Abstract
Background
The metalloprotease-disintegrin family, or ADAM, proteins, are implicated in cell-cell interactions, cell fusion, and cell signaling, and are widely distributed among metazoan phyla. Orthologous relationships have been defined for a few ADAM proteins including ADAM10 (Kuzbanian), and ADAM17 (TACE), but evolutionary relationships are not clear for the majority of family members. Human ADAM33 refers to a testis cDNA clone that does not contain a complete open reading frame, but portions of the predicted protein are similar to Xenopus laevis ADAM13.
Results
In a 48 kb region of mouse DNA adjacent to the Attractin gene on mouse chromosome 2, we identified sequences very similar to human ADAM33. A full-length mouse cDNA was identified by a combination of gene prediction programs and RT-PCR, and the probable full-length human cDNA was identified by comparison to human genomic sequence in the homologous region on chromosome 20p13. Mouse ADAM33 is 44% identical to Xenopus laevis ADAM13, however a phylogenetic alignment and consideration of functional domains suggests that the two genes are not orthologous. Mouse Adam33 is widely expressed, most highly in the adult brain, heart, kidney, lung and testis.
Conclusions
While mouse ADAM33 is similar to Xenopus ADAM13 in sequence, further examination of its embryonic expression pattern, catalytic activity and protein interactions will be required to assess the functional relationship between these two proteins. Adam33 is expressed in the mouse adult brain and could play a role in complex processes that require cell-cell communication.
Background
The metalloprotease-disintegrin family of proteins (called ADAMs, or MDC proteins) consists of over 30 members identified in various species. These proteins are membrane-anchored glycoproteins, named for two of the motifs they carry: adisintegrin domain, and ametalloprotease domain [1]. ADAM proteins also contain a single transmembrane domain, a C-rich region and an EGF-like repeat. These domains suggest roles in adhesive interactions, cell fusion, proteolysis and/or intracellular signaling and implicate this family of proteins in numerous biological processes including fertilization, neurogenesis, myoblast fusion, and protein-ectodomain shedding of cytokines and other cell surface proteins [2-5]. While some ADAM proteins, such as Kuzbanian (ADAM 10) and TACE (ADAM 17), have active metalloprotease domains and are thought to be required for proteolytic processing of other proteins [3,6,7], other ADAM proteins, including fertilin β, do not have active metalloprotease domains and appear to function primarily in adhesive interactions and/or cell-cell fusion events [8-12]. ADAM proteins are also implicated in several disease processes, including Alzheimer's disease [13,14].
We have identified a mouse ADAM gene that we have named Adam33 based on comparison to its likely human ortholog http://www.people.Virginia.EDU/~jag6n/Table_of_the_ADAMs.html) and a partial cDNA for mouse Adam 33 (Genbank AF386072). We find that Adam33 mRNA is expressed during embryonic development and in several adult tissues, including discrete regions of the adult brain. Based on sequence similarity, mouse and human ADAM33 may be mammalian homologs of Xenopus laevis X-ADAM13 (X-ADAM13), which is expressed in migrating neural crest cells, somitic mesoderm and specific regions of the central nervous system during embryonic development [15].
Results and Discussion
Mouse Adam33
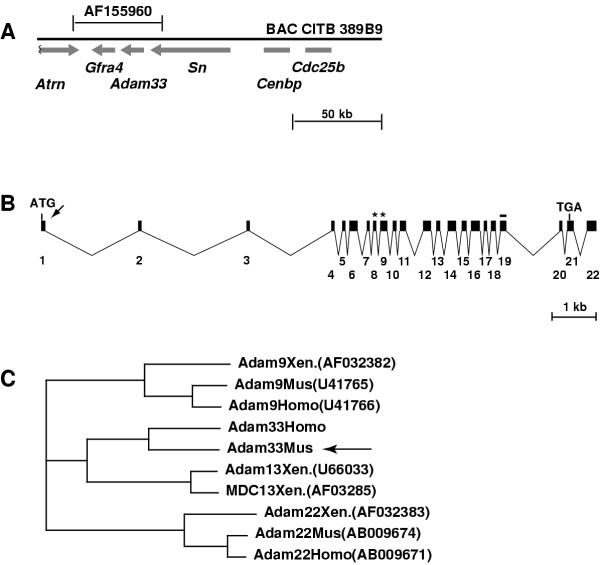
We identified Adam33 during the positional cloning of the mouse mahogany mutation, since Adam33 lies on the same bacterial artificial chromosome (BAC) that contains Attractin (Atrn), the gene mutated in mahogany[16]. This BAC, CITB 389B9, also contains Gfra4, Sialoadhesin (Sn), Cdc25b, Cenpb, and 7 clusters of expressed sequence tags (ESTs), represented by IMAGE clones 386736,388220,775311,614556,427645, 807697 and 514017.
