Abstract
Mountains create steep environmental gradients that are sensitive barometers of climate change. We calibrated 10 statistical models to formulate ensemble ecological niche models for 12 predominantly alpine, flightless grasshopper species in Aotearoa New Zealand, using their current distributions and current conditions. Niche models were then projected for two future global climate scenarios: representative concentration pathway (RCP) 2.6 (1.0°C rise) and RCP8.5 (3.7°C rise). Results were species specific, with two-thirds of our models suggesting a reduction in potential range for nine species by 2070, but surprisingly, for six species, we predict an increase in potential suitable habitat under mild (+1.0°C) or severe global warming (+3.7°C). However, when the limited dispersal ability of these flightless grasshoppers is taken into account, all 12 species studied are predicted to suffer extreme reductions in range, with a quarter likely to go extinct due to a 96–100% reduction in suitable habitat. Habitat loss is associated with habitat fragmentation that is likely to escalate stochastic vulnerability of remaining populations. Here, we present the predicted outcomes for an endemic radiation of alpine taxa as an exemplar of the challenges that alpine species, both in New Zealand and internationally, are subject to by anthropogenic climate change.
Keywords: alpine, climate change, ecological niche modelling, ensemble modelling, biomod2, fragmentation, FRAGSTATS
1. Introduction
Global climate change, whether driven by anthropogenic pollution or Milankovitch cycles, is expressed unevenly across the landscape. In particular, latitude and elevation influence the extent and rate of change in local conditions [1]. Where these gradients intersect in the alpine zone, organisms that endure diurnal and seasonal extremes in temperature and water availability, high winds and elevated UV-radiation during a contracted growth season are exposed to rapid environmental shifts. Among animals, the survival and reproduction of ectotherms is likely to be especially sensitive to displacement of climate isoclines [2,3].
Although most ecosystems will be impacted in some way, arctic, alpine and boreal biomes are predicted to be particularly vulnerable to the ecological impacts of anthropogenic climate change when compared with other systems [4–6]. Growing evidence suggests that global warming results in rapid temperature increases at higher elevations [7]. This is reflected by rising treelines and snowlines, and reduction in snowpack depth and longevity [8–10]. For alpine species, the consequences of such changes include upward shifts in both lower and upper elevational range limits where the landscape permits, increasing inter-species competition at higher elevation, reduction in richness at lower elevation and shifts in phenology [11–14].
Alpine habitat exists at increasing elevations from the poles towards the equator and in total is estimated to currently cover approximately 2.64% of Earth's total land area outside Antarctica (3.56 Mkm2) [15,16]. Temperature decreases by approximately 0.65°C for every 100 m of elevation subject to geographic, seasonal and diurnal variance [17,18]. In mountainous areas, this results in the stratification of conditions that is most evident in the abrupt zonation of alpine vegetation, including a clearly defined upper limit to trees on most mountains [19,20]. As a consequence, mountain environments are a sensitive barometer of global climate change, which can yield rapid shifts in the distribution and connectivity of alpine ecosystems [21,22].
The limited and patchy nature of montane topography limits opportunities for alpine adapted organisms to disperse and/or shift their ranges and for alpine environments to persist in the face of global warming [22,23]. For alpine specialists, habitat is typically isolated by an inhospitable low-elevation sea across which dispersal may be limited [24,25]. Fragmentation and patch size reduction, when coupled with poor dispersal ability, will undoubtedly negatively affect the ability of species to track and/or shift their range under future climatic change pressure.
Predicting species responses to global warming, while encompassing all relevant variables is impractical [26,27], but a tool that can be used to explore some facets is ecological niche modelling (ENM). Ecological niche models use data of known occurrences of a species (realized niche, RN) and its current environmental envelope to determine what environmental conditions are significant proxies for constraint of species' range [28,29]. The current environmental envelope of a species is estimated using environmental covariate data in a correlative statistical modelling framework. The model is then used to hindcast or forecast the potential distribution of the species of interest (potential niche, PN), providing an estimate of where these environmental conditions are most likely to occur in the past or future. Inferences from niche modelling will be limited if sampling of presence data is non-random and absence data is lacking [29], and is only useful for predicting future distributions if the environmental variables included are relevant and life-history characteristics are taken into account [28]. To improve model predictability, ensemble modelling is commonly used in ENM. Multiple statistical models are applied to datasets, and predictive outputs meeting an evaluation threshold are then combined in a single ensemble model [30]. Ensemble modelling allows the user to capture predictive signal from multiple models, reducing over-reliance on the assumptions of individual algorithms [30,31].
We examined environmental constraints on an endemic radiation of grasshoppers in Aotearoa New Zealand that belong to the acridid family Catantopinae [32]. Before the arrival of humans about 800 years ago [33], most of the New Zealand landscape was clothed in forest while open habitat was limited to high-elevation or semi-arid hinterland and braided riverbeds [34]. Though relatively scarce, these open areas support a rich biota with high species endemism [35]. The alpine zone in particular exerts strong selective pressure on the fauna and flora resulting in specialized adaptations. For example, many New Zealand alpine insects are freeze-tolerant, including the grasshoppers in this study, which can survive equilibrium freezing year round and at any life-history stage [36]. Grasshoppers are suitable representatives of the alpine fauna, because their distributions are closely associated with climatic gradients that predict the availability of open habitat [37,38], they display alpine adaptations and have limited dispersal capacity due to a lack of functional wings. They are therefore particularly vulnerable to the types of environmental change exerted by climate warming that is being documented around the globe [39]. We model the environmental envelope of 12 species of New Zealand flightless grasshoppers (10 alpine species and two lowland relatives) using presence and absence data in conjunction with environmental covariate data in a statistical modelling framework. We assess the potential impacts of climate change on projected distributions by transferring the ensemble models to future climate change scenarios to predict how their optimal habitat might grow, shrink and/or fragment over the next 50 years of climate change.
2. Material and methods
2.1. Collection of species location records
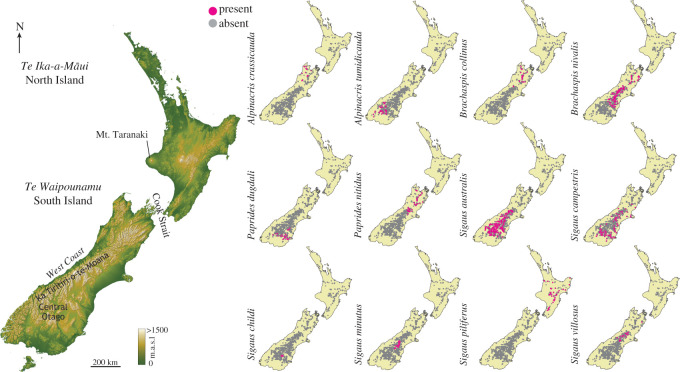
For 12 grasshopper species, location records were collated from insect collections, journal articles, theses, books and Crown Pastoral Lease Tenure Reviews (CPLTR) (electronic supplementary material S1) published between 1967 and 2016. These 12 endemic New Zealand grasshoppers consist of 10 high-elevation species: Alpinacris crassicauda, Alpinacris tumidicauda, Brachaspis collinus, Brachaspis nivalis, Paprides dugdali, Paprides nitidus, Sigaus australis, Sigaus campestris, Sigaus piliferus and Sigaus villosus, and two low-elevation relatives: Sigaus childi and Sigaus minutus. Given the range of sources used to obtain location data, our sample does not suffer from the common bias of only collecting from sites close to access roads. In particular, our location data benefit from the use of CPLTR that are produced by Land Information New Zealand and contain conservation reports and ecological surveys carried out by the Department of Conservation on pastures throughout Te Waipounamu (South Island), New Zealand. Additional records were retrieved from our specimen collections at Massey University. World Geodetic System 1984 (WGS84) coordinates (latitudes and longitudes) were obtained for each location record using NZ Topo Map [40]. Location records for two additional endemic lowland grasshopper species (Phaulacridium marginale and Phaulacridium otagoense) were included in the dataset, but not modelled in this study [37]. Both of these species have low-elevation distributions and provided additional location searches and absence data that increase the accuracy of the ENMs. Coordinates were reduced to two decimal places and duplicate records within species were removed, in order to ensure only one record per approximately 1 km2 for each species. Coordinates were also mapped for each species to ensure there were no outliers due to incorrect species identification. Following filtering, a total of 949 location records were available for our analysis (figure 1). All location records and their sources can be found in electronic supplementary material S1.
