anti-rabbit antibodies (Pierce, Rockford, IL), diluted 1:5,000 to 1:10,000 in blocking buffer. The membranes were developed with 0.33 mg mL−1 nitro blue tetrazolium and 0.165 mg mL−1 5-bromo-4-chloro-3-indolyl phosphate in alkaline phosphatase buffer (100 mm Tris [pH 9.5], 100 mm NaCl, and 5 mm MgCl2).
AGP Detection on Western Blots
β-Glucosyl Yariv reagent (Biosupplies, Melbourne, Australia), a diagnostic reagent for AGPs, was used to test for the presence of AGPs after electroblotting onto nitrocellulose as described by Baldwin et al. (1993).
Monosaccharide Analysis
Monosaccharides were either analyzed as their alditol acetates after trifluoroacetic acid hydrolysis (as described below after methylation) or as their trimethylsilylated methylglycosides following methanolysis. (Chaplin, 1982; Bacic et al., 1987), using 1 to 5 μg of purified PELP III. The trimethylsilyl methylglycosides were analyzed by gas-liquid chromatography-mass spectrometry as described by Bacic et al. (1987).
Carbohydrate Linkage Analysis
Purified PELPIII samples (5–20 μg) were methylated according to a modified procedure of Ciucanu and Kerek (1984) as described by Oxley and Bacic (1995). The methylated samples were hydrolyzed with 2 m trifluoroacetic acid at 100°C for 2 h, reduced with 1 m NaBD4 (sodium borodeuteride) in 2 m NH3 at room temperature for 2.5 h, and acetylated with acetic anhydride at 100°C for 2.5 h. After extracting the partially methylated alditol acetates with dichloromethane, they were identified and quantified by gas-liquid chromatography-mass spectrometry essentially as described by Sims and Bacic (1995).
Determination of Total Carbohydrate
The phenol-sulfuric acid assay (Dubois et al., 1956) was used to quantitatively determine the total amount of carbohydrates in a given sample using d(+) Gal and l(+) Ara as internal standards.
Determination of Protein Concentration
Protein concentrations were either determined from the amino acid analysis or with a protein assay (Bio-Rad) based on the Bradford (1976) dye-binding procedure using bovine serum albumin as the internal standard.
Amino Acid Sequencing
The two backbones obtained after chemical deglycosylation of purified PELPIII with TFMS were electrophoretically separated and electroblotted to polyvinylidene fluoride membrane using 10 mm 3-[cyclohexylamino]-1-propanesulfonic acid. Automated Edman degradation was performed using a protein sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA) by the Protein Analysis Laboratory of the University of Alabama (Birmingham) and by Midwest Analytical, Inc. (St. Louis).
ACKNOWLEDGMENTS
We gratefully acknowledge Professor Bacic for critically reviewing the manuscript. We thank Professor Bacic and colleagues (Plant Cell Biology Research Centre, School of Botany, University of Melbourne) for providing facilities and hospitality for the carbohydrate analysis, and the Dutch Foundation Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) for a travel grant to Maurice Bosch.
LITERATURE CITED
- Bacic A, Moody SF, McComb JA, Hinch JM, Clarke AE. Extracellular polysaccharides from shaken liquid cultures of Zea mays. Aust J Plant Physiol. 1987;14:633–641. [Google Scholar]
- Baldwin TC, McCann MC, Roberts K. A novel hydroxyproline-deficient arabinogalactan protein secreted by suspension-cultured cells of Daucus carota: purification and partial characterization. Plant Physiol. 1993;103:115–123. doi: 10.1104/pp.103.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chaplin MF. A rapid and sensitive method for the analysis of carbohydrate components in glycoproteins using gas-liquid chromatography. Anal Biochem. 1982;123:336–341. doi: 10.1016/0003-2697(82)90455-9. [DOI] [PubMed] [Google Scholar]
- Chen C, Cornish EC, Clarke AE. Specific expression of an extensin-like gene in the style of Nicotiana alata. Plant Cell. 1992;4:1053–1062. doi: 10.1105/tpc.4.9.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, May B, Kawata EE, Gu Q, Wu H. Characterization of cDNAs for stylar transmitting tissue-specific proline-rich proteins in tobacco. Plant J. 1993;3:151–160. [PubMed] [Google Scholar]
- Cheung AY, Wang H, Wu H. A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell. 1995;82:383–393. doi: 10.1016/0092-8674(95)90427-1. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wu HM. Arabinogalactan proteins in plant sexual reproduction. Protoplasma. 1999;208:87–98. [Google Scholar]
- Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 1984;131:209–217. [Google Scholar]
