Abstract
Nutrients are emerging regulators of adaptive immunity1. Selective nutrients interplay with immunological signals to activate mTORC1, a key driver of cell metabolism2-4, but how these environmental signals are integrated for immune regulation remains unclear. Here, we use genome-wide CRISPR screening combined with protein–protein interaction (PPI) networks to identify regulatory modules that mediate immune receptor- and nutrient-dependent signaling to mTORC1 in regulatory T (Treg) cells. Sec31a is identified to promote mTORC1 activation, by interacting with the GATOR2 component Sec13 to protect it from Skp1-dependent proteasomal degradation. Accordingly, loss of Sec31a impairs T cell priming and Treg suppressive function. In addition, the SWI/SNF complex restricts expression of the amino acid sensor Castor1, thereby enhancing mTORC1 activation. Moreover, we reveal the Ccdc101-associated SAGA complex as a potent inhibitor of mTORC1, which limits expression of glucose and amino acid transporters and maintains T cell quiescence in vivo. Specific deletion of Ccdc101 in Treg cells results in uncontrolled inflammation but improved antitumour immunity. Collectively, our results establish epigenetic and posttranslational mechanisms that underpin how nutrient transporters, sensors and transducers interplay with immune signals for three-tiered regulation of mTORC1 activity, and their pivotal roles in licensing T cell immunity and immune tolerance.
T cells are pivotal for adaptive immunity against pathogens and tumours. Activation of T cells requires conventional immune signals, namely T cell receptors (TCR), co-stimulation and cytokines. Moreover, nutrients are emerging as crucial regulators of T cell immunity, in part, by licensing mTORC1 activation1,4,5, but the regulation and signaling basis of nutrient transporters and sensors remain largely elusive. We also lack integrative, systems-level information of the regulatory circuits that transmit physiological signals to mTORC1. Here we address these questions by integrating CRISPR-based genetic screening, PPI network analysis, and in vivo functional dissection.
Integration of CRISPR screening with PPI network analysis
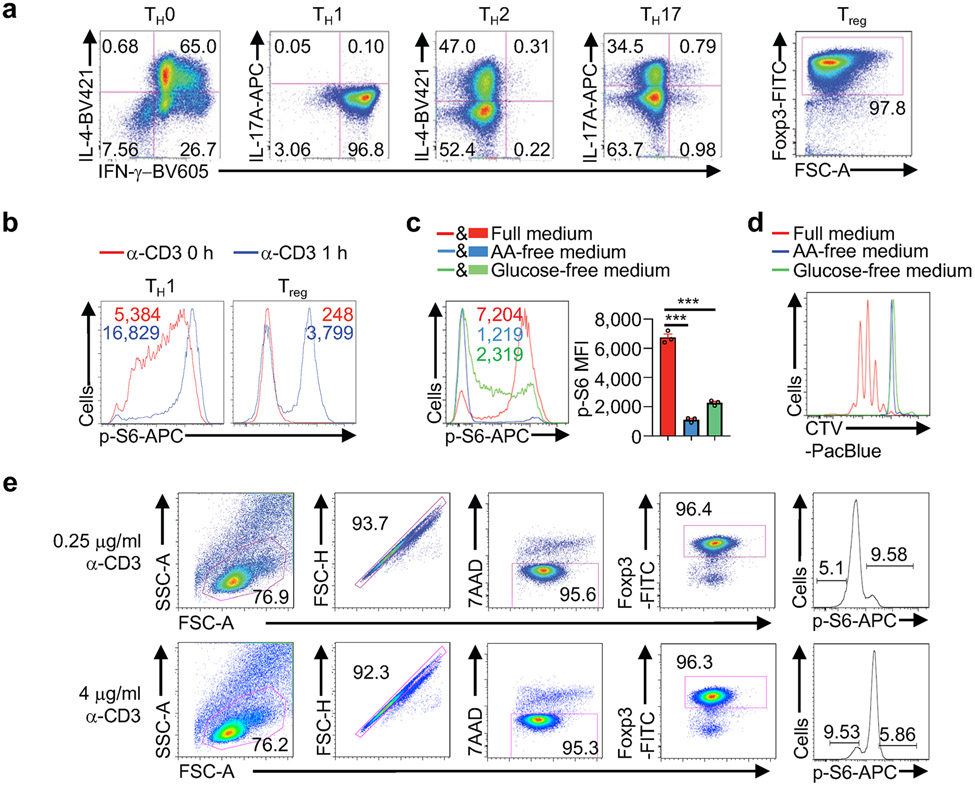
To establish a system for pooled screening of physiological regulators of mTORC1 activity, we measured mTORC1-dependent phosphorylation of S6 (p-S6)2-4 in effector and induced Treg cells following TCR stimulation under nutrient-replete conditions. Cells differentiated under TH1 and Treg conditions were selected for analysis, as they contained a more homogenous population than other effector cells (Extended Data Fig. 1a). Further, Treg cells showed TCR-mediated strong and bimodal induction of p-S6 (Extended Data Fig. 1b). Nutrients license TCR-dependent mTORC1 signaling1,5, and upon amino acid or glucose deprivation, TCR-induced mTORC1 activation and proliferation of Treg cells were severely impaired (Extended Data Fig. 1c, d). Therefore, Treg cells represent a faithful system for TCR- and nutrient-dependent mTORC1 activation.
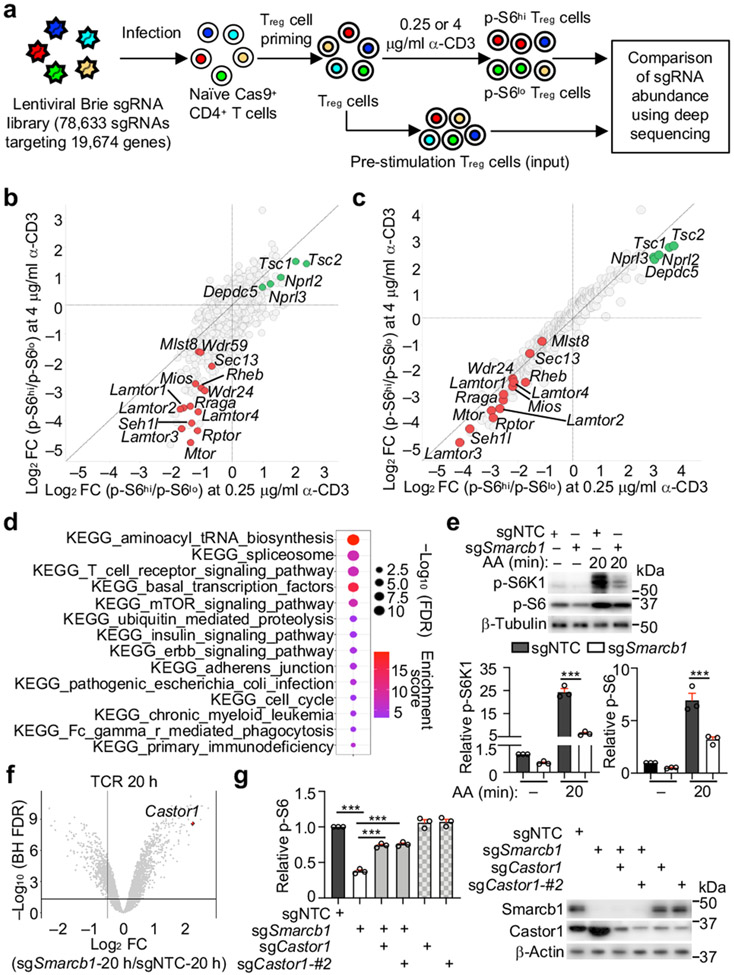
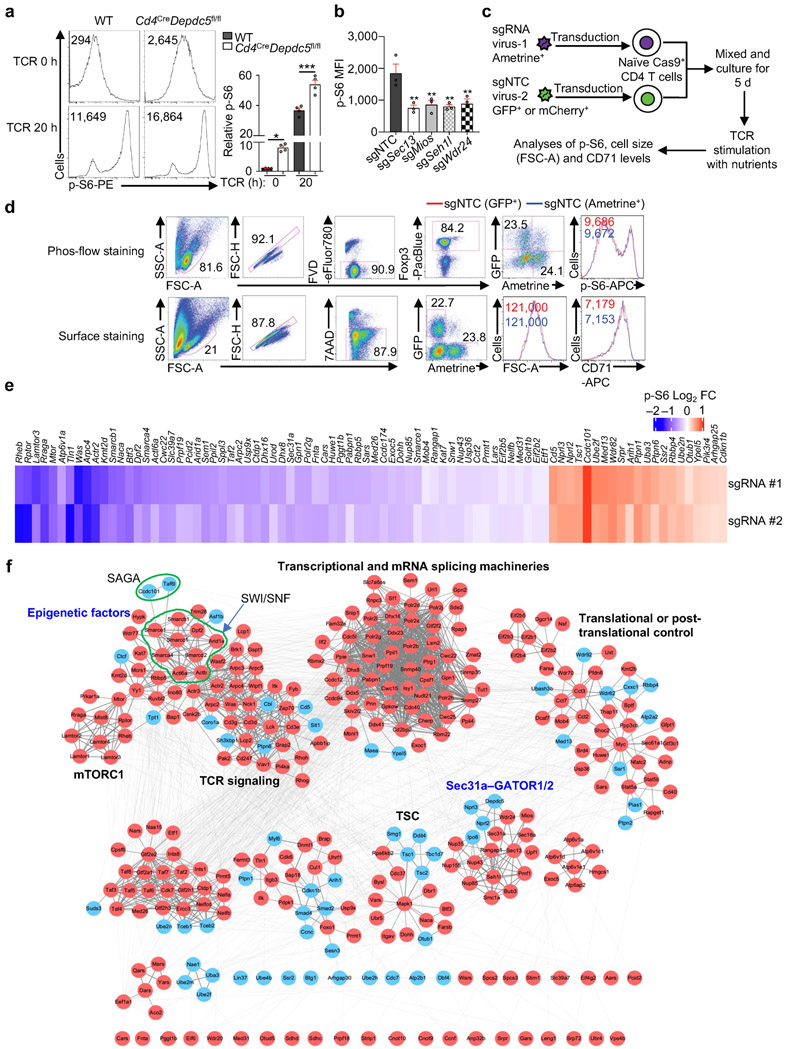
To perform pooled CRISPR screening (Fig. 1a), we transduced Cas9-expressing T cells activated with Treg-polarizing cytokines with the genome-wide lentiviral CRISPR library6, followed by stimulation with two separate doses of α-CD3 under nutrient-replete conditions. Treg cells with the highest (p-S6hi) and lowest (p-S6lo) levels of p-S6 were enriched for library preparation (Extended Data Fig. 1e). Fold change (FC)/FC plot analysis showed that both TCR stimulation conditions identified known mTORC1 positive regulators including GATOR2 (Sec13, Mios, Seh1l, Wdr24 and Wdr59)2,3, and negative regulators including Tsc1/2 and GATOR1 (Nprl2, Nprl3 and Depdc5)2,3 (Fig. 1b, Supplementary Table 1). Using two computational analyses (see Methods), we identified 292 positive and 125 negative mTORC1 regulators. Secondary CRISPR screening using a focused library targeting these 417 genes (Supplementary Table 2) revealed that 83% of the candidate genes (286 positive and 60 negative regulators) showed consistent enrichments between the two screens (Fig. 1c, Supplementary Table 3).
Figure 1. Genome-wide CRISPR screening uncovers mTORC1 regulatory networks in Treg cells.
(a) Genome-wide CRISPR screening approach. (b, c) Fold change (FC)/FC plot for primary (b) or secondary CRISPR screening (n = 6 sgRNAs per gene) (c). Positive (red) and negative (green) regulators. (d) Functional enrichment plot of genes validated by secondary CRISPR screening. (e) Immunoblot analysis and quantification of relative p-S6K1 and p-S6 in TCR-stimulated sgNTC- and sgSmarcb1-transduced cells after amino acid (AA) stimulation (n = 3 samples each group). (f) Gene expression profiles in sgNTC- or sgSmarcb1-transduced cells at 20 h of TCR stimulation (n = 4 samples each group). (g) Cells expressing indicated sgRNAs were stimulated with TCR for 3 h. Quantification of relative p-S6 level and immunoblot analysis of Smarcb1 and Castor1 expression (n = 3 samples each group). Mean ± s.e.m. (e, g). ***P < 0.001; one-way ANOVA (e, g). Data are representative of one (b–d, f) or two (g), or pooled from three (e) experiments.
We validated the respective inhibitory and stimulatory roles of GATOR1 and GATOR2 for mTORC1 activation in T cells (Extended Data Fig. 2a, b). We next developed a dual-color co-culture system with non-targeting control (NTC) ‘spike’ cells to examine cell-intrinsic effects of targeting a candidate gene on mTORC1 activation (Extended Data Fig. 2c), where expression of distinct fluorescent proteins did not impact mTORC1-dependent parameters (p-S6, cell size and CD71 expression)7 (Extended Data Fig. 2d). Using this strategy, we validated 63 positive and 21 negative candidate regulators for mTORC1 (Extended Data Fig. 2e), including previously uncharacterized regulators.
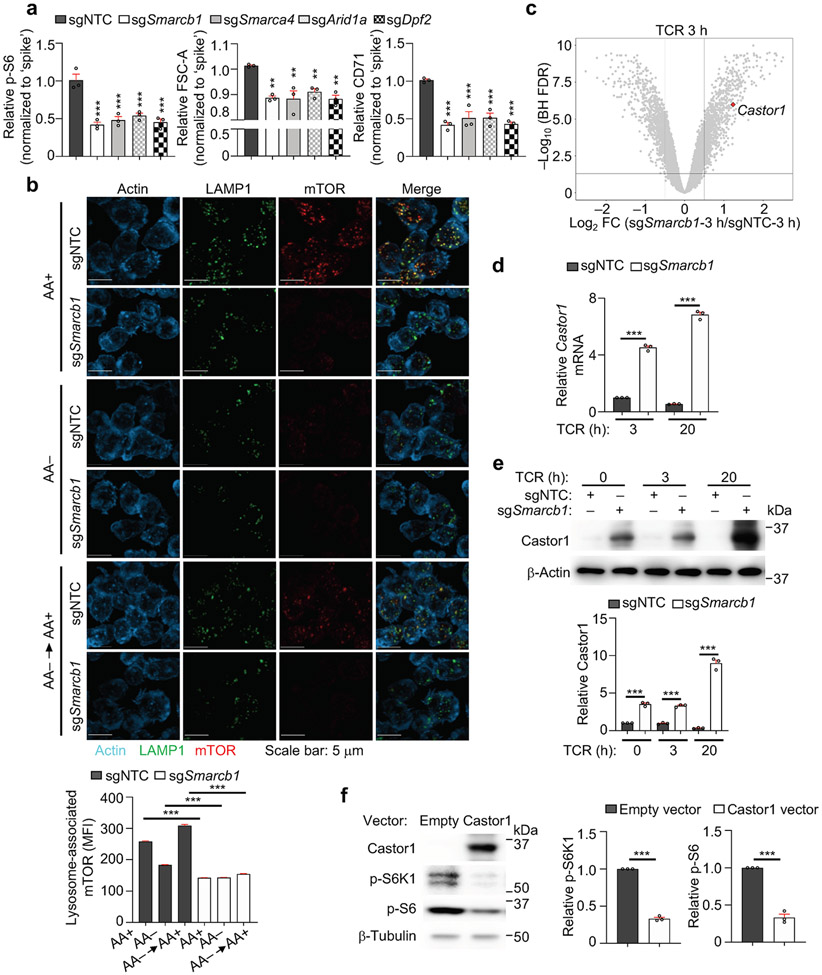
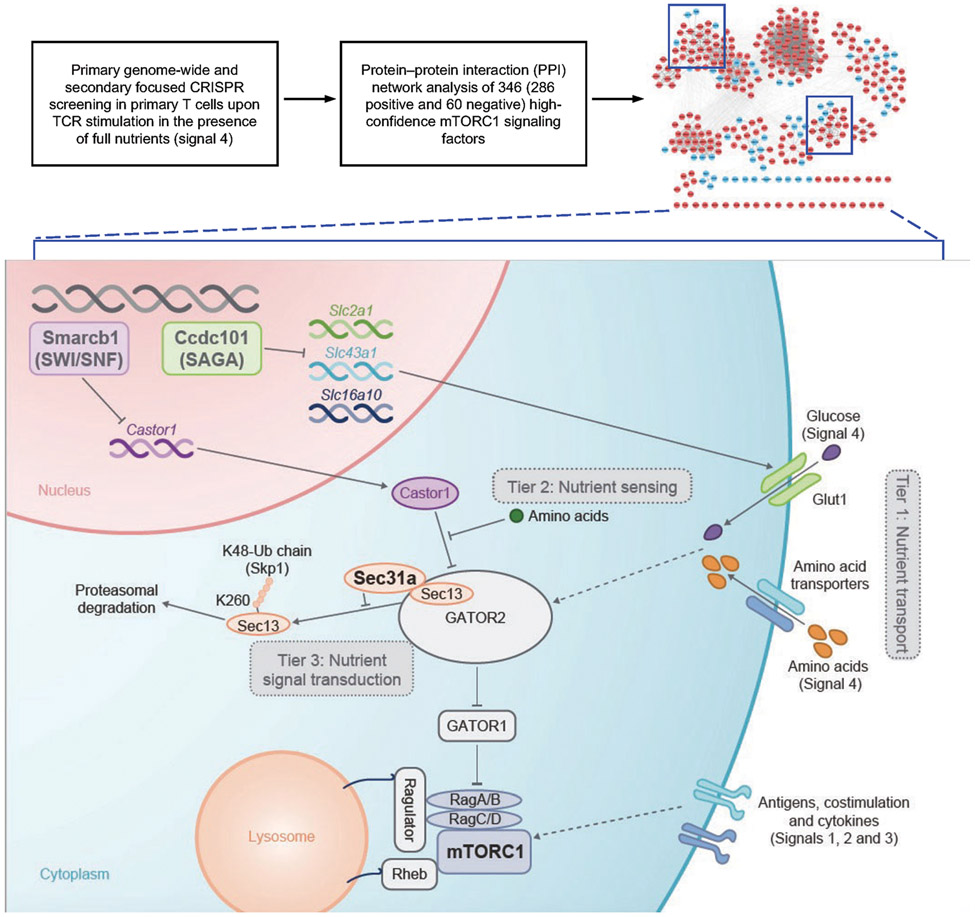
Functional enrichment analysis of the 346 high-confidence regulators of mTORC1 revealed the enrichment of diverse mTORC1-associated signaling processes (Fig. 1d). To reconstruct the mTORC1 signaling network, we integrated these high-confidence hits with composite PPI databases8,9, and identified multiple functional modules (Extended Data Fig. 2f). One module contained components of the SWI/SNF complex (Extended Data Fig. 2f, Supplementary Table 1), which regulates Foxp3 expression in Treg cells10. As the role of SWI/SNF in mTORC1 signaling is unknown, we validated that loss of Smarcb1, Smarca4, Arid1a, or Dpf2 significantly reduced mTORC1-dependent parameters (Extended Data Fig. 3a). Smarcb1-null cells also had reduced amino acid-induced mTORC1 activation and lysosome-associated mTOR5 (Fig. 1e, Extended Data Fig. 3b), indicating that Smarcb1 interplays with amino acids to regulate mTORC1.
