Abstract
The A-repeat region of the lncRNA Xist is critical for X inactivation and harbors several N6-methyladenosine (m6A) modifications. How the m6A modification affects the conformation of the conserved AUCG tetraloop hairpin of the A-repeats and how it can be recognized by the YTHDC1 reader protein is unknown. Here, we report the NMR solution structure of the (m6A)UCG hairpin, which reveals that the m6A base extends 5′ stacking of the A-form helical stem, resembling the unmethylated AUCG tetraloop. A crystal structure of YTHDC1 bound to the (m6A)UCG tetraloop shows that the (m6A)UC nucleotides are recognized by the YTH domain of YTHDC1 in a single-stranded conformation. The m6A base inserts into the aromatic cage and the U and C bases interact with a flanking charged surface region, resembling the recognition of single-stranded m6A RNA ligands. Notably, NMR and fluorescence quenching experiments show that the binding requires local unfolding of the upper stem region of the (m6A)UCG hairpin. Our data show that m6A can be readily accommodated in hairpin loop regions, but recognition by YTH readers requires local unfolding of flanking stem regions. This suggests how m6A modifications may regulate lncRNA function by modulating RNA structure.
Graphical Abstract
Graphical Abstract.
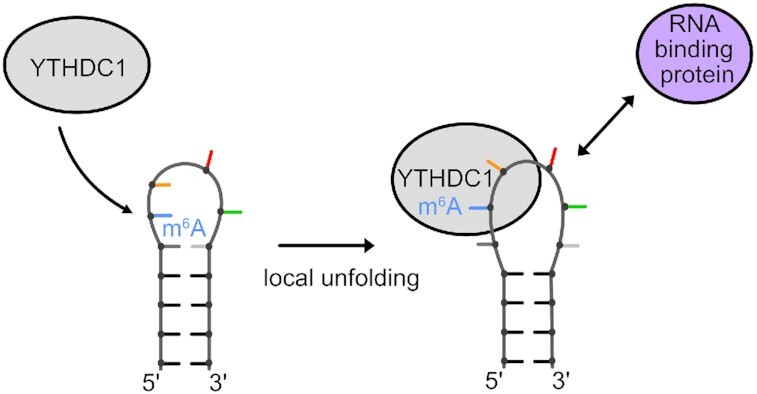
Recognition of a m6A modification in an Xist A-repeat RNA hairpin loop leads to local unfolding. This may modulate tertiary RNA contacts or interactions with RNA-binding proteins.
INTRODUCTION
N6-Methyladenosine (m6A) is the most abundant post-transcriptional base modification found in eukaryotic RNA (1). This modification, which is carried out by the ‘writer’ methylation machinery (i.e. the METTL3-METTL14 complex), serves as a regulatory factor in the gene expression of over 7000 human genes (2–4). The N6 methyl group in m6A can adopt syn and anti conformations (5,6), and may alter RNA secondary and tertiary structure: the presence of an N6-methyl group may destabilize base pairing in helical stem regions or may enhance stability when the m6A nucleotide is flanking the stem (7). The mechanism by which m6A regulates gene expression can involve the direct modulation of RNA recognition by RNA binding proteins, such as splicing factors (4), but in general depends on ‘reader’ proteins, which specifically recognize the m6A-base and flanking nucleotides. The recognition of m6A by reader proteins has been proposed to modulate RNA structure and accessibility and thereby regulate interactions with RNA binding proteins (7–10), although the underlying structural mechanisms are poorly characterized. The most prevalent reader proteins are the YT521-B homology (YTH) domain proteins (consisting of YTHDF1-3, YTHDC1 and YTHDC2), that function to regulate processes that include alternative splicing, mRNA translation and mRNA decay by specifically recognizing the m6A base (10–14).
While the regulation of mRNAs by YTH domain proteins is well-studied, other classes of RNAs, such as m6A-modified long non-coding RNAs (lncRNAs), are also ‘read’ by YTH proteins to influence m6A-linked biological functions (15–17). One of these lncRNAs is the X-inactive specific transcript (Xist), a 17kb RNA that is responsible for X-chromosome inactivation (XCI) in female placental mammals (18,19). Through six interspersed conserved repeat domains (A–F), the lncRNA Xist coats and transcriptionally silences the inactive X-chromosome. This involves interactions of numerous RBPs to the Xist lncRNA (20), including SHARP/SPEN which is required for Xist-mediated silencing (20–25). Critically, the A-repeat domain, which is located at the 5′ end of the transcript, is responsible for transcriptional silencing; in its absence, coating, but not silencing, occurs. This region is comprised of a 24-nucleotide conserved element that is repeated 8.5 times in human (26). The first 14 nucleotides of this conserved element fold into a thermodynamically stable AUCG tetraloop hairpin, while the remaining nucleotides promote duplex formation with other A-repeat elements (27–29).
Xist was recently found to harbor several hundred N6-methyladenosines along the length of the transcript, including the adenosine located within the conserved AUCG tetraloops of the A-repeats. (Figure 1A) (15). The m6A modification of Xist is facilitated by the RBPs RBM15/RBM15B. RBM15, which directly binds the A-repeats, associates and guides the ‘writer’ protein complex formed between WTAP (Wilms tumor-associated protein) and METTL3 to the lncRNA Xist, thereby facilitating the m6A-modification (Figure 1A) (15,22,30). Through co-immunoprecipitation assays, the m6A-modified nucleotides were found to be bound by the YTH domain of the reader protein YTHDC1 (15). While the degree to which m6A modification of Xist regulates XCI is disputed (15,24,30), it is widely accepted that recognition of m6A-modified bases of Xist by YTHDC1 promotes Xist-mediated transcriptional silencing (15).
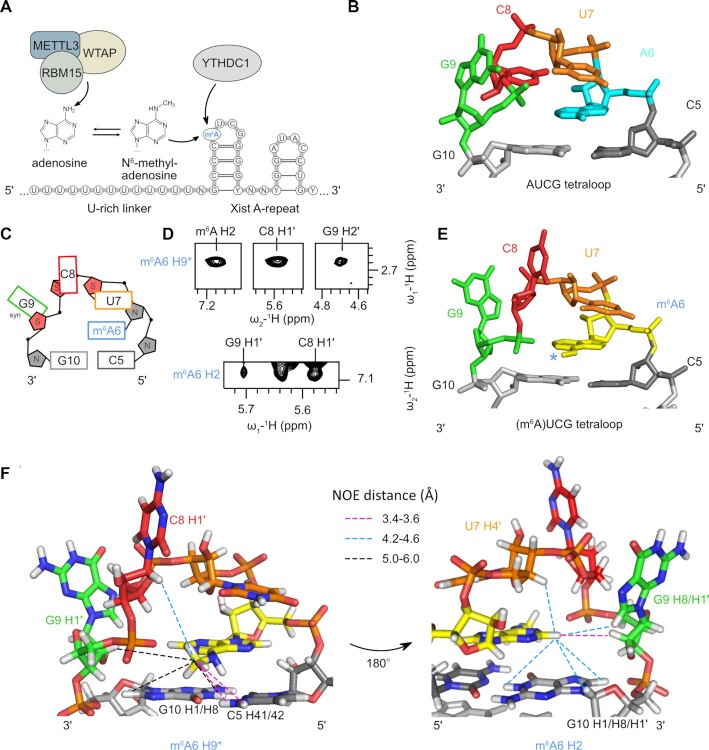
Figure 1.
Structure of the (m6A)UCG tetraloop in Xist A-repeats. (A) Schematic overview of the reader (RBM15) and writer proteins (METTL3 and WTAP) that facilitate m6A modification of adenosines located within the tetraloop of the Xist AUCG tetraloop hairpin. YTHDC1 recognizes and binds m6A-modified Xist RNA. (B) Overview of the non-methylated AUCG tetraloop structure (PDB ID: 2Y95). (C) Schematic of base orientation and sugar puckering of the (m6A)UCG tetraloop. (D) NOEs between the m6A H2 and methyl (H9*) protons, and inter-nucleotide NOEs between m6A H2 and methyl (H9*) protons to ribose protons of C8 and G9, as observed in the 1H–1H NOESY spectra at 150 ms mixing time. (E) Zoomed view of the (m6A)UCG tetraloop nucleotides in the lowest energy structure of the NMR ensemble of the (m6A)UCG tetraloop hairpin. The methyl group of m6A is marked with a blue asterisk. (F) Zoomed view of the (m6A)UCG tetraloop residues highlighting inter-residue NOEs between the m6A H2 and methyl (H9*) protons.
