Abstract
Background
The proteinase-activated receptor 4 (PAR4) is a G-protein-coupled receptor activated by proteases such as thrombin and trypsin. Although activation of PAR4 has been shown to modulate rat gastrointestinal motility, the rat PAR4 sequence was unknown until now. This study aimed to identify the rat PAR4 cDNA.
Results
The cDNA coding for the rat PAR4 homologue was cloned from the duodenum. Northern blots demonstrated a 3.0 kb transcript in the duodenum. Protein homology with mouse and human counterparts was 90% and 75% respectively. PAR4 is expressed predominantly in the esophagus, stomach, duodenum and the spleen. When expressed in COS cells, PAR4 is activated by trypsin (1 nM), thrombin (50 nM), mouse PAR4 specific peptide (500 μM) and a putative rat PAR4 specific activating peptide (100 μM), as measured by intracellular Ca2+-changes.
Conclusions
We have identified and characterized cDNA encoding the rat PAR4 homologue. PAR4 is expressed predominantly in the upper gastrointestinal tract. It is activated by trypsin, thrombin and its newly identified rat PAR4 specific activating peptide.
Background
Proteinase-activated receptors (PARs) comprise a sub-family of G-protein coupled receptors, which are activated by serine proteases such as trypsin and thrombin. Cleavage of PARs unmasks an extracellular N-terminal domain that subsequently binds and activates the receptor, thus acting as a tethered ligand. Four related receptors have been identified to date [1-7]. Most recently, both human and mouse proteinase-activated receptor 4 (AR4) have been cloned [5,7]. Although best known for their role in the vascular and clotting systems, PARs have also been implicated as potential modulators of gastrointestinal motility [8-11] and recent studies have suggested a possible similar role for PAR4 [10]. However, to date, the rat PAR4 homologue has not been cloned and the tissue distribution of rat PAR4 in the gastrointestinal tract is unknown. In this report, we describe the cloning of a functional rat PAR4 cDNA. When expressed transiently in COS-1 cells, the rat PAR4 was activated by thrombin, trypsin, mouse PAR4 activating peptide and its own newly identified rat PAR4 tethered ligand. The rat PAR4 is mainly expressed in the rat esophagus, stomach, duodenum and the spleen.
Results
Cloning of rat PAR4
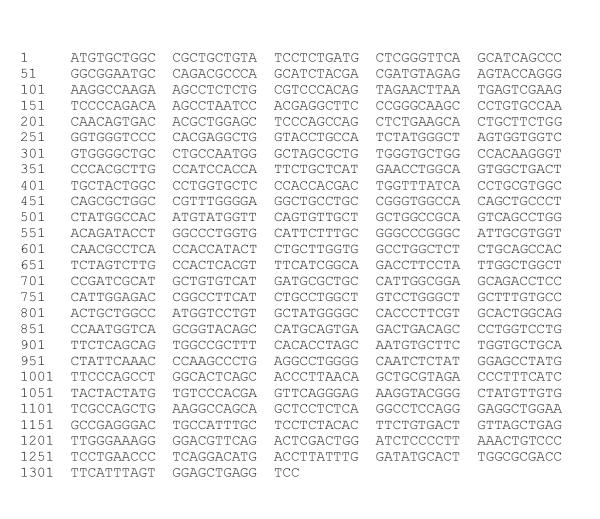
Primers, based upon the mouse PAR4 sequence, were designed to amplify the rat PAR4 coding region. We amplified a 1.1-kb fragment from rat duodenal RNA of which the nucleotide sequence shared 91 % and 82 % homology with mouse PAR4 and human PAR4 respectively. To establish the expression and corroborate the sequence of the PCR clone a rat duodenal cDNA library was screened. A partial clone was identified (0.7 kb), which contained part of the open reading frame (ORF) of the RT-PCR clone (nucleotides 596–1188) and a short segment of 3' untranslated region (nucleotides 1189–1323). To fill in the missing 5' end, 5' RACE was performed. This yielded a 0.6 kb fragment that represented the 5' end of the ORF (nucleotides 2–636). The ORF of the partial duodenal library clone and the 5' RACE fragments both shared 100% homology with the rat PAR4 PCR clone. Figure 1 shows the nucleotide sequence; figure 2 shows the deduced amino-acid sequence and its alignment with the human and mouse rat PAR4 homologue. The rat PAR4 nucleotide sequence is available in the GenBank under accession number AF310216.