The full-length Adam33 cDNA sequence was predicted using Genscan, GRAIL, and sequence data from EST clones, to identify exons within a ~48 kb contig of genomic DNA (Genbank AF155960). The predicted cDNA sequence was confirmed by sequencing RT-PCR products amplified from adult brain RNA. Adam33 lies centromere-distal to Atrn and Gfra4 and centromere-proximal to Sn and Cdc25b (Figure 1A). Beginning with the putative translational initiation site, the Adam33 cDNA consists of 22 exons spanning 12671 bp of genomic DNA and corresponds to a 2771 bp cDNA of which 2391 bp are protein-coding (Figure 1B, 2). The predicted 797 amino acid protein is most closely related to Xenopus laevis X-ADAM13 (44% identity, 58% similarity) and X-MDC13 (43% identity, 57% similarity), and to mouse ADAM12 (38% identity, 51% similarity) and ADAM19 (39 % identity, 53% similarity).
Figure 1.
Adam33 structure and phylogeny (A) Location and orientation of genes on BAC389B9. The entire Adam33 gene is contained within a 48 kb contig (AF155960) that also contains the Gfra4 gene and the 3' ends of Atrn and Sn. (B) Beginning with the translational start site, Adam33 is contained within 22 exons that span approximately 12.6 kb of genomic sequence. The putative signal sequence cleavage site (arrow), metalloprotease domain (asterisks) and transmembrane domain (line) are indicated. (C) ClustalW Phylogeny for ADAM proteins most closely related to Adam33 (arrow). For reference, human (Homo), mouse (Mus), and Xenopus laevis (Xen.) orthologs are shown for Adam9 and Adam22.
Figure 2.
Amino acid similarity among mouse (m) and human (h) ADAM33 proteins, and Xenopus laevis (X) ADAM13. Residues identical to the mouse protein are indicated in bold. Putative signal sequence cleavage sites (SS) are indicated by arrow. The metalloprotease site (MP), disintegrin loop (DL) and transmembrane domain (TM) are boxed and cysteine residues are highlighted by black boxes. The metalloprotease (MPD), disintegrin-like (DD), cysteine-rich (CRD), and EGF-like (EGF) domains are indicated.
The hallmark motifs of an ADAM protein include a signal sequence, pro-domain, metalloprotease, disintegrin-like, and cysteine-rich domains, an EGF-like repeat, a single transmembrane domain, and a cytoplasmic tail, which sometimes contains proline-rich regions implicated in SH3 binding. The mouse ADAM33 protein contains all of these motifs (Figure 2) and based on sequence, the metalloprotease domain of mouse ADAM33 is likely to be functional. The carboxy-terminal intracellular region contains one proline-rich region but no consensus sites for SH3-binding domains, as found in Xenopus ADAM 13.
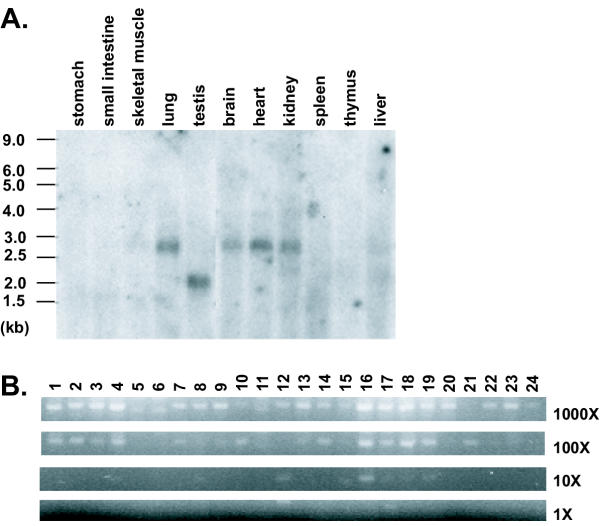
Northern blot hybridization using a probe corresponding to the end of the Adam33 coding region and the beginning of the 3'UTR detects a 2.7 kb RNA in adult brain, heart, kidney, and lung, and a 2 kb RNA in testis (Figure 3A). The size of the 2.7 kb RNA corresponds exactly to the prediction based on sequence, but the molecular basis for the testis-specific 2 kb transcript is not known (attempts to amplify the cDNA from testis using primers in exon 1 and the 3'UTR were unsuccessful). Using a more sensitive assay for expression based on semi-quantitative RT-PCR revealed expression in all samples examined (Figure 3B), including total RNA from mouse embryos on days 8.5, 9.5, 12.5 and 19 of development (Figure 3B).
