Abstract
Infectious diseases affect individual health and have widespread societal impacts. New ex vivo models are critical to understand pathogenesis, host response, and features necessary to develop preventive and therapeutic treatments. Pluripotent and tissue stem cell–derived organoids provide new tools for the study of human infections. Organoid models recapitulate many characteristics of in vivo disease and are providing new insights into human respiratory, gastrointestinal, and neuronal host–microbe interactions. Increasing culture complexity by adding the stroma, interorgan communication, and the microbiome will improve the use of organoids as models for infection. Organoid cultures provide a platform with the capability to improve human health related to infectious diseases.
Keywords: organoid, infection, stem cell, pathogenesis, therapies
INTRODUCTION
The coronavirus disease 2019 (COVID-19) pandemic has put a spotlight on infectious diseases and their potential for disastrous effects on humankind. Viruses like severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as well as bacteria, parasites, and fungi cause more than 10 million deaths worldwide each year, accounting for one-fifth of the overall global total (1). The top 10 causes of death include lower respiratory infections, diarrheal diseases, tuberculosis, HIV/AIDS, and malaria (2). In addition, tropical diseases such as Chagas and schistosomiasis simmer in less-developed areas, and as SARS-CoV-2 has demonstrated, there needs to be constant vigilance for emerging infectious diseases. Antimicrobial-resistant bacteria, such as methicillin-resistant Staphylococcus aureus (MRSA), present additional threats. Infectious diseases take a tremendous toll on individual health and well-being, with other confounding consequences affecting both widespread social and economic impacts (3). Finally, infectious diseases burden the capacity and management of the healthcare system (3). Although the development of highly effective vaccines and antimicrobials has reduced the burden of some infectious diseases, as SARS-CoV-2 has shown us, the threat will never disappear.
The continued presence of infectious pathogens drives the need to understand the mechanisms underlying disease development and progression so as to better design preventive measures, such as vaccines and therapeutic treatments, to combat illness once disease has been established. Both in vitro and in vivo models have been used to establish much of what is known about infectious diseases. Many of the in vitro standards, consisting predominantly of universally used transformed cell lines, do not recapitulate the complex untransformed cellular composition and microenvironment or the disease process that occurs at the whole-organism level. Animal models, due to species-specific differences, may not accurately reflect the physiology of human–pathogen interactions, and many results cannot be extrapolated to the human condition. Such classical approaches lack the ability to replicate human genetic variation or to study human-specific pathogens. Recent advances in stem cell and developmental biology have led to new approaches using cultures derived from human stem cells that bridge the gap between transformed cell lines and animal models by addressing aspects of in vivo complexity, offering an easy means of in vitro manipulation, and illuminating human genetic diversity and biology.
OVERVIEW OF ORGANOID MODELS
Promising new ex vivo models for infectious disease research are human stem cell–derived organoid cultures. Derived from either pluripotent or tissue-derived stem cells (Figure 1) and grown within a variety of extracellular matrices, such as Matrigel®, organoid cultures develop and self-assemble into three-dimensional structures that in some cases contain an underlying mesenchymal component. Mitogens, morphogens, and cytokines such as WNT are added to the surrounding media to promote growth. The cultures are genetically stable, can be propagated indefinitely, and can be frozen for storage in much the same manner as immortalized cells, thus providing ease of use, storage, and transfer. In contrast, other, nontransformed human ex vivo models, such as primary cells or tissue explants, have a finite replication capacity and rapidly senesce (i.e., reach the Hayflick limit). Manipulation of growth factors in the culture media induces further development of organoids from stem cells to differentiated cells that mimic functional epithelium. Initially, this three-dimensional structure limited access to the apical epithelial surface, where most pathogens initially attack, to labor-intensive microinjection (Figure 2). This technical challenge can be surmounted by using a two-dimensional monolayer organoid culture system (4) or by inverting the cell polarity of the three-dimensional organoids (5). This epithelial tissue recapitulates many aspects of the cellular heterogeneity and physiology of the native organ, such as cellular composition, region-specific features, and functional and physiological properties including circadian rhythm and polarization. Most importantly, organoids provide the means to investigate the role of individual genetic variability, including demographic factors such as age, sex, and ethnicity, at the cellular level. In this way, organoid cultures are a gateway to the development of personalized approaches to understanding infectious diseases.
Figure 1.
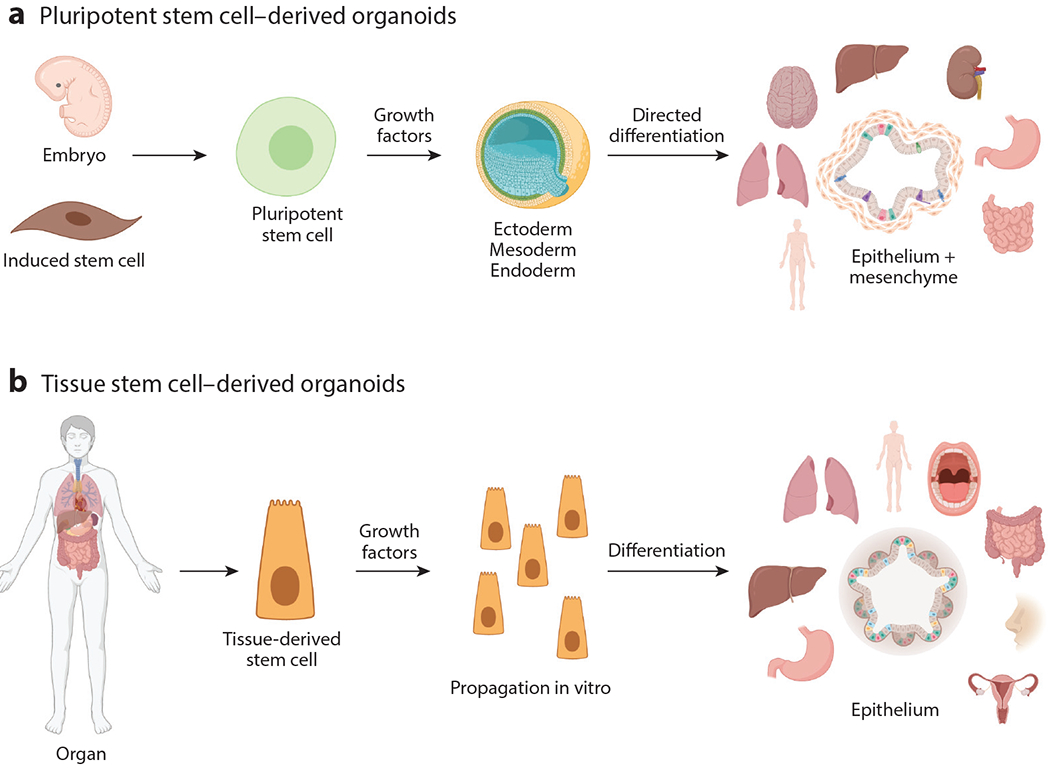
Establishment of pluripotent and tissue stem cell–derived organoids. (a) Pluripotent stem cell–derived organoids originate from embryonic stem cells or terminally differentiated cells that are induced or dedifferentiated synthetically to become stem cells. These stem cells are propagated with growth factors and respond to signals that result in the development of ectoderm, mesoderm, and endoderm. Further growth signals direct differentiation into organ-specific cells. In these organoids the mesenchymal component develops along with the epithelium. (b) Tissue stem cell–derived organoids originate from stem cells that reside in mature organs. Upon isolation and ex vivo culture with defined growth factors, the stem cells will divide indefinitely and, upon manipulation of the growth factors, will differentiate into organoids that consist of the epithelial layer of the tissue from which the stem cells were derived. Figure adapted from image created with BioRender.com.