The unmodified AUCG tetraloop of the Xist A-repeats adopts a unique fold, with the adenosine largely solvent inaccessible. Extended 5′ base stacking from C5 to U7 results in the first two nucleotides of the tetraloop (AU) adopting an A-form helical conformation (Figure 1B) (27). It is unclear how the addition of an m6A methyl group may affect the tetraloop conformation. Previous studies investigating the effect of m6A modification on RNA structure have revealed both stabilizing and destabilizing effects. In RNA duplexes, m6A modifications occurring at the termini of an A-form helix result in stabilization of base stacking, whereas N6 methylation of base-paired adenosines results in destabilization of duplex structure (7). While structures have been reported for the binding of YTH reader domains to m6A in single-stranded RNA regions (31–34), it is unknown if and how YTH domains can recognize the N6-methylated adenosine in the context of a hairpin structure.
Here, we combine NMR, crystallography and biochemical studies to study the effect of N6-methylation of the adenosine in the (m6A)UCG-tetraloop of Xist. We show that the N6 methylation is compatible with the tetraloop structure and retains base pairing in the helical stem. While the m6A base in the loop is stacked between C5 and U7, the remaining two nucleotides of the apical loop, C8 and G9 are solvent-exposed, and oriented perpendicular to the m6A base. This may contribute to the mechanism of how the YTH domain can access the m6A nucleotide. Our crystal structure of the YTH domain of YTHDC1 with the (m6A)UCG-tetraloop reveals that the m6A, U and C bases of the tetraloop are recognized in a single-stranded conformation. Strikingly, NMR and fluorescence experiments show that YTH binding leads to partial destabilization of the upper stem, while base-pairing in the lower region of the stem remains intact. The required local unfolding of the closing base pair is reflected in a slightly reduced affinity compared to an interaction with a single-stranded m6A RNA ligand. Our data provide insight into the structural mechanism of m6A recognition in the context of a hairpin structure to promote Xist-mediated X chromosome silencing.
MATERIALS AND METHODS
RNA transcription and purification
The single-stranded hexamer RNAs, 5′-CCAUCG-3′ and 5′-CC(m6A)UCG-3′, and the m6A-modified Xist A-repeat UG-mismatch RNA, 5′-GGCGU(m6A)UCGGCGCC-3′, were purchased from Dharmacon as HPLC- or PAGE-purified and desalted RNA oligonucleotides. The unlabeled and 13C–15N Xist A-repeat tetraloop RNAs, 5′-GGCGCAUCGGCGCC-3′ and 5′-GGCGC(m6A)UCGGCGCC-3′, were produced by in vitro transcription using in-house purified T7 polymerase and purified as described previously (27). DNA template (5′-GGCGCCGATGCGCCTATAGTGAGTCGTATTA-3′) containing the T7 promoter sequence (underlined) was purchased from Eurofins Genomics as a standard desalted DNA oligo. 13C-15N-labeled rCTP, rGTP and rUTP were purchased from Silantes, and N6-methyl-ATP, for the transcription of the m6A modified RNA, was purchased from Jena Bioscience. RNAs were concentrated to 1 mM in 25 mM NaCl and 25 mM sodium phosphate, pH 6.5.
Protein expression and purification
The pETM28 vector-containing residues 345–509 of H. sapiens YTHDC1 was purchased from Addgene (Addgene plasmid #64652). Nucleotides encoding an N-terminal GGGG-linker were cloned between the YTH domain and TEV cleavage site to improve cleavage efficiency. Escherichia coli BL21 (DE3) cells transformed with the vector were grown either in Lysogeny broth (LB) medium for unlabeled expression, or in M9 minimal medium supplemented with 0.5 g/l 15NH4Cl and 4.0 g/l unlabeled glucose or 2.0 g/l 13C-labeled glucose (for 15N- and 13C–15N-labeling, respectively). Expression and purification for all constructs was carried out as previously described (33). During the final purification by size-exclusion chromatography, the protein was eluted in NMR buffer (25 mM sodium phosphate pH 6.5, 150 mM NaCl and 5 mM dithiothreitol), and fractions containing the YTH domain were concentrated and stored at 4°C.
NMR spectroscopy
All NMR experiments were carried out on Bruker 500, 600, 900 and 950 MHz Avance III spectrometers equipped with TCI or QCI cryoprobes. Backbone assignments for 60% of the YTH domain of YTHDC1 were initially taken from Theler et. al (32) (PDB 2MTV); completion of backbone 1H, 15N, 13C’, 13Cα and 13Cβ assignments for the YTH domain were obtained from standard triple resonance experiments (CBCACONH, HNCACB, HNCO and HNCACO) at 298 K (35). Data were processed using NMRPipe (36) and analyzed using CCPN Analysis V2.5 (37).
RNA assignments for the (m6A)UCG tetraloop hairpin were partially facilitated using our previously reported chemical shifts for the AUCG Xist tetraloop hairpin (27,38). Due to chemical shift differences induced by the presence of the m6A base, further NMR experiments were collected and analyzed to unambiguously assign resonances corresponding to the m6A-modified RNA. All aromatic base (H2, H6, and H8) and anomeric ribose (H1′, H2′ and H3′) protons were assigned to completion. 60% of resonances corresponding to H4′, H5′ and H5″ were unambiguously assigned. The exchangeable 1H resonances of the (m6A)UCG tetraloop hairpin of Xist were assigned using homonuclear 1H–1H NOESY experiments at 280 K with graduated mixing times (50, 150 and 300 ms) in 90% H2O and 10% D2O. The nonexchangeable 1H resonances were assigned using homonuclear 1H-1H NOESY (50, 150, and 300 ms mixing times), homonuclear TOCSY (80 ms mixing time), and natural abundance 1H–13C HMQC experiments at 298 and 280 K on a sample dissolved in 99.99% D2O. Spectral overlap was resolved using 3D 13C-edited 1H–1H HMQC–NOESY–HMQC experiments implementing ultrashort broadband cooperative pulses (39). Hydrogen bond restraints of RNA base pairs were identified using HNN COSY experiments (40,41).
1H–15N SOFAST HMQC (42) titration experiments were recorded on a uniformly 15N-labeled YTH domain that was concentrated to 100 μM. Prior to measurement, all hairpin RNA constructs were snap-cooled by heating at 96°C for 5 min, followed by incubation on ice for 10 min. NMR experiments were carried out at 298 K in NMR buffer supplemented with 10% D2O. NMR spectra were processed in TopSpin 3.5 (Bruker) and analyzed in CCPN Analysis V2.5. The chemical shift perturbation plots were generated based on chemical shifts obtained from titration experiments using the equation:
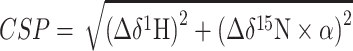 |
The scaling factor α is calculated according to the range of chemical shifts observed in bound states of each titration series and varies from 0.114 to 0.117 (43). Line broadening was evaluated as the change in peak intensity of the protein–RNA complex relative to the free protein.
1H 1D and homonuclear 1H–1H NOESY experiments with WATERGATE water flip-back were used for titration experiments. The NMR spectra of 200 μM unmodified and 160 μM m6A-modified Xist tetraloop hairpin RNA were acquired at 298 K in NMR buffer supplemented with 10% D2O. Before measurements, RNA samples were snap-cooled as described above. Protein to RNA molar ratios are indicated in the figure legends. NMR spectra were processed in TopSpin 3.5 and analyzed in CCPN Analysis V2.5.
Fluorescence quenching assays
The (m6A)UCG Xist tetraloop hairpin RNA construct with 6-FAM conjugated at the 5′-end and BhQ-1 at the 3′-end were purchased from Dharmacon and Eurofins Genomics, respectively, as PAGE-purified and desalted oligos. Prior to measurement, all RNA samples were diluted to 400 nM in NMR buffer (25 mM sodium phosphate pH 6.5, and 25 mM NaCl) and refolded by heating at 96°C for 5 min followed by incubation on ice for 10 min. A denaturing control was prepared by supplementing 400 nM RNA with 6 M urea and heating the RNA to 95°C prior to measurement. YTH protein was added at concentrations as indicated in the main figure. FAM fluorescence was excited at 495 nm with a slit of 2 nm. Emission was recorded at 517 nm with a slit of 3 nm for 0.5 s (integration time). The acquired data were normalized against buffer control and plotted. Three biological replicates were performed.