Figure 1.
Nucleotide sequence of the rat PAR4 gene; CD: 1–1188.
Figure 2.
Ammo-acid sequence of rat PAR4 and alignment with mouse and human PAR4 (Genbank accession number AF080215, mouse, and AF080214, human). The putative rat PAR4 tethered ligand (GFPGKP) sequence has 3 amino-acids (GPG) in common with the human (GYPGQV) and 4 amino-acids (GPGK) with the mouse ligand (GYPGKF). Activating peptide sequences are printed bold.
Analysis of the coded amino-acid sequence revealed an overall 90% and 75% protein identity of the rat PAR4 sequence with the mouse and human PAR4, respectively. Rat PAR4 shared 33% with rat PAR1 and 35% protein identity with rat PAR2. Identification of a protease cleavage site corresponding to the human PAR4 indicated that the putative rat PAR4 AP (GFPGKP) had 3 amino-acids (GPG) in common with the corresponding human PAR4 (GYPGQV) and 4 amino-acids (GPGK) in common with the mouse (GYPGKF) sequences [5,7].
Tissue distribution of rat PAR4
Northern blot analysis of poly(A)+ RNA from the duodenum detected a 3.0 kb band (Figure 3).
Figure 3.
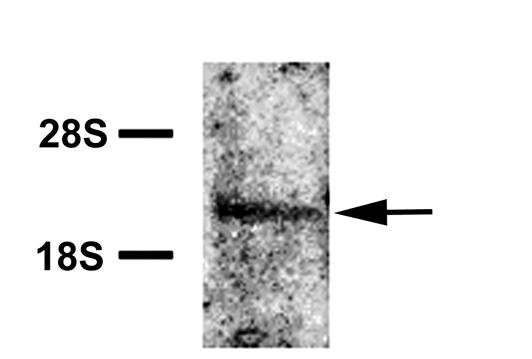
Northern blot of rat duodenal mRNA hybridized to a rat PAR4 probe. Left: ribosomal markers. The single hybridizing band corresponds to a mRNA of ≅ 3 kb.
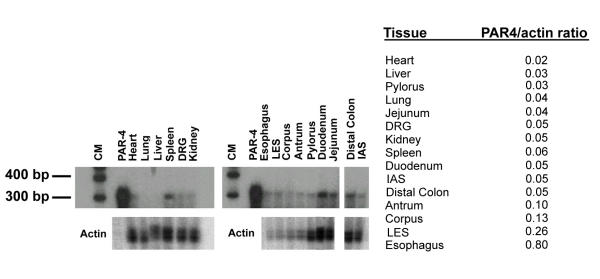
This corresponds in size to the predominant band previously described in human tissues. Expression in other tissues was measured by RPA (Figure 4).
Figure 4.
A Detection of PAR4 mRNA in rat tissues by RPA. Tissue sources are given at the top and the size of the protected fragment is given on the left. (CM, central marker; LES, lower esophageal sphincter; IAS, internal anal sphincter; DRG, dorsal root ganglia). B The PAR4/actin ratio suggests that PAR4 is predominantly expressed in the esophagus, LES, and the stomach.
Various amounts of total RNA were used based upon availability of the tissue, however analysis of the total amounts of PAR4 was expressed as the PAR4/actin ratio, thereby correcting for the different amounts of total RNA. Results showed that PAR4 is expressed at low levels in multiple gastrointestinal tissues including the esophagus, stomach, duodenum, jejunum and distal colon. It is also present in the spleen.