Figure 2.
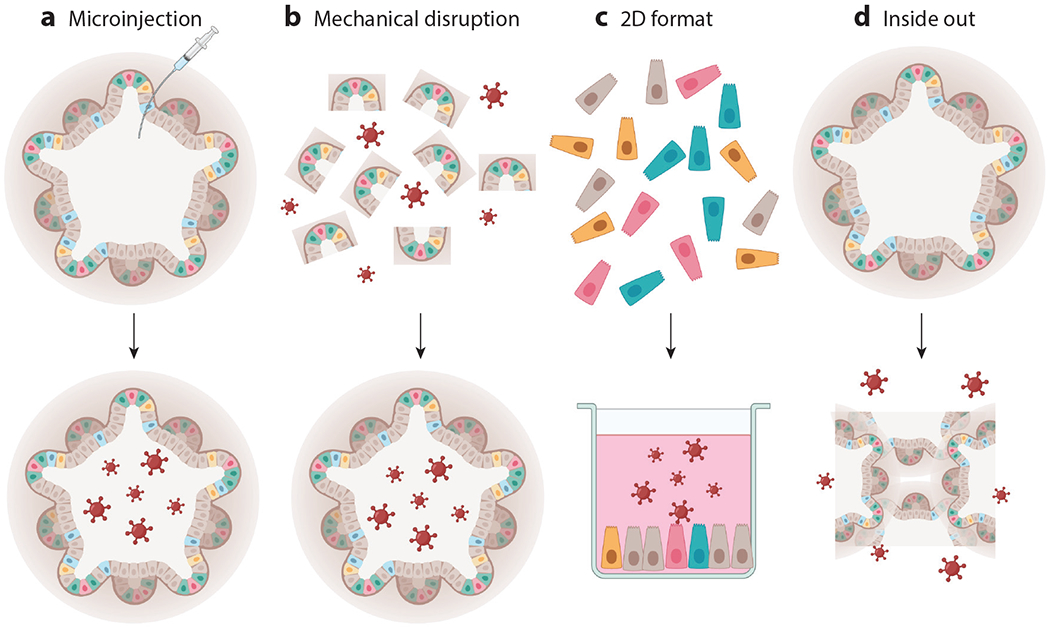
Modes of delivery of infectious organisms to organoid cultures. (a) Pathogens can be delivered to the lumen of an organoid via microinjection. (b) Organoids can be mechanically disrupted into fragments and incubated with pathogens. The fragments reassemble into intact organoids containing the pathogens within the lumen. (c) Organoids can be enzymatically treated, resulting in single cells. These single cells can be plated on plastic or on membranes to form two-dimensional (2D) monolayers that offer easy access to the apical side of the epithelium, which can be used for pathogen encounters. If 2D organoids are plated on Transwell® membranes, they can be exposed to pathogens either apically or basolaterally. (d) Three-dimensional organoids can also be inverted such that the apical side faces outward to interact with pathogens. Figure adapted from image created with BioRender.com.
HUMAN PLURIPOTENT STEM CELL–DERIVED ORGANOIDS
Human pluripotent stem cell–derived organoids have several advantages for investigations of infectious diseases. These cultures originate either from human blastocytes isolated from fetal tissue (embryonic stem cell lines) (6) or from fibroblasts or other differentiated somatic cells transduced with defined transcription factors (induced pluripotent stem cell lines) (7). Pluripotent stem cells have unlimited proliferative capacity and the ability to model all the cell types in the body through a multistep directed-differentiation process that takes typically 1–3 months to generate organoids (8). Using this approach, researchers have created organoids that model the intestine, brain, and airway, allowing investigations of previously unstudied interactions between pathogens and host organs. Organoids generated from pluripotent stem cells contain both epithelial and mesenchymal layers, allowing dissection of the role of the mesenchyme in protecting and defending the epithelium when damaged by infection. Although pluripotent stem cell–derived organoids contain a mesenchymal component, one of the challenges of using these organoids is their inability to mature sufficiently to completely model in vivo epithelium and stroma. Currently, many pluripotent stem cell–derived organoid cultures resemble developing tissue and may not fully recapitulate their adult counterparts. In some instances, implantation of organoid cultures into immunodeficient animals can result in increased development and induction of more mature organoids for further study (9–11). Despite this limitation, an advantage of pluripotent stem cell–derived organoid lines is that they can be easily genetically modified using approaches such as CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/caspase-9) (12), which allows molecular pathways such as innate immune response to infections to be interrogated relatively quickly. Additionally, lines can be generated from individuals with specific genetic mutations, yielding valuable tools for determining the susceptibility factors that influence infectious disease. These cultures are allowing modeling of human infectious diseases in ways that were not possible using transformed cell lines or animals.
HUMAN TISSUE STEM CELL–DERIVED ORGANOIDS
Tissue stem cell–derived organoid cultures contribute equally to the modeling of human infectious diseases. These cultures are derived from resident tissue stem cells that are isolated and recovered from the mature organ, and self-organizing organoids are usually established within a week. Manipulation of growth factors induces differentiation of the stem cells into mature epithelium that resembles the complex in vivo standard both cellularly and physiologically. As with pluripotent stem cell–derived organoids, tissue stem cell–derived organoids have unlimited proliferative capacity in vitro, can be genetically manipulated, and are generated from individuals to represent populations with specific single-nucleotide polymorphisms or diseases caused by specific mutations. Unlike pluripotent stem cell–derived organoids, tissue stem cell–derived organoids lack a mesenchymal component; instead, they have the advantage of retaining mature differentiation and region-specific features that are not yet achievable in pluripotent stem cell–derived organoids. Thus, tissue stem cell–derived organoids are crucial for advancing our understanding of the interactions between human tissue and infectious diseases, especially previously noncultivable human-specific organisms.
