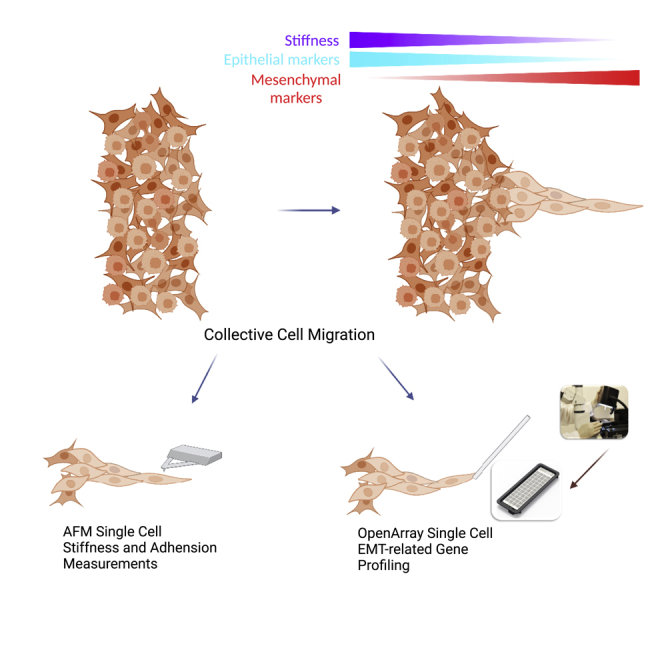
Summary
Collective cell migration is associated with cancer metastasis. Cancer fingers are formed when groups of migrating cancer cells follow the leader cells in the front. Epithelial to mesenchymal transition (EMT) is a critical process of cancer metastasis. However, the role of EMT in cancer finger formation remains unclear. In this work, we investigated the EMT-associated mechanical properties and gene expression at single-cell levels in non-small lung cancer fingers. We found that leader cells were more elastic and less sticky than follower cells. Spatial EMT-related gene expression profiling in cancer fingers revealed cellular heterogeneity. Particularly, SNAIL and VIM were found to be two key genes that positively correlated with leader cell phenotypes and controlled cancer finger formation. Silencing either SNAIL or VIM, decreased cancer cell elasticity, cancer finger formation and migration, and increased adhesiveness. These findings indicated that SNAIL and VIM are two driver genes for cancer finger formation.
Subject areas: Cell biology, Cancer, Transcriptomics
Graphical abstract
Highlights
-
•
Spatial mapping of EMT genes and mechanical properties of cancer finger at single cell level
-
•
Cancer cell elasticity and adhesiveness are two physical biomarkers for leader cells
-
•
SNAIL and VIM drive finger cell formation and are potential targets for therapy
Cell biology; Cancer; Transcriptomics
Introduction
Collective cancer cell migration and invasion, in which cancer cells invade cohesively as a multicellular unit, is a fundamental step in tumor progression (Friedl et al., 2012). A previous study based on a conventional cancer progression model proposed that tumor metastasis arose from single tumor cells, which became the foundation for epithelial-mesenchymal transition (EMT) and migratory cancer stem cells models (Irène Baccelli, 2012). Challenging the single cell metastasis model, experimental and clinical observations demonstrated that tumor cells could also migrate collectively during tumor metastasis (Cheung et al., 2016; Khalil and Friedl, 2010). Multiple genetically distinct tumor cell clones were found to be involved in tumor metastasis, such as breast, pancreas, and small cell carcinoma in the mouse model (Aceto et al., 2014; Maddipati and Stanger, 2015; McFadden et al., 2014), and in human metastatic prostate cancer patients (Gundem et al., 2015), shows the collective migration could involve multiple cell types with distinct properties. In sum, these observations provide accumulating evidence that collective cell migration contributes to metastasis.
Cellular heterogeneity was found in collective cancer cell migration and associated with the ability of cancer cell metastatic potential, leading to different therapeutic responses (Almendro et al., 2013; Fidler, 2003). Recent research showed that a small group of cancer cells led to collective cancer cell migration in the form of finger shape called “cancer finger” at the leading edge of the cluster trailed by follower cells from behind (Mayor and Etienne-Manneville, 2016). During migration, leader cells explored the tumor microenvironment and generated the migration and invasion path for follower cells (Gaggioli et al., 2007; Haeger et al., 2014). Unlike other normal epithelial cancer cells, the front of leader cells engaged with tissue substrate, and the other sides of leader cells contacted with the neighbor cancer cells. When groups of cancer cells migrate, the mechanotransduction between cells is going through integrins and cadherin-based cell-cell junctions (Weber et al., 2011). On the other hand, extracellular signals such as chemokines and growth factor signaling were received by the leader cells and transferred through the other cancer cells by intercellular communication in the group-like gap junction (Odenthal et al., 2016).
Collective migration, a critical process for tumor metastasis, is a result of incomplete epithelial-mesenchymal transition (EMT) (Yang et al., 2020). EMT increases actomyosin contractibility and weakens cell-cell junction because of lower expression of E-cadherin and desmosome, where mesenchymal-like cancer cells adopt mostly single cell movement. Nowadays, increasing evidence shows that EMT is a dynamic process instead of a binary one. Cancer cells can attain co-expression of both mesenchymal and epithelial markers that contribute to collective migration (Aiello and Kang, 2019; Lambert and Weinberg, 2021; Prieto-García et al., 2017). Although mesenchymal transition has been widely reported to facilitate tumor cell migration, the process may be incomplete (Vilchez Mercedes et al., 2021). For example, the H1975 non-small lung cancer cell is highly invasive and has stable expression of both epithelial markers and mesenchymal markers (Jolly et al., 2016). Although cell heterogeneity inside the collective cancer cell migration is well-known (Summerbell et al., 2020), the heterogeneity of EMT-associated features inside cancer fingers during collective migration remains unclear.
