Abstract
Extracellular vesicles (EVs) are secreted from all cell types and are intimately involved in tissue homeostasis. They are being explored as vaccine and gene therapy platforms, as well as potential biomarkers. As their size is below the diffraction limit of light microscopy, direct visualizations have been daunting and single‐particle studies under physiological conditions have been hampered. Here, direct stochastic optical reconstruction microscopy (dSTORM) was employed to visualize EVs in three‐dimensions and to localize molecule clusters such as the tetraspanins CD81 and CD9 on the surface of individual EVs. These studies demonstrate the existence of membrane microdomains on EVs. These were confirmed by Cryo‐EM. Individual particle visualization provided insights into the heterogeneity, structure, and complexity of EVs not previously appreciated
Keywords: biotechnology, exosome, extracellular vesicle, microvesicle, dSTORM, nanodomains, super‐resolution microscopy, tetraspanin
1. INTRODUCTION
Exosomes are a species of extracellular vesicles (EVs). They are differentiated from larger EVs by their size, typically measuring ∼30–200 nm in diameter, and their biogenesis pathway. Exosomes are believed to originate from inside the cell as a result of the inward budding of late endosomes into the multivesicular body (MVB) (Cocozza et al., 2020; Théry et al., 2002). The invaginated endosomes are enriched for proteins of the endosomal sorting complexes required for transport (ESCRT), for tetraspanin proteins, and for lipid raft‐associated proteins. The MVB then traffics to the plasma membrane to release the exosomes. Many of the proteins and characteristics ascribed to exosomes are shared with plasma membrane‐derived shedding microvesicles. These include size, and the presence of the tetraspanin proteins CD9, CD63, and CD81 (Pegtel & Gould, 2019; Pols & Klumperman, 2009; Verweij et al., 2019). As these studies use material from the cell supernatant, we will therefore refer to all vesicles discussed in this manuscript as EVs. Of note, recent studies have emphasized the heterogeneity of EVs in terms of protein composition and size (Kowal et al., 2016; Martin‐Jaular et al., 2021).
EVs are part of an evolutionarily conserved cell‐to‐cell communications network. They are critical for maintaining tissue and organismal homeostasis. The contents of an EV are modified by the state of the cell, with some of the most abrupt changes being observed upon virus infection and tumorigenesis (Raab‐Traub & Dittmer, 2017; Wortzel et al., 2019). The deposition of EV contents to neighbouring or distant recipient cells has been shown to influence cell differentiation, transcription, migration, proliferation, metabolic state, and disease progression (Chugh et al., 2013; Kamerkar et al., 2017; Mathieu et al., 2019; Mcnamara et al., 2019; Yogev et al., 2017). EVs transfer small metabolites, mRNAs, miRNAs, proteins, and even entire viruses between cells (Baglio et al., 2016; Feng et al., 2013; Gould et al., 2003; Kalamvoki et al., 2014; Melo et al., 2014; Valadi et al., 2007; Zhao et al., 2016). EVs are altered by pharmacological agents and human diseases (Demarino et al., 2018). Recent years have seen a rapidly expanding interest in EVs as delivery agents for small molecules, RNA, and proteins, as biomarkers for disease, and as vaccine platforms (Kamerkar et al., 2017; Mori et al., 2019). Yet, we know little about their structure.
Biological studies and pharmaceutical applications depend on thoroughly characterized and homogenous EV preparations. The most important EV measure is size. EVs, however, are too small to be visualized by conventional light microscopy. This represents a widely recognized barrier in the field (Verweij et al., 2021). Electron microscopy (EM) represents the standard in the field of nanoparticle research and has been applied to EVs as well. Many EM preparative methods, however, employ dehydration steps that potentially alter the three‐dimensional (3‐D) shape of any liquid vesicle. Techniques such as dynamic light scattering, nanoparticle tracking analysis (NTA), and more recently nanoscale flow cytometry frequently yield results as a single summary statistic (Dragovic et al., 2011). As such, they can be limited in identifying subpopulations and are unable to detect EV surface marker clustering (Arab et al., 2021). The majority of these methods are not as sensitive as dSTORM and may miss EV populations that only have a single surface marker molecule. To address this barrier, we adapted multi‐colour, 3‐D super‐resolution microscopy to measure individual EVs in solution and to localize proteins on their surface.
Super‐resolution microscopy was developed to visualize objects below the diffraction limit (∼ 250 nm) (Huang et al., 2008; Van De Linde et al., 2011). There are multiple technical modalities. Direct stochastic optical reconstruction microscopy (dSTORM) relies on the analysis of individual photoswitching events to bypass the diffraction limit. Notably, dSTORM does not require fixation. EV isolation, as well as the measurements, are performed under physiological conditions (osmolarity, pH, temperature). This study visualized hundreds of individual EVs in 3‐D per field of view, made precise measurements on individual particles, quantitated subpopulations based on tetraspanin markers, and localized tetraspanins complexes on the surface of individual, single particles. These data provide direct evidence for the existence of microdomains on the surface of EVs. The existence of these microdomains was confirmed by Cryo‐EM.
2. RESULTS
2.1. Cells expressing CD81‐mCherry and CD63‐GFP yield fluorescent and endocytosis‐competent EVs
The tetraspanin proteins, CD9, CD63, and CD81, are accepted markers for EVs and play functional roles in EV biogenesis (Hurwitz et al., 2018; Kowal et al., 2016; Lötvall et al., 2014). They have been used to generate fluorescent alleles to track EVs and to study EV cellular uptake and release (Verweij et al., 2018; York et al., 2021). We created clonal U‐2 OS cell lines expressing either green fluorescent protein (GFP)‐tagged CD63, or mCherry‐tagged CD81, or both. Stable expression was validated by flow cytometry (Figure 1A–D). Greater than 99% of the cells expressed the reporter proteins. The proteins were enriched along the cell membranes and vesicular compartments (Figure 1E–H), consistent with normal processing and trafficking (Hurwitz et al., 2018; Mathieu et al., 2021).
FIGURE 1.
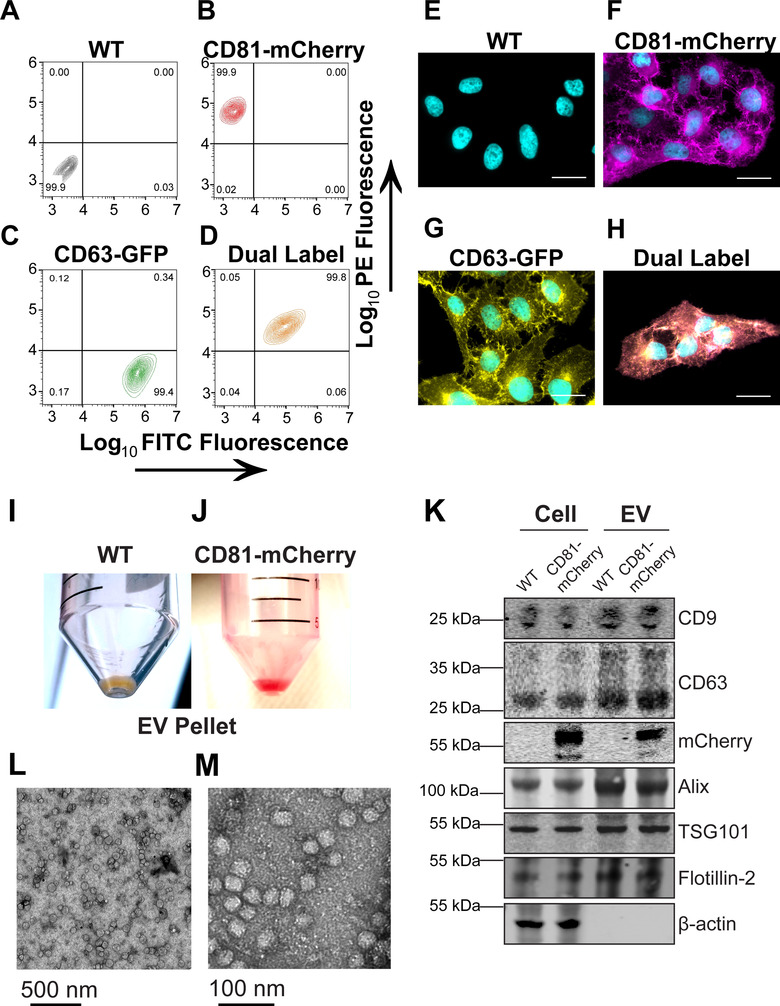
Generation of clonal CD81‐mCherry cells. (A–D) U‐2 OS cells were transfected with the indicated plasmid and selected via FACS and cell populations were analysed for clonal expansion via flow cytometry. (E–H) Fluorescence microscopy of cells sorted in A‐D (scale bar = 20 μm). (I) EV pellet from U‐2 OS WT cells. (J) EV pellet from U‐2 OS CD81‐mCherry cells. (K) Western blots of cell and EV pellets of U‐2 OS WT and CD81‐mCherry cells. (L) Transmission EM view of negatively stained affinity‐purified CD81+ EVs. (M) Zoomed‐in view of a cluster of CD81+ EVs imaged by transmission EM
EVs from WT and CD81‐mCherry expressing cells were isolated as previously described (Mcnamara et al., 2018). In brief, one liter of cell supernatant was concentrated by tangential flow filtration, EVs were precipitated by PEG, resuspended in 0.5 ml of PBS, treated with DNAse and RNAse, and passed through a CaptoCore700 column (1000x enrichment). This protocol does not use ultracentrifugation, thus avoiding exposing EVs to high g‐forces and minimizing rupture. By comparison to unlabelled EVs, the CD81‐mCherry EVs appeared bright red (Figure 1 I, J). The presence of the transgene did not affect EV protein composition (Figure 1K), demonstrating that the tagged tetraspanin did not interfere with EV biogenesis and was incorporated into EVs by the canonical pathways. It did not alter the particle size distribution of the EVs as determined by NTA (Figure S1A–D and Videos S1 and S2). The EVs exhibited the expected sphere‐like morphologies and size by transmission EM (Figure 1L‐M).
The EVs were either used directly or subjected to an additional purification step using anti‐CD81 affinity beads. The latter step increased purity and sped up image acquisition but was not required. The same results were obtained with total as with affinity‐purified EVs.