THE USE OF ORGANOIDS TO STUDY RESPIRATORY INFECTIONS
Respiratory infectious diseases enter the body through the epithelium that lines the respiratory tree, and airway organoid cultures represent significant advances in the study of human respiratory infections. Both pluripotent stem cell– and tissue stem cell–derived organoids have been created to model epithelial aspects of the respiratory tree (13–16) and contain various epithelial cell types, including basal, goblet, ciliated, club, and type I/type II alveolar cells. The organoids can be cultured under air–liquid interface conditions, which makes them more closely related to the physiology of the epithelium in vivo (17). Lung organoids are only beginning to be used to study respiratory bacterial, parasitic (18), and fungal infections. They are also a powerful new tool for the study of lung viral infections, such as parainfluenza virus 3 (19), influenza virus (20, 21), enterovirus (22), and respiratory syncytial virus (16). To date, investigators have examined a wide range of infectious properties, such as microbial growth; antiviral therapies; epithelial pathologies (or lack thereof), including apical extrusion of infected cells, cytoskeletal rearrangement, and syncytia formation; and susceptibility of the host epithelium to infection.
Perhaps the most exciting use of lung organoids is in studies of the novel SARS-CoV-2 coronavirus strains that are causing the current world pandemic. On preprint servers, yet-to-be-reviewed manuscripts describe the use of both pluripotent and tissue-derived stem cell lung organoid cultures to study the replication kinetics of the virus, tropism, and host epithelial response via techniques such as immunofluorescence, electron microscopy, flow cytometry, transcriptomics, and whole-genome sequencing. Of particular interest is the identification of specific cell populations within the lung that express receptors for SARS-CoV-2. ACE2 and TMPRSS2, which are thought to be critical for binding of the virus to its target cell, are expressed in human airway organoids (20, 23). In differentiated airway organoid cultures, ciliated cells appear to be a key target (24). Organoid studies have revealed that distal cells, such as club and type II alveolar cells, also express ACE2 and TMPRSS2, are infected by SARS-CoV-2, exhibit cytokine pathway induction similar to that of SARS-CoV-2 lung autopsy tissue, and may play a role in more severe disease pathology (25, 26). Furthermore, organoids are beginning to be used to screen for drugs that block viral binding and entry (26), an important first step in developing effective therapeutic treatments against severe disease. The vast increase in the use of organoids for SARS-CoV-2 research over the last year demonstrates the potential and power of human lung organoids to improve our understanding of both existing and emerging respiratory infectious diseases.
THE USE OF ORGANOIDS TO STUDY GASTROINTESTINAL INFECTIONS
Like airway organoids, gastric organoids can be generated either from directed differentiation of pluripotent stem cells or by propagating tissue-derived stem cells for use in investigating infectious diseases (8, 27, 28). Gastric organoids contain differentiated lineages of pit, mucous, neck, endocrine, chief, and parietal cells, recapitulating the extreme cellular heterogeneity that is present in the gastric pit and gland regions. Although the conditions in the stomach are the harshest among the epithelial surfaces, the gastric epithelium is still a target for some pathogens. The best-known organism that causes infections of the gastric epithelium is Helicobacter pylori, which is associated with gastritis, gastric ulcers, and gastric cancer. The use of gastric organoids to study H. pylori infection has advanced our understanding of the pathophysiology of human infection. Bacterial proteins and cellular receptors have been identified using organoid technology (29) and have revealed clues to the mechanisms that underlie the link between H. pylori and cellular proliferation that is the precursor to gastric cancer (see the sidebar titled Using Organoids to Study H. pylori Binding). This research has also led to the discovery of inhibitors to repress H. pylori–induced proliferation (30, 31), as these inhibitors may be useful clinically to prevent gastric cancer. Another area advanced by the use of organoid cultures concerns how H. pylori colonizes the gastric epithelium. An H. pylori chemoreceptor is induced that appears to recognize molecules, such as urea, produced by the gastric organoids. This recognition seems critical in the attraction of the organisms to the epithelium (32), providing some initial clues about mechanisms of colonization. In a vast improvement over earlier in vitro systems, gastric organoids have accelerated mechanistic insights into human gastric infectious diseases.
To date, intestinal organoids have contributed the most information to the biology of infectious diseases. This reflects the early development of intestinal organoids in 2009 and 2011, the relatively easy ability to access intestinal tissues to establish cultures, and the push to evaluate whether these novel, nontransformed, multicellular and physiologically active human organoids could overcome long-standing barriers to understand human-specific pathogens that lacked animal or in vitro culture models. Both pluripotent and tissue-derived stem cell intestinal organoids contain the multiple epithelial cell types that constitute the small and large intestinal epithelium (8, 33–35). Intestinal organoids exhibit physiological properties such as absorption, secretion, barrier function, and innate immune signals, all of which typify the epithelium of specific intestinal segments in vivo. The intestine is a site of attachment, invasion, and replication of many bacterial, viral, parasitic, and fungal pathogens that can lead to significant pathology and disease in the intestine. However, the pathogenesis of many of these organisms in the human host is not well understood. Intestinal organoid models have enhanced our understanding of several aspects of infection, including pathogen receptors, cellular entry pathways, mechanisms of epithelial barrier dysfunction, and responses to infection. In addition, many responses of the organoid cultures to infections reflect both clinical and epidemiological data. Studies involving bacterial pathogens such as Salmonella (5), Listeria monocytogenes (5), Escherichia coli (4, 36–38), Clostridioides difficile (39–41), Vibrio cholera (42), and Klebsiella pneumonia (43) have revealed novel interactions between bacterial pathogens and the intestinal epithelium as well as unique epithelial-intrinsic cellular responses that had not been previously appreciated (see the sidebar titled Pathogenesis of C. difficile Has Been Linked to Two Toxins). Intestinal organoids have met a key challenge in parasite infections by establishing the first ex vivo culture system that supports the complete life cycle of Cryptosporidium infection (18). The molecular mechanisms of the pathophysiology of this organism had previously been poorly understood, and organoids are currently being used to capitalize on these discoveries and develop treatments for this global infectious disease. Viral intestinal infections, such as those caused by rotavirus (44–47), enterovirus (48), echovirus (48), coxsackievirus (48), adenovirus (49), astrovirus (50), and coronavirus (24, 51–53), have all been found to model many features of in vivo infections, For example, human rotavirus infections of organoids recapitulate host restriction; induce organoid swelling, illustrating a physiological response mimicking fluid secretion and diarrhea induction; and can model novel calcium signaling involved in pathogenesis (46, 54). Studies in human organoid cultures have provided new details on pathogenesis, including in areas such as unexpected cellular tropisms, epithelial responses, and antiviral signaling.