Figure 1.
Relief map of Aotearoa New Zealand with locations referred to in the text (left). Distribution maps (right) show the locations of presence and absence data points input into ecological niche models for each of 12 New Zealand grasshopper species (Acrididae; Catantopinae).
2.2. Predictor variable layers
In order to define and project the PN of each grasshopper species, 19 bioclimatic variables were obtained from the Worldclim v. 1.4 database for two time periods—current (e.g. data averaged from 1960 to 1990) and future (e.g. data predicted and averaged from 2061 to 2080; electronic supplementary material, S2), at a resolution of 30 arc-seconds (approximately 1 km2) [41–43]. Two versions of predicted climate variables for the future were acquired, derived from representative concentration pathway (RCP) 2.6 and RCP8.5 models [44]. These represent potential low and high greenhouse gas concentration scenarios, respectively (electronic supplementary material, S3), and mean global temperature increases of 1.0°C (RCP2.6) and 3.7°C (RCP8.5). All climate layers were generated using the MIROC-ESM global climate model [45]. The bioclimatic layers were cropped in QGIS v. 2.16.1 [46] to the extent of New Zealand between latitudes −49° and −32° and longitudes 165° and 180°. To reduce variable collinearity and minimize model over-fitting, a variance inflation factor analysis was implemented in R using the ‘usdm’ package, applying a threshold of 0.85 [47]. This resulted in retention of seven independent ecologically informative bioclimatic variables.
Two additional environmental data layers were acquired from the New Zealand Land Resource Information System portal (https://lris.scinfo.org.nz/): fundamental soil layers New Zealand Soil Classification and Vegetative Cover Map of New Zealand [48,49]. The soil and vegetation data layers were rasterized, clipped and rescaled from the original files. These variables have previously been found to contribute significantly to models of grasshopper distribution [50,51]. These layers represent the current approximate state of vegetation and soil in New Zealand, and, as no future (e.g. 2070) GIS files exist for soil and vegetation cover in New Zealand, they were used as static layers throughout the modelling process. Ecological niche models that include such static layers are known to perform as well as, or better than, models where only dynamic variables are included [52].
2.3. Ecological niche models
We used ENM analyses implemented in the R package ‘biomod2’ v. 3.3–7 [53]. Ten different statistical modelling methods were applied to the nine environmental variables and presence/absence data of each individual grasshopper species: generalized linear model (GLM), generalized boosting model/boosted regression tree (GBM), generalized additive model (GAM), classification tree analysis (CTA), artificial neural network (ANN), surface range envelope (SRE), flexible discriminant analysis (FDA), multiple adaptive regression splines (MARS), random forest (RF) and maximum entropy (MAXENT). The Biomod2 BIOMOD_Modeling() function was used to initially run each statistical model, with all model parameters kept at default values. Eighty per cent of the input data were used to calibrate the models, with the remaining 20% used to test them. The Prevalence parameter was set at 0.5, allowing absences and presences to be weighted equally and VarImport was set to 3, allowing three permutations to estimate variable importance. Calibration and evaluation of each statistical model was repeated three times, resulting in 30 models per species. A summary of raw, clean, training and testing absence and presence points input into the models can be found in the electronic supplementary material, S4.
Model evaluation of the 30 ENMs for each species was carried out using two methods: receiver operating characteristic (ROC) and true skill statistic (TSS). Models with ROC values of 0.9–1 and TSS values of 0.8–1 are considered to have ‘excellent’ predictive power (accuracy) [54]. Following initial modelling and model evaluation, the Biomod2 BIOMOD_EnsembleModeling() function was used to formulate ensemble models for each species. Ensemble models were built using the ‘all’ total consensus model option, with the eval.metric set to ROC, and eval.metric.quality.threshold set to 0.9, so only models with ROC scores greater than 0.9 (or greater than 0.8 for three species) were used in building the ensembles. The BIOMOD_Projection() function was then applied, projecting ensemble forecasts for the two future climate scenarios. Plots were subsequently made for the three datasets (current, RCP2.6 and RCP8.5) using the ensemble model weighted mean model (EMwm) output. EMwm variable importance was calculated by applying the weights produced in the ensemble model to the variable importance results of the initial 30 model dataset. These were then averaged across the three model runs, and EMwm variable importance was calculated by summing the total of these averages for each predictor variable and dividing by the number of modelling methods used (10) [55]. Final scores of variable importance were converted into percentages, with higher percentages indicating a variable was more influential in enabling models to estimate the niche of a particular species [56].
2.4. Fragmentation
Binary raster layers of each ENM were generated for range change and fragmentation statistical analyses. These binary vectors were generated from the 30 niche model dataset, where each pixel that scored greater than a predetermined cut-off value in greater than 50% of models was ranked as a 1, and all other pixels as 0. When comparing binary vectors between current and future models, Biomod2 ranks pixels as either, ‘Never occupied’ (pixels were unoccupied and remain unoccupied between models), ‘Always occupied’ (pixels were occupied and remain occupied between models), ‘Lost’ (pixels were occupied but become unoccupied in the future RCP scenario) or ‘Gained’ (pixels that were unoccupied in the current model become occupied in the RCP model). From this information range change statistics were calculated with two contrasting models. The dispersal model assumed the species in question will occupy any pixels ‘gained’ in future models, implying no limitation on dispersal and/or establishment; the model essentially treats potential habitat as realized habitat. At the other extreme, the non-dispersal model precluded occupation of disconnected ‘gained’ pixels, which means nearby but separate habitat patches as well as more distant patches (e.g. on a separate island) are assumed to be uncolonized. We also applied FRAGSTATS, implemented in R package ‘SDM Tools’ v. 1.1-221 [57] (see McGarigal [58] for a detailed description of FRAGSTATS metrics), to the binary files to estimate fragmentation statistics that can be compared between different RCP scenarios from their current niche models. Pixels that are connected within each of the binary files are given unique patch identities, and the area and number of these patches calculated for each scenario. We excluded small patch fragments that were less than 0.1 km2 from the fragmentation statistics analysis. Methods and scripts implemented in this study can be found in the electronic supplementary material S5 and S6.
3. Results
After filtering, nine of the 12 grasshopper species had more than 30 location records as ‘present’ (table 1). A minimum of 30 occurrences are recommended for ENMs, below which model accuracy decreases, and variance of model accuracy increases [59–62]. The majority of evaluation scores averaged across the three model runs were greater than 0.9 ROC (65/120), but for the four most well-sampled and widespread species (B. nivalis, P. nitidus, S. australis and S. campestris), no individual model had a ROC score greater than 0.9 (electronic supplementary material, S7). Fewer models had TSS scores greater than 0.8 (55/120), with none gaining a TSS greater than 0.8 for B. nivalis, S. australis and S. campestris. The models that most commonly had the highest model evaluation scores across species were GBM, followed by MAXENT and RF, while CTA, ANN and SRE models consistently had low evaluation scores across species (electronic supplementary material, S7). For the species with ROC scores below the 0.9 threshold, we applied a ROC threshold of greater than 0.8 to formulate EMwm ensemble models. For most of the grasshopper species, the EMwm ensemble model had an improved ROC score compared with individual model outputs (electronic supplementary material, S7). The exception was the rare and localized grasshopper S. childi, whose RF model and EMwm model could not be improved upon (both had a ROC score of 1). The EMwm models for B. nivalis, S. australis and S. campestris had ROC scores greater than 0.9, improving on the predictive accuracy of their individual models. Ten species (A. crassicauda, A. tumidicauda, B. collinus, B. nivalis, P. dugdali, P. nitidus, S. childi, S. minutus, S. piliferus and S. villosus) had EMwm sensitivity scores (percentage correctly predicted presences) greater than 90%. Seven of these species also had specificity scores (proportion of correctly predicted absences) greater than 90% (electronic supplementary material, S7). The two species, S. australis and S. campestris, for which niche models had the poorest fit (i.e. scored less than 90% for both sensitivity and specificity) were both well-sampled, widespread, grasshoppers. In the case of S. campestris, this relatively low model fit can be explained by the availability of suitable habitat in North Island which is out of reach of this flightless insect (the assumption that realized distribution encompasses most of potential habitat is therefore invalid in this case). For S. australis, it is possible that the assumption of niche stability and conservatism has been invalidated by recent range shifts associated with anthropogenic land-use change [38].