Transcriptome profiling showed that Castor1, an arginine sensor that binds GATOR2 to inhibit mTORC1 upon nutrient deprivation2,3, was upregulated in Smarcb1-null cells (Fig. 1f, Extended Data Fig. 3c and Supplementary Table 5), which was verified by real-time PCR and immunoblot analyses (Extended Data Fig. 3d, e). The defective mTORC1 activation in Smarcb1-null cells is mediated by enhanced Castor1, as co-deletion of Smarcb1 and Castor1 reversed the defect (Fig. 1g). Furthermore, overexpression of Castor1 suppressed mTORC1 activation (Extended Data Fig. 3f). Therefore, the SWI/SNF complex represses Castor1 to promote mTORC1 activation in Treg cells.
Sec31a mediates nutrient and GATOR2-dependent mTORC1
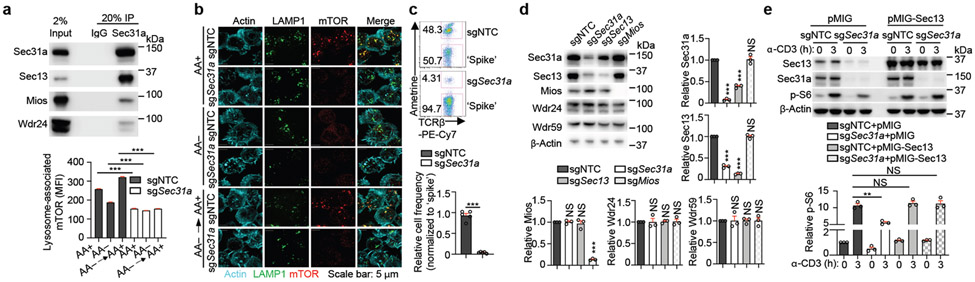
We identified a functional module containing multiple GATOR1 and GATOR2 components2,3, as well as Sec31a11 (Extended Data Fig. 2f). We found that Sec31a interacted with several GATOR2 components, especially Sec13, in T cells (Fig. 2a, Extended Data Fig. 4a). Further, deletion of Sec31a markedly impaired amino acid- or glucose-mediated mTORC1 activation (Extended Data Fig. 4b, c), and TCR-induced p-S6 level, cell size and CD71 expression (Extended Data Fig. 4d). Therefore, Sec31a is essential for integrating nutrient and immune signals to activate mTORC1.
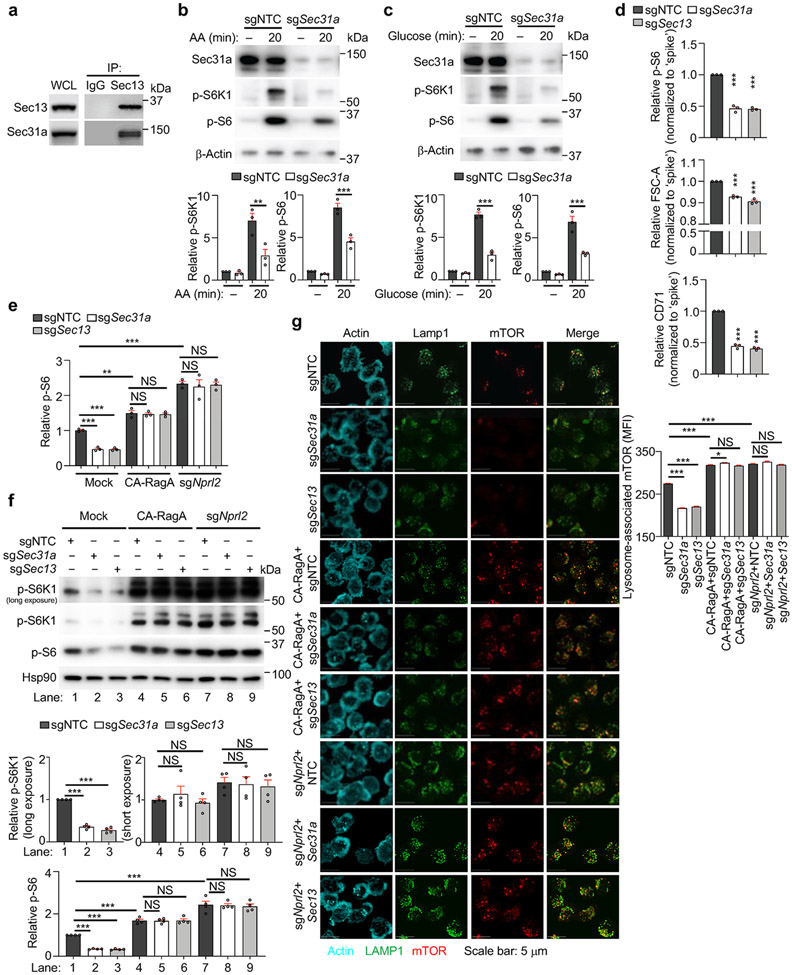
Figure 2. Sec31a is crucial for nutrient and GATOR2-dependent mTORC1 activation and the abundance of Sec13.
(a) Interaction of endogenous Sec31a with indicated proteins in induced Treg cells. (b) Images and quantification of lysosome-associated mTOR [calculated as mean fluorescence intensity (MFI)] for sgNTC- and sgSec31a-transduced cells after amino acid (AA) starvation and refeeding (n ≥ 700 cells per condition). (c) Naïve SMARTA–Cas9 T cells (Ametrine+CD45.1+) from sgNTC or sgSec31a-transduced ‘retrogenic’ mice were co-transferred with ‘spike’ (Ametrine−CD45.1+) naïve SMARTA cells (1:1 ratio) into naïve (CD45.2+) mice, followed by LCMV infection. Flow cytometry analysis and quantification of relative percentage of indicated cells (n = 5 mice per group). (d) Immunoblot analysis and quantification of indicated protein expression in indicated sgRNA-transduced cells (n = 3 samples each group). (e) sgNTC- or sgSec31a (both Ametrine+)-transduced cells were co-transduced with Sec13-overexpressing or empty vector (both GFP+) retrovirus. Immunoblot analysis of indicated proteins, and quantification of relative p-S6 level (n = 3 samples each group). Mean ± s.e.m. (b–e). NS, not significant; **P < 0.01; ***P < 0.001; two-tailed unpaired Student’s t-test (c) or one-way ANOVA (b, d, e). Data are representative of two (b, c) or five (a), or pooled from three (d, e) experiments.
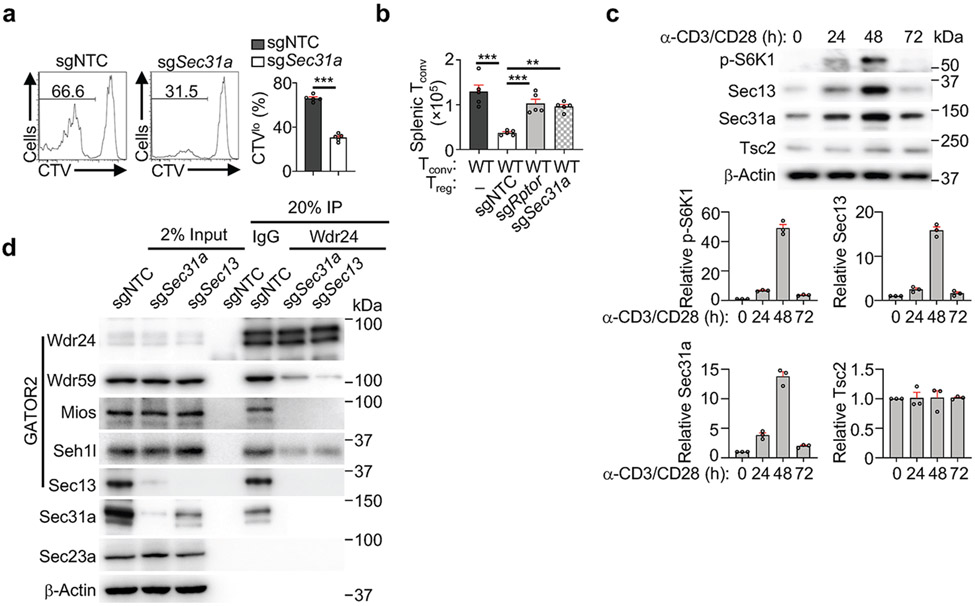
Immunofluorescence staining revealed that Sec31a-null cells had decreased lysosome-associated mTOR and were irresponsive to amino acid stimulation (Fig. 2b). We next performed epistasis assays between Sec31a and the established mTORC1 regulators. Expression of constitutively active Rag GTPase (RagAQ66L) or deletion of Nprl2 rectified the defective mTORC1 activation and localization in Sec31a- or Sec13-null cells (Extended Data Fig. 4e-g). Thus, Sec31a, like Sec13, functions upstream of GATOR1 and RagA to activate mTORC1. Accordingly, loss of Sec31a impaired in vivo Treg cell proliferation upon transfer into immunodeficient Rag1−/− mice (Extended Data Fig. 5a), and the ability of Treg cells to inhibit the accumulation of conventional T cells (Extended Data Fig. 5b). Antigen-specific T cell response following LCMV infection was also diminished (Fig. 2c), consistent with the roles of mTORC1 for activation and function of both Treg and conventional T cells1,7,12.
mTORC1 activity is dynamically regulated upon T cell activation via poorly understood mechanisms1,7. We found that mTORC1 activity, as indicated by p-S6K1, and expression of Sec13 and Sec31a (but not Tsc2), were increased up to 48 h after TCR stimulation but declined by 72 h (Extended Data Fig. 5c). Deletion of Sec31a or Sec13 led to a respective reduction of Sec13 or Sec31a expression, but not other GATOR2 components, while Mios deficiency did not have such effects (Fig. 2d). Moreover, loss of Sec31a or Sec13 disrupted integrity of GATOR2 complex, as shown by dampened interaction of Wdr24 with other GATOR2 components (Extended Data Fig. 5d). Thus, Sec31a and Sec13 are important for their reciprocal expression and contribute to the integrity of GATOR2. Further, overexpression of Sec13 fully restored defective mTORC1 activity in Sec31a-null cells, albeit without effects in the wild-type (WT) context (Fig. 2e), indicating that Sec13 is necessary but not sufficient for mTORC1 activation. Collectively, these results indicate a critical role of Sec31a in orchestrating Sec13 abundance and GATOR2 complex integrity.
Sec31a protects Sec13 from proteasomal degradation
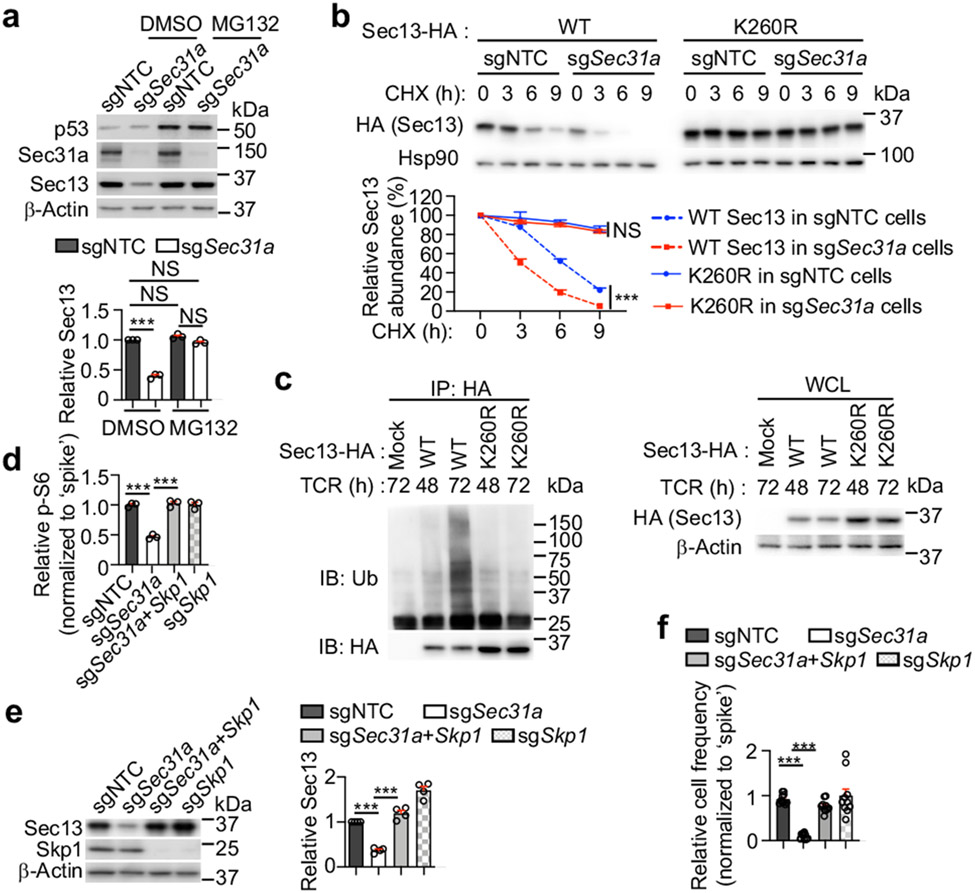
We determined the mechanisms for Sec31a-mediated regulation of Sec13 abundance. This effect was unlikely due to mRNA regulation, as Sec13 mRNA was slightly elevated in Sec31a-null cells (Extended Data Fig. 6a). Cycloheximide-chase analysis13 revealed that Sec13 half-life was reduced by half in Sec31a-null cells (Extended Data Fig. 6b), but blocking proteasomal degradation via MG132 restored Sec13 expression in these cells (Fig. 3a). In T cells with prolonged TCR stimulation, blocking proteasomal degradation also upregulated Sec13 and Sec31a levels (Extended Data Fig. 6c). To examine the mechanistic basis of proteasomal degradation, we expressed Sec13 together with wild-type ubiquitin (WT-Ub) or mutant ubiquitin that cannot undergo K48-linked (K48R-Ub) or K63-linked poly-ubiquitylation (K63R-Ub). Pulldown assay showed that Sec13 was efficiently labelled by WT-Ub or K63R-Ub, but had reduced labelling with K48R-Ub (Extended Data Fig. 6d), revealing that Sec13 mainly undergoes K48-linked poly-ubiquitylation. Upon loss of Sec31a, more Sec13 poly-ubiquitylation was observed (Extended Data Fig. 6e), suggesting that Sec31a prevents proteasomal degradation and poly-ubiquitylation of Sec13.
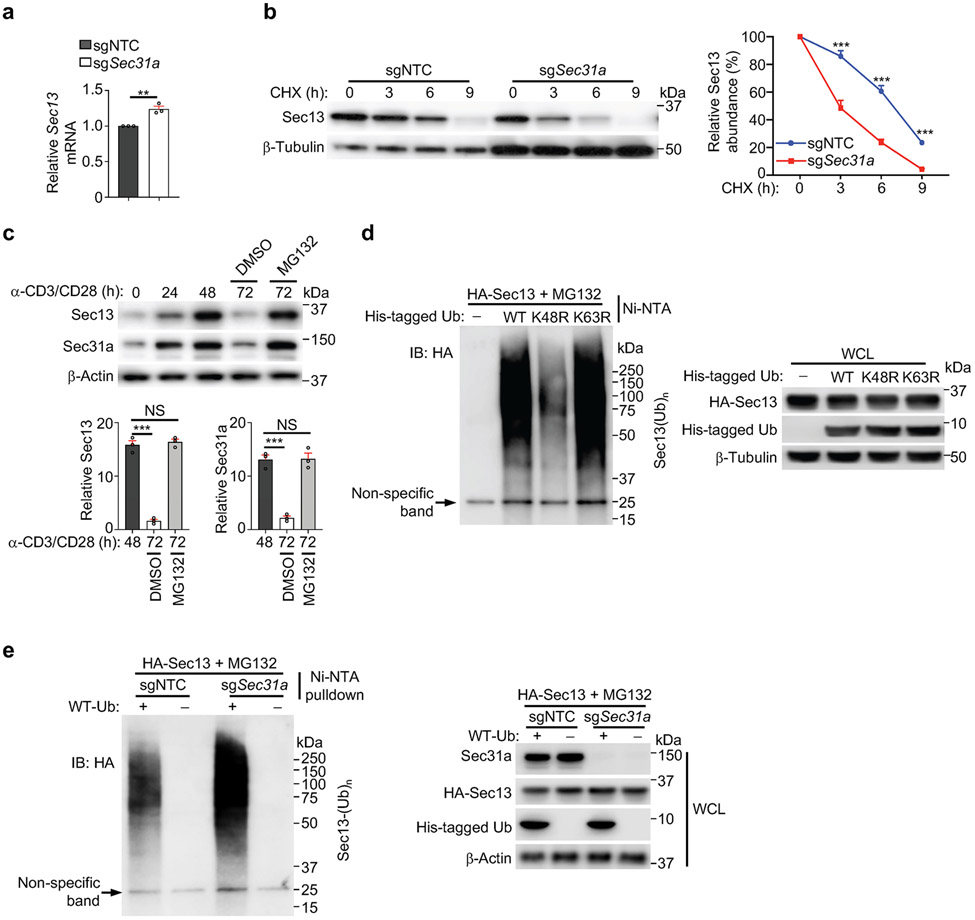
Figure 3. Sec31a protects Sec13 from Skp1-mediated proteasomal degradation.