At the physical level, EMT is generally accompanied by changing of cell deformability, which plays a critical role in the potency of collective cancer migration because of larger volume of cells passing through the confinements, like dense ECM matrix and blood capillaries with diameters smaller than tumor cells (Coughlin et al., 2013). Generally, nontumorigenic cells are stiffer than malignant cancerous cells (Rebelo et al., 2013). During EMT, cytoskeleton proteins transform to changes in mechanical properties of the cancer cells in contraction, stretchability, deformability, which are mesenchymal and viscoelastic parameters. How EMT signaling and related genes affect the mechanical properties inside the finger-like associated with collective cancer cells is not well understood.
In this work, we spatially profiled the mechanical properties, such as stiffness and adhesiveness, and biological properties, like the expression of 53 EMT-related genes in single cell resolution, in cancer fingers during collective migration. We demonstrated that leader cells exhibited lower stiffness and elasticity than follower cells in the collective migration. The expression of SNAIL and VIM, two EMT related genes, were significantly upregulated in leader cells and then decreased spatially from the tip to the rear on three lung cancer cell lines tested. After inhibiting the above two genes, collective cell fingers were diminished, and mechanical properties were regulated. In addition, EMT induction by TGF-β treatment led to higher levels of elasticity and stiffness with reduced migration ability. These findings showed the essential roles of SNAIL and VIM regulate the biological and mechanical properties in cancer fingers during collective migration and could act as the potential gene targets for cancer metastasis inhibition.
Results and discussion
Morphological characterization of leader cells in finger-like collective cancer cell migration
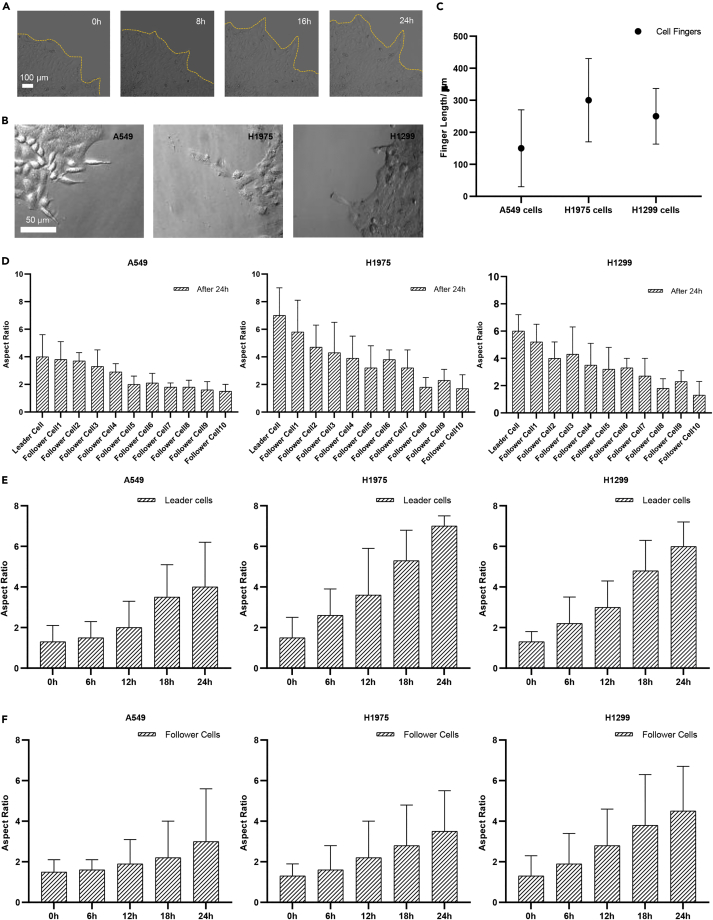
Lung cancer cells were cultured in Petri-dish for the cancer cell finger formation by using wound healing assays. A mass of cancer cells directed by several leader cells was observed to migrate unidirectionally for 24 h (Figure 1A and Video S1). In formed cell fingers, groups of cancer cells were led by one or two cancer cells at the tips of cancer cell fingers, which are so-called “leader cells”. The leader cells were found in all three non-small cell lung cancer cell lines, A549 cells, H1299 cells, and adenocarcinoma H1975 cells (Figure 1B). The displacement results of leader cells in migration assay for 24 h were analyzed. It represents H1975 cancer cells that formed much longer cancer cell fingers than A549 and H1299 cell lines (Figure 1C).
Figure 1.
The finger-like collective cancer cell migration and cancer cell morphology characterization
(A) finger-like collective cancer cell migration for 24 h.
(B) The finger formation of A549 cells, H1975 cells, and H1299 cells at 24 h time point.
(C)The average finger length of A549 cells, H1975 cells, and H1299 cells after 24 h migration.
(D) The aspect ratio of single cells in cancer cell fingers of A549 cells, H1975 cells, and H1299 cells under confinement and unconfinement.
(E) The leader cell’s morphology changes from 0 h to 24 h.
(F) The follower cell’s morphology changes from 0 h to 24 h (n=3 for each group)
Then, at single cell level, we observed that leader cells became much more elongated than follower cells inside the fingers by cell elongation length and width measurement. The aspect ratio of the cancer cells in the finger decreased gradually from the leader cell to follower cells (Figure 1D) based on the analysis of cell position ranking (Figure S1). The morphology of cancer leader cells was more elongated than follower cells with a larger aspect ratio. At time = 0 s, there were no cancer cell fingers, and the aspect ratio of cells remained at round shape with aspect ratio close to 1.0. During the collective cell migration and cancer cell finger formation, both leader cells and follower cells became elongated than before with the increasing cancer cell aspect ratio. After 24 h, the aspect ratio of leader cells increased more sharply than follower cells, which indicated that leader cells were more elongated than follower cells (Figures 1E and 1F). Leader cell morphology changing became more significant compared with follower cells after 12 h migration.