Figure 3.
Mouse Adam33 expression. (A) Northern hybridization assay of mouse Adam33 (using IMAGE clone 636599) on polyadenylated RNA from adult mouse tissues. (B) Expression profile of mouse Adam 33 in Origene Mouse Rapid-Scan cDNA panel. Panel consists of normalized cDNA from 24 mouse tissues or embryo stages, serially diluted over a 4-log range (1000X-1X), where the lowest concentration (IX) is approximately 1 pg. Adam33 expression was assayed by PCR amplification of a 368 bp fragment of the mouse Adam33 cDNA sequence (band shown). Lane 1: brain; 2: adrenal gland; 3: heart; 4: pancreas; 5: kidney; 6: uterus; 7: spleen; 8: prostate gland; 9: thymus; 10: day 8.5 embryo; 11: liver; 12: day 9.5 embryo; 13: stomach; 14: day 12.5 embryo; 15: small intestine; 16: day 19 embryo; 17: muscle; 18: virgin breast; 19: lung; 20: pregnant breast; 21: testis; 22: lactating breast; 23: skin; 24: involuting breast. Slight variation in the apparent mobility among lanes is due to sample differences; fragments of identical mobility among different tissue samples were produced when individual RT-PCR reactions were repeated. Absence of signal in the 1000× but presence in the 100× dilutions for two of the tissues are likely to be caused by contaminants in the RNA preparation that inhibited the RT-PCR reaction.
The expression pattern of Adam33 in the adult brain was also examined by in situ hybridization of coronal brain sections (Figure 4). A striking pattern of hybridization was observed in the granule later of the dentate gyrus, the pyramidal layer of the hippocampus (Figure 4B), and the granule layer of the cerebellum (Figure 4E). The expression of Adam33 in two of the primary locations of granule neurons in the brain is intriguing and it will be interesting to follow expression of this gene during neuronal migration; while Xenopus ADAM13 has been implicated in neural crest cell migration during embryonic development [18], perhaps ADAM33 plays a role in the migration of granule neurons in the mouse brain. In Drosophila, the ADAM protein kuzbanian (orthologous to mammalian ADAM 10) is required for proteolytic cleavage of notch and delta, associates with a number of molecules that have been implicated in axon guidance and migration, and has been shown to play a role in axon guidance [6,19-21].
Figure 4.
Expression pattern of Adam33 in adult mouse brain. (A, D) Photomicrographs of coronal sections of adult mouse brain, from the Mouse Brain Library [25],[26]. Boxes indicate approximate regions shown in panels B-C and E-F. (B-C and E-F) In situ hybridization of digoxigenin labeled antisense (B, E) and sense (C, F) RNA probes derived from IMAGE clone 636599. Adam33 expression was observed only in the dentate gyrus (DG) and pyramidal cell layer (Py) of the hippocampus (panel B) and the granule layer (Gr) of the cerebellum (panel E), indicated by arrows.
Human ADAM33
The human ortholog of mouse Adam33 was identified by comparing the mouse cDNA and predicted protein to genomic sequence in the public genome database at http://www.ensembl.org and in high throughput human genomic DNA sequence (Genbank AC055771, AL356755, AL109804, AC017113; *see note). Gene order and orientation are conserved between this region of mouse chromosome 2 and human Chromosome 20p13, and human ADAM33 has the same transcriptional orientation as its closest neighbors, SN, which lies immediately upstream, and GFRA4, which lies immediately downstream. A bona fide human cDNA for ADAM33 has not been isolated (see below), and only some of the exons have been annotated correctly in human genomic sequence. However, comparison of human genomic sequence to the mouse Adam33 cDNA reveals an identical exon-intron structure and a putative 814 aa protein with 71% identity and 78% similarity to mouse ADAM33 and 37% identity and 61% similarity to X-ADAM13 (Figure 1C, Figure 2).