- de Graaf BHJ. Pistil proline-rich proteins in Nicotiana tabacum, their involvement in pollen-pistil interaction. PhD thesis. The Netherlands: University of Nijmegen; 2000. [Google Scholar]
- Desai NN, Allen AK, Neuberger A. The properties of potato (Solanum tuberosum) lectin after deglycosylation by trifluoromethanesulphonic acid. Biochem J. 1983;211:273–276. doi: 10.1042/bj2110273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Zhu JK. A role for arabinogalactan-proteins in root epidermal cell expansion. Planta. 1997;203:289–294. doi: 10.1007/s004250050194. [DOI] [PubMed] [Google Scholar]
- Du H, Simpson RJ, Clarke AE, Bacic A. Molecular characterization of a stigma-specific gene encoding an arabinogalactan-protein (AGP) from Nicotiana alata. Plant J. 1996;9:313–323. doi: 10.1046/j.1365-313x.1996.09030313.x. [DOI] [PubMed] [Google Scholar]
- Du H, Simpson RJ, Moritz RL, Clarke AE, Bacic A. Isolation of the protein backbone of an arabinogalactan-protein from the styles of Nicotiana alata and characterization of a corresponding cDNA. Plant Cell. 1994;6:1643–1653. doi: 10.1105/tpc.6.11.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Gilles KH, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- Edge ASB, Faltynek CR, Hof L, Reichert LE, Jr, Weber P. Deglycosylation of glycoproteins by trifluo-romethanesulfonic Acid. Anal Biochem. 1981;118:131–137. doi: 10.1016/0003-2697(81)90168-8. [DOI] [PubMed] [Google Scholar]
- Fincher GB, Stone BA, Clarke AE. Arabinogalactan-proteins: structure, biosynthesis, and function. Annu Rev Plant Physiol. 1983;34:47–70. [Google Scholar]
- Fong C, Kieliszewski MJ, de Zacks R, Leykam JF, Lamport DTA. A gymnosperm extensin contains the serine-tetrahydroxyproline motif. Plant Physiol. 1992;99:548–552. doi: 10.1104/pp.99.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson PA, McNamara M, Wettenhall REH, Stone BA, Fincher GB. Characterization of the hydroxyproline-rich protein core of an arabinogalactan-protein secreted from suspension-cultured Lolium multiflorum (Italian ryegrass) endosperm cells. Biochem J. 1989;264:857–862. doi: 10.1042/bj2640857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB. Plants: novel developmental processes. Science. 1988;240:1460–1467. doi: 10.1126/science.3287622. [DOI] [PubMed] [Google Scholar]
- Goldman MHS, Pezotti M, Seurinck J, Mariani C. Developmental expression of tobacco pistil-specific genes encoding novel extensin-like proteins. Plant Cell. 1992;4:1041–1051. doi: 10.1105/tpc.4.9.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison Y, Heslop-Harrison J, Reger B. Pollen tube guidance and the regulation of tube number in Zea mays. Acta Bot Neerl. 1985;34:193–211. [Google Scholar]
- Kieliszewski MJ, Lamport DTA. Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 1994;5:157–172. doi: 10.1046/j.1365-313x.1994.05020157.x. [DOI] [PubMed] [Google Scholar]
- Knox JP. Developmentally regulated proteoglycans and glycoproteins of the plant cell surface. FASEB J. 1995;9:1004–1012. doi: 10.1096/fasebj.9.11.7544308. [DOI] [PubMed] [Google Scholar]
- Knox RB. Pollen-pistil interactions. In: Linskens HF, Heslop-Harrison J, editors. Cellular interactions: Encyclopedia of Plant Physiology. Vol. 17. New York, Berlin, Heidelberg: Springer; 1984. pp. 508–608. [Google Scholar]
- Kobata A. Structures and functions of the sugar chains of glycoproteins. Eur J Biochem. 1992;209:483–501. doi: 10.1111/j.1432-1033.1992.tb17313.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the heads of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lind JL, Bacic A, Clarke AE, Anderson MA. A style-specific hydroxyproline-rich glycoprotein with properties of both extensins and arabinogalactan proteins. Plant J. 1994;6:491–502. doi: 10.1046/j.1365-313x.1994.6040491.x. [DOI] [PubMed] [Google Scholar]
- Lind JL, Bönig I, Clarke AE, Anderson MA. A style-specific 120-kD glycoprotein enters pollen tubes of Nicotiana alata in vivo. Sex Plant Reprod. 1996;9:75–96. [Google Scholar]
- Lush WM, Opat A, Nie F, Clarke AE. An assay for assessing the effects of growth factors on N. alata pollen tubes in culture. Sex Plant Reprod. 1997;10:351–357. [Google Scholar]
- Majewska-Sawka A, Nothnagel EA. The multiple roles of arabinogalactan proteins in plant development. Plant Physiol. 2000;122:3–9. doi: 10.1104/pp.122.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mau SL, Chen C, Pu Z, Moritz RL, Simpson RJ, Bacic A, Clarke AE. Molecular cloning of cDNAs encoding the protein backbones of arabinogalactan-proteins from the filtrate of suspension-cultured cells of Pyrus communis and Nicotiana alata. Plant J. 1995;8:269–281. doi: 10.1046/j.1365-313x.1995.08020269.x. [DOI] [PubMed] [Google Scholar]