The above results showed that cancer cell fingers were formed during collective migration. For the cancer cell morphology, both leader cells and follower cells, especially leader cells, were elongated during collective migration and cancer cell finger formation. Usually, the acquisition of a mesenchymal-like elongated spindle morphology is associated with the development of the invasive phenotype in carcinomas and EMT (Kalluri and Weinberg, 2009; Odenwald et al., 2013).
Unique mechanical properties of leader cells in finger-like collective migration
During metastasis, tumor cells invade through the dense peritumoral stroma and intravasate into blood vessels that impose varying mechanical confinements to cancer cells. These mechanical confinements impede the collective migration stronger than single cells migration because of the larger volume of cells. Deformability is one of the critical factors that help cancer cells overcome this confinement. The microfluidic chip provides an efficient way to observe collective cell migration under a compressive space on a large scale in vitro. Based on the microfluidic chip study, we found that the collective cancer migration fingers (Figure 2A) have longer finger length under the confined space (device confinement height is 6 μm, cancer cell around 15 μm) than the ones in Petri-dish (Figures 2A, S2, and S3). In vivo leader cells were found to be the pioneers in tumor metastasis and invasion (Cheung and Ewald, 2014). The results also showed that physical confinement could stimulate and promote the collective cancer fingers formation.
Figure 2.
The stiffness of cells in cancer cell fingers
(A) cancer cell finger formation in confined and unconfined space with the channel height of 6 and 30μm.
(B) The Young’s Modulus of leader cells and follower cells in cancer fingers.
(C) The Young’s Modulus of normal cancer cells and mesenchymal cells.
(D) The Young’s Modulus of single cells in cancer fingers. (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ns not significantly, Two-tailed t-test)
Cell stiffness is a crucial mechanical property that is closely related to cell motility. Cell stiffness and adhesiveness are two important physical factors involved in tumor metastasis (Palmieri et al., 2015). Therefore, we first investigated the stiffness of the leader cells and follower cells (Figure S4). Atomic force microscope (AFM) is the most prevalent method used to determine cell stiffness by the quantitative parameter designated as Young's modulus, which is regarded as a marker of cell motility, especially in estimating the metastasis of cancer cells (Lekka, 2016). To study the mechanics of cancer cells during metastasis, we first determined the stiffness of cancer leader cells with an indentation method using an AFM and compared it with those of follower cells. We selected the cancer cells at the tip of cancer fingers as the leader cell and randomly picked follower cells behind to measure the Young’s modulus. Notably, in Figure 2B, it showed that the cancer leader cells had smaller Young’s Modulus values than the follower cells for A549, H1975, and H1299 cell lines. The results suggested that leader cells were significantly softer than follower cells. The Young’s modulus of cancer cells in normal condition is also higher than TGFβ induced lung cancer cells in the mesenchymal state, indicating that the leader cells may exhibit more mesenchymal property than epithelial property (Figure 2C) (Cascione et al., 2019).
We also measured the Young’s modulus of single cells within the cancer cell fingers according to the cell order based on their distance from the tip. We selected seven to nine cells from each cancer cell finger, and measured Young’s modulus of each selected cell. We found that cells at the front position had lower Young’s modulus values than their followers. The stiffness increased along with the cancer cell fingers from the leader cells to the follower cells for all A549, H1975, and H1299 cells (Figure 2D). In vivo, leader cells were in the front of the collective cancer cell migration, which were under the bio/chemical factor induction and physical space pressure (Clark and Vignjevic, 2015). The elasticity of cancer leader cells may provide them with the ability to invade through and survive in the confinement of the tissue and ECM, and the different cellular deformability with tumorigenic and metastatic potential may be underlined by the tumor heterogeneity (Swaminathan et al., 2011).
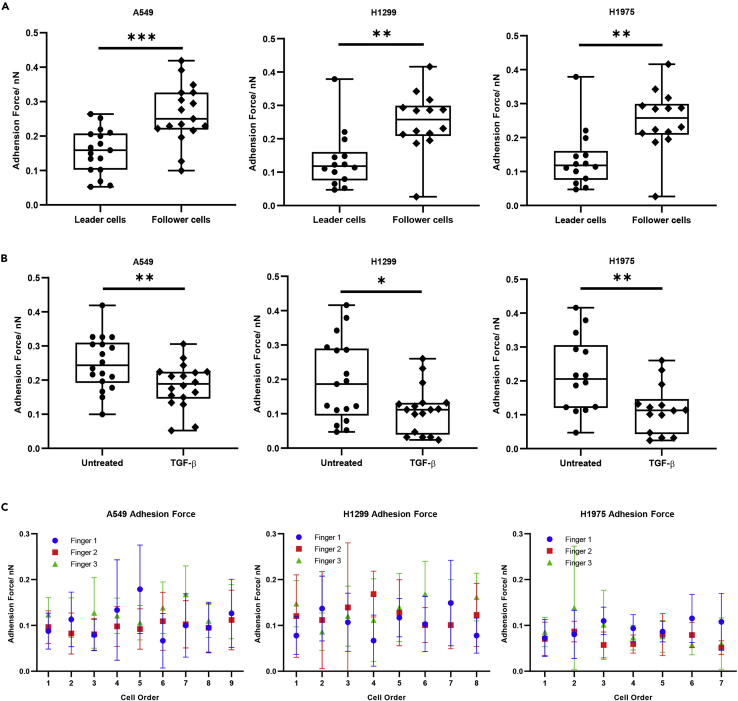
Other than stiffness, we also investigated the adhesiveness of each cancer cell during collective cancer cell migration by measuring the adhesion force. Studying cell adhesion is to measure how cells stick to and interact with each other and their environment. Some studies have suggested that the strength with which cells attach to the surrounding tumor tissue would indicate the likelihood of secondary tumor development (Fuhrmann et al., 2017). Tumor cells exhibit substantial variability in their adhesive strength. Understanding adhesive heterogeneity within an invasive population may provide another perspective on collective migration and finger formation.
