Abstract
Renal fibrosis contributes to progressive damage to renal structure and function. It is a common pathological process as chronic kidney disease develops into kidney failure, irrespective of diverse etiologies, and eventually leads to death. However, there are no effective drugs for renal fibrosis treatment at present. Lipid aggregation in the kidney and consequent lipotoxicity always accompany chronic kidney disease and fibrosis. Numerous studies have revealed that restoring the defective fatty acid oxidation in the kidney cells can mitigate renal fibrosis. Thus, it is an important strategy to reverse the dysfunctional lipid metabolism in the kidney, by targeting critical regulators of lipid metabolism. In this review, we highlight the potential “druggability” of lipid metabolism to ameliorate renal fibrosis and provide current pre-clinical evidence, exemplified by some representative druggable targets and several other metabolic regulators with anti-renal fibrosis roles. Then, we introduce the preliminary progress of noncoding RNAs as promising anti-renal fibrosis drug targets from the perspective of lipid metabolism. Finally, we discuss the prospects and deficiencies of drug targeting lipid reprogramming in the kidney.
Keywords: lipid metabolism, anti-renal fibrosis, drug targets, fatty acid oxidation, noncoding RNA
Introduction
Fibrosis is a pathological process that induces and aggravates multiple diseases, seriously impacting human health. The kidney is one of the major organs prone to suffering from fibrosis, with a strikingly high incidence—second only to the liver (on average, the annual incidence of renal fibrosis exceeds 10%) [1]. In the kidney, fibrosis often occurred in three parts: the glomeruli, renal interstitium, and intrarenal blood vessels [2]. The tubular tubule is most susceptible to fibrosis, and tubulointerstitial fibrosis may predict chronic kidney disease (CKD) [3, 4]. CKD resulting from different pathologies can involve fibrosis and progress to end-stage renal disease, therefore blocking fibrosis will delay CKD progression. Although the mechanism of renal fibrosis is being gradually elucidated, there are no available anti-renal fibrosis drugs in clinical practice. In recent years, growing evidence has revealed that lipid accumulation and its consequent lipotoxicity in renal cells are definite contributors to kidney injury, and the link between phenotype and fatty acid metabolism in advanced chronic kidney disease has been gradually clarified [5–7]. There is an intimate crosstalk between lipid metabolism and renal fibrosis, and the former is a direct pivotal cause of fibrosis, instead of being parallel to the fibrotic tendency. Moreover, restoring abnormal lipid metabolism in the kidney to a normal degree would effectively prevent and treat renal fibrosis, which has been confirmed by several in vitro and in vivo studies [8–10]. This has unveiled the therapeutic potential of intervening in lipid metabolism in renal fibrosis. Numerous groundbreaking studies have clarified the relationship between the transforming growth factor beta (TGF-β) signaling pathway and lipid metabolism. TGF-β1, one of the members of the TGF-β superfamily, induces metabolic reprogramming characterized by fatty acid oxidation (FAO) downregulation and lipid accumulation [8]. Hence, targeting molecules involved in the lipid metabolism pathway to resolve the metabolic dysregulation, such as FAO deficiency, emerges as a novel strategy against renal fibrosis.
Herein, we firstly introduce variously important mediators which take part in aberrant lipid metabolism in renal fibrosis and based on those possible targets, we summarize and discuss current experimental data and preclinical evidence around modulating lipid metabolism against renal fibrosis, and systemically assess the “druggability” of targeting lipid metabolism.
A brief overview of well-known mechanisms in renal fibrosis
Renal fibrosis is an evolutionary and dynamic process with complex mechanisms occurring in four inseparable stages: priming, activation, execution, and progression, which occur concomitantly and overlap mutually [11]. Inflammation and epithelial damage are often the two major initial characteristics in renal fibrosis development. Generally, the kidney can trigger an acutely protective inflammatory response when confronting stimulation or injury, but sustained chronic inflammation can recruit inflammatory cells convergence to the kidney, including but not limited to macrophages, mast cells, T cells, and extrarenal fibroblast cells [12–15]. These cells secrete fibrosis-related factors together with epithelial cells, podocytes, mesangial cells, and endothelial cells, eventually breaking the balance between extracellular matrix (ECM) production and degradation, generating irregularly gathering ECM [16] (Fig. 1). Myofibroblasts are the most important ECM secreting cells, and can rapidly produce substantial ECM, hence fostering renal fibrosis and corresponding structural and functional damage [17]. Their resource has not yet been clarified completely (Fig. 1), but transformation from different types of cells is important. For example, macrophages are recruited into the kidney by chemokines and activated by corresponding cytokines into the M1 subtype or M2 subtype. M1, the classically activated proinflammatory phenotype, will produce a series of cytokines, growth factors (e.g., TGF-β, PDGF, FGF, IL-1β, and TNF-α, etc.) and reactive oxygen species (ROS), to promote mesenchymal transformation of endothelial and epithelial cells, which is one approach to boost the surge of myofibroblasts [18]. Macrophages also undergo macrophage-to-myofibroblast transition to function as ECM producers [19]; myofibroblasts are the core executors devoted to fibrosis.
Fig. 1. The pathological progress in renal fibrosis.
Epithelial cells damage and inflammation that occur when the kidney is subjected to various harmful stimuli during renal fibrosis. In the inflammation branch, a series of immune cells outside the kidney are recruited into the kidney tissue and they secrete various cytokines, which will eventually promote the myofibroblasts activation, serving as the fibrosis executor enabling considerable extracellular matrix production rapidly. The main sources of myofibroblasts are epithelial cells, endothelial cells, pericytes, and fibrocytes (including those from the kidney itself and bone marrow). In the left branch, epithelial cell injury can trigger EMT and epithelial cell loss especially podocyte, which exacerbate renal fibrosis. Injured epithelial cell can also promote the activation of fibroblasts via secreting numerous fibrosis-related factors. Myofibroblast and its produced ECM are the dominantly executors in renal fibrosis, but the source of fibroblasts is not clarified yet, and the contribution of each cell type is still controversial. Wholly, the above epithelial cells damage and inflammation pathway are overlapping and dynamic, which are both regulated by complex molecular networks. Therefore, renal fibrosis is a particularly complicate process. The right gray part indicates the roles of lipid metabolism in fibrosis, and the contents in dotted ellipse are speculated mechanisms remaining to be verified.
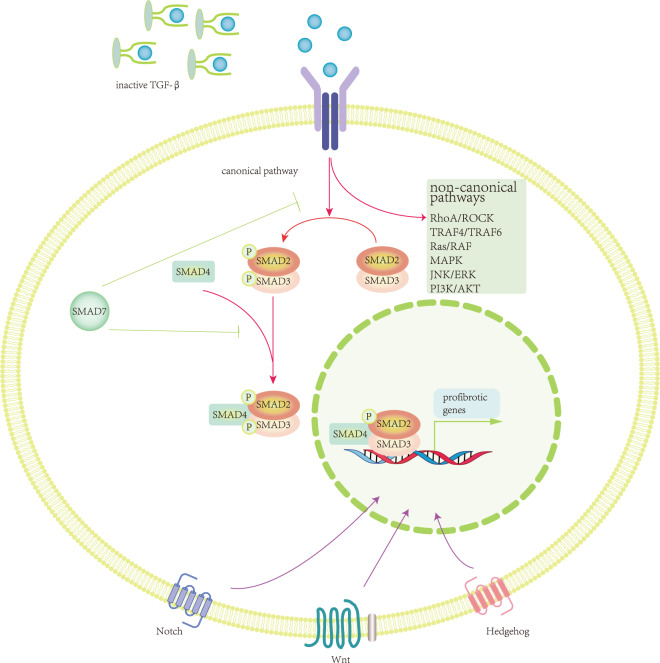
Since the TGF-β dominated pathway is the central mechanism in fibrosis [20–22], here we first introduce this pathway despite it being extensively reviewed elsewhere [23, 24]. In the canonical pathway, downstream biomolecules of TGF-β1 and smad2/smad3 [25, 26], can promote fibrosis and form a complex with smad4 upon phosphorylation to upregulate the transcription of profibrogenic genes, while smad2 and smad7 acts as an anti-fibrotic biomolecules, so the disrupted balance between smad3/smad4 and smad2/smad7 stimulates fibrogenesis [20] (Fig. 2). In addition to the conventional TGF-β/smad pathway, TGF-β also mediates fibrosis through signaling pathways such as RhoA/ROCK, TRAF4/6, Ras/RAF, MAPK, JNK/ERK, and PI3K/Akt, referred to as noncanonical pathways. Other well-known signaling pathways, such as WNT, Notch, and Hedgehog, have also been reported to crosstalk with TGF-β signals, increasing the complexity and obscurity of renal fibrosis mechanisms [27–30].
Fig. 2. Brief overview of classical TGF-β pathway in renal fibrosis.
Once released from the small latent complex, TGF-β binds to its receptor and activates the SMAD2/3 complex, which later forms complexes with SMAD4 and translocates into the nucleus to regulate gene regulation as a transcriptional factor. SMAD7 acts as an inhibitor of TGF-β mediated profibrotic role in canonical pathway. Several non-canonical pathways may regulate TGF-β signals to some extent. Other pathways such as Wnt, Notch, Hedgehog contribute into renal fibrosis and have a crosstalk with TGF-β signaling, portending the intricacy and intangibility of fibrogenesis mechanically.
According to clinical trials, anti-fibrotic druggable approaches in the TGF-β pathway include antibodies, small molecules, siRNA, and oligonucleotides [31]. These drugs are mostly designed for anti-fibrosis in the lungs, liver, skin, and digestive system (https://clinicaltrials.gov/). To date, there is no drug-specific for kidney fibrosis that has obtained an ideal outcome, and some clinical trials have already been withdrawn (eg, NCT00001959, NCT00878761, and NCT00464321). The major reason for this is that inhibition of isoforms or receptors engaged in the TGF-β signaling pathway in the long term can cause disastrous adverse effects, because TGF-β has pleiotropic effects on cellular activities, especially in the inflammatory response and cell proliferation. For instance, genetic knockout or pharmacological blockade of TGF-β using fresolimumab eventually leads to inflammation exacerbation [32] and multiple keratoacanthomas [33]. Accordingly, targeting intracellular TGF-β cascades or other profibrotic pathways may lead to development of relatively proper candidates [24]. For example, connective tissue growth factor (CTGF) is a stromal cell protein induced by TGF-β, and is recognized as a profibrotic element. There are clinical trials of CTGF antibodies for the treatment of idiopathic pulmonary fibrosis (NCT00074698 and NCT01890265), but whether CTGF is necessary for the development of fibrosis in the kidney is not yet clear [34, 35], and there has been no clinical trial to evaluate the efficacy of the CTGF antibody against renal fibrosis. Therefore, finding alternative efficient targets other than the TGF-β signaling pathway is indeed an important and urgent strategy for preventing and treating renal fibrosis. Tables 1 and 2 display the ongoing clinical trials of drugs for anti-renal fibrosis.
Table 1.
Clinical trials of drugs for the prevent/treatment of renal fibrosis.
| Drug | Condition or disease | Target or mechanism | stage | NCT number |
|---|---|---|---|---|
| MSCs | Renal Cirrhosis | Reducing the decompensated conditions | Phase 1 | NCT03460223 |
|
Renal Transplant Rejection Fibrosis |
Phase 2 | NCT02057965 | ||
| Pirfenidone | FSGS | TGF-β | Phase 2 | NCT00001959 |
| Fresolimumab | FGSC | TGF-β | Phase 2 | NCT01665391 |
| LY2382770 |
DN/ Glomerulosclerosis |
TGF-β | Phase 2 | NCT01113801 |
| Rapamycin | Kidney transplantation | mTOR | Phase 4 | NCT00493194 |
Table 2.