Table 1.
Environmental characteristics of 12 flightless, alpine New Zealand grasshopper species modelled in this study. The most important predictor of species distribution (from nine variables) is shown for each species (EMwms). Predictor variable importance scores for all models and all species are in electronic supplementary material, S8. Range-change percentages are based on the loss/gain/stability of pixels between vector binary maps (electronic supplementary material, S9). Predicted range changes include all gained pixels (with dispersal), while range change with no dispersal includes only gain of currently occupied habitat fragments.
| species | elevation range (m.a.s.l) | number of presence points | number of absence points | most important predictor variable | % range change from present (with dispersal) |
% range change from present (no dispersal) |
||
|---|---|---|---|---|---|---|---|---|
| RCP2.6 | RCP8.5 | RCP2.6 | RCP8.5 | |||||
| A. crassicauda | 975–1680 | 16 | 933 | mean diurnal range | −43 | 208 | −61 | −11 |
| A. tumidicauda | 600–1830 | 49 | 900 | precipitation of driest month | −74 | −35 | −74 | −37 |
| B. collinus | 1000–2000 | 30 | 919 | mean diurnal range | −95 | −66 | −96 | −89 |
| B. nivalis | 450–2000 | 93 | 856 | isothermality | 59 | 134 | −9 | −7 |
| P. dugdali | 400–1160 | 29 | 920 | precipitation seasonality | −75 | −96 | −82 | −97 |
| P. nitidus | 600–1830 | 91 | 858 | mean temp. of driest quarter | 12 | 17 | −61 | −35 |
| S. australis | 285–2020 | 278 | 671 | annual mean temp. | −74 | −93 | −75 | −93 |
| S. campestris | 0–1550 | 100 | 849 | soil | −45 | −67 | −45 | −67 |
| S. childi | 160–420 | 41 | 908 | precipitation of driest month | −47 | 154 | −100 | −89 |
| S. minutus | 500–1180 | 40 | 909 | mean diurnal range | 4 | 191 | −80 | −44 |
| S. piliferus | 725–1100 | 45 | 901 | mean temp. of wettest quarter | −15 | −58 | −45 | −77 |
| S. villosus | 1370–2130 | 23 | 926 | annual mean temp. | 168 | −77 | −20 | −94 |
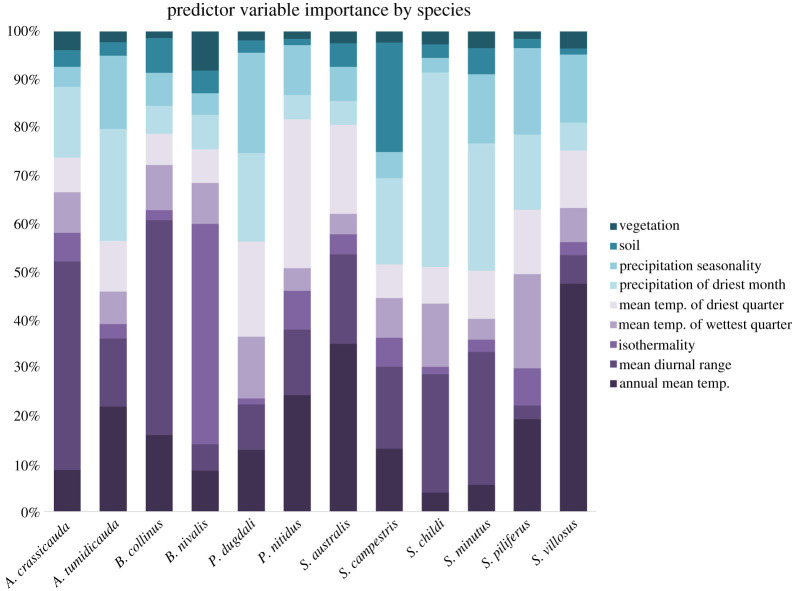
Considering the EMwm models only, eight of the nine predictor variables were important in explaining the distributions of the 12 grasshopper species (see electronic supplementary material, S8 for other models). Vegetation did not rank as an important predictor variable for any species and soil was influential for only one species, S. campestris (table 1 and figure 2). Temperature was an important predictor of habitat for most species (annual mean temperature, mean diurnal range and mean temperature of driest quarter). Water availability given as precipitation of driest month and precipitation seasonality were influential for three species (table 1). For one species, isothermality was the most important predictor (B. nivalis). Despite passing the plausibility criteria, it must be remembered that all of these variables are likely to be proxies for a combination of environmental traits.
Figure 2.
Environmental predictor variables differ in their importance in models of presence for 12 species of New Zealand alpine grasshoppers in the genera Alpinacris, Brachaspis, Paprides and Sigaus. Relative scores (percentages) are from the EMwm. See electronic supplementary material, S8 for results of other models.
3.1. Models for current conditions
Initial modelling using presence/absence records and current environmental conditions revealed where suitable habitat currently exists for each species (RN). This was compared with the known distribution of each species in order to determine if the models are ecologically plausible (figure 3). The modelled potential distribution of eight species (A. tumidicauda, B. collinus, B. nivalis, P. dugdali, P. nitidus, S. australis, S. childi, S. minutus and S. villosus) closely matched their current occupancy, indicating that their current distribution is probably constrained by the climate variables or their proxies in the models. For three species (A. crassicauda, S. campestris and S. piliferus), modelled potential distribution was larger than their current distributions. Of these three, all species had projected habitat that expanded onto the main island (North or South) where they are currently not found. Additionally, the projected suitable habitat of S. campestris and S. piliferus was found to be larger than their current distribution within their respective islands. Our model predicts suitable climatic environment to exist further west and north than these species have been recorded. Suitable climate for S. piliferus is suggested in the west (Mt Taranaki), and also along the southern east coast Te Ika-a-Māui (North Island), where the species has not been recorded.
Figure 3.
The realized and predicted niche space of 12 New Zealand alpine grasshopper species generated by ENM (EMwms). Maps show ecological niche models for current climate and as predicted for two future climate change scenarios (RCP2.6 (1°C rise) and RCP8.5(3.7°C rise) trajectories). Colours indicate probability (high to low) of suitable habitat under the model.
3.2. Future ecological niche models
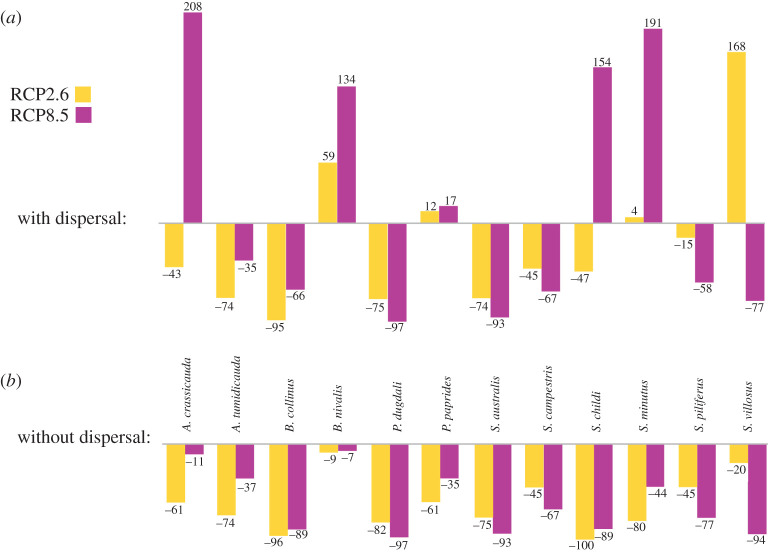
Changes in the size of each grasshopper projected niche (PN) were calculated for both RCP scenarios (electronic supplementary material, S9). Some species were predicted to lose habitat while other species were predicted to gain PN (table 1 and figure 4; electronic supplementary material, S9). With a mean temperature increase of 1°C by approximately 2070 (RCP2.6), eight species (A. crassicauda, A. tumidicauda, B. collinus, P. dugdali, S. australis, S. campestris, S. childi and S. piliferus) are predicted to lose PN, while four species are predicted to gain PN (B. nivalis, P. nitidus, S. minutus and S. villosus). Of those that will lose PN, four species (A. tumidicauda, B. collinus, P. dugdali and S. australis) will lose greater than 50%, with the greatest loss being 95% of PN for B. collinus. Sigaus villosus, however, is predicted to gain 168% of PN, while B. nivalis, P. nitidus and S. minutus gain 59%, 12% and 4% in our models, respectively. However, with a mean temperature increase of 3.7°C by approximately 2070 (RCP8.5), seven species (A. tumidicauda, B. collinus, P. dugdali, S. australis, S. campestris, S. piliferus and S. villosus) are predicted to lose PN, with six species (B. collinus, P. dugdali, S. australis, S. campestris, S. piliferus and S. villosus) predicted to lose over 50%, and one species (A. tumidicauda) predicted to lose 35% of its predicted PN. Five species (A. crassicauda, B. nivalis, P. nitidus, S. childi and S. minutus) gain PN in our models, with three species (A. tumidicauda, S. childi and S. minutus) gaining over 150%.