The ADAM33 designation was originally assigned to an ~3000 bp testis cDNA clone, DKFZp434K0521, whose full-length sequence (AL117415) was determined as part of a large-scale genome project. Comparison to the mouse cDNA and to human genomic DNA reveals that the testis cDNA clone represents residues 300–813 (with one frameshift) of our predicted human ADAM33 protein.
Is ADAM33 the mammalian ortholog of X-ADAM13?
The most closely related homolog of mouse and human ADAM33 is X-ADAM13, which encodes a 3200 bp cDNA with an ORF of 915 amino acids [15]. The overall level of similarity between mouse or human ADAM33 and X-ADAM13 is lower than that of other mammalian and Xenopus ADAM orthologs (Figure 1C). In addition, there are no putative SH3 binding sites in the cytoplasmic tail of the mouse or human proteins, whereas X-ADAM13 contains several. Like the mammalian ADAM33 proteins, X-ADAM13 has all the hallmark motifs of an ADAM protein, including potential metalloprotease, disintegrin and signaling activities. It is expressed during embryogenesis in somitic mesoderm and neural crest cells, suggesting a likely role in embryonic development. Mouse and human ADAM33 are also highly similar to X-MDC13, another Xenopus ADAM protein that is highly homologous to X-ADAM13 and is expressed in embryos and adult testes [22]. Characterization of the Adam33 embryonic expression pattern may provide additional insight into cross-species relationships, however, it should be noted that much of the work on X-ADAM13 to date has focused on its putative SH3 binding sites to identify interacting proteins [23], and the absence of these sites in mouse and human ADAM33 proteins indicates that such interactions are likely not conserved in mammals. Recent work does indicate that the metalloprotease domain of X-ADAM13 is active and that this protein is required for normal cranial neural crest cell migration [18]. As the metalloprotease domain of ADAM33 also matches the consensus sequence for an active metalloprotease, further study of this protein's potential catalytic activity and identification of its target proteins will be important in determining its biological role. In addition, availability of genomic sequence surrounding mouse Adam33 will facilitate cross-species comparisons to identify conserved regulatory sequences.
Conclusions
As complete genomic DNA sequence becomes available for more species, the number of putative genes identified at the sequence level will increase and more cross-species comparisons will become possible. While analyzing the sequence of a large BAC containing the Attractin locus, we identified a novel member of the ADAM family, Adam33, which was most similar to a partial human cDNA for ADAM33 and to Xenopus laevis ADAM13. Mouse Adam33 is expressed during embryogenesis and in a number of adult tissues, and preliminary characterization of its protein sequence predicts that ADAM33 contains an active metalloprotease domain. Despite overall sequence similarity to X-ADAM13, mouse ADAM33 does not contain putative SH3 binding sites in its cytoplasmic tail, suggesting that either these two proteins are not orthologs, or their functions have diverged during evolution. Further investigation is required to determine whether mouse ADAM33 has metalloprotease activity and, if it does, identification of its substrates would provide insight into its evolutionary and functional relationship to X-ADAM13 beyond what is possible by sequence analysis alone. Finally, mouse Adam33 is expressed in discrete regions of the adult brain, suggesting an important function for this gene in adult mice. Its expression in two regions of the brain critical for granule neuron migration and production suggests a possible role in neurogenesis and/or neuronal migration.
Materials and Methods
Random-insert Ml3 clones for mouse BAC clone 389B9 (Research Genetics) were generated and sequenced by the Stanford DNA Sequencing and Technology Center, following their standard protocols http://sequence-www.stanford.edu/group/techdev/inp.html. The Adam33 cDNA sequence was confirmed by sequencing RT-PCR products amplified from C3H/HeJ adult brain RNA (Titan RT-PCR kit, Roche Molecular Biochemicals, Indianapolis, IN). Mouse multiple tissue Northern blots and Mouse 24-tissue Rapid-Scan Gene Expression Panel were purchased from Origene Technologies, Inc. (Rockville, MD). Northern blots were hybridized with the insert of IMAGE clone 636599 (Research Genetics, Inc., Huntsville, AL). Primers (GCGTTCGCACCCTCCATCTCTTGA and ATCGGGCTCTGGGGTTCCTATGCT) that amplify a 368bp fragment corresponding to the end of the Adam33 coding region and the beginning of the 3'UTR were used to screen the Rapid-Scan panel. Sense and anti-sense digoxigenin-labeled RNA probes were generated from IMAGE clone 636599 using DIG RNA Labeling Kit (Roche Molecular Biochemicals, Indianapolis, IN) and hybridized to coronal sections of adult male C3H/HeJ brain, frozen in Tissue-Tek OCT compound and cut at 12 μm, following standard protocols [25]. The insert in IMAGE clone 636599 contains the last 155 bp of the coding region and continues into the 3'UTR, overlapping the 5' end of the PCR fragment used to screen the Rapid-Scan cDNA panel.
