PURPOSE
To establish a patient-specific polygenic score derived from cytarabine (ara-C) pathway pharmacogenomic evaluation to personalize acute myeloid leukemia (AML) treatment.
MATERIALS AND METHODS
Single nucleotide polymorphisms (SNPs) in the ara-C-pathway genes were analyzed with outcome in patients from the multicenter-AML02 trial (N = 166). Multi-SNP predictor modeling was used to develop 10-SNP Ara-C_SNP score (ACS10) using top SNPs predictive of minimal residual disease and event-free survival (EFS) from the AML02-cohort and four SNPs previously associated with ara-C triphosphate levels in the AML97 trial. ACS10 was evaluated for association with outcomes in each clinical trial arms: the standard low-dose ara-C (LDAC, n = 91) and augmented high-dose ara-C (HDAC, n = 75) arms of AML02 and the standard Ara-C, daunorubicin and etoposide (ADE) (n = 465) and the augmented ADE + gemtuzumab ozogamicin (GO; n = 466) arms of AAML0531 trial.
RESULTS
In the standard LDAC-arm of AML02 cohort, the low-ACS10 score group (≤ 0) had significantly worse EFS (ACS10 low v high hazard ratio [HR] = 2.81; 95% CI, 1.45 to 5.43; P = .002) and overall survival (OS; HR = 2.98; 95% CI, 1.32 to 6.75; P = .009) compared with the high-ACS10 group (score > 0). These results were validated in the standard-ADE arm of AAML0531, with poor outcome in the low-ASC10 group compared with the high-ACS10 group (EFS: HR = 1.35, 95% CI, 1.04 to 1.75, P = .026; OS: HR = 1.64, 95% CI, 1.2 to 2.22, P = .002). Within the augmented arms (AML02-HDAC and AAML0531-ADE + GO), EFS and OS did not differ between low- and high-ACS10 score groups. In both cohorts, patients with low-ACS10 consistently showed a 10-percentage point improvement in 5-year EFS with augmented therapy (AML02-HDAC or AAML0531-ADE + GO arms) than with standard therapy (AML02-LDAC or AAML0531-ADE arms).
CONCLUSION
Patients with low-ACS10 score experienced significantly poor outcome when treated on standard regimen. Augmentation with either high-dose ara-C or GO addition improved outcome in low-ACS10 group. A polygenic ACS10 score can identify patients with unfavorable pharmacogenetic characteristics and offers a potential for an elective augmented therapy option.
INTRODUCTION
For the past five decades, cytarabine (ara-C) has been the mainstay of acute myeloid leukemia (AML) chemotherapy regimen, primarily given in combination with anthracyclines. However, standard ara-C–containing chemotherapy fails to induce remission in roughly 10%-15% of children.1-4 Among those who achieve remission, approximately 40% relapse.5-7 This interpatient variation in treatment response, development of resistance, and high risk of relapse remain major hurdles to effective AML chemotherapy. Ara-C is a prodrug requiring activation to ara-C triphosphate (ara-CTP) by multiple phosphorylation steps, which upon incorporation in place of deoxycytidine triphosphate, results in chain termination, thereby blocking DNA/RNA synthesis and causing leukemic cell death. Thus, intracellular abundance of ara-CTP formation is one of the significant determinants of treatment response.8 We and others have previously sequenced multiple genes in the ara-C pathway and reported SNPs of functional and clinical relevance.9-19 Despite these efforts, a comprehensive evaluation of pharmacogenomic ara-C pathway genes for association with clinical outcome in AML is largely lacking. Our recent effort in this direction evaluated genetic variation in 16 ara-C metabolic pathway genes and developed a four SNP score predictive of leukemic cell intracellular levels of ara-CTP.9 Encouraged by these promising results, the current study was designed with the objective to perform a comprehensive pharmacogenomic evaluation of ara-C pathway and develop a polygenic score predictive of treatment outcome that also holds clinical utility for designing chemotherapeutic treatment regimens.
CONTEXT
Key Objective
Cytarabine (ara-C) is the mainstay of acute myeloid leukemia chemotherapy. The key objective of this work is to establish pharmacogenomics-based personalization of ara-C therapy to enhance treatment outcome.
Knowledge Generated
We developed a polygenic score consisting of 10 SNPs,designated as ACS10, that not only provides prognostic significance by predicting poor outcome in acute myeloid leukemia, but suggests that alternative treatment strategies with either high-dose ara-C or addition of gemtuzumab ozogamicin are more suitable strategies for patients with detrimental low-ACS10 score.
Relevance
Preemptive genotyping with quick turnaround time can allow for the most effective induction 1 chemotherapy combination regimen design to achieve maximum clinical benefit.
MATERIALS AND METHODS
Patient Cohorts
1. St Jude AML02 Discovery Cohort (NCT00136084): 166 pediatric, adolescent, and young adult patients with AML (2 days-21.4 years old) treated under the multicenter AML02 trial were included in the study. Details of study design and clinical outcome have been described elsewhere.3 Briefly, patients with de novo AML were randomly assigned to receive either high-dose (3 g/m2, every 12 hours on days 1, 3, and 5; high-dose ara-C [HDAC] arm, n = 75) or low-dose (100 mg/m2 every 12 hours on days 1-10; low-dose ara-C [LDAC] arm, n = 91) ara-C along with daunorubicin (50 mg/m2 over 6 hours on days 2, 4, and 6) and etoposide (100 mg/m2 over 4 hours on days 2-6) as a first course of chemotherapy.
2. COG-AAML0531 Validation Cohort (NCT00372593): 931 pediatric, adolescent and young adult patients with AML (1 month-29.9 years old) treated on the AAML0531 trial were included in this study. Patients were randomly assigned to receive ADE (ADE arm: ara-C 100 mg/m2/dose twice per day for 10 days alongside daunorubicin and etoposide—equivalent to the LDAC arm of St Jude AML02, n = 465) without or with the addition of two doses (at 3 mg/m2) of the CD33-targeting drug gemtuzumab ozogamicin (GO; ADE + GO arm, n = 466).2
The Data Supplement (online only) provides the summary of patient characteristics for the AML02 and COG-AAM05L31 cohorts. Patient risk assignments were as defined in the clinical trials reports published previously.2,3 St Jude Institutional Review Board approved the current research in the AML02 trial, and the COG ethics committee approved use of specimens and data for the current study. Written informed consent or assent as described in study protocols were obtained in accordance with Declaration of Helsinki and local institutional policies. University of Florida IRB approved the pharmacogenomic analysis for both studies.
Clinical End Point Definitions
Clinical end points used in this evaluation were defined as follows: (1) positive minimal residual disease at the end of induction I (MRD1), ≥ 1 leukemic cell per 1,000 mononuclear bone marrow cells (≥ 0.1%) determined using flow cytometry; (2) complete remission, trilineage hematopoietic recovery with < 5% blasts in the marrow after induction 1; (3) event-free survival (EFS), as the time from study enrollment to induction failure, relapse, secondary malignancy, death, or study withdrawal for any reason, with event-free patients censored on last follow-up for AML02 and as the time from study entry until death, induction failure, or relapse of any type with event-free patients censored on last follow-up for AAML0531; and (4) overall survival (OS) defined as the time from study enrollment to death, with living patients censored on the date of last follow-up.
Genotyping
Genomic DNA from the patients was genotyped for SNPs from both AML02 and AAML0531 clinical trials as described in the Data Supplement.
Statistical Analysis
1. Individual SNP analysis: SNP genotype groups in three different modes of inheritance (additive, dominant, and recessive) were tested for association with MRD1 using logistic regression models and with EFS and OS using Cox proportional hazard models. We performed outcome association analysis of SNPs with and without adjusting for risk group to identify SNPs associated with outcome independent of risk group.