Functional studies
COS-1 and COS-7 cells were transiently transfected with a pcDNA3.1+ expression vector containing the rat PAR4 1.1 kb amplimer. Cells were treated with trypsin, thrombin, mouse PAR4 AP (GYPGKF) or rat PAR4 AP (GFPGKP) and changes in intracellular Ca2+ were measured. The reversed sequence of the mouse PAR4 AP (FKGPYG) was used as a control peptide, as we did not have the rat PAR4 control peptide available. In addition, untransfected COS cells were used as controls.
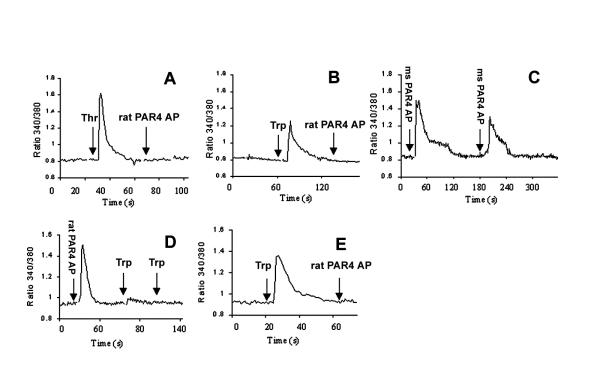
Transfected cells responded to mouse PAR4 AP 500 μM, trypsin 1 nM, thrombin 50 nM and the putative rat PAR4 AP 100 μM (Figure 5). Control COS cells showed no response to any agonists tested (data not shown) and transfected cells failed to respond to the mouse control peptide. After incubation with mouse PAR4 AP, the cells did respond to a second incubation with the mouse PAR4 AP 500 μM, although the magnitude of the response was less. Rat PAR4 AP caused an increase in intracellular Ca2+ at 100 μM. Subsequent stimulation of the receptor with trypsin did not demonstrate an increase in intracellular Ca2+. When these agonists were applied in reversed order, the same observations were made: the receptor was activated by trypsin but subsequently did not respond to stimulation with rat PAR4 AP.
Figure 5.
PAR4 mediated Ca2+ mobilization in transfected COS cells. (A) Rat thrombin 50 nM (1st arrow) followed by rat PAR4 AP 100 μM (2nd arrow), (B) Trypsin 1 nM (1st arrow) followed by rat PAR4 AP 100 μM (2nd arrow), (C) Mouse PAR4 AP 500 μM followed by repeat mouse PAR4 AP 500 μM, (D) Rat PAR4 AP 100 μM (1st arrow) followed by trypsin 1 nM (2nd and 3rd arrow), (E) Trypsin 1 nM (1st arrow) followed by rat PAR4 AP 100 μm (2nd arrow). Thr, thrombin; trp, trypsin; ms PAR4 AP, mouse PAR4 activating peptide.
Discussion
We have cloned a rat PAR cDNA from the rat duodenum. Based on its sequence homology with the mouse and human PAR4 and its functional characteristics we conclude that the amplimer represents the rat PAR4 homologue. Although our cloning strategy involved PCR techniques in both the initial PCR cloning of the rat PAR4 and in filling in the partial cDNA library clone with 5' RACE, we feel confident that the sequence of the rat receptor as reported here, is accurate. In both cases, we used the most accurate proofreading polymerase known, Pfu-Turbo, and in essence, we independently derived two PCR clones from our cloning experiments and found that their sequence was identical. In addition, the rat PAR4 is fully functional in a transient transfection assay, thus corroborating the accuracy of the sequence.
Synthetic peptides that mimic the tethered ligands of the PAR receptors have been used as specific agonists to study the function of these receptors. Site-directed mutagenesis studies by Xu et al., who simultaneously and independently cloned the human PAR4 receptor, suggested that the Arg-47/Gly-48 cleavage site is critical for subsequent receptor activation by thrombin and trypsin [5]. The rat PAR4 receptor contains such a cleavage site at position Arg-58/Gly-59. Indeed, both thrombin and trypsin function as a ligand for the rat PAR4 receptor.