Intestinal organoid cultures can transform infectious disease research, as demonstrated by the finding that they support human norovirus replication (55). These viruses are the leading cause of global epidemic and sporadic gastroenteritis in all age groups, characterized by profuse vomiting and diarrhea. Since their discovery in 1973, a major hurdle in studying this pathogen was the lack of in vitro platforms or animal model systems to culture and study the viruses. Human intestinal organoid cultures are permissive to infection and support replication of multiple human norovirus strains. Their power to dissect host genetic susceptibility was illustrated via replication of epidemiological and human volunteer studies in which cultures established from individuals who express a susceptibility gene (fucotransferase 2) support viral infection and replication while cultures from individuals who do not express this gene are not infectable by the virus. Additionally, investigators discovered strain-specific requirements for infection and replication of human noroviruses, including a need to mimic the intestinal luminal environment by the inclusion of certain bile acids in the media in order for some strains to replicate (56). Methods to inactivate viruses, and characterization of neutralizing antibodies induced by natural infection or vaccination, are enabling advances in infection prevention and therapies (55). These intestinal organoid breakthroughs highlight the potential for these remarkable cultures to stimulate advances in preventive and therapeutic treatments for gastrointestinal infectious diseases.
THE USE OF ORGANOIDS TO STUDY NEURONAL INFECTIONS
The development of cerebral organoids has led to the establishment of a previously nonexistent ex vivo model of nontransformed human brain cells that has important implications for the study of neurological infections. Unlike the respiratory and gastrointestinal models, where both pluripotent and tissue-derived stem cell organoid models are available, cerebral organoids have been established only from pluripotent stem cells (57–60). These cerebral organoid models contain proliferating neuronal progenitors, exhibit functional synapses and active neural networks, show features of dendritic spines, and exhibit a transcriptional profile that closely resembles that of developing brain tissue. Furthermore, they mimic the cellular arrangement and epigenomic signatures of human cerebral tissue.
Since little is known about the pathogenesis of many neuronal infectious pathogens, organoids provide a physiologically relevant system in which molecular features of infection can be studied. Viral infections that have been studied to date in the cerebral organoid model include West Nile virus (61), herpes simplex virus 1 (62), dengue virus (63), varicella zoster virus (64), cytomegalovirus (65), rabies lyssavirus (66), and HIV (67). The emergence of Zika virus in Central America in 2013 highlighted the importance of cerebral organoids for infectious disease research: Organoids played a central role in addressing the central nervous system (CNS) cellular targets of the virus and in defining the link between the infection and the clinical manifestation of microencephaly (68–70). Zika infection of cerebral organoids induced significant disorganization of cultures that exhibited growth defects due to reductions in progenitor cells. These findings could directly explain the neurological manifestations that were present in newborns. Multiple compounds capable of mitigating Zika-induced cytopathy have also been identified (71). The critical role of organoids in establishing the biology of Zika virus confirmed these cultures as an important technology for future studies of neurotropic infectious diseases. Although COVID-19 is considered primarily a respiratory disease, SARS-CoV-2 has neuroinvasive capacity, as shown by clear infection of neuronal and neural stem cells in brain organoids, which led to neuronal cell death in both target cells and neighboring cells (72). Further studies may help us understand the consequences of CNS infection on other organ systems or the long-term sequelae of infection.
OTHER ORGANOID MODELS AND THEIR APPLICATION TO INFECTIOUS DISEASES
Other organoid model systems used to study infectious diseases are only in their infancy. Both pluripotent (73) and tissue (74) stem cell–derived organoids have been developed to recapitulate some of the complexity of the renal architecture and composition that is lacking in more traditional cell lines. These lines have been used to study BK virus, which is associated with chronic infections and complications in renal transplantation recipients. Similarly, pluripotent (75) and tissue (76) stem cell–derived organoids have been generated to model the human liver and infections with hepatitis B (77) and hepatitis C (78) viruses, and ongoing studies are providing clues as to the links among these infectious pathogens, liver hepatitis, and hepatocellular carcinoma. Oral mucosal tissue stem cell–derived organoids have been developed to investigate infections with herpes simplex virus, which causes cold sores, and human papillomavirus, one of the causative agents for oral pharyngeal cancers (79). Female reproductive tract tissue stem cell–derived organoids have been developed from fallopian tube (80, 81) and endo- and ectocervical (82) tissues. Changes in differentiation of the epithelium are driven by Chlamydia trachomatis infection, and initial observations of lysis and collapse of organoids due to infection with herpes simplex virus 1 are stimulating future research to more fully explain the pathogenesis of these infections and how they relate to infertility. Pluripotent stem cell–derived skin organoids (83) were recently shown to model infection with Trichophyton rubrum, a fungus that causes athlete’s foot, and ringworm (84). Repression of cytokine signaling observed in the organoids may explain the chronic and recurrent nature of these infections. Expansion of available organoid cultures to encompass and represent more human organs will drive further in-depth studies involving infectious pathogens that will inform the prevention, management, and treatment of many infectious diseases.
LIMITATIONS OF ORGANOIDS IN THE STUDY OF INFECTIONS
Both pluripotent and tissue stem cell–derived organoids offer the opportunity to study human infectious disease in the context of nontransformed ex vivo cell cultures that are relatively simple and inexpensive to maintain, to investigate the relationship between genetics and infection, and to enable comparisons with epidemiological and clinical data. Results can be obtained rapidly and can more accurately predict in vivo human responses, in comparison to classical approaches such as transformed cell lines or animal models. Organoid genome editing provides tools to dissect the fine details of specific results. In addition, cultures offer enormous potential for future personalized approaches to the prevention, diagnosis, and treatment of infectious diseases. Although the use of organoid cultures has significantly advanced many aspects of research on the pathogenesis of infectious organisms and will continue to do so in the near future, there are several areas in which modification or enhancement of the cultures will substantially increase the importance of their role. The continuing development and evolution of organoid cultures will enhance their ability to dissect the molecular mechanisms underlying interactions between infectious pathogens and host tissue.