Figure 4.
Predicted loss of suitable habitat (as a percentage of current range) for 12 grasshopper species in the New Zealand alpine genera Alpinacris, Brachaspis, Paprides and Sigaus under two future climate scenarios: approximately 1.0°C rise by 2070(RCP2.6) and approximately 3.7°C rise by 2070(RCP8.5). Percentages are based on the loss/stability of pixels between vector binary maps (current models compared with future); (a) is the predicted loss/gain of all potential habitat patches and (b) is the predicted loss of space excluding fragments that would require dispersal by these flightless grasshoppers to newly available pixels (habitat).
As all grasshopper species in this study are flightless, dispersal between habitat patches will be limited. When niche space was recalculated to exclude newly available (gained) niche space that would require dispersal between fragments, all species were predicted to lose PN in the future (table 1). With a mean temperature increase of 1°C by approximately 2070 (RCP2.6), eight species (A. crassicauda, A. tumidicauda, B. collinus, P. dugdali, P. nitidus, S. australis, S. childi and S. minutus) will lose greater than 50% of PN if they are unable to disperse. Three other species (S. campestris, S. piliferus and S. villosus) will lose over 20% of their predicted PN. The greatest loss will be by S. childi (−100%), followed closely by B. collinus (−96%). With a mean temperature increase of 3.7°C by approximately 2070 (RCP8.5), seven species (B. collinus, P. dugdali, S. australis, S. campestris, S. childi, S. piliferus and S. villosus) will lose over 50% of their predicted PN, and three species (A. tumidicauda, P. nitidus and S. minutus) will lose over 35%. Paprides nitidus is predicted to have the greatest loss in this scenario (−97%), followed closely by S. villosus (−94%) and S. australis (−93%).
3.3. Fragmentation statistics
Fragmentation statistics were generated for current and the two future RCP scenarios based on vector binary maps (figure 5; electronic supplementary material, S10.1; S10.2; S11; S11; S12). Patches of connected ‘suitable habitat’ pixels were identified in each map and given unique IDs from which FRAGSTATS were calculated. The most common trend between current and future habitat fragmentation statistics was a decrease in the mean patch size associated with a decrease in suitable total habitat area. In some instances (11/24) of the scenarios across species: A. crassicauda (RCP2.6), A. tumidicauda (RCP2.6), B. collinus (RCP2.6 and RCP8.5), P. dugdali (RCP2.6 and RCP8.5), S. australis (RCP8.5), S. campestris (RCP2.6 and RCP8.5), S. piliferus (RCP8.5) and S. villosus (RCP8.5), a decrease in patch number was evident in an increase in the splitting index and/or decrease in the aggregation index in future scenarios (c.f. current conditions), indicating an increase in fragmentation and an increase in distance between patches, respectively. In three scenarios (A. tumidicauda (RCP8.5), S. australis (RCP2.6) and S. piliferus (RCP2.6)) where there was a decrease in the total area of patches, there was a decrease in mean patch size and increase in the number of patches. And for another three scenarios (P. nitidus (RCP2.6 and RCP8.5) and S. childi (RCP8.5)) where there was an increase in total patch area, there was a decrease in the mean area of patches, but an increase in the number of patches.
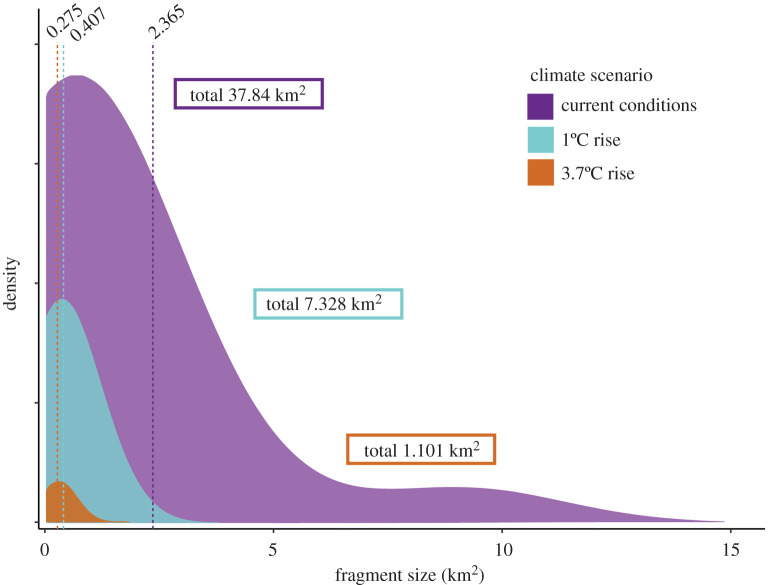
Figure 5.
Predicted habitat fragmentation for the endemic, flightless, alpine grasshopper Sigaus australis on Ka Tiritiri-o-te-Moana, Aotearoa (Southern Alps, New Zealand). Density distributions of habitat patches greater than 0.1 km2 in current conditions and two future scenarios under anthropogenic climate change (RCP2.6 and RCP8.5). Vertical dashed lines indicate means of each distribution with their values. Density plots are scaled to sum total of habitat greater than 0.1 km2 under each scenario (see electronic supplementary material, S12 for plots of other species).
4. Discussion
Patterns of biodiversity are intrinsically linked to changes in Earth's climate [63,64]. Long-term climate change is expected to result in species becoming more isolated, experiencing phenological mismatch, losing suitable habitat and encountering new competition and community assemblages as ranges shift [5]. Whether a species survives such changes or is extinguished depends on the severity and speed of change in climate, and the adaptive capacity of the species involved [65]. The pace of anthropogenic climate change [39] imposes intense selective pressure on many taxa, and those on steep climatic gradients are likely to be influenced most rapidly [4–6]. This is the situation for alpine grasshoppers endemic to Aotearoa New Zealand. Comparison of their current distributions with modelled suitable niche space reveals that climate is a primary factor controlling their distribution. Despite the ecological overlap of New Zealand's alpine grasshopper species, the environmental proxies/variables that best explain their current distributions were species specific. It is likely that changes in species distributions resulting from global warming will also be species specific, but most New Zealand grasshoppers will be negatively affected. Furthermore, for most species, we identified a decrease in the total area of suitable habitat patches and reduction in the mean size of these patches and the total number of patches, which will result in population fragmentation and vulnerability to stochastic loss and erosion of intra-specific diversity [66,67]. Despite New Zealand's alpine habitat being less than 10 million years old, it is home to an extensive array of specialized, endemic plants and insects [14,35], all of which will be vulnerable to climate change. Given our finding of idiosyncratic responses of individual grasshopper species, we recommend ENM be applied to other alpine specialists such as crickets Pharmacus spp. (Rhaphidophoridae) and penwiper plants Notothlaspi spp. (Brassicaceae) in order to identify whether species extinctions could be prevented with human-mediated dispersal of population samples to unoccupied alpine habitat.