In this work, we found that leader cells had lower adhesion force than the one of follower cells, which indicated that the leader cells were less adhesive than follower cells (Figure 3A). When compared with mesenchymal state (TGF-β treated) cells as a group, cancer cells show significantly higher adhesion force (Figure 3B). However, when we measured adhesion force in cancer fingers at single cell level, the adhesion force of each cancer cell does not show significant differences (Figure 3C). From the above results, we could identify intercellular heterogeneity in cancer fingers. Leader cells were more elastic and less adhesive than follower cells, which suggested that leader cells physically have more mesenchymal features than follower cells.
Figure 3.
The adhesion force of cancer cell in finger formation
(A) The adhesion force of leader cells and follower cells in cancer fingers.
(B) The adhesion force of cancer cells and mesenchymal state cells.
(C) The adhesion force of single cells in cancer fingers. (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ns not significantly, Two-tailed t-test)
Spatial EMT related gene expression profiling in cancer cell fingers
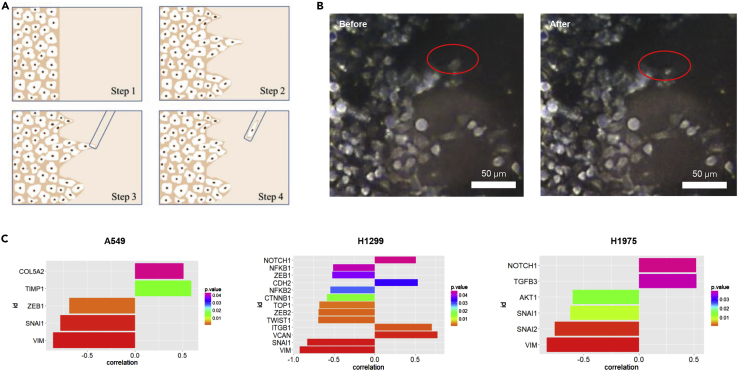
The above cell morphology and mechanical properties data indicated that the leader cells were more mesenchymal-like. To further investigate the EMT state in cancer fingers, we studied EMT related gene expressions at the single-cell level within the finger section. Individual leader cells and follower cells of the cancer fingers were picked up by capillary-based vacuum force (Figure 4A and 4B), which enabled the precise selection at single cell level for spatially mapping of gene expression. Figure 4B showed the result of how a leader cell was precisely picked up.
Figure 4.
EMT-related gene expression profiling of cancer cell fingers in single cell resolution
(A) schematic diagram of single cell picking from finger-like collective cancer migration.
(B) single cell picking by Kuckpick device.
(C) the significant correlation genes for A549, H1299, and H1975 cells
During collective cancer migration, leader cells led the group of cells to go through the extracellular matrix for tumor invasion while metastatic cancer cells undergo epithelial to mesenchymal transition. However, the role of EMT in physical properties and migratory behavior of leader cells remains unclear. Therefore, we then investigated the EMT related gene expression of cancer finger at single cell resolution. By single cell open array, we analyzed the expression profile of 53 EMT-related genes spatially for 16 single cells along each cancer cell finger of A549, H1975, and H1299, respectively, with three cancer fingers in total (Table S1). We found some genes were highly correlated with the spatial location of the cancer within the fingers. On the other hand, we also picked out spare single cells for each cell line from the Petri dish and performed an open array. Spare cells and leader cells separate into two different groups according to hierarchical clustering (Figure S5), which indicates the specialty of the leader cell.
Based on the results, we further analyzed the correlation of gene expression and cell location along the fingers of each cancer cell line. The significant genes in each cell line were shown in Figure 4C. In A549 cells, ZEB1, SNAI1, and VIM genes were negatively correlated with cancer cell locations, indicating the decreasing gene expression from leader cells to follower cells. COL5A2 and TIMP1 genes were positively correlated with cancer cell locations, indicating the increasing gene expression from leader cells to follower cells. For H1299 cells, SNAI1, VIM, TWIST1, ZEB2, TOP1, TNNB1, NFKB2, ZEB1, and NFKB1 showed negative correlation, whereas VCAN, ITGB1, CDH2, and NOTCH1 were positively correlated. For H1975 cells, VIM, SNAI1, SNAI2, and AKT1 genes were negatively correlated, whereas NOTCH1 and TGFB3 showed positive correlation. Among the significantly correlated genes, there were two common genes, SNAIL and VIM. Both genes were negatively correlated with cancer cell location in the cancer fingers.