Isothermal titration calorimetry
All ITC measurements were performed on a MicroCal PEAQ-ITC (Malvern Pananalytical Ltd., UK) at 25°C. The YTH domain and the unmodified and m6A-modified tetraloop hairpin RNA constructs were concentrated and dialyzed against NMR buffer (25 mM sodium phosphate, pH 6.5, 150 mM NaCl and 5 mM dithiothreitol) at 4°C overnight; the purchased single-stranded RNA oligos were diluted with dialysis buffer (25 mM sodium phosphate pH 6.5, 150 mM NaCl and 5 mM dithiothreitol or 1mM TCEP). Prior to measurement, all hairpin RNA samples were snap-cooled by heating at 96°C for 5 min, followed by incubation on ice for 10 min. For hexamer and tetraloop hairpin RNA series, 9.5–10.5-fold (or 290–315 μM) concentrated YTH domain was titrated into 30 μM RNA in the cell, both in dialysis buffer. The experimental settings include one 0.4 μl injection followed by 19 injections of 2 μl with 120 s spacing for equilibration. Collected data were analyzed with MicroCal PEAQ-ITC Analysis Software using a one-site binding model; KD and thermodynamic signatures of binding were determined based on three technical replicates. Note, that the distinct enthalpy and entropy contribution seen for the binding of the less stable RNA tetraloop with a closing UG base pair, might reflect unfolding of the hairpin upon complex formation.
CD-monitored thermal denaturation
CD measurements were performed on Jasco J-715 Spectropolarimeter using a quartz cuvette with 10 mm path length. 50 μM of RNA in NMR buffer (25 mM sodium phosphate buffer pH 6.4, 25 mM NaCl) was refolded by heating at 96°C for 5 min followed by incubating on ice for 10 min. The changes of CD as a function of wavelength for both unmodified and m6A-modified RNAs were recorded at 25°C and averaged based on five replicates. The specific wavelength values were derived for both unmodified (265 nm) and m6A-modified (265.5 nm) RNAs and further used to measure temperature-dependent changes in ellipticity from 10 to 100°C, with a 1°C/min slope and 5 min delay. Tm values were calculated as described previously (27) using the minima of first derivatives of the melting and refolding curves.
NMR structure calculations
The structure of the Xist (m6A)UCG tetraloop hairpin was calculated using restrained molecular dynamics followed by energy minimization in NIH-XPLOR using the RNA-ff1 force field (44). Briefly, an extended structure was generated and folded using NOE-derived distance restraints obtained from homonuclear 1H–1H NOESY experiments and unambiguous hydrogen bond and planarity restraints for base-paired nucleotides to generate 200 structures using the fold.py script. Based on qualitative assessment of homonuclear TOCSY and NOESY spectra, U7, C8 and G9 riboses were restrained to adopt C2′-endo sugar puckering (all other nucleotides adopt C3′-endo pucker conformation), and the G9 base was restrained to syn orientation. Refinement of the lowest energy structures was carried out as described (44) and included the aforementioned dihedral angle restraints, with final convergence being established when no NOE violations >0.5 Å or dihedral angle violations >5° occurred in the majority of calculated structures. Structural statistics are given in Table 1. Torsional violations listed in Table 1 arise from the sugar pucker of U7 in the apical loop.
Table 1.
NMR structural statistics for the ensemble of 10 lowest energy structures
| NMR restraints | |
| # restraints | 560 |
| NOEs | 202 |
| Intra-residue | 112 |
| Inter-residue | 90 |
| Torsional anglesa | 126 |
| Planarity | 15 |
| H-bonds | 15 |
| NMR ensemble | |
| R.M.S.D. NOE restraints (Å) | 0.037 ± 0.001 |
| NOE violations (>0.5 Å) | 0 |
| Torsion violations (>5°) | 1.2 |
| R.M.S.D. from the mean coordinates (Å)b | 0.34 ± 0.02 |
| R.M.S.D. from ideal geometry | |
| Bond lengths (Å) | 0.002 ± 0.000 |
| Bond angles (°) | 0.631 ± 0.061 |
| Average XPLOR energy (kcal/mol) | 1127.6 ± 33.46 |
aA-form duplex backbone torsion angle restraints derived from high-resolution crystal structures were used for the helical stem: α (300° ± 20°), β (180° ± 10°), ϒ (50° ± 10°), δ (80° ± 30°), ϵ (210° ± 10°), ζ (290° ± 20°).
ball heavy atoms.
X-ray crystallography
The 4:1 ratio of (m6A)UCG Xist tetraloop hairpin:YTH complex obtained from ITC was concentrated to 5.0 mg/ml and screening was carried out with the Qiagen Classics I Suite X-ray crystallography screening kit. A 400 nl drop volume, comprised of 200 nl of complex and 200 nl of crystallization solution, was used. Optimal crystals were formed using the sitting drop, vapor diffusion method after one week in 0.2 M lithium sulfate, 0.1M Tris–HCl pH 8.5, 30% (w/v) Peg4000. Crystals were supplemented with 30% ethylene glycol for cryogenic preservation prior to flash cooling. Diffraction datasets were collected on the beamlines at the Swiss Light Source (SLS, Villigen, Switzerland) at beamlines PXIII. Diffraction data were collected at cryogenic temperatures (100 K) at wavelengths of 1.0 Å. The data were indexed with the XDS package (45) before scaling with Aimless as part of the CCP4 package (46). The structure was solved by molecular replacement using two crystal structures from the Protein Data Bank (PDB), codes 4R3I and 4RCJ, as search models in Phaser (47); the missing residues and RNA were built into the visible electron density using Coot modeling building software (48). The models were refined using the Phenix suite (49). Figures were made using Pymol (Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 2.4, 2020). Structural statistics are given in Table 3.
Table 3.
Crystallographic data collection and refinement statistics
| Wavelength (Å) | 1.00 |
| Resolution range (Å) | 44.31–1.77 (1.833–1.77) |
| Space group | P 21 21 21 |
| Unit cell | 78.438, 85.119, 88.617, 90, 90, 90 |
| Unique reflections | 57 778 (5063) |
| Completeness (%) | 98.70 (87.53) |
| Wilson B-factor (Å2) | 29.26 |
| Reflections used in refinement | 57 723 (5027) |
| Reflections used for R-free | 2883 (251) |
| R-work | 0.1991 (0.4571) |
| R-free | 0.2313 (0.4734) |
| Number of non-hydrogen atoms | 4387 |
| macromolecules | 4131 |
| ligands | 106 |
| solvent | 234 |
| Protein residues | 495 |
| RMS (bonds) (Å) | 0.011 |
| RMS (angles) (°) | 1.22 |
| Ramachandran favored (%) | 98.16 |
| Ramachandran allowed (%) | 1.84 |
| Ramachandran outliers (%) | 0 |
| Rotamer outliers (%) | 0 |
| Clashscore | 3.14 |
| Average B-factor (Å2) | 64.87 |
| Macromolecules | 63.91 |
| Ligands | 84.67 |
| Solvent | 65.02 |
Statistics for the highest-resolution shell are shown in parentheses.
RESULTS
Structure of the m6A modified Xist A-repeat hairpin
We previously reported the NMR-resolved structure of the thermodynamically stable AUCG tetraloop hairpin of Xist (27). Here, the adenosine (A6) of the AUCG loops adopts an A-form helical conformation, and contributes to extended 5′ base stacking from C5 to U7. After U7, the phosphate backbone twists, causing the bases of C8 and G9 to be flipped out in solution (Figure 1B). We first monitored thermal denaturation with circular dichroism experiments to investigate potential effects of the N6 methyl group of m6A on the thermodynamic stability of the AUCG tetraloop hairpin. Notably, the (m6A)UCG tetraloop hairpin is thermodynamically stable, with a melting temperature of 85.25 ± 0.25°C (Supplementary Figure S1A), which is comparable to the melting temperature of the unmodified AUCG tetraloop hairpin (85.75 ± 0.25°C) (Supplementary Figure S1B), and consistent with measurements for a related tetraloop RNA (82.1 ± 0.4°C) (27).
