Abstract
It is currently unknown how many RNA transcripts are able to induce degradation of microRNAs (miRNA) via the mechanism known as target-directed miRNA degradation (TDMD). We developed TDMDfinder, a computational pipeline that identifies ‘high confidence’ TDMD interactions in the Human and Mouse transcriptomes by combining sequence alignment and feature selection approaches. Our predictions suggested that TDMD is widespread, with potentially every miRNA controlled by endogenous targets. We experimentally tested 37 TDMDfinder predictions, of which 17 showed TDMD effects as measured by RT-qPCR and small RNA sequencing, linking the miR-17, miR-19, miR-30, miR-221, miR-26 and miR-23 families to novel endogenous TDMDs. In some cases, TDMD was found to affect different members of the same miRNA family selectively. Features like complementarity to the miRNA 3′ region, bulge size and hybridization energy appeared to be the main factors determining sensitivity. Computational analyses performed using the multiomic TCGA platform substantiated the involvement of many TDMD transcripts in human cancer and highlighted 36 highly significant interactions, suggesting TDMD as a new potential oncogenic mechanism. In conclusion, TDMDfinder provides the first inventory of bona fide human and mouse TDMDs. Available as a free webtool, TDMDfinder allows users to search for any TDMD interaction of interest by customizing its selection criteria.
INTRODUCTION
MicroRNAs are a class of small non-coding RNAs that function as guide molecules in RNA silencing (1). The miRNA:target interaction is based on a limited base-pair complementarity, which usually involves nucleotides located at the 5′ end of the miRNA (the ‘seed’ region) and a complementary region typically located within the 3′ untranslated region of the target RNA (the miRNA responsive element, MRE). This interaction occurs within the RNA inducible silencing complex (RISC) and leads to mRNA destabilization and protein synthesis inhibition. The biological activity of a given miRNA is critically determined by its cellular copy number which must be precisely controlled (2,3). These cellular levels depend primarily on miRNA biogenesis, which involves several steps from the transcription of a primary transcript to the maturation of the latter into a small, single-stranded RNA molecule loaded onto the Argonaute (AGO) proteins, the catalytic components of the RISC (4).
In the last few years, it has emerged that miRNA degradation also plays a critical biological role and miRNA decay has been revealed to be a regulated process, modulating specific miRNAs in different biological contexts (5–8). The mechanism described to control miRNA degradation involves a specific class of target RNAs and has thus been termed ‘Target-Dependent MiRNA Degradation’ (TDMD) (9). At first, this mechanism was observed with artificial targets in vitro (10) and with viral RNA targets in vivo (11,12). TDMD was also shown to take place in the human and mouse brain in a peculiar fashion (9), although it was unclear which endogenous targets were involved. Cellular transcripts able to induce TDMD of their respective cognate miRNAs have been recently identified in three independent studies: Serpine1, shown to trigger miR-30b/c degradation in mouse fibroblasts (13); libra and Nrep, able to degrade miR-29b in zebrafish and mouse brain, respectively (14) and Cyrano, capable of inducing miR-7 decay in mouse (15). In each of these cases, the TDMD-transcript contained a miRNA degradation element (MDE) that, in addition to the usual seed pairing region, was characterized by a second region with complementarity to the 3′ end of the miRNA (named ‘3C pairing’) and by a central unpaired linker region (‘bulge’) between them. The structural bases of TDMD have been recently unveiled by determining the crystal structures of human Ago2 bound to either of two TDMD targets (16). The complementarity of the 3C pairing region forces the target:miRNA duplex to bend at the linker, pushing the miRNA 3′ end out of the PAZ domain of Ago2, where it becomes accessible to enzymatic degradation. The ZSWIM8 Cullin-RING E3 ubiquitin ligase has also been recently implicated in the mechanism (17,18). ZSWIM8 is recruited by the TDMD conformation and induces the degradation of AGO, thus resulting in miRNA decay. Importantly, these studies suggest that endogenous TDMD is widely involved in sculpting the levels of numerous miRNAs and that it acts in multiple contexts and animals, from flies and nematodes to mammals. Nonetheless, the endogenous TDMD interactions are still unknown and to date a genome-wide identification has not been attempted yet. In fact, commonly used target predictive algorithms, such as TargetScan (www.targetscan.org), miRanda (http://www.microrna.org) or miRDB (http://mirdb.org), that provide information about possible miRNA targets rely on a scoring system (e.g. context score) that focuses on the seed region and, therefore, does not allow to specifically search for TDMD interactions.
In this study, we present a computational pipeline called TDMDfinder that was developed to identify miRNA:target pairs with the potential to produce TDMD effects. We used TDMDfinder to investigate the human and mouse genomes and predicted TDMD interactions that gave a high accuracy score (47%) when assessed by experimental validation. Through the integration of multiple computational analyses performed on large cancer datasets, TDMDfinder also provided useful information on the involvement of TDMD in human cancer.
MATERIALS AND METHODS
mRNA:miRNA TDMD predictions
All the predicted target:miRNA interactions in human and mouse (both conserved (CS) and non-conserved (nCS), respectively, Nhuman_CS = 257 466, Nhuman_nCS = 14 279 806, Nmouse_CS = 193 264, Nmouse_nCS = 10 662 819) were downloaded from the TargetScan database Release 7.2 (http://www.targetscan.org/). Data were filtered to exclude alignments with non-canonical seed sites. The TargetScan database was also used to retrieve the exact position of each microRNA responsive element (MRE) within the corresponding 3′UTR region. Then, the ∼30 nucleotides upstream from the MRE were retrieved from Ensembl database (v75) and pairwise sequence alignments performed using the pairwiseAlignment function in the Bioconductor Biostrings package (v3.1). A global alignment of the reverse-complement of the mRNA and miRNA sequences converted to DNA strings, respectively pattern and subject, was performed using the gap penalty values and substitution scoring scheme provided by microRNA.org (http://www.microrna.org/). Only the alignments covering the entire length of the subject (i.e. miRNA) were further analyzed in order to find the ‘best alignment’, defined as the one having an alignment score falling between the 50th and the 100th percentile and the smallest number of gaps. For each aligned RNA duplex, information on the number of matches, mismatches, and consecutive aligned nucleotides was extracted using a custom implemented R function, together with additional information, such as dimension of the bulge (defined as the first block of at least two mismatching bases located beyond the seed region; only when this condition was not satisfied, the position of the first single mismatch beyond the seed was considered), and the presence/absence of base pairing both in positions 9, 10 and 11 and in the 3′ end four last positions of the miRNA. In order to select miRNAs whose 3′ complementary regions (3C) were compatible with TDMD, we initially identified all the pairs with seven or more (≥7) consecutive pairings (including G:U wobbles) and then we retained only 3C consecutive pairings with a limited number of G:U wobbles by applying the following rule: Number(3C consecutive_with_G:U) – Number(G:U_in_3C_consecutive_region) >5. For instance, in pairs with a stretch of seven consecutive pairings, only one G:U was allowed. Examples of the ‘3C types’ are shown in Figure 1A.
Figure 1.
Identification of TDMD pairs in human and mouse transcriptome. (A) Scheme summarizing the different types of miRNA:target interactions: canonical and TDMD. Bottom panel reports the nomenclature used to classify’3 complementarity (3C) types, based on number of consecutive pairings and the contribution of G:U wobbles. (B) MDEs of known TDMD targets are shown along with their structural and thermodynamic features: 3C type, bulge extension nucleotides (bulge), minimal free energy (mfe) of the interaction, reported as a ratio over an optimal TDMD duplex. (C–E) Schemes illustrating the workflow used to identify and investigate TDMD interactions. (C) TDMDfinder pipeline. (D) Criteria used to classify ‘Predicted TDMD’ interactions and, after evaluating experimental validation results, to isolate High-Confidence TDMD (HC). (E) Pan-cancer analysis, with schematics of the in silico tests used to support the involvement of TDMD interactions in human tumors.
Calculation of minimum free energy (MFE)
The RNAhybrid tool (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid) (19) was used to calculate the minimum free energy (MFE) of hybridization for each predicted target:miRNA duplex. For each target:miRNA alignment, the ratio between its MFE and the MFE of a ‘theoretically optimal TDMD’ was also calculated. For each of the annotated miRNAs, the corresponding ‘theoretically optimal TDMD’ was built using RNA sequences that were complementary to the miRNA sequence of interest in all positions except for position 9, 10 and 11 (i.e. bulge region).
Predicted and high confidence TDMD pairs selection and phylogenetic conservation
Out of the predicted TDMD pairs, we selected those that met the following criteria: alignments with seven or more (≥7) consecutive 3C matches, a bulge extension between 2 and 8 nucleotides long (≥2 and ≤8) and a MFE ratio greater than 0.7 (≥0.7). High Confidence (HC) TDMD pairs were selected from the list of predicted TDMD pairs by filtering those with six or more (≥6) consecutive 3C matches (considering canonical pairings only) and a MFE ratio greater than 0.8 (≥0.8).