- Mort AJ, Lamport DTA. Anhydrous hydrogen fluoride deglycosylates glycoproteins. Anal Biochem. 1977;82:289–309. doi: 10.1016/0003-2697(77)90165-8. [DOI] [PubMed] [Google Scholar]
- Nothnagel EA. Proteoglycans and related components in plant cells. Int Rev Cytol. 1997;174:195–288. doi: 10.1016/s0074-7696(08)62118-x. [DOI] [PubMed] [Google Scholar]
- Oxley D, Bacic A. Microheterogeneity of N-glycosylation on a stylar self-incompatibility glycoprotein of Nicotiana alata. Glycobiology. 1995;5:517–523. doi: 10.1093/glycob/5.5.517. [DOI] [PubMed] [Google Scholar]
- Oxley D, Bacic A. Structure of the glycosylphosphatidylinositol anchor of an arabinogalactan protein from Pyrus communis suspension-cultured cells. Proc Natl Acad Sci USA. 1999;96:14246–14251. doi: 10.1073/pnas.96.25.14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders LC, Lord EM. A dynamic role for the stylar matrix in pollen tube extension. Int Rev Cytol. 1992;140:297–318. [Google Scholar]
- Schultz C, Gilson P, Oxley D, Youl J, Bacic A. GPI-anchors on arabinogalactan-proteins: implications for signaling in plants. Trends Plant Sci. 1998;3:426–431. [Google Scholar]
- Schultz C, Hauser K, Lind JL, Atkinson AH, Pu Z, Anderson MA, Clarke AE. Molecular characterization of a cDNA sequence encoding the backbone of a style-specific 120kDa glycoprotein which has features of both extensins and arabinogalactan-proteins. Plant Mol Biol. 1997;35:833–845. doi: 10.1023/a:1005816520060. [DOI] [PubMed] [Google Scholar]
- Showalter AM. Structure and function of plant cell wall proteins. Plant Cell. 1993;5:9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM, Varner JE. Plant hydroxyproline-rich glycoproteins. In: Marcus A, editor. The Biochemistry of Plants: A Comprehensive Treatise. Vol. 15. New York: Academic Press; 1989. pp. 485–520. [Google Scholar]
- Sims IM, Bacic A. Characterization of extracellular polysaccharides from suspension cultures of Nicotiana plumbaginifolia. Phytochemistry. 1995;38:1397–1405. [Google Scholar]
- Sommer-Knudsen J, Bacic A, Clarke AE. Hydroxyproline-rich plant glycoproteins. Phytochemistry. 1998a;4:483–497. [Google Scholar]
- Sommer-Knudsen J, Clarke AE, Bacic A. A galactose-rich, cell-wall glycoprotein from styles of Nicotiana alata. Plant J. 1996;9:71–83. doi: 10.1046/j.1365-313x.1996.09010071.x. [DOI] [PubMed] [Google Scholar]
- Sommer-Knudsen J, Lush WM, Bacic A, Clarke AE. Re-evaluation of the role of a transmitting tract-specific glycoprotein on pollen tube growth. Plant J. 1998b;13:529–535. [Google Scholar]
- Stein WH, Moore S. Chromatography. Sci Am. 1951;81:1–10. [Google Scholar]
- Van Beeumen J, Van Driessche G, Huitema F, Duine JA, Canters GW. N-Terminal heterogeneity of methylamine dehydrogenase from Thiobacillus versutus. FEBS Lett. 1993;333:188–192. doi: 10.1016/0014-5793(93)80402-g. [DOI] [PubMed] [Google Scholar]
- Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogeler HP, Voigt J, Koenig WA. Polypeptide pattern of the insoluble wall component of Chlamydomonas reinhardii and its variation during the vegetative cell cycle. Plant Sci. 1990;71:119–128. [Google Scholar]
- Wu H, Wang H, Cheung AY. A pollen tube growth stimulatory glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell. 1995;82:395–403. doi: 10.1016/0092-8674(95)90428-x. [DOI] [PubMed] [Google Scholar]
- Wu H, Wong E, Ogdahl J, Cheung AY. A pollen tube growth-promoting arabinogalactan protein from Nicotiana alata is similar to the tobacco TTS protein. Plant J. 2000;22:165–176. doi: 10.1046/j.1365-313x.2000.00731.x. [DOI] [PubMed] [Google Scholar]
- Wu H, Zou J, May B, Gu Q, Cheung AY. A tobacco gene family for flower cell wall proteins with a proline-rich domain and a cysteine-rich domain. Proc Natl Acad Sci USA. 1993;90:6829–6833. doi: 10.1073/pnas.90.14.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yariv J, His H, Katchalski E. Precipitation of arabic acid and some seed polysaccharides by glycosylphenyl-azo dyes. Biochem J. 1967;105:10–20. doi: 10.1042/bj1050001c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yariv J, Rapport MM, Graf L. The interaction of glycosides and saccharides with antibody of the corresponding phenylazo glycosides. Biochem J. 1962;85:383–388. doi: 10.1042/bj0850383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youl JJ, Bacic A, Oxley D. Arabinogalactan-proteins from Nicotiana alata and Pyrus communis contain glycosylphoshatidylinositol membrane anchors. Proc Natl Acad Sci USA. 1998;95:7921–7926. doi: 10.1073/pnas.95.14.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]