4.1. Ecological niche modelling
ENM will only work well when species are limited by abiotic environmental factors rather than by species interactions and their dispersal ability to access suitable conditions. In cases where RN is close to fundamental niche, we can expect high performance of models if distribution data are adequately sampled and all relevant environmental drivers are included in the models. The predictive accuracy of ecological niche models is dependent on the number and distribution of presence records available for each species and the quality of absence information [59,60,62]. Here, each grasshopper species was represented by presence data that closely reflected the spatial ranges of each species, and the accumulation of observations of 14 New Zealand grasshopper species provided the advantages of accurate absence data. For some species (P. dugdali, S. piliferus and S. villosus), the potential for additional data points is limited because they have restricted and patchy occurrence in current conditions. In fact, it was those species with the highest numbers of presence data points (e.g. B. nivalis, P. nitidus, S. australis and S. campestris) that yielded models with the lowest sensitivity and specificity scores. Although seemingly contrariwise, the tendency for the distributions of rare or spatially restricted species to yield statistically more robust models than common or widely distributed species [56,68,69], reflects the detection of more environmental variance across wide-ranging locations. This might result from the inadvertent pooling of distinct lineages (cryptic taxa) adapted to different conditions, and with the potential to respond in different ways to environmental change. In such instances it may be beneficial to model phylogenetic lineages separately to improve the predictive accuracy of models [70–72].
With the available data, the individual statistical models generated were generally well supported by both ROC and TSS model evaluation metrics. Across species, a minimum of four statistical models that met evaluation thresholds were incorporated into ensemble models, although the combination of models varied by species. Evaluation metrics for the final ensemble models indicate the ensembles out-performed individual models (the exception being S. childi where they were equal), and this is consistent with other studies [31,59,73]. This emphasizes the importance and benefits of exploring a range of statistical models and applying the ensemble model approach. We did not need to rely on a single algorithm to model our predictions and different ensembles of statistical models could be applied depending on what was most relevant for each species.
Of the climate, vegetation and soil inputs applied in this study, climate variables were predominantly the most important at explaining the distribution of the grasshopper species. For most species, a single predictor variable tended to be a more important environmental proxy than any other. In the majority of instances, the current distribution of grasshopper species matched the ecological niche space predicted by the ENMs. A mismatch between the current distribution of a species and its predicted niche under current conditions indicates that other variables are limiting the distribution of these species [74]. This could involve abiotic variables not included in the analysis, or biological interactions such as competition, predation, disease and/or parasitism. Biotic variables are notoriously difficult to account for in ENM [74]. Climate is evidently a key driver in the distribution of New Zealand's endemic alpine grasshoppers. When transferring ecological niche models to future climate scenarios we assumed that environmental correlations remained the same as those determined for current New Zealand climate variables, but additional aspects including changes in anthropogenic land use will need consideration for protection of biodiversity.
4.2. Future climate
Predictions of how species may fare with future climate change varied, depending on the RCP scenario, and whether or not ability to disperse across habitat gaps was taken into account. Most species were predicted to lose at least 30% of suitable habitat under both RCP scenarios (table 1; electronic supplementary material, S8). In the last 20 years, global mean surface temperature has already increased by 0.66°C [75], suggesting that a rise of 1°C (RCP2.6) is an underestimate of the speed and extent of warming Earth's biota will experience. Even under this conservative scenario, the currently widespread alpine species B. collinus will lose 95% of its range, putting it at high risk of extinction. Other grasshopper species were predicted to gain PN from a rise of 1°C (RCP2.6), while also being predicted to lose PN from a rise of 3.7°C (RCP8.5), and vice versa. Furthermore, closely related species did not demonstrate similar responses to change. An example is the endangered species S. childi which currently has a narrow distribution in Central Otago of the South Island within the much wider range of its sister species S. australis. Sigaus childi is specialized to low-elevation semi-arid environments in Central Otago, while S. australis is primarily an alpine species with a range now extending to low elevations. With a rise of 3.7°C (RCP8.5), S. childi was predicted to gain substantial niche space, as semi-arid conditions expand in the Central Otago region, although only after losing 47% of its habitat at +1°C. By contrast, S. australis was predicted to lose projected niche space under all scenarios. Perhaps S. childi has the potential to expand its range and out-compete S. australis in the future (although hybridization might prevent this [76]). Of more immediate concern for this endangered species is that without the ability to colonize new habitat patches, S. childi is predicted to go extinct unless we invest in human-mediated dispersal. When no-dispersal was applied to the range-change calculations, no New Zealand grasshopper species maintained its range, with all species losing PN. Full dispersal and no dispersal are the extreme assumptions to apply to these models [77], and it is likely that even with their poor dispersal abilities, the grasshoppers have some potential to move short distances to colonize new patches. This would not, however, allow shifts between habitat on different mountain ranges or islands of Aotearoa, and the general pattern seen across grasshopper species in this study, was of negative range outcomes with climate change, as found for alpine invertebrates around the world [78–80].
The extent and consequences of PN loss will be species specific. The impact of a reduction of 10% of PN would probably be negligible for species that currently have wide distributions (e.g. S. australis, P. nitidus), compared with range-restricted species (e.g. S. childi). Despite being predicted to gain PN under various scenarios, species with lowland distributions (S. childi and S. minutus), will be at greatest risk of land-use change including conversion of native grasslands for agricultural land, invasion of exotic plants (e.g. Radiata pine) and fire, with the end result likely to be just as detrimental as for high-elevation species [37].
4.3. Fragmentation results and consequences
Habitat fragmentation typically results in reduced net population size and changes in metapopulation dynamics (e.g. [81]) that can result in a loss of genetic diversity through genetic drift and inbreeding depression [82–84]. By contrast, population fragmentation is also hypothesized as a mechanism generating biodiversity hotspots associated with small-island effects, including in alpine areas [85,86]. The outcomes of habitat fragmentation for inter- and intra-specific diversity are sensitive to many factors [87–89], but rapid, sustained fragmentation resulting in patch size reduction when coupled with limited dispersal ability will negatively affect the ability of species to track optimal conditions under future climatic change pressure.
As New Zealand's endemic alpine grasshoppers are flightless, and New Zealand is divided into two main islands, it is important to consider that even though new PN space might become available in the future, the grasshoppers may not be able to disperse into these new areas without human assistance. Evidence of this can be seen in models of current suitable niche space. For example, there is predicted to be suitable habitat for A. crassicauda and S. campestris in the North Island, but these species remain only distributed in the South Island. For S. piliferus, suitable habitat is available in the South Island, but this species is limited to the North Island. During the last glacial maximum (LGM), the North and South Islands were connected by lowland habitat across the Cook Strait—however, even this did not facilitate the movement of South Island species northwards or vice versa (or they have subsequently been extirpated) [90]. Even within a land area grasshopper ranges are restricted to suitable habitat, for example S. piliferus is predicted to exist on Mt Taranaki on the west of the North Island; however, this species has never been recorded there. The inability of these endemic New Zealand grasshoppers to readily disperse (i.e. via flight) is already a limiting factor in their current distributions.
The fragmentation statistics estimated from the ENMs predict that habitat for the grasshoppers will be increasingly fragmented in the future—regardless of predicted total habitat area. In most cases, future total area of patches was predicted to decrease, with patches either reducing in number as well as size, or reducing in size but increasing in number. Even when overall area was predicted to increase, this was accompanied by an increase in the number of patches (i.e. increased fragmentation) (figure 5). New Zealand's alpine environments have been more connected in the past—most recently during the LGM ending approximately 15 kya [38], with the ranges of many species subsequently receding and fragmenting. Around the world, many alpine ecosystems are recognized hotspots of endemism, hypothesized to be as a result of steady climate cycling, habitat fragmentation and species diversification [85,86]. Biodiversity outcomes from environmental fluctuation and species adaptation over tens or hundreds of thousands of years are not comparable with change occurring within 100 years. The rate of habitat loss and fragmentation predicted by 2070 in this study may be too fast for many alpine species to adapt. The alpine grasshoppers analysed here have approximately 50 generations to shift ranges, niches or adapt in situ to predicted changes. Furthermore, if humans are unable to curb carbon emissions, the consequences may be worse than what we have predicted here [39].