The putative rat PAR4 tethered ligand contains a phenylalanine in the second position, as do the human PAR1 and PAR3. This phenylalanine in the second position of the tethered ligand of the human PAR1 and PAR3 receptor is critical for function of this domain and we speculate that it is of equal importance in the rat PAR4 tethered ligand. By contrast, in the mouse PAR4, it is a tyrosine in the second position of the tethered ligand that is critical [7]. Future studies, requiring the construction of different receptor-related peptides using site-directed mutagenesis, are needed to confirm the role of the various amino-acids in conferring activity to the PAR4 ligand.
Although the mouse PAR4 tethered ligand (GYPGKF) differs from the putative rat PAR4 AP sequence (GFPGKP), it was nonetheless able to activate the rat PAR4 transfected COS cells. This may represent the ability of mouse PAR4 activating peptide to serve as a partial agonist, causing enough of an alteration in the conformation of the receptor to initiate transduction of a signal across the plasma membrane. In keeping with this theory, in vitro studies using rat tissue have previously reported functional responses to mouse PAR4 activating peptide [10,11].
When transfected COS cells were challenged with either rat PAR4 AP or trypsin, activation occurred as demonstrated by an increase in intracellular Ca2+. However, subsequent stimulation with rat PAR4 AP or trypsin did not evoke a biological response suggesting that desensitization had occurred. It is conceivable that activation of the receptor prevents subsequent stimulation by a second agonist, analogous to studies on PAR2 mechanisms of desensitization [12]. By contrast, mouse PAR4 AP did not completely abolish a response to a second exposure to mouse PAR4 AP, although the magnitude of the response was less. This may be due to a weaker agonistic effect, reflecting differences in its sequence.
Rat PAR4 message is found mainly in the esophagus, stomach, the duodenum and the spleen. The potential effect of PAR4 activation in the upper gastrointestinal tract and a hematopoietic organ like the spleen is an interesting topic for further study. Recently Andrade-Gordon et al partially cloned guinea-pig PAR4 and identified the guinea-pig activating peptide SFPGQA [14]. Guinea pig PAR4 appears to be involved in platelet aggregation. However, the exact physiological and pathophysiological roles of PARs in the gastrointestinal tract remains to be fully defined. Several studies have suggested a role for PARs in the modulation of gastrointestinal motility [8-10] and based on previous literature using cross-species experiments (mouse AP on rat GI tissue), a role for PAR4 has also been proposed in gastric contractility [11] and in the relaxation of esophageal muscularis mucosae [10]. The identification of the rat PAR4 homologue, its tethered ligand and its tissue distribution will now allow the testing of the specific PAR4 AP in various well established models of rat motility or other aspects of gastrointestinal function such as epithelial ion transport, as has been suggested by some studies [15].
In summary, we identified a rat PAR4 cDNA sequence from a rat duodenal cDNA library. A putative protease cleavage site (Arg-58/Gly-59) was identified. The rat PAR4 clone mobilized calcium in transiently transfected COS cells in response to thrombin, trypsin, mouse PAR4 AP and its newly identified rat PAR4 AP. The highest levels of expression of this receptor were observed in the esophagus and the stomach.
