Abstract
Background
Multidrug-resistant Klebsiella pneumoniae (MDR-KP) are becoming increasingly common over the world. The focus of this research was to get a quantitative assessment of K. pneumoniae and their multidrug resistance (MDR) profile in Nepal.
Methods
Three electronic databases: PubMed, Google Scholar, and Research4Life were used to search publications specifying K. pneumoniae infections and/or their MDR status from January 2015 to October 2021. Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines was followed for the review, and R language 4.1.1 was used for analysis. Depending upon heterogeneity of data, we used random model for pooled data to examine the prevalence of the organism and the multidrug resistance.
Results
Evaluation included 16 studies, and the pooled estimation of K. pneumoniae in total clinical samples was 3% (95% CI; 0.01–0.05). In the meta-analysis, 14 studies were combined for determining the prevalence of K. pneumoniae in total positive clinical isolates which was 16% (95% CI: 0.11–0.20), while from 12 research studies, MDR status in the pathogen was found to be 64% (95% CI, 0.53–0.74).
Conclusion
The MDR status of K. pneumoniae as well as the prevalence of the bacteria in Nepal was analyzed which showed alarming situation about administration of antibiotics and indicated choosing and developing reliable antibiotic strategies.
1. Introduction
Klebsiella pneumoniae is a Gram-negative bacteria that can be frequently found in the mouth, on the skin, and in the intestines, as well as in natural environments like water and soil [1–3]. The organism is one of the most common opportunistic bacteria linked to nosocomial and community-acquired infections, especially in immune-compromised patients responsible for causing infections in the urinary tract, respiratory tract, lower biliary duct, soft tissue, blood, surgical wounds, and liver [4–11]. K. pneumoniae has emerged as a major clinical and public health problem due to the rising prevalence of the infections caused by emerging multidrug-resistant strains [6, 7, 12].
The therapeutic options for infections caused by multidrug-resistant (MDR) K. pneumoniae are often limited. The prevalence of multidrug resistance bacterial species has risen significantly since the introduction and widespread use of new generation extended range antibiotics. By manufacturing enzymes like extended spectrum-lactamase (ESBLs), carbapenemase, and forming biofilms, the organism has been reported to develop antibiotic resistance faster than other bacteria [12, 13]. One of the primary causes in the production and spread of highly resistant bacteria for health-care-associated disorders is the intensive and continuous use of antibiotics in the hospital context [14]. The bacterium is resistant to a wide spectrum of medications, including fluoroquinolones and aminoglycosides [15–17]. As a result of increased resistance, choosing an effective antibiotic treatment for hospital-acquired infections is becoming increasingly difficult around the world [18].
Drug resistance in developing countries like Nepal have several reasons, including health-care professionals' behaviors and patients' attitudes toward antibiotic use, as well as antimicrobial supply networks in the population. This is the first meta-analysis so far according to our knowledge emphasizing in prevalence of K. pneumoniae infections and their multidrug status in Nepal. As a reason, the objective of this research was to explore at those characteristics in the organism isolated from Nepal in order to provide situation of the concerns. This analysis could create a deeper understanding about persistence of the infection and their MDR profile alerting the authorities locally and globally.
2. Methods
Following the PRISMA guidelines, this review was conducted using Medline/PubMed, Research4Life, and Google Scholar [19]. The terms used in the search were “MDR K. pneumoniae in Nepal” and “K. pneumoniae in Nepal.” The searches were restricted to articles published between 2015 and 2021, with work dates ranging from January 1, 2015, and October 20, 2021.
2.1. Eligibility Criteria
Each study's eligibility was chosen separately after the search results were examined, and any disagreements were resolved through discussion among the authors. Any discrepancies that arose during the review over whole articles were resolved by a majority vote. The title and abstract were used to evaluate the results of the initial search procedure. For inclusion and exclusion criteria, the whole texts of relevant papers were assessed.
2.2. Inclusion Criteria
Observational studies from Nepal that recorded the occurrence of K. pneumoniae in humans and/or their multidrug resistance profile were selected for quantitative synthesis. We considered all standard guidelines for antimicrobial therapy but only Clinical and Laboratory Standards Institute (CLSI) guidelines were found to be used in the included studies. Standard laboratory method included Kirby–Bauer disk diffusion method for antibiotic susceptibility test in all studies. In our study, MDR was defined as the organisms resistant to at least one antimicrobial agent in three or more antimicrobial categories.