(a) Immunoblot analysis of indicated protein expression in sgNTC- or sgSec31a-transduced cells treated with MG132 or DMSO for 9 h. Lower, quantification of relative Sec13 expression (n = 3 samples each group). (b) Control (sgNTC)- or Sec31a-null HEK293T expressing HA-tagged WT or K260R mutant Sec13 were treated with cycloheximide (CHX) for indicated times. Immunoblot analysis of HA and Hsp90 and quantification of relative Sec13 abundance (n = 3 samples each group). (c) T cells expressing HA-tagged WT or K260R mutant Sec13 were stimulated with α-CD3/CD28 for 48 or 72 h (with MG132 treatment for the last 6 h), followed by immunoprecipitation with anti-HA antibody. Immunoblot analysis for HA and β-Actin. (d) Indicated sgRNA-transduced cells were mixed with sgNTC (mCherry+; ‘spike’)-transduced cells, and stimulated with TCR for 3 h. Quantification of relative p-S6 level (n = 3 samples each group). (e) Immunoblot analysis of indicated protein expression and quantification of relative Sec13 abundance in indicated sgRNA-transduced cells (n = 4 samples each group). (f) Quantification of relative proportion of donor-derived (CD45.1+) T cells in the spleen of LCMV-infected mice (CD45.2+) at 7 d post-infection. See also Extended Data Fig. 7k (n = 10 mice per group). Mean ± s.e.m. (a, b, d–f). NS, not significant; ***P < 0.001; one-way ANOVA (a, d–f); two-way ANOVA (b). Data are representative of two (b–d) or three (e), or pooled from two (f) or three (a) experiments.
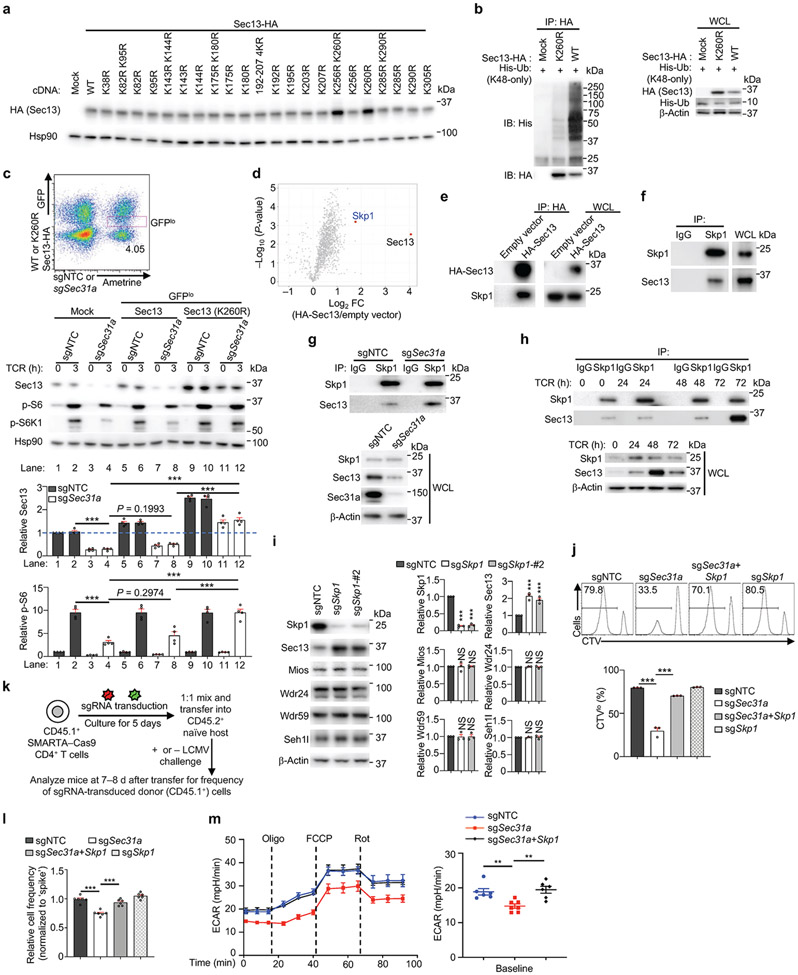
We determined the specific lysine residue(s) in Sec13 (16 in total) that mediate such poly-ubiquitylation. We found that the Sec13 K260R and K256R/K260R mutants had much higher abundance compared to WT Sec13 (Extended Data Fig. 7a), with the K260R mutant showing enhanced stability in both WT and Sec31a-null cells (Fig. 3b). Unlike WT Sec13, the Sec13 K260R mutant was not labelled by the K48-only Ub (Extended Data Fig. 7b). Further, poly-ubiquitylation of WT Sec13, but not Sec13 K260R mutant, was increased upon prolonged TCR stimulation, consistent with the elevated expression of Sec13 K260R mutant in T cells (Fig. 3c). Thus, lysine-260 mediates poly-ubiquitylation of Sec13 for degradation. We next tested the functional effects of the degradation-resistant Sec13 K260R mutant. Specifically, we transduced T cells with sgSec31a (or sgNTC) retrovirus (Ametrine+) together with WT or K260R mutant Sec13-expressing retrovirus (GFP+), and sorted the Ametrine+GFPlo population for TCR stimulation. While Sec13 WT protein expressed at the low level (i.e. GFPlo) had a very modest but trending rescue effect at restoring p-S6 and p-S6K1 in Sec31a-null cells, Sec13 K260R mutant fully rescued mTORC1 activity (Extended Data Fig. 7c), indicating a gain-of-function effect and the functional importance of Sec13 degradation.
Using quantitative interaction proteomics (interactome) analysis5,14, we identified Skp1, the essential adaptor protein of the SCF E3 ligase complex15, as among the top-enriched Sec13-interacting proteins (Extended Data Fig. 7d), which was subsequently validated by immunoprecipitation–immunoblot analysis (Extended Data Fig. 7e, f). Upon deletion of Sec31a, enhanced Sec13–Skp1 interaction was observed (Extended Data Fig. 7g), revealing Sec31a-dependent inhibition of this interaction. In α-CD3/CD28-stimulated T cells, the endogenous interaction between Skp1 and Sec13 was increased at 72 compared to 48 h (Extended Data Fig. 7h), in line with the upregulated Sec13 poly-ubiquitylation at 72 h (Fig. 3c). Thus, TCR stimulation leads to dynamic changes in Sec13 abundance and poly-ubiquitylation, and Sec13–Skp1 interaction.
Deletion of Skp1 increased the abundance of Sec13 but not other GATOR2 components (Extended Data Fig. 7i) or mTORC1 activity (Fig. 3d). However, co-deletion of Skp1 and Sec31a rectified the defective mTORC1 activation and Sec13 expression in Sec31a-null cells (Fig. 3d, e). Such co-deletion also restored the defective proliferation of Sec31a-null cells in vitro (Extended Data Fig. 7j), indicating the role of Skp1 in Sec13 degradation. Furthermore, upon adoptive transfer (Extended Data Fig. 7k), loss of Sec31a modestly reduced T cell accumulation (Extended Data Fig. 7l), but following LCMV infection, a pronounced defect in antigen-specific T cell expansion was observed (Fig. 3f). These effects of Sec31a deletion were largely rectified by co-deletion of Skp1 (Fig. 3f, Extended Data Fig. 7l). Moreover, Sec31a-null cells had reduced extracellular acidification rate (ECAR), a measure of aerobic glycolysis, which was also rescued by Skp1 co-deletion (Extended Data Fig. 7m). Collectively, these data indicate that Sec31a protects Sec13 from Skp1-dependent proteasomal degradation, thereby contributing to mTORC1 activation, glycolytic metabolism, and in vivo T cell priming.
Ccdc101 suppresses nutrient transporters and mTORC1
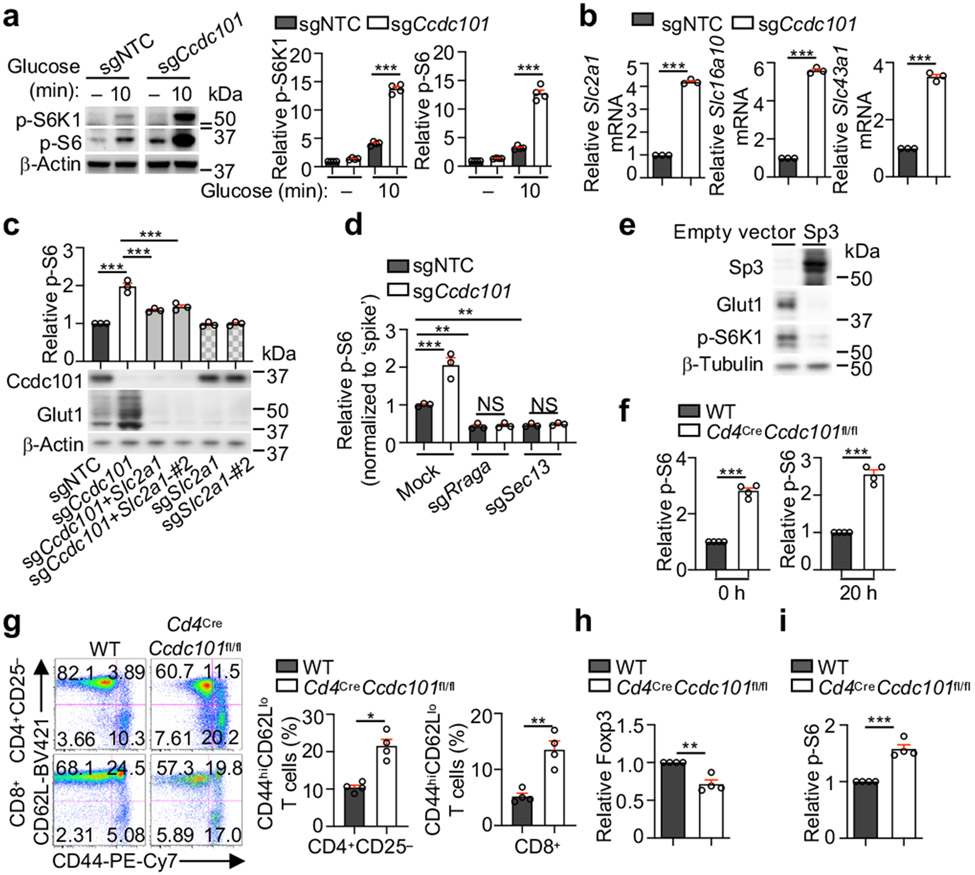
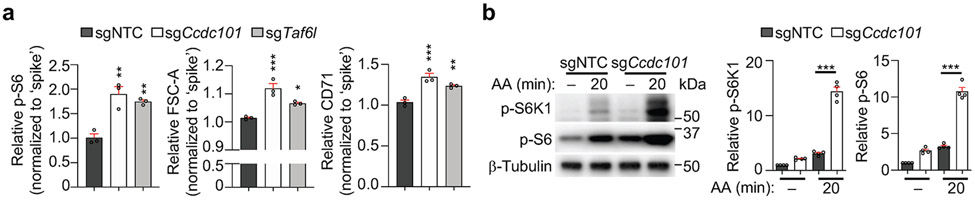
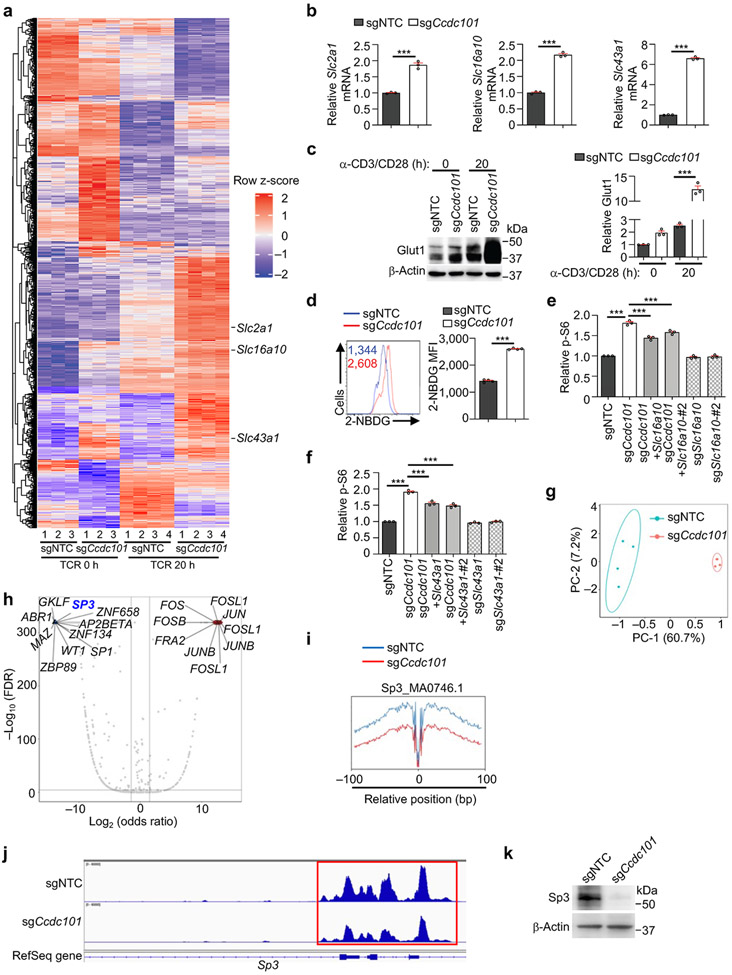
We also investigated mechanisms that suppress mTORC1 activity. Loss of Ccdc101 or Taf6l, components of chromatin-modifying SAGA complex10,16, increased TCR-induced mTORC1 activity (Extended Data Fig. 8a). Also, compared to WT cells, Ccdc101-null cells showed markedly elevated mTORC1 activity upon stimulation with glucose or amino acids (Fig. 4a, Extended Data Fig. 8b), indicating an inhibitory role of SAGA complex in TCR- and nutrient-induced mTORC1 activation. Transcriptome profiling of Ccdc101-null cells revealed significantly upregulated expression of transporters for glucose (Slc2a1, which encodes Glut1) and amino acids (Slc16a10 and Slc43a1) (Fig. 4b, Extended Data Fig. 9a, b and Supplementary Table 6). Glut1 protein expression was also increased in Ccdc101-null cells (Extended Data Fig. 9c), associated with enhanced glucose uptake (Extended Data Fig. 9d). To determine the functional effects, we deleted Slc2a1, Slc43a1 or Slc16a10 from Ccdc101-null cells. Co-deletion of Slc2a1 (Fig. 4c), and to a lesser extent, Slc16a10 or Slc43a1 (Extended Data Fig. 9e, f), partially blocked the elevated p-S6 levels in Ccdc101-null cells, indicating the functional importance of nutrient transporters, especially Glut1. Additionally, deletion of RagA or Sec13 substantially dampened the excessive mTORC1 activity in Ccdc101-null cells (Fig. 4d), suggesting that Ccdc101 functions upstream of GATOR2 and RagA. Therefore, Ccdc101 represses nutrient transporter expression and downstream GATOR2–RagA signaling to restrain mTORC1 activity.
Figure 4. SAGA complex suppresses nutrient transporter expression and mTORC1 activation.
(a) Immunoblot analysis and quantification of relative p-S6K1 and p-S6 in sgNTC- or sgCcdc101-transduced cells after glucose stimulation (n = 4 samples each group). (b) Slc2a1, Slc16a10 or Slc43a1 mRNA expression in sgNTC- and sgCcdc101-transduced cells after α-CD3/CD28 stimulation for 20 h (n = 3 samples each group). (c) Indicated sgRNA-transduced cells were stimulated with TCR for 3 h to quantify relative p-S6 level. Lower, immunoblot analysis of Ccdc101 and Glut1 expression (n = 3 samples each group). (d) sgNTC- or sgCcdc101 (both Ametrine+)-transduced cells were co-transduced with sgRraga or sgSec13 (both GFP+), mixed with sgNTC (mCherry+; ‘spike’)-transduced cells, and stimulated with TCR for 3 h to examine relative p-S6 level (n = 3 samples each group). (e) Immunoblot analysis of indicated protein expression in induced Treg cells transduced with indicated retrovirus. (f) Quantification of relative p-S6 level in freshly-isolated naïve (0 h) or α-CD3/CD28-stimulated (20 h) WT and Ccdc101-deficient CD4+ T cells (n = 4 mice per group). (g) Flow cytometry analysis and quantification of frequencies of effector/memory (CD44hiCD62Llo) subsets in splenic CD4+Foxp3− and CD8+ T cells from indicated mice (n = 4 mice per group). (h, i) Quantification of relative Foxp3 expression (h) or p-S6 levels (i) in splenic CD4+Foxp3+ Treg cells from indicated mice (n = 4 mice per group). Mean ± s.e.m. (a–d, f–i). NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed unpaired Student’s t-test (b, f–i) or one-way ANOVA (a, c, d). Data are representative of two (b–e), or pooled from two (a) or three (f–i) experiments.