The CD81‐mCherry EVs were uptake‐competent as determined by two approaches. First, purified EVs transferred the CD81‐mCherry protein to primary peripheral blood mononuclear cells (PBMC) at a similar efficiency to EVs labelled with the membrane dye 1,1′‐Dioctadecyl‐3,3,3′,3′‐tetramethylindocarbocyanine perchlorate (DiI) as measured by flow cytometry (Figure S1E and F). Second, cells expressing either CD63‐GFP or CD81‐mCherry were equally competent at generating EVs and transferring the fusion proteins to neighbouring cells using a coculture assay (Figure S1G and H). Coculture resulted in CD63‐GFP cells now also staining positive for CD81‐mCherry and vice versa. As expected, there were fewer transferred tetraspanins in the target cells than generated de novo, resulting in lower mean fluorescence of the transferred reporter. Direct cell‐to‐cell fusion, as measured by equal fluorescence of both transgenes amounted to < 0.2% of the cell population. These studies demonstrate that the modified EVs were physically and physiologically indistinguishable from normal EVs.
2.2. CD63 and CD81 colocalize intracellularly in the same vesicles
The first application of direct stochastic optical reconstruction microscopy (dSTORM) was to verify that tagged‐CD63 and CD81 utilize a common EV biogenesis pathway. Light microscopy is limited by the diffraction limit of ∼ 180 nm laterally and ∼500 nm axially (Fouquet et al., 2015), which is larger than the size of an exosome (30–200 nm). By contrast, the dSTORM system was able to achieve ± 16 nm resolution along the lateral (XY) axis, and ± 42 nm along the axial (Z) axis (Figure S2A–C). This constitutes a ten‐fold increase in resolution. Therefore, dSTORM is able to distinguish individual EVs from aggregates.
Typically, dSTORM uses optimized fluorophores (such as Alexa Fluor 647) that maximize photon output per switching event. Thus, the ability to detect mCherry and GFP was surprising at first. It was due to advancements in the oxygen‐buffering solution that was optimized for the detector and to extended recording times. Whereas dSTORM using optimized dyes requires a ∼10 millisecond exposure, dSTORM of mCherry and GFP required five times that exposure per frame.
At the 10 μm scale, CD63 and CD81 co‐localized intracellularly (Figure 2A‐C and Video S3). At the 0.5 μm scale, CD81 was enriched along the plasma membrane as compared to CD63, which was more often seen in punctate cytoplasmic regions. Large globular domains of CD81‐mCherry were also apparent and contained CD63 and CD81 together (Figure 2D–F, yellow colour). These results confirmed the existence of lipid rafts or tetraspanin enriched microdomains (TEMs) on cell membranes as previously observed by total internal reflection fluorescence microscopy (Espenel et al., 2008; Levental et al., 2020). By contrast, Flotillin‐2 stained in dense intracellular clusters but was not enriched on the cell surface of U2‐OS cells (Figure S2D–I). As a negative control, we used β‐actin. No co‐occupancy of CD81‐mCherry with β‐actin was observed (Figure 2G–L).
FIGURE 2.
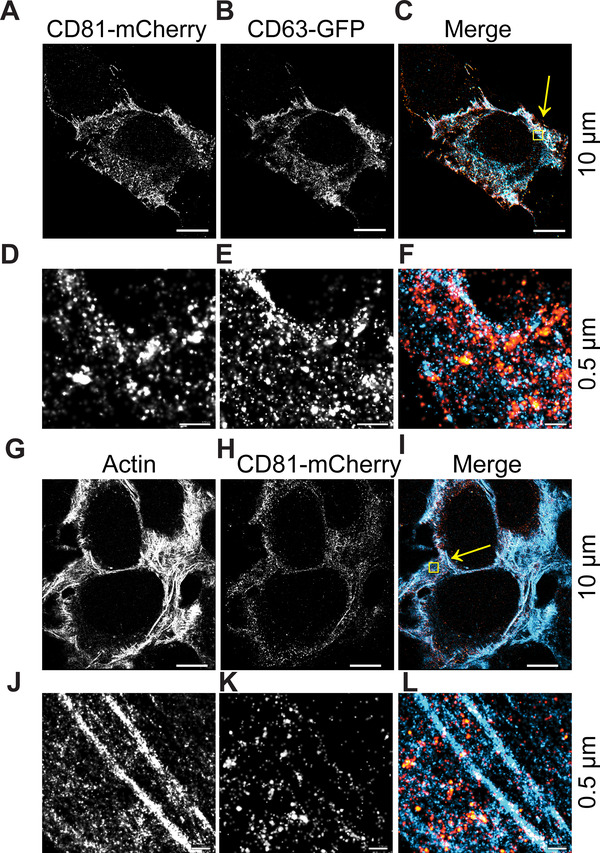
CD81 and CD63 co‐occupy intracellular regions. (A‐C) CD63‐GFP and CD81‐mCherry U‐2 OS expressing cells were visualized by dSTORM. (D–F) Zoomed‐in view of yellow box in C. (G–I) CD81‐mCherry does not co‐occupy regions with β‐actin, as visualized by dSTORM. (J–L) Zoomed‐in view of yellow box in I. Scale bars are shown with corresponding values on the right for each row of panels
Staining with fluorescently conjugated antibodies yielded the same localization as observed for tagged CD63‐GFP and CD81‐mCherry (Figure S3). This demonstrates the use of dSTORM for GFP and mCherry tagged proteins and it shows that these C‐terminal intracellular tags did not mislocalize the protein within the cell nor impair the tetraspanins’ ability to incorporate into TEMs.
2.3. Imaging of individual EV particles in solution by dSTORM
The diameter of EVs is at or below the wavelength (λ) of visible light, that is, below the diffraction limit. Therefore, individual EVs cannot be resolved by ordinary light microscopy. This barrier motivated the use of dSTORM. Purified EVs were incubated with the photoswitchable membrane dye CM Red (excitation 640 nm, peak emission ∼675 nm) and individual photoswitching events were captured throughout the exposure. Figure 3A shows the raw data, Figure 3B the data after super‐resolution filtering, and Figure 3C and D an individual EV. The experiment was repeated using the dye CM Green (excitation 473 nm, peak emission ∼520 nm). One‐hundred fifty individual dSTORM‐imaged EVs were used to generate a size distribution profile for each dye. The CM Green‐labelled EVs had a mean ± sd of 107 ± 38.6 (n = 150) and the CM Red‐labelled EVs had a mean ± sd of 126 ± 44.7 (n = 150). NTA population measurements yielded a mean ± sd of 117 ± 21.6 (n = 1500). These experiments demonstrated that EV size was neither excitation/ emission wavelength nor dye‐specific and comparable to NTA analysis (Figure 3E–F). Thus, dSTORM constitutes a novel, orthologous method to determine EV size under physiological conditions. Given a powerful enough computer, a dSTORM‐based workflow can be as fast as NTA or nano flow‐cytometry for EV size and marker determination.
FIGURE 3.
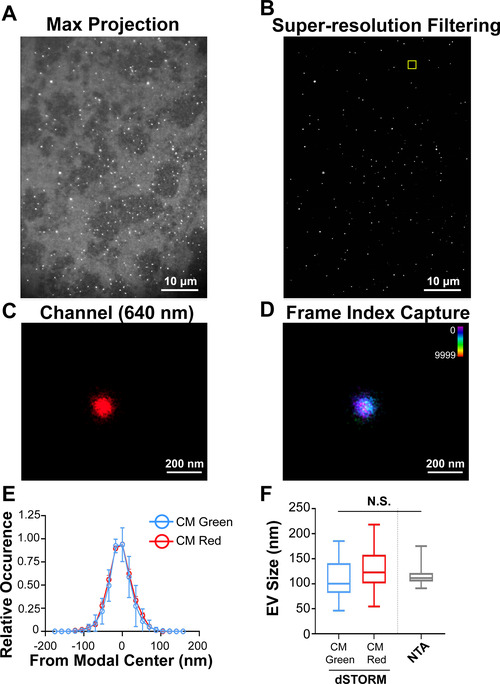
Super‐resolution (dSTORM) microscopy of a single EV in solution. (A) CD81+ EVs were affinity purified and labelled with the photoswitchable dye CM Red. Max projection image (i.e., pre‐dSTORM filtration) is shown. (B) dSTORM filtration of the image in A. (C) Zoomed‐in image of a single EV. (D) Frame‐index capture of CD81+ EVs in C showing photoswitching throughout the capture. (E) Size distribution analysis of CD81+ EVs viewed through dSTORM using CM Green or CM Red (n = 50 technical replicates, n = 3 biological replicates). (F) Box‐Whisker plot of CD81+ EV sizes stained with CM Green or CM Red as measured by dSTORM as compared to NTA
Next, EVs were stained simultaneously with CM Red and CM Green (Figure 4A–D). Panel A shows a representative image and panel B shows an artist rendering. Individual events are graphed in panel C and the distribution peaks scaled on the modal centres are shown in D. The two dyes co‐localized with a difference in modal distribution peaks of d = 9.5 nm (n > 400 photoswitching events post super‐resolution filtering per channel). The distance between modal centres can be attributed to sub‐optimal apochromatic correction, that is, the focal plane for the longer wavelength light was slightly different than that of the shorter wavelength light. This experiment established the limit of resolution for EV single particles analysis for two colours: two molecules will be considered co‐localized on the same EV if their modal peaks are within d ≤ 10 nm diameter of each other.
FIGURE 4.