2. Development of Multi-SNP Predictor Models for MRD, EFS, and OS: We developed models that use the genotypes of up to three SNPs to predict MRD at the completion of induction therapy, EFS, or OS. SNPs with P < .15 in the risk-adjusted association analyses described above were included as candidate predictor variables in logistic regression (for MRD1) or proportional hazards regression (for EFS) under a Bayesian information criterion (BIC) model selection framework that evaluated all models with up to three SNPs as predictors20 as described in depth in the Data Supplement Methods section.
3. Statistical analysis of ACS10 score with outcome: The SNPs identified in the multi-SNP predictor modeling were used to construct a multi-SNP score (ACS10 score) for each patient (see results and Fig 1 for detail). MRD1, EFS, and OS were evaluated for association with ACS10 score in the AML02 and AAML0531 cohorts using Kaplan-Meier and Cox regression models for EFS and OS and logistic regression for MRD1 as described in detail in the Data Supplement methods section. All statistical analyses were performed using R software.21
FIG 1.
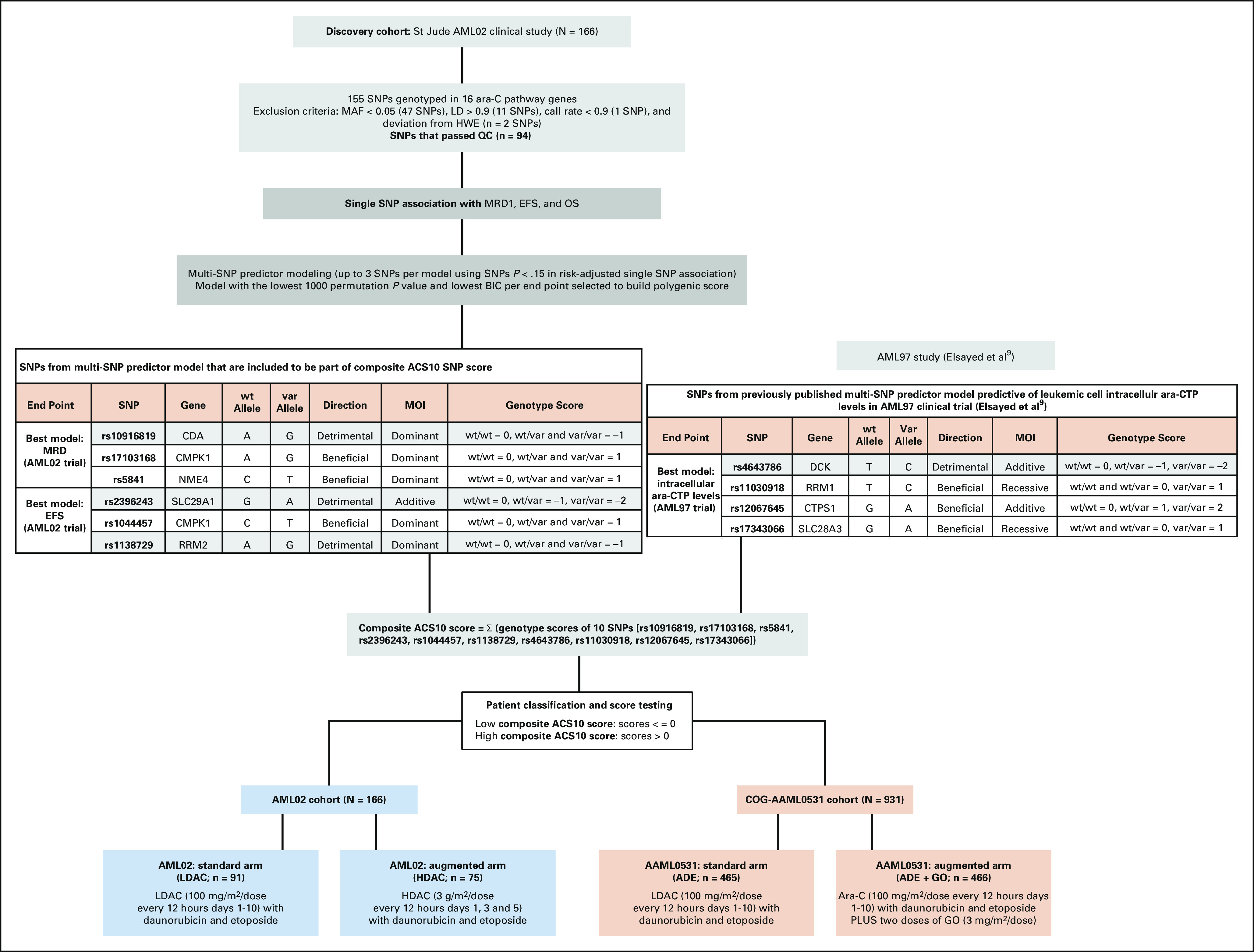
Overall study design. ACS10, ara-C pharmacogenomic 10-SNP; ADE, Ara-C, daunorubicin and etoposide; ara-C, cytarabine; ara-CTP, ara-C triphosphate; BIC, Bayesian information criterion; EFS, event-free survival; GO, gemtuzumab ozogamicin; HDAC, high-dose ara-C; HWE, Hardy-Weinberg Equilibrium; LD, linkage disequilibrium; LDAC, low-dose ara-C; MAF, minor allele frequency; MOI, mode of inheritance; MRD, minimal residual disease; MRD1, minimal residual disease at the end of induction I; OS, overall survival; QC, quality control; SNP, single nucleotide polymorphism; var, variant; wt, wild-type.
RESULTS
The overall study schema is shown in Figure 1, with details described below.
Ara-C SNPs and Outcome in the AML02 Discovery Cohort
Of the 155 SNPs genotyped in AML02 cohort, 94 passed initial quality control (SNPs excluded one with call rate < 90%; 47 with minor allele frequency < 5%; 11 high LD [r2 > 0.9] with other SNPs; and two deviated from Hardy-Weinberg equilibrium) and were included for further association analysis with multiple end points described above (Data Supplement). In unadjusted and diagnostic risk group–adjusted individual SNP analysis of the whole AML02 cohort, 34 SNPs within 12 genes were associated at the P < .05 level with at least one clinical outcome (MRD1, EFS, or OS; Data Supplement). Within treatment arms, 25 SNPs were significantly associated with at least one clinical end point in the LDAC arm and 15 SNPs within the HDAC arm (Data Supplement). Two SNPs, rs10916819 in CDA and rs507964 in SLC29A1, showed a consistent and significant detrimental association with MRD1 (LDAC arm: odds ratio [OR] = 1.75, P = .023, HDAC arm: OR = 1.87, P = .023) and EFS (LDAC arm: hazard ratio [HR] = 1.54, P = .03, HDAC arm: HR = 1.71, P = .013) in both arms, respectively.
Development of Three-SNP Predictor Models for MRD and EFS in AML02
The BIC model selection and permutation procedure found the model with a dominant mode of inheritance (MOI) of the CDA-rs10916819, a dominant MOI of the NME4-rs5841 and a dominant MOI of the CMPK1-rs17103168 to have smallest permutation P value (P = .047) and BIC model weight of 5.6% among predictor models for MRD1. This procedure also found the model with an additive MOI of the SLC29A1-rs2396243, a dominant MOI of the CMPK1-rs1044457, and a dominant MOI of the RRM2-rs1138729 to have the smallest P value (P = .059) and BIC model weight of 94.6% among predictor models for EFS. The top model for OS identified same SNPs as EFS model. The Data Supplement provides details and shows the association of individual SNPs with the respective end point.
