Summary
Background
Plasma extracellular vesicles (pEV) can harbor a diverse array of factors including active proteases and the amyloid-precursor-protein (APP) cleavage product Aβ, involved in plaque formation in Alzheimer`s diseases (AD). A potential role of such vesicles in AD pathology is unexplored.
Methods
In a case-control study of randomly selected patients with AD and other neurological diseases (n = 14), and healthy controls (n = 7), we systematically analyzed the content of pEV, using different assay systems. In addition, we determined their entry path into brain tissue, employing animal (mice) injection experiments with ex vivo generated EV that were similar to AD-pEV, followed by multi antigen analysis (MAA) of brain tissue (n = 4 per condition). The results were compared with an IHC staining of human brain tissue in a small cohort of AD patients (n = 3) and controls with no neurodegenerative diseases (n = 3).
Findings
We show that pEV levels are considerably upregulated in AD patients. Besides numerous inflammatory effectors, AD-pEV contained α-, β- and γ-secretases, able to cleave APP in in target cells. In vitro generated EV with similar characteristics as AD-pEV accumulated in the choroid plexus (CP) of injected animals and reached primarily hippocampal neurons. Corroborating findings were made in human brain samples. An inhibitor of hyaluronic-acid-synthetase (HAS) blocked uploading of proteases and Hyaluronan onto EV in vitro and abolished CP targeting in animal injection experiments.
Interpretation
We conclude that protease-containing pEV could be part of a communication axis between the periphery and the brain that could be become detrimental depending on pEV concentration and duration of target cell impact.
Keywords: Alzheimer's disease, Plasma extracellular vesicles, Choroid plexus, Hippocampus, Hyaluronan, Hyaluronic acid synthetase, Secretases, BACE, Aβ, APP
Research in context.
Evidence before this study
In 2002 it was firstly demonstrated that extracellular vesicles (EV), secreted by a cancer cell line, contain a protease, namely ADAM10, cleaving a substrate molecule in vesicles. In 2013 we detected the presence of ADAM10 and ADAM17 in plasma extracellular vesicles (pEV) from HIV and melanoma patients in a clinical stage-dependent manner, implying that the appearance of protease-containing pEV could correlate with disease development. In 2016 we demonstrated a potential function for pEV-contained proteases, showing that they can cleave precursor proteins in target cells. This suggested, that EV-associated proteases may serve to amplify cleavage events in the cellular environment, but also in distant target cells. Unrelated to these findings, the presence of the amyloid precursor protein (APP) cleavage product Aβ had been previously demonstrated in EV and pEV of Alzheimer's patients. These vesicles are currently assumed to derive from diseased brain tissue.
Added value of this study
In this study we extend and combine these previous observations, demonstrating a potential mechanism by which protease-containing pEV from the periphery may reach the brain, eventually contributing to the cleavage of substrate proteins like APP. By using an animal model, we demonstrate how and where these vesicles reach brain areas, previously associated with early Alzheimer pathology. Supporting this experimental findings, we present preliminary evidence for the presence of pEV-associated proteins in the limbic system of Alzheimer patients. Together our results suggests a hitherto unrecognized communication axis between the periphery and the brain through pEV that may have detrimental effects under non-physiological conditions.
Implications of all the available evidence
By demonstrating how protease-containing pEV can enter the limbic system, our findings delineate a possible pathway, by which peripheral inflammation can reach the brain, specifically the hippocampus. This is of importance as inflammation is believed to be a crucial factor in the development of many neurological and neurodegenerative diseases. However, additional studies have to confirm whether this mechanism has indeed a role in this context.
Alt-text: Unlabelled box
Introduction
Altered degradation of the amyloid precursor protein (APP) and/or the Aβ peptide may lead to the accumulation of amyloid plaques, a hallmark in the pathogenesis of AD.1, 2, 3 This process involves several proteases, including the aspartyl protease or β-secretase (BACE).4,5 Physiologically, APP is cleaved close to the outer plasma membrane by an α-secretase, likely ADAM10 or ADAM17.6,7 A second protease, γ-secretase, cleaves APP within the plasma membrane, liberating a small peptide in-between the cuts of both proteases. Under certain conditions the extracellular cleavage is made more distant from the membrane by BACE.8 Hence, the resulting fragment is somewhat longer (40–42 AA) and called amyloid-beta or Aβ, the main constituent of amyloid plaques.9,10 For reasons that are not clear, Aβ is also found in plasma extracellular vesicles (pEV) of AD patients.11
Secretases are membrane-associated enzymes. However, they are also packaged into extracellular vesicles (EV), as for example in Cancer- and HIV-infected cells, which appear as pEV in circulation.12, 13, 14, 15, 16, 17 For EV-associated ADAM17, we previously suggested a function, analyzing the processing of its substrate pro-TNF.12,18 After activation, ADAM17 first cleaved plasma membrane-associated pro-TNF, before it was endocytosed into endosomal compartments processing internal storage of pro-TNF. From there mature TNF and active ADAM17, both packaged into EV, were secreted. Such protease-containing EV/pEV could enter target cells and process more substrate. Hence, protease-containing EV may serve to maximize substrate processing locally and in distant tissue compartments, e.g. in a state of immune defense.
Model systems have demonstrated that EV/pEV can cross the blood brain barrier (BBB) by endocytosis after binding to endothelial cells, in part through receptors like integrins (α5 and α6) and CD46.19,20 However, their transition into brain tissue remains unclear. EV are also able to cross the BBB into the bloodstream,21 and brain-specific adhesion molecules (e.g. L1CAM) are used to isolate these neurally-derived pEV (NDEV) from plasma.22 However, this L1CAM-dependent procedure has been disputed recently.23 In diseased patients, NDEV contain typical biomarkers of Parkinson and AD, including α-synuclein, BACE, p-Tau and Aβ.24, 25, 26, 27 These findings supported assumptions that brain-derived EV may contribute to the dissemination of neurodegeneration to other brain areas.20,28,29 Likewise it was suggested that EV may disseminate inflammation in this manner. Inflammatory stimuli, like LPS, were shown to induce the transfer of pEV to the brain30 and stimulate choroid plexus epithelium cells for increased secretion of miRNA-containing EV.31 However, these mechanisms are currently not considered as being relevant for AD pathology.29,32
In this study, we speculated that active proteases would be present in AD-pEV of peripheral origin, able to reach brain tissue, eventually leading to the cleavage of APP and other substrates in target neurons. We provide evidence that supports this assumption, demonstrating that such pEV, containing a broad range of different proteases, can reach hippocampal neurons, potentially exerting detrimental effects when reaching target cells at high levels for extended time periods.
Methods
Study design and participants
The analysis of plasma samples and human tissue was carried out from July 2017 to December 2019. All samples from AD patients (n = 10), were obtained from the Department of Psychiatry after a standard set of diagnostic tests had been performed, collectively termed CERAD (consortium to establish registry for Alzheimer's disease). Additional samples were obtained from patients with established clinical diagnoses of depression, schizophrenia and cognitive impairment (n = 4). Since both, healthy and neurological controls had no pEV-associated protease activity (with one exception, Figure S1e), and only low levels of inflammatory factors (Figure 1f), we did not analyze more controls. The exclusion criteria for this study were: significant additional diseases as for example cancer, autoimmune diseases and acute or chronic infections, or patients where such information was not available. Age-matched healthy controls (n = 7) were obtained from the Department of Transfusion Medicine (Blood Bank) of the University clinic.
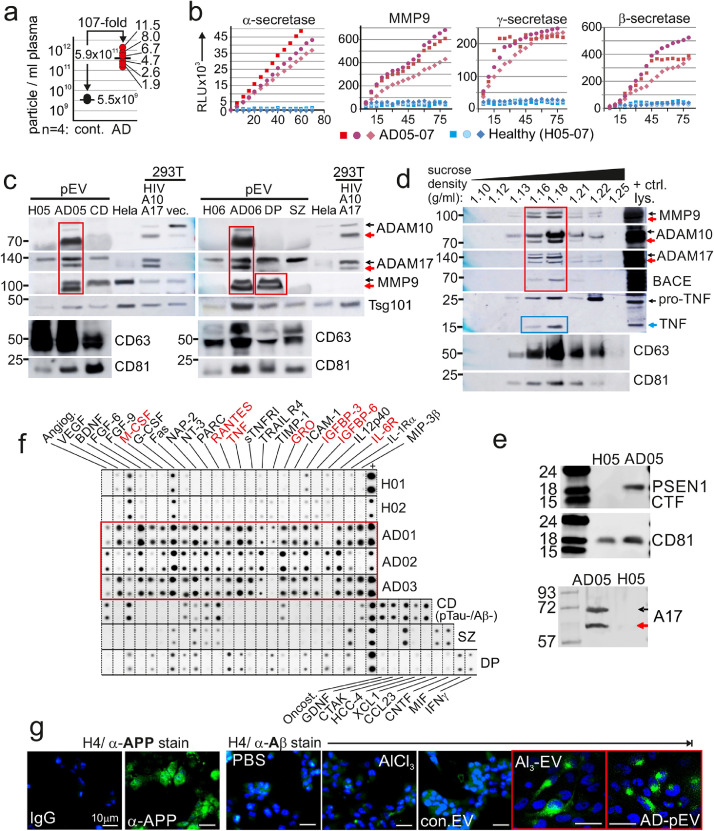
Figure 1.
Abundance of protease-containing pEV in patients with AD.