Representative studies on taking drug repurposing or targeting the key regulatory molecules involved in lipid metabolism to treat renal fibrosis.
| Target | Experimental model | Intervention manner | Involved lipid metabolism improvement | Fibrosis amelioration | Ref |
|---|---|---|---|---|---|
| CD36 | 5/6 nephrectomy with angiotensin II infusion & unilateral ureteral obstruction | Antagonist 5A peptide | Prevent lipid metabolism alteration | Y | [64] |
| CPT1/2 | FAN | CPT1 agonists, C75 | Block FA synthase while resrore CPT1 and ACOX1 expression, hence reinfore FAO | Y | [8] |
| FAN & UUO | CPT1A OE | Prevent mitochondrial function and facilitate FAO | Y | [67] | |
| DN | CPT2 OE | Inhibit AGEs induced FAO surppression | Y | [68] | |
| SREBP1/2 | UUO | Inhibitor, fatostatin | Undetected | Y | [72] |
| DN | Inhibitor, fatostatin | Undetected | N | [74] | |
| PPARs | UUO & remnant kidney model | PPAR-α agonist BAY PP1 | Undetected | Y | [81] |
| age-associated renal fibrosis model | PPARα/β dual agonist MHY2013 | Increase nuclear translocation and activity of PPARα/β and their target genes related to FAO | Y | [82] | |
| FAN | PPAR-α agonist fenofibrate | Restore FAO-related enzymes CPT1A/2 and ACOX1/2 | Y | [8] | |
| Renal Transplant Model | PPAR-α agonist fenofibrate | Undetected | Y | [83] | |
| UUO | PPAR-γ agonist lobeglitazone | Undetected | Y | [84] | |
| TGF-β1 induced renal fibrosis model | PPAR-γ agonist pioglitazone | Undetected | Y | [86] | |
| UUO | PPAR-γ agonist | Undetected | Y | [87] | |
| UUO | PPAR-γ agonist | Undetected | N | [88] | |
| PGC-1α | DKD | Use rosiglitazone as pharmaceutical inducers | Enhance mitochondrial function of energy metabolism | Y | [91] |
| FAN & UUO & APOL1-induced fibrosis models | Genetic re-expression of PGC-1α | Resist Notch induced FAO impairment and mitochondrial dysfunction | Y | [92] | |
| TGF-β1 induced in vitro fibrosis model | PGC-1α-siRNA | Undetected | Y | [94] | |
| PCSK9 | UUO & N-nitro-L-arginine methyl ester mice model | PCSK9Qβ-003 vaccine |
Upregulate FAO-related factors such as CD36, CPT1A, SREBP2, PPAR-α/γ, PGC-1, ACOX1, and LPL |
Y | [98] |
| MiR-21 | UUO & unilateral IRI | KD by Chemically modified ASO | Upregulate PPAR-α to enhance lipid metabolism | Y | [120] |
| Alport nephropathy | KD by Chemically modified ASO | Upregulate PPAR-α and its downstream FA oxidation and mitochondrial/peroxisomal biogenic functions | Y | [121] | |
| miR-9-5p | UUO | lentivirus carrying miR-9-5p mimics |
Regulate CPT1A and PGC-1α to enhance mitochondrial function |
Y | [122] |
| miR-33 | UUO & FAN | pHILP to carry miR33 PNA inhibitors | Increase CPT1A expression and improve FAO | Y | [124] |
| TUG1 | DN | Podocyte-specific OE of TUG1 | Upregulate PGC-1α and modulate mitochondrial bioenergetics | Undetected | [129] |
Y yes, N no, undetected the author detected other pathways such as TGF-β or reactive oxygen species rather than lipid metabolism pathway,
CD36 cluster of differentiation 36, CPT carnitine palmitoyl transferase, SREBP sterol regulatory element-binding protein regulatory element-binding protein 2, PPAR-α peroxisome proliferator-activated receptor α, PPAR-γ peroxisome proliferator-activated receptor γ, PGC-1 peroxisome proliferator-activated receptor gamma coactivator 1, AGEs advanced glycation end products, DN diabetic nephropathy, DKD diabetic kidney disease, UUO unilateral ureteral obstruction, FAN folic acid-induced nephropathy, IRI ischemia-reperfusion injury, ASO antisense oligonucleotide, pHLIP pH low insertion peptide, PNA peptide nucleic acid, KD knock down, OE overexpression, PCSK9 proprotein convertase subtilisin/kexin type 9.
Lipid metabolism involved in renal fibrosis
Unique pattern of lipid metabolism in kidney tissues
The kidney is a highly energy-consuming organ, which uses lipids instead of glucose as its primary energy source due to the function of reabsorption, especially for tubular epithelial cells (TECs). TECs primarily use fatty acids as fuel, and the mitochondrial β-oxidation of fatty acids is a major energy source for ATP generation, whereas glucose catabolism is less demanded and is not a key contributor to energy production in TECs [36]. Unlike adipocytes, which have massive intracellular lipid drops (LDs) storage under physiological conditions, TECs harbor much little number of LDs to maintain their energy homeostasis [37]. Lipid overload in TECs, which would be a result of excessive dietary intake or dysfunction in lipid consumption or degradation, exerts vicious effects termed lipotoxicity through the generation of reactive oxygen species, release of proinflammatory and profibrotic factors, as well as cell apoptosis [38]. This has been observed in obesity-associated kidney diseases, diabetic nephropathy, and kidney fibrosis progression [39]. The excess ectopic accumulation of lipid in TECs could be a subsequence of systemic dyslipidemia, but sometimes, it is an outcome of the imbalance of cellular lipid metabolism, because renal lipid accumulation occurred independent of the systemic lipid disorder in the contexts of hypertensive nephrosclerosis, focal segmental glomerulosclerosis, or minimal change disease [40]. Particularly, it has been manifested that this usually occurs in lower expression of key enzymes and regulators of fatty acid oxidation and in higher intracellular lipid deposition in humans and mouse models with tubulointerstitial fibrosis [41].
FAO is an especially critical metabolic step. Cluster of differentiation 36 (CD36), the most important FA transporter in the kidney responsible for FA uptake, is expressed on the membrane of most cell types, including renal tubular epithelial cells [42], podocytes [43], mesangial cells [44], and macrophages [45]. Its expression is increased as kidney diseases proceed, thus it induces lipid overload and lipotoxicity [46]. In addition, the fatty acid-binding protein (FABP) family and fatty acid transport proteins (FATP) family also participate in the cellular uptake of FA and other lipophilic substances [47]. After being transported into the cells, lipids are deposited into the cytoplasm or taken up into the peroxisome and mitochondria (FAO rate-limiting step) by carnitine palmitoyl-transferase 1 (CPT1) and CPT2 [48] (Figs. 3 and 4). Subsequently, β-oxidation of FA occurs, and ATPs are produced. Each molecule of FA can yield 106 ATPs, while one carbohydrate molecule can only offer 36 ATP maximally, so FAO is the optimal energy supply mode for cell activities in high-energy-consuming organs, especially the kidney [49]. Lipid uptake, synthesis, and degradation are regulated by a series of transcription factors involving peroxisome proliferator-activated receptors (PPARs), sterol regulatory element-binding transcription factor (SREBPs), farnesoid X receptor, and CCAAT enhancer-binding protein (CCAAT/Enhancer-binding Protein α, C/EBPα), and their gene expressions are also governed by some specific microRNAs (miRNAs) whose expressions change during renal fibrosis [50–53]. Therefore, targeting enzymes, transporters, transcription factors, or miRNAs responsible for lipid metabolism to artificially regulate renal lipid metabolism is possible in order to prevent or treat renal fibrosis.
Fig. 3. Critical lipid metabolism regulators with potential druggability in the kidney fibrosis discussed in the current review.
Here we use fatty acid (FA) as a schematic example. FA is transported into cells by CD36 primarily or other proteins such as FABP and FATP, then FA can be disposed into the following three directions, lipid droplet, peroxisome, and mitochondria, among which, mitochondrial β-oxidation is a fundamental process for FA metabolism, thus CPT1/2 downregulation or mitochondrial dysfunction will lead to FA deposition. PPARs and their co-activator PGC-1α promote lipid metabolism transcriptionally. While SREBP can enhance lipogenesis through its transcriptionally active N-terminal fragments cleaved in Golgi apparatus. Noncoding RNAs including miR21, miR-9-5p, and TUG1 can control renal fibrosis via lipid metabolism. Targeting these above regulators by utilizing nucleic, antibody, small molecule or phytochemical drug can yield anti-fibrosis effects in the kidney.
Fig. 4. MiRNA/lncRNA performs as upstream mediators to influence renal fibrosis in an epigenetic way.
By improvement of advanced delivery technique, specific target therapy for fibrosis within the kidney tissue can be achieved. Popular loading systems (including viral vector, polymeric vector, liposome, exosome, etc.) are employed to deliver exact miRNA, lncRNA, according siRNA or ASO to the kidney.
Important participants in lipid metabolism and renal fibrosis
Although preclinical and experimental studies are increasingly suggesting that targeting lipid metabolism can antagonize renal fibrosis (this section will be described in detail later), the exact mechanisms are still far from clear. As there are many commonalities in fibrosis in different organs, including TGF-β activation, myofibroblasts differentiation, and ECM excessive accumulation (Figs. 1 and 2), lipid metabolism markedly influences fibrosis within the kidney in a tissue-specific manner.
“Synthesis and degradation” of ECM equals “anabolism and catabolism” of ECM, thus, in other words, fibrosis may be controlled by ECM “metabolism”. Fibroblasts in radiation-induced skin fibrosis are characterized by an altered fuel-utilization balance, with enhanced glycolysis but compromised FAO. A new study claims that over-activation of PPARs/CD36 signaling of fibroblasts directly promotes ECM degradation by endowing fibroblasts with a catabolic phenotype with reduced ECM gene transcription. For CD36hi fibroblasts, the secretion ability of ECM-related proteins, including fibronectin, collagen-1, and plasminogen activator inhibitor type-1, is reduced [54]. This was further exemplified by the phenomenon that caffeic acid with a PPARs agonizing effect impedes SMAD3-dependent transcription and internalizes collagen-1 in dermal fibroblasts [54]. Thus, ECM homeostasis may be one major strategy for metabolic mediators to exert fibrosis regulation.
Several different types of cells contribute to chronic kidney disease, and they also have different metabolism characteristics.
Myofibroblast generation may be a striking element of the necessary downstream effect in the metabolism-fibrosis axis. Some literature has documented that augmented aerobic glycolysis initiates and sustains myofibroblast contractility and formation [55–58]. Succinate, a glycolysis intermediate, can directly promote myofibroblast differentiation through stabilizing hypoxia-inducible factor 1-α (HIF-1α) [55]. These conclusions encourage researchers to investigate whether myofibroblast quiescence and activation are also partly determined by a lipid metabolism switch, whereas this indispensable metabolic branch is relatively less investigated. As mentioned before, M2 macrophages are a major cell type prone to promoting myofibroblast differentiation (Fig. 1), and FAO is a fundamental characteristic of M2. Although detailed pathways remain elusive, given the discrepancy of immunometabolism between M1 and M2, it can be speculated that lipid metabolism may skew macrophage reprogramming to trigger fibrosis. However, recently, the question of whether M2 polarization is dependent on FAO has become controversial, because macrophages have been proven to use glucose metabolism for M2 phenotype transition in several separate studies [59]. Thus, more studies are needed to investigate whether and how FAO predominates in macrophage phenotype shift to control renal fibrosis.
Additionally, disordered lipid metabolism in the kidney will incur well-known stimulants of fibrogenesis, including cellular apoptosis, inflammation, and dedifferentiation. However, involved pathways connecting these pathogenic processes and the consequent metabolic changes are not understood well yet. There are relatively few studies on how lipid metabolism modifies kidney architecture and function. In the near future, we must pay more attention to the detailed mechanism by which lipid metabolism manipulates renal fibrosis. As we discussed above, there are several possible pathways, including but not limited to ECM “metabolism”, epithelial-mesenchymal transition (EMT) control, macrophage domestication, and other direct mechanisms such as TGF-β signaling.
Preclinical studies against renal fibrosis via interfering with lipid metabolism and hot-spot targets
Based on the above putative ideas, the feasibility and efficiency of targeting renal lipid metabolism pathways to ameliorate fibrosis have been explored in lots of preclinical experiments, although there are no clinical trials in humans currently. Here, we summarize some “star” lipid metabolism regulatory proteins, including CD36, CPT1/2, SREBP1/2, PPARs, peroxisome proliferator-activated receptor-γ coactivator (PGC-1α), and proprotein convertase subtilisin/kexin type 9 (PCSK9), to describe their latest research status as drug targets. We also discuss their advantages and disadvantages against renal fibrosis.
CD36
CD36, also known as scavenger receptor B2, is a transmembrane glycoprotein that performs as a receptor of long-chain FAs, oxidized lipids, advanced glycation end products, and oxidized protein products [60]. CD36 seems to play tissue-specific role in fibrosis. As mentioned earlier, CD36 is the most important transporter that is responsible for the entry of FA into cells, and upregulating CD36 reduces ECM accumulation in murine skin fibrosis [54]. However, its overexpression in different kidney pathophysiological models initiates lipid droplet formation and fibrosis development [61]. It was previously reported that reducing CD36 expression by gene intervention or drug treatment could ameliorate renal oxidative stress and inflammation, and inhibit TGF-β expression, suggesting that CD36 is a potential target for anti-renal fibrosis [62, 63]. In the 5/6 nephrectomy with angiotensin II perfusion mouse model and unilateral ureteral obstruction (UUO) model, a micropump containing CD36 antagonist 5A peptide reduced inflammatory cytokines and chemokines, delayed CKD progression, and inhibited renal interstitial fibrosis [64]. Human CD36 overexpressed transgenic mice had obviously more proinflammatory and profibrotic characteristics, as well as proteinuria, than wild mice during folic acid-induced (FAI) acute kidney injury. In a diabetic mice model, CD36 also mediated the renal lipotoxicity and profibrotic effects of advanced glycation end-products [46]. These results imply that CD36 regulates fibrosis in various disease conditions, in addition to CKD. Meanwhile, soluble CD36 in serum is positively correlated with CD36 in the kidney, thus can be used as a biomarker to predict CKD. Therefore, in-depth study of the role of CD36 in renal physiology and pathology is valuable for CKD diagnosis and treatment. However, there is a noteworthy problem that CD36 is highly expressed in various kidney cell types, and artificial changes of CD36 expression may cause unexpected consequences in phenotype. For example, interstitial fibrosis was significantly weakened in UUO mice after specific CD36 knockout in macrophages, while CD36 in proximal renal tubular cells is involved in the binding and uptake of albumin, its abnormally lower expression may lead to proteinuria, rather than fibrosis [65]. Therefore, the specific targeting of cell types should be considered when employing CD36 inhibitors. Of course, pharmacological application with CD36 target is also dependent on optional organs or tissues.