Acknowledgments
Acknowledgements
We thank Sue Kalman for help with sequence assembly and Bani Azari for technical assistance. This work was supported by grants to G.S.B. and R.W.D. from the National Institutes of Health, and by an American Heart Association Western States fellowship award to T.M.G. G.S.B. is an Associate Investigator of the Howard Hughes Medical Institute.
Mouse Adam 33 sequence has been deposited at GenBank under accession number AY054984. CITB BAC 389B9 sequence has been deposited as phase 1 HTGS sequence at GenBank under accession number AF415225.
*Note: During revisions of this manuscript, human ADAM33 sequence was deposited by NCBI to Genbank under accession number XM045437; this protein sequence is identical to 309 of the last 335 amino acids of the protein we predicted but appears to be missing 26 amino acids of the EGF-like repeat and is incomplete at the N-terminal end.
Contributor Information
Teresa M Gunn, Email: tmg25@cornell.edu.
Arezou Azarani, Email: aazarani@robsic.com.
Philip H Kim, Email: pkim24@yahoo.com.
Richard W Hyman, Email: Hyman@Genome.Stanford.Edu.
Ronald W Davis, Email: ronald.davis@stanford.edu.
Gregory S Barsh, Email: gbarsh@cmgm.stanford.edu.
References
- Wolfsberg TG, Primakoff P, Myles DG, White JM. ADAM, a novel family of membrane proteins containing A Disintegrin And Metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions. J Cell Biol. 1995;131:275–278. doi: 10.1083/jcb.131.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfsberg TG, Straight PD, Gerena RL, Huovila AP, Primakoff P, Myles DG, White JM. ADAM, a widely distributed and developmentally regulated gene family encoding membrane proteins with a disintegrin and metalloprotease domain. Dev Biol. 1995;169:378–383. doi: 10.1006/dbio.1995.1152. [DOI] [PubMed] [Google Scholar]
- Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- Howard L, Maciewicz RA, Blobel CP. Cloning and characterization of ADAM28: evidence for autocatalytic pro-domain removal and for cell surface localization of mature ADAM28. Biochem J. 2000;348 Pt 1:21–27. doi: 10.1042/0264-6021:3480021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Xu W, Loechel F, Wewer UM, Murphy LJ. ADAM 12, a disintegrin metalloprotease, interacts with insulin-like growth factor-binding protein-3. J Biol Chem. 2000;275:18574–18580. doi: 10.1074/jbc.M002172200. [DOI] [PubMed] [Google Scholar]
- Pan D, Rubin GM. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- Moss ML, Jin SL, Becherer JD, Bickett DM, Burkhart W, Chen WJ, Hassler D, Leesnitzer MT, McGeehan G, Milla M, et al. Structural features and biochemical properties of TNF-alpha converting enzyme (TACE). J Neuroimmunol. 1997;72:127–129. doi: 10.1016/S0165-5728(96)00180-4. [DOI] [PubMed] [Google Scholar]
- Huovila AP, Almeida EA, White JM. ADAMs and cell fusion. Curr Opin Cell Biol. 1996;8:692–699. doi: 10.1016/S0955-0674(96)80111-6. [DOI] [PubMed] [Google Scholar]
- Evans JP, Schultz RM, Kopf GS. Mouse sperm-egg plasma membrane interactions: analysis of roles of egg integrins and the mouse sperm homologue of PH-30 (fertilin) beta. J Cell Sci. 1995;108:3267–3278. doi: 10.1242/jcs.108.10.3267. [DOI] [PubMed] [Google Scholar]
- Evans JP, Schultz RM, Kopf GS. Characterization of the binding of recombinant mouse sperm fertilin alpha subunit to mouse eggs: evidence for function as a cell adhesion molecule in sperm-egg binding. Dev Biol. 1997;187:94–106. doi: 10.1006/dbio.1997.8612. [DOI] [PubMed] [Google Scholar]