(a) Concentration of particles in plasma of 6 AD patients and 4 age-matched controls by single particle tracking (ZetaView®). Measurements were repeated 3-times for each individual. (b) Representative plots of protease activities in pEV of AD patients (AD05-07) and healthy controls (H05-07). Aliquots corresponding to 0.3 ml plasma were analyzed at least twice. (c) Immunoblot of protease fragments in pEV (corresponding to 1 ml plasma) of AD patients and controls. HeLa and 293T cells transfected with HIVΔenv and ADAM17/10 (A17/A10)12,13 or vector (vec.) served as controls. (d) Immunoblot of proteases in sucrose gradient fractions of AD-pEV (from AD07). Control lysates (+ctrl. lys.) as in (c). (e) Immunoblot of Presenilin 1 (CTF: C-terminal fragment) on pEV lysates of AD05 and a healthy control. (f) Protein arrays assessing cytokines, chemokines and soluble factors (CCF) in pEV from indicated patients. (g) Representative staining of APP and Aβ in H4 glioma cells after incubation with Al3-EV and AD-pEV (AD09).
Patient material
Plasma
Blood samples from psychiatric patients were obtained from the Depatment of Psychiatry. Blood was drawn from patients after informed consent. In general, 20–30 ml of blood was obtained from each individual using 10 ml Vacutainers (BD). Blood samples were transferred to the lab within 1 h and Plasma was obtained after centrifugation of samples for 10 min at 1250 g. For clinical details of patients see Supplement Table S1. Human FFPE Tissue. Brain tissue samples (paraffin embedded tissue blocks (FFPE)) from deceased AD patients and controls were obtained from R. Coras (author) from the Department of Neuropathology of the University Clinic Erlangen. For clinical details see Supplement Table S2.
Ethics statement
The Erlangen University ethics committee approved the study in 2017 under the title (translated from German into English): “Immunological and cell-based diagnosis of dementia” (#3987). This included the analysis of tissue. Samples from healthy donors and were obtained from the local Blood Bank (Abteilung für Transfusionsmedizin und Hämostaseologie, Universitätsklinikum Erlangen), where all participants provided informed consent and permission for their blood to be used for scientific research. It was not possible for us to trace data from the donors (age, gender or any other). Mouse injection experiments. All experiments were performed in accordance with European Communities Council Directive (86/609/EEC), and were approved by the local ethics committee (Government of Middle Franconia, Germany).
Cell lines
All cell lines used are commercially available and hence were validated. Liver cell lines Huh7 (Human hepatocellular carcinoma cells, CLS Cat# 300156/p7178_HuH7, RRID: CVCL_0336) and Sk-Hep-1 (human hepatic adenocarcinoma cells, RRID:CVCL_0525) were kindly provided by P. Knolle (Technische Universität München). LX-2 (human hepatic stellate cells, RRID: CVCL_5792) were provided by SL. Friedman (Icahn School of Medicine). HEK293T (human embryonic kidney cells, RRID: CVCL_QW54), THP-1 (human monocytic cells, CLS Cat# 300356/p804_THP-1, RRID:CVCL_0006), Jurkat (human immortalized T lymphocyte cells, CLS Cat# 300223/p849_Jurkat_E61, RRID:CVCL_0367) and H4 (human brain neuroglioma cells were from ATCC (CLS Cat# 300184/p564_H4, RRID:CVCL_1239). For all cell lines were performed a mycoplasma PCR test at different times in order to detect mycoplasma contamination. The results were negative. Huh7, HEK293T, H4 cells were grown in DMEM (Sigma-Aldrich) supplemented with 10% Fetal calf serum (FCS, Sigma-Aldrich) and 1% penicillinstreptomycin (Lonza). Sk-Hep1 cells were additionally maintained in 40 µM β-mercaptoethanol (Carl Roth). THP-1 and Jurkat cells were maintained in Roswell Park Memorial Institute Medium (RPMI, Gibco) supplemented with 10% (v/v) FCS (Sigma-Aldrich), 2 mM GlutaMAX (Gibco), 1 mM sodium pyruvate (Gibco), and 100 U/mL penicillin-streptomycin (Lonza). LX-2 were cultured in DMEM high glucose (Life Technologies) supplemented with 2% FCS, 1% penicillin-streptomycin. FL83 cells were maintained in Ham's F-12K (Kaighn's) Medium supplemented with 10% FCS (Sigma-Aldrich) and 1% penicillinstreptomycin (Lonza). All cells were grown at 37 °C under 5% CO2.
Antibodies
Only commercially available and therefore validated antibodies were used. The following antibodies were used for immunostaining, flow cytometry or immunoblotting: anti-ADAM10 (mouse monoclonal, Abcam Cat# ab73402, RRID:AB_10562808), anti-ADAM10-PE (mouse monoclonal, BioLegend Cat# 352704, RRID:AB_10896790), anti-ADAM17 (rabbit polyclonal, Cell Signaling Technology Cat# 3976, RRID:AB_2242380), anti-MMP9 (mouse monoclonal, BioLegend Cat# 819701, RRID:AB_2564833), anti-BACE1 (rabbit polyclonal, Thermo Fisher Scientific Cat# PA1-757, RRID:AB_325863), anti-APP (mouse monoclonal, Millipore, 22C11, MAB348A4), anti-Abeta (mouse monoclonal, BioLegend Cat# 803013, RRID:AB_2564765), anti-HAS2 (mouse monoclonal, Santa Cruz Biotechnology Cat# sc-514737, RRID:AB_2753202), anti-HAS3 (mouse monoclonal, Thermo Fisher Scientific Cat# MA5-17088, RRID:AB_2538559), anti-HA (rabbit polyclonal, MyBioSource, MBS2003855), anti-pSrc-Y416 (rabbit polyclonal, Cell Signaling Technology Cat# 2101, RRID:AB_331697), anti-pFAK-Y397 (rabbit polyclonal, Cell Signaling Technology Cat# 3283, RRID:AB_2173659), anti-pAkt-S473 (rabbit monoclonal, Cell Signaling Technology Cat# 3787, RRID:AB_331170), anti-pMAPK/p44/42 (rabbit monoclonal, Cell Signaling Technology Cat# 7074, RRID:AB_2099233), anti-NeuN (mouse monoclonal, Millipore Cat# MAB377X, RRID:AB_2149209), anti-Bodipy (rabbit, polyclonal, Life Technologies, A5570), anti-alpha-smooth muscle actin (mouse monoclonal, Sigma-Aldrich Cat# F3777, RRID:AB_476977), anti-Tubulin (mouse monoclonal, Sigma-Aldrich Cat# F2043, RRID:AB_259461), anti-TNFα (rabbit monoclonal, Cell Signaling Technology Cat# 6945, RRID:AB_10859375), anti-CD31 (mouse monoclonal, Miltenyi Biotec Cat# 130-092-654, RRID:AB_871662), anti-CD63 (mouse monoclonal, BD Biosciences Cat# 556019, RRID:AB_396297), anti-CD81 (mouse monoclonal, BD Biosciences Cat# 555675, RRID:AB_396028), anti-Tsg101 (mouse monoclonal, Santa Cruz Biotechnology Cat# sc-7964, RRID:AB_671392), anti-CRP (rabbit polyclonal, Abcam Cat# ab32412, RRID:AB_726934), anti-Haptoglobin (rabbit polyclonal, GeneTex Cat# GTX112962, RRID:AB_1950507), Propidium iodide (Genaxxon bioscience, M3181.0010), DAPI (4′,6-diamidino-2-phenylindole, Biomol ABD-17510).
For FACS analysis of mouse cells the following antibodies were used: anti-Ly6C (rat monoclonal, BioLegend Cat# 128024, RRID:AB_10643270), anti-Ly6G (rat monoclonal, BioLegend Cat# 127659, RRID:AB_2819864), anti-CD11b (rat monoclonal, Novus Cat# NB 200-609, RRID:AB_577744), anti-B220 (rat monoclonal, BD Biosciences Cat# 612972, RRID:AB_2870244), anti-CD56 (NCAM-1) (rat monoclonal, BD Biosciences Cat# 562811, RRID:AB_2732006), anti-CD3 (rat monoclonal, BioLegend Cat# 100203, RRID:AB_312660), anti-CD4 (rat monoclonal R&D Systems, Cat# MAB554, RRID:AB_358492), anti-CD8 (rat monoclonal, BD Biosciences Cat# 553031, RRID:AB_394569).
Primary antibodies were used at 1–2 μg ml−1 for immunoblotting, 2 μg ml−1 for immunofluorescence, 1–10 μg ml−1 for Multi-antigen staining and 5–10 μg ml−1 for FACS analysis.
The following secondary antibodies were used: Alexa Fluor 488 goat anti-mouse (Thermo Fisher Scientific Cat# A-11001, RRID:AB_2534069 ), Alexa Fluor 555 goat anti-rabbit IgG (Thermo Fisher Scientific Cat# A-21428, RRID:AB_2535849), anti-mouse IgG-HRP (Cell Signaling Technology Cat# 7076, RRID:AB_330924) and anti-rabbit IgG-HRP (Cell Signaling Technology Cat# 7074, RRID:AB_2099233).
Reagents and drugs
All reagents used are commercially available and were validated by the vendor. 4MU: For inhibition experiments the sodium salt of 4-MU (Sigma, St. Louis, MO, USA) was dissolved in Hank's Balanced Salt Solution (HBSS, Lonza, Verviers, Belgium) at a concentration of 100 mM. Final concentration in culture medium was 1 mM. AlCl3 solution: Aluminum chloride hexahydrate was purchased from Sigma Aldrich (A3017). The powder was dissolved in distilled water at a concentration of 400 mM and filtered by 0.2 μm syringe filter.
DNA constructs and transfection
Expression plasmids for HIVΔenv, ADAM10, ADAM17 and GFP-proTNF-RFP were described previously.12,33 A BACE expression plasmid was obtained from the Department of Psychiatry, Erlangen. For immunoblotting experiments, plasmids were transfected with Lipofectamine® LTX with Plus™ Reagent (Invitrogen) according to the manufacturer's instructions, or using the classical calcium phosphate procedure. By this method a transfection efficiency of 60 to 80% was achieved. Cells were used for experiments 24–72 h after transfection.