CPT1/2
The CPT family proteins CPT1 and CPT2 are located on the outer and inner mitochondrial membranes, respectively. CPT1 has three subtypes: CPT1A, CPT1B, and CPT1C. CPT1/2 are rate-limiting enzymes that transport FAs into mitochondria and are indispensable for lipid catabolism. CPT1 downregulation or inactivation will block FAO, incur FAs accumulation in the cytoplasm and cell necrosis, and cause CKD as well as renal fibrosis [8].
The CPT1 activator C75, a synthetic chemical compound, can inhibit FA synthesis and improve renal function in a mouse model of FAI renal fibrosis. Thus, CPT1 agonists are effective in reversing metabolic reprogramming to hinder fibrosis progression [8]. CPT1A is a prevailing enzyme converting acyl-CoA to acylcarnitine. It is the most widely distributed subtype among CPT1 members and has been intensely studied in cardiovascular diseases and malignant tumors [66]. In patients with CKD, CPT1A expression is negatively correlated with the fibrosis degree. A series of experimental renal fibrosis models have shown that CPT1 overexpression can reduce inflammation and TGF-β-induced epithelial cell transformation, and remarkably mitigate renal fibrosis. Gain of function of FAO mediated by upregulating CPT1 restored the catastrophic lipid metabolic reprogramming, improving mitochondrial structure and metabolic activity, in three common animal models of renal fibrosis [67]. This evidence provides the opportunity to develop drugs for renal fibrosis, as well as CKD, based on CPT1. However, studies on CPT2 in renal fibrosis are relatively rare compared to CPT1. Recently, it was found that advanced glycation end products induce mitochondrial dysfunction through down-regulation of CPT2, impairing FAO and causing renal fibrosis and DKD [68]. Artificial overexpression of CPT2 could restore FAO and fibrosis-related genes to normal levels, suggesting that targeting CPT2 could help prevent diabetic renal fibrosis [68].
SREBP1/2
SREBPs (SREBP1a, SREBP1c, and SREBP2) belong to the membrane-binding transcription factors family, which is responsible for regulating lipid production. SREBP1a stimulates overall lipid synthesis, SREBP1c is responsible for FA and triglyceride synthesis, and SREBP2 specifically controls sterol production [69]. Under normal physiological conditions, SREBPs and their escort protein SREBP-cleavage activating protein (SCAP) are locked by a complex consisting of lipids and an endoplasmic reticulum transmembrane protein insulin-induced gene (INSIG), anchored on the endoplasmic reticulum, which prevents SREBP from entering the nucleus to guide the transcription of genes involved in lipogenesis. Once the SCAP/SREBP complex detaches from the endoplasmic reticulum, it will enter the Golgi apparatus freely, where SREBP is cleaved to release its transcriptionally active N-terminal fragments (Fig. 3). When SREBP is abnormally activated and ectopically located in the nucleus, it will lead to excessive lipid production and accumulation within the cells. Although the research on SREBPs is mainly concentrated in the field of cancer, in which the elemental functions of SREBPs in regulating tumor cell proliferation and escape have been clarified [70, 71]. Given their role in lipid homeostasis, we can speculate that they modulate the occurrence of renal fibrosis. In CKD and DKD patients, increased expression of SREBP1 and SREBP2 triggers kidney lipid deposition, lipotoxicity, and fibrosis. Moreover, SREBP1 can regulate renal fibrosis through the lipid independent pathway (directly acting on the TGF-β/Smad3 signal and inhibiting TGF-β receptor degradation)[10]. By giving fatostatin, an inhibitor of SREBP in UUO mice, Mustafa et al. demonstrated that blocking the signal transduction of SREBP1/2 to lipid production and TGF-β1 obviously reduced renal inflammation, necrosis, and fibrosis in UUO mice [72]. Recently, in another group, Richard Van Krieken et al. declared that fatostatin did not alleviate proteinuria and renal fibrosis in diabetic mice, and it also aggravated recruitment of pro-inflammatory cells (CD3+ T cells and macrophages) to kidneys in both nondiabetic and diabetic mice [73]. This discrepancy may be attributed to the different importance degree of SREBP in different kidney diseases. Sun et al. demonstrate that activation of renal SREBP-1 results in alterations in renal lipid metabolism and renal lipid accumulation plays an important role in the pathogenesis of diabetic nephropathy by transgenic mice [74], but failure of fatostatin against DN might suggest that SREBP-1 mediated lipid metabolism dysfunction might not be key element to induce fibrosis in DN. Therefore, the role of SREBPs in renal pathology and the renal protective function of fatostatin need to be evaluated in more nephrotic models.
Moreover, considering SREBP as a transcription factor involved in the regulation of many downstream genes, it might not be a suitable drug target considering its broad possible effects, also due to some of its effects were not lying on lipid metabolism. Therefore, it should be careful when using SREBP as a target to design strategy to treat renal fibrosis. Currently, several clinical trials of Oltipraz aiming to treat nonalcoholic steatohepatitis (NASH) were under investigation, such as NCT01373554, NCT02068339, and NCT04142749. Oltipraz is shown to inhibit fatty acid synthesis through AMPK-S6K1 pathway and LXRg-SREBP-1c pathway [75, 76]. These following clinical trial results are necessary to be explored to evaluate its real clinical value as an anti-fibrosis target.
PPARs
PPARs belong to the type II nuclear hormone receptor superfamily and are divided into three subtypes: PPAR-α, β (δ), and γ. They can serve as transcription factors binding to the response element within the promoter of genes related to glucose and lipid metabolism [77]. PPARs agonists, such as fibrates, are often used to treat metabolic diseases [78]. PPAR-α or PPAR-β activation enhances triglycerides and FAO decomposition, so PPAR-α agonists and PPAR-α/β dual agonists are mainly applied to reduce blood lipids [79]. PPAR-γ controls adipogenesis, adipocyte differentiation, insulin sensitivity, and glucose metabolism, and its agonists have become prevalent drugs for type 2 diabetes [80].
Robust evidence has manifested that intervening PPARs can regulate renal fibrosis. In an animal model of tubulointerstitial fibrosis, PPAR-α expression was significantly reduced, and PPAR-α agonist BAY PP1, but not fenofibrate, remarkably attenuated fibrosis through restoring the PPAR-α level in renal tubular cells without adverse effects [81]. During body aging, the degree of renal fibrosis gradually increases and renal function declines, and Ki Wung Chung et al. found that an impaired PPAR-α pathway accelerates fibrogenesis of aged kidneys in mice [9]. This research group later gave MHY2013, a PPARα/β dual agonist, to aging rats, to eliminate lipid aggregation and renal fibrosis in the rats’ kidneys [82]. These results confirmed PPARs agonists help delay renal dysfunction. Regarding renal fibrosis, recent studies have further shown that PPAR-α agonist fibrates have visible therapeutic effects. In the context of renal transplantation-induced fibrosis, administration of fenofibrate in kidney transplantation rat models suppressed EMT induced by oxidative stress, and relieved renal fibrosis through directly improving FA metabolism [83].
In parallel with this, PPAR-γ participates in the maintenance of renal metabolism homeostasis [53], and PPAR-γ agonists have also been reported to have similar effects in a few papers. Kim et al. found that lobelgitazone, a drug mainly used for diabetes, can inhibit the TGF-β/Smad3 pathway to slow down UUO-induced renal fibrosis, displaying kidney protective functions [84, 85]. Another two PPAR-γ agonists, including rosiglitazone, can inhibit EMT and tubulointerstitial fibrosis via antagonizing ROS in DKD [84]. Pioglitazone can reduce TGF-β1 induced renal fibrosis in mice by inhibiting EGR-1, STAT3, and AP-1 [86]. These findings indicate that PPAR-γ may be a potential target for the treatment of CKD and fibrosis in the future. Indeed, repositioning of the PPAR-γ agonist can decrease time cost and lower risk over the new drug development pipeline. However, in the UUO renal fibrosis model, two separate studies from Kawai et al. on the therapeutic effect of pioglitazone reached the opposite conclusion. Therefore, whether PPAR-γ has definite anti-renal fibrosis require further study [87, 88].
PGC-1α
PGC-1α belongs to PGC-1 family. The other two members of PGC-1 are PGC-1β and PGC-1-related coactivator. PGC-1α is a co-activator of PPARs and facilitates stimulating mitochondrial and peroxisomal biogenesis. PGC-1α can affect gene regulation of lipid synthesis and transport by interacting with PPARs, thus PGC-1α is also an important cellular lipid balancer and energy regulator [89]. PGC-1α expression is often reduced in aging or fibrotic kidneys, corresponding to weakened cellular metabolism [90]. Like PPARs, it is reasonable that PGC-1α also affects renal fibrosis. Administration of the PPAR-γ agonist rosiglitazone to DKD mice with fibrosis can increase PGC-1α expression, thus reducing ROS and protecting the kidney from injury [91]. Han et al. reported that PGC-1α is a target gene of Notch. After inhibition of Notch, PGC-1α transcription is blocked, and then kidney cells encounter FAO deficiency. Restoring PGC-1α gene expression can counteract Notch-mediated kidney damage and fibrosis [92]. Qin et al. also report that berberine protects against diabetic kidney fibrosis via promoting PGC-1α-regulated mitochondrial energy homeostasis [93]. However, Wang et al. silenced PGC-1α by siRNA, which broke the positive feedback loop between TGF-β and PGC-1α, unexpectedly decreasing the expression of fibronectin and collagen I [94]. This suggested that not PGC-1α inhibition but activation in renal tubular cells contributes to anti-fibrosis, contrary to the previous results. This was the first time that PGC-1α has been found to promote fibrosis by acting on TGFβ-RI, PI3K/Akt, and p38 MAPK. These studies imply that intervention in PGC-1α may change renal fibrosis, but the more recent opposite results obscure the role of PGC-1α in the kidney.
PCSK9
PCSK9 is a gene closely related to hypercholesterolemia, and its encoded enzyme PCSK9 mainly exists in the liver. In the kidney, PCSK9 is also expressed at a lower level. PCSK9 binds to the low-density lipoprotein receptor (LDL-R) on the cell surface to form a complex, which will undergo cellular internalization for lysosomal degradation, resulting in reduced LDC-R. Since LDC-R is necessary to clear low-density lipoprotein cholesterol (LDL-C), deficiency in LDC-R will elicit LDL-C overload. Accordingly, PCSK9 is a key molecule required to regulate blood LDL-C. PCSK9 inhibitor (PCSK9i) is principally used in lipid-lowering therapy to regulate blood lipid metabolism and improve cardiovascular function [95]. Alirocumab and evolocumab are currently available PCSK9i, both of which are monoclonal antibodies binding to PCSK9. PCSK9i has exhibited renal protective effects in several studies on CKD. The application of PCSK9i in kidney disease mainly focuses on lowering cholesterol levels and improving complications such as cardiovascular disease in CKD patients [96, 97]. There are few studies on PCSK9i employment in renal fibrosis. Through literature mining we found only one group documenting that the PCSK9Qβ-003 vaccine could enhance FAO via its own original lipid regulation effect, ameliorating hypercholesterolemia and renal fibrosis in UUO and N-nitro-L-arginine methyl ester mice model [98]. In more detail, the PCSK9Qβ-003 vaccine profoundly elevated CD36, CPT1A, PPARα/γ, PGC-1α, and SREBP2, while downregulating TGF-β/smad3 in the kidney, representing a possible anti-fibrosis mechanism through the FAO pathway. However, it is worthy to notice that the reduced hypercholesterolemia by PCSK9 vaccine in renal injury models is likely a result of improved LDL clearance in the liver due to PCSK9 antagonizing. Injury kidneys have increased circulating PCSK9 levels, thus, whether PCSK9 inhibition improves renal fibrosis via reducing circulating lipid levels rather than targeting renal lipid metabolism deserves further confirmation. This study provides an incentive to further confirm its effect on renal fibrosis. Exploring new indications of PCSK9i will circumvent the target confirmation stage and provide new drugs to this field faster and more economically. Of course, whether PCSK9i can treat renal fibrosis warrants further preclinical trials and clinical trials to determine. Additionally, whether the PCSK9 antibody mainly acts on the kidney cells themselves or on the PCSK9 highly expressed liver cells remains to be corroborated, as PCSK9 expression in the kidney is low.