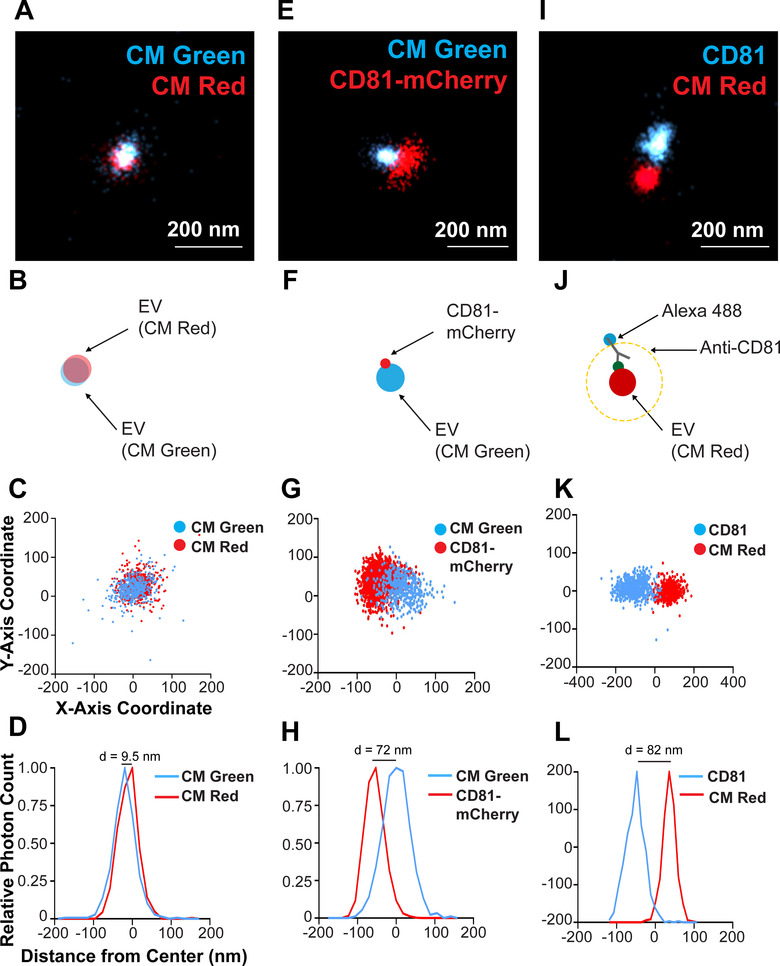
Two‐colour staining of a single CD81+ EV and visualization of CD81 localized to the membrane. (A) CD81+ EVs were dual stained with CM Green and CM Red and imaged using dSTORM. (B) Scheme of the experimental setup showing the overlap of membrane dyes. (C) X and Y‐axis scatter plot of photoswitching events of a small EV shown in B. (D) Photoswitching event distribution of the EV stained with CM Green and CM Red from the modal centre of the EV in the CM Red channel. The distance of modal centres between the channels is shown. (E) CD81‐mCherry EVs were stained with CM Green and imaged using dSTORM. (F) Scheme of the experimental setup showing the offset of the membrane dye with the tetraspanin. (G) X and Y‐axis scatter plot of photoswitching events of EV in E. (H) Photoswitching event distribution of the EV in F from the modal centre of the CM Green channel. (I) CD81+ EVs from WT cells were stained with CM Red and the endogenous CD81 was stained indirectly using Alex‐fluor antibody conjugates. (J) Scheme of the experimental setup showing the offset of the membrane dye with the antibody spacer directed against the tetraspanin. (K) X and Y‐axis scatter plot of photoswitching events of EV in I. (L) Photoswitching event distribution of the EV in I from the modal centre between signals
The CD81‐mCherry and CM Green did not colocalize on individual EVs. The tetraspanin was consistently offset from the modal centre of the EV as determined by CM Green (Figure 4E–H). The modal distribution peaks were separated by d = 72 nm (n > 400 photoswitching events post super‐resolution filtering per channel), that is, almost 10‐fold as large as the error attributable to sub‐optimal apochromatic correction. The offset direction between the membrane stain and the protein stain differed among different EVs. Sometimes the CD81‐mCherry was on the left and sometimes on the right of the EV centre. EVs harbouring two CD9 signals could also be observed (Figure S4). As tetraspanins are multimers (Min et al., 2006), each localization most likely represents multiple molecules. It demonstrates that CD81 molecules were not uniformly dispersed on the EV surface; rather CD81 appeared concentrated as a patch on one location of the EV as diagrammed in Figure 4F.
To validate this hypothesis, dSTORM was repeated on wild‐type EVs and the endogenous CD81 was detected using Alexa Fluor‐488‐conjugated anti‐CD81 antibody. Here, the anti‐CD81 antibody served as a spacer, with the fluorophore located further away from the centre of the EV. Consequently, the EV and antibody signals were cleanly separated with no overlap (Figure 4I, J). Quantitation is provided in Figure 4K, L. If CD81 was freely diffusible within the exposure time, one would expect a circular halo around the EV as indicated by the yellow line in diagram Figure 4J. This was not the case. These observations are consistent with the hypothesis of localized tetraspanin agglomeration on EV membranes.
To independently confirm this observation, we used a different tetraspanin, CD9 (Figure S5). Again, the anti‐CD9 antibody served as a spacer, with the high‐efficiency Alexa Fluor located firmly on the outside of the EV. As before, the signals from the membrane dye and the tetraspanin could be separated. These observations are inconsistent with tetraspanins being freely diffusible across the surface of the EV under physiological conditions at 37°C (∼250 s dSTORM signal acquisition for all frames). These experiments, thus, provide direct proof for the sub‐vesicular localization of multimeric protein complexes on an individual EV. In a manner of speaking these observations extend the fluid mosaic model from the cell membrane to EVs.
3. 3‐D DSTORM OF A SINGLE EV
A limitation of the initial experiments was that the signals represented projections of the three‐dimensional EV sphere onto the two‐dimensional focal plane of the microscope. To observe an individual EV in 3‐D, photoswitching events were collected at multiple axial (Z) depths (Figure 5A–C). Previously 3‐D dSTORM had been applied to large, fixed intracellular structures such as microtubules, clathrin‐coated pits, or viral episome tethering complexes (Grant et al., 2018; Huang et al., 2008), but never to individual particles. As in conventional microscopy, resolution along the Z‐axis was less than along the XY axes (Figure 5D–F). This 3‐D astigmatism was easily recognized by 2D scatter plots. The 2D projections showed a perfect circle, with a diameter of < 100 nm on the X/Y‐plane, but an ellipsis with the same short diameter in X and Y, but a long diameter of ∼170 nm along the Z‐axis (Figure 5G–I). The 3‐D dSTORM rendition of CD81+ EVs is shown in Video S4, where EVs look like elliptical rugby rather than spherical soccer balls. Nevertheless, 3‐D dSTORM affirmed the presence of distinct domains on individual EVs. The first series of experiments again used the hydrophobic membrane intercalating dyes but this time at saturating conditions. The axial (Z) distortion was eliminated computationally. The algorithm utilized the observation that at or near (dZ) the equator the short radius of an ellipsoid is the same as the inscribed sphere (Figure 6A). Having thus calculated the diameter, all points were projected onto a sphere with radius r = d / 2. Figure 6B shows the original data and Figure 6C the transformed data. Note, that this transformation preserves relative angles between the data and thus can be used to identify clusters of molecules on the EV surface. Sections along the Z‐axis showed a hollow sphere in 3‐D or a donut in 2 D. Figure 6D–F show 12 nm thick slices at different Z heights around the equator (Z = 0). A hollow sphere is consistent with the depth of the lipid bilayer, estimated at 6 nm plus headgroups or 1/10 of the diameter of a typical EV. The optical signal was unevenly distributed, with regions of higher density staining, reminiscent of nanodomains of denser lipids within an individual EV. The projected Anderson‐Darling uniformity test rejected a uniform distribution of signal (p≤ 0.05). A similarly skewed localization was evident for CD81‐mCherry sitting on the surface of an EV in 3‐D (Figure S6). These measurements establish proof‐of‐principle that single EV reconstruction can be accomplished using super‐resolution microscopy in three dimensions and multiple colour channels and that this direct visualization can be used to ascertain the existence of domains and protein complexes on the surface of EVs.
FIGURE 5.
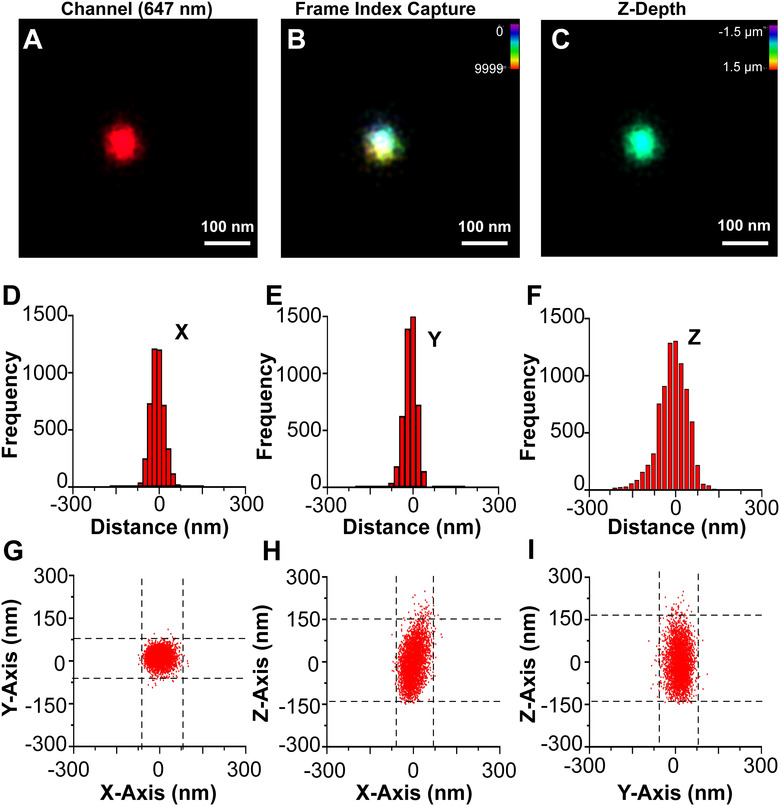
3‐D dSTORM of a single CD81+ EV. (A) A single CD81+ EV from WT cells stained with CM Red was visualized by dSTORM with Z‐axis astigmatism activated. (B) Frame index capture of the CD81+ EV shown in A. (C) Z‐depth information of the CD81+ EV in A. (D) X‐axis size histogram of EV with events plotted from the modal centre. (E) Y‐axis size histogram of EV with events plotted from the modal centre. (F) Z‐axis size histogram of EV with events plotted from the modal centre. (G) X and Y‐axis scatter plot of photoswitching events of an EV. (H) Same as G, but for the X‐Z axis. (I) Same as G, but for the Y‐Z axis
FIGURE 6.