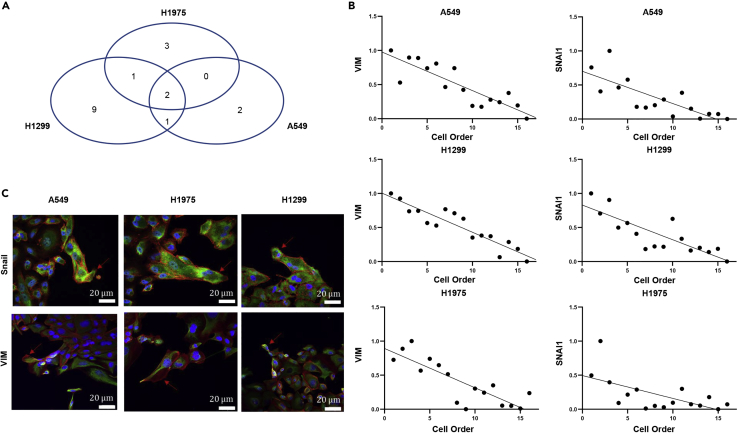
The above results demonstrated that for all the three lung cancer cell lines (A549 cells, H1975 cells, and H1299 cells), SNAIL and VIM, as two important EMT-related genes, are highly expressed in the leader cells. In addition, the expression of two genes both decreased from leader cells to follower cells (Figure 5A). The linear correlation of SNAIL and VIM with cancer cell locations in the cancer fingers were shown in Figure 5B for all three cancer cell lines. Both genes showed linear negative correlation with the cancer cell location. Leader cells expressed higher SNAIL and VIM than follower cells. The SNAIL and VIM expression results were verified by fluorescence immunostaining (Figure 5C). The fluorescence images showed that the leader cells expressed higher Snail and Vim proteins than the follower cells, showing that leader cells were with higher SNAIL and VIM gene expression than follower cells and the expressions were cell location dependent.
Figure 5.
SNAIL and VIM are upregulated in the leader cells of cancer fingers
(A) The significant expression genes of A549 cells, H1975 cells, and H1299 cells.
(B) The correlation of SNAIL and VIM gene expression in the cancer cell fingers of A549 cells, H1975 cells, and H1299 cells, respectively.
(C) The fluorescence immunostaining of SNAIL and VIM on cancer fingers (leader cell point out in red arrow).
SNAIL and VIM are two key EMT-related genes in cancer metastasis. SNAIL family members have been implicated in various important developmental processes, including neural differentiation, cell fate, and survival decisions, as well as left-right identity. Expression of SNAIL positively correlates with tumor grade, recurrence, metastasis, and poor prognosis in various tumors. The expression of SNAIL was found to be a critical regulator of EMT and closely associated with tumor metastasis (Wang et al., 2013). Many signaling molecules from tumor microenvironment have been shown to induce Snail expression (Jin et al., 2010). In addition, Snail-induced EMT accelerates metastasis through induction of immune-suppression (Kudo-Saito et al., 2009).
Meanwhile, VIM is now regarded as a canonical marker of EMT. After EMT, cells become more elongated and more dissociative with aggressive migration ability. The fact that leader cells from the three lung cancer cell lines had elongated morphology (Figure 1) and increased expression of mesenchymal markers (VIM and SNAIL) implies that mesenchymal transition is crucial for leader cells. However, we found that leader cells from A549 cells still have some CDH1 expression (Figure S6). These results are consistent with previous findings that incomplete EMT processes can also promote cell migration (Campbell et al., 2019). Moreover, the H1975 cells have been reported to have a stable hybrid epithelial/mesenchymal state, expressing both VIM and CDH1 (Jolly et al., 2016). But we found that the CDH1 was not detectable in finger cells from H1975, probably because of the loss of CDH1 expression finger cell formation. Previous cancer leader cell analysis mainly focused on the migration-related proteins, such as RhoA-Rock signaling which involves actin protein repolarization (Khalil and Friedl, 2010; Yamaguchi et al., 2015). In this work, we investigated the role of cancer cell EMT in cancer cell fingers single cell resolution. Our single cell analysis results suggest the heterogeneity of cancer cell fingers during collective cancer cell migration. Leader cell formation is regulated by SNAIL and VIM family genes. Therefore, the commonly upregulated genes SNAIL and VIM could be the potential target for inhibiting finger-like collective migration.
SNAIL and VIM are two key genes to determine collective migration by knockdown validation
To verify our finding, we knocked down the SNAIL and VIM genes on leader cells to investigate whether it can affect the cancer finger formation. SNAI1, SNAI2, and VIM genes were inhibited by siRNA transfection in cancer cell migration assay as shown by immunostaining and PCR methods (Figure S7). After genes inhibition, we characterized the finger formation during the collective cell migration. The time series images of collective cell migration were demonstrated in Figure 6A. In the same time frame, the cell migration without treatment is significantly faster than cells with SNAIL and VIM genes knocked down (Figure 6B). For A549, H1975, and H1299 cells, the cancer cell morphology was not significantly changed with time (Figure 6C). Cancer cell migration became substantially slower compared to the normal condition. We compared the collective cancer cell migration at normal condition, SNAIL inhibition condition and VIM inhibition condition together. The results showed that after SNAIL and VIM inhibition, the collective cancer cell migration boundaries were smoother with less finger formation than the normal condition, which indicated the SNAIL and VIM genes played important roles in cancer finger formation (Figure 6D). The cell cycle of cancer cells showed no significant changes after the inhibition of SNAIL and VIM (Figure S8).
Figure 6.
The characterization of collective cell migration after SNAIL and VIM genes inhibition
(A) the collective cancer cell migration after SNAIL and VIM gene expression.
(B) cell group migration speed. (n=3 for each group)
(C) the morphology changes of cancer cells in collective cancer migration in terms of aspect ratio.
(D) The cancer cell finger-like collective migration in A549 cells, H1299 cells, and H1975 cells.
(E) Young’s modulus of cancer cells in fingers under different conditions.
(F) The adhesion force of cancer cells in fingers under different conditions. (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ns not significantly, Two-tailed t-test).
