Abstract
Neurotrophin signaling is essential for normal nervous system development and adult function. Neurotrophins are secreted proteins that signal via interacting with two neurotrophin receptor types: the multifaceted p75 neurotrophin receptor and the tropomyosin receptor kinase receptors. In vivo, neurons compete for the limited quantities of neurotrophins, a process that underpins neural plasticity, axonal targeting, and ultimately survival of the neuron. Thirty years ago, it was discovered that p75 neurotrophin receptor and tropomyosin receptor kinase A form a complex and mediate high-affinity ligand binding and survival signaling; however, despite decades of functional and structural research, the mechanism of modulation that yields this high-affinity complex remains unclear. Understanding the structure and mechanism of high-affinity receptor generation will allow development of pharmaceuticals to modulate this function for treatment of the many nervous system disorders in which altered neurotrophin expression or signaling plays a causative or contributory role. Here we re-examine the key older literature and integrate it with more recent studies on the topic of how these two receptors interact. We also identify key outstanding questions and propose a model of inside-out allosteric modulation to assist in resolving the elusive high-affinity mechanism and complex.
Keywords: neurotrophin, p75 neurotrophin receptor, nerve growth factor, TrkA, Trk receptor, receptor tyrosine kinase, transmembrane domain, intracellular domain, high-affinity binding, receptor structure-function
Abbreviations: A293 cells, human embryonic kidney cell line (HEK293); ADAM10/17, a disintegrin and metalloprotease 10/17; Akt, protein kinase B; BDNF, brain-derived neurotrophic factor; c29, peptide mimetic of first 29 amino acids of Chopper domain; Chopper (domain), proximal intracellular juxtamembrane region of p75 (K273-S308, human numbering); D1–D5, Trk extracellular domains 1 to 5; ECD, extracellular domain; EGFR, epidermal growth factor receptor; EpoR, erythropoietin receptor; ErbB4, Erb-B2 receptor tyrosine kinase 4; ERK, extracellular signal–regulated kinase; GnRH, progonadoliberin-1; Grb2, growth factor receptor–bound protein 2; HeLa cells, Henrietta Lacks cervical cancer immortalized cell line; ICD, intracellular domain; Kd, dissociation constant; MAH cells, v-myc, adrenal-derived HNK1+ cell line; MAPK, mitogen-activated protein kinase; NGF, nerve growth factor; NT-3, neurotrophin-3; NT-4/5, neurotrophin-4/5; p75, p75 neurotrophin receptor; PC12 cell, cell line from pheochromocytoma of the rat adrenal medulla; PI3K, phosphoinositide 3-kinase; RIP, regulated intramembrane proteolysis; RTK, receptor tyrosine kinase; SCG, superior cervical ganglia; Shc, src homology 2 domain-containing transforming protein C1; TM, transmembrane; Trk, tropomyosin receptor kinase
Neurotrophin signaling regulates a diverse set of neural processes including differentiation, neurite outgrowth, axon pruning, apoptosis, and cell survival during development, adulthood, and following injury to the nervous system. Loss of, or aberrant, neurotrophic signaling is strongly implicated in multiple neurodegenerative and psychiatric disorders including Alzheimer’s and motor neuron diseases, chronic pain, and depression (1, 2). It is unsurprising, therefore, that manipulating protrophic neurotrophin signaling is of interest to pharmaceutical companies.
Introduction to neurotrophin signaling: Meet the players
There are four members of the human neurotrophin family: nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), neurotrophin-3 (NT-3), and neurotrophin-4/5 (NT-4/5). Each of these structurally related neurotrophins is synthesized as a proform prior to proteolytic cleavage to produce a mature neurotrophin (3). Proneurotrophin receptor binding initiates death signaling through interaction with the coreceptor complex of the p75 neurotrophin receptor (hereafter referred to as p75) and sortilin (4). p75 binding of mature neurotrophins can also result in death signaling and, as a member of the tumor necrosis receptor super family, the receptor is often referred to as a neural cell death receptor (5, 6). However, mature neurotrophins also bind to their cognate tropomyosin receptor kinase (Trk) family of receptors, with the result usually being prosurvival or trophic signaling that can also inhibit death signaling induced by p75 (3, 7, 8). Therefore, only in the absence of Trk receptor signaling does p75 typically promote cell death in response to binding mature neurotrophins.
Although both Trk and p75 can individually bind neurotrophins to elicit independent signaling events, there is substantial evidence that the two proteins work together to enhance trophic signaling during development and in the healthy brain by mediating at least 10-fold higher affinity of the Trk receptor for its cognate neurotrophin. Five decades of research have revealed that the neurotrophin signaling system is extremely complex, and the mechanism by which p75 and Trk receptors work together to enhance signaling is still not fully understood. This review will briefly describe the two receptors individually before outlining the evidence that Trk and p75 form a coreceptor complex and proposing how this interaction results in enhanced neurotrophin binding for optimal cellular responses.
Trk receptors
The Trk family has three members, namely TrkA, TrkB, and TrkC. Each of these has preferred neurotrophin binding partner(s); TrkA binds preferentially to NGF, TrkB binds BDNF and NT-4/5, and TrkC binds NT-3 (9, 10), although all three receptors bind NT-3 to some extent. The expression and functional activity of each Trk member in the nervous system is cell-type specific (albeit with some crossover and co-expression). For example, TrkB and TrkC are highly expressed in cortical and hippocampal pyramidal neurons and cochlear neurons, whereas TrkA is found in cholinergic basal forebrain neurons (11, 12, 13, 14). As such, although their signaling outputs and mechanisms of regulation have many commonalities, there are subtle differences in structure and function between Trk family members that influence how each Trk receptor and neurotrophin specifically maintains the health of the neuronal population in which it is expressed. In this review, we will focus on the interaction between p75, TrkA, and its cognate ligand NGF, calling attention to key known differences with TrkB and TrkC as appropriate.
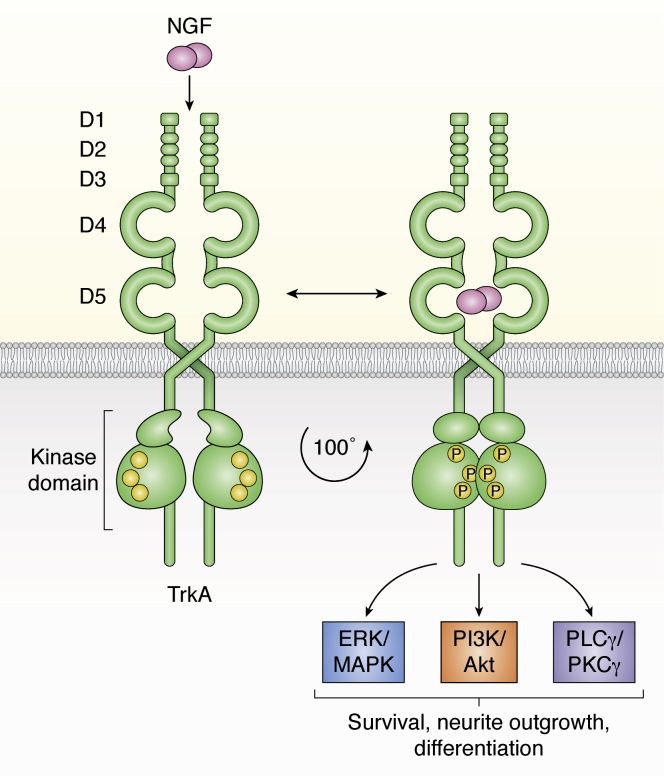
TrkA is a receptor tyrosine kinase (RTK) consisting of five extracellular domains (ECDs) (D1–D5) which resemble a “crab-claw” shape, an α-helical transmembrane (TM) domain, and an intracellular segment which includes a kinase domain that is structurally highly conserved between all RTKs (Fig. 1). D1 and D3 are “cap modules”, i.e., cysteine-rich domains which N- and C-terminally flank the three leucine-rich repeats that comprise D2. D4 and D5 are both Ig-like (commonly called Ig-C1 and Ig-C2, respectively) domains, the latter of which is responsible for NGF binding (15, 16, 17).
Figure 1.
TrkA receptor structure and signaling pathways. The TrkA receptor consists of five extracellular domains (D1–D5), an α-helical transmembrane domain, and an intracellular segment containing a kinase domain. The receptor can exist in preformed inactive dimers. NGF binding to TrkA activates the receptor, causing rotation and rearrangement of the dimers to an active state, autotransphosphorylation, and activation of multiple signaling pathways. TrkA receptor is shown in green, NGF dimers in pink, and phosphorylated sites of the active kinase domain as a “P”. PLC-γ, phosphoinositide phospholipase C-γ; PKC-γ, protein kinase C-γ; Trk, tropomyosin receptor kinase.
As is typical of RTKs, ligand binding at the cell surface to the ECD induces formation of active TrkA dimers or oligomers, autotransphosphorylation of the intracellular domain (ICD) via its kinase domain, and initiation of signaling via phosphorylation cascades (18, 19, 20). The most studied signaling pathways activated by Trk receptors are the extracellular signal–regulated kinase (ERK)/mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K)/ protein kinase B (Akt), and phosphoinositide phospholipase C-γ/ protein kinase C-γ pathways, which influence proliferation, neuronal survival, differentiation, and neurite outgrowth (19, 21, 22).
p75 neurotrophin receptor
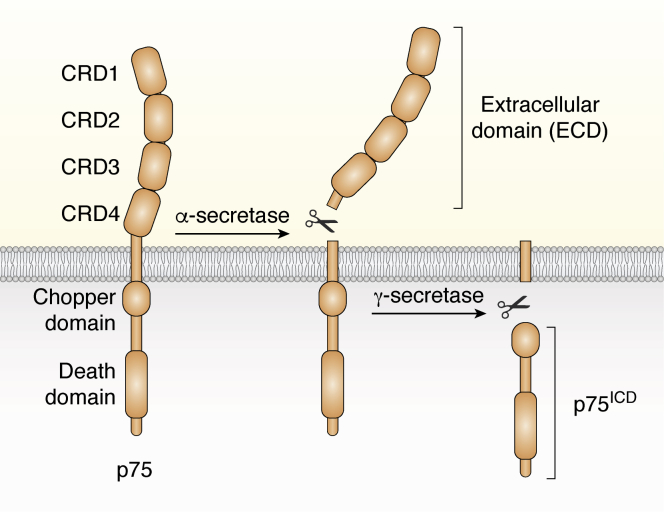
p75 is also a type 1 TM protein that was originally characterized as the NGF receptor but is now classified as the 16th member of the tumor necrosis factor receptor family (23). The extracellular segment comprises four cysteine-rich domains (CRD1–4) as well as sites for N- and O-linked glycosylations (24) (Fig. 2). An α-helical TM domain connects the ECD to the ICD of p75 (p75ICD) which is well known for including a death domain (25). Within the N-terminal half of p75ICD are multiple putative interacting protein binding sites, which are thought to be involved in p75 signal initiation as the receptor lacks any obvious intrinsic catalytic activity (26, 27, 28). The globular death domain of p75ICD is located in the latter C-terminal portion of the intracellular segment. While trimerization and higher-order oligomerization of tumor necrosis factor receptor family member death domains are usual for initiation of apoptotic signaling, experimental evidence remains inconclusive as to whether self-association of p75 death domains is required for its function (25, 29, 30, 31, 32, 33, 34). At any rate, p75 is known to (also) initiate cell death through several well-described apoptotic signaling pathways which are not dependent on death domain oligomerization (for reviews see (23, 35)). The focus of most of the recent studies on p75 has been on its role in mediating proapoptotic signaling in the nervous system during development and/or neurodegenerative conditions. However, the bifurcated function of the receptor is a significant feature, and it must be remembered that p75 was first described as a neurotrophin receptor involved in trophic signaling (30, 36, 37).