Other regulators of lipid metabolism with an emerging role in treating renal fibrosis
In addition to the typical modulators aforementioned, recently several other lipid metabolism regulators have also been preliminarily discovered, and they may deserve to be excavated as drug targets.
FABP4 is one of the subtypes of FABPs that is able to transport FAs and can downregulate its target gene PPAR-γ [99]. In vitro use of FABP4 siRNA or inhibitor, and in vivo FABP4 knockdown, can enhance PPAR-γ signaling and improve lipid metabolism, eventually leading to reduced inflammation and renal interstitial fibrosis through the PPAR-γ/ACOX1/CPT1 and PPAR-γ/NF-κB/ICAM pathways, respectively [100]. Analogously, FATP2 mediates the uptake of albumin-bound non-esterified FAs by distal renal tubular cells, avoiding the lipotoxicity of non-esterified FAs [101]. Activating transcription factor 6α (ATF6α) is a transcription factor reported to participate in lipid metabolism regulation. ATF6α may downregulate PPAR-α, mediate lipid aggregation and mitochondrial dysfunction, and even promote secretion or expression of fibrotic factors such as CTGF, alpha-SMA, and collagen I. With the help of a unilateral ischemia-reperfusion injury-induced kidney fibrosis model, researchers demonstrated that ATGFα in proximal renal tubular cells was activated and then moved into the nucleus, acting as an inhibitor of PPAR-α to reduce PPAR-α expression, then led to CPT2 (one target gene of PPAR-α) downregulation. Since CPT2 is a vital transporter mediating FAs entry into mitochondria, ATGα can influence FAO and mitochondrial functional homeostasis, inducing lipid aggregation/lipotoxicity and CTGF overexpression, followed by cell necrosis and finally fibrosis [102]. The subsequent use of fenofibrate to reverse ATF6α-mediated lipotoxicity and fibrosis in mouse kidneys further verified the pathway of ATF6α/PPAR-α/CPT2, but how ATFα was activated was not clarified. In addition, as the same study shown, the role of ATF6α in different disease models is very different, so its role needs further determination. Liver kinase B1 (LKB1) is a serine kinase regulating adipogenesis and differentiation. Seung Hyeok Han and others reported that phosphorylated LKB1 expression in tubule epithelial cells was decreased in a human fibrotic kidney, which indicated a link between LKB1 and renal fibrosis. Then they found that LKB1 deficiency-induced impaired renal glucose and FAs utilization through AMPK and PPAR-α [103]. Whether LKB1 can control fibrosis needs further verification. Feng et al report that activated NF-κB/Nrf2 and Wnt/β-catenin pathways are associated with lipid metabolism in CKD patients with microalbuminuria and macroalbuminuria, which provides several more possible targets for modulating renal lipid metabolism [104].
On the other hand, some molecules can indirectly regulate fibrosis by cross-talking with lipid metabolism. Numerous studies have proved that complement is a causative factor for various CKD and acute kidney diseases, and complement components including C3 and C5 are also involved in renal fibrosis [105–107]. In DKD, C5a inhibition can downregulate diacylglycerol O-Acyltransferase 1 and SREBP-1, and C5a inhibitor NOX-D21 reduced tubular sclerosis and improved lipid metabolism [108], suggesting drugs against complement may also prevent renal fibrosis through cross-talk with lipid metabolism. Autophagy is also tightly related to lipid metabolism. During renal fibrosis, autophagy activation often coexists with lipid aggregation. Preliminary studies have found that the autophagy protein beclin-1 can congregate in the endoplasmic reticulum, initiate autophagosomes generation, and promote the cellular deposition of lipid droplets, which may result from the accumulation of free FAs, so lipotoxicity caused by autophagy in the kidney will motivate the fibrotic phenotype [36], and axis “autophagy-metabolism-fibrosis” may offer an alternative pathway against renal fibrosis. HIF-1α is a key factor regulating cell lipid metabolism and lipid accumulation, thus provoking fibrosis by promoting ECM accumulation [109–111]. Mitochondrial uncoupling protein (UCP2) is also involved in regulation of mitochondrial functional activity and lipids metabolism, especially FAO. Ke et al. demonstrated that UCP2 and HIF-1α are involved in disorder of lipid metabolism-related to renal fibrosis in an acute ischemia-reperfusion induced renal fibrosis model. In renal tubular cells, UCP2 can stabilize HIF-1α by regulating mitochondrial respiration and oxygen content, thereby regulating lipid accumulation and ECM and controlling the initiation and development of fibrosis, which may be related to PPAR-α and CPT1A [112]. Other molecules, such as mitochondrial transcription factor A [113] and vascular endothelial growth factor B [114], have also been shown to influence nephropathy and fibrosis by regulating lipid metabolism.
Although in-depth studies on these emerging regulators are still lacking, they may represent a series of novel possible targets in the axis of “lipid metabolism-renal fibrosis”. Likewise, we can select candidates for anti-fibrosis from the repertoire of molecules with lipid metabolism regulation ability.
Lipid metabolism-related noncoding RNAs
Epigenetics and its derived gene therapy are hot spots at present, especially the miRNA-based and long noncoding RNA (lncRNA)-based agents with inspiring advancement. By virtue of cutting-edge sequencing techniques and effective gene intervening strategies, we can expedite our excavation on drugs based on epigenetics. As seen from nonalcoholic fatty liver disease (NAFLD), miRNAs and lncRNAs play crucial roles in the pathology of pulmonary lipid metabolism, metabolic inflammation, regeneration, and fibrogenesis [115]. These noncoding RNAs in the liver take part in cholesterol synthesis and FAO via targeting multiple metabolism-associated genes, thus control steatosis, NAFLD, and end-stage liver fibrosis, through lipid accumulation and lipotoxicity [115]. Likewise, targeting noncoding RNAs upstream of regulatory factors in the lipid metabolism pathway to treat renal fibrosis has also shown encouraging progress in preclinical studies.
miRNAs involved in lipid metabolism and renal fibrosis
miRNAs are a class of noncoding RNAs that regulate gene expression transcriptionally via binding to mRNAs, and play a key role in cellular physiology and disease pathology, including metabolism [116, 117] and CKD [118, 119] respectively. Although there are relatively fewer studies on miRNAs in renal fibrosis, miRNAs may be novel targets for anti-fibrosis, based on existing studies. Moreover, miRNAs may engage in kidney metabolism via checking specific genes, hence mediating fibrogenesis in a metabolic manner.
In this part, we solely focus on the axis of miRNAs-lipid metabolism-renal fibrosis. MiR-21 is the first miRNA that has been shown to link lipid metabolism to renal fibrosis. MiR-21 targets PPAR-α, Mpv17l (a gene that inhibits the production of ROS), and reck (inhibits metalloproteinases) to downregulate their expression, thus disrupting the balance of lipid metabolism and accelerating renal damage and fibrosis, while systematic knockout of miR-21 precluded the pathological role of miR-21 in a UUO and unilateral ischemia-reperfusion mice model [120]. Gomez et al. found that miR-21 can increase intracellular lipid aggregation and ROS production by silencing PPARα and inhibiting antioxidant proteins. Subsequently, they developed highly specific ASOs targeting miR-21, which was selectively positioned into kidney of mice and knocked down miR-21 after subcutaneous administration. MiR-21 ASO significantly improved proteinuria in an Alport nephropathy (a hereditary glomerular disease, with the clinical manifestation of chronic glomerulonephritis) model, and inhibited glomerular sclerosis and renal interstitial fibrosis. These therapeutic effects were related to enhanced mitochondrial functional activity, reinforced mitochondrial FAO activity, and reduced mitochondrial ROS production [121]. Note that nowadays the anti-miR21 oligonucleotide RG-012 has entered clinical trial phase I (NCT03373786) and is being tested in patients with Alport syndrome by subcutaneous injection. Although it is not specific for renal fibrosis, RG-012 may also be developed into a drug for the treatment of renal fibrosis considering that miR21 genuinely dictates renal fibrosis. On the other hand, miR-21 from serum or urine can reflect renal fibrosis, and there are clinical trials to assess its reliability as a biomarker (NCT03780101).
MiR-9-5p, a miRNA that functions to maintain the homeostasis of organ functions, can prevent fibrosis of organs, including the kidney. Compared with mice injected with only lentivirus vector, renal interstitial fibrosis in a UUO mouse model injected with lentivirus carrying miR-9-5p was profoundly alleviated, with a reverse of aberrant gene expressions mainly related to FAO, the tricarboxylic acid cycle, glycolysis, oxidative phosphorylation, and mitochondrial function, which are down-regulated during fibrosis. miR-9-5p prevented a decline of the CPT1A level, and it was further found that PGC-1α was necessary for miR-9-5p to exert renal protective effects. These findings illuminated the metabolic reprogramming effect of miR-9-5p in renal fibrosis, especially in regulation of the cellular lipid metabolism pathway [122].
Similarly, miR-33, an miRNA known to be encoded by the region within the SREBP gene and highly expressed in the kidney, can promote fibrosis and inflammation in DKD through the NF-κB/TGF-β pathway [123]. Based on this, Price et al. used a pH-low insertion peptide to carry miR-33 peptide nucleic acid inhibitors. This tool enabled miR-33 targeted delivery to the kidney, an acidic environment, then miR-33 downstream regulatory genes, CPT1A and carnitine O-octanoyltransferase (CROT, an enzyme catalyzing the reversible transfer of fatty acyl groups), showed increased expression. Finally, cellular lipid aggregation was eliminated in the kidneys, which effectively dampened renal fibrosis. This study reveals the possibility of targeting the kidney’s miRNAs to treat fibrosis [124].
LncRNAs in lipid metabolism and renal fibrosis
lncRNAs are a type of non-coding RNA with a length greater than 200 bps [125]. They are able to manipulate development of various diseases via regulating gene expression transcriptionally, post-transcriptionally, and epigenetically. Several lncRNAs have been characterized in renal fibrosis, such as Erbb4-IR [21], lnc-TSI [126], CYB4P1-PS1-001 [127], lncRNA ZEB1-AS1 [128], and lncRNA taurine upregulated gene 1 (TUG1). Among them, TUG1 could upregulate PGC-1α expression through interacting with the upstream region of the PGC-1α gene to promote the binding of PGC1-α to its promoter, which modulates multiple downstream genes of PGC-1α, including CPT1B, and partially reverses the high glucose-induced mitochondrial destruction in podocytes, facilitating energy metabolism [129]. The direct effect on PGC-1α endows TUG1 with the ability to regulate mitochondrial biosynthesis and energy metabolism activity, suggesting that engineered TUG1 is likely to treat renal fibrosis. For other lncRNAs listed here, metabolic reprogramming is not an essential component of their mode of action.
Due to the controversy over protein-level candidates (such as PPARs) as mentioned earlier, directly targeting the kidney’s miRNAs or lncRNAs may be a better strategy. At present, famous biotech companies (Synlogic, Miragen, Regulus Therapeutics and etc.) are vigorously propelling miRNA drug pipelines.
Obstacles of noncoding RNAs as drug targets
In contrast, it is difficult to define whether a miRNA or lncRNA is “good” or “bad” due to its different effects depending on the disease background or tissue localization. For example, miR-9-5p is “versatile”: it protects peripheral nerves [130] and the kidney [120] from pathological conditions and inhibits glioblastoma cell proliferation [131], while on the other hand, it incites cervical cancer angiogenesis and invasion [132]. Therefore, in order to avoid severe toxic effects, miRNAs or lncRNAs should be targeted as specifically as possible. To realize this purpose, drug delivery must be carefully designed. Viral vectors are widely used for loading nucleic acids in biological experiments, but they have some risk elements, including immunogenicity, limited packaging capacity, and the possibility of causing oncogenesis, so they have not been deemed as ideal biomaterial for in vivo drug delivery [133]. Nowadays, nanoparticles (NPs) are popular drug carriers. Nano-liposomes and other nanoparticles modified with specific peptide chains to achieve highly specific targeting of miRNAs and lncRNAs will help reduce undesirable effects within other organs and tissues [134]. Recently, an in vivo miRNA encapsulating system based on polymeric NPs was deemed as an efficient and sustainable delivery approach. Effective and safe infarcted myocardium restoration was achieved when locally injecting miRNA polymeric NPs with invasiveness [135]. Other types of NPs (bacterial minicells, gold NPs, dendrimers, etc.) are also exploited to encapsulate miRNA, especially for cancer therapy [136]. Exosomes are 30–200 nm membrane-bound vesicles naturally secreted by cells, containing lipids, proteins, DNA, mRNAs, miRNAs, and lncRNAs [137]. They are engineered to be delivery systems characterized by acquiring inspiring organotropism, a property that endows exosomes with specific organ distribution post systemic administration [138]. Delivering drugs for cancer is a cardinal hotspot in exosome research. miRNAs loaded into exosomes will target and enter into expected tissues through modification technologies, for example, exosomes carrying miRNA-126 are able to accomplish lung homing after intravenous administration, representing natural miRNA nano-carrier formation and successfully treating metastatic lung cancer both in vitro and in vivo. Mechanistically, exosomes express a high level of integrin β4 on their surface, which binds with surfactant protein C located on epithelial cells in the lungs [139].