Inspection of the imino proton NMR resonances of the (m6A)UCG tetraloop hairpin in 1H–1H NOESY spectra recorded in H2O revealed a complete ‘imino-walk’ (i.e., correlations based on NOEs between the imino protons of the guanosines involved in base-pairs) along the helical stem (G2-G12-G4-G10) (Supplementary Figure S2A). As expected, imino resonances corresponding to the first G–C base pair are not visible due to end-fraying. Strong imino-amino inter-residue NOEs between each guanosine H1 imino proton and its respective base-paired cytosine amino proton resonances are observed, confirming G–C base pair formation (Supplementary Figure S2A). These results show that the addition of the N6-methyl in m6A is compatible with the hairpin structure and does not alter stem-formation.
Assignment and analysis of aromatic H6/H8 and anomeric protons using homonuclear NOESY and TOCSY experiments (50) revealed that the N6-methyl of A6 is in the syn conformation and stacked between residues C5 and U7, following an A-form helix conformation. The intra- and inter-residual aromatic H6/H8 to anomeric H1′ proton NOE walk shows sequential connectivities from the 5′ end of the RNA through the H6 proton of U7 (Supplementary Figure S2B). The H8 proton of m6A is shifted downfield (7.68 ppm), which is characteristic of adenosines adopting an A-form conformation (27,50). NOEs are observed between the H5 and amino protons of C5 to the methyl protons of m6A (Supplementary Figure S2C). The H5/H6 protons of U7 also show NOEs to the H8 proton of the m6A base, supporting an orientation where the m6A base is sandwiched between the nucleobases of C5 and U7 (Supplementary Figure S2B). Further supporting this stacking arrangement, we find a weak-intensity NOE between the H1 proton of G10 and the N6-methyl group protons of m6A in the 1H–1H NOESY spectra of the RNA recorded in H2O (Supplementary Figure S2A). A qualitative analysis of the 1H–1H TOCSY spectra reveals strong H1′–H2′ and H1′–H3′ correlations for U7, C8 and G9, supporting C2′-endo sugar puckering for these nucleotides in the (m6A)UCG tetraloop (Figure 1C, Supplementary Figure S2D). All other riboses in the RNA stem region adopt a C3′-endo sugar pucker. The base of G9 adopts a syn conformation, as indicated by the strong NOE observed between its H8 base and H1′ ribose protons (Figure 1C, Supplementary Figure S2B). While these results are in good agreement with what was observed for the AUCG tetraloop hairpin (Supplementary Figure S2E), further inspection of the 1H–1H NOESY spectra revealed several medium- to weak-intensity NOEs between the H2 and methyl group protons of m6A to the C8 and G9 residues of the (m6A)UCG tetraloop that suggest a differential arrangement of these nucleobases relative to m6A. In particular, NOEs are observed between the C8 H1′ proton and the m6A methyl protons, and between the m6A H2 proton and the H2′ and H8 protons of G9 (Figure 1D).
To provide high-resolution structural insight, we determined the NMR solution structure of the (m6A)UCG tetraloop hairpin using molecular dynamics/simulated annealing in XPLOR-NIH (51,52). The structure is based on 202 NOE-derived distance restraints, and additional torsional angle and base planarity restraints (see Methods and Table 1). The ensemble of the 10 lowest energy structures is well converged (Supplementary Figure S2F, G) and shows that the stem region adopts an A-form helical conformation, with stacking extended at the 5′ strand from C8 base is solvent-exposed (Figure 1E, F). The H1′ proton of C8 is localized in close proximity to the methyl group of m6A. The base of G9 folds back into the minor groove with the H8 pointing toward the H2 of m6A (Figure 1F). Following G9, G10 is base paired with C5. Overall, the (m6A)UCG tetraloop hairpin forms a stable structure, with the UCG bases of the apical loop exposed to the solvent. Thus, the N6-methyl modification of A6 does not disrupt the stem formed by the RNA, but results in a rearrangement of nucleotides in the loop region.
The YTH domain recognizes m6A in the Xist A-repeat (m6A)UCG tetraloop
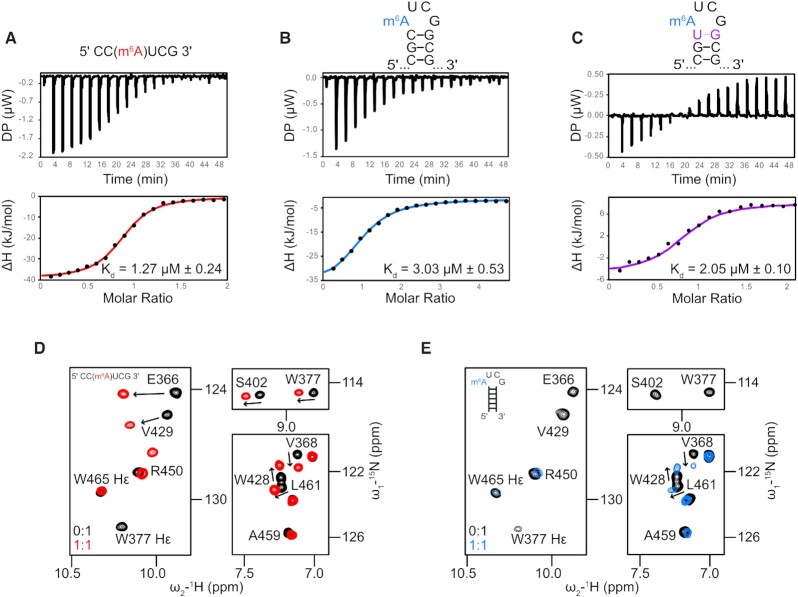
Previous studies have shown that the YTH domain of YTHDC1 binds single-stranded m6A-modified RNAs with nanomolar to micromolar binding affinity, while unmodified oligonucleotides of the same sequence are bound with considerably weaker binding affinity (i.e. 100 nM and 5 μM for 5'-UG(m6A)CAC-3' and 5'-UGACAC-3' RNAs, respectively) (32,33,53). We used isothermal calorimetry (ITC) to investigate the interactions of the YTH domain with unmodified and m6A-modified Xist AUCG tetraloop RNA, both as a single-stranded oligonucleotide and in the context of the structured hairpin (Figure 2A, B; Table 2). The YTH domain binds the single-stranded 5′-CC(m6A)UCG-3′ sequence with low micromolar affinity (dissociation constant KD = 1.27 ± 0.24 μM), while no binding is observed with the unmodified 5′-CCAUCG-3′ RNA (Supplementary Figure S3A). Notably, the YTH domain binds the (m6A)UCG tetraloop hairpin structure with reduced binding affinity (KD = 3.03 μM ± 0.53), while no binding is observable with the unmodified AUCG tetraloop hairpin in by ITC (Supplementary Figure S3B). The reduced binding affinity of the structured m6A-modified RNA relative to single-stranded m6A-modified RNA suggests that opening of base pairs in the upper stem region may be necessary for binding to the YTH domain. To investigate this, we thought to destabilize the upper region of the helical stem by modifying the closing C5-G10 base pair to a less stable U5–G10 base pair (UG base pairs are supported by two hydrogen bonds, whereas CG base pairs are supported by three). Indeed, the binding affinity of YTHDC1 is improved to 2.05 ± 0.1 μM for the UG Xist hairpin RNA relative to the native CG closing base pair RNA hairpin (Figure 2C, Table 2). As expected, the binding affinity is still somewhat reduced compared to what is observed with the single stranded m6A-modified hexamer. These results support the notion that the energetic penalty of breaking or destabilizing the upper stem base pair decreases the affinity to the hairpin versus the single-stranded RNA.
Figure 2.
Binding of m6A modified RNAs by the YTH domain. Isothermal titration calorimetry experiments for binding of the YTH domain (A) to 5′-CC(m6A)UCG-3′ RNA, (B) to the m6A-modified Xist A-repeat tetraloop hairpin and (C) to a variant of the m6A tetraloop with a destabilized closing base pair (CG to UG). (D, E) Zoomed-in regions of 1H–15N HSQC spectra overlays of the YTH domain free (black), (D) in complex with m6A-modified 5′-CC(m6A)UCG-3' hexameric oligo (red: 1:1), and (E) when bound to the m6A modified Xist A-repeat tetraloop hairpin (blue: 1:1).
Table 2.