Figure 2.
p75 structure and regulated intramembrane proteolysis (RIP). p75 consists of four extracellular cysteine-rich domains (CRD1–4), an α-helical TM domain, and an intracellular segment (ICD, p75ICD) which includes a death domain and the juxtamembrane Chopper domain. The receptor undergoes RIP; α-secretase/ADAM10/17–mediated cleavage in the extracellular domain (ECD) releases an ECD fragment, this is followed by γ-secretase cleavage in the transmembrane domain to yield a soluble p75ICD fragment. p75 receptor is shown in orange. p75, p75 neurotrophin receptor.
Following the discovery of the neurotrophin-binding Trk receptors with kinase activity, the lack of obvious catalytic domains in p75ICD relegated p75 to be considered an adaptor protein, incapable of activating signaling cascades and instead partnering with TrkA to enhance NGF trophic signaling (38, 39). There is now an increased understanding of signaling by protein multimerization, and more recent evidence supports the ability of p75 to initiate signal events in response to ligand binding in the absence of coreceptors such as TrkA (29). Indeed, many factors affect which of the bifurcated signaling outcomes p75 mediates; ligand type, coreceptor/adaptor protein presence, and relative expression levels can all alter neurotrophin–p75 signaling outcomes, leading to cell survival, apoptosis, growth cone collapse, neurite outgrowth, and/or proliferation (reviewed in (40, 41, 42)). To exemplify the complexity of p75 signaling regulation, one must consider that p75 binds each of the neurotrophins with similar affinity (43, 44) and as such, cell fate is influenced by ligand maturation, availability, and binding kinetics, as well as Trk receptor co-expression. NGF treatment of NGF-responsive sympathetic neurons or a neural progenitor–like cell line (PC12 cells) which endogenously expresses TrkA and p75 revealed that p75 participates in the survival signaling of the cells. Upon BDNF treatment of sympathetic neurons, p75 will instead mediate apoptotic signaling (45, 46, 47, 48). The same applies in reverse where treatment of BDNF-responsive, TrkB- and p75-expressing motor, or hippocampal neurons with NGF results in p75-mediated cell death (49, 50, 51, 52).
p75 regulated intramembrane proteolysis
p75 is one of a growing list of proteins, including ErbB4, Notch, and amyloid precursor protein, which undergo a two-step process of regulated intramembrane proteolysis (RIP) to yield a soluble cytoplasmic fragment with signaling capability (53, 54) (Fig. 2). First, α-secretase/ADAM10/17 cleaves within the juxtamembrane region of the ECD, thereby shedding the ECD to yield a membrane-bound C-terminal fragment (53, 55, 56). This is followed by presenilin-dependent γ-secretase cleavage within the TM domain to release a soluble p75ICD fragment into the cytoplasm (53). However, it was only after three decades of mechanistic study of p75 that this process came to be understood. As a result, the field must carefully consider the conclusions which were drawn in the earlier literature in light of more recent evidence that each p75 cleavage fragment could serve a distinct functional purpose (57).
Keep your friends close—how TrkA and p75 work together
It is well established that p75 potentiates Trk signaling events. It is also widely accepted that p75 directly influences Trk in such a way that the latter receptor forms a high-affinity binding site for its cognate ligand, for example TrkA with NGF. These high-affinity binding sites are frequently referenced in the neurotrophin literature as a requirement for prolonged cell survival and differentiation signaling. Individually, p75 and TrkA exhibit low affinity for NGF with Kd ≈ 1 × 10−9 M (58, 59, 60). However, in PC12 cells and sensory neurons that endogenously express both receptors, a TrkA high-affinity binding site for NGF is detectable with Kd ≈ 1 × 10−11 M (58, 60, 61). Of note, although published binding affinities of high- and low-affinity binding sites are very similar, differences in cell type (and therefore membrane composition), receptor clustering, and relative ratios of expressed receptors likely contribute to minor observed differences. Kinetic analysis of NGF binding to the individually expressed receptors has revealed that p75 has a rapid rate of ligand association and dissociation, whereas TrkA exhibits very slow association and dissociation (44, 62, 63). When the two are co-expressed, however, the association rate of NGF to TrkA increases 25-fold (62, 64) with still very slow dissociation rate—characteristic of the high-affinity binding site (62, 65, 66).
Subsequent experiments in which p75 and TrkA were co-expressed in cell lines lacking endogenous receptors demonstrated the formation of high-affinity binding sites only in the presence of both receptors, which correlated with the extent of TrkA phosphorylation increasing several-fold in response to NGF (27, 43, 61, 67, 68, 69, 70). Total TrkA expression and, where measured, expression at the plasma membrane remained unchanged. In vitro, PC12 cells have been shown to respond maximally to lower concentrations of NGF (67) than PC12 cells deficient in p75 (68). Mouse embryonic dorsal root ganglia and postnatal superior cervical ganglia (SCG) deficient in p75 also have decreased sensitivity to NGF, requiring 2- to 3-fold higher concentrations than WT neurons for survival (70). Furthermore, another sympathetic neural progenitor–like cell line (MAH) expressing both TrkA and p75 undergoes mitotic arrest and subsequent differentiation in response to NGF 2 to 3 days earlier than MAH cells expressing only TrkA (27). Similarly, NGF-stimulated axon outgrowth of sensory neurons during development is slower in cells that express only TrkA than those which co-express p75 (71). These studies have demonstrated that co-expression of TrkA and p75 yields a high-affinity binding site and that p75 is able to modulate TrkA such that is has increased sensitivity to lower concentrations of NGF. The importance of this for neurons is most evident during development where limited amounts of neurotrophin are present (72) and could be harnessed in rational design of therapeutics targeting disease where TrkA expression is limited.
In addition to increasing Trk sensitivity to neurotrophins, p75-mediated modulation results in increased specificity of ligand binding. In vitro, it is evident that p75 increases the ligand specificity of TrkA for its preferred ligand NGF and desensitizes the receptor to NT-3 (27, 61, 73, 74). For example, although NT-3 can bind to TrkA expressed by PC12 cells, it does not induce a differentiated phenotype such as that which occurs in response to NGF binding, suggesting that the high-affinity NGF receptor complex discriminates between the two structurally related neurotrophins (61). Further, very high concentrations of NT-3 are required to produce measurable TrkA activity, a marked difference to the heightened sensitivity to NGF. This was demonstrated in vivo in NGF-responsive sympathetic neurons which endogenously express p75 and TrkA (75). SCG neuron number was assessed in adult transgenic mice with mutations in p75, NGF, and/or NT-3. In p75−/− mice, the number of SCG neurons was normal, whereas in NGF+/− mice, there were 50% fewer sympathetic neurons. Remarkably, however, in p75−/− NGF+/− mice, the neuron number was restored to WT levels. This restoration was lost in p75−/− NGF+/− NT-3+/− mice, indicating that, when present, p75 increases TrkA ligand specificity for NGF, but when absent, NT-3 can substitute for NGF in activating a “nonspecific” (and low-affinity) TrkA. Similarly, Mischel et al. (74) found that co-expression of an intracellularly truncated p75 receptor inhibited NT-3 activation of TrkA, whereas co-expression of a chimeric ECDEGFR-p75TM-ICD receptor had no effect. The study concluded that p75 ECD is necessary to inhibit NT-3 signaling through TrkA. Importantly, the truncated p75 retained the first nine residues of the p75 intracellular juxtamembrane region, and the ECDEGFR-p75TM-ICD chimeric receptor is unlikely to have retained the α-secretase cleavage site. As discussed below, these two factors likely contributed to the observed results. Bibel et al. (9) also showed that in the absence of p75, TrkB is readily activated by BDNF, NT-3, and NT-4/5 in A293 cells, whereas when p75 is co-expressed, only BDNF can efficiently activate this receptor. To our knowledge, there is one report of p75 mediating specificity of TrkC ligand binding (66) and another detailing that p75 expression has no impact on TrkC activation or ligand specificity (76).
In summary, decades of research have revealed not only that p75 increases the affinity and specificity of TrkA and TrkB for their cognate ligands but also that this significantly impacts cell fate in vivo. Despite this knowledge and the clear importance of this signaling system in neurodevelopment, cell maintenance, and the response to disease (where neurotrophin or Trk receptor expression is altered), the mechanism by which p75 modulates Trk–neurotrophin interactions remains unknown. Clearly important questions remain unanswered: (i) what is the mechanism by which high-affinity binding is achieved, (ii) how does p75 promote this mechanism, and (iii) does the same or a different mechanism promote ligand-Trk receptor specificity?
Models of p75-Trk receptors
Several models of the mechanism by which p75 modulates Trk activity have been explored. Of these, three have been more intensely investigated: (i) the formation of heterocomplexes stabilized by p75, (ii) p75-mediated concentration of Trk ligand within the local microenvironment, and (iii) direct interaction between p75 and Trk in such a way this results in allosteric modulation for high-affinity receptor formation. However, as the first two models are not wholly supported by the experimental data and the third lacks direct structural evidence, they should not be considered as mutually exclusive. Rather, there is likely some interplay which accounts for the complexity and diversity of the signaling outcomes within the system. This is reviewed below.
Stabilized heterocomplex model
The first of the proposed models involved p75, Trk, and the neurotrophin ligand forming a simple 1:1:2 heterocomplex (77), that is highly stable and facilitates prolonged signaling (15, 78, 79). This model was consistent with other heterodimer receptor complexes described at the time, such as the ciliary neurotrophic factor receptor/glycoprotein 130 complex required for leukemia inhibitory factor binding and PI3K/Akt pathway activation (80). The existence of a neurotrophin coreceptor complex is supported by evidence of a direct interaction between p75 and TrkA receptors, including co-immunoprecipitation of TrkA and p75 (9), confirming previous reports of an association revealed by cross-linking and copatching techniques (64, 81, 82).
Structural evidence indicates that in the 1:1:2 stoichiometry, each receptor would bind the NGF dimer in antiparallel orientations. Although there are no steric clashes in this complex (15), the model includes only a TrkA monomer, thereby removing the possibility of Trk catalytic activity and making the functional value of such an interaction questionable. Furthermore, the structural data indicate that both NGF+p75 and NGF+TrkA make 2:2 complexes (15, 83). Of note, each receptor binds to the NGF dimer at sites on the same face, which, although not overlapping, precludes the other receptor from being able to bind simultaneously, thereby preventing any detectable or computationally obvious 2:2:2 complex of full-length receptors from forming (15, 79). Although He and Garcia (78) solved the p75 structure to bind in a 1:2 stoichiometry with an NGF homodimer, this was later explained as an artifact of solving the structure with unglycosylated p75 (84). Computational analysis demonstrated it is possible to form a 1:1:1 complex (78), though again the questionable functionality of such a complex makes it unlikely to offer any evolutionary advantage, and there have not yet been any further studies to support this model. Thus, it is improbable that simply forming a heterocomplex which interacts extracellularly is the mechanism whereby p75 modulates Trk receptor affinity.
Ligand-passing model
The second popular model, dubbed the “ligand-passing theory” (62, 67, 85), is based on affinity kinetics. It postulates that p75 is the first of the two receptors to bind NGF at the plasma membrane, thus increasing the local concentration of NGF which is available to bind to TrkA. p75 forms a temporary complex with TrkA by sandwiching the NGF dimer between the two receptors as it is passed from one receptor to the other, (for full review see (85)). This model is consistent with the findings that a mutant form of NGF, which is able to bind TrkA but not p75, binds only with low affinity (27, 67, 86, 87). However, the ligand-passing model is inconsistent with data which show that the p75 ECD (and therefore the ligand binding-domain) is not required for high-affinity receptor formation (88, 89). Rather, the results suggest that p75 modulates the ligand-binding site on TrkA, raising the possibility of an altered conformation of the Trk ECD in the presence of p75.