2.3. Exclusion Criteria
The articles that reported the pathogen from samples other than human samples were excluded to minimize heterogeneity and bias. Articles that did not apply established procedures for detecting drug resistance (according to guidelines), did not provide the sample size or had inappropriate data, were also eliminated.
2.4. Data Synthesis and Analysis
The screened publications contained variables like first author, publication date, study site, sample size, K. pneumoniae, and MDR-K. pneumoniae (supplementary file 1 (available here)). Statistical studies were performed using the R programming language (Meta package). Percentages were used to represent the distributions of category variables. The incidence of bacteria and the drug resistance in clinical settings was estimated as a proportion with a 95% confidence interval and shown as a forest plot using the random effects model. To identify study heterogeneity, the Cochran Q test was utilized, with a p value of less than 0.10 indicating significant heterogeneity. The I2 statistic was used to measure how much heterogeneity contributed to the total variation in research estimates. I2 values of 25%, 25–75%, and >75% indicate low, moderate, and high heterogeneity, respectively [20].
3. Results
3.1. Summary of Selected Study
The search approach yielded a total of 7081 important potential articles. After 644 duplicates were removed, the remaining 6437 papers were screened again by title and abstract, with 79 being chosen for full-text examination. A total of 16 papers were included in the quantitative meta-analysis for determining the prevalence of the pathogen, while only 12 papers were included for analyzing their MDR profile. In searching the relevant information, 63 articles were excluded due to a lack of complete information on MDR and the presence of the target organism, as well as investigations conducted outside Nepal. Among included studies, a majority of the investigations were from tertiary care hospitals in Kathmandu valley. In the specified work duration, total samples from all included studies were 29,741 in which 4099 showed positive growth upon culture. Among those positive isolates, 643 isolates were K. pneumoniae and 327 were determined to be MDR-KP. The samples were mostly from children and adults. Most of the patients were from the general section, while a study reported patients from ventilation. Patients suffering from UTI and neonatal sepsis were also included in this study. The flowchart for study selection is shown in Figure 1.
Figure 1.
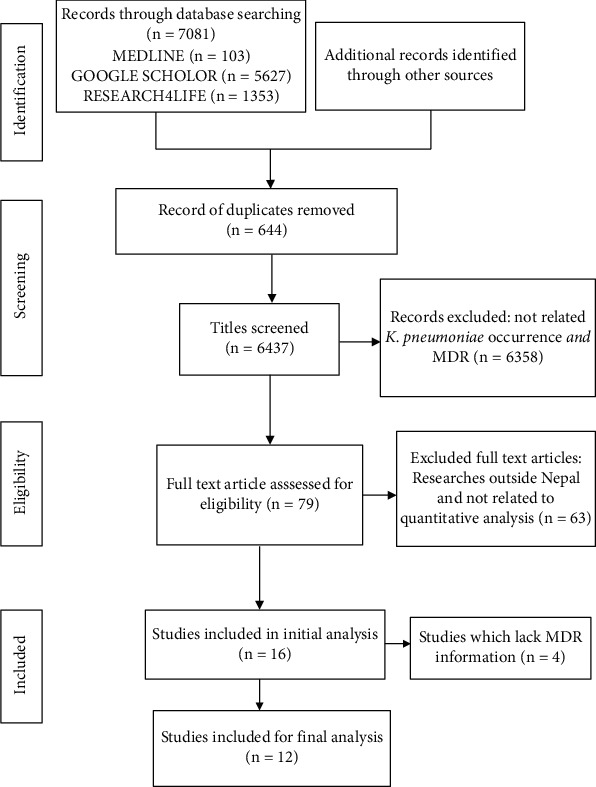
A flow diagram of the search strategy according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.
3.2. Meta-Analysis on Prevalence of K. pneumoniae
The pooled estimation of K. pneumoniae in clinical settings in various processed samples (29,741) that came in laboratory for investigations from 16 papers was 3% (95% CI; 0.01–0.05), with significant heterogeneity among studies (p < 0.01; I2 = 94%) (supplementary file 1 (available here)). The pooled estimation of the prevalence of the bacteria among the total positive isolates (4099) from 14 studies was 16% (95% CI; 0.11–0.20) with high heterogeneity (p < 0.01, I2 = 89%) (Figure 2).