Development of the Ara-C Pharmacogenomic 10 SNP Score
We developed the ara-C pharmacogenomic 10-SNP (ACS10) score by qualitatively combining the above results and our published study of the pharmacogenetics of intracellular ara-CTP levels9 in the AML97 clinical trial.22 These 10 unique SNPs included top three-SNP predictor model for EFS on AML02, top three-SNP predictor model for MRD1 on AML02, and four SNPs from previously published predictor model for intracellular ara-CTP levels on AML97.1 We chose to include the SNPs from all three of these models that each captured SNPs of distinct pharmacologic and clinical relevance. Each statistical model designated each SNP as having an additive, dominant, or recessive MOI and direction of association with its end point variable (EFS, MRD1, or ara-CTP levels). For each SNP, each genotype was assigned a point value of –2, –1, 0, +1, or +2 on the basis of direction of association and MOI.23 As illustrated in the table embedded within Figure 1, for each patient, the ACS10 score was defined and computed as the sum of genotype point values for the 10 SNPs as explained in equation below.
This qualitatively developed definition was fixed and then computed for each patient on the AML02 and AAML0531 clinical trials with available genotype data before performing the statistical analyses reported below (the Data Supplement shows score distribution in the two trials). Also, before subsequent analysis, we chose to dichotomize patients as low score (≤ 0) or high score (> 0), indicating whether they had more beneficial genotype points than detrimental genotype points. An example for ACS10 with hypothetical examples of patient score calculations and categorizations is shown in the Data Supplement.
Low-ACS10 Score Associates With Poor Outcome From Standard Therapy in Two Trials
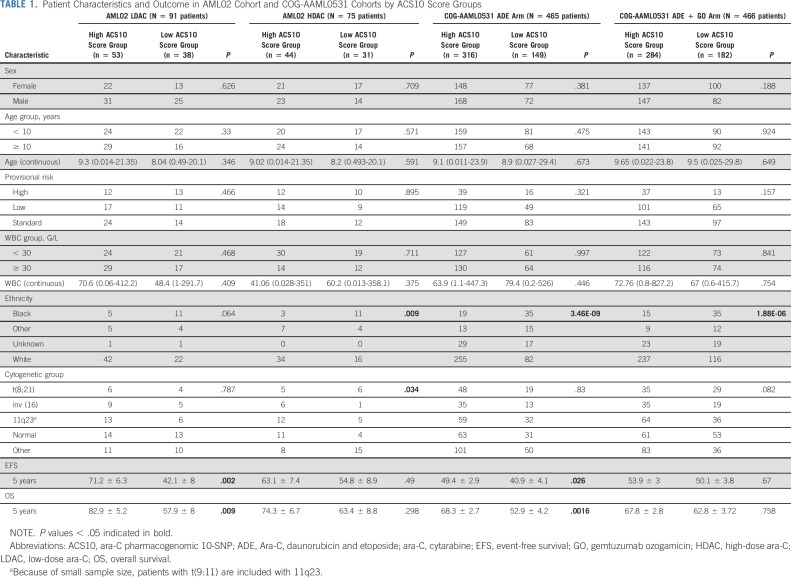
Patient characteristics and outcome end points within low-ACS10 and high-ACS10 groups from AML02 and AAML0531 cohorts are summarized in Table 1. Ethnicity was the only demographic factor found to be significantly different by score groups with approximately 70% (range, 65%-78%) of Black patients compared with approximately 30% (range, 24%-34%) White patients within low-ACS10 score group in both cohorts. A low-ACS10 score was significantly associated with poor outcome on the standard treatment arms of AML02 (LDAC arm) and the COG-AAML0531 cohorts (ADE arm). On the AML02-LDAC arm, complete response to induction I was significantly less common (OR = 3.77; 95% CI, 1.23 to 12.65; P = .012) and MRD+ rate at the end of induction I tended to be higher but was not statistically significant (ACS10 low v high OR = 1.87; 95% CI, 0.73 to 4.89; P = .19) among subjects with low-ACS10 score compared with high-ACS10 score group. Low-ACS10 score group within AML02-LDAC arm also had significantly worse EFS (ACS10 low v high HR = 2.81; 95% CI, 1.45 to 5.43; P = .002, Fig 2A) and OS (HR = 2.98; 95% CI, 1.32 to 6.75; P = .009, Fig 2B) than patients with high-ACS10 score. After adjustment for age, ethnicity, WBC, and diagnostic molecular risk, low-ACS10 score remained significantly associated with worse EFS (ACS10 low v high HR = 4.12; 95% CI, 1.83 to 9.33; P < .001, Fig 3A) and OS (HR = 4.04; 95% CI, 1.49 to 11.0; P = .006, Fig 3B) among patients on the AML02-LDAC treatment arm.
TABLE 1.
Patient Characteristics and Outcome in AML02 Cohort and COG-AAML0531 Cohorts by ACS10 Score Groups
FIG 2.
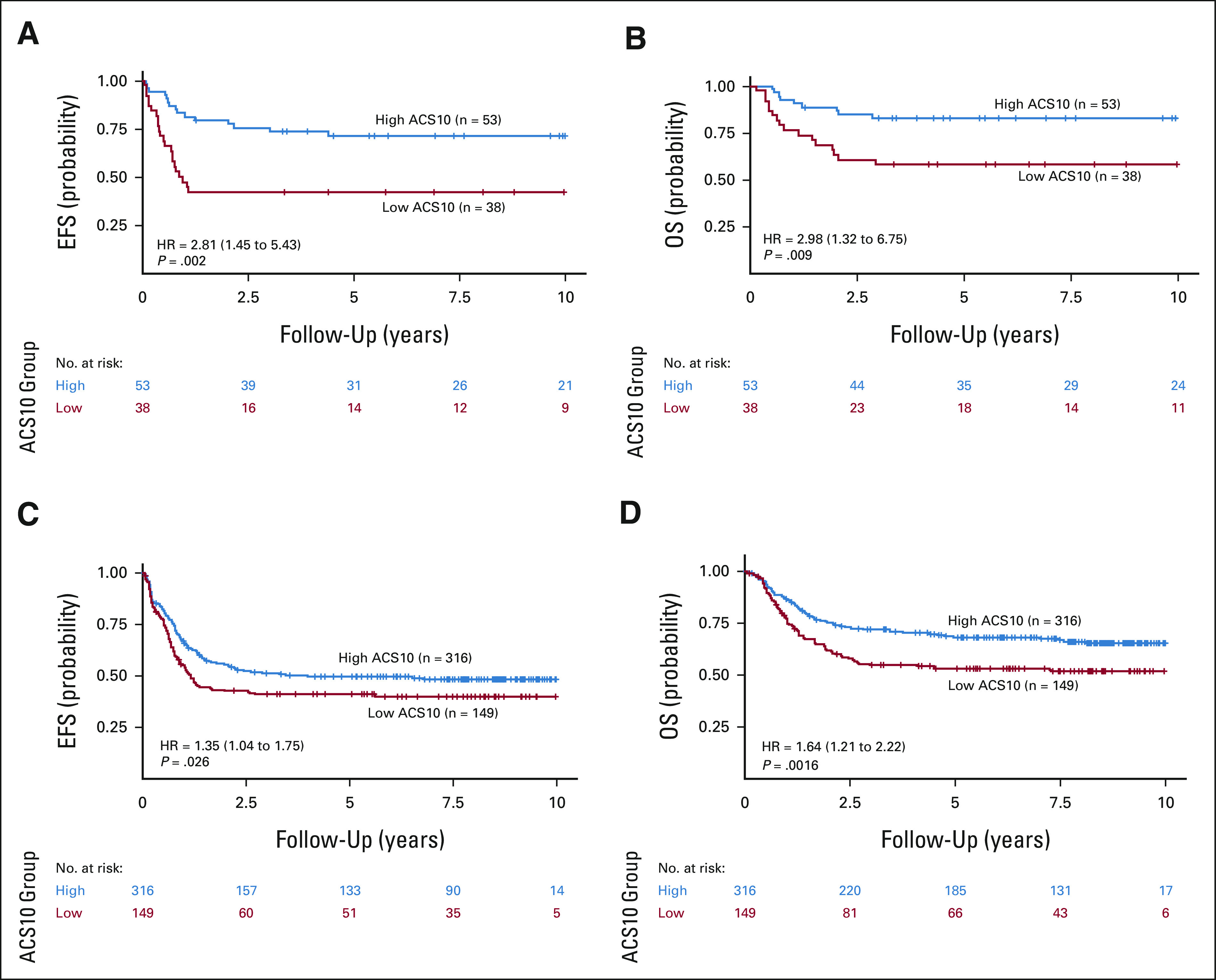
Patient outcomes by composite ACS10 score groups within standard treatment arms of AML02 and AAML0531 cohorts: (A) EFS in AML02 cohort-LDAC arm; (B) OS in AML02-LDAC arm; (C) EFS in COG AAML0531 ADE arm; and (D) OS in COG AAML0531 ADE arm. ACS10, ara-C pharmacogenomic 10-SNP; ADE, Ara-C, daunorubicin and etoposide; EFS, event-free survival; GO, gemtuzumab ozogamicin; HR, hazard ratio; OS, overall survival.