The most important aspect of infectious diseases that is lacking in current organoid cultures is the support cells that reside in the underlying stroma of most epithelial tissues. These cells include endothelial cells, neuronal cells, mesenchymal cells such as fibroblasts, and most importantly immune cells (Figure 3). Disease that occurs with infection results from a complex interaction among the human epithelium, the invading organisms, and the host immune system. Coculture models in which immune cells and organoids are examined together have already been established (36, 85–87) in an effort to understand these interactions. In some cases, these model systems are being applied to address questions concerning infectious diseases. A remarkable example is the use of tonsillar organoids as a tool to probe human humoral immune responses to infection (88). These cultures were able to reproduce germinal center responses, which were found to be variable across different patient-derived organoids, mimicking the variability in vaccine responses across human populations. This system has the potential to predict the quality and magnitude of the human humoral immune responses and might be a valuable tool for studies of novel prophylactic and therapeutic strategies. Understanding immune system responses will be crucial for understanding the physiology of disease as well as for designing approaches to both prevent and treat illness.
Figure 3.
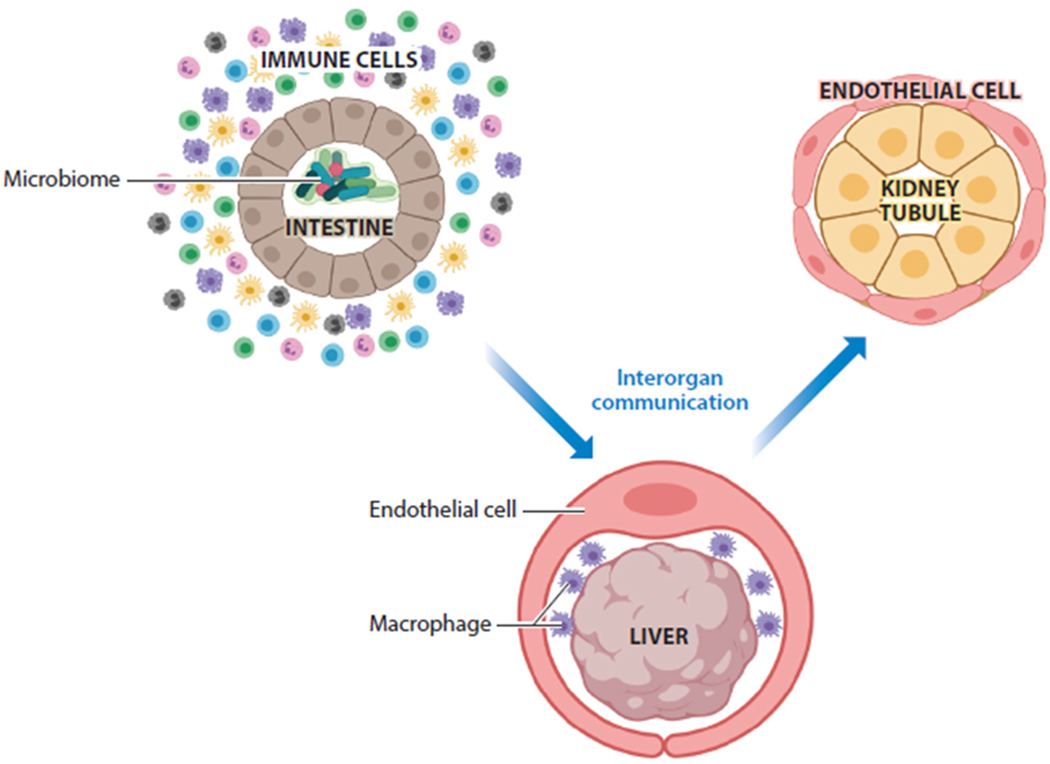
Key features of organoid cultures that will advance the use of organoids in infectious disease research. Incorporation of immune cells such as macrophages will be critical in establishing the interactions among the pathogen, the epithelium, and the immune system. Incorporation of the microbiome will also be critical. Interorgan communication will allow a more comprehensive view of the body’s response to infectious agents. Figure adapted from image created with BioRender.com.
In most cases, studies involving infectious diseases in organoid models reflect reductionist approaches that examine only epithelial intrinsic responses at the organ or tissue level and lack the ability to model interorgan communication. Rarely do infectious diseases remain isolated in a specific location or exert their effects only locally. Therefore, organoid models need to evolve to mimic more of the whole body. Out of this need has arisen the body-on-a-chip movement, with the goal of creating a mimic of the human body using organoid cultures that are interconnected on a single platform (89–91). Thus, the effects of perturbation of one organ could be studied not only on the directly affected organ but also on other organs that are connected via circulatory and lymphatic channels. These systems could be used to study host-pathogen interactions in addition to drug responses and vaccine efficacy. The interconnection among multiple organoids on the same platform would allow signals to pass between organoids and would enable investigation of a more global physiological response to insults such as infections.
Many epithelial surfaces that are modeled by organoid cultures are colonized by a microbiota. These organisms are now known to significantly influence epithelial biology in such areas as epithelial turnover and immune response. It is clear that the microbiota influences and competes with other pathogens and can influence disease outcome. Currently, most studies that use organoids to study infectious diseases lack a microbiota component. To comprehensively understand infectious diseases, studies must incorporate the role of the microbiota. In recognition of the important role the microbiota plays at epithelial surfaces, coculture systems that have both organoid and microbiota are being developed (92, 93).
Due to the lack of widely accepted standardized protocols and guidelines for generating and growing organoid cultures, as well as genetic differences among the individuals from whom cultures are derived, each culture that is generated is inherently variable. As a result, the heterogeneity of the system is reflected in the data analysis and interpretation of results obtained with each culture. Questions remain as to the best methods to assess both the quality and the validity of the cultures. With the advent of unbiased single-cell transcriptomic and epigenomic analyses that allow profiling of large numbers of cells, we can establish a better standard to which individual cultures can be measured. To this end, assembly of a comprehensive reference map of all human cells is underway (https://www.humancellatlas.org) and has the potential to affect every aspect of biology and medicine, including infectious diseases. Features such as cell hierarchy and cell identity are changing the landscape of what was previously known about human cells (94) and allowing these new cellular discoveries to be applied to microbe tropisms and intercellular signaling important for pathogenesis. This technology is already being applied to organoids, with the ultimate benchmark of comparison against native tissue (95–97). This technology has the potential to offer a deep view of the response of the epithelium to many infectious pathogens, and many new hypotheses are bound to emerge.