ATAC-seq revealed discrete chromatin accessibility profiles between sgNTC- and sgCcdc101-transduced cells (Extended Data Fig. 9g). Transcription factor motif searches on differentially accessible chromatin regions identified downregulated motifs corresponding to Sp3 binding sites in Ccdc101-null cells (Extended Data Fig. 9h, i). Also, the promoter region of Sp3 showed reduced accessibility (Extended Data Fig. 9j), associated with reduced Sp3 expression (Extended Data Fig. 9k). To test the functional effects, we overexpressed Sp3 and found decreased Glut1 levels and mTORC1 activity (Fig. 4e). Thus, Sp3, whose expression is contingent upon Ccdc101 function, limits Glut1 expression and mTORC1 signaling.
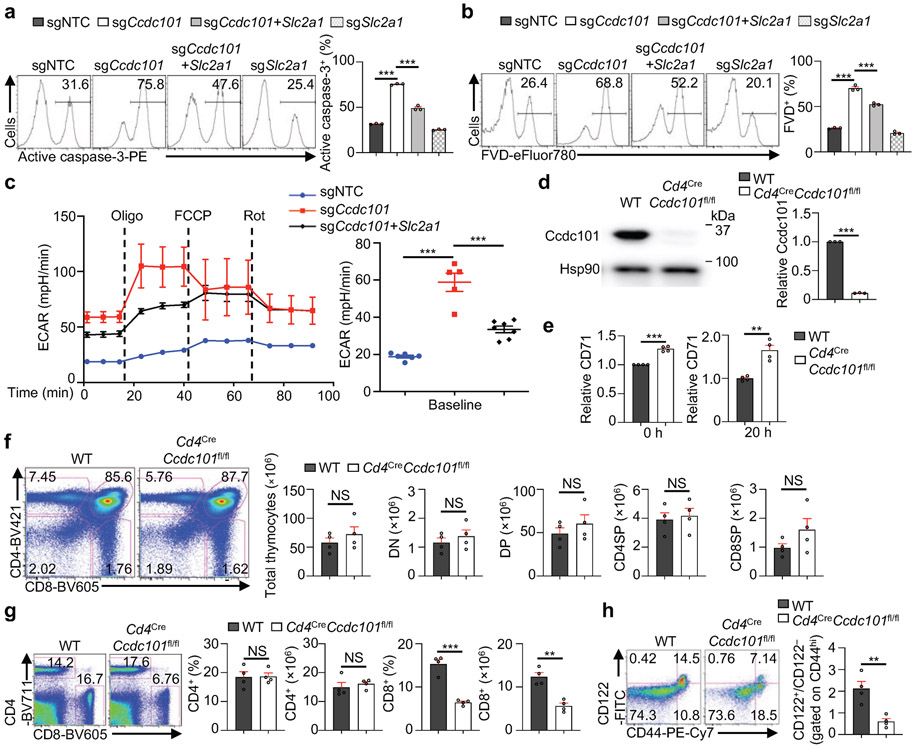
Excessive mTORC1 activation is associated with increased apoptosis and metabolism of T cells17. Accordingly, sgCcdc101-transduced cells had increased active caspase-3 and fixable viability dye (FVD) staining (Extended Data Fig. 10a, b), as well as elevated ECAR (Extended Data Fig. 10c), indicating increased apoptotic cell death and glycolytic metabolism. Consistent with the role of Ccdc101 in preventing mTORC1 hyperactivation through repressing Glut1 expression, co-deletion of Glut1 blocked the increased cell death and ECAR in Ccdc101-null cells (Extended Data Fig. 10a-c).
To further establish the in vivo physiological function of Ccdc101, we generated Cd4CreCcdc101fl/fl mice to delete Ccdc101 specifically in T cells (Extended Data Fig. 10d). Consistent with the results above, naïve T cells isolated from Cd4CreCcdc101fl/fl mice had higher expression of p-S6 (Fig. 4f) and CD71 (Extended Data Fig. 10e) than WT cells. Cd4CreCcdc101fl/fl mice had no overt defects in thymocyte development (Extended Data Fig. 10f). In contrast, T cells from Cd4CreCcdc101fl/fl mice had features of mTORC1 hyperactivation17, including a reduction of CD8+ but not CD4+ T cells in the spleen (Extended Data Fig. 10g), increased CD44hiCD62Llo effector/memory populations (Fig. 4g), and accumulation of semi-activated (CD44hiCD122hi) CD8+ T cells (Extended Data Fig. 10h). Thus, Ccdc101-deficient conventional T cells display loss of quiescence in vivo. Hyperactivation of mTORC1 is also associated with decreased lineage stability of Treg cells18. In line with this, Ccdc101-deficient Treg cells had reduced Foxp3 expression (Fig. 4h), associated with increased mTORC1 activity (Fig. 4i). Thus, Ccdc101 is a crucial negative regulator of mTORC1 to maintain T cell homeostasis and lineage stability.
Ccdc101 in immune tolerance and antitumour immunity
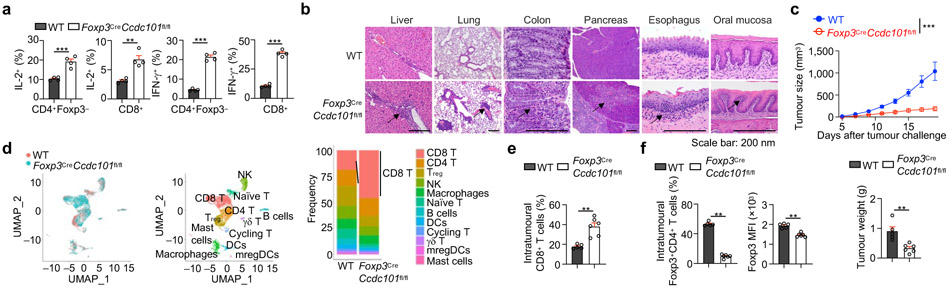
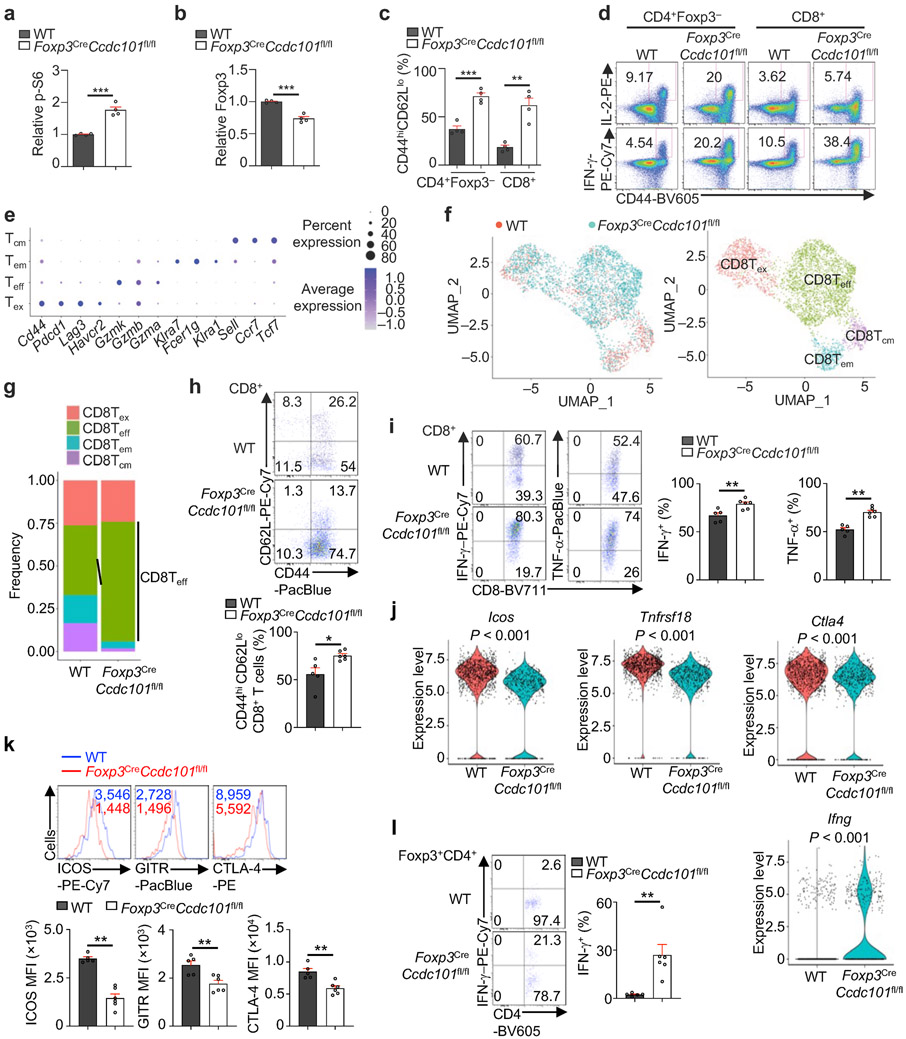
To investigate the specific role of Ccdc101 in Treg cells in vivo, we generated Foxp3CreCcdc101fl/fl mice, with specific deletion of Ccdc101 in Treg cells resulting in elevated p-S6 and reduced Foxp3 expression (Extended Data Fig. 11a, b). Foxp3CreCcdc101fl/fl mice had increased effector/memory populations and IL-2+ and IFN-γ+ cells (Fig. 5a, Extended Data Fig. 11c, d), and aged animals developed systemic inflammatory disorders, including inflammation in the periportal areas in the liver, perivascular/peribronchiolar locations in the lung, and interepithelial mucosal mast cells in the colon (Fig. 5b). Thus, Foxp3CreCcdc101fl/fl mice show altered immune homeostasis and spontaneous inflammation.
Figure 5. Steady state and tumour challenge phenotypes of Foxp3CreCcdc101fl/fl mice.
(a) Quantification of IL-2+ or IFN-γ+ populations of splenic CD4+Foxp3− and CD8+ T cells from indicated mice (~8 weeks old) (n = 4 mice each group). (b) H&E staining of indicated tissues from indicated mice (> 4 months old) (n = 5 mice each group). Arrows indicate various inflammatory features. (c–f) Indicated mice were inoculated with MC38 colon adenocarcinoma cells. (c) Tumour size and end point tumour weight (n ≥ 5 mice per group). (d) Intratumoural Treg cells, non-Treg immune cells, and myeloid cells were sorted at 19 d after tumour challenge, followed by scRNA-seq analysis (see Methods) (n = 2 replicates). UMAP embeddings of CD45+ immune cells grouped by genotype (left) and indicated immune cell subclusters (middle). Right, frequencies of indicated immune cell subclusters. (e, f) Quantification of the percentage of tumour-infiltrated CD8+ T (e) or Treg cells (f, left), and Foxp3 mean fluorescence intensity (MFI; gated on CD4+Foxp3+ cells; f, right) at 19 d after tumour challenge (n ≥ 5 mice per group). Mean ± s.e.m. (a, c, e, f); **P < 0.01; ***P < 0.001; two-tailed unpaired Student’s t-test (a, c, lower; e, f); two-way ANOVA (c, upper). Data are representative of one (b, d–f) or two (a, c) experiments.
Treg cells serve as a major barrier to antitumour immunity19. Indeed, tumour growth was strongly inhibited in Foxp3CreCcdc101fl/fl mice following MC38 tumour challenge (Fig. 5c), suggesting possibly impaired immunosuppressive function of Ccdc101-deficient Treg cells. To test this notion and the effects on the tumour microenvironment (TME), we used single-cell RNA-sequencing (scRNA-seq) to unbiasedly profile CD45+ immune cells in tumour-bearing mice (Fig. 5d). Foxp3CreCcdc101fl/fl mice contained increased proportions of total CD8+ T cells (Fig. 5d), especially effector-like CD8+ T cells, in the TME (Extended Data Fig. 11e-g). Flow cytometry analysis verified these results (Fig. 5e, Extended Data Fig. 11h). Also, tumours from Foxp3CreCcdc101fl/fl mice had higher percentages of IFN-γ+ and TNF-α+ CD8+ T cells (Extended Data Fig. 11i), indicating enhanced effector cell function.
Moreover, scRNA-seq and flow cytometry analyses of Ccdc101-null Treg cells from tumours revealed reduced expression of Treg cell suppressive markers Icos, Tnfrsf18 (encoding GITR) and Ctla4 (Extended Data Fig. 11j, k). In contrast, Ccdc101-null Treg cells had increased expression of IFN-γ (Extended Data Fig. 11j, l), consistent with the role of Treg cell-derived IFN-γ in antitumour immunity18,20. Moreover, Treg cell percentage and Foxp3 expression were dampened in intratumoural Ccdc101-null Treg cells (Fig. 5f). Collectively, these results reveal a crucial role of Ccdc101 in maintaining Treg cell fitness and suppressive function in tumours.
Here, we systemically identify the regulatory network for mTORC1 signaling in primary T cells, and uncover mechanisms of how nutrients license mTORC1 activity and adaptive immunity5,21. We establish the three-tier regulatory modules mediating nutrient transport, sensing and transduction that act in concert with immunological signals for shaping mTORC1 signaling in primary T cells (Extended Data Fig. 12). We also reveal TCR-mediated regulation of nutrient signaling processes, including the downregulation of mTORC1 activity and degradation of Sec13 upon prolonged TCR stimulation, and the active suppression of Glut1 expression by Ccdc101 in TCR-stimulated cells. Our findings support and expand upon the emerging concept of bidirectional metabolic signaling in adaptive immunity1 by establishing the connections between conventional immunological cues (antigens and costimulation or signals 1 and 2, and possibly cytokines or signal 3) and nutrient signals (emerging as signal 4) in programming immunometabolism and T cell function. Moreover, the integration of these signals by mTORC1 serves as a fundamental mechanism for connecting microenvironmental cues with T cell metabolism and activation state. The context-specific programming observed in primary T cells likely extends to other physiological processes. The identification of regulatory modules in mTORC1 signaling provides important insight into therapeutic targeting of nutrient signaling processes for infectious diseases and other immune-mediated disorders.
Methods
Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Cell lines and mice
HEK293T cell line was purchased from ATCC. The Plat-E cell line was kindly provided by Yun-Cai Liu. Cell lines used in this study were not independently authenticated or tested for mycoplasma contamination. Both cell lines were cultured in DMEM (Gibco) supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin-streptomycin. C57BL/6, Rag1−/−22, Cd4Cre23, Foxp3YFP-Cre24, SMARTA25 TCR transgenic, and Rosa26-Cas9 knockin mice26 were purchased from The Jackson Laboratory. We crossed Rosa26-Cas9 knockin mice with SMARTA TCR transgenic mice to express Cas9 in antigen-specific CD4+ T cells (SMARTA–Cas9 CD4+ T cells). Depdc5fl/fl and Ccdc101fl/fl mice were purchased from Infrafrontier. Cd4CreDepdc5fl/fl, Cd4CreCcdc101fl/fl and Foxp3CreCcdc101fl/fl mice were used at 7–10 weeks old unless otherwise noted. All genetic models were on the C57BL/6 background. Both male and female mice were used in experiments. To generate ‘retrogenic’ mice expressing sgNTC or sgSec31a, lineage-negative (Lin−) bone marrow (BM) cells from Cas9-expressing or SMARTA–Cas9-expressing mice were isolated with a lineage cell depletion kit (Miltenyi Biotec). Lin− BM cells were cultured in DMEM with 20% (vol/vol) FBS, 50 ng/ml recombinant stem cell factor (PeproTech), 20 ng/ml murine IL-3 (Thermo Fisher Scientific) and 50 ng/ml murine IL-6 (BD Biosciences) in non-tissue culture-treated plates. After 2 d of culture, retroviral transduction was performed by spin-infection at 900 × g at 25 °C for 3 h followed by additional cell culture for 48 h. Rag1−/− recipient mice were sub-lethally irradiated (5.5 Gy) 6 h prior to Lin− BM cell transfer. Transduced Lin− BM cells (5 × 105) were injected via tail vein. Mice were given antibiotic (Baytril) water for 3 weeks and reconstitution was determined by flow cytometry analysis of blood at 5–6 weeks post Lin− BM cell transfer. All mice were kept in a specific pathogen-free facility in the Animal Resource Center at St. Jude Children’s Research Hospital. Mice were kept with 12-hour light/dark cycles that coincide with daylight in Memphis, TN, USA. The St. Jude Children’s Research Hospital Animal Resource Center housing facility was maintained at 30–70 % humidity and 20–25 °C. Animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of St. Jude Children’s Research Hospital.
T cell purification and viral transduction
Naïve Cas9-expressing CD4+ T cells were isolated from the spleen and peripheral lymph nodes (PLNs) of Cas9 mice using naïve (CD25−CD44loCD62Lhi) CD4+ T cell isolation kit (Miltenyi Biotec) according to the manufacturer’s instructions. Purified naïve CD4+ T cells were activated in vitro for 20 h with 10 μg/ml α-CD3 (2C11; Bio X Cell, BE0001-1) and 5 μg/ml α-CD28 (37.51; Bio X Cell, BE0015-1) antibodies before viral transduction. Viral transduction was performed by spin-infection at 900 × g at 25 °C for 3 h with 10 μg/ml polybrene (Sigma), followed by continued culture under the polarizing conditions described below (see in vitro T cell differentiation). sgRNAs were designed by using the online tool (https://portals.broadinstitute.org/gppx/crispick/public) and subsequently cloned into retroviral or lentiviral vector27-29. sgRNAs used for validation in this study were listed in Supplementary Table 4 (two independent sgRNAs per gene; data for the second guide for certain genes are consistent with those from the first guide and are not shown). Retroviral sgRNA vector was co-transfected into Plat-E cells with the helper plasmid pCL-Eco (12371; Addgene) to produce retrovirus. Lentiviral sgRNA vector was co-transfected into HEK293T cells with the helper plasmid psPAX2 (12260; Addgene) and pCAG4-Eco (35617; Addgene) to produce lentivirus. The coding sequences of Sec13 (46332; Addgene) and Sp3 (NM_001018042.3) were subcloned into pMIG-II retroviral vector (52107; Addgene), which was co-transfected into Plat-E cells with the helper plasmid pCL-Eco (12371; Addgene) to produce retrovirus. Epistasis and rescue experiments were performed through double or triple transduction of Cas9-expressing T cells with Ametrine-, GFP-, and/or mCherry-expressing retroviral vectors (different color combinations were used to identify cells that had undergone successful dual or triple transduction), followed by flow cytometry or immunoblot analysis.