FIG 3.
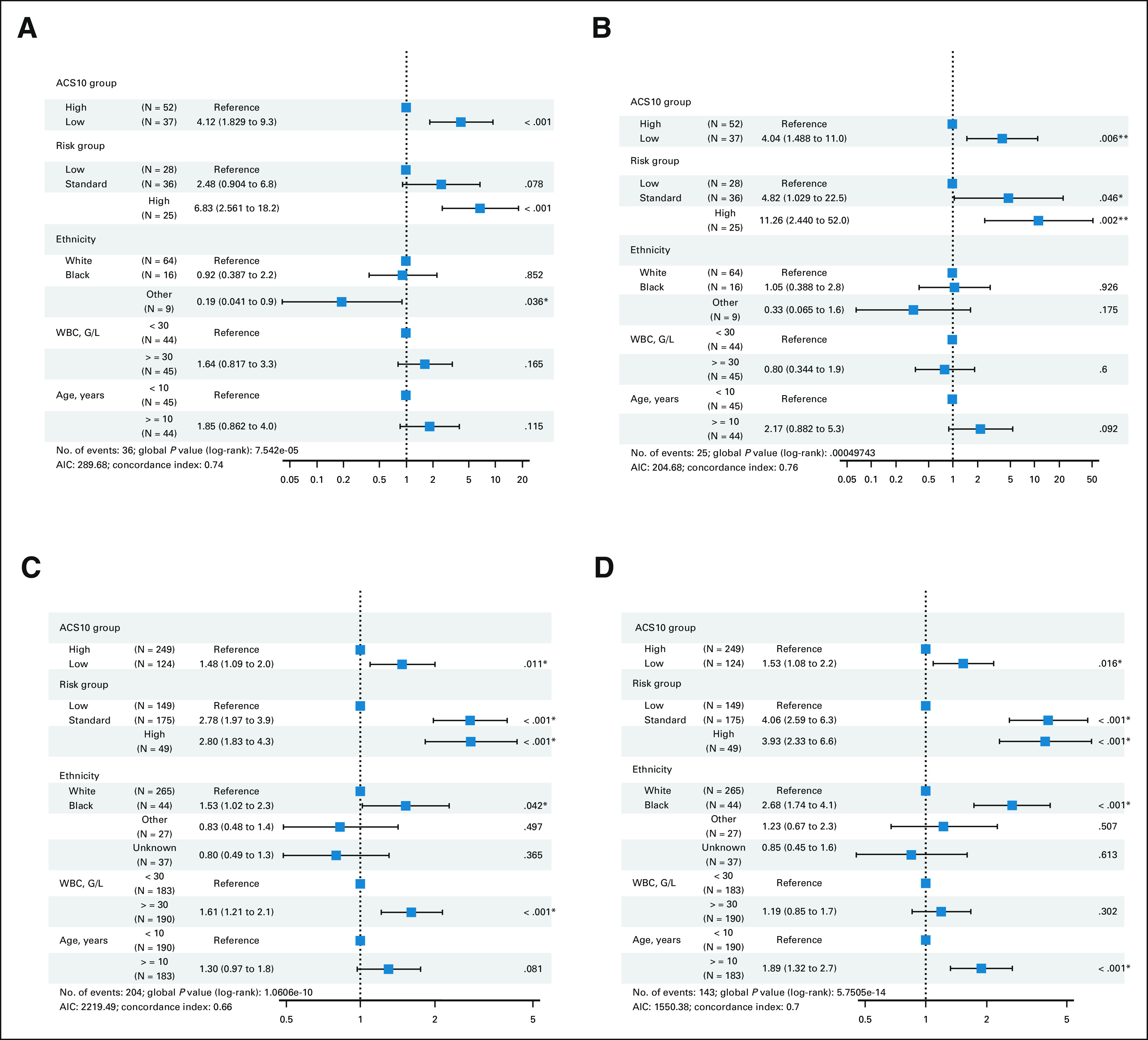
Forest plots of multivariable Cox proportional hazard models that includes ACS10 score groups, risk-group assignment, ethnicity, WBC count at diagnosis, and age for association with patient outcomes in standard treatment arms of the AML02 and AAML0531 cohorts. (A) EFS and (B) OS in the AML02 LDAC arm; (C) EFS and (D) OS in the COG-AAML0531-ADE arm. * indicates P values < 0.05. ACS10, ara-C pharmacogenomic 10-SNP; AIC, Akaike Information Criterion; EFS, event-free survival; HR, hazard ratio; GO, gemtuzumab ozogamicin; ADE, Ara-C, daunorubicin and etoposide; OS, overall survival.
Similarly, on the standard ADE arm of AAML0531, patients with low-ACS10 had significantly greater MRD1+ rate (ACS10 low v high OR = 1.69; 95% CI, 1.02 to 2.79; P = .038), worse EFS (ACS10 low v high HR = 1.35; 95% CI, 1.04 to 1.75; P = .026, Fig 2C), and worse OS (HR = 1.64; 95% CI, 1.2 to 2.22; P = .002, Fig 2D) than patients with high-ACS10 score. After adjustment for age, ethnicity, WBC, and diagnostic molecular risk, low-ACS10 score remained significantly associated with worse EFS (ACS10 low v high HR = 1.48; 95% CI, 1.09 to 2.0; P = .011, Fig 3C) and OS (HR = 1.53; 95% CI, 1.08 to 2.2; P = .016, Fig 3D). Because AAML0531 data were not used in any way to select ACS10 SNPs or define the ACS10 score, these results validate the ability of the ACS10 score to identify patients with poorer prognosis under standard therapy.
Given significant prognostic value of MRD after induction 1 chemotherapy, we evaluated ACS10 by MRD1 status. Overall, within both MRD1-negative and MRD-positive groups, ACS10-score groups demonstrated significant impact on outcome in both AML02-LDAC (Data Supplement) and AAML0531-ADE arms (Data Supplement).