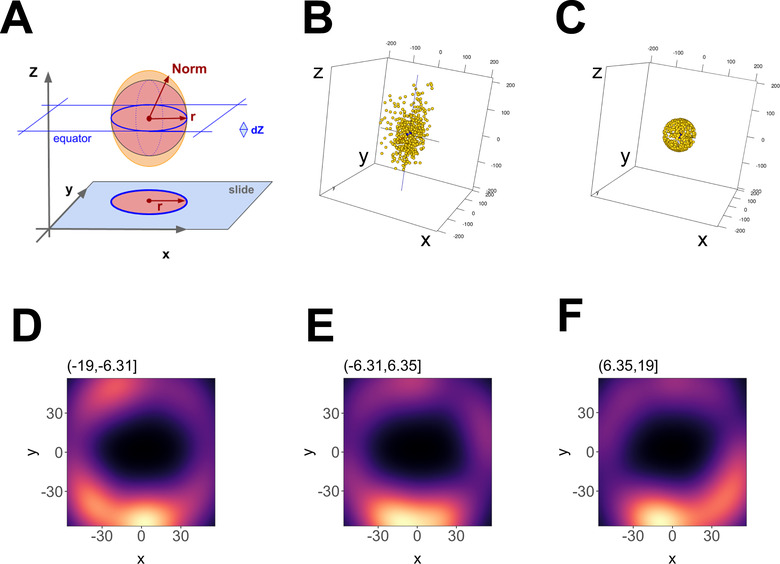
3‐D reconstruction of a single EV. (A) Outline of the geometric foundation, with “slide” indicating the focal plane at z = ‐max. The filled red circle and red radius depict how a 2D image would look like. Above, the red sphere indicates the ideal EV, and the orange ellipsoid indicates the actual data. The red arrow r = sqrt (x2 + y2) depicts the radius at the equator, whereas “Norm” indicates the point vector from the centre to any point on the ellipsoid surface. Norm = sqrt (x2 + y2 +z ), which at z = 0 equals r. (B) 3‐D representation of the data before transformation and (C) after transformation. (D) Principal component analysis (PCA) plot of an EV using 12 nm Z‐axis binning of photoswitching events, showing hollowed core. (E) Same as D, but for a bin at one Z‐axis increment shift. (F) Same as E, but for a bin at one Z‐axis increment shift
3.1. Clustering of tetraspanins on the surface of EVs
To provide further evidence for localized domains, two different tetraspanins on the surface of individual EVs were imaged in 3‐D. Total EVs, representing the physiological conditions, rather than affinity‐purified EVs were seeded onto glass slides and subjected to dSTORM using parameters described in Table S3 . At low resolution, clustering of three markers, CM Red, CD81‐mCherry, and CD9 (stained with Alexa‐488) within ≤200 nm2 space was apparent and used to select individual EVs. As before, the tetraspanins localized to CM Red but were never evenly distributed (Figure 7A). EVs were identified by the presence of CM Red using the thresholding in Table S2 . All CD81 and CD9 events occurring without a corresponding CM Red event were trimmed out from further analysis as they likely represented channel‐specific noise or non‐EV associated protein. By taking high‐confidence localizations (> 100 photoswitching events within a 50 nm radius filtration), we were able to perform multi‐channel clustering. EVs that were positive for CM Red and CD81‐mCherry were identified as double‐positive (Figure 7B). CD9 also colocalized with CM Red but was present on fewer EVs (Figure 7C). Finally, the EV signals for the two tetraspanins were plotted against each other for only those signals that were also positive for CM Red (Figure 7D). This identified CD81 only EVs, CD9 only EVs, and EVs carrying both CD81 and CD9. CD9 only EVs were present since this experiment used total EVs rather than affinity‐purified EVs. CD81‐mCherry, which was ectopically overexpressed, had higher levels of localization with total EVs (25.26% ± 2.58) than did CD9 (17.23% ± 3.94) (Figure 7E). The number of dual‐tetraspanin positive EVs (CD81 + CD9) was 8.2% ± 2.9 of the total EV pool, which is consistent with the established notion that total EVs are heterogeneous with regard to marker gene expression (Kowal et al., 2016).
FIGURE 7.
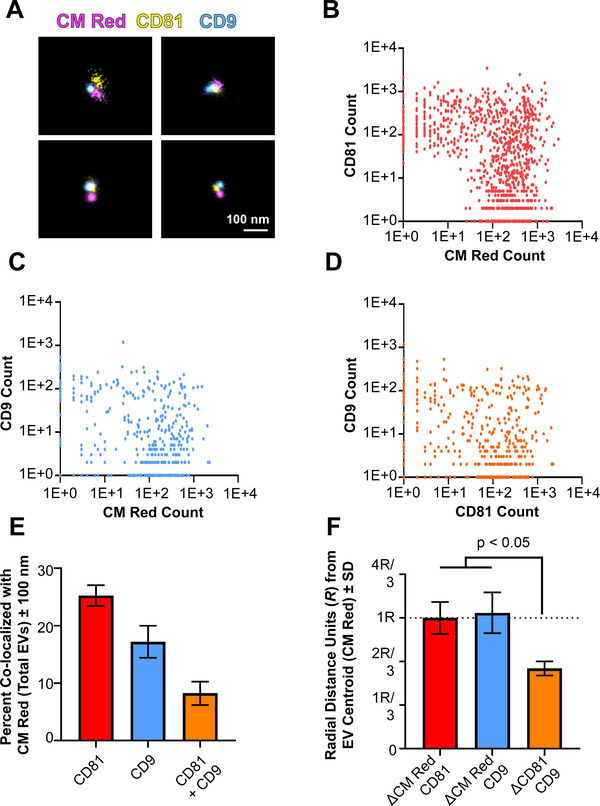
Tetraspanins cluster on the surface of single EVs. (A) Three‐colour dSTORM was performed on total EVs using emissions from CM Red, CD81‐mCherry, and anti‐CD9 Alexafluor‐488. Four representative images are shown: scale = 100 nm. (B) Clustering of photoswitching events of total EVs that were positive for CM Red and CD81‐mCherry. X‐ and Y‐axis show events collected post‐super‐resolution filtration in log scale. The max radius between modal centroids allowed was 150 nm (C) Same as B, but for co‐localizing events for CM Red and CD9. (D) Same as B, but for co‐localizing events for CD81‐mCherry and CD9. (E) The total number of EVs per exposure was quantified using the non‐specific membrane‐intercalating dye CM Red and co‐localizing frequencies were determined for CD81, CD9, or both. (F) The distance between modal centroids was determined between CM Red and CD81, and set to 1 radial measurement, R (dotted line). This was then compared to the distance between the centre of an EV and CD9, as well as between CD81 and CD9. For all experiments, three independent exposures were taken with > 900 individual EVs identified through CM Red clusters
In triple‐positive EVs, the two tetraspanins were frequently co‐localized near each other on the surface of an EV, like tetraspanin enriched microdomains on cellular surfaces (Perez‐Hernandez et al., 2013; Zuidscherwoude et al., 2015). As above, a distance‐based co‐localization analysis was performed using CM Red as control. The distance between the modal centroid of CM Red (EV) and CD81‐mCherry is set to R = 1, representing the normalized EV radius as above. We then compared distances between the centre of an EV and CD9 or CD81, as well as the distance between the tetraspanins CD81 and CD9 (Figure 7F). The CD81 and CD9 signals were significantly closer to each other than they were to the centre of the EV with R = 0.61 ± 0.051 (mean ± sd). If CD81 and CD9 were randomly and independently distributed on the surface of an EV, they would be on opposite poles some of the time with a distance of 2*R; they would be on opposite halves of the equator half the time resulting in a distance of ≥1*R. Hence, a mean of ≤ 1*R supports a model wherein, on average, the two tetraspanins CD81 and CD9 are clustered near to each other on one side of an EV.
The sample size (n≥900) was limited by experimental constraints. We are confident to image 900 EVs per slide without excess laser bleaching. Based on this, population averages were calculated. The distances between modal centroids between tetraspanins were significantly lower than the distance from the EV centroid to individual tetraspanins, which is the criterium used to establish clustering; however, the distances between the different tetraspanin signals varied, as these data follow a Poisson distribution, where the standard deviation equals the mean, rather than being a fraction of it as for normal distributed data. Improving accuracy by increasing sample size and more refined statistical analysis is the subject of ongoing studies. On the whole, though, the distances between modal centroids of tetraspanins were significantly lower than the distance from the EV centroid to individual tetraspanins.
3.2. Cryo‐EM of EVs confirms presence of EV microdomains
Cryogenic electron microscopy (Cryo‐EM) allows for angstrom‐level detail of molecular structures, particularly for larger biological entities such as viruses (Sevvana et al., 2018; Sirohi et al., 2016). We therefore asked if we could confirm the existence of distinct, protein‐rich domains on EVs through this method. EVs were purified under physiological conditions to prevent any artificial aggregation to one region of the EV prior to flash‐freezing. EVs were of the expected size, with a mean diameter of 60.3 ± 10.5 nm (mean ± sd, 95% CI = 51.75 – 68.93, n = 9 independent images from each of 3 biological replicates) (Figure 8E). Their membrane composition was too heterogenous for image reconstruction by particle averaging. Nevertheless, protein‐rich (rough) and lipid‐rich (smooth) regions on the surface of individual EVs could be readily identified, analogous, perhaps, to the rough and smooth ER. At times, multiple domains could be seen on a single EV depending on EV orientation and section (Figure 8A–D). The protein and lipid‐dense regions are shown with a yellow arrow, whereas regions without proteins or lipid‐dense patches are shown with white arrows. The lipid‐rich regions had a smooth appearance, while the protein microdomains showed extrusions consistent with membrane proteins. This result is consistent with 3‐D dSTORM and demonstrates directly the existence of different domains on the EV surface.
FIGURE 8.