Allosteric modulation model
The final prominent model describes a direct interaction between the two neurotrophin receptors, leading to allosteric modulation of the TrkA receptor such that NGF affinity and specificity is increased. This modulation may reflect that NGF has improved access to the D5 ligand-binding site or that other residues in the TrkA ECD are participating in the interaction. Either of these changes may also explain the inability of mutant NGF to bind TrkA with high affinity; should high-affinity binding be mediated through additional or alternate residues in either TrkA ECD or the NGF dimer, this may be prevented by NGF mutation. Challenges in solving the structure of full-length TrkA (as opposed to single domains), let alone solving the structure of Trk in the presence of p75, would first need to be overcome to thoroughly test the validity of this hypothesis. As noted above, biochemical evidence supports a direct interaction between Trk and p75. Previous work, where the p75 ECD, ICD, or TM domains were removed, demonstrated that for TrkA and TrkB to form the characteristic high-affinity receptor, based on measures of NGF or BDNF binding affinity, respectively, the ICD and/or TM domain of p75 is required for Trk receptor interaction (9, 61, 88, 89).
More recent studies have demonstrated that Trk signaling can be enhanced by promoting p75 RIP (50, 68, 90, 91) whereas an ECDEGFR-p75TM-ICD chimeric protein, in which the p75 extracellular α-secretase site (92) is replaced by the epidermal growth factor receptor portion of the chimera, failed to form a high-affinity complex (93). Indeed, PC12 cells expressing a mutant p75 construct unable to undergo RIP (30) displayed reduced NGF affinity and shorter neurite outgrowth upon NGF stimulation compared to cells expressing WT p75, and the p75ICD fragment alone could stimulate functional outcomes associated with high-affinity binding (68). Thus, endogenous RIP of p75 appears to be both required and sufficient for high-affinity Trk–p75 receptor complex generation. Although RIP of p75 to release the ECD could facilitate NGF passing to TrkA, this alone does not explain the requirement of the TM domain and/or p75ICD for high-affinity modulation and so cannot be the entire mechanism driving enhanced receptor-ligand affinity. As soluble p75ICD is unable to directly affect TrkA extracellular ligand binding, a mechanism involving allosteric modulation is most probable.
Evidence is mounting that the soluble proximal intracellular juxtamembrane region of p75, termed the Chopper domain (7), is sufficient to mediate the protrophic effects of p75/p75ICD fragment with Trk. Indeed, a soluble peptide mimetic of the juxtamembrane 29 amino acids of the Chopper domain (referred to herein as c29) has been demonstrated to be sufficient for the promotion of TrkA and TrkB trophic activity. c29 treatment of primary SCG neurons and PC12 cells has been shown to enhance their neurite outgrowth and trophic signaling responses to NGF 10-fold, with peptide treatment alone (i.e., no neurotrophin) demonstrating that c29 has no intrinsic activity (68). Similarly, c29 treatment of a motor neuron disease mouse model resulted in increased motor neuron survival and later onset of disease (50). These findings are consistent with earlier work which mapped the domains of p75 that are important for the high-affinity complex. Although not recognized at the time, others also demonstrated the requirement of the intracellular juxtamembrane segment when using truncated p75 constructs which contained intact TM and Chopper domains as well as intact secretase cleavage sites, reporting that they retained functional interaction and high-affinity ligand binding when complexed with TrkA and TrkB receptors (9, 58, 74, 89, 93); the conclusion at the time was that the TM domain was contributing to the interaction. Although recent evidence points to a possible role for the TM domain in high-affinity complex formation (88 and 128; see below), taken together, these data indicate that the 29 amino acid Chopper sequence is both sufficient and necessary for p75 to form a high-affinity complex with TrkA or TrkB, promoting trophic signaling in vivo and in vitro.
Further work is required to elucidate the mechanism by which p75ICD, and specifically the Chopper domain, influences the ligand-binding affinity of Trk. Although p75 does not directly affect the catalytic activity of the Trk kinase domain (58, 68, 73), to deduce how the allosteric modulation model by which p75ICD/c29 could facilitate an outwardly propagated TrkA conformational change nonetheless requires an understanding of RTK kinase domain activation and cis-autoinhibitory mechanisms.
Structural and steric contributions to Trk receptor activation
Kinase activation
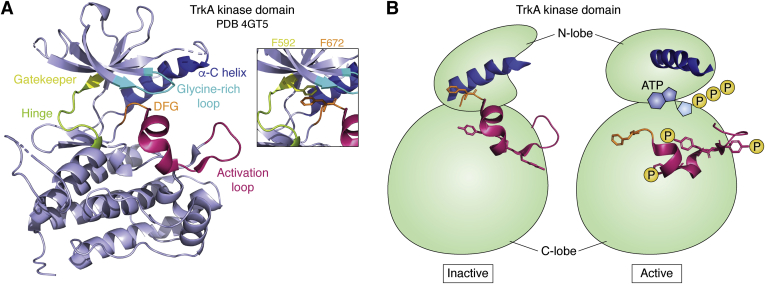
The overall structure and mechanism of activation of RTK kinase domains is highly conserved. Neurotrophin ligands are dimers, and binding to a Trk receptor ECD facilitates either dimerization of receptor monomers or rearrangement of preformed inactive dimers (94, 95). In doing so, the kinase domains are brought into close proximity in a suitable conformation for one of the kinase pair to autotransphosphorylate key tyrosine residues on the other’s kinase domain. This phosphorylation leads to major structural repositioning within the kinase domain, facilitating ATP binding and full catalytic activation (for in-depth review see (96, 97, 98)) (Fig. 3).
Figure 3.
Key TrkA kinase domain sites.A, TrkA kinase domain (PDB 4GT5) crystal structure with regions of importance for kinase activation indicated. Inset shows T-stacking interaction between the gatekeeper residue F592 and F672 of the DFG motif. B, schematic representation of the N-lobe and C-lobe shifts, repositioning of the α-C helix, DFG motif, and activation loop, as well as engagement of ATP and phosphorylation of activation loop tyrosine residues between the active and inactive TrkA kinase domains. Trk, tropomyosin receptor kinase.
Broadly speaking, the intracellular kinase domains of RTKs consist of two linked lobes. The N-terminal lobe (N-lobe) is made of five β-sheets and an α-C helix which is critical for the catalytic activity of the kinase domain. The C-terminal lobe (C-lobe) is considerably larger and comprises β-sheets and several α-helices (96). The N-lobe and C-lobe are connected by the hinge region which itself plays a role in the structural integrity of the inactive kinase domain. Additional key residues and motifs are found in each of the lobes. A large loop between β-sheet eight and β-sheet nine of the C-lobe is termed the activation loop, so named due to the presence of the critical tyrosine residues that are autotransphosphorylated during kinase activation. There is also an important alanine–phenylalanine–glycine (DFG) motif at the base of the activation loop that is positioned at the hydrophobic interface between the N-lobe and C-lobe where ATP will eventually bind. This interface abuts a glycine-rich loop of the N-lobe which is a flexible flap containing a conserved glycine-rich sequence motif GXGXФG (where Ф is an aromatic amino acid). The glycine-rich loop interacts closely with the adenine base, ribose sugar, and nonhydrolyzable phosphate groups of ATP for coordination of the nucleotide (97, 99). The conserved aromatic side chain forms a clamp around the site of γ-phosphate transfer (97). Simultaneous to this ATP engagement, the α-C helix is rotated into an active position that is stabilized by a salt bridge formed with β-sheet 3, which in turn facilitates α-C helix coordination with β-phosphate of the ATP molecule (99).
TrkA kinase activation
Trk family members adopt an inactive “DFG out” conformation (100) which positions the DFG motif of the activation loop within the ATP-binding site, blocking substrate access. The position of the loop is stabilized by a T-stacking interaction between the phenylalanine of the DFG motif and another phenylalanine at position 592 (rat TrkA numbering unless otherwise stated) which is at the start of the hinge region. F592 is therefore known as the gatekeeper residue. In this conformation, it is not possible for the crucial salt bridge between K547 (of β-sheet 3) and E563 (within the α-C helix) to form, as a result of which, the entire N-lobe is held in an inactive conformation in which the α-C helix is rotated and faces away from the ATP binding pocket (100). Indeed, mutation of K547 to either arginine or alanine, preventing β-sheet three interaction with the α-C helix, has provided a useful experimental tool by creating a kinase dead TrkA receptor (101).
TrkA has three sites of regulatory phosphorylation in the activation loop: Y679, Y683, and Y684. Upon autotransphosphorylation of these sites, the activation loop will adopt a new position, rotating the DFG motif such that the T-stacking interaction with the gatekeeper residue is broken and the ATP binding pocket becomes accessible (97). As ATP binds, the N-lobe “clamps” down, rotating the α-C helix into a position where interactions are now possible between ATP, the GXGXФG motif, and α-C helix (97). TrkA is then considered to have adopted a fully catalytic state and is ready for substrate binding and activation, e.g., Shc/Grb2 binding p-Y499 as adaptors for the ERK/MAPK and PI3K/Akt signaling pathways and phosphoinositide phospholipase C-γ binding p-Y794 for alternate ERK/MAPK activation (22).
The conformational switch from an inactive to active Trk receptor following ligand binding involves a sophisticated process of key residue repositioning and overcoming of various bonding constraints, which typically “hold” the inactive state. Interaction of Trk with p75, together with the corresponding changes to the Trk ECD that result in a high-affinity site, may also facilitate an inactive but more readily activated repositioning of the kinase domain. This is one possible argument for an allosteric modulation model in which ligand binding is still a requirement and p75 does not affect constitutive kinase activity, although ligand affinity is enhanced. This possibility is discussed in more detail below.
Multiple states of existence
Multiple groups have demonstrated that inactive TrkA exists in a monomer–dimer equilibrium, albeit with some disagreement on whether the majority of inactive receptors are monomeric or dimeric (95, 102). Maintaining a monomer–dimer equilibrium requires cis-autoinhibition to prevent inappropriate kinase activation. The proposed mechanisms of cis-autoinhibition for the RTK family are surprisingly diverse, implying that each receptor self-regulates in a unique manner (for review see (103)). How Trk receptors are cis-autoinhibited is as yet unknown, though inspiration may be drawn from other RTKs. The Ephrin type-B receptor 2 adopts a pseudo-closed confirmation, in which the highly ordered cytoplasmic juxtamembrane region interacts with both N-lobe and C-lobe of the kinase domain to kink the α-C helix, augmenting rearrangement of the lobes to prevent spontaneous kinase activation. Upon ligand binding, this distortion is relieved through initial phosphorylation of two tyrosine residues in the juxtamembrane region, which in turn allows phosphorylation of the activation loop tyrosines and full catalytic activity (99, 104). The erythropoietin receptor (EpoR) assumes at least two dimer orientations; interactions through one face of the TM α-helices results in stabilization of an inactive dimer, whereas an alternate interaction interface that is achieved through ∼100° rotation of the TM α-helices allows kinase activation. This rotation through the TM domain is achieved upon ligand binding and has been validated with experiments using chimeric mutants and cysteine-substitution mutants which form alternate relative TM orientations (105, 106, 107). These experiments also suggested that a partially active dimer, which exhibits specificity for signaling pathway activation, is possible through a third interaction interface, although whether this is achieved endogenously was not tested.