After the inhibition treatment, we also measured the stiffness and adhesiveness of the cancer cells in collective migration. For all A549, H1975, and H1299 cells, the Young’s modulus of cancer cells with VIM inhibition was significantly higher than the negative control, which indicated that cancer cell stiffness increased significantly after the VIM inhibition (Figure 6E). However, the Young’s modulus of cancer cells with SNAIL inhibition was no significant difference with negative control for A549 and H1975 cells. Only the H1299 cells' Young’s modulus was significantly higher than the negative control. Rather than VIM inhibition, the SNAIL inhibition showed no obvious effect on regulation of cancer cell stiffness. In terms of cancer cell stickiness, both SNAIL and VIM inhibition increased the adhesion force of cancer cells when compared with normal conditions (Figure 6F). The results showed that SNAIL and VIM, two cancer genes, can both regulate cancer cell stickiness.
Thus, for lung cancer cell lines, these findings suggested that SNAIL and VIM genes play an essential role in collective cancer migration by regulating cancer cell morphology, mechanical properties, and finger formation. They may serve as potential targets for preventing metastasis by inhibiting finger formation.
Conclusions
In this work, we investigated how EMT-associated genes and mechanical properties of single cells affect cancer fingers during collective migration. Experimentally, the stiffness and adhesiveness suggest that leader cells are less elastic and sticky than follower cells in cancer fingers. The cancer heterogeneity is involved in cancer fingers that can be demonstrated by the spatial EMT-related genes profiling. We find that SNAIL and VIM genes are highly expressed in the leader cells as demonstrated in maps. After the knockdown of these two genes, cancer cell finger formation was inhibited, and cancer cells stiffness and adhesiveness increased with the decreasing migration abilities. These findings confirmed that SNAIL and VIM play an important role in regulating the biological and mechanical properties of cancer fingers, and indicated that SNAIL and VIM could be potentially used as targets for inhibiting cancer finger formation and cancer metastasis.
Limitations of the study
The study reveals the difference and relationship between leader and follower cells based on mechanical and molecular properties. The current study can only show the relationship on the particular timepoint after leader cell and follower cell formed. The question of the leader cell formed with the natural potential of the cell or competition with other cells remains unclear. Picking single cells at different timepoints followed by the sequencing technique may help screen out the potential gene which causes leader cell formation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Vimentin | cell signalling technology | Cat#5741 |
| Snail | cell signalling technology | Cat#3879 |
| Anti-rabbit IgG (H+L), F(ab')2 Fragment (Alexa Fluor® 488 Conjugate) | cell signalling technology | Cat#4412 |
| Alexa Fluor® 647 Phalloidin | cell signalling technology | Cat#8940 |
| Experimental models: Cell lines | ||
| A549 | ATCC | CCL-185 |
| NCI-H1299 | ATCC | CRL-5803 |
| NCI-H1975 | ATCC | CRL-5908 |
| Oligonucleotides | ||
| Snail | Thermo Fisher Scientific | Cat#4392420 |
| Vim | Thermo Fisher Scientific | Cat#4390824 |
| TaqMan OpenArray | Thermo Fisher Scientific | Cat#4471125 |
Resource availability
Lead contact
Further information and requests for resources and data should be directed to and will be fulfilled by the lead contact, Mengsu Yang (bhmyang@cityu.edu.hk).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Non-small lung cancer cell lines, A549 cell line, H1975 cell line, H1299 cell line were cultured in DMEM/F12 (1:1, cat# 11039-021, GIBCO®, USA) in the incubator with 5% CO2 at 37 degree (CO2 water-jacketed incubator, Nuaire®, USA). The three cell lines were epithelial adenocarcinoma. The culture mediums were all supplementary with 10% fetal bovine serum (FBS) and 1% penicillin-Streptomycin (10000 Units/mL of Penicillin and 10000 μg/mL of Streptomycin, Life technologies, USA).
Method details
Non-scratching wound healing assay
Non-scratching wound healing assay was performed in the Ibidi culture insert (Ibidi®, Munich, Germany), which consists of two cell culture chambers separated by an interval with the width of 500 μm. 70 μL of cell suspension at a density of 5 × 105 cells/mL and 10% serum medium were seeded in each chamber. After cell adhesion and growth for 24 hours followed by removal of the insert. Fresh medium containing 2% FBS were prepared for collective cells migration and finger formation.
AFM instrumentation, imaging, surface stiffness and stickiness analysis
A BioScope Catalyst atomic force microscope (Bruker Nano, Santa Barbara, CA) was used to measure mechanical properties of live cells. A conically shaped silicon nitride AFM probe with a spring constant of 0.1 N/m (MLCT, Bruker-nano, Santa Barbara, CA) was used. Young’s modulus and adhesion force were derived by fitting curves extracted from 10-20 measurement on one single cells with Sneddon model.
Cell collection with Kuiqpick from cultures
Kuiqpick device was used according to the manual for collecting live cells. Single cell was carefully removed by using a syringe. The collected lung cancer cells were transferred onto a 6-well culture plate containing complete DMEM culture medium. Cells were maintained in PBS during the collection. Photographs were taken daily to record the growth and proliferation of cells.
Single cell analysis
Single cells from fingers were isolated by Kuiqpick with 30μm diameter of capillary tube. Cells were lysed, reverse transcribed and amplified with Single Cell-to-CT qRT-PCR Kit (Invitrogen) and a pool of Taqman-probe (please see the attachment). The amplified cDNA was then quantified by OpenArray.
SNAIL and VIM inhibitor transfection using lipofectamine
Prepare lipocomplex: add 10.56 ul lipofectamine 2000 to 220 ul OptiMEM and 2.5 ul (50 pmol) of Negative control/Snail mimics or 5 ul (100 pmol) Negative control/Snail inhibitors to 100 ul OptiMEM. Combine the 2 mixtures and incubate at room temperature for 20-30 minutes. Prepare cancer cell samples with the density of 5 x 104 cells in 1.76 mL OptiMEM medium. Add 50 ul lipocomplex containing RNAs to each well. Incubate for 5-6 hours at 37C. Change to complete medium and incubate for 2 more days.