ACS10 Does Not Associate With Prognosis With Augmented Therapy in Two Trials
Interestingly, ACS10 did not associate with prognosis on the augmented HDAC arm of AML02 nor the augmented ADE + GO arm of AAML0531. Within AML02-HDAC arm, though for patients with low-ACS10 score we observed greater MRD1 positivity at the end of induction I (ACS10 low v high, OR = 3.38; 95% CI, 1.13 to 10.58; P = .024), complete response to induction I did not differ (OR = 0.53; 95% CI, 0.14 to 1.91; P = .377) between low and high ACS10 score groups. In addition, within AML02-HDAC arm, low-ACS10 patients did not have significantly different EFS (ACS10 low v high HR = 1.28; 95% CI, 0.63 to 2.61; P = .49, Fig 4A) or OS (HR = 1.56; 95% CI, 0.68 to 3.6; P = .298, Fig 4B) compared with patients with high-ACS10 score. After adjustment for age, ethnicity, WBC, and diagnostic molecular risk, ACS10 score was still not associated with EFS (HR = 1.15; 95% CI, 0.53 to 2.5; P = .718) and OS (HR = 1.07; 95% CI, 0.427 to 2.7; P = .889) among the HDAC arm of AML02 (Data Supplement).
FIG 4.
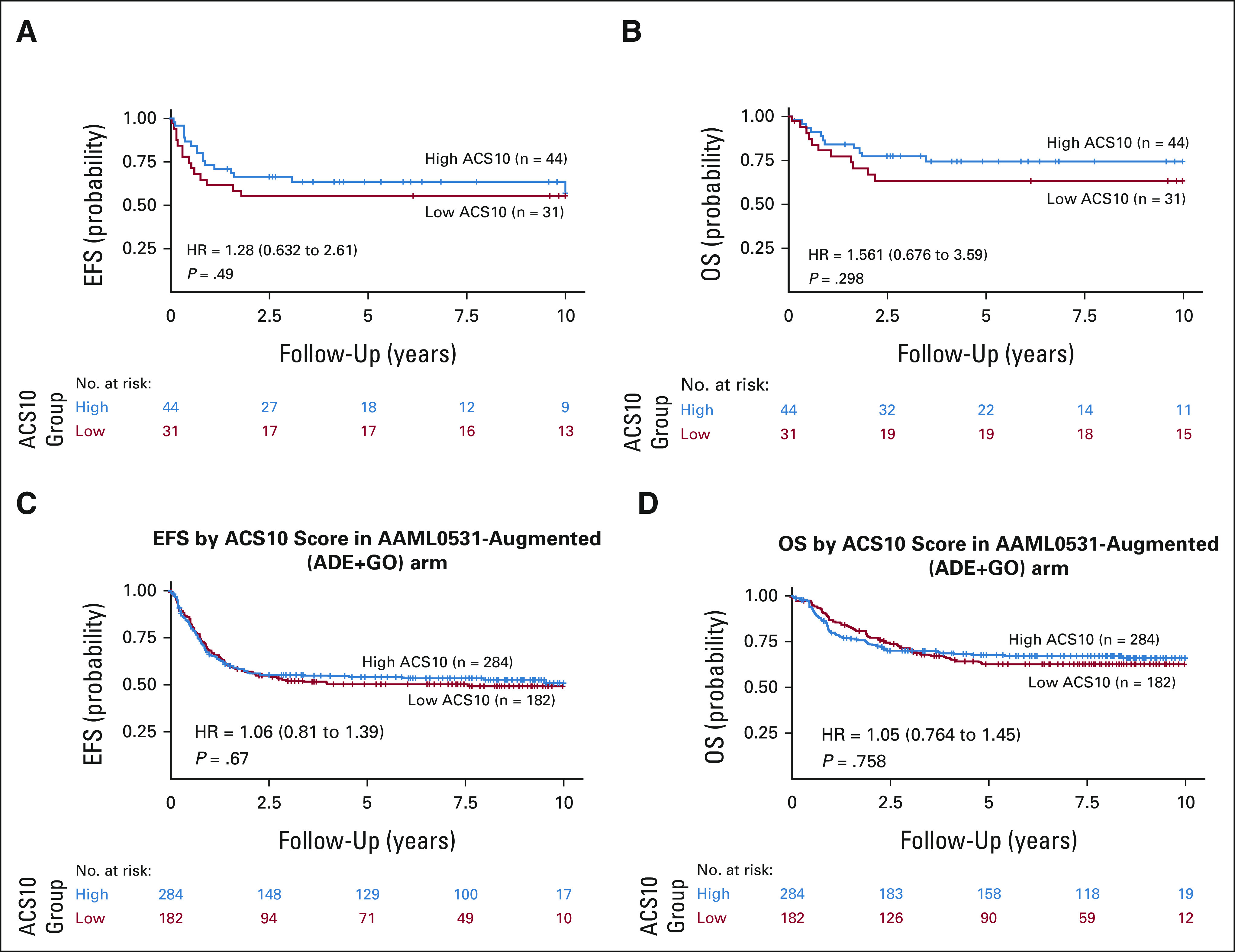
Patient outcomes by composite ACS10 score groups with augmented treatment arms: (A) EFS in the AML02 cohort-HDAC arm; (B) OS in the AML02-HDAC arm; (C) EFS in the COG AAML0531 ADE + GO arm; and (D) OS in the COG AAML0531 ADE + GO arm. ACS10, ara-C pharmacogenomic 10-SNP; ADE, Ara-C, daunorubicin and etoposide; EFS, event-free survival; GO, gemtuzumab ozogamicin; HR, hazard ratio; OS, overall survival.
On the augmented ADE + GO arm of AAML0531, patients with low-ACS10 did not have a significantly different MRD + rate at the end of induction I (ACS10 low v high OR = 1.22; 95% CI, 0.7 to 2.13; P = .499), EFS (ACS10 low v high HR = 1.06; 95% CI, 0.81 to 1.39; P = .67, Fig 4C), or OS (HR = 1.05; 95% CI, 0.76 to 1.45; P = .758, Fig 4D) compared with patients with high-ACS10 score. After adjustment for age, ethnicity, WBC, and diagnostic molecular risk, ACS10 score was still not significantly associated with EFS (HR = 1.1; 95% CI, 0.82 to 1.5; P = .479) or OS (HR = 1.1; 95% CI, 0.76 to 1.6; P = .613) among AAML0531 ADE + GO arm patients (Data Supplement).
Estimated Benefit of Augmented Therapy for Low-ACS10 Score Patients
Across the two trials, augmented therapy was consistently associated with a 10-percentage point improvement in 5-year EFS among low-ACS10 patients (Data Supplement). On the AAML0531 clinical trial, the 5-year EFS for low-ACS10 patients was 40.9% (95% CI, 33.6 to 49.7) on the standard ADE arm and 50.1% (95% CI, 43.2 to 58) with augmented ADE + GO therapy (ADE v ADE + GO HR = 1.39; 95% CI, 1.04 to 1.87; P = .027). Similarly, on the AML02 clinical trial, the 5-year EFS for low-ACS10 patients was 42.1% (95% CI, 29 to 61.1) with standard LDAC therapy and 54.8% (95% CI, 39.8 to 75.5) with augmented HDAC therapy (LDAC v HDAC HR = 1.39; 95% CI, 0.71 to 2.72; P = .337).