For each predicted TDMD duplex, the phylogenetic conservation of the region corresponding to the 3C pairing element was measured using the PhyloP scores retrieved from the UCSC (https://genome.ucsc.edu). We used PhyloP scores calculated from multiple sequence alignments of 99 vertebrate genomes with the human genome (hg19/GRCh37). In order to obtain a cumulative conservation measure for each 3C pairing element, the single positional values of the PhyloP scores, representing −log10P-values under a null hypothesis of neutral evolution, were averaged.
Pan-Cancer TCGA analysis
Target mRNA and miRNA expression data
Normalized Genomic Data Commons (GDC) harmonized RNA-seq data [both mRNA and miRNA, FPKM (Fragments Per Kilobase Million) and RPM (Reads Per Million) respectively], of 23 selected TCGA solid primary tumor types (BLCA, BRCA, CESC, COAD, ESCA, GBM, HNSC, KIRC, KIRP, LGG, LIHC, LUAD, LUSC, OV, PAAD, PCPG, PRAD, READ, SARC, SKCM, STAD, THCA, UCEC) were downloaded using the TCGAbiolinks R/Bioconductor package (20). Data were filtered to exclude studies with less than 100 samples and/or without available GISTIC2 copy number variation (CNV) data on the GDAC firehose portal (http://gdac.broadinstitute.org). A total of 21 datasets were retained for further analysis, including 8572 patient samples.
miRNA expression data were retrieved using TCGA biolinks and Loci-based isoform.quantification.txt files, which report counts (both raw and normalized to RPM) of every distinct sRNA-seq read observed. The level of expression of each mature miRNA was then calculated as the sum of all isoforms corresponding to the same unique miRBase MIMAT identifier, which was then translated into a miRBase V.21 name using the miRiadne tool (21).
Target:miRNA and target:activity correlations
The correlation between TDMD target expression and miRNA expression was measured for all HC TDMD pairs in each cancer type using the Spearman correlation test (hereafter referred to as ‘miR_Expression Test’). In each tumor sample, the activity of each miRNA of the human HC set was estimated with the ActMir algorithm (22), a computational method based on the expression levels of both miRNAs and their predicted target genes. Activity was measured using the list of predicted conserved targets but excluding candidate TDMD transcripts. The correlation between miRNA activity and the expression level of any of its predicted TDMD-targets was evaluated using the Spearman correlation test (hereafter referred to as ‘miR_Activity Test’). For the Pan-cancer interactions we calculated also the correlation between the TDMD target and the passenger strand of the targeted miRNAs in each tumor type. A ‘Normalized Spearman Correlation’ was then computed as (Rho.Guide - Rho.Passenger). For passenger miRNAs with multiple genomic loci (e.g. miR-24-1-5p and miR-24-2-5p), the average correlation was used.
CNV data analysis
For each selected TCGA cancer dataset, GISTIC2 all_thresholded.by_genes.txt output data, a gene-level table of discrete amplification and deletion indicators for all samples, was downloaded from the GDAC firehose web portal by using the RTCGA Toolbox package available in Bioconductor. Wild type samples (WT, table value = 0) and samples bearing deletions (DEL, table values = −1 or −2, possibly heterozygous and homozygous deletions respectively) were selected from the GISTIC gene-level table and used in the MDE CNV test. In the MDE CNV test, significantly different miRNA expression levels between WT and DEL groups were measured for each HC TDMD pair, using a cut-off of FDR-adjusted P-value (q value) < 0.05.
3′UTR alternative polyadenylation
The Cancer 3′UTR Atlas (TC3A) portal (23) was used to detect differential usage of 3′UTR alternative PolyA sites in TCGA samples for all the targets in the HC set. In TC3A, 3′UTR usage for each transcript is based on a two-PolyA model and the fraction of reads attributed to each isoform is estimated by calculating a Percentage of Distal PolyA site Usage Index (PDUI). First, we evaluated whether each MDE was potentially lost in the shorter 3′UTR variant. Then, we calculated the fraction of patients with a potential loss of the MDE, in each tumor type, as follows: [number of samples with >50% of RNA-seq reads attributable to the shorter PolyA]/[total number of samples where 3′UTR was modeled].
Data related to 3P-seq reads were retrieved from TargetScan7.1. MDE inclusion rate was evaluated using the clustered 3P-seq tags, which indicate the fraction of transcripts including a specific segment of the 3′UTR [expressed as affected isoform ratio, AIR, (24)]. The MDE was first attributed to a defined 3′UTR segment, then the AIR (0–100% scale) was used to classify the inclusion rate in classes.
TDMDfinder
TDMDfinder (http://213.82.215.117:9999/TDMDfinder/index.php) is a web tool consisting of a user interface frontend implemented in Javascript/Ajax/CSS and a computational backend implemented in PHP 5.5, R 3.3 and MySQL 5.5 for data storage. The user interface allows to search and visualize results in a meaningful way and to export results in standard formats, such as Excel/csv for tables and PNG for plots. The computational backend is designed to dynamically build SQL (Structured Query Language) queries for data retrieval and to perform statistical tests and draw plots using the analysis section.
Cell cultures and reagents
The Human cervical carcinoma HeLa cells (ECACC No. 96112022) were cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 mg/ml streptomycin. Cells were routinely checked for mycoplasma contamination and always tested negative.
TDMD-assay
In order to over-express the MDEs of putative TDMD transcripts, MDE sequences and controls (scrambled sequences and mutant seed sequences) were sub-cloned into the mammalian expression vector PCDNA 3.1, a gift from Doug Golenbock (Addgene plasmid, 13031), in the 3′UTR of an EGFP reporter gene. Two additional controls were designed for miR-106b and miR-26a, namely ‘Optimal TDMD’ MDEs (ad hoc sequences pairing with all bases of the miRNA sequence, except in position 9–10–11). HeLa cells were transfected (Lipo3000, Thermo Scientific) with the PCDNA EGFP-MRE-X constructs in six-wells (400 000 cells/well) and cells were collected 72 h after transfection. The full list of constructs used in the paper is provided in Supplementary Table S3.
Small RNA sequencing and data analysis
Total RNA was isolated with the miRNeasy Mini Kit (Qiagen). Small RNA sequencing (sRNA-seq) libraries were prepared using 1000 ng of total RNA with the TruSeq Small RNA Kit (Illumina), following the manufacturer's instructions. Sequencing was performed on an Illumina Novaseq 6000 (50 bp single-read mode at a 5 million read depth per sample). Sequencing quality was checked in the FASTQC report, and only experiments with Q30 or above were considered (Phred Quality Score). Raw data together with detailed description of the procedures are available in the GEO database (GSE168566). miRNA counting was performed with the Isomirage tool (25): after counting, miRNA reads were normalized based on the library size (reads-per-million, using the sum of all miRNA-matching reads). All the analyses were performed on canonical and templated miRNA reads, in order to exclude miRNA 3′ isoforms, which are typically modulated upon TDMD. Fold-change results were plotted as a function of miRNA levels in control cells.
miRNA, pri-miRNA and mRNA RT-qPCR expression analyses
Mature miRNAs were detected by RT-qPCR (5 ng of input cDNA) performed with the miScript system (Qiagen). miRNAs primary transcripts (pri-miRNAs) were evaluated by quantitative PCR on total RNA. RT-qPCR of pri-miRNAs (25ng of input cDNA) was performed using the SuperScript® VILO cDNA Synthesis Kit (Life Technologies cat. no. 11754050) and the Fast SYBR green master mix (Life Technologies). The Primer3 software was used to design primer pairs preferentially in the 500 bp region upstream from the sequence of the mature miRNA. A list of the RT-qPCR primers used in this study is provided in Supplementary Table S5.
RESULTS
Strategy for the identification of TDMD pairs in human and mouse transcriptome
We performed an integrative bioinformatics analysis on miRNA:target pairs that was specifically aimed at identifying TDMD interactions. We focused on the distinctive structural features shared by all TDMD pairs (viral and endogenous) so far reported (Figure 1A, B), which included: (i) the length of the 3C pairing region, (ii) the size of the central bulge and (iii) the miRNA:target hybridization energy (minimal free energy, MFE). We implemented a pipeline, based on a custom R function, to compute these parameters for miRNA:target pairs and identify predicted TDMD pairs at genome-wide level (Figure 1C). Briefly, we performed pairwise sequence alignments between miRNAs and targets based on a seed-anchoring approach, using as input all transcripts reported in the TargetScan7.2 database (human and mouse). For each miRNA:target pair, we then extracted structural information on its potential TDMD-like structure by computing all the relevant quantitative parameters of the bipartite duplex (seed-, 3C- pairing and central bulge, altogether forming the ‘miRNA Degradation Element’ or MDE, Figure 1A). More than 10 millions miRNA:target pairs were analyzed for each of the two genomes (Figure 1C – ‘TDMDfinder Pipeline’). Data are available online on the TDMDfinder webtool (http://213.82.215.117:9999/TDMDfinder/index.php), where users can freely explore and query the full dataset to make customized TDMD predictions starting from any of the available transcripts and/or microRNAs (see Supplemental Material).