Protease activity assays
The assay was performed essentially as described previously12 using a commercial Secretase Activity Assay Kits from AnaSpec, California: for α-secretases SensoLyte®520 (AnaSpec 72085), for γ-secretases γ-secretase fluorogenic substrate was purchased from Calbiochem (565764), for β-secretases SensoLyte ® 520 β - secretase assay (AS-71144), for MMP9 activity SensoLyte ® 490 MMP 9 assay (AS-71134) according to the manufacturer's instructions. Briefly, we placed sucrose gradient purified pEV (the equivalent of 1 ml plasma), or ADAM immunoprecipitates, on a 96-well, black, flat bottom plate (Greiner 655900) and added a 5-FAM (fluorophore) and QXL™ 520 (quencher) labeled FRET peptide substrate for continuous measurement of enzyme activity. Upon cleavage of the FRET peptide by the active enzyme, the fluorescence of 5-FAM is recovered and continuously monitored at excitation/emission = 490 nm/520 nm by a preheated (37 °C) TECAN infinite M200 Pro plate reader.
pEV stimulation assay, immunohistochemistry (IHC) and confocal analysis
In pEV incubation experiments, 3.0 × 105 H4 glioma cells were seeded in a 12 well plate and 10 µg of sucrose gradient purified AD pEV were added and incubated overnight at 37 °C under 5% CO2. Then cells were harvested and 1.0–2.5 × 105 cells were seeded on a Poly-(L)-Lysine (Sigma Aldrich) coated cover slips. The cells adhered for 2 h at 37 °C under 5% CO2 and were fixed thereafter (3% PFA for 30 min at room temperature followed with three washes with PBS/1% BSA). Then cells were permeabilized (0.1% Triton X-100/1% BSA) and immunostained by standard procedures (primary and secondary antibodies). GFP-proTNF-RFP containing samples were imaged with a laser scanning confocal microscope (LSM-780; Zeiss) equipped with a 63x objective. For Alexa488 the illumination was set at 488 nm and emissions were collected between 506 and 583 nm. For Alexa555 the illumination was at 561 nm and emission collected between 574 and 667 nm. Detecting DAPI, illumination was set to 405 nm and emission collected between 410 and 495 nm. For IHC of human FFPE samples tissue sections were deparaffinized and antigen retrieval was achieved in Tris-EDTA buffer pH 9 at 100 °C for 30 min. For antigen detection the LSAB method (Dako REAL Detection Systems) was used, performed by the Dako Autostainer Plus.
Flow cytometry analysis (FACS) of mouse cells
Single-cell suspension for FACS analysis from fresh mouse tissue were obtained by mechanical dissociation and enzymatic digestion with DNase and collagenase. Cells were stained with fluorochrome-conjugated antibodies and flow cytometric analysis was done using a FACS Canto II flow cytometer (BD Bioscience). Data were analyzed with the FCS Express 4 (De Novo Software) software. Percent of EV+/PKH26+ cells was determined with flow cytometry gating on Monocytes (Ly6chi, CD11b+, Ly6G−), B-cells (B220+, CD11b−, Ly6C−, Ly6G−), Granulocytes (SSChigh, Ly6G+, CD11b+, Ly6cint, B220−), NK cells (CD56+, CD3− ) and T cells (CD3, CD4/CD8, CD45).
Particle quantification
The procedure was performed essentially as described previously.13 Briefly, purified pEV (by sucrose gradient or differential centrifugation) were diluted 1:1000 for plasma samples (pEV) and 1:1000-5000 for culture-derived EV. Particles were quantified via particle tracking analysis by a commercially available ZetaView® particle tracker from ParticleMetrix (Germany) using a 1000µl aliquot of the diluted samples. The concentration of pEV was then calculated using the appropriate dilution factors.
EV depletion of FCS and human serum for cell culture
To assure that EV generated from cell culture were not contaminated by outside sources, heat inactivated FCS and human serum for medium supplementation were depleted of bovine EV by ultracentrifugation for 18 h at 110,000 g, 4 °C before use.
Isolation, purification, labelling and assessment of electron microscopy images of EV/pEV
EV purification was performed essentially as described previously,13 adhering to MISEV2018 guidelines. Briefly, cell supernatants were collected after 48 h of cell culture and centrifuged for 20 min at 2000 g, 30 min at 10,000 g (Beckmann Coulter AllegraX15R) and ultra-centrifuged for 1 h at 100,000 g (Beckmann Coulter AvantiJ-26XP). Pellets were resuspended in 35 ml PBS and centrifuged at 100,000 g for 1 h. Pellets were resuspended in 100 μl PBS and considered as EV preparations. For pEV purification from patient samples, 10–15 ml plasma was diluted with 1:1 with PBS and centrifuged for 30 min at 2000 g, 45 min at 12,000 g and ultra-centrifuged for 2 h at 110,000 g. Pellets were resuspended in 30 ml PBS and centrifuged at 110,000 g for 1 h. Pellets were again resuspended in 100 μl PBS and considered as pEV preparations. For gradient purification, pEV were diluted in 2 ml of 2.5 M sucrose, 20 mM Hepes/NaOH, pH 7.4 and a linear sucrose gradient (2–0,25 M sucrose, 20 mM Hepes/NaOH pH 7.4) was layered on top of the EV suspension or EV were diluted in 500 µl homogenizazion media (HM) of 0.25 M sucrose, 1 mM EDTA, 10 mM Tris-HCL and layered on top of linear OptiPrep (Axis Shield) gradient (40–5% OptiPrep, HM). The samples were then centrifuged at 210,000 g for 15 h. Gradient fractions were collected from top down and the refractive index was determined. Each fraction was diluted in 10 ml PBS and ultra-centrifuged for 1 h at 110,000 g. The yield and purity of exosomes were assessed by transmission electron microscopy (TEM) (Figure S1 and see below) and nanoparticle tracking analysis (see 2.11). EV were characterized by western blot analysis for Tetraspanins (CD63, CD9 and CD81) on EV lysates, as well as on individual fractions of sucrose gradients (Figures. 1d and S1f). The latter also confirmed the presence of vesicles. Proteins were blotted onto nitrocellulose membranes, and subsequently blocked with 5% fat-free milk before being incubated with primary and secondary antibodies (see 2.5). The immunoblots were visualized by enhanced chemiluminescence (ECL kit; Millipore). Pellets were solubilized in SDS sample buffer or resuspended in 100 μl PBS and analyzed by immunoblotting or Cytokine/Chemokine/ soluble Factor (CCF) protein array (see supplementary information). For labeling of EV with PKH we used the Mini26-1KT" PKH26 Red Fluorescent Cell Linker Mini Kit (Sigma) according to the manufacturers’ procedures. For labeling with Bodipy (BODIPY® TR, Invitrogen), EV (or nil for the stain control sample) were resuspended in 100 μL of PBS and 1 μL of the dye stock solution (1 mM DMSO stock) was added to obtain a final dye concentration of 10 μM. After incubate at 37 °C for 20 min excess dye was removed with an exosome spin column (MW 3000) following the standard protocol. The efficiency of exosome labeling was assessed using the Qubit® 2.0 Fluorometer (594 nm excitation for BODIPY® TR). Electron microscopy was done essentially as described previously.34 Briefly, pEV samples were fixed in 2% (w/v) paraformaldehyde in PBS at 4 °C overnight. Fixed pEV were spread on carbon-coated 400-square-mesh copper grids (Electron Microscopy Sciences, Hatfield, PA, USA). After 20 min of incubation grids were washed with PBS and post-fixed with 2% glutaraldehyde (w/v) in PBS for 5 min. After a series of washing steps using distilled water, grids were incubated in a 3% aqueous solution of uranyl acetate (pH 4.5) that had been filtered through a 0.22 μm filter for 5 min. Grids were dried at room temperature and examined with a transmission electron microscope (Leo 912; Zeiss, Oberkochen, Germany).
Immunoblots
Proteins separated by SDS-PAGE were transferred onto nitrocellulose filters (Schleicher & Schuell) using the wet blotting device “Mini-Protean II Cell and System” (BioRad) at 400 mA for 45 min. Filters were immersed in blocking buffer for 1 h at RT. After three washes with distilled water, primary antibody diluted 1:500–1:5000 in TBST was added and incubated for 1 h (RT) or overnight (4 °C). Thereafter, filters were washed 3 times for 5 min with PBS /0.02% Tween20 before being incubated for 1 h at 4 °C with a secondary horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit antibody diluted 1:2000–1:5000 in PBS /0.02% Tween20/5%. Finally, the filters were washed three times for 10 min with PBS /0.02% Tween20 and protein bands were visualized by enhanced chemiluminescence (ECL, PIERCE) according to the manufacturer's instructions.
Human cytokine/chemokine/soluble factor (CCF) array
Purified EV (through differential centrifugation, see below) were applied to the RayBiotec Human Cytokine Array C-S (Hölzel Diagnostika, AAH-CYT-1000-2) according to the manufacturer's instructions. For Figure 1f protein arrays AAH-CYT-6 and AAH-CYT-7 were used, containing the following factors: AAH-CYT-6: ANG, BDNF, BMP4, BMP6, CCL1, CCL11, CCL13, CCL15, CCL17, CCL18, CCL2, CCL20, CCL22, CCL23, CCL24, CCL26, CCL5, CCL7, CCL8, CNTF, CSF1, CSF2, CX3CL1, CXCL12, CXCL13, CXCL6, CXCL9, EGF, FGF6, FGF7, FLT3LG, GDNF, IFNG, IGF1, IGFBP1, IGFBP2, IGFBP4, IL10, IL13, IL15, IL16, IL1A, IL1B, IL1RN, IL2, IL3, IL4, IL5, IL6, IL7, KITLG, LEP, LTA, NTF3, PDGFB, PPBP, TGFB1, TGFB3, TNF, TNFSF14.