In vitro T cell differentiation
For TH0, TH1 or induced Treg cell differentiation, naïve CD4+ T cells were stimulated with 5 μg/ml each of plate-bound α-CD3/CD28 antibodies in the presence of human IL-2 (100 U/ml; PeproTech) for TH0 polarizing; human IL-2 (100 U/ml) plus mouse IL-12 p40 (0.5 ng/ml; BD Biosciences) for TH1 polarizing; or human IL-2 (100 U/ml) plus human TGF-β (0.5 ng/ml; PeproTech) for Treg polarizing for 2.5 d, and rested in complete Click’s medium (IrvineScientific) supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin-streptomycin and the cytokine combinations indicated above for 3 d. For TH2 or TH17 cell differentiation, naïve CD4+ T cells were stimulated with 2 μg/ml each of soluble α-CD3/CD28 antibodies and irradiated antigen presenting cells (T cell-depleted splenocytes) in the presence of human IL-2 (100 U/ml), mouse IL-4 (10 ng/ml; R&D systems) and anti-IFN-γ antibody (10 μg/mL; Bio X Cell) for TH2 polarizing; or human TGF-β (2 ng/ml; PeproTech) plus mouse IL-6 (20 ng/m; BD Biosciences) for TH17 polarizing in complete Click’s medium for 5.5 d.
Adoptive transfer and LCMV infection
For analysis of effects of Sec31a and Rptor deletion on antigen-specific T cell expansion in vivo, naïve SMARTA–Cas9 CD4+ T cells were isolated from ‘retrogenic’ mice as indicated above (CD45.1+Ametrine+). sgRNA-expressing SMARTA–Cas9 CD4+ T cells were co-adoptively transferred at a 1:1 ratio (5 × 105 each) with non-transduced SMARTA (CD45.1+Ametrine− ‘spike’) cells into naïve hosts (CD45.2+) followed by LCMV infection. Alternatively, SMARTA–Cas9 CD4+ T cells (CD45.1+; 5 × 105) that were transduced with sgNTC (mCherry+; ‘spike’) in vitro were mixed at a 1:1 ratio with SMARTA–Cas9 CD4+ T cells (CD45.1+) transduced with sgNTC (Ametrine+), sgSec31a (Ametrine+), sgSkp1 (Ametrine+), sgSec31a+Skp1 (GFP+ and Ametrine+), and adoptively transferred into naïve WT mice (CD45.2+) (n = 5 mice per group). The mice were left either uninfected or challenged with LCMV infection, and were analyzed at d 7–8 after transfer by flow cytometry analysis30. For LCMV infection, 2 × 105 plaque-forming units (PFU) of LCMV Armstrong strain virus were injected intraperitoneally.
Tumour model and tumour-infiltrating lymphocyte isolation
MC38 colon adenocarcinoma cells were cultured in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% (v/v) penicillin plus streptomycin. WT (included both Foxp3CreCcdc101+/+ or Foxp3CreCcdc101fl/+ mice) and Foxp3CreCcdc101fl/fl mice were injected subcutaneously with MC38 colon adenocarcinoma cells (5 × 105 cells) into the right flank. Tumours were measured every two days with digital calipers and tumour volumes were calculated by the formula: Length × Width × [(Length × Width) ^ 0.5] × π/6. Tumour size limits were approved to reach a maximum of 3,000 mm3 or ≤ 20% of body weight (whichever was less) by the IACUC at St. Jude Children’s Research Hospital. Tumour infiltrating lymphocytes (TILs) were prepared with mechanical dissociation and enzymatic [1 mg/ml Collagenase IV (Roche) + 200 U/ml Dnase I (Sigma)] disruption for 1 h at 37 °C, and then passed through 70 μm cell strainers to remove residual undigested tumour tissues. TILs were isolated by density-gradient centrifugation over Percoll (GE Healthcare).
Flow cytometry
For analysis of surface markers, cells were stained in PBS (Gibco) containing 2% (wt/vol) BSA (Sigma). Surface proteins were stained for 30 min on ice. For phospho-flow cytometry analysis, to preserve the fluorescent proteins, cells were fixed with 2% paraformaldehyde for 30 min at room temperature, followed by permeabilization with 90% ice-cold methanol for 30 min and then stained in 1× Permeabilization Buffer (eBioscience) for 30 min at room temperature. Transcription factor staining was performed with Foxp3/transcription factor staining buffers, according to the manufacturer’s instructions (eBioscience). Intracellular staining for cytokines was performed with a fixation/permeabilization kit (BD Biosciences). Active caspase-3 staining was performed using the instructions and reagents from the ‘Active Caspase-3 Apoptosis Kit (BD Biosciences). 7AAD (Sigma) or fixable viability dye (eBioscience) was used for dead cell exclusion. The following antibodies were used: α-CD4 (1:200, RM4-5, 100557), α-IFN-γ (1:200, XMG1.2, 505814), α-IL-4 (1:200, 11B11, 504109), α-IL-17A (1:200, TC11-18H10.1, 506916), α-CD25 (1:200, PC61.5, 102038), α-CD44 (1:200, IM7, 103047), α-CD71 (1:200, RI7217, 113808), α-CD122 (1:200, TM-b1, 123210), anti-TNF-α (1:200, MP6-XT22, 506314), α-CTLA-4 (1:200, UC10-4B9, 106310) (all from Biolegend); α-active caspase-3 (1:75, C92-605, 560626), α-Foxp3 (1:400, MF23, 560401) (all from BD Biosciences); α-TCRβ (1:400, H57-597, 1105961-82), α-ICOS (1:200, 7E.17G9, 17-1942-82), α-GITR (1:200, DTA-1, 12-5874-82); α-IL-2 (1:200, JES6-5H4, 11-7021-82) (all from eBioscience); α-p-S6 (1:200, D57.2.2E, 5316S) (from Cell Signaling Technology); α-CD8α (1:200, 53-6.7, 1103605) (from SONY); α-CD62L (1:400, MEL-14, 25-0621-U025) (both from TONBO Biosciences). To monitor cell division, cells were labelled with 5 μM CellTrace Violet (CTV; ThermoFisher; C34557). Glucose uptake was measured by staining with the glucose analog, 2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose (2-NBDG)30 (Thermo Fisher Scientific; N13195), which was dissolved in 100% ethanol and used at 10 μM in complete DMEM. Flow cytometry data were acquired on an LSR II or LSR Fortessa (BD Biosciences) via the BD FACSDiva software 8 and analyzed using Flowjo 9.9.4 (Tree Star).
Lentiviral sgRNA Brie library CRISPR–Cas9 mutagenesis screening
Lentiviral and retroviral sgRNA vector design
The lentiviral and retroviral sgRNA vectors were previously described27. Briefly, the lentiviral sgRNA vector was modified from lentiGuide-puro vector by replacing the EF-1α-PuroR fragment with a mouse PGK promoter-driven Ametrine (or GFP or mCherry) fluorescent protein. The retroviral sgRNA vector was modified from pLMPd-Amt vector31 by replacing the miR30 shRNA cassette with the U6 promoter-driven gRNA cassette from the lentiGuide-puro vector.
Primary genome-wide p-S6 CRISPR screening using Brie sgRNA library
Lentiviral Brie library plasmid6 was co-transfected into HEK293T cells with the helper plasmid psPAX2 (12260; Addgene) and pCAG4-Eco (35617; Addgene) to produce lentivirus. At 48 h after transfection, virus was harvested and frozen at −80 °C. 2 × 108 naïve CD4+ T cells were purified from Cas9 mice, transduced at a MOI of 0.3 to achieve ~20% transduction efficiency, followed by puromycin selection, and differentiated into Treg cells (two biological replicates were performed). At d 6 of differentiation, dead cells were removed by Percoll centrifugation, and 5 × 107 transduced cells were saved as input (~630× cell coverage per sgRNA), or were stimulated with 0.25 or 4 μg/ml of α-CD3 antibody for 3 h. Cells were incubated with the live/dead dye 7AAD during the last 20 min of α-CD3 stimulation, and then were fixed and permeabilized according to manufacturer’s (eBioscience) instructions, namely fixation in the Fixation/Permeabilization buffer for 30 min, followed by treatment with the permeabilization buffer for another 30 min. The cells were washed twice and then incubated with antibodies against Foxp3 and p-S6. For cell sorting, we first gated on singlets, and then 7AAD− cells, followed by gating of Foxp3+ cells and sorting of the top ≤ 10% of populations with the highest (p-S6hi) and lowest (p-S6lo) levels of p-S6 for library preparation by following a similar strategy used in a previous study (i.e. the top 10% or 5–15% of TNF-αhi and TNF-αlo populations)32. About 3–9 × 106 Treg cells per sample (38–113× cell coverage per sgRNA) were recovered for further analysis.
Sequencing library preparation
Genomic DNA was extracted by using the DNeasy Blood & Tissue Kits (69506; Qiagen). Primary PCR was performed by using the KOD Hot Start DNA Polymerase (71086; Millipore) and the following pair of Nextera NGS primers (Nextera NGS-F: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGTTGTGGAAAGGACGAAACACCG; Nextera NGS-R: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCCACTTTTTCAAGTTGATAACGG). Primary PCR products were purified using the AMPure XP beads (A63881; Beckman). A second PCR was performed to add adaptors and indexes to each sample. Hi-Seq 50-bp single-end sequencing (Illumina) was performed. To minimize false positive candidates, deep sequencing data were analyzed by two independent pipelines as detailed below. We superimposed the top-enriched genes (550 positive and 300 negative mTORC1 regulators) consistently captured by these two independent computational analyses and applied additional filters [∣z-score∣ > 1.96; ∣log2 fold change (FC) (p-S6hi/p-S6lo)∣ > 0.5; and log2 FC (p-S6lo/input) > 0.2 for positive regulator or log2 FC (p-S6hi/input) > 0.2 for negative regulator], resulting in the identification of a total of 292 positive and 125 negative mTORC1 regulators.
Data processing pipeline #1:
As described previously27, for data analysis, FASTQ read files obtained after sequencing were demultiplexed using the Hi-Seq analysis software (Illumina). The 50-bp single-end reads were trimmed and quality-filtered using the CLC Genomics Workbench v11 (Qiagen) and matched against sgRNA sequences from the genome-scale sgRNA Brie library (primary screening, 78,637 sgRNAs from 19,642 genes and 1,000 non-targeting control sgRNAs). Read counts for sgRNAs were normalized against total read counts across all samples for each screening. For each sgRNA, the fold change [log2 FC] for enrichment was calculated between each of the two (primary screening) biological replicates and the input experiment before the stimulation. The difference of log2 FC between p-S6hi and p-S6lo populations was then calculated, and the gene level difference was further summarized by averaging four sgRNAs for each gene.
Data processing pipeline #2:
As described previously33, for data analysis, we first demultiplexed, and quality-filtered all raw FASTQ read files using an in-house mapping pipeline. Single-end reads were trimmed and matched against sgRNA sequences from the Brie sgRNA library. Read counts were normalized to count per million across all samples, and then log2 transformed prior to the downstream analysis. The hit identification at the gene and sgRNA levels was analyzed by ScreenBEAM33, a Bayesian hierarchical modeling approach for meta-analysis of large-scale functional genomics analyses that minimizes the false positive rates of hit identification. Gene ranking was based on z-score and log2 FC outputted from the ‘DRAgeneLevel’ function in ScreenBEAM R package, comparing p-S6 altered samples with input samples, by integrating 4 sgRNAs across 2 biological replicates. Any sgRNA with median count less than 8 was filtered out when calculating the statistics for gene ranking. sgRNA ranking was also based on z-score and log2 FC calculated by the ‘DRAshRNALevel’ function in ScreenBEAM R package across 2 biological replicates.
Lentiviral sgRNA focused library CRISPR-Cas9 mutagenesis screening
Focused sgRNA library construction
To generate the focused library based on the candidates from the primary screening, a list of all sgRNA sequences found in the mouse Brie6 and GeCKO v234 whole genome sgRNA libraries was compiled. These libraries contain duplicate sgRNAs, as well as sgRNAs that bind to more than one site in the genome and may lead to off-target effects. Therefore, after duplicate sgRNAs were removed, a subsequent off-target analysis was performed, and up to six sgRNAs per gene, which have unique target sites in the genome, were selected. sgRNAs were manually designed for several genes for which no adequate sgRNAs could be found from the compiled libraries. The resulting oligo library was designed as described35 and synthesized (Twist Bioscience). Library amplification, Gibson Assembly into the pLentiPuro backbone (39481; Addgene) and library plasmid prep validation were performed as described35.
Secondary p-S6 CRISPR screening using focused sgRNA library and data processing
Lentiviral focused sgRNA plasmid was co-transfected into HEK293T cells with the helper plasmid psPAX2 (12260; Addgene) and pCAG4-Eco (35617; Addgene) to produce lentivirus. At 48 h after transfection, virus was harvested and frozen at −80 °C. 1 × 108 naïve CD4+ T cells from Cas9 mice were transduced at a MOI of 0.3 to achieve ~20% transduction efficiency, followed by puromycin selection, and differentiated into Treg cells (three biological replicates were performed). At d 6 of differentiation, dead cells were removed by Percoll centrifugation, and 3 × 106 transduced cells were saved as input (> 1,000× cell coverage per sgRNA), or were stimulated with 0.25 or 4 μg/ml of α-CD3 antibody for 3 h to sort the p-S6hi and p-S6lo populations (less than 10%). About 2–3 × 106 cells per sample (> 700× cell coverage per sgRNA) were recovered for further analysis. Data processing was performed as described above, using both pipelines #1 and #2.
Gene expression profiling
Induced Treg cells were analyzed using microarray analysis (Affymetrix Mouse Clariom S Assay), which were collected in the following three batches: (a) sgNTC- and sgCcdc101-transduced cells (n = 3 replicates each group) without stimulation (no α-CD3/CD28 antibodies); (b) sgNTC- and sgCcdc101-transduced cells (n = 4 replicates each group) stimulated with α-CD3/CD28 antibodies for 20 h; and (c) sgNTC- and sgSmarcb1-transduced cells (n = 4 replicates each group) stimulated with α-CD3/CD28 antibodies for 3 or 20 h. For microarrays, the expression signals were analyzed by Affymetrix Expression Console v1.1, followed by differential expression analysis performed using R package limma v.3.34.9. All the plots were generated using R packages ggplot2 v.2.2.1 and ComplexHeatmap v2.6.2. Differentially expressed transcripts were identified by ANOVA (Partek Genomics Suite version 6.5), and the Benjamini-Hochberg method was used to estimate the false discovery rate (FDR) as described12. Differentially expressed (DE) genes were defined by ∣log2 FC∣ > 0.5; FDR < 0.05. Microarray data have been deposited into the GEO series database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160598, token for access: ghojkuyyrhyphux) and will be made available to public upon manuscript publication.
ATAC-seq and data analysis
Library preparation
Induced Treg cells (n = 4 biological replicates each group) were stimulated with α-CD3/CD28 antibodies for 20 h. sgNTC- and sgCcdc101-transduced cells (n = 4 replicates each group) were collected. To prepare ATAC-seq library, a total of 5 × 104 cells were incubated in 50 μl ATAC-seq lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630) on ice for 10 min. Resulting nuclei were pelleted at 500 g for 10 min at 4 °C. Supernatant was carefully removed with a pipette and discarded. The pellet was resuspended in 50 μl transposase reaction mix (25 μl 2× TD buffer, 22.5 μl nuclease-free water, 2.5 μl Transposase) and incubated for 30 min at 37 °C. After the reaction, the DNA was cleaned up using the Qiagen MinElute kit. The barcoding reaction was run using the NEBNext HiFi kit based on manufacturer’s instructions and amplified for 5 cycles as described36 using the same primers. Ideal cycle numbers were determined from 5 μl (of 50 μl) from the previous reaction mix using KAPA SYBRFast (Kapa Biosystems) and 20 cycles of amplification on an Applied Biosystems 7900HT. Optimal cycles were determined from the linear part of the amplification curve, and the remaining 45 μl of PCR reaction was amplified in the same reaction mix using the optimal cycle number.
Data analysis
ATAC-seq analysis was performed as described previously27,36. Briefly, 2 × 50-bp paired-end reads obtained from NovaSeq were trimmed for Nextera adaptor by trimmomatic (v0.36, paired-end mode, with parameter LEADING:10 TRAILING:10 SLIDINGWINDOW:4:18 MINLEN:25) and aligned to mouse genome mm9 downloaded from gencode release M1 (https://www.gencodegenes.org/mouse/releases.html) by BWA (version 0.7.16, default parameters)36 . Duplicated reads were then marked with Picard (v2.9.4) and only non-duplicated proper paired reads have been kept by samtools (parameter ‘-q 1 -F 1804’ v1.9)36 . After adjustment of Tn5 shift (reads were offset by +4 bp for the sense strand and −5 bp for the antisense strand), we separated reads into nucleosome-free, mononucleosome, dinucleosome and trinucleosome as previously described37 by fragment size and generated ‘.bigwig’ files by using the centre 80-bp of fragments and scaled to 30 × 106 nucleosome-free reads. We observed reasonable nucleosome-free peaks and a pattern of mono-, di- and tri-nucleosomes on IGV (v2.4.13)36 . All samples in this study had approximately 1 × 107 nucleosome-free reads, indicative of good data quality. Next, peaks were called on nucleosome-free reads by MACS2 (v2.1.1.20160309, with default parameters with ‘–extsize 200–nomodel’)36. To assure reproducibility, we first finalized nucleosome-free regions for each sample and retained a peak only if it called with a higher cut-off (MACS2 -q 0.05). We further generated consensus peaks for each group by keeping peaks that were present in at least 50% of the replicates and discarding the remaining, non-reproducible peaks. The reproducible peaks were further merged between sgNTC- and sgCcdc101-transduced samples if they overlapped by 100-bp and then we counted nucleosome free reads from each of the 8 samples by bedtools (v.2.25.0)36. To identify the differentially accessible open chromatin regions (OCRs), we first normalized raw nucleosome-free read counts per million (CPM) followed by differential accessibility analysis by implementation of the negative binomial model in the DESeq2 R package36. FDR-corrected P value < 0.05, ∣log2 FC∣ > 0.5 were used as cut-offs for more- or less-accessible regions in sgCcdc101-transduced samples compared to their sgNTC-transduced ‘spike’ cells. Principal component analysis was performed using function prcomp in R. We then assigned the differentially accessible OCRs in the ATAC-seq data for the nearest genes to generate a list of DA genes using HOMER36. This analysis identified 25,646 open chromatin regions (OCRs) with differential expression in Ccdc101-null versus control cells (FDR < 0.05; ∣log2 FC (sgCcdc101/sgNTC)∣ > 0.5). Raw and processed ATAC-seq data have been deposited into the GEO series database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160598, token for access: ghojkuyyrhyphux) and will be made available to public upon manuscript publication.