Methods
RT-PCR cloning of rat PAR4
Total RNA was made from the duodenum of normal male Sprague-Dawley rats, using the Ultraspec™ RNA isolation system (Biotex Laboratories, Inc., Houston, TX) and treated with DNAse I. Using the ProSTAR Ultra HF RT-PCR System (Stratagene, La Jolla, CA), RT-PCR was performed with mouse PAR4 specific primers (Forward primer 5'-ATG TGC TGG CCG CTG CTG TA-3' and reverse primer 5'-CAG TCA CAG AAG TGT AGA GG-3', Genbank accession number AF080215) which were designed to amplify the entire coding sequence. PCR conditions were as follows: initial denaturation 95°C for 1 min (1 cycle), 95°C for 1 min, 56°C for 1 min, 68°C for 15 min (40 cycles), 68°C final extension for 10 minutes. The PCR products were run on a low melt agarose gel, excised and purified with the PCR prep kit (Promega, Madison, WI) and blunt-end ligated into a plasmid vector, pGEM 5Z(f)+ (Promega, Madison, WI). The PCR clone was completely sequenced using an automated ABI sequencer and its identity as a rat PAR4 was established by alignment of its sequence with the mouse and human PAR4 clones. To confirm the sequence of the rat PAR4 amplimer, a rat duodenal cDNA library was screened.
DNA library screening
Approximately 1 × 106 recombinant phage of a rat duodenal cDNA library constructed with the ZAP Express cDNA synthesis kit (Stratagene, La Jolla, CA) were screened using standard filter hybridization techniques with a 32P-labeled random primer (Prime-it II kit; Stratagene, La Jolla, CA) rat PAR4 PCR amplimer as a probe. The filter was washed at moderate stringency (20 min at 37°C in 2× SSC, 0.1% SDS and for 20 min at 55°C in 0.5× SSC, 0.1% SDS) and subsequently exposed to Biomax film with an intensifying screen overnight at -80°C. A single cDNA clone was plaque-purified after two more rounds of screening and subjected to in vivo excision as per manufacturer's protocol. The rat PAR4 cDNA was sequenced completely and found to be missing 595 bases at the 5' end.
5' RACE
Rapid amplification of cDNA ends (RACE) technique was used to complete the rat PAR4 cDNA sequence (Marathon RACE kit, Clontech, Palo Alto, CA). Double stranded rat duodenal cDNA was ligated to universal adapter primer and used as a template in 5' RACE reactions. The adapter-ligated cDNA was used in a PCR reaction with a sense primer corresponding to the adapter primer sequence and a PAR4 specific antisense primer (nucleotides 698–718 of the rat PAR4 sequence). RACE PCR conditions were similar as described under RT-PCR and also performed using a proofreading DNA polymerase (Pfu-Turbo-Stratagene La Jolla, CA). Fragments of the expected size were run on a low melt agarose gel, purified as described previously and blunt-end ligated into a plasmid vector, pGEM 5Z(f)+ (Promega, Madison, WI). The identity of the PCR product was confirmed by sequence analysis using an automated ABI sequencer.
Northern blot analysis
mRNA was purified using the mRNA Poly(A)Pure Purification Kit (Ambion, Austin, TX). Two μg of duodenal mRNA was run on a 1.0 % formaldehyde-agarose gel, transferred to a nylon membrane and UV-crosslinked. The Northern blot was pre-hybridized and hybridized in ExpressHyb (Clontech, Palo Alto, CA) at 68°C with the 32P-labeled random primer rat PAR4 PCR-amplified DNA clone. The membrane was washed at high stringency (20 min at 37°C in 2× SSC, 0.1% SDS, 20 min at 65°C in 0.1 × SSX, 0.1% SDS and exposed to Biomax film with an intensifying screen overnight at -80°C.
RNAse Protection assay
The levels of rat PAR4 transcripts in a variety of rat tissues were analyzed using a ribonuclease protection assay (RPA). The RPA template DNA was amplified by PCR incorporating a T7 promoter sequence before the antisense primer (5'CTCGGTAATACGACTCACTATAGGGATGGCAAGCGTGGCACCCTTGTG-3'). The antisense riboprobe was labeled with alpha-[32-P] uridine triphosphate (UTP) according to manufacturer's protocol (Maxiscript, Ambion Inc., Austin, TX). Various amounts of total RNA were precipitated and dissolved in 8 μl of hybridization buffer. One microliter each of rat PAR4 (80,000 counts/μl) and (β-actin (10,000 counts/μl) riboprobes were added to the hybridization mixture. Samples were heated to 90°C for 5 min and then hybridized at 56°C for 16 hours. After hybridization and RNase treatment, protected fragments were resolved on a 6.0 % sequencing gel. Radioactive bands were quantified with Cyclone Storage Phoshor Imaging System (Packard, Meriden, CT). PAR4 ratios were presented against the actin band for each tissue.