AAH-CYT-7: ADIPOQ, AGRP, ANGPT2, AREG, AXL, BTC, CCL16, CCL19, CCL25, CCL27, CCL28, CCL3, CCL4, CSF3, CXCL1, CXCL11, CXCL2, CXCL3, CXCL5, CXCL8, EGFR, FAS, FGF2, FGF4, FGF9, FIGF, HGF, ICAM1, ICAM3, IGF1R, IGFBP3, IGFBP6, IL11, IL12A, IL12B, IL17A, IL1R1, IL1RL1, IL2RA, IL6R, IL6ST, MIF, MST1, NGF, NTF4, OSM, PGF, PLAUR, THPO, TIMP1, TIMP2, TNFRSF10C, TNFRSF10D, TNFRSF11B, TNFRSF18, TNFRSF1A, TNFRSF1B, TNFSF18, TPO, TYRO3, VEGFA, XCL1. For Figure S1g the protein array AAH-MMP-1-2 was used: AAH-MMP-1-2: MMP1, MMP10, MMP13, MMP2, MMP3, MMP8, MMP9, TIMP1, TIMP2, TIMP4.
Mouse injection experiments
Healthy female C57BL/6 mice with standard weight (27–32 g) were bred in-house and kept at 4/cage at 24 ± 1 °C with a 12 h/day light cycle and free access to food and water. The weight did not change over the course of the experiment (72 h). Specific inclusion or exclusion criteria for animals, other than health, were not applied. During experiments animals were not excluded due to health or other conditions/criteria. The animals had been acclimated to their surroundings for 7 days prior to experiment initiation. Age-matched 12 week old mice were used for all experiments and randomly selected for each experimental condition (n = 4/condition/per experiment, total of 2 experiments; conditions: PBS+stain, cont. EV (not stimulated), Al3-EV, Al3-EV+4MU, 24 animals in total), n = 4/condition is a typical number/condition for the intended purpose (identification of target cells for EV). To minimize potential confounders no strategy was used or controlled for. Groups of 4 mice (a cage of animals) underwent one tail vein injection of purified cell culture (FL83 cells)-derived EV in 100 µl PBS, equivalent to a total amount of 100 µg double labelled EV (PKH26 and Bodipy, see above EV staining) per mouse. Control mice were injected with 100 µl PKH26-containing PBS. The injections were blinded and performed by an independent research group (E.Z. and L.S.). Only the first author was aware of the content of the injections (JH. L.). After 72 h mice were anesthetized with isoflurane and sacrificed, and blood, brains, lymph nodes and additional organs were collected, photos were taken, and tissue was embedded in Tissue Tek® O.C.D and frozen at -80 °C (C.O.) until samples were proceeded for immunohistochemistry (C.O.) and FACS analysis (S.S.).
Multi antigen staining technology (MAA)
Sample preparation
Tissue sections of 5 µm were prepared using a cryotome (Leica CM 3050S, Leica, Wetzlar, Germany), incubated in aceton (Carl Roth, Mannheim, Germany) for 10 min at -20 °C and air-dried for 5 min. For rehydration the slides were placed in PBS (PAA, Pasching, Austria) for 5 min at RT, followed by incubation with 5% NGS (Dako, Hamburg, Germany) in PBS for 30 min in order to block unspecific binding sites. MAA Data Generation. The MAA-technology has been described previously.35 The coverslip with the sample was positioned onto a motor-controlled XY stage of an inverted fluorescent microscope (Leica DM IRE2, Leica Microsystems, Wetzlar, Germany; × 20 air lens; numerical aperature, 0.7). The repetitive cyclic process of this method includes flowing steps: (a) antigen tagging by a fluorescence-coupled monoclonal antibody, (b) washing, (c) image assessment and (d) photo bleaching. After completion of the cycle, the next antibody was added to the same tissue sample. Two - four visual fields are recorded simultaneously during each MAA run. Data acquisition was achieved using imaging software developed by the former company MelTec GmbH (Magdeburg, Germany). For quantification and calculation of signal intensity the ROI (regions of interest) manager tool of the ImageJ software was used. MAA Data Analysis. Using the corresponding phase-contrast images, fluorescence images produced after each antibody stain were aligned pixel wise and were corrected for illumination faults using flat-field correction. The alignment reached a resolution of ±1 pixel. Post bleaching images were subtracted from the following fluorescence tag images. Superimposed images composed a n epitope expression in relation to each pixel (900 × 900 nm2 area) of a visual field (1024 × 1024 pixels). Protein expression quantification was performed using StrataQuest software (TissueGnostics, Austria) and is described in Figure S4c.
Statistical analysis
GraphPad Prism (Version 8, GraphPad Software, USA) was used for statistical analysis. For determination of the significance, multiple data sets were analyzed using one-way analysis of variance (ANOVA). Data were expressed as mean ± standard error of the mean unless otherwise stated. Statistical analysis was performed when the experiment was repeated three times or more. Significance was accepted for p-values less than 0.05. Heat maps and Principle component analysis. Spot signal intensities were quantified with the ImageJ plugin Protein Array Analyzer. For cross-blot normalization, subtraction of the average intensity of blank spots (n = 14) and division by the average of the positive controls was performed. A list of factors of interest was then selected and plotted in a heat map with Pearson correlation distance using the library pheatmap in R. Principal Component Analysis (PCA) was performed with function prcomp in R on the normalized values. The PCA result was visualized with the R library ggplot2.
Role of funders
The funding sponsors played no role in study design, data collection, data analysis, interpretation, writing of the report, and the decision to submit the paper for publication.
Results
Abundance of secretases in pEV of AD patients
Investigating a potential role of pEV in AD, we first measured numbers of purified particles (by sucrose gradient) from plasma of AD patients (Table S1) using single particle tracking (ZetaView®). Surprisingly, AD patients (n = 6) harbored pEV numbers/particles that were on average 107-fold higher compared to age matched healthy controls (n = 4) (Figure 1a). In melanoma and HIV patients, we had previously recorded 5 to 20-fold higher numbers.13,16 A different approach supported this finding. Protein concentrations in vesicle-containing sucrose gradient fractions (as in Figure 1d) were on average 91.9-fold higher using AD plasma as compared to controls (Figure S1a). The size, shape and electro-microscopic appearance of these pEV (Figure S1b, c) were similar to what we have described previously.34 The difference of this image compared to commonly seen TEM images (cup shaped vesicles) could be due to different reasons. Aside from the EM procedure, an origin from the plasma membrane and/or an unconventional secretion process seems possible, similar as proposed and shown for HIV-induced vesicle release.36,37 Alternatively, the vesicles shown here might have been released by a pearling effect, induced by increased glycocalyx expression, e.g. through HAS3 expression, as demonstrated recently38,39 (see also below).
A potential presence of protease activity in AD-pEV was analyzed using FRET peptides. As we have described recently for melanoma and HIV patients (Figure S1d).12,13 Different protease activities, including ADAM10/17 (α-secretases), MMP9, γ-secretase and β-secretases were recorded in all AD samples (n = 3), but not in healthy controls (Figure 1b), who, in general, harbor little to no protease activity in their pEV (see).13,16 For additional control, samples from patients with other neurological symptoms were analyzed accordingly, including schizophrenia (SZ), cognitive dysfunction (CD) and depression (DP). However, only the DP sample was positive for MMP9 activity (Figure S1e). To confirm the presence of active proteases in pEV, we performed immunoblots. In accordance with the protease assay results, only AD-pEV harbored faster migrating fragments of ADAM10/17 and MMP9, which likely represented processed/activated forms of the enzymes (red arrows in Figure 1c–e), as described previously12,36,40 (Figure 1c, red frames). Confirming an association with vesicles, these activated protease forms, and also BACE, co-migrated with typical vesicle proteins (CD63, CD81) in sucrose gradient fractions, (Figure 1d, red arrows and frame, healthy control in Figure S1f). In this analysis also mature TNF was detected, implying pro-TNF cleavage by ADAM17 before or during vesicle secretion (blue arrow and frame), similar as seen previously.41 Again, this suggested the presence of active proteases in AD-pEV. Finally, the presence of γ-secretase (Presenilin1 C-terminal fragment: CTF) was verified in AD-pEV (Figure 1e). In summary, increased pEV levels in AD patients contained an array of different active proteases.
The cellular origin of pEV is difficult to determine, as generally accepted organ/tissue-specific protein markers do not exist. However, pEV may contain patterns of effectors (hereafter referred to as CCF: Cytokines, Chemokines and soluble Factors), that could at least hint at their cellular source.13,34 CCFs of pEV from different neurological patients (AD, CD, SZ, DP) were assessed by protein array, revealing distinct patterns (Figure 1f). In AD the pattern was particularly rich in inflammatory factors (red frame), including Rantes, GROα, M-CSF, TNF, IGFBP3 and IL6-R (red letters) and several matrix metalloproteinases (MMP2, 3, 9 and 13; Figure S1g), while interleukins were largely absent. This pattern of factors hinted at hepatocytes as at least one possible source of AD-pEV,42, 43, 44, 45 similarly as we have demonstrated recently for pEV induced by HIV infection and Melanoma.34,46 Supporting this assumption, the factor content of EV from a stimulated hepatocyte cell line was almost identical to AD-pEV (see below Figure S2e, f). In summary, AD-pEV harbored a rich and seemingly disease-typical array of inflammatory factors, of which at least a fraction could have derived from hepatocytes.