Additionally, low-ACS10 patients had a statistically significant improvement in 5-year OS with augmented therapy on the AAML0531 clinical trial. In this trial, low-ACS10 score had a 5-year OS of 53% (95% CI, 45.4 to 61.8) with standard ADE therapy and 62.8% (95% CI, 55.9 to 70.6) with augmented ADE + GO therapy (ADE v ADE + GO HR = 1.52; 95% CI, 1.08 to 2.14; P = .016). Also, low-ACS10 patients demonstrated improvement in OS with augmented therapy on the AML02 trial; there was a 5-year OS of 57.9% (95% CI, 44.1 to 75.9) with standard LDAC therapy and 63.4% (95% CI, 48.3 to 83.2) with augmented HDAC therapy (LDAC v HDAC HR = 1.19; 95% CI, 0.55 to 2.57; P = .651). When comparing outcome by treatment arms within ACS10 score groups, the benefit of adding GO for low-ACS10 patients retained its statistical significance in a multivariable analysis adjusting for risk group, ethnicity, WBC, count, and age, but a similar impact was not seen in the high-ACS10 score group (Data Supplement).
Estimation of ACS10 Therapy Personalization Thresholds
We also evaluated ACS10 as a numeric variable by fitting statistical models to characterize the relationship of ACS10 with outcome on each therapy arm to estimate an appropriate threshold for personalization by ACS10 score (Fig 5). For AML02, we observed that 5-year EFS and OS improved with increasing ACS10 score more rapidly for the LDAC-arm than for the HDAC treatment regimens. Our models suggest that HDAC provides better EFS and OS than LDAC for patients with ACS10 < 0 but LDAC provides better EFS and OS than HDAC for patients with ACS10 > 0 (Figs 5A and 5B). The outcomes are comparable for patients with ACS10 = 0. The personalization threshold may be different for choosing between ADE and ADE + GO in AAML0531. As shown in Figures 5C and 5D, outcomes improve with increasing ACS10 score for ADE arm, but outcomes are not strongly associated with ACS10 score on the ADE + GO arm. For patients with ACS10 score < 2, outcomes are better with ADE + GO. For patients with higher ACS10 score, outcomes are better with ADE. For patients with ACS10 score 2-3, outcomes are similar (Figs 5C and 5D).
FIG 5.
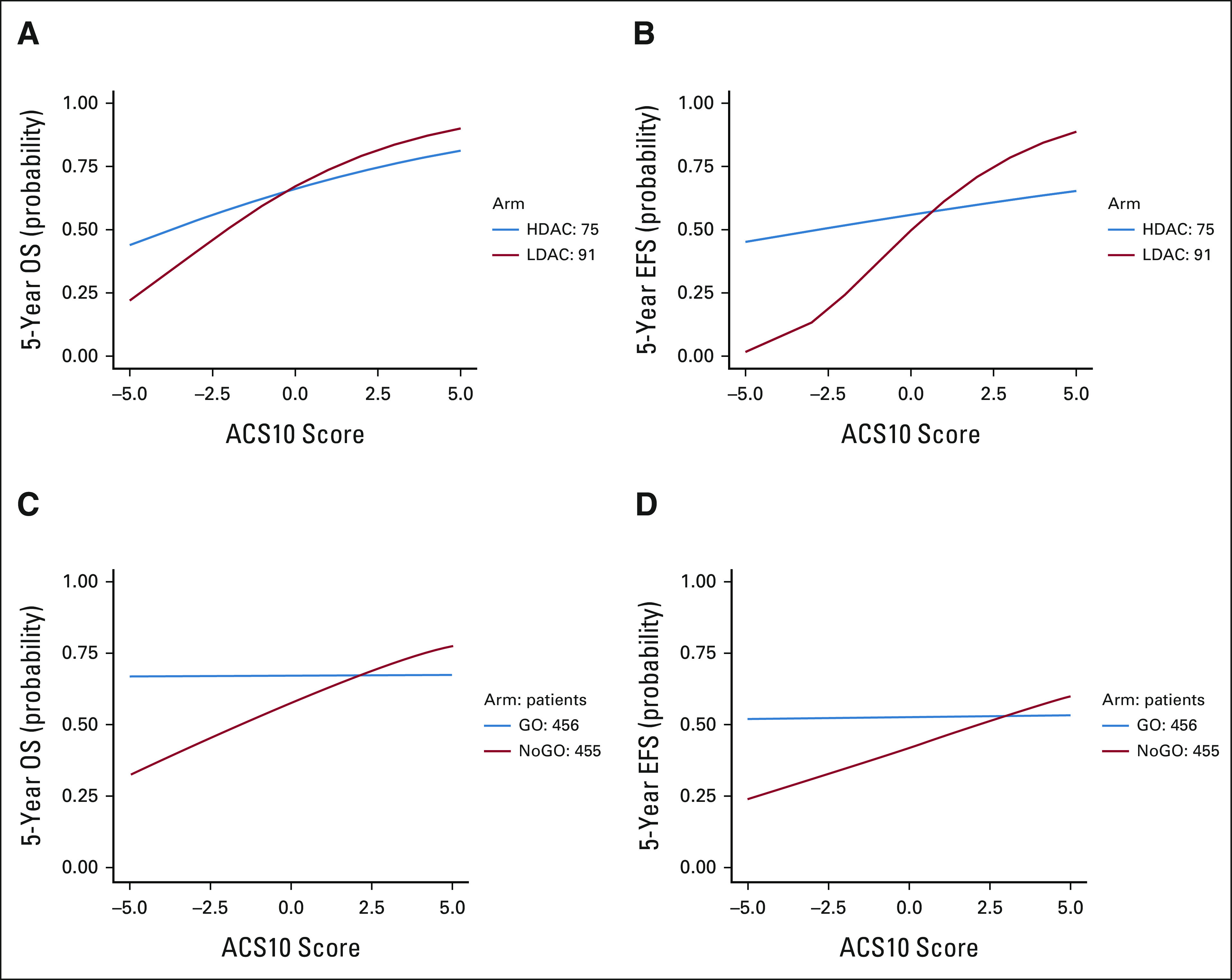
Impact of interaction between numerical ACS10 scores and treatment arms on 5-year EFS and OS in AML02 cohort and COG cohorts. (A) Five-year OS and (B) 5-year EFS in AML02 LDAC and HDAC treatment arms by ACS10 scores; (C) 5-year OS and (D) 5-year EFS in AAML0531 ADE and ADE + GO treatment arms by ACS10 scores. ACS10, ara-C pharmacogenomic 10-SNP; ADE, Ara-C, daunorubicin and etoposide; COG, Children's Oncology Group; EFS, event-free survival; GO, gemtuzumab ozogamicin; HDAC, high-dose ara-C; LDAC, low-dose ara-C; OS, overall survival.
Overall, in both cohorts, patients with low-ACS10 reflecting lower ara-CTP levels were associated with worse outcome when patients were treated with standard chemotherapy. Specifically, in AML02, low-ACS10 was detrimental in patients within the AML02-LDAC arm but not for the HDAC arm. In AAML0531, low-ACS10 was detrimental in patients within the AAML0531-ADE arm but not for the AAML0531-ADE + GO arm. These results demonstrate that low-ACS10 patients have a better outcome when treated with augmented therapy (AML02-HDAC or AAML0531-ADE + GO arms), presenting this as a personalized approach to improve outcome for this group of patients.