Therefore, considerable progress has been made in in vivo miRNA delivery systems, which is shown in Fig. 4, especially for tissue-targeted or cell-specific delivery. As for lncRNAs, their delivery is similar to that of miRNAs. These technologies have also been researched and preclinically applied in other fields besides carcinoma, including in cardiovascular diseases, wherein exosomes with cardiomyocyte-specific binding peptides on the surface lengthen their retention in the heart [140, 141]. In this scenario, designing kidney-specific anti-fibrotic drugs from noncoding RNA may be a promising area in the future, based on the premise that we constantly address challenges in pharmaceutical formulations, especially active delivery systems consisting of drug particles and specific molecules bound to cells of interest.
Recently, even lncRNA candidate drugs have entered into clinical trials (NCT03985072). Although among trials on miRNA/lncRNA, cancer rather than fibrosis is the mainstream field being evaluated, progress in nano-pharmacological techniques conducive to cancer treatment will also be able to be used to overcome renal fibrosis. Undoubtedly, adequate experiments are needed to clarify the regulatory network of a candidate miRNA or lncRNA, and their targeting security remains to be seen.
Other epigenetic targets in kidney lipid metabolism
Circular RNAs (circRNAs) are another group of noncoding RNAs, with closed-loop structures where the 3′ and 5′ RNA ends are covalently joined together [142]. Recently, circRNAs have been reported to participate in initiation and progression of renal diseases such as acute renal impairment, chronic nephritis, and diabetic glomerular injury [143]. circRNAs act as molecular sponges or scaffolds that bind with microRNAs or proteins and thus affect the intracorporeal processes of lipid metabolism, such as lipogenesis, lipolysis, lipophagy, lipid efflux, and degradation [142, 144], but there is rare research regarding how circRNAs mediate lipid metabolism within the kidney, as well as lipid-renal fibrosis axis. An increasing number of studies have demonstrated that circRNAs regulate multiple lipid-related genes and lipid metabolism processes in adipocytes, hepatocytes, and macrophages. For example, CircRNA_11897 promotes fatty acid synthesis through the miR-27b-3P/stearoyl-CoA desaturase pathway in adipocytes of Large White and Laiwu pigs [145]; and circSAMD4A promotes adipogenesis and preadipocyte differentiation through the miR-138-5p/EZH2 axis, resulting in obesity after bariatric surgery in humans [146]. Although currently there are no circRNAs can bridge lipid metabolism and kidney fibrosis, several novel circRNAs, especially circACTR2, have been reported to regulate high glucose-induced pyroptosis, inflammation, and fibrosis in proximal tubular cells [143]. There is a crosstalk between glucose and lipid metabolism, therefore, it is possible that more novel circRNAs directly involved in renal lipid metabolism will be identified in the future.
A number of recent studies revealed that another epigenetic modification, DNA methylation also plays a central role in the regulation of lipid metabolism [147]. DNA methylation modifications are important regulators of transcriptional networks that do not affect the DNA sequence and can translate genetic variants and environmental factors into phenotypic traits. Characterizing the genetic and environmental control of methylation and its contributions to the regulation of gene expression in lipid metabolism may help us understand the underlying biological mechanism and identify new biomarkers and treatment targets for renal fibrosis. For example, it is reported that methylation of one CpG upstream of a promoter region of CPT1A was strongly associated with triglyceride and negatively correlated with CPT1A expression in a clinical study [148], further gene expression changes in CPT1A explained 13.5% of the observed association of CPT1A DNA methylation at the CpG site with triglyceride [148, 149]. Other reports showed that PGC1a was differentially methylated in response to a 5-day high-fat diet in subcutaneous adipose tissues and skeletal muscle in a birth-weight specific manner [150, 151]. These studies suggested that DNA methylation is associated with gene expression, and the interaction explains related phenotype changes in lipid metabolism.
Summary and outlook
Aberrant intra-renal lipid metabolism is a universal characteristic and pathological factor in renal fibrosis. Therefore, developing drugs able to master renal lipid metabolism may help treat renal fibrosis. The rational of targeting lipid metabolism mainly includes regulating ER stress, ROS, fibroblast activation, ECM expression, tubule injury, pro-inflammatory microenvironment, and macrophage skewing/reprogramming (Fig. 1). In this article, we reviewed the advancement of representative lipid metabolism regulators in recent years, discussing the anti-fibrotic effects of existing drugs or inhibitors against these candidate targets in preclinical animal models. These evaluations, acquired from preliminary studies, have shown that “druggable” lipid metabolism has the potential to control renal fibrosis. Among other lipid metabolism-related molecules, gene therapy based on miRNAs has also been introduced, because studies are gradually emerging supporting that they connect kidney metabolism and fibrosis, and have the possibility to be anti-fibrotic drugs.
Equally of note, the resource of renal lipids is somewhat vague, as the kidney is an excretory organ with abounding blood and metabolites flowing through it, and suffers from countless exogenous invasions. To this end, although this review was focused on lipid metabolism within the kidney, we cannot exclude the possibility that kidney aberrant lipids originate from other tissues. Moreover, the exact mechanism based on the “lipid-fibrosis axis” is far from clear yet, albeit with phenotypic success, as announced in preclinical trials, which we discussed earlier.
In the field of oncology, metabolic research is undoubtedly a hot spot, and scientists are confident of developing new antineoplastic drugs from the perspective of metabolic reprogramming. In fibrosis, it is also likely to find new drugs targeting metabolic dysregulation and to achieve drug repositioning based on existing compounds with metabolic targets. The fibrosis mechanism is particularly complex and is far from having a clear conclusion, but preclinical experiments have shown promising aspects of lipid metabolism, such as the FAO molecular pathway, in treating renal fibrosis models. These should be valued by more researchers in the near future because many anti-renal fibrosis drugs targeting the traditional TGF-β pathway are ineffective and nonspecific. Since TGF-β is a fundamental conductor in many physiological processes, inhibition of TGF-β will lead to carcinogenesis [152], autoimmunity [153], compromised wound healing [154], and other unwanted effects. Thus, we must make a shift now. Metabolic imbalance may be the downstream reaction of or partly in parallel with the TGF-β pathway [1]; on the other hand, lipid metabolism affects the physiology of the kidney to a great extent, but research on finding a target in anti-renal fibrosis from this perspective is still lacking, and there has not been a suitable drug target determined yet. Therefore, it is urgent and essential to explore a set of weighting networks for the regulation of renal lipid metabolism, and to find the most critically targetable molecules in renal fibrosis.
From the point of view of practice, drugs targeting metabolism in cancer have been wedged into clinical trials. According to clinicals.gov (https://clinicaltrials.gov/), fatty acid synthase inhibitor TVB-2640, combined with paclitaxel and trastuzumab, was tested on patients with HER2 positive breast cancer to find out whether this metabolic drug can contribute to better anticancer effects (NCT03179904). Metformin, a well-known drug for metabolic diseases, has been examined in breast cancer clinically after the discovery of its anti-cancer effect in numerous animal model experiments (NCT04387630). Other drugs being designed to target glucose or amino acid metabolism include CPI-613 (α- ketoglutarate dehydrogenase complex, NCT04203160), leflunomide (dihydroorotate dehydrogenase 2, NCT03709446), CB-839 (glutaminase-1, NCT02861300), and ADI-PEG 20 (arginine deiminase, NCT03449901). These attempts will lay the foundation for clinical translation of metabolic strategies in fibrosis. Latest clinical trial has demonstrated that TVB-2640 has achieved satisfactory effect in reducing hepatic de novo lipogenesis and restoring liver function in NASH in clinical trial stage 2b, as well as NASH related fibrosis [155]. This excited result will encourage more anti-fibrosis trials on other organs, including kidney.
Considerable ongoing research on modifying metabolic reprogramming to overcome cancer will provide an opportune reference for fibrosis prevention and treatment, deepening our understanding of the molecular mechanisms. Advanced therapies for locally targeting kidney tissues, and even the exact cell types, will greatly depend on successful pharmaceutical preparations and delivery technology.
Funding
This paper was supported by the Drug Innovation Major Project of China [grant number 2018ZX09711001-002-010]; Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences [grant number 2016-I2M-3–011]; and Beijing Natural Science Foundation [grant numbers 7202138, 7181007].
Competing interests
The authors declare no competing interests.
Contributor Information
Xiao-guang Chen, Email: chxg@imm.ac.cn.
Sen Zhang, Email: zhangs@imm.ac.cn.