Motif analysis and footprinting of transcription factor binding sites
For motif analysis, we further selected 1,000 unchanged regions log2 FC < 0.5 and FDR-corrected P value > 0.5 as control regions. FIMO from MEME suite (v4.11.3, ‘–thresh 1e-4–motif-pseudo 0.0001’)38 was used for scanning motif (TRANSFAC database release 2019, only included Vertebrata and not 3D structure-based) matches in the nucleosome-free regions and two-tailed Fisher’s exact test was used to determine whether a motif was significantly enriched in differentially accessible compared to the control regions. To perform footprinting analysis of transcription factor binding site, the RGT HINT application was used to infer transcription factor activity and to plot the results39.
Single-cell RNA sequencing (scRNA-seq) and data analysis
Library preparation
WT (included a mixture of Foxp3CreCcdc101+/+ or Foxp3CreCcdc101fl/+ mice) and Foxp3CreCcdc101fl/fl mice were challenged with MC38 colon adenocarcinoma cells, and Treg cells (CD45+CD4+YFP+), non-Treg immune cells (CD45+YFP−CD11b−) and myeloid cells (CD45+CD11b+) in the tumour tissues were sorted on a Reflection cell sorter (iCyt) at 19 d after tumour challenge40 and mixed at 1:2:1 ratio (n = 2 biological replicates, pooled from 3-4 mice, per group). The cells were counted and examined for viability using a Luna Dual Florescence Cell Counter (Logos Biosystems). Samples were pelleted using centrifuge at 2,000 rpm for 5 min. The supernatant was discarded, and cells were resuspended in 100 μl of 1× PBS (Thermo Fisher Scientific) with 0.04% BSA (Amresco). A Luna Dual Florescence Cell Counter (Logos Biosystems) was then used to count the cell numbers and examine cell viability. Cell counts were approximately 1 × 106 cells/ml and cell viability was above 98%. Single-cell suspensions were loaded onto the Chromium Controller based on their respective cell counts to generate 8,000 single-cell gel beads in emulsion per sample. Each sample was loaded into a separate channel. Libraries were generated using the Chromium Next GEM Single Cell 3′ v.3.1 Library and Gel Bead Kit (10X Genomics). A high sensitivity D5000 ScreenTape with a TapeStation (Agilent Technologies) was used to quantify and quality-check the cDNA content of each sample after cDNA amplification of 12 cycles and determine the number of PCR amplification cycles for preparing a sufficient sequencing library. After library quantification and quality-checking using D5000 ScreenTape (Agilent Technologies), samples were diluted to 3.5 nM for loading onto the HiSeq 4000 (Illumina) with a 2 × 100-bp paired-end kit using the following read length: 28-bp read 1, 10-bp i7 index, 10-bp i5 index, and 90-bp read 2. An average of 1.8×108 reads per sample were obtained (approximately 5.8×104 reads per cell).
Alignment, barcode assignment and unique molecular identifier (UMI) counting
The Cell Ranger v6.0 Single-Cell software suite (10X Genomics) was implemented to process the raw sequencing data from the Illumina HiSeq run. This pipeline performed demultiplexing, alignment (using the mouse genome mm10 from ENSEMBL GRCm38) and barcode processing to generate gene–cell matrices. Seurat R package (v4.0) was used for downstream analysis. Specifically, data from CD45+YFP+ Treg cells, CD45+YFP−CD11b− non- Treg immune cells and CD45+CD11b+ myeloid cells in the tumour tissues, isolated from WT and Foxp3CreCcdc10 fl/fl mice (n = 2 biological replicates per group), were combined into one dataset for consistent filtering. Cells with low unique molecular identifier (UMI) counts (potentially dead cells with broken membranes) or high UMI counts (potentially two or more cells in a single droplet) were filtered. Potential dead cells with high percent (> 6%) of mitochondrial reads were also removed. Residual CD45− contaminants introduced by cell sorting and doublets were further removed. A total of 10,202 cells (WT mice, 5,281; Foxp3CreCcdc101fl/fl mice, 4,921) were captured, with an average of 2,266 mRNA molecules (UMIs, median: 6,862; range: 522–29,984). We normalized the expression level of each gene to 1e6 UMIs per cell and log-transformed them by adding 1 to the expression matrix.
Data visualization
To identify different clusters in tumour immune cells, data were further analyzed using Seurat and visualized by UMAP (Uniform Manifold Approximation and Projection), which partitioned cells into 14 unsupervised clusters based on their transcriptomes using resolution = 0.5 in FindClusters function. Twelve immune cell types were further identified based on prior knowledge and markers highly expressed in each unsupervised cluster. Curated violin plots that represent the expression level of genes were generated by VlnPlot function in Seurat R package. CD8+ T cells were further clustered into 4 subtypes, including central memory-like (Tcm), effector memory-like (Tem), effector-like (Teff) and exhausted-like (Tex) CD8+ T cells.
Immunoprecipitation and cell-based ubiquitylation assays
Immunoprecipitation–immunoblot analysis
For immunoprecipitation, 1 × 108 transduced cells were lysed in 100 μl CHAPS lysis buffer (0.3% CHAPS, 40 mM HEPES pH 7.4, 120 mM NaCl, 20 mM NaF, 1 mM EDTA). About 2–4 μg of the indicated antibodies were bound to 5 μl of protein A/G agarose (sc-2003; Santa Cruz) and then added to the cleared cellular lysates and incubated with rotation for 2 h. Immunoprecipitates captured with protein A/G agarose were washed three times with the CHAPS lysis buffer and eluted with 2× sample reducing buffer (39000; ThermoFisher), followed by immunoblot analysis. The following antibodies or antibody-conjugated beads were used for immunoprecipitation: α-Sec31a (17913-1-AP; ProteinTech), α-Sec13 (GTX101055; GeneTex), α-Skp1 (12248; Cell Signaling Technology), α-Wdr24 (20778-1-AP; ProteinTech), and α-HA magnetic beads (88837; ThermoFisher).
Cell-based ubiquitylation assay
As described41, HEK293T cells transfected with the indicated combination of plasmids for HA-tagged Sec13 together with (His-tagged wild-type (WT) ubiquitin (WT-Ub) or mutant ubiquitin that cannot undergo K48-linked poly-ubiquitylation (K48R-Ub) or K63-linked poly-ubiquitylation (K63R-Ub) were treated with MG132 for 6 h to facilitate the accumulation of ubiquitin-labelled target proteins. Cells were lysed in 1 ml denaturing buffer (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM imidazole pH 8.0) and passed through 23 G needle for 10 times, followed by sonification to shear the genomic DNA. After centrifugation at 20,000 × g for 30 min, cellular extracts were then incubated with 10 μl Ni-NTA His-Bind-Resin (70666; Millipore Sigma) for 3 h, washed 2 times with the denaturing buffer and buffer T1 (25 mM tris-Cl, 20 mM imidazole pH 6.8), and subjected to immunoblot analysis. Ni-NTA bead-based pulldown of His-tagged ubiquitin under denaturing conditions allows only for the pulldown of proteins that are covalently modified by ubiquitin (or unbound, free ubiquitin). To examine the K48-only ubiquitylation, HA-tagged WT or K260R mutant Sec13 were transfected into HEK293T cells together with the K48-only His-Ub, as described42. At 36 h after transfection, cells were treated with MG132 for 6 h and lysed in CHAPS buffer, followed by immunoprecipitation of Sec13-HA with anti-HA antibody. The immunoprecipitated proteins were assessed by immunoblot analysis for His-tagged Ub. To detect whether TCR triggered ubiquitylation of Sec13, T cells pre-activated with overnight TCR stimulation were retrovirally transduced with HA-tagged WT or K260R mutant Sec13, stimulated with α-CD3/CD28 for a total of 48 or 72 h (cells were treated with MG132 for the last 6 h of stimulation), followed by immunoprecipitation with anti-HA antibody and immunoblot analysis with an antibody against Ubiquitin.
Treg suppression and proliferation assays
In vivo Treg suppression assay
sgNTC-, sgSec31a-, and sgRptor-transduced induced Treg cells (CD45.1+; 0.5 × 106) were sorted and mixed with 2 × 106 conventional CD4+ T cells (CD45.2+), and transferred into Rag1−/− mice at a 1:4 ratio, as described43. At 7 d after transfer, recipients were euthanized for flow cytometry analysis to quantify the accumulation of conventional T cells in the spleen.
In vivo Treg cell proliferation
induced Treg cells transduced with retrovirus were sorted and labelled with CTV at 37 °C for 15 min, followed by 3× washes with complete Click’s medium and 1× wash with PBS. 2 × 106 cells were transferred into immunodeficient Rag1−/− mice, as described18. At 7 d after transfer, recipient mice were euthanized to assess the CTV dilution in Treg cells by flow cytometry analysis.
Protein extraction and immunoblot analysis
Cells were lysed in RIPA buffer (89900; ThermoFisher). Lysates were mixed with 5× Reducing Sample Buffer and resolved in 4–12% Criterion™ XT Bis-Tris Protein Gel (3450125; Bio-Rad) and transferred to PVDF membrane (1620177; Bio-Rad). Membranes were blocked using 5% BSA for 1 h and then incubated for overnight with α-p-S6 S235/236 (1: 5,000; 4858), α-S6 (1: 5,000; 2217), α-p-S6K1 T389 (1: 1,000; 9205), α-S6K1 (1: 1,000; 2708), α-Mios (1: 1,000; 13557), α-Skp1 (1: 1,000; 12248), α-Tsc2 (1: 1,000; 4308), α-β-Actin (1: 5,000; 3700), α-p53 (1: 1,000; 2524), α-HA (1: 5,000; 3724), α-β-Tubulin (1: 1,000; 2128), α-Wdr59 (1: 1,000; 53385), α-Sec23a (1: 1,000; 8162) (all from Cell Signaling Technology); α-Glut1 (1: 1,000; ab652), α-Seh1l (1: 5,000; ab218531) (from Abcam); α-Hsp90 (1: 10,000; 13171-1-AP), α-Sec31a (1: 3,000; 17913-1-AP), α-Ccdc101 (1: 1,000; 24061-1-AP), α-Wdr24 (1: 3,000; 20778-1-AP), α-His-tag (1: 1,000; 66005-1-Ig) (all from ProteinTech); α-Sec13 (1: 3,000; sc-514308), α-Sp3 (1: 1,000; sc-28305), α-Smarcb1 (1: 1,000; sc-166165), α-Castor1 (1: 1,000; sc-377385) and α-Ubiquitin (1: 1,000; sc-8017) (all from Santa Cruz). Membranes were washed 6 times with TBST and then incubated with 1:5,000 diluted HRP-conjugated α-mouse IgG (1: 10,000; W4021; Promega), HRP-conjugated α-rabbit IgG (1: 10,000; W4011; Promega), HRP-conjugated mouse α-rabbit IgG (conformation specific) (1: 3,000; 5127; Cell Signaling Technology) or HRP-conjugated α-mouse IgG (light chain specific) for immunoblotting after immunoprecipitation (1: 2,000; 115-035-174; Jackson Immuno Research) for 2 h. Following another 6 times of washes with TBST, the membranes were imaged using the ODYSSEY Fc Analyzer (LI-COR). To measure protein stability via cycloheximide (CHX) treatment, transfected HEK293T cells were treated with 30 μg/ml CHX for 0, 3, 6 or 9 h to block protein synthesis. Cells were then harvested for immunoblot analysis to examine the expression level of relevant proteins.
Immunofluorescence imaging
sgRNA-transduced cells were sorted and stimulated with surface-bound α-CD3 antibody using Ibidi chamber cover glasses (80426; Ibidi) in complete media for 3 h at 37 °C, followed by amino acid starvation for 40 min and full amino acid refeed for 20 min. Select lanes were washed 5× with amino acid free media, followed by amino acid starvation for 40 min at 37 °C. Selected lanes were subjected to refeeding with amino acid replete media for an additional 30 min at which time all samples were fixed with 4% paraformaldehyde in PBS. Cells were washed and permeabilized with 0.1% Triton X-100 for 3 min, and blocked with 1% BSA in PBS for 10 min at room temperature. Cells were subsequently stained with rabbit α-mTOR (7C10; Cell Signaling Technologies) and rat α-LAMP1 (1D4B; eBiosciences) antibodies overnight at 4 °C, followed by detection with AlexaFluor 555-conjugated donkey α-rabbit (A32794; ThermoFisher) and AlexaFluor 647-conjugated donkey α-rat (712-605-153; Jackson ImmunoResearch) secondary antibodies, while F-actin was detected with AlexaFluor 488-conjugated phalloidin (A12379; ThermoFisher). Samples were visualized using a Marianas confocal microscope (Intelligent Imaging Innovations) equipped with a CSU-W (Yokogawa) spinning disk to facilitate super resolution via optical reassignment (SoRA) imaging, in combination with a 100× 1.45 NA oil objective and Prime 95B camera detection. Images were subsequently quantified using Slidebook software (Intelligent Imaging Innovations), n > 700 cells per condition.
RNA isolation and real-time PCR
sgRNA-transduced cells (2 × 105) were sorted and RNA was isolated using the RNeasy Micro Kit (74004; Qiagen) following the manufacturer’s instructions. RNA was converted to cDNA using the High-Capacity cDNA Reverse Transcription Kit (4368813; ThermoFisher) according to manufacturer’s instructions. Real-time PCR was performed on the QuantStudio 7 Flex System (Applied Biosystems) using the PowerSYBR Green PCR Master Mix (4367659; ThermoFisher) and the following primers: Slc2a1-F: CAGTTCGGCTATAACACTGGTG, Slc2a1-R: GCCCCCGACAGAGAAGATG; Slc43a1-F: CTGAGAACAGGACCAATACCAC, Slc43a1-R: CAAGGCCATTAGAGTGCAGGA; Slc16a10-F: AGGTGCTCTTCATGTGCATTG, Slc16a10-R: TGGAGGTAGACCTTCTTCACAC; Castor1-F: CAGAACCGCTTTTGTGTCCTCAC, Castor1-R: GGAGAAAGCGAAGAACGGAATGG; Sec13-F: GAACACTGTGGACACCTCTCA, Sec13-R: CTCCATTCCGCACATCGAAAA.
Site-directed mutagenesis
We individually mutated all of the 16 lysine residues in Sec13 to arginine and ectopically expressed each of the lysine mutants of Sec13 (with the C-terminal HA tag, Sec13-HA) in HEK293T cells to screen for the lysine residue(s) that alter Sec13 expression or degradation. Site-Directed Mutagenesis of Sec13 was performed using the QuikChange XL Site-Directed Mutagenesis Kit (ThermoFisher), according to the manufacturer’s instructions using the primers listed in Supplementary Table 7.
Seahorse metabolic assay
Extracellular acidification rate (ECAR) was measured by following the manufacturer’s instructions of Seahorse XF Cell Mito Stress Test Kit (Agilent). In brief, 2.5 × 105 transduced induced Treg cells were stimulated with α-CD3/CD28 antibodies for 20 h and suspended in XF medium and then plated in a poly-L-lysine-coated XF96 plate. The ECAR under baseline condition or in response to 1 μM oligomycin, 1.5 μM fluoro-carbonyl cyanide phenylhydrazone (FCCP) and 500 nM rotenone was measured using an XF96 Extracellular Flux Analyzer (Seahorse Bioscience).
Affinity purification–mass spectrometry (AP–MS) and data analysis
AP–MS analysis combining tandem mass tag (TMT) labelling and two-dimensional liquid chromatography-tandem mass spectrometry was carried out as described previously to enable highly sensitive and unbiased analysis of PPI networks5,14. Briefly, immunoprecipitated proteins were digested by trypsin (V5113; Promega) and the resulting tryptic peptides were labelled by TMT pro reagents (A44520; ThermoScientific) and pooled together following the manufacturer’s instructions. Mixed TMT-labelled peptides were loaded on basic pH reversed phase LC for fractionation44. Each fraction was dried and reconstituted for online LC-MS/MS analysis using an optimized platform45. Protein database search, quantification and computational analyses were performed using the JUMP46,47 software (Version 1.13.003) suite as previously described48. In brief, spectra were searched against the Uniprot mouse database (52,490 protein entries; downloaded in February 2015) and filtered to achieve 1% FDR at unique protein (for whole proteome) level. To minimize redundancy, protein identifications from shared peptide sequences were grouped into unique proteins according to the principal of parsimony. TMT intensities were extracted, filtered, normalized and summarized into peptide and protein quantification. Contaminate proteins from the CRAPome database that are commonly detected by IP-MS were removed49. For Sec13 interactome analysis, TMT intensities were log2 transformed. Student’s t-test was used to identify DE events between HA-Sec13 versus empty vector control by the threshold of P value < 0.05, ∣log2 FC∣ > 1. Volcano plot depicting log2 FC and –log10 (P value) was plotted and top 20 enriched proteins by log2 FC were highlighted.