Figure 2.
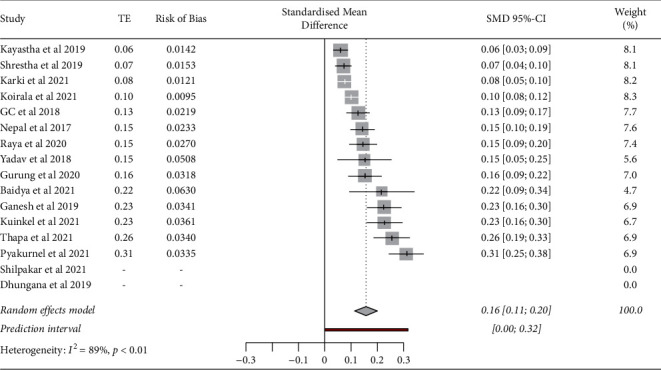
Prevalence of K. pneumoniae from clinical positive isolates in Nepal from 14 different studies ((I2 = 89%, pooled prevalence = 16%, 95% CI: 0.11–0.20, p < 0.01).
3.3. Multidrug Resistance
The pooled prevalence of multidrug resistance in total 643 K. pneumoniae from 12 researches was 64% (95% CI, 0.53–0.74). There was high heterogeneity among analyzed studies (p < 0.01, I2 = 97%) (Figure 3).
Figure 3.
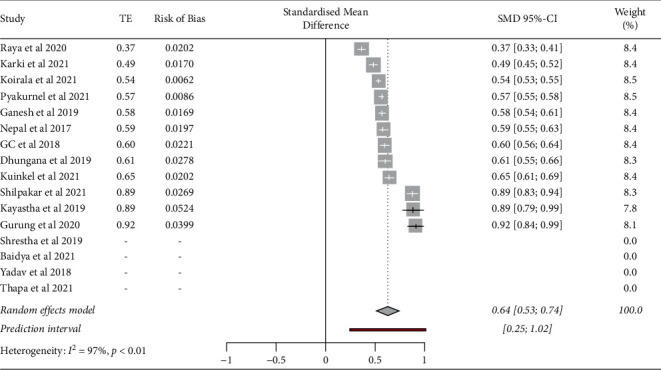
Prevalence of MDR-K. pneumoniae from clinical isolates in Nepal from 12 different studies (I2 = 97%, pooled prevalence = 64%, 95% CI: 0.53–0.74, p < 0.01).
4. Discussion
The prevalence of K. pneumoniae infections isolated from humans and their MDR status 2015 to 2021 were assessed in this meta-analysis. This is the first comprehensive meta-analysis on the prevalence of the pathogen in Nepal. We hope the information acquired thus far will help provide background aspects of drug resistance in order to avoid pan drug resistance in humans. The number of studies included in the meta-analysis was limited due to the study's search limitations. In Nepal, the occurrence of the pathogen to cause the infection ranges from about 7% to 37% displaying rise in the infection [21–36]. The results in this study determined the combined estimation of K. pneumoniae isolates in both overall suspected samples (3%) and positive isolates in the country (16%). In this investigation, a significant multidrug prevalence was found among K. pneumoniae (pooled prevalence, 64%). In various locations, the multidrug resistance was seen to vary from 6% to 91% [4, 23, 26, 27, 29–36].
Urinary tract infections, respiratory tract infections, and septicemia may be caused by K. pneumoniae, especially in immunocompromised people [8, 14, 37]. However, treatment options for infections caused by multidrug-resistant K. pneumoniae are frequently restricted [38]. Since the introduction, followed by unrestricted usage of new generation extended range antibiotics, the prevalence of multidrug resistant bacteria has increased substantially [39]. As a result of this resistance, there is a rising global difficulty with choosing an effective antibiotic treatment for hospital-acquired infections [8, 14, 18]. The pathogen is also involved in the transmission of antibiotic-resistant genes from bacteria in the environment to clinically significant pathogens [10, 40].
The meta-analysis has been important to determine the pathogen's pooled estimate and resistance to more than two classes of antibiotics. The antibiotic selective pressure in pathogen may develop the multiresistance to antibiotics, and to date, some strains of the organism have developed resistance to almost all currently available antimicrobial agents, including carbapenems, which were previously thought to be the drugs of choice for treating infection by this microorganism [31]. MDR infections are most common in complicated patients who require long-term antibiotic therapy and hospitalization and who frequently undergo invasive procedures [4, 5, 34, 38, 41]. Antimicrobial resistance risk factors may differ based on the type of organism and population analyzed [42].