References
- 1.Zhao X, Kwan JYY, Yip K, Liu PP, Liu FF. Targeting metabolic dysregulation for fibrosis therapy. Nat Rev Drug Discov. 2019:1–19:57–75. [DOI] [PubMed]
- 2.Humphreys BD. Mechanisms of renal fibrosis. Annu Rev Physiol. 2018;80:309–26. doi: 10.1146/annurev-physiol-022516-034227. [DOI] [PubMed] [Google Scholar]
- 3.Hewitson TD, Holt SG, Smith ER. Progression of tubulointerstitial fibrosis and the chronic kidney disease phenotype—role of risk factors and epigenetics. Front Pharmacol. 2017; 8:520. 10.3389/fphar.2017.00520. [DOI] [PMC free article] [PubMed]
- 4.Wilson PC, Kashgarian M, Moeckel G. Interstitial inflammation and interstitial fibrosis and tubular atrophy predict renal survival in lupus nephritis. Clin Kidney J. 2017;11:207–18. doi: 10.1093/ckj/sfx093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen DQ, Chen H, Chen L, Vaziri ND, Wang M, Li XR, et al. The link between phenotype and fatty acid metabolism in advanced chronic kidney disease. Nephrol Dial Transplant. 2017;32:1154–66. doi: 10.1093/ndt/gfw415. [DOI] [PubMed] [Google Scholar]
- 6.Dou F, Miao H, Wang JW, Chen L, Wang M, Chen H, et al. An integrated lipidomics and phenotype study reveals protective effect and biochemical mechanism of traditionally used alisma orientale juzepzuk in chronic kidney disease. Front Pharmacol. 2018;9:53. doi: 10.3389/fphar.2018.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Cao G, Chen DQ, Wang M, Vaziri ND, Zhang ZH, et al. Metabolomics insights into activated redox signaling and lipid metabolism dysfunction in chronic kidney disease progression. Redox Biol. 2016;10:168–78. doi: 10.1016/j.redox.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang HM, Ahn SH, Choi P, Ko Y-A, Han SH, Chinga F, et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med. 2015;21:37. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung KW, Lee EK, Lee MK, Oh GT, Yu BP, Chung HY. Impairment of PPAR alpha and the fatty acid oxidation pathway aggravates renal fibrosis during aging. J Am Soc Nephrol. 2018;29:1223–37. doi: 10.1681/ASN.2017070802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorotea D, Koya D, Ha H. Recent insights into SREBP as a direct mediator of kidney fibrosis via lipid-independent pathways. Front Pharmacol. 2020;11:265. doi: 10.3389/fphar.2020.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y. Cellular and molecular mechanisms of renal fibrosis. Nat Rev Nephrol. 2011;7:684–96. doi: 10.1038/nrneph.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato Y, Yanagita M. Functional heterogeneity of resident fibroblasts in the kidney. Proc Jpn Acad Ser B Phys Biol Sci. 2019;95:468–78. doi: 10.2183/pjab.95.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi R, Yang C. Renal tubular epithelial cells: the neglected mediator of tubulointerstitial fibrosis after injury. Cell Death Dis. 2018;9:1–11. doi: 10.1038/s41419-018-1157-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato Y, Yanagita M. Resident fibroblasts in the kidney: a major driver of fibrosis and inflammation. Inflamm Regen. 2017;37:17. 10.1186/s41232-017-0048-3. [DOI] [PMC free article] [PubMed]
- 15.Summers SA, Gan P-Y, Dewage L, Ma FT, Ooi JD, O’sullivan KM, et al. Mast cell activation and degranulation promotes renal fibrosis in experimental unilateral ureteric obstruction. Kidney Int. 2012;82:676–85. doi: 10.1038/ki.2012.211. [DOI] [PubMed] [Google Scholar]
- 16.Djudjaj S, Boor P. Cellular and molecular mechanisms of kidney fibrosis. Mol Asp Med. 2019;65:16–36. doi: 10.1016/j.mam.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Meng XM. Inflammatory mediators and renal fibrosis. Adv Exp Med Biol. 2019;1165:381–406. doi: 10.1007/978-981-13-8871-2_18. [DOI] [PubMed] [Google Scholar]
- 18.Meng X-M, Tang PM-K, Li J, Lan HY. Macrophage phenotype in kidney injury and repair. Kidney Dis. 2015;1:138–46. doi: 10.1159/000431214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang YY, Jiang H, Pan J, Huang XR, Wang YC, Huang HF, et al. Macrophage-to-myofibroblast transition contributes to interstitial fibrosis in chronic renal allograft injury. J Am Soc Nephrol. 2017;28:2053–67. doi: 10.1681/ASN.2016050573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Yang T, Lu D-W, Zhao H, Feng Y-L, Chen H, et al. Central role of dysregulation of TGF-β/Smad in CKD progression and potential targets of its treatment. Biomed Pharmacother. 2018;101:670–81. doi: 10.1016/j.biopha.2018.02.090. [DOI] [PubMed] [Google Scholar]
- 21.Feng M, Tang PM-K, Huang X-R, Sun S-F, You Y-K, Xiao J, et al. TGF-β mediates renal fibrosis via the Smad3-Erbb4-IR long noncoding RNA axis. Mol Ther. 2018;26:148–61. doi: 10.1016/j.ymthe.2017.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Cai H, Deng J, Tu X, Sun Y, Huang Z, et al. TGF-β-mediated upregulation of Sox9 in fibroblast promotes renal fibrosis. Biochim Biophys Acta Mol Basis Dis. 2018;1864:520–32. doi: 10.1016/j.bbadis.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 23.Györfi AH, Matei AE, Distler JHW. Targeting TGF-β signaling for the treatment of fibrosis. Matrix Biol. 2018;68:8–27. doi: 10.1016/j.matbio.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 24.Walton KL, Johnson KE, Harrison CA. Targeting TGF-β mediated SMAD signaling for the prevention of fibrosis. Front Pharmacol. 2017;8:461. doi: 10.3389/fphar.2017.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng XM, Huang XR, Chung AC, Qin W, Shao X, Igarashi P, et al. Smad2 protects against TGF-beta/Smad3-mediated renal fibrosis. J Am Soc Nephrol. 2010;21:1477–87. doi: 10.1681/ASN.2009121244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-beta: the master regulator of fibrosis. Nat Rev Nephrol. 2016;12:325–38. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 27.Edeling M, Ragi G, Huang S, Pavenstädt H, Susztak K. Developmental signalling pathways in renal fibrosis: the roles of Notch, Wnt and Hedgehog. Nat Rev Nephrol. 2016;12:426–39. doi: 10.1038/nrneph.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He W, Dai C, Li Y, Zeng G, Monga SP, Liu Y. Wnt/beta-catenin signaling promotes renal interstitial fibrosis. J Am Soc Nephrol. 2009;20:765–76. doi: 10.1681/ASN.2008060566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bielesz B, Sirin Y, Si H, Niranjan T, Gruenwald A, Ahn S, et al. Epithelial Notch signaling regulates interstitial fibrosis development in the kidneys of mice and humans. J Clin Invest. 2010;120:4040–54. doi: 10.1172/JCI43025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabian SL, Penchev RR, St-Jacques B, Rao AN, Sipilä P, West KA, et al. Hedgehog-Gli pathway activation during kidney fibrosis. Am J Pathol. 2012;180:1441–53. doi: 10.1016/j.ajpath.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rauchman M, Griggs D. Emerging strategies to disrupt the central TGF-β axis in kidney fibrosis. Transl Res. 2019;209:90-104. [DOI] [PMC free article] [PubMed]
- 32.Chen Y, Shi‐Wen X, Eastwood M, Black CM, Denton CP, Leask A, et al. Contribution of activin receptor–like kinase 5 (transforming growth factor β receptor type I) signaling to the fibrotic phenotype of scleroderma fibroblasts. Arthritis Rheum. 2006;54:1309–16. doi: 10.1002/art.21725. [DOI] [PubMed] [Google Scholar]
- 33.Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, et al. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–9. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montford JR, Furgeson SB. A new CTGF target in renal fibrosis. Kidney Int. 2017;92:784–6. doi: 10.1016/j.kint.2017.04.042. [DOI] [PubMed] [Google Scholar]
- 35.Nagai Y, Matoba K, Kawanami D, Takeda Y, Akamine T, Ishizawa S, et al. ROCK2 regulates TGF-β-induced expression of CTGF and profibrotic genes via NF-κB and cytoskeleton dynamics in mesangial cells. Am J Physiol-Ren Physiol. 2019;317:F839–F51. doi: 10.1152/ajprenal.00596.2018. [DOI] [PubMed] [Google Scholar]
- 36.Yan Q, Song Y, Zhang L, Chen Z, Yang C, Liu S, et al. Autophagy activation contributes to lipid accumulation in tubular epithelial cells during kidney fibrosis. Cell Death Discov. 2018;4:39. doi: 10.1038/s41420-018-0065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welte MA. Expanding roles for lipid droplets. Curr Biol. 2015;25:R470–81. doi: 10.1016/j.cub.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Jr., Ory DS, et al. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci USA. 2003;100:3077–82. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Izquierdo-Lahuerta A, Martínez-García C, Medina-Gómez G. Lipotoxicity as a trigger factor of renal disease. J Nephrol. 2016;29:603–10. doi: 10.1007/s40620-016-0278-5. [DOI] [PubMed] [Google Scholar]
- 40.Bobulescu IA. Renal lipid metabolism and lipotoxicity. Curr Opin Nephrol Hypertens. 2010;19:393–402. doi: 10.1097/MNH.0b013e32833aa4ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang HM, Ahn SH, Choi P, Ko YA, Han SH, Chinga F, et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med. 2015;21:37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou Y, Wu M, Wei J, Ren Y, Du C, Wu H, et al. CD36 is involved in high glucose-induced epithelial to mesenchymal transition in renal tubular epithelial cells. Biochem Biophys Res Commun. 2015;468:281–6. doi: 10.1016/j.bbrc.2015.10.112. [DOI] [PubMed] [Google Scholar]
- 43.Hua W, Huang HZ, Tan LT, Wan JM, Gui HB, Zhao L, et al. CD36 mediated fatty acid-induced podocyte apoptosis via oxidative stress. PLoS One. 2015;10:e0127507. [DOI] [PMC free article] [PubMed]
- 44.Hughes J, Liu Y, Van Damme J, Savill J. Human glomerular mesangial cell phagocytosis of apoptotic neutrophils: mediation by a novel CD36-independent vitronectin receptor/thrombospondin recognition mechanism that is uncoupled from chemokine secretion. J Immunol. 1997;158:4389–97. [PubMed] [Google Scholar]
- 45.Pennathur S, Pasichnyk K, Bahrami NM, Zeng L, Febbraio M, Yamaguchi I, et al. The macrophage phagocytic receptor CD36 promotes fibrogenic pathways on removal of apoptotic cells during chronic kidney injury. Am J Pathol. 2015;185:2232–45. doi: 10.1016/j.ajpath.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Zhang T, Geng J, Wu Z, Xu L, Liu J, et al. Advanced oxidation protein products promote lipotoxicity and tubulointerstitial fibrosis via CD36/β-catenin pathway in diabetic nephropathy. Antioxid Redox Signal. 2019;31:521–38. doi: 10.1089/ars.2018.7634. [DOI] [PubMed] [Google Scholar]
- 47.Abumrad N, Coburn C, Ibrahimi A. Membrane proteins implicated in long-chain fatty acid uptake by mammalian cells: CD36, FATP and FABPm. Biochim Biophys Acta. 1999;1441:4–13. doi: 10.1016/s1388-1981(99)00137-7. [DOI] [PubMed] [Google Scholar]
- 48.Wanders RJA, Waterham HR, Ferdinandusse S. Metabolic interplay between peroxisomes and other subcellular organelles including mitochondria and the endoplasmic reticulum. Front Cell Dev Biol. 2016;3:83. 10.3389/fcell.2015.00083. [DOI] [PMC free article] [PubMed]
- 49.Meyer C, Nadkarni V, Stumvoll M, Gerich J. Human kidney free fatty acid and glucose uptake: evidence for a renal glucose-fatty acid cycle. Am J Physiol-Endocrinol Metab. 1997;273:E650. doi: 10.1152/ajpendo.1997.273.3.E650. [DOI] [PubMed] [Google Scholar]
- 50.Wang M. Kidney miR-33 controls fatty acid oxidation. Nat Rev Nephrol. 2020;16:66. doi: 10.1038/s41581-019-0228-2. [DOI] [PubMed] [Google Scholar]
- 51.Hou X, Tian J, Geng J, Li X, Tang X, Zhang J, et al. MicroRNA-27a promotes renal tubulointerstitial fibrosis via suppressing PPARγ pathway in diabetic nephropathy. Oncotarget. 2016;7:47760. doi: 10.18632/oncotarget.10283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang XX, Jiang T, Shen Y, Adorini L, Pruzanski M, Gonzalez FJ, et al. The farnesoid X receptor modulates renal lipid metabolism and diet-induced renal inflammation, fibrosis, and proteinuria. Am J Physiol Ren Physiol. 2009;297:F1587–F96. doi: 10.1152/ajprenal.00404.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyu Z, Mao Z, Li Q, Xia Y, Liu Y, He Q, et al. PPARγ maintains the metabolic heterogeneity and homeostasis of renal tubules. EBioMedicine. 2018;38:178–90. doi: 10.1016/j.ebiom.2018.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao X, Psarianos P, Ghoraie LS, Yip K, Goldstein D, Gilbert R, et al. Metabolic regulation of dermal fibroblasts contributes to skin extracellular matrix homeostasis and fibrosis. Nat Metab. 2019;1:147–57. doi: 10.1038/s42255-018-0008-5. [DOI] [PubMed] [Google Scholar]