Microfluidic chip fabrication
The confined microfluidic device was fabricated according to the protocol we published previously using soft lithography technique by molding poly(dimethylsiloxane) (PDMS) (10:1 silicone elastomer with curing agent, Sylgard 184, Dow Corning®, Midland, MI.) against a master made of printed circuit board (PCB,75×125×1.6mm, Kinsten® Pty. Ltd.). The microfluidic device consisted of two layers molded from the PCB masters etched by ferric trichloride solution (1.56×10-3 mol/mL) for different times: the top layer was etched for 45 minutes for the generation of master with the height of 21 μm, and the bottom layer was etched for 16 minutes such that the height of the microchannels was about 6 μm, respectively. Inlets and outlets on the PDMS replica were drilled by circular holes puncher with the diameter of 1.22 mm. Two PDMS layers were treated by air plasma (Plasma cleaner/sterilizer, PPC-3XG, Harrick®, NY, US) and bonded together with precise alignment under the microscope.
Quantification and statistical analysis
Statistical analysis
For both conventional assays and on-chip analysis, at least triplicated experiments were performed for each cell line. In each condition, at least 25 cells were measured, and all quantitative data were presented as means ± s.d. Single cell gene analysis was dependent on the t-test significant results.
Acknowledgments
We gratefully acknowledge fruitful discussion with Dr. Zhang Liang of City University of Hong Kong Department of Biomedical Science. The author gratefully acknowledges funding support by a grant from the National Key Research and Development Program of China (2018YFA0901104), Hong Kong Research Grant Council (CityU_11319516) (M.Y), Basic Research Projects of Shenzhen Knowledge Innovation Program (JCYJ20170818095453642 and JCYJ20180307123759162) (MY), and Research Impact Fund (R1020-18F) of the Hong Kong Research Grant Council.
Author contributions
H.Z. and M.Y. conceived the initial idea for this research. M.Y. guided the work. H.Z., Z.Y., and Y.C. conducted experiment, data analysis, and drafted the manuscript. S.A.Y., M.K.A., T.S., and T.X. helped single cell picking, manuscript revision, and data handling. H.Z., Z.Y., and Y.C. contributed equally. All the authors were responsible for writing the paper and have given approval to the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103917.
Supplemental information
Data and code availability
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aceto N., Bardia A., Miyamoto D.T., Donaldson M.C., Wittner B.S., Spencer J.A., Yu M., Pely A., Engstrom A., Zhu H. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello N.M., Kang Y. Context-dependent EMT programs in cancer metastasis. J. Exp. Med. 2019;216:1016–1026. doi: 10.1084/jem.20181827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almendro V., Marusyk A., Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu. Rev. Pathol. Mech. Dis. 2013;8:277–302. doi: 10.1146/annurev-pathol-020712-163923. [DOI] [PubMed] [Google Scholar]
- Campbell K., Rossi F., Adams J., Pitsidianaki I., Barriga F.M., Garcia-Gerique L., Batlle E., Casanova J., Casali A. Collective cell migration and metastases induced by an epithelial-to-mesenchymal transition in Drosophila intestinal tumors. Nat. Commun. 2019;10:2311. doi: 10.1038/s41467-019-10269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascione M., Leporatti S., Dituri F., Giannelli G. Transforming growth factor-β promotes morphomechanical effects involved in epithelial to mesenchymal transition in living hepatocellular carcinoma. Int. J. Mol. Sci. 2019;20:7–9. doi: 10.3390/ijms20010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.J., Ewald A.J. Invasive leader cells: metastatic oncotarget. Oncotarget. 2014;5:1390. doi: 10.18632/oncotarget.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.J., Padmanaban V., Silvestri V., Schipper K., Cohen J.D., Fairchild A.N., Gorin M.A., Verdone J.E., Pienta K.J., Bader J.S. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl. Acad. Sci.U S A. 2016;113:E854–E863. doi: 10.1073/pnas.1508541113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.G., Vignjevic D.M. Modes of cancer cell invasion and the role of the microenvironment. Curr. Opin. Cell Biol. 2015;36:13–22. doi: 10.1016/j.ceb.2015.06.004. [DOI] [PubMed] [Google Scholar]
- Coughlin M.F., Bielenberg D.R., Lenormand G., Marinkovic M., Waghorne C.G., Zetter B.R., Fredberg J.J. Cytoskeletal stiffness, friction, and fluidity of cancer cell lines with different metastatic potential. Clin. Exp. Metastasis. 2013;30:237–250. doi: 10.1007/s10585-012-9531-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler I.J. The pathogenesis of cancer metastasis: the’seed and soil’hypothesis revisited. Nat. Rev. Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- Friedl P., Locker J., Sahai E., Segall J.E. Classifying collective cancer cell invasion. Nat. Cell Biol. 2012;14:777–783. doi: 10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- Fuhrmann A., Banisadr A., Beri P., Tlsty T.D., Engler A.J. Metastatic state of cancer cells may be indicated by adhesion strength. Biophys. J. 2017;112:736–745. doi: 10.1016/j.bpj.2016.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggioli C., Hooper S., Hidalgo-Carcedo C., Grosse R., Marshall J.F., Harrington K., Sahai E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- Gundem G., Van Loo P., Kremeyer B., Alexandrov L.B., Tubio J.M.C., Papaemmanuil E., Brewer D.S., Kallio H.M.L., Högnäs G., Annala M. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeger A., Krause M., Wolf K., Friedl P. Cell jamming: collective invasion of mesenchymal tumor cells imposed by tissue confinement. Biochim. Biophys. Acta. 2014;1840:2386–2395. doi: 10.1016/j.bbagen.2014.03.020. [DOI] [PubMed] [Google Scholar]