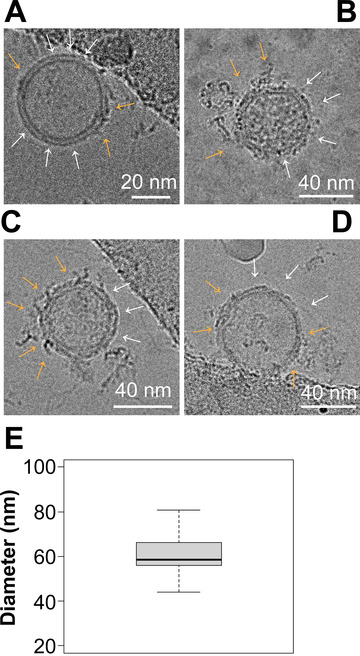
Cryo‐EM of EVs shows protein‐rich clusters on the surface. (A–D) Representative images of EVs viewed under Cryo‐EM. Surface nanodomains are shown with an orange arrow, highlighting the protein and/or lipid‐dense regions of an individual EV. Empty regions are shown with white arrows. (E) Size analysis of EVs viewed through Cryo‐EM. EVs were measured in diameter in nanometers based on their lipid bilayer and do not include protruding proteins from the surface
4. DISCUSSION
EVs are crucial mediators of cellular physiology, viral pathogenesis, and cancer pathologies (Mcnamara & Dittmer, 2020; Pegtel & Gould, 2019; Wortzel et al., 2019). This class of biological response mediators had been understudied because of the difficulty in visualizing EVs. Like viruses, EVs fall beneath the limit of diffraction for visible light and thus had been studied indirectly or as collectives of underdefined composition (Hyenne et al., 2019).
There is no single gold‐standard technique that can fulfil the characterization needs of the field for EV analysis. One method historically used to visualize individual particles is EM. Transmission and scanning EM, however, include dehydration steps, which make it difficult to confidently characterize the native state of EVs. Detecting specific cargo associated with EVs through Cryo‐EM requires considerable technical expertise. Cryo‐EM has outstanding resolution with regard to EV visualization as shown in Figure 8, but it is time‐, resource‐, and labour‐intensive (Arraud et al., 2014).
Super‐resolution microscopy allows for the rapid and direct visualization of individual EVs and their associated surface protein markers in a dilute, aqueous solution where EVs retain their native structure. With proper image processing, it can be used to inspect thousands of particles per hour. Prior studies applied dSTORM to EVs (Chen et al., 2016; Lennon et al., 2019; Maire et al., 2021; Nizamudeen et al., 2018). These relied on extraneous chemical labelling and were limited to imaging in two‐dimensions only, or only with two colours. Our approach differed in that we employed dSTORM using three colours in 3‐D, allowing for the identification of surface‐marker subpopulations within heterogenous EV mixtures, as well as detection of membrane microdomains on individual EVs.
We performed multi‐colour, 3‐D dSTORM on EVs by first marking EVs with the dyes CM Red or CM Green, which preferably partition into membranes. We chose this as the first step in image trimming as non‐EV‐associated tetraspanins may exist and may aggregate in solution given their hydrophobic properties. While membrane‐staining added an extra step in the labelling process, we consider membrane‐staining as imperative for downstream identification of EV surface marker subpopulations.
Due to the low quantum efficiency of GFP/mCherry as opposed to quantum dots, we can only make a statement about the composition of EVs that have at least one (GFP or mCherry) positive signal. It remains a possibility that EVs that do not stain positive, nevertheless have tetraspanins, but at a level that is below the detection limit of this system.
Image quality was not correlated with EV size up to the resolution limit of dSTORM. Particles as small as 20 nm can be imaged by dSTORM using a membrane dye or fluorescent tag with sufficient quantum yield. On the one hand, even a 20 nm particle incorporates enough membrane‐dye to allow detection under saturating dye concentrations. On the other hand, GFP and mCherry are not as sensitive as directly labelled high‐quantum yield fluorophores, for which fluorophores others achieved single protein subunit detection and resolution (Grant et al., 2018).
There are limitations to this study. For instance, apochromatic compensation will need to be improved and better algorithms for axial compensation are needed. This work is ongoing.
While this study demonstrates that GFP and mCherry are dSTORM‐compatible, the same concerns apply to dSTORM as to other imaging studies: tagged, exogenously expressed proteins may behave differently than unmodified proteins expressed at native levels. For tagged‐CD9 and CD81 localization by dSTORM exactly mirror prior work with untagged proteins. In contrast, for tagged‐CD63, which is normally localized exclusively to the late endosome and not on the plasma membrane, we were able to detect some plasma membrane localization by dSTORM. Confirmation of tetraspanin clustering in additional cell lines and primary cells is warranted. Antibody‐based detection is limited by the specificity profile of the particular reagents used, for instance monoclonal antibodies only recognize a single epitope and it is assumed that this epitope is exposed on all molecules in the experiment. This may not be true for heavily glycosylated proteins such as tetraspanins. In addition, using multiple antibodies on small structures such as EVs may yield steric hindrance, whereby one antibody occludes binding of a second antibody to a nearby molecule. Therefore, the approach presented here used an orthogonal experimental design, that is, a combination of direct protein labelling and antibody detection.
A multitude of purification strategies for EVs exist, each with their intrinsic pros and cons. In our study, we built upon our previously validated approach of tangential flow filtration followed by PEG precipitation, CaptoCore filtration, and affinity selection (Mcnamara et al., 2018). We elected to use this approach as it minimizes structural damage to the membrane of EVs during purification, maximizes yield, and selects for specifically marked EVs – in our case those with CD81. PEG can cause aggregation of EVs if used as the sole purification step. We do not think that EV aggregation due to PEG carried through to imaging, as we used additional purification steps (column and beads) afterwards and obtained a pure population as ascertained by NTA and EM. This has also been the experience of others in the field that use PEG as the first, but never as the only step to isolate EVs (Ludwig et al., 2018; Rider et al., 2016).
Lastly, the present study does not attempt to address components inside of an EV by dSTORM. Intra‐EV visualization is extremely technically challenging due to the intrinsically small and heterogeneous nature of EVs. Detergents can create pore sizes larger than an EV itself (Zhao & Keswani, 2016) and may induce fusion. We exclusively imaged non‐detergent treated EVs and, therefore, this study is limited to characterizing EV composition at the surface level. Of course EV composition is not limited to proteins; other groups have been able to recently link nucleic acid modifications to specific cancers including glioblastoma (Maire et al., 2021), which was used as a classification tool in addition to EV subpopulation clustering from surface markers. Improvements upon, and/or the development of, more techniques that can classify EVs as the single vesicle level is of high interest in the biotechnology and liquid biopsy fields.
We hope that the experimental principles derived here will be useful for the study of EVs, viruses, and nanoparticles in the 30–200 nm size range. Direct visualization of EVs in multiple fluorescent channels and three dimensions represents a fundamental step forward towards our understanding of their biophysical properties, composition, and function.
As an appropriate first application for 3‐D dSTORM on EVs, this study set out to uncover and document the structural heterogeneity of EVs and study EV membrane microdomains. Structural surface organization and compositional complexity represent the key distinction between EVs and artificial liposomes, which comprise only a limited number of different lipids, and no proteins. By dSTORM, tetraspanin clusters on the surface of single and differently sized EVs could be discerned. This was verified by Cryo‐EM. The two orthogonal approaches demonstrate that EVs including exosomes contain distinct domains on their surfaces. A similar phenomenon has been previously described on cellular membranes and has been termed TEMs (De Gassart et al., 2003; Hurwitz et al., 2017; Perez‐Hernandez et al., 2013; Verweij et al., 2011; Yáñez‐Mó et al., 2009; Yáñez‐Mó et al., 2015). Future studies will utilize chemical inhibitors or shRNA/CRISPR‐based knockdown of individual tetraspanins and other possible components to understand these TEMs in detail. In sum, our study shows that EVs contain previously unrecognized surface structures and spatial organization. Further investigation into EV surface assembly could yield novel insights into their intracellular assembly, specific packing, and tissue‐specific destinations.
5. METHODS AND MATERIALS
5.1. Cell lines
Human osteosarcoma (U‐2 OS) cells were obtained from the ATCC (HTB‐96) and grown in Dulbecco's Modified Eagle Medium (DMEM) (Thermo Fisher 21013024) supplemented with 10% exosome‐free Foetal Bovine Serum (FBS) (VWR 97068–085) and 100 units/ml of Penicillin, 100 μg/ml of Streptomycin solution (Gibco 15140‐122). Cells were maintained at 37°C and 5% CO2. To develop clonal cell lines, 106 U‐2 OS cells (ATCC HTB‐96) were plated onto a 10‐cm tissue culture plate (Fisher 430167) and transfected with 1 μg of plasmid with 5 μl of Lipofectamine‐2000 (ThermoFisher 11668019) diluted in DMEM. Cells were selected using 500 μg/ml of Geneticin (Thermo Fisher 10131035). Single‐cell fluorescence‐assisted cell sorting (FACS) was done using the FACS Aria II, maintained and operated by the UNC Flow Cytometry Core. Cells were grown in selective media and verified for continual fluorescence activity using an BD Accuri 6 Plus flow cytometer (BD Biosciences) equipped with a 488 and 640 laser, along with the emission filters FITC (488–519 nm), PE (546–578 nm), PerCP (640–678 nm), and APC (650–670 nm). mCherry‐CD81‐10 was a gift from Michael Davidson (Addgene plasmid # 55012). CD63‐pEGFP C2 was a gift from Paul Luzio (Addgene plasmid # 62964).
5.2. EV isolation and purification
U‐2 OS cells were grown to confluency, and supernatant containing EVs was processed as previously described (Mcnamara et al., 2018). In short, 0.5–1.0 L of cell culture supernatant was passaged through a 0.45 and 0.22 μm vacuum filtration apparatus (Genesee 25–230 and 25–227, respectively). Clarified supernatant was concentrated and equilibrated with 1X phosphate‐buffered saline (PBS) via tangential‐flow filtration using the AKTA Flux S (GE Healthcare 29038437) equipped with a 750 kDa cut‐off filter (GE Healthcare 29‐0142‐95). The concentrated/equilibrated solution was further concentrated by precipitating with 40 mg/ml of PEG‐8000 overnight and centrifugation at 1200 * g at 4°C for 1 h. EV pellets were resuspended in 0.5 ml of 1X PBS, DNAse and RNAse‐treated, and incubated with 50 μg/ml of CellMask Red and/or CellMask Green (CM Red or CM Green, respectively, Thermo Fisher C10046 and C37608) for full EV saturation as imaged by dSTORM (see below), and RNase A (50 μg/ml, Thermo Scientific, EN0531) at 4°C for 1 h and fractionated on the AKTA Start equipped with a HiTrap Capto Core 700 column (GE Healthcare 17548151). Affinity selection for CD81+ EVs was done using anti‐CD81 beads (Thermo Fisher 10616D) overnight. Beads were washed three times with 1X PBS and CD81+ EVs were eluted using 0.2 M Glycine pH = 2.0 at 37 °C for 30 min. The EV solution was then transferred to another tube containing an equal volume of 100 mM Tris‐HCl pH = 7.5 in 1X PBS.