Due to the small number of reports, the current study had limitations. Many of the investigations were excluded from the study due to a lack of relevant information. Out of 16 papers, only 12 studies were eligible to determine the status of multidrug resistance in the pathogen. In the meta-analysis, another limitation was the type of patient was not considered. More research into the prevalence of pathogens as well as multidrug resistance patterns in clinical settings based on infection site and patient type is required. Such research could provide a more comprehensive picture of MDR patterns in clinical settings, as well as help in controlling those resistant bacteria that have a high risk of disease development.
5. Conclusion and Recommendation
In Nepal, the prevalence of K. pneumoniae has been shown to be high (16%), with MDR patterns in the pathogen reaching up to 64%. This is a concerning scenario, and relevant authorities must remain vigilant in order to prevent worsening of the situation. This study suggests that more research into the process and reasons of antibiotic resistance in the organism, as well as the development of new antibiotics, is required. Therefore, antimicrobials used in treatment should be carefully managed.
Acknowledgments
The authors like to express their gratitude to all of the researchers of the studies included in this meta-analysis.
Data Availability
The data used to support the finding of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
The study design and the data analysis was performed by PD. RO searched for the literature and drafted the report. The manuscript was revised by PD. Both authors evaluated and approved the final version of the paper.
Supplementary Materials
Data on studies, occurrence of K. pneumoniae, MDR pattern, and publication bias are present in supplementary file 1.
References
- 1.Abd El-Rahman A. H. H. Leuven, Belgium: ATMIRE; 2017. Sensitivity of Escherichia coli, Klebsiella sp, Pseudomonas sp and Staphylococcus aureus to aqueous and alcoholic extracts of four medicinal plants. Master Thesis. [Google Scholar]
- 2.Hurst C. J. Opportunistic bacteria associated with mammalian livestock disease. In: Hurst C. J., editor. The Connections between Ecology and Infectious Disease. Advances in Environmental Microbiology . Berlin, Germany: Springer International Publishing; 2018. pp. 185–238. [DOI] [Google Scholar]
- 3.Siddhardha B., Dyavaiah M., Syed A. Biofilm Formation and Antimicrobial Drug Discovery . Berlin, Germany: Springer Nature; 2020. Model Organisms for Microbial Pathogenesis. [Google Scholar]
- 4.Holt K. E., Wertheim H., Zadoks R. N., et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proceedings of the National Academy of Sciences . 2015;112(27):E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caneiras C., Lito L., Melo-Cristino J., Duarte A. Community-and hospital-acquired Klebsiella pneumoniae urinary tract infections in Portugal: virulence and antibiotic resistance. Microorganisms . 2019;7(5):p. 138. doi: 10.3390/microorganisms7050138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu Y., Lin D., Xu Y., et al. Invasive Klebsiella pneumoniae infections in community-settings and healthcare settings. Infection and Drug Resistance . 2021;14:2647–2656. doi: 10.2147/IDR.S315871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gipson K. S., Nickerson K. P., Drenkard E., et al. The great ESKAPE: exploring the crossroads of bile and antibiotic resistance in bacterial pathogens. Infection and Immunity . 2020;88(10) doi: 10.1128/IAI.00865-19.e00865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paczosa M. K., Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiology and Molecular Biology Reviews . 2016;80(3):629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh N., Limaye A. P. Infections in solid-organ transplant recipients. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases . 2015:3440–3452. doi: 10.1016/B978-1-4557-4801-3.00313-1. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7151835/ [DOI] [Google Scholar]
- 10.Bassetti M., Righi E., Carnelutti A., Graziano E., Russo A. Multidrug-resistant Klebsiella pneumoniae: challenges for treatment, prevention and infection control. Expert Review of Anti-Infective Therapy . 2018;16(10):749–761. doi: 10.1080/14787210.2018.1522249. [DOI] [PubMed] [Google Scholar]