Criteria used in predicting TDMD pairs
With the aim of compiling a list of potential TDMD interactions, we leveraged previous studies (9,13,14,16) and selected the optimal criteria to obtain ‘Predicted TDMD pairs’ (Figure 1D), as: (i) the presence of a 3C pairing region including at least seven consecutive bases and tolerating only a minimal proportion of G:U wobbles (‘3C type’ as shown in Figure 1A; see Materials and Methods for G:U filtering criteria), (ii) the presence of a bulge from 2 to 8 nt in size and (iii) an miRNA:target hybridization energy at least 0.7 times greater than that of a theoretically perfect TDMD duplex (MFE ratio ≥ 0.7) (Supplementary Figure S1A, B; see Materials and Methods for ‘MFE ratio’ definition). This last requirement was added in order to exclude any energetically unstable pair generated by the alignment process. To verify our predictions, we performed the thorough experimental tests described in the following paragraphs (‘Experimental Validation’ – details are in Figure 3–6). We used the validation results to define more stringent criteria that would lead to the isolation of a set of ‘High Confidence TDMD pairs’, hereafter named as ‘HC set’ (Figure 1D). Finally, we exploited large TCGA datasets to further investigate HC TDMD pairs in the context of human cancer and find evidence of their possible relevance at endogenous levels (Figure 1E, ‘Pan-Cancer Analysis’ – details are in Figures 7 and 8). Data from the Pan-Cancer Analysis on all the HC pairs are also available in the TDMDfinder online tool, with tables, graphical outputs and statistics (see Supplemental Material).
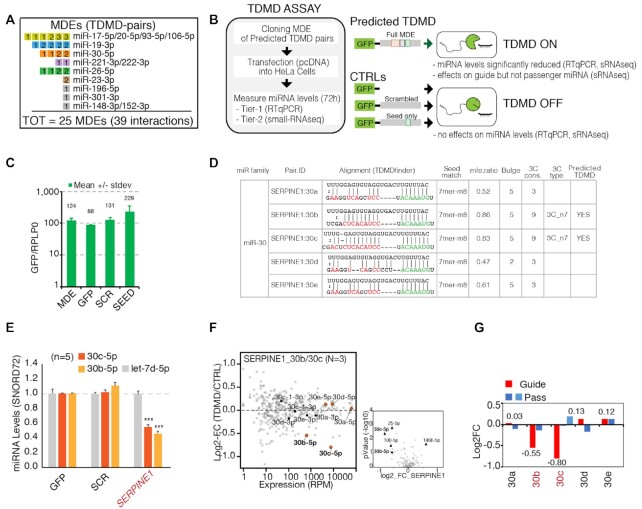
Figure 3.
Strategy for the validation of predicted TDMD pairs: the TDMD-Assay. (A) List of the predicted TDMD pairs that were experimentally validated. Each miRNA degradation element (MDE) is represented by a single square. MDEs that target the same miRNA family are grouped together. Inscribed in each square is the total number of interactions. Colored squared indicate that at least one of the corresponding TDMD interactions has been successfully validated. (B) Schematics of the TDMD-assay. For each candidate transcript, a region (∼100 bp or ∼400 bp long, depending on the construct) containing the predicted MDE was cloned into a GFP vector and transiently over-expressed in HeLa cells. (C) Expression levels of the different constructs are reported as ratios to the housekeeping gene RPLP0 expression level. (D) TDMDfinder predictions for SERPINE1 transcript and miR-30 family. Alignment and features are reported as in the online tool. (E–G) TDMD assays performed on SERPINE1 MDE. (E) Results by RT-qPCR. Mature miRNAs levels are shown as mean values with s.e.m. calculated on different biological replicates (indicated as N), normalized over CTRL_GFP. An unrelated miRNA, Let-7d, was also measured. SNORD72 was used as housekeeping gene. (n.a., not assayed). P-values were calculated by Dunnet's t-Test, using the SCR/SEED group as reference. (F) Results by small RNA sequencing (sRNA-seq). On the left, the scatter plot shows the fold changes (MDE vs CTRL, y axis) over the expression levels (reads per million, x-axis) of all the detected miRNAs. N, number of replicates. The members of the targeted miR-family (guide and passenger miRNAs) are highlighted and colored. On the right, the volcano plot highlights those miRNAs that are regulated in a statistically significant manner (P-values by Student's t-test <0.05 and log2FC >|0.5|). SERPINE1:30b is also highlighted, as it was predicted by TDMDfinder, close to significance in the sRNA-seq (P-value = 0.08) and validated by RT-qPCR in independent samples (see E). (G) Mature miRNAs levels for each individual member of the miR-30 family (with guide and passenger miRs) as measured by sRNA-seq (N, number of replicates).
Figure 6.
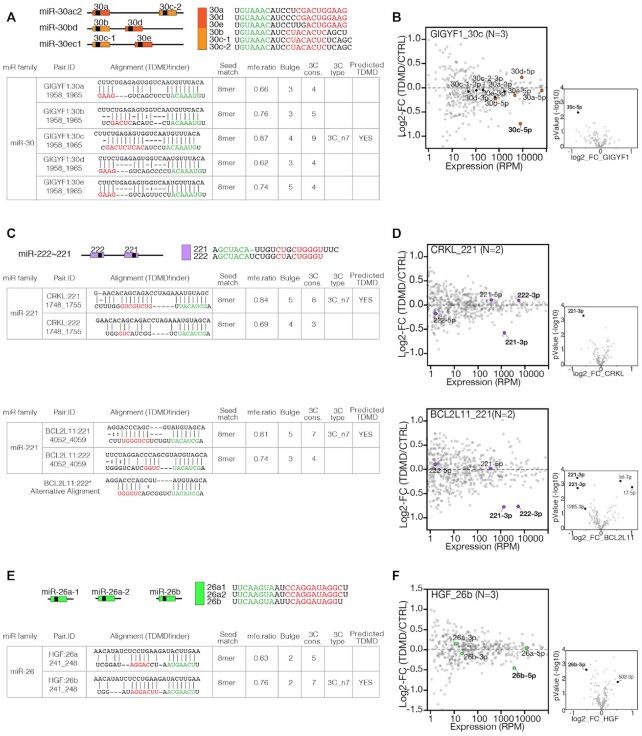
TDMD can discriminate between miRNAs from the same family. (A, C, E) miR-30, miR-222 and miR-26 family predictions that were subjected to experimental validation. Guide miRNAs sequences (seed and 3C pairing regions are highlighted) and genomic cluster organization are shown. The tables report TDMDfinder predictions of miRNA:target alignment and the scores for the different features used to select for TDMD (MFE ratio, Bulge extension, 3C and 3C type). (A) miR-30 family. (C) miR-221/222 family. (E) miR-26 family. (B, D, F) Small RNA sequencing results by TDMD assay. The scatter plot shows the fold changes (log2 of MDE versus CTRL, y axis) over the expression levels (reads per million, x-axis) of all the detected miRNAs. Above each panel, predicted interactions (target_miRNA) and number of replicates (N) are reported. The members of the targeted miR-family (guide and passenger miRNAs) and co-clustered miRNAs (if any) are highlighted and colored (color-code as in A-E). On the lower right of each panel, volcano plots highlight miRNAs regulated in a statistically significant manner (P-values by Student's t-test <0.05 and log2FC >|0.5|).
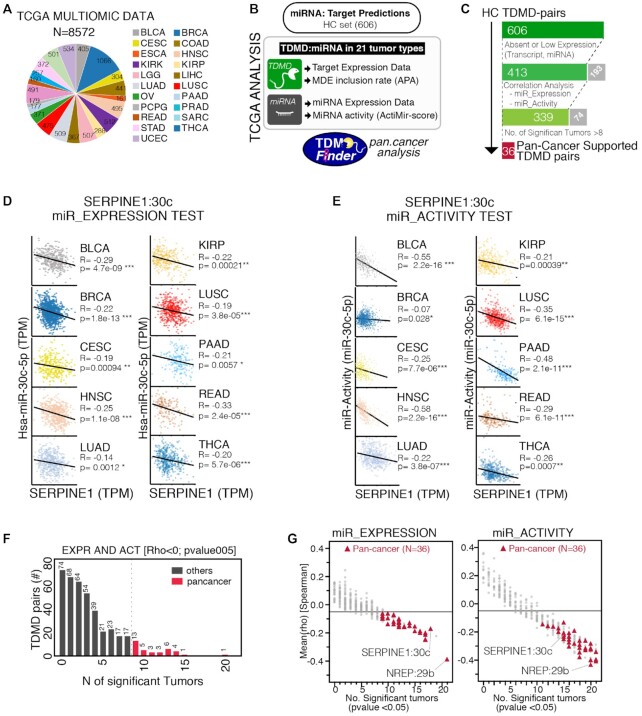
Figure 7.