5. Conclusion
By modelling the environmental envelopes of New Zealand's endemic alpine grasshopper fauna, and the likely changes to local environmental conditions under future climate change scenarios, we witness the complexities in predicting the distributions of species in the future, despite shared ecosystems and history. We have determined that the overall trend for this group will be a decrease in projected niche space by 2070; however, there are some exceptions, and sensitivity of models to different warming scenarios varies. We demonstrate the importance of considering the dispersal ability of species when inferring predictions, in addition to fragmentation and total area of PN, as this could greatly impact the chances of species surviving future climate change and land-use changes.
Acknowledgements
We would like to thank the New Zealand Department of Conservation for granting access and collecting permits (Authorization no.: 49878-RES). We would also like to thank all those who have contributed Acrididae specimens to the Phoenix Lab collection at Massey University, including: Edwina Dowle, Julia Goldberg, Danilo Hegg, Sam King, Ben Anderson, Louisa Sivyer, Briar Taylor-Smith, Ted & Bee Trewick.
Data accessibility
Specimen locality data are presented in the electronic supplementary file. Environmental input and coding information are available in file Biomod2_Archive at the website: evolves.massey.ac.nz. Link: http://evolves.massey.ac.nz/Data/Biomod2_Archive.zip.
The data are provided in the electronic supplementary material [91].
Authors' contributions
E.M.K.: formal analysis, investigation, methodology and writing—original draft; M.M.-R.: conceptualization, project administration, supervision and writing—revision, review and editing; S.A.T.: conceptualization, funding acquisition, investigation, project administration, supervision, resources, figures and writing—revision, review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This research was supported by a doctoral scholarship from Massey University and a grant from the Miss E. L. Hellaby Indigenous Grasslands Research Trust.
References
- 1.Clark JS, Bell DM, Hersh MH, Nichols L. 2011. Climate change vulnerability of forest biodiversity: climate and competition tracking of demographic rates. Glob. Change Biol. 17, 1834-1849. ( 10.1111/j.1365-2486.2010.02380.x) [DOI] [Google Scholar]
- 2.Ordonez A, Williams JW, Svenning JC. 2016. Mapping climatic mechanisms likely to favour the emergence of novel communities. Nat. Clim. Change 6, 1104-1109. ( 10.1038/nclimate3127) [DOI] [Google Scholar]
- 3.Loarie SR, Duffy PB, Hamilton H, Asner GP, Field CB, Ackerly DD. 2009. The velocity of climate change. Nature 462, 1052-1055. ( 10.1038/nature08649) [DOI] [PubMed] [Google Scholar]
- 4.Sala OE, et al. 2000. Global biodiversity scenarios for the year 2100. Science 287, 1770-1774. ( 10.1126/science.287.5459.1770) [DOI] [PubMed] [Google Scholar]
- 5.Parmesan C. 2006. Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37, 637-669. ( 10.1146/annurev.ecolsys.37.091305.110100) [DOI] [Google Scholar]
- 6.Turner C. 2018. Climate change and biodiversity. Waltham Abbey, UK: ED Tech Press. [Google Scholar]
- 7.Pepin N, et al. 2015. Elevation-dependent warming in mountain regions of the world. Nat. Clim. Change 5, 424-430. ( 10.1038/nclimate2563) [DOI] [Google Scholar]
- 8.Kullman L, Öberg L. 2009. Post-Little Ice Age tree line rise and climate warming in the Swedish Scandes: a landscape ecological perspective. J. Ecol. 97, 415-429. ( 10.1111/j.1365-2745.2009.01488.x) [DOI] [Google Scholar]
- 9.Gatti RC, Callaghan T, Velichevskaya A, Dudko A, Fabbio L, Battipaglia G, Liang J. 2019. Accelerating upward treeline shift in the Altai Mountains under last-century climate change. Sci. Rep. 9, 1-13. ( 10.1038/s41598-019-44188-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein G, Vitasse Y, Rixen C, Marty C, Rebetez M. 2016. Shorter snow cover duration since 1970 in the Swiss Alps due to earlier snowmelt more than to later snow onset. Clim. Change 139, 637-649. ( 10.1007/s10584-016-1806-y) [DOI] [Google Scholar]
- 11.Wilson RJ, Gutierrez D, Gutierrez J, Monserrat VJ. 2007. An elevational shift in butterfly species richness and composition accompanying recent climate change. Glob. Change Biol. 13, 1873-1887. ( 10.1111/j.1365-2486.2007.01418.x) [DOI] [Google Scholar]
- 12.Tafani M, Cohas A, Bonenfant C, Gaillard JM, Allainé D. 2013. Decreasing litter size of marmots over time: a life history response to climate change? Ecology 94, 580-586. ( 10.1890/12-0833.1) [DOI] [PubMed] [Google Scholar]
- 13.Parmesan C, Yohe G. 2003. A globally coherent fingerprint of climate change impacts across natural systems. Nature 421, 37-42. ( 10.1038/nature01286) [DOI] [PubMed] [Google Scholar]
- 14.Chinn W, Chinn T. 2020. Tracking the snow line: responses to climate change by New Zealand alpine invertebrates. Arctic Ant. Alpine Res. 52, 361-389. ( 10.1080/15230430.2020.1773033) [DOI] [Google Scholar]
- 15.Testolin R, Attorre F, Jiménez-Alfaro B. 2020. Global distribution and bioclimatic characterization of alpine biomes. Ecography 43, 779-788. ( 10.1111/ecog.05012) [DOI] [Google Scholar]
- 16.Körner C, Paulsen J, Spehn EM. 2011. A definition of mountains and their bioclimatic belts for global comparisons of biodiversity data. Alpine Bot. 121, 73. ( 10.1007/s00035-011-0094-4) [DOI] [Google Scholar]
- 17.Lute A, Abatzoglou JT. 2020. Best practices for estimating near-surface air temperature lapse rates. Int. J. Climatol. 41, E110-E125. ( 10.1002/joc.6668) [DOI] [Google Scholar]
- 18.Minder JR, Mote PW, Lundquist JD. 2010. Surface temperature lapse rates over complex terrain: lessons from the Cascade Mountains. J. Geophys. Res. 115, 1-13. ( 10.1029/2009JD013493) [DOI] [Google Scholar]
- 19.Körner C, Paulsen J. 2004. A world-wide study of high altitude treeline temperatures. J. Biogeogr. 31, 713-732. ( 10.1111/j.1365-2699.2003.01043.x) [DOI] [Google Scholar]
- 20.Körner C. 1998. A re-assessment of high elevation treeline positions and their explanation. Oecologia 115, 445-459. ( 10.1007/s004420050540) [DOI] [PubMed] [Google Scholar]
- 21.MacDonald GM, Velichko AA, Kremenetski CV, Andreev A. 2000. Holocene treeline history and climate change across northern Eurasia. Quat. Res. 53, 302-311. ( 10.1006/qres.1999.2123) [DOI] [Google Scholar]
- 22.Greenwood S, Jump AS. 2014. Consequences of treeline shifts for the diversity and function of high altitude ecosystems. Arctic Ant. Alpine Res. 46, 829-840. ( 10.1657/1938-4246-46.4.829) [DOI] [Google Scholar]
- 23.Boggs CL, Murphy DD. 1997. Community composition in mountain ecosystems: climatic determinants of montane butterfly distributions. Glob. Ecol. Biogeogr. Lett. 6, 39-48. ( 10.2307/2997525) [DOI] [Google Scholar]