- 11.Frieri M., Kumar K., Boutin A. Antibiotic resistance. Journal of Infection and Public Health . 2017;10(4):369–378. doi: 10.1016/j.jiph.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Donelli G., Vuotto C. Biofilm-based infections in long-term care facilities. Future Microbiology . 2014;9(2):175–188. doi: 10.2217/fmb.13.149. [DOI] [PubMed] [Google Scholar]
- 13.Munita J. M., Arias C. A. Mechanisms of antibiotic resistance. Microbiology Spectrum . 2016;4(2) doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prestinaci F., Pezzotti P., Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathogens and Global Health . 2015;109(7):309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fair R. J., Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspectives in Medicinal Chemistry . 2014;6 doi: 10.4137/PMC.S14459.PMC.S14459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dsouza R., Pinto N. A., Hwang I., et al. Panel strain ofKlebsiella pneumoniaefor beta-lactam antibiotic evaluation: their phenotypic and genotypic characterization. PeerJ . 2017;5 doi: 10.7717/peerj.2896.e2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferreira R. L., da Silva B. C. M., Rezende G. S., et al. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-lactamase encoding genes in a Brazilian intensive care unit. Frontiers in Microbiology . 2019;9:p. 3198. doi: 10.3389/fmicb.2018.03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies J., Davies D. Origins and evolution of antibiotic resistance. Microbiology and Molecular Biology Reviews . 2010;74(3):417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Page M. J., McKenzie J. E., Bossuyt P. M., et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ . 2021;372 doi: 10.1136/BMJ.N71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins J. P. T., Thompson S. G. Quantifying heterogeneity in a meta-analysis. Statistics in Medicine . 2002;21(11):1539–1558. doi: 10.1002/SIM.1186. [DOI] [PubMed] [Google Scholar]
- 21.Yadav N. S., Sharma S., Chaudhary D. K., et al. Bacteriological profile of neonatal sepsis and antibiotic susceptibility pattern of isolates admitted at Kanti Children’s Hospital, Kathmandu, Nepal. BMC Research Notes . 2018;11(1):p. 301. doi: 10.1186/s13104-018-3394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuinkel S., Acharya J., Dhungel B., et al. Biofilm formation and phenotypic detection of ESBL, MBL, KPC and AmpC enzymes and their coexistence in Klebsiella spp. isolated at the national reference laboratory, Kathmandu, Nepal. Microbiology Research . 2021;12(3):683–697. doi: 10.3390/microbiolres12030049. [DOI] [Google Scholar]
- 23.Baidya S., Sharma S., Mishra S. K., Kattel H. P., Parajuli K., Sherchand J. B. Biofilm formation by pathogens causing ventilator-associated pneumonia at intensive care units in a tertiary care hospital: an armor for refuge. BioMed Research International . 2021;2021:10. doi: 10.1155/2021/8817700.8817700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shrestha L. B., Baral R., Poudel P., Khanal B. Clinical, etiological and antimicrobial susceptibility profile of pediatric urinary tract infections in a tertiary care hospital of Nepal. BMC Pediatrics . 2019;19(1):p. 36. doi: 10.1186/s12887-019-1410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Binod G. C., Sapkota N. R., Rayamajhee B. Detection of blaNDM-1 gene among the carbapenem resistant Escherichia coli and Klebsiella pneumoniae isolates from a children’s hospital in Nepal. Novel Research in Microbiology Journal . 2018;2(5):65–74. doi: 10.21608/NRMJ.2018.17862. [DOI] [Google Scholar]
- 26.Dhungana K., Krishna Awal B., Dhungel B., Sharma S., Banjara M. R., Rijal K. R. Detection of Klebsiella pneumoniae carbapenemase (KPC) and metallo-beta lactamase (MBL) producing gram negative bacteria isolated from different clinical samples in A transplant center, Kathmandu, Nepal. Acta Scientific Microbiology . 2019;2(12):60–69. doi: 10.31080/ASMI.2019.02.0432. [DOI] [Google Scholar]
- 27.Thapa S., Adhikari N., Shah A. K., et al. Detection of NDM-1 and VIM genes in carbapenem-resistant Klebsiella pneumoniae isolates from a tertiary health-care center in Kathmandu, Nepal. Chemotherapy . 2021;66(5-6):199–209. doi: 10.1159/000518256. [DOI] [PubMed] [Google Scholar]