Identification of ‘Pan-cancer’ TDMD pairs. (A) Number of samples from the TGCA database that were used in the correlation analysis, divided by tumor type. (B) Schematics of the strategy used to analyze TDMD pairs (HC set) in human cancer using data from A. (C) Steps followed to identify Pan-cancer supported TDMD pairs, with number of pairs that passed each filtering step. Excluded pairs are shown in grey. (D, E) Bivariate analysis, with linear fitting and Spearman correlation results (R and P-values), between the TDMD-transcript and the levels (D) or the activity (E) of its cognate miRNA. Shown are the tumor types with significant anti-correlation for both tests. TPM, transcripts per million. (F) Bar chart with the distribution of those pairs that, at least in one tumor, scored positively in both the miRNA Expression and the miRNA Activity test (Rho←0.05 and p<0.05). 36 pairs scored positively for both tests in at least nine different tumor types (Pan-cancer supported, in red). (G) Scatter plots showing, for each TDMD-pair, the Spearman correlation across all tumors (mean, y-axis) against the total number of tumor types with a significative anti-correlation (x-axis) by using the miRNA Expression (left) or Activity test (right). Pan-cancer supported TDMD pairs are in red.
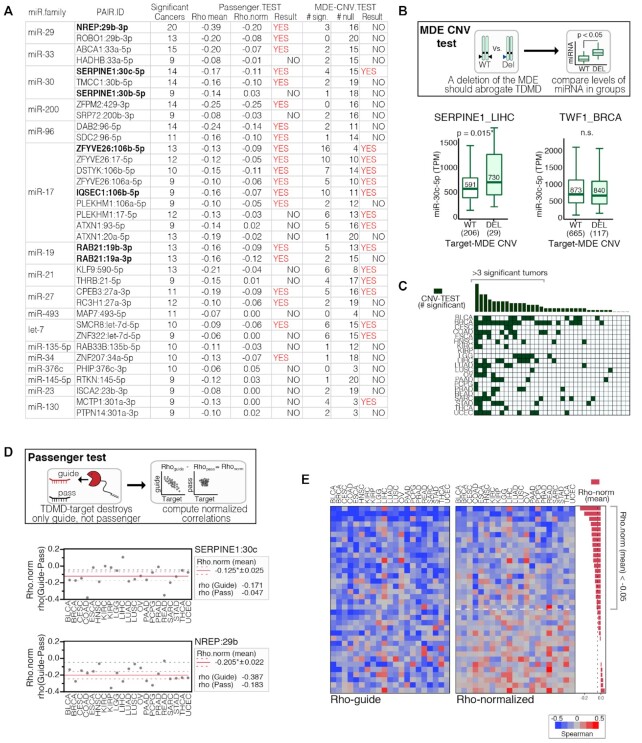
Figure 8.
Additional tests in support of Pan-cancer TDMD pairs. (A) Table reporting Pan-Cancer supported pairs and MDE CNV and Passenger test results. Experimentally validated interactions are reported in bold. (B) Top: schematics of the ‘MDE CNV test’, with miRNA expression levels computed in patients both with and without a deletion in the TDMD target. Bottom: MDE CNV test results for miR-30c and either its TDMD target SERPINE1 (left) or its canonical target TWF1 (right). Results with a ratio ≥1.1 (miRNA.in.DEL/ miRNA.in.WT) and a P-value <0.05 by Welch's t-test, were considered significant. (C) Heatmap showing the results of the CNV test in each tumor type for the 36 Pan-cancer-supported TDMD pairs. Highlighted are the pairs with a score ≥4. (D) Top: schematics of the ‘Passenger test’: a normalized Spearman correlation coefficient for each TDMD-pair was calculated as (Rho-norm = Rho-guide – Rho-pass); the normalization was performed at the level of each individual tumor type and only if the Rho of the passenger was <0, to avoid artifacts. Bottom: Passenger-test results for SERPINE1:miR-30c and NREP:miR-29b. (E) Heatmaps showing the results of the Passenger test (Rho-guide and Rho-norm) in each tumor type for the Pan-cancer supported TDMD pairs.
Features of the predicted TDMD pairs
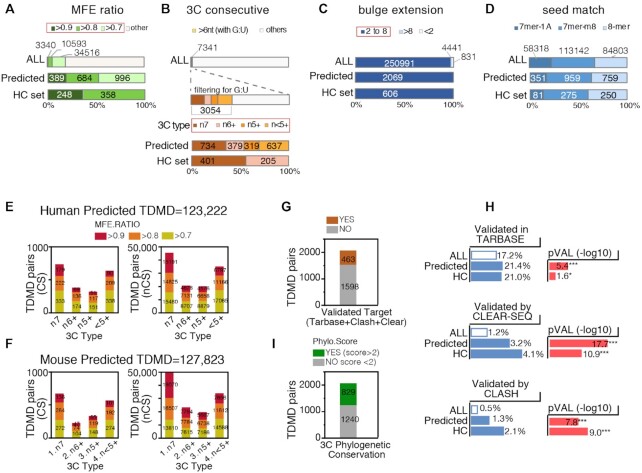
Overall, both in human and in mouse, the Predicted TDMD pairs represent 0.8% of all the interactions found, with a total of ∼2,000 conserved (CS) and over 100,000 non conserved (nCS) interactions. Of the criteria used to select for TDMD, the MFE ratio and, in particular, the 3C consecutive pairing type resulted to be the more stringent (Figure 2A–D). In the case of the conserved human targets, 19% of pairs showed a MFE ratio >0.7, while only 2.9% of pairs met the 3C consecutive match requirement (Figure 2A, B). Although TDMDs were selected regardless of what type of match their seed regions formed, predicted TDMDs were slightly enriched for 8mer and 7mer-m8 types and depleted for 7mer-1A (Figure 2D). In both the human and the mouse predicted set, the most represented (∼25% of the predicted pairs) 3C type had at least seven consecutive bases with no G:U (n7) (Figure 2E, F). We annotated experimentally validated direct interactions either reported in the Human TarBase (26) or obtained from intramolecular ligation-based approaches, namely CLEAR-SEQ and CLASH (27,28). Overall, >20% of the predicted TDMDs corresponded to validated interactions (Figure 2G) and the overlap with each of the three validated datasets was significantly enriched (Figure 2H).
Figure 2.
Features of genes and miRNAs involved in predicted endogenous TDMD. (A–D) Bar plots showing the distribution, across all human conserved TargetScan predictions (ALL), the Predicted and the HC TDMD pairs, of the TDMD pair features TDMDfinder predictions were based on: (A) MFE ratio class, (B) 3C type, (C) bulge size, (D) seed type. (E, F) Number of Predicted-TDMD pairs identified in the conserved (CS) and non-conserved (NCS) target groups, stratified according to their 3C-type and MFE ratio class. (G) Human Conserved Predicted TDMDs were stratified according to their target validation status (according to TarBase or assessed by CLEAR-SEQ or CLASH approaches); (H) The frequency of the validated targets (as in G) was calculated in the TargetScan database (ALL) and in the Predicted and High Confidence (HC) TDMD pairs sets. P-values by Chi-test. (I) Phylogenetic Conservation of Predicted TDMDs as measured by PhyloP scores (average conservation over the whole 3C pairing region).
Phylogenetic conservation of the 3C pairing regions
Phylogenetic conservation is a key feature of biologically functional miRNA target sites, and it is usually limited to the seed pairing region (1). We noticed that in previously identified endogenous TDMD [such as Serpine1 and Nrep, (13,14)], also the 3C pairing regions were significantly conserved at phylogenetic level (Supplementary Figure S1C). Hence, a phylogenetic conservation analysis was performed on all predicted TDMD alignments. As a whole, the 3C pairing regions in the MDEs of both Predicted and HC TDMDs were only slightly more phylogenetically conserved when compared to canonical MREs (Supplementary Figure S1D). When a score was individually assigned to predicted MDEs, a number of them (829/2069, 40%) displayed significant phylogenetic conservation (PhyloScore ≥2, which corresponds to P-value ≤0.01 in –log10 scale), possibly hinting at some sort of selective pressure (Figure 2I and Supplementary Table S1). Phylogenetically Conserved predicted TDMDs are also annotated in the TDMDfinder online tool.