Isothermal titration calorimetry
| RNA | K D | N | ΔG (kcal/mol) | ΔH (kcal/mol) | –TΔS (kcal/mol) |
|---|---|---|---|---|---|
| CC(m6A)UCG single-stranded | 1.27 ± 0.24 μM | 0.88 ± 0.02 | −33.77 ± 0.52 | −41.37 ± 2.02 | 7.64 ± 2.39 |
| CCAUCG single-stranded | No binding | N/A | N/A | N/A | N/A |
| GGCGC(m6A)UCGGCGCC hairpin | 3.03 ± 0.53 μM | 0.827 ± 0.09 | −31.57 ± 0.42 | −40.07 ± 4.45 | 8.47 ± 4.89 |
| GGCGCAUCGGCGCC hairpin | No binding | N/A | N/A | N/A | N/A |
| GGCGU(m6A)UCGGCGCC hairpin | 2.05 ± 0.1 μM | 0.93 ± 0.07 | −32.5 ± 0.1 | −13 ± 1.7 | −19.5 ± 1.7 |
Altogether, these data demonstrate that the N6-methyl is required, as expected, for recognition by the YTH domain. The 3-fold reduced affinity of the recognition of m6A in the hairpin fold compared to a single-stranded RNA sequence, and the increased binding to a weakened hairpin compared to a more stable one, suggest that binding is accompanied with some energetic penalty due to required conformational rearrangements.
NMR spectroscopy was used to assess which regions of the YTH domain interact with the Xist A-repeat (m6A)UCG tetraloop hairpin and the single-stranded 5′-CC(m6A)UCG-3′ sequence comprising the tetraloop nucleotides (Figure 2D, E, Supplementary Figure S4A–F). In 1H–15N HSQC NMR spectra of 15N-labeled YTH, large chemical shift perturbations (CSPs) are observed for amide resonances upon titration of the single-stranded 5′-CC(m6A)UCG-3′ RNA, while severe line-broadening is induced in the presence of the structured (m6A)UCG tetraloop hairpin (Figure 2D and E, Supplementary Figure S4A–F). The line-broadening observed in the NMR spectra likely reflects a combination of increased relaxation rates of the RNA–protein complex (due to the significantly increased molecular weight) and distinct binding kinetics of the complex with the hairpin RNA, which, in contrast to the single-stranded RNA interaction, require conformation changes in the hairpin. Consistent with the binding affinities determined by ITC, the single-stranded m6A RNA shows slow binding kinetics on the NMR time scale, while the tetraloop exhibits fast-to-intermediate binding kinetics associated with line-broadening (Supplementary Figure S4A, D). Importantly, the YTH amide resonances exhibiting CSPs and line-broadening comprise residues reported to mediate m6A-nucleotide recognition (31–33) (Figure 2D, E), including W377, W428 and N367, which are a part of the aromatic cage (see also Figure 3). We find very minor spectral changes upon addition of the unmodified Xist A-repeat AUCG tetraloop or the corresponding single-stranded RNA, consistent with the specificity of YTH binding to m6A-modified RNAs (Supplementary Figure S3C–F).
Figure 3.
Structure of YTH/(m6A)UCG tetraloop complex. (A) Crystal structure of YTH bound to the (m6A)UCG tetraloop of a Xist A-repeat. (B) The aromatic cage that recognizes the m6A is formed by W428, W377 and L439 side chains. (C) Hydrogen bonds (dashed lines) formed between S378, N367 and N363 mediate specific contacts to the m6A base. (D) A positively charged surface area interacts with the backbone of (m6A)UCG tetraloop and (E) mediates electrostatic interactions with the RNA (dashed lines). (F) π/cation stacking interaction between R475 and the U7 base.
Altogether, the ITC and NMR data show the specificity of YTHDC1 to m6A-modified Xist sequences over unmodified Xist RNA, and demonstrate the YTH domain is able to recognize and bind m6A, despite its stacking onto the A-form helical stem in the tetraloop hairpin. However, the reduced binding affinity to the hairpin RNA compared to single-stranded RNA, suggests that an energetic penalty is associated with the recognition of the m6A in the hairpin RNA, presumably due to conformational changes that are required.
Crystal structure of the complex of YTH with the (m6A)UCG tetraloop
To gain insight into the structural basis that defines YTH recognition of the Xist (m6A)UCG tetraloop, we crystallized the YTH domain of YTHDC1 with the 14-mer (m6A)UCG tetraloop hairpin of the Xist A-repeats. We obtained crystals of the complex that diffracted to 1.7 Å in space group of P21 21 21 (Table 3). Nucleotides of the stem flanking each end of the (m6A)UCG tetraloop (G1-C5, and G10-C14) are missing in the electron density map, despite there being sufficient space in the crystal lattice for these nucleotides. This is likely due to local destabilization of the upper stem of the RNA upon binding by YTHDC1, which results in increased flexibility. However, the (m6A)UCG tetranucleotides are clearly visible and structurally well-defined (Supplementary Figure S5A). Consistent with previously determined structures of the YTH domains with RNA (31–33), the protein adopts an open α/β fold, consisting of five helices (α0-α4), six strands (β1-β6) and 310 helices (η1-η3) (Figure 3A). The (m6A)UCG tetraloop of the Xist A-repeat hairpin RNA is bound by an arc-like surface of the YTH domain (Figure 3A; Supplementary Figure S5B). The N6-methyl-adenosine adopts a syn conformation and is buried in a pocket that is formed by residues of α1, β1 and β2 (amino acids 355 – 378), and the loop region connecting β4 and β5 (amino acids 418-440). Three conserved aromatic/hydrophobic residues, W377, W428 and L439, form the aromatic cage that recognizes the m6A methyl group by stacking of the m6A base with the aromatic rings of the two tryptophan residues, W377 and W428 (Figure 3B). The specificity for N6-methyl adenine is achieved by base-specific hydrogen bonds of the nitrogen atoms of the m6A base with the main chain amide of N363, the side chain NH2 of N367, and the carbonyl oxygen of S378 (Figure 3C). The bases of the other three nucleotides of the (m6A)UCG tetraloop, U7, C8 and G9, are stacked with each other while the phosphodiester backbone is bound by a charged surface pocket formed by K361, R404, K469, K472 and R475 in the YTH domain (Figure 3D,E). The 3′ phosphate of the m6A residue interacts with the R404 side chain (Figure 3E), while the U7 base forms a π-cation interaction with the guanidino group of R475 (Figure 3F). This binding interface is in excellent agreement with the residues that show significant chemical shift changes and line-broadening in our NMR titration experiments (Figure 2D, Supplementary Figure S4B, E). Taken together these interactions provide specific recognition of the loop region of the (m6A)UCG tetraloop.
Overall, the structure of the YTH/(m6A)UCG complex tetraloop structure resembles the recognition of m6A seen in other reported structures of YTH domains with single-stranded RNA ligands. A structural alignment of the YTH/(m6A)UCG complex and the YTH/(m6A)CU RNA in PDB 6RT4 (34) shows that the two YTH domain structures are nearly identical, with a root mean square deviation (RMSD) of 0.146 Å for heavy atoms (Supplementary Figure S5C). This indicates a conserved mode of m6A recognition in a single-stranded RNA context and implies that the YTH binding requires at least a local conformational change of the (m6A)UCG tetraloop, which avoids steric clashes of the remaining helical stem region with the YTH domain (Supplementary Figure S5D). The helical stem is thus oriented away from the YTH domain but with a dynamic orientation as indicated by the lack of observable electron density in our crystal structure.
Binding of YTHDC1 to the Xist (m6A)UCG tetraloop leads local unfolding of the RNA stem
In our NMR-derived structure of the (m6A)UCG tetraloop hairpin RNA, the m6A base is stacked with C5 and U7 and is not fully solvent-exposed. The recognition of m6A seen in our crystal structure shows that the m6A base is no longer stacked, and the remaining residues of the tetraloop (U7, C8 and G9, Figure 3A) are stacked with each other orientated away from the protein surface. Notably, no electron density is visible for the remaining residues of the RNA stem region. Thus, the conformation of the tetraloop residue within the (m6A)UCG tetraloop hairpin and when bound to the YTH domain is drastically different and involves substantial conformational rearrangements that likely affect destabilization of the helical stem region to enable m6A recognition. We therefore sought to determine whether the YTH domain fully or only partially unwinds the stem-loop upon binding.