- 28.Gurung S., Kafle S., Dhungel B., et al. Detection of OXA-48 gene in carbapenem-resistant Escherichia coli and Klebsiella pneumoniae from urine samples. Infection and Drug Resistance . 2020;13:2311–2321. doi: 10.2147/IDR.S259967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganesh R., Shrestha D., Bhattachan B., Rai G. Epidemiology of urinary tract infection and antimicrobial resistance in a pediatric hospital in Nepal. BMC Infectious Diseases . 2019;19(1):p. 420. doi: 10.1186/s12879-019-3997-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nepal K., Pant N. D., Neupane B., et al. Extended spectrum beta-lactamase and metallo beta-lactamase production among Escherichia coli and Klebsiella pneumoniae isolated from different clinical samples in a tertiary care hospital in Kathmandu, Nepal. Annals of Clinical Microbiology and Antimicrobials . 2017;16(1):p. 62. doi: 10.1186/s12941-017-0236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kayastha K., Pant D., Neupane B., et al. Extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella species in pediatric patients visiting international friendship children’s hospital, Kathmandu, Nepal. Infectious Diseases: Research and Treatment . 2020;13 doi: 10.1177/1178633720909798.117863372090979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raya G. B., Dhoubhadel B. G., Shrestha D., et al. Multidrug-resistant and extended-spectrum beta-lactamase-producing uropathogens in children in Bhaktapur, Nepal. Tropical Medicine and Health . 2020;48(1):p. 65. doi: 10.1186/s41182-020-00251-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pyakurel S., Ansari M., Kattel S., et al. Prevalence of carbapenemase-producing Klebsiella pneumoniae at a tertiary care hospital in Kathmandu, Nepal. Tropical Medicine and Health . 2021;49(1):p. 78. doi: 10.1186/s41182-021-00368-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koirala S., Khadka S., Sapkota S., et al. Prevalence of CTX-M β-lactamases producing multidrug resistant Escherichia coli and Klebsiella pneumoniae among patients attending bir hospital, Nepal. BioMed Research International . 2021;2021:11. doi: 10.1155/2021/9958294.9958294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shilpakar A., Ansari M., Rai K. R., Rai G., Rai S. K. Prevalence of multidrug-resistant and extended-spectrum beta-lactamase producing Gram-negative isolates from clinical samples in a tertiary care hospital of Nepal. Tropical Medicine and Health . 2021;49(1):p. 23. doi: 10.1186/s41182-021-00313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kashefieh M., Hosainzadegan H., Baghbanijavid S., Ghotaslou R. The molecular epidemiology of resistance to antibiotics among Klebsiella pneumoniae isolates in Azerbaijan, Iran. Journal of Tropical Medicine . 2021;2021:9. doi: 10.1155/2021/9195184.9195184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khalili H., Izadpanah M. Antibiotic regimens for treatment of infections due to multidrug-resistant gram-negative pathogens: an evidence-based literature review. Journal of Research in Pharmacy Practice . 2015;4(3):105–114. doi: 10.4103/2279-042X.162360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aslam B., Wang W., Arshad M. I., et al. Antibiotic resistance: a rundown of a global crisis. Infection and Drug Resistance . 2018;11:1645–1658. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam M. M. C., Wick R. R., Wyres K. L., et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microbial Genomics . 2018;4(9) doi: 10.1099/mgen.0.000196.e000196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karki D., Dhungel B., Bhandari S., et al. Antibiotic resistance and detection of plasmid mediated colistin resistance mcr-1 gene among Escherichia coli and Klebsiella pneumoniae isolated from clinical samples. Gut Pathogens . 2021;13(1):p. 45. doi: 10.1186/s13099-021-00441-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters L., Olson L., Khu D. T. K., et al. Multiple antibiotic resistance as a risk factor for mortality and prolonged hospital stay: a cohort study among neonatal intensive care patients with hospital-acquired infections caused by gram-negative bacteria in Vietnam. PLoS One . 2019;14(5) doi: 10.1371/journal.pone.0215666.e0215666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allcock S., Young E. H., Holmes M., et al. Antimicrobial resistance in human populations: challenges and opportunities. Global Health, Epidemiology and Genomics . 2017;2:p. e4. doi: 10.1017/gheg.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data on studies, occurrence of K. pneumoniae, MDR pattern, and publication bias are present in supplementary file 1.
Data Availability Statement
The data used to support the finding of this study are available from the corresponding author upon request.