CONCLUSION
Organoid models have great potential for the study of human infectious diseases at both cellular and environmental levels. The technology is rapidly advancing in complexity as well as in techniques for standardization and validation. Cocultures of organoids with viruses, bacteria, parasites, and fungi have accelerated knowledge at the interface of infectious diseases and human biology. Key molecules that are critical for cell recognition, pathogen entry and replication, and host response are already being identified and applied to the development of preventive and therapeutic modalities. These tools have already provided an unprecedented opportunity to improve human health.
USING ORGANOIDS TO STUDY H. PYLORI BINDING.
McCracken et al. (29) microinjected pathogenic and attenuated (lacking CagA) H. pylori into the lumen of human gastric organoids. After 24 h, the bacteria were found tightly adhered to the apical surface of the gastric epithelium within the organoid. Immunoprecipitation of the bacteria indicated that they were bound to and influenced the tyrosine-dependent phosphorylation status of c-Met (an oncogene). This interaction was dependent on expression of CagA by H. pylori and resulted in increased proliferation of the gastric organoids. These results strengthened the link between CagA and its role in H. pylori–linked gastric cancer pathogenesis.
PATHOGENESIS OF C. DIFFICILE HAS BEEN LINKED TO TWO TOXINS.
The contribution of C. difficile toxins TcdA and TcdB to overall disease is unknown. The use of colonic organoids and recombinant toxins showed that both toxins induced changes in crypt architecture, reduced goblet cell numbers, and induced the loss of tight junctions over adherens junctions—all of which reduced organoid epithelial permeability (39). These changes replicate those found in pseudomembranous colitis, which is associated with severe C. difficile infection. A transcriptional analysis of the organoids demonstrated the induction of inflammatory, actin-regulation, and junctional complex pathways, which could explain the detrimental effects of both toxins.
SUMMARY POINTS.
Organoids are transformative new tools for the study of human infectious diseases.
Organoids are grown from either pluripotent stem cells (embryonic or induced) or tissue-derived stem cells.
Lung organoids have been used to study respiratory viral infections including SARS-CoV-2.
Intestinal organoids have been used to study gastrointestinal infections with viruses, bacteria, and parasites.
Viral infections have been established in cerebral organoids.
Other organoids, such as liver, skin, and kidney, are beginning to be used to investigate infectious diseases.
Areas of focus that are improving organoid models include the addition of stromal components, improved methods of validation, establishment of interorgan communication, and incorporation of the microbiome.
Organoids provide an opportunity to improve human health in relation to infectious diseases through a better understanding of the mechanisms of pathogenesis and the development of new methods to prevent and treat infections.
ACKNOWLEDGMENTS
Research in our laboratories related to organoids and infectious diseases has been supported by Public Health Service grants P01AI057788, U19 AI116497, P30 DK056338, U01DK103168, and U19AI144297, and NASA grant NNX16A069.
Glossary
- In vitro
refers to the study of something outside the living organism in an artificial environment
- Ex vivo
refers to something extracted from a living organism and studied with minimal changes to the native environment
- Organoid
simplified version of an organ produced in vitro that models representative microanatomy
- Matrigel®
gelatinous protein mixture that mimics the extracellular matrix and is used to culture cells
- Mesenchymal
refers to a type of connective tissue found underlying most epithelia
- WNT
highly conserved signaling molecule that plays a role in axis patterning, cell fate, cell proliferation, and cell migration
- Epithelial
refers to one of the basic types of tissue that lines the outer and inner surfaces of organs
- CRISPR/Cas9
method used to edit genes within a cell
- Air-liquid interface
a cell culture method in which the basal surface of the cells is in contact with media and the apical surface is exposed to air
- Body on a chip
multiorgan in vitro system that models interactions and connections between different human tissues
- Microbiota
communities of organisms that live on multicellular organisms and play a role in homeostasis of the organism
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.GBD 2016 DALYs and HALE Collab. 2017. Global, regional, and national disability-adjusted life-years (DALYs) for 333 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390(10100):1260–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michaud CM. 2009. Global burden of infectious diseases. Encycl. Microbiol 2009:444–54 [Google Scholar]
- 3.Colzani E 2019. Beyond morbidity and mortality: the burden of infectious diseases on healthcare services. Epidemiol. Infect 147:e251 [Google Scholar]
- 4.VanDussen KL, Marinshaw JM, Shaikh N, et al. 2015. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 64(6):911–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Co JY, Margalef-Catala M, Li X, et al. 2019. Controlling epithelial polarity: a human enteroid model for host–pathogen interactions. Cell Rep. 26(9):2509–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282(5391):1145–47 [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, Tanabe K, Ohnuki M, et al. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131(5):861–72 [DOI] [PubMed] [Google Scholar]
- 8.McCracken KW, Howell JC, Wells JM, Spence JR. 2011. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat. Protoc 6(12):1920–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Berg CW, Ritsma L, Avramut MC, et al. 2018. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell Rep. 10(3):751–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mansour AA, Gonçalves JT, Bloyd CW, et al. 2018. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol 36(5):432–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson CL, Mahe MM, Munera J, et al. 2014. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med 20(11):1310–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artegiani B, Hendriks D, Beumer J, et al. 2020. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat. Cell Biol 22(3):321–31 [DOI] [PubMed] [Google Scholar]
- 13.Wong AP, Bear CE, Chin S, et al. 2012. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol 30(9):876–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang SX, Islam MN, O’Neill J, et al. 2014. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol 32(1):84–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dye BR, Hill DR, Ferguson MA, et al. 2015. In vitro generation of human pluripotent stem cell derived lung organoids. eLife 4:e05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sachs N, Papaspyropoulos A, Zomer-van Ommen DD, et al. 2019. Long-term expanding human airway organoids for disease modeling. EMBO J 38(4):e100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konishi S, Gotoh S, Tateishi K, et al. 2016. Directed induction of functional multi-ciliated cells in proximal airway epithelial spheroids from human pluripotent stem cells. Stem Cell Rep. 6(1):18–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heo I, Dutta D, Schaefer DA, et al. 2018. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol 3(7):814–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porotto M, Ferren M, Chen YW, et al. 2019. Authentic modeling of human respiratory virus infection in human pluripotent stem cell–derived lung organoids. mBio 10(3):00723–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J, Li C, Sachs N, et al. 2018. Differentiated human airway organoids to assess infectivity of emerging influenza virus. PNAS 115(26):6822–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hui KPY, Ching RHH, Chan SKH, et al. 2018. Tropism, replication competence, and innate immune responses of influenza virus: an analysis of human airway organoids and ex-vivo bronchus cultures. Lancet Respir. Med 6(11):846–54 [DOI] [PubMed] [Google Scholar]