Features of the miRNAs predicted to be involved in TDMD
Human Predicted TDMD pairs comprised a total of 1457 different TDMD-genes (i.e. 11.8% of the genes with at least one conserved MRE) and 324 different miRNAs (87.6% of the 370 miRNAs with at least one CS target). Thus, most miRNAs are possibly susceptible to degradation by TDMD, while a relatively small set of genes is potentially able to regulate miRNA stability. On average, each miRNA showed five CS Predicted MDEs and 36 NCS Predicted MDEs (Supplementary Figure S2A). A correlation analysis was performed to compare the number of canonical vs predicted TDMD pairs. For most miRNAs, a positive correlation was observed between the number of conserved MREs (canonical targets, predicted by TargetScan) and that of MDEs (TDMD targets, predicted by TDMDfinder) (R = 0.50, Supplementary Figure S2B). For some of the miRNA families, such as miR-15, miR-17, miR-302, miR-28, known to comprise multiple members and/or to have a complex genomic organization, the number of conserved predicted TDMD pairs was larger (Supplementary Figure S2B). As for the non-conserved TDMD interactions, they were found to be unusually over-represented in the miR-17 family (Supplementary Figure S2C, D).
Features of the genes predicted to be involved in TDMD
The genes predicted to establish TDMD interactions (TDMD-genes) showed some peculiar features. In comparison to canonical target genes, they had significantly longer 3′ UTRs and considerably more MREs, which were also at a higher density (Supplementary Figure S3A). Within the 3′UTRs, MREs and MDEs were found to be similarly positioned (Supplementary Figure S3B) and to have comparable rates of inclusion in alternative transcript isoforms (Supplementary Figure S3C). As for their biological functions, TDMD-genes were enriched for (i) ontologies related to development, with neuronal functions particularly over-represented and (ii) pathways and upstream regulators that are frequently altered in human cancer, such as estrogen, P53, WNT and TGF-beta signaling (Supplementary Figure S4A–C and Table S2). Tissue-specific expression analysis revealed that most TDMD-genes were expressed in multiple tissues, but a subset of them (99 genes) resulted highly specific for the brain (Supplementary Figure S4D). This result might suggest higher propensity for TDMD to occur in the brain, the tissue where this mechanism was originally described as being particularly active (9).
Strategy for the experimental validation of the predicted TDMD pairs
In order to experimentally verify the accuracy of the TDMDfinder predictions, some of the TDMD targets predicted by our pipeline were selected and used in an assay (henceforth named ‘TDMD assay)’ that verifies their ability to promote the degradation of the cognate miRNAs (9,13,14,16). Candidates were picked based on the following criteria: (i) the targeted miRNA family should be, according to a broad literature, biologically relevant; (ii) the expression level of the miRNA should be above detection limit in the cell line used for the assay (i.e. HeLa), iii) the seed matching region should be phylogenetically conserved. A subset of seven pairs was additionally included because an analysis presented below (see Figures 7 and 8) suggested that they could be cancer relevant. As a positive control, we used the Serpine1:30c interaction, previously identified in mouse fibroblasts (9,13,14,16). In total, 39 TDMD predicted interactions, corresponding to 25 different TDMD-genes and nine miRNA families were considered (Figure 3A and Supplementary Table S3).
Usually, the occurrence of TDMD can be directly verified by overexpressing the MDE of interest and using appropriate mutants designed to prove that the effect under observation is specific (9,13,14,16,29). Hence, a ‘TDMD assay’ was set up, in which a GFP reporter plasmid, containing in its 3′UTR the MDE of the predicted TDMD transcript of interest, was ectopically over-expressed in HeLa cells at high transfection efficiency (>90%) (Figure 3B, C). To check if the effect was specific, the following negative controls were used: (i) an empty GFP vector (CTRL_GFP) and (ii) a GFP vector encoding either a scramble sequence (SCR) or a mutant form of the MDE of interest, obtained by deleting the 3C pairing region while leaving the seed pairing sequence (SEED) intact. The expression level of the target miRNA was measured by either RT-qPCR o small RNA sequencing (sRNA-seq). As predicted, expression of the positive control, i.e. the MDE of SERPINE1, significantly reduced the levels of both its targets, i.e. miR-30c and 30b (Figure 3D–F). sRNA-seq was employed to further verify specific repression effects on predicted miRNA(s) and monitor the expression levels of guide and passenger strands (Figure 3F, G). It is known that changes in the expression levels of guide and passenger strands generated from the same miRNA precursor molecule are very highly correlated. By acting post-transcriptionally and specifically targeting only one of them, TDMD is predicted to uncouple the expression of these two miRNA strands, as confirmed both when using the positive control SERPINE1 and when employing two artificial ‘optimal TDMDs’ (Figure 3G and Supplementary Figure S5A, B). In conclusion, these data demonstrate that the ‘TDMD assay’ is an effective strategy for testing TDMDfinder predictions.
Validation of the predicted TDMD pairs
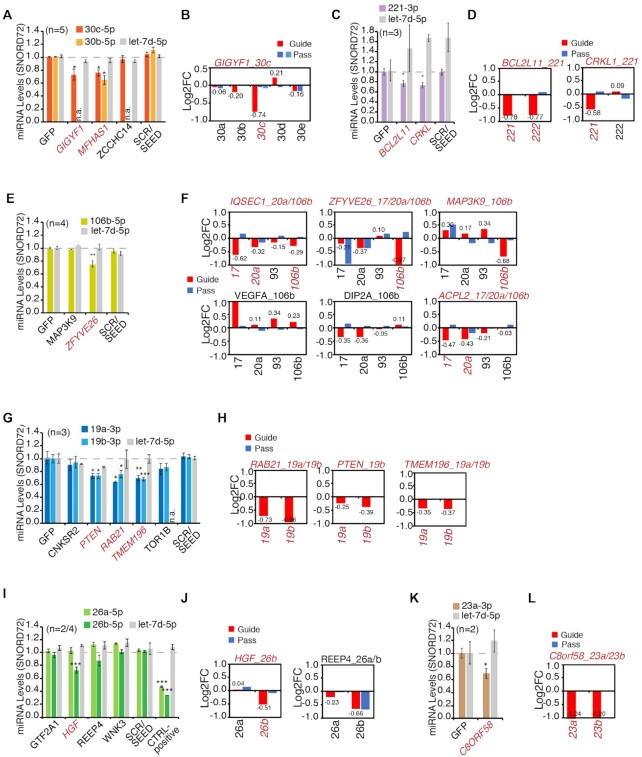
In our TDMD assay, using either RT-qPCR or sRNA-seq, we observed a significant reduction in the levels of the predicted guide miRNA in 20 of the 37 tested pairs (54.1%, Figure 4 and Supplementary Table S3). Six distinct miRNA families were shown to be responsive to TDMD: (i) miR-30, already known to be regulated by TDMD (GIGYF1:30c, Figure 4A, B) and five families, (ii) miR-221 (CRKL:221, BCL2L11:221, Figure 4C, D), (iii) miR-17 (IQSEC1:20a/106b, ZFYVE26:106b, MAP3K9:106b, ACPL2:17 Figure 4E, F), (iv) miR-19 (RAB21:19a/b, PTEN:19b, TMEM196:19a/b, Figure 4G, H), (v) miR-26 (HGF:26b, Figure 4I, J) and (vi) miR-23 (C8ORF58:23a/b, Figure 4K, L), whose susceptibility to endogenous TDMD was observed here for the first time. Three other families (miR-196 miR-148, miR-301) were tested, but their predicted MDEs did not induce significant effects (Supplementary Figure S5C, D). As already discussed in the case of SERPINE1:miR-30, we accepted as experimentally validated only those MDEs that selectively affected the guide but not the passenger miRNA. In most cases (17/20), sRNA-seq analysis clearly showed negligible differences in the levels of the passenger miRNAs, while for three pairs (REEP4:26b, ACPL2:20a and ZFYVE26:20a) repression of the passenger strand was significant (Figure 4B, D, F, J, L). To further check that the decrease in miRNA levels was due to a post-transcriptional mechanism, we measured the levels of primary transcripts (pri-miRs) for two of the targeted miRNA families (miR-30s and miR-19s – Supplementary Figure S5E, F). In both instances, the pri-miRs expression levels could not explain the observed change in miRNA levels. We can, therefore, conclude that in total 17 of the 37 tested pairs (among which 4 of the 7 pairs taken from those potentially cancer relevant) and 13 of the 24 tested MDEs are capable of TDMD (Supplementary Table S3).
Figure 4.
Experimental validation of predicted TDMD pairs. (A–L) Results of TDMD assays grouped by miRNA family. The plots (A, C, E, G, I, K) show mature miRNAs levels measured by RT-qPCR and normalized over CTRL_GFP, as in Figure 3E. P-values were calculated by Dunnet's t-Test, using the SCR/SEED group as reference. The plots (B, D, F, H, J, L) show mature miRNAs levels for the entire miRNA family (with guide and passenger miRs) measured by small RNA sequencing and normalized over CTRL_GFP (N, number of replicates). Above each bar graph, predicted pairs are reported (gene_miRNA/s, in red if the prediction was confirmed).