- 55.Xie N, Tan Z, Banerjee S, Cui H, Ge J, Liu R-M, et al. Glycolytic reprogramming in myofibroblast differentiation and lung fibrosis. Am J Respir Crit Care Med. 2015;192:1462–74. doi: 10.1164/rccm.201504-0780OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Para R, Romero F, George G, Summer R. Metabolic reprogramming as a driver of fibroblast activation in pulmonary fibrosis. Am J Med Sci. 2019;357:394–8. doi: 10.1016/j.amjms.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernard K, Logsdon NJ, Ravi S, Xie N, Persons BP, Rangarajan S, et al. Metabolic reprogramming is required for myofibroblast contractility and differentiation. J Biol Chem. 2015;290:25427–38. doi: 10.1074/jbc.M115.646984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gibb AA, Lazaropoulos MP, Elrod JW. Myofibroblasts and fibrosis: mitochondrial and metabolic control of cellular differentiation. Circ Res. 2020;127:427–47. doi: 10.1161/CIRCRESAHA.120.316958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Batista-Gonzalez A, Vidal R, Criollo A, Carreño LJ. New insights on the role of lipid metabolism in the metabolic reprogramming of macrophages. Front Immunol. 2019;10:2993. doi: 10.3389/fimmu.2019.02993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang X, Okamura DM, Lu X, Chen Y, Moorhead J, Varghese Z, et al. CD36 in chronic kidney disease: novel insights and therapeutic opportunities. Nat Rev Nephrol. 2017;13:769. doi: 10.1038/nrneph.2017.126. [DOI] [PubMed] [Google Scholar]
- 61.Susztak K, Ciccone E, McCue P, Sharma K, Böttinger EP. Multiple metabolic hits converge on CD36 as novel mediator of tubular epithelial apoptosis in diabetic nephropathy. PLoS Med. 2005;2:e45. 10.1371/journal.pmed.0020045. [DOI] [PMC free article] [PubMed]
- 62.Yang YL, Lin SH, Chuang LY, Guh JY, Liao TN, Lee TC, et al. CD36 is a novel and potential anti‐fibrogenic target in albumin‐induced renal proximal tubule fibrosis. J Cell Biochem. 2007;101:735–44. doi: 10.1002/jcb.21236. [DOI] [PubMed] [Google Scholar]
- 63.Okamura DM, Pennathur S, Pasichnyk K, López-Guisa JM, Collins S, Febbraio M, et al. CD36 regulates oxidative stress and inflammation in hypercholesterolemic CKD. J Am Soc Nephrol. 2009;20:495–505. doi: 10.1681/ASN.2008010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Souza ACP, Bocharov AV, Baranova IN, Vishnyakova TG, Huang YG, Wilkins KJ, et al. Antagonism of scavenger receptor CD36 by 5A peptide prevents chronic kidney disease progression in mice independent of blood pressure regulation. Kidney Int. 2016;89:809–22. doi: 10.1016/j.kint.2015.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baines RJ, Chana RS, Hall M, Febbraio M, Kennedy D, Brunskill NJ. CD36 mediates proximal tubular binding and uptake of albumin and is upregulated in proteinuric nephropathies. Am J Physiol-Ren Physiol. 2012;303:F1006–F14. doi: 10.1152/ajprenal.00021.2012. [DOI] [PubMed] [Google Scholar]
- 66.Schlaepfer IR, Joshi M. CPT1A-mediated fat oxidation, mechanisms, and therapeutic potential. Endocrinology. 2020;161:bqz046. 10.1210/endocr/bqz046. [DOI] [PubMed]
- 67.Miguel V, Tituaña J, Herrero JI, Herrero L, Serra D, Cuevas P, et al. Renal tubule Cpt1a overexpression mitigates kidney fibrosis by restoring mitochondrial homeostasis. J Clin Invest. 2021;131:e140695. [DOI] [PMC free article] [PubMed]
- 68.Lee J, Hyon J-Y, Min JY, Huh YH, Kim HJ, Lee H, et al. Mitochondrial carnitine palmitoyltransferase 2 is involved in Nε-(carboxymethyl)-lysine-mediated diabetic nephropathy. Pharmacol Res. 2020;152:104600. doi: 10.1016/j.phrs.2019.104600. [DOI] [PubMed] [Google Scholar]
- 69.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–31. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wen YA, Xiong X, Zaytseva YY, Napier DL, Vallee E, Li AT, et al. Downregulation of SREBP inhibits tumor growth and initiation by altering cellular metabolism in colon cancer. Cell Death Dis. 2018;9:265. doi: 10.1038/s41419-018-0330-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen J, Wu Z, Ding W, Xiao C, Zhang Y, Gao S, et al. SREBP1 siRNA enhance the docetaxel effect based on a bone-cancer dual-targeting biomimetic nanosystem against bone metastatic castration-resistant prostate cancer. Theranostics. 2020;10:1619. doi: 10.7150/thno.40489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mustafa M, Wang TN, Chen X, Gao B, Krepinsky JC. SREBP inhibition ameliorates renal injury after unilateral ureteral obstruction. Am J Physiol-Ren Physiol. 2016;311:F614–F25. doi: 10.1152/ajprenal.00140.2016. [DOI] [PubMed] [Google Scholar]
- 73.Van Krieken R, Marway M, Parthasarathy P, Mehta N, Ingram AJ, Gao B, et al. Inhibition of SREBP with fatostatin does not attenuate early diabetic nephropathy in male mice. Endocrinology. 2018;159:1479–95. doi: 10.1210/en.2018-00093. [DOI] [PubMed] [Google Scholar]
- 74.Sun L, Halaihel N, Zhang W, Rogers T, Levi M. Role of sterol regulatory element-binding protein 1 in regulation of renal lipid metabolism and glomerulosclerosis in diabetes mellitus. J Biol Chem. 2002;277:18919–27. doi: 10.1074/jbc.M110650200. [DOI] [PubMed] [Google Scholar]
- 75.Kim TH, Eom JS, Lee CG, Yang YM, Lee YS, Kim SG. An active metabolite of oltipraz (M2) increases mitochondrial fuel oxidation and inhibits lipogenesis in the liver by dually activating AMPK. Br J Pharmacol. 2013;168:1647–61. doi: 10.1111/bph.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hwahng SH, Ki SH, Bae EJ, Kim HE, Kim SG. Role of adenosine monophosphate-activated protein kinase-p70 ribosomal S6 kinase-1 pathway in repression of liver X receptor-alpha-dependent lipogenic gene induction and hepatic steatosis by a novel class of dithiolethiones. Hepatology. 2009;49:1913–25. doi: 10.1002/hep.22887. [DOI] [PubMed] [Google Scholar]
- 77.Tyagi S, Gupta P, Saini AS, Kaushal C, Sharma S. The peroxisome proliferator-activated receptor: a family of nuclear receptors role in various diseases. J Adv Pharm Technol Res. 2011;2:236–40. doi: 10.4103/2231-4040.90879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han L, Shen W-J, Bittner S, Kraemer FB, Azhar S. PPARs: regulators of metabolism and as therapeutic targets in cardiovascular disease. Part I: PPAR-α. Future Cardiol. 2017;13:259–78. doi: 10.2217/fca-2016-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lakhia R, Yheskel M, Flaten A, Quittner-Strom EB, Holland WL, Patel V. PPARα agonist fenofibrate enhances fatty acid β-oxidation and attenuates polycystic kidney and liver disease in mice. Am J Physiol-Ren Physiol. 2018;314:F122–F31. doi: 10.1152/ajprenal.00352.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bermúdez V, Finol F, Parra N, Parra M, Pérez A, Peñaranda L, et al. PPAR-gamma agonists and their role in type 2 diabetes mellitus management. Am J Ther. 2010;17:274–83. doi: 10.1097/MJT.0b013e3181c08081. [DOI] [PubMed] [Google Scholar]
- 81.Boor P, Celec P, Martin IV, Villa L, Hodosy J, Klenovicsova K, et al. The peroxisome proliferator-activated receptor-alpha agonist, BAY PP1, attenuates renal fibrosis in rats. Kidney Int. 2011;80:1182–97. doi: 10.1038/ki.2011.254. [DOI] [PubMed] [Google Scholar]
- 82.Chung KW, Ha S, Kim SM, Kim DH, An HJ, Lee EK, et al. PPAR alpha/beta activation alleviates age-associated renal fibrosis in Sprague Dawley rats. J Gerontol A Biol Sci Med Sci. 2020;75:452–8. doi: 10.1093/gerona/glz083. [DOI] [PubMed] [Google Scholar]
- 83.Wang Y, Pang L, Zhang Y, Lin J, Zhou H. Fenofibrate improved interstitial fibrosis of renal allograft through inhibited epithelial-mesenchymal transition induced by oxidative stress. Oxid Med Cell Longev. 2019; 2019:8936856. 10.1155/2019/8936856. [DOI] [PMC free article] [PubMed]
- 84.Bae KH, Seo JB, Jung YA, Seo HY, Kang SH, Jeon HJ, et al. Lobeglitazone, a novel peroxisome proliferator-activated receptor γ agonist, attenuates renal fibrosis caused by unilateral ureteral obstruction in mice. Endocrinol Metab. 2017;32:115–23. doi: 10.3803/EnM.2017.32.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim HS, Bae KH, Jung GS, Ham HJ, Park BY, Choi YK, et al. Proceedings of the 19th European Congress of Endocrinology. (BioScientifica).
- 86.Németh Á, Mózes MM, Calvier L, Hansmann G, Kökény G. The PPARγ agonist pioglitazone prevents TGF-β induced renal fibrosis by repressing EGR-1 and STAT3. BMC Nephrol. 2019;20:245. doi: 10.1186/s12882-019-1431-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kawai T, Masaki T, Doi S, Arakawa T, Yokoyama Y, Doi T, et al. PPAR-γ agonist attenuates renal interstitial fibrosis and inflammation through reduction of TGF-β. Lab Invest. 2009;89:47–58. doi: 10.1038/labinvest.2008.104. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Y, Wang J, Zhou QD, Zhang CH, Li Q, Huang S, et al. Peroxisome proliferator-activated receptor-γ agonist pioglitazone fails to attenuate renal fibrosis caused by unilateral ureteral obstruction in mice. J Huazhong Univ Sci Technol [Med Sci] 2016;36:41–7. doi: 10.1007/s11596-016-1539-1. [DOI] [PubMed] [Google Scholar]
- 89.Liang H, Ward WF. PGC-1alpha: a key regulator of energy metabolism. Adv Physiol Educ. 2006;30:145–51. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- 90.Lee G, Uddin MJ, Kim Y, Ko M, Yu I, Ha H. PGC-1alpha, a potential therapeutic target against kidney aging. Aging Cell. 2019;18:e12994. doi: 10.1111/acel.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang L, Liu J, Zhou F, Wang W, Chen N. PGC-1alpha ameliorates kidney fibrosis in mice with diabetic kidney disease through an antioxidative mechanism. Mol Med Rep. 2018;17:4490–8. doi: 10.3892/mmr.2018.8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Han SH, Wu MY, Nam BY, Park JT, Yoo TH, Kang SW, et al. PGC-1α protects from notch-induced kidney fibrosis development. J Am Soc Nephrol. 2017;28:3312–22. doi: 10.1681/ASN.2017020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qin X, Jiang M, Zhao Y, Gong J, Su H, Yuan F, et al. Berberine protects against diabetic kidney disease via promoting PGC-1alpha-regulated mitochondrial energy homeostasis. Br J Pharmacol. 2020;177:3646–61. doi: 10.1111/bph.14935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang JL, Chen CW, Tsai MR, Liu SF, Hung TJ, Yu Ju H, et al. Antifibrotic role of PGC-1alpha-siRNA against TGF-beta1-induced renal interstitial fibrosis. Exp Cell Res. 2018;370:160–7. doi: 10.1016/j.yexcr.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 95.Kaufman TM, Warden BA, Minnier J, Miles JR, Duell PB, Purnell JQ, et al. Application of PCSK9 inhibitors in practice. Circ Res. 2019;124:32–7. doi: 10.1161/CIRCRESAHA.118.314191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Charytan DM, Sabatine MS, Pedersen TR, Im K, Park JG, Pineda AL, et al. Efficacy and safety of evolocumab in chronic kidney disease in the FOURIER trial. J Am Coll Cardiol. 2019;73:2961–70. doi: 10.1016/j.jacc.2019.03.513. [DOI] [PubMed] [Google Scholar]
- 97.González Sanchidrián S, Labrador Gómez PJ, Aguilar Aguilar JC, Davin Carrero E, Gallego Domínguez S, Gómez-Martino Arroyo JR. Evolocumab for the treatment of heterozygous familial hypercholesterolemia in end-stage chronic kidney disease and dialysis. Nefrologia. 2019;39:218–20. doi: 10.1016/j.nefro.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 98.Wu D, Zhou Y, Pan Y, Li C, Wang Y, Chen F, et al. Vaccine against PCSK9. J Am Heart Assoc. 2020;9:e014358. doi: 10.1161/JAHA.119.014358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hotamisligil GS, Bernlohr DA. Metabolic functions of FABPs—mechanisms and therapeutic implications. Nat Rev Endocrinol. 2015;11:592. doi: 10.1038/nrendo.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qiao Y, Liu L, Yin L, Xu L, Tang Z, Qi Y, et al. FABP4 contributes to renal interstitial fibrosis via mediating inflammation and lipid metabolism. Cell Death Dis. 2019;10:1–12. doi: 10.1038/s41419-019-1610-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 101.Khan S, Cabral PD, Schilling WP, Schmidt ZW, Uddin AN, Gingras A, et al. Kidney proximal tubule lipoapoptosis is regulated by fatty acid transporter-2 (FATP2) J Am Soc Nephrol. 2018;29:81–91. doi: 10.1681/ASN.2017030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jao TM, Nangaku M, Wu CH, Sugahara M, Saito H, Maekawa H, et al. ATF6 alpha downregulation of PPARalpha promotes lipotoxicity-induced tubulointerstitial fibrosis. Kidney Int. 2019;95:577–89. doi: 10.1016/j.kint.2018.09.023. [DOI] [PubMed] [Google Scholar]