5.3. EV quantitation and biophysical characterizations
Size and particle concentrations were determined using the ZetaView (Particle Metrix PMX‐120BASIC). EVs were diluted in nanopure water until approximately 50–200 particles were in a field of view. Size distribution profiles and concentrations were taken using eleven technical replicates per sample. A total of ≥ three complete biological replicates from independent cultures at different dates were done to ensure reproducibility.
5.4. Animal care
All experiments using rhesus macaques were approved by the Tulane Institutional Animal Care and Use Committee. The Tulane National Primate Research Centre (TNPRC) is an Association for Assessment and Accreditation of Laboratory Animal Care International‐accredited facility (AAALAC #000594). The NIH Office of Laboratory Animal Welfare assurance number for the TNPRC is A3071‐01. All clinical procedures, including administration of anaesthesia and analgesics, were carried out under the direction of a laboratory animal veterinarian. All possible measures are taken to minimize the discomfort of all the animals used in this study. Tulane University complies with NIH policy on animal welfare, the Animal Welfare Act, and all other applicable federal, state, and local laws.
5.5. Fluorescence microscopy
Cells were seeded onto glass coverslips previously sterilized with 1% HCl diluted in 70% ethanol inside of a 6‐well plate (Fisher 07‐200‐83). Cells were imaged as previously described (Mcnamara et al., 2018) using the DM55008 microscope (Leica) equipped with the Leica HCX PL Apo 63x Oil Objective and the Leica HCX PL Apo 100x Oil Objective lenses (both with numerical aperture = 1.40). Coverslips were mounted onto Frosted Micro Slides (Corning 2948–75 × 25) using 30 μl of ProLong Gold Antifade Reagent (Cell Signalling 9071S). Z‐stacks were captured and deconvoluted using MetaMorph V 7.8.12.0 (Molecular Devices), and multi‐plane images were visualized using Imaris V 9.2.0 (Bitplane).
5.6. Super‐resolution microscopy
U‐2 OS cell lines were grown in Glass Bottom 15 μ‐Slide 8 well plates (Ibidi 80827). Cells were fixed and permeabilized as above. Cells were blocked with 5% bovine serum albumin (BSA) (Thermo Fisher BP9706‐100) diluted in 1X PBS at 4°C for 1 h and incubated with Phalloidin‐488 (Thermo Fisher A12379) at a dilution of 1:100 for 1 h at room temperature. Cells were overlaid with 200 μl of B‐cubed buffer (Oxford Nanoimaging) for 30 min at room temperature before visualization.
The Nanoimager (Oxford Nanoimaging) was calibrated for dSTORM using 100 nm Tetraspek microspheres (Invitrogen T7279) diluted in water. Calibration beads were viewed under the Nanoimager using a 405/473/561/640 nm laser configuration with a 100X oil‐objective lens. X, Y, and Z axes errors were obtained after the 3‐D mapping calibration was completed. Parameters for super‐resolution are summarized in Tables S1 and S 2 . The software analysis program used was the Nanoimager Software v 1.4.8740 (Oxford Nanoimaging). Channels were pseudocoloured (Channel 0 = light blue #55aaff; Channel 1 = bright red #ff0011) for RG‐colourblind individuals. Videos of the 3‐D CD81+ EV were created by drawing a 3‐D visualization box around a region of interest. The video was recorded using ONI software and then converted into an MPEG4 or WMV file using HandBrake v 1.3.3.
5.7. EV tracing using super‐resolution images
For image localization points, .csv files were obtained by using the Nanoimager Software XYZ Plane View Tool (X by Y; X by Z; Y by Z). For size‐distribution analysis, individual EVs identified through dSTORM were identified and the Line Histogram Tool (ONI) was used to bisect the EV. Events were binned into 31 nm fragments away from the modal centre, allowing for size distribution analysis to be conducted. A total of 50 manual traces were done, each across three separate biological experiments, yielding n = 150. Individual particle tracking was done on 3‐D images using the Tracking Tool. Z‐position was used, and point tracking was done to give an estimated concentration and mean diameter of 3‐D objects.
5.8. EV lysis and immunoblotting
EVs were lysed in EV‐lysis buffer (1% NP‐40, 5% Glycerol, 0.5% sodium dodecyl sulphate (SDS), 0.5% sodium deoxycholate, 150 mM NaCl, and 1 mM PMSF) in two separate tubes. To one tube, dithiothreitol (DTT) was added to a final concentration of 1 mM DTT. Cellular and EV fractions were run on a 4–12% NuPAGE Bis‐Tris 15‐well gel (ThermoFisher NP0336BOX) using 1X Bolt MES buffer (ThermoFisher B0002) at constant voltage for 60 min and transferred to nitrocellulose membranes (Bio‐Rad 1620115) at 250 mA for 90 min. Primary antibodies at dilutions listed in Table S4 in 8% milk in Tris‐buffered saline + tween‐20 (TBS‐T) were incubated with the membrane for > 1 h. Membranes were vigorously washed 3X with tris‐buffered saline + tween‐20 (TBS‐T) for 10 min per wash and then secondary antibodies at dilutions shown in Table S4 in 8% milk in TBS‐T were incubated with the membrane for > 1 h. Images were taken on the Li‐Cor Odyssey and analysed using Image Studio V. 5.2. Tetraspanins were detected using non‐reducing conditions, whereas other proteins were detected under reducing conditions.
5.9. Transmission Electron Microscopy (EM) preparation
EV samples were adsorbed to glow‐discharged carbon‐coated 400‐mesh copper grids for 3 min and then stained with 2% (weight/volume) uranyl acetate in water, rinsed briefly with water, and air‐dried. The grids were visualized in a FEI Tecnai 12 transmission EM at 80 kV. Images were captured on a Gatan Orius CCD camera with Gatan Digital Micrograph software.
5.10. Cryogenic electron microscopy (Cryo‐EM)
The Cryo‐EM core operated by UNC houses a 200 KV Thermo Fisher Scientific Talos Arctica G3 TEM equipped with a Gatan K3 direct electron detector. EVs purified in the absence of any acidic elution steps were added onto a Quantifoil grid (R 1.2/1.3, 400 Mesh, Copper) from EMS (ID: Q425CR1.3) and snap‐frozen in ethane and propane mixture prechilled by liquid nitrogen to ‐165°C at a concentration of 1010 particles/ml. Individual Cryo‐EM snapshots were obtained in .mrc format and exported to high‐resolution .tiff images using IMOD 4.11 (University of Colorado).
AUTHOR CONTRIBUTIONS
Yijun Zhou: Data curation; Methodology; Validation. Smaranda Willcox: Data curation; Formal analysis; Methodology; Visualization. Blake Schouest: Data curation; Formal analysis; Investigation; Visualization; Conceptualization, Ryan P. McNamara and Dirk P. Dittmer; Methodology, Ryan P. McNamara, Anthony B. Eason, Yijun Zhou, Tiffany A. Peterson, Justin T. Landis, Blake Schouest, Nicholas J. Maness, Andrew G. MacLean, Smaranda Willcox, and Dirk P. Dittmer; Software, Justin T. Landis, B.Y., Dirk P. Dittmer; Validation Ryan P. McNamara, and Dirk P. Dittmer; Formal Analysis, Ryan P. McNamara, Justin T. Landis, and Dirk P. Dittmer; Investigation Ryan P. McNamara, Anthony B. Eason, Yijun Zhou, Tiffany A. Peterson, Justin T. Landis, B.Y., Blake Schouest, Nicholas J. Maness, Andrew G. MacLean, and Dirk P. Dittmer; Resources, Nicholas J. Maness, Andrew G. MacLean, and Dirk P. Dittmer; Data Curation Ryan P. McNamara, Justin T. Landis, and Dirk P. Dittmer, Writing, Ryan P. McNamara, Anthony B. Eason, and Dirk P. Dittmer; Visualization, Ryan P. McNamara, Anthony B. Eason, Justin T. Landis, B.Y., Smaranda Willcox, Jack D. Griffith, and Dirk P. Dittmer; Supervision, Dirk P. Dittmer; Project Administration Dirk P. Dittmer; Funding Acquisition, Ryan P. McNamara, Nicholas J. Maness, Andrew G. MacLean, Jack D. Griffith, and Dirk P. Dittmer.
CONFLICTS OF INTEREST
Ryan P. McNamara and Dirk P. Dittmer received material support from Oxford Nanoimaging (ONI) Inc. and Cytiva Inc. (formerly GE Healthcare). Ryan P. McNamara and Dirk P. Dittmer declare competing interests for possible commercialization of some of the information presented. These are managed by the University of North Carolina. The other authors declare no relevant conflicts of interest. Funding sources were not involved in the interpretations or writing of this manuscript.