Of these and other proposed models of cis-autoinhibition, it seems most likely that domain rotation is involved in Trk family RTK regulation and/or may explain how p75 promotes Trk function. Crystal structures of the TrkA kinase domain have proven notably stable in the absence of cocrystallizing of the intracellular juxtamembrane region, indicating that this domain is not required for the stability of the inactive kinase conformation (100, 108, 109, 110). Furthermore, early functional studies revealed that, unlike EpoR, the Trk activation loop tyrosines are phosphorylated prior to those tyrosines outside the kinase domain; the latter serve as docking units for signaling pathway activation and are not involved in cis-autoinhibition (111, 112, 113, 114, 115, 116).
Intracellularly propagated rotation as a requirement for kinase domain activation has been previously described in multiple kinase receptors, including epidermal growth factor receptor, EpoR, tyrosine kinase-type cell surface receptor HER2 (ErbB2), growth hormone receptor, and platelet-derived growth factor receptor-β (105, 117, 118, 119). Recent structural evidence suggests that inactive TrkA dimers are stabilized by interaction of the receptor TM domains, specifically residues belonging to an LXXFAXXF motif (spanning L424-F431, human numbering) (95, 102). Franco et al. (95) also demonstrated that a different dimerization interface, still within the TM domain although slightly N-terminal to the aforementioned motif, is engaged by active TrkA, with residues located on the opposite side of the α-helices. The authors propose that A428 (human numbering) is a central point of interaction between the dimeric Trk TM interfaces during receptor activation. As the TrkA dimer transitions from an inactive to active state, the TM regions pivot through A428 to allow the receptors to interact more closely. Franco et al. (95) proposed that a conformational change in the TrkA ECD is induced when NGF binds to TrkA, bringing the extracellular proximal juxtamembrane region into close proximity. This leads to the pivoting movement around A428 and propagates an allosteric shift of the TrkA ICD equivalent to a 100° rotation of the kinase domains, which then allows autotransphosphorylation to occur. However, whether this 100° rotation is the only mechanism required to overcome cis-autoinhibition requires further investigation.
The ECDs of TrkA have also been identified as key regions for regulating cis-autoinhibition of the receptor (120). In the absence of neurotrophin and upon deletion of either or both Ig-like domains (D4 and D5), active Trk dimers formed readily, a phenomenon not observed in WT receptors or full ECD truncated mutants. Furthermore, although deletion of these Ig-like domains did not impact cell proliferation, differentiation was stimulated. Evidently, the relative conformation of the ECD influences signaling specificity and cellular outcomes, possibly by altering ligand binding site accessibility and therefore ligand affinity and/or intracellularly transmitted information that affects the longevity of signaling or subsequent fate of the receptor (121). This again suggests that extracellular and intracellular structural changes do not occur independent of each other.
Numerous agonists and activators have been identified as possible therapeutics based on their ability to modulate neurotrophic signaling, thereby providing additional mechanistic insights. Of these molecules, the vast majority target the ligand-binding domain of the Trk receptor, or at least some portion of its ECD (for reviews see (122, 123)). Although some activators act as ligand mimetics—including monoclonal antibody 5C3 and the small molecule 7,8-dihydroxyflavone that bind D5 of TrkA and TrkB, respectively—others do not interact with the known neurotrophin binding domains yet are still capable of promoting receptor dimerization and/or autotransphosphorylation (124, 125). Examples include the small molecule D3 noncompetitive TrkA agonist which binds D5 (IgG-C2) although not at the same epitope as NGF, amitriptyline hydrochloride which binds to the TrkA and TrkB D2, and mAb 1D7 that binds the D2–D3 domains of TrkB (126, 127, 128). These examples endorse the idea that alternate extracellular conformations are compatible with inducing kinase domain activation to produce neurotrophic outcomes; the agonist–Trk complexes are sufficiently stable to promote activation of downstream signaling for cell survival and differentiation, as is seen with NGF–TrkA–p75 complexes, despite the absence of neurotrophin. Interestingly, recent computational simulations and in vitro binding and kinase activation assays have revealed an antidepressant-drug binding pocket within the TM domain of TrkB (only) (129), demonstrating that the interaction between TrkB and fluoxetine stabilizes TrkB dimerization and promotes increased receptor trafficking to cholesterol-rich subdomains of the cell surface of dendritic spines to facilitate BDNF-dependent receptor activation (129). p75 can be palmitoylated (130) and could similarly regulate Trk subcellular localization and function (131). To our knowledge, the only published agonist apart from the c29 peptide that targets the ICD of Trk receptors is gambogic amide. Gambogic amide specifically activates TrkA, but not TrkB or TrkC, inducing robust neurotrophic signaling, which both protects against cell death and promotes neurite outgrowth in PC12 cells (132). The mechanism driving gambogic amide-induced TrkA activation, and whether this is additive or identical to NGF binding to TrkA, remains unclear.
Insight into structural differences in high- and low-affinity TrkA receptors can also be inferred from a TrkA mutant that has been characterized as constitutively high affinity (133). This mutant TrkA has a P203A substitution located within a linker region between D2 and D4. When stimulated with NGF, TrkA P203A binds with high affinity (133). However, this substitution also promotes constitutive dimerization and near-maximal activation of the kinase domain even in the absence of NGF (133). Although D5 is considered the primary domain involved in NGF binding (17), it has also been suggested that D4 and D1 participate in neurotrophin activation through unknown mechanisms (95, 134). It has been proposed that the P203A substitution compromises the integrity of the “crab-claw”-like structure of the TrkA ECD; however, exactly how this impacts cis-autoinhibition or translates to high-affinity binding is currently unclear. There are at least three possibilities for how P203A might impact NGF binding affinity. The first is that in WT TrkA, NGF access to D5 is obstructed by D1 and D4 interacting within the TrkA pair or with NGF. P203A may therefore disrupt these interactions and permit NGF to gain easier access to D5 (faster association rate). The second possibility is that in WT TrkA, NGF binding to D5 is stabilized by D1 and D4, with P203A changing the ECD conformation to promote easier secondary interactions between NGF and D1/D4, thereby forming high-affinity binding sites (slower dissociation rate). The third possibility is that NGF binds through two separate interfaces in the low- and high-affinity receptors. D1 and D4 may mediate the second interface through which NGF binds when the receptor is in a high-affinity binding conformation. These are experimentally testable models which we believe should be investigated.
Of interest to this issue is the observation that c29 peptide treatment of TrkA P203A-expressing cells does not further enhance (above the already enhanced effect of P203A) NGF binding to the cells, as it does in WT TrkA-expressing cells (68). This result suggests that the actions of c29, which induces a faster NGF–TrkA association rate, and the change mediated by the P203A mutation are acting by the same or a similar mechanism, rather than causing additive structural changes. However, given that Arevalo et al. (133) reported that P203A produces a slower off rate, it is also possible that the changes to the ICD induced by P203A prevent c29 from interacting with or influencing the Trk ICD structure. Further affinity studies would clarify this matter. In either case, the notion that conformational changes to the Trk ECD are propagated to the ICD, and vice versa, is well supported by the current evidence.
Very recent NMR and molecular dynamics simulations of p75 and TrkA TM domains support the idea of heterodimerization occurring through these domains (135). Notably, p75 constitutively interacted with the TrkA inactive interface, leaving the active interface available for dimerization with another Trk monomer. Mutation of three p75 TM domain residues key to the simulated TrkA interaction was shown to abolish p75-promoted TrkA high-affinity receptor function in PC12 and HeLa cells. Franco et al. (135) proposed that p75 binding to the TrkA-inactive TM interface to “hold” the receptor in an active-dimer position is the underlying mechanism by which high-affinity NGF binding to TrkA is promoted, fitting with the stabilized heterocomplex model. However, two of these three residues A262 (rat p75 numbering) and G266 have previously been identified to be critical for p75 γ-secretase cleavage (136), allowing for these results to be alternatively explained by a loss of p75ICD generation and therefore its affinity-promoting function. Further, this positioning of TrkA to readily dimerize through the active interface does not promote constitutive kinase activity, suggesting that merely rotating to interact through the active TM interface is insufficient to relieve cis-autoinhibition of the kinase, at least in the presence of p75 (where Trk ECD has presumably already changed to a high-affinity binding conformation). Moreover, the inactive interface residues identified in TrkA are only weakly conserved in TrkB and are not conserved in TrkC. As this receptor family has co-evolved and displays substantial functional similarities, Trk TM domain positioning by p75 is unlikely to wholly underpin how the receptors form high-affinity heterocomplexes. However, the ability of the p75 TM to affect TrkA in addition to the effect of the p75ICD could be part of a two-step activation process unique to TrkA.
We know that the ECD conformation of a high-affinity Trk receptor is different from that of a low-affinity receptor, but what of the TM and ICDs? Do these domains also adopt multiple conformations in accordance with the state of the ECD? If so, such changes may contribute to signaling specificity to produce different cellular outcomes. An important unanswered question is whether the conformational changes brought about by any of the agonists mirror those induced by the interaction between p75 and Trk. This can only be robustly tested experimentally once there is a suitable model for the allosteric modulation of Trk. Clarifying the structural changes induced by P203A and extracellular agonists and those produced by c29 and gambogic amide should assist in determining the mechanisms by which TrkA structural change and activation can be induced and modulated therapeutically.
Inside-out activation of TrkA by p75: a new model
As discussed above, the binding of ligands (including agonists) to a range of sites within the ECD of TrkA causes an ECD structural change that relieves cis-autoinhibition and propagates intracellularly to permit and/or facilitate an allosteric shift of the TrkA ICD equivalent to a 100° rotation of the kinase domain(s), orientating them to allow for autotransphosphorylation. Conversely, p75ICD-mediated modulation of Trk activation/binding-affinity likely involves rotation of the TM domain to make the ECD high-affinity binding site accessible but without relief of cis-autoinhibition of the kinase. This may be in addition to the modulation by p75 TM domain of TrkA or may follow as a reinforcement to this modulation. Importantly, it is possible that TrkA has several regulated points of rotation which, rather than (or as well as) regulating kinase activation status, regulate conformational changes to the ECD.
Compelling evidence supports two hypotheses where either the p75 TMD and/or p75ICD interact with TrkA to promote high-affinity receptor activity. Perhaps two independent mechanisms allow p75 to potentiate TrkA activity, and the occurrence of either depends on context or subcellular differences (e.g., local membrane lipid content). Alternatively, they may both be involved in a multistep process whereby TrkA is first stabilized (or localized to a specific microdomain of the plasma membrane) by p75 TM domain interaction, allowing low-affinity Trk activation which promotes p75 RIP while the receptors are still proximal. The proximally localized p75ICD can then bind through a second interaction site on Trk ICD to promote conformational changes for high-affinity binding. This particular process may be unique to TrkA as the TM domain residues involved are not highly conserved with TrkB or TrkC, and likewise TrkB TM domain modulation by cholesterol is not translatable to TrkA/C (129). However, p75ICD promotes activity in both TrkA and TrkB and is likely to be working through a conserved mechanism of action.