Hepatocytes secrete AD pEV-like EV
In order to identify target cells of AD-pEV, we attempted to generate similar vesicles in vitro for animal injection experiments. We had noticed that a certain concentration range of AlCl347 (hereafter Al3) stimulated the processing of ADAM10 and 17 in Huh7 hepatocytes. In addition, BACE expression increased. Conversely, liver endothelial cells (Sk-Hep1) were unresponsive and stimulation with other metal ions (Zn2, Fe3) had no effect, except for a slight activation of ADAM10 by Fe3 (Figure S2a–d). We therefore explored whether Al3-treatment of Huh7 would produce EV similar to AD-pEV.
The CCF profile of such EV was very similar to that found in AD-pEV, as for example MMP2, 3, 9 and 13 were also present (Figure S2e, f). In addition, Al3-EV contained α-secretase-, and MMP9- β-secretase activities and proteins (Figure S2g, h). Using an anti-Aβ mouse monoclonal, we also detected Aβ in Al3-EV, similar as reported previously for AD-pEV11 (Figure S2j). No Aβ was detected in EV derived from stimulated Sk-Hep1 cells or primary astrocytes, although APP and protein fragments thereof were seemingly present in all EV/pEV.
The presence of APP and Aβ in AD-pEV/Al3-EV implied that co-secreted proteases cleaved APP in a manner yielding Aβ. To test this assumption, we used APP-expressing H4 Glioma cells as EV targets. Stimulation of H4 with Al3 or control EV had no effect, while incubation with Al3-EV, and more so AD-pEV, led to the appearance of Aβ (Figure 1g, red frames, quantification in Figure S3a), seemingly packaged into vesicular structures. These vesicular compartments, better recognizable in H4 cells stably transfected with APP, were transported to the cell periphery, potentially to be secreted (Figure S3b). This appeared similar to what we had observed for pro-TNF cleavage through ADAM17-containing EV.18 For clarification, we employed our indicator construct for pro-TNF cleavage (GFP-proTNF-RFP).18 Transfected into H4 cells, Al3-EV incubation cleaved intra-endosomal GFP-proTNF-RFP (yellow) into GFP-pro (green) and TNF-RFP (red), the latter packaged into vesicular compartments (Figure S3c). Taken together, Al3-EV appeared to be similar to AD-pEV in factor content and were able to cleave different substrates in target cells.
EV reach the brain via the choroid plexus
For animal experiments, we first confirmed that Al3-stimulated murine hepatocytes (FL83) uploaded α-secretases and MMP9 into EV similar as Huh7 (Figure S4a; see also below Figure S6d). Then double-labeled EV (BodipyFL-C5 Ceramide (green), PKH26 (red)) from Al3-/non-stimulated FL83 cells, or the staining dyes diluted in PBS, were injected into tail veins of mice (n = 4/condition; Figure 2a). No behavioral changes of the animals were noticed during the experiment. Only animals receiving Al3-EV (4 of 4) presented large reddish lymph nodes (at 72h) with roughly 24% of monocytes (Ly6C+) detected therein harboring labeled EV (Figure 2b, red frames). Only 3.9% of such cells were found in lymph nodes of controls. This was determined by FACS analysis on tissue-isolated B-, T- and NK-cells, monocytes and granulocytes. Aside from monocytes in lymph nodes, the percentage of EV-containing immune cells was low in all tissues (brain, liver, lymph node, lung and spleen) and not significantly different between Al3-EV and control EV (Figure S4b).
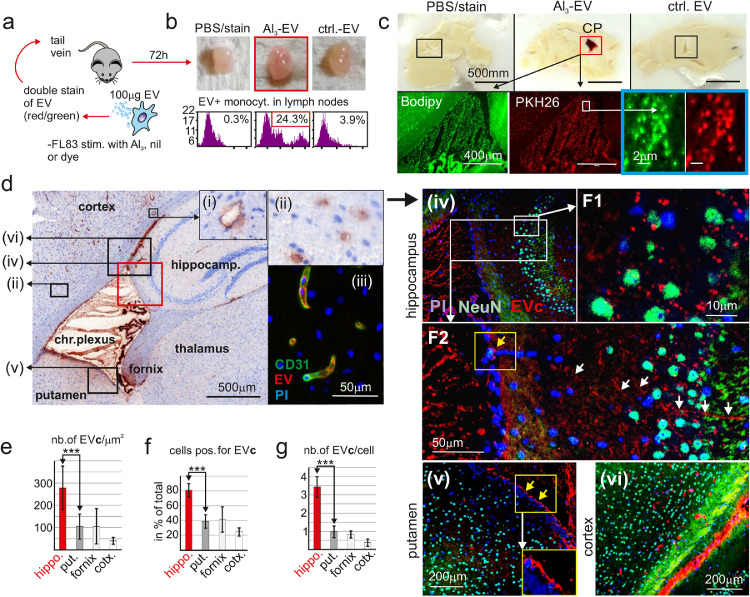
Figure 2.
Plasma EV reach hippocampal neurons via the choroid plexus (CP).
(a) Cartoon depicting the injection of Al3-induced EV into mice. (b) Representative images of lymphnodes 72 h after EV injection (n = 4/condition) before analysis for EV-containing monocytes by FACS. (c) Sagital brain section with accumulation of labelled Al3-induced EV in the choroid plexus (CP). (d) Representative image showing Al3-EV accumulate in hippocampal neurons and blood vessel endothelia. Boxed areas were magnified (i),(ii), or analyzed by multi-antigen IHC (iii)–(vi). (e–g) Quantification of EV clusters (EVc) in different brain areas and neurons, using imaging analysis. Statistical significance for (e–g) by one-way ANOVA and Tukey's test ***p < 0.005. Data are means ± SEM from 3 visual fields of 3 different animals.
In sagittal brain sections we noticed a prominent red staining of the ventricle area/choroid plexus (CP); however, only Al3-EV injected mice (Figure 2c, red frame; 4 of 4, one presented). Fluorescence microscopy revealed countless 0.5–2 μm sized double stained vesicular structures (blue frame), apparently clusters of labeled EV (EVc). A counter stain with an anti-Bodipy-FL antibody supported this conclusion and revealed additional EV-positive areas around the CP (Figure 2d), and in blood vessels (2d (i) and (ii)). The latter was confirmed by multi-antigen IHC staining,33,35 showing a co-localization of endothelial marker CD31 with EV (Figure 2d (iii)).
The same IHC technology was used to localize and quantify neurons, EV and nuclei in areas around the ventricle/CP (Figure 2d (iv)–(vi); quantification explained in Figure S4c). We found that the number of EVc /μm2 tissue (Figure 2e), the percentage of neurons positive for EVc (Figure 2f) and the average number of EVc in neurons (Figure 2g) was highest in the hippocampus (Figure 2d (iv), F1), followed by the putamen (Figure 2d (v)), the fornix (IHC not shown), the cortex (Figure 2d (vi)) and the thalamus (IHC not shown). Supporting this finding, EV/EVc appeared to move along axons or conduit-like structures into the hippocampus (Figure 2d (iv), F2, white arrows), but not into other brain areas. Furthermore, whereas EVc seemingly crossed the brain ependyma towards the hippocampus (yellow box/arrow in Figs. 2d, F2), they accumulated at the ependyma bordering to other brain nuclei and the cortex (yellow boxes/arrows in Figure 2d (v)). Taken together, peripheral Al3-EV reached the brain, and particularly neurons of the hippocampus, through the CP.
pEV components in human CP/brain
To confirm a potential concentration of pEV in the CP, we analyzed AD CP/brain tissue samples (n = 3) (Table S2). Assuming that at least a fraction of AD-pEV were of hepatocyte origin, we speculated to detect liver acute phase proteins, commonly not found in brain tissue. Supporting this assumption, both haptoglobin and CRP (C-reactive protein) were prominent in lysates of AD-pEV, but were also present in pEV from healthy controls, albeit at lower concentrations (Figure S5a). Brain tissue samples from age-matched controls had few CP cells positive for haptoglobin and CRP. Conversely, CP tissue from AD patients gave a strong staining for both markers (Figs. 3a, S5b) in the lumen of vessels (Figure 3a, yellow arrows), and in sub-membrane areas of CP cells (black arrows). The latter was more prominent for CRP (quantified in Figure 3b), whereas haptoglobin was prominent in vessel lumens. Notably, the sub-membrane CRP staining was similar to the appearance of labelled Al3-EV in the CP of injected animals (Figure 3c, white arrows). Confirming the presence of vesicles in the CP of an AD patient (AD-13), we could demonstrate a co-localization of CD63 and CRP using the multi-antigen analysis technology (Figure 3d). Furthermore we detected CRP in up to 20% of hippocampal neurons, but not in other brain areas, as for example the cortex. Conversely, in controls only around 4% of hippocampal neurons revealed a weak staining for CRP (Figs. 3e; S5c). Consistent with our findings in animals, AD-pEV seemingly reached hippocampal neurons via the CP.
Figure 3.
Liver-derived pEV factors in AD brain tissue.
(a) Staining of CP brain sections for CRP and haptoglobin from one age-matched control (H-08) and 3 AD patients (AD-12-14). (b) Quantification of CP cell staining (pixel/cell) for CRP and haptoglobin by imaging software. SDM based on 3 visual areas analyzed (40 cells/area). (c) Representative image of mouse CP cells incorporating Al3-induced EV. (d) Representative images of a multiantigen analysis on human tissue (AD13), showing that pEV markers and CRP co-localize in the CP. (e) Representative images of CRP staining in hippocampal brain sections of one control (Con-01) and one AD patient (AD-13) and quantification of positive cells in hippocampal and cortical areas (3 controls and 3 AD patients (see also Fig. S5b). SDM based on 100 cells analyzed in each sample. Statistical significance for (d) and (f) by two-way ANOVA and Dunnett's test, ***p < 0.005.