DISCUSSION
ara-C–based regimens have been the mainstay of AML therapy for more than five decades and are likely to remain the backbone of therapy in coming years as newly approved agents24,25 are primarily given in sequence or in combination with ara-C. Thus, efforts to incorporate newly approved agents into clinical care by genomics-guided stratification of patients can have a significant impact in AML treatment strategies. In this study, we used a multistep approach to develop a polygenic 10 SNP score (ACS10) of relevance to ara-C pharmacology. Our results show that: (1) Patients with low-ASC10 score had worse outcome when compared with those in the high-ACS10 score group in AML02 and AAML0531 trials when given standard chemotherapy regimen. However, such an impact of score was not observed in the augmented arms of the both trials (AML02-HDAC arm) and AAML0531 (ADE + GO arm). The results from AML02 trial indicate that patients with low-ACS10 score benefit from high-dose of ara-C. Further patients within high-ACS10 score had similar outcome in LDAC and HDAC arms implying that treatment with low dose ara-C might reduce the risk of toxicity. In the AAML0531 cohort, patients with low-ACS10 score demonstrated improvement in outcome with addition of GO, indicating this as an alternate treatment strategy for these patients. Thus, consistently across two trials, augmenting standard therapy with HDAC or GO mitigates the poor prognostic impact of low-ACS10 score. (2) Although sample size of our study cohorts limited our ability to perform any subgroup analysis by ethnicity or risk groups, we observed a greater abundance of the low-ACS10 score in Black patients within both AML02 and AAML0531 cohorts (approximately 30% White patients v approximately 70% Black patients had low-ACS10 score). Three of the 10 SNPs in the score contribute toward these racial differences and included a DCK SNP-rs4643786 with detrimental impact (variant allele frequency 0.038 in White v 0.48 v Black patients) and SNPs within CMPK1 (rs1044457: allele frequency 0.5 in White v 0.11 in Black patients) and SLC28A3 (rs17343066: allele frequency 0.53 in White v 0.15 in Black patients) with beneficial impact. Previous studies have reported worse outcomes in Black patients compared with White patients.26-28 Although validation in larger cohorts is required, higher abundance of low-ACS10 score within Black patients might contribute to the observed racial disparity in outcome. (3) Given MRD1 is considered a prognostic factor in predicting outcome, our results show that ACS10 score holds potential for classification of patients beyond MRD1 stratification to predict outcome and design downstream treatment strategies.
In conclusion, our comprehensive approach not only provides a unique ACS10 score of prognostic significance that can predict poor outcome in AML, but suggests that alternative treatment strategies with either high-dose ara-C or addition of GO are more suitable strategies for patients with detrimental low-ACS10 score. Our results also warrant the need to further test whether patients within low-ACS10 score groups would benefit from augmentation with other existing or newly approved agents such as clofarabine (studied in AML08 trial), glasdegib, venetoclax etc. In fact, we have reported a splicing-SNP (rs12459419C>T) in CD33 that is predictive of response to GO.29 Future studies are focused on in-depth evaluation of interaction between ACS10 score and CD33 pharmacogenomics to more accurately identify patients most likely to benefit from ADE-GO combination. Recently, a systemic review protocol on the basis of PRISMA guidelines for evaluation of ara-C–related SNPs associated with response and toxicity in AML has been published30; our results significantly contribute to this ongoing effort to accelerate evidence-based practice for better patient management. Although our results are in pediatric AML, we anticipate that the impact of ACS10 score will be observed in adult AML, warranting the need to expand this investigation in adults. Given that genotyping assays for SNPs within ACS10 score are readily available, preemptive genotyping with a rapid turnaround time can be accomplished using a wide range of samples (blood, buccal swab, or skin) in most of the clinical settings. Thus, prospective investigation of these germline polymorphisms in clinical laboratories is highly feasible. Development of a web-based tool that could be integrated into the electronic health records for easy calculation and categorization of the patient on the basis of ACS10 would accelerate clinical translation of ACS10 for making treatment decisions. Our results also open up opportunities to further refine the guidelines for customizing regimens on the basis of the pharmacogenomic evaluation of patient. Success of such examples as TPMT and NUDT15 pharmacogenomics-guided regimens to guide therapeutic decisions are already in clinics for the treatment of acute lymphoblastic leukemia.
ACKNOWLEDGMENT
NIH R01-CA132946 (J.K.L. and S.P.), ALSAC and American Society of Hematology Bridge funding (J.K.L.), UF Opportunity seed grant AGR DTD 04-26-2018 (J.K.L.), NCTN Operations Center Grant U10CA180886, American Cancer Society, SAP-21-061-01 - SBF-ACS (J.K.L.), and St Baldrick's Foundation funded the study. The authors thank Drs Campana and Coustan-Smith for minimal residual disease (MRD) data, and Egyptian Drug Authority (EDA), Egypt, for supporting A.H.E.
Abdelrahman H. Elsayed
Patents, Royalties, Other Intellectual Property: I have a patent pending application T18496 titled Pharmacogenomics Score to Make Decisions on Therapy Augmentation in AML
Christopher Cogle
Consulting or Advisory Role: Bristol Myers Squibb
Alan Gamis
Consulting or Advisory Role: Novartis
Edward Anders Kolb
Travel, Accommodations, Expenses: Roche/Genentech
Richard Aplenc
Expert Testimony: Vorys
Jeffrey Rubnitz
Consulting or Advisory Role: Kura Oncology, Biomea, Pinotbio
Research Funding: Abbvie (Inst)
Stanley Pounds
Patents, Royalties, Other Intellectual Property: I have pending patents for the six-gene pediatric leukemia stem cell score (https://pubmed.ncbi.nlm.nih.gov/31645648/) and other omics scores predictive of pediatric AML outcomes
Jatinder K. Lamba
Patents, Royalties, Other Intellectual Property: Title Status Application No Methods for Predicting AML Outcome Published PCT/US2020/051961 Development of Novel CD33 Antibodies Filed 63/078,686 CD33-Targeted Cancer Therapy Nationalized PCT/US2017/026369
No other potential conflicts of interest were reported.
See accompanying article on page 784
DISCLAIMER
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
PRIOR PRESENTATION
Presented at 2021 American Society of Clinical Pharmacology and Therapeutics annual meeting, March 15, 2021, virtual.
AUTHOR CONTRIBUTIONS
Conception and design: Abdelrahman H. Elsayed, Xueyuan Cao, Amit K. Mitra, Alan Gamis, Edward Anders Kolb, Soheil Meshinchi, Stanley Pounds, Jatinder K. Lamba
Financial support: Jatinder K. Lamba
Administrative support: Jatinder K. Lamba
Provision of study materials or patients: Susana Raimondi, Raul C. Ribeiro, Alan Gamis, Richard Aplenc, Jeffrey Rubnitz, Jatinder K. Lamba
Collection and assembly of data: Susana Raimondi, Edward Anders Kolb, Richard Aplenc, Todd A. Alonzo, Jeffrey Rubnitz, Jatinder K. Lamba
Data analysis and interpretation: Abdelrahman H. Elsayed, Xueyuan Cao, Amit K. Mitra, Huiyun Wu, Christopher Cogle, Zeina Al-Mansour, Raul C. Ribeiro, Alan Gamis, Edward Anders Kolb, Richard Aplenc, Todd A. Alonzo, Soheil Meshinchi, Stanley Pounds, Jatinder K. Lamba
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Polygenic Ara-C Response Score Identifies Pediatric Patients With Acute Myeloid Leukemia in Need of Chemotherapy Augmentation
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Abdelrahman H. Elsayed
Patents, Royalties, Other Intellectual Property: I have a patent pending application T18496 titled Pharmacogenomics Score to Make Decisions on Therapy Augmentation in AML
Christopher Cogle
Consulting or Advisory Role: Bristol Myers Squibb
Alan Gamis
Consulting or Advisory Role: Novartis
Edward Anders Kolb
Travel, Accommodations, Expenses: Roche/Genentech
Richard Aplenc
Expert Testimony: Vorys
Jeffrey Rubnitz
Consulting or Advisory Role: Kura Oncology, Biomea, Pinotbio
Research Funding: Abbvie (Inst)
Stanley Pounds
Patents, Royalties, Other Intellectual Property: I have pending patents for the six-gene pediatric leukemia stem cell score (https://pubmed.ncbi.nlm.nih.gov/31645648/) and other omics scores predictive of pediatric AML outcomes
Jatinder K. Lamba
Patents, Royalties, Other Intellectual Property: Title Status Application No Methods for Predicting AML Outcome Published PCT/US2020/051961 Development of Novel CD33 Antibodies Filed 63/078,686 CD33-Targeted Cancer Therapy Nationalized PCT/US2017/026369
No other potential conflicts of interest were reported.