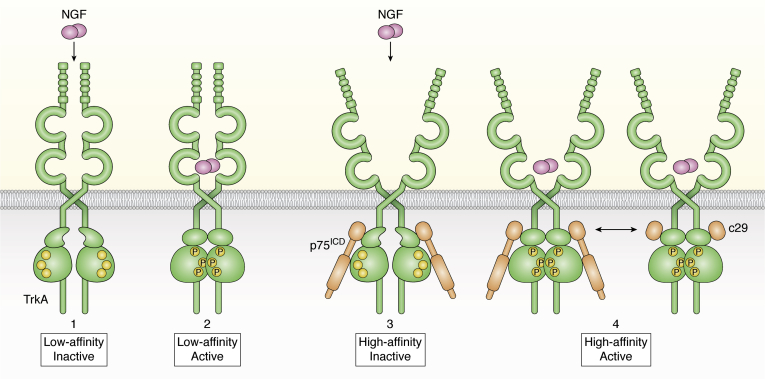
We therefore propose an “inside-out” modulation model in which the p75ICD interacts with the Trk kinase domain to propagate an allosteric conformational change to the Trk ECD, which does not cause constitutive kinase activation but rather promotes the formation of inactive receptors which are primed for high-affinity binding. This suggests that there are four possible conformations of the Trk receptor: low-affinity inactive, high-affinity primed inactive, low-affinity active, and high-affinity active. This model does not preclude the other proposed models but rather focuses particular attention on the structural aspects and ability of the juxtamembrane portion of p75ICD to promote Trk receptor allosteric modulation. Far from being a radical idea, this type of allosteric modulation has previously been reported for GnRH, Fc, Epo, and integrin receptors (98, 137, 138, 139). Reiteration of key known structural and steric contributions to TrkA activation will help to illustrate how our proposed inside-out modulation model applies to the neurotrophin receptors (Fig. 4).
-
1.
Low-affinity inactive. Inactive TrkA receptors are in a monomer–dimer equilibrium in the absence of NGF binding. Dimers interact through the LXXFAXXF motif in the TM domain, and the kinase domain occupies the DFG-out inactive conformation.
-
2.
Low-affinity active. TrkA dimers bind NGF with low affinity through D5 and assume an active dimer conformation. TrkA extracellular juxtamembrane residues are located proximally and TM domains pivot through A428, inducing a 100° rotation of the kinase domains to positions which are conducive to autotransphosphorylation. The DFG motif and activation loop are reorganized to allow for ATP and substrate binding. It appears that the active interaction interface in the TM domain now stabilizes the dimer, likely due to the rotation of the entire C-terminal portion of the receptor downstream of the proximal juxtamembrane residues.
-
3.
High-affinity inactive (primed). TrkA is complexed with p75ICD and adopts a high-affinity binding ECD conformation but remains unliganded. The stoichiometry of the p75–TrkA complex is uncertain. Trk dimers may be stabilized through the inactive LXXFAXXF motif of the TM domain, the active interaction interface or an alternate “high-affinity-receptor” interaction interface. The proximity of Trk extracellular juxtamembrane residues and the rotational status of the TrkA kinase domains in the presence of p75ICD are unknown. It is probable that the kinase domains are in an inactive arrangement, reflecting the reported lack of constitutive activity. The c29 peptide is sufficient to promote this TrkA receptor state. This structure would be an intermediatory between inactive and fully active dimers, primed for high-affinity activation.
-
4.
High-affinity active. The p75ICD–TrkA (or c29–TrkA) active complex binds NGF through high-affinity ECD sites. Allosteric modulation to the Trk ECD may (i) improve ligand access to D5, (ii) position D1 and D4 appropriately to promote ligand-D5 interaction, and/or (iii) reveal a second NGF-binding site. We postulate that TrkA extracellular juxtamembrane residues are in close proximity, an active or alternate TM interaction interface is engaged, and the kinase domain is rotated into a catalytically viable position.
Figure 4.
Inside-out model of p75ICD/c29 allosteric modulation of TrkA. In the absence of p75, TrkA exists either as monomers (not shown) or preformed yet inactive dimers (shown in 1), appropriately positioned to bind NGF with low affinity (shown in 2). Upon low-affinity NGF binding, the TrkA C-terminal domains of inactive preformed dimers rotate 100° to facilitate autotransphosphorylation at key tyrosine residues within the kinase domain of the dimer partner. Co-expression of TrkA with either the p75ICD fragment or c29 peptide is sufficient to promote allosteric modulation of TrkA to produce a primed, high-affinity receptor (shown in 3 and 4). However, this interaction does not render the receptor constitutively active. TrkA receptors are depicted in green, p75ICD and c29 in orange, NGF dimers in pink, and phosphorylated sites of the active kinase domain as a “P”. NGF, nerve growth factor; p75, p75 neurotrophin receptor; Trk, tropomyosin receptor kinase.
Having summarized the evidence for an inside-out modulation model, it is important to highlight that many questions remain. (i) What is the actual mechanism by which p75 promotes high-affinity ligand-to-Trk-receptor binding? and (ii) does the same mechanism also promote ligand-Trk receptor specificity? Identifying the intracellular p75–TrkA interaction interface will enable the structural ramifications of this interaction to be inferred. This would also provide a foundation for understanding if an allosteric modulation by p75 is required for Trk to bind ligand(s) with high affinity and allow functional questions about promoting Trk signaling in situations other than development, e.g., in the treatment of neurodegenerative diseases, to be answered. (iii) Is Trk affinity influenced by p75 TM domain and p75ICD in an additive manner? This can be tested with side-by-side comparisons of ligand affinity for TrkA when the various forms of p75 are co-expressed. (iv) Do the TM and ICD of Trk as well as the ECD adopt different conformations in the high-affinity binding receptor from that of the low-affinity Trk receptor? This particular question is challenging in light of the limitations of X-ray crystallography and would be more suitably addressed with cryo-electron microscopy. However, the TrkA structure(s) will need to be solved in the presence and absence of p75 (or suitable mimetic peptides), and solving a TM heterocomplex is not without challenges. (v) Are signaling strength and cellular outcomes influenced only by the Trk–ligand complex or also by its subcellular localization (e.g., lipid-rich domains (121, 129))? Various methods now exist to facilitate targeting of proteins to specific subcellular locations. These, in combination with super-resolution microscopy of signaling complexes, will provide correlative evidence of the localization–function relationship of Trk–ligand complexes.
We submit to the field our testable model of inside-out allosteric modulation of Trk, with the challenge that the process of verifying (or rejecting) the validity of this model will assist in answering the above outstanding questions. Given the importance of neurotrophin signaling for correct nervous system development and maintenance, there will have been significant evolutionary pressure to both maintain function of the receptor while also optimizing function of the organism. Our model suggests that having regulated steps that produce alternate Trk conformations allows for fine-tuned regulation of signaling and cellular responses to minute concentrations of neurotrophin. This information will also be crucial for future drug targeting and design to permit precise modulation of neurotrophin receptors in order to target pathologies to which disrupted neurotrophin signaling contributes, including neurodegenerative disease, chronic pain, and cancers.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank other members of the Coulson group and Dr Benjamin Luo for interesting discussions and Rowan Tweedale for critical reading and editing of the manuscript.
Author contributions
J. N. C. and E. J. C. conceptualization; J. N. C. investigation; J. N. C. data curation; J. N. C. writing-original draft; J. N. C. and E. J. C. writing-review and editing.
Funding and additional information
This work was supported by the Australian Federal Government Research Training Program Scholarships, Australia to J. N. C. and National Health and Medical Research Council, Australia funding (GRP1162505) to E. J. C.
Edited by Paul Fraser
References
- 1.Lu B., Nagappan G., Lu Y. BDNF and synaptic plasticity, cognitive function, and dysfunction. Handb. Exp. Pharmacol. 2014;220:223–250. doi: 10.1007/978-3-642-45106-5_9. [DOI] [PubMed] [Google Scholar]
- 2.Nagappan G., Lu B. Activity-dependent modulation of the BDNF receptor TrkB: Mechanisms and implications. Trends Neurosci. 2005;28:464–471. doi: 10.1016/j.tins.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Huang E.J., Reichardt L.F. Trk receptors: Roles in neuronal signal transduction. Annu. Rev. Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 4.Nykjaer A., Lee R., Teng K.K., Jansen P., Madsen P., Nielsen M.S., Jacobsen C., Kliemannel M., Schwarz E., Willnow T.E., Hempstead B.L., Petersen C.M. Sortilin is essential for proNGF-induced neuronal cell death. Nature. 2004;427:843–848. doi: 10.1038/nature02319. [DOI] [PubMed] [Google Scholar]
- 5.Cheema S.S., Barrett G.L., Bartlett P.F. Reducing p75 nerve growth factor receptor levels using antisense oligonucleotides prevents the loss of axotomized sensory neurons in the dorsal root ganglia of newborn rats. J. Neurosci. Res. 1996;46:239–245. doi: 10.1002/(SICI)1097-4547(19961015)46:2<239::AID-JNR12>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 6.Vicario A., Kisiswa L., Tann J.Y., Kelly C.E., Ibáñez C.F. Neuron-type-specific signaling by the p75NTR death receptor is regulated by differential proteolytic cleavage. J. Cell Sci. 2015;128:1507–1517. doi: 10.1242/jcs.161745. [DOI] [PubMed] [Google Scholar]
- 7.Coulson E.J., Reid K., Baca M., Shipham K.A., Hulett S.M., Kilpatrick T.J., Bartlett P.F. Chopper, a new death domain of the p75 neurotrophin receptor that mediates rapid neuronal cell death. J. Biol. Chem. 2000;275:30537–30545. doi: 10.1074/jbc.M005214200. [DOI] [PubMed] [Google Scholar]
- 8.Yoon S.O., Casaccia-Bonnefil P., Carter B., Chao M.V. Competitive signaling between TrkA and p75 nerve growth factor receptors determines cell survival. J. Neurosci. 1998;18:3273–3281. doi: 10.1523/JNEUROSCI.18-09-03273.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bibel M., Hoppe E., Barde Y.A. Biochemical and functional interactions between the neurotrophin receptors Trk and p75NTR. EMBO J. 1999;18:616–622. doi: 10.1093/emboj/18.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbacid M. The Trk family of neurotrophin receptors. J. Neurobiol. 1994;25:1386–1403. doi: 10.1002/neu.480251107. [DOI] [PubMed] [Google Scholar]
- 11.Cellerino A., Maffei L., Domenici L. The distribution of brain-derived neurotrophic factor and its receptor TrkB in parvalbumin-containing neurons of the rat visual cortex. Eur. J. Neurosci. 1996;8:1190–1197. doi: 10.1111/j.1460-9568.1996.tb01287.x. [DOI] [PubMed] [Google Scholar]