Identification of High-Confidence TDMD pairs
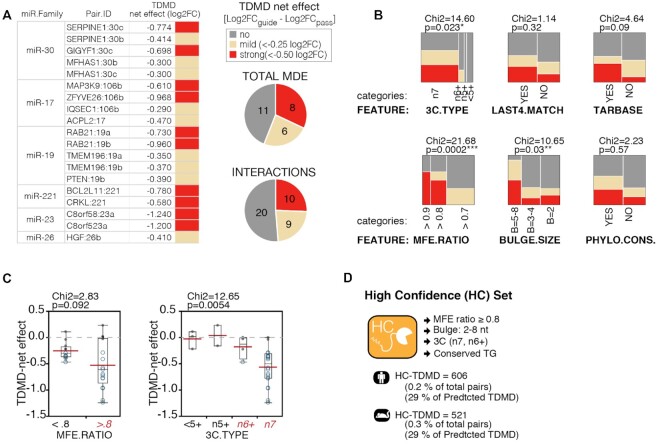
In order to isolate the MDEs responsible for the strongest TDMD effects, we used what we called the ‘TDMD net effect’ to correct for any transcriptional effect that the assay might have introduced. For each miRNA, the ‘TDMD net effect’ was calculated as the ratio between the reduction in the expression of the guide strand over the passenger strand. Eight MDEs (corresponding to 10 different interactions) displayed a strong TDMD net effect (–0.5 log2 normalized decrease; Figure 5A): GIGYF1 and SERPINE1 (miR-30c), MAP3K9 and ZFYVE26 (miR-106b), RAB21 (miR-19a/b), BCL2L11 and CRKL (miR-221), and C8orf58 (miR-23a/b). We used contingency analysis to correlate TDMD net effects with predicted TDMD pairs features and observed that stronger effects were significantly associated with MDEs characterized by extensive 3C pairings (n7), high binding energy (MFE ratio ≥0.8) and bulges >4 nt in size (Figure 5B). No significant association was found with other features, such as the level of phylogenetic conservation, the number of matches at the miRNA 3′ end (last four nts), or with whether or not the miRNA:target interaction of interest had been experimentally verified (according to Tarbase) (Figure 5B). However, a positive trend was found in all of the above cases. Of note, all the strongest interactions belonged to either the n7 or the n6+ 3C type and had a MFE ratio of at least 0.8 (Figure 5C), thus prompting us to group them together into a ‘High Confidence TDMD’ subset (HC TDMD), including miRNA:target pairs with the highest probability of being actual TDMD interactions. The HC set includes ∼0.2% of all interactions, with 606 and 521 CS pairs in human and mouse, respectively. About 29% of the Human Predicted TDMD set are found also in the HC set. HC TDMD pairs have been annotated in the TDMDfinder tool.
Figure 5.
Features associated to experimentally validated TDMD. (A) The table and pie charts summarize validation results by sRNA-seq. reported is the ‘TDMD net effect’, normalizing the log2 fold change (MDE versus CTRL) of the guide strand over the corresponding effect on the passenger strand. (B) Contingency analysis, correlating the different ‘TDMD net effect’ classes with the main features related to TDMD predictions. P-values by Chi-test. (C) Box plots of the TDMD net effects, with TDMD pairs grouped according to their MFE ratio (left) or 3C-type (right). P-values and Chi-square by Wilcoxon test. (D) List of the criteria used in classifying human and mouse TDMD pairs as High-Confidence.
TDMDfinder predicts selective TDMD effects for miRNA families
We used the sRNA-seq data to evaluate if TDMDfinder could selectively predict the effect of different MDEs on miRNA families (Figure 6 and Supplementary Figure S6). miRNAs having the same seed sequence are organized into families of miRNAs and associated with similar sets of canonical targets. However, as their 3′ ends differ from each other, miRNAs from the same family are theoretically susceptible to post-transcriptional regulation by different sets of MDEs. For instance, TDMDfinder predicts that the different members of the miR-30 family have various degrees of susceptibility to TDMD, due to the different pairing conformations they adopt when binding their targets (Figure 3D and Figure 6A). sRNA-seq data from the TDMD assay confirmed that validated miR-30 TDMD targets (SERPINE1, GIGYF1) significantly changed the expression levels of only a few miRNAs and displayed a clear specificity. In particular, SERPINE1 repressed both 30b and 30c (Figure 3F) while GIGYF1 had effect only on 30c (Figure 6B). In the case of miRNAs of the miR-221 and miR-26 families, TDMDfinder also predicted different susceptibility to TDMD as the result of a very different type of 3C pairing and a different energetic stability (Figure 6C, E). Accordingly, CRKL1 discriminated between miR-221 and miR-222 (Figure 6D) and HGF distinguished miR-26b from miR-26a (Figure 6F). Conversely, BCL2L11, which had been predicted to target only miR-221 (Figure 6C), could significantly repress miR-222 too (Figure 6D). Of note, a different alignment with a 3C pairing made up of six consecutive nucleotides at the miRNA 3′ end, is also possible and might explain this effect (Figure 6C). The miRNAs belonging to each of the remaining families (miR-17, miR-19 and miR-23) have similar 3′ end sequences (Supplementary Figure S6A, D, G) so that TDMDfinder predictions were less selective, with the same MDE similarly targeting different family members (Supplementary Figure S6B, E, H). Accordingly, results from sRNA-seq were more widely distributed, with multiple members of the predicted miRNA families being repressed (Supplementary Figure S6C, F, I). For some MDEs (i.e. ZFYVE26, RAB21, TMEM196 and PTEN) unrelated miRNAs were also regulated, possibly as the results of secondary effects in these TDMD assays (30). Interestingly, miR-106b, whose 3C region is the most divergent within the miR-17 family, was found, as predicted by TDMDfinder, to be specifically repressed by MAP3K9 (Supplementary Figure S6B, C). In conclusion, TDMDfinder predictions could spot if TDMD affects different members of the same miRNA family selectively. This might be particularly the case with miRNAs whose 3C regions are sufficiently different.
Analysis of TDMD pairs in cancer datasets
The TDMD mechanism also depends on quantitative parameters, primarily the expression levels of both the miRNA and the target (13,15,17), which cannot be recapitulated by a predictive tool based on sequence alignment and structural features calculations. Vast cancer datasets are available, containing miRNA and gene expression data from thousands of samples and different tissues. We used the multi-omic molecular landscape of human tumors provided by The Cancer Genome Atlas (TCGA) to retrieve quantitative information, perform in silico analyses and infer which TDMD interactions might be relevant in human cancer. This investigation was also prompted by the fact that predicted TDMD genes were found enriched for cancer genes (see Supplementary Figure S4B). We retrieved matched miRNA and mRNA expression data in 21 cancer datasets for a total of 8572 tumor samples, along with their corresponding genomic data (i.e. copy number variations) (Figure 7A and Supplementary Table S4). We focused on the most reliable set of predictions, the ‘HC set’, which contained 606 miRNA:target pairs. For each of these pairs we retrieved information on miRNA expression, target expression and MDE inclusion by alternative polyadenylation analysis (APA) (Figure 7B, C). APA, which has frequently been observed in cancer (31), could change 3′UTR sequences by either including or excluding an MDE and therefore affecting the possibility of TDMD to occur (Supplementary Table S4). To investigate which TDMD interactions might be relevant in human cancer, we performed a series of correlation analysis. We hypothesized that if TDMD occurs, then there should be a negative correlation in cancer samples between the TDMD-transcript and its cognate miRNA (‘miR_Expression Test’). For example, in the case of the SERPINE1:miR-30c pair, a significant Spearman negative correlation was observed in multiple (N = 18) cancer types (Figure 7D and Supplementary Table S4). The reduction of miRNA levels by TDMD should also produce effects on miRNA activity (i.e. the ability of a given miRNA to repress its canonical targets), as previously observed for endogenous TDMD (7). We, therefore, exploited the ‘ActMiR score’ (22), a computational method for inferring the activity of miRNAs based on the changes in the expression levels of their predicted target genes in cancer datasets, and measured the correlation between TDMD-transcript and miRNA activity (‘miR_Activity Test’). Indeed, in many cancers (N = 15), we observed a significant negative Spearman correlation between SERPINE1 expression and miR-30 activity (Figure 7E and Supplementary Table S4), strongly suggesting that this TDMD interaction does occur in human cancer. In the pan-cancer module of the TDMDfinder webtool, MDE inclusion, ‘miR_Expression Test’ and ‘miR_Activity Test’ are provided for all HC TDMD pairs.