HAS3 is required for CP targeting
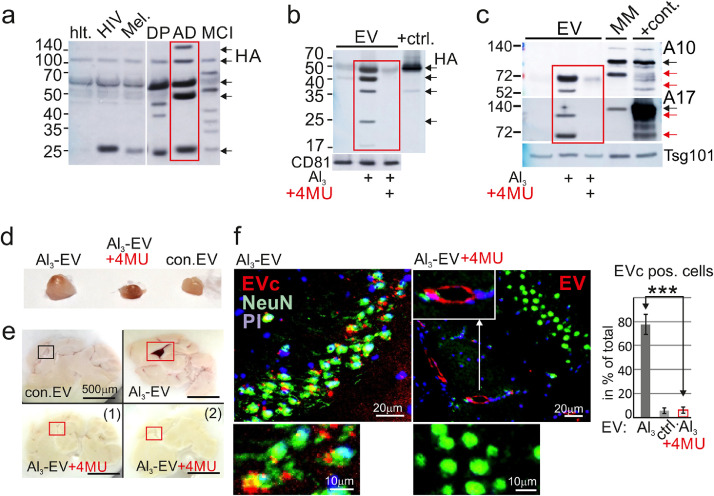
We asked why Al3-EV but not control EV accumulated in the CP. Previously hyaluronic acid synthetase 3 (HAS3) expression was found to induce the uploading of Hyaluronan (HA) onto EV,48 a docking ligand for receptors like CD44 or LYVE-1.49,50 Indeed, AD-pEV harbored different fragments of HA (Figure 4a, red frame), that is usually represented by multiple cleavage products.51 This HA fragment pattern seemed different or absent in other diseases/conditions, including HIV infection, Melanoma, DP, MCI and a healthy control. In vitro, HA upload could be mimicked by Al3 stimulation, and this was blocked by the addition of the HAS inhibitor 4-methylumbelliferone (4MU)52,53 (Figure 4b, red frame). Surprisingly, 4MU also blocked the uptake of other factors, including ADAM10/17 (Figure 4c, red frame) and Hck (Figure S6a). We demonstrated previously that Hck was required for packaging of ADAM protease onto EV/pEV.34,36 Supporting this assumption, Hck was present in AD-pEV (Figure S6b). Finally, and in line with these findings, Al3 stimulated the expression of HAS3 and HAS2 (Figure S6c). In summary, Al3-stimulated HAS3 expression induced the uploading of different factors onto EV, and this could be blocked by 4MU.
Figure 4.
4-methylumbelliferone (4MU) blocks EV upload and CP targeting.
(a) Plasma EV from different patients blotted for Hyaluronic acid (HA) fragments: hlt.: healthy; Mel.: Melanoma; DP: Depression; MCI: mild cognitive impairment, AD (AD10) (2 ml plasma analyzed). (b) 4MU blocks HA uptake into EV. +ctrl: Huh7 lysate. (c) 4MU blocks upload of ADAM10/17 (A10, A17) into EV. Red arrows indicate processed protease forms. MM: melanoma cell line harboring ADAM10 (12). +cont.: 293T lysates as in Fig. 1c. (d,e) Representative images of lymph nodes (d) and brain sections (e) 72 h after injection of 4MU treated EV (n = 4/condition). (f) IHC of a hippocampal section and quantification of EVc pos. cells as in Fig. 2f. Statistical significance by one-way ANOVA and Tukey's test, ***p < 0.005.
To test whether 4MU prevented EV from docking to the CP, we generated FL83 hepatocyte-derived EV in the presence of 4MU. We verified that their protease content and HA level were reduced (Figure S6d, e) and injected the EV as described above. Those EV did not cause a lymph node swelling, although a strong reddish color was noticed (Figure 4d). Furthermore, the CPs of these mice did not capture any pEV (Figure 4e, compare red frames). IHC staining confirmed that labelled EV (EVc) did not reach the hippocampus (Figure 4f); however, they still attached to the endothelium of blood vessels (white arrows and insert). Together, this implied that HA was involved in docking pEV to the CP, although we could not exclude that 4MU also blocked other ligands.
Discussion
We report the presence of protease-containing pEV in plasma of AD patients. In vitro copies of these pEV reached hippocampal neurons via trafficking through the CP, potentially cleaving substrate proteins like APP and pro-TNF as shown in vitro (Figs. 1j, S3c). This could lead to the secretion of TNF,18 Aβ (see model in Figure 5) and potentially additional amyloidogenic peptides54 from brain cells. By the same route, pEV-associated Aβ could accumulate in the brain, similar as demonstrated here for liver acute phase proteins (Figs. 3 and S5). Plasma EV may connect with brain cells for different physiological reasons. For example, in the wake of a systemic infection, vesicles may transfer effectors to establish an immune defense. Supporting this assumption, Aβ was suggested to be an antimicrobial peptide.55
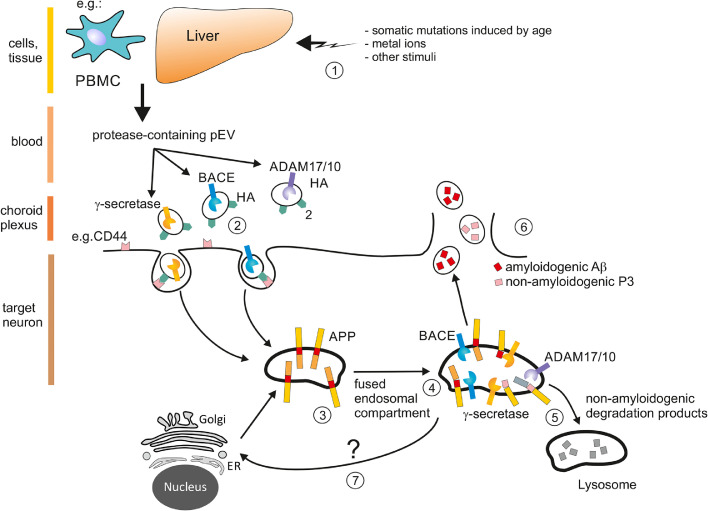
Figure 5.
Speculative model of Aβ generation by protease-containing pEV
(1) Somatic mutations induced by age, metal ions or other stress stimuli may stimulate liver and/or cells of other organs or the immune system (e.g. PBMC)46 for an inflammatory response that includes the secretion of pEV containing CCF and active proteases. These pEV possibly function to alert or activate the immune defense in the periphery, for example by differentiating precursor cells.65(2) Depending on their ligands on the vesicle surface, in this case Hyaluronan (HA) and/or HA fragments, pEV are incorporated by specific target cells like monocytes, CP cells or neurons in the hippocampus (Figs. 2,3 and S4c). A single vesicles may contain only one type of protease or inflammatory factor. (3) Once incorporated, these incoming vesicles may fuse with early endosomes (18) containing various substrates including APP. (4) Endosomal compartments may fuse with vesicles containing different proteases, including ADAM10/17, cleaving APP in a physiological (non-amyloidogenic) manner, but also non-physiological (amyloidogenic) manner that includes BACE activity. (5) Non-amyloidogenic peptides, like P3, may be secreted or degraded in lysosomes. Conversely, 6) amyloidogenic Aβ withstands degradation, may accumulate (not shown) and/or be released by secretion to function as an anti-microbial peptide. (7) Likely, the cleavage of APP leads to a feedback loop to the ER/Golgi inducing the translation of more APP.
There are numerous publications suggesting a role of EV in neurodegenerative diseases, although many of these studies were done in vitro or in transgenic animals.29 Most current models envision EV to be part of the intercellular communication between brain cells. In that function they could also cause the spreading of disease-associated proteins like PrPSc, α-synuclein and Aβ.56 EV detected in plasma with markers of neurodegeneration are assumed to have crossed the BBB into the periphery,24,26,27 potentially as a consequence of barrier damage, for example in the course of cancer growth.57 Conversely, pEV may reach the brain in a state of inflammation.30,31 In line with these latter studies, our results point to a pEV-mediated communication axis between the periphery and the brain through the CP. In a state of inflammation, pEV-transported proteases could lead to the activation of inflammatory cytokines in target cells, as demonstrated here for pro-TNF (Figure S3c). A prolonged or chronic transfer of such pEV into brain cells could turn harmful, particularly when regeneration and compensation mechanisms fail or decline with age.
The broad array of effectors and unusually high levels of AD-pEV could hint at a strong non-physiological stimulus. The biggest risk factor in AD is age. In the process of aging, the mutational load in regenerative tissues increases remarkably.58 Mutated somatic cells may show behavior of immortalized cell clones59 that could include a higher secretory activity. Supporting this idea, we recently demonstrated that BRAFV600E increased the secretion of ADAM10 and other factors.33 This assumed non-physiological stimulus also led to the release of α-secretases, considered to be protective in AD. The presence of both, Aβ and TNF in pEV and target cells (Figs. 1d, S2j, S3), implies stochastic cleavage events, and could lead to a fluctuating appearance of different cleavage products in target cells. While non-amyloidogenic cleavage products are expected to be degraded, amyloidogenic peptides, particularly when functioning as an anti-microbial peptide like Aβ, would be secreted, undergoing a slower degradation as compared to other peptides. Over time, this would lead to an accumulation of Aβ, even if alpha-, beta- and gamma secretases are entering neurons in comparable quantities (Figure 5).
The origin of inflammatory pEV in AD is a central issue. Plasma vesicles presumably have a short life span of about 30 min.60 Therefore the high levels of pEV recorded here (Figure 1a) may originate from a larger secretory organ. In our previous work, we provided circumstantial evidence for liver cells being at least one possible source of pEV.13,34,46 Indeed, this large organ could constantly replenish factor-loaded vesicles. In line with this assumption we found high levels of liver acute phase proteins in pEV and human tissue sections of the CP and the hippocampus. Furthermore, the factor profile of AD-pEV and Al3-stimulated EV from a hepatocyte line (Figure S2) was almost identical. Conversely, it seems unlikely that the here reported high amounts of vesicles originated from brain cells, for example as a consequence of AD. That would require a very high secretory activity of diseased/dying brain cells, for which there has been no evidence put forward so far. Supporting our assumption we found that injected EV accumulated in astonishing concentrations in the choroid plexus, implying a physiological target mechanism from the periphery. However, additional evidence is necessary to confirm that peripheral cells, like liver cells, secrete protease-containing pEV in AD.