Supporting information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information ‐ Table S1
Supplementary Information ‐ Table S2
Supplementary Information ‐ Table S3
Supplementary Information ‐ Table S4
ACKNOWLEDGEMENTS
We would like to thank Oxford Nanoimaging for constructive feedback and guidance. We thank Ruotian Wu and Steve Marron for statistical discussions. This work was funded by 5UM1CA121947‐10 to Ryan P. McNamara, 1R01DA040394 to Dirk P. Dittmer, R21‐MH113517 to Andrew G. MacLean and Dirk P. Dittmer, the 5T32OD011124‐13 to Tiffany A. Peterson, and 2P01CA01901438 to Jack D. Griffith, and ESO‐31635 to Jack D. Griffith. Research reported in this publication was supported in part by the North Carolina Biotech Institutional Support Grant 2012‐IDG‐1006. The Tulane National Primate Research Centre is supported by the base grants P51‐OD11104, U42‐OD024282, U42‐OD010568. The UNC Flow Cytometry Core is supported by the NCI Centre Core Support Grant CA16086. The UNC Cryo‐EM Core is supported by the National Cancer Institute of the National Institutes of Health under award number P30CA016086. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
McNamara, R. P. , Zhou, Y. , Eason, A. B. , Landis, J. T. , Chambers, M. G. , Willcox, S. , Peterson, T. A. , Schouest, B. , Maness, N. J. , MacLean, A. G. , Costantini, L. M. , Griffith, J. D. , & Dittmer, D. P. (2022). Imaging of Surface Microdomains on Individual Extracellular Vesicles in 3‐D. Journal of Extracellular Vesicles, 11, e12191. 10.1002/jev2.12191
REFERENCES
- Cocozza, F. , Grisard, E. , Martin‐Jaular, L. , Mathieu, M. , & Théry, C. (2020).SnapShot: Extracellular vesicles. Cell 182, 262–262.e1 e1 [DOI] [PubMed] [Google Scholar]
- Théry, C. , Zitvogel, L. , & Amigorena, S. (2002).Exosomes: Composition, biogenesis and function. Nature Reviews Immunology 2, 569–579 [DOI] [PubMed] [Google Scholar]
- Pegtel, D. M. , & Gould, S. J. (2019).Exosomes Annual Review of Biochemistry 88, 487–514 [DOI] [PubMed] [Google Scholar]
- Pols, M. S. , & Klumperman, J. (2009).Trafficking and function of the tetraspanin CD63. Experimental Cell Research 315, 1584–1592 [DOI] [PubMed] [Google Scholar]
- Verweij, F. J. , Revenu, C. , Arras, G. , Dingli, F. , Loew, D. , Pegtel, D. M. , Follain, G. , Allio, G. , Goetz, J. G. , Zimmermann, P. , Herbomel, P. , Del Bene, F. , Raposo, G. , & Van Niel, G. (2019).Live tracking of inter‐organ communication by endogenous exosomes in vivo. Developmental Cell 48, 573–589.e4 e4 [DOI] [PubMed] [Google Scholar]
- Kowal, J. , Arras, G. , Colombo, M. , Jouve, M. , Morath, J. P. , Primdal‐Bengtson, B. , Dingli, F. , Loew, D. , Tkach, M. , & Théry, C. (2016).Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. PNAS 113, E968–E977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Jaular, L. , Nevo, N. , Schessner, J. P. , Tkach, M. , Jouve, M. , Dingli, F. , Loew, D. , Witwer, K. W. , Ostrowski, M. , Borner, G. H. H. , & Théry, C. (2021).Unbiased proteomic profiling of host cell extracellular vesicle composition and dynamics upon HIV‐1 infection. Embo Journal 40, e105492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortzel, I. , Dror, S. , Kenific, C. M. , & Lyden, D. (2019).Exosome‐mediated metastasis: Communication from a distance. Developmental Cell 49, 347–360 [DOI] [PubMed] [Google Scholar]
- Raab‐Traub, N. , & Dittmer, D. P. (2017).Viral effects on the content and function of extracellular vesicles. Nature Reviews Microbiology 15, 559–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcnamara, R. P. , Chugh, P. E. , Bailey, A. , Costantini, L. M. , Ma, Z. , Bigi, R. , Cheves, A. , Eason, A. B. , Landis, J. T. , Host, K. M. , Xiong, J. , Griffith, J. D. , Damania, B. , & Dittmer, D. P. (2019).Extracellular vesicles from Kaposi Sarcoma‐associated herpesvirus lymphoma induce long‐term endothelial cell reprogramming. Plos Pathogens 15, e1007536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev, O. , Henderson, S. , Hayes, M. J. , Marelli, S. S. , Ofir‐Birin, Y. , Regev‐Rudzki, N. , Herrero, J. , Enver, T. Herpesviruses shape tumour microenvironment through exosomal transfer of viral microRNAs. Plos Pathogens 13, e1006524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh, P. E. , Sin, S. H. , Ozgur, S. , Henry, D. H. , Menezes, P. , Griffith, J. , Eron, J. J. , Damania, B. , & Dittmer, D. P. (2013).Systemically circulating viral and tumor‐derived microRNAs in KSHV‐associated malignancies. Plos Pathogens 9, e1003484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu, M. , Martin‐Jaular, L. , Lavieu, G. , Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell‐to‐cell communication. Nature Cell Biology 21, 9–17 (2019). [DOI] [PubMed] [Google Scholar]
- Kamerkar, S. , Lebleu, V. S. , Sugimoto, H. , Yang, S. , Ruivo, C. F. , Melo, S. A. , Lee, J. J. , & Kalluri, R. (2017).Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 546, 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, S A. , Sugimoto, H. , O'connell, J T. , Kato, N. , Villanueva, A. , Vidal, A. , Qiu, Le , Vitkin, E. , Perelman, L T. , Melo, C A. , Lucci, A. , Ivan, C. , Calin, G A. , & Kalluri, R. (2014).Cancer exosomes perform cell‐independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H. , Yang, L. , Baddour, J. , Achreja, A. , Bernard, V. , Moss, T. , Marini, J. C. , Tudawe, T. , Seviour, E. G. , San Lucas, F. A. , Alvarez, H. , Gupta, S. , Maiti, S. N. , Cooper, L. , Peehl, D. , Ram, P. T. , Maitra, A. , Nagrath, D. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 5, e10250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Z. , Hensley, L. , Mcknight, K. L. , Hu, F. , Madden, V. , Ping, L. , Jeong, S. ‐. H. , Walker, C. , Lanford, R. E. , & Lemon, S. M. (2013).A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496, 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi, H. , Ekström, K. , Bossios, A. , Sjöstrand, M. , Lee, J. J. , & Lötvall, J. O. (2007).Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology 9, 654–659 [DOI] [PubMed] [Google Scholar]
- Baglio, S. R. , Van Eijndhoven, M. A. J. , Koppers‐Lalic, D. , Berenguer, J. , Lougheed, S. M. , Gibbs, S. , Léveillé, N. , Rinkel, R. N. P. M. , Hopmans, E. S. , Swaminathan, S. , Verkuijlen, S. A. W. M. , Scheffer, G. L. , Van Kuppeveld, F. J. M. , De Gruijl, T. D. , Bultink, I. E. M. , Jordanova, E. S. , Hackenberg, M. , Piersma, S. R. , Knol, J. C. … Pegtel, D. M. (2016).Sensing of latent EBV infection through exosomal transfer of 5'pppRNA. PNAS 113, E587–E596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamvoki, M. , Du, Te , Roizman, B. (2014). Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. PNAS 111, E4991–E4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, S. J. , Booth, A. M. , & Hildreth, J. E. K. (2003).The Trojan exosome hypothesis. PNAS 100, 10592–10597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarino, C. , Pleet, M. L. , Cowen, M. , Barclay, R. A. , Akpamagbo, Y. , Erickson, J. , Ndembi, N. , Charurat, M. , Jumare, J. , Bwala, S. , Alabi, P. , Hogan, M. , Gupta, A. , Noren Hooten, N. , Evans, M. K. , Lepene, B. , Zhou, W. , Caputi, M. , Romerio, F. … Kashanchi, F. (2018).Antiretroviral drugs alter the content of extracellular vesicles from HIV‐1‐infected cells. Science Reports 8, 7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, M A. , Ludwig, R G. , Garcia‐Martin, R. , Brandão, B B. , Kahn, C. R Extracellular miRNAs: From biomarkers to mediators of physiology and disease. Cell Metabolism 30, 656–673 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij, F. J. , Balaj, L. , Boulanger, C. M. , Carter, D. R. F. , Compeer, E. B. , D'angelo, G. , El Andaloussi, S. , Goetz, J. G. , Gross, J. C. , Hyenne, V. , Krämer‐Albers, E. ‐. M. , Lai, C. P. , Loyer, X. , Marki, A. , Momma, S. , Nolte‐‘T Hoen, E. N. M. , Pegtel, D. M. , Peinado, H. , Raposo, G. … Van Niel, G. (2021).The power of imaging to understand extracellular vesicle biology in vivo. Nature Methods 18, 1013–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovic, R A. , Gardiner, C. , Brooks, A S. , Tannetta, D S. , Ferguson, D J.P. , Hole, P. , Carr, B. , Redman, C W.G. , Harris, A L. , Dobson, P J. , Harrison, P. , Sargent, I L. Sizing and phenotyping of cellular vesicles using nanoparticle tracking analysis. Nanomedicine 7, 780–788 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab, T. , Mallick, E. R. , Huang, Y. , Dong, L. , Liao, Z. , Zhao, Z. , Gololobova, O. , Smith, B. , Haughey, N. J. , Pienta, K. J. , Slusher, B. S. , Tarwater, P. M. , Tosar, J. P. , Zivkovic, A. M. , Vreeland, W. N. , Paulaitis, M. E. , & Witwer, K. W. (2021).Characterization of extracellular vesicles and synthetic nanoparticles with four orthogonal single‐particle analysis platforms. Journal of Extracellular Vesicles 10, e12079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Linde, S. , Löschberger, A. , Klein, T. , Heidbreder, M. , Wolter, S. , Heilemann, M. , Sauer, M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nature Protocols 6, 991–1009 (2011). [DOI] [PubMed] [Google Scholar]