Protein–protein interaction (PPI) network analysis
The analysis was performed using our in-house JUMPn software (Version 0.19.006) as previously described8,9,44. Briefly, the 346 mTORC1 regulatory candidates were superimposed onto a composite PPI database [combines STRING (v10)50, BioPlex51, and InWeb_IM52] with edge confidence scores filtered by best fitting the scale-free network property53. The PPI network was then visualized by Cytoscape V3.7.254. Main protein modules were then identified by MCODE algorithm55 in Cytoscape app clusterMaker256 and annotated by core enriched categories.
Histology
Mouse tissues were fixed by 10% (vol/vol) neutral buffered formalin solution, embedded in paraffin, sectioned and stained with hematoxylin and eosin; the clinical signs of inflammation were analyzed by an experienced pathologist (P. Vogel)40.
Statistical analysis for biological experiments
For biological experiment (non-omics) analyses, data were analyzed using Prism 8 software (GraphPad) by two-tailed unpaired Student’s t-test, Two-sided Wilcoxon rank sum test, one-way ANOVA with Newman-Keuls’s test, or two-way ANOVA. P < 0.05 was considered significant. Data are presented as mean ± s.e.m.
Extended Data
Extended Data Figure 1 (to Figure 1). Two rounds of pooled CRISPR screening to identify novel regulators of nutrient and mTORC1 signaling.
(a) Flow cytometry analysis of IFN-γ, IL-4, IL-17A or Foxp3 expression in cells cultured in TH0-, TH1-, TH2-, TH17- or induced Treg-polarizing condition (n = 3 samples each group). (b) Flow cytometry analysis of p-S6 in TH1 and Treg cells with TCR stimulation for 0 or 1 h (n = 3 samples each group). (c) Induced Treg cells were stimulated with α-CD3/CD28 antibodies in the presence or absence of amino acids (AA) or glucose for 3 h followed by flow cytometry analysis and quantification of p-S6 level [based on mean fluorescence intensity (MFI)] (n = 3 samples each group). (d) Induced Treg cells were labelled with CellTrace Violet (CTV) and stimulated with α-CD3/CD28 antibodies in the presence or absence of AA or glucose for 3 d, followed by flow cytometry analysis of CTV dilution (n = 3 samples each group). (e) Gating strategy used for sorting cells with the ≥ 10% highest (p-S6hi) and ≤ 10% lowest (p-S6lo) levels after stimulation with 0.25 or 4 μg/ml of α-CD3 antibody for 3 h (n = 2 samples each group). Mean ± s.e.m. (c). ***P < 0.001; one-way ANOVA (c). Data are representative of two (b–d) or three (a) experiments.
Extended Data Figure 2 (to Figure 1). Validation of individual candidate mTORC1 regulators.
(a) Flow cytometry analysis and quantification of p-S6 level [based on mean fluorescence intensity (MFI)] in naïve or activated WT and Depdc5-deficient CD4+Foxp3− T cells (n = 4 samples each group). Naïve CD4+ T cells among freshly isolated splenocytes from WT and Cd4CreDepdc5fl/fl mice were gated (indicated as TCR 0 h), or naïve CD4+ T cells were sorted and stimulated with α-CD3/CD28 antibodies overnight for flow cytometry analysis of p-S6 level. (b) Quantification of relative p-S6 level in induced Treg cells transduced with sgNTC, sgSec13, sgMios, sgSeh1l, or sgWdr24 (all Ametrine+) were stimulated with TCR for 3 h (n = 3 samples each group). (c) Diagram of dual-color co-culture system to examine cell-intrinsic effects of deletion of a candidate gene on TCR-induced p-S6, cell size and CD71 expression. Specifically, Cas9+ cells transduced with sgNTC (mCherry+ or GFP+; ‘spike’) were mixed with those transduced with those targeting a specific gene (Ametrine+), and stimulated with α-CD3 antibody for 3 h (for p-S6) or with α-CD3/CD28 antibodies for 20 h (for cell size and CD71). (d) Validation of dual-color co-culture system by using two sgNTC-expressing vectors with different fluorophores. Cells transduced with sgNTC (GFP+; ‘spike’) were mixed with those transduced with sgNTC (Ametrine+), and stimulated with α-CD3 antibody for 3 h to examine p-S6 (see phos-flow staining), or stimulated with α-CD3/CD28 antibodies for 20 h (n = 3 samples each group) to measure cell size and CD71 expression (see surface staining). (e) Heatmap summary of log2 FC (p-S6hi/p-S6lo) for individually validated candidate genes (63 positive and 21 negative regulators) including positive (Rheb, Rptor, Lamtor3, Rraga and Mtor) and negative (Cd5, Nprl3, Nprl2 and Tsc1) control genes (2 sgRNAs for each candidate). Specifically, Cas9-expressing CD4+ T cells transduced with sgRNA for target genes (Ametrine+) or non-targeting control sgRNA (sgNTC) (mCherry+; ‘spike’) were mixed and differentiated into induced Treg cells. These cells were then stimulated with α-CD3 antibody for 3 h (n = 3 samples each group). Relative p-S6 level (normalized to ‘spike’) was analyzed by flow cytometry. (f) Analysis of protein–protein interaction (PPI) networks of high-confidence regulators. Specifically, 286 positive and 60 negative high-confidence hits were integrated with the composite PPI databases that encompass STRING, BioPlex and InWeb_IM databases for the inference of functional modules. Red and blue circles represent genes whose deletion represses and promotes mTORC1 activity, respectively. Mean ± s.e.m. (a, b). *P < 0.05; **P < 0.01; ***P < 0.001; one-way ANOVA (a, b). Data are representative of one (f) or three (d, e), or pooled from two (a) or three (b) experiments.
Extended Data Figure 3 (to Figure 1). SWI/SNF complex represses expression of nutrient sensor Castor1 to support mTORC1 activation.
(a) Quantification (normalized to ‘spike’) of relative p-S6 level, cell size (FSC-A) and CD71 expression in induced Treg cells transduced with indicated sgRNAs followed by stimulation with α-CD3 antibody for 3 h to measure p-S6 level, or with α-CD3/CD28 antibodies for 20 h to measure cell size (FSC-A) and CD71 expression by flow cytometry (n = 3 samples each group). (b) Imaging analysis and quantification of lysosome-associated mTOR [based on mean fluorescence intensity (MFI)] in sgNTC- or sgSmarcb1-transduced cells that were stimulated with α-CD3 antibody for 3 h, starved of amino acids (AA), and refed AA for 20 min (n > 230 cells per condition). (c) Volcano plots of expression levels of transcripts, including Castor1, in sgNTC or sgSmarcb1 (both Ametrine+)-transduced cells that were stimulated with CR for 3 h (n = 4 samples each group). (d) Castor1 mRNA expression in sgNTC- or sgSmarcb1 (both Ametrine+)-transduced cells were stimulated with α-CD3 antibody for 3 h or with α-CD3/CD28 antibodies for 20 h (n = 3 samples per group). (e) sgNTC or sgSmarcb1 (both Ametrine+)-transduced cells were left unstimulated (indicated by 0 h) or stimulated with α-CD3 antibody for 3 h or α-CD3/CD28 antibodies for 20 h. Immunoblot analysis and quantification of relative Castor1 expression (n = 3 samples each group). (f) Immunoblot analysis and quantification of relative p-S6K1 and p-S6 levels in cells transduced with empty vector or vector expressing Castor1, followed by stimulation with α-CD3/CD28 antibodies for 2 d (n = 3 samples each group). Mean ± s.e.m. (a, b, d–f). **P < 0.01; ***P < 0.001; two-tailed unpaired Student’s t-test (f); one-way ANOVA (a, b, d, e). Data are representative of one (c) or two (b), or pooled from two (d, e) or three (a, f) experiments.
Extended Data Figure 4 (to Figure 2). Sec31a is required for mTORC1 activation.
(a) Interaction of endogenous Sec13 with Sec31a in induced Treg cells as assessed by immunoprecipitation (IP)–immunoblot analysis. (b, c) sgNTC- or sgSec31a-transduced cells were starved of and refed amino acids (AA, b) or glucose (c) for 20 min, followed by immunoblot analysis of Sec31a, p-S6K1, p-S6 and β-Actin. Lower, quantification of relative p-S6K1 and p-S6 levels (n = 3 samples each group). (d) Cells transduced with the indicated sgRNAs (all Ametrine+) were mixed with sgNTC (mCherry+; ‘spike’)-transduced cells, and stimulated with α-CD3 antibody for 3 h to measure p-S6 level or with α-CD3/CD28 antibodies for 20 h to measure cell size (FSC-A) and CD71 expression by flow cytometry (normalized to ‘spike’) (n = 3 samples each group). (e–g) sgNTC-, sgSec31a- or sgSec13 (all Ametrine+)-transduced cells were co-transduced with constitutively active RagAQ66L (CA-RagA)-expressing retrovirus (GFP+) or sgNprl2 (GFP+), followed by stimulated with TCR for 3 h to examine relative p-S6 level by flow cytometry (n = 3 samples each group) (e), p-S6K1 and p-S6 levels by immunoblot analysis (n = 4 samples each group) (f), or lysosomal localization of mTOR [based on mean fluorescence intensity (MFI)] (n > 700 cells per condition) (g). In f, two different exposures for p-S6K1 were included to account for the differential intensities between mock and CA-RagA or sgNprl2 conditions, and relative p-S6K1 level was quantified from long exposure for mock and short exposure for CA-RagA or sgNprl2 conditions (middle). Lower, quantification of p-S6 level. Mean ± s.e.m. (b–g). NS, not significant; **P < 0.01; ***P < 0.001; one-way ANOVA (b–g). Data are representative of two (e–g) or four (a), or pooled from three (b–d) experiments.
Extended Data Figure 5 (to Figure 2). Sec31a–Sec13 axis promotes mTORC1 activation and cell proliferation in vivo.
(a) Cells transduced with sgNTC or sgSec31a (both Ametrine+) were labelled with CellTrace Violet (CTV) and transferred into Rag1−/− mice. Flow cytometry analysis of CTV dilution, and quantification of percentage of proliferated (CTVlo) cells at 7 d after transfer (n = 5 samples each group). (b) WT, Raptor- and Sec31a-null Treg cells (all CD45.1+Ametrine+) were mixed with conventional CD4+ T cells (Tconv; CD45.2+) at a 1:4 ratio and transferred into Rag1−/− mice. Quantification of the accumulation of conventional T cells in the spleen at 7 d after transfer (n = 5 samples each group). (c) Naïve CD4+ T cells were stimulated with α-CD3/CD28 antibodies for 0, 24, 48 or 72 h followed by immunoblot analysis of the indicated protein expression, and quantification of p-S6K1, Sec13, Sec31a, and Tsc2 (n = 3 samples each group). (d) sgNTC-, sgSec31a- and sgSec13-transduced cells were sorted and lysed with CHAPS buffer for immunoprecipitation (IP) with an antibody against Wdr24. The immunoprecipitated proteins were analyzed by immunoblot for Wdr24, Wdr59, Mios, Seh1l, Sec13, Sec31a, Sec23a and β-Actin. Mean ± s.e.m. (a–c). **P < 0.01; ***P < 0.001; two-tailed unpaired Student’s t-test (a); one-way ANOVA (b). Data are representative of one (a, b) or two (c, d) experiments.
Extended Data Figure 6 (to Figure 3). Sec31a protects Sec13 from proteasomal degradation to sustain mTORC1 activation.
(a) Sec13 mRNA expression in sgNTC- or sgSec31a-transduced induced Treg cells (n = 3 samples each group). (b) sgNTC- or sgSec31a-transduced induced Treg cells were sorted and treated with cycloheximide (CHX) for the indicated times. Total protein extracts of cells transduced with sgNTC (5 μg) or sgSec31a (12.5 μg; more protein was loaded to equalize basal Sec13 amount between these cells) were used for immunoblot analysis and quantification of relative Sec13 abundance (n = 4 samples each group). (c) Naïve CD4+ T cells were stimulated with α-CD3/CD28 antibodies for 0, 24, or 48 h and then treated with DMSO or MG132 at 48–72 h of stimulation, followed by immunoblot analysis and quantification of Sec13 and Sec31a expression (n = 3 samples each group). (d) HEK293T cells were transfected with HA-tagged Sec13 and 6× His-tagged WT-, K48R- or K63R-ubiquitin and treated with MG132 for 6 h. Ni-nitrilotriacetic acid (Ni-NTA) bead-based pulldown and immunoblot analysis of HA-Sec13. Lower, expression of indicated proteins in whole cell lysates (WCL). (e) sgNTC- or sgSec31a-transduced HEK293T cells were transfected with HA-tagged Sec13 and 6× His-tagged WT ubiquitin (His-tagged Ub), and treated with MG132 for 6 h. Left, Ni-NTA bead-based pulldown of His-tagged Ub-labelled proteins followed by immunoblot analysis for HA-Sec13. Right, immunoblot analysis of WCL for expression of endogenous Sec31a or HA-Sec13, His-tagged Ub, and β-Actin. Mean ± s.e.m. (a–c). NS, not significant; **P < 0.01; ***P < 0.001; two-tailed unpaired Student’s t-test (a); two-way ANOVA (b); one-way ANOVA (c). Data are representative of two (d, e) or three (a), or pooled from two (b) or three (c) experiments.
Extended Data Figure 7 (to Figure 3). Sec31a protects Sec13 from Skp1-mediated proteasomal degradation and supports T cell functional fitness.
(a) HA-tagged WT or the indicated lysine mutant constructs of Sec13 were transfected into HEK293T cells individually. Immunoblot analysis of HA and Hsp90. (b) HA-tagged WT or K260R mutant Sec13 was transfected into HEK293T cells together with the K48-only His-Ub followed by MG132 treatment and anti-HA immunoprecipitation (IP). Immunoblot analysis for HA, His-Ub and β-Actin. WCL, whole cell lysate. (c) Cas9-expressing CD4+ T cells were transduced with sgNTC or sgSec31a retrovirus (Ametrine+) together with WT or K260R mutant Sec13-expressing retrovirus (GFP+). Ametrine+GFPlo cells (see gating on flow cytometry plot, upper) were stimulated with TCR for 0 or 3 h. Immunoblot analysis and quantification of relative Sec13 and p-S6 levels (n = 4 samples each group). (d) Volcano plot of proteins, including Skp1, that interact with HA-Sec13 in induced Treg cells as identified by mass spectrometry (n = 3 samples each group). (e) Induced Treg cells transduced with HA-tagged-Sec13- or empty vector-expressing retrovirus were lysed with CHAPS buffer followed by α-HA immunoprecipitation (IP) and immunoblot analysis of HA and Skp1. (f) Induced Treg cells were lysed with CHAPS buffer followed by immunoprecipitation of endogenous Skp1 and immunoblot analysis of Skp1 and Sec13. (g) Interaction of endogenous Skp1 with Sec13 in sgNTC- or sgSec31a-transduced cells. (h) Naïve CD4+ T cells were stimulated with α-CD3/CD28 antibodies for 0, 24, 48 or 72 h. Immunoblot analysis of anti-Skp1 immunoprecipitants and WCL for Skp1, Sec13, and β-Actin (n = 2 samples per group). (i) Immunoblot analysis and quantification of relative expression of indicated proteins in sgNTC- or sgSkp1 (both Ametrine+)-transduced cells (n = 3 samples per group). (j) Indicated sgRNA-transduced cells were labelled with CellTrace Violet (CTV), and stimulated with α-CD3/CD28 antibodies for 72 h, followed by flow cytometry analysis and quantification of CTV dilution (n = 3 samples each group). (k) Diagram of SMARTA T cell transfer and LCMV infection system. Briefly, SMARTA–Cas9 CD4+ T cells (CD45.1+) transduced with sgRNA for candidate genes (CD45.1+Ametrine+) and mixed with sgNTC (CD45.1+mCherry+; ‘spike’)-transduced cells at a 1:1 ratio, and adoptively transferred into naïve (unchallenged; CD45.2+) mice that were left uninfected (see l) or challenged with LCMV infection (see Fig. 3f). (l) Quantification of the relative proportion (normalized to ‘spike’) of donor-derived (CD45.1+) T cells in the spleen of uninfected mice at 7 d after transfer (n =6 mice per group). (m) Cells transduced with sgNTC (Ametrine+), sgSec31a (Ametrine+) or sgSec31a-Skp1 (GFP+ and Ametrine+) were sorted and stimulated with α-CD3/CD28 antibodies for 20 h (n = 6-7 samples per group), followed by the measurement of extracellular acidification rate (ECAR). Oligo, oligomycin; FCCP, fluoro-carbonyl cyanide phenylhydrazone; Rot, rotenone. Mean ± s.e.m. (c, i, j, l, m). **P < 0.01; ***P < 0.001; one-way ANOVA (c, i, j, l, m, right); two-way ANOVA (m, left). Data are representative of one (d, j, l, m) or two (a, b, e–h), or pooled from two (c) or three (i) experiments.
Extended Data Figure 8 (to Figure 4). SAGA complex suppresses mTORC1 activation.
(a) Quantification (normalized to ‘spike’) of relative p-S6 level, cell size (FSC-A) and CD71 expression in induced Treg cells transduced with sgRNA for Ccdc101 or Taf6l, followed by stimulation with α-CD3 antibody for 3 h to measure p-S6 level (left) or with α-CD3/CD28 antibodies for 20 h to measure cell size (FSC-A; middle) and CD71 (right) expression by flow cytometry (n = 3 samples each group). (b) Immunoblot analysis and quantification of relative p-S6K1 and p-S6 expression in sgNTC- or sgCcdc101-transduced cells that were starved of and refed amino acids (AA) for 20 min (n = 4 samples each group). Mean ± s.e.m. (a, b). *P < 0.05; **P < 0.01; ***P < 0.001; one-way ANOVA (a, b). Data are representative of three (a), or pooled from three (b) experiments.
Extended Data Figure 9 (to Figure 4). SAGA complex represses the expression of nutrient transporters and mTORC1 activation.