- Evans JP, Kopf GS, Schultz RM. Characterization of the binding of recombinant mouse sperm fertilin beta subunit to mouse eggs: evidence for adhesive activity via an egg betal integrin-mediated interaction. Dev Biol. 1997;187:79–93. doi: 10.1006/dbio.1997.8611. [DOI] [PubMed] [Google Scholar]
- Myles DG, Kimmel LH, Blobel CP, White JM, Primakoff P. Identification of a binding site in the disintegrin domain of fertilin required for sperm-egg fusion. Proc Natl Acad Sci U SA. 1994;91:4195–4198. doi: 10.1073/pnas.91.10.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss ML, White JM, Lambert MH, Andrews RC. TACE and other ADAM proteases as targets for drug discovery. Drug Discov Today. 2001;6:417–426. doi: 10.1016/S1359-6446(01)01738-X. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Higuchi Y, Yoshiyama K, Shimizu E, Kataoka M, Hijiya N, Matsuura K. ADAM family proteins in the immune system. Immunol Today. 1999;20:278–284. doi: 10.1016/S0167-5699(99)01464-4. [DOI] [PubMed] [Google Scholar]
- Alfandari D, Wolfsberg TG, White JM, DeSimone DW. ADAM 13: a novel ADAM expressed in somitic mesoderm and neural crest cells during Xenopus laevis development. Dev Biol. 1997;182:314–330. doi: 10.1006/dbio.1996.8458. [DOI] [PubMed] [Google Scholar]
- Gunn TM, Miller KA, He L, Hyman RW, Davis RW, Azarani A, Schlossman SF, Duke-Cohan JS, Barsh GS. The mouse mahogany locus encodes a transmembrane form of human attractin. Nature. 1999;398:152–156. doi: 10.1038/18217. [DOI] [PubMed] [Google Scholar]
- Gould E, Tanapat P. Stress and hippocampal neurogenesis. Biol Psychiatry. 1999;46:1472–1479. doi: 10.1016/S0006-3223(99)00247-4. [DOI] [PubMed] [Google Scholar]
- Alfandari D, Cousin H, Gaultier A, Smith K, White JM, Darribere T, DeSimone DW. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr. 2001;11:918–930. doi: 10.1016/S0960-9822(01)00263-9. [DOI] [PubMed] [Google Scholar]
- Qi H, Rand MD, Wu X, Sestan N, Wang W, Rakic P, Xu T, Artavanis-Tsakonas S. Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- Schimmelpfeng K, Gogel S, Klambt C. The function of leak and kuzbanian during growth cone and cell migration. Mech Dev. 2001;106:25–36. doi: 10.1016/S0925-4773(01)00402-6. [DOI] [PubMed] [Google Scholar]
- Hattori M, Osterfield M, Flanagan JG. Regulated cleavage of a contact mediated axon repellant. Science. 2000;289:1360–1365. doi: 10.1126/science.289.5483.1360. [DOI] [PubMed] [Google Scholar]
- Cai H, Kratzschmar J, Alfandari D, Hunnicutt G, Blobel CP. Neural crest-specific and general expression of distinct metalloprotease-disintegrins in early Xenopus laevis development. Dev Biol. 1998;204:508–524. doi: 10.1006/dbio.1998.9017. [DOI] [PubMed] [Google Scholar]
- Cousin H, Gaultier A, Bleux C, Darribere T, Alfandari D. PACSIN2 is a regulator of the metalloprotease/disintegrin ADAM13. Dev Biol. 2000;227:197–210. doi: 10.1006/dbio.2000.9871. [DOI] [PubMed] [Google Scholar]
- Storm EE, Kingsley DM. Joint patterning defects caused by single and double mutations in members of the bone morphogenetic protein (BMP) family. Development. 1996;112:3969–3979. doi: 10.1242/dev.122.12.3969. [DOI] [PubMed] [Google Scholar]
- Rosen GD, Williams AG, Capra JA, Connolly MT, Cruz B, Lu L, Airey DC, Kulkarni K, Williams RW. The Mouse Brain Library. Int Mouse Genome Conference. 2000;14:166. http://www.mbl.org [Google Scholar]
- Williams RW. Mapping genes that modulate mouse brain development: a quantitative genetic approach. In: Goffinet AF, Rakic P, editor. Mouse Brain Development. Springer, New York; 2000. pp. 21–49. [DOI] [PubMed] [Google Scholar]