Because 4MU abolished the ability of pEV to dock at CP cells (Figure 4e, f), Hyaluronan, or cleavage products thereof, may function as tissue entry ligands for pEV, engaging receptors like LYVE-1 or CD44.49,50 The strong presence of labelled pEV in the CP, but not other regions of the brain, implies such a mechanism. Interestingly, Hyaluronan synthesis involves chitin oligomers,61,62 which are toxic for neurons and are found in amyloid plaques in AD.63 Furthermore, levels of these oligomers are increased in plasma of patients with AD.64 These findings would be in line with a model in which excess production of Hyaluronan-coated pEV could be involved in the pathogenesis of AD. However this hypothesis requires further analysis.
A limitation of the study is the insufficient number of plasma and tissue samples that were analyzed in order to substantiate our findings. Furthermore, we do not have definite proof of the producer cells of AD-pEV and whether they really concentrate in the CP due to a ligand-receptor interaction involving HA. Finally, aside from Aβ, additional APP-derived peptides in pEV54 could play a role in the here described mechanism, that were not explored. Nevertheless we conclude that both, pEV and 4MU might be further explored with respect to the pathogenesis, diagnosis and potentially treatment of neurological disorders.
Contributors
The project was designed and coordinated by A.S.B. Substantial contributions to the work were made by J-H.L. (pEV purification, transfections, protein assays, Western blots and protein arrays); C.O. (tissue analysis and staining, MELC analysis); S.Sch. (immune cell analysis by FACS); M.E., and J.G-V. (heat map clustering and bioinformatics analysis); K.B. (pEV array analysis); M.M. T.O. (clinical diagnostic, patient selection, blood sampling, and advice), E.Z. and L.S. (animal injections and handling), H.B. (electron microscopy), B.P. (MAA software analysis). All authors read and approved the final version of the manuscript. All data were controlled and verified by both first authors (JH.L., C.O.). Data produced by members from collaborating labs were confirmed by the respective contributors.
Data sharing statement
Data, materials and software information supporting the conclusions of this article are included within the article and its additional file.
Declaration of interests
B. Plosnita is employed by TissueGnostics GmbH, Vienna. Dr Baur considers to submit a patent. Dr. Eberhardt and Prof. Vera-Gonzalez reports grants from German Ministry of Education and Research (BMBF), during the conduct of the study. The other authors have declared no conflicts of interest.
Acknowledgments
This work was supported by funds from the German Science Foundation (DFG): SFB 643 BA961/3-1 and the German Federal Ministry of Education and Research (BMBF) under grant 01GU1107A, 031L0073A (MelEVIR) and 161L0244A (KI-VesD). JH. L. as well as S.Sch. and E.A. were supported from the IZKF (Interdisziplinäres Zentrum für Klinische Forschung) of the Fredrich-Aleander-University of Erlangen-Nürnberg. (S. Sch. and E.A.). C.O. was supported by the Comprehensive Cancer Center (CCC) Erlangen and the German Science Foundation (DFG): (DFG; OS 578). J.V-G. and M.E. were supported by the BMBF as part of the project eBio:miRSys (0316175A). J.V-G. is also funded by the Elan Funds of the Medical Faculty of the Friedrich-Alexander-University of Erlangen-Nürnberg (FAU) and the DFG through the project SPP 1757/1 (VE 642/1-1). T.H. was supported by the Emerging Fields Program of the Friedrich-Alexander-University of Erlangen-Nürnberg. L.Z. and L.S. from the CCC Erlangen (CRC1181). We are grateful to TissueGnostics for providing the StrataQuest Software. We are greatful to the The Netherlands Brain Bank in Amsterdam for providing brain tissue samples. The study has not received any funds from industrial corporations.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103903.
Appendix. Supplementary materials
References
- 1.Braak H., Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 2.Esch F.S., Keim P.S., Beattie E.C., et al. Cleavage of amyloid beta peptide during constitutive processing of its precursor. Science. 1990;248(4959):1122–1124. doi: 10.1126/science.2111583. [DOI] [PubMed] [Google Scholar]
- 3.Masters C.L., Bateman R., Blennow K., Rowe C.C., Sperling R.A., Cummings J.L. Alzheimer's disease. Nat Rev Dis Prim. 2015;1:15056. doi: 10.1038/nrdp.2015.56. [DOI] [PubMed] [Google Scholar]
- 4.Vassar R., Bennett B.D., Babu-Khan S., et al. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286(5440):735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 5.Haass N.K., Ripperger D., Wladykowski E., et al. Melanoma progression exhibits a significant impact on connexin expression patterns in the epidermal tumor microenvironment. Histochem Cell Biol. 2010;133(1):113–124. doi: 10.1007/s00418-009-0654-5. [DOI] [PubMed] [Google Scholar]
- 6.Vincent B., Checler F. α-Secretase in Alzheimer's disease and beyond: mechanistic, regulation and function in the shedding of membrane proteins. Curr Alzheimer Res. 2012;9(2):140–156. doi: 10.2174/156720512799361646. [DOI] [PubMed] [Google Scholar]
- 7.Lammich S., Kojro E., Postina R., et al. Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci U S A. 1999;96(7):3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haass C. Take five–BACE and the gamma-secretase quartet conduct Alzheimer's amyloid beta-peptide generation. EMBO J. 2004;23(3):483–488. doi: 10.1038/sj.emboj.7600061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glenner G.G. Alzheimer's disease: its proteins and genes. Cell. 1988;52(3):307–308. doi: 10.1016/s0092-8674(88)80021-7. [DOI] [PubMed] [Google Scholar]
- 10.Glenner G.G., Wong C.W. Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120(3):885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 11.Rajendran L., Honsho M., Zahn T.R., Keller P., Geiger K.D., Verkade P., et al. Alzheimer's disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci U S A. 2006;103(30):11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J.H., Wittki S., Brau T., et al. HIV Nef, paxillin, and Pak1/2 regulate activation and secretion of TACE/ADAM10 proteases. Mol Cell. 2013;49(4):668–679. doi: 10.1016/j.molcel.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Lee J.H., Schierer S., Blume K., et al. HIV-Nef and ADAM17-containing plasma extracellular vesicles induce and correlate with immune pathogenesis in chronic HIV infection. EBioMedicine. 2016;6:103–113. doi: 10.1016/j.ebiom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimoda M., Khokha R. Proteolytic factors in exosomes. Proteomics. 2013;13(10-11):1624–1636. doi: 10.1002/pmic.201200458. [DOI] [PubMed] [Google Scholar]
- 15.Stoeck A., Keller S., Riedle S., et al. A role for exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem J. 2006;393(Pt 3):609–618. doi: 10.1042/BJ20051013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J.H., Dindorf J., Eberhardt M., et al. Innate extracellular vesicles from melanoma patients suppress beta-catenin in tumor cells by miRNA-34a. Life Sci Alliance. 2019;2(2) doi: 10.26508/lsa.201800205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller M.D., Ching K.L., Liang F.X., et al. Decoy exosomes provide protection against bacterial toxins. Nature. 2020;579(7798):260–264. doi: 10.1038/s41586-020-2066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostalecki C., Wittki S., Lee J.H., et al. HIV Nef- and Notch1-dependent endocytosis of ADAM17 induces vesicular TNF secretion in chronic HIV infection. EBioMedicine. 2016;13:294–304. doi: 10.1016/j.ebiom.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuroda H., Tachikawa M., Yagi Y., et al. Cluster of differentiation 46 is the major receptor in human blood-brain barrier endothelial cells for uptake of exosomes derived from brain-metastatic melanoma cells (SK-Mel-28) Mol Pharm. 2019;16(1):292–304. doi: 10.1021/acs.molpharmaceut.8b00985. [DOI] [PubMed] [Google Scholar]
- 20.Saint-Pol J., Gosselet F., Duban-Deweer S., Pottiez G., Karamanos Y. Targeting and crossing the blood-brain barrier with extracellular vesicles. Cells. 2020;9(4) doi: 10.3390/cells9040851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Romero N., Carrion-Navarro J., Esteban-Rubio S., et al. DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients. Oncotarget. 2017;8(1):1416–1428. doi: 10.18632/oncotarget.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi M., Liu C., Cook T.J., et al. Plasma exosomal alpha-synuclein is likely CNS-derived and increased in Parkinson's disease. Acta Neuropathol. 2014;128(5):639–650. doi: 10.1007/s00401-014-1314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norman M., Ter-Ovanesyan D., Trieu W., et al. L1CAM is not associated with extracellular vesicles in human cerebrospinal fluid or plasma. Nat Methods. 2021;18(6):631–634. doi: 10.1038/s41592-021-01174-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pulliam L., Sun B., Mustapic M., Chawla S., Kapogiannis D. Plasma neuronal exosomes serve as biomarkers of cognitive impairment in HIV infection and Alzheimer's disease. J Neurovirol. 2019;25(5):702–709. doi: 10.1007/s13365-018-0695-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loov C., Scherzer C.R., Hyman B.T., Breakefield X.O., Ingelsson M. α-Synuclein in extracellular vesicles: functional implications and diagnostic opportunities. Cell Mol Neurobiol. 2016;36(3):437–448. doi: 10.1007/s10571-015-0317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiandaca M.S., Kapogiannis D., Mapstone M., et al. Identification of preclinical Alzheimer's disease by a profile of pathogenic proteins in neurally derived blood exosomes: a case-control study. Alzheimers Dement. 