- 22.van der Sanden SMG, Sachs N, Koekkoek SM, et al. 2018. Enterovirus 71 infection of human airway organoids reveals VP1-145 as a viral infectivity determinant. Emerg. Microbes Infect 7(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuyama S, Nao N, Shirato K, et al. 2020. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. PNAS 117(13):7001–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamers MM, Beumer J, van der Vaart J, et al. 2020. SARS-CoV-2 productively infects human gut enterocytes. Science 369(6499):50–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salahudeen AA, Choi SS, Rustagi A, et al. 2020. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 588(7839):670–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han Y, Duan X, Yang L, et al. 2021. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589(7841):270–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartfeld S, Bayram T, van de Wetering M, et al. 2015. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148(1):126–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlaermann P, Toelle B, Berger H, et al. 2016. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut 65(2):202–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCracken KW, Catá EM, Crawford CM, et al. 2014. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516(7531):400–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wroblewski LE, Piazuelo MB, Chaturvedi R, et al. 2015. Helicobacter pylori targets cancer-associated apical-junctional constituents in gastroids and gastric epithelial cells. Gut 64(5):720–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertaux-Skeirik N, Feng R, Schumacher MA, et al. 2015. CD44 plays a functional role in Helicobacter pylori–induced epithelial cell proliferation. PLOS Pathog 11(2):e1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang JY, Sweeney EG, Sigal M, et al. 2015. Chemodetection and destruction of host urea allows Helicobacter pylori to locate the epithelium. Cell Host Microbe 18(2):147–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spence JR, Mayhew CN, Rankin SA, et al. 2011. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470(7332):105–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato T, Vries RG, Snippert HJ, et al. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459(7244):262–65 [DOI] [PubMed] [Google Scholar]
- 35.Sato T, Stange DE, Ferrante M, et al. 2011. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141(5):1762–72 [DOI] [PubMed] [Google Scholar]
- 36.Noel G, Baetz NW, Staab JF, et al. 2017. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep 7:45270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.In J, Foulke-Abel J, Zachos NC, et al. 2016. Enterohemorrhagic Escherichia coli reduce mucus and intermicrovillar bridges in human stem cell–derived colonoids. Cell Mol. Gastroenterol. Hepatol 2(1):48–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajan A, Vela L, Zeng XL, et al. 2018. Novel segment- and host-specific patterns of enteroaggregative Escherichia coli adherence to human intestinal enteroids. mBio 9(1):02419–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leslie JL, Huang S, Opp JS, et al. 2015. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect. Immun 83(1):138–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engevik MA, Yacyshyn MB, Engevik KA, et al. 2015. Human Clostridium difficile infection: altered mucus production and composition. Am. J. Physiol. Gastrointest. Liver Physiol 308(6):G510–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Yamamoto Y, Wilson LH, et al. 2015. Cloning and variation of ground state intestinal stem cells. Nature 522(7555):173–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuhlmann FM, Santhanam S, Kumar P, et al. 2016. Blood group O–dependent cellular responses to cholera toxin: parallel clinical and epidemiological links to severe cholera. Am. J. Trop. Med. Hyg 95(2):440–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamoto N, Sasaki N, Aoki R, et al. 2019. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat. Microbiol 4(3):492–503 [DOI] [PubMed] [Google Scholar]
- 44.Yin Y, Bijvelds M, Dang W, et al. 2015. Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antivir. Res 123:120–31 [DOI] [PubMed] [Google Scholar]
- 45.Finkbeiner SR, Zeng XL, Utama B, et al. 2012. Stem cell–derived human intestinal organoids as an infection model for rotaviruses. mBio 3(4):e00159–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saxena K, Blutt SE, Ettayebi K, et al. 2016. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J. Virol 90(1):43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang-Graham AL, Danhof HA, Engevik MA, et al. 2019. Human intestinal enteroids with inducible neurogenin-3 expression as a novel model of gut hormone secretion. Cell Mol. Gastroenterol. Hepatol 8(2):209–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drummond CG, Bolock AM, Ma C, et al. 2017. Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage–specific manner. PNAS 114(7):1672–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holly MK, Smith JG. 2018. Adenovirus infection of human enteroids reveals interferon sensitivity and preferential infection of goblet cells. J. Virol 92(9):e00250–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kolawole AO, Mirabelli C, Hill DR, et al. 2019. Astrovirus replication in human intestinal enteroids reveals multi-cellular tropism and an intricate host innate immune landscape. PLOS Pathog. 15(10):e1008057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou J, Li C, Zhao G, et al. 2017. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv 3(11):eaao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zang R, Gomez Castro MF, McCune BT, et al. 2020. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol 5(47):eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou J, Li C, Liu X, et al. 2020. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med 26(7):1077–83 [DOI] [PubMed] [Google Scholar]
- 54.Chang-Graham AL, Perry JL, Engevik MA, et al. 2020. Rotavirus induces intercellular calcium waves through ADP signaling. Science 370(6519):eabc3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ettayebi K, Crawford SE, Murakami K, et al. 2016. Replication of human noroviruses in stem cell–derived human enteroids. Science 353(6306):1387–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murakami K, Tenge VR, Karandikar UC, et al. 2020. Bile acids and ceramide overcome the entry restriction for GII.3 human norovirus replication in human intestinal enteroids. PNAS 117(3):1700–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lancaster MA, Renner M, Martin CA, et al. 2013. Cerebral organoids model human brain development and microcephaly. Nature 501(7467):373–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quadrato G, Nguyen T, Macosko EZ, et al. 2017. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545(7652):48–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Camp JG, Badsha F, Florio M, et al. 2015. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. PNAS 112(51):15672–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dezonne RS, Sartore RC, Nascimento JM, et al. 2017. Derivation of functional human astrocytes from cerebral organoids. Sci. Rep 7:45091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang B, West N, Vider J, et al. 2019. Inflammatory responses to a pathogenic West Nile virus strain. BMC Infect. Dis 19(1):912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lafaille FG, Pessach IM, Zhang SY, et al. 2012. Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature 491(7426):769–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Desole G, Sinigaglia A, Riccetti S, et al. 2019. Modelling neurotropic flavivirus infection in human induced pluripotent stem cell–derived systems. Int. J. Mol. Sci 20(21):5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Markus A, Lebenthal-Loinger I, Yang IH, et al. 2015. An in vitro model of latency and reactivation of varicella zoster virus in human stem cell–derived neurons. PLOS Pathog. 11(6):e1004885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cosset E, Martinez Y, Preynat-Seauve O, et al. 2015. Human three-dimensional engineered neural tissue reveals cellular and molecular events following cytomegalovirus infection. Biomaterials 53:296–308 [DOI] [PubMed] [Google Scholar]