‘Pan-cancer’ TDMD pairs
After filtering out those pairs whose either mRNA or miRNA was poorly or not-at-all expressed (Figure 7C), the remaining 413 TDMD pairs were analyzed with the aim of identifying a set of TDMD interactions potentially relevant in human cancer. We considered as relevant those TDMD pairs that, in the same cancer type, showed significant anti-correlation in both the miR_Expression and miR_Activity tests (Supplementary Figure S7A, B). For most TDMD pairs (339/413, 82.1%), we observed co-occurrence of significance in at least one type of cancer (Supplementary Figure S7B), suggesting that most of the high-confidence TDMDs may potentially occur in human cancer. To avoid any possible bias from individual tumor datasets (e.g. tumor purity, molecular subtypes, genetic events, etc.), we focused on TDMD pairs that had given significant and consistent scores in 9 or more cancer types (Supplementary Figure S7B and Figure 7F, G). These TDMD interactions, named as ‘Pan-cancer supported’, comprised 36 pairs, 28 transcripts and 30 MDEs. They included previously known endogenous TDMDs, such as SERPINE1 and NREP (Figure 8A), and others that were validated in this study by the TDMD assay (ZFYVE26, RAB21, PTEN, IQSEC1, BLC2L11). The miRNAs predicted to be targeted included well-known oncogenic (miR-19, miR-17) and tumor-suppressor families (let-7, miR-30, miR-34, Figure 8A) (32).
Additional tests in support of Pan-cancer TDMD pairs
Anti-correlation between the target and the miRNA is in itself necessary but not sufficient to prove that TDMD is taking place. Therefore, to provide further evidence of TDMD occurring, we devised additional tests based on: (i) the effect that variations in the TDMD gene copy number (CNV) have on miRNA expression (‘MDE CNV test’) and (ii) the correlation between either the guide or the passenger miRNA and the TDMD-transcript (‘Passenger Test’). The ‘MDE CNV test’ is based on the assumption that, in the miRNA:target interaction, the TDMD target plays an epistatic role (the target's genetic status influences miRNA expression) rather than the hypostatic role typical of canonical targets (the miRNA’s genetic status influences the target expression) (Figure 8B). We used GISTIC (33) to find possible deletions of TDMD genes across all TCGA tumors and checked whether these were associated with higher expression of the targeted miRNA. Several TDMD pairs scored positively in multiple tumors (Figure 8A, C). The ‘Passenger Test’ is based on the hypothesis that the TDMD mechanism uncouples the expression of guide and passenger miRNAs, as discussed previously (see Figure 3). Hence, the correlation between miRNA and target in tumors was normalized for the passenger correlation, to get rid of the transcriptional effects that guide and passenger have in common (Figure 8D). As expected, and proving the validity of our approach, known TDMD pairs, such as SERPINE1:30c and NREP: 29b, were modestly affected by normalization (Figure 8D). Overall, 20 out of the 36 TDMD pairs of Pan-cancer supported group retained a negative anti-correlation in multiple tumor types after the normalization by the ‘Passenger Test’ (Figure 8A, E). For the other TDMD pairs, both guide and passenger strands were found to be similarly correlated with the TDMD target, suggesting that the correlation measured in cancer datasets mostly occurred at the transcriptional level. The results of additional tests are also included in TDMDfinder (pan.cancer module) and can help selecting TDMD pairs potentially relevant in human cancer and therefore worthy of further investigation by functional studies.
DISCUSSION
An inventory of bona fide human and mouse TDMD pairs
Here, we provide the first inventory of miRNA:target interactions that are prone to TDMD. TDMDfinder is a computational pipeline that, using both quantitative and qualitative parameters, enables its users to find miRNA:target interactions with the potential of producing TDMD effects. Applied to the human and mouse genomes, our pipeline has identified thousands of pairs with structural features that are compatible with TDMD. According to our predictions, ∼1% of all miRNA:target interactions could induce miRNA degradation. Therefore, as virtually every miRNA could be regulated by multiple independent transcripts via TDMD, this mechanism might have a pervasive role in miRNA biology.
We performed extensive experimental validations in HeLa cells in order to support the soundness of our predictions. When validating a set of candidate TDMD pairs related to nine different miRNA families, we measured an overall accuracy of 45.9% (17 validated out of 37), in the same range with the computational tools usually employed to predict canonical miRNA targets (34). As previously said, some of the tested pairs were selected from the Pan-cancer set, with four out of seven validated by the TDMD assay. While this result confirms that cancer-relevant TDMD interactions might exist, the peculiar nature of these pairs might have inflated the validation rate. Of note, even removing the Pan-cancer supported pairs from the calculation, TDMD predictions were still found correct in >40% of cases (13 validated out of 30, 43.3%). Two parameters, namely the thermodynamic stability (MFE ratio) and the number of consecutive pairings in the miRNA 3′ region (3C type), were found to be significantly associated with validated pairs and were used to select a set of high-confidence TDMD pairs. All the predictions (‘Predicted’ or ‘HC’ TDMD) can be easily retrieved from TDMDfinder, a user-friendly webtool that provides all the relevant structural information on any TDMD-like interaction involving a given miRNA and its targets. It should be pointed out that the criteria we imposed to decide if a miRNA:target interaction actually forms a TDMD pair are not rigid and could be adapted in case of MDE:miRNA pairings with peculiar features. Therefore, we encourage users to customize TDMDfinder selection criteria as needed. Although TDMDfinder predictions had a high validation rate in our assays, we do emphasize that it will be important to experimentally validate individual predictions of interest. For instance, TDMD predictions could be validated through the ‘TDMD assay’ that we used here, which in principle is similar to the reporter assay used for canonical miRNA targets and based on the ectopic expression of the MRE of interest. In the TDMD assay, the levels of miRNAs are measured following the overexpression of the selected MDE, in a procedure widely used for analyzing TDMD effects (9,13,16,29). The specificity for TDMD was assessed by using MDE mutants whose seed regions had been left intact but whose 3C pairing sequences had been deleted (basically, MDEs converted into MREs). As TDMD acts post-transcriptionally and affects selectively just one of the two miRNAs, a further verification test was performed by measuring the levels of guide and passenger miRNAs.
TDMD interactions in miRNA biology
TDMD is a post-transcriptional mechanism capable of introducing differences in the expression of miRNAs that are transcriptionally linked. We used small RNA sequencing to experimentally verify that MDEs can discriminate between different members of the same miRNA family provided that sufficiently different 3C pairings are present. Hence, TDMD can finely tune the expression of miRNAs that are organized in clusters or families. In addition, TDMD is a mechanism that, by impacting on shared targets, ultimately influences miRNA activity (13–15). As a matter of fact, it can be considered to be a non-coding regulatory mechanism that involves specific RNA molecules, regardless of their translational potential, and that regulates gene expression in trans (35).
It must be said that a transcript's intrinsic ability to trigger miRNA degradation, which can be predicted to some extent by TDMDfinder, does not necessarily imply that a significant degradation of the miRNA also occurs in a physiological or pathological context, even in the case of interactions validated by the TDMD assay, which has also the limitation of expressing MDE sites outside of their endogenous context (i.e. full 3′UTR). For example, as part of its native transcript, an MDE might not be accessible to miRNA binding, because of RNA binding proteins (RBPs) masking the site or of a peculiar 3′UTR structure. This is also true for canonical targets. The identification and experimental validation of a miRNA responsive element (MRE) is not sufficient to demonstrate that regulation by miRNA occurs in vivo and at endogenous level, as other factors, such as availability and expression levels of both miRNA and of the targets, also play a part. So far, the quantitative requirements for TDMD have been investigated only to a limited extent (13,15) and cannot be easily accounted for by a predictive tool. Further and specific studies are needed to demonstrate in which specific contexts each of the predicted MDEs works. In this regard, it is important to underline that the recent discovery of the role played by the ZSWIM8 Cullin-RING E3 ubiquitin ligase in the TDMD mechanism, has also revealed that TDMD is more widely implicated than previously thought, as it acts on many miRNAs and in multiple biological contexts (17,18). Looking ahead, in order to further study the TDMD mechanism and its role in miRNA biology, we envision an effective strategy based on the combined use of predictive and experimental tools. This will entail inhibiting ZSWIM8 function to highlight the involvement of TDMD in a given biological context, while using TDMDfinder predictions to identify potentially involved TDMD interactions.
TDMD interactions in human cancer
Human cancer is an intriguing biological context for evaluating the implications of the TDMD interactions predicted by our tool. From a number of TCGA datasets we extracted quantitative information on both miRNAs and targets across thousands of samples and investigated the relevance of TDMD interactions at endogenous levels. Based on a series of correlation analyses that inferred how TDMD affects both miRNA levels and miRNA activity, we speculated that TDMD might be occurring in human cancer. Indeed, the dysregulation of miRNA levels often observed in human cancer (32) cannot be always explained by alterations in miRNA biogenesis. Modulation of cancer genes and pathways or chromosomal rearrangements (including amplifications or deletions) may cause sudden changes in the expression of TDMD transcripts, contributing to miRNA alterations in cancer. Of relevance, we isolated a number of solidly supported interactions (‘Pan-cancer supported’ TDMD) that passed additional tests (e.g. ‘Passenger Test’) and thus reinforced the idea that TDMD could indeed take place in such cases. These ‘Pan-cancer supported’ TDMD pairs include endogenous transcripts (NREP, SERPINE1) that have been previously shown to cause TDMD, as well as many of those that have been experimentally validated by the TDMD-assay in the present study, here considered at their endogenous levels of expression. This might be just the tip of the iceberg. In-depth characterization studies on various TDMD pairs, tumor types and cancer phenotypes are required to fully appreciate the importance of TDMD in cancer initiation and/or evolution. miRNAs are key molecules in cancer biology and have tremendous potential as therapeutic tools. We foresee that further elucidation of the TDMD mechanism in cancer will have an impact on both basic and translational research.