- Irène Baccelli A.T. The evolving concept of cancer and metastasis stem cells. J. Cell Biol. 2012;198:281–293. doi: 10.1083/jcb.201202014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Yu Y., Zhang T., Zhou X., Zhou J., Jia L., Wu Y., Zhou B.P., Feng Y. Snail is critical for tumor growth and metastasis of ovarian carcinoma. Int. J. Cancer. 2010;126:2102–2111. doi: 10.1002/ijc.24901. [DOI] [PubMed] [Google Scholar]
- Jolly M.K., Tripathi S.C., Jia D., Mooney S.M., Celiktas M., Hanash S.M., Mani S.A., Pienta K.J., Ben-Jacob E.L.H. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget. 2016;7:27067–27084. doi: 10.18632/oncotarget.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A.A., Friedl P. Determinants of leader cells in collective cell migration. Integr. Biol. 2010;2:568–574. doi: 10.1039/c0ib00052c. [DOI] [PubMed] [Google Scholar]
- Kudo-Saito C., Shirako H., Takeuchi T., Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Lambert A.W., Weinberg R.A. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat. Rev. Cancer. 2021;21:325–338. doi: 10.1038/s41568-021-00332-6. [DOI] [PubMed] [Google Scholar]
- Lekka M. Discrimination between normal and cancerous cells using AFM. Bionanoscience. 2016;6:65–80. doi: 10.1007/s12668-016-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddipati R., Stanger B.Z. Pancreatic cancer metastases harbor evidence of polyclonality. Cancer Discov. 2015;5:1086–1097. doi: 10.1158/2159-8290.CD-15-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R., Etienne-Manneville S. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 2016;17:97. doi: 10.1038/nrm.2015.14. [DOI] [PubMed] [Google Scholar]
- McFadden D.G., Papagiannakopoulos T., Taylor-Weiner A., Stewart C., Carter S.L., Cibulskis K., Bhutkar A., McKenna A., Dooley A., Vernon A. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. 2014;156:1298–1311. doi: 10.1016/j.cell.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenthal J., Takes R., Friedl P. Plasticity of tumor cell invasion: governance by growth factors and cytokines. Carcinogenesis. 2016;37:1117–1128. doi: 10.1093/carcin/bgw098. [DOI] [PubMed] [Google Scholar]
- Odenwald M.A., Prosperi J.R., Goss K.H. APC/β-catenin-rich complexes at membrane protrusions regulate mammary tumor cell migration and mesenchymal morphology. BMC Cancer. 2013;13:12. doi: 10.1186/1471-2407-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri V., Lucchetti D., Maiorana A., Papi M., Maulucci G., Calapà F., Ciasca G., Giordano R., Sgambato A., De Spirito M. Mechanical and structural comparison between primary tumor and lymph node metastasis cells in colorectal cancer. Soft Matter. 2015;11:5719–5726. doi: 10.1039/c5sm01089f. [DOI] [PubMed] [Google Scholar]
- Prieto-García E., Díaz-García C.V., García-Ruiz I., Agulló-Ortuño M.T. Epithelial-to-mesenchymal transition in tumor progression. Med. Oncol. 2017;34:122. doi: 10.1007/s12032-017-0980-8. [DOI] [PubMed] [Google Scholar]
- Rebelo L.M., de Sousa J.S., Mendes Filho J., Radmacher M. Comparison of the viscoelastic properties of cells from different kidney cancer phenotypes measured with atomic force microscopy. Nanotechnology. 2013;24:55102. doi: 10.1088/0957-4484/24/5/055102. [DOI] [PubMed] [Google Scholar]
- Summerbell E.R., Mouw J.K., K Bell J.S., Knippler C.M., Pedro B., Arnst J.L., Khatib T.O., Commander R., Barwick B.G., Konen J., et al. Epigenetically heterogeneous tumor cells direct collective invasion through filopodia-driven fibronectin micropatterning. Sci. Adv. 2020;6:1–15. doi: 10.1126/sciadv.aaz6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan V., Mythreye K., O’Brien E.T., Berchuck A., Blobe G.C., Superfine R. Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res. 2011;71:5075–5080. doi: 10.1158/0008-5472.CAN-11-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez Mercedes S.A., Bocci F., Levine H., Onuchic J.N., Jolly M.K., Wong P.K. Decoding leader cells in collective cancer invasion. Nat. Rev. Cancer. 2021;21:592–604. doi: 10.1038/s41568-021-00376-8. [DOI] [PubMed] [Google Scholar]
- Wang Y., Shi J., Chai K., Ying X., Zhou B.P. The role of snail in EMT and tumorigenesis. Curr. Cancer Drug Targets. 2013;13:963–972. doi: 10.2174/15680096113136660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G.F., Bjerke M.A., DeSimone D.W. Integrins and cadherins join forces to form adhesive networks. J. Cell Sci. 2011;124:1183–1193. doi: 10.1242/jcs.064618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N., Mizutani T., Kawabata K., Haga H. Leader cells regulate collective cell migration via Rac activation in the downstream signaling of integrin β1 and PI3K. Sci. Rep. 2015;5:1–8. doi: 10.1038/srep07656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Antin P., Berx G., Blanpain C., Brabletz T., Bronner M., Campbell K., Cano A., Casanova J., Christofori G., et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020;21:341–352. doi: 10.1038/s41580-020-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.