REFERENCES
- 1.Tierens A, Bjorklund E, Siitonen S, et al. : Residual disease detected by flow cytometry is an independent predictor of survival in childhood acute myeloid leukaemia; results of the NOPHO-AML 2004 study. Br J Haematol 174:600-609, 2016 [DOI] [PubMed] [Google Scholar]
- 2.Gamis AS, Alonzo TA, Meshinchi S, et al. : Gemtuzumab ozogamicin in children and adolescents with de novo acute myeloid leukemia improves event-free survival by reducing relapse risk: Results from the randomized phase III Children’s Oncology Group trial AAML0531. J Clin Oncol 32:3021-3032, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubnitz JE, Inaba H, Dahl G, et al. : Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: Results of the AML02 multicentre trial. Lancet Oncol 11:543-552, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wells RJ, Adams MT, Alonzo TA, et al. : Mitoxantrone and cytarabine induction, high-dose cytarabine, and etoposide intensification for pediatric patients with relapsed or refractory acute myeloid leukemia: Children's Cancer Group Study 2951. J Clin Oncol 21:2940-2947, 2003 [DOI] [PubMed] [Google Scholar]
- 5.Bose P, Vachhani P, Cortes JE: Treatment of relapsed/refractory acute myeloid leukemia. Curr Treat Options Oncol 18:17, 2017 [DOI] [PubMed] [Google Scholar]
- 6.Döhner H, Weisdorf DJ, Bloomfield CD: Acute myeloid leukemia. N Engl J Med 373:1136-1152, 2015 [DOI] [PubMed] [Google Scholar]
- 7.Rowe JM, Tallman MS: How I treat acute myeloid leukemia. Blood 116:3147-3156, 2010 [DOI] [PubMed] [Google Scholar]
- 8.Plunkett W, Iacoboni S, Estey E, et al. : Pharmacologically directed ara-C therapy for refractory leukemia. Semin Oncol 12:20-30, 1985. (2 suppl 3) [PubMed] [Google Scholar]
- 9.Elsayed AH, Cao X, Crews KR, et al. : Comprehensive Ara-C SNP score predicts leukemic cell intracellular ara-CTP levels in pediatric acute myeloid leukemia patients. Pharmacogenomics 19:1101-1110, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao X, Mitra AK, Pounds S, et al. : RRM1 and RRM2 pharmacogenetics: Association with phenotypes in HapMap cell lines and acute myeloid leukemia patients. Pharmacogenomics 14:1449-1466, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamba JK: Genetic factors influencing cytarabine therapy. Pharmacogenomics 10:1657-1674, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamba JK, Crews K, Pounds S, et al. : Pharmacogenetics of deoxycytidine kinase: Identification and characterization of novel genetic variants. J Pharmacol Exp Ther 323:935-945, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Mitra AK, Crews KR, Pounds S, et al. : Genetic variants in cytosolic 5'-nucleotidase II are associated with its expression and cytarabine sensitivity in HapMap cell lines and in patients with acute myeloid leukemia. J Pharmacol Exp Ther 339:9-23, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amaki J, Onizuka M, Ohmachi K, et al. : Single nucleotide polymorphisms of cytarabine metabolic genes influence clinical outcome in acute myeloid leukemia patients receiving high-dose cytarabine therapy. Int J Hematol 101:543-553, 2015 [DOI] [PubMed] [Google Scholar]
- 15.Braunagel D, Schaich M, Kramer M, et al. : The T_T genotype within the NME1 promoter single nucleotide polymorphism -835 C/T is associated with an increased risk of cytarabine induced neurotoxicity in patients with acute myeloid leukemia. Leuk Lymphoma 53:952-957, 2012 [DOI] [PubMed] [Google Scholar]
- 16.Hyo Kim L, Sub Cheong H, Koh Y, et al. : Cytidine deaminase polymorphisms and worse treatment response in normal karyotype AML. J Hum Genet 60:749-754, 2015 [DOI] [PubMed] [Google Scholar]
- 17.Kim JH, Lee C, Cheong HS, et al. : SLC29A1 (ENT1) polymorphisms and outcome of complete remission in acute myeloid leukemia. Cancer Chemother Pharmacol 78:533-540, 2016 [DOI] [PubMed] [Google Scholar]
- 18.Kim SR, Saito Y, Maekawa K, et al. : Twenty novel genetic variations and haplotype structures of the DCK gene encoding human deoxycytidine kinase (dCK). Drug Metab Pharmacokinet 23:379-384, 2008 [DOI] [PubMed] [Google Scholar]
- 19.Zhang DY, Yuan XQ, Yan H, et al. : Association between DCK 35708 T>C variation and clinical outcomes of acute myeloid leukemia in South Chinese patients. Pharmacogenomics 17:1519-1531, 2016 [DOI] [PubMed] [Google Scholar]
- 20.Schwartz G: Estimating the dimension of a model. Ann Statist 6:461-464, 1978 [Google Scholar]
- 21.www.r-project.org
- 22.Rubnitz JE, Crews KR, Pounds S, et al. : Combination of cladribine and cytarabine is effective for childhood acute myeloid leukemia: Results of the St Jude AML97 trial. Leukemia 23:1410-1416, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsepilov YA, Shin SY, Soranzo N, et al. : Nonadditive effects of genes in human metabolomics. Genetics 200:707-718, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jen EY, Ko CW, Lee JE, et al. : FDA approval: Gemtuzumab ozogamicin for the treatment of adults with newly diagnosed CD33-positive acute myeloid leukemia. Clin Cancer Res 24:3242-3246, 2018 [DOI] [PubMed] [Google Scholar]
- 25.Lancet JE, Uy GL, Cortes JE, et al. : CPX-351 (cytarabine and daunorubicin) liposome for injection versus conventional cytarabine plus daunorubicin in older patients with newly diagnosed secondary acute myeloid leukemia. J Clin Oncol 36:2684-2692, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Pan J, Wang S, et al. : The epidemiological trend of acute myeloid leukemia in childhood: A population-based analysis. J Cancer 10:4824-4835, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Children's Oncology G, Aplenc R, Alonzo TA, et al. : Ethnicity and survival in childhood acute myeloid leukemia: A report from the Children's Oncology Group. Blood 108:74-80, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubnitz JE, Lensing S, Razzouk BI, et al. : Effect of race on outcome of white and black children with acute myeloid leukemia: The St. Jude experience. Pediatr Blood Cancer 48:10-15, 2007 [DOI] [PubMed] [Google Scholar]
- 29.Lamba JK, Chauhan L, Shin M, et al. : CD33 splicing polymorphism determines gemtuzumab ozogamicin response in de novo acute myeloid leukemia: Report from randomized phase III Children's Oncology Group trial AAML0531. J Clin Oncol 35:2674-2682, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puty TC, Sarraf JS, Do Carmo Almeida TC, et al. : Evaluation of the impact of single-nucleotide polymorphisms on treatment response, survival and toxicity with cytarabine and anthracyclines in patients with acute myeloid leukaemia: A systematic review protocol. Syst Rev 8:109, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]