- 12.Xu B., Gottschalk W., Chow A., Wilson R.I., Schnell E., Zang K., Wang D., Nicoll R.A., Lu B., Reichardt L.F. The role of brain-derived neurotrophic factor receptors in the mature hippocampus: Modulation of long-term potentiation through a presynaptic mechanism involving TrkB. J. Neurosci. 2000;20:6888–6897. doi: 10.1523/JNEUROSCI.20-18-06888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szobota S., Mathur P.D., Siegel S., Black K., Saragovi H.U., Foster A.C. BDNF, NT-3 and Trk receptor agonist monoclonal antibodies promote neuron survival, neurite extension, and synapse restoration in rat cochlea ex vivo models relevant for hidden hearing loss. PLoS One. 2019;14 doi: 10.1371/journal.pone.0224022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sobreviela T., Clary D.O., Reichardt L.F., Brandabur M.M., Kordower J.H., Mufson E.J. TrkA-immunoreactive profiles in the central nervous system: Colocalization with neurons containing p75 nerve growth factor receptor, choline acetyltransferase, and serotonin. J. Comp. Neurol. 1994;350:587–611. doi: 10.1002/cne.903500407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wehrman T., He X., Raab B., Dukipatti A., Blau H., Garcia K.C. Structural and mechanistic insights into nerve growth factor interactions with the TrkA and p75 receptors. Neuron. 2007;53:25–38. doi: 10.1016/j.neuron.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 16.Ultsch M.H., Wiesmann C., Simmons L.C., Henrich J., Yang M., Reilly D., Bass S.H., de Vos A.M. Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC. J. Mol. Biol. 1999;290:149–159. doi: 10.1006/jmbi.1999.2816. [DOI] [PubMed] [Google Scholar]
- 17.Wiesmann C., Ultsch M.H., Bass S.H., de Vos A.M. Crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor. Nature. 1999;401:184–188. doi: 10.1038/43705. [DOI] [PubMed] [Google Scholar]
- 18.Lamballe F., Klein R., Barbacid M. The trk family of oncogenes and neurotrophin receptors. Princess Takamatsu Symp. 1991;22:153–170. [PubMed] [Google Scholar]
- 19.Vetter M.L., Martin-Zanca D., Parada L.F., Bishop J.M., Kaplan D.R. Nerve growth factor rapidly stimulates tyrosine phosphorylation of phospholipase C-gamma 1 by a kinase activity associated with the product of the trk protooncogene. Proc. Natl. Acad. Sci. U. S. A. 1991;88:5650–5654. doi: 10.1073/pnas.88.13.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan D.R., Martin-Zanca D., Parada L.F. Tyrosine phosphorylation and tyrosine kinase activity of the trk proto-oncogene product induced by NGF. Nature. 1991;350:158–160. doi: 10.1038/350158a0. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan D.R., Miller F.D. Neurotrophin signal transduction in the nervous system. Curr. Opin. Neurobiol. 2000;10:381–391. doi: 10.1016/s0959-4388(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 22.Arévalo J.C., Wu S.H. Neurotrophin signaling: Many exciting surprises! Cell. Mol. Life Sci. 2006;63:1523–1537. doi: 10.1007/s00018-006-6010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skeldal S., Coulson E.J. In: Encyclopedia of Cell Biology, 3. Bradshaw R.A., Stahl PD, editors. Vol. 3. Academic Press; Waltham, MA: 2016. Signaling and function of death receptors of the tumor necrosis factor receptor superfamily; pp. 67–75. [Google Scholar]
- 24.Underwood C.K., Coulson E.J. The p75 neurotrophin receptor. Int. J. Biochem. Cell Biol. 2008;40:1664–1668. doi: 10.1016/j.biocel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 25.Liepinsh E., Ilag L.L., Otting G., Ibáñez C.F. NMR structure of the death domain of the p75 neurotrophin receptor. EMBO J. 1997;16:4999–5005. doi: 10.1093/emboj/16.16.4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gentry J.J., Barker P.A., Carter B.D. The p75 neurotrophin receptor: Multiple interactors and numerous functions. Prog. Brain Res. 2004;146:25–39. doi: 10.1016/S0079-6123(03)46002-0. [DOI] [PubMed] [Google Scholar]
- 27.Verdi J.M., Birren S.J., Ibáñez C.F., Persson H., Kaplan D.R., Benedetti M., Chao M.V., Anderson D.J. p75LNGFR regulates Trk signal transduction and NGF-induced neuronal differentiation in MAH cells. Neuron. 1994;12:733–745. doi: 10.1016/0896-6273(94)90327-1. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka K., Kelly C.E., Goh K.Y., Lim K.B., Ibáñez C.F. Death domain signaling by disulfide-linked dimers of the p75 neurotrophin receptor mediates neuronal death in the CNS. J. Neurosci. 2016;36:5587–5595. doi: 10.1523/JNEUROSCI.4536-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roux P.P., Barker P.A. Neurotrophin signaling through the p75 neurotrophin receptor. Prog. Neurobiol. 2002;67:203–233. doi: 10.1016/s0301-0082(02)00016-3. [DOI] [PubMed] [Google Scholar]
- 30.Underwood C.K., Reid K., May L.M., Bartlett P.F., Coulson E.J. Palmitoylation of the C-terminal fragment of p75(NTR) regulates death signaling and is required for subsequent cleavage by gamma-secretase. Mol. Cell. Neurosci. 2007;37:346–358. doi: 10.1016/j.mcn.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Li Z., Lin Z., Ibáñez C.F. Evidence for the equilibrium between monomers and dimers of the death domain of the p75 neurotrophin receptor. bioRxiv. 2021 doi: 10.1101/2021.05.02.442373. [preprint] [DOI] [PubMed] [Google Scholar]
- 32.Qu Q., Chen J., Wang Y., Gui W., Wang L., Fan Z., Jiang T. Structural characterization of the self-association of the death domain of p75(NTR) PLoS One. 2013;8 doi: 10.1371/journal.pone.0057839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goncharuk S.A., Artemieva L.E., Nadezhdin K.D., Arseniev A.S., Mineev K.S. Revising the mechanism of p75NTR activation: Intrinsically monomeric state of death domains invokes the “helper” hypothesis. Sci. Rep. 2020;10:13686. doi: 10.1038/s41598-020-70721-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Z., Tann J.Y., Goh E.T., Kelly C., Lim K.B., Gao J.F., Ibanez C.F. Structural basis of death domain signaling in the p75 neurotrophin receptor. Elife. 2015;4 doi: 10.7554/eLife.11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sankorrakul K., Qian L., Thangnipon W., Coulson E.J. Is there a role for the p75 neurotrophin receptor in mediating degeneration during oxidative stress and after hypoxia? J. Neurochem. 2021;158:1292–1306. doi: 10.1111/jnc.15451. [DOI] [PubMed] [Google Scholar]
- 36.Ibáñez C.F., Simi A. p75 neurotrophin receptor signaling in nervous system injury and degeneration: Paradox and opportunity. Trends Neurosci. 2012;35:431–440. doi: 10.1016/j.tins.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Herrup K., Shooter E.M. Properties of the beta nerve growth factor receptor of avian dorsal root ganglia. Proc. Natl. Acad. Sci. U. S. A. 1973;70:3884–3888. doi: 10.1073/pnas.70.12.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang M.S., Arevalo J.C., Chao M.V. Ternary complex with Trk, p75, and an ankyrin-rich membrane spanning protein. J. Neurosci. Res. 2004;78:186–192. doi: 10.1002/jnr.20262. [DOI] [PubMed] [Google Scholar]
- 39.Hempstead B.L. The many faces of p75NTR. Curr. Opin. Neurobiol. 2002;12:260–267. doi: 10.1016/s0959-4388(02)00321-5. [DOI] [PubMed] [Google Scholar]
- 40.Barker P.A. p75NTR: A study in contrasts. Cell Death Differ. 1998;5:346–356. doi: 10.1038/sj.cdd.4400375. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y., Zeng J., Cen L., Wang X., Yao G., Wang W., Qi W., Kong K. Multiple roles of the p75 neurotrophin receptor in the nervous system. J. Int. Med. Res. 2009;37:281–288. doi: 10.1177/147323000903700201. [DOI] [PubMed] [Google Scholar]
- 42.Lee F.S., Kim A.H., Khursigara G., Chao M.V. The uniqueness of being a neurotrophin receptor. Curr. Opin. Neurobiol. 2001;11:281–286. doi: 10.1016/s0959-4388(00)00209-9. [DOI] [PubMed] [Google Scholar]
- 43.Davies A.M., Lee K.-F., Jaenisch R. p75-deficient trigeminal sensory neurons have an altered response to NGF but not to other neurotrophins. Neuron. 1993;11:565–574. doi: 10.1016/0896-6273(93)90069-4. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez-Tébar A., Dechant G., Barde Y.A. Binding of brain-derived neurotrophic factor to the nerve growth factor receptor. Neuron. 1990;4:487–492. doi: 10.1016/0896-6273(90)90107-q. [DOI] [PubMed] [Google Scholar]
- 45.Troy C.M., Friedman J.E., Friedman W.J. Mechanisms of p75-mediated death of hippocampal neurons. Role of caspases. J. Biol. Chem. 2002;277:34295–34302. doi: 10.1074/jbc.M205167200. [DOI] [PubMed] [Google Scholar]
- 46.Kenchappa R.S., Tep C., Korade Z., Urra S., Bronfman F.C., Yoon S.O., Carter B.D. p75 neurotrophin receptor-mediated apoptosis in sympathetic neurons involves a biphasic activation of JNK and up-regulation of tumor necrosis factor-alpha-converting enzyme/ADAM17. J. Biol. Chem. 2010;285:20358–20368. doi: 10.1074/jbc.M109.082834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bamji S.X., Majdan M., Pozniak C.D., Belliveau D.J., Aloyz R., Kohn J., Causing C.G., Miller F.D. The p75 neurotrophin receptor mediates neuronal apoptosis and is essential for naturally occurring sympathetic neuron death. J. Cell Biol. 1998;140:911–923. doi: 10.1083/jcb.140.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casaccia-Bonnefil P., Carter B.D., Dobrowsky R.T., Chao M.V. Death of oligodendrocytes mediated by the interaction of nerve growth factor with its receptor p75. Nature. 1996;383:716–719. doi: 10.1038/383716a0. [DOI] [PubMed] [Google Scholar]
- 49.Friedman W.J. Neurotrophins induce death of hippocampal neurons via the p75 receptor. J. Neurosci. 2000;20:6340–6346. doi: 10.1523/JNEUROSCI.20-17-06340.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matusica D., Alfonsi F., Turner B.J., Butler T.J., Shepheard S.R., Rogers M.L., Skeldal S., Underwood C.K., Mangelsdorf M., Coulson E.J. Inhibition of motor neuron death in vitro and in vivo by a p75 neurotrophin receptor intracellular domain fragment. J. Cell Sci. 2016;129:517–530. doi: 10.1242/jcs.173864. [DOI] [PubMed] [Google Scholar]
- 51.Wiese S., Metzger F., Holtmann B., Sendtner M. The role of p75NTR in modulating neurotrophin survival effects in developing motoneurons. Eur. J. Neurosci. 1999;11:1668–1676. doi: 10.1046/j.1460-9568.1999.00585.x. [DOI] [PubMed] [Google Scholar]
- 52.Domeniconi M., Hempstead B.L., Chao M.V. Pro-NGF secreted by astrocytes promotes motor neuron cell death. Mol. Cell. Neurosci. 2007;34:271–279. doi: 10.1016/j.mcn.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanning K.C., Hudson M., Amieux P.S., Wiley J.C., Bothwell M., Schecterson L.C. Proteolytic processing of the p75 neurotrophin receptor and two homologs generates C-terminal fragments with signaling capability. J. Neurosci. 2003;23:5425–5436. doi: 10.1523/JNEUROSCI.23-13-05425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee H.J., Jung K.M., Huang Y.Z., Bennett L.B., Lee J.S., Mei L., Kim T.W. Presenilin-dependent gamma-secretase-like intramembrane cleavage of ErbB4. J. Biol. Chem. 2002;277:6318–6323. doi: 10.1074/jbc.M110371200. [DOI] [PubMed] [Google Scholar]