- 66.Sundaramoorthy V, Godde N, Farr RJ, et al. 2020. Modelling lyssavirus infections in human stem cell-derived neural cultures. Viruses 12(4):359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ryan SK, Gonzalez MV, Garifallou JP, et al. 2020. Neuroinflammation and EIF2 signaling persist despite antiretroviral treatment in an hiPSC tri-culture model of HIV infection. Stem Cell Rep. 14(5):991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qian X, Nguyen HN, Song MM, et al. 2016. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165(5):1238–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garcez PP, Correia Loiola E, Madeiro da Costa R, et al. 2016. Zika virus impairs growth in human neurospheres and brain organoids. Science 352(6287):816–18 [DOI] [PubMed] [Google Scholar]
- 70.Cugola FR, Fernandes IR, Russo FB, et al. 2016. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534(7606):267–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watanabe M, Buth JE, Vishlaghi N, et al. 2017. Self-organized cerebral organoids with human-specific features predict effective drugs to combat Zika virus infection. Cell Rep. 21(2):517–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song E, Zhang C, Israelow B, et al. 2021. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med 218(3):e20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takasato M, Er PX, Chiu HS, et al. 2015. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526(7574):564–68 [DOI] [PubMed] [Google Scholar]
- 74.Schutgens F, Rookmaaker MB, Margaritis T, et al. 2019. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat. Biotechnol 37(3):303–13 [DOI] [PubMed] [Google Scholar]
- 75.Takebe T, Sekine K, Enomura M, et al. 2013. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499(7459):481–84 [DOI] [PubMed] [Google Scholar]
- 76.Huch M, Gehart H, van Boxtel R, et al. 2015. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160(1–2):299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shlomai A, Schwartz RE, Ramanan V, et al. 2014. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell–derived hepatocellular systems. PNAS 111(33):12193–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baktash Y, Madhav A, Coller KE, Randall G. 2018. Single particle imaging of polarized hepatoma organoids upon hepatitis C virus infection reveals an ordered and sequential entry process. Cell Host Microbe 23(3):382–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Driehuis E, Kolders S, Spelier S, et al. 2019. Oral mucosal organoids as a potential platform for personalized cancer therapy. Cancer Discov. 9(7):852–71 [DOI] [PubMed] [Google Scholar]
- 80.Kessler M, Hoffmann K, Fritsche K, et al. 2019. Chronic Chlamydia infection in human organoids increases stemness and promotes age-dependent CpG methylation. Nat. Commun 10(1):1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kopper O, de Witte CJ, Lohmussaar K, et al. 2019. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med 25(5):838–49 [DOI] [PubMed] [Google Scholar]
- 82.Lõhmussaar K, Oka R, Espejo Valle-Inclan J, et al. 2021. Patient-derived organoids model cervical tissue dynamics and viral oncogenesis in cervical cancer. Cell Stem Cell 28(8):1380–96 [DOI] [PubMed] [Google Scholar]
- 83.Lee J, Rabbani CC, Gao H, et al. 2020. Hair-bearing human skin generated entirely from pluripotent stem cells. Nature 582(7812):399–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang X, Wang S, Guo B, et al. 2021. Human primary epidermal organoids enable modeling of dermatophyte infections. Cell Death Dis. 12(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nozaki K, Mochizuki W, Matsumoto Y, et al. 2016. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. J. Gastroenterol 51(3):206–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rogoz A, Reis BS, Karssemeijer RA, Mucida D. 2015. A 3-D enteroid-based model to study T-cell and epithelial cell interaction. J. Immunol. Methods 421:89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cook L, Stahl M, Han X, et al. 2019. Suppressive and gut-reparative functions of human type 1 T regulatory cells. Gastroenterology 157(6):1584–98 [DOI] [PubMed] [Google Scholar]
- 88.Wagar LE, Salahudeen A, Constantz CM, et al. 2021.Modeling human adaptive immune responses with tonsil organoids. Nat. Med 27(1):125–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maschmeyer I, Lorenz AK, Schimek K, et al. 2015. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 15(12):2688–99 [DOI] [PubMed] [Google Scholar]
- 90.Materne EM, Maschmeyer I, Lorenz AK, et al. 2015. The multi-organ chip—a microfluidic platform for long-term multi-tissue coculture. J. Vis. Exp 98:e52526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Herland A, Maoz BM, Das D, et al. 2020. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat. Biomed. Eng 4(4):421–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim HJ, Huh D, Hamilton G, Ingber DE. 2012. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12(12):2165–74 [DOI] [PubMed] [Google Scholar]
- 93.Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, et al. 2019. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng 3(7):520–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Han X, Zhou Z, Fei L, et al. 2020. Construction of a human cell landscape at single-cell level. Nature 581(7808):303–9 [DOI] [PubMed] [Google Scholar]
- 95.Subramanian A, Sidhom EH, Emani M, et al. 2019. Single cell census of human kidney organoids shows reproducibility and diminished off-target cells after transplantation. Nat. Commun 10(1):5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kanton S, Boyle MJ, He Z, et al. 2019. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574(7778):418–22 [DOI] [PubMed] [Google Scholar]
- 97.Andor N, Simonds EF, Czerwinski DK, et al. 2019. Single-cell RNA-seq of follicular lymphoma reveals malignant B-cell types and coexpression of T-cell immune checkpoints. Blood 133(10):1119–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
RELATED RESOURCES
- Bhatia SN, Ingber DE. 2014. Microfluidic organs-on-chips. Nat. Biotechnol 32(8):760–72 [DOI] [PubMed] [Google Scholar]
- Bar-Ephraim YE, Kretzschmar K, Clevers H. 2020. Organoids in immunological research. Nat. Rev. Immunol 20(5):279–93 [DOI] [PubMed] [Google Scholar]
- Brazovskaja A, Treutlein B, Camp JG. 2019. High-throughput single-cell transcriptomics on organoids. Curr. Opin. Biotechnol 55:167–71 [DOI] [PubMed] [Google Scholar]
- May S, Evans S, Parry L. 2017. Organoids, organs-on-chips and other systems, and microbiota. Emerg. Top. Life Sci 1(4):385–400 [DOI] [PMC free article] [PubMed] [Google Scholar]