- 24.Gifford ME, Kozak KH. 2012. Islands in the sky or squeezed at the top? Ecological causes of elevational range limits in montane salamanders. Ecography 35, 193-203. ( 10.1111/j.1600-0587.2011.06866.x) [DOI] [Google Scholar]
- 25.Lyons MP, Kozak KH. 2020. Vanishing islands in the sky? A comparison of correlation-and mechanism-based forecasts of range dynamics for montane salamanders under climate change. Ecography 43, 481-493. ( 10.1111/ecog.04282) [DOI] [Google Scholar]
- 26.Piao S, et al. 2019. Plant phenology and global climate change: current progresses and challenges. Glob. Change Biol. 25, 1922-1940. ( 10.1111/gcb.14619) [DOI] [PubMed] [Google Scholar]
- 27.Thuiller W, et al. 2008. Predicting global change impacts on plant species' distributions: future challenges. Perspect. Plant Ecol. Evol. Syst. 9, 137-152. ( 10.1016/j.ppees.2007.09.004) [DOI] [Google Scholar]
- 28.Regos A, Gagne L, Alcaraz-Segura D, Honrado JP, Domínguez J. 2019. Effects of species traits and environmental predictors on performance and transferability of ecological niche models. Sci. Rep. 9, 1-14. ( 10.1038/s41598-019-40766-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sillero N, Barbosa AM. 2021. Common mistakes in ecological niche models. Int. J. Geogr. Inf. Sci. 35, 213-226. ( 10.1080/13658816.2020.1798968) [DOI] [Google Scholar]
- 30.Hao T, Elith J, Guillera-Arroita G, Lahoz-Monfort JJ. 2019. A review of evidence about use and performance of species distribution modelling ensembles like BIOMOD. Divers. Distrib. 25, 839-852. ( 10.1111/ddi.12892) [DOI] [Google Scholar]
- 31.Araújo MB, New M. 2007. Ensemble forecasting of species distributions. Trends Ecol. Evol. 22, 42-47. ( 10.1016/j.tree.2006.09.010) [DOI] [PubMed] [Google Scholar]
- 32.Koot EM, Morgan-Richards M, Trewick SA. 2020. An alpine grasshopper radiation older than the mountains, on Kā Tiritiri o te Moana (Southern Alps) of Aotearoa (New Zealand). Mol. Phylogenet. Evol. 147, 106783. ( 10.1016/j.ympev.2020.106783) [DOI] [PubMed] [Google Scholar]
- 33.Wilmshurst JM, Anderson AJ, Higham TF, Worthy TH. 2008. Dating the late prehistoric dispersal of Polynesians to New Zealand using the commensal Pacific rat. Proc. Natl Acad. Sci. USA 105, 7676-7680. ( 10.1073/pnas.0801507105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Argiriadis E, et al. 2018. Lake sediment fecal and biomass burning biomarkers provide direct evidence for prehistoric human-lit fires in New Zealand. Sci. Rep. 8, 12113. ( 10.1038/s41598-018-30606-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winkworth RC, Wagstaff SJ, Glenny D, Lockhart PJ. 2005. Evolution of the New Zealand mountain flora: origins, diversification and dispersal. Org. Divers. Evol. 5, 237-247. ( 10.1016/j.ode.2004.12.001) [DOI] [Google Scholar]
- 36.Hawes TC. 2015. Canalization of freeze tolerance in an alpine grasshopper. Cryobiology 71, 356-359. ( 10.1016/j.cryobiol.2015.07.008) [DOI] [PubMed] [Google Scholar]
- 37.Sivyer L, Morgan-Richards M, Koot E, Trewick SA. 2018. Anthropogenic cause of range shifts and gene flow between two grasshopper species revealed by environmental modelling, geometric morphometrics and population genetics. Insect Conserv. Divers. 11, 415-434. ( 10.1111/icad.12289) [DOI] [Google Scholar]
- 38.Carmelet-Rescan D, Morgan-Richards M, Koot EM, Trewick SA. 2021. Climate and ice in the last glacial maximum explain patterns of isolation by distance inferred for alpine grasshoppers. Insect Conserv. Divers. 14, 568-581. ( 10.1111/icad.12488) [DOI] [Google Scholar]
- 39.Masson-Delmotte V, et al. 2021. Climate Change 2021: The physical science basis. In Contribution of Working Group I to the sixth assessment report of the Intergovernmental Panel on Climate Change. IPCC. [Google Scholar]
- 40.LINZ. 2015. NZ Topo Map. See https://www.topomap.co.nz/.
- 41.Hijmans RJ, Cameron S, Parra J. 2015. WorldClim – Global Climate Data. See https://www.worldclim.com/.
- 42.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965-1978. [Google Scholar]
- 43.Taylor KE, Stouffer RJ, Meehl GA. 2012. An overview of CMIP5 and the experiment design. Bull. Am. Meteorol. Soc. 93, 485-498. ( 10.1175/BAMS-D-11-00094.1) [DOI] [Google Scholar]
- 44.Pachauri RK, et al. 2014. Climate change 2014: synthesis report. In Contribution of Working Groups I, II and III to the fifth assessment report of the Intergovernmental Panel on Climate Change. IPCC. [Google Scholar]
- 45.Watanabe S, et al. 2011. MIROC-ESM 2010: model description and basic results of CMIP5-20c3m experiments. Geosci. Model Dev. 4, 845. ( 10.5194/gmd-4-845-2011) [DOI] [Google Scholar]
- 46.QGIS Development Team. 2015. QGIS geographic information system.
- 47.Naimi B, Araújo M. 2016. sdm: a reproducible and extensible R platform for species distribution modelling. Ecography 39, 368-375. ( 10.1111/ecog.01881) [DOI] [Google Scholar]
- 48.Leathwick J, McGlone M, Walker S, Briggs C. 2004. New Zealand's potential vegetation pattern. Lincoln, New Zealand: Manaaki Whenua Press. [Google Scholar]
- 49.Newsome P. 1987. Vegetative cover of New Zealand. Wellington, New Zealand: Water and Soil Directorate, Ministry of Works and Development. [Google Scholar]
- 50.Nattier R, Grandcolas P, Pellens R, Jourdan H, Couloux A, Poulain S, Robillard T. 2013. Climate and soil type together explain the distribution of microendemic species in a biodiversity hotspot. PLoS ONE 8, e80811. ( 10.1371/journal.pone.0080811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss N, Zucchi H, Hochkirch A. 2013. The effects of grassland management and aspect on Orthoptera diversity and abundance: site conditions are as important as management. Biodivers. Conserv. 22, 2167-2178. ( 10.1007/s10531-012-0398-8) [DOI] [Google Scholar]
- 52.Stanton JC, Pearson RG, Horning N, Ersts P, Reşit Akçakaya H. 2012. Combining static and dynamic variables in species distribution models under climate change. Methods Ecol. Evol. 3, 349-357. ( 10.1111/j.2041-210X.2011.00157.x) [DOI] [Google Scholar]
- 53.Thuiller W, Georges D, Engler R, Breiner F. 2016. biomod2: Ensemble Platform for Species Distribution Modeling. See http://CRAN.R-project.org/package=biomod2.
- 54.Thuiller W, Lafourcade B, Araujo M. 2009. ModOperating manual for BIOMOD. In, Thuiller W, Lafourcade B (2010) BIOMOD: species/climate modelling functions. R package version, 1.1-5.
- 55.Fletcher D, Gillingham P, Britton J, Blanchet S, Gozlan RE. 2016. Predicting global invasion risks: a management tool to prevent future introductions. Sci. Rep. 6, 1-8. ( 10.1038/srep26316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allouche O, Tsoar A, Kadmon R. 2006. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 43, 1223-1232. ( 10.1111/j.1365-2664.2006.01214.x) [DOI] [Google Scholar]
- 57.VanDerWal J, et al. 2014. Package ‘SDMTools’. R package (R Foundation for Statistical Computing).