DATA AVAILABILITY
Raw and processed data together with detailed description of the procedures are available in the Gene Expression Omnibus database, under accession number GSE168566. The TDMDfinder web tool is available at http://213.82.215.117:9999/TDMDfinder/index.php.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Claudia Crovace for manuscript editing. The results published here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Author contributions: the TDMDfinder pipeline was developed by I.S. with the supervision of M.J.M and F.N. Bioinformatics analyses were performed by I.S., T.M.R.N., M.J.M. and F.N. Validation experiments (presented in Figure 3, 5 and 6 and Supplementary Figures S5 and S6) were performed by C.R. and D.F. The TDMDfinder webtool was developed in collaboration with L.C. M.J.M. and F.N. supervised the project and wrote the manuscript.
Contributor Information
Ines Simeone, Center for Genomic Science of IIT@SEMM, Istituto Italiano di Tecnologia (IIT)—Via Adamello 16, 20139 Milan, Italy.
Carmela Rubolino, Center for Genomic Science of IIT@SEMM, Istituto Italiano di Tecnologia (IIT)—Via Adamello 16, 20139 Milan, Italy.
Teresa Maria Rosaria Noviello, Department of Electrical Engineering and Information Technology, University of Naples “Federico II”, Via Claudio 21, Naples 80128, Italy; BioGeM, Institute of Genetic Research “Gaetano Salvatore”, Via Camporeale, Ariano Irpino (AV) 83031, Italy.
Diego Farinello, Center for Genomic Science of IIT@SEMM, Istituto Italiano di Tecnologia (IIT)—Via Adamello 16, 20139 Milan, Italy.
Luigi Cerulo, Dep. of Science and Technology, University of Sannio, via de Sanctis, 11, Benevento 82100, Italy; BioGeM, Institute of Genetic Research “Gaetano Salvatore”, Via Camporeale, Ariano Irpino (AV) 83031, Italy.
Matteo Jacopo Marzi, Center for Genomic Science of IIT@SEMM, Istituto Italiano di Tecnologia (IIT)—Via Adamello 16, 20139 Milan, Italy.
Francesco Nicassio, Center for Genomic Science of IIT@SEMM, Istituto Italiano di Tecnologia (IIT)—Via Adamello 16, 20139 Milan, Italy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Associazione Italiana per la Ricerca sul Cancro (AIRC) [IG18774, IG22851 to F.N.]; Fondazione Cariplo [2015-0590 to F.N., 2016-0615 to M.J.M.]; C.R. was supported by a FIRC-AIRC fellowship for Italy [22438]. Funding for open access charge: Core funding from IIT.
Conflict of interest Statement. None declared.
REFERENCES
- 1. Bartel D.P. Metazoan microRNAs. Cell. 2018; 173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mukherji S., Ebert M.S., Zheng G.X.Y., Tsang J.S., Sharp P.A., van Oudenaarden A.. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 2011; 43:854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mullokandov G., Baccarini A., Ruzo A., Jayaprakash A.D., Tung N., Israelow B., Evans M.J., Sachidanandam R., Brown B.D.. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods. 2012; 9:840–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Treiber T., Treiber N., Meister G.. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019; 20:5–20. [DOI] [PubMed] [Google Scholar]
- 5. Ruegger S., Grosshans H.. MicroRNA turnover: when, how, and why. Trends Biochem. Sci. 2012; 37:436–446. [DOI] [PubMed] [Google Scholar]
- 6. Duffy E.E., Rutenberg-Schoenberg M., Stark C.D., Kitchen R.R., Gerstein M.B., Simon M.D.. Tracking distinct RNA populations using efficient and reversible covalent chemistry. Mol. Cell. 2015; 59:858–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marzi M.J., Ghini F., Cerruti B., de Pretis S., Bonetti P., Giacomelli C., Gorski M.G., Kress T., Pelizzola M., Muller H.et al.. Degradation dynamics of microRNAs revealed by a novel pulse-chase approach. Genome Res. 2016; 26:554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kingston E.R., Bartel D.P.. Global analyses of the dynamics of mammalian microRNA metabolism. Genome Res. 2019; 29:1777–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de la Mata M., Gaidatzis D., Vitanescu M., Stadler M.B., Wentzel C., Scheiffele P., Filipowicz W., Grosshans H.. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep. 2015; 16:500–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ameres S.L., Horwich M.D., Hung J.H., Xu J., Ghildiyal M., Weng Z., Zamore P.D.. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010; 328:1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cazalla D., Yario T., Steitz J.A.. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010; 328:1563–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marcinowski L., Tanguy M., Krmpotic A., Radle B., Lisnic V.J., Tuddenham L., Chane-Woon-Ming B., Ruzsics Z., Erhard F., Benkartek C.et al.. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 2012; 8:e1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ghini F., Rubolino C., Climent M., Simeone I., Marzi M.J., Nicassio F.. Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. Nat. Commun. 2018; 9:3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bitetti A., Mallory A.C., Golini E., Carrieri C., Carreno Gutierrez H., Perlas E., Perez-Rico Y.A., Tocchini-Valentini G.P., Enright A.J., Norton W.H.J.et al.. MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat. Struct. Mol. Biol. 2018; 25:244–251. [DOI] [PubMed] [Google Scholar]
- 15. Kleaveland B., Shi C.Y., Stefano J., Bartel D.P.. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018; 174:350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sheu-Gruttadauria J., Pawlica P., Klum S.M., Wang S., Yario T.A., Schirle Oakdale N.T., Steitz J.A., MacRae I.J.. Structural basis for target-directed MicroRNA degradation. Mol. Cell. 2019; 75:1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi C.Y., Kingston E.R., Kleaveland B., Lin D.H., Stubna M.W., Bartel D.P.. The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science (New York, NY). 2020; 370:eabc9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han J., LaVigne C.A., Jones B.T., Zhang H., Gillett F., Mendell J.T.. A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science (New York, NY). 2020; 370:eabc9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kruger J., Rehmsmeier M.. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006; 34:W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I.et al.. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016; 44:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bonnal R.J., Rossi R.L., Carpi D., Ranzani V., Abrignani S., Pagani M.. miRiadne: a web tool for consistent integration of miRNA nomenclature. Nucleic Acids Res. 2015; 43:W487–W492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee E., Ito K., Zhao Y., Schadt E.E., Irie H.Y., Zhu J.. Inferred miRNA activity identifies miRNA-mediated regulatory networks underlying multiple cancers. Bioinformatics. 2016; 32:96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feng X., Li L., Wagner E.J., Li W.. TC3A: the cancer 3′ UTR atlas. Nucleic Acids Res. 2018; 46:D1027–D1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Agarwal V., Bell G.W., Nam J.W., Bartel D.P.. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015; 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Muller H., Marzi M.J., Nicassio F.. IsomiRage: from functional classification to differential expression of miRNA isoforms. Front. Bioeng. Biotechnol. 2014; 2:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karagkouni D., Paraskevopoulou M.D., Chatzopoulos S., Vlachos I.S., Tastsoglou S., Kanellos I., Papadimitriou D., Kavakiotis I., Maniou S., Skoufos G.et al.. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018; 46:D239–D245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Helwak A., Kudla G., Dudnakova T., Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013; 153:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moore M.J., Scheel T.K., Luna J.M., Park C.Y., Fak J.J., Nishiuchi E., Rice C.M., Darnell R.B.. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015; 6:8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pawlica P., Moss W.N., Steitz J.A.. Host miRNA degradation by Herpesvirus saimiri small nuclear RNA requires an unstructured interacting region. RNA (New York, N Y). 2016; 22:1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mogilyansky E., Rigoutsos I.. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013; 20:1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gruber A.J., Zavolan M.. Alternative cleavage and polyadenylation in health and disease. Nat. Revi. Genet. 2019; 20:599–614. [DOI] [PubMed] [Google Scholar]
- 32. Lujambio A., Lowe S.W.. The microcosmos of cancer. Nature. 2012; 482:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G.. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011; 12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P.. The impact of microRNAs on protein output. Nature. 2008; 455:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kopp F., Mendell J.T.. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018; 172:393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data together with detailed description of the procedures are available in the Gene Expression Omnibus database, under accession number GSE168566. The TDMDfinder web tool is available at http://213.82.215.117:9999/TDMDfinder/index.php.