- Huang, Bo , Wang, W. , Bates, M. , & Zhuang, X. (2008).Three‐dimensional super‐resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz, S. N. , Cheerathodi, M. R. , Nkosi, D. , York, S. B. , & Meckes, D. G. (2018).Tetraspanin CD63 bridges autophagic and endosomal processes to regulate exosomal secretion and intracellular signaling of epstein‐barr virus LMP1. Journal of Virology 92(5), 10.1128/JVI.01969-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötvall, J. , Hill, A F. , Hochberg, F. , Buzás, E I. , Di Vizio, D. , Gardiner, C. , Gho, Y. S. , Kurochkin, I V. , Mathivanan, S. , Quesenberry, P. , Sahoo, S. , Tahara, H. , Wauben, M H. , Witwer, K W. , Théry, C. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. Journal of Extracellular Vesicles 3, 26913 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij, F. J. , Bebelman, M. P. , Jimenez, C. R. , Garcia‐Vallejo, J. J. , Janssen, H. , Neefjes, J. , Knol, J. C. , De Goeij‐De Haas, R. , Piersma, S. R. , Baglio, S. R. , Verhage, M. , Middeldorp, J. M. , Zomer, A. , Van Rheenen, J. , Coppolino, M. G. , Hurbain, I. , Raposo, G. , Smit, M. J. , Toonen, R. F.G. … Pegtel, D. M. (2018).Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. Journal of Cell Biology 217, 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York, S. B. et al. Zika virus hijacks extracellular vesicle tetraspanin pathways for cell‐to‐cell transmission. mSphere, e0019221 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu, M. , Névo, N. , Jouve, M. , Valenzuela, J. I. , Maurin, M. , Verweij, F. J. , Palmulli, R. , Lankar, D. , Dingli, F. , Loew, D. , Rubinstein, E. , Boncompain, G. , Perez, F. , & Théry, C. (2021).Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nature Communication 12, 4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcnamara, R. P. , Caro‐Vegas, C. P. , Costantini, L. M. , Landis, J. T. , Griffith, J. D. , Damania, B. A. , & Dittmer, D. P. (2018).Large‐scale, cross‐flow based isolation of highly pure and endocytosis‐competent extracellular vesicles. Journal of Extracellular Vesicles 7, 1541396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquet, C. , Gilles, J. F. , Heck, N. , Dos Santos, M. , Schwartzmann, R. , Cannaya, V. , Morel, M. P. , Davidson, R. S. , Trembleau, A. , Bolte, S. Improving axial resolution in confocal microscopy with new high refractive index mounting media. Plos One 10, e0121096 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental, I. , Levental, K. R. , & Heberle, F. A. (2020).Lipid rafts: Controversies resolved, mysteries remain. Trends in Cell Biology 30, 341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenel, C. , Margeat, E. , Dosset, P. , Arduise, Cé , Le Grimellec, C. , Royer, C. A. , Boucheix, C. , Rubinstein, E. , & Milhiet, P. E. (2008).Single‐molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. Journal of Cell Biology 182, 765–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, G. , Wang, H. , Sun, T. T. , & Kong, X. P. (2006).Structural basis for tetraspanin functions as revealed by the cryo‐EM structure of uroplakin complexes at 6‐A resolution. Journal of Cell Biology 173, 975–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, M. J. , Loftus, M. S. , Stoja, A. P. , Kedes, D. H. , & Smith, M. M. (2018).Superresolution microscopy reveals structural mechanisms driving the nanoarchitecture of a viral chromatin tether. PNAS 115, 4992–4997, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Hernandez, D. , Gutiérrez‐Vázquez, C. , Jorge, I. , López‐Martín, S. , Ursa, A. , Sánchez‐Madrid, F. , Vázquez, J. , Yáñez‐Mó, M. The intracellular interactome of tetraspanin‐enriched microdomains reveals their function as sorting machineries toward exosomes. Journal of Biological Chemistry 288, 11649–11661 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuidscherwoude, M. , Göttfert, F. , Dunlock, V. M. E. , Figdor, C. G. , Van Den Bogaart, G. , & Spriel, A. B. V. (2015).The tetraspanin web revisited by super‐resolution microscopy. Science Reports 5, 12201 https://www.nature.com/articles/srep12201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevvana, M. , Long, F. , Miller, A. S. , Klose, T. , Buda, G. , Sun, L. , Kuhn, R. J. , & Rossmann, M. G. (2018).Refinement and analysis of the mature zika virus Cryo‐EM structure at 3.1 Å resolution. Structure (London, England) 26, 1169–1177.e3.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohi, D. , Chen, Z. , Sun, L. , Klose, T. , Pierson, T. C. , Rossmann, M. G. , & Kuhn, R. J. (2016).The 3.8 Å resolution cryo‐EM structure of Zika virus. Science 352, 467–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcnamara, R. P. , & Dittmer, D. P. (2020).Extracellular vesicles in virus infection and pathogenesis. Current Opinion in Virology 44, 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyenne, V. , Ghoroghi, S. , Collot, M. , Bons, J. , Follain, G. , Harlepp, S. , Mary, B. , Bauer, J. , Mercier, L. , Busnelli, I. , Lefebvre, O. , Fekonja, N. , Garcia‐Leon, M. J. , Machado, P. , Delalande, F. , López, A. A. , Silva, S. G. , Verweij, F. J. , Van Niel, G. … Goetz, J. G. (2019).Studying the fate of tumor extracellular vesicles at high spatiotemporal resolution using the zebrafish embryo. Developmental Cell 48, 554–572.e7 e7 [DOI] [PubMed] [Google Scholar]
- Arraud, N. , Linares, R. , Tan, S. , Gounou, C. , Pasquet, J. ‐ M. , Mornet, S. , Brisson, A. R. Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. Journal of Thrombosis and Haemostasis 12, 614–627 (2014). [DOI] [PubMed] [Google Scholar]
- Nizamudeen, Z. , Markus, R. , Lodge, R. , Parmenter, C. , Platt, M. , Chakrabarti, L. , & Sottile, V. (2018).Rapid and accurate analysis of stem cell‐derived extracellular vesicles with super resolution microscopy and live imaging. Biochimica et Biophysica Acta (BBA) ‐ Molecular Cell Research 1865, 1891–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Zong, S. , Wang, Z. , Lu, Ju , Zhu, D. , Zhang, Y. , & Cui, Y. (2016).Imaging and intracellular tracking of cancer‐derived exosomes using single‐molecule localization‐based super‐resolution microscope. ACS Applied Materials & Interfaces 8, 25825–25833 [DOI] [PubMed] [Google Scholar]
- Lennon, K. M. , Wakefield, D. L. , Maddox, A. L. , Brehove, M. S. , Willner, A. N. , Garcia‐Mansfield, K. , Meechoovet, B. , Reiman, R. , Hutchins, E. , Miller, M. M. , Goel, A. , Pirrotte, P. , Van Keuren‐Jensen, K. , & Jovanovic‐Talisman, T. (2019).Single molecule characterization of individual extracellular vesicles from pancreatic cancer. Journal of Extracellular Vesicles 8, 1685634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maire, C. L. , Fuh, M. M. , Kaulich, K. , Fita, K. D. , Stevic, I. , Heiland, D. H. , Welsh, J. A. , Jones, J. C. , Görgens, A. , Ricklefs, T. , Dührsen, L. , Sauvigny, T. , Joosse, S. A. , Reifenberger, G. , Pantel, K. , Glatzel, M. , Miklosi, A. G. , Felce, J. H. , Caselli, M. … Ricklefs, F. L. Genome‐wide methylation profiling of glioblastoma cell‐derived extracellular vesicle DNA allows tumor classification. Neuro‐oncology 23, 1087–1099 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, A. K. , De Miroschedji, K. , Doeppner, T. R. , Börger, V. , Ruesing, J. , Rebmann, V. , Durst, S. , Jansen, S. , Bremer, M. , Behrmann, E. , Singer, B. B. , Jastrow, H. , Kuhlmann, J. D. , El Magraoui, F. , Meyer, H. E. , Hermann, D. M. , Opalka, B. , Raunser, S. , Epple, M. … Giebel, B. (2018).Precipitation with polyethylene glycol followed by washing and pelleting by ultracentrifugation enriches extracellular vesicles from tissue culture supernatants in small and large scales. Journal of Extracellular Vesicles 7, 1528109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider, M A. , Hurwitz, S N. , Meckes, D G. ExtraPEG: A polyethylene glycol‐based method for enrichment of extracellular vesicles. Science Reports 6, 23978 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M. , & Keswani, M. (2016).Fabrication of radially symmetric graded porous silicon using a novel cell design. Science Reports 6, 24864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij, F. J. , Van Eijndhoven, M. A. J. , Hopmans, E. S. , Vendrig, T. , Wurdinger, T. , Cahir‐Mcfarland, E. , Kieff, E. , Geerts, D. , Van Der Kant, R. , Neefjes, J. , Middeldorp, J. M. , & Pegtel, D. M. (2011).LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF‐kappaB activation. Embo Journal 30, 2115–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gassart, A. , Géminard, C. , Février, B. , Raposo, Graça , & Vidal, M. (2003).Lipid raft‐associated protein sorting in exosomes. Blood 102, 4336–4344 [DOI] [PubMed] [Google Scholar]
- Hurwitz, S. N. , Nkosi, D. , Conlon, M. M. , York, S. B. , Liu, X. , Tremblay, D. C. , & Meckes, D. G. (2017).CD63 regulates epstein‐barr virus LMP1 exosomal packaging, enhancement of vesicle production, and noncanonical NF‐kappaB signaling. Journal of Virology 91(5):e02251‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez‐Mó, M. , Barreiro, O. , Gordon‐Alonso, M. , Sala‐Valdés, M. , Sánchez‐Madrid, F. (2009). Tetraspanin‐enriched microdomains: a functional unit in cell plasma membranes. Trends in Cell Biology 19, 434–446. [DOI] [PubMed] [Google Scholar]
- Yáñez‐Mó, M. , Siljander, P. R.‐M. , Andreu, Z. , Bedina Zavec, A. , Borràs, F. E. , Buzas, E. I. , Buzas, K. , Casal, E. , Cappello, F. , Carvalho, J. , Colás, E. , Cordeiro‐Da Silva, A. , Fais, S. , Falcon‐Perez, J. M. , Ghobrial, I. M. , Giebel, B. , Gimona, M. , Graner, M. , Gursel, I. … De Wever, O. (2015).Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles 4(1), 27066. https://www.tandfonline.com/doi/full/10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcnamara, R P. , Costantini, L M. , Myers, T. A , Schouest, B. , Maness, N J. , Griffith, J D. , Damania, B A. , Maclean, A G. , Dittmer, D P(2018) Nef secretion into extracellular vesicles or exosomes is conserved across human and simian immunodeficiency viruses. MBio 9(1):e02344‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information ‐ Table S1
Supplementary Information ‐ Table S2
Supplementary Information ‐ Table S3
Supplementary Information ‐ Table S4