- 58.McGarigal K. 1995. FRAGSTATS: spatial pattern analysis program for quantifying landscape structure. General Technical Report PNW-351. Corvallis, OR: USDA Forest Service. [Google Scholar]
- 59.Breiner FT, Guisan A, Bergamini A, Nobis MP. 2015. Overcoming limitations of modelling rare species by using ensembles of small models. Methods Ecol. Evol. 6, 1210-1218. ( 10.1111/2041-210X.12403) [DOI] [Google Scholar]
- 60.Hernandez PA, Graham CH, Master LL, Albert DL. 2006. The effect of sample size and species characteristics on performance of different species distribution modeling methods. Ecography 29, 773-785. ( 10.1111/j.0906-7590.2006.04700.x) [DOI] [Google Scholar]
- 61.Stockwell DR, Peterson AT. 2002. Effects of sample size on accuracy of species distribution models. Ecol. Model 148, 1-13. ( 10.1016/S0304-3800(01)00388-X) [DOI] [Google Scholar]
- 62.Wisz MS, et al. 2008. Effects of sample size on the performance of species distribution models. Divers. Distrib. 14, 763-773. ( 10.1111/j.1472-4642.2008.00482.x) [DOI] [Google Scholar]
- 63.Gillespie RG, Roderick GK. 2014. Evolution: geology and climate drive diversification. Nature 509, 297-298. ( 10.1038/509297a) [DOI] [PubMed] [Google Scholar]
- 64.Nogués-Bravo D, Rodríguez-Sánchez F, Orsini L, de Boer E, Jansson R, Morlon H, Fordham DA, Jackson ST. 2018. Cracking the code of biodiversity responses to past climate change. Trends Ecol. Evol. 33, 765-776. ( 10.1016/j.tree.2018.07.005) [DOI] [PubMed] [Google Scholar]
- 65.Visser ME. 2008. Keeping up with a warming world; assessing the rate of adaptation to climate change. Proc. R. Soc. B 275, 649-659. ( 10.1098/rspb.2007.0997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burkey TV. 1999. Extinction in fragmented habitats predicted from stochastic birth–death processes with density dependence. J. Theor. Biol. 199, 395-406. ( 10.1006/jtbi.1999.0967) [DOI] [PubMed] [Google Scholar]
- 67.Pflüger FJ, Signer J, Balkenhol N. 2019. Habitat loss causes non-linear genetic erosion in specialist species. Glob. Ecol. Conserv. 17, e00507. ( 10.1016/j.gecco.2018.e00507) [DOI] [Google Scholar]
- 68.Bouska KL, Whitledge GW, Lant C. 2015. Development and evaluation of species distribution models for fourteen native central US fish species. Hydrobiologia 747, 159-176. ( 10.1007/s10750-014-2134-8) [DOI] [Google Scholar]
- 69.Syphard AD, Franklin J. 2010. Species traits affect the performance of species distribution models for plants in southern California. J. Veg. Sci. 21, 177-189. ( 10.1111/j.1654-1103.2009.01133.x) [DOI] [Google Scholar]
- 70.Leaché AD, Koo MS, Spencer CL, Papenfuss TJ, Fisher RN, McGuire JA. et al. 2009. Quantifying ecological, morphological, and genetic variation to delimit species in the coast horned lizard species complex (Phrynosoma). Proc. Natl Acad. Sci. USA 106, 12 418-12 423. ( 10.1073/pnas.0906380106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith AB, Godsoe W, Rodríguez-Sánchez F, Wang HH, Warren D. 2019. Niche estimation above and below the species level. Trends Ecol. Evol. 34, 260-273. ( 10.1016/j.tree.2018.10.012) [DOI] [PubMed] [Google Scholar]
- 72.Ikeda DH, Max TL, Allan GJ, Lau MK, Shuster SM, Whitham TG. 2017. Genetically informed ecological niche models improve climate change predictions. Glob. Change Biol. 23, 164-176. ( 10.1111/gcb.13470) [DOI] [PubMed] [Google Scholar]
- 73.Marmion M, Parviainen M, Luoto M, Heikkinen RK, Thuiller W. 2009. Evaluation of consensus methods in predictive species distribution modelling. Divers. Distrib. 15, 59-69. ( 10.1111/j.1472-4642.2008.00491.x) [DOI] [Google Scholar]
- 74.Pearson RG, et al. 2006. Model-based uncertainty in species range prediction. J. Biogeogr. 33, 1704-1711. ( 10.1111/j.1365-2699.2006.01460.x) [DOI] [Google Scholar]
- 75.Valipour M, Bateni SM, Jun C. 2021. Global Surface Temperature: A New Insight. Climate 9,81..
- 76.Dowle EJ, Morgan-Richards M, Trewick SA. 2014. Morphological differentiation despite gene flow in an endangered grasshopper. BMC Evol. Biol. 14, 1-5. ( 10.1186/s12862-014-0216-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thuiller W, Lavorel S, Araújo MB, Sykes MT, Prentice IC. 2005. Climate change threats to plant diversity in Europe. Proc. Natl Acad. Sci. USA 102, 8245-8250. ( 10.1073/pnas.0409902102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Descombes P, Pradervand JN, Golay J, Guisan A, Pellissier L. 2016. Simulated shifts in trophic niche breadth modulate range loss of alpine butterflies under climate change. Ecography 39, 796-804. ( 10.1111/ecog.01557) [DOI] [Google Scholar]
- 79.Biella P, Bogliani G, Cornalba M, Manino A, Neumayer J, Porporato M, Rasmont P, Milanesi P. 2017. Distribution patterns of the cold adapted bumblebee Bombus alpinus in the Alps and hints of an uphill shift (Insecta: Hymenoptera: Apidae). J. Insect Conserv. 21, 357-366. ( 10.1007/s10841-017-9983-1) [DOI] [Google Scholar]
- 80.Xue Z, Zhang Z, Lu X, Zou Y, Lu Y, Jiang M, Tong S, Zhang K. 2014. Predicted areas of potential distributions of alpine wetlands under different scenarios in the Qinghai-Tibetan Plateau, China. Glob. Planetary Change 123, 77-85. ( 10.1016/j.gloplacha.2014.10.012) [DOI] [Google Scholar]
- 81.Kahilainen A, van Nouhuys S, Schulz T, Saastamoinen M. 2018. Metapopulation dynamics in a changing climate: increasing spatial synchrony in weather conditions drives metapopulation synchrony of a butterfly inhabiting a fragmented landscape. Glob. Change Biol. 24, 4316-4329. ( 10.1111/gcb.14280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pavlova A, et al. 2017. Severe consequences of habitat fragmentation on genetic diversity of an endangered Australian freshwater fish: a call for assisted gene flow. Evol. Appl. 10, 531-550. ( 10.1111/eva.12484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rehnus M, Bollmann K, Schmatz DR, Hackländer K, Braunisch V. 2018. Alpine glacial relict species losing out to climate change: the case of the fragmented mountain hare population (Lepus timidus) in the Alps. Glob. Change Biol. 24, 3236-3253. ( 10.1111/gcb.14087) [DOI] [PubMed] [Google Scholar]
- 84.Crooks KR, Sanjayan M. 2006. Connectivity conservation, vol. 14. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 85.Chen JH, et al. 2019. Genome-wide analysis of Cushion willow provides insights into alpine plant divergence in a biodiversity hotspot. Nat. Commun. 10, 5230. ( 10.1038/s41467-019-13128-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang P, Yao H, Gilbert KJ, Lu Q, Hao Y, Zhang Z, Wang N. 2018. Glaciation-based isolation contributed to speciation in a Palearctic alpine biodiversity hotspot: evidence from endemic species. Mol. Phylogenet. Evol. 129, 315-324. ( 10.1016/j.ympev.2018.09.006) [DOI] [PubMed] [Google Scholar]
- 87.Fahrig L, et al. 2019. Is habitat fragmentation bad for biodiversity? Biol. Conserv. 230, 179-186. ( 10.1016/j.biocon.2018.12.026) [DOI] [Google Scholar]
- 88.Fletcher RJ Jr, et al. 2018. Is habitat fragmentation good for biodiversity? Biol. Conserv. 226, 9-15. ( 10.1016/j.biocon.2018.07.022) [DOI] [Google Scholar]
- 89.Fahrig L. 2017. Ecological responses to habitat fragmentation per se. Ann. Rev. Ecol. Evol. Syst. 48, 1-23. ( 10.1146/annurev-ecolsys-110316-022612) [DOI] [Google Scholar]
- 90.Trewick S, Bland K. 2012. Fire and slice: palaeogeography for biogeography at New Zealand's North Island/South Island juncture. J. R. Soc. New Zealand 42, 153-183. ( 10.1080/03036758.2010.549493) [DOI] [Google Scholar]
- 91.Koot EM, Morgan-Richards M, Trewick SA. 2022. Climate change and alpine-adapted insects: modelling environmental envelopes of a grasshopper radiation. Figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Koot EM, Morgan-Richards M, Trewick SA. 2022. Climate change and alpine-adapted insects: modelling environmental envelopes of a grasshopper radiation. Figshare. [DOI] [PMC free article] [PubMed]
Data Availability Statement
Specimen locality data are presented in the electronic supplementary file. Environmental input and coding information are available in file Biomod2_Archive at the website: evolves.massey.ac.nz. Link: http://evolves.massey.ac.nz/Data/Biomod2_Archive.zip.
The data are provided in the electronic supplementary material [91].