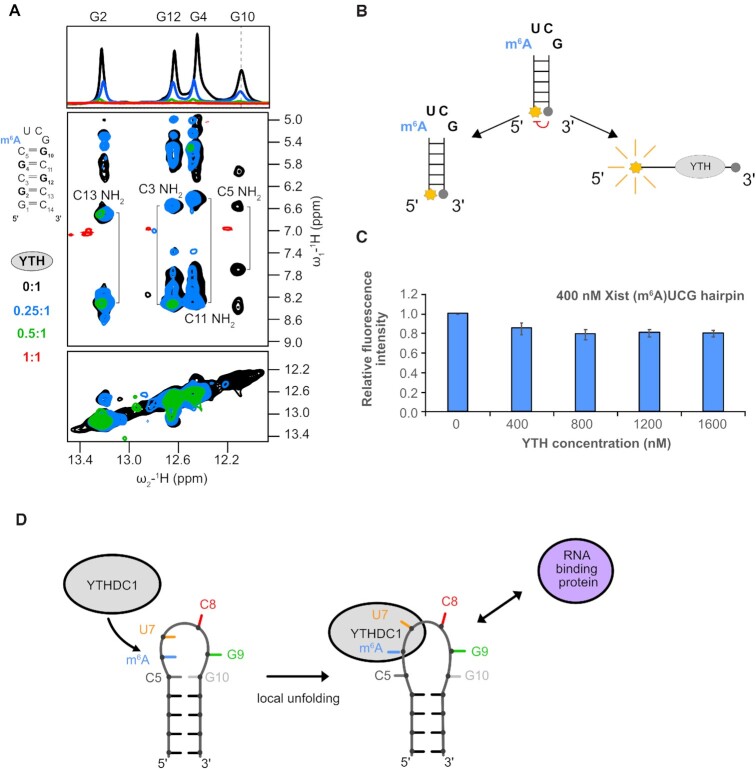
To investigate the conformation of the (m6A)UCG tetraloop hairpin and potential changes induced by YTH binding we used NMR and fluorescence quenching experiments. First, we followed intensity changes of imino resonances of the (m6A)UCG tetraloop stem by in 1D 1H NMR and 1H-1H NOESY experiments upon addition of YTH to the RNA. While the guanine imino proton resonances of the lower stem experience modest line-broadening upon addition of YTH protein, the G10 imino signal of the closing base pair becomes severely broadened already at a 0.25:1 ratio of YTH to RNA (Figure 4A). Furthermore, inspection of the 1H-1H NOESY spectra reveals that at this substoichiometric ratio, NOEs observed between the amino protons of C5 (H41 and H42) and the imino proton of G10 (H1) are no longer present, while amino-imino NOE correlations for other G-C base pairs are still observed. At a 1:1 ratio, imino proton resonances in both spectra are broadened (due to increased relaxation associated with complex formation and conformational dynamics). However, while the diagonal peaks representing the imino protons of the base pairs in the lower stem region remain visible, the G10 imino is no longer observed (Figure 4A). These results indicate that the closing C5-G10 base pair in the upper stem of the (m6A)UCG tetraloop hairpin is disrupted upon YTH binding, whereas the lower stem remains folded.
Figure 4.
YTH domain binding requires opening of the closing base pair of the (m6A)UCG tetraloop stem. (A) 1H 1D and 1H–1H NOESY spectra showing broadening imino proton resonances upon addition of YTH domain protein (black: apo RNA, blue: 0.25:1, green: 0.5:1 and red: 1:1). The G10 imino proton resonance (as indicated by the dashed line) disappears even in the presence of 0.25 molar ratio of YTH protein. (B) Schematic representation of the (m6A)UCG tetraloop hairpin with a fluorophore conjugated to the 5′ end and a quencher to the 3′ end. When the two probes are separated, there is emission of fluorescence intensity. (C) Bar plot showing the effect the YTH domain has on unwinding of the lower stem of the (m6A)UCG tetraloop hairpin at increasing concentration of protein. The RNA concentration was held constant at 400 nM. (D) Model illustrating the effects of m6A modification and YTHDC1 binding on the upper region of the Xist (m6A)UCG stem–loop. The conformational changes induced in the RNA upon YTH binding might modulate interactions with RNA-binding proteins or tertiary contacts involving the hairpin RNA.
To further support this conclusion, we performed fluorescence quenching experiments using an (m6A)UCG tetraloop hairpin with a 6-FAM fluorophore and a Black Hole Quencher 1 (54) conjugated to the 5′ and 3′ ends of the RNA, respectively (Figure 4B). Formation of the helical stem in the absence and presence of increasing amounts of the YTH domain of YTHDC1 is assessed based on the intensity of the fluorescence emission of the RNA. Relative to an RNA-only control, which shows significant quenching of fluorescence emission due to the spatial proximity of fluorescent dye and quencher attached to the two arms of the helical stem, addition of YTH protein does not notably increase the fluorescence emission, even at a 4:1 protein to RNA ratio (Figure 4C). As a positive control, denaturing of the RNA leads to more than a 5-fold increase in fluorescence emission (Supplementary Figure S6). Taken together the NMR and fluorescence quenching data indicate that binding of the YTH domain to the (m6A)UCG hairpin leads to local opening of the closing base pair, while the lower part of the helical stem remains base paired.
DISCUSSION
To date, >150 different RNA modifications have been identified across the RNA transcriptome (55). These modifications are not limited to protein-encoding mRNA, but are also prevalent in noncoding RNA transcripts (2,3,56). As has been demonstrated in several studies, RNA modifications increase the diversity of RNA function, influencing the regulation of numerous biological processes that include pre-mRNA splicing, mRNA translation, and RNA export and stability (1,4,10,11,13,57,58). The involvement of RNA modifications in each of these cellular processes thus highlights their integral role in the regulation of gene expression. However, the effects of the m6A modification to RNA structure and molecular interactions are still poorly understood.
Here, we have explored how the m6A modification affects the structure of the conserved AUCG tetraloop hairpin structure located within the A-repeats of the lncRNA Xist, and the structural mechanisms that enable m6A recognition by the YTH domain of YTHDC1 reader protein. The ensemble of structures of the unmodified AUCG tetraloop hairpin structure shows that G9 in the tetraloop is solvent exposed and that C8, G9 and G10, are conformationally dynamic, while the A6 base is stabilized by extended stacking at the 5′ arm of the RNA duplex (27). The final ensemble of structures of the (m6A)UCG tetraloop presented here reveals that the presence of the methylamino group on A6 results in a minor but notable conformational rearrangement of nucleotides of the tetraloop. The C8 nucleobase is more solvent exposed and in closer proximity to U7, with its O2 group positioned down over the inside of the major groove and H5 and H6 protons exposed to solution. On the other hand, our structure reveals that the N6-methyl group the m6A base remains stacked with the 5′ arm of the RNA stem, demonstrating that the N6-methyl group does not destabilize the Xist AUCG tetraloop structure. This is distinct from the reported destabilization of RNA duplex regions upon introducing an m6A base within an RNA helical stem (5,9). This is attributed to the requirement of an anti conformation of the N6-methyl to sustain base pairing within an RNA helical region, while the energetically more favorable syn conformation of the N6-methyl in m6A is favored in the single-stranded regions in the absence of base pairing. Indeed, our structure of the (m6A)UCG tetraloop shows that the m6A base is not base-paired, but stacked on top of the A-form helical stem. This structural arrangement of the m6A is compatible with the thermodynamically more favorable syn conformation of the N6-methyl and does not affect the stability of the Xist AUCG hairpin.