- 55.DiStefano P.S., Johnson E.M., Jr. Identification of a truncated form of the nerve growth factor receptor. Proc. Natl. Acad. Sci. U. S. A. 1988;85:270–274. doi: 10.1073/pnas.85.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown M.S., Ye J., Rawson R.B., Goldstein J.L. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 57.Skeldal S., Matusica D., Nykjaer A., Coulson E.J. Proteolytic processing of the p75 neurotrophin receptor: A prerequisite for signalling?: Neuronal life, growth and death signalling are crucially regulated by intra-membrane proteolysis and trafficking of p75(NTR) Bioessays. 2011;33:614–625. doi: 10.1002/bies.201100036. [DOI] [PubMed] [Google Scholar]
- 58.Berg M.M., Sternberg D.W., Hempstead B.L., Chao M.V. The low-affinity p75 nerve growth factor (NGF) receptor mediates NGF-induced tyrosine phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 1991;88:7106–7110. doi: 10.1073/pnas.88.16.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greene L.A., Tischler A.S. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. U. S. A. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Landreth G.E., Shooter E.M. Nerve growth factor receptors on PC12 cells: Ligand-induced conversion from low- to high-affinity states. Proc. Natl. Acad. Sci. U. S. A. 1980;77:4751–4755. doi: 10.1073/pnas.77.8.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hempstead B.L., Martin-Zanca D., Kaplan D.R., Parada L.F., Chao M.V. High-affinity NGF binding requires coexpression of the trk proto-oncogene and the low-affinity NGF receptor. Nature. 1991;350:678–683. doi: 10.1038/350678a0. [DOI] [PubMed] [Google Scholar]
- 62.Mahadeo D., Kaplan L., Chao M.V., Hempstead B.L. High affinity nerve growth factor binding displays a faster rate of association than p140trk binding. Implications for multi-subunit polypeptide receptors. J. Biol. Chem. 1994;269:6884–6891. [PubMed] [Google Scholar]
- 63.Radeke M.J., Misko T.P., Hsu C., Herzenberg L.A., Shooter E.M. Gene transfer and molecular cloning of the rat nerve growth factor receptor. Nature. 1987;325:593–597. doi: 10.1038/325593a0. [DOI] [PubMed] [Google Scholar]
- 64.Gargano N., Levi A., Alema S. Modulation of nerve growth factor internalization by direct interaction between p75 and TrkA receptors. J. Neurosci. Res. 1997;50:1–12. doi: 10.1002/(SICI)1097-4547(19971001)50:1<1::AID-JNR1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 65.Sutter A., Riopelle R.J., Harris-Warrick R.M., Shooter E.M. Nerve growth factor receptors. Characterization of two distinct classes of binding sites on chick embryo sensory ganglia cells. J. Biol. Chem. 1979;254:5972–5982. [PubMed] [Google Scholar]
- 66.Rodríguez-Tébar A., Dechant G., Götz R., Barde Y.A. Binding of neurotrophin-3 to its neuronal receptors and interactions with nerve growth factor and brain-derived neurotrophic factor. EMBO J. 1992;11:917–922. doi: 10.1002/j.1460-2075.1992.tb05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barker P.A., Shooter E.M. Disruption of NGF binding to the low affinity neurotrophin receptor p75LNTR reduces NGF binding to TrkA on PC12 cells. Neuron. 1994;13:203–215. doi: 10.1016/0896-6273(94)90470-7. [DOI] [PubMed] [Google Scholar]
- 68.Matusica D., Skeldal S., Sykes A.M., Palstra N., Sharma A., Coulson E.J. An intracellular domain fragment of the p75 neurotrophin receptor (p75(NTR)) enhances tropomyosin receptor kinase A (TrkA) receptor function. J. Biol. Chem. 2013;288:11144–11154. doi: 10.1074/jbc.M112.436469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hantzopoulos P.A., Suri C., Glass D.J., Goldfarb M.P., Yancopoulos G.D. The low affinity NGF receptor, p75, can collaborate with each of the Trks to potentiate functional responses to the neurotrophins. Neuron. 1994;13:187–201. doi: 10.1016/0896-6273(94)90469-3. [DOI] [PubMed] [Google Scholar]
- 70.Lee K.F., Davies A.M., Jaenisch R. p75-deficient embryonic dorsal root sensory and neonatal sympathetic neurons display a decreased sensitivity to NGF. Development. 1994;120:1027–1033. doi: 10.1242/dev.120.4.1027. [DOI] [PubMed] [Google Scholar]
- 71.Yamashita T., Tucker K.L., Barde Y.A. Neurotrophin binding to the p75 receptor modulates Rho activity and axonal outgrowth. Neuron. 1999;24:585–593. doi: 10.1016/s0896-6273(00)81114-9. [DOI] [PubMed] [Google Scholar]
- 72.Deppmann C.D., Mihalas S., Sharma N., Lonze B.E., Niebur E., Ginty D.D. A model for neuronal competition during development. Science. 2008;320:369–373. doi: 10.1126/science.1152677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benedetti M., Levi A., Chao M.V. Differential expression of nerve growth factor receptors leads to altered binding affinity and neurotrophin responsiveness. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7859–7863. doi: 10.1073/pnas.90.16.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mischel P.S., Smith S.G., Vining E.R., Valletta J.S., Mobley W.C., Reichardt L.F. The extracellular domain of p75NTR is necessary to inhibit neurotrophin-3 signaling through TrkA. J. Biol. Chem. 2001;276:11294–11301. doi: 10.1074/jbc.M005132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brennan C., Rivas-Plata K., Landis S.C. The p75 neurotrophin receptor influences NT-3 responsiveness of sympathetic neurons in vivo. Nat. Neurosci. 1999;2:699–705. doi: 10.1038/11158. [DOI] [PubMed] [Google Scholar]
- 76.Vesa J., Kruttgen A., Shooter E.M. p75 reduces TrkB tyrosine autophosphorylation in response to brain-derived neurotrophic factor and neurotrophin 4/5. J. Biol. Chem. 2000;275:24414–24420. doi: 10.1074/jbc.M001641200. [DOI] [PubMed] [Google Scholar]
- 77.Chao M.V., Hempstead B.L. p75 and Trk: A two-receptor system. Trends Neurosci. 1995;18:321–326. [PubMed] [Google Scholar]
- 78.He X.L., Garcia K.C. Structure of nerve growth factor complexed with the shared neurotrophin receptor p75. Science. 2004;304:870–875. doi: 10.1126/science.1095190. [DOI] [PubMed] [Google Scholar]
- 79.Wiesmann C., de Vos A.M. Nerve growth factor: Structure and function. Cell. Mol. Life Sci. 2001;58:748–759. doi: 10.1007/PL00000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Auernhammer C.J., Kopp F.B., Vlotides G., Dorn F., Isele N.B., Spöttl G., Cengic N., Weber M.M., Senaldi G., Engelhardt D. Comparative study of gp130 cytokine effects on corticotroph AtT-20 cells--redundancy or specificity of neuroimmunoendocrine modulators? Neuroimmunomodulation. 2004;11:224–232. doi: 10.1159/000078440. [DOI] [PubMed] [Google Scholar]
- 81.Huber L.J., Chao M.V. A potential interaction of p75 and trkA NGF receptors revealed by affinity crosslinking and immunoprecipitation. J. Neurosci. Res. 1995;40:557–563. doi: 10.1002/jnr.490400415. [DOI] [PubMed] [Google Scholar]
- 82.Ross A.H., Daou M.C., McKinnon C.A., Condon P.J., Lachyankar M.B., Stephens R.M., Kaplan D.R., Wolf D.E. The neurotrophin receptor, gp75, forms a complex with the receptor tyrosine kinase TrkA. J. Cell Biol. 1996;132:945–953. doi: 10.1083/jcb.132.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aurikko J.P., Ruotolo B.T., Grossmann J.G., Moncrieffe M.C., Stephens E., Leppänen V.M., Robinson C.V., Saarma M., Bradshaw R.A., Blundell T.L. Characterization of symmetric complexes of nerve growth factor and the ectodomain of the pan-neurotrophin receptor, p75NTR. J. Biol. Chem. 2005;280:33453–33460. doi: 10.1074/jbc.M503189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gong Y., Cao P., Yu H.J., Jiang T. Crystal structure of the neurotrophin-3 and p75NTR symmetrical complex. Nature. 2008;454:789–793. doi: 10.1038/nature07089. [DOI] [PubMed] [Google Scholar]
- 85.Barker P.A. High affinity not in the vicinity? Neuron. 2007;53:1–4. doi: 10.1016/j.neuron.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 86.Rydén M., Hempstead B., Ibáñez C.F. Differential modulation of neuron survival during development by nerve growth factor binding to the p75 neurotrophin receptor. J. Biol. Chem. 1997;272:16322–16328. doi: 10.1074/jbc.272.26.16322. [DOI] [PubMed] [Google Scholar]
- 87.Horton A., Laramee G., Wyatt S., Shih A., Winslow J., Davies A.M. NGF binding to p75 enhances the sensitivity of sensory and sympathetic neurons to NGF at different stages of development. Mol. Cell. Neurosci. 1997;10:162–172. doi: 10.1006/mcne.1997.0650. [DOI] [PubMed] [Google Scholar]
- 88.Hempstead B.L., Patil N., Thiel B., Chao M.V. Deletion of cytoplasmic sequences of the nerve growth factor receptor leads to loss of high affinity ligand binding. J. Biol. Chem. 1990;265:9595–9598. [PubMed] [Google Scholar]
- 89.Esposito D., Patel P., Stephens R.M., Perez P., Chao M.V., Kaplan D.R., Hempstead B.L. The cytoplasmic and transmembrane domains of the p75 and Trk A receptors regulate high affinity binding to nerve growth factor. J. Biol. Chem. 2001;276:32687–32695. doi: 10.1074/jbc.M011674200. [DOI] [PubMed] [Google Scholar]
- 90.Ceni C., Kommaddi R.P., Thomas R., Vereker E., Liu X., McPherson P.S., Ritter B., Barker P.A. The p75NTR intracellular domain generated by neurotrophin-induced receptor cleavage potentiates Trk signaling. J. Cell Sci. 2010;123:2299–2307. doi: 10.1242/jcs.062612. [DOI] [PubMed] [Google Scholar]
- 91.Kommaddi R.P., Thomas R., Ceni C., Daigneault K., Barker P.A. Trk-dependent ADAM17 activation facilitates neurotrophin survival signaling. FASEB J. 2011;25:2061–2070. doi: 10.1096/fj.10-173740. [DOI] [PubMed] [Google Scholar]
- 92.Zampieri N., Xu C.F., Neubert T.A., Chao M.V. Cleavage of p75 neurotrophin receptor by alpha-secretase and gamma-secretase requires specific receptor domains. J. Biol. Chem. 2005;280:14563–14571. doi: 10.1074/jbc.M412957200. [DOI] [PubMed] [Google Scholar]
- 93.Yan H., Schlessinger J., Chao M.V. Chimeric NGF-EGF receptors define domains responsible for neuronal differentiation. Science. 1991;252:561–563. doi: 10.1126/science.1850551. [DOI] [PubMed] [Google Scholar]
- 94.Shen J., Maruyama I.N. Nerve growth factor receptor TrkA exists as a preformed, yet inactive, dimer in living cells. FEBS Lett. 2011;585:295–299. doi: 10.1016/j.febslet.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 95.Franco M.L., Nadezhdin K.D., Goncharuk S.A., Mineev K.S., Arseniev A.S., Vilar M. Structural basis of the transmembrane domain dimerization and rotation in the activation mechanism of the TRKA receptor by nerve growth factor. J. Biol. Chem. 2020;295:275–286. doi: 10.1074/jbc.RA119.011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hubbard S.R., Till J.H. Protein tyrosine kinase structure and function. Annu. Rev. Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- 97.Huse M., Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 98.Jiang G., Hunter T. Receptor signaling: When dimerization is not enough. Curr. Biol. 1999;9:R568–R571. doi: 10.1016/s0960-9822(99)80357-1. [DOI] [PubMed] [Google Scholar]
- 99.Wybenga-Groot L.E., Baskin B., Ong S.H., Tong J., Pawson T., Sicheri F. Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region. Cell. 2001;106:745–757. doi: 10.1016/s0092-8674(01)00496-2. [DOI] [PubMed] [Google Scholar]