- 103.Cui J, Wu X, Song Y, Chen Y, Wan J. Deletion of Lkb1 in renal tubular epithelial cells leads to CKD by altering metabolism. J Am Soc Nephrol. 2016;27:439–53. doi: 10.1681/ASN.2014121181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Feng YL, Chen H, Chen DQ, Vaziri ND, Su W, Ma SX, et al. Activated NF-kappaB/Nrf2 and Wnt/beta-catenin pathways are associated with lipid metabolism in CKD patients with microalbuminuria and macroalbuminuria. Biochim Biophys Acta Mol Basis Dis. 2019;1865:2317–32. doi: 10.1016/j.bbadis.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 105.Rangan GK, Pippin JW, Couser WG. C5b-9 regulates peritubular myofibroblast accumulation in experimental focal segmental glomerulosclerosis. Kidney Int. 2004;66:1838–48. doi: 10.1111/j.1523-1755.2004.00957.x. [DOI] [PubMed] [Google Scholar]
- 106.Cui J, Wu X, Song Y, Chen Y, Wan J. Complement C3 exacerbates renal interstitial fibrosis by facilitating the M1 macrophage phenotype in a mouse model of unilateral ureteral obstruction. Am J Physiol Renal Physiol. 2019;317:F1171–F1182. [DOI] [PubMed]
- 107.Liu Y, Wang K, Liang X, Li Y, Zhang Y, Zhang C, et al. Complement C3 produced by macrophages promotes renal fibrosis via IL-17A secretion. Front Immunol. 2018;9:2385. doi: 10.3389/fimmu.2018.02385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yiu WH, Li RX, Wong DWL, Wu HJ, Chan KW, Chan LYY, et al. Complement C5a inhibition moderates lipid metabolism and reduces tubulointerstitial fibrosis in diabetic nephropathy. Nephrol Dialysis Transplant. 2017;33:1323–32. doi: 10.1093/ndt/gfx336. [DOI] [PubMed] [Google Scholar]
- 109.Higgins DF, Kimura K, Bernhardt WM, Shrimanker N, Akai Y, Hohenstein B, et al. Hypoxia promotes fibrogenesis in vivo via HIF-1 stimulation of epithelial-to-mesenchymal transition. J Clin Invest. 2007;117:3810–20. doi: 10.1172/JCI30487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kimura K, Iwano M, Higgins DF, Yamaguchi Y, Nakatani K, Harada K, et al. Stable expression of HIF-1α in tubular epithelial cells promotes interstitial fibrosis. Am J Physiol-Ren Physiol. 2008;295:F1023–F9. doi: 10.1152/ajprenal.90209.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bensaad K, Favaro E, Lewis CA, Peck B, Lord S, Collins JM, et al. Fatty acid uptake and lipid storage induced by HIF-1α contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014;9:349–65. doi: 10.1016/j.celrep.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 112.Ke Q, Yuan Q, Qin N, Shi C, Luo J, Fang Y, et al. UCP2-induced hypoxia promotes lipid accumulation and tubulointerstitial fibrosis during ischemic kidney injury. Cell Death Dis. 2020;11:1–13. doi: 10.1038/s41419-019-2219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huang S, Park J, Qiu C, Chung KW, Li SY, Sirin Y, et al. Jagged1/Notch2 controls kidney fibrosis via Tfam-mediated metabolic reprogramming. PLoS Biol. 2018;16:e2005233. doi: 10.1371/journal.pbio.2005233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Falkevall A, Mehlem A, Palombo I, Sahlgren BH, Ebarasi L, He L, et al. Reducing VEGF-B signaling ameliorates renal lipotoxicity and protects against diabetic kidney disease. Cell Metab. 2017;25:713–26. doi: 10.1016/j.cmet.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 115.Su Q, Kumar V, Sud N, Mahato RI. MicroRNAs in the pathogenesis and treatment of progressive liver injury in NAFLD and liver fibrosis. Adv Drug Deliv Rev. 2018;129:54–63. doi: 10.1016/j.addr.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 116.Yu Y, Du H, Wei S, Feng L, Li J, Yao F, et al. Adipocyte-derived exosomal MiR-27a induces insulin resistance in skeletal muscle through repression of PPARγ. Theranostics. 2018;8:2171. doi: 10.7150/thno.22565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Castaño C, Kalko S, Novials A, Párrizas M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc Natl Acad Sci. 2018;115:12158–63. doi: 10.1073/pnas.1808855115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bai M, Chen H, Ding D, Song R, Lin J, Zhang Y, et al. MicroRNA-214 promotes chronic kidney disease by disrupting mitochondrial oxidative phosphorylation. Kidney Int. 2019;95:1389–404. doi: 10.1016/j.kint.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 119.Rysz J, Gluba-Brzózka A, Franczyk B, Jabłonowski Z, Ciałkowska-Rysz A. Novel biomarkers in the diagnosis of chronic kidney disease and the prediction of its outcome. Int J Mol Sci. 2017;18:1702. doi: 10.3390/ijms18081702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chau BN, Xin C, Hartner J, Ren S, Castano AP, Linn G, et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med. 2012;4:121ra18-ra18. doi: 10.1126/scitranslmed.3003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gomez IG, MacKenna DA, Johnson BG, Kaimal V, Roach AM, Ren S, et al. Anti-microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J Clin Invest. 2015;125:141–56. doi: 10.1172/JCI75852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fierro‐Fernández M, Miguel V, Márquez‐Expósito L, Nuevo‐Tapioles C, Herrero JI, Blanco-Ruiz E, et al. MiR-9-5p protects from kidney fibrosis by metabolic reprogramming. FASEB J. 2020;34:410–31. doi: 10.1096/fj.201901599RR. [DOI] [PubMed] [Google Scholar]
- 123.Cao M, Bai L, Wang D, Zhai Q, Li Y, Hai J, et al. miRNA-33 expression and its mechanism in patients and model rats with type 2 diabetic nephropathy. Int J Clin Exp Med. 2018;11:1661–8. [Google Scholar]
- 124.Price NL, Miguel V, Ding W, Singh AK, Malik S, Rotllan N, et al. Genetic deficiency or pharmacological inhibition of miR-33 protects from kidney fibrosis. JCI Insight. 2019;4:e131102. 10.1172/jci.insight.131102. [DOI] [PMC free article] [PubMed]
- 125.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang P, Luo M-L, Song E, Zhou Z, Ma T, Wang J, et al. Long noncoding RNA lnc-TSI inhibits renal fibrogenesis by negatively regulating the TGF-β/Smad3 pathway. Sci Transl Med. 2018;10:eaat2039. doi: 10.1126/scitranslmed.aat2039. [DOI] [PubMed] [Google Scholar]
- 127.Wang M, Wang S, Yao D, Yan Q, Lu W. A novel long non-coding RNA CYP4B1-PS1-001 regulates proliferation and fibrosis in diabetic nephropathy. Mol Cell Endocrinol. 2016;426:136–45. doi: 10.1016/j.mce.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 128.Wang J, Pan J, Li H, Long J, Fang F, Chen J, et al. lncRNA ZEB1-AS1 was suppressed by p53 for renal fibrosis in diabetic nephropathy. Mol Ther-Nucleic Acids. 2018;12:741–50. doi: 10.1016/j.omtn.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Long J, Badal SS, Ye Z, Wang Y, Ayanga BA, Galvan DL, et al. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J Clin Invest. 2016;126:4205–18. doi: 10.1172/JCI87927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li Z, Li Y, Li Q, Zhang Z, Jiang L, Li X. Role of miR-9-5p in preventing peripheral neuropathy in patients with rheumatoid arthritis by targeting REST/miR-132 pathway. In Vitro Cell Dev Biol Anim. 2019;55:52–61. doi: 10.1007/s11626-018-0310-2. [DOI] [PubMed] [Google Scholar]
- 131.Zhang H, Li Y, Tan Y, Liu Q, Jiang S, Liu D, et al. MiR-9-5p inhibits glioblastoma cells proliferation through directly targeting FOXP2 (Forkhead Box P2). Front Oncol. 2019;9:1176. 10.3389/fonc.2019.01176. [DOI] [PMC free article] [PubMed]
- 132.Wei Y, Jiao X, Zhang S, Xu Y, Li S, Kong B. MiR-9-5p could promote angiogenesis and radiosensitivity in cervical cancer by targeting SOCS5. Eur Rev Med Pharmacol Sci. 2019;23:7314–26. doi: 10.26355/eurrev_201909_18837. [DOI] [PubMed] [Google Scholar]
- 133.Lee SWL, Paoletti C, Campisi M, Osaki T, Adriani G, Kamm RD, et al. MicroRNA delivery through nanoparticles. J Control Release. 2019;313:80–95. doi: 10.1016/j.jconrel.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nastase MV, Zeng-Brouwers J, Wygrecka M, Schaefer L. Targeting renal fibrosis: mechanisms and drug delivery systems. Adv Drug Deliv Rev. 2018;129:295–307. doi: 10.1016/j.addr.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 135.Yang H, Qin X, Wang H, Zhao X, Liu Y, Wo HT, et al. An in vivo miRNA delivery system for restoring infarcted myocardium. ACS Nano. 2019;13:9880–94. doi: 10.1021/acsnano.9b03343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.O’Neill CP, Dwyer RM. Nanoparticle-based delivery of tumor suppressor microRNA for cancer therapy. Cells. 2020;9:521. doi: 10.3390/cells9020521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9:19-. doi: 10.1186/s13578-019-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang B, Chen Y, Shi J. Exosome biochemistry and advanced nanotechnology for next-generation theranostic platforms. Adv Mater. 2019;31:e1802896. doi: 10.1002/adma.201802896. [DOI] [PubMed] [Google Scholar]
- 139.Nie H, Xie X, Zhang D, Zhou Y, Li B, Li F, et al. Use of lung-specific exosomes for miRNA-126 delivery in non-small cell lung cancer. Nanoscale. 2020;12:877–87. doi: 10.1039/c9nr09011h. [DOI] [PubMed] [Google Scholar]
- 140.Mentkowski KI, Lang JK. Exosomes engineered to express a cardiomyocyte binding peptide demonstrate improved cardiac retention in vivo. Sci Rep. 2019;9:10041-. doi: 10.1038/s41598-019-46407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen Y, Li Z, Chen X, Zhang S. Long non-coding RNAs: from disease code to drug role. Acta Pharm Sin B. 2020;11:340-54. [DOI] [PMC free article] [PubMed]
- 142.Jin J, Sun H, Shi C, Yang H, Wu Y, Li W, et al. Circular RNA in renal diseases. J Cell Mol Med. 2020;24:6523–33. doi: 10.1111/jcmm.15295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wen S, Li S, Li L, Fan Q. circACTR2: a novel mechanism regulating high glucose-induced fibrosis in renal tubular cells via pyroptosis. Biol Pharm Bull. 2020;43:558–64. doi: 10.1248/bpb.b19-00901. [DOI] [PubMed] [Google Scholar]
- 144.Yu G, Yang Z, Peng T, Lv Y. Circular RNAs: rising stars in lipid metabolism and lipid disorders. J Cell Physiol. 2021;236:4797-806. [DOI] [PubMed]
- 145.Li A, Huang W, Zhang X, Xie L, Miao X. Identification and characterization of CircRNAs of two pig breeds as a new biomarker in metabolism-related diseases. Cell Physiol Biochem. 2018;47:2458–70. doi: 10.1159/000491619. [DOI] [PubMed] [Google Scholar]
- 146.Liu Y, Liu H, Li Y, Mao R, Yang H, Zhang Y, et al. Circular RNA SAMD4A controls adipogenesis in obesity through the miR-138-5p/EZH2 axis. Theranostics. 2020;10:4705–19. doi: 10.7150/thno.42417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.He Z, Zhang R, Jiang F, Hou W, Hu C. Role of genetic and environmental factors in DNA methylation of lipid metabolism. Genes Dis. 2018;5:9–15. doi: 10.1016/j.gendis.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hidalgo B, Irvin MR, Sha J, Zhi D, Aslibekyan S, Absher D, et al. Epigenome-wide association study of fasting measures of glucose, insulin, and HOMA-IR in the Genetics of Lipid Lowering Drugs and Diet Network study. Diabetes. 2014;63:801–7. doi: 10.2337/db13-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gillberg L, Jacobsen SC, Rönn T, Brøns C, Vaag A. PPARGC1A DNA methylation in subcutaneous adipose tissue in low birth weight subjects–impact of 5 days of high-fat overfeeding. Metab Clin Exp. 2014;63:263–71. doi: 10.1016/j.metabol.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 151.Brøns C, Jacobsen S, Nilsson E, Rönn T, Jensen CB, Storgaard H, et al. Deoxyribonucleic acid methylation and gene expression of PPARGC1A in human muscle is influenced by high-fat overfeeding in a birth-weight-dependent manner. J Clin Endocrinol Metab. 2010;95:3048–56. doi: 10.1210/jc.2009-2413. [DOI] [PubMed] [Google Scholar]
- 152.Principe DR, Doll JA, Bauer J, Jung B, Munshi HG, Bartholin L, et al. TGF-β: duality of function between tumor prevention and carcinogenesis. J Natl Cancer Inst. 2014;106:djt369. 10.1093/jnci/djt369. [DOI] [PMC free article] [PubMed]
- 153.Kelly A, Houston SA, Sherwood E, Casulli J, Travis MA. Regulation of innate and adaptive immunity by TGFβ. Adv Immunol. 2017;134:137-233. [DOI] [PubMed]
- 154.Shah M, Foreman DM, Ferguson M. Neutralising antibody to TGF-beta 1, 2 reduces cutaneous scarring in adult rodents. J Cell Sci. 1994;107:1137–57. doi: 10.1242/jcs.107.5.1137. [DOI] [PubMed] [Google Scholar]
- 155.Syed-Abdul MM, Parks EJ, Gaballah AH, Bingham K, Hammoud GM, Kemble G, et al. Fatty acid synthase inhibitor TVB-2640 reduces hepatic de novo lipogenesis in males with metabolic abnormalities. Hepatology. 2020;72:103–18. doi: 10.1002/hep.31000. [DOI] [PubMed] [Google Scholar]