Our crystal structure of the YTH domain bound to the (m6A)UCG tetraloop shows that the m6A base is specifically recognized by the aromatic cage of the YTH domain. The U7, C8 and G9 bases are stacked with each other, and their sugar-phosphate backbone interacts with a charged surface area (Figure 3D). These interactions are highly similar to the recognition of single-stranded m6A containing RNA sequences (31–34). However, the fact that the m6A base is not readily accessible in the (m6A)UCG tetraloop structure raises the question of how the YTH domain can induce conformational changes in the hairpin RNA to enable recognition of the m6A. It has been suggested that the interaction formed between the two nucleotides immediately 3′ of m6A nucleotides (in our case U7 and C8) with the charged pocket may facilitate the recognition of m6A by the YTH domain through a two-step mechanism (34,59). Following a zipper-like mechanism of binding, the charged pocket on the surface of the YTH domain may first interact with the sugar-phosphate backbone of the two nucleotides 3′ of the m6A base, thus guiding the m6A base into the aromatic pocket. Our NMR structure of the m6A modified AUCG hairpin indeed shows that the U7, C8 and G9 nucleotides are solvent-exposed in the apo-RNA, which could enable an initial interaction by the charged surface in the YTH domain (Figure 3D). The stacking of R475 with U7 and charged interactions with the phosphate-ribose backbone may then alter the backbone conformation of the RNA and thereby trigger a reorientation of the m6A base into a position that enables its recognition by the aromatic cage of the YTH domain. This binding mechanism may rationalize the conformational rearrangement of the hairpin loop that is required for YTH binding. Indeed, the orientation of the RNA in the crystal structure does not align with the tetraloop residues of our NMR-resolved RNA structure (Supplementary Figure S5D). Furthermore, this conformational change may be reflected in energetic penalty that leads to the reduced binding affinity of the (m6A)UCG RNA tetraloop compared to the binding to single-stranded m6A RNA, and an improved binding to the m6A-modified tetraloop when the closing base pair is destabilized while the rest of the stem remains base-paired (Figure 2, Table 2). Our ITC data indicate that the differences in binding affinity result from distinct enthalpic and entropic contributions. These likely reflect the more complex binding mechanisms and conformational changes that are coupled with the recognition of m6A in a hairpin RNA by the YTH domain. As expected the binding enthalpy with the hairpin RNA is slightly reduced compared to the single-stranded m6A RNA due to the local unfolding of the closing base pair. Interestingly, recognition of m6A in the context of both the single-stranded or the hairpin RNA ligand by the YTH domain shows slightly unfavorable binding entropy (consistent with previous reports), although the release of solvent molecules from the pocket of the protein that accommodates the N6 methyl is expected to show favorable entropic contributions for binding (60,61).
The crystal structure of the YTH complex with the (m6A)UCG RNA and our NMR data indicate that the closing C5-G10 base pair must be opened to enable recognition of the m6A sequence by YTH. Consistent with this, a recent analysis of m6A/YTH domain interactions suggests that residues located upstream, i.e. 5′, of the m6A residue do not form stable interactions with the protein and remain flexible (34). Strikingly, our NMR and fluorescence data demonstrate that the lower region of the stem remains base paired, and thus, the RNA hairpin is not completely unfolded. Considering that the stem is comprised only of stable G-C base pairs, it is reasonable that the fold of the helical stem is partially maintained even in the complex with the YTH domain.
What are the consequences of the m6A of a loop residue in the Xist tetraloop hairpin? The binding of the YTH domain and the conformational changes induced in the hairpin structure may modulate molecular interactions of the RNA as a consequence of m6A modification in multiple ways. (i) The binding of YTH domain to the (m6A)UCG tetraloop may sterically block interactions with RNA binding proteins (i.e. SHARP, which has been shown to bind to the Xist A-repeat region) (23,25) and thereby modulate the A-repeat mediated silencing activity. (ii) Local unfolding of the RNA hairpin, i.e. by opening of the closing base pair, may expose single-stranded RNA sequence motifs that can then be recognized by an RNA binding protein to further modulate the fate of the RNA (9). (iii) YTH binding can attract additional RNA binding proteins (i.e. SRSF3) via protein-protein interactions to modulate biological activity, such as alternative splicing (10). (iv) Finally, conformational changes of the RNA structure may modulate tertiary contacts, which, in the case of the A-repeat regions are expected to be required for its biological function (29). In general, the specific effects of m6A will depend on whether the modification occurs in a single-stranded RNA region, in the middle of an RNA duplex, or flanking a helical stem as seen with the Xist AUCG tetraloop hairpin.
In conclusion, our data demonstrate that m6A modification of lncRNAs are compatible with retaining overall structural features but may increase the potential and complexity of fine-tuning and regulating their functional activity by modulating local structure and accessibility, for example, for RNA binding proteins.
DATA AVAILABILITY
Atomic coordinates and structural restraints for the NMR structure of the (m6A)UCG tetraloop RNA and the crystal structure of the complex with the YTHDC1 YTH domain have been deposited with the Protein Data Bank under accession numbers 7POF and 7PO6, respectively. Assignments and chemical shifts for the reported NMR structure have been deposited in the BMRB with accession number 51079.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are thankful to Arie Geerlof for preparation of T7 polymerase, Sam Asami and Gerd Gemmecker for assistance with NMR experiments, Grzegorz Popowicz for advice with crystallography, Charles Schwieters for help with m6A parameter and topology file generation, and Winfried Meining for assistance with the computational setup. The authors thank Giovanni Bussi, Hyun-Seo Kang, and Santiago Martinez-Lumbreras for comments and discussions. E.T. is grateful for support by the International Max-Planck Research School for Molecular Life Sciences (IMPRS-LS) and Ludwig-Maximilians-Universität Graduate School Life Science Munich.
Contributor Information
Alisha N Jones, Institute of Structural Biology, Helmholtz Zentrum München, Ingolstädter Landstr. 1, 85764, Neuherberg, Germany; Bavarian NMR Center, Department of Chemistry, Technical University of Munich, Lichtenbergstr. 4, 85747, Garching, Germany.
Ekaterina Tikhaia, Institute of Structural Biology, Helmholtz Zentrum München, Ingolstädter Landstr. 1, 85764, Neuherberg, Germany; Bavarian NMR Center, Department of Chemistry, Technical University of Munich, Lichtenbergstr. 4, 85747, Garching, Germany.
André Mourão, Institute of Structural Biology, Helmholtz Zentrum München, Ingolstädter Landstr. 1, 85764, Neuherberg, Germany; Bavarian NMR Center, Department of Chemistry, Technical University of Munich, Lichtenbergstr. 4, 85747, Garching, Germany.
Michael Sattler, Institute of Structural Biology, Helmholtz Zentrum München, Ingolstädter Landstr. 1, 85764, Neuherberg, Germany; Bavarian NMR Center, Department of Chemistry, Technical University of Munich, Lichtenbergstr. 4, 85747, Garching, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (German Research Foundation) [SFB 1309 – 325871075 to M.S.]. Funding for open access charge: internal budget.
Conflict of interest statement. None declared.