Rat PAR4 functional studies
Two oligonucleotide primers were designed to amplify the entire rat PAR4 coding region from rat duodenal RNA. The 5' primer contained a Bam HI site and a Kozak sequence (5'-GCTGGTACCGGATCCGCCATGTGCTGGCCGCTGCTGTATCCTCTGATG-3') and the antisense primer contained a Xba1 site (5'-GCGTCTAGAGCGGCCGCATCACAGAAGTGTAGAGGAGCAAATGGCAGT-3'). The amplified PAR4 cDNA was cleaved with the restriction enzyme Bam H1 and Xba1, directionally cloned into the expression vector pcDNA3.1+ (Invitrogen, Carlsbad, CA) and its sequence verified. For the tranfection, COS-7 cells and COS-1 cells were grown in DMEM with 10% fetal bovine serum (FBS) supplemented with penicillin/streptomycin. Cells were plated 1 day before transfection. PAR4 DNA (1 μg) was cotransfected with green fluorescent protein (GFP) according to manufacturer's protocol (FuGene™ 6 Transfection Reagent; Roche, Indianapolis, IN) and the cells were incubated for 36–48 hours. The cells were then loaded with the calcium indicator fura-2 and [Ca2+]i-mobilization was measured using the 340/380 ratio. Specific PAR4 agonists were used including trypsin (Sigma, St. Louis, MO), rat thrombin (Sigma, St. Louis, MO) and rat and mouse PAR4 AP. (rat PAR4, GFPGKP-NH2; mouse PAR4, GYPGKF-NH2; reversed mouse PAR4, FKGPYG-NH2. All peptides were synthesized at the UTMB Core Protein Facility using standard solid phase synthesis procedures).
Abbreviations
PAR, proteinase-activated receptor; RPA, ribonuclease protection assay; AP, activating peptide; ORF, open reading frame
Acknowledgments
Acknowledgement
We thank Brenda Kenworthy for excellent technical assistance in the preparation of this manuscript.
Contributor Information
Willemijntje A Hoogerwerf, Email: wahooger@utmb.edu.
Helen Lee Hellmich, Email: hhellmic@utmb.edu.
Maria Adelaide Micci, Email: mmicci@utmb.edu.
John H Winston, Email: lzou@utmb.edu.
Lei Zou, Email: jhwinsto@utmb.edu.
Pankaj J Pasricha, Email: jpasrich@utmb.edu.