2015;11(6):600–607. doi: 10.1016/j.jalz.2014.06.008. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goetzl E.J., Mustapic M., Kapogiannis D., et al. Cargo proteins of plasma astrocyte-derived exosomes in Alzheimer's disease. FASEB J. 2016;30(11):3853–3859. doi: 10.1096/fj.201600756R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Candelario K.M., Steindler D.A. The role of extracellular vesicles in the progression of neurodegenerative disease and cancer. Trends Mol Med. 2014;20(7):368–374. doi: 10.1016/j.molmed.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill A.F. Extracellular vesicles and neurodegenerative diseases. J Neurosci. 2019;39(47):9269–9273. doi: 10.1523/JNEUROSCI.0147-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridder K., Keller S., Dams M., et al. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol. 2014;12(6) doi: 10.1371/journal.pbio.1001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balusu S., Van Wonterghem E., De Rycke R., et al. Identification of a novel mechanism of blood-brain communication during peripheral inflammation via choroid plexus-derived extracellular vesicles. EMBO Mol Med. 2016;8(10):1162–1183. doi: 10.15252/emmm.201606271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long J.M., Holtzman D.M. Alzheimer disease: an update on pathobiology and treatment strategies. Cell. 2019;179(2):312–339. doi: 10.1016/j.cell.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ostalecki C., Lee J.H., Dindorf J., et al. Multiepitope tissue analysis reveals SPPL3-mediated ADAM10 activation as a key step in the transformation of melanocytes. Sci Signal. 2017;10(470) doi: 10.1126/scisignal.aai8288. [DOI] [PubMed] [Google Scholar]
- 34.Lee J.H., Ostalecki C., Zhao Z., et al. HIV activates the tyrosine kinase Hck to secrete ADAM protease-containing extracellular vesicles. EBioMedicine. 2018;28:151–161. doi: 10.1016/j.ebiom.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schubert W., Bonnekoh B., Pommer A.J., et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat Biotechnol. 2006;24(10):1270–1278. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Z., Kesti T., Ugurlu H., Baur A.S., Fagerlund R., Saksela K. Tyrosine phosphorylation directs TACE into extracellular vesicles via unconventional secretion. Traffic. 2019;20(3):202–212. doi: 10.1111/tra.12630. [DOI] [PubMed] [Google Scholar]
- 37.Muratori C., Cavallin L.E., Kratzel K., et al. Massive secretion by T cells is caused by HIV Nef in infected cells and by Nef transfer to bystander cells. Cell Host Microbe. 2009;6(3):218–230. doi: 10.1016/j.chom.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Shurer C.R., Kuo J.C., Roberts L.M., et al. Physical principles of membrane shape regulation by the glycocalyx. Cell. 2019;177(7):1757–1770. doi: 10.1016/j.cell.2019.04.017. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rilla K. Diverse plasma membrane protrusions act as platforms for extracellular vesicle shedding. J Extracell Vesicles. 2021;10(11):e12148. doi: 10.1002/jev2.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adrain C., Zettl M., Christova Y., Taylor N., Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012;335(6065):225–228. doi: 10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Escrevente C., Morais V.A., Keller S., Soares C.M., Altevogt P., Costa J. Functional role of N-glycosylation from ADAM10 in processing, localization and activity of the enzyme. Biochim Biophys Acta. 2008;1780(6):905–913. doi: 10.1016/j.bbagen.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Rowell D.L., Eckmann L., Dwinell M.B., et al. Human hepatocytes express an array of proinflammatory cytokines after agonist stimulation or bacterial invasion. Am J Physiol. 1997;273(2 Pt 1):G322–GG32. doi: 10.1152/ajpgi.1997.273.2.G322. [DOI] [PubMed] [Google Scholar]
- 43.Arany E., Afford S., Strain A.J., Winwood P.J., Arthur M.J., Hill D.J. Differential cellular synthesis of insulin-like growth factor binding protein-1 (IGFBP-1) and IGFBP-3 within human liver. J Clin Endocrinol Metab. 1994;79(6):1871–1876. doi: 10.1210/jcem.79.6.7527416. [DOI] [PubMed] [Google Scholar]
- 44.Desgeorges A., Gabay C., Silacci P., et al. Concentrations and origins of soluble interleukin 6 receptor-alpha in serum and synovial fluid. J Rheumatol. 1997;24(8):1510–1516. [PubMed] [Google Scholar]
- 45.Calabro S.R., Maczurek A.E., Morgan A.J., et al. Hepatocyte produced matrix metalloproteinases are regulated by CD147 in liver fibrogenesis. PLoS One. 2014;9(7):e90571. doi: 10.1371/journal.pone.0090571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J.H., Eberhardt M., Blume K., Vera J., Baur A.S. Evidence for liver and peripheral immune cells secreting tumor-suppressive extracellular vesicles in melanoma patients. EBioMedicine. 2020;62 doi: 10.1016/j.ebiom.2020.103119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darbre P.D., Bakir A., Iskakova E. Effect of aluminium on migratory and invasive properties of MCF-7 human breast cancer cells in culture. J Inorg Biochem. 2013;128:245–249. doi: 10.1016/j.jinorgbio.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 48.Rilla K., Pasonen-Seppanen S., Deen A.J., et al. Hyaluronan production enhances shedding of plasma membrane-derived microvesicles. Exp Cell Res. 2013;319(13):2006–2018. doi: 10.1016/j.yexcr.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 49.Jackson D.G. Hyaluronan in the lymphatics: the key role of the hyaluronan receptor LYVE-1 in leucocyte trafficking. Matrix Biol. 2019;78–79:219–235. doi: 10.1016/j.matbio.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 50.Skandalis S.S., Karalis T.T., Chatzopoulos A., Karamanos N.K. Hyaluronan-CD44 axis orchestrates cancer stem cell functions. Cell Signal. 2019;63 doi: 10.1016/j.cellsig.2019.109377. [DOI] [PubMed] [Google Scholar]
- 51.Stern R., Asari A.A., Sugahara K.N. Hyaluronan fragments: an information-rich system. Eur J Cell Biol. 2006;85(8):699–715. doi: 10.1016/j.ejcb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 52.Nakamura T., Funahashi M., Takagaki K., et al. Effect of 4-methylumbelliferone on cell-free synthesis of hyaluronic acid. Biochem Mol Biol Int. 1997;43(2):263–268. doi: 10.1080/15216549700204041. [DOI] [PubMed] [Google Scholar]
- 53.Kultti A., Pasonen-Seppanen S., Jauhiainen M., et al. 4-Methylumbelliferone inhibits hyaluronan synthesis by depletion of cellular UDP-glucuronic acid and downregulation of hyaluronan synthase 2 and 3. Exp Cell Res. 2009;315(11):1914–1923. doi: 10.1016/j.yexcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 54.Laulagnier K., Javalet C., Hemming F.J., et al. Amyloid precursor protein products concentrate in a subset of exosomes specifically endocytosed by neurons. Cell Mol Life Sci. 2018;75(4):757–773. doi: 10.1007/s00018-017-2664-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar D.K., Choi S.H., Washicosky K.J., et al. Amyloid-β peptide protects against microbial infection in mouse and worm models of Alzheimer's disease. Sci Transl Med. 2016;8(340):340ra72. doi: 10.1126/scitranslmed.aaf1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aguzzi A., Rajendran L. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron. 2009;64(6):783–790. doi: 10.1016/j.neuron.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 57.Tominaga N., Kosaka N., Ono M., et al. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat Commun. 2015;6:6716. doi: 10.1038/ncomms7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alexandrov L.B., Jones P.H., Wedge D.C., et al. Clock-like mutational processes in human somatic cells. Nat Genet. 2015;47(12):1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alcolea M.P., Greulich P., Wabik A., Frede J., Simons B.D., Jones P.H. Differentiation imbalance in single oesophageal progenitor cells causes clonal immortalization and field change. Nat Cell Biol. 2014;16(6):615–622. doi: 10.1038/ncb2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rand M.L., Wang H., Bang K.W., Packham M.A., Freedman J. Rapid clearance of procoagulant platelet-derived microparticles from the circulation of rabbits. J Thromb Haemost. 2006;4(7):1621–1623. doi: 10.1111/j.1538-7836.2006.02011.x. [DOI] [PubMed] [Google Scholar]
- 61.Stern R. Go fly a chitin: the mystery of chitin and chitinases in vertebrate tissues. Front Biosci (Landmark Ed) 2017;22:580–595. doi: 10.2741/4504. [DOI] [PubMed] [Google Scholar]
- 62.Weigel P.H., West C.M., Zhao P., Wells L., Baggenstoss B.A., Washburn J.L. Hyaluronan synthase assembles chitin oligomers with -GlcNAc(alpha1–>)UDP at the reducing end. Glycobiology. 2015;25(6):632–643. doi: 10.1093/glycob/cwv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castellani R.J., Siedlak S.L., Fortino A.E., Perry G., Ghetti B., Smith M.A. Chitin-like polysaccharides in Alzheimer's disease brains. Curr Alzheimer Res. 2005;2(4):419–423. doi: 10.2174/156720505774330555. [DOI] [PubMed] [Google Scholar]
- 64.Sotgiu S., Musumeci S., Marconi S., Gini B., Bonetti B. Different content of chitin-like polysaccharides in multiple sclerosis and Alzheimer's disease brains. J Neuroimmunol. 2008;197(1):70–73. doi: 10.1016/j.jneuroim.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 65.Schierer S., Ostalecki C., Zinser E., et al. Extracellular vesicles from mature dendritic cells (DC) differentiate monocytes into immature DC. Life Sci Alliance. 2018;1(6) doi: 10.26508/lsa.201800093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.