- 100.Bertrand T., Kothe M., Liu J., Dupuy A., Rak A., Berne P.F., Davis S., Gladysheva T., Valtre C., Crenne J.Y., Mathieu M. The crystal structures of TrkA and TrkB suggest key regions for achieving selective inhibition. J. Mol. Biol. 2012;423:439–453. doi: 10.1016/j.jmb.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 101.Hanks S.K., Quinn A.M., Hunter T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 102.Ahmed F., Hristova K. Dimerization of the trk receptors in the plasma membrane: Effects of their cognate ligands. Biochem. J. 2018;475:3669–3685. doi: 10.1042/BCJ20180637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hubbard S.R. Theme and variations: Juxtamembrane regulation of receptor protein kinases. Mol. Cell. 2001;8:481–482. doi: 10.1016/s1097-2765(01)00350-1. [DOI] [PubMed] [Google Scholar]
- 105.Seubert N., Royer Y., Staerk J., Kubatzky K.F., Moucadel V., Krishnakumar S., Smith S.O., Constantinescu S.N. Active and inactive orientations of the transmembrane and cytosolic domains of the erythropoietin receptor dimer. Mol. Cell. 2003;12:1239–1250. doi: 10.1016/s1097-2765(03)00389-7. [DOI] [PubMed] [Google Scholar]
- 106.Remy I., Wilson I.A., Michnick S.W. Erythropoietin receptor activation by a ligand-induced conformation change. Science. 1999;283:990–993. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- 107.Lu X., Gross A.W., Lodish H.F. Active conformation of the erythropoietin receptor: Random and cysteine-scanning mutagenesis of the extracellular juxtamembrane and transmembrane domains. J. Biol. Chem. 2006;281:7002–7011. doi: 10.1074/jbc.M512638200. [DOI] [PubMed] [Google Scholar]
- 108.Tanaka H., Sase H., Tsukaguchi T., Hasegawa M., Tanimura H., Yoshida M., Sakata K., Fujii T., Tachibana Y., Takanashi K., Higashida A., Hasegawa K., Ono Y., Oikawa N., Mio T. Selective TRK inhibitor CH7057288 against TRK fusion-driven cancer. Mol. Cancer Ther. 2018;17:2519–2529. doi: 10.1158/1535-7163.MCT-17-1180. [DOI] [PubMed] [Google Scholar]
- 109.Bagal S.K., Andrews M., Bechle B.M., Bian J., Bilsland J., Blakemore D.C., Braganza J.F., Bungay P.J., Corbett M.S., Cronin C.N., Cui J.J., Dias R., Flanagan N.J., Greasley S.E., Grimley R., et al. Discovery of potent, selective, and peripherally restricted pan-Trk kinase inhibitors for the treatment of pain. J. Med. Chem. 2018;61:6779–6800. doi: 10.1021/acs.jmedchem.8b00633. [DOI] [PubMed] [Google Scholar]
- 110.Choi H.S., Rucker P.V., Wang Z., Fan Y., Albaugh P., Chopiuk G., Gessier F., Sun F., Adrian F., Liu G., Hood T., Li N., Jia Y., Che J., McCormack S., et al. (R)-2-Phenylpyrrolidine substituted imidazopyridazines: A new class of potent and selective pan-TRK inhibitors. ACS Med. Chem. Lett. 2015;6:562–567. doi: 10.1021/acsmedchemlett.5b00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Watson F.L., Porcionatto M.A., Bhattacharyya A., Stiles C.D., Segal R.A. TrkA glycosylation regulates receptor localization and activity. J. Neurobiol. 1999;39:323–336. doi: 10.1002/(sici)1097-4695(199905)39:2<323::aid-neu15>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 112.Segal R.A., Bhattacharyya A., Rua L.A., Alberta J.A., Stephens R.M., Kaplan D.R., Stiles C.D. Differential utilization of Trk autophosphorylation sites. J. Biol. Chem. 1996;271:20175–20181. doi: 10.1074/jbc.271.33.20175. [DOI] [PubMed] [Google Scholar]
- 113.Obermeier A., Halfter H., Wiesmüller K.H., Jung G., Schlessinger J., Ullrich A. Tyrosine 785 is a major determinant of Trk--substrate interaction. EMBO J. 1993;12:933–941. doi: 10.1002/j.1460-2075.1993.tb05734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Obermeier A., Lammers R., Wiesmüller K.H., Jung G., Schlessinger J., Ullrich A. Identification of Trk binding sites for SHC and phosphatidylinositol 3'-kinase and formation of a multimeric signaling complex. J. Biol. Chem. 1993;268:22963–22966. [PubMed] [Google Scholar]
- 115.Loeb D.M., Stephens R.M., Copeland T., Kaplan D.R., Greene L.A. A Trk nerve growth factor (NGF) receptor point mutation affecting interaction with phospholipase C-gamma 1 abolishes NGF-promoted peripherin induction but not neurite outgrowth. J. Biol. Chem. 1994;269:8901–8910. [PubMed] [Google Scholar]
- 116.Stephens R.M., Loeb D.M., Copeland T.D., Pawson T., Greene L.A., Kaplan D.R. Trk receptors use redundant signal transduction pathways involving SHC and PLC-gamma 1 to mediate NGF responses. Neuron. 1994;12:691–705. doi: 10.1016/0896-6273(94)90223-2. [DOI] [PubMed] [Google Scholar]
- 117.Bell C.A., Tynan J.A., Hart K.C., Meyer A.N., Robertson S.C., Donoghue D.J. Rotational coupling of the transmembrane and kinase domains of the Neu receptor tyrosine kinase. Mol. Biol. Cell. 2000;11:3589–3599. doi: 10.1091/mbc.11.10.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brown R.J., Adams J.J., Pelekanos R.A., Wan Y., McKinstry W.J., Palethorpe K., Seeber R.M., Monks T.A., Eidne K.A., Parker M.W., Waters M.J. Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat. Struct. Mol. Biol. 2005;12:814–821. doi: 10.1038/nsmb977. [DOI] [PubMed] [Google Scholar]
- 119.Maruyama I.N. Activation of transmembrane cell-surface receptors via a common mechanism? The “rotation model”. Bioessays. 2015;37:959–967. doi: 10.1002/bies.201500041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Arevalo J.C., Conde B., Hempstead B.L., Chao M.V., Martin-Zanca D., Perez P. TrkA immunoglobulin-like ligand binding domains inhibit spontaneous activation of the receptor. Mol. Cell. Biol. 2000;20:5908–5916. doi: 10.1128/mcb.20.16.5908-5916.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Matusica D., Coulson E.J. Local versus long-range neurotrophin receptor signalling: Endosomes are not just carriers for axonal transport. Semin. Cell Dev. Biol. 2014;31:57–63. doi: 10.1016/j.semcdb.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 122.Obianyo O., Ye K. Novel small molecule activators of the Trk family of receptor tyrosine kinases. Biochim. Biophys. Acta. 2013;1834:2213–2218. doi: 10.1016/j.bbapap.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saragovi H.U., Galan A., Levin L.A. Neuroprotection: Pro-survival and anti-neurotoxic mechanisms as therapeutic strategies in neurodegeneration. Front. Cell. Neurosci. 2019;13:231. doi: 10.3389/fncel.2019.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jang S.W., Liu X., Yepes M., Shepherd K.R., Miller G.W., Liu Y., Wilson W.D., Xiao G., Blanchi B., Sun Y.E., Ye K. A selective TrkB agonist with potent neurotrophic activities by 7,8-dihydroxyflavone. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2687–2692. doi: 10.1073/pnas.0913572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.LeSauteur L., Maliartchouk S., Le Jeune H., Quirion R., Saragovi H.U. Potent human p140-TrkA agonists derived from an anti-receptor monoclonal antibody. J. Neurosci. 1996;16:1308–1316. doi: 10.1523/JNEUROSCI.16-04-01308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maliartchouk S., Feng Y., Ivanisevic L., Debeir T., Cuello A.C., Burgess K., Saragovi H.U. A designed peptidomimetic agonistic ligand of TrkA nerve growth factor receptors. Mol. Pharmacol. 2000;57:385–391. [PubMed] [Google Scholar]
- 127.Jang S.W., Liu X., Chan C.B., Weinshenker D., Hall R.A., Xiao G., Ye K. Amitriptyline is a TrkA and TrkB receptor agonist that promotes TrkA/TrkB heterodimerization and has potent neurotrophic activity. Chem. Biol. 2009;16:644–656. doi: 10.1016/j.chembiol.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bai Y., Xu J., Brahimi F., Zhuo Y., Sarunic M.V., Saragovi H.U. An agonistic TrkB mAb causes sustained TrkB activation, delays RGC death, and protects the retinal structure in optic nerve axotomy and in glaucoma. Invest. Ophthalmol. Vis. Sci. 2010;51:4722–4731. doi: 10.1167/iovs.09-5032. [DOI] [PubMed] [Google Scholar]
- 129.Casarotto P.C., Girych M., Fred S.M., Kovaleva V., Moliner R., Enkavi G., Biojone C., Cannarozzo C., Sahu M.P., Kaurinkoski K., Brunello C.A., Steinzeig A., Winkel F., Patil S., Vestring S., et al. Antidepressant drugs act by directly binding to TRKB neurotrophin receptors. Cell. 2021;184:1299–1313. doi: 10.1016/j.cell.2021.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Barker P.A., Barbee G., Misko T.P., Shooter E.M. The low affinity neurotrophin receptor, p75LNTR, is palmitoylated by thioester formation through cysteine 279. J. Biol. Chem. 1994;269:30645–30650. [PubMed] [Google Scholar]
- 131.Fukata Y., Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 2010;11:161–175. doi: 10.1038/nrn2788. [DOI] [PubMed] [Google Scholar]
- 132.Jang S.W., Okada M., Sayeed I., Xiao G., Stein D., Jin P., Ye K. Gambogic amide, a selective agonist for TrkA receptor that possesses robust neurotrophic activity, prevents neuronal cell death. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16329–16334. doi: 10.1073/pnas.0706662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Arevalo J.C., Conde B., Hempstead B.I., Chao M.V., Martín-Zanca D., Pérez P. A novel mutation within the extracellular domain of TrkA causes constitutive receptor activation. Oncogene. 2001;20:1229–1234. doi: 10.1038/sj.onc.1204215. [DOI] [PubMed] [Google Scholar]
- 134.Zaccaro M.C., Ivanisevic L., Perez P., Meakin S.O., Saragovi H.U. p75 co-receptors regulate ligand-dependent and ligand-independent Trk receptor activation, in part by altering Trk docking subdomains. J. Biol. Chem. 2001;276:31023–31029. doi: 10.1074/jbc.M104630200. [DOI] [PubMed] [Google Scholar]
- 135.Franco M.L., Nadezhdin K.D., Light T.P., Goncharuk S.A., Soler-Lopez A., Ahmed F., Mineev K.S., Hristova K., Arseniev A.S., Vilar M. Interaction between the transmembrane domains of neurotrophin receptors p75 and TrkA mediates their reciprocal activation. J. Biol. Chem. 2021;297:100926. doi: 10.1016/j.jbc.2021.100926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sykes A.M., Palstra N., Abankwa D., Hill J.M., Skeldal S., Matusica D., Venkatraman P., Hancock J.F., Coulson E.J. The effects of transmembrane sequence and dimerization on cleavage of the p75 neurotrophin receptor by γ-secretase. J. Biol. Chem. 2012;287:43810–43824. doi: 10.1074/jbc.M112.382903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Margadant C., Monsuur H.N., Norman J.C., Sonnenberg A. Mechanisms of integrin activation and trafficking. Curr. Opin. Cell Biol. 2011;23:607–614. doi: 10.1016/j.ceb.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 138.Millar R.P., Pawson A.J. Outside-in and inside-out signaling: The new concept that selectivity of ligand binding at the gonadotropin-releasing hormone receptor is modulated by the intracellular environment. Endocrinology. 2004;145:3590–3593. doi: 10.1210/en.2004-0461. [DOI] [PubMed] [Google Scholar]
- 139.Koenderman L. Inside-out control of fc-receptors. Front. Immunol. 2019;10:544. doi: 10.3389/fimmu.2019.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]