References
- Vu TK, Hung DT, Wheaton VI, Coughlin SR. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell, 1991;64:1057–1068. doi: 10.1016/0092-8674(91)90261-V. [DOI] [PubMed] [Google Scholar]
- Nystedt S, Emilsson K, Wahlestedt C, Sundelin J. Molecular cloning of a potential proteinase activated receptor. Proc Natl Acad Sci, 1994;91:9208–9212. doi: 10.1073/pnas.91.20.9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt S, Emilsson K, Larsson AK, Strömbeck B, Sundelin J. Molecular cloning and functional expression of the gene encoding the human proteinase-activated receptor 2. Eur J Biochem, 1995;232:84–89. doi: 10.1111/j.1432-1033.1995.tb20784.x. [DOI] [PubMed] [Google Scholar]
- Saifeddine M, Al-Ani B, Cheng C-H, Wang L, Hollenberg MD. Rat proteinase-activated receptor-2 (PAR-2): cDNA sequence and activity of receptor-derived peptides in gastric and vascular tissue. Br J Pharmacol, 1996;118:521–530. doi: 10.1111/j.1476-5381.1996.tb15433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Andersen H, Whitmore TE, Presnell SR, Yee DP, Ching A, Gilbert T, Davie EW, Foster DC. Cloning and characterization of human protease-activated receptor 4. Proc Natl Acad Sci, 1998;95:6642–6646. doi: 10.1073/pnas.95.12.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H, Connolly AJ, Zeng D, Kahn ML, Zheng YW, Tommons C, Tram T, Coughlin SR. Protease-activated receptor 3 is a second thrombin receptor in humans. Nature, 1997;386:502–506. doi: 10.1038/386502a0. [DOI] [PubMed] [Google Scholar]
- Kahn ML, Zheng YW, Huang W, Bigornia V, Zen D, Moff S, Farese JR RV, Tam C, Coughlin SR. A dual thrombin receptor system for platelet activation. Nature, 1998;394:690–694. doi: 10.1038/29325. [DOI] [PubMed] [Google Scholar]
- Cocks TM, Sozzi V, Moffatt JD, Selemidis S. Protease-activated receptors mediate Apamin-sensitive relaxation of mouse and guinea pig gastrointestinal smooth muscle. Gastroenterology. 1999;116:586–592. doi: 10.1016/s0016-5085(99)70180-0. [DOI] [PubMed] [Google Scholar]
- Kawabata A, Kuroda R, Nishikawa H, Kawai K. Modulation by protease-activated receptors of the rat duodenal motility in vitro: possible mechanisms underlying the evoked contraction and relaxation. Br J Pharmacol, 1999;128:865–872. doi: 10.1038/sj.bjp.0702755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata A, Kuroda R, Kuroki N, Nishikawa H, Kawai K. Dual modulation by thrombin of the motility of rat oesophageal muscularis mucosae via two distinct protease-activated receptors (PARs): a novel role for PAR-4 as opposed to PAR-1. Br J Pharmacol, 2000;131:578–584. doi: 10.1038/sj.bjp.0703590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg MD, Saifeddine M, Al-Ani B, Gui Y. Proteinase-activated receptor 4 (PAR4): action of PAR-4 activating peptides in vascular and gastric tissue and lack of cross-reactivity with PAR-1 and PAR-2. Can J Physiol, 1999;77:459–464. [PubMed] [Google Scholar]
- Bohm SK, Khitin LM, Grady EF, Aponte G, Payan DG, Bunnett NW. Mechanisms of desensitization and resensitization of proteinase-activated receptor-2. J Biol Chem, 1996;271:22003–22016. doi: 10.1074/jbc.271.36.22003. [DOI] [PubMed] [Google Scholar]
- Déry O, Corvera CU, Steinhoff M, Bunnett NW. Proteinase-activated receptors: novel mechanisms of signaling by serine proteases. Am J Physiol, 1998;274:C1429–C1452. doi: 10.1152/ajpcell.1998.274.6.C1429. [DOI] [PubMed] [Google Scholar]
- Andrade-Gordon P, Derian CK, Maryanoff BE, Zhang HC, Addo MF, Cheung W, Damiano BP, D'Andrea MR, Darrow AL, de Garavilla L, Eckardt AJ, Giardino EC, Haertlein BJ, McComsey DF. Administration of a potent antagonist of protease-activated receptor-1 (PAR-1) attenuates vascular restenosis following balloon angioplasty in rats. J Pharmacol Exp Ther, 2001;298:34–42. [PubMed] [Google Scholar]
- Vergnolle N, MacNaughton WK, Al-Ani B, Saifeddine M, Wallace JL, Hollenberg MD. Proteinase-activated receptor 2 (PAR-2)-activating peptides: Identification of a receptor distinct from PAR-2 that regulates intestinal transport. Proc Natl Acad Sci, 1998;95:7766–7771. doi: 10.1073/pnas.95.13.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]