REFERENCES
- 1. Zhao B.S., Roundtree I.A., He C.. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017; 18:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M.et al.. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 3. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mendel M., Delaney K., Pandey R.R., Chen K.M., Wenda J.M., Vagbo C.B., Steiner F.A., Homolka D., Pillai R.S.. Splice site m(6)A methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell. 2021; 184:3125–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roost C., Lynch S.R., Batista P.J., Qu K., Chang H.Y., Kool E.T.. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc. 2015; 137:2107–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi H., Liu B., Nussbaumer F., Rangadurai A., Kreutz C., Al-Hashimi H.M.. NMR chemical exchange measurements reveal that N(6)-Methyladenosine slows RNA annealing. J. Am. Chem. Soc. 2019; 141:19988–19993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou K.I., Parisien M., Dai Q., Liu N., Diatchenko L., Sachleben J.R., Pan T.. N(6)-Methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J. Mol. Biol. 2016; 428:822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zaccara S., Ries R.J., Jaffrey S.R.. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019; 20:608–624. [DOI] [PubMed] [Google Scholar]
- 9. Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T.. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015; 518:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiao W., Adhikari S., Dahal U., Chen Y.S., Hao Y.J., Sun B.F., Sun H.Y., Li A., Ping X.L., Lai W.Y.et al.. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 2016; 61:507–519. [DOI] [PubMed] [Google Scholar]
- 11. Li A., Chen Y.S., Ping X.L., Yang X., Xiao W., Yang Y., Sun H.Y., Zhu Q., Baidya P., Wang X.et al.. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017; 27:444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roundtree I.A., He C.. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Trends Genet. 2016; 32:320–321. [DOI] [PubMed] [Google Scholar]
- 13. Roundtree I.A., Luo G.Z., Zhang Z., Wang X., Zhou T., Cui Y., Sha J., Huang X., Guerrero L., Xie P.et al.. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017; 6:e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C.. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017; 27:315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Patil D.P., Chen C.K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R.. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016; 537:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X., Liu C., Zhang S., Yan H., Zhang L., Jiang A., Liu Y., Feng Y., Li D., Guo Y.et al.. N(6)-methyladenosine modification of MALAT1 promotes metastasis via reshaping nuclear speckles. Dev. Cell. 2021; 56:702–715. [DOI] [PubMed] [Google Scholar]
- 17. Liu H., Xu Y., Yao B., Sui T., Lai L., Li Z.. A novel N6-methyladenosine (m6A)-dependent fate decision for the lncRNA THOR. Cell Death. Dis. 2020; 11:613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Avner P., Heard E.. X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet. 2001; 2:59. [DOI] [PubMed] [Google Scholar]
- 19. Brockdorff N., Bowness J.S., Wei G.. Progress toward understanding chromosome silencing by Xist RNA. Genes Dev. 2020; 34:733–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chu C., Zhang Q.C., da Rocha S.T., Flynn R.A., Bharadwaj M., Calabrese J.M., Magnuson T., Heard E., Chang H.Y.. Systematic discovery of xist RNA binding proteins. Cell. 2015; 161:404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Monfort A., Di Minin G., Postlmayr A., Freimann R., Arieti F., Thore S., Wutz A.. Identification of spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 2015; 12:554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moindrot B., Cerase A., Coker H., Masui O., Grijzenhout A., Pintacuda G., Schermelleh L., Nesterova T.B., Brockdorff N.. A pooled shRNA screen identifies rbm15, spen, and wtap as factors required for Xist RNA-Mediated silencing. Cell Rep. 2015; 12:562–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dossin F., Pinheiro I., Zylicz J.J., Roensch J., Collombet S., Le Saux A., Chelmicki T., Attia M., Kapoor V., Zhan Y.et al.. SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature. 2020; 578:455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nesterova T.B., Wei G., Coker H., Pintacuda G., Bowness J.S., Zhang T., Almeida M., Bloechl B., Moindrot B., Carter E.J.et al.. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat. Commun. 2019; 10:3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McHugh C.A., Chen C.K., Chow A., Surka C.F., Tran C., McDonel P., Pandya-Jones A., Blanco M., Burghard C., Moradian A.et al.. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015; 521:232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wutz A., Rasmussen T.P., Jaenisch R.. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002; 30:167. [DOI] [PubMed] [Google Scholar]
- 27. Duszczyk M.M., Wutz A., Rybin V., Sattler M.. The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. 2011; 17:1973–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maenner S., Blaud M., Fouillen L., Savoye A., Marchand V., Dubois A., Sanglier-Cianferani S., Van Dorsselaer A., Clerc P., Avner P.et al.. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol. 2010; 8:e1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jones A.N., Sattler M.. Challenges and perspectives for structural biology of lncRNAs-the example of the Xist lncRNA A-repeats. J. Mol. Cell Biol. 2019; 11:845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Coker H., Wei G., Moindrot B., Mohammed S., Nesterova T., Brockdorff N.. The role of the Xist 5′ m6A region and RBM15 in X chromosome inactivation. Wellcome Open Res. 2020; 5:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Luo S., Tong L.. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:13834–13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Theler D., Dominguez C., Blatter M., Boudet J., Allain F.H.. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 2014; 42:13911–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu C., Wang X., Liu K., Roundtree I.A., Tempel W., Li Y., Lu Z., He C., Min J.. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014; 10:927–929. [DOI] [PubMed] [Google Scholar]
- 34. Li Y., Bedi R.K., Wiedmer L., Huang D., Sledz P., Caflisch A.. Flexible binding of m(6)A reader protein YTHDC1 to its preferred RNA motif. J. Chem. Theory Comput. 2019; 15:7004–7014. [DOI] [PubMed] [Google Scholar]
- 35. Sattler M., Schleucher J., Griesinger C.. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 1999; 34:93–158. [Google Scholar]
- 36. Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.. NMRpipe - a multidimensional spectral processing system based on Unix pipes. J. Biomol. NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 37. Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D.. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005; 59:687–696. [DOI] [PubMed] [Google Scholar]
- 38. Duszczyk M.M., Sattler M.. (1)H, (1)(3)C, (1)(5)N and (3)(1)P chemical shift assignments of a human Xist RNA A-repeat tetraloop hairpin essential for X-chromosome inactivation. Biomol. NMR Assign. 2012; 6:75–77. [DOI] [PubMed] [Google Scholar]
- 39. Asami S., Kallies W., Gunther J.C., Stavropoulou M., Glaser S.J., Sattler M.. Ultrashort broadband cooperative pulses for multidimensional biomolecular NMR experiments. Angew. Chem. Int. Ed. Engl. 2018; 57:14498–14502. [DOI] [PubMed] [Google Scholar]
- 40. Dallmann A., Simon B., Duszczyk M.M., Kooshapur H., Pardi A., Bermel W., Sattler M.. Efficient detection of hydrogen bonds in dynamic regions of RNA by sensitivity-optimized NMR pulse sequences. Angew. Chem. Int. Ed. Engl. 2013; 52:10487–10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dingley A.J., Grzesiek S.. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc. 1998; 120:8293–8297. [Google Scholar]
- 42. Schanda P., Brutscher B.. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J. Am. Chem. Soc. 2005; 127:8014–8015. [DOI] [PubMed] [Google Scholar]
- 43. Williamson M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013; 73:1–16. [DOI] [PubMed] [Google Scholar]
- 44. Bermejo G.A., Clore G.M., Schwieters C.D.. Improving NMR structures of RNA. Structure. 2016; 24:806–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kabsch W Xds. Acta. Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A.et al.. Overview of the CCP4 suite and current developments. Acta. Crystallogr. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta. Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J.et al.. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol. 2019; 75:861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Varani G., Aboul-ela F., Allain F.H.. NMR investigation of RNA structure. Prog. Nucl. Magn. Reson. Spectrosc. 1996; 29:51–127. [Google Scholar]
- 51. Schwieters C.D., Kuszewski J.J., Marius Clore G.. Using Xplor–NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 2006; 48:47–62. [Google Scholar]
- 52. Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M.. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003; 160:65–73. [DOI] [PubMed] [Google Scholar]
- 53. Zhang Z., Theler D., Kaminska K.H., Hiller M., de la Grange P., Pudimat R., Rafalska I., Heinrich B., Bujnicki J.M., Allain F.H.et al.. The YTH domain is a novel RNA binding domain. J. Biol. Chem. 2010; 285:14701–14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tyagi S., Kramer F.R.. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996; 14:303–308. [DOI] [PubMed] [Google Scholar]
- 55. Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crecy-Lagard V., Ross R., Limbach P.A., Kotter A.et al.. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic. Acids. Res. 2018; 46:D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Coker H., Wei G., Brockdorff N.. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim. Biophys. Acta Gene Regul. Mech. 2019; 1862:310–318. [DOI] [PubMed] [Google Scholar]
- 57. Pandolfini L., Barbieri I., Bannister A.J., Hendrick A., Andrews B., Webster N., Murat P., Mach P., Brandi R., Robson S.C.et al.. METTL1 promotes let-7 MicroRNA processing via m7G methylation. Mol. Cell. 2019; 74:1278–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Solomon O., Oren S., Safran M., Deshet-Unger N., Akiva P., Jacob-Hirsch J., Cesarkas K., Kabesa R., Amariglio N., Unger R.et al.. Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR). RNA. 2013; 19:591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhu T., Roundtree I.A., Wang P., Wang X., Wang L., Sun C., Tian Y., Li J., He C., Xu Y.. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014; 24:1493–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Krepl M., Damberger F.F., Schroetter C., Theler D., Pokorna P., Allain F.H., Sponer J.. Recognition of N6-methyladenosine by the YTHDC1 YTH domain studied by molecular dynamics and NMR spectroscopy: the role of hydration. J. Phys. Chem. B. 2021; 125:7691–7705. [DOI] [PubMed] [Google Scholar]
- 61. Li Y., Bedi R.K., Wiedmer L., Sun X., Huang D., Caflisch A.. Atomistic and thermodynamic analysis of N6-methyladenosine (m(6)A) recognition by the reader domain of YTHDC1. J. Chem. Theory Comput. 2021; 17:1240–1249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structural restraints for the NMR structure of the (m6A)UCG tetraloop RNA and the crystal structure of the complex with the YTHDC1 YTH domain have been deposited with the Protein Data Bank under accession numbers 7POF and 7PO6, respectively. Assignments and chemical shifts for the reported NMR structure have been deposited in the BMRB with accession number 51079.