(a) Heatmap of differentially expressed genes in Ccdc101-null Treg cells stimulated with α-CD3/CD28 antibodies for 0 (n = 3 samples each group) or 20 h (n = 4 samples each group). (b) Slc2a1, Slc16a10 or Slc43a1 mRNA expression in sgNTC- or sgCcdc101-transduced cells at steady state (n = 3 samples each group). (c) Immunoblot analysis and quantification of relative Glut1 expression in sgNTC- or sgCcdc101-transduced cells that were stimulated with α-CD3/CD28 antibodies for 0 or 20 h (n = 3 samples each group). (d) Flow cytometry analysis and quantification of 2-NBDG uptake in sgNTC or sgCcdc101 (both Ametrine+)-transduced cells were stimulated with α-CD3/CD28 antibodies for 20 h (n = 4 samples each group). (e, f) Quantification of relative p-S6 level in cells transduced with the indicated sgRNAs that were stimulated with TCR for 3 h (n = 3 samples each group). (g) Principal component analysis (PCA) of ATAC-seq for cells transduced with sgNTC (n = 4 samples) or sgCcdc101 (both Ametrine+) (n = 3 samples) and stimulated with α-CD3/CD28 antibodies for 20 h. (h) Motif enrichment analysis of ATAC-seq of sgNTC- and sgCcdc101-transduced cells (n = 4 samples each group). (i) Footprinting analysis of Sp3 binding in ATAC-seq. (j) Accessibility of the Sp3 locus in sgNTC- and sgCcdc101-transduced cells as identified by ATAC-seq. Highlighted peaks in the red box indicate differential accessible regions. (k) Immunoblot analysis of Sp3 expression in sgNTC- or sgCcdc101 (both Ametrine+)-transduced cells. Mean ± s.e.m. (b–f). ***P < 0.001; two-tailed unpaired Student’s t-test (b, d); one-way ANOVA (c, e, f). Data are representative of one (a, g–j), two (b, e, f, k), or pooled from two (c, d) experiments.
Extended Data Figure 10 (to Figure 4). SAGA complex prevents mTORC1 hyperactivation to enforce immune homeostasis in vivo.
(a, b) Induced Treg cells transduced with indicated sgRNAs were stimulated with α-CD3/CD28 antibodies for 20 h (n = 3 samples each group). Flow cytometry analysis and quantification of staining with active caspase-3 (a) and fixable viability dye (FVD, b) (n = 3 per group). (c) Cells transduced with indicated sgRNAs were stimulated with α-CD3/CD28 antibodies for 20 h (n = 5–7 samples per group), followed by the measurement of extracellular acidification rate (ECAR). Oligo, oligomycin; FCCP, fluoro-carbonyl cyanide phenylhydrazone; Rot, rotenone. (d) Immunoblot analysis and quantification of Ccdc101 expression in naïve CD4+ T cells from WT and Cd4CreCcdc101fl/fl mice (n = 3 mice per group). (e) Quantification of CD71 expression on naïve or activated WT and Ccdc101-deficient CD4+ T cells. Naïve CD4+ T cells among freshly isolated splenocytes from WT and Cd4CreCcdc101fl/fl mice (n = 4 mice per group) were gated (indicated as 0 h), or naïve CD4+ T cells were stimulated with α-CD3/CD28 antibodies for 20 h. (f) Flow cytometry analysis and quantification of numbers of total, double-negative (DN), double-positive (DP), CD4 single-positive (CD4SP), and CD8 single-positive (CD8SP) thymocytes from WT and Cd4CreCcdc101fl/fl mice (n = 4 mice each group). (g) Flow cytometry analysis and quantification of proportions and numbers of splenic CD4+ and CD8+ T cells from WT and Cd4CreCcdc101fl/fl mice (n = 4 mice each group). (h) Flow cytometry analysis and normalized ratio of CD122+ versus CD122− cells among CD44hi populations (gated on splenic CD8+ T cells) from indicated mice (n = 4 mice each group). Mean ± s.e.m. (a–h). NS, not significant; **P < 0.01; ***P < 0.001; two-tailed unpaired Student’s t-test (d–h); one-way ANOVA (a, b, c, right); two-way ANOVA (c, left). Data are representative of one (c) or two (a, b), or pooled from three (d–h) experiments.
Extended Data Figure 11 (to Figure 5). Treg-specific deletion of Ccdc101 disrupts immune homeostasis and boosts antitumour response.
(a) Quantification of relative p-S6 level in splenic CD4+Foxp3+ cells from WT and Foxp3CreCcdc101fl/fl (~8 weeks old) mice (n = 4 mice per group). (b) Quantification of relative Foxp3 expression (gated on splenic Foxp3+CD4+ T cells) from WT and Foxp3CreCcdc101fl/fl mice (n = 4 mice per group). (c) Quantification of percentages of effector/memory (CD44hiCD62Llo) subsets in splenic CD4+Foxp3− and CD8+ T cells from WT and Foxp3CreCcdc101fl/fl (~8 weeks old) mice (n = 4 mice each group). (d) Representative flow cytometry analysis of IL-2+ or IFN-γ+ population of splenic CD4+Foxp3− and CD8+ T cells from WT and Foxp3CreCcdc101fl/fl (~8 weeks old) mice. (e–g) WT and Foxp3CreCcdc101fl/fl mice were inoculated with MC38 colon adenocarcinoma cells. Treg cells (CD45+CD4+YFP+), non-Treg immune cells (CD45+YFP−CD11b−) and myeloid cells (CD45+CD11b+) were isolated and sorted from tumours, and mixed at a 1:2:1 ratio for scRNA-seq analysis (2 biological replicates, pooled from 3-4 mice each, per group) at 19 d after tumour inoculation. Dot plot showing the differentially expressed marker genes for 4 subclusters of CD8+ T cells in the MC38 tumours (e; see Methods for details). UMAP embeddings of CD8+ T cells grouped by genotype (f, left) and indicated subclusters (f, right). Frequencies of the indicated subclusters were quantified for each genotype (g). Teff-like, effector-like CD8+ T cells; Tex-like, exhaustion-like CD8+ T cells; Tcm-like, central memory-like CD8+ T cells; Tem-like, effector/memory-like CD8+ T cells. (h) Flow cytometry analysis and quantification of the percentages of CD44hiCD62Llo intratumoural CD8+ T cells from WT and Foxp3CreCcdc101fl/fl mice (n ≥ 5 per group). (i) Flow cytometry analysis and quantification of IFN-γ+ and TNF-α+ cells among intratumoural CD8+ T cells from WT and Foxp3CreCcdc101fl/fl mice (n ≥ 5 per group). (j) Violin plots of scRNA-seq data depicting Icos, Tnfrsf18, Ctla4 and Ifng expression in intratumoural Treg cells. (k) Flow cytometry analysis and quantification of ICOS, GITR and CTLA-4 for intratumoural Treg cells from WT and Foxp3CreCcdc101fl/fl mice (n ≥ 5 per group). (l) Flow cytometry analysis and quantification of IFN-γ expression in intratumoural Treg cells from WT and Foxp3CreCcdc101fl/fl mice (n ≥ 5 per group). Mean ± s.e.m. (a–c, h, i, k, l). *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed unpaired Student’s t-test (a–c, h, i, k, l); Two-sided Wilcoxon rank sum test in j. Data are representative of one (e–l) or two (d), or pooled from two (a–c) experiments.
Extended Data Figure 12 (to Figure 5). Schematic of applying CRISPR screening and integrative analyses to dissect nutrient and mTORC1 signaling in primary T cells.
With two rounds of genome-wide and focused CRISPR screenings, we identify 346 high-confidence mTORC1 signaling factors, including many novel activators and inhibitors, as well as known regulators (identified in other systems) that have not been studied in primary T cells. Notably, using analysis of protein–protein interaction (PPI) networks and unbiased functional and proteomic approaches, we further establish the epigenetic and posttranslational mechanisms underpinning the three-tier regulatory modules of nutrient signaling, composed of nutrient transporters (e.g. via affecting expression of Glut1 and other transporters by SAGA complex), sensors (e.g. via epigenetic regulation of Castor1 expression by SWI/SNF complex) and transducers (e.g. via shaping GATOR2 complex stability by Sec31a; regulating Sec13 ubiquitination at lysine 260), which transmit immunological and nutrient cues to mTORC1 signaling for proper regulation of T cell activity in vivo and in vitro.
Supplementary Material
Acknowledgements
The authors acknowledge Melissa Hendren and Sherri Rankin for animal colony management, Jing Wen for help with phenotypic studies, Chunliang Li and Sheng Zhou for technical and scientific insights, Geoffrey Neale and Scott Olsen for assistance with sequencing, St. Jude Immunology FACS core facility for cell sorting, and St. Jude CSMCO for artwork. This work was supported by ALSAC and by US National Institutes of Health grants R01AG053987 (to J.P.), AI105887, AI131703, AI140761, AI150241, AI150514, CA250533 and CA253188 (to H.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interests
H. Chi is a consultant for Kumquat Biosciences, Inc.
Supplementary information The online version contains supplementary material available.
Data availability
The authors declare the data supporting the findings of this study are available within the paper and its Supplementary Information files. All microarray, ATAC-seq and scRNA-seq data described in the manuscript have been deposited in the NCBI Gene Expression Omnibus (GEO) database and are accessible through the GEO SuperSeries accession number: 160598 (accessible link, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160598). Source data provided with this paper. CRAPome database (https://reprint-apms.org/); Uniprot mouse database (https://www.uniprot.org/); STRING (v10) (https://string-db.org/); BioPlex (https://bioplex.hms.harvard.edu/).
References
- 1.Chapman NM, Boothby MR & Chi H Metabolic coordination of T cell quiescence and activation. Nat Rev Immunol 20, 55–70, doi: 10.1038/s41577-019-0203-y (2020). [DOI] [PubMed] [Google Scholar]
- 2.Kim J & Guan KL mTOR as a central hub of nutrient signalling and cell growth. Nat Cell Biol 21, 63–71, doi: 10.1038/s41556-018-0205-1 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Liu GY & Sabatini DM mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 21, 183–203, doi: 10.1038/s41580-019-0199-y (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang H, Long L, Zhou P, Chapman NM & Chi H mTOR signaling at the crossroads of environmental signals and T-cell fate decisions. Immunol Rev 295, 15–38, doi: 10.1111/imr.12845 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi H et al. Amino Acids License Kinase mTORC1 Activity and Treg Cell Function via Small G Proteins Rag and Rheb. Immunity 51, 1012–1027 e1017, doi: 10.1016/j.immuni.2019.10.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34, 184–191, doi: 10.1038/nbt.3437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang K et al. T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity 39, 1043–1056, doi: 10.1016/j.immuni.2013.09.015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan H et al. Integrative Proteomics and Phosphoproteomics Profiling Reveals Dynamic Signaling Networks and Bioenergetics Pathways Underlying T Cell Activation. Immunity 46, 488–503, doi: 10.1016/j.immuni.2017.02.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai B et al. Deep Multilayer Brain Proteomics Identifies Molecular Networks in Alzheimer's Disease Progression. Neuron 105, 975–991 e977, doi: 10.1016/j.neuron.2019.12.015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loo CS et al. A Genome-wide CRISPR Screen Reveals a Role for the Non-canonical Nucleosome-Remodeling BAF Complex in Foxp3 Expression and Regulatory T Cell Function. Immunity 53, 143–157 e148, doi: 10.1016/j.immuni.2020.06.011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang BL et al. Mammalian homologues of yeast sec31p. An ubiquitously expressed form is localized to endoplasmic reticulum (ER) exit sites and is essential for ER-Golgi transport. J Biol Chem 275, 13597–13604, doi: 10.1074/jbc.275.18.13597 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Zeng H et al. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature 499, 485–490, doi: 10.1038/nature12297 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P Determining protein half-lives. Methods Mol Biol 284, 67–77, doi: 10.1385/1-59259-816-1:067 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Shi H et al. Hippo Kinases Mst1 and Mst2 Sense and Amplify IL-2R-STAT5 Signaling in Regulatory T Cells to Establish Stable Regulatory Activity. Immunity 49, 899–914 e896, doi: 10.1016/j.immuni.2018.10.010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skaar JR, Pagan JK & Pagano M SCF ubiquitin ligase-targeted therapies. Nat Rev Drug Discov 13, 889–903, doi: 10.1038/nrd4432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortez JT et al. CRISPR screen in regulatory T cells reveals modulators of Foxp3. Nature 582, 416–420, doi: 10.1038/s41586-020-2246-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang K, Neale G, Green DR, He W & Chi H The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol 12, 888–897, doi: 10.1038/ni.2068 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei J et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat Immunol 17, 277–285, doi: 10.1038/ni.3365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakaguchi S et al. Regulatory T Cells and Human Disease. Annu Rev Immunol 38, 541–566, doi: 10.1146/annurev-immunol-042718-041717 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Overacre-Delgoffe AE et al. Interferon-gamma Drives Treg Fragility to Promote Anti-tumor Immunity. Cell 169, 1130–1141 e1111, doi: 10.1016/j.cell.2017.05.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su W et al. Protein Prenylation Drives Discrete Signaling Programs for the Differentiation and Maintenance of Effector Treg Cells. Cell Metab 32, 996–1011 e1017, doi: 10.1016/j.cmet.2020.10.022 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Online References
- 22.Mombaerts P et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877, doi: 10.1016/0092-8674(92)90030-g (1992). [DOI] [PubMed] [Google Scholar]
- 23.Lee PP et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774, doi: 10.1016/s1074-7613(01)00227-8 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Rubtsov YP et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558, doi: 10.1016/j.immuni.2008.02.017 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Oxenius A, Bachmann MF, Zinkernagel RM & Hengartner H Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur J Immunol 28, 390–400, doi: (1998). [DOI] [PubMed] [Google Scholar]
- 26.Platt RJ et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455, doi: 10.1016/j.cell.2014.09.014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei J et al. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature 576, 471–476, doi: 10.1038/s41586-019-1821-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang H et al. In vivo CRISPR screening reveals nutrient signaling processes underpinning CD8(+) T cell fate decisions. Cell 184, 1245–1261 e1221, doi: 10.1016/j.cell.2021.02.021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu G et al. Metabolic control of TFH cells and humoral immunity by phosphatidylethanolamine. Nature 595, 724–729, doi: 10.1038/s41586-021-03692-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng H et al. mTORC1 and mTORC2 Kinase Signaling and Glucose Metabolism Drive Follicular Helper T Cell Differentiation. Immunity 45, 540–554, doi: 10.1016/j.immuni.2016.08.017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen R et al. In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity 41, 325–338, doi: 10.1016/j.immuni.2014.08.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parnas O et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell 162, 675–686, doi: 10.1016/j.cell.2015.06.059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J, Silva J & Califano A ScreenBEAM: a novel meta-analysis algorithm for functional genomics screens via Bayesian hierarchical modeling. Bioinformatics 32, 260–267, doi: 10.1093/bioinformatics/btv556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784, doi: 10.1038/nmeth.3047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joung J et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc 12, 828–863, doi: 10.1038/nprot.2017.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karmaus PWF et al. Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity. Nature 565, 101–105, doi: 10.1038/s41586-018-0806-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218, doi: 10.1038/nmeth.2688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailey TL et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37, W202–208, doi: 10.1093/nar/gkp335 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z et al. Identification of transcription factor binding sites using ATAC-seq. Genome biology 20, 45, doi: 10.1186/s13059-019-1642-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim SA et al. Lipid signalling enforces functional specialization of Treg cells in tumours. Nature 591, 306–311, doi: 10.1038/s41586-021-03235-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim KL et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci 25, 2002–2009, doi: 10.1523/JNEUROSCI.4474-04.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wertz IE et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430, 694–699, doi: 10.1038/nature02794 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Collison LW et al. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature 450, 566–569, doi: 10.1038/nature06306 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Wang H et al. Deep multiomics profiling of brain tumors identifies signaling networks downstream of cancer driver genes. Nat Commun 10, 3718, doi: 10.1038/s41467-019-11661-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H et al. Integrated analysis of ultra-deep proteomes in cortex, cerebrospinal fluid and serum reveals a mitochondrial signature in Alzheimer's disease. Mol Neurodegener 15, 43, doi: 10.1186/s13024-020-00384-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X et al. JUMP: a tag-based database search tool for peptide identification with high sensitivity and accuracy. Mol Cell Proteomics 13, 3663–3673, doi: 10.1074/mcp.O114.039586 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y et al. JUMPg: An Integrative Proteogenomics Pipeline Identifying Unannotated Proteins in Human Brain and Cancer Cells. J Proteome Res 15, 2309–2320, doi: 10.1021/acs.jproteome.6b00344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart E et al. Identification of Therapeutic Targets in Rhabdomyosarcoma through Integrated Genomic, Epigenomic, and Proteomic Analyses. Cancer Cell 34, 411–426 e419, doi: 10.1016/j.ccell.2018.07.012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mellacheruvu D et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods 10, 730–736, doi: 10.1038/nmeth.2557 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szklarczyk D et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–452, doi: 10.1093/nar/gku1003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huttlin EL et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440, doi: 10.1016/j.cell.2015.06.043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li T et al. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat Methods 14, 61–64, doi: 10.1038/nmeth.4083 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barabasi AL & Oltvai ZN Network biology: understanding the cell's functional organization. Nat Rev Genet 5, 101–113, doi: 10.1038/nrg1272 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Shannon P et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504, doi: 10.1101/gr.1239303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bader GD & Hogue CW An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4, 2, doi: 10.1186/1471-2105-4-2 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morris JH et al. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics 12, 436, doi: 10.1186/1471-2105-12-436 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare the data supporting the findings of this study are available within the paper and its Supplementary Information files. All microarray, ATAC-seq and scRNA-seq data described in the manuscript have been deposited in the NCBI Gene Expression Omnibus (GEO) database and are accessible through the GEO SuperSeries accession number: 160598 (accessible link, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160598). Source data provided with this paper. CRAPome database (https://reprint-apms.org/); Uniprot mouse database (https://www.uniprot.org/); STRING (v10) (https://string-db.org/); BioPlex (https://bioplex.hms.harvard.edu/).